Abstract
The prevalence of physiological and pathological left ventricular hypertrophy (LVH) among young adults is about 5%. A use of electrocardiographic (ECG) voltage criteria and machine learning for the ECG parameters to identify the presence of LVH is estimated only 20-30% in the general population. The aim of this study is to develop an ECG system with anthropometric data using machine learning to increase the accuracy and sensitivity for a screen of LVH. In a large sample of 2,196 males, aged 17–45 years, the support vector machine (SVM) classifier is used as the machine learning method for 31 characteristics including age, body height and body weight in addition to 28 ECG parameters such as axes, intervals and voltages to link the output of LVH. The diagnosis of LVH is based on the echocardiographic criteria for young males to be 116 gram/meter2 (left ventricular mass (LVM)/body surface area) or 49 gram/meter2.7 (LVM/body height2.7). On the purpose of increasing sensitivity, the specificity is adjusted around 70-75% and all data tested in proposed model reveal high sensitivity to 86.7%. The area under curve (AUC) of the Precision-Recall (PR) curve is 0.308 in the proposed model which is better than 0.109 and 0.077 using Cornell and Sokolow-Lyon voltage criteria for LVH, respectively. Our system provides a novel screening tool using age, body height, body weight and ECG data to identify most of the LVH among young adults. It provides a fast, accurate and practical diagnosis tool to identify LVH.
Keywords: Anthropometrics, electrocardiographic system, left ventricular hypertrophy, machine learning, young adults
The aim of this study was to develop an ECG system with anthropometric data using machine learning to increase the accuracy and sensitivity for a screen for physiological and pathological left ventricular hypertrophy (LVH) among young adults. In a large sample of 2,196 males aged 17-45 years, the support vector machine (SVM) classifier was used as the machine learning method for 31 characteristics, including age, body height and body weight, in addition to 28 ECG parameters such as axes, intervals and voltages to link the output of LVH. Our system provides a novel screening tool using age, body height, body weight and ECG data to identify most of the LVH among young adults. It provides a fast, accurate and practical diagnosis tool to identify LVH.
I. Introduction
Artificial intelligence (AI) grows fast with the improvement of technology and the availability of various kinds of big data. Machine learning, an AI of the computational statistics, has been introduced in clinical medicine which could provide accurate diagnosis of disease and prediction of the risk [1]–[12]. For example, [12] utilizes the random survival forest technique identifying the top-20 risk factors of cardiovascular events and the performance is superior to the traditional risk calculators. In the modern era, using machine learning techniques has become an efficient and reliable tool for clinical practice by physicians globally.
Left ventricular hypertrophy (LVH), which is clinically considered as a sign of end-organ damage related to long-term hypertension, has been associated with heart failure and cardiovascular disease events among middle and old-aged individuals [13], [14]. In contrast, the prevalence of LVH in young adults is low, accounting for approximately 5% [15], and the phenotypes are usually caused by physiologic adaptions to intense physical training [16] and congenital hypertrophic cardiomyopathy [17]. A prior population research also shows that the presence of LVH at young ages is associated with higher risk of incident cardiovascular disease events [18]. The 12-lead surface electrocardiography (ECG) is the currently most used tool for screening the presence of LVH in the general population [19]. Several ECG-based criteria such as the Cornell and Sokolow-Lyon formulas have been proposed for more than 30 years [20], [21]; however, the performance of the ECG-based criteria for LVH consistently yields high specificity (>95%) but low sensitivity (20%-30%). Over the past 5 years, a few population studies were presented by machine learning and deep learning for the ECG characteristics to detect presence of LVH [1]–[3]. For hypertrophic cardiomyopathy (HCM), one of the most common pathological LVH, Rahman et al. firstly used the random forest and the support vector machine (SVM) techniques, and 5-fold cross validations, for hundreds of ECG characteristics training, where showed excellent results regarding the sensitivity, specificity and precision up to 90% in a hospital-based population study [2]. Subsequently, Tison et al. used the deep learning of convolutional neural network for numerous ECG parameters to identify HCM, consistently showing excellent results in a hospital-based population study [3]. However, the sensitivity using the specific ECG criteria for HCM has approached up to 90% [22], [23]. By contrast, a community-based population study using machine learning for the ECG to screen any unspecific LVH was proposed by Sparapani et al. [1]. Despite this study utilized the tree-based, Bayesian nonparametric machine learning technique for a number of ECG characteristics alone, the sensitivity for detecting any unspecific LVH phenotype in a general population of middle and old aged individuals is increased up to 29.0%, merely a little improvement compared with the other ECG criteria for LVH [1]. Accordingly, computerized training of the ECG alone might not be adequate in screening for unspecific LVH in the general population level.
In this paper, we aim to develop a clinically accessible ECG-based system which uses a large sample of the military young personnel taking age, anthropometric data and several ECG characteristics into account for machine learning by the method of SVM to predict the presence of unspecific LVH as shown in Fig. 1. The rest of this paper is organized as follows. The materials are presented in Section II. In Section III, the proposed algorithm regarding the ECG system for unspecific LVH detection is described in detail. Section IV displays the experimental results. Section V concludes this paper.
FIGURE 1.
Schematic diagram of proposed system.
II. Data Collection and Features Selection
A. Data Collection
This study uses a population of 2,196 military males aged 17–45 years from the ancillary cardiorespiratory fitness and hospitalization events in armed forces (CHIEF) substudy performed in the Hualien Armed Forces General Hospital in Hualien city, Taiwan, R.O.C. Each participant received an ECG and an echocardiography at the same clinic visit in a health examination prior to the annual exercise tests for the military rank promotions and awards. The study design has been described in detail previously [24]–[33]. The raw data of 12-lead ECG parameters are interpreted by the software products of two ECG manufacturers: one is the CARDIOVIT MS-2015 (Schiller AG, Baar, Switzerland), and another one is the TC70 CARDIOGRAPH (Philips, Amsterdam, Netherlands). The transthoracic echocardiography is performed via the IE33 (Philips, Amsterdam, Netherlands). All the echocardiography and ECG procedures are implemented by the same technician who has been certificated with plenty of experiences for longer than 20 years. The 28 ECG characteristics adopted in the proposed method include heart rate, the durations of P wave, PR interval, QRS interval, QT interval and QTc interval in Lead II, and the axes of P, QRS, and T waves in Lead II, and the voltages of R waves in all Limb Leads I, II, III, aVR, aVL, aVF and S wave in Lead aVL, and the voltages of R and S waves in all precordial Leads V1-V6, where the voltage of 1 mV indicates 10 mm. In addition, a population of 203 military females aged 17–42 from the ancillary CHIEF substudy is utilized as an additional test set using the male model of machine learning by age, anthropometric data and ECG parameters. The comparison methods are the Sokolow-Lyon voltage criterion for LVH [20] and the Cornell voltage criteria for males and females [21], which are revealed in Table 1, respectively.
TABLE 1. Electrocardiographic and Echocardiographic Criteria for Left Ventricular Hypertrophy.
| Criteria | Sex-specific | Description | |
|---|---|---|---|
| Electrocardiographic | Sokolow-Lyon voltage | Both Sexes | (S-V1 or S-V2+ R-V5 or R-V6) ≥ 35 mm |
| Cornell voltage | Male Adults | R-aVL+S-V3 ≥ 28 mm | |
| Female Adults | R-aVL+S-V3 ≥ 20 mm | ||
| Echocardiographic | CARDIA-based | Young Male Adults | LVM/BSA ≥ 116 gram/meter2 or LVM/height2.7≥ 49 gram/meter2.7 |
| CHIEF-based | Young Female Adults | LVM/BSA ≥ 88 gram/meter2 or LVM/ height2.7 ≥ 41 gram/meter2.7 |
Abbreviations: CARDIA, coronary artery risk development in young adults; CHIEF, cardiorespiratory fitness and hospitalization events in armed forces
The diagnosis of LVH is based on the recommendations of the American Society of Echocardiography [34]. Quantification of left ventricular internal dimension (LVIDd) and left ventricular wall thickness including interventricular septum (IVSd) and left ventricular posterior wall (LVPWd) is measured by M-mode and 2-dimensional methods at the mitral valve tips and at the onset of the QRS complex in ECG of end diastole in echocardiographic parasternal long axis view. Left ventricular mass (LVM) is calculated on the basis of the corrected echocardiographic formula proposed by Devereux et al. [35] as shown in (1).
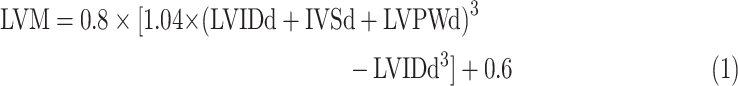 |
LVM is respectively indexed for body surface area (BSA) and for height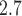 based on the Dubois and Dubois [36] and de Simone et al. formula [37], respectively. Echocardiographic LVH for young males is defined to be the
based on the Dubois and Dubois [36] and de Simone et al. formula [37], respectively. Echocardiographic LVH for young males is defined to be the 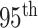 percentile of the military males and according to the finding of a prior Coronary Artery Risk Development in Young Adults study (CARDIA) [18]. In addition, echocardiographic LVH for young females is defined according to the
percentile of the military males and according to the finding of a prior Coronary Artery Risk Development in Young Adults study (CARDIA) [18]. In addition, echocardiographic LVH for young females is defined according to the 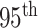 percentile of the military females in the CHIEF study and the results of a prior study for Southeastern Asia young females [38]. The sex-specific echocardiographic criteria for LVH are listed in Table 1. To develop the proposed machine learning method, the data are partitioned into 80% for cross validation and 20% for test for the male samples. The study protocol was approved by the Institutional Review Broad of Mennonite Christian Hospital (No. 16-05-008) in Hualien City, Taiwan.
percentile of the military females in the CHIEF study and the results of a prior study for Southeastern Asia young females [38]. The sex-specific echocardiographic criteria for LVH are listed in Table 1. To develop the proposed machine learning method, the data are partitioned into 80% for cross validation and 20% for test for the male samples. The study protocol was approved by the Institutional Review Broad of Mennonite Christian Hospital (No. 16-05-008) in Hualien City, Taiwan.
B. Pre-Test for Input Features
To select the proper features, at initial stage, we stepwise add several biological parameters on the 28 ECG parameters, as input features for SVM machine learning to determine the most clinically efficient system. These biological parameters include age, body height, body weight, body mass index (BMI), waist circumference, systolic blood pressure (SBP), diastolic blood pressure (DBP) and body surface area (BSA). The preliminary performances of additional biological parameters and adopted 28 ECG parameters are listed in Table 2. For the stepwise pre-test of input features, we only take training set and test set without cross validation for SVM model to compare the results of various ECG-based combinations. As shown in Table 2, when more input parameters are trained, there are larger area under curves (AUCs) of the Receiver Operating Characteristic (ROC) curves and Precision-Recall (PR) curves in the test set. A significant improvement in AUCs of the ROC and PR curves is observed when using age, body height, body weight with the 28 ECG parameters as inputs to relate to the output of LVH. Additional inputs of BMI, waist circumference, BSA, SBP and DBP are neutral or merely increase a little in performance. Thus, the 31 features including age, body height, body weight, and the 28 ECG parameters are determined as the input features of our machine learning model. The average values in each parameter of the participants are revealed in Table 3. The label of LVH is by the echocardiographic LVM/BSA ≥ 116 gram/meter2 or LVM/height2.7 ≥ 49 gram/meter2.7 for young males. As shown in Table 3, the characteristics in those with and those without LVH are continuous data which are expressed as mean ± standard deviation and compared by two samples t-test. A p-value < 0.05 is considered significant. Notably, older age, lower body height and greater body weight are observed in those with echocardiographic LVH.
TABLE 2. Preliminary Performance of Additional Biological Parameters and Adopted 28 ECG Parameters.
| ECG (28) | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| Age | – | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| Height and Weight | – | – | ✔ | ✔ | ✔ | ✔ | ✔ |
| BMI | – | – | – | ✔ | ✔ | ✔ | ✔ |
| Waist Circumference | – | – | – | – | ✔ | – | ✔ |
| SBP and DBP | – | – | – | – | – | ✔ | ✔ |
| BSA | – | – | – | – | – | – | ✔ |
| Number of input features | 28 | 29 | 31 | 32 | 33 | 34 | 36 |
| ROC AUC | 0.765 | 0.772 | 0.855 | 0.858 | 0.860 | 0.857 | 0.866 |
| PR AUC | 0.215 | 0.214 | 0.303 | 0.311 | 0.314 | 0.322 | 0.339 |
TABLE 3. Characteristics of Study Participants (Males).
| Features | Total N=2196 | Non-LVH N=2046 | LVH N=150 | p-value |
|---|---|---|---|---|
| Age (years) | 25.93± 6.77 | 25.68± 6.67 | 29.41± 7.18 | < 0.001 |
| Height (cm) | 172.01± 5.92 | 172.19± 5.89 | 169.55± 5.68 | < 0.001 |
| Weight (kg) | 72.47± 12.03 | 71.86± 11.73 | 80.73± 13.06 | < 0.001 |
| Heart rate (bpm) | 67.38± 11.83 | 67.47± 11.69 | 66.03± 13.53 | 0.206 |
| P-II(ms) | 105.85± 15.26 | 105.80± 15.17 | 106.57± 16.49 | 0.551 |
| PR-II(ms) | 157.91± 21.02 | 157.40± 20.97 | 164.91± 20.45 | < 0.001 |
| QRS-II(ms) | 97.26± 10.75 | 97.11± 10.70 | 99.28± 11.24 | 0.017 |
| QT-II(ms) | 372.21± 29.64 | 371.21± 29.28 | 385.82± 31.24 | < 0.001 |
| QTc-II(ms) | 390.14± 24.56 | 389.71± 24.30 | 396.01± 27.34 | 0.007 |
| P axis-II(degree) | 44.78± 27.21 | 45.41± 27.20 | 36.09± 25.99 | < 0.001 |
| QRS axis-II(degree) | 65.22± 32.23 | 65.92± 31.96 | 55.71± 34.34 | < 0.001 |
| T axis-II(degree) | 35.87± 21.37 | 36.52± 21.14 | 27.03± 22.65 | < 0.001 |
| R-I(mm) | 5.81± 3.00 | 5.65± 2.90 | 7.99± 3.43 | < 0.001 |
| R-II(mm) | 12.86± 4.93 | 12.97± 4.91 | 11.35± 5.02 | < 0.001 |
| R-III(mm) | 8.53± 5.83 | 8.71± 5.86 | 6.04± 4.81 | < 0.001 |
| R-aVR(mm) | 1.62± 2.31 | 1.58± 2.26 | 2.14± 2.79 | 0.019 |
| R-aVL(mm) | 2.34± 2.23 | 2.24± 2.10 | 3.73± 3.24 | < 0.001 |
| S-aVL(mm) | 2.21± 2.98 | 2.28± 3.02 | 1.31± 2.23 | < 0.001 |
| R-aVF(mm) | 10.50± 5.34 | 10.67± 5.33 | 8.20± 4.80 | < 0.001 |
| R-V1(mm) | 3.54± 2.28 | 3.55± 2.27 | 3.37± 2.45 | 0.347 |
| S-V1(mm) | 10.25± 5.30 | 10.27± 5.25 | 9.96± 6.01 | 0.542 |
| R-V2(mm) | 8.58± 4.24 | 8.55± 4.19 | 9.08± 4.90 | 0.199 |
| S-V2(mm) | 16.01± 6.89 | 16.08± 6.92 | 15.04± 6.52 | 0.075 |
| R-V3(mm) | 13.26± 6.23 | 13.20± 6.22 | 14.02± 6.26 | 0.119 |
| S-V3(mm) | 8.45± 5.34 | 8.39± 5.30 | 9.29± 5.80 | 0.045 |
| R-V4(mm) | 19.73± 6.84 | 19.80± 6.87 | 18.78± 6.44 | 0.076 |
| S-V4(mm) | 5.15± 4.10 | 5.10± 4.09 | 5.82± 4.28 | 0.039 |
| R-V5(mm) | 19.86± 5.86 | 19.85± 5.83 | 19.95± 6.31 | 0.848 |
| S-V5(mm) | 3.17± 2.93 | 3.14± 2.91 | 3.49± 3.18 | 0.168 |
| R-V6(mm) | 16.35± 5.08 | 16.32± 5.06 | 16.84± 5.32 | 0.223 |
| S-V6(mm) | 1.86± 1.99 | 1.85± 1.98 | 1.92± 2.20 | 0.721 |
III. Proposed Method
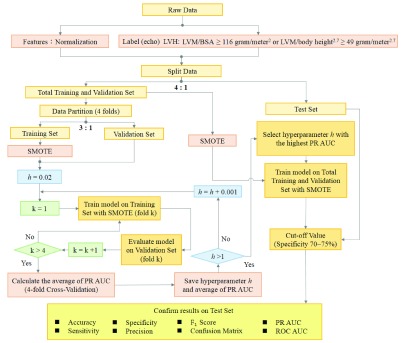
According to the preliminary pre-test outcomes, the 31 input factors for machine learning are age, body height, body weight and the adopted 28 ECG parameters. This paper uses the SVM for predicting the presence of LVH among young adults. The reason for selecting SVM as the model is due to the advantages of SVM classifier which are effective in high dimensional spaces and memory efficient, and could provide successful discriminative models in many fields [2], [39]–[41]. In addition, the training time and running time of SVM are extremely short. Therefore, we utilize the SVM machine learning technique which can be feasible in an ECG equipment to achieve practical application. The flowchart of the proposed method is illustrated in Fig.2.
FIGURE 2.
Flowchart of the proposed method.
A. Data Normalization
Firstly, we use the normalization of Min-Max scaling [42], [43] to individually normalize the original data of 31 input features into the interval between 0~1 for solving the problem of different dynamic ranges of various input features. Min-Max normalization performs a linear transformation on the original data. Each of the actual data 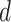 of feature
of feature 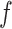 is mapped to a normalized value which lies in the range of 0 to 1. The Min-Max normalization is calculated by using (2).
is mapped to a normalized value which lies in the range of 0 to 1. The Min-Max normalization is calculated by using (2).
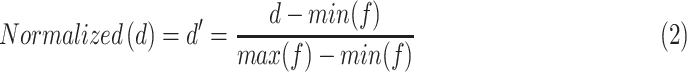 |
where 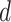 indicates the original data of feature
indicates the original data of feature 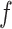 among the 31 input features, min(
among the 31 input features, min(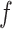 ) and max(
) and max(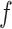 ) represent the minimum and maximum values of the input feature
) represent the minimum and maximum values of the input feature 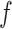 , respectively.
, respectively. 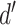 denotes the normalized data.
denotes the normalized data.
B. Cross Validation
The data of 2,196 military males are segmented into a total training and validation set and a test set with 4:1 ratio. The total training and validation set is partitioned into four equal size groups. Among the four groups, one group is treated as the validation set for validating the model, and the remaining three groups are taken as the training set. Each of the four groups is used once as the validation set. The proportions of non-LVH and LVH cases are similar across each group. The cross validation process is then repeated four times. Four AUCs of PR curves from the four folds are averaged as a single performance of the results. By using a 4-fold cross validation, a better generalization assessment of the performance for training can be obtained.
C. Application of Smote
The data numbers illustrated by four folds are described in detail in Table 4. Our datasets are predominately composed of non-LVH cases with only a small percentage of LVH cases since the prevalence of LVH in young adults is approximately 5%. For example, in Table 4, the 1st cross validation, the numbers of the training set and validation set are 1,317 (Non-LVH: 1,225, LVH: 92) and 439 (Non-LVH: 408, LVH: 31), respectively. This imbalance in sample sizes between the Non-LVH and LVH cases is obvious. The solution for this issue is to increase LVH cases in pre-processing by applying the synthetic minority over-sampling technique (SMOTE) [44]. The main idea of SMOTE is to create new minority class samples by choosing a near minority class neighbor randomly and interpolating as described as follows. Firstly, for each minority class sample 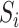 , its
, its 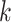 nearest neighbors from other minority class samples are taken. Secondly, minority class sample
nearest neighbors from other minority class samples are taken. Secondly, minority class sample 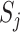 among the
among the 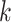 neighbors is randomly selected. Finally, the
neighbors is randomly selected. Finally, the 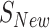 is generated as the synthetic sample by interpolating between
is generated as the synthetic sample by interpolating between 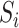 and
and 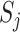 as (3).
as (3).
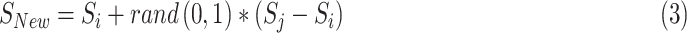 |
where rand (0, 1) stands for a random number between 0 and 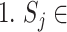 {
{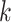 neighbors of
neighbors of 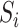 }. The process of applying SMOTE can be treated as interpolating between two LVH samples in the viewpoint of geometry. The decision space for the LVH samples is expanded. Thus, it allows the SVM method to have a higher prediction performance on unknown LVH samples. The SMOTE is used in the process of 4-fold cross validation. The training data of the LVH group are pre-processed and augmented by SMOTE to be the same numbers with those of the Non-LVH group as 1,225, 1,230, 1,229 and 1,215, respectively, for the four folds as shown in Table 4. We also compare the performances of the test set for the proposed model with and without using SMOTE.
}. The process of applying SMOTE can be treated as interpolating between two LVH samples in the viewpoint of geometry. The decision space for the LVH samples is expanded. Thus, it allows the SVM method to have a higher prediction performance on unknown LVH samples. The SMOTE is used in the process of 4-fold cross validation. The training data of the LVH group are pre-processed and augmented by SMOTE to be the same numbers with those of the Non-LVH group as 1,225, 1,230, 1,229 and 1,215, respectively, for the four folds as shown in Table 4. We also compare the performances of the test set for the proposed model with and without using SMOTE.
TABLE 4. Data Numbers in the Training and Validation Set.
| Fold | Data | Non-LVH | LVH | Total |
|---|---|---|---|---|
| 1st | Training Set | 1225 | 92 | 1317 |
| Pre-processed by SMOTE | 1225 | 1225 | 2450 | |
| Validation Set | 408 | 31 | 439 | |
| 2nd | Training Set | 1230 | 87 | 1317 |
| Pre-processed by SMOTE | 1230 | 1230 | 2460 | |
| Validation Set | 403 | 36 | 439 | |
| 3rd | Training Set | 1229 | 88 | 1317 |
| Pre-processed by SMOTE | 1229 | 1229 | 2458 | |
| Validation Set | 404 | 35 | 439 | |
| 4th | Training Set | 1215 | 102 | 1317 |
| Pre-processed by SMOTE | 1215 | 1215 | 2430 | |
| Validation Set | 418 | 21 | 439 |
D. Machine Learning Model
Our proposed method utilizes SVM [45]–[48] as a binary classifier for machine learning. In our method, a 31-dimensional vector represents a data point and we employ the linear kernel (Linear SVM) to separate such points by a 30-dimesional hyperplane. Maximum-margin is constructed by SVM so that the distance from the hyperplane to the nearest subset of the training data points (support vectors) of Non-LVH or LVH class is maximized. The soft-margin SVM is adopted in our method. Soft-margin SVM allows the wide decision margin and some outliers are inside or on the wrong side of the margin.
Let 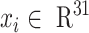 denote a 31-dimensional training vector with associated label
denote a 31-dimensional training vector with associated label 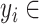 {1, −1}.
{1, −1}. 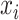 also includes the synthetic samples of LVH group applied by SMOTE and all the data of
also includes the synthetic samples of LVH group applied by SMOTE and all the data of 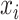 are processed by Min-Max normalization.
are processed by Min-Max normalization. 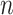 indicates the number of training vectors. The weight vector
indicates the number of training vectors. The weight vector 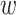 , which is related to the construction of hyperplane for SVM, is obtained by solving the objective function as shown in (4)
[41], [47]. The second term in (4) is the squared hinge loss (L2 loss) function for the soft-margin SVM evaluated on the training data and weighted by hyperparameter
, which is related to the construction of hyperplane for SVM, is obtained by solving the objective function as shown in (4)
[41], [47]. The second term in (4) is the squared hinge loss (L2 loss) function for the soft-margin SVM evaluated on the training data and weighted by hyperparameter 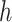 . The soft-margin formulation can help in avoiding over-fitting.
. The soft-margin formulation can help in avoiding over-fitting.
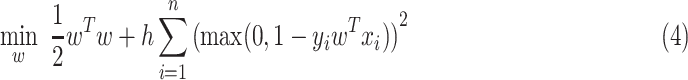 |
where 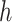 is a hyperparameter which decides the trade-off between maximizing the margin and minimizing the training error. When
is a hyperparameter which decides the trade-off between maximizing the margin and minimizing the training error. When 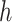 is large, avoiding misclassification is emphasized at the expense of maintaining the margin small, whereas when
is large, avoiding misclassification is emphasized at the expense of maintaining the margin small, whereas when 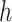 is small, classification errors are presented less importance and focus is more on maximizing the margin. The optimized hyperparameter
is small, classification errors are presented less importance and focus is more on maximizing the margin. The optimized hyperparameter 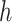 is chosen by grid search according to the average AUC of the PR curves of the cross validation in our algorithm. As demonstrated in Fig.2, the hyperparameter
is chosen by grid search according to the average AUC of the PR curves of the cross validation in our algorithm. As demonstrated in Fig.2, the hyperparameter 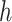 is initialized to 0.02. The training processes with the increment 0.001 of
is initialized to 0.02. The training processes with the increment 0.001 of 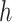 for grid search is iterated until
for grid search is iterated until 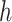 reaches to 1. The optimized hyperparameter is chosen based on the highest AUC of the PR curves among the candidates of
reaches to 1. The optimized hyperparameter is chosen based on the highest AUC of the PR curves among the candidates of 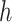 .
.
After selecting the optimized hyperparameter, the SVM training model will be determined by the data in the total training and validation set. As shown in Table 5, the data of total training and validation set for the LVH group are pre-processed by SMOTE, and the number is increased to 1,633.
TABLE 5. Data Numbers of Total Data.
| Data | Non-LVH | LVH | Total |
|---|---|---|---|
| Total Training and Validation Set | 1633 | 123 | 1756 |
| Pre-processed by SMOTE | 1633 | 1633 | 3266 |
| Test Set | 413 | 27 | 440 |
| Total Data | 2046 | 150 | 2196 |
IV. Results and Discussion
Our proposed method is implemented using the software scikit learn v0.20.2 with Python programming language [49]. In addition, the optimal weight vector 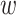 for hyperplane is obtained by LIBLINEAR (A Library for Large Linear Classification) [47], an open source library for large linear classification. The optimized hyperparameter
for hyperplane is obtained by LIBLINEAR (A Library for Large Linear Classification) [47], an open source library for large linear classification. The optimized hyperparameter 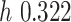 is chosen when the highest AUC of the PR curve averaged from the 4-fold cross validation is found from the values of 981 trials.
is chosen when the highest AUC of the PR curve averaged from the 4-fold cross validation is found from the values of 981 trials.
A. Performance Measurement
The specificity 70-75% is the criterion to decide the most appropriate test cut-off probability [50] for our SVM method. The performance is assessed by several standard measurements including accuracy, specificity, sensitivity (recall), precision, F1 score, the AUC of the ROC curve and the AUC of the PR curve [51], [52].
The definitions of accuracy, specificity, sensitivity and precision are calculated by true positive (TP), true negative (TN), false positive (FP), and false negative (FN) as denoted in (5) - (8). The F1 score, which is the harmonic average of the precision and recall, is described in (9).
 |
B. Results
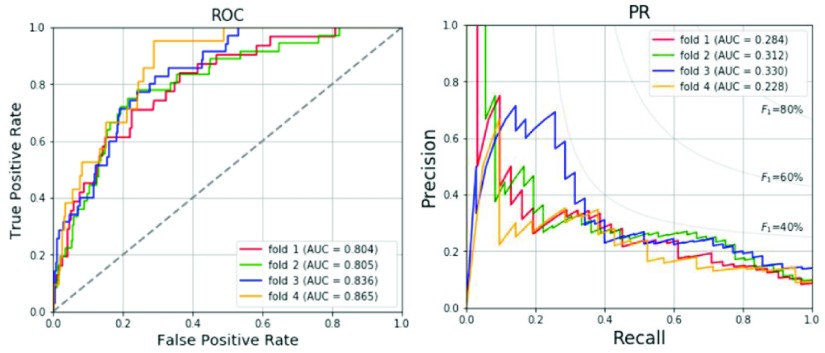
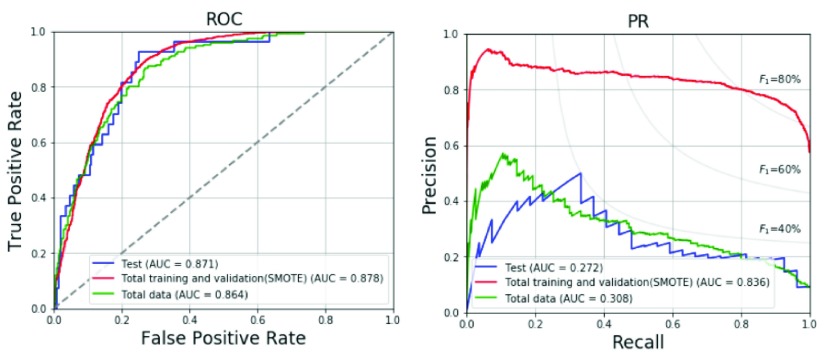
The results of the 4-fold cross validation for the validation set with the optimized hyperparameter are shown in Table 6. The prevalence of LVH in the validation set is range from 4.8% to 8.2% as shown in Table 6. Average accuracy, specificity, sensitivity, precision and F1 score are 73.3%, 72.9%, 78.8%, 18.3% and 29.3%, respectively. The ROC and PR curves for the four folds are compared in Fig. 3. The average AUC of the ROC curve is 0.828 and the average AUC of the PR curve is 0.289. Table 7 shows the prediction results of the total training and validation set, test set and total data. In the total training and validation set, the SMOTE is applied for the LVH group to increase the prevalence rate to 50%. Thus, the precision, F1 score and AUC of PR curves are much better than the other two datasets. In the test set and total data, the prevalence of LVH is generally distributed around 6-7% in the population of young adults. The results of the test set for the model with SMOTE regarding the accuracy, specificity, sensitivity, precision and F1 score are 76.1%, 75.1%, 92.6%, 19.5% and 32.2%, respectively, which are in line with the results of the total data for the model with SMOTE, and better than 74.6%, 73.9%, 85.2%, 17.6%, and 29.2%, respectively, for the model without using SMOTE. The ROC and PR curves for various datasets are compared in Fig. 4. The AUC values for the three datasets are similar in ROC curves. The AUC values of ROC and PR curves of the proposed method with SMOTE for test set are 0.871 and 0.272, respectively, and larger than 0.841 and 0.259, respectively, for the model without using SMOTE.
TABLE 6. Data Numbers and Performances for the 4-Fold Cross Validation.
| Validation set 1st fold | Validation set 2nd fold | Validation set 3rd fold | Validation set 4th fold | Average | |
|---|---|---|---|---|---|
| Non-LVH Group | 408 | 403 | 404 | 418 | – |
| LVH Group | 31 | 36 | 35 | 21 | – |
| Total | 439 | 439 | 439 | 439 | – |
| Prevalence Rate | 7.1% | 8.2% | 8.0% | 4.8% | – |
| Accuracy | 67.2% | 77.9% | 75.9% | 72.0% | 73.3% |
| Specificity | 66.4% | 78.2% | 75.7% | 71.3% | 72.9% |
| Sensitivity | 77.4% | 75.0% | 77.1% | 85.7% | 78.8% |
| Precision | 14.9% | 23.5% | 21.6% | 13.0% | 18.3% |
| F1-score | 25.0% | 35.8% | 33.7% | 22.6% | 29.3% |
| ROC AUC | 0.804 | 0.805 | 0.836 | 0.865 | 0.828 |
| PR AUC | 0.284 | 0.312 | 0.330 | 0.228 | 0.289 |
| True Negative | 271 | 315 | 306 | 298 | – |
| False Negative | 7 | 9 | 8 | 3 | – |
| False Positive | 137 | 88 | 98 | 120 | – |
| True Positive | 24 | 27 | 27 | 18 | – |
FIGURE 3.
ROC and PR curves for the 4-fold cross validation.
TABLE 7. Predicted Results of the Proposed Method for Various Datasets.
| Total Training and validation set (SMOTE) | Test set | Total data | |
|---|---|---|---|
| Non-LVH Group | 1633 | 413 | 2046 |
| LVH Group | 1633 | 27 | 150 |
| Total | 3266 | 440 | 2196 |
| Prevalence Rate | 50.0% | 6.1% | 6.8% |
| Accuracy | 81.2% | 76.1% | 74.2% |
| Specificity | 72.8% | 75.1% | 73.3% |
| Sensitivity | 89.6% | 92.6% | 86.7% |
| Precision | 76.7% | 19.5% | 19.2% |
| F1-score | 82.6% | 32.2% | 31.4% |
| ROC AUC | 0.878 | 0.871 | 0.864 |
| PR AUC | 0.836 | 0.272 | 0.308 |
| True Negative | 1189 | 310 | 1499 |
| False Negative | 170 | 2 | 20 |
| False Positive | 444 | 103 | 547 |
| True Positive | 1463 | 25 | 130 |
FIGURE 4.
ROC and PR curves of the proposed method for various datasets.
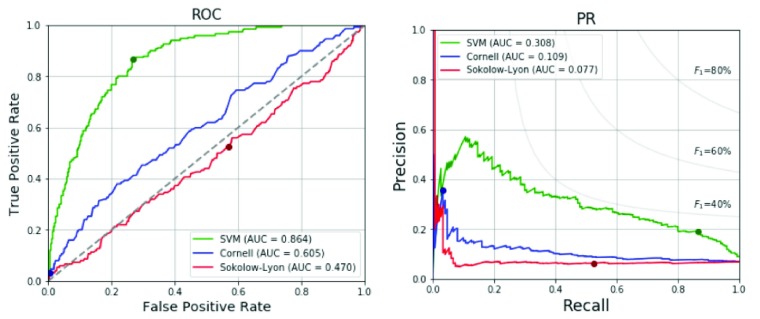
Our proposed SVM machine learning method is also compared with the Sokolow-Lyon voltage and Cornell voltage criteria for LVH as shown in Table 8. All data of the 2,196 military males are tested in the model. With the specificity of 73.3%, intended to be set between 70-75%, our SVM technique provides much better sensitivity 86.7% compared to 3.3% and 52.7% regarding the Cornell and Sokolow-Lyon voltage criteria, respectively. The ROC and PR curves for the three approaches are shown in Fig. 5. It is obvious that the proposed method is much superior to the other two traditional ECG voltage criteria.
TABLE 8. Performance Comparison of Proposed Method and Traditional ECG Voltage Criteria.
| SVM | Cornell voltage | Sokolow-Lyon voltage | |
|---|---|---|---|
| Cut-off Value | 0.422 | 28 mm | 35 mm |
| Accuracy | 74.2% | 93.0% | 43.8% |
| Specificity | 73.3% | 99.6% | 43.2% |
| Sensitivity | 86.7% | 3.3% | 52.7% |
| Precision | 19.2% | 35.7% | 6.4% |
| F1-score | 31.4% | 6.0% | 11.4% |
| ROC AUC | 0.864 | 0.605 | 0.470 |
| PR AUC | 0.308 | 0.109 | 0.077 |
| True Negative | 1499 | 2037 | 883 |
| False Negative | 20 | 145 | 71 |
| False Positive | 547 | 9 | 1163 |
| True Positive | 130 | 5 | 79 |
FIGURE 5.
ROC and PR curves of the proposed method and traditional ECG voltage criteria.
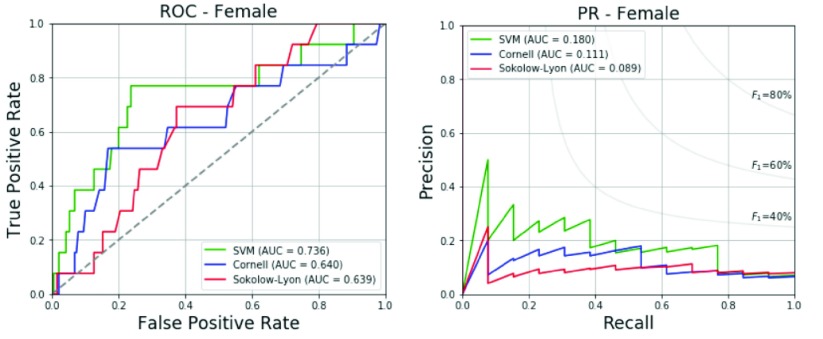
In addition, we also test the CHIEF military female subcohort data with the label of echocardiographic LVH by the definitions of LVM/BSA ≥ 88 gram/meter2 or LVM/height2.7 ≥ 41 gram/meter2.7 for young females using the proposed SVM model trained by the military young males. The baseline data with an average for each adopted biological and ECG features of the female participants with and without echocardiographic LVH are revealed in Table 9. It is contrary to the findings of the male participants that there are no significant differences in the adopted biological parameters including age, body height and body weight, and there are only two significant differences in the ECG characteristics including the amplitudes of R waves in chest Leads V1 and V3. In addition, the results of the female test set with regard to the accuracy, specificity, sensitivity, precision and F1 score are 76.4%, 76.3%, 76.9%, 18.2% and 29.4%, respectively, and shown in detail in Table 10. Compared to the conventional Sokolow-Lyon voltage and Cornell voltage criteria specifically for females [21], the proposed SVM method can provide superior performance evaluated by F1 score, and the AUCs of the ROC and PR curves. The ROC and PR curves obtained from the female’s test data are shown in Fig. 6.
TABLE 9. Characteristics of Study Participants (Females).
| Features | Total N=203 | Non-LVH N=190 | LVH N=13 | p-value |
|---|---|---|---|---|
| Age (years) | 25.42± 5.38 | 25.47± 5.31 | 24.69± 6.56 | 0.6162 |
| Height (cm) | 160.55± 4.81 | 160.63± 4.75 | 159.35± 5.72 | 0.3570 |
| Weight (kg) | 58.90± 9.24 | 58.55± 8.83 | 64.00± 13.34 | 0.1711 |
| Heart rate (bpm) | 68.40± 11.72 | 68.23± 11.24 | 71.00± 17.77 | 0.5884 |
| P-II(ms) | 100.07± 15.99 | 99.90± 15.60 | 102.54± 21.48 | 0.5661 |
| PR-II(ms) | 148.78± 23.52 | 149.41± 23.63 | 139.62± 20.53 | 0.1470 |
| QRS-II(ms) | 89.93± 12.10 | 89.55± 11.23 | 95.46± 21.00 | 0.3338 |
| QT-II(ms) | 383.96± 34.32 | 383.16± 32.74 | 395.54± 52.76 | 0.4193 |
| QTc-II(ms) | 410.01± 29.14 | 408.98± 27.24 | 425.08± 48.41 | 0.2576 |
| P axis-II(degree) | 42.16± 28.33 | 42.83± 28.22 | 32.31± 29.16 | 0.1957 |
| QRS axis-II(degree) | 71.02± 31.76 | 71.65± 30.85 | 61.92± 43.47 | 0.2866 |
| T axis-II(degree) | 33.94± 20.82 | 34.18± 20.06 | 30.31± 30.80 | 0.6621 |
| R-I(mm) | 4.19± 2.16 | 4.11± 2.08 | 5.36± 3.02 | 0.1647 |
| R-II(mm) | 11.38± 3.75 | 11.32± 3.70 | 12.22± 4.48 | 0.4014 |
| R-III(mm) | 8.34± 4.61 | 8.36± 4.57 | 8.07± 5.37 | 0.8273 |
| R-aVR(mm) | 0.90± 1.17 | 0.93± 1.18 | 0.57± 0.83 | 0.2879 |
| R-aVL(mm) | 1.46± 1.18 | 1.42± 1.15 | 1.93± 1.47 | 0.1330 |
| S-aVL(mm) | 2.69± 2.80 | 2.72± 2.81 | 2.38± 2.70 | 0.6813 |
| R-aVF(mm) | 9.83± 3.92 | 9.81± 3.92 | 10.12± 4.03 | 0.7816 |
| R-V1(mm) | 2.75± 1.60 | 2.67± 1.53 | 3.92± 2.10 | 0.0059 |
| S-V1(mm) | 7.03± 3.52 | 6.91± 3.51 | 8.72± 3.37 | 0.0732 |
| R-V2(mm) | 6.42± 2.88 | 6.34± 2.80 | 7.70± 3.70 | 0.0983 |
| S-V2(mm) | 9.78± 4.82 | 9.70± 4.76 | 11.02± 5.76 | 0.3387 |
| R-V3(mm) | 9.20± 4.40 | 9.03± 4.30 | 11.68± 5.14 | 0.0348 |
| S-V3(mm) | 5.50± 3.76 | 5.41± 3.68 | 6.83± 4.79 | 0.1872 |
| R-V4(mm) | 13.24± 4.45 | 13.16± 4.42 | 14.38± 4.91 | 0.3411 |
| S-V4(mm) | 3.64± 2.94 | 3.61± 2.91 | 4.21± 3.46 | 0.4768 |
| R-V5(mm) | 13.52± 3.96 | 13.45± 3.98 | 14.45± 3.55 | 0.3824 |
| S-V5(mm) | 2.53± 2.19 | 2.51± 2.16 | 2.70± 2.60 | 0.7664 |
| R-V6(mm) | 12.09± 3.80 | 12.01± 3.78 | 13.15± 4.08 | 0.2956 |
| S-V6(mm) | 1.61± 1.57 | 1.62± 1.55 | 1.54± 1.87 | 0.8512 |
TABLE 10. Performance Comparison of Proposed Method and Traditional ECG Voltage Criteria for Female’s Test Data.
| SVM | Cornell voltage | Sokolow-Lyon voltage | |
|---|---|---|---|
| Cut-off Value | 0.355 | 20 mm | 35 mm |
| Accuracy | 76.4% | 93.1% | 91.6% |
| Specificity | 76.3% | 99.5% | 97.4% |
| Sensitivity | 76.9% | 0% | 7.7% |
| Precision | 18.2% | 0% | 16.7% |
| F1-score | 29.4% | 0% | 10.5% |
| ROC AUC | 0.736 | 0.640 | 0.639 |
| PR AUC | 0.180 | 0.111 | 0.089 |
| True Negative | 145 | 189 | 185 |
| False Negative | 3 | 13 | 12 |
| False Positive | 45 | 1 | 5 |
| True Positive | 10 | 0 | 1 |
FIGURE 6.
ROC and PR curves of the proposed method and traditional ECG voltage criteria for the female’s test data.
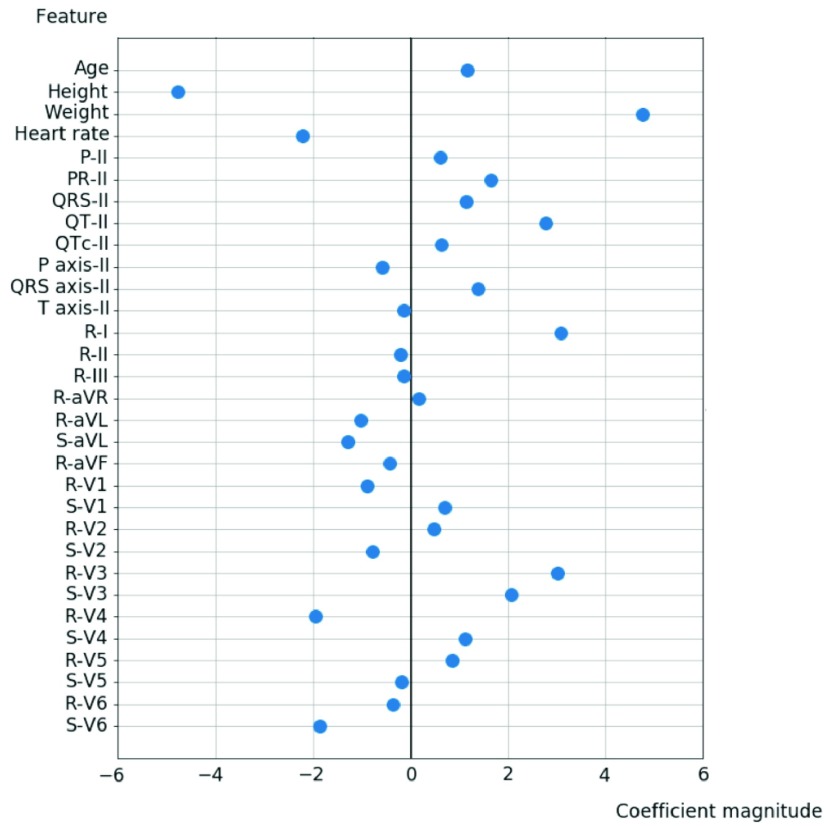
Fig. 7 shows the feature importance with regard to the overall 31 input characteristics. We can see that body height and body weight are the most important factors of echocardiographic LVH with a coefficient magnitude greater than 4 in our SVM model. The other significant predictors of LVH with greater coefficient magnitude include age, heart rate, PR interval, uncorrected QT interval, QRS axis in Lead II, R amplitudes in Lead I, Lead V3, V4, and S amplitudes in Lead V3, V6.
FIGURE 7.
Feature importance of the 31 input parameters.
C. Discussion
A few studies have utilized machine learning or deep learning techniques for ECG characteristics training to predict the LVH presence [1]–[3], [53]–[55]. However, the disadvantages are that most of the studies include a small sample size of participants [53]–[55], or the output is aimed merely for HCM [2], [3] but not for all kinds of LVH phenotypes in the general population. To our knowledge, the Multi-Ethnic Study of Atherosclerosis (MESA) study might be the only one research in screening for the presence of any unspecific LVH based on the definition of cardiac magnetic resonance imaging in a large sample size of middle and old aged general population [1]. As compared with MESA, our study has superior results as both the ECG characteristics and simple biological features are trained by the SVM. On the basis of feature importance analysis of the proposed SVM model, it is obvious that body height and body weight are the strongest predictors of LVH among young adults. We also notice that an addition of systolic and diastolic blood pressures on the currently used SVM model can improve merely a little or is similar in the detection of LVH. It is reasonable that elevated levels of blood pressure are highly correlated with greater body mass index among young adults [56] and the effect time on cardiac remodeling is relatively short. Therefore, blood pressure may not play a critical role on the development of LVH in young adults.
V. Conclusion
This study develops a clinically effective ECG-based system with age and simple anthropometric data through the SVM machine learning technique in screening for unspecific LVH among young adults, which improves much in the sum of sensitivity and specificity as compared with the traditional ECG criteria for LVH or using the ECG parameters alone for the machine learning. The sensitivity of our proposed method achieves up to 92.6%. In addition, since the test performances regarding accuracy, specificity, sensitivity, precision, F1 score, and the AUCs of the ROC and PR curves for the female samples are not optimal by adopting the SVM model trained by male samples, future studies should be done to clarify the validity of our system operated specifically for young females.
Funding Statement
This work was supported by Hualien Armed Forces General Hospital, Taiwan, under Grant 805-C109-07.
References
- [1].Sparapani R.et al. , “Detection of left ventricular hypertrophy using Bayesian additive regression trees: The MESA (Multi-Ethnic Study of Atherosclerosis),” J. Am. Heart Assoc., vol. 8, no. 9, May 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Rahman Q. A., Tereshchenko L. G., Kongkatong M., Abraham T., Abraham M. R., and Shatkay H., “Utilizing ECG-based heartbeat classification for hypertrophic cardiomyopathy identification,” IEEE Trans. Nanobiosci., vol. 14, no. 5, pp. 505–512, Jul. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Tison G. H., Zhang J., Delling F. N., and Deo R. C., “Automated and interpretable patient ECG profiles for disease detection, tracking, and discovery,” 2018, arXiv:1807.02569. [Online]. Available: http://arxiv.org/abs/1807.02569 [DOI] [PMC free article] [PubMed]
- [4].Teijeiro A. E., Shokrekhodaei M., and Nazeran H., “The conceptual design of a novel workstation for seizure prediction using machine learning with potential eHealth applications,” IEEE J. Transl. Eng. Health Med., vol. 7, 2019, Art. no. 2900110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Yadav D. P., Sharma A., Singh M., and Goyal A., “Feature extraction based machine learning for human burn diagnosis from burn images,” IEEE J. Transl. Eng. Health Med., vol. 7, 2019, Art. no. 1800507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Decaro C.et al. , “Machine learning approach for prediction of hematic parameters in hemodialysis patients,” IEEE J. Transl. Eng. Health Med., vol. 7, 2019, Art. no. 4100308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Fan X., Yao Q., Cai Y., Miao F., Sun F., and Li Y., “Multiscaled fusion of deep convolutional neural networks for screening atrial fibrillation from single lead short ECG recordings,” IEEE J. Biomed. Health Informat., vol. 22, no. 6, pp. 1744–1753, Nov. 2018. [DOI] [PubMed] [Google Scholar]
- [8].Yang G.et al. , “An IoT-enabled stroke rehabilitation system based on smart wearable armband and machine learning,” IEEE J. Transl. Eng. Health Med., vol. 6, 2018, Art. no. 2100510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lin G.-M.et al. , “Transforming retinal photographs to entropy images in deep learning to improve automated detection for diabetic retinopathy,” J. Ophthalmol., vol. 2018, Sep. 2018, Art. no. 2159702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chen M.-J.et al. , “Machine learning to relate PM2.5 and PM10 concentrations to outpatient visits for upper respiratory tract infections in taiwan: A nationwide analysis,” World J. Clin. Cases, vol. 6, no. 8, pp. 200–206, Aug. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Tao R.et al. , “Magnetocardiography-based ischemic heart disease detection and localization using machine learning methods,” IEEE Trans. Biomed. Eng., vol. 66, no. 6, pp. 1658–1667, Jun. 2019. [DOI] [PubMed] [Google Scholar]
- [12].Ambale-Venkatesh B.et al. , “Cardiovascular event prediction by machine learning: The multi-ethnic study of atherosclerosis,” Circulat. Res., vol. 121, no. 9, pp. 1092–1101, Oct. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Levy D., “Echocardiographically detected left ventricular hypertrophy: Prevalence and risk factors,” Ann. Internal Med., vol. 108, no. 1, pp. 7–13, Jan. 1988. [DOI] [PubMed] [Google Scholar]
- [14].Ruilope L. M. and Schmieder R. E., “Left ventricular hypertrophy and clinical outcomes in hypertensive patients,” Amer. J. Hypertension, vol. 21, no. 5, pp. 500–508, May 2008. [DOI] [PubMed] [Google Scholar]
- [15].Su F.-Y.et al. , “A comparison of cornell and sokolow-lyon electrocardiographic criteria for left ventricular hypertrophy in a military male population in taiwan: The cardiorespiratory fitness and HospItalization events in armed forces study,” Cardiovascular Diagnosis Therapy, vol. 7, no. 3, pp. 244–251, Jun. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Rawlins J., Bhan A., and Sharma S., “Left ventricular hypertrophy in athletes,” Eur. J. Echocardiography, vol. 10, no. 3, pp. 350–356, 2009. [DOI] [PubMed] [Google Scholar]
- [17].Georgakopoulos D. and Tolis V., “Hypertrophic cardiomyopathy in children, teenagers and young adults,” Hellenic J. Cardiol., vol. 48, no. 4, pp. 228–233, Jul.-Aug. 2007. [PubMed] [Google Scholar]
- [18].Armstrong A. C.et al. , “Framingham score and LV mass predict events in young adults: CARDIA study,” Int. J. Cardiol., vol. 172, no. 2, pp. 350–355, Mar. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Okin P. M.et al. , “Ethnic differences in electrocardiographic criteria for left ventricular hypertrophy: The LIFE study,” Amer. J. Hypertension, vol. 15, no. 8, pp. 663–671, Aug. 2002. [DOI] [PubMed] [Google Scholar]
- [20].Sokolow M. and Lyon T. P., “The ventricular complex in left ventricular hypertrophy as obtained by unipolar precordial and limb leads,” Amer. Heart J., vol. 37, no. 2, pp. 161–186, Feb. 1949. [DOI] [PubMed] [Google Scholar]
- [21].Casale P. N., Devereux R. B., Alonso D. R., Campo E., and Kligfield P., “Improved sex-specific criteria of left ventricular hypertrophy for clinical and computer interpretation of electrocardiograms: Validation with autopsy findings,” Circulation, vol. 75, no. 3, pp. 565–572, Mar. 1987. [DOI] [PubMed] [Google Scholar]
- [22].Corrado D. and McKenna W. J., “Appropriate interpretation of the athlete’s electrocardiogram saves lives as well as money,” Eur. Heart J., vol. 28, no. 16, pp. 1920–1922, Aug. 2007. [DOI] [PubMed] [Google Scholar]
- [23].Potter S. L. P.et al. , “Detection of hypertrophic cardiomyopathy is improved when using advanced rather than strictly conventional 12-lead electrocardiogram,” J. Electrocardiol., vol. 43, no. 6, pp. 713–718, Nov. 2010. [DOI] [PubMed] [Google Scholar]
- [24].Lin G.-M.et al. , “Rationale and design of the cardiorespiratory fitness and hospitalization events in armed forces study in eastern taiwan,” World J. Cardiol., vol. 8, no. 8, pp. 464–471, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Chen Y.-J.et al. , “Chronic hepatitis B, nonalcoholic steatohepatitis and physical fitness of military males: CHIEF study,” World J. Gastroenterology, vol. 23, no. 25, pp. 4587–4594, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Chen K.-W.et al. , “Sex-specific association between metabolic abnormalities and elevated alanine aminotransferase levels in a military cohort: The CHIEF study,” Int. J. Environ. Res. Public Health, vol. 15, no. 3, p. 545, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Tsai K.-Z.et al. , “Association of betel nut chewing with exercise performance in a military male cohort: The CHIEF study,” J. Roy. Army Med. Corps, vol. 164, no. 6, pp. 399–404, Nov. 2018. [DOI] [PubMed] [Google Scholar]
- [28].Lin J.-W.et al. , “Sex-specific association between serum uric acid and elevated alanine aminotransferase in a military cohort: The CHIEF study,” Endocrine, Metabolic Immune Disorders—Drug Targets, vol. 19, no. 3, pp. 333–340, Apr. 2019. [DOI] [PubMed] [Google Scholar]
- [29].Chao W.-H., Su F.-Y., Lin F., Yu Y.-S., and Lin G.-M., “Association of electrocardiographic left and right ventricular hypertrophy with physical fitness of military males: The CHIEF study,” Eur. J. Sport Sci., vol. 19, no. 9, pp. 1214–1220, Oct. 2019. [DOI] [PubMed] [Google Scholar]
- [30].Meng F.-C., Lin Y.-P., Su F.-Y., Yu Y.-S., and Lin G.-M., “Association between electrocardiographic and echocardiographic right ventricular hypertrophy in a military cohort in taiwan: The CHIEF study,” Indian Heart J., vol. 69, no. 3, pp. 331–333, May 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Tsai K.-Z.et al. , “Association between mild anemia and physical fitness in a military male cohort: The CHIEF study,” Sci. Rep., vol. 9, no. 1, Dec. 2019, Art. no. 11165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lu S.-C.et al. , “Quantitative physical fitness measures inversely associated with myopia severity in military males: The CHIEF study,” Amer. J. Men’s Health, vol. 13, no. 5, Oct. 2019, Art. no. 155798831988376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Su F.-Y., Wang S.-H., Lu H. H.-S., and Lin G.-M., “Association of tobacco smoking with physical fitness of military males in taiwan: The CHIEF study,” Can. Respiratory J., vol. 2020, pp. 1–6, Jan. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lang R. M.et al. , “Recommendations for cardiac chamber quantification by echocardiography in adults: An update from the American society of echocardiography and the European association of cardiovascular imaging,” J. Amer. Soc. Echocardiography, vol. 28, no. 1, pp. 1–39, Jan. 2015. [DOI] [PubMed] [Google Scholar]
- [35].Devereux R. B., “Detection of left ventricular hypertrophy by M-mode echocardiography. Anatomic validation, standardization, and comparison to other methods,” Hypertension, vol. 9, no. 2, pp. 19–26, Feb. 1987. [DOI] [PubMed] [Google Scholar]
- [36].Dubois D. and Dubois E. F., “A formula to estimate the approximate surface area if height and weight be known,” Arch. Intern. Med., vol. 17, pp. 863–871, 1916. [Google Scholar]
- [37].de Simone G.et al. , “Strong heart study investigators: Normalization for body size and population-attributable risk of left ventricular hypertrophy,” Am. J. Hypertens., vol. 18, pp. 191–196, Feb. 2005. [DOI] [PubMed] [Google Scholar]
- [38].Wong R. C.-C., Yip J. W.-L., Gupta A., Yang H., and Ling L. H., “Echocardiographic left ventricular mass in a multiethnic southeast Asian population: Proposed new gender and age-specific norms,” Echocardiography, vol. 25, no. 8, pp. 805–811, Sep. 2008. [DOI] [PubMed] [Google Scholar]
- [39].Mukkamala S., Janoski G., and Sung A., “Intrusion detection: Support vector machines and neural networks,” in Proc. Int. Jt. Conf. Neural Netw. (IJCNN), May 2002, pp. 1702–1707. [Google Scholar]
- [40].Ç. Çöltekin and Rama T., “Discriminating similar languages: Experiments with linear SVMs and neural networks,” in Proc. VarDialogy, Dec. 2016, pp. 15–24. [Google Scholar]
- [41].Cumani S. and Laface P., “Analysis of large-scale SVM training algorithms for language and speaker recognition,” IEEE Trans. Audio, Speech, Language Process., vol. 20, no. 5, pp. 1585–1596, Jul. 2012. [Google Scholar]
- [42].Xu S. and Lai S., “Fast Fourier transform based data perturbation method for privacy protection,” in Proc. IEEE Intell. Secur. Informat., May 2007, pp. 45–50. [Google Scholar]
- [43].Suresh Y., Kumar L., and Rath S. K., “Statistical and machine learning methods for software fault prediction using CK metric suite: A comparative analysis,” ISRN Softw. Eng., vol. 2014, pp. 1–15, 2014. [Google Scholar]
- [44].Chawla N. V., Bowyer K. W., Hall L. O., and Kegelmeyer W. P., “SMOTE: Synthetic minority over-sampling technique,” J. Artif. Intell. Res., vol. 16, pp. 321–357, Jun. 2002. [Google Scholar]
- [45].Scholkopf B., Learning With Kernels: Support Vector Machines, Regularization, Optimization, and Beyond. Cambridge, MA, USA: MIT Press, 2001. [Google Scholar]
- [46].Chang Y. W. and Lin C. J., “Feature ranking using linear SVM,” J. Mach. Learn. Res., vol. 3, pp. 53–64, Dec. 2008. [Google Scholar]
- [47].Fan R.-E., Chang K.-W., Hsieh C.-J., Wang X.-R., and Lin C.-J., “LIBLINEAR: A library for large linear classification,” J. Mach. Learn. Res., vol. 9, pp. 1871–1874, Jun. 2008. [Google Scholar]
- [48].Burges C. J. C., “A tutorial on support vector machines for pattern recognition,” Data Mining Knowl. Discovery, vol. 2, no. 2, pp. 121–167, 1998. [Google Scholar]
- [49].Pedregosa F.et al. , “Scikit-learn: Machine learning in Python,” J. Mach. Learn. Res., vol. 12, pp. 2825–2830, Oct. 2011. [Google Scholar]
- [50].Habibzadeh F., Habibzadeh P., and Yadollahie M., “On determining the most appropriate test cut-off value: The case of tests with continuous results,” Biochemia Medica, pp. 297–307, 2016. [DOI] [PMC free article] [PubMed]
- [51].Davis J. and Goadrich M., “The relationship between precision-recall and ROC curves,” in Proc. 23rd Int. Conf. Mach. Learn. (ICML), Jun. 2006, pp. 233–240. [Google Scholar]
- [52].Hajian-Tilaki K., “Receiver operating characteristic curve analysis for medical diagnostic test evaluation,” Caspian J. Intern. Med., vol. 4, no. 2, pp. 627–635, Mar. 2013. [PMC free article] [PubMed] [Google Scholar]
- [53].Afkhami R. G. and Tinati M. A., “ECG based detection of left ventricular hypertrophy using higher order statistics,” in Proc. 23rd Iranian Conf. Electr. Eng., May 2015, pp. 1–5. [Google Scholar]
- [54].Deng Y., Duan X., Jiao B., Zhu T., and Wang Z., “Electrocardiographic criteria for the diagnosis of left ventricular hypertrophy based on data mining,” in Proc. 10th Biomed. Eng. Int. Conf. (BMEiCON), Aug. 2017, pp. 1–5. [Google Scholar]
- [55].Sengupta S., Ghosh U., Sarkar S., and Das S., “Application of fractional calculus to distinguish left ventricular hypertrophy with normal ECG,” in Proc. 4th Int. Conf. Recent Adv. Inf. Technol. (RAIT), Mar. 2018, pp. 1–6. [Google Scholar]
- [56].Linderman G. C.et al. , “Association of body mass index with blood pressure among 1.7 million Chinese adults,” JAMA Netw. Open, vol. 1, no. 4, Aug. 2018, Art. no. e181271. [DOI] [PMC free article] [PubMed] [Google Scholar]