Abstract
It remains largely unknown how Zika virus (ZIKV) infection causes severe microcephaly in human newborns. We examined an Asian lineage ZIKV, SZ01, which similarly infected and demonstrated comparable growth arrest and apoptotic pathological changes in human neuroprogenitors (NPCs) from forebrain dorsal, forebrain ventral as well as hindbrain and spinal cord brain organoids derived from human pluripotent stem cells. Transcriptome profiling showed common overactivated antiviral response in all regional NPCs upon ZIKV infection. ZIKV infection directly activated a subset of IFN-stimulated genes (ISGs) in human NPCs, which depended on the presence of IRF3 and NF-κB rather than IFN production and secretion, highlighting a key role of IFN-independent acute antiviral pathway underlying ZIKV infection-caused neuropathy. Our findings therefore reveal that overactivated antiviral response is detrimental rather than protective in human NPCs, and the IFN-independent acute antiviral pathway may serve as a potential target to ameliorate ZIKV infection-triggered neuropathy.
Subject terms: Neuroscience, Cell biology
Introduction
Zika virus (ZIKV), an enveloped, single-stranded RNA virus of the flavivirus family, has re-emerged and spread throughout many countries recently [1]. Microcephaly is the most devastating birth defect associated with ZIKV-infected newborns. Epidemiological statistics reported a 20-fold increase in the number of newborn microcephaly cases during the outbreak of ZIKV in Brazil in 2015 [2]. Clinical case-control studies have also established solid connections between maternal ZIKV infection in the first trimester of pregnancy and microcephaly [2, 3]. The neurotropism of ZIKV and the microcephaly phenotype caused by ZIKV infection have also been demonstrated in mouse models [4–8] and recently in rhesus monkeys [9].
The innate immune system defends the host from viral infections through eliciting type I interferons (IFNs), which exert autocrine or paracrine functions to induce hundreds of IFN-stimulated genes (ISGs) [10–12]. Infection of RNA viruses or their production of RNA intermediates can also directly activate a subset of ISGs independent of type I IFN signaling through intrinsic expression or activation of IFN-regulatory factor 3 (IRF3) or nuclear factor-κB signaling pathway (NF-κB) [12–14]. The induction of ISGs through both IFN-dependent and -independent pathways is thought to be the key for host innate immunity against viruses [14].
Though mouse models can partially resemble microcephaly caused by ZIKV infection in the fetal human brain, a human-based system is of broad interest to study the pathological processes underpinning ZIKV infection. Human embryonic stem cells (hESCs) and human induced pluripotent stem cells (hiPSCs) can be efficiently specified into neural progenitor cells (NPCs), which closely mimics human embryonic neural development [15–19]. Infection with ZIKV in these pluripotent stem cell-derived human NPCs causes severe growth arrest and cell death by apoptosis and autophagy, effectively mirroring the neurotropism of ZIKV and characteristic pathological microcephaly [8, 20–23].
It is currently unknown whether ZIKV infects human NPCs with different regional identities equally or if bias exists. How ZIKV causes cell growth arrest and apoptosis in human NPCs also remains elusive. Here, we show that an Asian lineage, ZIKV strain SZ01, isolated from a patient infected in Samoa in early 2016 [24], efficiently infects the human forebrain dorsal (FD), forebrain ventral (FV), as well as hindbrain and spinal cord (H&S) NPCs derived from hESCs. The ZIKV infection leads to reduced cell viability and proliferation rate, elevated apoptosis, and triggers robust ISG activation. Unexpectedly, the acutely activated ISGs is extremely detrimental for the normal proliferation of human NPCs, which might account for the microcephaly pathology upon ZIKV infection.
Results
ZIKV efficiently infects hESC-derived NPCs of different regional identities
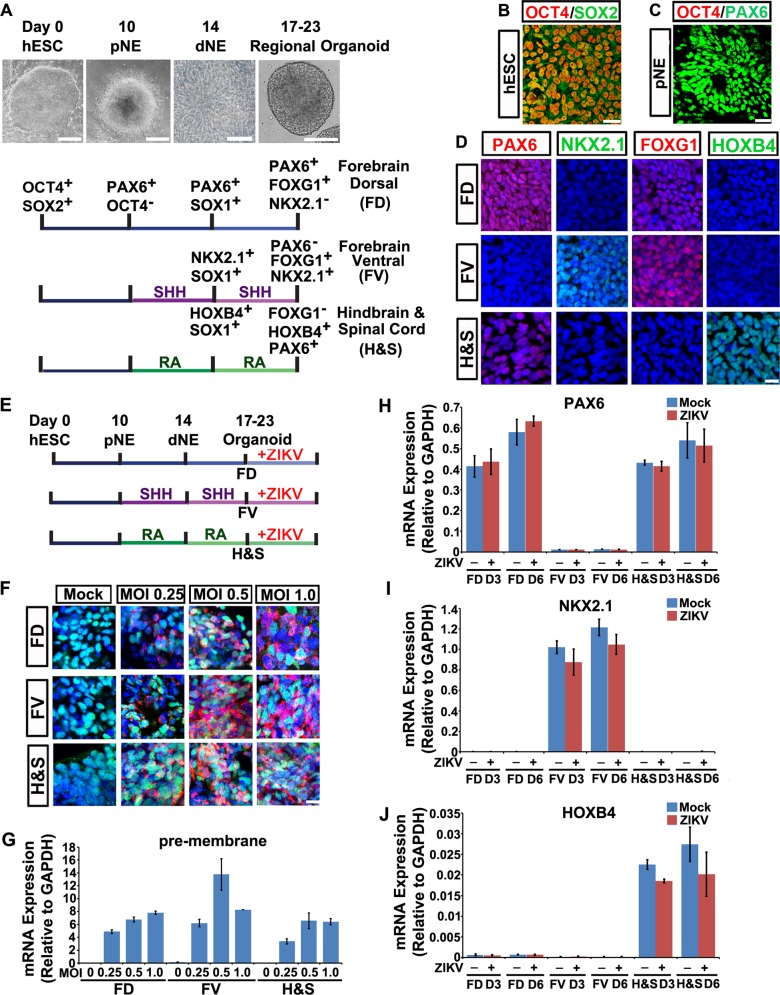
We and others established a sophisticated procedure to specify hESCs to primitive neuroectoderm cells (pNE), definitive neuroepithelial cells (dNE), and NPCs with committed regional identities formed organoid structures in the absence or presence of patterning morphogens (Fig. 1a) [16, 25–27]. At days 14–17 of neural differentiation under chemically defined conditions, hESC-derived dNE cells presented typical columnar neuroepithelial cell morphology and organized into neural tube-like rosettes (Fig. 1a). The hESCs uniformly expressed Oct4 and Sox2, hallmark genes of pluripotency (Fig. 1b). At the pNE stage, cells lost Oct4 and began to express Pax6 (Fig. 1c) [25]. Without patterning morphogens, NPCs within the organoid adopted an FD identity, which exclusively expressed FoxG1 and Pax6 (Fig. 1a, d, h–J). Sonic hedgehog (Shh) and retinoid acid (RA) are two potent patterning morphogens that ventralize and caudalize pNE along the dorsoventral and anteroposterior axes, respectively. The yielded FV NPCs patterned through Shh expressed FoxG1 and Nkx2.1, whereas the H&S NPCs patterned through RA expressed HoxB4 and Pax6 (Fig. 1a, d, h–j) [16, 28]. Confocal images further showed that brain organoids at day 23 of FD, FV and H&S regional identities mostly composed of Sox1-expressing NPCs, while the population of differentiated neurons were less than 0.05% as revealed by a small fraction of Map2, Tuj1 and NeuN immune-labeled cells at the periphery of the organoids (Fig. S1A).
Fig. 1.
ZIKV efficiently infects hESC-derived brain organoids of different regional identities. a Schematic representation of hESCs patterning to regional brain organoids in vitro. Without patterning morphogens, human ESCs differentiated into forebrain dorsal (FD) NPCs at day 17. When Shh was applied from day 10 to day 17, hESCs were patterned into forebrain ventral (FV) NPCs. To generate hindbrain & spinal cord (H&S) NPCs, retinoid acid (RA) was added from day 10 to day 17. Light images show typical hESC clone at D0, columnar pNE at D10 (Scale bar, 200 μm), organized neurotube-like rosette of dNE at D14 (Scale bar, 25 μm) and regional organoid at D20 (Scale bar, 100 μm). b Confocal image showing undifferentiated hESCs were Oct4+/Sox2+. Scale bar, 50 μm. c Confocal image showing day 10 pNE cells were Pax6+/Oct4−. Scale bar, 50 μm. d FD, FV and H&S organoids were dissociated and plated onto coverslips for immunostaining. FD NPCs were Pax6+/Nkx2.1−/Foxg1+. FV NPCs were Pax6−/Nkx2.1+/Foxg1+. H&S NPCs were Foxg1−/Hoxb4+/Pax6+. Scale bar, 50 μm. e Schematic representation of ZIKV infection of FD, FV and H&S organoids from day 17 to day 23. f Confocal images stained for viral envelope proteins (red), Sox2 (green) and nuclei (blue) of ZIKV-infected FD, FV, H&S NPCs at different viral MOI (0.25, 0.5, 1.0) or mock control at 3 dpi. Sox2 is a hallmark protein expressed in NPCs. Scale bar, 50 μm. g qRT-PCR analyses of ZIKV pre-membrane mRNAs relative to GAPDH extracted from FD, FV, H&S NPCs infected with ZIKV at different MOI (0.25, 0.5, 1.0) or mock control at 2 dpi. Quantification data are presented as mean ± SEM. n = 3. h–j qRT-PCR analyses of regional marker genes Pax6, Nkx2.1 and Hoxb4 mRNAs relative to GAPDH extracted from FD, FV, H&S NPCs infected with ZIKV at 0.5 MOI or mock control at 3 or 6 dpi. Quantification data are presented as mean ± SEM. n = 3
It is unclear whether human NPCs with various regional identities are equally vulnerable to ZIKV infection. We then exposed FD, FV and H&S organoids to ZIKV SZ01 from day 17 to day 23 (Fig. 1e). Immunostaining with an antiserum that recognized ZIKV envelope protein (ZIKV E) confirmed abundant viral envelope proteins clustered round the nucleus in all three organoids, especially at multiplicity of infection (MOI) = 0.5 and 1.0 (Fig. 1f). Quantitative analysis showed increased transcription of viral pre-membrane RNA (Fig. 1g), envelope RNA, and NS5 RNA (Data not shown) in the three regional organoids, which plateaued at a MOI =0.5–1. Western blot showed ZIKV E protein increased with similar patterns to ZIKV RNAs (Fig. S1B, S1C). Infection of three regional organoids with ZIKV did not affect the regional identities of all three NPCs (Fig. 1h–j). Thus, ZIKV showed similar infection sensitivities in human NPCs with all regional identities.
ZIKV causes growth arrest and programmed cell death in all regional NPCs
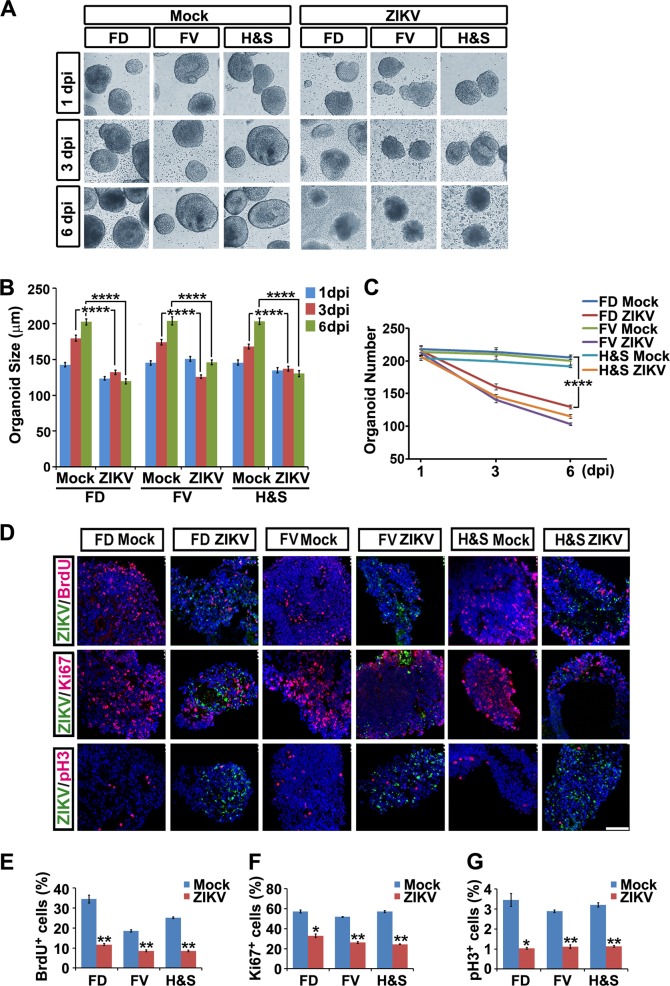
We photo recorded FD, FV, and H&S organoids at 1, 3, and 6 days post infection (dpi) of ZIKV at an MOI = 0.5. Mock-infected organoids presented expected round and intact shape. Remarkably, ZIKV-infected organoids of all regional identities stopped growing at 3 and 6 dpi (Fig. 2a–c). Two hours of BrdU (50 mM, 1:5000) incorporation analysis and immune-labeling of mitotic markers Ki67 and pH3 all showed significant reduction at 3 dpi in all three regional organoids (Fig. 2d–g). These results indicate that ZIKV infection leads to cell growth arrest and phenocopies microcephaly in all spectra of the regional NPCs.
Fig. 2.
ZIKV abrogates growth of hESC-derived brain organoids of different regional identities. a Light images of FD, FV, H&S organoids exposed to mock conditions or ZIKV at MOI =0.5 at 1, 3 or 6 dpi. b Size of FD, FV and H&S organoids exposed to mock or ZIKV at MOI = 0.5 at 1, 3 or 6 dpi. Quantification data of organoids diameters are presented as mean ± SEM. n = 200 from three independent experiments. Unpaired two-tailed Student’s t-test. ****p < 0.0001 compared to mock controls. c Numbers of FD, FV and H&S organoids remaining after mock or ZIKV infection at MOI = 0.5 at 1, 3 or 6 dpi. Quantification data are presented as mean ± SEM. n = 3. Unpaired two-tailed Student’s t-test. ****p < 0.0001 compared to mock controls. d Confocal images stained with ZIKV envelope proteins in green, or BrdU, Ki67 and pH3 in red of mock or ZIKV-infected FD, FV, H&S organoids. Scale bar, 100 μm. e–g Quantitative analyses of percentage of BrdU, Ki67 and pH3 positive cells in mock or ZIKV groups. Data are presented as mean ± SEM. n = 3. Unpaired two-tailed Student’s t-test. *p < 0.05, **p < 0.01 compared to corresponding mock controls
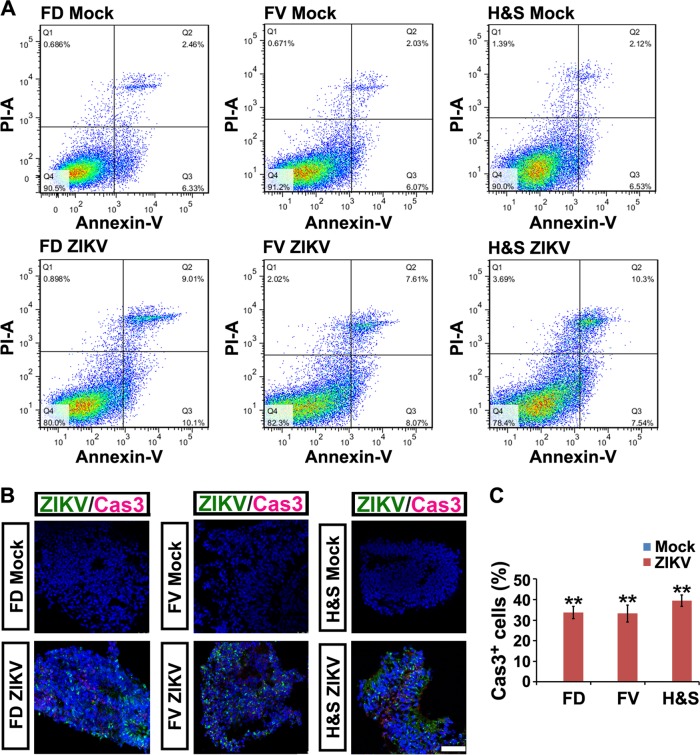
ZIKV infection induced extensive cell death, as observed by the irregular organoid morphology and cell debris detached from the organoids in all three groups (Fig. 2a). The numbers of organoids were also remarkably reduced at 3 and 6 dpi with an MOI = 0.5 (Fig. 2c). Fluorescein isothiocyanate-conjugated annexin V and propidium iodide were used for assessing early and late apoptotic cells with MOI = 0.006 to avoid immediate cell death at higher MOI induced by ZIKV. The percentage of apoptotic NPCs increased 3- to 4-fold at 6 dpi in all three groups (Fig. 3a). This was further confirmed by immunostaining of cleaved caspase 3, a marker of programmed cell death (Fig. 3b, c).
Fig. 3.
ZIKV induces programmed cell death of hESC-derived NPCs. a Annexin V-FITC/PI staining followed by flow cytometry analyses of cell apoptosis in mock and ZIKV groups (MOI=0.006 for 6 days). Q1, dead cells and cell debris. Q2, late apoptotic cells. Q3, early apoptotic cells. Q4, living cells. b Confocal images stained with ZIKV envelope proteins in green and cleaved Caspase 3 in red of mock or ZIKV-infected FD, FV and H&S organoids at 3 dpi. Scale bar, 100 μm. c Percentage of cleaved Caspase 3 positive cells in mock and ZIKV-infected groups. Data are presented as mean ± SEM. n = 3. Unpaired two-tailed Student’s t-test. **p<0.01 compared to mock controls
ZIKV infection strongly induces antiviral immune responses in human NPCs
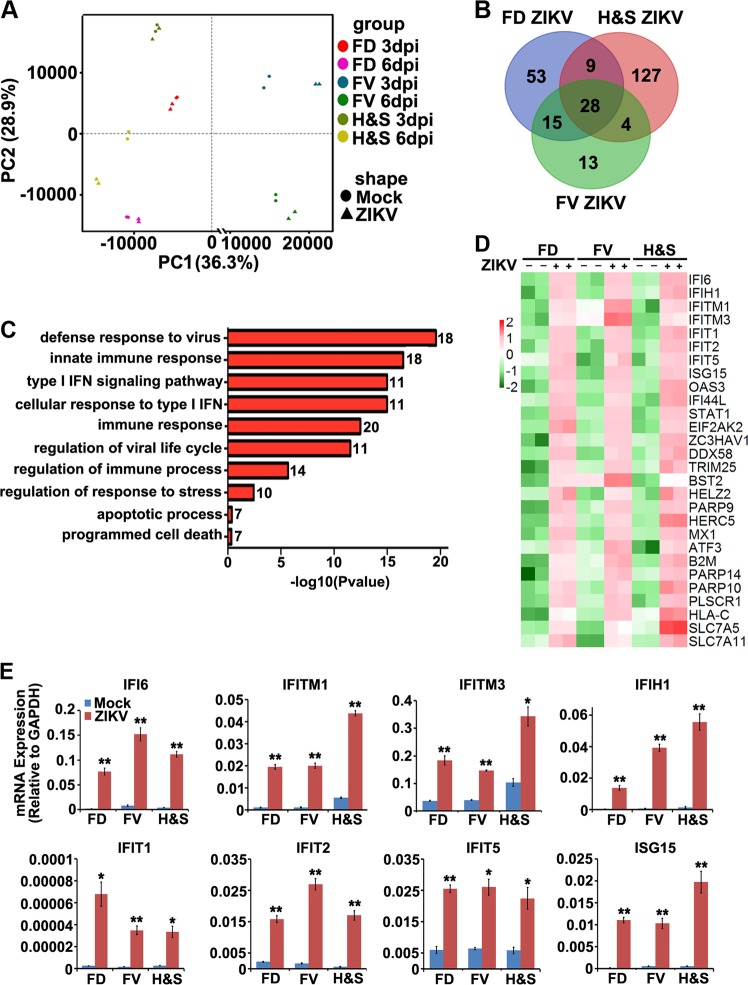
To investigate the cellular pathologies in human NPCs after ZIKV infection, we performed global transcriptome profiling of the FD, FV, and H&S organoids at 3 and 6 dpi. Principal component analysis (PCA) plots showed that samples were primarily clustered by their regional identities and differentiation stages; suggesting ZIKV infection did not significantly affect neural developmental programs. ZIKV infection caused mild, but consistent gene expression changes, which were mostly upregulated and became predominant at 6 dpi (Fig. 4a). We found 105 upregulated genes in the FD NPCs, 60 genes in the FV NPCs, and 168 genes in the H&S NPCs, with at least 1.5-fold differences found between ZIKV-infected groups and the control groups at 6 dpi. Among them, 28 genes overlapped in all three regional NPCs (Fig. 4b). Gene Ontology (GO) analyses of these 28 common genes exclusively referred to the pathways associated with immune responses to viral infection, especially the type I IFN signaling pathway (Fig. 4c). Heatmap analyses further indicated that these 28 genes were strongly upregulated in all three groups (Fig. 4d). While these 28 genes could be clustered to the ISG family [14], we did not identify IFN mRNA induction in all three groups under ZIKV infection (Fig. S2A), nor the type I IFN, indicating the acute antiviral response is mediated by the IFN-independent ISG activation pathway. ZIKV infection-induced ISG activation was also validated by quantitative RT-PCR analyses in all three regional organoids (Fig. 4e). Similar results were observed in a mouse study by infection of ZIKV in the fetal mouse brain at E13.5–16.5 (Fig. S2B and S2C) [4].
Fig. 4.
ZIKV infection induces antiviral immune response through activation of ISGs. a Principal component analysis of global RNA sequencing from mock (round) and MOI = 0.5 ZIKV-infected (triangle) FD, FV, H&S organoids at 3 dpi or 6 dpi. b Upregulated genes in ZIKV-infected organoids at 6 dpi compared to mock groups. 28 genes were significantly upregulated in all three ZIKV-treated regional organoids. c List of Gene Ontology (GO) classification of the common 28 upregulated genes and the corresponding number. d Heatmaps of 28 upregulated genes in ZIKV-infected FD, FV and H&S organoids. The RPKM values were log2-transformed and row-scaled. e qRT-PCR analysis of mRNA expression of 8 ISGs of MOI = 0.5 ZIKV infected vs mock infected FD, FV and H&S organoids at 3 dpi. Data are presented as mean ± SEM. n = 3. Unpaired two-tailed Student’s t-test. *p < 0.05, **p < 0.01 compared to mock controls
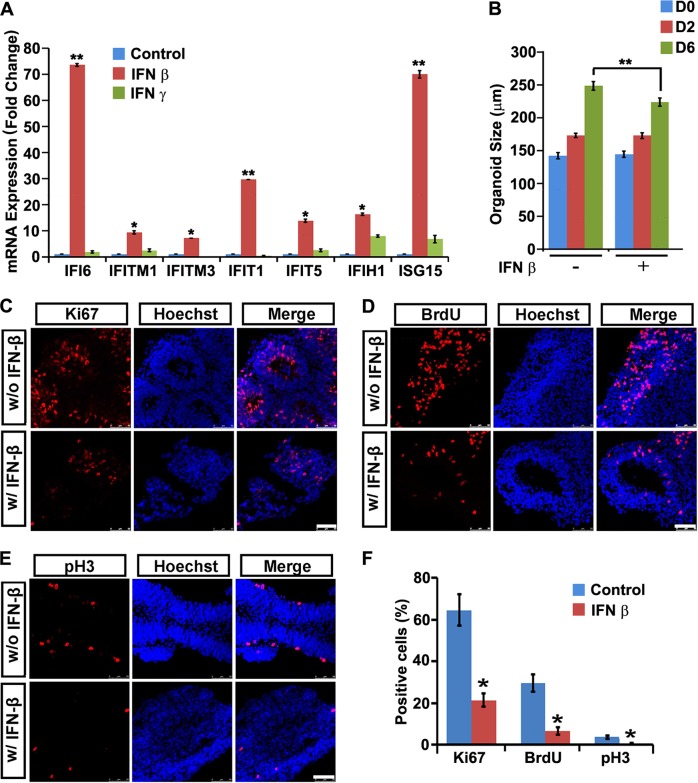
Activation of ISGs through IFNβ administration causes growth arrest in human NPCs
All three regional NPCs expressed abundant type I and type II IFN receptors (Fig. S2A). The FD, FV, and H&S organoids were then treated with IFNβ or IFNγ for 6 d. Quantitative RT-PCR analyses revealed IFNβ, but not IFNγ administration, induced the expression of those key ISGs efficiently (Fig. 5a, S3A, S3B). Strikingly, IFNβ treatment for 6 d hindered the expansion of the FD, FV, and H&S NPCs, as demonstrated by the reduced organoids diameters (Fig. 5b, S3C). BrdU incorporation studies and immune-labeling of Ki67 and pH3 showed a dramatic reduction in positively labeled cells in all three regional organoids after IFNβ treatment (Fig. 5c–f, S3D, S3E). FACS analysis for apoptotic cells of regional organoids with IFNβ administration showed that the percentage of apoptotic NPCs remained unchanged upon IFNβ administration (Fig. S4A,S4B). Thus, IFNβ could not trigger NPCs apoptosis, but dampen NPCs growth. These data indicate that overactivated ISG pathways might be detrimental for human NPCs of various regional identities.
Fig. 5.
Activation of ISGs by IFNβ administration causes NPC growth arrest. a qRT-PCR analysis of ISG mRNA expression of FD organoids treated with 40 ng/ml IFNβ or IFNγ or untreated control for 6 days. Data are presented as mean ± SEM. n=3. Unpaired two-tailed Student’s t-test. *p < 0.05, **p < 0.01 compare to untreated controls. b Size of FD organoids treated with or without 40 ng/ml IFNβ for 0, 2 and 6 days. Data are presented as mean ± SEM. n = 200 from three independent experiments. Unpaired two-tailed Student’s t-test. **p < 0.01 compared to the untreated controls. c–e Confocal images stained with Ki67, BrdU or pH3 in red and Hoechst nuclei staining in blue of untreated or 40 ng/ml IFNβ−treated FD organoids for 3 days. Scale bar, 100 μm. f Percentage of Ki67, BrdU and pH3 positive cells. Data are presented as mean ± SEM. n = 3. Unpaired two-tailed Student’s t-test. *p < 0.05 compared to untreated controls
Protection of ZIKV infection-induced neuropathy by abrogating ISG induction
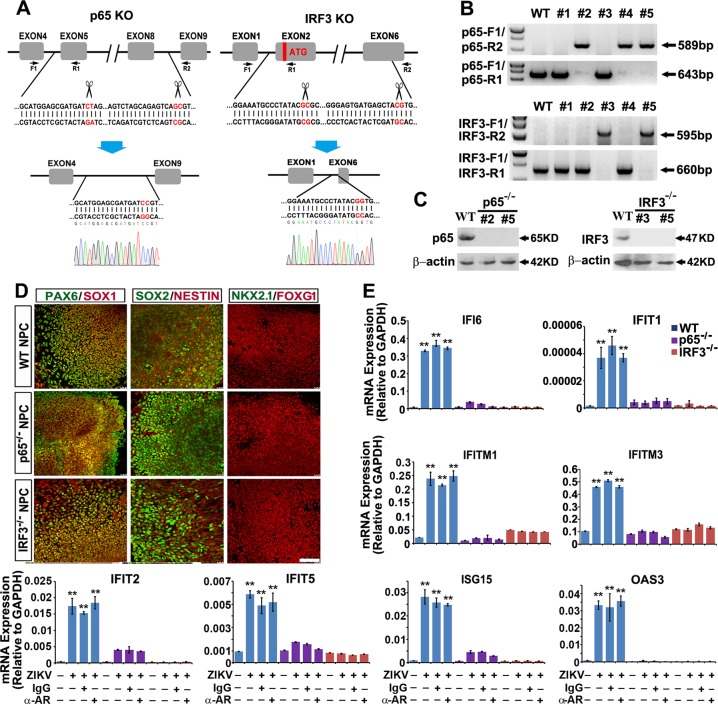
Infection of RNA viruses or their production of RNA intermediates can activate a subset of ISGs through activation of IRF3, IRF7 and NF-κB [12–14]. NPCs expressed high levels of IRF3 and NF-κB subunit p65 (RELA) [29], and basal level of IRF7 according to our profiling data (data not shown). We therefore ablated p65 and IRF3 in hESCs through donor-free paired gRNA-guided CRISPR/Cas9 knockout (paired-KO) system we established (Fig. 6a) [30]. The KO genotype was verified via genomic DNA PCR analyses (Fig. 6b). Western blotting further confirmed the KO efficacy (Fig. 6c). Ablation of p65 or IRF3 in hESCs did not interfere regular neural differentiation programs and ZIKV infection (Fig. 6d, S1D). Quantitative RT-PCR analyses revealed that ZIKV infection-triggered ISGs expression was almost completely abrogated in p65 or IRF3 KO organoids (Fig. 6e). In all ISGs tested, administration of IFNAR blocking antibody could not ameliorated ZIKV-induced ISGs activation, supporting an IFN-independent ISG induction via activation of IRF3 and NF-κB upon ZIKV infection in NPCs.
Fig. 6.
Knockout of p65 or IRF3 abrogates ISG activation upon ZIKV infection. a Schematic representation of the paired-KO strategy for NF-κB p65 or IRF3 knockout. The cleavage sites are pointed out by the scissors. Genomic DNA sequences before and after the paired-gRNA mediated cleavage and repair are shown in red. A representative Sanger sequencing peak map verifies precise re-joining of the double blunt ends. The positions of the designed primer sets for genomic PCR are shown as arrows. b Genomic DNA PCR results showing 5 colonies retrieved from p65 or IRF3 paired-KO in H9 hESCs. Clones #2, #4 and #5 are p65 KO (upper). Clones #3 and #5 are IRF3 KO (lower). c Western blot results further verify complete lack of target protein expression of clones #2, #5 of p65 KO, and #3, #5 of IRF3 KO. d Confocal images showing NPCs within FD organoids from WT, p65 KO or IRF3 KO are all Pax6+/Sox1+/Sox2+/Foxg1+/Nestin+/Nkx2.1−. Scale bar, 100 μm. e qRT-PCR analysis of ISG mRNA expression of WT, p65 KO and IRF3 KO FD organoids infected with ZIKV at MOI = 0.5 alone or with IFNAR blocking antibody (2 μg/ml, mouse IgG2a, pbi assay science), or control mouse IgG2a (2 μg/ml, mouse IgG2a, R&D) at 5 dpi as well as each mock groups. Data are presented as mean ± SEM. n = 3. Unpaired two-tailed Student’s t-test. **p < 0.01 compared to mock controls
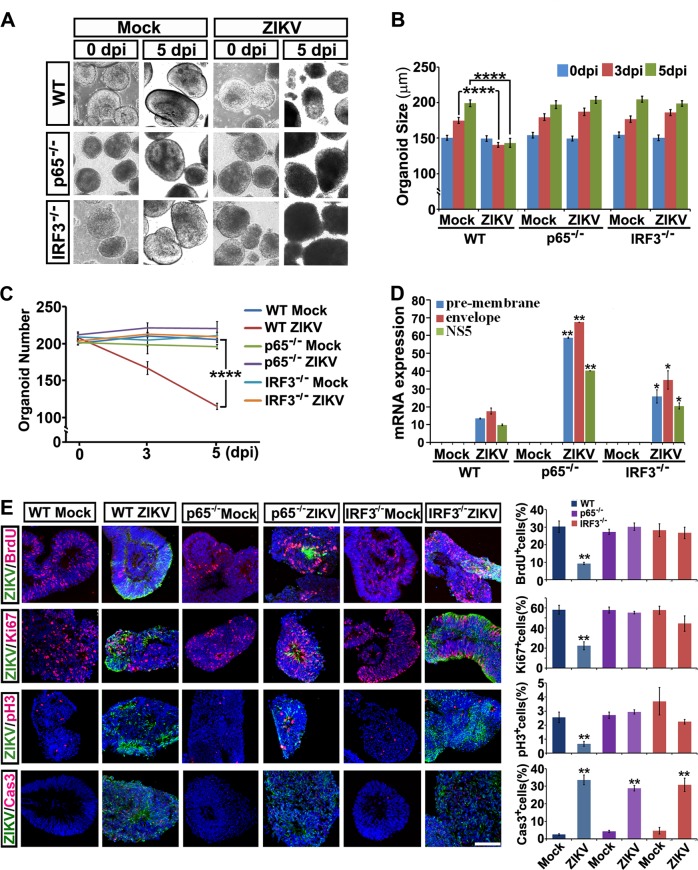
In striking contrast to the wild type (WT) control, ZIKV-infected p65 or IRF3 KO organoids showed similar growth rates as that of their uninfected groups (Fig. 7a–c). Quantitative RT-PCR analyses and Western blot showed significant increase of viral transcripts and envelope protein in p65 or IRF3 KO organoids compared to the WT control, indicating the KO cells are more sustainable for a higher viral burden (Fig. 7d and S1D). Of note, BrdU incorporation studies and immune-labeling of Ki67 and pH3 showed a significant restoration in KO organoids with ZIKV infection (Fig. 7e). These data suggest that ZIKV infection-induced growth arrest of human NPCs could be largely protected by abrogation of the IFN-independent ISG pathway. Interestingly, cleaved caspase 3 staining experiments and FACS analysis showed that the apoptosis of the ZIKV-infected NPCs were not equally protected by ablation of p65 or IRF3 (Fig. 7e, S4C, S4D), possibly because of the severer viral burden in these NPCs lack of ISG activation.
Fig. 7.
Knockout of p65 or IRF3 in hESCs protects brain organoids from ZIKV infection-induced neuropathy. a Light images of FD organoids differentiated from WT, p65 or IRF3 KO hESCs exposed to mock conditions or ZIKV at MOI = 0.5 at 0 or 5 dpi. b Size of WT, p65 or IRF3 KO FD organoids exposed to mock or ZIKV at MOI = 0.5 at 0, 3 or 5 dpi. Quantification data are presented as mean ± SEM. n = 200 from three independent experiments. Unpaired two-tailed Student’s t-test. ****p < 0.0001 compared to mock controls. c Number of WT, p65 or IRF3 KO organoids remaining with mock or ZIKV infection at MOI = 0.5 at 0, 3 or 5 dpi. Quantification data are presented as mean ± SEM. n = 3. Unpaired two-tailed Student’s t-test. ****p < 0.0001 compared to the mock control. d qRT-PCR analyses of ZIKV pre-membrane, envelope and NS5 mRNAs relative to GAPDH extracted from WT, p65 or IRF3 KO organoids infected with ZIKV at 0.5 MOI or mock control at 5 dpi. Quantification data are presented as mean ± SEM. n = 3. Unpaired two-tailed Student’s t-test. *p < 0.05, **p < 0.01 compared to the corresponding WT controls. e Confocal images stained with ZIKV envelope proteins in green, or BrdU, Ki67, pH3 and cleaved Caspase 3 in red of mock or ZIKV-infected WT, p65 or IRF3 KO organoids at 5 dpi. Scale bar, 100 μm (left). Quantitative analyses of percentage of BrdU, Ki67, pH3 and cleaved Caspase 3 positive cells of WT, p65 or IRF3 KO organoids in mock or ZIKV groups (right). Data are presented as mean ± SEM. n = 3. Unpaired two-tailed Student’s t-test. **p < 0.01 compared to corresponding mock controls
Activation of the innate immune receptor TLR3 leads to nuclear accumulation of NF-κB and IRF3 and subsequently boosts ISGs expression. To investigate the link between ZIKV-mediated overactivation of antiviral responses and TLR3 pathway [31], qRT-PCR analyses were performed to check the basal TLR3 mRNA level of WT, p65 or IRF3 KO organoids, or after stimulation with TLR3 agonist poly (I:C), or combined with TLR3 competitive inhibitor, thiophenecarboxamidopropionate compound. Compared to the positive control of PH5CH8 cells, TLR3 agonist poly(I:C) failed to robustly induce TLR3 mRNA expression in human NPCs (Fig. S5A), nor did ZIKV infection (Fig. S5B). To further test the role of TLR3 pathway in ZIKV-mediated neuropathy, WT, p65 or IRF3 KO organoids were challenged with TLR3 agonist poly (I:C) or combined with TLR3 competitive inhibitor. Activation or blockage of TLR3 signaling for 3 or 6 days showed no obvious effects in the growth rates of human NPCs (Fig. S5C). Moreover, TLR3 competitive inhibitor could not ameliorate ZIKV-mediated neuropathy of NPC organoids (Fig. S5D). Therefore, in our system, ZIKV infection-mediated microcephaly phenotype results from IFN-independent ISG overactivation rather than boosting TLR3 signaling.
Discussion
The mechanisms underlying ZIKV infection-triggered microcephaly in human newborns remain poorly understood. Our results demonstrated that ZIKV directly invaded and robustly replicated in the brain and spinal cord organoids derived from human pluripotent stem cells. Moreover, ZIKV infection in human NPCs led to cell growth arrest and apoptosis, as evidenced by reduced expression of Ki67 and pH3, lower BrdU incorporation, and increased cleavage of Caspase 3. It has been reported that severe degeneration of the lumbar spinal cord occurs in ZIKV-infected fetuses, which is in line with our conclusion that ZIKV unbiasedly infects all spectra of human FD, FV, and H&S NPCs [20–22, 32, 33].
ZIKV infection during the first trimester of pregnancy causes much severer fetal outcomes than infections occurring in later developmental stages [34–36]. Our results showed that FD, FV, and H&S regional NPCs at day 17 post-initiation of human pluripotent stem cell differentiation were similar to the regional NPCs of 5- to 10-week-old developing embryos [26]. Pathogenesis related to ZIKV infection of in vitro differentiated human NPCs at this stage will thus maximally mirror in vivo neural abnormalities, which model microcephaly in progress. Indeed, transcriptome profiling analyses revealed that ZIKV-infected FD, FV, and H&S regional NPCs had similar molecular signatures on the differentially expressed genes. These results support the effectiveness of in vitro neural differentiation of human pluripotent stem cells in yielding developmentally relevant NPCs for studying neural malformation caused by ZIKV infection.
Type I IFNs (IFN α, β, ε, κ, and ω) elicit a potent antiviral effect in cells by stimulating the IFNα/β receptors (IFNAR1 and IFNAR2) to activate Janus kinase (JAK)-signal transducers and activators of transcription (STAT) signals, leading to the upregulation of hundreds of ISGs for antagonizing viral infection and replication [37, 38]. Type II IFN (IFNγ) is released by immune cells and signals through the IFNγ receptor (IFNGR) to regulate immune responses [39, 40]. Intrauterine inoculation with ZIKV in a mouse model demonstrated that ZIKV infection of the placenta increases production of IFNβ, which, in turn, stimulates ISG expression to limit viral infection [41]. In the brain, astrocytes are also capable of producing IFNs [42]. Interestingly, ZIKV infection in human NPCs failed to induce IFNs. Similar results were observed in the mouse study that ZIKV infection showed under detectable IFN vs strongly induced ISGs (Fig. S2). These observations together with ours demonstrate that intracellular processes responsible for viral infection vary significantly among different cell types, which might account for the tissue specific pathology in ZIKV-infected individuals [41, 43–46].
As a key component of innate immune system, TLR3 mediates antiviral response including IFNβ secretion and NFκB signaling activation. By generating cerebral organoids from hESCs, Dang et al. efficiently mimicked microcephaly pathology after ZIKV infection in vitro [31]. In their study, TLR3 was upregulated after ZIKV infection and blocking TLR3 signaling ameliorated the phenotypic effects of ZIKV infection. This is to some extent in line with our current research that ZIKV infection-induced activation of innate immune responses are detrimental for neural cells. However, in our system, TLR3 agonist poly(I:C) and ZIKV infection failed to robustly induce TLR3 mRNA expression (Fig. S5). Moreover, poly(I:C) treatment for 3 or 6 days did not affect normal growth of brain organoids. TLR3 competitive inhibitor also failed to block the phenotypic effects of ZIKV infection. Therefore, in our system, ZIKV infection-mediated microcephaly phenotype results from IFN-independent ISG overactivation rather than boosting TLR3 signaling. We reason that the discrepancy may come from the different neural cell types residing within the brain organoids. We used brain organoids which comprise almost pure NPCs, while in Dang’s study, they used cerebral organoids of a later developmental stage, which are a mixture of NPCs and differentiated neurons. It will be of great interest to investigate whether NPCs, neurons and glial cells have different mechanisms in responding to ZIKV infection.
IRF3 and NFκB transcription factors have strong consensus binding sequences and Chip-seq studies reveal that their binding sites have high co-occurrence. However, IRF3 is more competent than NFκB in recruiting transcriptional machineries to the promoters of target genes, whereas NF-κB is more like an effector for transcription pause release [47]. An open question in our current study is whether IRF3 and NF-κB have equal or differential effects upon ZIKV infection. In our qRT-PCR analyses, the ZIKV pre-membrane, envelope and NS5 mRNAs under the p65 KO are higher than those under IRF3 KO conditions, suggesting a higher viral loads in p65 KO NPCs. Future studies are apparently needed to investigate whether there is a difference of virus entry, virus replication or cell survival between IRF3 and NFκB KO cells upon ZIKV infection.
In a recently published paper, Wu and others showed that stem cells express a subset of ISGs to protect stem cells and their differentiation potential during viral infection [46]. In our current study, ISG activation either by ZIKV infection or IFNβ administration interfered with normal proliferation of human NPCs. Our data thus suggest a detrimental rather than protective role of ISGs, no matter induced via the IFN-dependent or -independent pathways in early human fetal brain upon viral infection. Indeed, among the ISG family, IFIT proteins lead to the accumulation of cells at the G1-S phase transition, or bind and sequester the ribosomal protein to contribute to the antiproliferative capacity [48, 49]. ISG15 has also been reported to inhibit cancer cell growth and promote apoptosis [50].
Taken together, our current study reveals IFN-independent ISG overactivation in human NPCs upon ZIKV infection, and their detrimental role in causing cell growth arrest in human NPCs. We also demonstrate that abrogation of intrinsic ISGs activation might serve a way to mitigate ZIKV infection-induced neuropathy.
Methods
Human ESC culture
Human ESC line H9 (WA09, passages 25–45, WiCell Agreement No. 14-W0377) was cultured on a feeder layer of irradiated mouse embryonic fibroblasts (MEFs) as previously described [51]. Cells were passaged every 5 days through dispase (Gibco, 17105) digestion. The components of the human ESC culture medium (hESCM) are: DMEM/F12, 20% knockout serum replacer, 1× MEM non-essential amino acids solution, 1× l-glutamine solution, 0.1 mM β-mercaptoethanol and 4 ng/ml FGF2.
Construction of the gRNA plasmids
The blank gRNA vector with two BbsI restriction sites was described as before [52]. The gRNA vector was digested with BbsI, gel purified, and ligated to the annealed oligoes containing targeting sequences of designed gRNAs. Sequences of double gRNAs used for NF-κB p65 (RELA) KO were:
p65#1: GCATGGAGCGATGATCTAG,
p65#2: AGTCTAGCAGAGTCAGCGT.
Sequences of double gRNAs used for IRF3 KO were:
IRF3#1: GGAAATGCCCTATACGCGC,
IRF3#2: GGGAGTGATGAGCTACGTG.
Genomic DNA PCR primer sets for validation of NF-κB KO were:
p65 F1: AGTGAAGGGACAGGGTTCGTTG,
p65 R1 AATGGGCACCAAGATTCCAG,
p65 R2: CCAAATCTGCTCCTGTCACCTCT.
Genomic DNA PCR primer sets for validation of IRF3 KO were:
IFR3 F1: TATACTGGCGGAATTGAGGGAG,
IRF3 R1: AAGATTCCGAAATCCTCCTGCTGT,
IRF3 R2: TCCTGAACTTGTCATCTGCCCAC.
Generation of knockout hESC lines
hESCs were cultured in hESCM with Y-27632 (1 mM, Calbiochem, Y27632), a Rho kinase inhibitor, for 3 h prior to electroporation. Cells were then digested by trypsin for 3 min into single cells and rinsed with PBS twice. 1 × 107 cells were electroporated with appropriate combination of Cas9 plasmids (5 μg) (addgene#44719), two gRNAs (5 μg) and CAG promoter-driven puromycin plasmid (5 μg) in 200 μl of electroporation buffer using the Gene Pulser Xcell System (Bio-Rad) at 250V, 500 μF in a 0.4 cm cuvettes (Phenix Research Products). Cells were treated with puromycin (0.5 μg/ml) from day 2–5 post electroporation. After puromycin selection, MEF-conditioned hESCM was supplied, and drug-resistant colonies could be picked up for genotyping analyses after 10 days post electroporation [30].
Neural differentiation and ZIKV infection
The procedure for the neural differentiation of human pluripotent stem cells were described previously [51, 53–56]. Briefly, human ESCs were detached from the MEF layers after dispase digestion. ESC aggregates were then pipetted up and down against the bottom of a 50 ml conical centrifuge tube to break up the colonies into 100–200 μm pieces and suspended in hESCM for 4 days to form embryoid bodies (EBs). EBs were then switched to the neural induction medium (NIM) to guide the cells toward the pNE fate for 2 days. At day 6, cell aggregates were plated on laminin-coated culture surface and neural tube-like rosettes could be seen at days 14–17. For regional patterning, Shh (500 ng/ml, 130095730, Miltenyi Biotechnology) or RA (0.1 μM, 302794, Sigma) was added to the pNE cells from day 10–17 to generate FV or H&S regional NPCs, respectively [16, 53]. NPCs were maintained in suspension culture as organoids in NIM. The recipe for NIM is as follows: DMEM/F12, 1× N2 supplement, 1× MEM non-essential amino acids solution and 2 μg/ml heparin. For ZIKV infection, quantified virus were added into the organoid culture for 2 h, and then washed with fresh NIM medium. Culture medium was refreshed every 2 days.
Virus
ZIKV strain SZ01/2016 (GenBank number: KU866423) was isolated from a patient who returned from Samoa [24]. Virus stocks were prepared by inoculation onto a confluent monolayer of C6/36 mosquito cells. Virus stocks were titrated by plaque forming units (PFU) on BHK-21 cells.
Plaque assay
ZIKV quantification was completed by plaque assay on BHK-21 cell cultures. Duplicate wells were infected with 0.1 ml aliquots from serial 10-fold dilutions in growth medium and virus was adsorbed for 2 h. Following incubation, the inoculum was removed, and monolayers were overlaid with 3 ml medium containing a 1:1 mixture of 1.2% oxoid agar and 2× DMEM (Gibco, Carlsbad, CA) with 10% (vol/vol) FBS and 2% (vol/vol) penicillin/streptomycin. Cells were incubated at 37 °C in 5% CO2 for 4–6 d for plaque development. Cell monolayers then were stained with 2 ml crystal violet formaldehyde solution, and incubated overnight at room temperature before counting the number of plaques.
mRNA extraction and RT-PCR analysis
Total cellular RNA was isolated using the Trizol kit (Invitrogen) and RNA concentration was determined by NanoDrop 2000c (Thermo Scientific). 1 μg of total RNA from each sample was reverse transcribed into cDNA using SuperScript III (Invitrogen). Real-time PCR was performed (Bio-Rad, CFX Connect Real-Time System) using the Ssofast EvaGreen kit (Bio-Rad). Primer oligonucleotides used for qRT-PCR were as follows:
| Genes | Forward primers | Reverse primers |
|---|---|---|
| GAPDH | ATGACATCAAGAAGGTGGTG | CATACCAGGAAATGAGCTTG |
| IFI6 | CTGGTCTGCGATCCTGAATG | AGAGGTTCTGGGAGCTGCTG |
| IFIM1 | ACTCCGTGAAGTCTAGGGACA | TGTCACAGAGCCGAATACCAG |
| IFIM3 | GGTCTTCGCTGGACACCAT | TGTCCCTAGACTTCACGGAGTA |
| IFIT1 | GAAAGCCTCAGTCTTGCAGC | CATCACCATTTGTACAAGAGCCT |
| IFIT2 | AGCGAAGGTGTGCTTTGAGA | GAGGGTCAATGGCGTTCTGA |
| IFIT5 | CGTCCTTCGTTATGCAGCCAAG | CCCTGTAGCAAAGTCCCATCTG |
| IFIH1 | GGGGCATGGAGAATAACTCA | TGCCCATGTTGCTGTTATGT |
| ISG15 | GCCTTCAGCTCTGACACC | CGAACTCATCTTTGCCAGTACA |
| OAS3 | TCTGAGACTCACGTTTCCTGA | CACTGTTGAGGAGGGTAGAGTA |
| ZIKV prM | CGGAGATCTAGAAGACTGTGAC | ATGACTTTTTGGCTCGTTGAGC |
| ZIKV E | GATTGAAGGGCGTGTCATACTCC | TCAGTGGAACGGGGTTAGCGG |
| ZIKV NS5 | CTGGTATATGTGGCTAGGGGC | TGGTGATTAAGAGCTTCATTCTCC |
| TLR3 | TTGCCTTGTATCTACTTTTGGGG | TCAACACTGTTATGTTTGTGGGT |
Western blot
Cells were washed three times with PBS and were solubilized in 1×SDS-PAGE loading buffer at 50 oC for 20 min and separated by SDS-PAGE. Primary antibodies used were anti-ZIKV E proteins (1:5000, mouse IgG, Biofront), p65 (1:1000, rabbit IgG, Abcam), IRF3 (1:1000, rabbit IgG, Abcam) and β-actin (1:10,000, Mouse IgG, sigma). Images were developed and semi-quantified by Image J.
Immunocytochemistry and imaging
Coverslip cultures were fixed in 4% paraformaldehyde for 10 min at room temperature. After adequate washing with PBS, cells were incubated in a blocking buffer (10% donkey serum plus 0.2% Triton X-100 in PBS) for 60 min at room temperature followed by primary antibody incubation at 4 oC overnight. On the next day, coverslips were washed with PBS and stained with the fluorescently conjugated secondary antibodies (1:1000, Jackson, West Grove, PA). Nuclei were counterstained with Hoechst 33258. Coverslips were visualized with Leica TSC SP5 (Leica Microsystems, Bensheim, Germany) confocal laser-scanning microscope. Antibodies used in this study included Pax6 (1:1000, rabbit IgG, Covance), Nkx2.1 (1:400, mouse IgG, Chemicon), FoxG1 (1:1000, rabbit IgG, Abcam), Sox1 (1:500, goat IgG, R&D), Sox2 (1:1000, goat IgG, R&D), Oct4 (1:1000, mouse lgG, Santa Cruz), HoxB4 (1:50, mouse IgG, DSHB), Nestin (1:500, mouse IgG, millipore), Map2 (1:10,000, chicken IgY, abcam), Tuj1 (1:5000, mouse IgG, sigma), cleaved caspase 3 (1:500, rabbit IgG, CST), BrdU (1:200, rat IgG, Abcam), Ki67 (1:500, rabbit IgG, Abcam), pH3 (1:500, rabbit IgG, CST) and ZIKV (1:500, mouse anti-flavivirus group antigen antibody, millipore). Light images were visualized with Leica DMI3000 (Leica Microsystems, Bensheim, Germany) microscope and organoid size was measured with Leica application Suite softwares.
Flow cytometry analysis
Annexin V-fluorescein isothiocyanate (FITC) and pro-pidium iodide (PI) were used for identification of early apoptotic cells and late apoptotic cells. The organoids were trypsinized into single cells and washed with PBS twice. NPCs were stained by the Annexin V-FITC/PI kit (Beyotime institute of biotechnology, China) for 15 min in the dark. After incubation, the samples were immediately analyzed by FACSCalibur Flow Cytometer (BD Biosciences, San Jose, CA).
Transcriptome analysis
Three regional organoids were infected with ZIKV at MOI = 0.5 or mock infection, and total RNAs were extracted at 3 dpi or 6 dpi and used for global transcriptome analysis. RNA-seq libraries were generated from duplicated samples per condition using the Next Illumina Ultra RNA library prep kit (NEB) following manufacturer’s protocols. RNA concentration of library was measured using Qubit RNA Assay Kit in Qubit 2.0. The insert size was assessed using the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA) and then accurate quantification was performed with Taqman fluorescence probe of AB Step One Plus Real-Time PCR system and sequenced by an Illumina Hiseq 2500 platform. RNA-seq reads were aligned using tophat v2.1.1. Significantly differentially expressed genes were identified using DESeq2 by comparing reads per kilobase of transcript per million mapped reads (RPKMs) between all pairs of samples with p value <0.05 and log2 Fold change >1.5. Principle component analysis (PCA) is performed with R package ggpolot. Gene Ontology analyses on biological process were performed by The Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.8. The gene expression profiles of different samples were displayed as heatmap, the RPKM values were log2-transformed and row-scaled for better visualization. The raw data were deposited in the NCBI’s Sequence Read Archive (accession number, GSE129180).
Statistical analysis
Data were analyzed using Student’s t-test for comparison of independent means with pooled estimates of common variances.
Supplementary information
Acknowledgements
The present study was supported by the National Key Research and Development Program of China (grant no. 2018YFA0108000), the National Natural Science foundation of China (31771132, 31872760, 31301157, 31400934, 81471397 and 31601908), the National Science and Technology Major Project (2016YFD0500208, 2017ZX10304402001012, 2017ZX10304402001006); the Shanghai Municipal Education Commission (C120114), the Fundamental Research Funds for the Central Universities, and the Major Program of Development Fund for Shanghai Zhangjiang National Innovation Demonstration Zone (Stem Cell Strategic Biobank and Clinical Translation Platform of Stem Cell Technology, ZJ2018-ZD-004).
Author contributions
LL, JG-H, ZX-H and ZX-Q conceived the study. CZ, LL, HY, ZX and LS performed most of the experiments. LS, LL and ZX-H provided ZIKVs. LL, ZX and FH performed the bioinformatics analyses. LL, CZ, HY, DY-H and ZX collected and analyzed the data. LL and ZX-Q wrote the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by M. Deshmukh
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ling Liu, Zhenyu Chen, Xin Zhang, Shun Li
Contributor Information
Guohua Jin, Email: jguohua@ntu.edu.cn.
Xiaohui Zhou, Email: zhouxiaohui@shphc.org.cn.
Xiaoqing Zhang, Email: xqzhang@tongji.edu.cn.
Supplementary information
The online version of this article (10.1038/s41418-019-0324-7) contains supplementary material, which is available to authorized users.
References
- 1.Wikan N, Smith DR. Zika virus: history of a newly emerging arbovirus. Lancet Infect Dis. 2016;16:e119–126. doi: 10.1016/S1473-3099(16)30010-X. [DOI] [PubMed] [Google Scholar]
- 2.Marrs C, Olson G, Saade G, Hankins G, Wen T, Patel J, et al. Zika virus and pregnancy: a review of the literature and clinical considerations. Am J Perinatol. 2016;33:625–39. doi: 10.1055/s-0036-1580089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ventura CV, Maia M, Bravo-Filho V, Gois AL, Belfort R., Jr Zika virus in Brazil and macular atrophy in a child with microcephaly. Lancet. 2016;387:228. doi: 10.1016/S0140-6736(16)00006-4. [DOI] [PubMed] [Google Scholar]
- 4.Li C, Xu D, Ye Q, Hong S, Jiang Y, Liu X, et al. Zika virus disrupts neural progenitor development and leads to microcephaly in mice. Cell Stem Cell. 2016;19:120–6. doi: 10.1016/j.stem.2016.04.017. [DOI] [PubMed] [Google Scholar]
- 5.Lazear HM, Govero J, Smith AM, Platt DJ, Fernandez E, Miner JJ, et al. A mouse model of Zika virus pathogenesis. Cell Host Microbe. 2016;19:720–30. doi: 10.1016/j.chom.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rossi SL, Tesh RB, Azar SR, Muruato AE, Hanley KA, Auguste AJ, et al. Characterization of a novel murine model to study Zika virus. Am J Trop Med Hyg. 2016;94:1362–9. doi: 10.4269/ajtmh.16-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu KY, Zuo GL, Li XF, Ye Q, Deng YQ, Huang XY, et al. Vertical transmission of Zika virus targeting the radial glial cells affects cortex development of offspring mice. Cell Res. 2016;26:645–54. doi: 10.1038/cr.2016.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cugola FR, Fernandes IR, Russo FB, Freitas BC, Dias JL, Guimaraes KP, et al. The Brazilian Zika virus strain causes birth defects in experimental models. Nature. 2016;534:267–71. doi: 10.1038/nature18296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirsch AJ, Smith JL, Haese NN, Broeckel RM, Parkins CJ, Kreklywich C, et al. Zika Virus infection of rhesus macaques leads to viral persistence in multiple tissues. PLoS Pathog. 2017;13:e1006219. doi: 10.1371/journal.ppat.1006219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Veer MJ, Holko M, Frevel M, Walker E, Der S, Paranjape JM, et al. Functional classification of interferon-stimulated genes identified using microarrays. J Leukoc Biol. 2001;69:912–20. [PubMed] [Google Scholar]
- 11.Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;95:15623–8. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol. 2014;32:513–45. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diamond MS, Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol. 2013;13:46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–5. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang SC, Wernig M, Duncan ID, Brustle O, Thomson JA. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat Biotechnol. 2001;19:1129–33. doi: 10.1038/nbt1201-1129. [DOI] [PubMed] [Google Scholar]
- 16.Li XJ, Hu BY, Jones SA, Zhang YS, Lavaute T, Du ZW, et al. Directed differentiation of ventral spinal progenitors and motor neurons from human embryonic stem cells by small molecules. Stem Cells. 2008;26:886–93. doi: 10.1634/stemcells.2007-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang XQ, Zhang SC. Differentiation of neural precursors and dopaminergic neurons from human embryonic stem cells. Methods Mol Biol. 2009;584:355–66. doi: 10.1007/978-1-60761-369-5_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li XJ, Du ZW, Zarnowska ED, Pankratz M, Hansen LO, Pearce RA, et al. Specification of motoneurons from human embryonic stem cells. Nat Biotechnol. 2005;23:215–21. doi: 10.1038/nbt1063. [DOI] [PubMed] [Google Scholar]
- 19.Lupo G, Bertacchi M, Carucci N, Augusti-Tocco G, Biagioni S, Cremisi F. From pluripotency to forebrain patterning: an in vitro journey astride embryonic stem cells. Cell Mol Life Sci. 2014;71:2917–30. doi: 10.1007/s00018-014-1596-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcez PP, Loiola EC, Madeiro da Costa R, Higa LM, Trindade P, Delvecchio R, et al. Zika virus impairs growth in human neurospheres and brain organoids. Science. 2016;352:816–8. doi: 10.1126/science.aaf6116. [DOI] [PubMed] [Google Scholar]
- 21.Tang H, Hammack C, Ogden SC, Wen Z, Qian X, Li Y, et al. Zika virus infects human cortical neural progenitors and attenuates their growth. Cell Stem Cell. 2016;18:587–90. doi: 10.1016/j.stem.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165:1238–54. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Souza BS, Sampaio GL, Pereira CS, Campos GS, Sardi SI, Freitas LA, et al. Zika virus infection induces mitosis abnormalities and apoptotic cell death of human neural progenitor cells. Sci Rep. 2016;6:39775. doi: 10.1038/srep39775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deng YQ, Zhao H, Li XF, Zhang NN, Liu ZY, Jiang T, et al. Isolation, identification and genomic characterization of the Asian lineage Zika virus imported to China. Sci China Life Sci. 2016;59:428–30. doi: 10.1007/s11427-016-5043-4. [DOI] [PubMed] [Google Scholar]
- 25.Pankratz MT, Li XJ, Lavaute TM, Lyons EA, Chen X, Zhang SC. Directed neural differentiation of human embryonic stem cells via an obligated primitive anterior stage. Stem Cells. 2007;25:1511–20. doi: 10.1634/stemcells.2006-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X, Huang CT, Chen J, Pankratz MT, Xi J, Li J, et al. Pax6 is a human neuroectoderm cell fate determinant. Cell Stem Cell. 2010;7:90–100. doi: 10.1016/j.stem.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chi L, Fan B, Zhang K, Du Y, Liu Z, Fang Y, et al. Targeted differentiation of regional ventral neuroprogenitors and related neuronal subtypes from human pluripotent stem cells. Stem Cell Rep. 2016;7:941–54. doi: 10.1016/j.stemcr.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pattyn A, Morin X, Cremer H, Goridis C, Brunet JF. Expression and interactions of the two closely related homeobox genes Phox2a and Phox2b during neurogenesis. Development. 1997;124:4065–75. doi: 10.1242/dev.124.20.4065. [DOI] [PubMed] [Google Scholar]
- 29.Ruben SM, Dillon PJ, Schreck R, Henkel T, Chen CH, Maher M, et al. Isolation of a rel-related human cDNA that potentially encodes the 65-kD subunit of NF-kappa B. Science. 1991;251:1490–3. doi: 10.1126/science.2006423. [DOI] [PubMed] [Google Scholar]
- 30.Liu Z, Hui Y, Shi L, Chen Z, Xu X, Chi L, et al. Efficient CRISPR/Cas9-mediated versatile, predictable, and donor-free gene knockout in human pluripotent stem cells. Stem Cell Rep. 2016;7:496–507. doi: 10.1016/j.stemcr.2016.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dang J, Tiwari SK, Lichinchi G, Qin Y, Patil VS, Eroshkin AM, et al. Zika virus depletes neural progenitors in human cerebral organoids through activation of the innate immune receptor TLR3. Cell Stem Cell. 2016;19:258–65. doi: 10.1016/j.stem.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mlakar J, Korva M, Tul N, Popovic M, Poljsak-Prijatelj M, Mraz J, et al. Zika virus associated with microcephaly. N Engl J Med. 2016;374:951–8. doi: 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- 33.Cui Liyuan, Zou Peng, Chen Er, Yao Hao, Zheng Hao, Wang Qian, Zhu Jing-Ning, Jiang Shibo, Lu Lu, Zhang Jiayi. Visual and Motor Deficits in Grown-up Mice with Congenital Zika Virus Infection. EBioMedicine. 2017;20:193–201. doi: 10.1016/j.ebiom.2017.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Calvet G, Aguiar RS, Melo AS, Sampaio SA, de Filippis I, Fabri A, et al. Detection and sequencing of Zika virus from amniotic fluid of fetuses with microcephaly in Brazil: a case study. Lancet Infect Dis. 2016;16:653–60. doi: 10.1016/S1473-3099(16)00095-5. [DOI] [PubMed] [Google Scholar]
- 35.Martines RB, Bhatnagar J, Keating MK, Silva-Flannery L, Muehlenbachs A, Gary J, et al. Notes from the field: evidence of Zika virus infection in brain and placental tissues from two congenitally infected newborns and two fetal losses--Brazil, 2015. MMWR Morb Mortal Wkly Rep. 2016;65:159–60. doi: 10.15585/mmwr.mm6506e1. [DOI] [PubMed] [Google Scholar]
- 36.Sarno M, Sacramento GA, Khouri R, do Rosario MS, Costa F, Archanjo G, et al. Zika virus infection and stillbirths: a case of hydrops fetalis, hydranencephaly and fetal demise. PLoS Negl Trop Dis. 2016;10:e0004517. doi: 10.1371/journal.pntd.0004517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu YJ. IPC: professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Annu Rev Immunol. 2005;23:275–306. doi: 10.1146/annurev.immunol.23.021704.115633. [DOI] [PubMed] [Google Scholar]
- 38.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–86. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 39.Husband AJ. Overview of the mammalian immune system. Adv Nutr Res. 2001;10:3–14. doi: 10.1007/978-1-4615-0661-4_1. [DOI] [PubMed] [Google Scholar]
- 40.Parkin J, Cohen B. An overview of the immune system. Lancet. 2001;357:1777–89. doi: 10.1016/S0140-6736(00)04904-7. [DOI] [PubMed] [Google Scholar]
- 41.Vermillion MS, Lei J, Shabi Y, Baxter VK, Crilly NP, McLane M, et al. Intrauterine Zika virus infection of pregnant immunocompetent mice models transplacental transmission and adverse perinatal outcomes. Nat Commun. 2017;8:14575. doi: 10.1038/ncomms14575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aschner M. Astrocytes as mediators of immune and inflammatory responses in the CNS. Neurotoxicology. 1998;19:269–81. [PubMed] [Google Scholar]
- 43.Bayer A, Lennemann NJ, Ouyang Y, Bramley JC, Morosky S, Marques ET, Jr., et al. Type III interferons produced by human placental trophoblasts confer protection against Zika virus infection. Cell Host Microbe. 2016;19:705–12. doi: 10.1016/j.chom.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miner JJ, Cao B, Govero J, Smith AM, Fernandez E, Cabrera OH, et al. Zika virus infection during pregnancy in mice causes placental damage and fetal demise. Cell. 2016;165:1081–91. doi: 10.1016/j.cell.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frumence E, Roche M, Krejbich-Trotot P, El-Kalamouni C, Nativel B, Rondeau P, et al. The South Pacific epidemic strain of Zika virus replicates efficiently in human epithelial A549 cells leading to IFN-beta production and apoptosis induction. Virology. 2016;493:217–26. doi: 10.1016/j.virol.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 46.Wu X, Dao Thi VL, Huang Y, Billerbeck E, Saha D, Hoffmann HH, et al. Intrinsic immunity shapes viral resistance of stem cells. Cell. 2018;172:423–38 e425. doi: 10.1016/j.cell.2017.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Freaney JE, Kim R, Mandhana R, Horvath CM. Extensive cooperation of immune master regulators IRF3 and NFkappaB in RNA Pol II recruitment and pause release in human innate antiviral transcription. Cell Rep. 2013;4:959–73. doi: 10.1016/j.celrep.2013.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiao S, Li D, Zhu HQ, Song MG, Pan XR, Jia PM, et al. RIG-G as a key mediator of the antiproliferative activity of interferon-related pathways through enhancing p21 and p27 proteins. Proc Natl Acad Sci USA. 2006;103:16448–53. doi: 10.1073/pnas.0607830103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hsu YA, Lin HJ, Sheu JJ, Shieh FK, Chen SY, Lai CH, et al. A novel interaction between interferon-inducible protein p56 and ribosomal protein L15 in gastric cancer cells. DNA Cell Biol. 2011;30:671–9. doi: 10.1089/dna.2010.1149. [DOI] [PubMed] [Google Scholar]
- 50.Swaim CD, Scott AF, Canadeo LA, Huibregtse JM. Extracellular ISG15 signals cytokine secretion through the LFA-1 integrin receptor. Mol Cell. 2017;68:581–90 e585. doi: 10.1016/j.molcel.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–7. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 52.Chen Y, Cao J, Xiong M, Petersen AJ, Dong Y, Tao Y, et al. Engineering human stem cell lines with inducible gene knockout using CRISPR/Cas9. Cell Stem Cell. 2015;17:233–44. doi: 10.1016/j.stem.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chi L, Fan B, Feng D, Chen Z, Liu Z, Hui Y, et al. The dorsoventral patterning of human forebrain follows an activation/transformation model. Cereb Cortex 2016;27:2941–54. [DOI] [PubMed]
- 54.Liu L, Liu X, Ren X, Tian Y, Chen Z, Xu X, et al. Smad2 and Smad3 have differential sensitivity in relaying TGFbeta signaling and inversely regulate early lineage specification. Sci Rep. 2016;6:21602. doi: 10.1038/srep21602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen Z, Ren X, Xu X, Zhang X, Hui Y, Liu Z, et al. Genetic engineering of human embryonic stem cells for precise cell fate tracing during human lineage development. Stem Cell Rep. 2018;11:1257–71. doi: 10.1016/j.stemcr.2018.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Du Y, Liu Z, Cao X, Chen X, Chen Z, Zhang X, et al. Nucleosome eviction along with H3K9ac deposition enhances Sox2 binding during human neuroectodermal commitment. Cell Death Differ. 2017;24:1121–31. doi: 10.1038/cdd.2017.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.