Abstract
Background
Changes in platelet count are common in COVID-19 patients. The platelet count reflects the thrombocyte turnover, acting as a sensitive indicator of illness severity that is of great clinical utility to monitor a quickly changing health condition of patients affected by aggressive viral infections. This study aims to investigate the significance of platelet count during the progression of the disease in COVID-19 patients.
Methods
A total of 532 COVID-19 patients were involved in the cohort study from the First People’s Hospital of Jiangxia District in Wuhan from January 7, 2020, to February 28, 2020. We collected the clinical characteristics and laboratory data of patients. Patients still hospitalized before February 29, 2020, died on admission, with malignant tumors, previous gastrointestinal surgery, missing baseline platelet count, or platelet count detected only once, were excluded. We used a generalized additive model and generalized additive mixed model to compare trends in platelet count over time among survivors and non-survivors, with an adjustment for potential confounders.
Results
During the follow-up, twenty-nine subjects died (mortality rate, 5.45%). The platelets among non-survivors decreased and among survivors increased gradually within 1 week after admission. In addition, the difference between the two groups showed an increasing trend during 1 week after admission. This difference increased by an average of 5.3 × 10^9/L daily.
Conclusions
In the early stage, platelet count can dynamically reflect the pathophysiological changes in COVID-19 patients. Early decrease in platelet count was associated with mortality in patients with COVID-19. Causality, however, cannot be deduced from our data.
Keywords: Platelet count, COVID-19, SARS-CoV-2, Early prognosis, Predictive preventive personalized medicine (PPPM), Clinical knowledge mining, Generalized additive mixed model (GAMM)
Introduction
The “2019 novel coronavirus” was discovered due to unexplained pneumonia cases in Wuhan at the end of 2019 [1–3]. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) is a new coronavirus strain that has never been found in humans before, causing widespread worldwide concern [2, 4, 5]. On February 11, 2020, the World Health Organization officially named SARS-CoV-2-induced diseases as coronavirus disease 2019 (COVID-19) [6]. Until March 4, 2020, 80,422 people have been diagnosed with COVID-19, among them 6416 are severe cases, leading to the death of 2984 patients in mainland China.
The incubation period of COVID-19 is as long as 2 weeks or even longer, and it is highly contagious [7]. The main target organ is lung tissue. Some patients rapidly develop into acute respiratory failure, acute respiratory distress syndrome (ARDS), or multiple organ failure, losing the best time to be rescued [8, 9]. Therefore, it is important to identify critical patients early, which is useful for good prognosis.
The White Paper of the“European Association for Predictive, Preventive and Personalised Medicine”(EPMA) emphasized that a central component of preventive strategies is to identify individuals at risk of developing emergent and severe cases [10]. In the position paper of the 2016 EPMA, it is also suggested that individualization of the diagnostic and treatment process is a key to future success [11]. Therefore, it is necessary to find prognostic indicators to identify potential and severe cases of COVID-19, which help to apply personalized approaches in treatment.
At present, the relevant diagnosis and treatment guidelines are continuously updated. So far, the main risk factors of COVID-19-related deaths include oldness, increasing counts of neutrophil, D-dimer, and decreasing counts of lymphocyte [12, 13]. In fact, several clinical observational studies have found that COVID-19 patients had lower counts of platelet [8, 9, 12, 14–16]. Recently, a meta-analysis reported that thrombocytopenia was associated with severe COVID-19 [17]. However, the relationship between decreasing counts of platelet and prognosis in COVID-19 patients is unknown. This study aimed to investigate the difference in platelet count between survivors and non-survivors among COVID-19 patients.
Methods
Study design and participants
In this cohort study, we recruited patients from January 7, 2020, to February 28, 2020, at the First People’s Hospital of Jiangxia District in Wuhan, China. In this period, a total of 1066 patients were diagnosed as COVID-19 according to the World Health Organization interim guidance. Patients meeting the following criteria were excluded: (1) those still hospitalized before February 29, 2020 (n = 345); (2) those died on admission (n = 5); (3) those with malignant tumors (n = 6); (4) those with previous gastrointestinal surgery (n = 7); (5) those missing baseline platelet count (n = 31); (6) those platelet count detected only once (n = 143). Finally, a total of 532 eligible patients were included (as shown in Fig. 1). This case series was approved by the Medical Ethics Committee of the First People’s Hospital of Jiangxia District. Oral consent was obtained from patients and was recorded by the physician. Written informed consent was not required because the data were anonymous, and the study was an observational one [18].
Fig. 1.
Study population. Asterisk indicates that subjects met combined conditions
Variables measured
We obtained demographic, medical history, and outcome data from patients’ medical records. After an overnight fast, all venous blood samples were obtained from patients. A complete blood count analysis (including white blood cell count, neutrophil count, lymphocyte count, and platelet count) performed within 2 h of collection using BC-3000 auto hematology analyzer (Mindray Medical International, Inc.), following the principle of electrical impedance counting and the hospital laboratory policy. The laboratory follows the quality assurance system in China. The reference range for platelet count in a healthy person is 100–300 × 10^9/L (male, 108–273 × 10^9/L; female, 148–257 × 10^9/L). Repeated measurements of platelet count were performed. The repeated measurements of platelet count were irregularly spaced over time. Two physicians checked all data.
Definition of COVID-19
The upper respiratory tract specimens were positive for SARS-CoV-2 nucleic acid by real-time RT-PCR in Wuhan Institute of Virology, Chinese Academy of Sciences, which excluded other viruses, such as influenza A virus (H1N1, H3N2, H7N9), influenza B virus, respiratory syncytial virus, parainfluenza virus, adenovirus, Severe Acute Respiratory Syndrome coronavirus (SARSCoV), and Middle Eastern Respiratory Syndrome coronavirus (MERSCoV). The method of virus RNA detection was described previously [9].
Statistical analysis
All analyses were performed using R (http://www.R-project.org) and EmpowerStats software (www.empowerstats.com, X&Y solutions, Inc., Boston, MA, USA). We presented continuous measurements as mean (SD) and categorical variables as count (%). A two-sided P value < 0.05 was considered statistically significant.
We first showed the difference in platelet count between survivors and non-survivors stratified by gender (Table 1). Next, baseline characteristics of all patients at admission were examined by a t test or Kruskal–Wallis rank sum test for continuous variables and chi-square tests for categorical data (Table 2). We also illustrated the age distribution of 29 non-survivors (Fig. 2). Then, the generalized additive model (GAM) was used to investigate changes of platelet count over time between groups (Fig. 3). Finally, the relationship between early (0–7 days) changes in platelet count and death in 532 COVID-19 patients derived from a generalized additive mixed model (GAMM) was analyzed (Table 3). The GAM and GAMM are particularly suitable for analyzing the results of repeated measures, especially when some data are missing, the interval between repeated measures is irregular, and the sample size is moderate [19, 20].
Table 1.
The difference in platelet count between survivors and non-survivors stratified by gender
| Time | Platelet count, 10^9/L, mean(SD) median (25th–75th percentile) | ||
|---|---|---|---|
| Male | |||
| Survivors,n = 232 | Non-survivors,n = 14 | Pvalue | |
| On admission | 176.0 (69.2) 165.0 (128.0–212.0) | 171.7 (93.7) 168.0 (121.2–196.0) | 0.826 |
| On 5–6th day | 218.5 (93.0) 195.0 (152.0–265.2) | 142.3 (97.7) 115.0 (75.2–209.0) | 0.003 |
| On 14–15th day | 239.9 (84.7) 232.0 (187.0–291.0) | 114.3 (95.0) 109.5 (42.2–128.5) | <0.001 |
| Female | |||
| Survivors,n = 271 | Non-survivors,n = 15 | Pvalue | |
| On admission | 194.7 (74.0) 180.0 (143.0–225.0) | 180.3 (87.4) 148.0 (125.0–233.5) | 0.466 |
| On 5–6th day | 227.1 (85.1) 212.0 (165.0–274.0) | 194.0 (86.7) 169.0 (136.2–263.0) | 0.157 |
| On 14–15th day | 250.7 (85.8) 247.0 (191.0–300.2) | 151.1 (58.8) 157.0 (137.0–196.0) | 0.001 |
p value: as for the difference between survivors and non-survivors; Kruskal–Wallis test was applied for the variables with a skewed distribution. On the 5–6th day: including 1 patient with missing data; on the 14–15th day: including 113 patients with missing data
Table 2.
Baseline characteristics and clinical outcomes of patients infected with SARS-CoV-2
| Characteristic | Survivors | Non-survivors | p value |
|---|---|---|---|
| N | 503 | 29 | |
| Age, years | 48.2 ± 14.4 | 64.7 ± 13.1 | < 0.001 |
| White blood cell count, 10^9/L | 5.4 ± 3.1 | 7.6 ± 3.3 | < 0.001 |
| Neutrophil count, 10^9/L | 3.8 ± 2.9 | 6.3 ± 3.3 | < 0.001 |
| Lymphocyte count, 10^9/L | 1.1 ± 0.5 | 0.8 ± 0.4 | < 0.001 |
| Platelet count, 10^9/L | 186.1 ± 72.3 | 176.1 ± 89.0 | 0.478 |
| Sex | 0.821 | ||
| Male | 232 (46.1%) | 14 (48.3%) | |
| Female | 271 (53.9%) | 15 (51.7%) | |
| History of hypertension | < 0.001 | ||
| No | 411 (81.7%) | 13 (44.8%) | |
| Yes | 92 (18.3%) | 16 (55.2%) | |
| History of diabetes | < 0.001 | ||
| No | 453 (90.1%) | 20 (69.0%) | |
| Yes | 50 (9.9%) | 9 (31.0%) |
Data were mean ± SD, n (%). p values comparing groups were from Student’s t test for continuous data and chi-squared tests for categorical variables
Fig. 2.
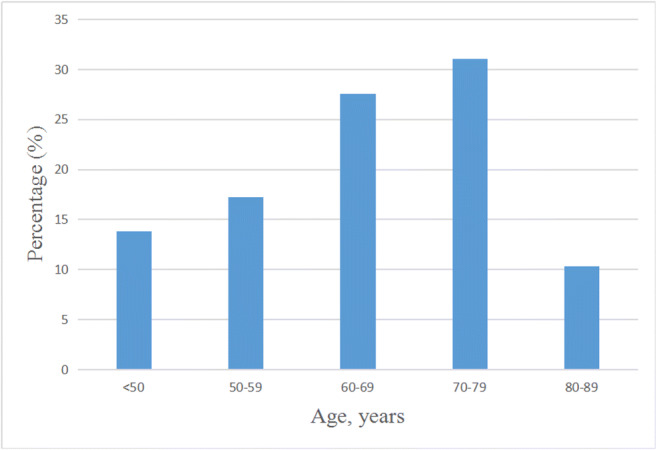
Age distribution of 29 non-survivors
Fig. 3.
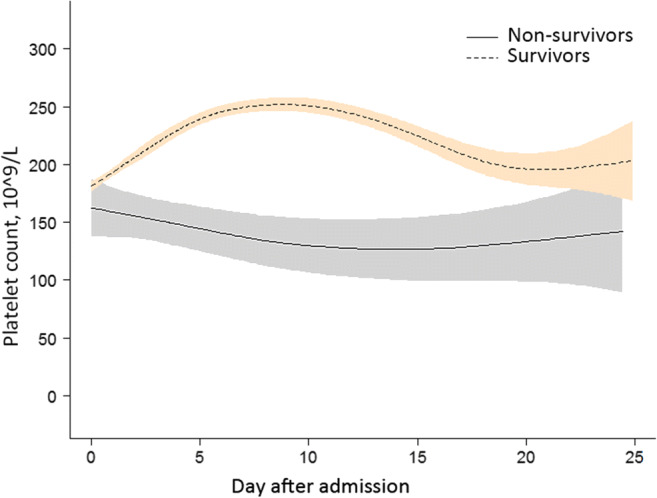
Association between changes in platelet count and mortality. A nonlinear association between changes in platelet count and mortality was found in a generalized additive model (GAM). Smooth curve fitting graph illustrated the platelet count in 532 COVID-19 patients (29 non-survivors and 503 survivors) based on the days after admission to hospital. The dotted line represented the survivors. The solid line represented the non-survivors. The blue and red-colored areas around the particular lines represented the 95% credible intervals. All adjusted for sex; age, years; white blood cell count, 10^9/L; neutrophil count, 10^9/L; lymphocyte count, 10^9/L; baseline platelet count, 10^9/L; history of hypertension; history of diabetes
Table 3.
Relationship between early (0–7 days) changes in platelet count (10^9/L) and death in 532 COVID-19 patients derived from a generalized additive mixed model (GAMM)
| Outcome | Model I | Model II | ||
|---|---|---|---|---|
| β (95%CI) | p value | β (95%CI) | p value | |
| Intercept | 167.9 (141.5, 194.2) | < 0.0001 | 168.9 (142.1, 195.74) | < 0.0001 |
| Day | 2.9 (2.4, 3.5) | < 0.0001 | 2.9 (2.4, 3.5) | < 0.0001 |
| Death | − 27.9 (− 55.7, − 0.1) | 0.05 | − 27.9 (− 56.1, 0.1) | 0.0509 |
| Day × death | − 5.3 (− 7.1, − 3.4) | < 0.0001 | − 5.3 (− 7.1, − 3.4) | < 0.0001 |
CI, confidence interval; Intercept, the mean of platelet count at day = 0 and death = 0; Day, the mean of the increasing of platelet count at death = 0 over time (daily); Death, the difference of platelet count at day = 0 between the group of death = 1 and the group of death = 0; Day × death, the average increasing in platelet count daily under the condition of the group of death = 1 compared with the group of death = 0; Model I: adjusted for Sex; Age, years; Neutrophil count, 10^9/L; Lymphocyte count, 10^9/L; Model II: adjusted for Sex; Age, years; White blood cell count, 10^9/L; Neutrophil count, 10^9/L; Lymphocyte count, 10^9/L; History of hypertension; History of diabetes
Results
In Table 1, we compared the difference in platelet count between survivors and non-survivors of different gender on admission, on the 5–6th day and the 14–15th day. There was one patient and 113 patients with missing platelet count on the 5–6th day and on the 14–15th day, respectively. The results showed that the mean value of platelet count of survivors was significantly higher than that of non-survivors at the above mentioned time points, especially on the 5–6th day (p = 0.003) and the 14–15th day (p < 0.001) of male patients and on the 14–15th day (p = 0.001) of female patients.
Baseline data of the 532 COVID-19 patients were shown in Table 2. The sample included 503 survivors and 29 non-survivors. No significant difference in sex composition between the two groups. The white blood cell count and neutrophil count on admission of the surviving group were significantly lower than those of the non-surviving group (p < 0.001). The baseline value of lymphocyte count in the surviving group was significantly higher than that in the non-surviving group (p < 0.001). The platelet count on admission in the surviving group was not significantly different from that in the non-surviving group. In addition, the non-surviving group had a higher proportion of hypertension (55.2% vs. 18.3%) and diabetes (31.0% vs. 9.9%) than the surviving group. The mean age of the non-surviving group was 64.7 ± 13.1 years, which was significantly higher than that of the surviving group (48.2 ± 14.4 years, p < 0.001). In this case, the average age at SD of 13.1 is not sufficient from the personalization point of view. The age of each non-survivor is of great importance for the scientific community, together with the changes in platelet count over time since admission time to the time of death. Therefore, we illustrated the age distribution of 29 non-survivors for further research (Fig. 2).
In Fig. 3, we can observe changes in platelet count over time between the two groups. We found that platelet count first decreased and then increased as time went on in the non-surviving group. However, the trend was the opposite in the other group. In addition, it can be seen that the changes in platelet count in 1 week after admission were significantly different by comparing the changes in platelet count between the two groups.
In Table 3, we revealed the relationship between early (0–7 days) changes in platelet count and death in 532 COVID-19 patients. Based on GAMM, we draw a conclusion that the platelet count in the non-surviving group was significantly lower than that in the surviving group. What’s more, the difference between the two groups showed an increasing trend within 1 week after admission, which increased by an average of 5.3 × 10^9/L daily. The increasing value remained 5.3 × 10^9/L with an adjustment for diverse variables, indicating that the result was stable.
Discussion
Here, we report the retrospective cohort study that has investigated the change in platelet count among non-survivors and survivors suffering from COVID-19. We found that platelet count in non-survivors decreased more than in survivors in 1 week after admission. This trend persisted after adjustment for potential confounders. It might be considered that early decrease in platelet count is associated with poor prognosis in COVID-19 patients. This study further expanded the effect of the change of platelet count.
Consistent with the other study, we also found that the group of non-survivors was older than the surviving group [12]. The majority of non-survivors were those aged 60–69 and 70–79 years. This may be related to the fact that older patients are more inclined to have more risk factors for diseases. Compared with survivors, non-survivors had higher white blood cell count, neutrophil count, and lower lymphocyte count, which may be related to disorders of inflammatory and immune responses in non-survivors. The non-surviving group had a higher proportion of hypertension and diabetes than the surviving group. The history of hypertension was a risk indicator in the Multilobular infiltration, hypo-Lymphocytosis, Bacterial coinfection, Smoking history, hyper-Tension and Age (MuLBSTA) score, a clinically predictive tool for assessing mortality risk of viral pneumonia [21]. SARS-CoV-2 can bind to its target cells through angiotensin-converting enzyme 2 (ACE2) [22], which would facilitate infection with COVID-19, while hypertension and diabetes mellitus were linked to ACE2 polymorphisms, specifically in Asian populations [23].
Our results contrasted with previous reports based on the clinical characteristics of affected patients, which concluded that platelets were out of the normal range in many COVID-19 patients [8, 9, 12, 14–16]. However, our study extended previous reports in several ways. First, we showed variation trend of the platelet count over time. Second, we compared the variation trend of platelets among survivors and non-survivors in COVID-19 patients. Third, our results helped to clarify the relation between variation trend of the platelet count and prognosis among COVID-19 patients. Fourth, by using the GAM and GAMM to analyze the collected data on COVID-19 patients, we also highlighted the complementary power of this method for the mining of the useful clinical knowledge, which might otherwise be lost if the data were analyzed only by the traditional methods based on the analytical performance and the concept of reference change value (RCV), total error, bias, intraindividual biological variation, and biological variation in group [24, 25]. It is important to keep this observation in mind when analyzing other data sets in relation to another laboratory test.
As well known, the platelet count shows the production and decay of thrombocytes, which also reflects the relation of the consumption and generation of platelets in the body to some degree [26]. Therefore, platelet count can act as a sensitive indicator of illness severity and shows early changes in conditions, indirectly reflecting the body’s ability to resist the disease. Recent studies reported that the lungs of COVID-19 patients showed bilateral diffuse alveolar damage, which was very similar to those found in SARS-CoV and MERS-CoV infections [27]. There were several reasons why the platelets of COVID-19 patients declined in the early stages. On the one hand, viral infection induced lung tissue damage, resulting in activation, aggregation, and entrapment of the platelet. This led to thrombosis at the lung injury site, which increased the consumption of platelet. On the other hand, mature megakaryocytes may release platelets in the lungs. Therefore, when the damaged lungs caused pulmonary fibrosis and pathological changes, production of platelet might be affected [28]. Furthermore, virus infection may bring about immune damages to platelets by inducing auto-antibodies and immune complexes. The use of immunomodulators and short courses of corticosteroids at low-to-moderate dose may potentially be helpful for critically ill patients with COVID-19, using, essentially, very personalized attitude towards each particular patient.
Our results have several potential clinical implications. Platelets have a short life cycle and there are very few platelets stored in the bone marrow. When platelet count changes in the early stages of the disease, it can sensitively reflect the severity of the patient’s condition. Progression of disease is related to the risk of lowering the platelet count, and reduced platelet count may also aggravate the disease. They are interrelated and each subtly influencing one another. What’s more, a downward trend of platelet count is more informative than whether it is within the normal range. The blood routine test is easy to carry out and it is inexpensive. We believe that frequent measurements of the platelet count may be helpful for the treatment of COVID-19, leading to more effective measures much earlier.
This study has several limitations. First, only 503 patients were included. However, the sample size was sufficient to get a conclusion. More populations in other cities in China or in other countries should be preferably included. Second, although we have shown a relationship between the downward trend of platelet count and prognosis among COVID-19 patients, whether platelets have effects on COVID-19, and what specific effects those are, it all needs to be confirmed by further basic research. Third, multiple potential confounding variables have been adjusted, but the residual confounding issues may not be fully resolved.
Conclusions and expert recommendations
Platelet count dynamically shows pathophysiological changes in body, which can act as an early warning index for the early treatment of COVID-19. It can also serve as a simple and repeatable clinical indicator. Compared with clinical symptoms, platelet count changes occur earlier and can be easily determined, so clinicians should pay more attention to the changes to personalize the therapeutic interventions and treatment procedures.
The 2012 EPMA white paper and the 2016 EPMA position paper both highlight the significance of suitable biomarker to early and predictive diagnostics and targeted prevention [10, 11]. Our findings conform with the predictive, preventive, and personalized medicine (PPPM) concepts of the above two papers and further refine their content, showing the practical application of PPPM in particular case of worldwide epidemic of SARS-CoV-2. Our data indicate that platelet count has important information, which could be used for improved prediction, patient stratification (moderate vs high severity of the disease), and personalized prevention needed for the COVID-19-infected individuals, in order to save lives and economical resources. We recommend adding platelet count as an additional biomarker in the current management process of COVID-19 patients. Consequently, it is important to observe the daily changes of platelet count and regard it as a sign of COVID-19 treatment.
Acknowledgments
We thank all patients and families who agreed their data to support this study. We thank Juan Feng and Haibin Han (Department of Information Center, The First People’s Hospital of Jiangxia District, Wuhan City, and Union Jiangnan Hospital, Huazhong University of Science and Technology, Wuhan, China) for their assistance in establishing the clinical database.
Abbreviations
- ACE2
angiotensin-converting enzyme 2
- ARDS
acute respiratory distress syndrome
- COVID-19
corona virus disease 2019
- EPMA
European Association for Predictive, Preventive and Personalised Medicine
- GAM
generalized additive model
- GAMM
generalized additive mixed model
- MERSCoV
Middle Eastern Respiratory Syndrome coronavirus
- MuLBSTA
Multilobular infiltration, hypo-Lymphocytosis, Bacterial coinfection, Smoking history, hyper-Tension and Age
- PPPM
predictive, preventive, and personalized medicine
- RCV
reference change value
- SARSCoV
Severe Acute Respiratory Syndrome coronavirus
- SARS-CoV-2
Severe Acute Respiratory Syndrome Coronavirus 2
Authors’ contributions
X.Z. and C.L. designed the study. X.Z., P.Z., Y.L., M.Z., S.X., and H.Z. collected the data. X.Z., K.W., and X.C. performed the analytical work. X.Z., K.W., P.Z., and Y.L. wrote the paper. All authors approved the final manuscript.
Funding information
This work was supported by the National Natural Science Foundation of China (No. 81671386 and No. 81974222).
Data availability
The datasets generated in the current research can be obtained from the corresponding author upon reasonable request.
Compliance with ethical standards
Competing interests
The authors declare that they have no competing interests.
Ethics approval and consent to participate
The study was approved by the Medical Ethics Committee of the First People’s Hospital of Jiangxia District and complied with the Declaration of Helsinki. Verbal informed consent was obtained from all the patients.
Consent for publication
We verbally informed the patients that their data would be used anonymously for medical studies and had their permission. No written informed consent, because the data were anonymous, and the study was an observational one.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KSM, Lau EHY, Wong JY, Xing X, Xiang N, Wu Y, Li C, Chen Q, Li D, Liu T, Zhao J, Liu M, Tu W, Chen C, Jin L, Yang R, Wang Q, Zhou S, Wang R, Liu H, Luo Y, Liu Y, Shao G, Li H, Tao Z, Yang Y, Deng Z, Liu B, Ma Z, Zhang Y, Shi G, Lam TTY, Wu JT, Gao GF, Cowling BJ, Yang B, Leung GM, Feng Z. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu JT, Leung K, Leung GM. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020;395(10225):689–697. doi: 10.1016/S0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bajema KL, Oster AM, McGovern OL, Lindstrom S, Stenger MR, Anderson TC, et al. Persons evaluated for 2019 novel coronavirus - United States, January 2020. MMWR Morb Mortal Wkly Rep. 2020;69(6):166–170. doi: 10.15585/mmwr.mm6906e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang F, Zhang C. What to do next to control the 2019-nCoV epidemic? Lancet. 2020;395(10222):391–393. doi: 10.1016/S0140-6736(20)30300-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lum L, Tambyah PA. Outbreak of COVID-19 - an urgent need for good science to silence our fears? Singapore Med J. 2020;61(2):55–7. [DOI] [PMC free article] [PubMed]
- 7.Backer JA, Klinkenberg D, Wallinga J. Incubation period of 2019 novel coronavirus (2019-nCoV) infections among travellers from Wuhan, China, 20-28 January 2020. Euro Surveill. 2020;25(5):2000062. doi: 10.2807/1560-7917.ES.2020.25.5.2000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Xia J', Yu T, Zhang X, Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Wei Y, Cheng Z, Yu T, Xia J, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Golubnitschaja O, Costigliola V. General report & recommendations in predictive, preventive and personalised medicine 2012: white paper of the European Association for Predictive, Preventive and Personalised Medicine. EPMA J. 2012;3(1):14. doi: 10.1186/1878-5085-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Golubnitschaja O, Baban B, Boniolo G, Wang W, Bubnov R, Kapalla M, Krapfenbauer K, Mozaffari MS, Costigliola V. Medicine in the early twenty-first century: paradigm and anticipation - EPMA position paper 2016. EPMA J. 2016;7(1):23. doi: 10.1186/s13167-016-0072-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, Xiang H, Cheng Z, Xiong Y, Zhao Y, Li Y, Wang X, Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang N, Li D, Wang X, Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J Thromb Haemost. 2020;18(4):844–847. doi: 10.1111/jth.14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guan W, Ni Z, Hu Y, Liang W, Ou C, He J, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020; NEJMoa2002032. [DOI] [PMC free article] [PubMed]
- 15.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, Yang Y, Zhang C, Huang F, Wang F, Yuan J, Wang Z, Li J, Li J, Feng C, Zhang Z, Wang L, Peng L, Chen L, Qin Y, Zhao D, Tan S, Yin L, Xu J, Zhou C, Jiang C, Liu L. Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci China Life Sci. 2020;63(3):364–374. doi: 10.1007/s11427-020-1643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lippi G, Plebani M, Henry BM. Thrombocytopenia is associated with severe coronavirus disease 2019 (COVID-19) infections: a meta-analysis. Clin Chim Acta. 2020;506:145–148. doi: 10.1016/j.cca.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Filion KB, Azoulay L, Platt RW, Dahl M, Dormuth CR, Clemens KK, Hu N, Paterson JM, Targownik L, Turin TC, Udell JA, Ernst P. A multicenter observational study of incretin-based drugs and heart failure. N Engl J Med. 2016;374(12):1145–1154. doi: 10.1056/NEJMoa1506115. [DOI] [PubMed] [Google Scholar]
- 19.Gueorguieva R, Krystal JH. Move Over ANOVA: Progress in analyzing repeated-measures data and its reflection in papers published in the archives of general psychiatry. Arch Gen Psychiatry. 2004;61(3):310–317. doi: 10.1001/archpsyc.61.3.310. [DOI] [PubMed] [Google Scholar]
- 20.Najjar SS, Scuteri A, Shetty V, Wright JG, Muller DC, Fleg JL, Spurgeon HP, Ferrucci L, Lakatta EG. Pulse wave velocity is an independent predictor of the longitudinal increase in systolic blood pressure and of incident hypertension in the Baltimore Longitudinal Study of aging. J Am Coll Cardiol. 2008;51(14):1377–1383. doi: 10.1016/j.jacc.2007.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo L, Wei D, Zhang X, Wu Y, Li Q, Zhou M, Qu J. Clinical features predicting mortality risk in patients with viral pneumonia: the MuLBSTA score. Front Microbiol. 2019;10:2752. doi: 10.3389/fmicb.2019.02752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94(7):e00127–e00120. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang L, Karakiulakis G, Roth M. Are patients with hypertension and diabetes mellitus at increased risk for COVID-19 infection? Lancet Respir Med. 2020;8(4):e21. doi: 10.1016/S2213-2600(20)30116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fraser CG. Reference change values. Clin Chem Lab Med. 2011;50(5):807–812. doi: 10.1515/CCLM.2011.733. [DOI] [PubMed] [Google Scholar]
- 25.Coşkun A, Carobene A, Kilercik M, Serteser M, Sandberg S, Aarsand AK, Fernandez-Calle P, Jonker N, Bartlett WA, Díaz-Garzón J, Huet S, Kızıltaş C, Dalgakıran I, Ugur E, Unsal I, European Biological Variation Study of the EFLM Working Group on Biological Variation Within-subject and between-subject biological variation estimates of 21 hematological parameters in 30 healthy subjects. Clin Chem Lab Med. 2018;56(8):1309–1318. doi: 10.1515/cclm-2017-1155. [DOI] [PubMed] [Google Scholar]
- 26.Holinstat M. Normal platelet function. Cancer Metastasis Rev. 2017;36(2):195–198. doi: 10.1007/s10555-017-9677-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, Liu S, Zhao P, Liu H, Zhu L, Tai Y, Bai C, Gao T, Song J, Xia P, Dong J, Zhao J, Wang FS. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang J, Yang M, Xu F, Li K, Lee SKM, Ng P, et al. Effects of oxygen-induced lung damage on megakaryocytopoiesis and platelet homeostasis in a rat model. Pediatr Res. 2003;54(3):344–352. doi: 10.1203/01.PDR.0000079186.86219.29. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated in the current research can be obtained from the corresponding author upon reasonable request.



