Abstract
Most caspases can be positioned unambiguously within the regulated cell death networks of apoptosis and pyroptosis, but the role of caspase-2, a highly conserved protease within the family, remains enigmatic. This is mainly due to lack of selective chemical and biochemical tools for the investigation of this protease. In this study, we used our hybrid combinatorial substrate library (HyCoSuL) approach to broadly profile caspase-2 substrate specificity using peptide scanning libraries. This screen uncovered previously unknown caspase-2 peptidyl substrate preferences, which were further used to develop caspase-2 selective fluorogenic substrates and covalent, irreversible AOMK inhibitors. Finally, we used the champion inhibitor (NH-23-C2) in reversine-treated HCT-116 colon cancer cells to selectively block caspase-2 activity and caspase-2-mediated MDM-2 cleavage. In addition, we showed that NH-23-C2 does not block caspase-3 or caspase-8, which makes it a powerful chemical tool to dissect the true role of caspase-2 in various biological setups.
Subject terms: Proteases, Chemical tools, Proteolysis
Introduction
Caspases are proteases that play dominant roles in apoptosis and inflammation, but also participate in cell differentiation [1]. Generally, these enzymes can be divided into three major groups: initiators of apoptosis (caspases-8, -9, and -10), executioners of apoptosis (caspases-3, -6, -7), and proinflammatory caspases (-1, -4, and -5 in humans, -1 and -11 in mice) [2, 3]. Caspase-2 (formerly known as NEDD-2 [4] or Ich-1 [5]) has been reported to act as an apoptosis initiator or executioner as well as participant in non-apoptotic processes [6–8]. Caspase-2 is highly conserved in vertebrates; however, despite over two decades of research, its biological role remains far less understood compared to other caspases [6]. The scope of caspase-2 action is much broader than for other caspases, however, there is still some controversy as several caspase-2-related studies failed to be replicated [6, 9, 10]. It is reported that caspase-2 participates in multiple forms of cell death caused by various stimulants, including endoplasmic reticulum (ER) stress [11], metabolic stress [12], heat shock response [13, 14], response to DNA damage [15, 16], senescence [17], mitotic catastrophe [18], and others (see reviews [6, 9, 19–21]). This enzyme can also be involved in cancer by acting as a tumor suppressor [4, 22].
Difficulties in dissecting the role of caspase-2 in physiology and disease arise from the lack of selective tools that may distinguish between caspase-2 and other family members. Most of the experiments rely on the use of antibodies, mRNA knockdowns [23], caspase-2-deficient mouse models [24], or measuring caspase-2 activation by induced proximity assays with protein fluorescent probes [25]. However, since the lack of one caspase can be compensated by another [26, 27], it would be beneficial to the field to develop highly selective caspase-2 substrates and inhibitors that can be used to interrogate various systems where caspase-2 might be involved.
Caspase-2 has an almost absolute requirement for Asp at the P1 position [2]. Positional scanning synthetic combinatorial library (PS-SCL) strategies using short substrates revealed that this enzyme has a narrow specificity at P4 with high preferences for Asp, and broader preferences at P3–P2 region, which overlaps with caspases-3 and -7 specificity [2]. Similarities in specificity between caspases-2, -3, and -7 reported for short substrates are in line with recent proteomic data demonstrating that these enzymes cleave almost the same pool of protein substrates, but with significantly different catalytic efficiency [28, 29], and it has been hypothesized that the substrates cleavage rates rather than overall specificity might be important to understand the true role of caspase-2 [28]. Another unique feature of this enzyme is the requirement of at least five amino acids in the non-prime region for the efficient catalysis, with the VDVAD peptide being the most active caspase-2 substrate [30, 31]. However, it has been demonstrated that substrates and inhibitors based on VDVAD sequence are even more potent toward caspase-3 [32, 33]. The discrimination between caspases-2 and -3 is difficult, as amino acids that build the substrate pockets in both enzymes are very similar, with the exception of the S2 pocket, which was exploited by Maillard et al. to develop caspase-2 reversible inhibitors with improved selectivity [33].
The lack of selective chemical tools is a bottleneck in studying caspase-2 activation and function, and this field could certainly benefit from a potent and selective caspase-2 probe that bypasses caspase-3 cross-reactivity [34]. We have recently demonstrated that the use of unnatural amino acids is a good strategy to develop selective inhibitors for initiator caspases [35]. To challenge this problem, we used our hybrid combinatorial substrate library (HyCoSuL) strategy to in-depth profile caspase-2 specificity in the P5–P2 positions [36, 37]. This analysis performed with a broad spectrum of unnatural amino acids revealed some unexpected caspase-2 preferences that were used to develop selective substrates and inhibitors. In order to further validate the usefulness of this approach, we further interrogated most selective inhibitors in paradigms of apoptosis and reversine checkpoint assembly inhibition with MDM2 cleavage. The latter paradigm, demonstrated genetically to depend on caspase-2, allowed us to validate the use of a highly selective and cell penetrant caspase-2 inhibitor, which we propose can be used in other cell-based studies to elucidate caspase-2 activation and function upon various stimulants.
Methods and materials
Reagents
All chemicals were obtained from commercial sources and used without further purification. Fmoc- and Boc-protected amino acids were purchased from Iris Biotech GmbH (Marktredwitz, Germany), Sigma-Aldrich (Poznan, Poland), Bachem (Torrance, CA, USA), Creosalus (Louisville, KY, USA), and PE Biosciences Limited (Hong Kong, China). Fmoc-ACC fluorescent dye was synthesized according to the procedure published previously by Maly et al. Rink Amide AM resin (200–300 mesh, loading 0.48 mmol/g), 2-chlorotrityl chloride resin (100–200 mesh, loading 1.59 mmol/g), biotin, (2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU), 1-[Bis(dimethylamino) methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxid hexafluorophosphate (HATU), piperidine (PAP), diisopropylcarbodiimide (DICI), 2,2,2-trifluoroethanol (TFE), and trifluoroacetic acid (TFA) were purchased from Iris Biotech GmbH. Anhydrous HOBt was purchased from Creosalus. 2,4,6-Trimethylcollidine, acetonitrile (ACN, HPLC gradient grade), triisopropylsilane (TIPS), hydrobromic acid solution (30% HBr wt. in acetic acid (AcOH)), N-methylmorpholine (NMM), tetrahydrofuran (THF, anhydrous), isobutylchloroformate (IBCF), and 2,6-dimethylbenzoic acid (2,6-DMBA) were purchased from Sigma-Aldrich. N,Nʹ-dimethylformamide (DMF, pure for analysis), methanol (MeOH), dichloromethane (DCM), AcOH, diethyl ether (Et2O), and phosphorus pentoxide (P2O5) were from POCh (Gliwice, Poland). Diazomethane for the acyloxymethyl ketone (AOMK) probe/inhibitor synthesis was generated according to the Aldrich Technical Bulletin (AL-180) protocol. All compounds (peptides, ACC fluorescent substrates, inhibitors, and biotinylated probes) were purified by reverse phase HPLC on Waters system (Waters M600 solvent delivery module and Waters M2489 detector system) using semi-preparative Discovery® C8 column (particle size 10 μm). The purity of the compounds were confirmed on the above HPLC system using analytical Discovery® C8 column (particle size 10 μm). The solvent composition was as follows: phase A (water/0.1% TFA), phase B (ACN/0.1% TFA). For the purification and compounds analysis, the assay was run for 30 min in a linear gradient (from 100% phase A to 0% phase A). The molecular weight of each compound was confirmed on High Resolution Mass Spectrometer WATERS LCT Premier XE with electrospray ionization (ESI) and time-of-flight (TOF) module.
P4–P2 HyCoSuL
The design and synthesis of three fluorescent substrate sub-libraries (P4, P3, and P2) containing natural and unnatural amino acids is described elsewhere [36].
Synthesis of the P5 substrate library
P5 library with the general formula of Ac-P5-Mix-Glu-Mix-Asp-ACC, where the P5 is one of the 169 amino acids (19 natural omitting Cys and 150 unnatural). Mix is an equimolar mixture of natural amino acids, and ACC is a fluorescent tag, was synthesized on a solid support according to the general methods described previously. About 16 g (1 eq., 7.68 mmol) of Fmoc-protected Rink Amide resin (0.48 mol/g) was placed into 250 mL glass cartridge for solid-phase synthesis and swollen in DCM for 30 min. Then the DCM was drained and the resin was washed three times with DMF. Next, the Fmoc-protecting group was removed using 20% piperidine in DMF in three cycles (5 min, 5 min, and 25 min), and the resin was washed six times with DMF. About 2.5 eq. of Fmoc-ACC-OH (19.2 mmol, 8.5 g) was pre-activated with 2.5 eq. of HOBt (19.2 mmol, 2.9 g) and 2.5 eq. of DICI (19.2 mmol, 2.5 mL) in a minimal amount of DMF for 3 min and poured onto the resin. The cartridge was gently agitated for 30 min, and more of the DMF was added as the mixture became too dense. The reaction was carried out for 24 h, followed by filtration and washing the resin three times with DMF. The Fmoc-ACC-OH coupling was repeated to improve the coupling yield using 1.5 eq. of the above reagents. After 24 h, the resin was washed three times with DMF and the ninhydrin test was performed to confirm the complete Fmoc-ACC-OH coupling. Then the Fmoc group was removed from ACC by using 20% piperidine in DMF, followed by washing the resin six times with DMF. Next, 2.5 eq. of Fmoc-l-Asp(tBu)-OH (19.2 mmol, 7.9 g) was pre-incubated with 2.5 eq. of HATU (19.2 mmol, 7.3 g) and 2.5 eq. of 2,4,6-collidine (19.2 mmol, 2.5 mL) in DMF for 3 min and poured onto the H2N-ACC-resin. The reaction was carried out for 24 h and repeated for another 24 h with the use of 1.5 eq. of the Fmoc-l-Asp(tBu)-OH/HATU/2,4,6-collidine reagents. Then the Fmoc-l-Asp(tBu)-ACC-resin was washed three times with DMF, followed by Fmoc de-protection with 20% piperidine in DMF, and washing the resin six times with DMF. An isokinetic mixture of 19 natural amino acids (excluding cysteine, and with norleucine mimicking methionine) was coupled into P2 position. To do this, 5 eq. of the isokinetic mixture, 5 eq. of HOBt (38.5 mmol, 5.76 g), and 5 eq. of DICI (38.5 mmol, 5 mL) were diluted in DMF and pre-activated for 3 min. Then the mixture was poured onto the resin and the cartridge was agitated for 3 h. The slurry was filtrated and washed three times with DMF. The P2 complete coupling was confirmed by a ninhydrin test. Next, the Fmoc group was removed with 20% piperidine in DMF and the Fmoc-l-Glu(tBu)-OH was coupled into the P3 position. About 2.5 eq. of this amino acid (19.2 mmol, 8.2 g), 2.5 eq. of HOBt (19.2 mmol, 2.9 g), and 2.5 eq. of DICI (19.2 mmol, 2.5 mL) was diluted into DMF, pre-activated for 3 min and poured onto H2N-mix-Asp(tBu)-ACC-resin. The reaction was carried out for 3 h, followed by washing the slurry with DMF (six times), and performing a ninhydrin test to confirm complete coupling. The Fmoc group was removed with 20% piperidine in DMF, and the resin was washed six times with DMF. An isokinetic mixture was coupled into the P4 position in the same manner as for P2 position: 5 eq. of mixture/HOBt/DICI in DMF for 3 h, followed by a ninhydrin test. Subsequently, Fmoc group was removed with 20% piperidine in DMF, and the resin was washed three times with DMF, three times with DCM, and three times with MeOH. Finally, the resin was dried in a desiccator over P2O5 overnight and divided into 170 portions (100 mg each, 0.048 mmol). The P5 coupling was performed using a MultiChem 48-well synthesis apparatus (FlexChem from SciGene, CA, USA) in four cycles. Cycle 1: amino acids 1 to 48, cycle 2: 49–96, cycle 3: 97–144, cycle 4: 145–170. For the first cycle, 48 portions of resin were placed in the 48-well cartridge and swollen with DCM for 30 min. Then the DCM was filtrated, and resin was washed three times with DMF. In 48 1.5-mL tubes, 2.5 eq. of various amino acids (0.12 mmol) was mixed with 1 mL of DMF containing 2.5 eq. of HOBt (0.12 mmol, 18 mg) and 2.5 eq. of DICI (0.12 mmol, 16 μL) and poured onto the resin. The P5 coupling reaction was carried out for 4 h, followed by a ninhydrin test. Finally, the Fmoc group was removed with 20% piperidine in DMF, and the free N-terminal end of the substrates was acetylated with 5 eq. of AcOH (0.24 mmol, 14 μL), 5 eq. of HBTU (91 mg), and 5 eq. of DIPEA (42 μL) in DMF for 30 min, followed by a ninhydrin test. Next the acetylated substrates on resin were washed three times with DMF, three times with DCM, three times with MeOH, and dried in a desiccator over P2O5 overnight. In the same manner, amino acids from groups second (49–96), third (97–144), and fourth (145–169) were coupled to the resin. Substrate no. 170 lacked P5 position and served as a control (Ac-P4-P3-P2-P1-ACC). All substrates were cleaved from the resin using ice-cold TFA/TPS/water (% v/v/v, 95/2.5/2.5) mixture for 2 h (shaking once per 15 min). The solution from each well was collected separately and the remaining resin was washed with TFA. Substrates were then precipitated with ice-cold Et2O for 30 min and centrifuged. Next, the supernatant was discarded, and the pellet was re-suspended in ice-cold Et2O and centrifuged again. The supernatant was then discarded and the pellet was allowed to dry out, and was dissolved in 5 mL of 1:1 water:ACN mixture, frozen at −80 °C and lyophilized. The final product (white powder) was then dissolved in dimethyl sulfoxide (DMSO) to the final concentration of 20 mM and stored at −80 °C until use.
Preparation of recombinant caspases
The detailed protocol for the expression and purification of human apoptotic caspases can be found elsewhere [38].
Enzymatic kinetic studies
All the kinetic studies were performed using a CLARIOstar plate reader (BMG LABTECH, Germany) operating in fluorescence kinetic mode using 96-well plates. The ACC-fluorescence was monitored using 355 nm (excitation) and 460 nm (emission) wavelengths. Before kinetic analysis, all caspases were active site-titrated using the zVAD-fmk inhibitor (Cayman Chemical Company, Cat No. 14467) according to the protocol described by Stennicke and Salvesen [38]. For the library screens, substrate and inhibitor kinetics, the caspase assay buffer was 10% w/v sucrose, 20 mM Pipes, 10 mM NaCl, 1 mM EDTA, and 10 mM DTT (pH = 7.2). The buffer for caspases-2, −8, -9, and -10 was supplemented with 0.75 M sodium citrate to ensure maximal enzymatic activity. Buffers were prepared at room temperature, and all kinetic assays were performed at 37 °C. All enzymes were preincubated for 15 min before any activity assays. All the results (library screens, substrate and inhibitor kinetics) were performed at least three times and average values with SD were presented. The kinetic data were analyzed using Graph Pad Prism software.
Characterization of caspase-2 specificity using P4–P2 HyCoSuL
The specificity of caspase-2 was determined in a similar manner as described previously for other caspases [36]. The reaction volume was 100 μL, substrate library concentration was 100 μM, and caspase-2 concentration was 100 nM. The total assay time was 30 min, but only the linear portion of the fluorescence progress curve was taken for the analysis. The best amino acid from each library was set as 100% and the activity of other amino acids was adjusted accordingly.
Characterization of apoptotic caspase specificity at P5 position
The P5 preference of caspases-2, -3, and -8 was determined using Ac-P5-Mix-Glu-Mix-Asp-ACC library. Each caspase was tested in its assay buffer (with or without 0.75 M sodium citrate). All screens were made on 96-well plates in 100 μL volume. Library concentration was 100 μM, and the caspase concentrations were as follows: caspase-2 50 nM, caspase-3 10 nM, and caspase-8 50 nM. The total assay time was 30 min, but only the linear portion of the fluorescence progress curve was taken for the analysis. For each caspase, the best P5 amino acid was set as 100%, and the activity of other amino acids was adjusted accordingly. As a control, a tetrapeptide substrate (Ac-Mix-Glu-Mix-Asp-ACC) lacking P5 position was used.
Synthesis and kinetic analysis of individual optimized substrates
The procedure for the synthesis, purification, HPLC/HRMS, and kinetic (kcat/KM, kcat, KM) analysis of the ACC-labeled substrates can be found elsewhere [36, 39].
Synthesis of AOMK inhibitors and activity-based probes
The detailed protocol for the synthesis of P1 Asp inhibitor and probe with AOMK electrophile warhead has been recently described by Poreba et al. [37, 40]. In brief, Boc-l-Asp(Bzl)-OH amino acid was transformed into Boc-l-Asp(Bzl)-CH2N2 through the use of diazomethane solution in Et2O. Next, the product was transformed into Boc-l-Asp(Bzl)-CH2Br with the use of a solution of HBr (30% wt. in AcOH) and water. The crude product, a pale yellow oil, was then incubated with 2,6-DMBA in the presence of potassium fluoride (KF) in DMF, to obtain Boc-l-Asp(Bzl)-AOMK. Then the Boc group was removed using 50% TFA in DCM and the final product (H2N-l-Asp(Bzl)-AOMK) was used for further synthesis without any purification. In a separate synthesis, caspase-2 selective peptide fragments containing appropriate protecting groups were synthesized (HN-Idc-hGlu(tBu)-Thr(Bzl)-Ser(tBu)-OH and Ac-Idc-hGlu(tBu)-Thr(Bzl)-Ser(tBu)-OH for inhibitors, and biotin-Idc-hGlu(tBu)-Thr(Bzl)-Ser(tBu)-OH and HN-Idc-hGlu(tBu)-Lys(Cy5)-Ser(tBu)-OH for activity-based probes (ABPs)). All peptide fragments were synthesized using 2-chlorotrityl chloride resin and used without further purification. Next, the peptides were coupled with H2N-l-Asp(Bzl)-AOMK in DMF using HATU/DIPEA coupling reagents. The crude product was purified on HPLC, lyophilized, and de-protected using 50% TFA in DCM, followed by removing the TFA/DCM under reduced pressure. Finally, the Bzl group from Asp was hydrolyzed via hydrogenolysis (Pd/C, H2, DMF). The final product was purified on HPLC, lyophilized, and dissolved in DMSO to the final concentration of 10 mM.
Determination of kobs/I inhibition parameter for inhibitors and probes toward caspases and legumain
Caspases-2, -3, -8, -9, and -10 were active site titrated using zVAD-fmk inhibitor. All enzymes were assayed in caspase buffer supplemented with 0.75 M sodium citrate. Human recombinant prolegumain was kindly provided by Dr. Dusan Turk. Prolegumain was first activated (pH 4.5) and then transferred into assay buffer (pH 5.5) as described previously [40]. The kobs/I parameter was measured under pseudo-first-order kinetic conditions. Inhibitor/probe was diluted in a 96-well plate and mixed with the appropriate substrate (NH-Idc-hGlu-Thr(Bzl)-Ser-Asp-ACC for caspase-2, Ac-DEVD-ACC for caspase-3, Ac-LEHD-ACC for caspases-8, -9, and -10, and Ac-DTyr-Tic-Ser-Asp-ACC for legumain); final substrate concentration was 100 μM) and pre-incubated at 37 °C for 15 min. At the same time in a separate tube, the caspases and legumain were pre-activated in assay buffer at 37 °C, and after 15 min, the enzymes were added and fluorescence was monitored for 30 min, starting immediately. Second-order rate of inhibition (kobs/I) was determined in at least three independent experiments and presented as an average value. All assays were performed using a CLARIOstar plate reader (BMG Labtech) operating in kinetic mode (355 nm excitation, 460 nm emission, gain 650) using 96-well plates (Corning, NY, USA).
Inhibition profile of NH-23-C2 and Ac-23-C2 toward cysteine cathepsins and proteasome
Human recombinant cysteine cathepsins were kindly provided by Dr. Boris Turk and active-site titrated with E-64 inhibitor (Bachem). Kinetic analysis was performed in cathepsin assay buffer (100 mM sodium acetate, 100 mM sodium chloride, 10 mM DTT, and 1 mM EDTA, pH 5.8 “buffer 1” or pH 7.2 “buffer 2”). Human proteasome was purchased from BostonBiochem (h20S proteasome, E-360) and assayed in buffer containing 20 mM HEPES and 0.5 mM EDTA (pH 7.2). Cysteine cathepsins (cat B 1 nM, cat L 1 nM, cat V 3 nM, cat S 10 nM, cat K 8 nM) or proteasome (1 nM) were incubated with inhibitors (concentration range 50 μM–4.4 μM) for 15 min, followed by substrate addition (10 μM, Cbz-Phe-Arg-AMC for cathepsins and 100 μM WRP23, Ac-hArg-Pro-Phe(2F)-Asp-ACC selective for caspase-like subunit [41]). The fluorescence signal was recorded for 15 min (excitation 355 nm, emission 460 nm) and the percentage of inhibition was calculated based on the rate of substrate hydrolysis (relative fluorescence units (RFU/s)).
Indirect competition of NH-23-C2, Ac-23-C2, and zVAD-fmk with active caspases
To assess inhibitor selectivity, 50 nM of each active site-titrated recombinant caspase (-2, -3, -8, -9, and -10) were incubated with inhibitor at various concentration ranges (10 nM—3 μM) in caspase assay buffer supplemented with 0.75 M sodium citrate for 30 min at 37 °C. Next, the residual caspase activity was quenched with 1 μM of broad-spectrum caspase biotin-labeled inhibitor biotin-LEHD-AOMK for another 30 min, and the reaction products were precipitated as follows: 100 μL of ice-cold 30% trichloroacetic acid (TCA) was added to each sample, incubated on ice for 30 min and centrifuged for 30 min (20,000×g, 4 °C). Supernatants were gently aspirated using a vacuum device, the pellet was re-suspended in 200 μL of ice-cold 10% TCA, and the samples were re-centrifuged for 15 min (20,000×g, 4 °C). Supernatants were again removed, and the pellet re-suspended in 200 μL of acetone. Samples were centrifuged for 10 min (20,000×g, 4 °C) and the supernatant was removed. Samples were dried at 37 °C for 15 min and then 80 μL of 1× SDS/DTT buffer was added to each, followed by denaturation at 95 °C for 5 min. Samples (20 μL) were subjected to SDS-PAGE (30 min, 200 V), followed by transfer to a nitrocellulose membrane (60 min, 10 V) and blocking with 2% BSA in TBS-T (60 min, room temperature (RT)). The biotin-labeled probe/caspase complex was detected by probing the membrane with fluorescently labeled streptavidin (30 min, RT, 1:10,000, IRDye® 800CW, LI-COR), followed by scanning in LI-COR system (800 nm channel, Odyssey® CLx, Lincoln, NE, USA). Data were analyzed using Image Studio Lite software (Lincoln, NE, USA).
Caspase-2 inhibition in reversine-treated HCT116 cells
About 100,000 low passage HCT116 cells were seeded on a 12-well plate (1 mL per well) with fresh McCoy’s medium supplemented with 10% fetal bovine serum (FBS, Atlas Biologicals), 2 mM l-glutamine (Omega Scientific), 100 units/mL penicillin, and 10 μg/mL streptomycin (both Gibco Life Technologies) in a humidified 5% CO2 atmosphere at 37 °C. The next day, the medium was changed, and caspase-2 inhibitors were added at various concentrations to the wells and pre-incubated for 4 h. After this time, 1 μM of reversine (Cayman Chemical) was added to the cells, followed by 24 h incubation. The next day, medium was removed, washed with 1× PBS, and cells were harvested with trypsin-EDTA (0.5 mL per well), quenched with fresh medium (0.5 mL), transferred to 1.5-mL Eppendorf tubes and centrifuged for 5 min at 500×g. The supernatant was aspirated, cell pellet was washed with 1× PBS (1 mL), and the cells were centrifuged for 5 min at 500×g. Then, PBS was aspirated, and the cell pellets were suspended in 50 μL of 1× SDS/DTT and boiled at 95 °C for 5 min. Samples (20 μL) were subjected to SDS-PAGE (30 min, 200 V, 4–12% Bis-Tris 15-well gels), followed by transfer to a nitrocellulose membrane (60 min, 10 V) and blocking with 2% BSA in TBS-T (60 min, RT). Proteins were detected by probing the membrane with primary antibodies (overnight, +4 °C) followed by the incubation with fluorescently labeled secondary antibodies (30 min, RT). The membrane was finally scanned in LI-COR system (700 nm/red and 800 nm/green channels, Odyssey® CLx, Lincoln, NE, USA). Protein bands were quantified using Image Studio Lite software (Lincoln, NE, USA). The following antibodies were used in immunoblotting experiments: mouse anti-MDM-2 (IF2, Merck Millipore, MABE340, 1:100), mouse anti-PARP (BD Pharmigen, 556362, form mouse ascites, 1:2000), rabbit anti-p53 (Cell Signaling, 9282, 1:1000), goat anti-actin (Abcam, 8229, 1:500), rabbit anti-caspase-3 (Cell Signaling, 9662S, 1:1000), mouse anti-caspase-2 (Cell Signaling, 2224, 1:1000). Monoclonal caspase-8 antibody C15 raised against an epitope of the large subunit was a kind gift of Dr. Marcus Peter, University of Chicago (dilution 1:200) [42]. As secondary antibodies, we used appropriate IRDye® 680RD and IRDye® 800CW fluorescent labeled antibodies from LI-COR (dilution 1:10,000).
Toxicity of NH-23-C2 and Ac-23-C2
HCT-116 cells were seeded on a 96-well plate (20,000 cells per well) and allowed to attached overnight. The following day, cells were incubated with NH-23-C2 and Ac-23-C2 (concentration range 50 μM–4.4 μM) for 24 h. After this time, cell viability was determined using MTS assay, according to the manufacturer’s protocol (CellTiter 96® Aqueous One Solution Cell Proliferation Assay, G3582, Promega). The number of living cells was reported as a survival ratio, calculated based on the number of non-treated control cells.
Apoptosis initiation and inhibition in TRAIL-stimulated HCT116 cells
About 100,000 low passage HCT116 cells were seeded on a 12-well plate (1 mL per well) as described in the above section. The next day, the medium was changed, and caspase-2 inhibitors were added at various concentrations to the wells and pre-incubated for 4 h. The cells were then stimulated 24 h with KillerTRAIL (Enzo Life Sciences, 100 ng/mL). The next day, floating cells were collected into 1.5 mL Eppendorf tubes, and adherent cells were harvested as described above. Then, floating and harvested cells were combined, and centrifuged for 5 min at 500×g. The supernatant was aspirated, cell pellet was washed with 1× PBS (1 mL) and cells were centrifuged for 5 min at 500×g. Then, PBS was aspirated, and the cell pellets were suspended into 50 μL of 1× SDS/DTT and boiled at 95 °C for 5 min. Samples (20 μL) were subjected to SDS-PAGE (30 min, 200 V, 4–12% Bis-Tris 15-well gels), followed by transfer to a nitrocellulose membrane (60 min, 10 V) and blocking with 2% BSA in TBS-T (60 min, RT). Proteins were detected by probing the membrane with primary antibodies (overnight, +4 °C) followed by incubation with fluorescently labeled secondary antibodies (30 min, RT). The membranes were finally scanned in LI-COR system (700 nm/red and 800 nm/green channels, Odyssey® CLx, Lincoln, NE, USA). The protein bands were quantified using Image Studio Lite software (Lincoln, NE, USA).
Results
P4-P2 HyCoSuL
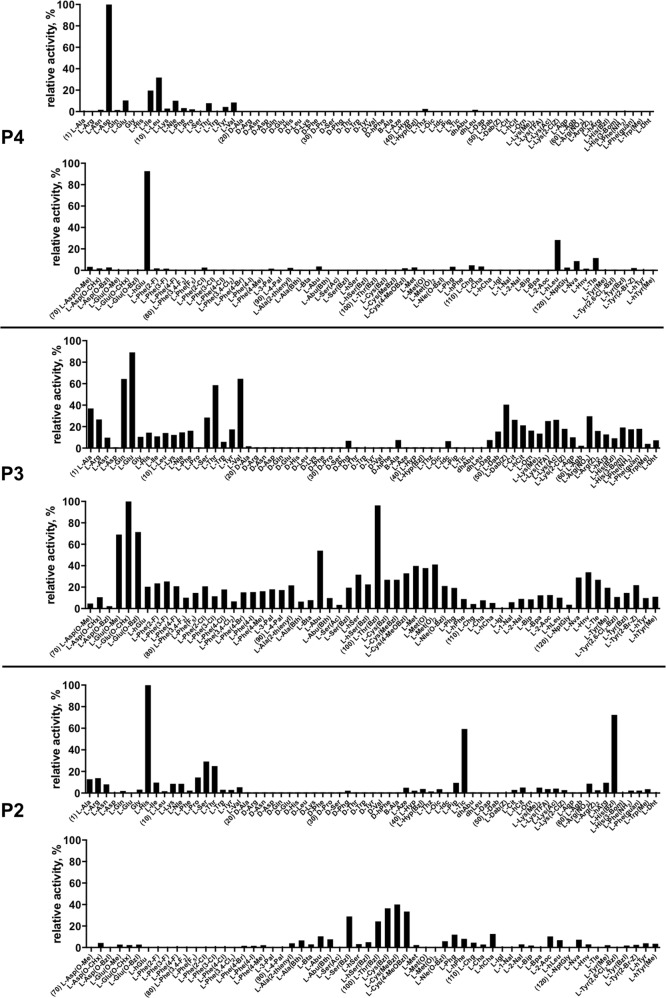
Caspase-2 substrate specificity was determined using a substrate combinatorial library containing Asp at P1 position, as this amino acids is recognized by caspases with high stringency [36]. The general library architecture was Ac-P4-P3-P2-Asp-ACC, and the library was composed of three sub-libraries: P4, Ac-X-Mix-Mix-Asp-ACC; P3, Ac-Mix-X-Mix-Asp-ACC; and P2, Ac-Mix-Mix-X-Asp-ACC, where Mix is an equimolar mixture of 18 natural amino acids (minus methionine and cysteine) and norleucine. In this library, we used over 100 unnatural amino acids that allows us to explore far more chemical space than conventional PS-SCL methods. All three sub-libraries were screened with caspase-2, and the results were presented as specificity matrixes, one for each tested position (Fig. 1). We found that caspase-2 displays very narrow specificity at P4 position, preferring Asp and hGlu (homoGlutamic acid) (100% and 90%, respectively). Interestingly, glutamic acid, which is intermediate between Asp and hGlu in carbon chain length, was poorly recognized by this enzyme. Caspase-2 can also accommodate branched, hydrophilic amino acids like Ile, Leu, and hLeu, while all other amino acids are almost completely ignored. The high tolerance for hGlu in the S4 pocket has a potential to be used to discriminate between caspase-2 and -3, as the latter one recognizes hGlu very poorly [36]. This is somehow unexpected, as in both enzymes, the amino acids forming the S4 pocket are well conserved (caspase-2: Asn-232, Trp-238, Tyr-273; caspase-3: Asn-208, Trp-214, Phe-250) [33]. P3 position appears to be more liberal, as almost all amino acids, except D-isomers, were recognized. The most active ones were unnatural Glu(Chx) (glutamic acid cyclohexyl ester), Thr(Bzl) (threonine benzyl ether), followed by natural Glu and its analogs. Finally, the P2 position shows narrow preference for His, His(Bzl), Tic, and Cys-benzyl derivatives. All remaining amino acids were only poorly accommodated in caspase-2 S2 pocket. The analysis of P4-P2 HyCoSuL provided us a detailed picture of caspase-2 preferences, which can be further used to develop caspase-2 selective peptides. Indeed, when comparing these data with other caspase specificity profiles, it appears that the strong preference of caspase-2 for hGlu at P4 is unique across the caspase family [36]. The differences between caspase-2 and other apoptotic caspases in P3 and P2 positons are less pronounced; however, we still were able to flush out some amino acids that can discriminate between these enzymes.
Fig. 1.
Human caspase-2 preferences at the P4, P3, and P2 positions. Human caspase-2 preferences were determined using three sub-libraries, Ac-P4-Mix-Mix-Asp-ACC, Ac-Mix-P3-Mix-Asp-ACC, Ac-Mix-Mix-P2-Asp-ACC, all containing natural and unnatural amino acids. The x axis shows abbreviated names of amino acids and the y axis displays relative activity presented as a percentage of the best-recognized amino acid. Standard deviations calculated from three screening were 15% of values shown in the figure. The order of the amino acids on the x axis corresponds to the order in Table S4. Every tenth amino acid is marked with a number
P5 library combinatorial substrate library
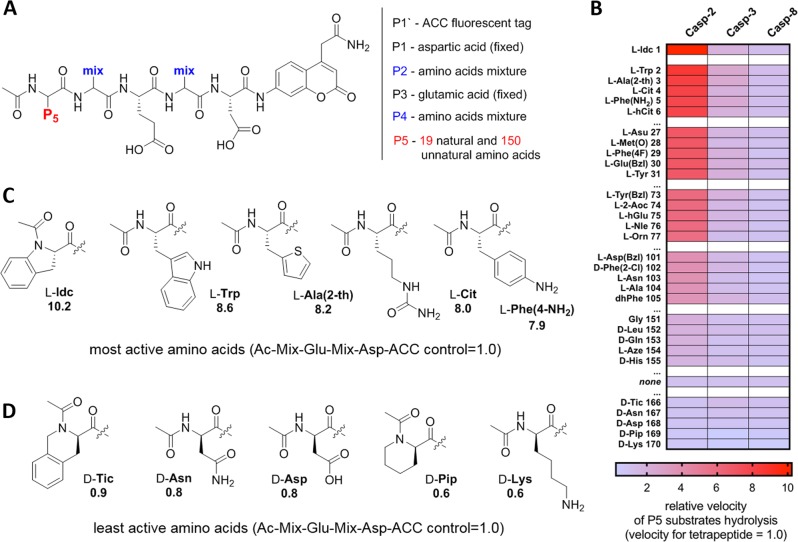
Since caspase-2 is the only caspase that hydrolyzes pentapeptides much more efficiently than tetrapeptides, we found it reasonable to screen caspase preferences at the P5 position [30, 43]. To do so, we synthesized a combinatorial fluorogenic substrate library of the general formula Ac-P5-Mix-Glu-Mix-Asp-ACC, where P5 is fixed natural or unnatural amino acids, and Mix is an equimolar mixture of 18 natural amino acids and norleucine (Fig. 2a). Analysis of the P5 library revealed that indeed, caspase-2 uniquely prefers pentapeptides over tetrapeptides, as 155 out of 169 substrates were more active than a tetrapeptide substrate lacking a P5 moiety (Ac-Mix-Glu-Mix-Asp-ACC) (Fig. 2b–d, Fig. S1). The most active P5 amino acid was the unnatural Idc, which was over tenfold faster hydrolyzed than a reference tetrapeptide substrate. Other amino acids, including natural Trp, and unnatural Ala(2th), Cit, and Phe(4-NH2), were also much better hydrolyzed than the control substrate, with the pentapeptide/tetrapepide ratio of around 8.0. We also investigated caspases-3 and -8, as these enzymes are known to share some substrate similarities with caspase-2, but the kinetic analysis showed that for these caspases P5 has no significant impact on the overall rate of substrate hydrolysis (Fig. 2b, Fig. S1). This broad caspase screen at the P5 position with 169 natural and unnatural amino acids, allowed us to make a very precise map of caspase–substrate interactions.
Fig. 2.
Analysis of caspase-2 P5 preferences. a General architecture of the P5 combinatorial fluorogenic substrate library, Ac-P5-Mix-Glu-Mix-Asp-ACC. The structure of this library allows for versatile screening of all caspases. b Substrate specificity of three apoptotic caspases at P5 position. The data are presented as a heat-map, where the velocity of substrate hydrolysis is expressed as ratio between P5 substrate (Ac-P5-Mix-Glu-Mix-Asp-ACC) and a tetrapeptide substrate lacking the P5 amino acid (Ac-Mix-Glu-Mix-Asp-ACC), which serves as a control. The results are sorted according to caspase-2 preferences, from the most to least active. The full P5 specificity profiles of these caspases can be found in Fig. S1. “none”—activity of tetrapeptide substrate lacking P5 amino acid. c, d The structures of the best (c) and worst (d) caspase-2 amino acids at P5 position. The numbers in c and d represent the ratio between cleavage rates of pentapeptides and Ac-Mix-Glu-Mix-Asp-ACC tetrapeptide, which served as a control
Design of caspase-2 selective substrates
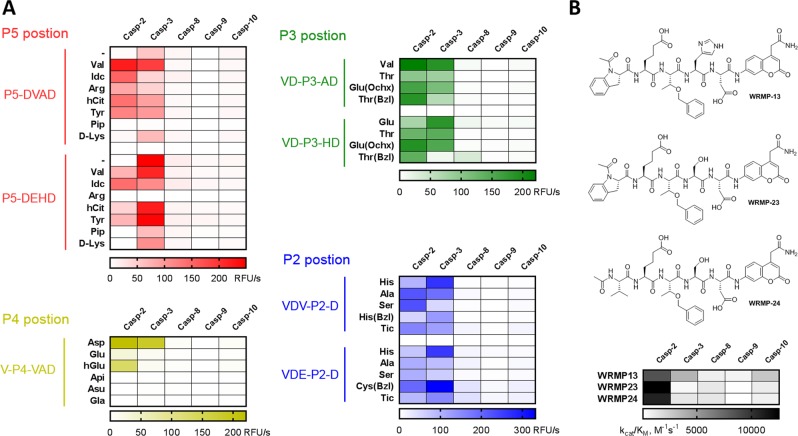
Despite a long history of developing caspase-specific substrates, inhibitors, and ABPs, caspase-2 studies have been largely neglected and to date no reasonably selective substrate or inhibitor has been developed [34]. The most heavily used caspase-2 substrate, the pentapeptide VDVAD sequence, was demonstrated to be efficiently hydrolyzed by caspases-3 and -7, which significantly impedes its application in biological systems where other caspases are active [32]. To develop a caspase-2 selective substrate lacking off-target activity, we performed in depth analysis of P5–P2 caspase specificity profiles. We first determined whether the HyCoSuL screens reflect the caspase-2 preferences toward individual substrates, an effect called subsite cooperativity. To do this, we used two pentapeptides as scaffolds for the synthesis of our substrates: (1) the reference VDVAD and (2) HyCoSuL-derived VDEHD. Then for each position tested (P5–P2), we synthesized several analogs and hydrolyzed them with a panel of five apoptotic caspases (-2, -3, -8, -9, -10) (Fig. 3a, Table S1). The kinetic analysis showed that the most caspase-2-selective amino acids were: P5–Idc and Val, P4–hGlu, P3–Thr(Bzl), and P2–Ser. Based on this dataset, we synthesized two new substrates (WRMP23 and WRMP24) and determined their second-order rate of hydrolysis (kcat/KM) against five caspases (Fig. 3b), demonstrating that both substrates are active and selective toward caspase-2. Moreover, the exchange of P2 Ser into His (WRMP13 substrate) resulted in a decrease of both activity and selectivity, which is in line with the data obtained from our validation screen (Fig. 3a). This two-step validation revealed that although according to HyCoSuL (and PS-SCL [2]) screening, VDEHD seems to be the most active natural substrate, the VDVAD is hydrolyzed much better. This might be explained by the formerly mentioned enzyme subsite cooperativity, as caspase-2 specificity is slightly different when screened against individual substrates or combinatorial mixtures. Nevertheless, the HyCoSuL approach allowed us to detect some key amino acids that can differentiate caspase-2 from its paralogs (Fig. 3).
Fig. 3.
Validation of combinatorial library screens and the design of caspase-2 selective substrates. a The rate of hydrolysis (expressed as RFU/s, relative fluorescence units per second) of a panel of pentapeptide substrates (10 μM) with five human caspases (50 nM). b A detailed kinetic analysis of three most selective substrates for caspase-2 (top panel: chemical structures of pentapeptide substrates; lower panel: kcat/KM matrix presented as heat-map)
Optimization of the P5 position in caspase-2 substrate
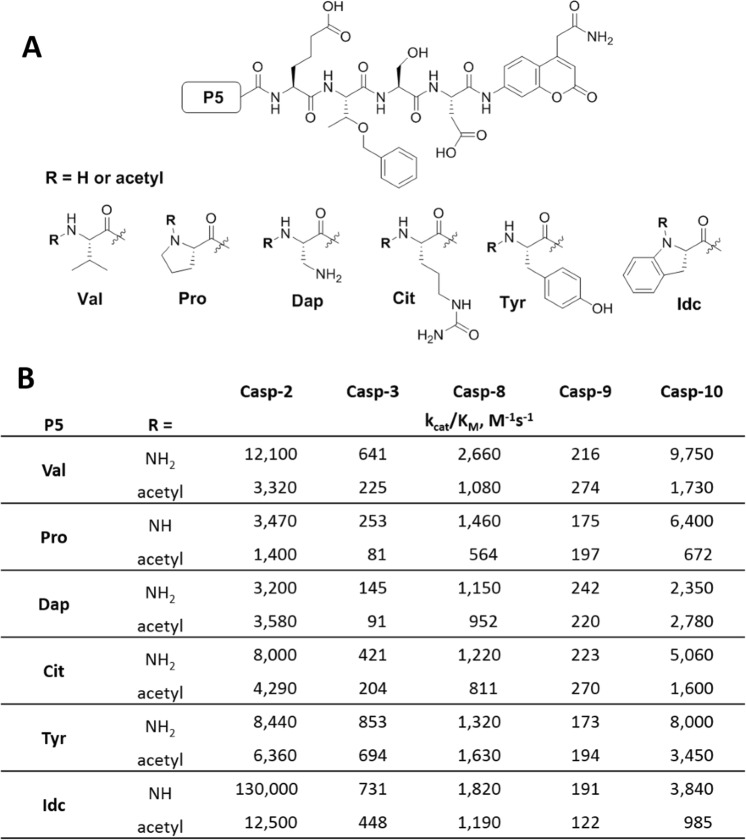
As we demonstrated above, the most selective caspase-2 substrate we found using VDVAD and VDEHD peptide scaffolds was Ac-Idc-hGlu-Thr(Bzl)-Ser-Asp (WRMP-23). In this step, we decided to optimize our champion substrate by testing other caspase-2 selective amino acids at P5. In addition, given that the P5 position has a strong impact on substrate binding and hydrolysis, we also checked whether the N-terminal acetylation may affect the rate of substrate hydrolysis. Thus, we synthesized twelve hGlu-Thr(Bzl)-Ser-Asp-based derivatives with six various acetylated or non-acetylated amino acids and determined their kinetic parameters toward five caspases (-2, -3, -8, -9, and -10) (Fig. 4). Surprisingly, we found that the N-terminal acetylation has a significant impact on caspase-2 kinetics. The most active substrate (Idc at P5) was hydrolyzed almost ten times more efficiently when the N-terminus lacked a blocking group (kcat/KM 130,000 M−1 s−1 vs. 12,500 M−1 s−1). Other substrates also displayed pronounced increase of cleavage rates when non-acetylated. This indicates that the caspase-2 S5 pocket may stabilize the substrate of pentapeptides by binding to its N-terminal amine group. Indeed, the detailed kinetic analysis of Ac/NH-Idc-hGlu-Thr(Bzl)-Ser-Asp-ACC substrate showed that the non-acetylated substrate had much lower KM (7.6 μM) than its acetylated counterpart (50.6 μM) (Table S2). Finally, we showed that NH-WRMP23-ACC displayed a 178-fold kcat/KM preference for caspase-2 over caspase-3 (Table S2).
Fig. 4.
Analysis of N-terminal modifications of caspase-2 selective substrates. a General structure of caspase-2 selective substrate that served as a scaffold for the optimization of the N-terminus. b Kinetic analysis (kcat/KM) of 12 caspase-2-selective substrates N-capped with various acetylated and non-acetylated amino acids
Design and kinetic characteristic of selective inhibitor and ABP
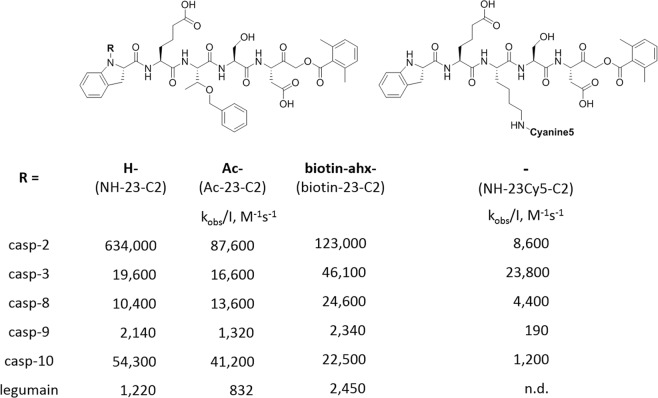
Profiling of protease substrate specificity with combinatorial fluorogenic substrate mixtures provides extensive information on the architecture of protease active sites. The information obtained in this manner of optimal subsite preferences is frequently utilized to produce very selective fluorogenic substrates, or inhibitors that contain strong electrophilic binding group at the C terminus. Moreover such substrate, if equipped with a biotin or fluorescent moiety on the N-terminus, can be used as ABP for enzyme localization. Having optimized a caspase-2 selective substrate, we aimed to use the champion pentapeptide to develop the first, selective and potent inhibitor, as currently available VDVAD-based inhibitors lack selectivity [32, 44]. To do this, we conjugated the most selective peptide we obtained with AOMK, an electrophile that is known to covalently react with the catalytic Cys of caspases. To get better insight into caspase-2 inhibition requirements, we synthesized two inhibitors, acetylated (Ac-23-C2) or non-acetylated (NH-23-C2) at the P5 position, and measured the second-order rate constant of enzyme inactivation (kobs/I, M−1 s−1) (Fig. 5). Similar to the champion substrates, the non-acetylated inhibitor was more potent (by sevenfold) than the acetylated analog. The differences between inhibitor kinetics for other caspases (-3, -8, -9, and -10) were insignificant. In the next step, we aimed to convert our caspase-2-selective inhibitor into a biotin-labeled ABP. To do this, we attached a biotin moiety to the N-terminus of the inhibitor. However, since the new compound (biotin-23-C2) lacked the key terminal amine group, it was five times less potent than parental NH-23-C2. In our next attempt to develop caspase-2 ABP, we decided to synthesize a Cy5-labeled probe with a free N-terminal. Since our HyCoSuL analysis indicated that caspase-2 has a relatively broad specificity at P3 position, we replaced Thr(Bzl) with Lys(Cy5) to obtain NH-23Cy5-C2 (Fig. 5). Unfortunately, this ABP almost completely lost potency against caspase-2, indicating that there might be steric hindrance in the enzyme S3 pocket.
Fig. 5.
Structures and kinetic parameters of caspase-2 inhibitor and Cy5-labeled activity-based probe. NH-Idc-hGlu-Thr(Bzl)-Ser-Asp-AOMK (NH-23-C2) is the most potent and selective caspase-2 inhibitor. Acetylation (Ac-23-C2) or biotinylation (b-23-C2) of the parental NH-23-C2 inhibitor significantly reduce potency of these compounds toward caspase-2. Substitution of Thr(Bzl) at P3 for Lys(Cy5) in NH-MP-C2 inhibitor dramatically decreases its caspase-2 potency and selectivity. Enzymes were assayed in the optimal buffers (pH 7.2 for caspases and pH 5.8 for legumain). n.d.—not determined
Analysis of the selectivity of NH-23-C2 and Ac-23-C2 in cancer cells
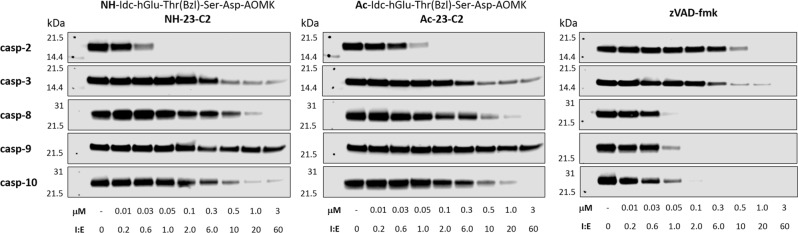
Although the caspase-2 ABP is still to be developed, we were able to obtain a very potent and selective inhibitor for this enzyme. To test their utility, we first compared their selectivtiy toward five purified recombinant caspases (2, 3, 8, 9, and 10). To do this, we incubated each inhibitor with a panel of caspases, and after 30 min, we labeled caspase residual activity using a broad-spectrum biotinylated probe (biotin-LEHD-AOMK) (Fig. 6). Our analysis showed that the non-acetylated inhibitor blocks caspase-2 activity even at 1:1 inhibitor–enzyme ratio. In addition, it displays weak potency toward other caspases, which reflects the kobs/I analysis. The acetylated inhibitor is slightly less potent, but still very selective. Finally, as expected, zVAD-fmk efficiently blocks caspases-8, -9, and -10, and displays only moderate activity versus caspases-2 and -3.
Fig. 6.
Inhibition profiles of three covalent inhibitors versus five human apoptotic caspases. NH-23-C2, Ac-23-C2, and zVAD-fmk inhibitors at various concentration ranges (10 nM—3 μM) were incubated with 50 nM of active caspases for 30 min, followed by addition of a broad-spectrum caspase probe (biotin-LEHD-AOMK, 1 μM) to label all residual caspase activity. Samples were then subjected for SDS-PAGE analysis, transferred to a PVDF membrane, and biotin-LEHD-caspases were detected with streptavidin
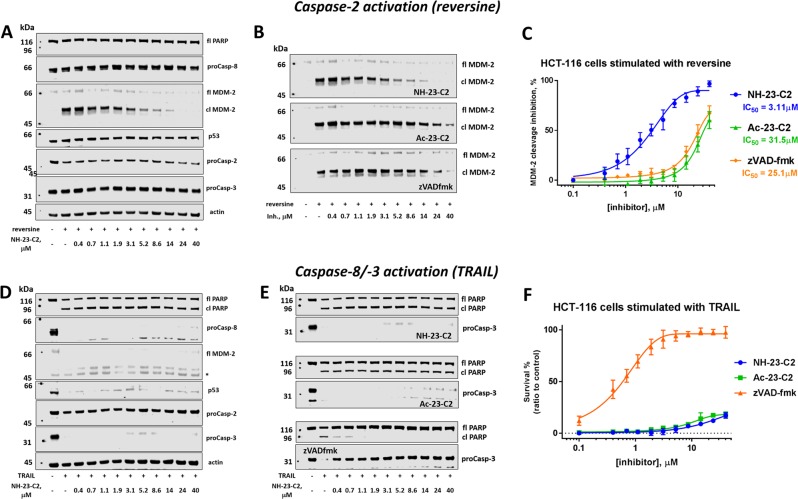
To validate the usefulness of our small molecule inhibitors, we sought a cell-based system where caspase-2 provides a reliable phenotype in the absence of apoptotic caspases. We investigated our inhibitors a paradigm of MDM-2-cleavage following chromosome miss-segregation and DNA damage, since this has been demonstrated to serve the roles we sought: involvement of caspase-2 without apoptotic caspases. Stimulation of proliferating cells with reversine, a potent Mps1 kinase inhibitor, leads to chromosome mis-segregation followed by caspase-2 activation and MDM-2 cleavage [45, 46]. MDM-2 is one of a very few bona fide caspase-2 substrates, and is cleaved in response to DNA damage [45] into MDM-2 p60 fragment. Previously published reports demonstrated that caspase-2 cleaves MDM-2 in HCT116 cells upon reversine treatment, thus we decided to use the same colon cancer cell model in our study [47]. To analyze the potency and selectivity of our inhibitors, we treated HCT116 cells with reversine in the presence of NH-23-C2, Ac-23-C2, and zVAD-fmk at various concentration ranges (0–40 μM) (Fig. 7 top panel, Fig. S2). In the absence of inhibitors, reversine induced the rapid accumulation of a cleaved (p60) MDM-2 fragment and decrease in full length caspase-2, which indicates the proteolytic activation of this enzyme (Fig. 7a). No evidence of apoptosis was observed, as neither caspase-8 nor caspase-3 was cleaved, and PARP, a caspase-3 substrate, was not processed. However, when HCT116 cells were pre-incubated with NH-23-C2 inhibitor, MDM-2 cleavage was inhibited in a concentration-dependent manner (IC50 = 3.11 μM), indicating that endogenous caspase-2 was inactivated (Fig. 7a–c). Ac-23-C2 and zVAD-fmk inhibitors were also able to block MDM-2 processing; however, they were both much less effective than NH-23-C2 with IC50 values of 31.5 μM and 25.1 μM, respectively (Fig. 7b, c). This data demonstrate that NH-23-C2 is a cell penetrant, potent caspase-2 inhibitor. To dissect its selectivity over other caspases, we induced apoptosis in HCT116 cells via TRAIL stimulation (Fig. 7 bottom panel, Fig. S3) [48]. In the absence of inhibitors, HCT116 cells rapidly underwent extrinsic apoptosis, which was observed by the processing of upstream procaspase-8 and downstream procaspase-3, and cleavage of PARP protein (Fig. 7d). No MDM-2 cleavage product was detected, which might indicate that the activation of caspase-2 in TRAIL-mediated apoptosis is negligible. More importantly, increasing concentration of NH-23-C2 and Ac-23-C2 (up to 40 μM) did not affect the apoptosis events (Fig. 7d–f). This strongly demonstrates that both compounds do not inhibit caspase-8 and caspase-3 in the apoptosis paradigm. In contrast, zVAD-fmk appeared as an excellent caspase-8 inhibitor, as even at very low concentration (0.4 μM), procaspases-8 and -3 processing and PARP cleavage were completely blocked. Taken together, these experiments revealed that NH-23-C2 is a potent, selective and cell-permeable inhibitor of caspase-2.
Fig. 7.
NH-23-C2 is a potent and selective inhibitor of endogenous caspase-2. Top panel: Caspase-2 activation. a Caspase activation and protein processing pattern in HCT116 cells stimulated with reversine in the presence of various concentrations of NH-23-C2. b Processing of MDM-2 protein in HCT116 cells stimulated with reversine (1 μM, 24 h) in the presence of various concentration of caspase-2 inhibitors (NH-23-C2 and Ac-23-C2) and a broad-spectrum caspase inhibitor, zVAD-fmk. c Caspase-2 inhibition in HCT116 cells expressed as IC50 for three inhibitors: NH-23-C2, Ac-23-C2, and zVAD-fmk. Bottom panel: Caspases-3/-8 activation. d Caspase activation and protein processing pattern in HCT116 cells undergoing TRAIL-mediated apoptosis. e PARP processing and caspase-3 activation in TRAIL-stimulated HCT116 cells in the presence of various concentrations of NH-23-C2, Ac-23-C2, and zVAD-fmk. f zVAD-fmk, but not NH-23-C2 or Ac-23-C2, inhibits apoptosis in TRAIL-stimulated HCT116 cells; fl—full length, cl—cleaved form
NH-23-C2 and Ac-23-C2 off-reactivity with cysteine and threonine proteases
Since our inhibitors contain an Asp-AOMK structural motif, they might also display some cross-reactivity with other proteases. To evaluate this, we decided to test the selectivity of NH-23-C2 and Ac-23-C2 inhibitors toward lysosomal cathepsins (K, B, L, V, and S), legumain, and the constitutive proteasome. Our experiments demonstrated that Ac-23-C2 displays moderate cross-reactivity with cathepsins B, L, V, S, and legumain at pH 5.5 and only slight reactivity at pH 7.2 (Fig. S4A, Fig. 5). On the other hand, the non-acetylated inhibitor (NH-23-C2) showed significantly lower potency toward cathepsins (pH 5.5 and 7.2) and legumain at pH 5.5 (Fig. S4B, Fig. 5), thus it is highly unlikely that this inhibitor would block these proteases when tested in reversine-treated HCT116 cells at the concentrations used (0.4 μM–40 μM). Finally, we also tested constitutive 20S proteasome, as this threonine protease displays cytosolic distribution and its β1 catalytic subunit prefers Asp or Glu at the P1 position. Our analysis revealed that both Ac-23-C2 and NH-23-C2 showed negligible reactivity with the proteasome even at high concentrations (50 μM). With these experiments, we demonstrated that inhibitor NH-23-C2 does not target proteases that are the most likely off-target candidate enzymes. However, since we could not test the off-target reactivity against all proteases, we determined the toxicity of both inhibitors toward HCT116 cells via cell viability assay. Since even at high inhibitor concentrations no cell death was observed, we concluded that neither Ac-23-C2 nor NH-23-C2 is toxic for this cancer cell line (Fig. S4B, D).
Discussion
Caspase-2 was the second human caspase to be discovered; however, despite significant research efforts, its role in physiology and disease is not always clear. One reason for this is its dual biochemical character, as depending on the environment caspase-2 can either act as an initiator or executioner of apoptosis [20]. Importantly, since procaspase-2 is a substrate for caspases-3 and -8, procaspase-2 in vitro processing does not necessarily reflect caspase-2 activation. In order to monitor caspase-2 activity, it is crucial to develop selective substrates and inhibitors. Unfortunately, the optimal caspase-2 substrate with the VDVAD sequence used in many studies is efficiently hydrolyzed by caspase-3, and to a lesser extent by caspase-7, which significantly impedes the functional characterization of this protease. Some current biochemical techniques rely on the use of caspase broad spectrum biotin-labeled probes in combination with caspase selective antibodies [13], or the parallel application of several substrates with improved selectivity by taking advantage of differences in caspase recognition preferences [32, 49]. Nevertheless, the development of truly selective substrate and small inhibitor for caspase-2 is still outstanding. To challenge this problem, we applied our HyCoSuL approach and explored chemical space in the caspase-2 active site surrounding the S5-S2 pockets. This analysis allowed us to find several natural and unnatural amino acids that can distinguish between caspase-2 from other apoptotic caspases. The best structure–activity feature we can distill from this work is that although caspase-2 displays narrow selectivity at the P4 position by preferring Asp, it can tolerate hGlu at almost the same level, which is a unique feature among caspases. This is somehow unexpected, as caspase-3 which has very similar S4 architecture recognizes hGlu very poorly. Another caspase-2-distinguishing feature is a very high preference for unnatural Idc at P5 position, but in order to obtain high catalytic efficiency, the N-terminal end must lack the commonly used acetyl cap. Dissecting caspase-2 preferences in P5–P2 positions allowed us to develop very active and selective fluorogenic substrate that most importantly almost completely bypasses caspase-3-mediated hydrolysis. Further, we used the optimal sequence and developed potent and selective caspase-2 covalent irreversible inhibitor. To check whether this compound would be useful in cell-based studies, we induced caspase-2 activation in HCT116 colon cancer cells by reversine treatment. In this scenario, activated caspase-2 cleaves MDM-2 protein, which can be monitored by Western blotting. Indeed, pre-incubation of HCT116 cells with our best caspase-2 inhibitor in low micromolar range completely prevented MDM-2 cleavage (IC50 = 3.11 μM), demonstrating its potency. To further validate this inhibitor, we induced caspase-8 and caspase-3 activation in HCT116 cells by TRAIL stimulation. We observed only slight apoptosis inhibition when NH-23-C2 was used at high concentration (40 μM). This is the first report demonstrating that endogenous caspase-2 can be selectively blocked by a small molecule inhibitor. Moreover, this approach can be used for other cell-based systems to dissect the authentic role of caspase-2.
Supplementary information
Acknowledgements
This project received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement no. 661187 (to MP) and was supported by the National Science Centre grant 2011/03/B/ST5/01048 in Poland (to MD), the “TEAM/2017-4/32” project, which is carried out within the TEAM programme of the Foundation for Polish Science cofinanced by the European Union under the European Regional Development Fund (to MD), and the NIH grant R01 GM099040 to (GSS). Sanford Burnham Prebys NCI Cancer Center Support Grant P30 CA030199.
Author contributions
MP and MD conceived the project; MP, WR, GS, and MD designed and MP, WR, and KG performed the experiments; SJS contributed new reagents and tools. MP drafted the manuscript and all authors edited the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by A. Ashkenazi
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Marcin Poreba, Email: marcin.poreba@pwr.edu.pl.
Marcin Drag, Phone: +48 71 320 24 55, Email: marcin.drag@pwr.edu.pl.
Supplementary information
The online version of this article (10.1038/s41418-019-0329-2) contains supplementary material, which is available to authorized users.
References
- 1.Shalini S, Dorstyn L, Dawar S, Kumar S. Old, new and emerging functions of caspases. Cell Death Differ. 2015;22:526–39. doi: 10.1038/cdd.2014.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thornberry NA, Rano TA, Peterson EP, Rasper DM, Timkey T, Garcia-Calvo M, et al. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J Biol Chem. 1997;272:17907–11. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 3.Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–5. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- 4.Kumar S, Kinoshita M, Noda M, Copeland NG, Jenkins NA. Induction of apoptosis by the mouse Nedd2 gene, which encodes a protein similar to the product of the Caenorhabditis elegans cell death gene ced-3 and the mammalian IL-1 beta-converting enzyme. Genes Dev. 1994;8:1613–26. doi: 10.1101/gad.8.14.1613. [DOI] [PubMed] [Google Scholar]
- 5.Wang L, Miura M, Bergeron L, Zhu H, Yuan J. Ich-1, an Ice/ced-3-related gene, encodes both positive and negative regulators of programmed cell death. Cell. 1994;78:739–50. doi: 10.1016/s0092-8674(94)90422-7. [DOI] [PubMed] [Google Scholar]
- 6.Bouchier-Hayes L, Green DR. Caspase-2: the orphan caspase. Cell Death Differ. 2012;19:51–57. doi: 10.1038/cdd.2011.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tiwari M, Sharma LK, Vanegas D, Callaway DA, Bai Y, Lechleiter JD, et al. A nonapoptotic role for CASP2/caspase 2: modulation of autophagy. Autophagy. 2014;10:1054–70. doi: 10.4161/auto.28528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slee EA, Harte MT, Kluck RM, Wolf BB, Casiano CA, Newmeyer DD, et al. Ordering the cytochrome c-initiated caspase cascade: hierarchical activation of caspases-2, -3, -6, -7, -8, and -10 in a caspase-9-dependent manner. J Cell Biol. 1999;144:281–92. doi: 10.1083/jcb.144.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fava LL, Bock FJ, Geley S, Villunger A. Caspase-2 at a glance. J Cell Sci. 2012;125(Pt 24):5911–5. doi: 10.1242/jcs.115105. [DOI] [PubMed] [Google Scholar]
- 10.Krumschnabel G, Sohm B, Bock F, Manzl C, Villunger A. The enigma of caspase-2: the laymen’s view. Cell Death Differ. 2009;16:195–207. doi: 10.1038/cdd.2008.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheung HH, Lynn Kelly N, Liston P, Korneluk RG. Involvement of caspase-2 and caspase-9 in endoplasmic reticulum stress-induced apoptosis: a role for the IAPs. Exp Cell Res. 2006;312:2347–57. doi: 10.1016/j.yexcr.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 12.Machado MV, Michelotti GA, Jewell ML, Pereira TA, Xie G, Premont RT, et al. Caspase-2 promotes obesity, the metabolic syndrome and nonalcoholic fatty liver disease. Cell Death Dis. 2016;7:e2096. doi: 10.1038/cddis.2016.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tu S, McStay GP, Boucher LM, Mak T, Beere HM, Green DR. In situ trapping of activated initiator caspases reveals a role for caspase-2 in heat shock-induced apoptosis. Nat Cell Biol. 2006;8:72–7. doi: 10.1038/ncb1340. [DOI] [PubMed] [Google Scholar]
- 14.Bonzon C, Bouchier-Hayes L, Pagliari LJ, Green DR, Newmeyer DD. Caspase-2-induced apoptosis requires bid cleavage: a physiological role for bid in heat shock-induced death. Mol Biol Cell. 2006;17:2150–7. doi: 10.1091/mbc.E05-12-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manzl C, Peintner L, Krumschnabel G, Bock F, Labi V, Drach M, et al. PIDDosome-independent tumor suppression by Caspase-2. Cell Death Differ. 2012;19:1722–32. doi: 10.1038/cdd.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fava LL, Schuler F, Sladky V, Haschka MD, Soratroi C, Eiterer L, et al. The PIDDosome activates p53 in response to supernumerary centrosomes. Genes Dev. 2017;31:34–45. doi: 10.1101/gad.289728.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dorstyn L, Puccini J, Wilson CH, Shalini S, Nicola M, Moore S, et al. Caspase-2 deficiency promotes aberrant DNA-damage response and genetic instability. Cell Death Differ. 2012;19:1288–98. doi: 10.1038/cdd.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castedo M, Perfettini JL, Roumier T, Valent A, Raslova H, Yakushijin K, et al. Mitotic catastrophe constitutes a special case of apoptosis whose suppression entails aneuploidy. Oncogene. 2004;23:4362–70. doi: 10.1038/sj.onc.1207572. [DOI] [PubMed] [Google Scholar]
- 19.Kumar S. Caspase 2 in apoptosis, the DNA damage response and tumour suppression: enigma no more? Nat Rev Cancer. 2009;9:897–903. doi: 10.1038/nrc2745. [DOI] [PubMed] [Google Scholar]
- 20.Vakifahmetoglu-Norberg H, Zhivotovsky B. The unpredictable caspase-2: what can it do? Trends Cell Biol. 2010;20:150–9. doi: 10.1016/j.tcb.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 21.Miles MA, Kitevska-Ilioski T, Hawkins CJ. Old and novel functions of caspase-2. Int Rev Cell Mol Biol. 2017;332:155–212. doi: 10.1016/bs.ircmb.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 22.Ho LH, Taylor R, Dorstyn L, Cakouros D, Bouillet P, Kumar S. A tumor suppressor function for caspase-2. Proc Natl Acad Sci USA. 2009;106:5336–41. doi: 10.1073/pnas.0811928106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahmed Z, Kalinski H, Berry M, Almasieh M, Ashush H, Slager N, et al. Ocular neuroprotection by siRNA targeting caspase-2. Cell Death Dis. 2011;2:e173. doi: 10.1038/cddis.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jesenberger V, Procyk KJ, Yuan J, Reipert S, Baccarini M. Salmonella-induced caspase-2 activation in macrophages: a novel mechanism in pathogen-mediated apoptosis. J Exp Med. 2000;192:1035–46. doi: 10.1084/jem.192.7.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bouchier-Hayes L, Oberst A, McStay GP, Connell S, Tait SW, Dillon CP, et al. Characterization of cytoplasmic caspase-2 activation by induced proximity. Mol Cell. 2009;35:830–40. doi: 10.1016/j.molcel.2009.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng TS, Hunot S, Kuida K, Momoi T, Srinivasan A, Nicholson DW, et al. Deficiency in caspase-9 or caspase-3 induces compensatory caspase activation. Nat Med. 2000;6:1241–7. doi: 10.1038/81343. [DOI] [PubMed] [Google Scholar]
- 27.Parrish AB, Freel CD, Kornbluth S. Cellular mechanisms controlling caspase activation and function. Cold Spring Harb Perspect Biol. 2013;5:a008672. doi: 10.1101/cshperspect.a008672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Julien O, Zhuang M, Wiita AP, O’Donoghue AJ, Knudsen GM, Craik CS, et al. Quantitative MS-based enzymology of caspases reveals distinct protein substrate specificities, hierarchies, and cellular roles. Proc Natl Acad Sci USA. 2016;113:E2001–10. doi: 10.1073/pnas.1524900113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wejda M, Impens F, Takahashi N, Van Damme P, Gevaert K, Vandenabeele P. Degradomics reveals that cleavage specificity profiles of caspase-2 and effector caspases are alike. J Biol Chem. 2012;287:33983–95. doi: 10.1074/jbc.M112.384552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Talanian RV, Quinlan C, Trautz S, Hackett MC, Mankovich JA, Banach D, et al. Substrate specificities of caspase family proteases. J Biol Chem. 1997;272:9677–82. doi: 10.1074/jbc.272.15.9677. [DOI] [PubMed] [Google Scholar]
- 31.Benkova B, Lozanov V, Ivanov IP, Mitev V. Evaluation of recombinant caspase specificity by competitive substrates. Anal Biochem. 2009;394:68–74. doi: 10.1016/j.ab.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 32.McStay GP, Salvesen GS, Green DR. Overlapping cleavage motif selectivity of caspases: implications for analysis of apoptotic pathways. Cell Death Differ. 2008;15:322–31. doi: 10.1038/sj.cdd.4402260. [DOI] [PubMed] [Google Scholar]
- 33.Maillard MC, Brookfield FA, Courtney SM, Eustache FM, Gemkow MJ, Handel RK, et al. Exploiting differences in caspase-2 and -3 S(2) subsites for selectivity: structure-based design, solid-phase synthesis and in vitro activity of novel substrate-based caspase-2 inhibitors. Bioorg Med Chem. 2011;19:5833–51. doi: 10.1016/j.bmc.2011.08.020. [DOI] [PubMed] [Google Scholar]
- 34.Poreba M, Szalek A, Kasperkiewicz P, Rut W, Salvesen GS, Drag M. Small molecule active site directed tools for studying human caspases. Chem Rev. 2015;115:12546–629. doi: 10.1021/acs.chemrev.5b00434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poreba M, Groborz K, Navarro M, Snipas SJ, Drag M, Salvesen GS. Caspase selective reagents for diagnosing apoptotic mechanisms. Cell Death Differ. 2018;26:229–44. doi: 10.1038/s41418-018-0110-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poreba M, Kasperkiewicz P, Snipas SJ, Fasci D, Salvesen GS, Drag M. Unnatural amino acids increase sensitivity and provide for the design of highly selective caspase substrates. Cell Death Differ. 2014;21:1482–92. doi: 10.1038/cdd.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poreba M, Salvesen GS, Drag M. Synthesis of a HyCoSuL peptide substrate library to dissect protease substrate specificity. Nat Protoc. 2017;12:2189–214. doi: 10.1038/nprot.2017.091. [DOI] [PubMed] [Google Scholar]
- 38.Stennicke HR, Salvesen GS. Caspases: preparation and characterization. Methods. 1999;17:313–9. doi: 10.1006/meth.1999.0745. [DOI] [PubMed] [Google Scholar]
- 39.Poreba M, Szalek A, Kasperkiewicz P, Drag M. Positional scanning substrate combinatorial library (PS-SCL) approach to define caspase substrate specificity. Methods Mol Biol. 2014;1133:41–59. doi: 10.1007/978-1-4939-0357-3_2. [DOI] [PubMed] [Google Scholar]
- 40.Poreba M, Solberg R, Rut W, Lunde NN, Kasperkiewicz P, Snipas SJ, et al. Counter selection substrate library strategy for developing specific protease substrates and probes. Cell Chem Biol. 2016;23:1023–35. doi: 10.1016/j.chembiol.2016.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rut W, Poreba M, Kasperkiewicz P, Snipas SJ, Drag M. Selective substrates and activity-based probes for imaging of the human constitutive 20s proteasome in cells and blood samples. J Med Chem. 2018;61:5222–34. doi: 10.1021/acs.jmedchem.8b00026. [DOI] [PubMed] [Google Scholar]
- 42.Scaffidi C, Medema JP, Krammer PH, Peter ME. FLICE is predominantly expressed as two functionally active isoforms, caspase-8/a and caspase-8/b. J Biol Chem. 1997;272:26953–8. doi: 10.1074/jbc.272.43.26953. [DOI] [PubMed] [Google Scholar]
- 43.Tang Y, Wells JA, Arkin MR. Structural and enzymatic insights into caspase-2 protein substrate recognition and catalysis. J Biol Chem. 2011;286:34147–54. doi: 10.1074/jbc.M111.247627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chauvier D, Ankri S, Charriaut-Marlangue C, Casimir R, Jacotot E. Broad-spectrum caspase inhibitors: from myth to reality? Cell Death Differ. 2007;14:387–91. doi: 10.1038/sj.cdd.4402044. [DOI] [PubMed] [Google Scholar]
- 45.Oliver TG, Meylan E, Chang GP, Xue W, Burke JR, Humpton TJ, et al. Caspase-2-mediated cleavage of Mdm2 creates a p53-induced positive feedback loop. Mol Cell. 2011;43:57–71. doi: 10.1016/j.molcel.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fava LL, Schuler F, Sladky V, Haschka MD, Soratroi C, Eiterer L, et al. The PIDDosome activates p53 in response to supernumerary centrosomes. Genes Dev. 2017;31:34–45. doi: 10.1101/gad.289728.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lopez-Garcia C, Sansregret L, Domingo E, McGranahan N, Hobor S, Birkbak NJ, et al. BCL9L dysfunction impairs caspase-2 expression permitting aneuploidy tolerance in colorectal cancer. Cancer Cell. 2017;31:79–93. doi: 10.1016/j.ccell.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.LeBlanc H, Lawrence D, Varfolomeev E, Totpal K, Morlan J, Schow P, et al. Tumor-cell resistance to death receptor--induced apoptosis through mutational inactivation of the proapoptotic Bcl-2 homolog Bax. Nat Med. 2002;8:274–81. doi: 10.1038/nm0302-274. [DOI] [PubMed] [Google Scholar]
- 49.Kitevska T, Roberts SJ, Pantaki-Eimany D, Boyd SE, Scott FL, Hawkins CJ. Analysis of the minimal specificity of caspase 2 and identification of Ac-VDTTD-AFC as a caspase 2 selective peptide substrate. Biosci Rep. 2014;34:127. doi: 10.1042/BSR20140025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.