Abstract
Mobile communications are propagated by electromagnetic fields (EMFs), and since the 1990s, they operate with pulse-modulated signals such as the GSM-1800 MHz. The biological effects of GSM-EMF in humans affected by neuropathological processes remain seldom investigated. In this study, a 2-h head-only exposure to GSM-1800 MHz was applied to (i) rats undergoing an acute neuroinflammation triggered by a lipopolysaccharide (LPS) treatment, (ii) age-matched healthy rats, or (iii) transgenic hSOD1G93A rats that modeled a presymptomatic phase of human amyotrophic lateral sclerosis (ALS). Gene responses were assessed 24 h after the GSM head-only exposure in a motor area of the cerebral cortex (mCx) where the mean specific absorption rate (SAR) was estimated to be 3.22 W/kg. In LPS-treated rats, a genome-wide mRNA profiling was performed by RNA-seq analysis and revealed significant (adjusted p value < 0.05) but moderate (fold changes < 2) upregulations or downregulations affecting 2.7% of the expressed genes, including genes expressed predominantly in neuronal or in glial cell types and groups of genes involved in protein ubiquitination or dephosphorylation. Reverse transcription-quantitative PCR analyses confirmed gene modulations uncovered by RNA-seq data and showed that in a set of 15 PCR-assessed genes, significant gene responses to GSM-1800 MHz depended upon the acute neuroinflammatory state triggered in LPS-treated rats, because they were not observed in healthy or in hSOD1G93A rats. Together, our data specify the extent of cortical gene modulations triggered by GSM-EMF in the course of an acute neuroinflammation and indicate that GSM-induced gene responses can differ according to pathologies affecting the CNS.
Keywords: Electromagnetic fields, Mobile phone, Neuroinflammation, Lipopolysaccharide, Amyotrophic lateral sclerosis, RNA-seq
Introduction
The general use of mobile phone communications results in partial or whole-body exposures to electromagnetic field (EMF). Since the 1990s, these communications use pulse-modulated signals, among which is the global system for mobile communication (GSM) operating in the 900-MHz or 1800-MHz bands. Concerns about possible health hazard resulting from these EMF exposures have generated a wealth of investigation, many of which aimed at determining the impact of EMF at cellular or molecular levels (Narayanan et al. 2019). This issue was approached experimentally using cell cultures or animals that were submitted to calibrated exposures to EMF (Gherardini et al. 2014; Marino et al. 2011; McNamee and Chauhan 2009). Mobile phone communications commonly bring a source of EMF in contact with the ear, which favors the exposure of brain cells to EMF. Studies carried out with rodents have searched for effects of GSM-related EMF in brain cells, such as changes in the expression of genes involved in cell stress responses or that mark brain tissue alterations. For instance, investigations have focused on the expression of heat shock proteins (Fritze et al. 1997; Miyakoshi et al. 2005; Watilliaux et al. 2011), immediate early genes including Fos (Finnie 2005), synaptic components (Kim et al. 2017), astroglial GFAP marker, or proinflammatory mediators (Bouji et al. 2012; Hirose et al. 2010; Lu et al. 2014; Su et al. 2018; Thorlin et al. 2006). However, gene responses to GSM-EMF were inconstant from one study to another. In particular, astroglial reactions marked by increased expression of GFAP were observed in response to a unique head-only exposure to GSM-900 MHz (Brillaud et al. 2007; Mausset-Bonnefont et al. 2004), but was not observed in other studies using either single head-only exposures (Bouji et al. 2012; Fritze et al. 1997; Watilliaux et al. 2011) or repeated head- or whole-body exposures (Court-Kowalski et al. 2015; Petitdant et al. 2016). The reasons for such discrepancies remain unclear but can result from differences in experimental setting used in the different studies. Key parameters include the number, duration, and the power of EMF exposures absorbed by the tissues, which is determined by the specific absorption rate (SAR) expressed in W/kg.
In an effort to identify molecular targets of mobile phone exposures, recent studies have used high-throughput screening techniques, such as whole genome microarray analyses, to perform transcriptome profiling in rodent brains following in vivo exposure to GSM-related EMF (Belyaev et al. 2006; Fragopoulou et al. 2018; McNamee et al. 2016; Nittby et al. 2008; Paparini et al. 2008). Two of these studies (McNamee et al. 2016; Paparini et al. 2008) have carried out their microarray analyses using statistical procedures incorporating an appropriate false discovery rate (FDR) adjustment for multiple comparison testing. Under these stringent conditions, Paparini et al. (2008) could not detect any EMF-induced modulation of gene expression assessed in the whole brain of mice after a 1-h whole-body exposure at an average whole-body SAR value of 1.1 W/kg. Also, McNamee et al. (2016) found no significant gene responses to EMF in different brain regions that were analyzed following 4-h daily whole-body exposures repeated for 5 days at (whole-body) average SAR values up to 1.45 W/kg. In both studies, the relevance of FDR adjustments was emphasized, by showing that gene responses detected in the absence of FDR adjustment could not be confirmed in semi-quantitative RT-PCR analyses.
So far, most of the reported in vivo molecular analyses including transcriptome profiling studies were carried out applying GSM-EMF to healthy rodents. However, recent investigations indicate that exposure to EMF influences preexisting neuropathological processes. In a rat model of epilepsy induced by pilocarpine treatment, proconvulsive activity of pilocarpine and its stimulating effect on Fos gene expression were enhanced when animals were submitted to a single whole-body exposure to GSM-900 MHz at an average SAR of 0.15 W/kg (Lopez-Martin et al. 2006). In transgenic mice modeling human Alzheimer diseases, chronic whole-body exposures to pulsed 918 MHz EMF (estimated SAR between 0.25 and 1.05 W/kg) or 1950 MHz (average SAR of 5 W/kg) could alleviate the neurodegenerative process, as indicated by improvement in cognitive behaviors and brain mitochondrial function, or reduction in reactive gliosis and ß-amyloid deposits (Arendash et al. 2010; Dragicevic et al. 2011; Jeong et al. 2015; Son et al. 2018). Glioblastoma-bearing rats submitted to daily head exposures to GSM 900 MHz, over a month (brain-averaged SAR of 0.5 W/kg), showed reductions in the numbers of apoptotic tumor cell and infiltrating immune cells albeit with no impact on animal survival (Ouadah et al. 2018).
Transient neuroinflammation associated to a sickness syndrome is most frequently observed in human populations, as a consequence of peripheral immune challenges triggered by an acute infection (Dantzer et al. 2008). This pathological condition is mimicked in rodents by lipopolysaccharide (LPS) injection (Dantzer 2001; Hoogland et al. 2015). Investigating brain cell responses triggered by RF in LPS-treated rats, we previously reported that a single 2-h head-only exposure to GSM-1800 MHz led to alterations in the neuroinflammatory process (Lameth et al. 2017; Occelli et al. 2018). These alterations involved modifications in the morphologies of proinflammatory effector cells such as microglia and reduced expression of the interleukin-1ß (Il1ß) gene. The effect could be detected as late as 24 h after the GSM exposure in the dorso-medial cerebral cortex where the average SAR value was estimated as 2.94 W/kg. GSM signals also impacted on the evoked activity of cortical neurons and neurotransmission, as indicated by the reduction of action potential emission evoked by sounds in the auditory cortex (Occelli et al. 2018) and by reduced phosphorylation of glutamatergic ionotropic alpha-amino-3-hydroxy-5 methyl-4-isoxazole propionic acid receptors (AMPARs) (Lameth et al. 2017). Importantly, these GSM-1800 MHz cell responses were not observed in the brain of healthy rats that were submitted to the same head-only exposure, indicating that the brain cell response to GSM signals was promoted by the LPS-induced neuroinflammation.
Our previous analyses of gene expression were limited to a set of proinflammatory genes that were assessed by reverse transcription-quantitative PCR (RT-qPCR). In the present study, we performed whole genome mRNA profiling of the cerebral cortex to further determine cortical gene responses to a single head-only exposure to GSM-1800 MHz, in the context of an acute neuroinflammation triggered by LPS. To get further insight into the effect of GSM-1800 MHz according to neuroinflammatory conditions, we also investigated the cortical gene response to the EMF in genetically modified rats expressing a mutated form of human (h) SOD1 gene (G93A mutation, hSOD1G93A) (Howland et al. 2002). The mutation is responsible for inherited human amyotrophic lateral sclerosis (ALS), a neurodegenerative disease characterized by a progressive loss of motor neurons associated with chronic inflammation in the spinal cord, the brainstem, and in the cerebral cortex (Al-Chalabi et al. 2017). Rodent expressing mutated hSOD1 are well-established models of human ALS (Philips and Rothstein 2015). They were used to uncover the role of neuroinflammation in the degeneration of motor neurons (Beers et al. 2006; Boillee et al. 2006; Chiot et al. 2019; Mesci et al. 2015) and to investigate how the progression of the disease could be affected by environmental factors, including extremely low-frequency EMF (Blacher et al. 2019; Poulletier de Gannes et al. 2009). A recent epidemiological study provides evidence for increased risk of ALS in association to exposure to RF emitted by GSM antennas (Luna et al. 2019).
We here report the modification of the coding transcriptome induced by GSM signals in the cerebral cortex of LPS-treated rats by mean of a RNA-seq analysis that reveals molecular signaling and CNS cell types affected by the EMF exposure. We used RT-qPCR to probe the effect of GSM signals in healthy rats, LPS-treated rats, and in presymptomatic ALS (hSOD1 G93A) rats (before clinical signs marked by a progressive paralysis) and show that the gene responses to RF vary according to the health status and the neuroinflammatory context.
Methods
Animals
Wild-type (WT) Sprague-Dawley rats obtained from Janvier Laboratories and transgenic hSOD1G93A rats (Sprague-Dawley background) grown up in our animal facility were housed in a humidity (50–55%) and temperature (22–24 °C) controlled facility on a 12-h/12-h light/dark cycle (light on at 7:30 a.m.) with free access to food and water. Adult male hSOD1G93A rats were obtained from male hSOD1G93A × female WT breeding. All procedures and experiments were conducted in accordance with the guidelines established by the European Communities Council Directive (2010/63/EU Council Directive Decree), with approval of the ethical committees Ile de France (CEAA N°5, approval number 01419.03) and Paris-Sud and Centre (CEEA N°59, approval number 03729.02). Health status of SOD1G93A rats was monitored by weekly assessment of weight and motor behavior from the age of 2 months. All exposure or sham exposure to GSM-EMF were carried out with 3-month-old male rats. In hSOD1G93A rats, this age falls in the presymptomatic phase, before the onset of the earliest clinical sign of the disease, which is characterized by weight peak before the animal starts to lose body weight due to muscle wasting linked to progressive motor neuron degeneration.
WT rats obtained from Janvier Laboratories were habituated to the colony rooms for at least 1 week before exposure (or sham exposure) to GSM-EMF with or without pretreatment with LPS. Rats were injected intraperitoneally (i.p.) with Escherichia coli LPS (250 μg/kg, serotype 0127:B8, Sigma) diluted in sterile endotoxin-free isotonic saline or with saline only (WT controls) 24 h before GSM or sham exposure.
EMF Exposure System and Procedure
The exposure system was identical to that used previously (Watilliaux et al. 2011) and has been described in detail elsewhere (Leveque et al. 2004). Briefly, a radiofrequency generator emitting a GSM electromagnetic field (RFPA, 1/8 duty factor) pulsed at 217 Hz (pulse emission every 4.6 ms for 546 μs) was connected to a four-output divider, allowing simultaneous exposure of four animals. Each output was connected to a loop antenna (Sama-Sistemi srl; Roma) enabling local exposure of the animal’s head. The loop antenna consisted of a printed circuit, with two metallic lines engraved in a dielectric epoxy resin substrate (dielectric constant εr = 4.6). At one end, this device consisted of a 1-mm-wide line forming a loop placed close to the animals’ head. As in previous studies (Lameth et al. 2017; Watilliaux et al. 2011), the incident power of the GSM signals was set to 0.2 W and specific absorption rates (SARs) were determined numerically using a numerical rat model with the finite difference time domain (FDTD) method (Yee 1966; Kunz and Luebbers 1993; Taflove 1995; Leveque et al. 2004). They were also determined experimentally in homogeneous rat model using a Vitek or Luxtron probe for measurement of temperature variation (Collin et al. 2016). In this case, SARs, expressed in W/kg, were calculated using the following equation: SAR = CΔT / Δt with C being the calorific capacity in J/(kg K), ΔT the temperature change in °K, and Δt the time in seconds. Numerically determined SAR values were compared with experimental SAR values obtained using homogenous models, especially in the equivalent rat brain area. The difference between the numerical SAR determinations and the experimentally detected SAR values was less than 30%.
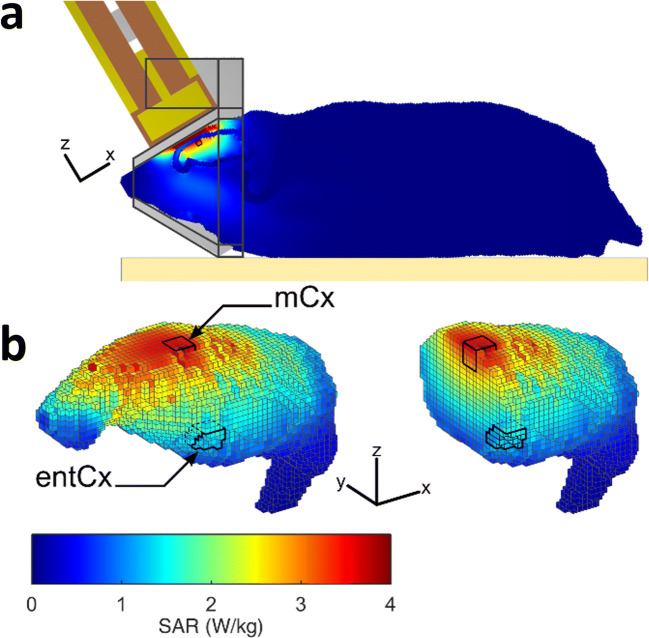
Figure 1 shows the SAR distribution in the rat brain with a rat model, which matched that of the rats used in our study in terms of weight and size. SAR values were the highest in cortical areas located directly below the loop antenna: they were 3.22 ± 0.27 W/kg (volume-averaged value ± SD among the SAR voxel values over the defined volume) in the motor cortex and dropped to 1.21 ± 0.07 W/kg in the entorhinal cortex, two cortical areas selected for biological analyses. The whole brain-averaged SAR was 1.3 ± 0.82 W/kg. As the weight of the exposed rats was homogenous, differences in tissue thickness at the level of the head were probably negligible, so the actual SAR in the cerebral cortex was probably very similar from one exposed animal to another.
Fig. 1.
Dosimetric analysis of specific absorption rates (SARs) in the rat brain during exposure to GSM-1800 MHz. The heterogeneous model of phantom rat and loop antenna described by Leveque et al. (2004) was used to evaluate the local SAR in the brain with a 0.5-mm3 cubic mesh. a Global view of the rat phantom in the exposure setup with the loop antenna above its head and the metal-free heat pad (yellow) under the body. b Distribution of SAR values in the adult brain at 0.5-mm3 spatial resolution. The cortical regions delimited by black contours in 3D whole brain view, or the sagittal section, correspond to the motor cortex (mCx) and the entorhinal cortex (entCx) where gene expression was analyzed. The color-coded scale of SAR values shown applies to all numerical simulations shown in the figure
Six groups of rats including two for hSOD1G93A, two for LPS-treated WT, and two for WT control rats (n = 6 to 7 animals per group) were used to analyze the effects of RF exposures (combined or not with LPS treatment or hSOD1G93A expression). Half of these groups were submitted to sham exposures. As in previous studies (Lameth et al. 2017), exposure of LPS-treated rats to GSM signals was carried out 24 h after the LPS injection. The animals were lightly anesthetized with ketamine/xylazine (ketamine 80 mg/kg, i.p.; xylazine 10 mg/kg, i.p.) before exposure to prevent movements and ensure reproducible position of the animal’ heads below the loop antenna emitting the GSM signals. Half the dose of the anesthetics was reinjected subcutaneously during the second hour of exposure to maintain the level of anesthesia. All the animals were awake less than 1 h following the end of the exposure. Half of the rats from the same cages (20 rats, WT treated or not with LPS and hSOD1G93A) were placed below the loop antenna with the energy of the GSM signals set to zero. The average weight of the exposed and sham-exposed animals was similar (exposed 511 ± 10 g; sham-exposed 530 ± 10 g; mean ± SEM, unpaired t test p = 0.19, ns). All the anesthetized animals were placed on a metal-free heating pad to maintain their body temperature around 37 °C throughout the experiment. As in previous experiments, the exposure duration was set at 2 h.
Tissue Preparation for Biochemical Analyses
Rats were killed by decapitation under isoflurane anesthesia; the brains were quickly removed from the skulls and frozen. Tissue samples were extracted from the motor cortex (mCx) and from the entorhinal cortex (entCx) in 100-μm-thick coronal sections cut on a cryostat and distributed over a cortical region starting 1.9-mm rostral to the bregma and extending over 2 mm in the rostro-caudal axis. Lumbar spinal cord was dissected out from hSOD1G93A rats and age-matched WT rats. Tissues were stored frozen at − 80 °C until use. RNAs were extracted from cortical tissues collected from the right and left hemispheres and from spinal cords.
RNA-seq and Data Analysis
Total RNA were extracted from the mCx collected from the right and left hemispheres of LPS-injected rats sham-exposed (n = 6) or exposed to GSM (n = 7) using Nucleospin miRNA kit (Macherey Nagel). Quality of RNA was confirmed on Agilent TapeStation (RIN > 8). RNA-seq libraries were prepared using TruSeq Stranded mRNA LT kit (Illumina) and sequenced with Illumina HiSeq 2500 platform (double-stranded 2 × 30′000 reads with 2 × 75 nt length per sample).
RNA-seq data analysis was performed by GenoSplice technology (www.genosplice.com). Sequencing, data quality, reads repartition (e.g., for potential ribosomal contamination), and insert size estimation were performed using FastQC, Picard-Tools, Samtools, and rseqc. Reads were mapped using STARv2.4.0 (Dobin et al. 2013) on the rn6 rat genome assembly. Gene expression regulation study was performed as already described (Noli et al. 2015). Briefly, for each gene present in the Rat FAST DB v2016_1 annotations, reads aligning on constitutive regions (that are not prone to alternative splicing) were counted. Based on these read counts, normalization and differential gene expression were performed using DESeq2 (Love et al. 2014) on R (v.3.2.5). Only genes expressed in at least one of the two compared experimental conditions were further analyzed. Genes were considered as expressed if their rpkm value was greater than 90% of the background rpkm value based on intergenic regions. Results were considered statistically significant for adjusted p values ≤ 0.05 and fold changes ≥ 1.2. Clusterings have been performed using “dist” and “hclust” functions in R, using Euclidean distance and ward agglomeration method. Analyses for enriched gene ontology (GO) terms were performed using DAVID Functional Annotation Tool (v6.8). GO terms and pathways were considered as enriched if fold enrichment ≥ 2.0, uncorrected p value ≤ 0.05, and minimum number of regulated genes in pathway/term ≥ 2.0. Analysis was performed three times: using all regulated genes, using upregulated genes, and using downregulated genes only. Union of these three analyses was made to provide a single list of results.
Reverse Transcription-Quantitative PCR
cDNA was synthesized from 400 ng of total RNA extracted from mCx or entCx (GSM- and sham-exposed rats) or from spinal cords (SOD1G93A tg and WT rats), using Maxima First-Strand cDNA synthesis kit (Thermo Scientific, Waltham, USA). Quantitative PCRs were performed with LightCycler 480 SYBR Green I Master Mix (Roche Diagnostics, France, Maylan). cDNA was amplified with primers for genes listed in Table 1, which were designed to span introns. Specificity of designed primers was checked by Ensembl BLAST analyses. The samples were amplified by 45 cycles (15 s at 95 °C, 1 min at 60 °C). Each reaction was performed in triplicate. Single peak dissociation curves with expected Tm were verified for each sample. Gene expression was normalized to TATA-box binding protein Tbp mRNA expression and analyzed by calculating 2−ΔCt values. Ct values for the assessed genes (Table 1) were all < 33. Differences between groups of animals (n = 6 or 7 rats) were analyzed using non-parametric Mann–Whitney U tests with Benjamini Hochberg multiple testing corrections. The FDR set at 0.05 was applied to determine significant differences.
Table 1.
List of the primers used in RT-qPCR analyses
| Foward primer (5′-3′) | Reverse primer (5′-3′) | |
|---|---|---|
| Tbp | TGGGATTGTACCACAGCTCCA | CTCATGATGACTGCAGCAAAC |
| Il1-β | CAACAAAAATGCCTCGTGC | TGCTGATGTACCAGTTGGG |
| Tnf-α | GCCAATGGCATGGATCTCAAAG | CAGAGCAATGACTCCAAA |
| Ccl5 | CCTTGCAGTCGTCTTTGTCA | ATCCCCAGCTGGTAAGGACT |
| Nos2 | CCTTGTTCAGCTACGCCTTC | GGTATGCCCGAGTTCTTTCA |
| Nox2 (gp91phox) | CCGTATTGTGGGAGACTGGA | CTTGAGAATGGAGGCAAAGG |
| Cox2 | CTACACCAGGGCCCTTCC | TCCAGAACTTCTTTTGAATCAGG |
| Parvg | AGAAGGTGCTGATGGAATGG | AGCCCATCAAACATGTCCTC |
| Akna | TGATGAGGAGCTGGATGAGA | GGCTGAGATCTCCCTGGTCT |
| Litaf | TGGGAAGGGCATGAATCCAC | GCGGTCGTAGAAGGAGATGG |
| Clu | TGAAGTTCTACGCACGCG | GGTCCCCGTTCATCCAGAAG |
| Fam107a | GGGCTGACATCGAGGGAC | GGGTTTAGCAGCTTCTTGGG |
| Adamts4 | GTGGACCAGCTCAAGGACTT | CAAGTCCTGGAGCAGTCACC |
| Micu3 | TGTGTGCTACCAGCTGTATGG | AAACGAAACCGTCTCTCTCG |
| Crlf1 | CTGACGTGCTCACACTGGAT | GTCCTCCAGGCCCCCAAC |
| Ppp4r3a | TGCATTTTCTCAAACGCTACA | TCCTGCATGACAAACTCTCG |
| Atf3 | GCTGGAGTCAGTCACCATCA | CCGCCTCCTTTTTCTCTCAT |
| Fos | TGGAGCCGGTCAAGAACATT | GCCGGAAACAAGAAGTCATCA |
| Cd11b | ATGCCGCGAAGATCCTAGTTGTCA | AATGGCAGCGCTAAAGCCTTCTTG |
Results
Gene Responses to GSM-1800 MHz Exposure in LPS-Treated Rats
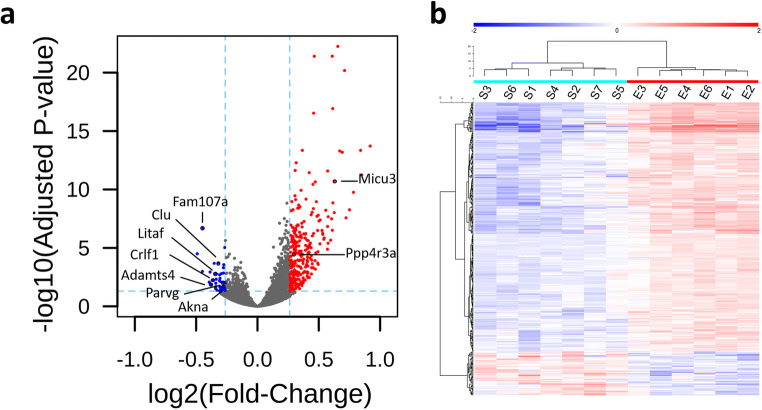
As in previous studies (Lameth et al. 2017; Occelli et al. 2018), adult male rats were exposed head only to GSM-1800 MHz for 2 h and the exposure was performed 24 h after the LPS injection. Based on previous evidence of gene responses in the dorso-medial region of the cerebral cortex (Lameth et al. 2017), we focused our investigation on an area of this brain region in which SAR values were maximal, making the assumption that local gene responses increase with local SAR values. Important for our study, this area corresponded to a sub region of the motor cerebral cortex (mCx, Fig. 1b) that is affected not only by LPS-triggered neuroinflammation but also in ALS. Dosimetric analyses showed that the mean SAR value reached 3.22 W/kg in the mCx that was selected for a whole genome mRNA analysis. Considering that altered molecular signaling could be observed 1 day after the GSM exposure (Lameth et al. 2017) and to determine gene responses that did not fade out within a few hours, gene expressions were assessed 24 h after the end of the GSM (n = 6 rats) or sham (n = 7) exposure, by means of RNA-seq analyses. We detected the expression of up to 11,971 genes including 10,476 annotated and 1495 unknown genes in this cortical area. Differential gene expression of LPS-treated/GSM-1800 MHz exposed rats relative to the LPS/sham control–exposed group was carried out applying a fold change (FC) cutoff of 1.2. Statistical analyses using an FDR-adjusted p value cutoff (p ≤ 0.05) showed that among the 11,971 expressed genes, a total of 321 genes were differentially expressed between GSM- and sham-exposed animals (Fig. 2a). FC values did not exceed 1.89 and averaged 1.29. Unsupervised hierarchical clustering of these RNA-seq data confirmed RF-induced modulations of the transcriptome, showing segregation without overlap between the sham and exposed groups (Fig. 2b). The differentially expressed genes (DEGs) consisted in 272 upregulated and 49 downregulated genes in response to the GSM exposure (Fig. 2a). Unknown genes were over-represented in the upregulated genes (42%, 115 unknown versus 157 annotated genes), whereas all but one (48) of the downregulated genes were annotated (Table 2).
Fig. 2.
RNA-seq analysis of the mCx in LPS-treated rats after sham or GSM exposure. RNA was extracted 24 h after GSM or sham exposures. a Volcano plot of differential gene expression between GSM and sham-exposed animals (FC cutoff 1.2). Genes that were significantly upregulated or downregulated in response to the GSM exposure are indicated by red or blue dots, respectively. Gray dots correspond to genes that were not significantly modulated. Genes selected for qPCR analyses are indicated. b Heat map showing color-coded normalized expression levels of the 321 genes, which were modulated by the GSM exposure (FC ≥ 1.2, adjusted p ≤ 0.05). Gene clustering was performed using Euclidean distance and ward agglomeration method. S(1–7) correspond to each of the sham-exposed rats; E(1–6) indicate GSM-exposed rats
Table 2.
Genes differentially expressed in the mCx of LPS-treated rats and GSM- and sham-exposed rats
| Gene symbol | Gene name | Regulation | Fold change | Adjusted p value |
|---|---|---|---|---|
| Bet1 | Bet1 golgi vesicular membrane trafficking protein | Up | 1.59 | 5.14E−14 |
| Micu3 | Mitochondrial calcium uptake family, member 3 | Up | 1.55 | 1.93E−11 |
| Pter | Phosphotriesterase related | Up | 1.53 | 3.09E−07 |
| Ttpa | Alpha tocopherol transfer protein | Up | 1.52 | 3.26E−08 |
| Ppp4r3b | Protein phosphatase 4, regulatory subunit 3B | Up | 1.49 | 2.17E−08 |
| Tmsbl1 | – | Up | 1.49 | 1.46E−07 |
| Usp34 | Ubiquitin-specific peptidase 34 | Up | 1.41 | 3.20E−07 |
| Sh3bgrl | SH3 domain binding glutamate-rich protein like | Up | 1.40 | 1.97E−08 |
| Mgat4c | MGAT4 family, member C | Up | 1.38 | 1.23E−04 |
| Dynlt3 | Dynein light chain Tctex-type 3 | Up | 1.37 | 3.64E−11 |
| Mcts1 | MCTS1, reinitiation and release factor | Up | 1.36 | 9.44E−07 |
| Marcks | Myristoylated alanine-rich protein kinase C substrate | Up | 1.36 | 8.95E−09 |
| Zfp458 | Zinc finger protein 458 | Up | 1.35 | 9.58E−06 |
| Katnbl1 | Katanin regulatory subunit B1 like 1 | Up | 1.35 | 3.87E−06 |
| Bcap29 | B cell receptor-associated protein 29 | Up | 1.35 | 7.50E−05 |
| Tmem126a | Transmembrane protein 126A | Up | 1.34 | 5.52E−06 |
| Cggbp1 | CGG triplet repeat binding protein 1 | Up | 1.33 | 2.38E−06 |
| Orc4 | Origin recognition complex, subunit 4 | Up | 1.33 | 3.55E−04 |
| Olfm3 | Olfactomedin 3 | Up | 1.33 | 7.60E−06 |
| Lrif1 | Ligand-dependent nuclear receptor interacting factor 1 | Up | 1.33 | 1.16E−03 |
| Trim59 | Tripartite motif-containing 59 | Up | 1.33 | 5.98E−05 |
| Ppp2r3c | Protein phosphatase 2, regulatory subunit B″, gamma | Up | 1.33 | 1.38E−04 |
| Zfp347 | Zinc finger protein 347 | Up | 1.33 | 1.61E−04 |
| Gtpbp8 | GTP-binding protein 8 (putative) | Up | 1.32 | 2.54E−03 |
| Dbp | D-box binding PAR bZIP transcription factor | Up | 1.32 | 2.74E−04 |
| Wif1 | Wnt inhibitory factor 1 | Up | 1.32 | 1.18E−03 |
| Ccdc34 | Coiled-coil domain containing 34 | Up | 1.31 | 4.10E−04 |
| Eif3j | Eukaryotic translation initiation factor 3, subunit J | Up | 1.31 | 1.56E−05 |
| Rpp30 | Ribonuclease P/MRP 30 subunit | Up | 1.31 | 6.68E−07 |
| Trim33 | Tripartite motif-containing 33 | Up | 1.31 | 1.06E−03 |
| Tmem47 | Transmembrane protein 47 | Up | 1.31 | 1.41E−06 |
| Ccng2 | Cyclin G2 | Up | 1.30 | 5.40E−08 |
| Gpr22 | G protein-coupled receptor 22 | Up | 1.30 | 6.33E−07 |
| Arhgap5 | Rho GTPase activating protein 5 | Up | 1.30 | 8.99E−05 |
| Sh2d1a | SH2 domain containing 1A | Up | 1.30 | 6.38E−03 |
| Syce2 | Synaptonemal complex central element protein 2 | Up | 1.30 | 9.73E−03 |
| Zfp40 | Zinc finger protein 40 | Up | 1.29 | 1.28E−04 |
| Plcxd3 | Phosphatidylinositol-specific phospholipase C, X domain containing 3 | Up | 1.29 | 3.58E−05 |
| Tmem65 | Transmembrane protein 65 | Up | 1.29 | 3.72E−05 |
| H2afz | H2A histone family, member Z | Up | 1.29 | 4.52E−14 |
| Spdya | Speedy/RINGO cell cycle regulator family member A | Up | 1.29 | 1.20E−02 |
| Ptpn4 | Protein tyrosine phosphatase, non-receptor type 4 | Up | 1.28 | 7.66E−08 |
| Spcs3 | Signal peptidase complex subunit 3 | Up | 1.28 | 5.32E−05 |
| Serp1 | Stress-associated endoplasmic reticulum protein 1 | Up | 1.28 | 2.94E−05 |
| Ptges3 | Prostaglandin E synthase 3 | Up | 1.28 | 1.19E−10 |
| Id2 | Inhibitor of DNA binding 2, HLH protein | Up | 1.28 | 8.06E−07 |
| Pp2d1 | Protein phosphatase 2C-like domain containing 1 | Up | 1.28 | 1.92E−03 |
| Mal2 | Mal, T cell differentiation protein 2 | Up | 1.28 | 1.05E−04 |
| Coq10b | Coenzyme Q10B | Up | 1.27 | 4.46E−04 |
| Ccdc126 | Coiled-coil domain containing 126 | Up | 1.27 | 3.63E−05 |
| Calm2 | Calmodulin 2 | Up | 1.27 | 2.41E−06 |
| Zfp192 | Zinc finger protein 192 | Up | 1.27 | 4.52E−07 |
| Fam172a | Family with sequence similarity 172, member A | Up | 1.27 | 2.50E−05 |
| Usp14 | Ubiquitin specific peptidase 14 | Up | 1.27 | 1.19E−06 |
| Cops2 | COP9 signalosome subunit 2 | Up | 1.27 | 1.28E−04 |
| Rnf152 | Ring finger protein 152 | Up | 1.27 | 4.97E−03 |
| Alg6 | ALG6, alpha-1, 3-glucosyltransferase | Up | 1.26 | 1.27E−03 |
| Prpf39 | pre-mRNA processing factor 39 | Up | 1.26 | 6.77E−06 |
| Taf1d | TATA-box binding protein associated factor, RNA polymerase I subunit D | Up | 1.26 | 1.85E−03 |
| Epm2aip1 | EPM2A interacting protein 1 | Up | 1.26 | 3.98E−09 |
| Rnf103 | Ring finger protein 103 | Up | 1.26 | 2.16E−09 |
| Cited2 | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 | Up | 1.26 | 1.15E−07 |
| Mblac2 | Metallo-beta-lactamase domain containing 2 | Up | 1.26 | 2.47E−03 |
| Taf13 | TATA-box binding protein associated factor 13 | Up | 1.26 | 1.06E−07 |
| Usmg5 | Upregulated during skeletal muscle growth 5 homolog (mouse) | Up | 1.26 | 1.10E−04 |
| Bcl2l11 | BCL2 like 11 | Up | 1.26 | 1.70E−03 |
| Ell2 | Elongation factor for RNA polymerase II 2 | Up | 1.25 | 2.68E−05 |
| RGD1307621 | Hypothetical LOC314168 | Up | 1.25 | 1.21E−02 |
| Wee1 | WEE1 G2 checkpoint kinase | Up | 1.25 | 3.50E−04 |
| Rpl22l1 | Ribosomal protein L22 like 1 | Up | 1.25 | 4.95E−03 |
| Tmed7 | Transmembrane p24 trafficking protein 7 | Up | 1.25 | 3.36E−06 |
| Med7 | Mediator complex subunit 7 | Up | 1.25 | 1.05E−03 |
| Ube2g1 | Ubiquitin-conjugating enzyme E2G 1 | Up | 1.25 | 1.33E−07 |
| Cmpk1 | Cytidine/uridine monophosphate kinase 1 | Up | 1.25 | 1.05E−06 |
| Rap2c | RAP2C, member of RAS oncogene family | Up | 1.25 | 6.93E−06 |
| Arl8b | ADP-ribosylation factor like GTPase 8B | Up | 1.24 | 5.18E−13 |
| Zfp367 | Zinc finger protein 367 | Up | 1.24 | 2.85E−05 |
| Pank3 | Pantothenate kinase 3 | Up | 1.24 | 8.17E−07 |
| Chm | CHM, Rab escort protein 1 | Up | 1.24 | 8.21E−06 |
| Lin7c | lin-7 homolog C, crumbs cell polarity complex component | Up | 1.24 | 3.09E−07 |
| Dcun1d1 | Defective in cullin neddylation 1 domain containing 1 | Up | 1.24 | 1.23E−03 |
| Pkia | Protein kinase (cAMP-dependent, catalytic) inhibitor alpha | Up | 1.24 | 2.27E−07 |
| Yipf4 | Yip1 domain family, member 4 | Up | 1.24 | 1.47E−05 |
| Gmfb | Glia maturation factor, beta | Up | 1.24 | 7.38E−10 |
| Slc35a3 | Solute carrier family 35 member A3 | Up | 1.24 | 1.85E−03 |
| LOC500584 | Similar to casein kinase 1, gamma 3 isoform 2 | Up | 1.24 | 3.06E−02 |
| RGD1306941 | Similar to CG31122-PA | Up | 1.24 | 2.42E−05 |
| Rnf138 | Ring finger protein 138 | Up | 1.24 | 1.67E−02 |
| Snx10 | Sorting nexin 10 | Up | 1.24 | 2.74E−06 |
| Cript | CXXC repeat containing interactor of PDZ3 domain | Up | 1.24 | 2.10E−04 |
| RGD1564541 | Similar to hypothetical protein FLJ22965 | Up | 1.24 | 4.55E−06 |
| Sub1 | SUB1 homolog, transcriptional regulator | Up | 1.24 | 2.36E−12 |
| Ube3a | Ubiquitin protein ligase E3A | Up | 1.24 | 4.36E−03 |
| Eif1ax | Eukaryotic translation initiation factor 1A, X-linked | Up | 1.24 | 3.44E−10 |
| Dnajb9 | DnaJ heat shock protein family (Hsp40) member B9 | Up | 1.23 | 3.17E−05 |
| Tceb1 | Transcription elongation factor B subunit 1 | Up | 1.23 | 3.09E−07 |
| Ppp3cb | Protein phosphatase 3 catalytic subunit beta | Up | 1.23 | 5.41E−07 |
| Dcaf12l1 | DDB1 and CUL4 associated factor 12-like 1 | Up | 1.23 | 5.67E−07 |
| Fgf13 | Fibroblast growth factor 13 | Up | 1.23 | 7.63E−04 |
| Kbtbd3 | Kelch repeat and BTB domain containing 3 | Up | 1.23 | 1.43E−03 |
| Sms | Spermine synthase | Up | 1.23 | 3.47E−04 |
| Osbpl8 | Oxysterol binding protein-like 8 | Up | 1.23 | 1.41E−05 |
| Pds5b | PDS5 cohesin associated factor B | Up | 1.23 | 2.16E−03 |
| Rnf139 | Ring finger protein 139 | Up | 1.23 | 3.54E−06 |
| Ppp4r3a | Protein phosphatase 4, regulatory subunit 3A | Up | 1.23 | 3.72E−05 |
| Gnpnat1 | Glucosamine-phosphate N-acetyltransferase 1 | Up | 1.23 | 1.16E−03 |
| Kctd12 | Potassium channel tetramerization domain containing 12 | Up | 1.23 | 3.96E−02 |
| Tdp2 | Tyrosyl-DNA phosphodiesterase 2 | Up | 1.23 | 1.63E−04 |
| Ccnc | Cyclin C | Up | 1.23 | 3.42E−03 |
| Ndufc2 | NADH:ubiquinone oxidoreductase subunit C2 | Up | 1.23 | 3.62E−07 |
| C1d | C1D nuclear receptor co-repressor | Up | 1.23 | 3.12E−02 |
| Ppp1r2 | Protein phosphatase 1, regulatory (inhibitor) subunit 2 | Up | 1.22 | 1.83E−07 |
| LOC499331 | Similar to hypothetical protein D030056L22 | Up | 1.22 | 3.01E−02 |
| Sar1b | Secretion associated, Ras-related GTPase 1B | Up | 1.22 | 1.33E−06 |
| Fancm | Fanconi anemia, complementation group M | Up | 1.22 | 4.17E−03 |
| Tmem41b | Transmembrane protein 41B | Up | 1.22 | 2.15E−03 |
| Trnt1 | tRNA nucleotidyl transferase 1 | Up | 1.22 | 3.75E−04 |
| Arl5b | ADP-ribosylation factor like GTPase 5B | Up | 1.22 | 8.98E−03 |
| RGD1309995 | Similar to CG13957-PA | Up | 1.22 | 1.83E−07 |
| Tnxa-ps1 | Tenascin XA, pseudogene 1 | Up | 1.22 | 3.63E−02 |
| Herc6 | HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 | Up | 1.22 | 1.82E−02 |
| Tbc1d19 | TBC1 domain family, member 19 | Up | 1.22 | 1.68E−03 |
| Slc7a11 | Solute carrier family 7 member 11 | Up | 1.22 | 1.40E−03 |
| Emc2 | ER membrane protein complex subunit 2 | Up | 1.22 | 1.41E−06 |
| Zfc3h1 | Zinc finger, C3H1-type containing | Up | 1.22 | 1.80E−06 |
| Naa30 | N(alpha)-acetyltransferase 30, NatC catalytic subunit | Up | 1.21 | 1.67E−04 |
| Ppp3r1 | Protein phosphatase 3, regulatory subunit B, alpha | Up | 1.21 | 1.09E−09 |
| Crebrf | CREB3 regulatory factor | Up | 1.21 | 1.67E−04 |
| Kcna4 | Potassium voltage-gated channel subfamily A member 4 | Up | 1.21 | 1.19E−02 |
| Ccne2 | Cyclin E2 | Up | 1.21 | 1.02E−03 |
| Herc4 | HECT and RLD domain containing E3 ubiquitin protein ligase 4 | Up | 1.21 | 1.92E−02 |
| Arl6ip6 | ADP ribosylation factor like GTPase 6 interacting protein 6 | Up | 1.21 | 1.66E−02 |
| Plpp6 | Phospholipid phosphatase 6 | Up | 1.21 | 5.79E−03 |
| Scamp1 | Secretory carrier membrane protein 1 | Up | 1.21 | 6.47E−09 |
| RGD1561931 | Similar to KIAA2022 protein | Up | 1.21 | 5.01E−04 |
| Rab9b | RAB9B, member RAS oncogene family | Up | 1.21 | 1.79E−04 |
| Triqk | Triple QxxK/R motif containing | Up | 1.21 | 1.95E−02 |
| Wwp1 | WW domain containing E3 ubiquitin protein ligase 1 | Up | 1.21 | 1.10E−04 |
| Hnmt | Histamine N-methyltransferase | Up | 1.21 | 3.06E−02 |
| Naa15 | N(alpha)-acetyltransferase 15, NatA auxiliary subunit | Up | 1.21 | 1.45E−04 |
| Fmr1 | Fragile X mental retardation 1 | Up | 1.21 | 1.53E−04 |
| Vps4b | Vacuolar protein sorting 4 homolog B | Up | 1.21 | 1.13E−07 |
| Ssr3 | Signal sequence receptor, gamma | Up | 1.21 | 5.18E−05 |
| Slf1 | SMC5-SMC6 complex localization factor 1 | Up | 1.21 | 8.87E−03 |
| Snap25 | Synaptosomal-associated protein 25 | Up | 1.21 | 8.67E−07 |
| Chka | Choline kinase alpha | Up | 1.21 | 1.88E−02 |
| Fam133b | Family with sequence similarity 133, member B | Up | 1.21 | 8.80E−06 |
| Zfp280d | Zinc finger protein 280D | Up | 1.21 | 6.20E−03 |
| Npy1r | Neuropeptide Y receptor Y1 | Up | 1.21 | 2.32E−02 |
| Dpy19l4 | dpy-19-like 4 (C. elegans) | Up | 1.20 | 3.61E−03 |
| Ccdc88a | Coiled coil domain containing 88A | Up | 1.20 | 3.40E−03 |
| Mob1b | MOB kinase activator 1B | Up | 1.20 | 5.17E−03 |
| Rock1 | Rho-associated coiled-coil containing protein kinase 1 | Up | 1.20 | 1.38E−02 |
| Rngtt | RNA guanylyltransferase and 5′-phosphatase | Up | 1.20 | 1.57E−02 |
| Dleu7 | Deleted in lymphocytic leukemia, 7 | Up | 1.20 | 1.68E−02 |
| Klhl7 | Kelch-like family member 7 | Up | 1.20 | 5.39E−05 |
| Ap3s1 | Adaptor-related protein complex 3, sigma 1 subunit | Up | 1.20 | 1.83E−03 |
| Shroom3 | Shroom family member 3 | Down | 1.41 | 3.16E−05 |
| Hes5 | hes family bHLH transcription factor 5 | Down | 1.37 | 1.04E−03 |
| Fam107a | Family with sequence similarity 107, member A | Down | 1.36 | 2.04E−07 |
| Akap17b | A kinase (PRKA) anchor protein 17B | Down | 1.31 | 8.54E−03 |
| Mob3a | MOB kinase activator 3A | Down | 1.31 | 1.15E−03 |
| Parvg | Parvin, gamma | Down | 1.30 | 1.45E−02 |
| Crlf1 | Cytokine receptor-like factor 1 | Down | 1.29 | 5.62E−03 |
| Fxyd1 | FXYD domain-containing ion transport regulator 1 | Down | 1.28 | 2.08E−04 |
| Litaf | Lipopolysaccharide-induced TNF factor | Down | 1.27 | 1.63E−03 |
| Sox9 | SRY box 9 | Down | 1.27 | 2.60E−02 |
| Creb3l1 | cAMP responsive element binding protein 3-like 1 | Down | 1.27 | 2.96E−02 |
| Htra3 | HtrA serine peptidase 3 | Down | 1.27 | 2.01E−02 |
| Dap | Death-associated protein | Down | 1.27 | 1.05E−02 |
| Bag3 | Bcl2-associated athanogene 3 | Down | 1.26 | 4.76E−03 |
| Rhpn2 | Rhophilin, Rho GTPase binding protein 2 | Down | 1.26 | 1.65E−03 |
| Cfap52 | Cilia and flagella associated protein 52 | Down | 1.26 | 4.17E−02 |
| Clu | Clusterin | Down | 1.25 | 2.02E−04 |
| Stat5a | Signal transducer and activator of transcription 5A | Down | 1.24 | 2.00E−03 |
| Shc1 | SHC adaptor protein 1 | Down | 1.24 | 6.42E−03 |
| Snx22 | Sorting nexin 22 | Down | 1.24 | 4.88E−02 |
| Meltf | Melanotransferrin | Down | 1.24 | 2.20E−02 |
| Zfp516 | Zinc finger protein 516 | Down | 1.24 | 4.01E−03 |
| Flnc | Filamin C | Down | 1.23 | 7.38E−03 |
| Gna13 | G protein subunit alpha 13 | Down | 1.23 | 2.07E−02 |
| Tnfrsf11b | TNF receptor superfamily member 11B | Down | 1.23 | 3.80E−02 |
| Mas1 | MAS1 proto-oncogene, G protein-coupled receptor | Down | 1.23 | 4.88E−02 |
| RGD1305464 | Similar to human chromosome 15 open reading frame 39 | Down | 1.23 | 2.82E−02 |
| Arhgap15 | Rho GTPase activating protein 15 | Down | 1.23 | 4.02E−02 |
| Sytl5 | Synaptotagmin-like 5 | Down | 1.23 | 2.58E−02 |
| Ston2 | Stonin 2 | Down | 1.23 | 1.58E−03 |
| Ikbke | Inhibitor of kappa light polypeptide gene enhancer in B cells, kinase epsilon | Down | 1.23 | 3.28E−02 |
| Lipe | Lipase E, hormone-sensitive type | Down | 1.22 | 3.92E−02 |
| Sod3 | Superoxide dismutase 3, extracellular | Down | 1.22 | 2.97E−02 |
| Hps1 | HPS1, biogenesis of lysosomal organelles complex 3 subunit 1 | Down | 1.22 | 3.30E−02 |
| Adamts4 | ADAM metallopeptidase with thrombospondin type 1 motif, 4 | Down | 1.22 | 9.01E−03 |
| Tor4a | Torsin family 4, member A | Down | 1.21 | 2.94E−02 |
| Cntrob | Centrobin, centriole duplication and spindle assembly protein | Down | 1.21 | 2.33E−02 |
| Clic4 | Chloride intracellular channel 4 | Down | 1.21 | 4.94E−04 |
| Szrd1 | SUZ RNA binding domain containing 1 | Down | 1.21 | 1.40E−03 |
| Akna | AT-hook transcription factor | Down | 1.21 | 1.90E−02 |
| Rlbp1 | Retinaldehyde binding protein 1 | Down | 1.21 | 2.71E−04 |
| Pmepa1 | Prostate transmembrane protein, androgen induced 1 | Down | 1.21 | 1.93E−02 |
| Bmpr1b | Bone morphogenetic protein receptor type 1B | Down | 1.21 | 2.84E−02 |
| Egfl7 | EGF-like domain, multiple 7 | Down | 1.20 | 2.89E−02 |
| Phospho1 | Phosphoethanolamine/phosphocholine phosphatase 1 | Down | 1.20 | 9.34E−06 |
| Gstt3 | Glutathione S-transferase, theta 3 | Down | 1.20 | 3.37E−02 |
| Scamp2 | Secretory carrier membrane protein 2 | Down | 1.20 | 4.37E−02 |
| Fam181b | Family with sequence similarity 181, member B | Down | 1.20 | 9.84E−03 |
Gene Ontology–Based Analysis of Differentially Expressed Genes Following RF Exposure
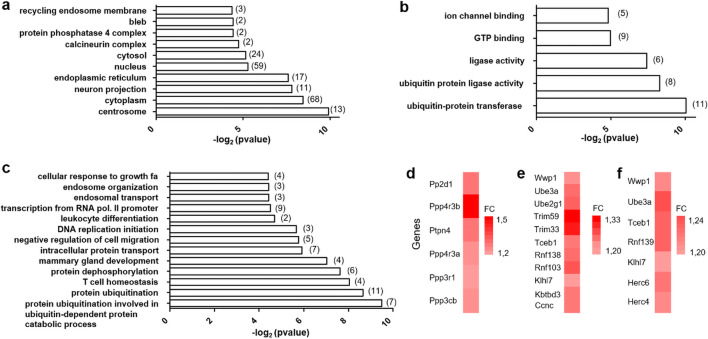
To further specify the transcriptional remodeling triggered by RF exposure, we performed a GO-based enrichment analysis to map DEG with respect to cellular components, biological processes, or molecular functions (Fig. 3).
Fig. 3.
Gene ontology (GO) annotation of the differentially expressed genes (DEGs) between GSM and sham-exposed rats. All the animals were treated with LPS. Significantly enriched GO terms and enrichment p values are indicated, with respect to cellular component (a), molecular function (b), and biological process (c). The number of DEG related to the GO terms is in brackets. Color-coded bars (d–f) indicates the names and fold changes (FCs) of the DEG associated to three GO terms (biological process): protein dephosphorylation (d), protein ubiquitination (e), and protein ubiquitination involved in ubiquitin-dependent protein catabolic process (f)
Besides the cytoplasm that contain proteins encoded by the largest DEG groups (68 genes), significant enrichment in GO terms related to “cellular component” corresponded to a limited subset of organelles including the nucleus, the endoplasmic reticulum, the centrosomes, or other cellular compartments such as the endosomal recycling system or neuronal projections. Other enriched GO terms corresponded to molecular complexes endowed with serine/threonine phosphatase activities such as calcineurin complex and protein phosphatase 4 complex (Fig. 3a). In keeping with the cellular distribution of DEG products, DEG mapping according to “biological process” led to significant enrichments in GO terms covering molecular activities localized in the nucleus (DNA transcription or replication) or related to endosomes or phosphatase complexes (“protein dephosphorylation,” Fig. 3c, d). However, the most significant enrichments corresponded to protein ubiquitination and suggested that RF exposure not only affected mRNA synthesis or degradation but also protein stability (Fig. 3c, e, f). Consistently, enriched GO terms related to molecular functions included enzymatic processes involved in protein ubiquitination but also highlighted molecular mechanism in signal transduction such as GTP or ion channel binding (Fig. 3b).
RT-qPCR Assessment of Gene Responses to GSM-1800 MHz in LPS-Treated Rats
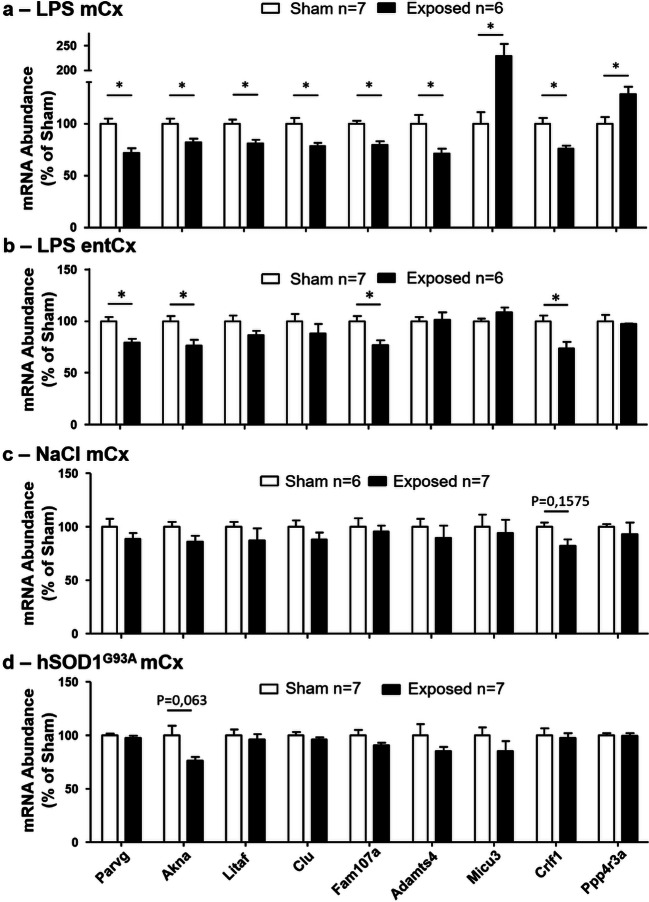
Cortical RNA used for RNA-seq analyses originated from the different cortical cell types including neurons and glial cells such as oligodendrocytes, oligodendrocyte progenitors, astrocytes, and microglia. To gain insight into how these different cell types contributed to the gene response to RF, the cellular distribution of DEG was analyzed using gene expression databases established from defined cell types or single cells derived from the mouse cerebral cortex (Zhang et al. 2014; Zeisel et al. 2015). Part of the DEGs appeared to be preferentially or selectively expressed in glia or in glial subtypes, among which Adamts4 (oligodendrocytes), Clu (astrocytes), Litaf (oligodendrocytes, microglia), and Fam107a (astrocytes, oligodendrocytes), whereas other genes including Micu3, Crlf1, and Ppp4r3a were expressed in neurons or in both neurons and glia. Although microglia account for less than 15% of the cortical non-neuronal cells (Ling and Leblond 1973), Parvg, a gene selectively and strongly expressed by microglia, was identified as a DEG whose expression was reduced in response to RF exposure. Akna, another microglia-expressed gene, was also affected by RF exposure. These genes were selected to verify, by RT-qPCR, the effect of RF on the level of transcripts. RT-qPCR analyses of the mCx from sham or GSM-exposed animals confirmed that the levels of transcripts from the selected genes were significantly enhanced or reduced as in our RNA-seq analysis, in response to the GSM exposure (Fig. 4a). Consistent with the range of FC values determined in RNA-seq analyses (Fig. 2a, Table 2), the optimal RF effect detected in RT-qPCR analyses corresponded to Micu3 gene (Fig. 4a).
Fig. 4.
RT-qPCR analyses of nine genes selected from RNA-seq data. RNA were extracted from the mCx (a, c, d) or the entCx (b) 24 h after sham or GSM exposure in LPS-injected rats (a, b), control saline-injected rats (c), and in hSOD1G93A rats (d). The mRNA levels were normalized to mean values from sham-exposed animals. Open bars correspond to sham rats, and black bars correspond to GSM-exposed rats. Data are means ± SEM from six or seven animals. *p < 0.05, Mann–Whitney test with Benjamini–Hochberg multiple testing corrections
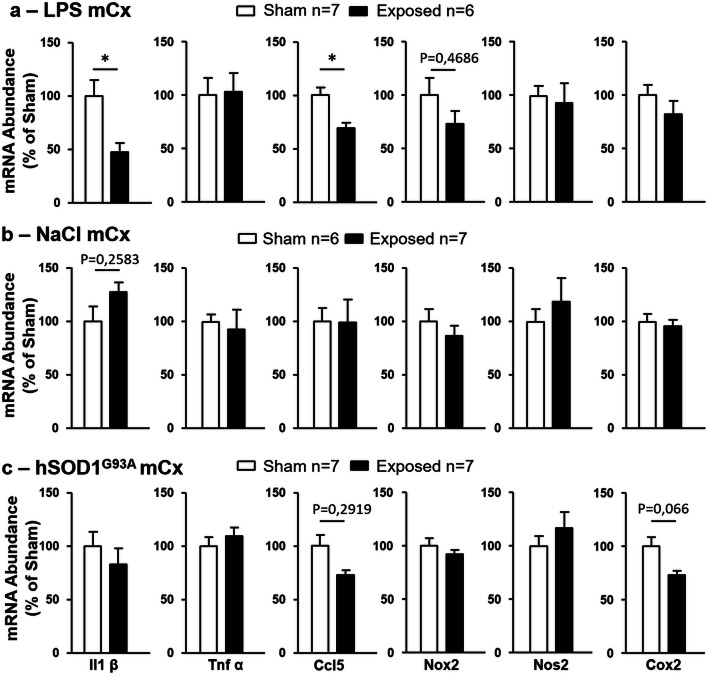
To determine whether gene responses to GSM-EMF could also be detected in cortical regions in which SAR values was below those determined at the level of the mCx, RT-qPCR analyses of the selected DEG were performed in the entorhinal cortex (entCx) where the mean SAR value was reduced to 1.21 W/kg. Comparison of the level of entCx transcripts in sham- and GSM-exposed animals showed that the expression of four of the nine selected DEG (Parvg, Akna, Fam107a, and Crlf1) was significantly reduced in response to GSM exposure (Fig. 4b).
LPS injection swiftly but transiently stimulates microglia expression of proinflammatory genes encoding cytokines (Hoogland et al. 2015). We have shown previously that in developing (15-day old) or young adult Wistar rats (2-month old) treated with LPS, a 2-h head exposure to GSM 1800 MHz at local mean SAR values of 2.9 W/kg resulted in reductions in cortical levels of IL-1ß mRNA. In the current study carried out with 3-month-old Sprague-Dawley rats, the expression of Il1ß or Ccl5 genes assessed in the mCx 48 h after LPS injection (e.g., 24 h after sham or GSM exposure) had already dropped to a level that precluded quantification in our RNA-seq analysis. However performing RT-qPCR, we could detect Il1ß and Ccl5 transcripts in the mCx of both sham and GSM-exposed rats and we found that GSM exposure resulted in significant reduction in the abundance of these transcripts, whereas the EMF had no effect on the local expression of other proinflammatory genes encoding Tnf-alpha or enzymes catalyzing the production of bioactive lipids or oxygen free radicals (Nox2, Nos2, Cox2) (Fig. 5a). RT-qPCR assessment of Il-1ß and Ccl5 genes in the entCx (1.21 W/kg) showed no significant differences between GSM- and sham-exposed animals (not shown).
Fig. 5.
RT-qPCR analyses of proinflammatory gene responses to GSM exposure in LPS-injected (a), control saline-injected (b), and hSOD1G93A rats (c). Data are means ± SEM from six to seven animals. *p < 0.05, Mann–Whitney test with multiple testing corrections
Effect of GSM-1800 MHz on Gene Expression in Healthy Rats
To address the importance of the LPS-triggered inflammation in gene responses uncovered by our RNA-seq analysis, healthy rats (untreated with LPS) were submitted to GSM or sham head exposure before RT-qPCR assessment of gene expressions in the mCx (SAR, 3.22 W/Kg). In these healthy rats, GSM signals had no effect on the level of expression of the nine selected genes that were responsive to GSM under LPS-triggered neuroinflammation (Fig. 4c). As expected from our previous study (Lameth et al. 2017), GSM signals had also no significant effect on the expression of proinflammatory genes including Il1ß and CCL5 in the healthy rats (Fig. 5b). These results indicate that LPS-triggered acute neuroinflammation promotes cortical gene responses to GSM-1800 MHz.
Effect of GSM Signals in hSOD1G93A Rats
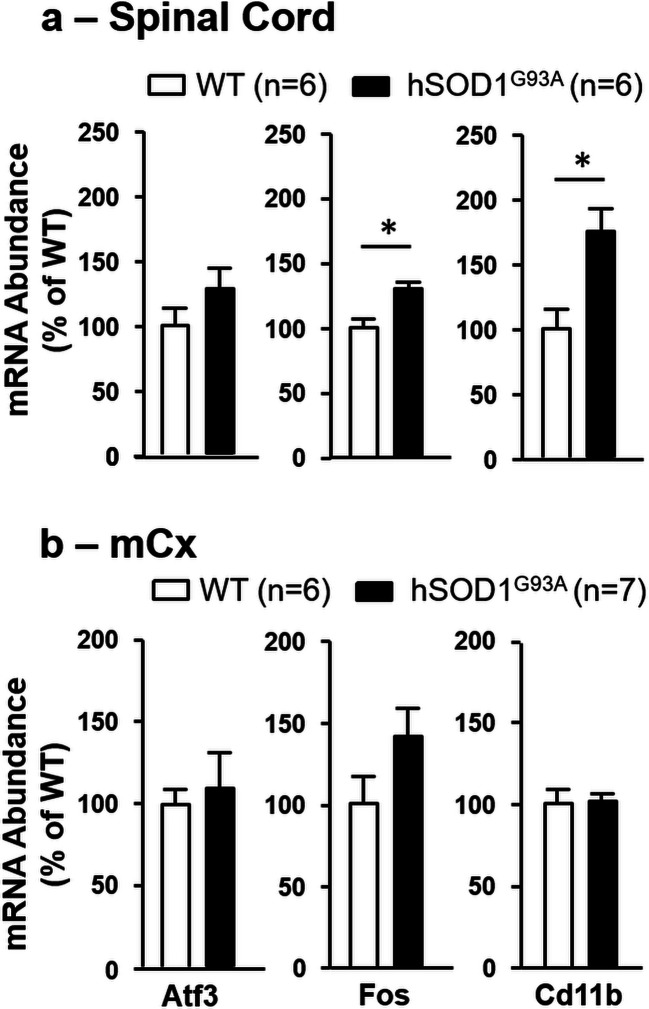
To further explore the effect of GSM-1800 MHz according to proinflammatory neuropathologies, we used the hSOD1G93A rat model to look for cortical gene response to GSM signals in rats primed for the development of a chronic neuroinflammatory process affecting CNS motor area including the mCx. As for LPS-treated or healthy rats, hSOD1G93A rats were exposed to GSM-EMF at the age of 3 months, before the onset of clinical signs of ALS. RT-qPCR assessment of three early markers of the disease (Atf3, Fos, Cd11b) (Lobsiger et al. 2007; Vlug et al. 2005) showed that despite the absence of clinical signs, hSOD1G93A rats submitted to the GSM exposure were already affected by the neuropathological process, as indicated by increased levels of Fos, and Cd11b transcripts in the spinal cord compared with WT controls (Fig. 6a). These early alterations in gene expression were not observed at the level of the mCx (Fig. 6b).
Fig. 6.
RT-qPCR analyses of ALS early stimulated genes in the lumbar spinal cord and the mCx from hSOD1G93A and age-matched (3-month old) WT rats. RNA was extracted from the spinal cord (a) or the mCx (b) 24 h after sham exposure. mRNA levels were normalized to mean values from WT animals. The data are means ± SEM from six or seven animals. *p < 0.05, Mann–Whitney test with multiple testing corrections
RT-qPCR analyses of the mCx of sham and GSM-exposed hSOD1G93A rats showed no significant effects of the GSM signals on the expression of the nine genes selected from RNA-seq analyses. Notably however, there was a trend toward a reduction in the expression Akna in the GSM-exposed animals with a p value (p = 0.063) close to the threshold for statistical significance (Fig. 4d). Assessment of genes encoding proinflammatory mediators showed no statistically significant effect of the GSM-EMF, albeit with a trend toward reduction in the expression of the Ccl5 (p = 0.212) and Cox2 (p = 0.066) genes (Fig. 5c).
Discussion
In this study, we have investigated gene responses to GSM-1800 MHz signal in the cerebral cortex of rats affected or not by neuropathological processes. Our RNA-seq analysis provides a comprehensive determination of coding transcriptome modifications in the context of an acute neuroinflammation and highlights molecular signaling that can be modified following a GSM-EMF exposure. Our study also shows that an acute neuroinflammation promotes gene responses to GSM signals.
GSM-Induced Modulations in Cortical Gene Expression: Implications in Cell Signaling
Our previous studies have shown that in adult rats undergoing a LPS-triggered neuroinflammation, a single 2-h exposure to GSM-1800 MHz resulted in reduced expression of the Il1ß gene in the dorso-medial cerebral cortex (Lameth et al. 2017). Focusing on a motor area of the dorso-medial cerebral cortex in which the mean SAR value was 3.22 W/kg, our RNA-seq analysis specifies the extent of the gene response assessed 24 h after the end exposure. Applying a FC threshold of 1.2, significant changes were limited to 2.7% of the expressed genes detected in our RNA-seq analysis. The EMF-induced gene modulations were moderated considering that all FC values were < 2. However, GO analyses uncovered potential modifications in biological processes that impact on proteostasis and intracellular signaling. The most significant modification (p < 0.005) corresponded to GO terms related to protein ubiquitination including proteasome-dependent catabolic processes. Ubiquitination is a multistep posttranslational modification that involves an enzymatic cascade leading to the attachment of a 15-kd ubiquitin peptide or polyubiquitin chains to lysine residues of protein substrates. According to the residues involved in ubiquitin-to-ubiquitin linkages, polyubiquitin chains act as proteolytic signal by targeting substrates for proteasome-mediated degradation, or control other processes like protein interaction signaling, endocytosis of membrane receptors, or DNA repair (Chen and Sun 2009; Swatek and Komander 2016). Our study reveals GSM-induced upregulation of several genes encoding E3 ubiquitin ligases or components of E3 ubiquitin ligase complexes that catalyze the final step of protein ubiquitination and determine the specificity of ubiquitinated substrates (Zheng and Shabek 2017). We also found enhanced expression of transcripts encoding two ubiquitin-specific proteases (Table 2), including USP14 that suppresses protein degradation through deubiquitination of proteasome substrates, non-catalytic inhibition of proteasome activity, and by regulating autophagy (de Poot et al. 2017; Xu et al. 2016).
In addition to ubiquitination and protein catabolism, our GO analysis has highlighted significant impact of RF on genes encoding phosphatase subunits, indicating that the cerebral effects of GSM signals could rely on EMF-induced posttranslational modification of the proteome. In keeping with this hypothesis, we previously found (Lameth et al. 2017) that in LPS-treated rats, a same head exposure to GSM 1800 MHz impacted on glutamate-mediated excitatory neurotransmission, as indicated by a reduced phosphorylation of serine residues (Ser 845 and 831) in the gluA1 subunit of cortical AMPAR, which are known to affect the conductance of AMPAR channel and synaptic plasticity. Alterations in excitatory (glutamate-mediated) neurotransmission were further documented by electrophysiological recording in the primary auditory cortex, showing GSM-induced reduction in spontaneous and sound-evoked firing rate (Occelli et al. 2018). Our current study strongly suggests that the reduced phosphorylation of gluA1 subunit implicates GSM-induced upregulation of protein phosphatase 3 (also called calcineurin). This Ca2+-calmodulin–dependent serine/threonine phosphatase catalyzes dephosphorylation of gluA1 Ser 845 (Hell 2016; Sanderson et al. 2012; Sanderson et al. 2018) whereas our RNA-seq analysis shows that the GSM exposure triggered increased expression of genes encoding catalytic and regulatory subunits (Ppp3cb, Ppp3r1) of this enzyme and its calmodulin activator (Calm2 in Table 2).
Changes in ubiquitin-mediated signaling could also contribute to GSM-triggered alterations in cortical excitatory neurotransmission. In line with this assumption, we observed that GSM exposure led to significant increases in the level of Ube3a transcripts. Ube3a encodes an ubiquitin E3 ligase that is expressed to high levels in cortical neurons and promotes ubiquitin-mediated degradation of synaptic proteins (Greer et al. 2010). Over-expression of Ube3a was found to impair excitatory synaptic transmission at both presynaptic and postsynaptic levels (Smith et al. 2011).
GSM-1800 MHz Affects Genes Expressed by Glial Cells
Microglia play key role in the intracerebral propagation of the LPS-triggered neuroinflammation through upregulation of genes encoding proinflammatory cytokines and chemokines (Hoogland et al. 2015). This microglial gene response peaks within hours following a single injection with LPS and was significant in the rat cerebral cortex at the time of GSM or sham exposure (e.g., 24 h after LPS injection) (Lameth et al. 2017). Despite rapid decay of this LPS-triggered microglial response, our RT-qPCR analysis shows that exposure to GSM-1800 MHz led to reduced expression of two genes encoding proinflammatory cytokines (Il1ß and Ccl5), providing evidence that GSM signals impact on the microglia-mediated neuroinflammatory reaction. Furthermore, our RNA-seq analysis uncovered other microglial genes, which are downregulated by exposure to GSM-EMF, including Akna and Litaf that encode transcription factors. While their role in LPS-mediated neuroinfammation remain to be specified, Litaf promotes expression of CCL5 and other cytokines in LPS-treated peripheral macrophages (Tang et al. 2006) whereas expression of proinflammatory cytokines was found to be under control of Akna in the lungs or in hypophysis cells (Liu et al. 2017; Ma et al. 2011).
We have shown previously that exposure of LPS-treated rats to GSM-1800 MHz signal resulted in the extension of microglia cell processes. The mechanism underlying this cell response is not determined but could possibly involve integrin signaling that was found to promote elongation of processes in microglia (Meller et al. 2017; Ohsawa et al. 2010). In line with this hypothesis, our RNA-seq analysis reveals that the GSM exposure reduces expression of Parvg, a microglia-expressed gene that is known to serve in integrin outside-in signal to actin cytoskeleton (Yoshimi et al. 2006).
Besides microglial DEG, our results provide evidence for GSM-induced alteration in other glial cell types. We found no increase in the expression of GFAP, a gene response that hallmarks astrocyte reactions in many pathological contexts and which have been inconstantly detected in the brain of rodents exposed to GSM signals (see “Introduction” section for references). However in our study, GSM exposure resulted in reduced expression of genes encoding other astrocyte markers including Sox9 (Table 2) (Sun et al. 2017). We also found a significant downregulation of Adamts 4 that is primarily expressed in oligodendrocytes (Levy et al. 2015). Thus, our data provide evidence that a 2-h exposure of cortical tissues (mCx) at a SAR level of 3.22 W/kg trigger gene responses in neurons as well as in the three major glial cell types. While our RNA-seq analysis was limited to mCx, RT-qPCR analysis of nine selected DEG showed that four of these genes (Fam107a, Parvg, Akna, and Crlf1) remained responsive to GSM-1800 MHz in the entCx where the mean SAR level was reduced to 1.21 W/kg. Consistent with previous demonstration that cortical microglia morphologically react to SAR values in the range of 1.5 W/kg (Occelli et al. 2018), two of the four responsive DEG corresponded to microglia-expressed genes (Akna and Parvg), emphasizing the sensitivity of microglia to a GSM exposure in LPS-treated rats.
Cortical Gene Responses to GSM-1800 MHz Vary According to Pathological States
Considering the broad incidence of acute neuroinflammatory episodes caused by infections or other diseases in human populations, it is important to determine if these pathological states modify responsiveness of brain cells to a GSM exposure. In former studies, applying a same head-only GSM exposure to healthy developing or adult rats, we could not detect any significant cortical cell responses when assessing heat shock proteins, proinflammatory mediators, microglia or astroglial cell markers, molecular effectors involved in excitatory neurotransmission, or electrophysiological activities of neurons (Lameth et al. 2017; Occelli et al. 2018; Watilliaux et al. 2011). Our current investigation extends these observations showing no effect of GSM exposure on the expression of proinflammatory genes or RNA-seq–related DEG in the mCx (3.22 W/kg) of healthy rats. These negative results indicate that LPS-triggered neuroinflammation can promote gene responses to GSM signals and supports our contention that the gene responses contribute to the GSM-induced changes on neuronal activity, neurotransmission, or microglial cell morphologies, which were observed in LPS-treated rats (Lameth et al. 2017; Occelli et al. 2018).
Besides acute pathological states, a wealth of evidence indicates that neuroinflammation contributes importantly to the progression of chronicle neurodegenerative diseases, including ALS (Chiot et al. 2019; Heneka et al. 2014). ALS rodents expressing the hSOD1G93A mutation display a progressive neuroinflammation marked by microglial activation profiles that differ from those triggered by LPS (Chiu et al. 2013; Hirbec et al. 2018). As a first investigation of the gene response to GSM-1800 MHz in ALS rats, we chose to avoid any bias due to manipulation of rodents with reduced body weight and altered skeletal muscles. In hSOD1G93A rats, the head exposure to GSM-EMF was carried out at a presymptomatic phase of the disease, albeit near to the weight peak timepoint that marks the onset of clinical signs. At this stage of the disease, we could detect altered expressions of genes including enhanced levels of Cd11b transcripts in the spinal cord, which were indicative of an early microglial activation. However, these gene modifications were not detected at the level of the mCx, and we found no significant effect of the GSM exposure on the cortical expression of RNA-seq–related DEG or proinflammatory genes. Interestingly however, our qPCR analysis indicates a trend toward reduction in the cortical expression of Cox2 (p = 0.066) and Akna (p = 0.063), two genes shown to be expressed in microglia purified from WT or hSOD1G93A mice (Chiu et al. 2013). This trend was not observed in age-matched healthy rats exposed to GSM-EMF, suggesting that significant gene responses to GSM exposure could be observed in the mCx at more advanced stages of the neuroinflammatory process in ALS rats. Considering the altered expression of genes, including a microglial gene in the spinal cord of ALS rats (Fig. 6), our data raise the possibility that whole-body GSM exposures at presymptomatic stages may affect the transcriptome and the nascent inflammation in the spinal cord, with or without impact on the onset of the clinical disease.
Limitations and Concluding Remarks
In this study, we have addressed the extent of transcriptome modifications induced by a 2-h head-only exposure to GSM-1800 MHz in the mCx of LPS-treated rats. As an experimental constraint, the exposure was carried out under (ketamine/xylazine) anesthesia to prevent head movement relative to the loop antenna, which could strongly and unpredictably modify the levels and distribution of cortical SAR values (Fig. 1). Ketamine was shown to affect neuronal activity and expression of genes, during the sedation period (Jacobson et al. 2019). Whether anesthesia alters brain cell responsiveness to GSM-1800 MHz could not be ruled out. However, anesthesia avoided acute stress that would have been generated by the 2-h restrained conditions necessary to expose awake animals, which could also interfere with the effects of the EMF on the brain. Our data show moderate but significant modulation of genes triggered by the GSM signals and reveal molecular signaling that can be affected by the GSM-1800 MHz exposure. The molecular responses were detected in the motor cortex where the local SAR level reached 3.22 W/kg. This value is clearly above the upper limit set for exposure of the general public, which is 2 W/kg in tissues as close as possible to the emitting device (ICNIRP 1998). Our study shows that the gene response is attenuated in a cortical region (entCx) where the local SAR value dropped to 1.21 W/kg. Further investigation will be needed to evaluate gene responses under power conditions that match the common uses of mobile phones. Considering the time lag between the EMF exposure and the gene assessment, our results reveal gene responses that are delayed or that last up to at least 24 h after the GSM exposure, but their sustainability remains unknown. We found that the LPS-triggered acute inflammation promotes significant gene responses to GSM signals, which were not detected in healthy rats or in ALS rats exposed to GSM signals at a presymptomatic stage of the disease. However, our results provide some evidence that gene responses could be promoted at a more advanced stage of ALS. The potentiation of GSM-induced gene responses, according to the stages or the types of neuroinflammatory reaction, remains to be further explored. Understanding of how pathological conditions affect CNS cell response to GSM-EMF should help to better appraise noxious, or potentially beneficial, effects due to the EMF environment.
Acknowledgments
We thank the ICM core facility IGENSEQ, which received funding from the program “Investissements d’avenir” ANR-10-IAIHU-06, and the technical staff from CEF (Centre d’expérimentation fonctionnelle). We also thank Noémie Robil and Pierre de la Grange from GenoSplice for RNA-seq analyses; Delphine Bouteiller, Emeline Mundwiller, and Yannick Marie from IGENSEQ for RNA-seq sample preparations; and Annie Gervais, Catherine Colin, Jean-Baptiste Hure, and Carlos Parras for technical advices and stimulating discussions.
Funding Information
This research was supported by the French National Research Program for Environmental and Occupational Health of Anses (grants 2015/2 RF/12 and 2018/2 RF/16).
Compliance with Ethical Standards
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethical Approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. All procedures and experiments were conducted in accordance with the guidelines established by the European Communities Council Directive (2010/63/EU Council Directive Decree). All protocols were approved by the institutional ethics committees Ile de France (CEAA N°5, approval number 01419.03) and Paris-Sud and Centre (CEEA N°59, approval number 03729.02).
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Al-Chalabi A, van den Berg LH, Veldink J. Gene discovery in amyotrophic lateral sclerosis: implications for clinical management. Nat Rev Neurol. 2017;13:96–104. doi: 10.1038/nrneurol.2016.182. [DOI] [PubMed] [Google Scholar]
- Arendash GW, Sanchez-Ramos J, Mori T, Mamcarz M, Lin X, et al. Electromagnetic field treatment protects against and reverses cognitive impairment in Alzheimer’s disease mice. J Alzheimers Dis. 2010;19:191–210. doi: 10.3233/JAD-2010-1228. [DOI] [PubMed] [Google Scholar]
- Beers DR, Henkel JS, Xiao Q, Zhao W, Wang J, et al. Wild-type microglia extend survival in PU.1 knockout mice with familial amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2006;103:16021–16026. doi: 10.1073/pnas.0607423103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyaev IY, Koch CB, Terenius O, Roxstrom-Lindquist K, Malmgren LO, et al. Exposure of rat brain to 915 MHz GSM microwaves induces changes in gene expression but not double stranded DNA breaks or effects on chromatin conformation. Bioelectromagnetics. 2006;27:295–306. doi: 10.1002/bem.20216. [DOI] [PubMed] [Google Scholar]
- Blacher E, Bashiardes S, Shapiro H, Rothschild D, Mor U, et al. Potential roles of gut microbiome and metabolites in modulating ALS in mice. Nature. 2019;572:474–480. doi: 10.1038/s41586-019-1443-5. [DOI] [PubMed] [Google Scholar]
- Boillee S, Yamanaka K, Lobsiger CS, Copeland NG, Jenkins NA, et al. Onset and progression in inherited ALS determined by motor neurons and microglia. Science. 2006;312:1389–1392. doi: 10.1126/science.1123511. [DOI] [PubMed] [Google Scholar]
- Bouji M, Lecomte A, Hode Y, de Seze R, Villegier AS. Effects of 900 MHz radiofrequency on corticosterone, emotional memory and neuroinflammation in middle-aged rats. Exp Gerontol. 2012;47:444–451. doi: 10.1016/j.exger.2012.03.015. [DOI] [PubMed] [Google Scholar]
- Brillaud E, Piotrowski A, de Seze R. Effect of an acute 900MHz GSM exposure on glia in the rat brain: a time-dependent study. Toxicology. 2007;238:23–33. doi: 10.1016/j.tox.2007.05.019. [DOI] [PubMed] [Google Scholar]
- Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33:275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- Chiot A, Lobsiger CS, Boillee S. New insights on the disease contribution of neuroinflammation in amyotrophic lateral sclerosis. Curr Opin Neurol. 2019;32:764–770. doi: 10.1097/WCO.0000000000000729. [DOI] [PubMed] [Google Scholar]
- Chiu IM, Morimoto ET, Goodarzi H, Liao JT, O’Keeffe S, et al. A neurodegeneration-specific gene-expression signature of acutely isolated microglia from an amyotrophic lateral sclerosis mouse model. Cell Rep. 2013;4:385–401. doi: 10.1016/j.celrep.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin A, Perrin A, Cretallaz C, Pla S, Arnaud-Cormos D, et al. In vivo set-up characterization for pulsed electromagnetic field exposure at 3 GHz. Phys Med Biol. 2016;61:5925–5941. doi: 10.1088/0031-9155/61/16/5925. [DOI] [PubMed] [Google Scholar]
- Court-Kowalski S, Finnie JW, Manavis J, Blumbergs PC, Helps SC, et al. Effect of long-term (2 years) exposure of mouse brains to global system for mobile communication (GSM) radiofrequency fields on astrocytic immunoreactivity. Bioelectromagnetics. 2015;36:245–250. doi: 10.1002/bem.21891. [DOI] [PubMed] [Google Scholar]
- Dantzer R. Cytokine-induced sickness behavior: where do we stand? Brain Behav Immun. 2001;15:7–24. doi: 10.1006/brbi.2000.0613. [DOI] [PubMed] [Google Scholar]
- Dantzer R, O’Connor JC, Freund GG, Johnson RW, Kelley KW. From inflammation to sickness and depression: when the immune system subjugates the brain. Nat Rev Neurosci. 2008;9:46–56. doi: 10.1038/nrn2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Poot SAH, Tian G, Finley D. Meddling with fate: the proteasomal deubiquitinating enzymes. J Mol Biol. 2017;429:3525–3545. doi: 10.1016/j.jmb.2017.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragicevic N, Bradshaw PC, Mamcarz M, Lin X, Wang L, Cao C, Arendash GW. Long-term electromagnetic field treatment enhances brain mitochondrial function of both Alzheimer’s transgenic mice and normal mice: a mechanism for electromagnetic field-induced cognitive benefit? Neuroscience. 2011;185:135–149. doi: 10.1016/j.neuroscience.2011.04.012. [DOI] [PubMed] [Google Scholar]
- Finnie JW. Expression of the immediate early gene, c-fos, in mouse brain after acute global system for mobile communication microwave exposure. Pathology. 2005;37:231–233. doi: 10.1080/00313020500098942. [DOI] [PubMed] [Google Scholar]
- Fragopoulou AF, Polyzos A, Papadopoulou MD, Sansone A, Manta AK, et al. Hippocampal lipidome and transcriptome profile alterations triggered by acute exposure of mice to GSM 1800 MHz mobile phone radiation: an exploratory study. Brain Behav. 2018;8:e01001. doi: 10.1002/brb3.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritze K, Wiessner C, Kuster N, Sommer C, Gass P, et al. Effect of global system for mobile communication microwave exposure on the genomic response of the rat brain. Neuroscience. 1997;81:627–639. doi: 10.1016/S0306-4522(97)00228-5. [DOI] [PubMed] [Google Scholar]
- Gherardini L, Ciuti G, Tognarelli S, Cinti C. Searching for the perfect wave: the effect of radiofrequency electromagnetic fields on cells. Int J Mol Sci. 2014;15:5366–5387. doi: 10.3390/ijms15045366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer PL, Hanayama R, Bloodgood BL, Mardinly AR, Lipton DM, et al. The Angelman syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell. 2010;140:704–716. doi: 10.1016/j.cell.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell JW. How Ca2+−permeable AMPA receptors, the kinase PKA, and the phosphatase PP2B are intertwined in synaptic LTP and LTD. Sci Signal. 2016;9:e2. doi: 10.1126/scisignal.aaf7067. [DOI] [PubMed] [Google Scholar]
- Heneka MT, Kummer MP, Latz E. Innate immune activation in neurodegenerative disease. Nat Rev Immunol. 2014;14:463–477. doi: 10.1038/nri3705. [DOI] [PubMed] [Google Scholar]
- Hirbec H, Marmai C, Duroux-Richard I, Roubert C, Esclangon A, et al. The microglial reaction signature revealed by RNAseq from individual mice. Glia. 2018;66:971–986. doi: 10.1002/glia.23295. [DOI] [PubMed] [Google Scholar]
- Hirose H, Sasaki A, Ishii N, Sekijima M, Iyama T, et al. 1950 MHz IMT-2000 field does not activate microglial cells in vitro. Bioelectromagnetics. 2010;31:104–112. doi: 10.1002/bem.20532. [DOI] [PubMed] [Google Scholar]
- Hoogland IC, Houbolt C, van Westerloo DJ, van Gool WA, van de Beek D. Systemic inflammation and microglial activation: systematic review of animal experiments. J Neuroinflammation. 2015;12:114. doi: 10.1186/s12974-015-0332-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howland DS, Liu J, She Y, Goad B, Maragakis NJ, et al. Focal loss of the glutamate transporter EAAT2 in a transgenic rat model of SOD1 mutant-mediated amyotrophic lateral sclerosis (ALS) Proc Natl Acad Sci U S A. 2002;99:1604–1609. doi: 10.1073/pnas.032539299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ICNIRP (International Commission on Non Ionizing Radiation Protection) Guidelines for limiting exposure to time-varying electric, magnetic and electromagnetic fields (up to 300 GHz) Health Phys. 1998;74:494–522. [PubMed] [Google Scholar]
- Jacobson GM, Voss LJ, Klockars A, Bird S, Dimitrov I, et al. Transcriptional changes in response to ketamine ester-analogs SN 35210 and SN 35563 in the rat brain. BMC Genomics. 2019;20:281. doi: 10.1186/s12864-019-5649-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong YJ, Kang GY, Kwon JH, Choi HD, Pack JK, et al. 1950 MHz electromagnetic fields ameliorate abeta pathology in Alzheimer’s disease mice. Curr Alzheimer Res. 2015;12:481–492. doi: 10.2174/156720501205150526114448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Kim HJ, Yu DH, Kweon HS, Huh YH, et al. Changes in numbers and size of synaptic vesicles of cortical neurons induced by exposure to 835 MHz radiofrequency-electromagnetic field. PLoS One. 2017;12:e0186416. doi: 10.1371/journal.pone.0186416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz KS, Luebbers RJ. The finite difference time domain method for electromagnetics. Boca Raton: CRC Press; 1993. [Google Scholar]
- Lameth J, Gervais A, Colin C, Leveque P, Jay TM, et al. Acute neuroinflammation promotes cell responses to 1800 MHz GSM electromagnetic fields in the rat cerebral cortex. Neurotox Res. 2017;32:444–459. doi: 10.1007/s12640-017-9756-3. [DOI] [PubMed] [Google Scholar]
- Leveque P, Dale C, Veyret B, Viart J. Dosimetric analysis of a 900 MHz rat head exposure system. IEE Trans Microw Theory Tech. 2004;52:2076–2083. doi: 10.1109/TMTT.2004.831984. [DOI] [Google Scholar]
- Levy C, Brooks JM, Chen J, Su J, Fox MA. Cell-specific and developmental expression of lectican-cleaving proteases in mouse hippocampus and neocortex. J Comp Neurol. 2015;523:629–648. doi: 10.1002/cne.23701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling EA, Leblond CP. Investigation of glial cells in semithin sections. II. Variation with age in the numbers of the various glial cell types in rat cortex and corpus callosum. J Comp Neurol. 1973;149:73–81. doi: 10.1002/cne.901490105. [DOI] [PubMed] [Google Scholar]
- Liu X, Huang D, Guo P, Wu Q, Dai M, et al. PKA/CREB and NF-kappaB pathway regulates AKNA transcription: a novel insight into T-2 toxin-induced inflammation and GH deficiency in GH3 cells. Toxicology. 2017;392:81–95. doi: 10.1016/j.tox.2017.10.013. [DOI] [PubMed] [Google Scholar]
- Lobsiger CS, Boillee S, Cleveland DW. Toxicity from different SOD1 mutants dysregulates the complement system and the neuronal regenerative response in ALS motor neurons. Proc Natl Acad Sci U S A. 2007;104:7319–7326. doi: 10.1073/pnas.0702230104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Martin E, Relova-Quinteiro JL, Gallego-Gomez R, Peleteiro-Fernandez M, Jorge-Barreiro FJ, et al. GSM radiation triggers seizures and increases cerebral c-Fos positivity in rats pretreated with subconvulsive doses of picrotoxin. Neurosci Lett. 2006;398:139–144. doi: 10.1016/j.neulet.2005.12.082. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, He M, Zhang Y, Xu S, Zhang L, et al. Differential pro-inflammatory responses of astrocytes and microglia involve STAT3 activation in response to 1800 MHz radiofrequency fields. PLoS One. 2014;9:e108318. doi: 10.1371/journal.pone.0108318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna J, Leleu JP, Preux PM, Corcia P, Couratier P, Marin B, Boumediene F, Fralim Consortium Residential exposure to ultra high frequency electromagnetic fields emitted by Global System for Mobile (GSM) antennas and amyotrophic lateral sclerosis incidence: a geo-epidemiological population-based study. Environ Res. 2019;176:108525. doi: 10.1016/j.envres.2019.108525. [DOI] [PubMed] [Google Scholar]
- Ma W, Ortiz-Quintero B, Rangel R, McKeller MR, Herrera-Rodriguez S, et al. Coordinate activation of inflammatory gene networks, alveolar destruction and neonatal death in AKNA deficient mice. Cell Res. 2011;21:1564–1577. doi: 10.1038/cr.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino C, Lagroye I, Scarfi MR, Sienkiewicz Z. Are the young more sensitive than adults to the effects of radiofrequency fields? An examination of relevant data from cellular and animal studies. Prog Biophys Mol Biol. 2011;107:374–385. doi: 10.1016/j.pbiomolbio.2011.09.002. [DOI] [PubMed] [Google Scholar]
- Mausset-Bonnefont AL, Hirbec H, Bonnefont X, Privat A, Vignon J, de Sèze R. Acute exposure to GSM 900-MHz electromagnetic fields induces glial reactivity and biochemical modifications in the rat brain. Neurobiol Dis. 2004;17:445–454. doi: 10.1016/j.nbd.2004.07.004. [DOI] [PubMed] [Google Scholar]
- McNamee JP, Bellier PV, Konkle AT, Thomas R, Wasoontarajaroen S, et al. Analysis of gene expression in mouse brain regions after exposure to 1.9 GHz radiofrequency fields. Int J Radiat Biol. 2016;92:338–350. doi: 10.3109/09553002.2016.1159353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamee JP, Chauhan V. Radiofrequency radiation and gene/protein expression: a review. Radiat Res. 2009;172:265–287. doi: 10.1667/RR1726.1. [DOI] [PubMed] [Google Scholar]
- Meller J, Chen Z, Dudiki T, Cull RM, Murtazina R et al (2017) Integrin-Kindlin3 requirements for microglial motility in vivo are distinct from those for macrophages. JCI Insight 2. 10.1172/jci.insight.93002 [DOI] [PMC free article] [PubMed]
- Mesci P, Zaidi S, Lobsiger CS, Millecamps S, Escartin C, et al. System xC- is a mediator of microglial function and its deletion slows symptoms in amyotrophic lateral sclerosis mice. Brain. 2015;138:53–68. doi: 10.1093/brain/awu312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakoshi J, Takemasa K, Takashima Y, Ding GR, Hirose H, et al. Effects of exposure to a 1950 MHz radio frequency field on expression of Hsp70 and Hsp27 in human glioma cells. Bioelectromagnetics. 2005;26:251–257. doi: 10.1002/bem.20077. [DOI] [PubMed] [Google Scholar]
- Narayanan Sareesh Naduvil, Jetti Raghu, Kesari Kavindra Kumar, Kumar Raju Suresh, Nayak Satheesha B., Bhat P. Gopalakrishna. Radiofrequency electromagnetic radiation-induced behavioral changes and their possible basis. Environmental Science and Pollution Research. 2019;26(30):30693–30710. doi: 10.1007/s11356-019-06278-5. [DOI] [PubMed] [Google Scholar]
- Nittby H, Widegren B, Krogh M, Grafström G, Berlin H, Rehn G, Eberhardt JL, Malmgren L, Persson BRR, Salford LG. Exposure to radiation from global system for mobile communications at 1800 MHz significantly changes gene expression in rat hippocampus and cortex. Environmentalist. 2008;28:458–465. doi: 10.1007/s10669-008-9170-8. [DOI] [Google Scholar]
- Noli L, Capalbo A, Ogilvie C, Khalaf Y, Ilic D. Discordant growth of monozygotic twins starts at the blastocyst stage: a case study. Stem Cell Reports. 2015;5:946–953. doi: 10.1016/j.stemcr.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Occelli F, Lameth J, Adenis V, Huetz C, Leveque P, et al. A single exposure to GSM-1800MHz signals in the course of an acute Neuroinflammatory reaction can Alter neuronal responses and microglial morphology in the rat primary auditory cortex. Neuroscience. 2018;385:11–24. doi: 10.1016/j.neuroscience.2018.06.002. [DOI] [PubMed] [Google Scholar]
- Ohsawa K, Irino Y, Sanagi T, Nakamura Y, Suzuki E, et al. P2Y12 receptor-mediated integrin-beta1 activation regulates microglial process extension induced by ATP. Glia. 2010;58:790–801. doi: 10.1002/glia.20963. [DOI] [PubMed] [Google Scholar]
- Ouadah NS, Lecomte A, Robidel F, Olsson A, Deltour I, et al. Possible effects of radiofrequency electromagnetic fields on in vivo C6 brain tumors in Wistar rats. J Neuro-Oncol. 2018;140:539–546. doi: 10.1007/s11060-018-03012-y. [DOI] [PubMed] [Google Scholar]
- Paparini A, Rossi P, Gianfranceschi G, Brugaletta V, Falsaperla R, et al. No evidence of major transcriptional changes in the brain of mice exposed to 1800 MHz GSM signal. Bioelectromagnetics. 2008;29:312–323. doi: 10.1002/bem.20399. [DOI] [PubMed] [Google Scholar]
- Petitdant N, Lecomte A, Robidel F, Gamez C, Blazy K, et al. Cerebral radiofrequency exposures during adolescence: impact on astrocytes and brain functions in healthy and pathologic rat models. Bioelectromagnetics. 2016;37:338–350. doi: 10.1002/bem.21986. [DOI] [PubMed] [Google Scholar]
- Philips T, Rothstein JD. Rodent models of amyotrophic lateral sclerosis. Curr Protoc Pharmacol. 2015;69:567. doi: 10.1002/0471141755.ph0567s69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulletier de Gannes F, Ruffie G, Taxile M, Ladeveze E, Hurtier A, et al. Amyotrophic lateral sclerosis (ALS) and extremely-low frequency (ELF) magnetic fields: a study in the SOD-1 transgenic mouse model. Amyotroph Lateral Scler. 2009;10:370–373. doi: 10.3109/17482960802320396. [DOI] [PubMed] [Google Scholar]
- Sanderson JL, Gorski JA, Gibson ES, Lam P, Freund RK, et al. AKAP150-anchored calcineurin regulates synaptic plasticity by limiting synaptic incorporation of Ca2+−permeable AMPA receptors. J Neurosci. 2012;32:15036–15052. doi: 10.1523/JNEUROSCI.3326-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson JL, Scott JD, Dell’Acqua ML. Control of homeostatic synaptic plasticity by AKAP-anchored kinase and phosphatase regulation of Ca(2+)-permeable AMPA receptors. J Neurosci. 2018;38:2863–2876. doi: 10.1523/JNEUROSCI.2362-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SE, Zhou YD, Zhang G, Jin Z, Stoppel DC, et al. Increased gene dosage of Ube3a results in autism traits and decreased glutamate synaptic transmission in mice. Sci Transl Med. 2011;3:103ra197. doi: 10.1126/scitranslmed.3002627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son Y, Kim JS, Jeong YJ, Jeong YK, Kwon JH, et al. Long-term RF exposure on behavior and cerebral glucose metabolism in 5xFAD mice. Neurosci Lett. 2018;666:64–69. doi: 10.1016/j.neulet.2017.12.042. [DOI] [PubMed] [Google Scholar]
- Su L, Yimaer A, Xu Z, Chen G. Effects of 1800 MHz RF-EMF exposure on DNA damage and cellular functions in primary cultured neurogenic cells. Int J Radiat Biol. 2018;94:295–305. doi: 10.1080/09553002.2018.1432913. [DOI] [PubMed] [Google Scholar]
- Sun W, Cornwell A, Li J, Peng S, Osorio MJ, et al. SOX9 is an astrocyte-specific nuclear marker in the adult brain outside the neurogenic regions. J Neurosci. 2017;37:4493–4507. doi: 10.1523/JNEUROSCI.3199-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swatek KN, Komander D. Ubiquitin modifications. Cell Res. 2016;26:399–422. doi: 10.1038/cr.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taflove A. Computational electrodynamics: the finite-difference time domain method. Norwood, MA: Artech House; 1995. [Google Scholar]
- Tang X, Metzger D, Leeman S, Amar S. LPS-induced TNF-alpha factor (LITAF)-deficient mice express reduced LPS-induced cytokine: evidence for LITAF-dependent LPS signaling pathways. Proc Natl Acad Sci U S A. 2006;103:13777–13782. doi: 10.1073/pnas.0605988103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorlin T, Rouquette JM, Hamnerius Y, Hansson E, Persson M, et al. Exposure of cultured astroglial and microglial brain cells to 900 MHz microwave radiation. Radiat Res. 2006;166:409–421. doi: 10.1667/RR3584.1. [DOI] [PubMed] [Google Scholar]
- Vlug AS, Teuling E, Haasdijk ED, French P, Hoogenraad CC, Jaarsma D. ATF3 expression precedes death of spinal motoneurons in amyotrophic lateral sclerosis-SOD1 transgenic mice and correlates with c-Jun phosphorylation, CHOP expression, somato-dendritic ubiquitination and Golgi fragmentation. Eur J Neurosci. 2005;22:1881–1894. doi: 10.1111/j.1460-9568.2005.04389.x. [DOI] [PubMed] [Google Scholar]
- Watilliaux A, Edeline JM, Leveque P, Jay TM, Mallat M. Effect of exposure to 1,800 MHz electromagnetic fields on heat shock proteins and glial cells in the brain of developing rats. Neurotox Res. 2011;20:109–119. doi: 10.1007/s12640-010-9225-8. [DOI] [PubMed] [Google Scholar]
- Xu D, Shan B, Sun H, Xiao J, Zhu K, et al. USP14 regulates autophagy by suppressing K63 ubiquitination of Beclin 1. Genes Dev. 2016;30:1718–1730. doi: 10.1101/gad.285122.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee KS. Numerical solution of initial boundary value problems involving Maxwell’s equations in isotropic media. IEEE Trans Antennas Propag. 1966;14(3):302–307. doi: 10.1109/TAP.1966.1138693. [DOI] [Google Scholar]
- Yoshimi R, Yamaji S, Suzuki A, Mishima W, Okamura M, et al. The gamma-parvin-integrin-linked kinase complex is critically involved in leukocyte-substrate interaction. J Immunol. 2006;176:3611–3624. doi: 10.4049/jimmunol.176.6.3611. [DOI] [PubMed] [Google Scholar]
- Zheng N, Shabek N. Ubiquitin ligases: structure, function, and regulation. Annu Rev Biochem. 2017;86:129–157. doi: 10.1146/annurev-biochem-060815-014922. [DOI] [PubMed] [Google Scholar]
- Zeisel A., Munoz-Manchado A. B., Codeluppi S., Lonnerberg P., La Manno G., Jureus A., Marques S., Munguba H., He L., Betsholtz C., Rolny C., Castelo-Branco G., Hjerling-Leffler J., Linnarsson S. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347(6226):1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Chen K., Sloan S. A., Bennett M. L., Scholze A. R., O'Keeffe S., Phatnani H. P., Guarnieri P., Caneda C., Ruderisch N., Deng S., Liddelow S. A., Zhang C., Daneman R., Maniatis T., Barres B. A., Wu J. Q. An RNA-Sequencing Transcriptome and Splicing Database of Glia, Neurons, and Vascular Cells of the Cerebral Cortex. Journal of Neuroscience. 2014;34(36):11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]