Abstract
Purpose
The basophil activation test (BAT) has been reported to be useful for the diagnosis of various food allergies, such as allergy to peanut, but not to fish. This study aimed to evaluate the diagnostic performance of the BAT for fish allergy.
Methods
We performed a retrospective review of patients with fish allergy who underwent the BAT using a panel of fish extracts (15 kinds) to examine the differential reactivity to several species of fish. The BAT score for each extract was expressed as the ratio of CD203chigh% with the extract to that with anti-IgE antibody. Clinical reactivity to each fish was confirmed by positive oral food challenge or a typical history of fish-induced immediate allergy symptoms. Receiver-operating-characteristic (ROC) analysis was performed to evaluate the diagnostic performance.
Results
Fifty-one patients with fish allergy were analyzed. Using extracts of 15 species of fish, the BAT was performed a total of 184 times on the patients. Clinical allergy to each species of fish was confirmed in 90 (48.9%) of those tests. ROC analysis yielded high areas under the curve for the BAT scores for the 5 most common fish species (0.72–0.88). The diagnostic accuracy ranged from 0.74 to 0.86. Using a tentative cutoff value of 0.3 deduced from the ROC analyses of the 5 fish species, the accuracy for other fish allergic reactions was generally high (0.6–1.0), except the fish tested in a small number of patients.
Conclusions
The BAT score based on CD203c expression may be useful for fish allergy diagnosis, especially since a large variety of fish can be tested by the BAT using fish extracts prepared by a simple method.
Keywords: Basophil activation test, CD203c, fish allergy, allergen extract
INTRODUCTION
Consumption of fish is increasing due to a better understanding of the nutritional benefits of fish such as reduced risk of chronic inflammatory disorders, including allergic diseases.1,2,3,4 On the other hand, fish allergy patients are increasing worldwide, not only in major fish-eating countries such as Japan and Korea, but in many Western countries.5 Parvalbumin, a major fish allergen, shows a wide range of cross-reactivity among various fish species.6,7,8 Collagen is also a pan-allergen in fish in countries where raw fish consumption is common.9,10 Thus, patients with allergy to one kind of fish often react to other kinds of fish.11 As a result, physicians often instruct fish allergy patients to eliminate all kinds of fish. However, this can result in nutritional and social disadvantages because fish have considerable nutritional value in preventing various diseases12,13 and a variety of species are recently utilized in many nations' cuisine. Oral food challenge (OFC), the gold standard for food allergy diagnosis, can alleviate the problem by correctly distinguishing between symptom-eliciting and “safe” fish, permitting the patient to be selective. However, the performance of OFC in clinics is hindered by the fact that it is quite time-consuming and might induce severe symptoms such as anaphylaxis.
Specific IgE (sIgE) and skin prick tests are used to predict OFC results.14 However, sIgE was reported to show high rates of false-negative and false-positive results for fish allergy,15 and the number of fish species that can be tested is limited. The allergen extracts available for skin prick tests are also limited. Meanwhile, prick-to-prick tests using fish meat itself may cause false-positive results because of histamine generated in the fish meat, and the histamine may cause anaphylaxis.16
The basophil activation test (BAT) has been shown to be a useful in vitro diagnostic method for the evaluation of food allergy, especially for allergens for which there is no commercially available sIgE test.17,18,19,20,21 The BAT requires only a small amount of allergen extract, which can be easily prepared in a standard laboratory. However, there have been few reports on the performance of the BAT for fish allergy. The purpose of this study was to assess the usefulness of BAT for the diagnosis of fish allergy.
MATERIALS AND METHODS
Study design and study population
Retrospective analysis of clinical records was performed. We enrolled patients suspected of manifesting fish allergy and on whom the BAT was performed at Mie National Hospital between October 2010 and March 2018. In each patient, the BAT was performed for several fish species, including not only the suspect fish but also other edible fish as neutral controls. Patients were diagnosed as being allergic to a fish species based on a positive OFC test or a history of an immediate allergic reaction after eating that fish. Patients were diagnosed as being non-allergic on the basis of a negative OFC test or regular eating of that fish without any symptoms. The OFC was considered positive if an objective clinical reaction was noted, such as urticaria, angioedema, rhinoconjunctivitis, cough, wheezing, vomiting, diarrhea and/or a decrease in blood pressure. The OFC was also considered positive if the patient reported intense abdominal pain even if no objective signs were present. The OFC was considered negative if no symptoms were observed 2 hours after ingesting a regular serving amount of fish meat at challenge. Full emergency equipment and medications were readily at hand during the course of all procedures. Antihistamine use was suspended 72 hours before the OFC. Blood sampling for the BAT was performed within 6 months after the diagnosis of fish allergy.
Extraction of fish allergens
Fish extracts were prepared as previously reported.22 In brief, fresh fish meat from salmon (Oncorhynchus keta), mackerel (Scomber japonicus), tuna (Thunnus obesus), red sea bream (Pagrus major), yellowtail (Seriola quinqueradiata), flounder (Paralichthys olivaceus), bonito (Katsuwonus pelamis), pacific ocean perch (Sebastes alutus), pacific saury (Cololabis saira), horse mackerel (Trachurus japonicus), spanish mackerel (Scomberomorus niphonius), barracuda (Sphyraena pinguis), anchovy (Engraulis japonicus), atka mackerel (Pleurogrammus azonus) or smelt (Spirinchus lanceolatus) was mixed with an equal volume of phosphate-buffered saline (PBS), followed by heating in a microwave oven for 1 to 2 minutes after coming to a boil. Each mixture was minced and pureed in a food processor. Particles were eliminated twice by filter centrifugation for 5 minutes at 20,000 g, and the supernatant was collected for use as an extract and dialyzed against PBS overnight. The extracts were freeze-dried and stored at −80°C until use. The frozen extracts were reconstituted in PBS, and then the protein concentration in each was measured with a spectrophotometer (NanoDrop ND-1000®, Wilmington, DE, USA) and diluted to 100 μg/mL with PBS at the time of use.
BAT and sIgE test
A commercial kit (Allergenicity Kit; Beckman Coulter, Fullerton, CA, USA) was used to quantify basophil CD203c expression.20 Briefly, EDTA-containing whole blood was incubated with each fish extract at concentrations of 1 and 10 µg/mL for 15 minutes at 37ºC after addition of a sufficient amount of calcium solution to neutralize the chelating capacity of EDTA. Anti-human IgE antibody at 4 µg/mL and PBS served as positive and negative controls, respectively. PC7-conjugated anti-CD3, fluorescein isothiocyanate–conjugated anti–chemoattractant receptor-homologous molecules expressed on TH2 lymphocytes (CRTH2), and phycoerythrin-conjugated anti-CD203c antibodies were added during the reaction. The samples were then analyzed on a flow cytometer (Gallios®, Beckman Coulter). Basophils were detected on the basis of the forward- and side-scatter characteristics, and negative CD3 and positive CRTH2 results. Up-regulation of CD203c on basophils was determined using a threshold that was defined by the fluorescence of unstimulated cells (negative control) and expressed as the CD203chigh percentage (CD203chigh%). The ratio of CD203chigh% induced by a fish allergen to that induced by anti-human IgE antibody was calculated21, 23, 24 and expressed as the BAT score. Patients were defined as low responders and excluded from the analysis if CD203chigh% in response to anti-human IgE antibody was <10%.
Allergen-sIgE was measured with ImmunoCAP® (ThermoFisher Diagnostics, Uppsala, Sweden). In Japan, there are commercially available sIgE tests for tuna, salmon, mackerel and horse mackerel. We also measured IgE specific for cod parvalbumin (Gad c1) and carp parvalbumin (Cyp c1).
Statistical analysis
The χ2 test was used to compare the proportions of categorical variables, and the Mann-Whitney U test was used to compare 2 groups. Receiver-operating-characteristic (ROC) curve analysis was employed to evaluate the predictive performances of the BAT score and sIgE. Optimal cutoff values were determined as the best balance between sensitivity and specificity, which correspond to the highest Youden index. The area under the ROC curve (AUC), sensitivity, specificity, positive predictive value, negative predictive value and accuracy were calculated. GraphPad Prizm 7.0® (GraphPad Software, Inc., San Diego, CA, USA) was used for all statistical analyses.
Ethics
This study was approved by the Ethics Committee of Mie National Hospital (#24-01). Since it was a retrospective study and no private information was collected, the opportunity to opt out was made known to the public.
RESULTS
Characteristics of the patients
Sixty-seven patients were enrolled, 16 of whom were excluded from the analyses: 10 due to ambiguity of symptoms after ingesting fish and 6 who were low responders. The remaining 51 patients were considered to have fish allergy and analyzed. Table 1 shows the demographics of the subjects analyzed. There were no significant differences between the analyzed and excluded subjects in sex, age, comorbid allergy (including other food allergy) or total serum IgE level (data not shown). The BAT was performed a total of 184 times on the 51 analyzed subjects using extracts of 15 species of fish, and 90 (48.9%) of those tests were positive. The prevalence of allergy to individual fish species ranged from 0% to 80%. A total of 83 sIgE tests were performed for 4 fish species (Table 2).
Table 1. Demographics of the patients.
| Patient background | Analyzed patients (n = 51) |
|---|---|
| Male | 33 (65) |
| Age (yr) | 6 (3–11) |
| Other food allergy | 22 (43) |
| Atopic dermatitis | 20 (39) |
| Bronchial asthma | 8 (16) |
| Allergic rhinitis | 42 (82) |
| Total IgE (IU/mL) | 487 (118–1,156) |
Data are presented as number (%) or median (interquartile range).
Table 2. List of the fish species tested with the BAT.
| Generic name | Scientific name | Patients tested* | Fish allergy† |
|---|---|---|---|
| Salmon | Oncorhynchus keta | 31 (29) | 16 (52) |
| Mackerel | Scomber japonicus | 29 (28) | 12 (41) |
| Tuna | Thunnus obesus | 24 (21) | 5 (21) |
| Red sea bream | Pagrus major | 22 | 11 (50) |
| Yellowtail | Seriola quinqueradiata | 19 | 13 (68) |
| Flounder | Paralichthys olivaceus | 15 | 6 (40) |
| Bonito | Katsuwonus pelamis | 13 | 7 (54) |
| Pacific ocean perch | Sebastes alutus | 9 | 7 (78) |
| Pacific saury | Cololabis saira | 5 | 3 (60) |
| Horse mackerel | Trachurus japonicus | 5 (5) | 4 (80) |
| Spanish mackerel | Scomberomorus niphonius | 4 | 3 (75) |
| Barracuda | Sphyraena pinguis | 3 | 2 (67) |
| Anchovy | Engraulis japonicus | 2 | 0 (0) |
| Atka mackerel | Pleurogrammus azonus | 2 | 1 (50) |
| Smelt | Spirinchus lanceolatus | 1 | 0 (0) |
Data are presented as *number (number of tested with ImmunoCAP) and †number (%) of patients diagnosed with allergy to indicated fish.
Diagnostic performance of the BAT
Since the fish extracts at 10 μg/mL, but not at 1 μg/mL, induced statistically significant CD203c expression in fish-allergy patients (data not shown), we employed the extracts at 10 μg/mL for the BAT in this study.
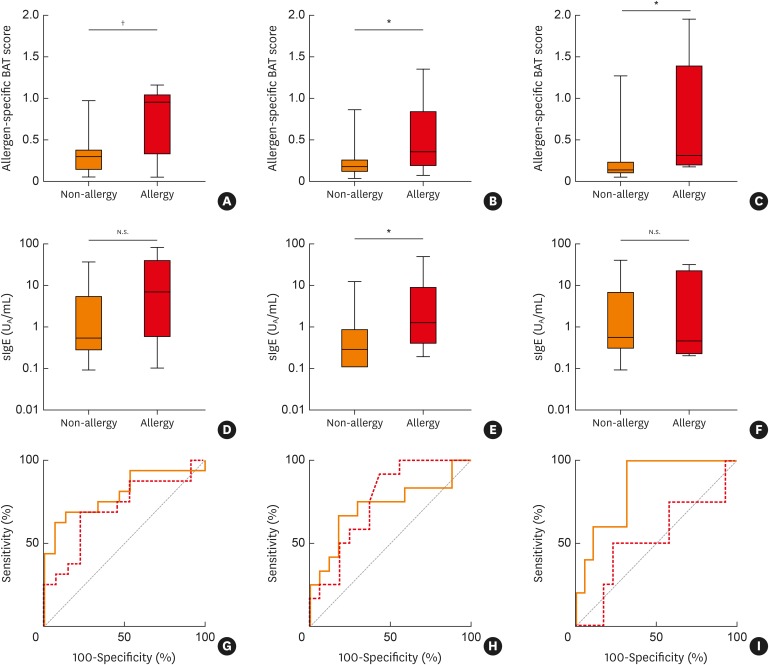
First, to achieve sufficient statistical power, the results for 5 species of fish—namely, salmon, mackerel, tuna, red sea bream and yellowtail—for which 125 subjects underwent the BAT were analyzed for the predictive performance of the BAT score. For each of the 5 fish species, the BAT score for patients with allergy was significantly higher than for the non-allergic patients (Table 3; Fig. 1A, B and C). ROC analysis revealed that the AUC ranged from 0.72 to 0.88. At the optimal cutoff value, the specificity ranged from 0.82 to 1.0. Although the sensitivity was not high for some fish, the accuracy of the test was sufficiently high, ranging from 0.74 to 0.91 (Table 3; Fig. 1G, H and I).
Table 3. Diagnostic performance of BAT scores in the diagnosis of allergies to 5 most common fish species.
| Fish | BAT score (mean ± SD) | AUC | Cut-off | Sensitivity | Specificity | PPV | NPV | Accuracy | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Allergy | No. | Non-allergy | No. | P value | ||||||||
| Salmon | 0.75 ± 0.39 | 16 | 0.32 ± 0.27 | 15 | 0.004 | 0.80 | 0.54 | 0.69 | 0.87 | 0.85 | 0.72 | 0.77 |
| Mackerel | 0.50 ± 0.40 | 12 | 0.24 ± 0.23 | 17 | 0.048 | 0.72 | 0.30 | 0.67 | 0.82 | 0.73 | 0.78 | 0.76 |
| Tuna | 0.70 ± 0.76 | 5 | 0.21 ± 0.27 | 19 | 0.019 | 0.84 | 0.30 | 0.60 | 0.90 | 0.60 | 0.90 | 0.83 |
| Red sea bream | 0.86 ± 0.41 | 11 | 0.18 ± 0.09 | 11 | 0.002 | 0.88 | 0.38 | 0.82 | 1.0 | 1.0 | 0.85 | 0.91 |
| Yellowtail | 0.59 ± 0.51 | 13 | 0.18 ± 0.08 | 6 | 0.017 | 0.85 | 0.30 | 0.69 | 0.83 | 0.90 | 0.56 | 0.74 |
BAT, basophil activation test; SD, standard deviation; AUC, area under the receiver-operating-characteristic curve; PPV, positive predictive value; NPV, negative predictive value.
Fig. 1. Allergen-specific BAT scores in patients allergic/non-allergic to (A) salmon, (B) mackerel and (C) tuna. Specific IgE levels in patients allergic-/non-allergic to (D) salmon, (E) mackerel and (F) tuna. ROC curves of BAT (solid line) and sIgE (dotted line) for (G) salmon, (H) mackerel and (I) tuna are shown. The areas under the ROC curve were 0.80, 0.72 and 0.84 for the BAT score and 0.73, 0.75 and 0.51 for sIgE.
BAT, basophil activation test; sIgE, specific IgE; ROC, receiver-operating-characteristic.
*P < 0.05; †P < 0.005.
Next, to evaluate the BAT score for each of the fish species, including those for which only a small number of patients was tested, the predictive performance was calculated by employing a common cutoff value of 0.3, which was deduced from the initial analysis for the 5 fish species (Table 4). Although there were some variations, the diagnostic accuracy was at least 0.6 for fish that were tested in 5 or more patients. For fish species tested in fewer than 5 subjects, the accuracy ranged widely from 0.33 to 1.0.
Table 4. Predictive performance of BAT scores using a cutoff value of 0.3.
| Fish species | Sensitivity | Specificity | PPV | NPV | Accuracy | |
|---|---|---|---|---|---|---|
| No. of subjects ≥ 5 | ||||||
| Salmon | 0.75 | 0.53 | 0.63 | 0.67 | 0.65 | |
| Mackerel | 0.67 | 0.82 | 0.73 | 0.78 | 0.76 | |
| Tuna | 0.60 | 0.89 | 0.60 | 0.89 | 0.83 | |
| Red sea bream | 0.82 | 0.91 | 0.90 | 0.83 | 0.86 | |
| Yellowtail | 0.69 | 0.83 | 0.90 | 0.56 | 0.74 | |
| Flounder | 0.67 | 0.56 | 0.50 | 0.71 | 0.60 | |
| Bonito | 1.00 | 0.50 | 0.70 | 1.00 | 0.77 | |
| Pacific ocean perch | 0.71 | 0.50 | 0.83 | 0.33 | 0.67 | |
| Pacific saury | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
| Horse mackerel | 0.75 | 0.00 | 0.75 | 0.00 | 0.60 | |
| No. of subjects < 5 | ||||||
| Spanish mackerel | 0.67 | 0.00 | 0.67 | 0.000 | 0.50 | |
| Barracuda | 0.00 | 1.00 | NA | 0.33 | 0.33 | |
| Anchovy | NA | 1.00 | NA | 1.00 | 1.00 | |
| Atka mackerel | 0.00 | 1.00 | NA | 0.50 | 0.50 | |
| Smelt | NA | 0.00 | 0.000 | NA | 0.00 | |
BAT, basophil activation test; PPV, positive predictive value; NPV, negative predictive value; NA, not applicable.
Comparison with sIgE
We next compared the sIgE results with the BAT scores. Since commercially available sIgE products for fish allergens are limited, we examined the sIgE values only for salmon, mackerel and tuna, for which sufficient numbers of patients were tested. The sIgE levels were significantly higher in patients allergic to mackerel than in the non-allergic subjects (Fig. 1D and E), whereas the difference was not significant for salmon and tuna allergy (Fig. 1F). The AUCs were 0.70, 0.75 and 0.51 for salmon, mackerel and tuna, respectively (Fig. 1G, H and I). In contrast, the differences in the BAT scores between the allergic and non-allergic subjects for the 3 fish species were all statistically significant, and the AUCs were 0.80, 0.72 and 0.84, respectively (Fig. 1A, B, C, G, H and I). These results suggest that for certain fish the diagnostic performance of the BAT score is at least comparable to, or even better than, the diagnostic performance of the sIgE level.
Since parvalbumin is reported to be a fish pan-allergen, the overall diagnostic performance of sIgE for Gad c1 and Cyp c1 was compared with the BAT score (Supplementary Fig. S1). Both the BAT score and the sIgE levels for Gad c1 and Cyp c1 were significantly higher in patients with allergy to a specific fish species than in those with no allergy. The AUCs were 0.79, 0.70 and 0.70 for the BAT score, anti-Gad c1- and anti-Cyp c1-sIgE, respectively; the AUC was thus highest for BAT, although the difference was not statistically significant. For the diagnosis of individual fish allergy, the predictive performance of anti-parvalbumin-sIgE for salmon, mackerel and tuna allergies was not as high as the predictive performance of the BAT score (Supplementary Fig. S1).
Possible influence of anisakis sensitization
Anisakis allergy is sometimes misdiagnosed as fish allergy because the induced symptoms are indistinguishable and a definitive diagnosis of anisakis allergy is difficult because no standardized anisakis allergen for OFC is commercially available. We thus investigated the influence of anisakis sensitization in our study population and found that there was no difference in the BAT score, sIgE levels for Gad c1 and Cyp c1, or the prevalence of fish allergy diagnosis between anisakis-sensitized and non-sensitized subjects (Supplementary Table S1). The only difference was that the median age was significantly higher in the anisakis-sensitized subjects than in the non-sensitized subjects.
DISCUSSION
In this study, we found that BAT quantification of CD203c expression by using in-house fish extracts showed a good predictive performance in the diagnosis of fish allergy. Although the available sIgE tests showed similar performance, the BAT has an advantage because a theoretically unlimited number of fish species can be tested by using fish extracts that can be easily prepared in a standard laboratory.
The BAT utilizes flow cytometry to measure expression of activation markers, such as CD203c25 and CD63,26 on the surface of basophils following cross-linking of high-affinity IgE receptor-bound IgE antibodies by allergen molecules or anti-IgE antibody molecules. The BAT can be performed using whole blood, which allows not only allergen-sIgE but also other plasma factors like blocking IgG4 to modulate the reaction and may better reflect in vivo allergies.27 The BAT has been reported to be a good biomarker for clinical allergies to various foods such as peanuts,24 milk,23 eggs28,29,30 and wheat.17,20 It can also be applied to identify rare allergens such as lotus root18 and erythritol.19 However, there have been few reports on the performance of the BAT for fish allergy. In this study, we for the first time demonstrated the utility of the BAT for identifying clinical allergies to 15 fish species. For 5 common fish species, namely, salmon, mackerel, tuna, red sea bream and yellowtail, the BAT showed high specificity (0.8 to 0.91) or moderately high sensitivity. Using a cutoff value of 0.3, which was deduced from the results for the 5 common fish species, the diagnostic performance for other fish species was also good.
Detection of IgE reactivity in fish allergy would not be expected to show high specificity because of putative cross-reactivity among fish allergens. Clinically, many patients who start to react to 1 fish species often experience clinical reactions to a multitude of other fish species. Several IgE-based analyses reported extensive cross-reactivity among a variety of fish species.6,11,15 However, a recent double-blind, placebo-controlled food challenge study of different fish species showed that more than 50% of 35 fish allergy patients had no objective symptoms and that approximately 30% had subjective tolerance to at least 1 of the 3 investigated fish species.31 Although that study involved only a small number of subjects, individual diagnosis of fish allergy appears to be important. In this context, our finding of high specificity of the BAT is significant.
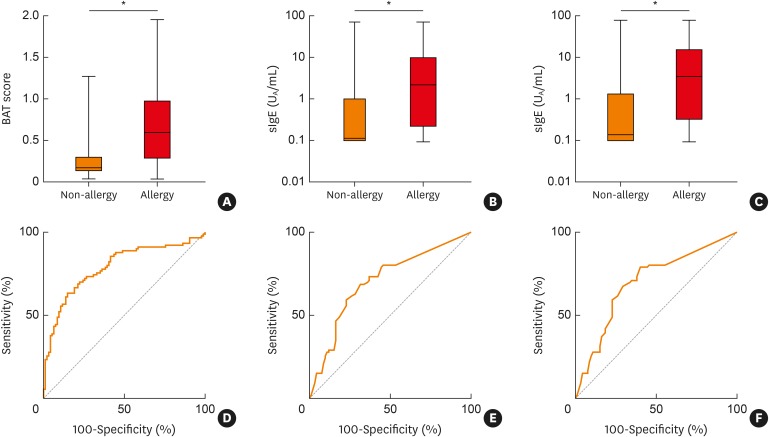
What accounts for the high specificity of the BAT? Fish allergens belong to 4 protein families, namely, parvalbumins,32 enolases, aldolases33 and collagen.10,34 Cross-reactivity has been shown for these proteins from commonly consumed fish species such as salmon and tuna.11,33,35 Recently, however, species-specific epitopes have been identified on parvalbumins,31 and differences in the parvalbumin content have been postulated to influence clinical symptoms.36 In this study, we found that the levels of sIgE for parvalbumins from cod and carp were significantly higher in fish allergy patients than in non-allergic patients, with an AUC of 0.7. Although the BAT showed a higher AUC of 0.79, the difference was not statistically significant (Fig. 2). In addition, the diagnostic performance of sIgE for parvalbumins was not satisfactory for the diagnosis of individual fish allergies. The fish extracts utilized in the BAT possibly contain various species-specific epitopes, while the parvalbumin concentrations in the extracts reflect the actual content in fish meat. It is believed that these factors contributed to the high specificity of BAT.
Fig. 2. Allergen-specific BAT score (A), and sIgE levels for Gad c1 (B) and Cyp c1 (C) in patients who were allergic or non-allergic to any fish species. ROC curves of allergen-specific BAT score (D) and sIgE levels for Gad c1 (E) and Cyp c1 (F) are shown. The AUCs were 0.79 (D), 0.70 (E) and 0.70 (F).
BAT, basophil activation test; sIgE, specific IgE; ROC, receiver-operating-characteristic.
*P < 0.0001.
This study has several limitations. First, anisakis allergy37 may have confounded the results. Anisakis simplex is a parasitic nematode that infests crustaceans, fish and marine mammals. L3 larvae of anisakis reside in the intestine and liver of fish, and they migrate to muscle upon the host's death.38,39 Humans become incidental hosts through eating infested raw/undercooked fish or can become sensitized even through eating cooked fish if sensitizing antigens remain.40 In this study, we prepared the fish extracts from very fresh fish, with the meat separated immediately upon sacrifice to prevent migration of A. simplex larvae to muscle tissue. Thus, anisakis antigen contamination of the extracts seems unlikely. We also confirmed that there was no difference in the BAT scores or other sensitization parameters between anisakis-sensitized and -unsensitized patients, suggesting that the influence of anisakis allergy was minimal. Secondly, not all patients underwent OFC to definitively diagnose their suspected fish allergy. However, a detailed history of allergy symptoms after eating a suspect fish was carefully taken in terms of the nature and reproducibility of the symptoms. Thirdly, lipopolysaccharide (LPS) may have contaminated the fish extracts and influenced basophil reactivity. We did not measure LPS in the extracts. However, it was reported that LPS did not cause basophil activation by itself and only enhanced allergen-induced basophil responses at a high concentration,41,42 namely, 1,000 EU/mL. Such a high concentration is unlikely in our reaction system. Although the LPS content in edible fish meat has not been reported, its level in a probiotic product containing viable gram-negative bacteria (Escherichia coli), for example, was reported to be 900-1700 EU/mL.43 If the above mentioned probiotic products were used as allergens in our system, the final concentration of LPS would be only 90–170 EU/mL, which is far less than the basophil-stimulating concentration.41 We used fresh and edible fish that may contain commensal bacteria, but it is unlikely that there was overgrowth of pathogenic bacteria or even commensal bacteria in fish meat present more than the “concentrated” probiotic product. Fourthly, there is lack of dose-response tests in this study. All the tests were performed at 2 concentrations, not at 5 different concentrations. Since there are individual variations in basophil responses, other parameters, such as the EC50 (the concentration at which 50% of the maximal basophil response occurs) when performing all the measurements at multiple concentrations, may be better. However, since such methods require more time and cost, single measurement at a given concentration is utilized in many studies. Fifthly, for several fish species the sample size was too small, so a larger-scale study is needed to confirm our findings. Finally, this study is of retrospective design. The key factors and characteristics defining the patients' fish allergy were retrieved from clinical records prepared by others, whose accuracy cannot be confirmed. Prospective studies need to be performed in order to establish the utility of BAT for diagnosis of fish allergy.
In conclusion, the BAT based on CD203c expression may be a reliable method for predicting fish allergy, especially for fish species for which an sIgE test is not available.
ACKNOWLEDGMENTS
This research was supported by a grant-in-aid from the Japan Agency for Medical Research and Development (AMED). The authors would like to thank Ms. Kyoko Nishinaka, Ms. Sumiko Yoshikawa and Ms. Manami Negoro for their excellent technical assistance.
Footnotes
Disclosure: TF has received lecture fees from Maruho Co., Ltd. and MSD K.K. All other authors declare that they have no conflicts of interest.
SUPPLEMENTARY MATERIALS
Characteristics of patients who were sensitized or not sensitized to anisakis
The sIgE levels for Gad c1 in patients who were allergic-/non-allergic to (A) salmon, (B) mackerel and (C) tuna. ROC curves of sIgE levels for Gad C1 for (C) salmon, (D) mackerel and (E) tuna are shown. The AUCs were 0.74 (D), 0.73 (E) and 0.53 (F). Specific IgE levels for Cyp c1 in patients who were allergic/non-allergic to (G) salmon (H), mackerel and (I) tuna. ROC curves of sIgE levels for Gad C1 for (J) salmon, (K) mackerel and (L) tuna are shown. The AUCs were 0.73 (D), 0.71 (E) and 0.53 (F).
References
- 1.Clausen M, Jonasson K, Keil T, Beyer K, Sigurdardottir ST. Fish oil in infancy protects against food allergy in Iceland-results from a birth cohort study. Allergy. 2018;73:1305–1312. doi: 10.1111/all.13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Papamichael MM, Shrestha SK, Itsiopoulos C, Erbas B. The role of fish intake on asthma in children: a meta-analysis of observational studies. Pediatr Allergy Immunol. 2018;29:350–360. doi: 10.1111/pai.12889. [DOI] [PubMed] [Google Scholar]
- 3.Vasileiadou S, Wennergren G, Strömberg Celind F, Åberg N, Pettersson R, Alm B, et al. Eating fish and farm life reduce allergic rhinitis at the age of twelve. Pediatr Allergy Immunol. 2018;29:283–289. doi: 10.1111/pai.12875. [DOI] [PubMed] [Google Scholar]
- 4.Hemmings O, Kwok M, McKendry R, Santos AF. Basophil activation test: old and new applications in allergy. Curr Allergy Asthma Rep. 2018;18:77. doi: 10.1007/s11882-018-0831-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moonesinghe H, Mackenzie H, Venter C, Kilburn S, Turner P, Weir K, et al. Prevalence of fish and shellfish allergy: a systematic review. Ann Allergy Asthma Immunol. 2016;117:264–272.e4. doi: 10.1016/j.anai.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 6.Perez-Gordo M, Cuesta-Herranz J, Maroto AS, Cases B, Ibáñez MD, Vivanco F, et al. Identification of sole parvalbumin as a major allergen: study of cross-reactivity between parvalbumins in a Spanish fish-allergic population. Clin Exp Allergy. 2011;41:750–758. doi: 10.1111/j.1365-2222.2011.03721.x. [DOI] [PubMed] [Google Scholar]
- 7.Kobayashi A, Kobayashi Y, Shiomi K. Fish allergy in patients with parvalbumin-specific immunoglobulin E depends on parvalbumin content rather than molecular differences in the protein among fish species. Biosci Biotechnol Biochem. 2016;80:2018–2021. doi: 10.1080/09168451.2016.1189318. [DOI] [PubMed] [Google Scholar]
- 8.Sharp MF, Stephen JN, Kraft L, Weiss T, Kamath SD, Lopata AL. Immunological cross-reactivity between four distant parvalbumins-Impact on allergen detection and diagnostics. Mol Immunol. 2015;63:437–448. doi: 10.1016/j.molimm.2014.09.019. [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi Y, Kuriyama T, Nakagawara R, Aihara M, Hamada-Sato N. Allergy to fish collagen: Thermostability of collagen and IgE reactivity of patients' sera with extracts of 11 species of bony and cartilaginous fish. Allergol Int. 2016;65:450–458. doi: 10.1016/j.alit.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 10.Hamada Y, Nagashima Y, Shiomi K, Shimojo N, Kohno Y, Shibata R, et al. Reactivity of IgE in fish-allergic patients to fish muscle collagen. Allergol Int. 2003;52:139–147. [Google Scholar]
- 11.Kobayashi Y, Huge J, Imamura S, Hamada-Sato N. Study of the cross-reactivity of fish allergens based on a questionnaire and blood testing. Allergol Int. 2016;65:272–279. doi: 10.1016/j.alit.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 12.Ghasemi Fard S, Wang F, Sinclair AJ, Elliott G, Turchini GM. How does high DHA fish oil affect health? A systematic review of evidence. Crit Rev Food Sci Nutr. 2019;59:1684–1727. doi: 10.1080/10408398.2018.1425978. [DOI] [PubMed] [Google Scholar]
- 13.Dale HF, Madsen L, Lied GA. Fish-derived proteins and their potential to improve human health. Nutr Rev. 2019;77:nuz016. doi: 10.1093/nutrit/nuz016. [DOI] [PubMed] [Google Scholar]
- 14.Ebisawa M, Ito K, Fujisawa T Committee for Japanese Pediatric Guideline for Food Allergy, The Japanese Society of Pediatric Allergy and Clinical Immunology, The Japanese Society of Allergology. Japanese guidelines for food allergy 2017. Allergol Int. 2017;66:248–264. doi: 10.1016/j.alit.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Schulkes KJ, Klemans RJ, Knigge L, de Bruin-Weller M, Bruijnzeel-Koomen CA, Marknell deWitt A, et al. Specific IgE to fish extracts does not predict allergy to specific species within an adult fish allergic population. Clin Transl Allergy. 2014;4:27. doi: 10.1186/2045-7022-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haktanir Abul M, Orhan F. Anaphylaxis after prick-to-prick test with fish. Pediatr Int. 2016;58:503–505. doi: 10.1111/ped.12856. [DOI] [PubMed] [Google Scholar]
- 17.Chinuki Y, Kaneko S, Dekio I, Takahashi H, Tokuda R, Nagao M, et al. CD203c expression-based basophil activation test for diagnosis of wheat-dependent exercise-induced anaphylaxis. J Allergy Clin Immunol. 2012;129:1404–1406. doi: 10.1016/j.jaci.2012.02.049. [DOI] [PubMed] [Google Scholar]
- 18.Hiraguchi Y, Tokuda R, Gen M, Shingaki T, Yoshino S, Kumagai Y, et al. Identification of a novel food allergen in lotus root. Allergol Int. 2018;67:141–143. doi: 10.1016/j.alit.2017.05.006. [DOI] [PubMed] [Google Scholar]
- 19.Shirao K, Inoue M, Tokuda R, Nagao M, Yamaguchi M, Okahata H, et al. “Bitter sweet”: a child case of erythritol-induced anaphylaxis. Allergol Int. 2013;62:269–271. doi: 10.2332/allergolint.12-LE-0517. [DOI] [PubMed] [Google Scholar]
- 20.Tokuda R, Nagao M, Hiraguchi Y, Hosoki K, Matsuda T, Kouno K, et al. Antigen-induced expression of CD203c on basophils predicts IgE-mediated wheat allergy. Allergol Int. 2009;58:193–199. doi: 10.2332/allergolint.08-OA-0023. [DOI] [PubMed] [Google Scholar]
- 21.Mehlich J, Fischer J, Hilger C, Swiontek K, Morisset M, Codreanu-Morel F, et al. The basophil activation test differentiates between patients with alpha-gal syndrome and asymptomatic alpha-gal sensitization. J Allergy Clin Immunol. 2019;143:182–189. doi: 10.1016/j.jaci.2018.06.049. [DOI] [PubMed] [Google Scholar]
- 22.Kondo Y, Kakami M, Koyama H, Yasuda T, Nakajima Y, Kawamura M, et al. IgE cross-reactivity between fish roe (salmon, herring and pollock) and chicken egg in patients anaphylactic to salmon roe. Allergol Int. 2005;54:317–323. [Google Scholar]
- 23.Rubio A, Vivinus-Nébot M, Bourrier T, Saggio B, Albertini M, Bernard A. Benefit of the basophil activation test in deciding when to reintroduce cow's milk in allergic children. Allergy. 2011;66:92–100. doi: 10.1111/j.1398-9995.2010.02432.x. [DOI] [PubMed] [Google Scholar]
- 24.Santos AF, Du Toit G, Douiri A, Radulovic S, Stephens A, Turcanu V, et al. Distinct parameters of the basophil activation test reflect the severity and threshold of allergic reactions to peanut. J Allergy Clin Immunol. 2015;135:179–186. doi: 10.1016/j.jaci.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bühring HJ, Seiffert M, Giesert C, Marxer A, Kanz L, Valent P, et al. The basophil activation marker defined by antibody 97A6 is identical to the ectonucleotide pyrophosphatase/phosphodiesterase 3. Blood. 2001;97:3303–3305. doi: 10.1182/blood.v97.10.3303. [DOI] [PubMed] [Google Scholar]
- 26.Knol EF, Mul FP, Jansen H, Calafat J, Roos D. Monitoring human basophil activation via CD63 monoclonal antibody 435. J Allergy Clin Immunol. 1991;88:328–338. doi: 10.1016/0091-6749(91)90094-5. [DOI] [PubMed] [Google Scholar]
- 27.Santos AF, James LK, Bahnson HT, Shamji MH, Couto-Francisco NC, Islam S, et al. IgG4 inhibits peanut-induced basophil and mast cell activation in peanut-tolerant children sensitized to peanut major allergens. J Allergy Clin Immunol. 2015;135:1249–1256. doi: 10.1016/j.jaci.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sato S, Tachimoto H, Shukuya A, Kurosaka N, Yanagida N, Utsunomiya T, et al. Basophil activation marker CD203c is useful in the diagnosis of hen's egg and cow's milk allergies in children. Int Arch Allergy Immunol. 2010;152(Suppl 1):54–61. doi: 10.1159/000312126. [DOI] [PubMed] [Google Scholar]
- 29.Berin MC, Grishin A, Masilamani M, Leung DY, Sicherer SH, Jones SM, et al. Egg-specific IgE and basophil activation but not egg-specific T-cell counts correlate with phenotypes of clinical egg allergy. J Allergy Clin Immunol. 2018;142:149–158.e8. doi: 10.1016/j.jaci.2018.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Itoh-Nagato N, Inoue Y, Nagao M, Fujisawa T, Shimojo N, Iwata T, et al. Desensitization to a whole egg by rush oral immunotherapy improves the quality of life of guardians: a multicenter, randomized, parallel-group, delayed-start design study. Allergol Int. 2018;67:209–216. doi: 10.1016/j.alit.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 31.Sørensen M, Kuehn A, Mills EN, Costello CA, Ollert M, Småbrekke L, et al. Cross-reactivity in fish allergy: a double-blind, placebo-controlled food-challenge trial. J Allergy Clin Immunol. 2017;140:1170–1172. doi: 10.1016/j.jaci.2017.03.043. [DOI] [PubMed] [Google Scholar]
- 32.Bugajska-Schretter A, Elfman L, Fuchs T, Kapiotis S, Rumpold H, Valenta R, et al. Parvalbumin, a cross-reactive fish allergen, contains IgE-binding epitopes sensitive to periodate treatment and Ca2+ depletion. J Allergy Clin Immunol. 1998;101:67–74. doi: 10.1016/S0091-6749(98)70195-2. [DOI] [PubMed] [Google Scholar]
- 33.Kuehn A, Hilger C, Lehners-Weber C, Codreanu-Morel F, Morisset M, Metz-Favre C, et al. Identification of enolases and aldolases as important fish allergens in cod, salmon and tuna: component resolved diagnosis using parvalbumin and the new allergens. Clin Exp Allergy. 2013;43:811–822. doi: 10.1111/cea.12117. [DOI] [PubMed] [Google Scholar]
- 34.Kobayashi Y, Akiyama H, Huge J, Kubota H, Chikazawa S, Satoh T, et al. Fish collagen is an important panallergen in the Japanese population. Allergy. 2016;71:720–723. doi: 10.1111/all.12836. [DOI] [PubMed] [Google Scholar]
- 35.Matricardi PM, Kleine-Tebbe J, Hoffmann HJ, Valenta R, Hilger C, Hofmaier S, et al. EAACI molecular allergology user's guide. Pediatr Allergy Immunol. 2016;27(Suppl 23):1–250. doi: 10.1111/pai.12563. [DOI] [PubMed] [Google Scholar]
- 36.Kuehn A, Scheuermann T, Hilger C, Hentges F. Important variations in parvalbumin content in common fish species: a factor possibly contributing to variable allergenicity. Int Arch Allergy Immunol. 2010;153:359–366. doi: 10.1159/000316346. [DOI] [PubMed] [Google Scholar]
- 37.Nieuwenhuizen NE, Lopata AL. Allergic reactions to Anisakis found in fish. Curr Allergy Asthma Rep. 2014;14:455. doi: 10.1007/s11882-014-0455-3. [DOI] [PubMed] [Google Scholar]
- 38.Berger SA, Marr JS. Human parasitic diseases sourcebook. Burlington: Jones & Bartlett Learning; 2006. [Google Scholar]
- 39.Murphy TM, Berzano M, O'Keeffe SM, Cotter DM, McEvoy SE, Thomas KA, et al. Anisakid larvae in Atlantic salmon (Salmo salar L.) grilse and post-smolts: molecular identification and histopathology. J Parasitol. 2010;96:77–82. doi: 10.1645/GE-2194.1. [DOI] [PubMed] [Google Scholar]
- 40.Vidacek S, de las Heras C, Solas MT, Mendizábal A, Rodriguez-Mahillo AI, Tejada M. Antigenicity and viability of Anisakis larvae infesting hake heated at different time-temperature conditions. J Food Prot. 2010;73:62–68. doi: 10.4315/0362-028x-73.1.62. [DOI] [PubMed] [Google Scholar]
- 41.Gyimesi E, Gönczi F, Szilasi M, Pál G, Baráth S, Sipka S. The effects of various doses of bacterial lipopolysaccharide on the expression of CD63 and the release of histamine by basophils of atopic and non-atopic patients. Inflamm Res. 2013;62:213–218. doi: 10.1007/s00011-012-0569-9. [DOI] [PubMed] [Google Scholar]
- 42.Smith TF, Aelvoet M, Morrison DC. The effect of bacterial lipopolysaccharide (LPS) on histamine release from human basophils. I. Enhancement of immunologic release by LPS. Clin Immunol Immunopathol. 1985;34:355–365. doi: 10.1016/0090-1229(85)90184-9. [DOI] [PubMed] [Google Scholar]
- 43.Wassenaar TM, Zimmermann K. Lipopolysaccharides in food, food supplements, and probiotics: should we be worried? Eur J Microbiol Immunol (Bp) 2018;8:63–69. doi: 10.1556/1886.2018.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characteristics of patients who were sensitized or not sensitized to anisakis
The sIgE levels for Gad c1 in patients who were allergic-/non-allergic to (A) salmon, (B) mackerel and (C) tuna. ROC curves of sIgE levels for Gad C1 for (C) salmon, (D) mackerel and (E) tuna are shown. The AUCs were 0.74 (D), 0.73 (E) and 0.53 (F). Specific IgE levels for Cyp c1 in patients who were allergic/non-allergic to (G) salmon (H), mackerel and (I) tuna. ROC curves of sIgE levels for Gad C1 for (J) salmon, (K) mackerel and (L) tuna are shown. The AUCs were 0.73 (D), 0.71 (E) and 0.53 (F).