Abstract
A key goal of cancer systems biology is to use big data to elucidate the molecular networks by which cancer develops. However, to date there has been no systematic evaluation of how far these efforts have progressed. In this Analysis, we survey six major systems biology approaches for mapping and modelling cancer pathways with attention to how well their resulting network maps cover and enhance current knowledge. Our sample of 2,070 systems biology maps captures all literature-curated cancer pathways with significant enrichment, although the strong tendency is for these maps to recover isolated mechanisms rather than entire integrated processes. Systems biology maps also identify previously underappreciated functions, such as a potential role for human papillomavirus-induced chromosomal alterations in ovarian tumorigenesis, and they add new genes to known cancer pathways, such as those related to metabolism, Hippo signalling and immunity. Notably, we find that many cancer networks have been provided only in journal figures and not for programmatic access, underscoring the need to deposit network maps in community databases to ensure they can be readily accessed. Finally, few of these findings have yet been clinically translated, leaving ample opportunity for future translational studies. Periodic surveys of cancer pathway maps, such as the one reported here, are critical to assess progress in the field and identify underserved areas of methodology and cancer biology.
Much as the Human Genome Project made it possible to quickly sequence all the genes in the genome, a big focus of systems biology has been to determine how these genes functionally interrelate, resulting in the generation of large molecular network and pathway maps. Over the past decade, pathway mapping has been markedly accelerated by a series of groundbreaking technologies, including gene editing1,2, nucleic acid sequencing3, proteomics4 and machine learning5. Fuelled by these advances, systems biology pathway maps have impacted nearly all facets of biology, including diverse subject areas such as developmental biology, neuroscience and immunity.
As with many modes of biological inquiry, many systems biology studies have been directed at cancer research. This focus is owing to the high cancer death rates in the general population (~16% of deaths worldwide6) and, more recently, to the large resources of systematic and quantitative data that have been generated to characterize patients with cancer, cell lines and animal models over multiple layers of molecular and phenotypic information. In particular, organizations such as The Cancer Genome Atlas (TCGA)7 and the International Cancer Genome Consortium (ICGC)8 have publicly released exomewide somatic mutations, copy number aberrations and in some cases methylomes, transcriptomes and proteomes for thousands of tumours spanning dozens of tumour types. Cancer systems biology has also grown to encompass numerous collaborative research efforts and institutes in most developed countries worldwide. For instance, the Cancer Systems Biology Consortium, supported by the US National Cancer Institute, has brought together dozens of universities and research institutes to comprehensively understand the complexity of cancer and to advance cancer diagnosis and treatment.
Despite all of this activity, to date there has been no systematic reckoning of how far cancer systems biology pathway mapping and modelling efforts have progressed, with respect to either classical molecular biology approaches or understanding cancer at large. To what extent have pathways derived from systems biology studies been able to recapitulate previous knowledge of major cancer signalling pathways? Have these same systems approaches allowed us to discover new cancerrelated pathways, and what are these pathways? Have the advances and discoveries in systems biology had meaningful impacts for patients with cancer?
To approach these questions, we focused on six major types of systems biology approaches that have been recently and repeatedly applied to map and model cancer pathways (Box 1). These approaches include genetic and protein interaction mapping, inference of gene regulatory networks, subnetwork identification methods and pathway modelling using constraintbased models or differential equations. For each type of systems approach, we surveyed published studies, identified their collections of cancer pathway maps and, when possible, downloaded their network representations (henceforth called ‘systems biology maps’ (SBmaps), Fig. 1). We then conducted an analysis to compare and contrast the extent to which these SBmaps agree with, or are divergent from, cancer pathways curated from the literature (literaturecurated pathways (LCpathways)). Next we evaluated whether these systems biology pathway mapping approaches have uncovered novel areas of biology or therapeutic opportunities. Lastly, we evaluated the extent to which systems biology methods and discoveries have been translated to the clinic. We apologize in advance to those authors whose work may have been omitted from this survey should their research fall outside the scope of this analysis or because of space limitations.
Box 1 | A diversity of methods in cancer systems biology included in this analysis.
Mapping of epistatic genetic interaction networks
a primary means of identifying functional relationships between genes, and the pathways in which those genes are active, is through mapping of epistatic genetic interactions104. when epistatic interactions are measured systematically across a panel of genes, these interactions tend to cluster genes with very similar functions, which allows the hierarchical organization of these genes into functional complexes and pathways105. the use of genetic interactions to map cellular wiring has been shown most powerfully in model systems such as Saccharomyces cerevisiae, in which systematic genetic manipulations have long been tractable106,107. with the advent of CrisPr–Cas9 gene editing technology, large-scale epistasis interaction mapping has begun to elucidate the architecture of pathways in human cancer cells28–32 (Fig. 2a).
Systematic mapping of protein–protein interactions
systematic measurement of biophysical protein interactions is often performed as a first step in understanding the structure and function of protein complexes or protein signalling pathways. widely used methods for protein interaction mapping include affinity purification–tandem mass spectrometry (AP–Ms/Ms), co-elution–mass spectrometry108, yeast two-hybrid screening109 and the protein-fragment complementation assay110. For example, in AP–Ms/Ms, a protein of interest is fused to one or more affinity tags. this ‘bait’ is then expressed in cancer cell lines and interrogated to identify physically interacting ‘prey’ proteins111,112 (Fig. 2b).
Inference of gene regulatory networks and upstream master regulators
A gene regulatory network is a collection of master regulators (for example, transcription factors and kinases) that interact to control downstream gene expression. A number of methods have been developed to infer these networks from genome-wide expression profiles75,113–116. ARACNE (algorithm for the reconstruction of accurate cellular networks), for example, analyses the gene expression profiles of tumours to reconstruct cancer-specific transcriptional interaction networks on the basis of the pairwise mutual information of each gene pair113,116. It then infers an optimal causal path through these gene pairs, which removes the vast majority of indirect interaction candidates (Fig. 2c). These inferred regulatory networks can then be used to identify critical transcriptional regulators driving a phenotype of interest (for example, metastasis), novel driver genes and potential mechanisms of action for drugs.
Heat diffusion for integration of protein networks with tumour mutations
Heat-diffusion subnetwork identification is a method used to map the genetic alterations of a cancer onto a protein interaction network to find driver pathways. Genetic alterations for each protein in the network are designated as a ‘heat source’, and this heat is then allowed to diffuse across the edges of the network, spreading the influence of each genetic alteration to neighbouring proteins. Cancer subnetworks are those subnetworks in which the nodes both send and receive a significant amount of heat (Fig. 2d). a number of network diffusion methods have been developed (for example, HotNet, HotNet2, ReMiC, VarWalker and NBS) for identification of cancer pathways117–121. the presence or absence of a mutation in each pathway can be used to stratify a tumour cohort to reveal distinct cancer subtypes and prognoses, as well as to predict differential drug response121,122.
Flux balance analysis for integration of metabolic networks with gene expression
Flux balance analysis (extensively reviewed by Bordbar et al.123) uses a model of metabolic pathways, encoded by a mathematical matrix, to estimate the fluxes through the enzymatic reactions of a cell (that is, their rates of substrate-to-product conversion in the steady state) (Fig. 2e). Fluxes are typically computed under the assumption that the cell population is growing optimally given the available metabolic precursors, an assumption that may not always apply in tumours. Fluxes may be additionally constrained by genome-wide expression data, which place reasonable upper and lower bounds on the rate of each reaction given the mrNa or protein levels of its enzymes. Given the metabolic network and gene expression profile of a tumour, this approach can indicate which metabolic reactions and pathways are most and least active across different conditions.
Mechanistic modelling of signalling networks using gene expression data
Mechanistic models such as ordinary differential equations or fuzzy logic have long been used to simulate the components of a biological system (for example, protein kinases of a cancer signalling network) and their dynamics, given their functional interrelationships. Ordinary differential equations, for example, specify the set of rules that govern how the states of interest vary over time and are most useful when the structural and functional connections of the system are well known. Once the structure has been specified, experimentally measured values for some of the states may be used to estimate the values of unknown states (for example, using measured phosphoprotein levels to estimate activities of upstream kinases). These simulations can then be compared with, or validated by, experimentally derived values. Mechanistic models are often used to simulate cancer signalling pathways (Fig. 2f).
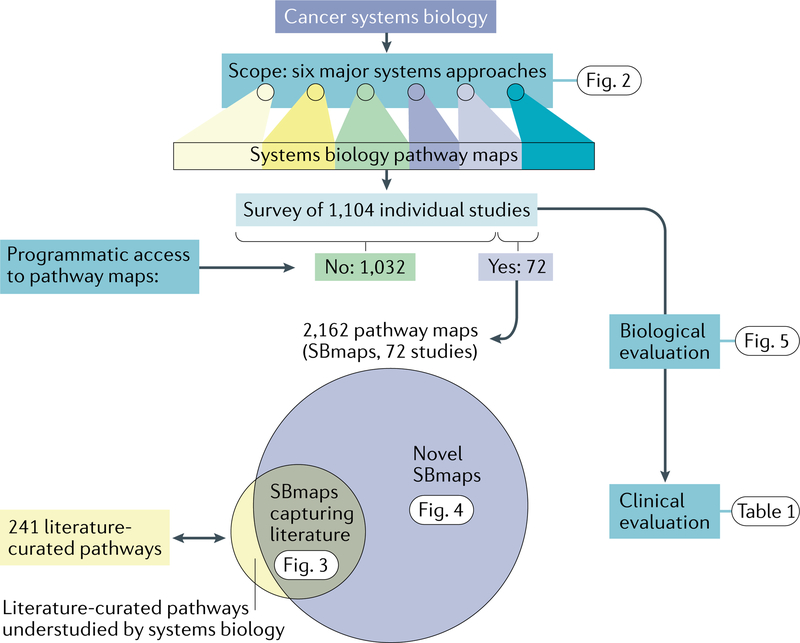
Fig. 1|. Structure of the analysis.
In the analysis presented here, we defined a scope of six major systems approaches used to map and model cancer signalling pathways. For these approaches, we identified publications referencing each of these six approaches. We then retrieved programmatically accessible pathway maps derived from systems biology studies, compared these maps with literature-curated cancer pathways and assessed the novel mechanisms emerging from these studies. Finally, we evaluated the extent to which systems biology methods and discoveries have been translated to the clinic. SBmaps, systems biology maps.
Collection of network and pathway maps
Our survey of cancer systems biology included approaches for direct measurement or inference of protein, genetic or transcriptional networks specific to cancer processes (Fig. 2a–c) as well as distinct methods to identify cancer networks through integration of general network databases with tumour molecular profiles (Fig. 2d–f; Supplementary methods). We surveyed literature in PubMed and identified 1,104 research articles that applied these systems biology approaches to cancer (Supplementary methods; Supplementary Table 1). We next sought to download the network maps from these studies, provided their network results were readily available in a format that could be parsed. We considered a network to be accessible if both gene/protein and interaction information was included (for example, adjacency matrix, interaction list or Cytoscape file). This step posed an unexpected challenge as we found that ~96% of studies insufficiently reported their resulting networks. For example, networks were provided only as static network images in figures, were summarized only in pathway or Gene Ontology9 enrichment tables, were summarized in difficulttoparse PDF, Microsoft Word or Excel formats or had only a small portion of the network available. In other cases, the list of genes, proteins and other molecules in the network was sufficiently reported but without machinereadable information about which pairs of these entities interconnect (we nonetheless chose to include these lists in our analysis, despite the absence of interactions). All identified networks were downloaded directly from supplementary material or reconstructed from shared raw data and standardized as gene lists representing the network. In total, our survey identified 2,162 cancer systems biology network maps from 72 publications (Supplementary Table 2; the data in network format are available in the Network Data Exchange10 (NDEx), an online commons where scientists can upload, store and share biological networks at all stages of development and publication11). We removed all networks with fewer than three genes or proteins, yielding 2,042 SBmaps for further analysis.
Fig. 2|. Cancer systems biology approaches covered in this analysis.
Six different approaches are discussed in this article. For additional details, see Box 1. a | Discovery of epistatic and functional gene interaction networks using genetic perturbation technologies such as CRISPR–Cas9. b | Discovery of protein–protein interactions, complexes and signalling networks in cancer-relevant contexts. An example of tandem affinity purification of a protein of interest coupled with liquid chromatography–tandem mass spectrometry (LC–MS/MS)-based proteomics is shown. Additional techniques are discussed in Box 1. c | Inference of gene regulatory networks and upstream master regulators. One example using ARACNE (algorithm for the reconstruction of accurate cellular networks)113 to assemble gene regulatory networks from tumour mRNA expression data is shown, where direct regulatory interactions between transcriptional regulators and target genes are inferred from gene expression data followed by the removal of many potential indirect interactions. Many other techniques to identify master regulators exist. Parts d–f show examples of integration of existing networks with tumour molecular profiles to identify molecular pathways and complexes altered in cancer. d | Integration of protein networks with tumour mutations using heat diffusion. Node colour corresponds to mutation frequency, modelled as heat. Black arrows represent heat diffusion to interacting proteins in the network. e | Integration of the glycolysis metabolic network with tumour gene expression using flux balance analysis. The metabolic solution space is shown with two example reactions, glucose (Gluc) to glucose 6-phosphate (G6P) (V1) and phosphoenolpyruvate (PEP) to pyruvate (PYR) (V2), with tumour biomass production as the objective function (Vobj). The constrained solution space represents the potential values of V1 and V2 that can be applied to maximize Vobj. f | Integration of signalling networks with tumour gene expression using ordinary differential equations (ODEs). Shown is an example ODE describing changes in cell cycle gene expression over time. F6P, fructose 6-phosphate; G3P, glyceraldehyde 3-phosphate; gRNA, guide RNA. Part d adapted from Leiserson et al.117, Springer Nature Limited.
To evaluate the degree to which these SBmaps covered extant knowledge of cancer processes, we collected a list of 241 LCpathways and made them available in NDEx12. These LCpathways included entries documented by the National Cancer Institute Pathway Interaction Database effort, the NetPath collection of cancer signalling pathways and the Signalling Network Open Resource (SIGNOR); overlapping pathways were merged where appropriate13–15 (Supplementary Table 3). Functional enrichment analysis indicated that both SBmaps and LCpathways were broadly representative of cancer processes, including antiapoptotic and proliferative signalling, DNA replication and repair, immune response, metabolism, migration and metastasis, and the tumour microenvironment (Fig. 3a). Relative to LCpathways, SBmaps showed higher coverage of cell proliferation and DNA repair functions and substantially lower coverage of immune pathways, likely reflecting the relative ease or difficulty, respectively, of interrogating these processes in systems biology studies.
Fig. 3|. Coverage of LCpathways by SBmaps.
a | Functional enrichment was performed for each systems biology map (SBmap) (left pie chart) using the Gene Ontology biological process branch. Significant terms (hypergeometric test, P < 10−8) were sorted by the number of SBmaps enriched. The top 300 were retained and organized under six broad process categories (colours). Each pie slice represents the number of SBmaps enriched within each category (note a map with multiple functional enrichments can be counted multiple times). A separate but identical analysis was performed for literature-curated pathways (LCpathways) (right pie chart). b | Analysis of overlap between SBmaps and LCpathways. Blue fill represents the proportion of SBmaps (right) that were significantly enriched for the genes of an LCpathway (left) (hypergeometric test, P < 10−8). Grey fill represents the proportion of SBmaps that were not enriched for an LCpathway. Lines connect each SBmap to the significantly overlapping LCpathway for which it had the highest recall. c | Precision–recall plot showing the best SBmap recovery of each LCpathway (points). The best-matching SBmap is selected according to the F score (point colours and contours), a combined measure of precision and recall. d | Specific precision and recall analysis for an example SBmap (Zhang118_27559151)18. Precision and recall are computed against each LCpathway (points). The best recall of an LCpathway is for vascular endothelial growth factor (VEGF) and VEGF receptor (VEGFR) signalling, displayed at the bottom. Red nodes indicate genes/proteins covered by both the SBmap and the matching LCpathway. Blue or green nodes indicate genes/proteins specific to the SBmap or LCpathway, respectively. High-confidence interactions (0.7 or greater) between genes in this network were obtained from STriNg124. e | Histograms of the number of genes/proteins belonging to each SBmap (top) or LCpathway (bottom). For display purposes, this analysis is limited to SBmaps with 200 or fewer genes (representing more than 95% of SBmaps). f | Network diagram of the LCpathway ‘β−3-integrin signalling’ and SBmap Park318_26635139 (reF.19). The colour mapping is the same as in part d.
Comparative evaluation of systems biology maps versus literature curation.
We next evaluated the overlap between the sets of genes assigned to each LCpathway and SBmap. Strikingly, SBmaps were enriched for all 241 LCpathways (P < 1 × 10–8 by the hypergeometric test, minimum two overlapping genes), with each LCpathway matching to 32 SBmaps on average (Fig. 3b). To understand the amount of overlap represented by this significant enrichment, each LCpathway was paired with the matching SBmap using the highest F score16, an accuracy measure combining precision (the fraction of SBmap genes present in the LCpathway) and recall (the fraction of LCpathway genes present in the SBmap). Indeed, certain LCpathways had been recapitulated by cancer systems biology approaches with both high precision and high recall (F > 0.5; Fig. 3c). An example is the vascular endothelial growth factor (VEGF) and VEGF receptor (VEGFR) pathway, which governs angiogenesis and plays a key role in development and metastasis of tumour types such as colorectal, breast and lung cancer, where VEGFR inhibitors are currently approved for treatment17. The VEGF and VEGFR signalling pathway was closely captured by an SBmap, Zhang118_27559151, emerging from the integration of mutation data with protein interaction networks18 (Fig. 3d). This same SBmap also covered a highly related LCpathway, VEGFR1specific signalling (Fig. 3d). Other LCpathways well captured by SBmaps included pololike kinase 3 (PLK3) signalling (SBmap Park263_26635139 (reF.19)), ERBB growth factor receptor signalling (SBmap Creighton2_23792563 (reF.20)), autophagy (SBmap Zhang155_27559151 (reF.18)), Hippo signalling (SBmap Xiong1_29983373 (reF.21)) and circadian rhythms (SBmap Grasso7_22722839 (reF.22)) (Fig. 3c).
Beyond these anecdotes, we found that more than 70% of LCpathways were matched by SBmaps with relatively low recall (less than 0.3) (Fig. 3c). This result was readily explained by the further observation that LCpathways tend to be much larger than SBmaps produced by systems biology approaches (median size 36 genes versus 8 genes; Fig. 3e). For example, the β3 integrin cell surface interaction pathway was recovered with high precision by an SBmap (Park318_26635139, precision 0.81)19; however, as this SBmap had 15 genes versus 43 genes in the LCpathway, this recovery was of relatively low recall (0.2; Fig. 3f).
We also observed LCpathways for which the best matching SBmap had both low precision and low recall, despite this overlap having been scored significant by the hypergeometric test. These poorly covered pathways included those involving GTPases: the RHOA signalling pathway, which is important in many cellular processes, including cytoskeletal organization and cell adhesion, regulation of CDC42 activity, which is an important regulator of cell cycle progression, and regulation of RAC1 activity, which controls many cancerrelated processes, including glucose transport and activation of kinase signalling23. Together, these results suggest that many SBmaps capture focused or isolated mechanisms within LCpathways, but they do not comprehensively recapitulate all genes involved in these LCpathways.
Biases in systems biology pathway maps
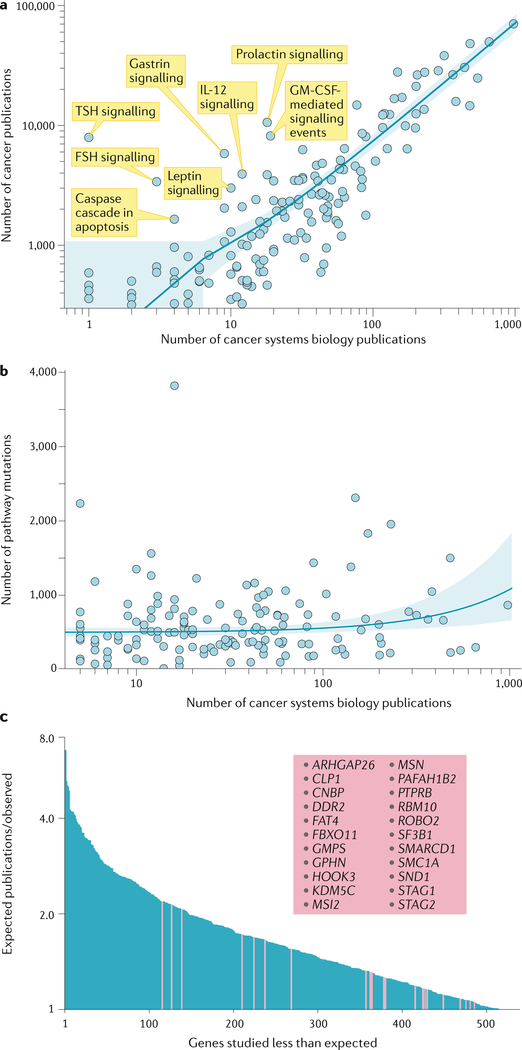
As expected, the number of systems biology publications exhibited a strong correlation with the total number of cancer publications for each pathway overall (Fig. 4a). To gain insights into the reasons why certain pathways are studied more than others, we next compared the number of cancer systems biology publications for each LCpathway with the aggregate mutation frequency of each gene in the LCpathway based on mutational data from TCGA. Strikingly, the amount an LCpathway was studied did not correlate with the aggregate mutation frequency (pancancer; Fig. 4b). These results suggest that the focus of cancer systems biology has been influenced less by the clinical importance of a pathway, in terms of the number of mutations observed across cancer types, than by issues such as the presence of prior publications, funding support for hot topics and the availability of relevant experimental reagents (for example, specific antibodies and inhibitors needed to study a pathway). Such factors may influence researchers to focus on bettercharacterized pathways and away from riskier novel leads.
Fig. 4|. Assessment of relative research coverage of cancer pathways by systems biology.
a | Scatterplot showing, for each literature-curated pathway (LCpathway), the number of publications related to that pathway overall versus the number of cancer publications specifically related to cancer systems biology. The number of publications was retrieved using custom PubMed search terms for both cancer systems biology and cancer publications for each LCpathway (listed in Supplementary Table 3). Selected pathways with few systems biology publications relative to overall cancer publications are labelled. The line represents the fit of the linear regression model, with the 95% confidence interval shown as the shaded area (note the log–log axes warp the linear fit). b | Scatterplot showing, for each LCpathway, the number of cancer systems biology publications related to that pathway versus the aggregate mutation count for that pathway (determined with The Cancer Genome Atlas Pan-Cancer Atlas7). The line represents the fit of the linear regression model, with the 95% confidence interval shown as the shaded area (note the log–log axes warp the linear fit). c | Waterfall plot of the number of expected publications divided by the number of observed publications for experimentally accessible genes (that is, there are antibodies, inhibitors and/or assays available to study them). The expected number of publications is based on a statistical model using chemical, physical and biological features of each gene24. Cancer genes are highlighted in pink, with these gene names listed. FSH, follicle- stimulating hormone; GM-CSF, granulocyte–macrophage colony-stimulating factor ; IL-12, interleukin-12; TSH, thyroid-stimulating hormone.
In addition to these biases, we found that a number of hormonerelated LCpathways (for example, folliclestimulating hormone and thyroidstimulating hormone signalling pathways) were more widely studied by traditional methods than by systems biology (Fig. 4a), which was also reflected by their relatively low F score (mean 0.14 ± 0.08). This bias is likely because much of cancer systems biology research is performed in cell line models, whereas the role of many hormone signalling pathways becomes clear only in vivo, where there is an intact endocrine system. To further quantify the extent to which some cancer genes are understudied despite having the appropriate tools and technology, we consulted a statistical model of the number of expected publications for a gene based on its individual chemical and biological characteristics24. Using this model, we identified 22 experimentally accessible cancer genes/proteins (that is, with antibodies, inhibitors and/or assays available to study them) that had been studied significantly less than expected, including genes that are highly mutated in cancer with known roles in oncogenesis (for example, the genes encoding the discoidin domain receptor tyrosine kinase DDR2 (reF.25) and the RNAbinding protein RBM10 (reF.26); Fig. 4c).
Potential new mechanisms
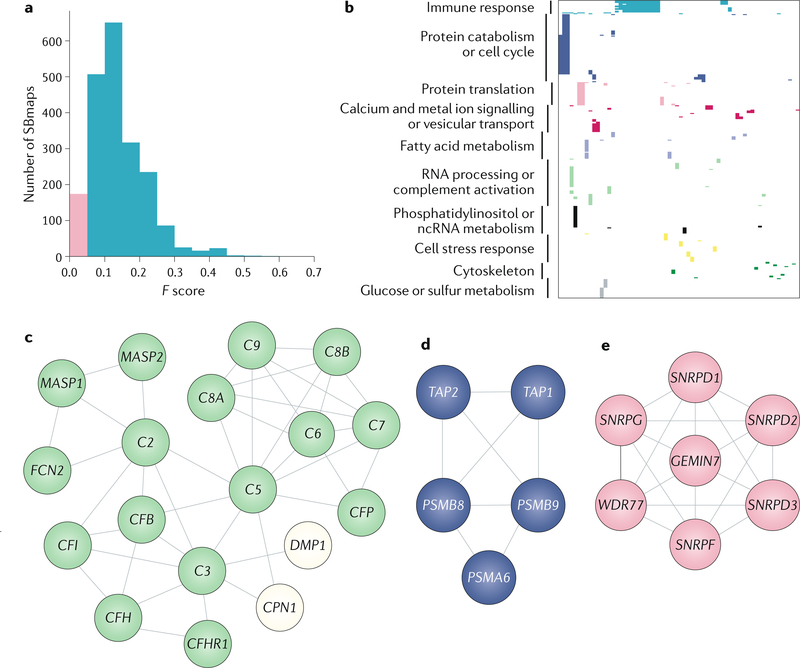
Approximately 9% (190/2,120) of the SBmaps were novel in that they did not significantly map to any LCpathways (F < 0.05; Fig. 5a,b). Most of these maps also were not enriched for any known biological processes with use of Gene Ontology9 (63%, 120/190; Supplementary Table 4). The remaining novel SBmaps (37%) were enriched for processes including numerous metabolic pathways, protein catabolism, cell cycle, vesicular transport, RNA processing, complement activation and protein translation (Fig. 5c–e). In the following sections we discuss some of the novel findings uncovered from our analysis of SBmaps as well as the findings from select studies that did not have an SBmap explicitly available.
Fig. 5|. representative SBmaps not previously reported in the literature.
a Histogram of best F score for each systems biology map (SBmap). The pink area indicates SBmaps with F < 0.05 selected for further analysis. b | Clustering of enriched Gene Ontology (GO) processes for SBmaps with F < 0.05. SBmaps without GO enrichment are not shown. Cluster names were determined by the most prevalent GO term enrichments; as such the processes listed together are not necessarily connected biologically. Parts c–e show example heat diffusion-derived SBmaps from the analysis in part b. c | Olcina27_30590044 (reF.125). d | Babaei9_23343428 (REF126.). e | Zhang133_27559151 (reF.18). Node colour corresponds to genes with functions shown in part b. Green nodes map to RNA processing and complement activation functions. Blue nodes map to protein catabolism and cell cycle functions. Pink nodes map to protein translation functions. White nodes do not map to a function. Edges show high-confidence (0.7 or greater) interactions as scored by STRING124. ncRNA, non-coding RNA.
Potential new mechanisms in cancer metabolism.
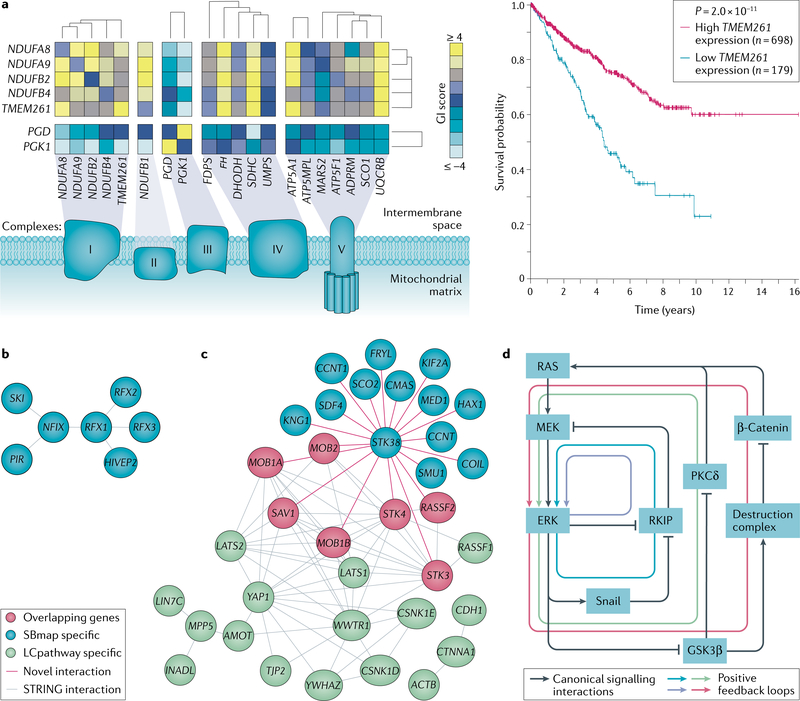
Dysregulated cellular metabolism is one of the longest studied phenomena in cancer research, beginning with the seminal work of Otto Warburg nearly 100 years ago27. Intriguingly, systemslevel studies continue to identify novel interactions and aspects of cancer metabolism. For example, recent advances in CRISPR–Cas9 gene editing technology and largescale epistasis interaction mapping have begun to elucidate the architecture of metabolic pathways in human cancer cells28–32. A study mapping epistatic genetic interactions in leukaemia cells using CriSPr interference28 has helped define a number of pathways and protein complexes, including one potentially important in human chronic lymphocytic leukaemia metabolism. Here the authors constructed a large lossoffunction genetic interaction map by testing the pairwise silencing of 222,784 gene pairs (472 × 472 genes) previously shown to be important for cell proliferation. By clustering the identified genetic interactions, the authors were able to reconstruct a number of pathways and protein complexes such as mTOR signalling, the proteasome complex and nucleotide excision repair. In addition, the authors were able to identify novel components of pathways and protein complexes such as assignment of the protein encoded by the largely uncharacterized gene TMEM261 as a potential component of the electron transport chain (Horlbeck28_30033366; Fig. 6a)28. The electron transport chain controls oxidative phosphorylation, which drives aerobic ATP production and has been suggested to be a potential target for cancer treatment33. TMEM261 expression has been shown to correlate with favourable prognosis in endometrial and renal cancer (Human Protein Atlas34), suggesting a potentially important role in several cancer types (Fig. 6a).
Fig. 6|. Potential new mechanisms emerging from cancer systems biology studies.
a | Example genetic interaction map demonstrating the recovery of complexes I, II, III, IV and V of the electron transport chain (left). Colour corresponds to the genetic interaction score (GI score) upon CRISPR knockdown of a pair of genes (row × column). The systems biology map (SBmap) is available: Horlbeck28_30033366. TMEM261 was found to be involved in the electron transport chain from this genetic interaction map. A Kaplan–Meier plot is shown demonstrating the prognosis of a patient with renal cancer based on high or low TMEM261 mRNA expression based on the median fragments per kilobase of exon per million reads (right). The logrank P value is shown. Data for the Kaplan–Meier plot were retrieved from the Human Protein Atlas. Similar results exist for endometrial cancer. b | Example SBmap, Bell16_21720365 (reF.48), which was not enriched for any known biological processes and may have a potential role in human papillomavirus-related ovarian oncogenesis. c | Merged network diagram of the literature-curated pathway (LCpathway) ‘Hippo signalling’ and SBmap Xiong1_29983373 (reF.21). Pink nodes indicate genes covered by both the SBmap and the matching LCpathway. Blue or green nodes indicate genes specific to the SBmap or LCpathway, respectively. Grey edges represent high-confidence (0.7 or greater) gene interactions from STRING124. Pink edges represent novel interactions identified from affinity purification–tandem mass spectrometry. d | Wiring diagram showing positive feedback loops involving members of the RAS signalling pathway (RAS, MEK and ERK), WNT signalling pathway (glycogen synthase kinase 3β (GSK3β), destruction complex and β-catenin) and epithelial–mesenchymal transition pathway (Snail, RAF kinase inhibitor protein (RKIP) and protein kinase Cδ (PKCδ)), where the presence of feedback was identified through mathematical modelling. Black arrows represent canonical signalling interactions. Coloured arrows represent each of the four identified positive feedback loops. Top left panel of part a adapted with permission from Horlbeck et al.28, Elsevier. Part d adapted with permission from the American Association for Cancer Research: Shin, S.-Y. et al. Functional Roles of Multiple Feedback Loops in Extracellular Signal-Regulated Kinase and Wnt Signaling Pathways That Regulate Epithelial-Mesenchymal Transition. Cancer Res. 70, 6715–6724, https://doi.org/10.1158/0008–5472.CAN-10–1377 (2010)60.
Genomewide metabolic models have also begun to provide insights into how metabolic networks influence cancer processes such as immune activation or evasion. For example, Bordbar et al.35 developed a genomescale metabolic model of macrophages using flux balance analysis to determine the metabolites and metabolic pathways important for macrophage activation. Not only was this model able to identify metabolites associated with immune activation (for example, glucose and arginine) and immune suppression (for example, tryptophan and vitamin D3), but these model predictions were also supported by multiomic analysis (transcriptomics, proteomics and metabolomics) of macrophage activation. This study suggested that the metabolic capabilities of effector immune cells (for example, production and excretion of metabolites) may act as remote sensors that mediate the local immune activation status of macrophages and other immune cells. This finding has been supported by a number of other studies demonstrating that alterations of the concentrations of specific metabolites in the tumour microenvironment reduce detection of tumours by the immune system36. There have been numerous attempts at similar genomescale models of human metabolism, spanning thousands of genes, metabolic reactions and metabolites37–46. These models have been used to identify novel associations between cancer proliferation and the Warburg effect, to stage tumours on the basis of metabolic states and to identify novel oncometabolites and drug opportunities, such as targeting recurrently mutated metabolic enzymes (for example, isocitrate dehydrogenase 1 (IDH1) or fatty acid synthase (FASN)), whose gain of function can result in promiscuous catalytic activities, thereby perturbing the metabolic network43–46. Other models have focused on the bestknown metabolic pathways in cancer and were able to accurately predict cancer cell growth rates, global metabolic activity during proliferation and response to metabolicbased therapy47.
Potential new mechanisms in cancer initiation and progression.
While many SBmaps recapitulate portions of LCpathways, some capture fundamental new pathway directions for cancer research. Largescale systemslevel analyses are uncovering a number of novel routes for the initiation and progression of cancer. For instance, the SBmap Bell16_21720365 (reF.48), derived from network diffusion of ovarian tumour mutation and DNA copy number alteration profiles, was among those that were not enriched for any known functions in our systematic comparison of SBmaps and LCpathways. We found that this SBmap interconnected genes within chromosomal regions that have been shown to have copy number alterations (for example, RFX1, RFX2, RFX3, SKI and NFIX) or expression changes (for example, PIR) from chromosomal alterations induced by human papillomavirus (HPV) infection49–53 (Fig. 6b). These genetic alterations are a result of deregulated E6 and E7 HPV proteins, which lead to chromosomal instability through inhibition of p53 and the induction of oxidative stress leading to the accumulation of specific (epi)genetic changes in the genome54,55. This SBmap may therefore be associated with HPVinduced ovarian carcinogenesis, a littlestudied and controversial phenomenon56–58.
Some SBmaps capture only a portion of an LCpathway but add new interactions between genes to welldescribed pathways. For example, a recent study of the interactome of the serine/threonine kinase NDR1 (also known as STK38), which is involved in cell proliferation, using a FLAGtagged NDR1 for affinity purification of NDR1binding proteins coupled with liquid chromatography–tandem mass spectrometry identification, significantly recapitulated the Hippo signalling pathway (Xiong1_29983373, F = 0.49)21. This SBmap was thus enriched for a number of major Hippo pathway proteins, including MOB1A, STK4, STK3, SAV1 and RASSF2 (reFS21,59). In addition to recapitulating these major Hippo signalling components, the study authors identified and validated FRYL as a novel interactor of NDR1 and potential pathway component (Fig. 6c). Intriguingly, FRYL is an undercharacterized transcriptional coactivator (only 12 publications in PubMed referencing FRYL at the time of this writing), the gene of which is mutated in ~12% of patients with cutaneous melanoma (TCGA PanCancer Atlas7), suggesting a potentially important role in cancer.
Potential new mechanisms in feedback loops and therapeutic resistance.
Mechanistic models of cancer signalling have been extensively used to study complex phenomena such as feedback loops60, crosstalk between pathways61–65, oncogene addiction66 and response to therapy67–71. For example, Shin et al.60 showed that the influence of ERK and WNT signalling pathways on epithelial–mesenchymal transition is greatly affected by multiple positive feedback loops. Through extensive ordinary differential equation modelling of combinations of feedback loops, they found that activated ERK counteracts protein kinase Cδ (PKCδ) inhibition by glycogen synthase kinase 3β (GSK3β), leading to decreased Ecadherin expression and a more mesenchymal phenotype. Conversely, a RAF kinase inhibitor protein (RKIP) feedback loop had the opposite effect (SBmap and model code not available; Fig. 6d). Together these simulations have demonstrated that a small number of signalling components can encode biological specificity in epithelial–mesenchymal transition, a finding supported by other modelling approaches60,72.
Similarly, a highthroughput protein interaction screen identified the kinase NUAK2 as a novel YAP/ TAZ activator (SBmap not available), which represents a positive feedforward loop driving Hippo signalling and cancer progression73. Loss of NUAK2 activity further decreased cell proliferation and tumour growth, suggesting NUAK2 as a potential therapeutic target for tumours driven by Hippo signalling. Interactome profiling has even begun to reveal mutationspecific rewiring of protein complexes resulting in altered therapeutic efficacy. A recent study of protein phosphatase 2A (PP2A) using affinity purification–mass spectrometry found that PP2A with the recurrent tumourpromoting R183W mutation rewires the PP2A interactome towards striatin-interacting phosphatase and kinase (STriPAK) and integrator complex components (SBmap not available)74. These interactions lead to MAPK pathway activation and reduced MEK inhibitor sensitivity.
Similarly, novel driver genes75 as well as mechanisms of response and resistance to therapy76–78 have been uncovered by studying the underlying gene regulatory networks. For example, master regulator computational analysis (Box 1) following treatment with the BET bromodomain inhibitor JQ1 in castrationresistant prostate cancer (SBmap not available) found 23 altered master regulators that contribute to JQ1 activity76. The study authors subsequently validated three of these (CBX3, MCM2 and MCM5) but the full activity of JQ1 likely involves a combination of all 23. Use of BET bromodomain inhibitors is a promising strategy to treat aggressive subtypes of castrationresistant prostate cancer that no longer respond to androgen receptortargeting agents. Moving forward, resistance to BET inhibitors is inevitable; however, a deep understanding of these master regulators could provide strategies to prevent BET inhibitor resistance. A similar study was performed to find potential mechanisms of resistance involving master regulators following treatment with a PI3K and mTOR inhibitor or a MEK inhibitor in basallike breast cancer77. The study authors found increases in activity of MAPK, BCL2 and nuclear factorκB (NFκB) signalling pathways in PI3K and mTOR inhibitortreated cells and increased PI3K, integrin and JUN Nterminal kinase (JNK) signalling pathway activation in MEK inhibitortreated cells. Inhibition of these master regulator pathways in combination with either the PI3K and mTOR inhibitor or the MEK inhibitor enhanced the efficacy of these drugs and could therefore help to prevent or overcome resistance to these therapies.
Progress to clinical systems biology
Although clinical translation of systems biology has arguably been slow, this pace parallels the slow progress of translation seen for molecular and ‘omics’ research more generally79,80. For instance, technological advances in proteomics, genome sequencing and gene expression profiling have resulted in more than 150,000 articles documenting thousands of biomarkers; however, fewer than 100 of these biomarkers have been validated for routine clinical practice79.
Despite these caveats, there has indeed been some very recent progress in implementing systems biology methods and discoveries in clinical practice (TABle 1). For example, the investigational VeriStrat test scores patient prognosis in lung cancer using mass spectrometrybased proteomics and a k-nearest neighbours model. The model assigns patients to either good or poor clinical outcomes on the basis of features of mass spectral peaks81,82. Similarly, there have been a number of efforts to create gene expression classifiers for breast cancer diagnosis and prognosis, such as the Genomic Grade Index83, PAM50 (reF.84), Breast Cancer Index85,86, EndoPredict87, RxPonder88, Oncotype DX89 and the US Food and Drug Administration (FDA)approved breast cancer prognostic test MammaPrint90. While many of the abovementioned advances have been for breast cancer, similar tests are being developed for lung cancer91 and acute lymphoblastic leukaemia92, among other cancers. The overall efficacy of such tests is not entirely characterized, as prognostic tests such as Oncotype DX and MammaPrint have been shown to have lowtomoderate concordance with one another93. For this reason, some consider prognostic gene signatures to have limited clinical utility at present and have suggested revised clinical trial designs for their effective evaluation94. Nonetheless, two multigene diagnostic tests measuring cancerassociated mutations have also recently been approved by the FDA (MSKIMPACT and FoundationOne CDx), which are the first multigene diagnostic tests to be approved for clinical use95,96.
Table 1 |.
example clinical trials implementing systems biology methods or discoveries
| Systems approach | Cancer type | Trial name | Phase or type of trial | Year started | Current status | Clinical trial identifiera |
|---|---|---|---|---|---|---|
| Gene expression profiling | ||||||
| Oncotype DX | Breast | TAILORx | Phase 3 | 2006 | Active, not recruiting | NCT00310180 |
| MammaPrint | Breast | MINDACT | Phase 3 | 2007 | Active, not recruiting | NCT00433589 |
| Oncotype DX or MammaPrint | Breast | OPTIMA | Phase 3 | 2012 | Recruiting | ISRCTN42400492 |
| Transcriptome and FoundationOne CDx | Multiple | WINTHER | NAb | 2013 | Suspended | NCT01856296 |
| Gene expression signatures | Prostate | PROVENGE | Prospective | 2014 | Complete | NCT02237170 |
| 10-gene expression signature | Thyroid | ThyroidPrint | Prospective | 2017 | Recruiting | NCT03309631 |
| 11-gene expression signature | Sarcoma | PREDISARC | Prospective | 2018 | Recruiting | NCT03625791 |
| Mutation profiling | ||||||
| MSK-IMPACT | Multiple | MSK-IMPACT | Prospective | 2013 | Recruiting | NCT01775072 |
| Transcriptome and FoundationOne CDx | Multiple | WINTHER | NA | 2013 | Suspended | NCT01856296 |
| 38-gene mutation profiling | Colorectal cancer | GENESIS | NA | 2015 | Complete | NCT02595645 |
| Gene mutation profiling and radiomics | Hepatocellular carcinoma | Med-HCC-1 | Observational | 2015 | Active, not recruiting | NCT02372162 |
| FoundationOne CDx | Multiple | I-PREDICT | Observational | 2015 | Recruiting | NCT02534675 |
| Other systems approaches and discoveries | ||||||
| Gene mutation profiling and radiomics | Hepatocellular carcinoma | Med-HCC-1 | Observational | 2015 | Active, not recruiting | NCT02372162 |
| Single-cell analysis | Melanoma | PEMSYS | Phase 2 | 2018 | Recruiting | NCT03534635 |
| Polypharmacologyc | Non-small cell lung cancer | Ceritinib Plus Docetaxel | Phase 1 | 2018 | Recruiting | NCT03611738 |
Clinical trials were selected to highlight ongoing trials that cover various tissue types and to highlight additional efforts beyond those explicitly discussed. NA, not applicable.
Clinical trials with ‘NCT’ identifiers can be accessed in the ClinicalTrials.gov database. The ISRCTN42400492 trial can be accessed in the ISRCTN registry.
Not applicable is used to describe trials without FDA-defined phases.
Polypharmacology is the design or use of a single drug to target multiple pathways and an extension of network medicine. This trial is the first trial implementing a polypharmacology compound to target multiple pathways simultaneously.
Currently, many clinical trials are designed to test a single marker (for example, efficacy of inhibitors of ALK receptor tyrosine kinase in patients harbouring ALK rearrangements)97. Such has been the paradigm for advancing individual targeted therapies. In contrast, biomarkers emerging from systems biology are generally at the pathway level and thus not amenable to a single marker trial design. A typical systems biologybased predictive algorithm will use a large amount of data (for example, mutational panel and gene expression profile) to determine whether a patient will respond to therapy. To fit such algorithms into the clinical trial framework, fundamentally different trial designs are needed. Adaptive trial designs such as basket trials and umbrella trials may therefore be more appropriate to evaluate systems biology platforms98,99 (reviewed by Senft et al.100). As the data from these newer types of trials become available, the field can begin to evaluate predictive treatment models against standard practice in retrospective studies. Similarly, new predictive models could then be developed by pooling clinical data sources and training models on mutation panels (such as the MSKIMPACT or FoundationOne CDx panels) using k-fold cross validation to tune and evaluate the model94. Collaborative research networks such as the Oncology Research Information Exchange Network (ORIEN), the American Association for Cancer Research project Genomics Evidence Neoplasia Information Exchange (GENIE) and the Quantum Immuno-oncology Lifelong Trial (QUILT) programme will likely be critical for these types of studies. In the future, one could imagine a scenario in which all patients are profiled against large panels (for example, mutation, epigenetic and immune) that inform a predictive algorithm to match each patient to a combination of therapies predicted to be most effective.
Conclusions
Over the past two decades, systems biology has matured considerably and has been transformed by powerful new experimental and computational technologies. These improvements, along with the continued efforts of a rapidly growing systems biology community, have proven reasonably effective at recapitulating knowledge of cancer pathways. In particular, publicly accessible SBmaps wereable to recover the vast majority of LCpathways with significant overlap, although further investigation showed that this overlap is generally characterized by high precision but relatively low recall. One reason for the generally low recall is that SBmaps tend to be small relative to cancer pathways assembled through literature curation, with the consequence that they can never achieve full recovery of the LCpathway. Whether this size difference reflects an inherent difference between systems biology and other modes of biological investigation is unclear, since SBmaps are the products of individual studies, whereas LCpathways are the synthesized products of many studies.
Systems biology has identified a number of novel SBmaps that were not well represented in the literature, at least as captured by LCpathways. It is interesting that many of these SBmaps also were not enriched for genes with known biological processes or functions. Either these pathways are false positives or there is a sizable opportunity for identifying new cancer pathway associations; these two opposing possibilities are currently not easily distinguished. Certainly the novel SBmaps advance some noteworthy hypotheses, for instance HPV involvement in ovarian carcinogenesis and potential new genes in the Hippo pathway. Moving forward, a greater emphasis on focused followup experiments is needed to systematically validate pathway hypotheses that have arisen from systems approaches.
It is important to note that the cancer literature reference used in this survey, LCpathways, combines information curated by noble but imperfect human editors from multiple tissue types and cellular contexts. It is likely that the definition and meaning of each LCpathway varies across tumour tissue types, akin to the context specificity observed for genetic interaction networks across cancer cell lines29 Given the myriad interconnections among cellular factors, whether cancer pathways can truly be described according to a discrete set of independent pathway maps seems unlikely. In this light, our comparison of SBmaps and LCpathways should not be viewed so much as a validation of experimental results against a gold standard as a contrast between two distinct and complementary modes by which cancer pathways have been studied.
Moving forward, an enormous opportunity for cancer systems biology and its pathway maps is in the interpretation of cancer genotype and somatic tumour alterations. Large-scale genomic studies continue to catalogue new mutations and single-nucleotide variants associated with disease. However, our knowledge of how genomic context dictates gene or pathway function, and ultimately a cancer phenotype, remains incomplete. For this reason, functional genomic studies (for example, genetic interaction mapping) are becoming much more prevalent to understand how individual genetic variants and mutations influence larger biological networks as a whole. Functional genomic maps of cancer across genotypes thus provide the wiring diagrams necessary to understand how individual genotypes drive diverse cancer phenotypes.
Clearly, a major limitation of our census of cancer pathways was its restriction to SBmaps accessible for download. Working to ensure that current and future SBmaps are truly available for public access will undoubtedly improve recall of the cancer literature and the discovery of new cancer pathways. Indeed, while great strides have been made in establishing standards for publication and sharing of genomic101, transcriptomic102 and proteomic data103, publication and data-sharing standards for networks are still evolving. This lack of standardized network storage and sharing clearly hinders the ability to use and compare cancer networks identified by systems biology studies, as well as the ability of researchers to reproduce the work of others, slowing progress towards understanding cancer at the network level. To improve the availability of networks in the future, we urge journals to examine their standards for network content and investigators working with networks to either share their networks in the supporting material of their articles or upload their network models to network sharing repositories. As one solution to this problem we recently developed NDEx11, which is integrated with the Cytoscape environment for network visualization and analysis, allowing seamless movement of networks to or from the cloud. Similarly, repositories exist to store and host quantitative ordinary differential equation models of biological networks, such as BioModels and CellML. The models in these repositories are often stored as code (for example, C++ and MATLAB), which allows researchers to readily reproduce the results from these network studies. GitHub, which is commonly used to distribute code, could potentially also be used to share network models.
While cancer systems biology has moved more slowly clinically, the application of systems biology to medical practice has the potential to become the future of medicine. This vision includes individualized cancer treatment that is based on altered cancer networks, immune landscapes and evolutionary trajectories rather than on tumour pathology alone. Continued efforts to profile the therapeutic courses of patients and their genomic, transcriptomic and proteomic information, along with improvements in machine learning, are poised to yield huge advances in the years to come.
Supplementary Material
Acknowledgements
The authors gratefully acknowledge the support for this work provided by grants from the US National Institutes of Health to T.I. (CA209891, CA184427, ES014811) and B.M.K. (CA212456).
Glossary
- Fuzzy logic
A predictive model that attempts to use vague or imprecise information to obtain accurate predictions and solve complex problems.
- Adjacency matrix
A square matrix used to represent the structure of a finite network in which rows and columns represent nodes in the network and the binary elements of the matrix represent the edges.
- Interaction list
A simple, tabular network representation containing two columns (source and target) detailing the edges of a network.
- Cytoscape
An open-source software platform for visualizing complex networks and integrating these with any type of attribute data for further analyses.
- Functional enrichment analysis
A method to identify collections of genes or proteins (often disease-associated pathways) that are over represented or under represented in a large set of genes or proteins.
- Hypergeometric test
A statistical test used to calculate the statistical significance of having drawn specific successes from a given population, often used to identify subpopulations that are over represented or under represented in that population.
- F score
A measure of a test’s accuracy that takes into account both the precision and the recall of the test to compute the score. Similarly to precision and recall, the F score has a highest value of 1 and a lowest value of 0.
- STRING
A database of known and predicted protein–protein interactions that includes both direct (physical) and indirect (functional) interactions.
- Epistasis
The phenomenon whereby genetic alterations at two or more genetic loci (for example, mutations or deletions in different genes) produce a phenotype that is unexpected on the basis of the phenotypes of each of the single genetic alterations.
- CRISPR interference
A genetic perturbation technique that allows sequence specific repression of gene expression in prokaryotic and eukaryotic cells.
- Network diffusion
A method to analyse how the topology of a network impacts how information spreads across a given network.
- Striatin-interacting phosphatase and kinase (STRIPAK) and integrator complex
An evolutionarily conserved supramolecular protein complex which regulates the phosphorylation status and therefore activation status of various pathways.
- k-nearest neighbours model
A non-parametric machine learning method used for classification and regression tasks that learns to classify new cases on the basis of a similarity measure (for example, distance functions).
- Basket trials
Trials designed to test the effects of a single drug, or a combination of drugs, in a variety of cancer types on the basis of the presence of a specific biomarker.
- Umbrella trials
Trials designed to test the effect of different drugs on the basis of the presence of different biomarkers within a single cancer type.
- k-fold cross validation
A resampling procedure used to evaluate machine learning models on a limited data sample by repeatedly splitting the data into training and test sets.
Footnotes
Competing interests
The authors declare no competing interests.
Peer review information
Nature Reviews Cancer thanks A. Mardinoglu, M. Vidal, D. Hill and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information is available for this paper at https://doi.org/10.1038/s41568–020-0240–7.
Related links
American Association for Cancer Research project Genomics Evidence neoplasia information Exchange: http://www.aacr.org/Research/Research/Pages/aacr-projectgenie.aspx
BioModels: https://www.ebi.ac.uk/biomodels/
Cancer systems Biology Consortium: http://csbconsortium.org/
CellMl: https://www.cellml.org/
ClinicalTrials.gov: https://clinicaltrials.gov/
Gene Ontology: http://geneontology.org/
GitHub: https://github.com/
Human Protein Atlas: http://proteinatlas.org/
isRCTn: https://www.isrctn.com/
network Data Exchange: http://www.ndexbio.org
Oncology Research information Exchange network: http://oriencancer.org
Quantum immuno-oncology lifelong Trial programme: https://clinicaltrials.gov/ct2/results?term=QUILT
References
- 1.Cong L et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mali P et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goodwin S, McPherson JD & McCombie WR Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet. 17, 333–351 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cox J & Mann M Quantitative, high-resolution proteomics for data-driven systems biology. Annu. Rev. Biochem. 80, 273–299 (2011). [DOI] [PubMed] [Google Scholar]
- 5.LeCun Y, Bengio Y & Hinton G Deep learning. Nature 521, 436–444 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Wang H et al. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 388, 1459–1544 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoadley KA et al. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell 173, 291–304 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J et al. International Cancer Genome Consortium Data Portal — a one-stop shop for cancer genomics data. Database 2011, bar026 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ashburner M et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuenzi BM et al. Nature Reviews Cancer - SBmaps. NDEx.org http://www.ndexbio.org/#/networkset/7cd9b57c-8322-11e9-848d-0ac135e8bacf (2019). [Google Scholar]
- 11.Pratt D et al. NDEx, the Network Data Exchange. Cell Syst. 1, 302–305 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuenzi BM et al. Nature Reviews Cancer - LCpathways. NDEx.org http://www.ndexbio.org/#/networkset/d01d40d4-fcdd-11e8-8438-0ac135e8bacf (2019). [Google Scholar]
- 13.Kandasamy K et al. NetPath: a public resource of curated signal transduction pathways. Genome Biol. 11, R3 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaefer CF et al. PID: the Pathway Interaction Database. Nucleic Acids Res. 37, D674–D679 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perfetto L et al. SIGNOR: a database of causal relationships between biological entities. Nucleic Acids Res. 44, D548–D554 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chinchor N MUC-4 evaluation metrics in Proc. of the Fourth Message Understanding Conference 22–29 (Morgan Kaufmann, 1992). [Google Scholar]
- 17.Shibuya M Vascular endothelial growth factor (VEGF) and its receptor (VEGFR) signaling in angiogenesis. Genes Cancer 2, 1097–1105 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang F et al. A network medicine approach to build a comprehensive atlas for the prognosis of human cancer. Brief. Bioinform. 17, 1044–1059 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park S et al. An integrative somatic mutation analysis to identify pathways linked with survival outcomes across 19 cancer types. Bioinformatics 32, 1643–1651 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 499, 43–49 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiong S et al. structural basis for auto-inhibition of the NDR1 kinase domain by an atypically long activation segment. Structure 26, 1101–1115.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grasso CS et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 487, 239–243 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sit S-T & Manser E Rho GTPases and their role in organizing the actin cytoskeleton. J. Cell Sci. 124, 679–683 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Stoeger T, Gerlach M, Morimoto RI & Amaral LAN Large-scale investigation of the reasons why potentially important genes are ignored. PLoS Biol. 16, e2006643 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bai Y et al. Adaptive responses to dasatinib-treated lung squamous cell cancer cells harboring DDR2 mutations. Cancer Res. 74, 7217–7228 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seiler M et al. Somatic mutational landscape of splicing factor genes and their functional consequences across 33 cancer types. Cell Rep. 23, 282–296 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Warburg O & Minami S Versuche an Überlebendem Carcinom-gewebe. Klin. Wochenschr. 2, 776–777 (1923). [Google Scholar]
- 28.Horlbeck MA et al. Mapping the genetic landscape of human cells. Cell 174, 953–967.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen JP et al. Combinatorial CRISPR-Cas9 screens for de novo mapping of genetic interactions. Nat. Methods 14, 573–576 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang T et al. Gene essentiality profiling reveals gene networks and synthetic lethal interactions with oncogenic Ras. Cell 168, 890–903 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han K et al. Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nat. Biotechnol. 35, 463–474 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao D et al. Combinatorial CRISPR-Cas9 metabolic screens reveal critical redox control points dependent on the KEAP1-NRF2 regulatory axis. Mol. Cell 69, 699–708 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ashton TM, McKenna WG, Kunz-Schughart LA & Higgins GS Oxidative phosphorylation as an emerging target in cancer therapy. Clin. Cancer Res. 24, 2482–2490 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Uhlén M et al. Tissue-based map of the human proteome. Science 347, 1260419 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Bordbar A et al. Model‐driven multi‐omic data analysis elucidates metabolic immunomodulators of macrophage activation. Mol. Syst. Biol. 8, 558 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Domblides C, Lartigue L & Faustin B Control of the antitumor immune response by cancer metabolism. Cells 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duarte NC et al. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc. Natl Acad. Sci. USA 104, 1777–1782 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma H et al. The Edinburgh human metabolic network reconstruction and its functional analysis. Mol. Syst. Biol. 3, 135 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thiele I et al. A community-driven global reconstruction of human metabolism. Nat. Biotechnol. 31, 419–425 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mardinoglu A et al. Integration of clinical data with a genome-scale metabolic model of the human adipocyte. Mol. Syst. Biol. 9, 649 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mardinoglu A et al. Genome-scale metabolic modelling of hepatocytes reveals serine deficiency in patients with non-alcoholic fatty liver disease. Nat. Commun. 5, 3083 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Yizhak K, Chaneton B, Gottlieb E & Ruppin E Modeling cancer metabolism on a genome scale. Mol. Syst. Biol. 11, 817 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shlomi T, Benyamini T, Gottlieb E, Sharan R & Ruppin E Genome-scale metabolic modeling elucidates the role of proliferative adaptation in causing the Warburg effect. PLoS Comput. Biol. 7, e1002018 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jerby L et al. Metabolic associations of reduced proliferation and oxidative stress in advanced breast cancer. Cancer Res. 72, 5712–5720 (2012). [DOI] [PubMed] [Google Scholar]
- 45.Nam H et al. A systems approach to predict oncometabolites via context-specific genome-scale metabolic networks. PLoS Comput. Biol. 10, e1003837 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Agren R et al. Identification of anticancer drugs for hepatocellular carcinoma through personalized genome-scale metabolic modeling. Mol. Syst. Biol. 10, 721 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Resendis-Antonio O, Checa A & Encarnación S Modeling core metabolism in cancer cells: surveying the topology underlying the Warburg effect. PLoS One 5, e12383 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baldwin A, Pirisi L & Creek KE NFI-Ski interactions mediate transforming growth factor beta modulation of human papillomavirus type 16 early gene expression. J. Virol. 78, 3953–3964 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilting SM et al. Genomic profiling identifies common HPV-associated chromosomal alterations in squamous cell carcinomas of cervix and head and neck. BMC Med. Genomics 2, 32 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bodelon C et al. Chromosomal copy number alterations and HPV integration in cervical precancer and invasive cancer. Carcinogenesis 37, 188–196 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu Q-J et al. Detection of human papillomavirus-16 in ovarian malignancy. Br. J. Cancer 89, 672 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jeannot E, Harlé A, Holmes A & Sastre-Garau X Nuclear factor I X is a recurrent target for HPV16 insertions in anal carcinomas. Genes Chromosomes Cancer 57, 638–644 (2018). [DOI] [PubMed] [Google Scholar]
- 54.zur Hausen H Papillomaviruses causing cancer: evasion from host-cell control in early events in carcinogenesis. J. Natl Cancer Inst. 92, 690–698 (2000). [DOI] [PubMed] [Google Scholar]
- 55.Marullo R et al. HPV16 E6 and E7 proteins induce a chronic oxidative stress response via NOX2 that causes genomic instability and increased susceptibility to DNA damage in head and neck cancer cells. Carcinogenesis 36, 1397–1406 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roos P, Orlando PA, Fagerstrom RM & Pepper JW In North America, some ovarian cancers express the oncogenes of preventable human papillomavirus HPV-18. Sci. Rep. 5, 8645 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ingerslev K et al. High-risk HPV is not associated with epithelial ovarian cancer in a Caucasian population. Infect. Agent. Cancer 11, 39 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosa MI et al. The prevalence of human papillomavirus in ovarian cancer: a systematic review. Int. J. Gynecol. Cancer 23, 437–441 (2013). [DOI] [PubMed] [Google Scholar]
- 59.Meng Z, Moroishi T & Guan K-L Mechanisms of Hippo pathway regulation. Genes. Dev. 30, 1–17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shin S-Y et al. Functional roles of multiple feedback loops in extracellular signal-regulated kinase and Wnt signaling pathways that regulate epithelial-mesenchymal transition. Cancer Res. 70, 6715–6724 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aldridge BB, Saez-Rodriguez J, Muhlich JL, Sorger PK & Lauffenburger DA Fuzzy logic analysis of kinase pathway crosstalk in TNF/EGF/ insulin-induced signaling. PLoS Comput. Biol. 5, e1000340 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kolch W, Calder M & Gilbert D When kinases meet mathematics: the systems biology of MAPK signalling. FEBS Lett. 579, 1891–1895 (2005). [DOI] [PubMed] [Google Scholar]
- 63.Orton RJ et al. Computational modelling of the receptor-tyrosine-kinase-activated MAPK pathway. Biochem. J. 392, 249–261 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heinrich R, Neel BG & Rapoport TA Mathematical models of protein kinase signal transduction. Mol. Cell 9, 957–970 (2002). [DOI] [PubMed] [Google Scholar]
- 65.Pan S Modeling the mitogen activated protein (MAP)-kinase pathway using ordinary differential equations. Comput. Biol. Bioinf. 1, 6–9 (2013). [Google Scholar]
- 66.Tran PT et al. Survival and death signals can predict tumor response to therapy after oncogene inactivation. Sci. Transl Med. 3, 103ra99 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Claas AM, Atta L, Gordonov S, Meyer AS & Lauffenburger DA Systems modeling identifies divergent receptor tyrosine kinase reprogramming to MAPK pathway inhibition. Cell. Mol. Bioeng. 11, 451–469 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morris MK, Clarke DC, Osimiri LC & Lauffenburger DA Systematic analysis of quantitative logic model ensembles predicts drug combination effects on cell signaling networks. CPT Pharmacomet. Syst. Pharmacol. 5, 544–553 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gierut JJ et al. Network-level effects of kinase inhibitors modulate TNF-α–induced apoptosis in the intestinal epithelium. Sci. Signal. 8, ra129 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lorz A, Botesteanu D-A & Levy D Modeling cancer cell growth dynamics in vitro in response to antimitotic drug treatment. Front. Oncol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Palacios-Moreno J et al. Neuroblastoma tyrosine kinase signaling networks involve FYN and LYN in endosomes and lipid rafts. PLoS Comput. Biol. 11, e1004130 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Choudhary KS et al. EGFR signal-network reconstruction demonstrates metabolic crosstalk in EMT. PLoS Comput. Biol. 12, e1004924 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gill MK et al. A feed forward loop enforces YAP/TAZ signaling during tumorigenesis. Nat. Commun. 9, 3510 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.O’Connor CM et al. Inactivation of PP2A by a recurrent mutation drives resistance to MEK inhibitors. Oncogene 39, 703–717 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alvarez MJ et al. Functional characterization of somatic mutations in cancer using network-based inference of protein activity. Nat. Genet. 48, 838–847 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Coleman DJ et al. BET bromodomain inhibition blocks the function of a critical AR-independent master regulator network in lethal prostate cancer. Oncogene 38, 5658–5669 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Risom T et al. Differentiation-state plasticity is a targetable resistance mechanism in basal-like breast cancer. Nat. Commun. 9, 3815 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Echeverria GV et al. Resistance to neoadjuvant chemotherapy in triple-negative breast cancer mediated by a reversible drug-tolerant state. Sci. Transl Med. 11, eaav0936 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Parker LA et al. Diagnostic biomarkers: are we moving from discovery to clinical application? Clin. Chem. 64, 1657–1667 (2018). [DOI] [PubMed] [Google Scholar]
- 80.Poste G Bring on the biomarkers. Nature 469, 156–157 (2011). [DOI] [PubMed] [Google Scholar]
- 81.Carbone DP et al. Prognostic and predictive role of the VeriStrat plasma test in patients with advanced non-small-cell lung cancer treated with erlotinib or placebo in the NCIC Clinical Trials Group BR.21 trial. J. Thorac. Oncol. 7, 1653–1660 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Amann JM et al. Genetic and proteomic features associated with survival after treatment with erlotinib in first-line therapy of non-small cell lung cancer in Eastern Cooperative Oncology Group 3503. J. Thorac. Oncol. 5, 169–178 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Filho OM, Ignatiadis M & Sotiriou C Genomic Grade Index: an important tool for assessing breast cancer tumor grade and prognosis. Crit. Rev. Oncol. Hematol. 77, 20–29 (2011). [DOI] [PubMed] [Google Scholar]
- 84.Parker JS et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 27, 1160–1167 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jerevall P-L et al. Prognostic utility of HOXB13:IL17BR and molecular grade index in early-stage breast cancer patients from the Stockholm trial. Br. J. Cancer 104, 1762–1769 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ma X-J et al. A five-gene molecular grade index and HOXB13:IL17BR are complementary prognostic factors in early stage breast cancer. Clin. Cancer Res. 14, 2601–2608 (2008). [DOI] [PubMed] [Google Scholar]
- 87.Filipits M et al. A new molecular predictor of distant recurrence in ER-positive, HER2-negative breast cancer adds independent information to conventional clinical risk factors. Clin. Cancer Res. 17, 6012–6020 (2011). [DOI] [PubMed] [Google Scholar]
- 88.Sparano JA et al. Prospective validation of a 21-gene expression assay in breast cancer. N. Engl. J. Med. 373, 2005–2014 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cronin M et al. Analytical validation of the Oncotype DX genomic diagnostic test for recurrence prognosis and therapeutic response prediction in node-negative, estrogen receptor-positive breast cancer. Clin. Chem. 53, 1084–1091 (2007). [DOI] [PubMed] [Google Scholar]
- 90.van ‘t Veer LJ et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 415, 530–536 (2002). [DOI] [PubMed] [Google Scholar]
- 91.Silvestri GA et al. A bronchial genomic classifier for the diagnostic evaluation of lung cancer. N. Eng. J. Med. 373, 243–251 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yeoh E-J et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell 1, 133–143 (2002). [DOI] [PubMed] [Google Scholar]
- 93.Stein RC et al. OPTIMA prelim: a randomised feasibility study of personalised care in the treatment of women with early breast cancer. Health Technol. Assess. Winch. Engl. 20, 1–201 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Michiels S, Ternès N & Rotolo F Statistical controversies in clinical research: prognostic gene signatures are not (yet) useful in clinical practice. Ann. Oncol. 27, 01–09 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cheng DT et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT). J. Mol. Diagn. 17, 251–264 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Harris J FDA approves FoundationOne CDx, CMS agrees to cover. OncLive, November (2017). [Google Scholar]
- 97.Dacic S & Nikiforova MN Present and future molecular testing of lung carcinoma. Adv. Anat. Pathol. 21, 94–99 (2014). [DOI] [PubMed] [Google Scholar]
- 98.Rashdan S & Gerber DE Going into BATTLE: umbrella and basket clinical trials to accelerate the study of biomarker-based therapies. Ann. Transl Med. 4, 529 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Biankin AV, Piantadosi S & Hollingsworth SJ Patient-centric trials for therapeutic development in precision oncology. Nature 526, 361–370 (2015). [DOI] [PubMed] [Google Scholar]
- 100.Senft D, Leiserson MDM, Ruppin E & Ronai ZA Precision oncology: the road ahead. Trends Mol. Med. 23, 874–898 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mailman MD et al. The NCBI dbGaP database of genotypes and phenotypes. Nat. Genet. 39, 1181–1186 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Clough E & Barrett T The Gene Expression Omnibus database. Methods Mol. Biol. Clifton NJ 1418, 93–110 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jones P et al. PRIDE: a public repository of protein and peptide identifications for the proteomics community. Nucleic Acids Res. 34, D659–D663 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mani R, St.Onge RP, Hartman JL, Giaever G. & Roth FP Defining genetic interaction. Proc. Natl. Acad. Sci. 105, 3461–3466 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Collins SR et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446, 806–810 (2007). [DOI] [PubMed] [Google Scholar]
- 106.Costanzo M et al. A global genetic interaction network maps a wiring diagram of cellular function. Science 353, aaf1420 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Costanzo M et al. The genetic landscape of a cell. Science 327, 425–431 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Havugimana PC et al. A census of human soluble protein complexes. Cell 150, 1068–1081 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Brückner A, Polge C, Lentze N, Auerbach D & Schlattner U Yeast two-hybrid, a powerful tool for systems biology. Int. J. Mol. Sci. 10, 2763–2788 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Remy I & Michnick SW Application of protein- fragment complementation assays in cell biology. BioTechniques 42, 137–145 (2007). [DOI] [PubMed] [Google Scholar]
- 111.Bürckstümmer T et al. An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat. Methods 3, 1013–1019 (2006). [DOI] [PubMed] [Google Scholar]
- 112.Dunham WH, Mullin M & Gingras A-C Affinity-purification coupled to mass spectrometry: basic principles and strategies. Proteomics 12, 1576–1590 (2012). [DOI] [PubMed] [Google Scholar]
- 113.Margolin AA et al. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics 7, S7 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lefebvre C et al. A human B‐cell interactome identifies MYB and FOXM1 as master regulators of proliferation in germinal centers. Mol. Syst. Biol. 6, 377 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Olow A et al. An atlas of the human kinome reveals the mutational landscape underlying dysregulated phosphorylation cascades in cancer. Cancer Res. 76, 1733–1745 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lachmann A, Giorgi FM, Lopez G & Califano A ARACNe-AP: gene network reverse engineering through adaptive partitioning inference of mutual information. Bioinformatics 32, 2233–2235 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Leiserson MDM et al. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat. Genet. 47, 106–114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Vandin F, Upfal E & Raphael BJ Algorithms for detecting significantly mutated pathways in cancer. J. Comput. Biol. 18, 507–522 (2011). [DOI] [PubMed] [Google Scholar]
- 119.Jia P & Zhao Z VarWalker: personalized mutation network analysis of putative cancer genes from next-generation sequencing data. PLoS Comput. Biol. 10, e1003460 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Park J et al. AF1q is a novel TCF7 co-factor which activates CD44 and promotes breast cancer metastasis. Oncotarget 6, 20697–20710 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hofree M, Shen JP, Carter H, Gross A & Ideker T Network-based stratification of tumor mutations. Nat. Methods 10, 1108–1115 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bidkhori G et al. Metabolic network-based stratification of hepatocellular carcinoma reveals three distinct tumor subtypes. Proc. Natl Acad. Sci. 115, E11874–E11883 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bordbar A, Monk JM, King ZA & Palsson BO Constraint-based models predict metabolic and associated cellular functions. Nat. Rev. Genet. 15, 107–120 (2014). [DOI] [PubMed] [Google Scholar]
- 124.Szklarczyk D et al. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Olcina MM et al. Mutations in an innate immunity pathway are associated with poor overall survival outcomes and hypoxic signaling in cancer. Cell Rep. 25, 3721–3732.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Babaei S, Hulsman M, Reinders M & de Ridder J Detecting recurrent gene mutation in interaction network context using multi-scale graph diffusion. BMC Bioinformatics 14, 29 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.