Abstract
Background and aims
Genetic variants involved in vitamin D metabolism have been associated with diabetes and related syndromes/diseases. We wanted to investigate possible associations of polymorphisms in genes involved in vitamin D metabolism with indices of insulin resistance and insulin secretion, and also with development of diabetes after gestational diabetes mellitus (GDM).
Materials and methods
We have studied 376 women with previous GDM. Eight single nucleotide polymorphisms (SNPs) in the genes for vitamin D receptor (VDR) [rs731236, rs7975232, rs10735810, and rs1544410], vitamin D binding protein (DBP) [rs7041 and rs4588], and cytochrome P450 family 27 subfamily B member 1 (CYP27B1) [rs10877012 and rs4646536] were genotyped by TaqMan Allelic Discrimination Assay using the Quantstudio 7 Flex system. A 75-g oral glucose tolerance test (OGTT) was performed 1–2 years postpartum. The homeostasis model assessment of insulin resistance (HOMA-IR) and the disposition index [(insulinogenic index: I30/G30)/HOMA-IR] were used to calculate insulin resistance and insulin secretion, respectively. Serum samples for determination of 25(OH)D3 were collected at the time of the OGTT. Manifestation of diabetes was followed up to five years postpartum.
Results
After adjustment for BMI, age, and ethnicity, the A-allele of the VDR rs1544410 polymorphism was found to be associated with increased disposition index (difference per allele = 3.56, 95% CI: 0.4567–6.674; p = 0.03). The A-allele of the DBP rs7041 polymorphism was found to be associated with 25(OH)D3 levels (difference [in nmol/L] per allele = −5.478, 95% CI: -8.315 to −2.641; p = 0.0002), as was the T-allele of the DBP rs4588 polymorphism (OR = −6.319, 95% CI: −9.466 to −3.171; p = 0.0001). None of the SNPs were significantly associated with HOMA-IR or postpartum diabetes.
Conclusions
This study provides evidence that the rs1544410 polymorphism of the VDR gene may be associated with increased insulin secretion in women after pregnancy complicated by GDM. Further studies in other populations are needed to confirm the results.
Introduction
Gestational diabetes mellitus (GDM) is defined as any degree of glucose intolerance with onset or first recognition during pregnancy [1]. The prevalence of GDM is increasing [2], in parallel with the increasing prevalence of type-2 diabetes (T2D) worldwide [3]. Women with a history of GDM have an increased risk of developing T2D relative to those with a normoglycemic pregnancy [4]. GDM is characterized by insulin resistance and beta-cell dysfunction, and the same is true for T2D [5].
Vitamin D has been implicated in several medical conditions [6]. It has been hypothesized that it may have a positive effect on insulin secretion and sensitivity. The mechanisms may be mediated by activation of the vitamin D receptor (VDR) on pancreatic beta cells and insulin-sensitive organs and also by regulation of calcium homeostasis [7, 8]. Studies have suggested that circulating concentrations of vitamin D may be inversely associated with the risk of diabetes, metabolic syndrome, insulin secretion, and insulin resistance [8–10]. Malik et al., in addition to a meta-analysis, have found associations between polymorphisms in the VDR gene and both diabetes and insulin resistance-related diseases [11, 12]. Moreover, single nucleotide polymorphisms (SNPs) in the vitamin D binding protein gene (DBP) affect 25(OH)D3 levels [13, 14] and increase the risk of T2D and the metabolic syndrome [15, 16]. There is also evidence of associations between polymorphisms in the CYP27B1 gene and type-1 diabetes [17, 18].
We have recently reported from the “Mamma Study” that vitamin D deficiency/insufficiency in women with a history of GDM appeared to be associated with beta-cell dysfunction and insulin resistance, but not with postpartum diabetes, after adjustment for factors that are well known to influence T2D [19]. We therefore hypothesized that 8 SNPs in genes known to be involved in the vitamin D metabolism, diabetes, and related traits (4 SNPs in VDR, 2 SNPs in DBP, and 2 SNPs in CYP27B1) [8–18] might be associated with insulin resistance and/or beta-cell function at 1–2 years postpartum and also development of diabetes after up to 5 years of follow-up in women with a history of GDM.
Material and methods
Patients
The design of the “Mamma Study” has been described in detail elsewhere [20]. Briefly, women delivering between 2003–2005 were invited to participate in the study, covering 86% of all pregnancies in the County of Skane in southern Sweden, including four of five delivery departments. The diagnosis of GDM was made using a 75-g oral glucose tolerance test (OGTT) at the twenty-eighth and/or the twelfth week of gestation. In the original evaluation, GDM was defined as two-hour capillary plasma glucose ≥10.0 mmol/l, gestational IGT as two-hour capillary plasma glucose 8.6–9.9 mmol/l, and normal glucose tolerance (NGT) during pregnancy as two-hour capillary plasma glucose <8.6 mmol/l. Among those who accepted to participate in the study and after exclusion of those who had already been diagnosed as having diabetes, 160 women had GDM, 309 had Gestational IGT and 167 had NGT. Women were followed for the development of diabetes using an OGTT at 1–2 years and 5 years after pregnancy, or until the diagnosis of diabetes. The World Health Organization (WHO) 1999 diagnostic criteria were used for classification of diabetes during follow-up [21]. Measurements of both glucose and insulin concentrations at 0, 30, and 120 min during OGTT at 1–2 years postpartum were performed to calculate indices of beta-cell function and insulin resistance, as previously reported [22]. Serum samples for determination of 25(OH)D3 were collected during the OGTT at 1–2 years postpartum. In the present investigation, we used the diagnostic criteria for GDM recommended by the WHO in 1999 [21]. Based on these criteria and on successful measurements of 25OHD3 concentrations, we identified 376 women who had previously had GDM and who formed the basis of the present study. All women gave their written informed consent and the Ethics Committee of Lund University approved the study (LU 259–00, 2002-04-17), which was performed in accordance with the Declaration of Helsinki.
Chemical analysis
Homeostasis model assessment was used to estimate insulin resistance [(HOMA-IR): (fasting serum insulin × fasting plasma glucose)/22.5] [23, 24]. Insulin secretory capacity was estimated using the insulinogenic index (I/G30): [insulin 30 min−insulin 0 min)/(glucose 30 min − glucose 0 min] [25]. As insulin resistance modulates insulin secretion, the disposition index was used to adjust insulin secretion for the degree of insulin resistance, obtained by dividing I/G30 by HOMA-IR [26]. The serum concentration of 25(OH)D3 (in nmol/L) was determined by liquid chromatography mass spectrophotometry (LC-MS/MS). Assays were performed according to accredited methods at the Department of Clinical Chemistry, Skåne University Hospital, Malmö, Sweden.
Genetic analysis
The SNPs were chosen for analysis based on previous associations with circulating vitamin D levels, diabetes, and related traits [8–18]. The following SNPs were genotyped: rs731236 (TaqI), rs7975232 (ApaI), rs10735810 (FokI), and rs1544410 (BsmI) in VDR; rs7041 and rs4588 in DBP; and rs10877012 and rs4646536 in CYP27B1. DNA was extracted from whole blood using the MaxiPrep Kit (QIAGEN, Sweden). SNPs were genotyped by TaqMan Allelic Discrimination Assay using the Quantstudio 7 Flex system. The genotyping results were confirmed by using positive controls during the genotyping as well as by re-genotyping of about 20% of the samples using the same genotyping method. The minor allele frequency for all SNPs was > 0.05. No deviation from Hardy-Weinberg equilibrium in the whole group or in the any of the subgroups was found. The success rate of genotyping was ≥96% (2 samples were not genotyped because of no available DNA). The genotyping protocol has been deposited in protocols.io. The DOI is: dx.doi.org/10.17504/protocols.io.bcjbiuin.
eQTL lookups
The association of the rs1544410 polymorphism with gene expression in human pancreatic islets was looked up in data from RNAseq data from 191 donors [https://www.biorxiv.org/content/10.1101/435743v2.full]. The data are uploaded with the following EGA accession numbers: RNAseq: EGAS00001004042, GWAS: EGAS00001004044 and Phenotype: EGAS00001004056.
Statistical analysis
All statistical analyses were performed using IBM SPSS Statistics for Windows, version 22.0 (IBM Corp., Armonk, NY). All genetic analyses were performed using PLINK version 1.07 (http://pngu.mgh.harvard.edu/~purcell/plink/index.shtml). Logistic and linear regressions with age, ethnicity, and body mass index (BMI) as covariates were used to estimate SNP associations and the data are presented as difference per allele or odds ratios (ORs) with 95% confidence intervals (CIs). The p-values are based on additive models for the genetic variants. Correction for multiple testing was performed using permutations.
Results
The clinical characteristics of the study subjects are presented in a previous publication [19]. The study included 376 women who were diagnosed with GDM during pregnancy in southern Sweden. At five years postpartum, 253 (67.3%) had normal glucose tolerance, 57 (15.2%) had diabetes, and for 66 (17.6%) there were missing data. The women were of European origin (n = 287, 76.3%) or non-European origin (n = 78, 20.8%) but 11 women (2.9%) with GDM were unclassifiable. The non-European group consisted of the subgroups Arab women (n = 33), Asian women (n = 35), and women of other origins (n = 10).
Of the SNPs tested, the A-allele of the rs1544410 polymorphism was associated with increased disposition index (difference per allele = 3.56, CI: 0.46–6.67; p = 0.03) using linear regression analysis with age, ethnicity, and BMI as covariates (Table 1).
Table 1. The association between the SNPs studied and disposition index.
| SNP | Associated allele | Frequency of associated allele | Difference in disposition index per allele (95% CI) | p-value |
|---|---|---|---|---|
| rs7041 | A | 0.41 | −0.47 (−2.71 to 1.76) | 0.68 |
| rs4588 | T | 0.38 | −0.62 (−3.14 to 1.91) | 0.63 |
| rs731236 | G | 0.38 | 0.25 (−1.90 to 2.40) | 0.82 |
| rs7975232 | C | 0.40 | −1.19 (−3.31 to 0.92) | 0.27 |
| rs10735810 | A | 0.38 | 0.62 (−1.59 to 2.82) | 0.58 |
| rs1544410 | A | 0.12 | 3.56 (0.46 to 6.67) | 0.03 |
| rs10877012 | T | 0.34 | 0.97 (−1.36 to 3.30) | 0.42 |
| rs4646536 | G | 0.34 | 1.05 (−1.30 to 3.37) | 0.38 |
The associations were tested by using linear regression analysis with age, ethnicity, and BMI as covariates.
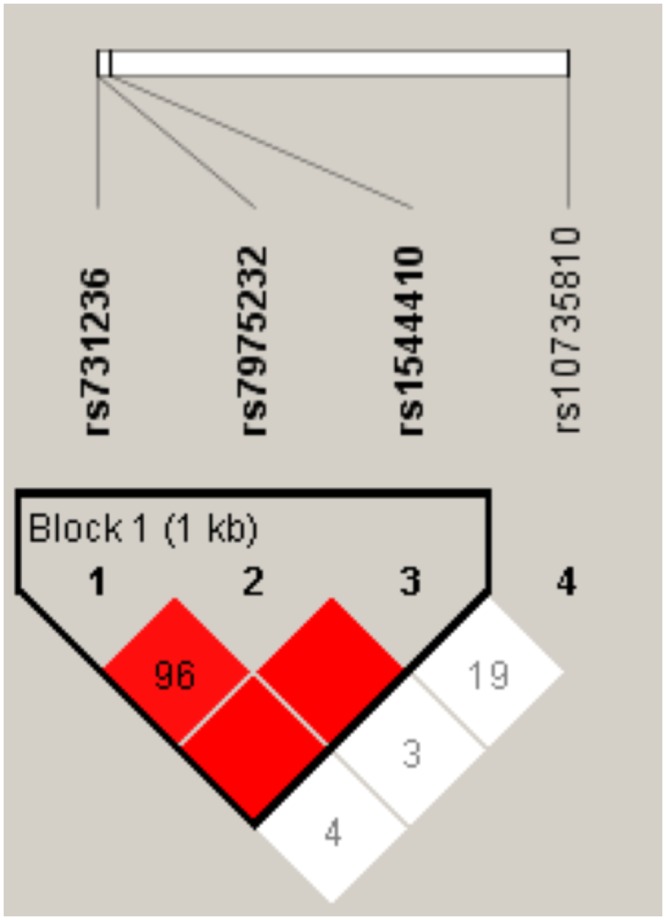
The rs731236 (Taql), rs7975232 (Apal), and rs1544410 (Bsml) polymorphisms of the VDR gene were in complete linkage disequilibrium (Fig 1), as previously shown [27]. However, the rs10735810 (FokI) polymorphism was not in linkage disequilibrium with the SNPs mentioned.
Fig 1. Linkage disequilibrium between the rs731236, rs7975232, rs10735810, and rs1544410 polymorphisms of the VDR gene.
However, we found no effect of the SNPs in question on the HOMA-IR, using linear multiple regression analysis (Table 2).
Table 2. The association between the SNPs studied and HOMA-IR.
| SNP | Allele | Allele Frequency | Difference in HOMA-IR per allele (95% CI) | p-value |
|---|---|---|---|---|
| rs7041 | A | 0.41 | 0.05 (−0.14 to 0.24) | 0.62 |
| rs4588 | T | 0.38 | 0.01 (−0.20 to 0.22) | 0.93 |
| rs731236 | G | 0.38 | −0.02 (−0.20 to 0.16) | 0.83 |
| rs7975232 | C | 0.40 | 0.01 (−0.16 to 0.18) | 0.91 |
| rs10735810 | A | 0.38 | −0.07 (−0.26 to 0.12) | 0.45 |
| rs1544410 | A | 0.12 | −0.07 (−0.34 to 0.20) | 0.60 |
| rs10877012 | T | 0.34 | −0.05 (−0.25 to 0.15) | 0.61 |
| rs4646536 | G | 0.34 | −0.004 (−0.20 to 0.20) | 0.97 |
The associations were tested by using linear regression analysis with age, ethnicity, and BMI as covariates.
In logistic regression analysis with age, ethnicity, and BMI as covariates, the SNPs studied showed no associations with postpartum diabetes in women with a history of GDM (Table 3).
Table 3. The association between the SNPs studied and postpartum diabetes.
| SNP | Associated allele | Frequency of associated allele | OR (95% CI) | p-value |
|---|---|---|---|---|
| rs7041 | A | 0.41 | 1.01 (0.66–1.54) | 0.98 |
| rs4588 | G | 0.62 | 1.42 (0.89–2.26) | 0.14 |
| rs731236 | A | 0.62 | 1.42 (0.95–2.12) | 0.09 |
| rs7975232 | C | 0.40 | 1.15 (0.77–1.72) | 0.50 |
| rs10735810 | G | 0.62 | 1.16 (0.76–1.78) | 0.48 |
| rs1544410 | A | 0.12 | 1.64 (0.81–3.30) | 0.17 |
| rs10877012 | G | 0.66 | 1.12 (0.73–1.73) | 0.60 |
| rs4646536 | A | 0.66 | 1.10 (0.71–1.68) | 0.68 |
The associations were tested using logistic regression analysis with age, ethnicity, and BMI as covariates.
The A-allele of the rs7041 polymorphism of the DBP gene was associated with a reduction in circulating 25(OH)D3 (difference [in nmol/L] per allele = −5.48, 95% CI: −8.32 to −2.64; p = 0.0002). The same was observed for the T-allele of the rs4588 polymorphism (difference [in nmol/L] per allele = −6.32, 95% CI: −9.47 to −3.17; p = 0.0001). The other SNPs did not show any effect on circulating levels of 25(OH)D3 (Table 4).
Table 4. The association between the SNPs tested and circulating 25(OH)D3.
| SNP | Associated allele | Frequency of associated allele | Difference in 25(OH)D3 (in nmol/L) per allele (95% CI) | p-value |
|---|---|---|---|---|
| rs7041 | A | 0.41 | −5.48 (−8.32 to −2.64) | 0.0002 |
| rs4588 | T | 0.38 | −6.32 (−9.47 to −3.17) | 0.0001 |
| rs731236 | G | 0.38 | 0.20 (−2.57 to −2.97) | 0.89 |
| rs7975232 | C | 0.40 | 1.01 (−1.71 to 3.73) | 0.47 |
| rs10735810 | A | 0.38 | −0.87 (−3.78 to 2.05) | 0.56 |
| rs1544410 | A | 0.12 | −0.66 (−4.75 to 3.42) | 0.75 |
| rs10877012 | T | 0.34 | −1.16 (−4.14 to 1.82) | 0.45 |
| rs4646536 | G | 0.34 | −1.14 (−4.08 to 1.81) | 0.45 |
The associations were tested using linear regression analysis with age, ethnicity, and BMI as covariates
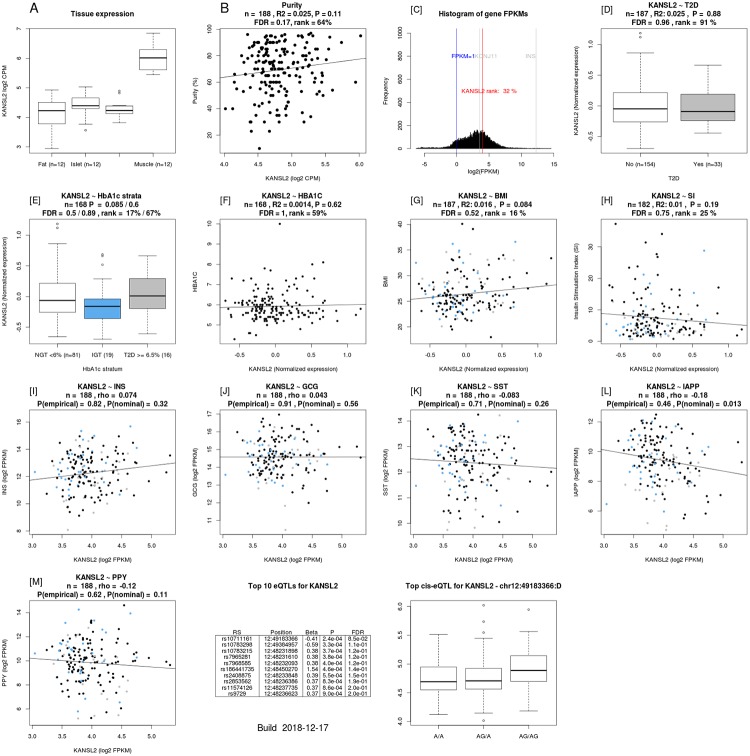
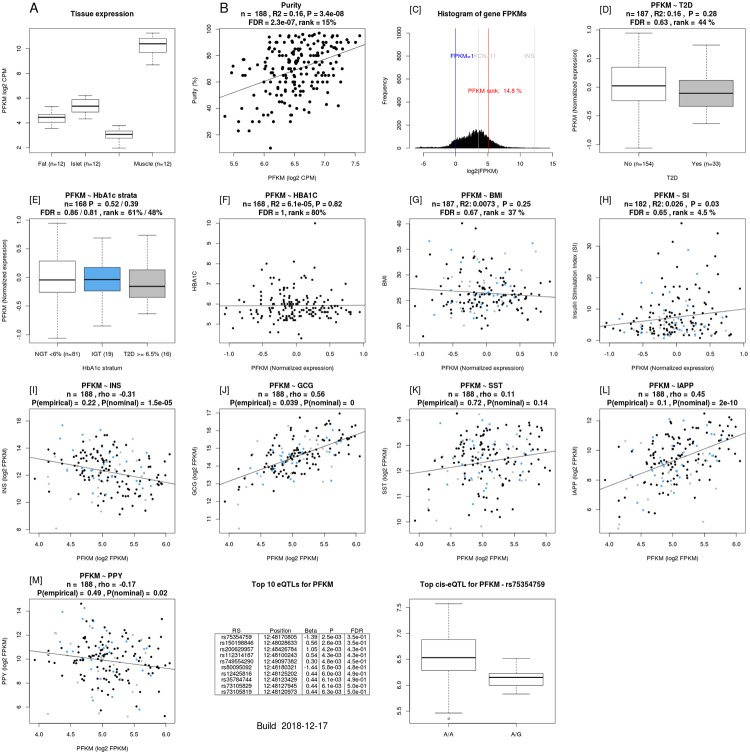
The rs1544410 polymorphism was an eQTL for the KAT8 regulatory NSL complex subunit 2 (KANSL2) and phosphofructokinase, muscle (PFKM) genes in human pancreatic islets [https://www.biorxiv.org/content/10.1101/435743v2.full] (Table 5). KANSL2 gene expression correlated with that of Islet Amyloid Polypeptide (IAPP) gene (Fig 2). PFKM expression correlated with stimulatory index (a measure of insulin secretion in islets), as well as with expression of insulin, glucagon, and IAPP genes (Fig 3) [https://www.biorxiv.org/content/10.1101/435743v2.full].
Table 5. The rs1544410 polymorphism was an eQTL for the KANSL2 and PFKM genes in human pancreatic islets.
| SNP | SNP_rsid | Gene name | beta | t-stat | p-value |
|---|---|---|---|---|---|
| 12:48239835 | rs1544410 | KANSL2 | 0,29472031 | 2,864151456 | 0,004661904 |
| 12:48239835 | rs1544410 | PFKM | 0,223429837 | 2,167193122 | 0,031491255 |
“Lookup” of significant associations in human pancreatic islets from 191 donors
Fig 2. KANSL2 gene expression is correlated with that of IAPP gene.
Fig 3. The correlation of the PFKM expression with stimulatory index as well as with expression of insulin, glucagon, and IAPP genes.
Discussion
We have recently shown that vitamin D deficiency/insufficiency in women with a history of GDM is associated with beta-cell dysfunction and insulin resistance [19]. The key finding of the current study was that the A-allele of the rs1544410 polymorphism of the VDR gene was associated with increased insulin secretion, measured as disposition index in women with a history of GDM. VDR is a member of the nuclear receptor superfamily of transcriptional regulators. It forms a heterodimer with a retinoid X receptor (RXR) and is widely distributed throughout the tissues, including pancreatic beta cells [28]. The 25(OH)D3 is widely used in assessment of vitamin D status. 25(OH)D3 is converted in the liver to 1,25(OH)2D, which is the main ligand for VDR [29]. When 1,25(OH)2D binds to this complex, it leads to the transcription of several vitamin D-regulated genes [23]. A relatively recent study has shown increased expression and production of major renin-angiotensin system components in isolated islets from VDR knockout mice incubated under high-glucose conditions [30]. This was prevented and reversed by 1,25(OH)2D in parallel with an increase in glucose-stimulated insulin secretion [30]. Ogunkolade et al. have shown that the TaqI (rs731236) genotype of the VDR gene was an independent determinant of the insulin secretion index in healthy Bangladeshi adults, who have a high risk of developing T2D [27]. The FokI (rs10735810) polymorphism of the VDR gene was found to be an additional independent determinant of insulin secretion in individuals with vitamin D insufficiency [27]. Furthermore, VDR mRNA expression has been shown to be a determinant of insulin secretory capacity [27]. Unlike our study, there was no direct association of the BsmI (rs1544410) polymorphism with insulin secretion. Interestingly, the TaqI t allele (i.e. the G allele in the present study) and the BsmI b (G) allele may be susceptibility alleles for diabetes in the Kashmiri population [11]. This is consistent with our present finding that the opposite allele, the A-allele, of the rs1544410 polymorphism was associated with increased disposition index, and therefore would prevent the development of diabetes. On the other hand, the TaqI t homozygotes were found to be associated with the highest levels of insulin secretion index compared to TT and Tt genotypes in healthy Bangladeshi adults, who have a high risk of developing T2D [27]. However, it should be noted that there was no adjustment for the degree of insulin resistance (i.e. disposition index), as we have done in the present study. Another explanation might be that the susceptibility alleles differ in different populations. Previous studies have shown tight linkage disequilibrium between the ApaI, BsmI, and TaqI genotypes [27, 31], and the same was found in our study (Fig 1). Our finding that the rs1544410 polymorphism was an eQTL for the PFKM gene in human pancreatic islets (Table 4) and that the PFKM expression correlated with stimulatory index (a measure of insulin secretion in islets), as well as with expression of INS, GCG, and IAPP genes (Fig 3) supports the role of this SNP in the insulin secretory capacity.
In the Rancho Bernardo study, the BsmI bb (GG) genotype was found to be associated with HOMA-IR after adjustment for age, sex, BMI, calcium, and vitamin D use in older adults without diabetes [32]. In the present study, we found no association between the polymorphisms tested and the HOMA-IR. Interestingly, a recent meta-analysis provided evidence of an association between VDR BsmI polymorphism and metabolic syndrome, supporting the idea that the VDR BsmI variant G allele may be a susceptibility marker of metabolic syndrome [12]. On the other hand, gene carriers with the VDR BsmI BB (AA) genotype have been found to have significantly higher levels of fasting glucose than gene carriers with the genotype Bb or bb in young men from Germany with low physical activity, but the effect was absent in men with a high degree of physical activity [33]. This discrepancy in the risk allele perhaps represents a gender difference.
In addition, we found that the A-allele of the rs7041 polymorphism and the T-allele of the rs4588 polymorphism of the DBP gene were associated with lower levels of circulating 25(OH)D3. The composite genotype of these 2 SNPs (rs7041 and rs4588) results in different DBP isotypes (Gc1f, Gc1s, and Gc2). These isotypes are known to influence the binding affinity of DBP and circulating vitamin D levels [34, 35]. Our results are consistent with previous findings that T-allele carriers (i.e. A-allele carriers in this study) of the rs7041 polymorphism had less circulating 25(OH)D3, regardless of reproductive state among third-trimester pregnant, lactating, and non-pregnant/non-lactating women [13]. Moreover, the TT genotype of the rs7041 polymorphism was found to be associated with metabolic syndrome in PCOS women, and with significantly lower 25(OH)D3 levels in both PCOS and control groups [15]. This was also shown in Jordanian subjects, where the genotypes containing the variant allele of rs7041 (TT, TG) and rs4588 (AA, AC) (i.e. T-allele in this study) were found to be associated with lower 25(OH)D3 levels than the wild-type genotypes (GG and CC, respectively) [14]. Interestingly, Rahman et al. found that the Glu/Glu genotype of the rs7041 and the Lys/Lys genotype of the rs4588 variants of DBP gene were significantly higher in subjects with type 2 diabetes than controls [16].
We have not observed any association between the SNPs studied and postpartum diabetes in the present study, which could be due to the small number of women who developed diabetes, to differences in ethnic background, to differences in study design, or simply to absence of an effect of these SNPs on the development of postpartum diabetes.
The main strength of the study was that we used the disposition index derived from the OGTT as a measure of beta-cell function, adjusted for insulin resistance. This is a more accurate measure of insulin secretory capacity than the HOMA-b [25]. However, the study still lacked enough statistical power (i.e. > 80%) to detect small potential effects of the SNPs in question on prediction of postpartum diabetes.
In conclusion, our study provides evidence that the rs1544410 polymorphism of the VDR gene may be associated with insulin secretion. Further studies in other populations will be needed to confirm the results.
Supporting information
(SAV)
Abbreviations
- 25(OH)D3
25-hydroxyvitamin D3
- BMI
body mass index
- CI
confidence interval
- CYP27B1
cytochrome P450 family 27 subfamily B member 1
- DBP
vitamin D binding protein
- GDM
gestational diabetes mellitus
- HOMA-IR
homeostasis model assessment of insulin resistance
- OGTT
oral glucose tolerance test
- OR
odds ratio
- SNP
single nucleotide polymorphism
- VDR
vitamin D receptor gene
Data Availability
All relevant data are within the manuscript and its Supporting Information files. EGA accession numbers are: RNAseq: EGAS00001004042 GWAS: EGAS00001004044 Phenotype: EGAS00001004056.
Funding Statement
The study was supported by grants from the Research Funds of Skåne University Hospital, the Skåne County Council Research and Development Foundation and ALF Region Skåne. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Metzger BE, Coustan DR. Summary and recommendations of the Fourth International Workshop-Conference on Gestational Diabetes Mellitus. The Organizing Committee. Diabetes Care. 1998;21 Suppl 2:B161–7. . [PubMed] [Google Scholar]
- 2.Fadl HE, Simmons D. Trends in diabetes in pregnancy in Sweden 1998–2012. BMJ Open Diabetes Res Care. 2016;4(1):e000221 10.1136/bmjdrc-2016-000221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collaboration NCDRF. Worldwide trends in diabetes since 1980: a pooled analysis of 751 population-based studies with 4.4 million participants. Lancet. 2016;387(10027):1513–30. 10.1016/S0140-6736(16)00618-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellamy L, Casas JP, Hingorani AD, Williams D. Type 2 diabetes mellitus after gestational diabetes: a systematic review and meta-analysis. Lancet. 2009;373(9677):1773–9. 10.1016/S0140-6736(09)60731-5 . [DOI] [PubMed] [Google Scholar]
- 5.Ben-Haroush A, Yogev Y, Hod M. Epidemiology of gestational diabetes mellitus and its association with Type 2 diabetes. Diabet Med. 2004;21(2):103–13. 10.1046/j.1464-5491.2003.00985.x . [DOI] [PubMed] [Google Scholar]
- 6.Wang H, Chen W, Li D, Yin X, Zhang X, Olsen N, et al. Vitamin D and Chronic Diseases. Aging Dis. 2017;8(3):346–53. 10.14336/AD.2016.1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forouhi NG, Luan J, Cooper A, Boucher BJ, Wareham NJ. Baseline serum 25-hydroxy vitamin d is predictive of future glycemic status and insulin resistance: the Medical Research Council Ely Prospective Study 1990–2000. Diabetes. 2008;57(10):2619–25. 10.2337/db08-0593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiu KC, Chu A, Go VL, Saad MF. Hypovitaminosis D is associated with insulin resistance and beta cell dysfunction. Am J Clin Nutr. 2004;79(5):820–5. 10.1093/ajcn/79.5.820 . [DOI] [PubMed] [Google Scholar]
- 9.Scragg R, Sowers M, Bell C, Third National H, Nutrition Examination S. Serum 25-hydroxyvitamin D, diabetes, and ethnicity in the Third National Health and Nutrition Examination Survey. Diabetes Care. 2004;27(12):2813–8. 10.2337/diacare.27.12.2813 . [DOI] [PubMed] [Google Scholar]
- 10.Knekt P, Laaksonen M, Mattila C, Harkanen T, Marniemi J, Heliovaara M, et al. Serum vitamin D and subsequent occurrence of type 2 diabetes. Epidemiology. 2008;19(5):666–71. 10.1097/EDE.0b013e318176b8ad . [DOI] [PubMed] [Google Scholar]
- 11.Malik R, Farooq R, Mehta P, Ishaq S, Din I, Shah P, et al. Association of Vitamin D Receptor Gene Polymorphism in Adults With Type 2 Diabetes in the Kashmir Valley. Can J Diabetes. 2018;42(3):251–6. 10.1016/j.jcjd.2017.06.003 . [DOI] [PubMed] [Google Scholar]
- 12.Han FF, Lv YL, Gong LL, Liu H, Wan ZR, Liu LH. VDR Gene variation and insulin resistance related diseases. Lipids Health Dis. 2017;16(1):157 10.1186/s12944-017-0477-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ganz AB, Park H, Malysheva OV, Caudill MA. Vitamin D binding protein rs7041 genotype alters vitamin D metabolism in pregnant women. FASEB J. 2018;32(4):2012–20. 10.1096/fj.201700992R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lafi ZM, Irshaid YM, El-Khateeb M, Ajlouni KM, Hyassat D. Association of rs7041 and rs4588 Polymorphisms of the Vitamin D Binding Protein and the rs10741657 Polymorphism of CYP2R1 with Vitamin D Status Among Jordanian Patients. Genet Test Mol Biomarkers. 2015;19(11):629–36. 10.1089/gtmb.2015.0058 . [DOI] [PubMed] [Google Scholar]
- 15.Santos BR, Lecke SB, Spritzer PM. Genetic variant in vitamin D-binding protein is associated with metabolic syndrome and lower 25-hydroxyvitamin D levels in polycystic ovary syndrome: A cross-sectional study. PLoS One. 2017;12(3):e0173695 10.1371/journal.pone.0173695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rahman MM, Hosen MB, Faruk MO, Hasan MM, Kabir Y, Howlader MZH. Association of vitamin D and vitamin D binding protein (DBP) gene polymorphism with susceptibility of type 2 diabetes mellitus in Bangladesh. Gene. 2017;636:42–7. 10.1016/j.gene.2017.09.008 . [DOI] [PubMed] [Google Scholar]
- 17.Bailey R, Cooper JD, Zeitels L, Smyth DJ, Yang JH, Walker NM, et al. Association of the vitamin D metabolism gene CYP27B1 with type 1 diabetes. Diabetes. 2007;56(10):2616–21. Epub 2007/07/04. 10.2337/db07-0652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cooper JD, Smyth DJ, Walker NM, Stevens H, Burren OS, Wallace C, et al. Inherited variation in vitamin D genes is associated with predisposition to autoimmune disease type 1 diabetes. Diabetes. 2011;60(5):1624–31. Epub 2011/03/29. 10.2337/db10-1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaat N, Ignell C, Katsarou A, Berntorp K. Glucose homeostasis, beta cell function, and insulin resistance in relation to vitamin D status after gestational diabetes mellitus. Acta Obstet Gynecol Scand. 2017;96(7):821–7. 10.1111/aogs.13124 . [DOI] [PubMed] [Google Scholar]
- 20.Anderberg E, Landin-Olsson M, Kalen J, Frid A, Ursing D, Berntorp K. Prevalence of impaired glucose tolerance and diabetes after gestational diabetes mellitus comparing different cut-off criteria for abnormal glucose tolerance during pregnancy. Acta Obstet Gynecol Scand. 2011;90(11):1252–8. 10.1111/j.1600-0412.2011.01214.x . [DOI] [PubMed] [Google Scholar]
- 21.Surveillance WHODoND. Definition, diagnosis and classification of diabetes mellitus and its complications: report of a WHO consultation. Part 1, Diagnosis and classification of diabetes mellitus. Geneva: World Health Organization. 1999.
- 22.Ignell C, Shaat N, Ekelund M, Berntorp K. The impact of ethnicity on glucose homeostasis after gestational diabetes mellitus. Acta Diabetol. 2013;50(6):927–34. 10.1007/s00592-013-0484-8 . [DOI] [PubMed] [Google Scholar]
- 23.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28(7):412–9. 10.1007/BF00280883 . [DOI] [PubMed] [Google Scholar]
- 24.Muniyappa R, Lee S, Chen H, Quon MJ. Current approaches for assessing insulin sensitivity and resistance in vivo: advantages, limitations, and appropriate usage. Am J Physiol Endocrinol Metab. 2008;294(1):E15–26. 10.1152/ajpendo.00645.2007 . [DOI] [PubMed] [Google Scholar]
- 25.Phillips DI, Clark PM, Hales CN, Osmond C. Understanding oral glucose tolerance: comparison of glucose or insulin measurements during the oral glucose tolerance test with specific measurements of insulin resistance and insulin secretion. Diabet Med. 1994;11(3):286–92. 10.1111/j.1464-5491.1994.tb00273.x . [DOI] [PubMed] [Google Scholar]
- 26.Jensen CC, Cnop M, Hull RL, Fujimoto WY, Kahn SE, American Diabetes Association GSG. Beta-cell function is a major contributor to oral glucose tolerance in high-risk relatives of four ethnic groups in the U.S. Diabetes. 2002;51(7):2170–8. 10.2337/diabetes.51.7.2170 . [DOI] [PubMed] [Google Scholar]
- 27.Ogunkolade BW, Boucher BJ, Prahl JM, Bustin SA, Burrin JM, Noonan K, et al. Vitamin D receptor (VDR) mRNA and VDR protein levels in relation to vitamin D status, insulin secretory capacity, and VDR genotype in Bangladeshi Asians. Diabetes. 2002;51(7):2294–300. 10.2337/diabetes.51.7.2294 . [DOI] [PubMed] [Google Scholar]
- 28.Bouillon R, Carmeliet G, Verlinden L, van Etten E, Verstuyf A, Luderer HF, et al. Vitamin D and human health: lessons from vitamin D receptor null mice. Endocr Rev. 2008;29(6):726–76. 10.1210/er.2008-0004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marquina C, Mousa A, Scragg R, de Courten B. Vitamin D and cardiometabolic disorders: a review of current evidence, genetic determinants and pathomechanisms. Obes Rev. 2019;20(2):262–77. 10.1111/obr.12793 . [DOI] [PubMed] [Google Scholar]
- 30.Cheng Q, Li YC, Boucher BJ, Leung PS. A novel role for vitamin D: modulation of expression and function of the local renin-angiotensin system in mouse pancreatic islets. Diabetologia. 2011;54(8):2077–81. 10.1007/s00125-011-2100-1 . [DOI] [PubMed] [Google Scholar]
- 31.Hitman GA, Mannan N, McDermott MF, Aganna E, Ogunkolade BW, Hales CN, et al. Vitamin D receptor gene polymorphisms influence insulin secretion in Bangladeshi Asians. Diabetes. 1998;47(4):688–90. 10.2337/diabetes.47.4.688 . [DOI] [PubMed] [Google Scholar]
- 32.Oh JY, Barrett-Connor E. Association between vitamin D receptor polymorphism and type 2 diabetes or metabolic syndrome in community-dwelling older adults: the Rancho Bernardo Study. Metabolism. 2002;51(3):356–9. 10.1053/meta.2002.29969 . [DOI] [PubMed] [Google Scholar]
- 33.Ortlepp JR, Metrikat J, Albrecht M, von Korff A, Hanrath P, Hoffmann R. The vitamin D receptor gene variant and physical activity predicts fasting glucose levels in healthy young men. Diabet Med. 2003;20(6):451–4. 10.1046/j.1464-5491.2003.00971.x . [DOI] [PubMed] [Google Scholar]
- 34.Arnaud J, Constans J. Affinity differences for vitamin D metabolites associated with the genetic isoforms of the human serum carrier protein (DBP). Hum Genet. 1993;92(2):183–8. 10.1007/BF00219689 . [DOI] [PubMed] [Google Scholar]
- 35.Lauridsen AL, Vestergaard P, Hermann AP, Brot C, Heickendorff L, Mosekilde L, et al. Plasma concentrations of 25-hydroxy-vitamin D and 1,25-dihydroxy-vitamin D are related to the phenotype of Gc (vitamin D-binding protein): a cross-sectional study on 595 early postmenopausal women. Calcif Tissue Int. 2005;77(1):15–22. 10.1007/s00223-004-0227-5 . [DOI] [PubMed] [Google Scholar]