Abstract
The loss of imprinting of MEST has been linked to certain types of cancer by promoter switching. However, MEST-mediated regulation of tumorigenicity and metastasis are yet to be understood. Herein, we reported that MEST is a key regulator of IL-6/JAK/STAT3/Twist-1 signal pathway-mediated tumor metastasis. Enhanced MEST expression is significantly associated with pathogenesis of breast cancer patients. Also, MEST induces metastatic potential of breast cancer through induction of the EMT-TFs-mediated EMT program. Moreover, MEST leads to Twist-1 induction by STAT3 activation and subsequently enables the induction of activation of the EMT program via the induction of STAT3 nuclear translocation. Furthermore, the c-terminal region of MEST was essential for STAT3 activation via the induction of JAK2/STAT3 complex formation. Finally, MEST is required for metastasis in an experimental metastasis model. These observations suggest that MEST is a promising target for intervention to prevent tumor metastasis.
Subject terms: Tumour-suppressor proteins, Metastasis, Tumour biomarkers
Introduction
Mesoderm-specific transcript (MEST) has been shown to maternally imprint on parthenogenetic embryos, and only the paternally inherited allele is preferentially expressed in the placenta of humans. Also, MEST expressed in extraembryonic tissues during development is paternally transmitted, and the offspring exhibit severe intrauterine growth retardation after a null allele, suggesting that MEST may have a role in development [1, 2].
Recently, several studies have described the frequent loss of imprinting of MEST, which has been linked to certain types of cancer. In choriocarcinoma, trophoblast tumors, breast cancer, lung adenocarcinomas, uterine leiomyoma, colon cancer, human adult blood lymphocytes, and lymphoblastoid cell lines, MEST is expressed from both the paternal allele and the maternal allele via promoter switching from isoform 1 to isoform 2 of MEST [1, 3–7]. Although MEST is expressed in many tumors by promoter switching, it is currently unclear how MEST regulates tumorigenicity and metastasis of the tumor. Moreover, the signaling mechanism that induces and maintains tumorigenicity and tumor metastasis by MEST has remained unclear.
In this report, we first identified a new network involved in tumor metastasis and EMT that regulates and coordinates MEST. We found that a high level of MEST expression is significantly correlated with pathogenesis in breast cancer patients and that MEST acts as a key regulator of tumor metastasis through IL-6/JAK/STAT3/Twist-1 signal pathway-mediated EMT induction.
Results
Elevated MEST expression in human breast tumors and cells
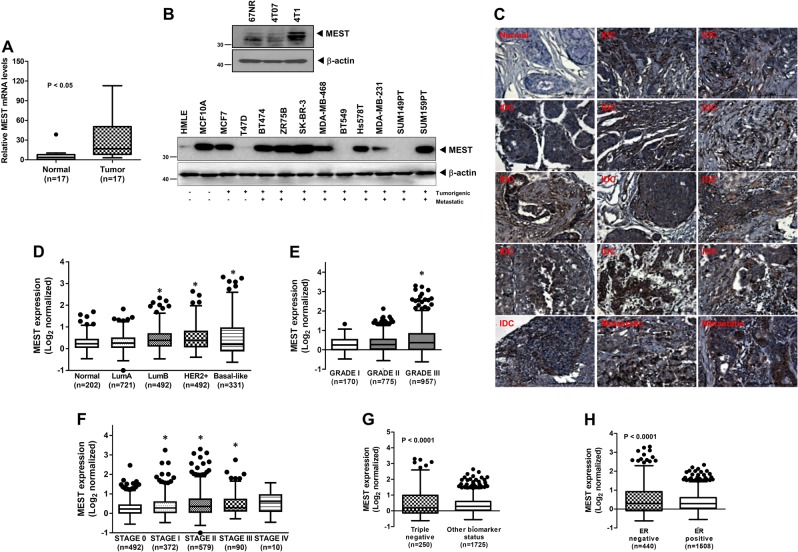
Although the loss of MEST imprinting is linked to certain types of cancer, MEST expression have not been well characterized in human breast cancer. To evaluate its potential involvement in breast cancer, we first assessed MEST expression in clinical breast tumors. Strikingly, the level of expression of MEST was significantly enhanced in 16 out of 17 tumor tissues of breast cancer patients (94%) compared to those in corresponding patient-matched normal tissue samples (Fig. 1a). On the basis of these observations, we next examined MEST expression in a panel of mouse and human breast cancer cells. MEST was highly expressed in human breast cancer cells compared to the level in human mammary epithelial cells (HMLE) and non-tumorigenic MCF10A cells. In addition, MEST was upregulated in 4T1 cells, the most aggressive of the three mouse mammary tumor cells and the only one capable of forming macroscopic lung metastases [8] (Fig. 1b and S1). Furthermore, in immunohistochemistry analysis, normal breast tissue exhibited low expression of MEST. However, elevated MEST expression was detected in the infiltrating ductal (IDC) and metastatic breast carcinoma samples (Fig. 1c and S2).
Fig. 1.
Expression of MEST is correlated with breast pathological phenotypes. a The expression of the mRNA encoding MEST in 17 samples of individual human breast cancer patient relative to that of the healthy tissue, as determined by quantitative RT-PCR. The 18S mRNA level was used to normalize the variability in template loading. P < 0.05, t-test. b Western blot analysis of MEST expression in tumorigenic and metastatic human or mouse breast cancer cells. β-actin was used as a loading control. c Representative immunohistochemical images of MEST staining in normal human breast tissue, infiltrating duct carcinoma (IDC), and metastatic breast carcinoma (magnification: ×200). d Box-and-whisker (Tukey) plots are shown for the expression of MEST in 2137 human breast cancer patients by subtypes of breast cancer. The MEST level was extracted from the METABRIC dataset and averaged for each tumor. (Asterisks) P < 0.05 relative to normal; P-value is based on a post-hoc test after one-way ANOVA. e Box-and-whisker (Tukey) plots are shown for the expression of MEST on the grade of 2137 human breast cancer patients. (Asterisks) P <0.05 relative to GRADE I; P-value is based on a post-hoc test after one-way ANOVA. f Box-and-whisker (Tukey) plots are shown for the expression of MEST on the stage of 2137 human breast cancer patients. (Asterisks) P < 0.05 relative to SATGE 0; P-value is based on a post-hoc test after one-way ANOVA. g Box-and-whisker (Tukey) plots are shown for the expression of MEST on triple-negative or other biomarkers from 2137 human breast cancer patients. P < 0.0001, t-test. h Box-and-whisker (Tukey) plots are shown for the expression of MEST on ERα negative or ERα positive samples from 2137 human breast cancer patients. The MEST level from d–h was extracted from the METABRIC dataset and averaged for each tumor. Points below and above the whiskers are drawn as individual dots. P < 0.05, t-test
We next investigated the clinical relevance using publicly available microarray datasets of breast cancer patients from the METABRIC and the UNC dataset (GSE26338) [9, 10]. Consistent with the above observations, the level of MEST expression was significantly higher in both basal-like and claudin-low breast cancer subtypes, and moreover, MEST expression was correlated with the grade and stage of breast cancer patients (Fig. 1d–f, S3A, S3B, and S3C). Furthermore, MEST expression was significantly upregulated in triple-negative breast cancer (TNBC) relative to the luminal and HER2-positive markers (Fig. 1g and S3D), and in particular, MEST expression was strongly negatively correlated with ER-α expression (Fig. 1h and S3E). In addition, we examined a potential linkage of the MEST expression levels with nine clinical parameters. Multivariate statistical analysis based on an orthogonal projection in latent structure (OPLS) was applied to explore the latent association (Figure S4). First, the multivariate analysis validated MEST expression levels were highly correlated with survival time (VIP > 2). The analysis further showed the moderate levels of association with survival status, stage, and grade (VIP > 1). In addition, lymph nodes positivity and high cellularity were positively correlated with the MEST levels. Of particular interest, the OPLS model proposed the expression levels were strongly linked to the variability within other breast types rather than triple-negative type although the absolute expression levels were significantly higher in triple-negative type. Our observations indicated that MEST might have an important role in the metastatic dissemination of breast cancer.
MEST is required for transformation and migration of triple-negative breast cancer cells
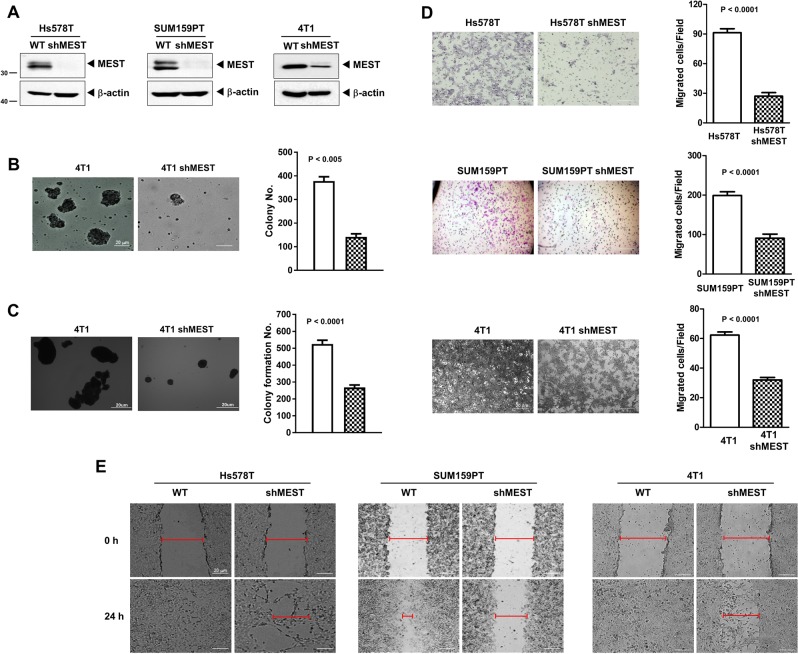
The invasion-metastasis cascade is driven by the acquisition of anchorage independence, a gain of motility, invasion in and out of circulation, and colonization of distant organs [11–13]. So, we speculated that MEST might contribute to tumorigenicity and the metastasis process of breast cancer. To test this notion, we selected highly metastatic Hs578T, SUM159PT, and 4T1 cells and engineered stably expressing MEST-shRNAs into the cell lines.
By an anoikis assay measuring the anchorage-independent survival [14], we examined the clonal formation abilities of 4T1 control and 4T1 MEST-shRNA cells in suspension. 4T1 control-shRNA cells proliferated as large spheroid aggregates in suspension relative to MEST-shRNA cells (Fig. 2b).
Fig. 2.
MEST is required for transformation and migration of triple-negative breast cancer cells. a Western blot analysis of MEST expression in Hs578T, SUM159PT, and 4T1 control-shRNA or MEST-shRNA cells. β-actin was used as a loading control. b Formation of spheroid colonies of 4T1 control-shRNA or MEST-shRNA cells. Spheroid colonies in suspension were then photographed at ×200 magnification. c The soft agar colony-forming assay of the 4T1 control-shRNA or MEST-shRNA cells (n = 3). P < 0.0001, t-test. d The migration assay of in Hs578T, SUM159PT, and 4T1 control-shRNA or MEST-shRNA cells (n = 3). P < 0.0001, t-test. e Wound healing assay of Hs578T, SUM159PT, and 4T1 control-shRNA or MEST-shRNA cells. Wound closures were photographed at 0 and 24 h after wounding (magnification: ×200)
We next examined whether the loss of MEST affected the colony-forming ability of 4T1 cells. 4T1 control-shRNA cells exhibited almost twofold higher colony-forming activity compared to the MEST-shRNA cells (Fig. 2c). Also, we conducted cell motility and wound healing assays. Control-shRNA and MCF10A-MEST cells had significantly increased cell migration, whereas MEST-shRNA cells and MCF10A-control cells inhibited the migration of breast cancer cells, indicating that the increased cell migration was correlated with the level of MEST expression (Fig. 2d, e, S5, and S6). These findings suggest that MEST accelerates the secondary tumor forming ability to distant organ sites.
MEST regulates the invasion-metastasis cascade through induction of a Twist-mediated EMT program
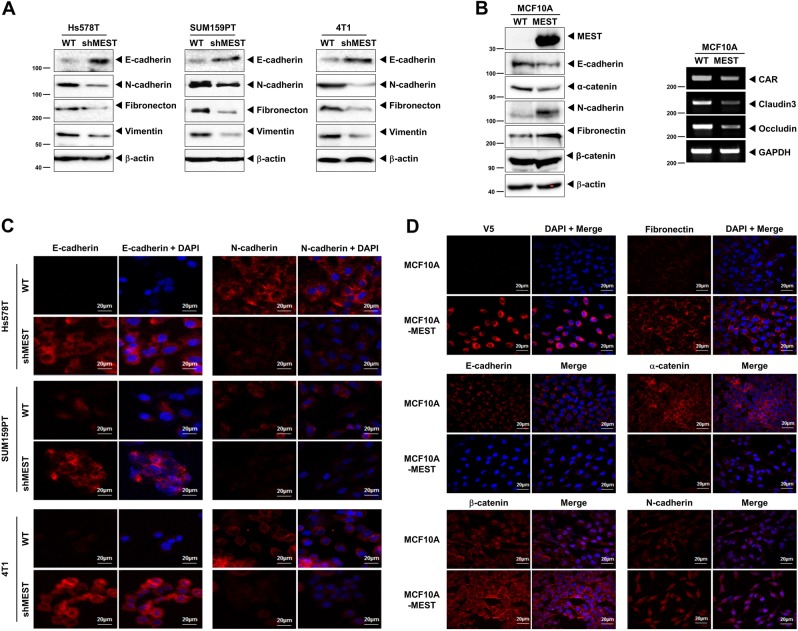
Since the epithelial–mesenchymal transition (EMT) broadly regulates invasion and metastasis by acquiring cellular traits associated with high-grade malignancy [11, 12], we speculated that the acquisition of tumorigenic and metastatic potential by MEST might depend on induction of the EMT program. To address the issue, we examined whether MEST regulates an EMT program in breast cancer. Specifically, MEST-shRNA or MCF10A-control cells exhibited increased mRNA or protein levels of E-cadherin and significant decreases in expression of mesenchymal markers relative to control-shRNA or MCF10A-MEST cells (Fig. 3a, b, S7A, and S7B).
Fig. 3.
High MEST expression induces EMT program. a Western blot analysis of the expression of E-cadherin, N-cadherin, fibronectin, and vimentin proteins in Hs578T, SUM159PT, and 4T1 control-shRNA or MEST-shRNA cells. β-actin was used as a loading control. b Western blot (left) and RT-PCR (Right) analysis of the expression of CAR, caludin3, occludin, E-cadherin, α-catenin, N-cadherin, fibronectin, and β-catenin proteins in MCF10A-control or MCF10A-MEST cells. GAPDH and β-actin were used as a loading control. c Immunofluorescence images of E-cadherin and N-cadherin in Hs578T, SUM159PT, and 4T1 control-shRNA or MEST-shRNA cells. The red signal represents staining of the corresponding protein, while the blue signal represents DAPI staining. d Immunofluorescence images of V5, E-cadherin, β-catenin, fibronectin, α-catenin, and N-cadherin in MCF10A-control or MCF10A-MEST cells. The red signal represents the staining of the corresponding protein, while the blue signal represents DAPI staining
We also performed immunostaining to further determine whether MEST expression induced the EMT program. The immunostaining intensity of epithelial or mesenchymal markers in control-shRNA cells corresponded to the level of protein in the cells (Fig. 3c). These results indicating that the depletion of MEST prevents the conversion of mesenchymal traits on epithelial cells. In addition, the expression levels of E-cadherin and α-catenin in the MCF10A-MEST cells were significantly decreased, while N-cadherin, fibronectin, and β-catenin expression levels were markedly increased (Fig. 3d), compared to the expression level in MCF10A-control cells.
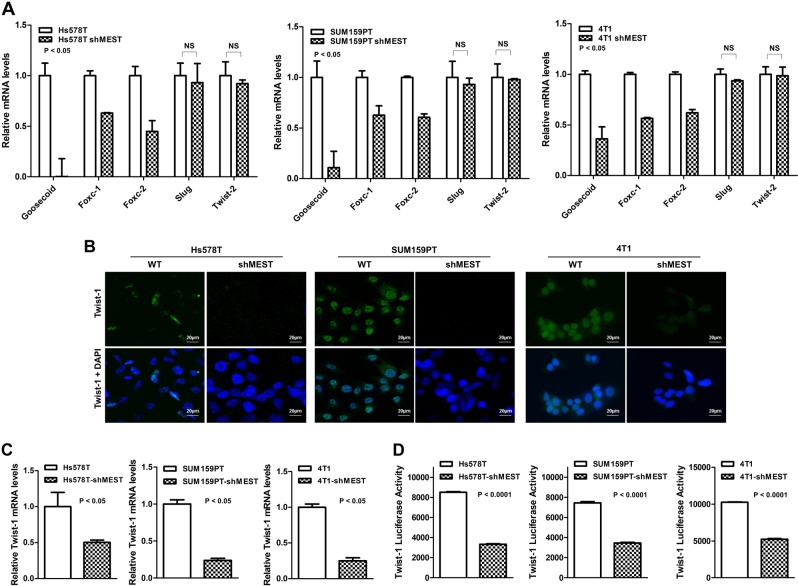
During the process of tumor metastasis, a set of EMT-TFs orchestrate the EMT program and related migratory processes. In addition, these EMT-TFs are expressed in various combinations in a number of malignant tumor types [12, 15]. These recent studies and the above observation led us to speculate that MEST might contribute to the induction of EMT via the upregulation of EMT-TFs. To test this possibility, we examined the expression of a set of EMT-TFs. There was a considerable increase in the expression of EMT-TFs, specifically Goosecoid, Foxc-1, Foxc-2, and Twist-1, in control-shRNA cells but not Slug, and Twist-2, compared to the expression in MEST-shRNA cells (Fig. 4a–c). The difference was also confirmed by quantitative RT-PCR (RT-qPCR). MCF10A-MEST cells overexpressed EMT-TFs (Figure S8A), compared to the MCF10A-control cells. These data indicate that the MEST upregulates the expression of multiple genes in EMT-induced cells and MEST may be a key mediator of twist gene trans-activation, resulting in the loss of epithelial characteristics and the gain of mesenchymal properties.
Fig. 4.
MEST knockdown is associated with loss of EMT transcription factors. a The relative expression levels of mRNA encoding Goosecoid, Foxc-1, Foxc-2, Slug, Twist-1, and Twist-2 in Hs578T, SUM159PT, and 4T1 control-shRNA or MEST-shRNA cells, as determined by quantitative RT-PCR. 18S was used as a loading control. P < 0.05, t-test. b Immunofluorescence images of Twist-1 in Hs578T, SUM159PT, and 4T1 control-shRNA or MEST-shRNA cells. The green signal represents the staining of the corresponding protein, while the blue signal represents DAPI staining. c The relative expression levels of mRNA encoding Twist-1 in Hs578T, SUM159PT, and 4T1 control-shRNA or MEST-shRNA cells, as determined by quantitative RT-PCR. 18S was used as a loading control. P < 0.05, t-test. d Promoter activity of Twist-1 gene in Hs578T, SUM159PT, and 4T1 control-shRNA or MEST-shRNA cells were measured by the Luciferase reporter assay. Each bar represents the mean ± SEM of three experiments. P < 0.0001, t-test
Because MEST more significantly regulates Twist-1 expression compared to other EMT-inducing transcription factors (Fig. 4c), we initially focused on the upregulation of Twist-1 by MEST. Immunofluorescence studies confirmed that Twist in control-shRNA cells and MCF10A-MEST cells were expressed primarily as a nuclear form of Twist (Fig. 4b and S8B). The intensity of overlap between Twist immunofluorescence and DAPI significantly decreased in MEST-shRNA cells and MCF10A-control cells, compared to those in control-shRNA cells, and MCF10A-MEST cells. Also, we tested the possibility of the role of MEST in Twist regulation at the transcriptional level. A significant reduction of Twist promoter activity was also observed in MEST knockdown cells (Fig. 4d). Collectively, these data imply that MEST is a positive mediator of twist gene trans-activation, resulting in the loss of epithelial characteristics and the gain of mesenchymal properties.
MEST upregulates Twist-1 expression through activation of STAT3
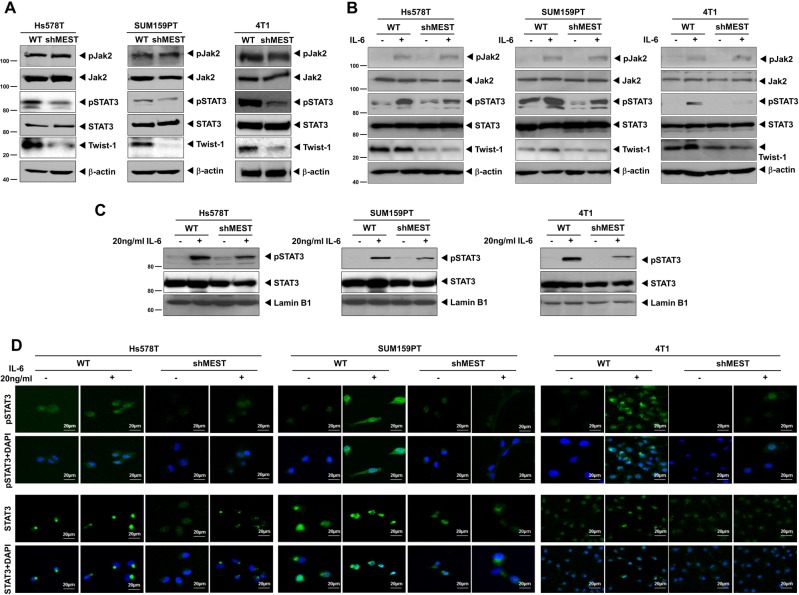
Although gene ontology (GO) analysis from the UniProtKB/Swiss-Prot database proposed that MEST localizes to the endoplasmic reticulum (ER), MEST has not been shown to localize to or be associated with organelles and specialized subcellular compartments. As subcellular localization is important for functionality, we examined the subcellular localization of MEST. It was found that the majority of MEST was present in the membrane fraction, including the plasma/ER/Golgi/mitochondrial membranes, and a small fraction of MEST protein was found in the nucleus where the Twist protein was primarily located. It is worth noting that cytokeratin 18 expression is well-known as a luminal epithelial marker and was markedly increased in the MEST-shRNA cells compared to the control-shRNA cells (Figures S9A and S9B). This result supports that MEST regulates the invasion-metastasis cascade through induction of the Twist-mediated EMT program. However, the distinction in the subcellular localization of MEST and Twist led us to hypothesize that MEST might have a role as a linker or scaffold protein having characteristics of both nuclear and cytoplasmic signaling molecules. Recently, Cheng et al. [16] demonstrated that the active form of STAT3 was able to directly bind to the Twist promoter and promote its transcriptional activity. These observations led us to speculate that MEST might be involved in the regulation of STAT3 activation and that it was functionally linked to the regulation of Twist-1. To test the effect of MEST in STAT3 activation, we initially examined whether MEST knockdown affected both the total and active forms of STAT3 protein. STAT3 activation, as well as Twist-1 expression, was markedly decreased in the MEST-shRNA cells relative to control-shRNA cells; however, STAT3 expression was not affected in the knockdown of MEST. Moreover, the levels of phosphorylated and total Jak2 were not altered upon MEST knockdown (Fig. 5a). In addition, similar results were obtained with MCF10A-MEST cells. STAT3 phosphorylation and Twist-1 expression were significantly increased, but JAK2 phosphorylation, total JAK2, and STAT3 expression were not changed (Figure S9C).
Fig. 5.
MEST led to Twist-1 upregulation through activation of STAT3. a Western blot analysis of the expression of phospho-JAK2, JAK2, phospho-STAT3, STAT3, and Twist-1 proteins in Hs578T, SUM159PT, and 4T1 control-shRNA or MEST-shRNA cells. β-actin was used as a loading control. b Western blot analysis of the expression of phospho-JAK2, JAK2, phospho-STAT3, STAT3, and Twist-1 proteins in Hs578T, SUM159PT, and 4T1 control-shRNA or MEST-shRNA cells treated with or without 20 ng/ml IL-6. β-actin was used as a loading control. c Western blot analysis of the expression of phospho-STAT3 and STAT3 proteins in Hs578T, SUM159PT, and 4T1 control-shRNA or MEST-shRNA cells after nuclear fractionation. Lamin B1 was used as a nuclear loading control. d Immunofluorescence images of phospho-STAT3 and STAT3 in Hs578T, SUM159PT, and 4T1 control-shRNA or MEST-shRNA cells. The green signal represents the staining of the corresponding protein, while the blue signal represents DAPI staining
As Jak2 expression and activation were similar between control and MEST knockdown cells, we examined whether there was any difference in terms of ligand-induced Jak2-STAT3 activation between the control and MEST knockdown cells or the levels of Twist were affected by ligand-induced STAT3 activation in MEST knockdown cells. To test this notion, we assessed the expression level and activation of JAK2 of control-shRNA and MEST-shRNA cells with IL-6 activation. Interestingly, there was no difference in the expression or activation of JAK2 or STAT3 expression between the control and MEST knockdown cell in response to IL-6. However, IL-6-induced STAT3 activation was markedly reduced in MEST knockdown cells, relative to the control-shRNA cells, which was strongly correlated with the level of Twist-1 expression (Fig. 5b).
As activated STAT3 can translocate into the nucleus and act as a transcription factor, we consequently tested whether MEST could modulate IL-6-dependent subcellular localization of STAT3. In control-shRNA cells, we observed an increase in the nuclear translocation and nuclear expression of STAT3 and phospho-STAT3 with or without IL-6 treatment, compared to those in MEST-shRNA cells (Fig. 5c, d). These results suggest that treatment of IL-6 could induce STAT3 phosphorylation in control cells; however, the loss of MEST seemed to have inhibitory effects on STAT3 phosphorylation after IL-6 treatment. Taken together, these observations indicate that MEST regulates expression of Twist-1, possibly mediated through JAK2 activation-independent STAT3 activation.
In addition, the STAT3 mRNA expression and STAT3 promoter activity were not affected by IL-6 treatment in both the control and MEST knockdown cells (Figures S10A and S10B). To more elucidate the mechanistic basis for regulation of IL-6/STAT3 signaling by MEST, we examined whether the level of STAT3 activation by IL-6 is affected by MEST expression by performing a dose–response or time-course experiment. The level of Twist-1 or phospho-STAT3 in control-shRNA cells were significantly increased in IL-6 dose-dependent or time-course manner relative to the MEST-shRNA cells but the level of serine phosphorylation of STAT3 in control or MEST-shRNA cells does not changed by treatment of IL-6 (Figure S11A and S11B). Interestingly, the basal level of phospho-stat1 was significantly higher in control-shRNA cells than in MEST-shRNA cells and phospho-STAT1 levels in both of control and MEST-shRNA cells markedly decreased by IL-6 dose-dependent or time-course manner (Figure S11A and S11B). Interestingly, the basal level of STAT3 activation and Twist-1 expression were markedly increased in MCF10A-MEST cells compared to that in control cells (Figure S14A). Also, the activation of STAT3 and Twist-1 expression were significantly increased in MCF10A-MEST cells by treatment of IL-6 (Figure S14A). These findings suggest that MEST alone is sufficient to induce Twist-1 induction by STAT3 activation.
To further examined whether EMT induction via upregulation of Twist-1 expression by MEST is mediated through STAT3, we evaluated the change of Twist-1 expression by inhibition of JAK2/STAT3 signaling via treatment of JAK2 inhibitor (AG490) or STAT3 inhibitor (S32-201). The inhibition of JAK2 activation in Hs578T and SUM149 cells by AG490 significantly reduced JAK2 and STAT3 phosphorylation and Twist-1 expression (Figure S12A). Also, treatment of S32-201 significantly reduced STAT3 phosphorylation and Twist-1 expression. However, phospho-STAT3 levels were not significantly different after treatment with STAT3 inhibitor (Figure S12B). Moreover, we investigated whether the change of STAT3 activation by STAT3 kinase-dead mutant (STAT3YF) or STAT3 constitutive activated mutant (STAT3CA) may affect the induction of EMT program by upregulation of Twist-1. As shown in Figure S13, the introduction of STAT3 wild type or STAT3CA mutant in MEST-shRNA cells significantly induced Twist-1 and downregulated the expression of E-cadherin, but increased expression of mesenchymal markers. However, the introduction of STAT3YF mutant induced high levels of E-cadherin but significantly reduced expression of mesenchymal markers relative to STAT3 wild type or STAT3CA mutant. Also, we found that MCF10A-MEST cell had significantly upregulated basal expression of mesenchymal markers but downregulated expression of E-cadherin. Furthermore, by treatment of IL-6, the expression of E-cadherin was more reduced but more induced expression of mesenchymal markers (Figure S14B). These findings suggest that activation of STAT3 by MEST is required EMT induction through Twist-1 induction.
MEST induces STAT3 activation through MEST–STAT3 complex formation
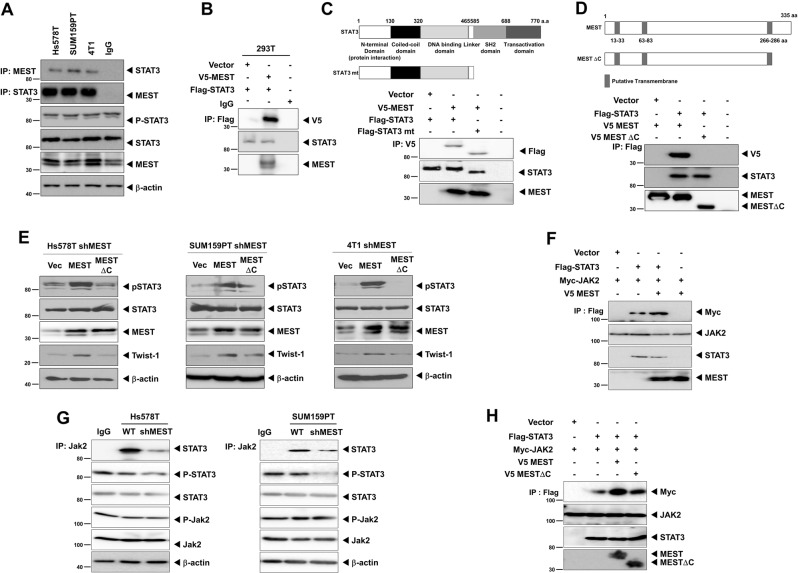
To test the regulatory role of MEST in STAT3 activation, we attempted to determine whether MEST regulates STAT3 activation through MEST–STAT3 complex formation by investigating the complex formation of MEST and STAT3 endogenously. An interaction between these two proteins was readily detected. (Fig. 6a). Moreover, MEST directly interacts with STAT3 under physiological conditions (Fig. 6b). To identify the STAT3 functional domain responsible for its interaction with MEST, we used a series of deletion constructs of STAT3. A STAT3 mutant containing the N-terminal domain (NTD: protein interaction domain) interacted with MEST, whereas MEST did not interact with SH2 or the trans-activation domain of STAT3 (Fig. 6c and S15A). MEST protein is annotated as a putative multi-pass membrane protein by sequence prediction (Fig. 6d). To identify the MEST functional regions responsible for its interaction with STAT3, we used a deletion construct of MESTΔC and full-length MEST. Full-length MEST interacts with STAT3, whereas MESTΔC with a C-terminal deletion after the 287th amino acid did not interact with STAT3 (Fig. 6d).
Fig. 6.
MEST induces STAT3 activation through induction of JAK2–STAT3 complex formation. a Identification of the complex formation of endogenous MEST/STAT3 in Hs578T, SUM159PT and 4T1 cells. b Western blot analysis of the whole-cell lysates and immunoprecipitates derived from 293T cells transfected with the V5-MEST and Flag-STAT3 constructs, as indicated. c Identification of the STAT3 region that interacted with MEST. Western blot analysis of whole-cell lysates and immunoprecipitates derived from 293T cells transfected with V5-MEST, Flag-STAT3 wild, and the STAT3 deletion construct (STAT3 mt) as indicated. d Western blot analysis of the whole-cell lysates and immunoprecipitates derived from 293T cells transfected with the V5-MEST, V5-MESTΔC, and Flag-STAT3 constructs, as indicated. e Western blot analysis of the expression of phospho-STAT3 and STAT3 proteins in Hs578T, SUM159PT, and 4T1 MEST-shRNA cells after transient transfection of MEST or MESTΔC constructs. β-actin was used as a loading control. f Western blot analysis of whole-cell lysates and immunoprecipitates derived from 293T cells after transfection of JAK2, STAT3, or MEST constructs. β-actin was used as a loading control. g Identification of the complex formation of endogenous JAK2/STAT3 in Hs578T and SUM159PT control-shRNA or MEST-shRNA cells. h Western blot analysis of whole-cell lysates and immunoprecipitates derived from 293T cells after transfection of JAK2, STAT3, MEST, or MESTΔC constructs. β-actin was used as a loading control
On the basis of the above observation, we speculated that MESTΔC might contribute to the activation of STAT3. To test this notion, we assessed STAT3 expression and STAT3 activation in MEST knockdown cells after transient transfection of MEST or MESTΔC. Compared to those in the MEST knockdown cells expressing the control or MESTΔC, the introduction of MEST to the MEST knockdown cells showed significantly increased activation and expression of STAT3 or Twist-1 (Fig. 6e and S15B). These results indicated that the C-terminal region (amino acid 287–335) of MEST is essential for STAT3 activation.
Next, we attempted to determine whether MEST regulates JAK2/STAT3 complex formation through the interaction of MEST and STAT3. Interestingly, MEST induces JAK2/STAT3 complex formation (Fig. 6f, g). To identify the MEST functional region responsible for the induction of JAK2/STAT3 complex formation, we tested JAK2/STAT3 complex formation by MEST and MESTΔC. JAK2/STAT3 complex formation by MESTΔC still remained, but the level of JAK2/STAT3 interaction was significantly decreased relative that of MEST (Fig. 6h). These findings suggest that MEST knockdown decreases the efficiency of this complex.
MEST expression significantly increases metastasis in vivo
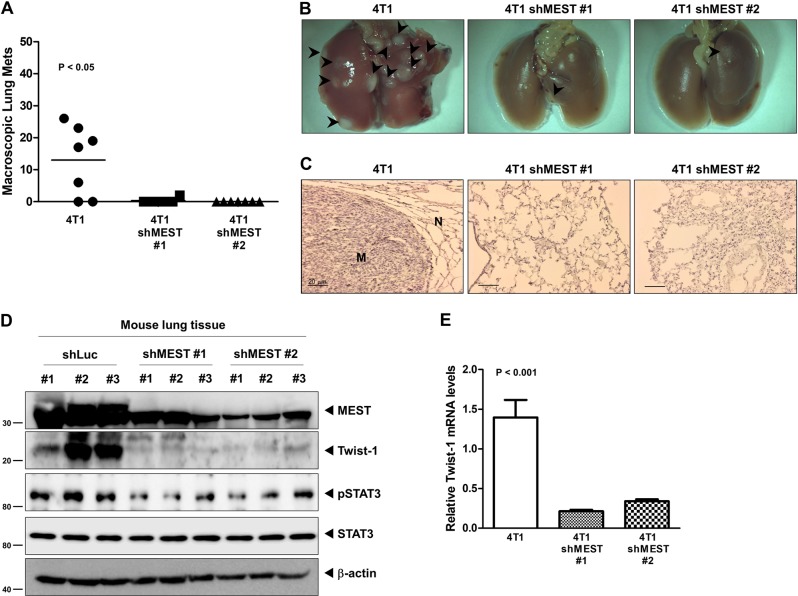
The 4T1 MEST-shRNA cells strongly reduced the number of metastatic nodules relative to the 4T1 control-shRNA cells (Fig. 7a). In addition, a few metastatic nodules in the lungs of mice carrying MEST-shRNA cells retained MEST expression, but MEST expression in the lungs of mice with MEST-shRNA cells was greatly reduced relative to the lungs of mice carrying control-shRNA cells (Fig. 7b–d). Moreover, histological analyses confirmed that the number of micrometastatic lesions was drastically reduced in the lungs of mice with 4T1 MEST-shRNA cells (Fig. 7c).
Fig. 7.
Suppression of MEST inhibited the ability of 4T1 cells to metastasize to the lung. a Lung metastasis by 4T1 control-shRNA or 4T1 MEST-shRNA cells. The total numbers of lung metastatic nodules from each mouse harboring 4T1 tumors expressing control-shRNA or MEST-shRNA were counted using a dissection scope (n = 6 mice per group). P < 0.05, ANOVA. b Representative images of the lungs from the mice harboring 4T1 control-shRNA or 4T1 MEST-shRNA cells for 30 days after implantation of mouse mammary fat pads of mice. c Representative H&E staining in the sections of the lungs from Fig. 7b. N normal lung tissue, M metastatic nodule. d Western blot analysis of MEST, phospho-STAT3, STAT3, and Twist-1 in tumor cells recovered from the lungs of individual mice expressing either 4T1 control-shRNA or 4T1 MEST-shRNA. β-actin was used as a loading control. e Quantitative RT-PCR of Twist-1 in tumor cells recovered from the lungs of individual mice expressing either 4T1 control-shRNA or 4T1 MEST-shRNA. 18S mRNA was used as a loading control. P < 0.001, t-test
We speculated that the presence of small numbers of nodules in the lungs of mice carrying 4T1 MEST-shRNA cells was caused by incomplete knockdown of MEST. To test this notion, we examined the level of MEST expression in the lungs of mice carrying 4T1 control-shRNA or MEST-shRNA cells. MEST expression in the lungs of mice with 4T1 MEST-shRNA cells was significantly lower than that of the 4T1 control-shRNA cells. Moreover, STAT3 activation and Twist-1 expression were markedly reduced in the lungs of mice with 4T1 MEST-shRNA cells relative to that of 4T1 control-shRNA cells (Fig. 7d). Furthermore, RT-qPCR analysis of Twist-1 revealed reduced expression in the lungs of mice injected with 4T1 MEST-shRNA cells compared to their control counterparts (Fig. 7e). These results indicated that expression of MEST is required for the full metastatic ability of highly metastatic 4T1 breast cancer cells.
Discussion
In this study, we found that MEST is frequently overexpressed in highly metastatic breast cancer cells and clinical breast tumor samples relative to normal epithelial cells and tissue. Moreover, MEST expression was significantly associated with the subtypes, grade, stage, and triple-negative phenotype of breast cancer patients. Our observations are consistent with other previous studies, which showed that Slug, HOXB13, HER2/neu, TrkA, TrkC, Foxc-2, and Goosecoid, which promote metastatic behaviors and poor prognosis in breast cancer, are already overexpressed in breast cancer patients and breast cancer cells before the appearance of the malignant tumor phenotype [17–21]. In addition, our results suggest that MEST expression may correlate with the characteristics of metastatic breast cancer.
Metastatic breast cancer is an aggressive, chemoresistant tumor and forms a distinct subtype that is closely related to receptor-negative breast cancer, such as basal and claudin-low cells characterized by increased motility, poor survival outcome, and the resistance of apoptosis [22]. In our results, suppression of MEST in highly metastatic breast cancer cells results in increased anoikis and the loss of motility and invasion potential. These observations indicate that high MEST expression enables breast cancer to acquire cellular traits associated with high-grade malignancy.
Recent studies demonstrated that metastatic breast cancer is closely linked to the EMT program. Metastatic breast cancer has an enriched EMT core signature that is associated with invasion, migration, resistance to chemotherapy, as well as the acquisition of mesenchymal and cancer stem cell (CSC) traits [12, 15, 23, 24]. In our present work, high MEST expression induces the EMT program, which correlates strongly with the poor survival of breast cancer. Our observations suggest that MEST expression is critical to EMT induction through EMT-TFs and governs the conversion to a high degree of plasticity.
The IL-6/JAK2/STAT3 pathway induces mesenchymal traits via the induction of Twist-1 and resistance to chemotherapy [25–30]. The molecular mechanisms of MEST-mediated IL-6/JAK2/STAT3 modulation in breast cancer are unclear, and none of the findings reported to date hinted at a link between MEST expression and IL-6/JAK2/STAT3 modulation. We found that upregulation of Twist-1 and the activation of the IL-6/JAK2/STAT3 pathway through MEST–STAT3 complex formation induced tumorigenicity and metastatic potential of breast cancer.
STAT3 activation by IL-6 leads to CSC self-renewal traits, induction of EMT, and cell transformation of a number of oncogenes [31–33]. Our studies further uncovered MEST function as a key regulator of Twist-1 via the activation of IL-6/JAK2/STAT3 signaling. In the presence of IL-6, the induction of STAT3 nuclear translocation by MEST promotes EMT through the increased expression of Twist-1.
MEST protein may have a putative multi-pass membrane protein according to gene ontology (GO). In our hands, the MESTΔC deletion mutant containing amino acids 1–286 did not interact with STAT3 and lead to STAT3 deactivation. In addition, MEST regulates JAK2/STAT3 complex formation through the interaction of MEST and STAT3. Moreover, high MEST expression promotes the metastasis of breast cancer in vivo.
Overall, we first identified a new molecular and functional network present in cancer metastasis that regulates and coordinates MEST. These results suggest that MEST seems to represent a new potential target for improving the treatment efficacy of metastatic cancer and the development of novel anticancer therapeutics.
Methods
Cell lines and cell culture
Mouse (67NR, 4T07, and 4T1) and human breast cancer cells (MCF10A, MCF7, T47D, BT474, ZR75B, MDA-MB-468, SK-BR-3, BT549, Hs578T, and MDA-MB-231) and 293T cells obtained from American Type Culture Collection (Manassas, VA, USA); SUM149PT and SUM159PT breast cells were obtained from Dr. Stephen Ethier (Kramanos Institute) and were maintained as described previously [34, 35]. HMLE were obtained from Dr. Robert A. Weinberg (Massachusetts Institute of Technology) and maintained as described previously [24]. Human IL-6 were purchased from PeproTech.
Human breast tumor samples
RNA from normal and tumorous human breast samples was obtained from the Gangnam Severance Hospital after approval (IRB approval number: 3-2011-0191) as previously described [36].
Plasmids and viral production
pLKO.1 lentiviral plasmids encoding shRNAs targeting the MEST gene were obtained from Sigma-Aldrich (SHCLNG-NM_002402-human and SHCLNG-NM_008590-mouse). Hs578T, SUM159PT, and 4T1 cells were selected for 15 days with 1 mg/ml puromycin after infection with lentivirus as previously described [36]. The cDNA encoding human MEST was subcloned into pEF6/V5-His-TOPO and plenti6.3/V5-TOPO vectors, respectively (Invitrogen). The cDNA encoding MESTΔC was produced via amplification of residues 1–286 of MEST using primers (Table S1). STAT3 deletion constructs [37] were provided by Dr. Kong Y-Y. Also, Plasmid STAT3CA and STAT3YF [38] were provided from Dr. Hong S, and Dr. Song MR.
Chemical, antibodies, western blotting, immunoprecipitation, and immunofluorescence
We performed western blotting, immunoprecipitation, and immunofluorescence analysis as previously described [14]. Antibodies were obtained from the following sources: anti-MEST (SAB2501254) and anti-Flag (F3165) were from Sigma-Aldrich; anti-V5 (R960-CUS) was from Invitrogen; anti-JAK2 (ab108596), anti-Twist-1 (ab50887), anti-phosphoserine/threonine (ab17464), and anti-phosphotyrosine (ab10321) were from Abcam; anti-STAT3 (9139), anti-phospho-STAT3 (4113), and anti-phospho-JAK2 (3771) were from Cell Signaling Technology; and anti-E-cadherin (610405), anti-fibronectin (610078), anti-N-cadherin (610920), anti-alpha-catenin (610194), anti-vimentin (550513) and anti-beta-catenin (610154) were from BD PhamingenTM. JAK2 activity inhibitor (AG490), and STAT3 inhibitor (S32-201) were from Santa Cruz biotechnology.
Luciferase reporter assay
Cells that were 50% confluent in 12-well dishes were transfected using Lipofectamine 2000 (Invitrogen). A total of 0.5 μg STAT3 or Twist-1 reporter gene constructs and 0.5 μg of pCMV-β-gal were co-transfected per well. The cell extracts were prepared 48 h after transfection, and the luciferase activity was quantified using the Enhanced Luciferase Assay Kit (BD Biosciences). All experiments were performed in triplicate.
Anoikis assay, soft agar assays, wound healing assays, anchorage-independent cell growth, RT-PCR, and Matrigel invasion assays
All the assays were performed as previously described [14, 36]. For anchorage-independent cell growth and soft agar assays, 1 × 103 cells were seeded into 6 well cell culture plates. For the wound healing assay, 1 × 106 cells were seeded into 6 well cell culture plates. For the invasion assay, 1 × 104 cells were seeded into BD Matrigel invasion chambers with 8 μm pores (Corning, 62405–744). For anoikis assay, 4T1 cells infected with recombinant lentiviruses carrying either an empty vector or an MEST-shRNA were seeded into an Ultra Low Cluster plate (Corning Life Sciences, Acton, MA) at 1 × 10 5 cells per well in a six-well plate and photographed at 7 days. In addition, the primer sequences used to amplify the genes are listed in the supplemental experimental procedures (Table S1).
RNA preparation and quantitative RT-PCR
Total RNA was isolated using RNeasy Mini Kits (Qiagen) according to the manufacturer’s instructions and reverse transcribed with hexa-nucleotide mix (Roche). The resulting cDNA was subjected to PCR using the SYBR-Green Master PCR mix and the Taqman master PCR mix (Applied Biosystems) in triplicate. PCR and data collection were performed using the 7900HT Fast Real-Time PCR System (Applied Biosystems). All quantitations were normalized to the 18S RNA endogenous control. Specific Human MEST (Hs00853380_g1), Mouse MEST (Mm00485003_m1), mouse 18S (Mm04277571_S1), and human 18S (Hs99999901_s1) quantitative probes for Taqman RT-PCR were obtained from Applied Biosystems.
Animal studies
Animal studies were performed as previously described [36]. For extravasation studies, 4T1 control-shRNA or MEST-shRNA cells (1 × 106 cells) were injected into the tail vein of female BALB/c mice (7 weeks old, nic7) and handled in compliance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) of Gachon University (Approval No. LCDI-2010-0074). After 29 days, the lungs were excised, fixed in 10% formalin, paraffin embedded, and sectioned for analysis.
Microarray data analysis
MEST expression signatures in 2136 breast cancer patients of the METABRIC dataset [9] and 855 breast cancer patients of the UNC dataset (GSE26338) [10] were extracted and averaged, and then in silico analysis was performed. The boxplot graphs were plotted with gene expression using GraphPad Prism v 5.0 (GraphPad Software, Inc.). P < 0.0001 was considered statistically significant.
Statistical analysis
Data are expressed as the means ± SEM. Statistical analyses of the data were conducted via the Student’s t-test (two-tailed) and ANOVA. Differences were considered statistically significant at P < 0.05, P < 0.001, or P < 0.0001.
Multivariate statistics was performed using OPLS analysis using SIMCA 14 (Umetrics AB, Umea, Sweden) to examine the correlation between clinical parameters and MEST expression levels. Nine clinical parameters were assigned to x-variable (stage, subtype, survival time, grade, survival status, triple-negative breast type, tumor size, lymph node positivity, and cellularity), whereas MEST was assigned to y-variable. Variable importance in projection (VIP) analysis was conducted by the OPLS model, which presented the importance of variables (clinical variables) in association with MEST expression (VIP > 1). The OPLS coefficients were computed as regression model for y-variable against scaled and centered x-variable. The quantity of the coefficient represented the change level and direction of the y-variable with the changes of the X-variables from 0 to 1. Each Y variable, and the coefficients for the first Y variable are displayed above. By default, the regression coefficients relate to the centered and scaled data and are computed from all extracted components. These coefficients are not independent (as in an MLR model), as the X variables are not independent. The coefficients are displayed with a confidence interval computed by jack-knifing.
Supplementary information
Acknowledgements
This work was supported by a National Research Foundation of Korea grant (NRF-2012R1A2A2A01002728 to WJ and 2015R1D1A1A01059406, 2018R1C1B6008372 to MSK). This work was supported by the Cooperative Research Program for Agriculture Science & Technology Development (Project no. PJ0132772019 to WJ), Rural Development Administration. This research was a part of the project entitled ‘Development of Biomedical materials based on marine proteins’, funded by the Ministry of Oceans and Fisheries, Korea.
Author contributions
MSK, SKK, and WJ designed research; MSK, HSL, YJK, and DYL performed research; MSK, SKK, and WJ analyzed data; and SKK and WJ wrote the main manuscript text.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by S. Fulda
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Sung Gyun Kang, Email: sgkang@kiost.ac.kr.
Wook Jin, Email: jinwo@gachon.ac.kr.
Supplementary information
The online version of this article (10.1038/s41418-019-0322-9) contains supplementary material, which is available to authorized users.
References
- 1.Mayer W, Hemberger M, Frank HG, Grummer R, Winterhager E, Kaufmann P, et al. Expression of the imprinted genes MEST/Mest in human and murine placenta suggests a role in angiogenesis. Dev Dyn. 2000;217:1–10. doi: 10.1002/(SICI)1097-0177(200001)217:1<1::AID-DVDY1>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 2.Lefebvre L, Viville S, Barton SC, Ishino F, Surani MA. Genomic structure and parent-of-origin-specific methylation of Peg1. Hum Mol Genet. 1997;6:1907–15. doi: 10.1093/hmg/6.11.1907. [DOI] [PubMed] [Google Scholar]
- 3.Kosaki K, Kosaki R, Craigen WJ, Matsuo N. Isoform-specific imprinting of the human PEG1/MEST gene. Am J Hum Genet. 2000;66:309–12. doi: 10.1086/302712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pedersen IS, Dervan PA, Broderick D, Harrison M, Miller N, Delany E, et al. Frequent loss of imprinting of PEG1/MEST in invasive breast cancer. Cancer Res. 1999;59:5449–51. [PubMed] [Google Scholar]
- 5.Nakanishi H, Suda T, Katoh M, Watanabe A, Igishi T, Kodani M, et al. Loss of imprinting of PEG1/MEST in lung cancer cell lines. Oncol Rep. 2004;12:1273–8. [PubMed] [Google Scholar]
- 6.Moon YS, Park SK, Kim HT, Lee TS, Kim JH, Choi YS. Imprinting and expression status of isoforms 1 and 2 of PEG1/MEST gene in uterine leiomyoma. Gynecol Obstet Invest. 2010;70:120–5. doi: 10.1159/000301555. [DOI] [PubMed] [Google Scholar]
- 7.Nishihara S, Hayashida T, Mitsuya K, Schulz TC, Ikeguchi M, Kaibara N, et al. Multipoint imprinting analysis in sporadic colorectal cancers with and without microsatellite instability. Int J Oncol. 2000;17:317–22. doi: 10.3892/ijo.17.2.317. [DOI] [PubMed] [Google Scholar]
- 8.Jin W, Yun C, Kwak MK, Kim TA, Kim SJ. TrkC binds to the type II TGF-beta receptor to suppress TGF-beta signaling. Oncogene. 2007;26:7684–91. doi: 10.1038/sj.onc.1210571. [DOI] [PubMed] [Google Scholar]
- 9.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–52. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harrell JC, Prat A, Parker JS, Fan C, He X, Carey L, et al. Genomic analysis identifies unique signatures predictive of brain, lung, and liver relapse. Breast Cancer Res Treat. 2012;132:523–35. doi: 10.1007/s10549-011-1619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147:275–92. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 13.Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003;3:453–8. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 14.Jin W, Kim GM, Kim MS, Lim MH, Yun C, Jeong J, et al. TrkC plays an essential role in breast tumor growth and metastasis. Carcinogenesis. 2010;31:1939–47. doi: 10.1093/carcin/bgq180. [DOI] [PubMed] [Google Scholar]
- 15.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–15. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng GZ, Zhang WZ, Sun M, Wang Q, Coppola D, Mansour M, et al. Twist is transcriptionally induced by activation of STAT3 and mediates STAT3 oncogenic function. J Biol Chem. 2008;283:14665–73. doi: 10.1074/jbc.M707429200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim MS, Kim GM, Choi YJ, Kim HJ, Kim YJ, Jin W. TrkC promotes survival and growth of leukemia cells through Akt-mTOR-dependent up-regulation of PLK-1 and Twist-1. Mol Cells. 2013;36:177–84. doi: 10.1007/s10059-013-0061-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma XJ, Salunga R, Tuggle JT, Gaudet J, Enright E, McQuary P, et al. Gene expression profiles of human breast cancer progression. Proc Natl Acad Sci USA. 2003;100:5974–9. doi: 10.1073/pnas.0931261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartwell KA, Muir B, Reinhardt F, Carpenter AE, Sgroi DC, Weinberg RA. The Spemann organizer gene, Goosecoid, promotes tumor metastasis. Proc Natl Acad Sci USA. 2006;103:18969–74. doi: 10.1073/pnas.0608636103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim MS, Kim GM, Choi YJ, Kim HJ, Kim YJ, Jin W. c-Src activation through a TrkA and c-Src interaction is essential for cell proliferation and hematological malignancies. Biochem Biophys Res Commun. 2013;441:431–7. doi: 10.1016/j.bbrc.2013.10.082. [DOI] [PubMed] [Google Scholar]
- 21.Mani SA, Yang J, Brooks M, Schwaninger G, Zhou A, Miura N, et al. Mesenchyme Forkhead 1 (FOXC2) plays a key role in metastasis and is associated with aggressive basal-like breast cancers. Proc Natl Acad Sci USA. 2007;104:10069–74. doi: 10.1073/pnas.0703900104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hennessy BT, Gonzalez-Angulo AM, Stemke-Hale K, Gilcrease MZ, Krishnamurthy S, Lee JS, et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res. 2009;69:4116–24. doi: 10.1158/0008-5472.CAN-08-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taube JH, Herschkowitz JI, Komurov K, Zhou AY, Gupta S, Yang J, et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci USA. 2010;107:15449–54. doi: 10.1073/pnas.1004900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scheel C, Eaton EN, Li SH, Chaffer CL, Reinhardt F, Kah KJ, et al. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell. 2011;145:926–40. doi: 10.1016/j.cell.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu S, Wicha MS. Targeting breast cancer stem cells. J Clin Oncol. 2010;28:4006–12. doi: 10.1200/JCO.2009.27.5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosen JM, Jordan CT. The increasing complexity of the cancer stem cell paradigm. Science. 2009;324:1670–3. doi: 10.1126/science.1171837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frank NY, Schatton T, Frank MH. The therapeutic promise of the cancer stem cell concept. J Clin Invest. 2010;120:41–50. doi: 10.1172/JCI41004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci USA. 2009;106:13820–5. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X, Lewis MT, Huang J, Gutierrez C, Osborne CK, Wu MF, et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst. 2008;100:672–9. doi: 10.1093/jnci/djn123. [DOI] [PubMed] [Google Scholar]
- 30.Marotta LL, Almendro V, Marusyk A, Shipitsin M, Schemme J, Walker SR, et al. The JAK2/STAT3 signaling pathway is required for growth of CD44(+)CD24(−) stem cell-like breast cancer cells in human tumors. J Clin Invest. 2011;121:2723–35. doi: 10.1172/JCI44745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–62. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 32.Buettner R, Mora LB, Jove R. Activated STAT signaling in human tumors provides novel molecular targets for therapeutic intervention. Clin Cancer Res. 2002;8:945–54. [PubMed] [Google Scholar]
- 33.Haura EB, Turkson J, Jove R. Mechanisms of disease: Insights into the emerging role of signal transducers and activators of transcription in cancer. Nat Clin Pract Oncol. 2005;2:315–24. doi: 10.1038/ncponc0195. [DOI] [PubMed] [Google Scholar]
- 34.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–39. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 35.Fillmore CM, Kuperwasser C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. 2008;10:R25. doi: 10.1186/bcr1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim MS, Lee WS, Jeong J, Kim SJ, Jin W. Induction of metastatic potential by TrkB via activation of IL6/JAK2/STAT3 and PI3K/AKT signaling in breast cancer. Oncotarget. 2015;6:40158–71. doi: 10.18632/oncotarget.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kwon MC, Koo BK, Moon JS, Kim YY, Park KC, Kim NS, et al. Crif1 is a novel transcriptional coactivator of STAT3. EMBO J. 2008;27:642–53. doi: 10.1038/sj.emboj.7601986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hong S, Song MR. STAT3 but not STAT1 is required for astrocyte differentiation. PLoS ONE. 2014;9:e86851. doi: 10.1371/journal.pone.0086851. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.