Abstract
Brahman cattle (Bos indicus) are well adapted to thrive in tropical environments. Since their introduction to Australia in 1933, Brahman’s ability to grow and reproduce on marginal lands has proven their value in the tropical beef industry. The poll phenotype, which describes the absence of horns, has become desirable in the cattle industry for animal welfare and handler safety concerns. The poll locus has been mapped to chromosome one. Four alleles, each a copy number variant, have been reported across this locus in B. indicus and Bos taurus. However, the causative mutation in Brahman cattle has not been fully characterized. Oxford Nanopore Technologies’ minION sequencer was used to sequence four homozygous poll (PcPc), four homozygous horned (pp), and three heterozygous (Pcp) Brahmans to characterize the poll allele in Brahman cattle. A total of 98 Gb were sequenced and an average coverage of 3.33X was achieved. Read N50 scores ranged from 9.9 to 19 kb. Examination of the mapped reads across the poll locus revealed insertions approximately 200 bp in length in the poll animals that were absent in the horned animals. These results are consistent with the Celtic poll allele, a 212-bp duplication that replaces 10 bp. This provides direct evidence that the Celtic poll allele is segregating in the Australian Brahman population.
Keywords: bovine, long read sequencing, Oxford Nanopore, poll, structural variant, whole genome sequencing
Introduction
The poll phenotype, a desirable trait in both the dairy and beef industries, describes a lack of horns in cattle (Drögemüller et al., 2005; Mariasegaram et al., 2012; Medugorac et al., 2012; Allais-Bonnet et al., 2013). Dehorning has been widely adopted in both the industries to decrease the risk horns pose to the animals and handlers (Medugorac et al., 2012). The bruising of carcasses from horns is also estimated to cost the Australian beef industry AUD $20 million annually (Prayaga, 2005, 2007; Misch et al., 2007). Dehorning is labor-intensive, painful for the animal, and leaves animals prone to secondary infection and may lead to mortality (Prayaga, 2005). Goonewardene et al. (1999) observed a 4.3% difference in average daily weight gain between steers that were dehorned and naturally poll. This equated to a loss of 530 kg per 100 steers, meaning a noninvasive genetic solution to horns would be highly advantageous (Winks et al., 1977; Prayaga, 2005).
The poll phenotype is controlled at the poll locus on bovine chromosome one (Long and Gregory, 1978; Georges et al., 1993; Drögemüller et al., 2005) by at least four known autosomal dominant poll (P) alleles (Medugorac et al., 2012; Crystal Ketel, 2019). The Celtic allele (Pc), a 212-bp duplication replacing a 10-bp segment at position Chr1:2,429,326-2,429,335 bp according to the Bos taurus reference ARS-UCD1.2, is found predominantly in breeds originating from Scandinavia and Great Britain (Medugorac et al., 2012; Crystal Ketel, 2019). An 80-kb duplication characterizes the Friesian allele (Rothammer et al., 2014). Medugorac et al. (2017) later discovered a third allele associated with the polled phenotype in Mongolian taurine cattle, and a fourth was identified as a 110-kb duplication by Utsunomiya et al. (2019).
Koufariotis et al. (2018a) used short reads to sequence 46 Australian Brahman bulls. They reported an increase in coverage at the location of the Celtic allele for homozygous polled animals, consistent with a duplication. Their results suggest that the poll allele in the Brahman breed is likely of Celtic origin; however, the identification of structural variants using short-read data can be misleading and inaccurate (Couldrey et al., 2017; Merker et al., 2018; Mahmoud et al., 2019). Long reads, however, are able to identify structural variants accurately by sequencing the variant, including break points, with enough flanking sequence to still be accurately placed within the genome. This study aimed to characterize the poll allele in a small sample Australian Brahman population using long reads from Oxford Nanopore’s minION (Oxford Nanopore Technologies, Oxford).
Materials and Methods
Ethics statement
All samples were obtained from a commercial genotyping facility. Samples were originally taken to undergo genomic evaluation of the animals’ genetic merit for commercial purposes. As such, no animals were directly involved in this study, so Animal Care and Use Committee approval was not obtained for this study.
DNA extraction and library preparation
The Gentra Puregene (Qiagen, Hilden, Germany) DNA extraction kit was used for the extraction of DNA from both semen and tail hair from four homozygous poll (PcPc), three heterozygous (Pcp), and four homozygous horned (pp) animals. The following amendments were made for the semen extraction: 24 μL of 1 M dithiothreitol was added during the overnight proteinase K digestion and the final DNA pellet was resuspended in EB buffer; 20 to 30 hairs were used for the tail hair DNA extraction and all Gentra Puregene reagents were scaled up accordingly. The DNA pellet was also dissolved in a pre-warmed DNA hydration solution. Oxford Nanopore’s 1D genomic DNA by ligation (SQK-LSK 109) library preparation protocol was used. Pipette tips were cut and all vortexing steps were replaced with flicking the samples to decrease shearing of the DNA during library preparation. The DNA input into the library preparation was increased from the recommended 2 to 6 μg to increase pore occupancy on the flow cells during sequencing.
Sequencing
Oxford Nanopore’s MinION sequencer (version R9.4.1 minION flow cells) was used to sequence 11 semen and tail hair samples (four homozygous poll [PcPc], three heterozygous [Pcp], and four homozygous horn [pp]) from registered Brahmans. Each sample was sequenced on a single Oxford Nanopore flow cell using a runtime of 48 h. Oxford Nanopore’s minKNOW software was used for base calling in Fast5 and Fastq outputs.
Data clean-up was done using NanoFilt (version 2.3.0; De Coster et al., 2018) to remove reads shorter than 200 bp and PoreChop (version 0.2.4) to remove adapter sequences from the ends of reads. Minimap2 (version 2.14; Heng, 2018) was used to align the reads to the B. taurus reference ARS-UCD1.2. The inbuilt “map-ont” setting was used along with a matching score of 3 and a gap open penalty of 1, 0; alignments were output in the SAM format. These settings were used so that large insertions/deletions were favored over smaller insertions/deletions. All alignments were visualized at the poll locus using the Integrative Genomics Viewer (IGV; version 2.8.0; Robinson et al., 2011), and dot plots were generated using Gepard (version 1.3; Krumsiek et al., 2007).
Results
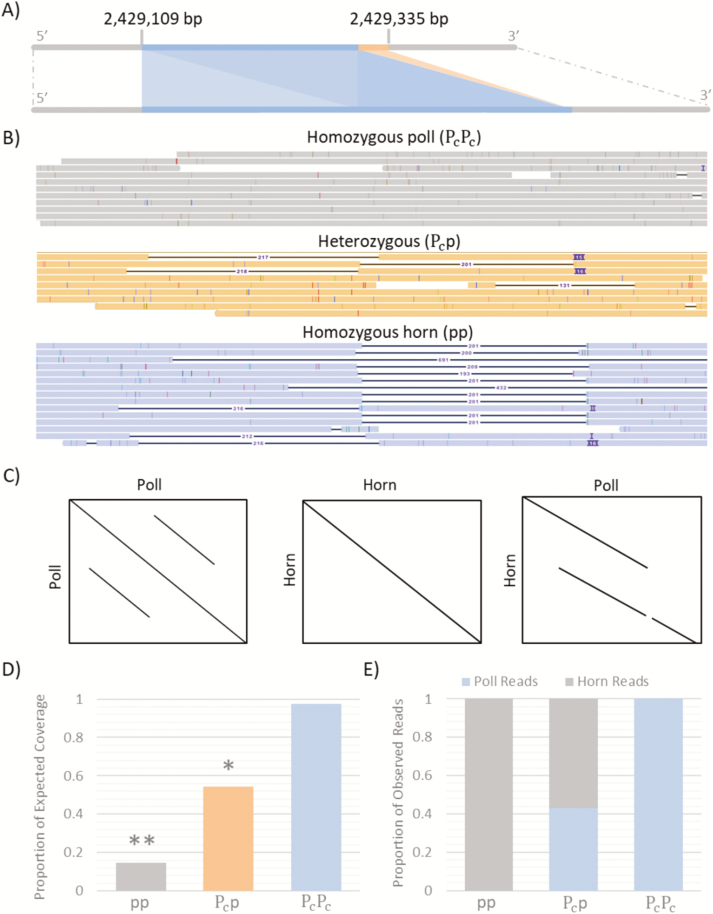
A whole-genome average coverage of 3.33X was achieved across all 11 samples. The N50 from each run varied between 9.5 and 19 kb with the longest single sequence being 234 kb. The average Phred quality score for the bases was 10. At the poll locus, an average coverage of 3.8X per sample was achieved, with a total of 34 reads spanning the Celtic locus. From visualization of the alignments against the B. taurus reference (ARS-UCD1.2) using IGV, insertions are clearly evident in the homozygous poll samples at the Celtic locus (Figure 1 and Supplementary Material 2). The insertions vary in size between 181 and 206 bp, slightly shorter than the reported 212-bp duplication, and occur at the exact location of the duplication reported in the Celtic allele (Chr1:2,429,109-2,429,320). No insertions were seen in the Celtic region in any of the four homozygous horn samples (Supplementary Material 2. This provides support that the insertions observed in the homozygous poll animals are the Celtic allele.
Figure 1.
Sequencing the poll allele in Brahman cattle using Oxford Nanopore sequencing. (A) Stylized representation of the Celtic allele on bovine chromosome one. The location of the Celtic allele in the B. taurus reference ARS-UCD1.2 is indicated. (B) IGV sequence alignments against the poll consensus sequence (Supplementary Material 3) created using minimap2. The poll consensus sequence was used here instead of the horn reference for clearer visualization of the Celtic allele. Each sample has been grouped based on poll/horn genotype: homozygous poll, homozygous horn, and heterozygous. (C) Stylized summary of dotplots created using Gepard across the Celtic locus. (D) Poll locus coverage for the three genotype groups as a proportion of the expected coverage, where the expected coverage was calculated based on an average depth at three random nearby loci. Both the heterozygous (*P < 0.01) and homozygous horn (**P < 0.001) had a poll locus coverage statistically different to the expected coverage. (E) Proportion of horn and Celtic reads observed at the Celtic locus for each of the three genotype groups.
Evidence of both the Celtic and horned alleles were found in the heterozygous samples. Five reads demonstrated duplications similar in size to those observed in the homozygous poll animals at the Celtic locus. While the remaining three reads spanning the Celtic locus in the heterozygous group, all mapped without a duplication to ARS-UCD1.2, suggesting they are the horn alleles. Dot plots of reads thought to be carrying the Celtic allele aligned to ARS-UCD1.2 also demonstrate clear duplications at the reported Celtic locus (Supplementary Material 1). No duplications were observed in dot plots in the homozygous horned samples (Supplementary Material 1).
The Friesian locus was also examined for structural variations between the poll and horned animals. Despite adequate sequencing coverage at the Friesian locus, no evidence of the allele was observed in this data set. There was no increase in coverage across the locus, which is often characteristic of duplications nor was any soft clipping observed at the reported structural variant break points.
Discussion
This is the first publication of the application of Nanopore sequencing for whole-genome sequencing in beef cattle. The average sequencing depth achieved in this study was slightly less than that achieved by long-read studies on other species (Xie and Tammi, 2009; Li et al., 2011; Fumagalli, 2013; Kukekova et al., 2018). This is a reflection of the other studies examining smaller genomes or using multiple flow cells per individual, while here the focus was to obtain long-read data on multiple animals. Across the three groups (PP, Pp, and pp), the coverage achieved in this study at the Celtic locus is similar to the minimum coverage used in two genome-wide structural variant calling studies (Sedlazeck et al., 2018; Kosugi et al., 2019). As this study focused on the detection of only two known structural variants (the Friesian and Celtic alleles), the low coverage still provided strong evidence that the Celtic allele is present in the Australian Brahman population.
Both the Friesian and Celtic loci were examined for structural variants. Within the Friesian locus, no significant variation was observed between the three groups. Koufariotis et al. (2018b) also found no evidence of the Friesian allele in 46 Brahman bulls they sequenced using Illumina short reads. They reported no increase in coverage across the Friesian locus (Koufariotis et al. 2018b). Increases in coverage are used by short-read sequencing technologies to identify genome duplications, which they often are unable to span. A more recent study by Randhawa et al. (2019) found that the Friesian allele was present but very rare in Australian Brahmans (frequency Friesian: 0.001; Celtic: 0.135). The average read lengths achieved here likely affected the ability to characterize the Friesian locus. However, reads flanking significant segments of the duplication breakpoint still did not indicate any structural variation, and no increase in coverage was observed.
Clear insertions matching the description of the Celtic allele were evident in the poll animals at the Celtic locus. The variation in the size of the duplication reported by Allais-Bonnet et al. (2013) and those seen in the homozygous samples may be attributed to the indel error profile of the minION. The majority of the variation observed on IGV across the genome were small insertions and deletions; therefore, it is likely a number of small deletions within the inserted sequences have shortened its length. Using Samtools, the error rate of the mapped reads was approximately one error per 10 bases matched, which was also reported by Cretu Stancu et al. (2017). Given that an error of 21 bp is expected in the 212-bp region, the average insertion length of 194 bp is within the expected size range for the Celtic allele and is unlikely to reflect a new variant.
This study has provided strong direct evidence that the Celtic allele is present in Australian Brahmans, and that emerging long-read technology is a useful tool to identify economically important structural variants that were previously difficult to accurately characterize en masse. It is important to note that other known or novel poll alleles may exist and will only be captured by increasing the sample size. Characterization of the Celtic allele in this Australian Brahman population will help Australia’s northern beef industry progress toward a genetic solution to horns and move away from the practice of dehorning.
Supplementary Material
Acknowledgments
We acknowledge financial contributions from Meat and Livestock Australia (Project P.PSH.0868 and P.PSH.0833). We would also like to acknowledge contributions from all members of the Centre for Animal Science at the Queensland Alliance for Agriculture and Food Innovation, in particular Dr. Roy Costilla Monteagudo and Dr. Bailey Engle. We are thankful to Dr. Brian Burns for the collection of some of the samples, and Dr. Burns contributions to the field of beef cattle genomics will be keenly missed.
Glossary
Abbreviations
- IGV
Integrative Genomics Viewer
- p
the wild-type horn allele at the poll locus
- P
the polled allele at the poll locus
- Pc
the Celtic poll allele at the poll locus
Conflicts of interest statement
The authors declare no actual or potential conflicts of interest that affect their ability to objectively present or review research or data.
Literature Cited
- Allais-Bonnet A., Grohs C., Medugorac I., Krebs S., Djari A., Graf A., Fritz S., Seichter D., Baur A., Russ I., . et al. 2013. Novel insights into the bovine polled phenotype and horn ontogenesis in Bovidae. PLoS One. 8:1–14. doi: 10.1371/journal.pone.0063512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couldrey C., Johnson T., Lopdell T., Zhang I. L., Littlejohn M. D., Keehan M., Sherlock R. G., Tiplady K., Scott A., Davis S. R., . et al. 2017. Bovine mammary gland X chromosome inactivation. J. Dairy Sci. 100:5491–5500. doi: 10.3168/jds.2016-12490 [DOI] [PubMed] [Google Scholar]
- Cretu Stancu M., van Roosmalen M. J., Renkens I., Nieboer M. M., Middelkamp S., de Ligt J., Pregno G., Giachino D., Mandrile G., Espejo Valle-Inclan J., . et al. 2017. Mapping and phasing of structural variation in patient genomes using nanopore sequencing. Nat. Commun. 8:1326. doi: 10.1038/s41467-017-01343-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crystal Ketel M. A.-C. 2019. Investigating candidate scur genes in Bos taurus breeds. J. Anim. Sci. 97:226–227. doi: 10.1093/jas/skz258.461 [DOI] [Google Scholar]
- De Coster W., D’Hert S., Schultz D. T., Cruts M., and Van Broeckhoven C.. . 2018. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics 34:2666–2669. doi: 10.1093/bioinformatics/bty149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drögemüller C., Wöhlke A., Mömke S., and Distl O.. . 2005. Fine mapping of the polled locus to a 1-Mb region on bovine chromosome 1q12. Mamm. Genome 16:613–620. doi: 10.1007/s00335-005-0016-0 [DOI] [PubMed] [Google Scholar]
- Fumagalli M. 2013. Assessing the effect of sequencing depth and sample size in population genetics inferences. PLoS One. 8:e79667. doi: 10.1371/journal.pone.0079667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georges M., Drinkwater R., King T., Mishra A., Moore S. S., Nielsen D., Sargeant L. S., Sorensen A., Steele M. R., and Zhao X.. . 1993. Microsatellite mapping of a gene affecting horn development in Bos taurus. Nat. Genet. 4:206–210. doi: 10.1038/ng0693-206 [DOI] [PubMed] [Google Scholar]
- Goonewardene L. A., Pang H., Berg R. T., and Price M. A.. . 1999. A comparison of reproductive and growth traits of horned and polled cattle in three synthetic beef lines. Can. J. Anim. Sci. 79:123–127. doi: 10.4141/A98-096 [DOI] [Google Scholar]
- Heng L. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34:6. doi: 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S., Momozawa Y., Liu X., Terao C., Kubo M., and Kamatani Y.. . 2019. Comprehensive evaluation of structural variation detection algorithms for whole genome sequencing. Genome Biol. 20:117. doi: 10.1186/s13059-019-1720-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koufariotis L., Hayes B. J., Kelly M., Burns B. M., Lyons R., Stothard P., Chamberlain A. J., and Moore S.. 2018a. Extensive sequencing of a tropically adapted breed - the Brahman Sequencing Project. Proceedings from the 22nd conference of the Association for the Advancement of Animal Breeding and Genetics, Vol. 22; July 2, 2018; Townsville, Australia; p. 167–170. [Google Scholar]
- Koufariotis L., Hayes B. J., Kelly M., Burns B. M., Lyons R., Stothard P., Chamberlain A. J., and Moore S.. . 2018b. Sequencing the mosaic genome of Brahman cattle identifies historic and recent introgression including polled. Sci. Rep. 8:17761. doi: 10.1038/s41598-018-35698-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumsiek J., Arnold R., and Rattei T.. . 2007. Gepard: a rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics 23:1026–1028. doi: 10.1093/bioinformatics/btm039 [DOI] [PubMed] [Google Scholar]
- Kukekova A. V., Johnson J. L., Xiang X., Feng S., Liu S., Rando H. M., Kharlamova A. V., Herbeck Y., Serdyukova N. A., Xiong Z., . et al. 2018. Author Correction: Red fox genome assembly identifies genomic regions associated with tame and aggressive behaviours. Nat. Ecol. Evol. 2:1514. doi: 10.1038/s41559-018-0664-6 [DOI] [PubMed] [Google Scholar]
- Li Y., Sidore C., Kang H. M., Boehnke M., and Abecasis G. R.. . 2011. Low-coverage sequencing: implications for design of complex trait association studies. Genome Res. 21:940–951. doi: 10.1101/gr.117259.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long C. R., and Gregory K. E.. . 1978. Inheritance of the horned, scurred, and polled condition in cattle. J. Hered. 69:395–400. doi: 10.1093/oxfordjournals.jhered.a108980 [DOI] [Google Scholar]
- Mahmoud M., Gobet N., Cruz-Dávalos D. I., Mounier N., Dessimoz C., and Sedlazeck F. J.. . 2019. Structural variant calling: the long and the short of it. Genome Biol. 20:246. doi: 10.1186/s13059-019-1828-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariasegaram M., Harrison B. E., Bolton J. A., Tier B., Henshall J. M., Barendse W., and Prayaga K. C.. . 2012. Fine-mapping the POLL locus in Brahman cattle yields the diagnostic marker CSAFG29. Anim. Genet. 43:683–688. doi: 10.1111/j.1365-2052.2012.02336.x [DOI] [PubMed] [Google Scholar]
- Medugorac I., Graf A., Grohs C., Rothammer S., Zagdsuren Y., Gladyr E., Zinovieva N., Barbieri J., Seichter D., Russ I., . et al. 2017. Whole-genome analysis of introgressive hybridization and characterization of the bovine legacy of Mongolian yaks. Nat. Genet. 49:470–475. doi: 10.1038/ng.3775 [DOI] [PubMed] [Google Scholar]
- Medugorac I., Seichter D., Graf A., Russ I., Blum H., Göpel K. H., Rothammer S., Förster M., and Krebs S.. . 2012. Bovine polledness–an autosomal dominant trait with allelic heterogeneity. PLoS One. 7:e39477. doi: 10.1371/journal.pone.0039477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merker J. D., Wenger A. M., Sneddon T., Grove M., Zappala Z., Fresard L., Waggott D., Utiramerur S., Hou Y., Smith K. S., . et al. 2018. Long-read genome sequencing identifies causal structural variation in a Mendelian disease. Genet. Med. 20:159–163. doi: 10.1038/gim.2017.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misch L. J., Duffield T. F., Millman S. T., and Lissemore K. D.. . 2007. An investigation into the practices of dairy producers and veterinarians in dehorning dairy calves in Ontario. Can. Vet. J. 48:1249–1254. [PMC free article] [PubMed] [Google Scholar]
- Prayaga K. C. 2005. Genetic options to replace dehorning of beef cattle in Australia. North Sydney (NSW, Australia): Meat and Livestock; Available from https://www.mla.com.au/CustomControls/PaymentGateway/ViewFile.aspx?D4fKfu8wWlktaPvZX8YDc2yLIauPA9FB9/d3AoKI7coyx2uU4/cmamUgTn+Mp5E/3EYMKKAfsht7d1Tnt3BqiA== [Accessed January 12, 2020]. [Google Scholar]
- Prayaga K. C. 2007. Genetic options to replace dehorning in beef cattle – a review. Aust. J. Agric. Res. 58:1–8. doi: 10.1071/AR06044 [DOI] [Google Scholar]
- Randhawa I. A. S., Burns B. M., McGowan M. R., Porto-Neto L. R., Hayes B. J., Ferretti R., Schutt K. M., and Lyons R. E.. . 2019. Optimized genetic testing for polledness in multiple breeds of cattle. G3 (Bethesda). 10:539:544. doi: 10.1534/g3.119.400866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E. S., Getz G., and Mesirov J. P.. . 2011. Integrative genomics viewer. Nat. Biotechnol. 29:24–26. doi: 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothammer S., Kremer P. V., Bernau M., Fernandez-Figares I., Pfister-Schär J., Medugorac I., and Scholz A. M.. . 2014. Genome-wide QTL mapping of nine body composition and bone mineral density traits in pigs. Genet. Sel. Evol. 46:68. doi: 10.1186/s12711-014-0068-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlazeck F. J., Rescheneder P., Smolka M., Fang H., Nattestad M., von Haeseler A., and Schatz M. C.. . 2018. Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods 15:461–468. doi: 10.1038/s41592-018-0001-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsunomiya Y. T., Milanesi M., Fortes M. R. S., Porto-Neto L. R., Utsunomiya A. T. H., Silva M. V. G. B., Garcia J. F., and Ajmone-Marsan P.. . 2019. Genomic clues of the evolutionary history of Bos indicus cattle. Anim. Genet. 50:557–568. doi: 10.1111/age.12836 [DOI] [PubMed] [Google Scholar]
- Winks L., Holmes A. E., and Orourke P. K.. . 1977. Effect of dehorning and tipping on liveweight gain of mature Brahman crossbred steers. Aust. J. Exp. Agr. 17:16–19. doi: 10.1071/EA9770016 [DOI] [Google Scholar]
- Xie C., and Tammi M. T.. . 2009. CNV-seq, a new method to detect copy number variation using high-throughput sequencing. BMC Bioinformatics 10:80. doi: 10.1186/1471-2105-10-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.