Summary Statement
Skeletal muscle fusion occurs during development, growth, and regeneration. To investigate how muscle fusion compares among different muscle cell types and developmental stages, we studied muscle cell fusion over time in wild-type, myomaker (mymk), and jam2a mutant zebrafish. Using live imaging, we show that embryonic myoblast elongation and fusion correlate tightly with slow muscle cell migration. In wild-type embryos, only fast muscle fibers are multinucleate, consistent with previous work showing that the cell fusion regulator gene mymk is specifically expressed throughout the embryonic fast muscle domain. However, by 3 weeks post-fertilization, slow muscle fibers also become multinucleate. At this late-larval stage, mymk is not expressed in muscle fibers, but is expressed in small cells near muscle fibers. Although previous work showed that both mymk and jam2a are required for embryonic fast muscle cell fusion, we observe that muscle force and function is almost normal in mymk and jam2a mutant embryos, despite the lack of fast muscle multinucleation. We show that genetic requirements change post-embryonically, with jam2a becoming much less important by late-larval stages and mymk now required for muscle fusion and growth in both fast and slow muscle cell types. Correspondingly, adult mymk mutants perform poorly in sprint and endurance tests compared to wild-type and jam2a mutants. We show that adult mymk mutant muscle contains small mononucleate myofibers with average myonuclear domain size equivalent to that in wild type adults. The mymk mutant fibers have decreased Laminin expression and increased numbers of Pax7-positive cells, suggesting that impaired fiber growth and active regeneration contribute to the muscle phenotype. Our findings identify several aspects of muscle fusion that change with time in slow and fast fibers as zebrafish develop beyond embryonic stages.
Keywords: Myomaker, Jam2a, Slow-twitch muscle, Type I muscle fiber, Fast-twitch muscle
Introduction
Cell-cell fusion merges individual cells, generating syncytial tissue, and is critical for development of organs such as placenta, bone, and skeletal muscle (Aguilar et al., 2013). As a genetically programmed process, cell-cell fusion employs three characteristic stages: 1) cellular differentiation and fusion competence, 2) cell recognition and adhesion that commits cells to fusion, and 3) lipid destabilization and membrane merging resulting in fusion (Aguilar et al., 2013; Kim et al., 2015). Each process uses key regulators to transition to the next fusion phase. Each of these stages is observed during myoblast fusion (Kim et al., 2015), generating the syncytial tissue required for skeletal muscle development, growth, and regeneration (Goh and Millay, 2017; Millay et al., 2014, 2013; White et al., 2010). Although many molecules are known to regulate muscle cell fusion, it remains unclear how muscle cell fusion mechanisms change over the course of development.
Myoblast fusion in Drosophila is well-characterized and uses two transcriptionally-distinct types of myoblasts: founder cells (FCs) and fusion-competent myoblasts (FCMs) (Abmayr and Pavlath, 2012; Kim et al., 2015; Rochlin et al., 2010). The asymmetric fusion process requires cell adhesion molecule-mediated recognition and adhesion, actin cytoskeleton-directed FCM protrusions into FCs, and membrane destabilization to allow cell merging (Kim et al., 2015; Ruiz-Gomez et al., 2000). FC asymmetry is maintained by endocytic recycling of the FCM membrane protein, Sticks and Stones, enabling multiple fusion events (Haralalka et al., 2014). Thus, asymmetric cell-cell interactions lead to fusion in fruit fly muscles.
Several conserved Drosophila fusion genes have been implicated in vertebrate muscle fusion (Baas et al., 2012; Moore et al., 2007; Pajcini et al., 2008; Sampath et al., 2018; Sohn et al., 2009; Srinivas et al., 2007; Tamir-Livne et al., 2017). Vertebrate muscle fusion also requires several genes not found in flies (Bi et al., 2017; Landemaine et al., 2014; Powell and Wright, 2011; Quinn et al., 2017; Zhang et al., 2017). For instance, the vertebrate gene Myomaker (Mymk) is a muscle-specific multi-pass transmembrane protein required for skeletal muscle fusion in mouse, chick, and zebrafish (Di Gioia et al., 2017; Landemaine et al., 2014; Luo et al., 2015; Millay et al., 2013; Zhang and Roy, 2017). If expressed in a non-muscle cell like a fibroblast, Mymk can facilitate fusion with a myoblast (Millay et al., 2013) and Mymk overexpression causes hyper-fusion, indicating that it is sufficient to induce skeletal muscle fusion (Millay et al., 2013; Zhang and Roy, 2017). A recent report shows that Mymk initiates membrane fusion by facilitating mixing between the outer cell membrane leaflet, in a process called hemifusion (Leikina et al., 2018). After Mymk initiates hemifusion, another fusogen (called Myomerger, Minion, or Myomixer) generates fusion pores, acting in trans with Mymk, leading to fusion (Bi et al., 2017; Leikina et al., 2018; Quinn et al., 2017; Shi et al., 2017; Zhang et al., 2017). It remains unclear how membranes are disposed of after pore formation, and whether these fusion mechanisms are consistent during different phases of myogenesis.
Skeletal muscle fusion contributes not only to embryonic muscle formation, but also to muscle growth and regeneration (Goh and Millay, 2017; Millay et al., 2014). Muscle growth is generally categorized as either the fusion-dependent addition of new fibers (hyperplasia) or the growth of existing fibers (hypertrophy). In mammalian embryos both modes contribute to muscle growth but fiber addition stops shortly after birth and thereafter occurs only by expansion of existing muscle fibers (Fiorotto, 2012; Matsakas et al., 2010; White et al., 2010). In early postnatal development, mammalian myofibers grow both by expanding the myofiber volume supported by each nucleus (the myonuclear domain) and by fusion-dependent addition of new nuclei (Mantilla et al., 2008; White et al., 2010). After the third week of postnatal development, muscle continues to grow through adulthood by expanding the myonuclear domain size without addition of new nuclei (White et al., 2010). In the adult mouse, stem cell-mediated muscle fusion occurs only after muscle injury or intense exercise. Mymk is transiently expressed in muscle progenitors during fusion-dependent muscle growth (Goh and Millay, 2017) and is required during muscle growth and regeneration after injury (Goh and Millay, 2017; Millay et al., 2014). Because Mymk is essential for fusion-dependent processes in both embryonic and adult muscle (Di Gioia et al., 2017; Goh and Millay, 2017; Landemaine et al., 2014; Millay et al., 2013; Shi et al., 2018; Zhang and Roy, 2017), we use zebrafish mymk mutants to investigate how fusion influences skeletal muscle development, growth, and regeneration.
In zebrafish embryos, only fast-twitch muscle precursors fuse, and fast muscle cell fusion is coordinated with slow-twitch fiber morphogenesis (Cortés et al., 2003; Devoto et al., 1996; Henry and Amacher, 2004; Roy et al., 2001; Snow et al., 2008). Fast-twitch and slow-twitch fibers (hereafter, “fast fibers” and “slow fibers”) are not intermingled but are spatially segregated (Devoto et al., 1996; Roy et al., 2001). Slow muscle identity is specified in medially-located cells that then migrate laterally through the fast muscle precursors (Cortés et al., 2003; Devoto et al., 1996; Henry and Amacher, 2004). After migration, the slow fibers comprise a thin layer of superficially-localized muscle that overlays the much larger fast muscle mass (Devoto et al., 1996). This migration is thought to trigger fast muscle morphogenesis including myoblast fusion (Cortés et al., 2003; Devoto et al., 1996; Henry and Amacher, 2004; Roy et al., 2001; Snow et al., 2008). Fusion appears symmetric in zebrafish embryos, because all known fusogens are expressed uniformly in fast muscle precursors, and because mosaic experiments using fusion-deficient cells demonstrated that any fast-type myoblast can initiate fusion (Powell and Wright, 2012). Heterophilic interaction between vertebrate-specific junction adhesion molecules Jam2a and Jam3b is required for zebrafish myoblast fusion (Powell and Wright, 2011), though, the mammalian orthologs, Jam2 and Jam3, are not required for muscle fusion at the developmental stages examined (Arcangeli et al., 2014; Gliki et al., 2004; Powell and Wright, 2011). When embryonic muscle fusion is blocked, as in zebrafish jam2a mutants, jam3b mutants (Powell and Wright, 2011; Sawamiphak et al., 2017; Zhang and Roy, 2016), and mymk mutants (Shi et al., 2018), muscle progenitors form functional mononucleate muscle fibers. Zebrafish muscle undergoes continuous fusion-dependent hypertrophic and hyperplastic growth from embryonic through adult stages (Barresi et al., 2001; Gurevich et al., 2015; Nguyen et al., 2017; Patterson et al., 2008; Roy et al., 2017). Post-embryonic growth phases are thought to employ Pax7-positive satellite-like cells, which are also used during muscle regeneration (Berberoglu et al., 2017; Gurevich et al., 2016; Nguyen et al., 2017; Pipalia et al., 2016; Seger et al., 2011). However, little is known about fusion mechanisms in late-larval or adult zebrafish.
In this study, we characterize zebrafish muscle fusion at different developmental stages. Using live imaging we visualize cell-cell fusion in zebrafish embryos, revealing membrane breakdown and dispersal events that produce multinucleate myofibers. We investigate genetic requirements for fiber fusion and find that while mymk is required throughout development, jam2a is essential only through the first week of development. While myofibers become multinucleate in jam2a mutants by the second week of development, they have fewer nuclei than wild-type adults, suggesting thatjam2a plays a nominal role in fusion at later stages. In adult jam2a fibers with reduced myonuclei number and in mononucleate adult mymk mutant fibers, the total fiber volume per nucleus closely matches that of wild-type fibers, suggesting that myonuclear domain is established independent of fusion capacity. Even though mymk is required for fusion at all developmental stages, mymk expression becomes more restricted by the late-larval stage, consistent with the idea that cell fusion is differentially regulated over developmental time. Surprisingly, we show that slow fibers, which are strictly mononucleate during embryonic stages, become multinucleate by the late-larval stage, and that this process is mymk-dependent. Lastly, proliferation and regeneration markers are expressed in mymk mutant adult muscle, implying that regenerative pathways are activated. Our findings reveal that fusion mechanisms change after embryonic stages and indicate that skeletal muscle growth and maintenance require skeletal muscle fusion.
Results
2.1. Time-lapse imaging reveals cell-cell interactions leading to myoblast fusion
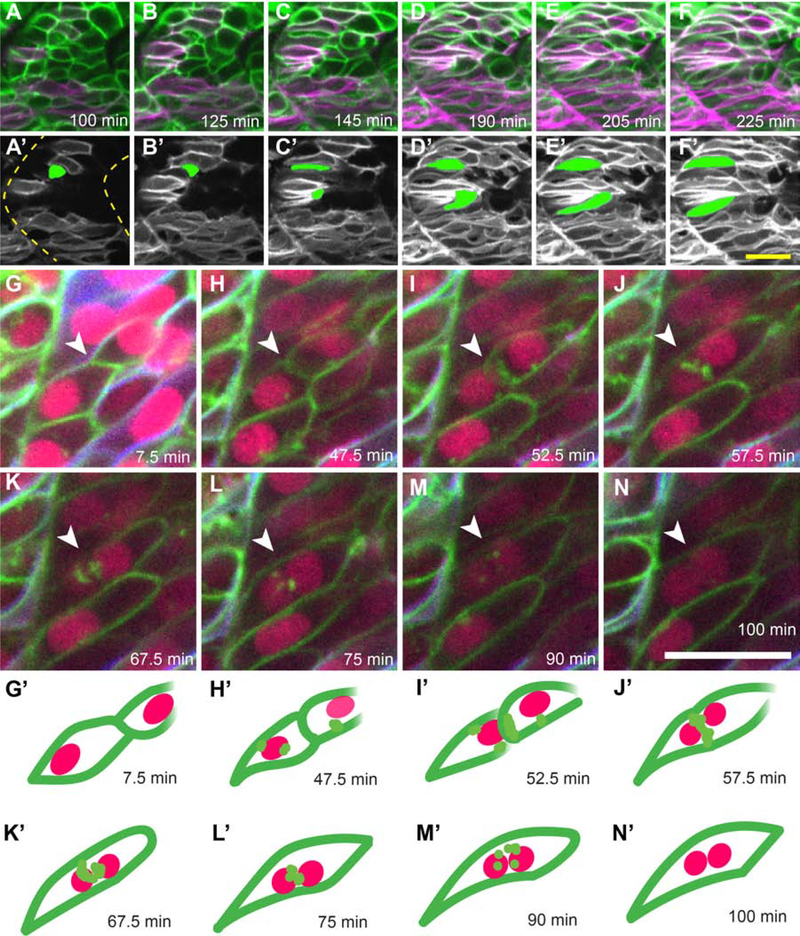
The zebrafish model has long been established for studying embryonic skeletal muscle fusion (Cortés et al., 2003; Devoto et al., 1996; Henry and Amacher, 2004; Roy et al., 2001; Snow et al., 2008). To capture myoblast fusion events in vivo, we performed confocal time-lapse imaging at 20 hours post fertilization (hpf) of embryos injected with mRNA encoding nuclear-localized CFP and carrying two transgenes: six1b:lyn-GFP that marks the plasma membrane of fast muscle precursors and smyhc1:lyn-tdTomato that marks the cytoplasm of slow muscle cell precursors (Talbot et al., In press; Wang et al., 2011). As expected, slow muscle remains mononucleate during early embryogenesis (Roy et al., 2001). However, fast muscle precursors often begin to elongate and almost simultaneously begin fusing when slow muscle cells migrate past them (Cortés et al., 2003; Henry and Amacher, 2004; Roy et al., 2001) (Fig. 1; Supplemental Movies 1–3). While interacting with slow muscle cells, fast muscle precursors elongate anteriorly and attach to the somite boundary (termed boundary capture, (Snow et al., 2008)) (Fig. 1 A–F’). Following boundary capture, fast muscle myoblasts fuse posteriorly until the growing muscle fiber spans the somite (Supplemental movie 1). Fusion begins between neighboring fast myoblasts after they contact slow muscle cells (Fig. 1 G, H). Then, small gaps appear in membranes at the interface between the two fusing cells (Fig. 1 I–J). Lastly, the plasma membrane breaks down and disperses into small GFP-positive punctae (Fig. 1 K–M), creating a binucleate fast muscle cell (Fig. 1 N). From membrane coalescence to binucleate cell formation, the fusion process takes approximately 1 hour (62.5 ± 3.15 min, n=4; ± indicates standard deviation). Thus, migrating slow muscle cells appear to trigger fast muscle precursor elongation towards the anterior edge of the somite, after which, myoblast fusion occurs from anterior to posterior within the somite.
Figure 1: Live imaging of slow muscle cell migration concurrent with fast muscle fusion events.
(A-F) Frames from a time-lapse movie (Supplemental Movie 1) of a 19 hpf six1b:lyn-GFP (green); smyhc1:lyn-tdTomato (magenta) double transgenic embryo. (A’-F’) Pseudo-colored images show the slow muscle channel (white) from frames (A-F) overlaid with the shape of two fast muscle precursors (green) as they interact with slow muscle cells and extend anteriorly. Dotted outlines in A’ show somite boundaries (yellow). (G-N) Frames from a time-lapse movie of a 20 hpf six1b:lyn-GFP (green); smyhc1:lyn-tdTomato (blue) double transgenic embryo injected with mRNA encoding H2B-CFP (fuchsia). The movie (Supplemental Movie 2), which spans 92.5 minutes, was taken when fusion is actively occurring. (G’-N’) Illustrations depict a pair of fusing cells from Supplemental Movie 2 (G-N; white arrowhead). The time stamp of each frame is indicated. Scale bars in F’ (for A-F’) and N (for G-N) are 20 μm.
2.2. mymk is expressed in fast muscle cells
In zebrafish, the fusogen gene mymk is expressed specifically in the fast muscle domain (Di Gioia et al., 2017; Landemaine et al., 2014; Zhang and Roy, 2017) beginning as early as the 10 somite stage (Fig. S1 A–B’). As expected, when slow muscle fate is blocked by treatment with the Smoothened inhibitor Cyclopamine, mymk expression appears normal, consistent with fast muscle-specific expression (Fig. S1 C–D’). In prdm1a mutants, where slow muscle precursors (adaxial cells) adopt a fast muscle identity (von Hofsten et al., 2008), mymk expression expands into adaxial cells (Fig. S1 E–F”). These data provide additional evidence that mymk is expressed specifically in the embryonic fast muscle domain.
2.3. Fast myoblasts do not fuse in mymk mutant embryos
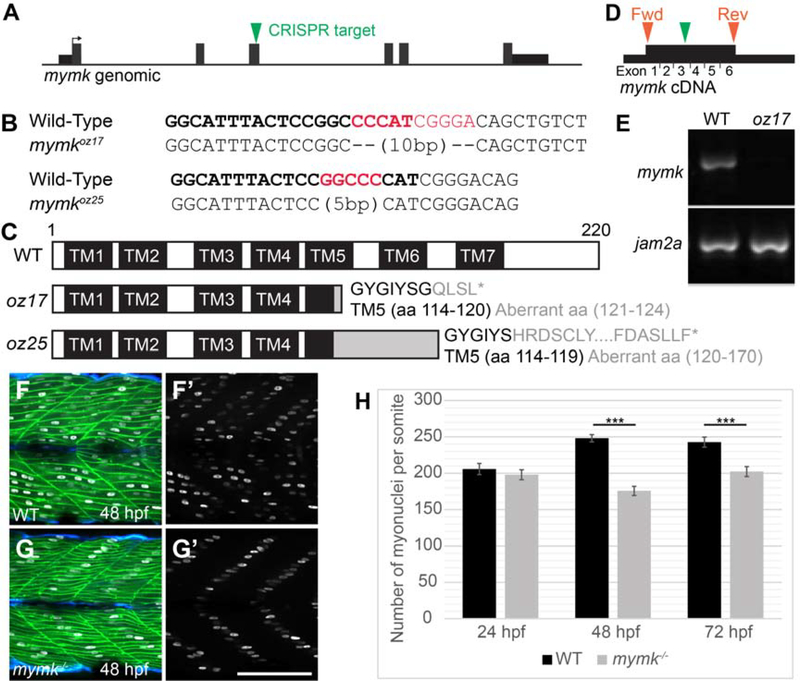
To genetically characterize skeletal muscle fusion in zebrafish, we used CRISPR/Cas9 technology to generate mutations in mymk, a gene encoding a muscle-specific transmembrane protein necessary and sufficient for skeletal muscle cell fusion in mammals (Millay et al., 2013) and chick (Luo et al., 2015). mymk is also required for fast muscle cell fusion in zebrafish, as recently shown using genetic mutants (Di Gioia et al., 2017; Shi et al., 2018; Si et al., 2019; Zhang and Roy, 2017). We targeted the third mymk exon to generate two additional alleles: mymkoz17, a 10 bp deletion and mymkoz25, a 5 bp deletion (Fig. 2 A, B). Both lesions are predicted to frameshift mymk after amino acid 170, before the last two transmembrane domains and the intracellular C-terminal domain (Fig. 2 C). Because the C-terminal domain is essential for mouse Mymk function (Millay et al., 2016), both zebrafish mymk mutant alleles are predicted to encode nonfunctional proteins. mymk transcript is not detected at 48 hpf in mymkozr7 mutants, suggesting that the mutant transcript is subject to nonsense-mediated decay and providing additional evidence that mymkoz17 is a strong loss of function allele (Fig. 2 D, E). Consistent with recent studies, we find that embryonic fast muscle fusion fails in zebrafish mymkoz17 mutants (Fig. 2 F–G) and in mymkoz25 mutant embryos (data not shown). By 48 hpf, the single myonucleus in each muscle cell characteristically aligns about halfway between fiber tips (Fig. 2 F–G’), similar to other fusion-deficient mutants like jam2a, jam3b, and myog (Ganassi et al., 2018; Powell and Wright, 2011). Although fusion is compromised in mymk mutants, myonuclei number is normal at 24 hpf, suggesting that most mymk mutant fast muscle precursors generate mononucleate myofibers at this stage (Fig. 2 G, H). After 24 hpf, myonuclei number increases in wild-type siblings, likely due to secondary fusion events. In mymk mutants, myonuclei number does not increase between 24 and 72 hpf, suggesting that fusion is needed for myonuclei addition during this time interval (Fig. 2H). Together, these data are consistent with previous work indicating that mymk is required for embryonic myoblast fusion (Di Gioia et al., 2017; Shi et al., 2018; Si et al., 2019; Zhang and Roy, 2017) and suggest that fusion is needed for a normal rate of myonuclei addition.
Figure 2: Embryonic skeletal muscle fusion requires mymk.
(A) The mymkCRISPR target site is within exon 3. (B) DNA lesions for mymkoz17 and mymkoz25 induced at the CRISPR target sequence (bold text), with the corresponding wild-type (WT) sequences (red text is deleted in mutant). (C) The resulting frame-shifting mutations are predicted to introduce 4 and 30 aberrant amino acids respectively (grey text), followed by a premature stop codon (*) that would truncate the protein in transmembrane (TM) domain 5 and eliminate C-terminal fusogenic elements (Millay et al. 2016). (D) Forward (fwd) and reverse (rev) primers (orange arrowheads) for RT-PCR amplify exons 1 through 6 of the cDNA including the mymkoz17 lesion site in exon 3 (green arrowhead). (E) RT-PCR of mymk and jam2a transcripts in wild-type (WT) and mymkoz17 mutant individuals at 48 hpf. (F-G’) Wild-type embryos (F, F’) contain multinucleated fast fibers at 48 hpf, while fast fibers in mymkoz17 mutant embryos (G, G’) are mononucleate, with the single nucleus in each fiber located about midway between fiber tips. myog:H2B-mRFP (white) marks myonuclei, mylfpa:lyn-cyan (green) labels fast muscle cells, and smyhc1:EGFP (aqua) labels slow muscle cells. (H) The total number of myonuclei per somite is reduced in mymk mutants compared to WT siblings beginning at 48 hpf. Myonuclei in somites 12 and 15 of each embryo were counted and averaged to get the number per somite (n= 6 for 24 hpf, n=8 for 48 hpf and 72 hpf). (Student’s t-test, p*** < 0.001). Scale bar in E’ (for D-E’) is 100 μm.
2.4. Fast muscle growth and maintenance differentially requires zebrafish embryonic fusion regulators mymk and jam2a
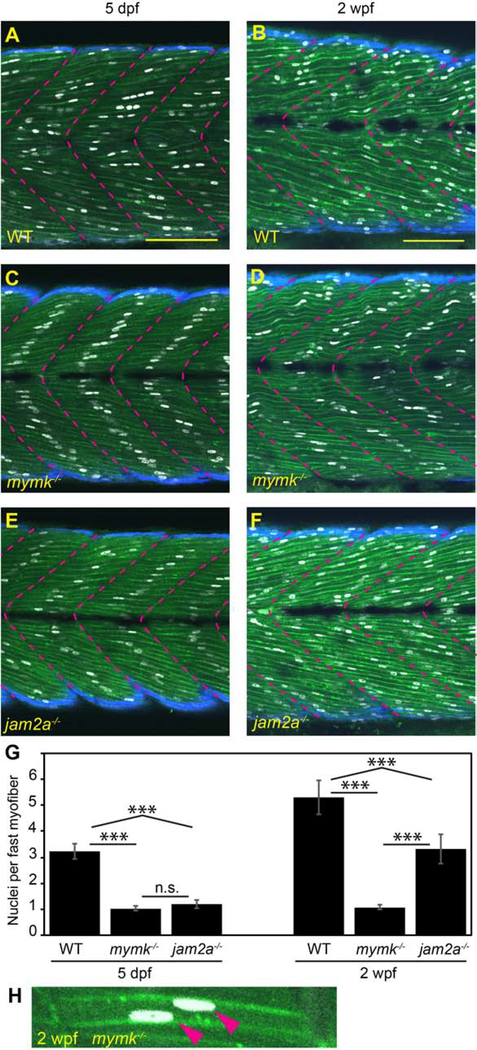
While mymk is thought to be required for fusion throughout development, recent findings have indicated that jam2a, which in embryonic stages is essential for fusion, is not needed by adulthood (Si et al., 2019). To learn when fusion phenotypes diverge between these mutants, we used a “3MuscleGlow” (3MG) transgenic combination, Tg(smyhc1:EGFP)i104 (Elworthy et al., 2008), Tg(mylfpa:lyn-cyan)fb122 (Ignatius et al., 2012), and Tg(myog:H2B-mRFP)fb121 (Tang et al., 2016), to visualize slow muscle cells, fast muscle cells, and myonuclei, respectively (Fig. 3 A–F). When examined at 5 dpf and 2 weeks post fertilization (wpf), wild-type fish are multinucleate and mymk mutants are essentially mononucleate (Fig 3 A–D, G). Only 3% (6/200) of mymk fast fibers were binucleate at 5 dpf and 7% (14/200) were binucleate at 2 wpf (Fig 3G, H). In comparison, jam2a mutant fibers, which are typically mononucleate at 5 dpf, become multinucleate by 2 wpf, having about 3 nuclei per cell (Fig 3 E, F, G). These findings reveal that the requirements for jam2a and mymk in muscle fusion diverge during early larval development.
Figure 3: jam2a is no longer required for multinucleation by 2 weeks post fertilization.
(A-F) Sagittal confocal sections of 3MuscleGlow transgenic fish at 5 dpf (A, C, E) and 2 wpf (B, D, F). myog:H2B-mRFP (white) labels myonuclei, mylfpa:lyn-cyan (green) marks fast muscle cell membranes, and smyhc1:EGFP (aqua) marks slow muscle cells; myotomal boundaries are indicated by dotted lines (magenta). (G) Quantitative analysis reveals that mymk mutant myofibers are typically mononucleate at both stages. jam2a mutant myofibers, which are typically mononucleate at 5 dpf, are consistently multinucleate at 2 wpf. (H) An example of a rare mymk mutant fast myofiber with two nuclei (red arrows). Scale bars in A (for A, C, E) and B (for B, D, E) are 100 μm. Significance determined by ANOVA with Tukey-Kramer post-hoc analysis (p*** < 0.001 compared to WT; n.s. indicates not significant).
2.5. Myofiber growth and organization requires mymk
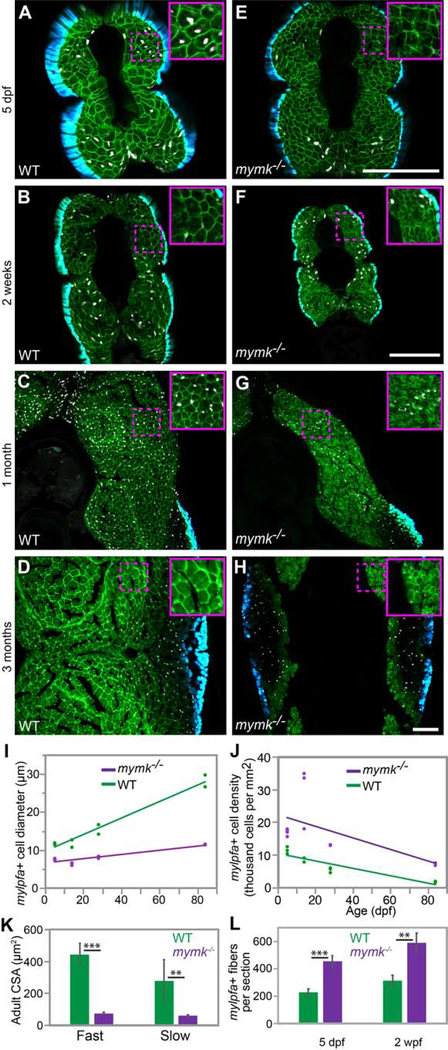
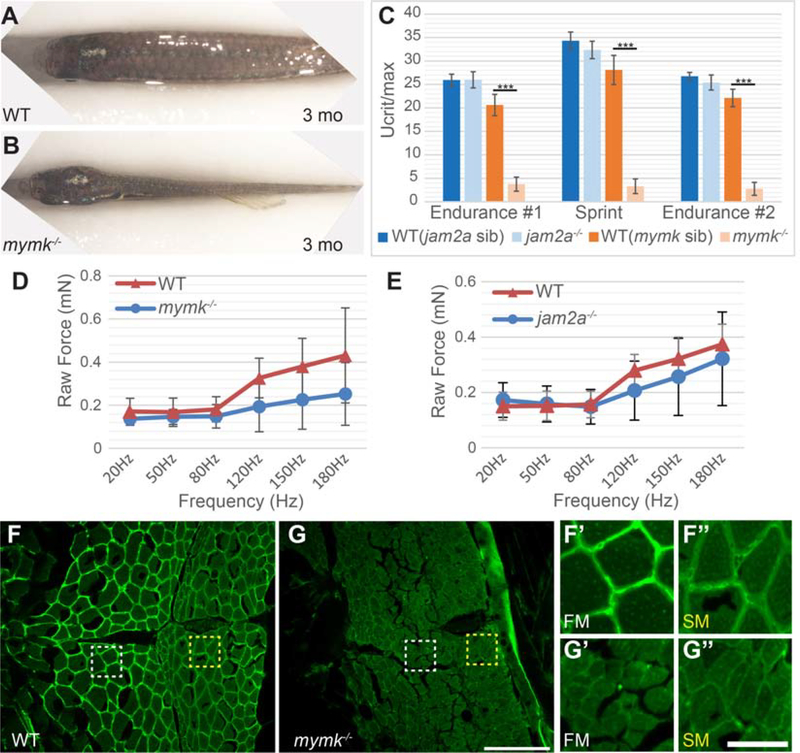
Because muscle volume correlates with myonuclei number, we hypothesized that mymk function is key to muscle growth. We examined mymk mutants from larval through adult stages using the 3MuscleGlow transgenes (Figure 4 A–H) and observe that mymk mutant fast muscle cells, including myoblasts and myofibers, are smaller in diameter and more densely packed than wild-type siblings at every time point examined (Fig. 4 I, J), consistent with the idea that fusion is important for muscle growth. Myofiber cross-sectional area is also smaller in mymk mutant versus wild-type adults (Fig. 4 K). Consistent with the mymk cell fusion defect, the total number of fibers per section is higher in 5 dpf and 2 wpf mymk mutants than wild-type animals (Figure 4 L). As expected, fast muscle cell diameter increases over time in both genotypes, though the mymk mutants take 1 month to achieve a cell diameter that wild-type animals attain in only 5 days (Fig. 4 I). In mymk adults, we find small diameter muscle fibers throughout both slow and fast muscle regions (Fig. S2). These mutant fibers may be small due to growth defects, fiber wasting, or because they are newly formed. Additionally, the distinct boundary between fast and slow muscle domains that is present in wild-type muscle is irregular in adult mymk mutants; in mymk mutants, we find slow muscle fibers within the fast muscle domain (Fig. S2 B) and vice versa (Fig. S2 B”’). These data reveal that mymk is required for larval and adult muscle fiber growth and organization.
Figure 4: mymk is required for normal myofiber growth.
Transverse sections through the mid-trunk region of wild-type (WT) (A-D) and mymkoz17 (E-H) 3MuscleGlow transgenic fish at 5 days, 2 weeks, 1 month, and 3 months post-fertilization. myog:H2B-mRFP (white) labels myonuclei, mylfpa:lyn-cyan (green) marks fast muscle cell membranes, and smyhc1:EGFP (aqua) marks slow muscle cells. Insets show magnified images of regions indicated by the dotted box. (I) Diameter of mylpfa-expressing fast muscle cells in WT and mymkoz17 individuals at the same stages, with trends over time determined by linear regression (model R2 is 0.97). (J) Fast muscle cell density is shown for the same WT (green) and mymkoz17 (purple) individuals as in (I) and regression lines show trends through time (model R2 is 0.63). Linear regression was performed in JMP using standard least squares modeling. (K) Adult myofiber cross-sectional area is higher in WT than mymkoz17 in both slow and fast myofiber types. (L) Total myofiber counts per section are lower in WT than in mymkoz17 at 5 dpf and 2 wpf. Scale bars in E (for A, E), in F (for B, F, C, G), and in H (for D, H) are 100 μm. Student’s t-test, p**<0.01, p***< 0.001.
2.6. mymk is necessary for normal adult muscle size and strength
mymkoz17 mutant adults are viable but have severe defects. Adult mymk mutants have phenotypes consistent with muscle reduction, including narrow bodies (Fig. 5 A, B) and often an open jaw (Fig. S3) (Di Gioia et al., 2017). Even though mymk and jam2a mutant embryos have remarkably similar embryonic muscle fusion defects, adult mymk and jam2a mutants appear strikingly different, with jam2a mutants indistinguishable from wild-type (data not shown; Si et al., 2018; Zhang and Roy, 2016), suggesting that myofiber number and or size differs in adult mymk and jam2a mutants. Because skeletal muscle cell size correlates with force and power (Russell et al., 2000), we tested skeletal muscle strength in adult mymk and jam2a mutants using a swim tunnel assay. We find that mymk mutants perform poorly compared to wild-type siblings and jam2a mutants in both endurance and sprint assays (Fig. 5 C). jam2a mutant performance is comparable to that of wild-type siblings suggesting that adult muscle strength does not require jam2a function (Fig. 5 C). We also measured larval muscle force at 3 dpf using an in vivo contractile strength assay (Martin et al., 2015). The assay records the force generated by whole body contraction after pulsed electrical stimulation; these pulses begin at low frequency (20 Hz) and then are applied at increasing frequencies until the muscle reaches a fused tetanic contraction (180 Hz). At 3 dpf, raw contractile force at frequencies higher than 50 Hz trends lower in jam2a and mymk mutants compared to wild-type siblings (Fig. 5 D, E), but the difference (calculated by ANOVA) is not statistically significant. Together, these data indicate that larval muscle can function normally when mononucleate, but adult muscle cannot. Since adult mymk mutant muscle fusion defects correlate with reduced muscle strength, we investigated whether mymk mutant fibers show signs of structural weakness or damage by assessing Laminin expression in the extracellular matrix, a commonly used measure of muscle integrity (Acharyya et al., 2005; He et al., 2013). We find that Laminin expression is substantially decreased in both fast and slow mymk mutant adult muscle domains compared to wild-type muscle (Fig. 5 F–G”). Our findings suggest that mymk mutant fusion defects lead to marked reductions in zebrafish muscle function and integrity by adult stages.
Figure 5: Muscle performance is severely compromised in adult mymk mutants.
(A,B) Dorsal view of wild-type (WT) (A) and mymk mutant (B) adult fish showing the difference in muscle size. jam2a individuals are indistinguishable from wild-type (Si et al., 2018; Zhang and Roy, 2017; and data not shown). (C) Swim tunnel performance of adult fish (3–6 months) reveals a dramatic difference in endurance and sprint capacity of mymk mutant and wild-type siblings (n=5 each) but no difference between jam2a mutant and wild-type siblings (n=6 each). (D,E) Assays of raw contractile force in stimulated 3 dpf mymk mutant and sibling control larvae (D) (n=3–10 tested per frequency) and jam2a mutant and sibling control larvae (E) (n=4 WT and n=6 jam2a mutant embryos tested per frequency) indicate no significant force difference among comparable genotypes. At higher frequencies, muscle-specific force trends lower in both mutants but the difference is not statistically significant. (F-G”) Transverse sections of adult WT (F-F”) and mymk mutants (G-G”), near the horizontal myoseptum, immunolabeled to detect Laminin (green). Magnified views of boxed regions in F and G show fast muscle (FM; white box) (F’ and G’) and slow muscle (SM; yellow box) (F” and G”) regions of WT and mymk mutant individuals respectively. Scale bar in G (for F and G) is 100 μm and scale bar in G” (for F’-G”) is 25 μm. Student’s t-test, p*** < 0.001.
2.7. mymk mutant muscle expresses markers of nascent myofibers
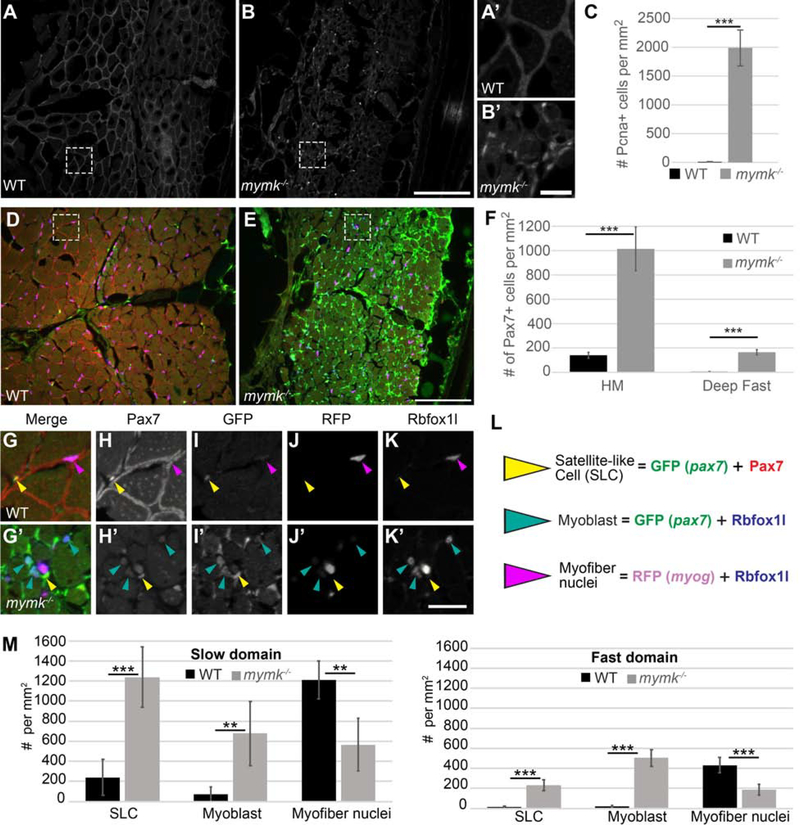
Adult mymk mutant muscle fibers weakly express Laminin (Fig. 5 G) and are small in diameter (Fig 4 H, I), suggesting that they may be immature. To investigate this possibility, we assessed typical hallmarks of regenerating muscle (Berberoglu et al., 2017; Gurevich et al., 2016; Pipalia et al., 2016; Saera-Vila et al., 2016, 2015; Tee et al., 2012). To assess proliferation, we quantified cells expressing Pcna, an established proliferation marker (Fig. 6 A–C) (Dray et al., 2015; Katz et al., 2016; Panza et al., 2015). We observe > 100-fold more proliferating cells within adult mymk mutant muscle than in wild-type siblings (Fig. 6 C). To assess whether satellite cells are activated in mymk mutant adults, we quantified the number of cells expressing Pax7 which, when strongly expressed, is a reliable satellite cell marker (Berberoglu et al., 2017). In mammals, Pax7 is required for satellite cell specification, maintenance, and muscle regeneration (Günther et al., 2013; Maltzahn et al., 2013; Seale et al., 2000). In zebrafish, Pax7 is expressed in cells that appear and behave like satellite cells (termed satellite-like cells, or SLCs) (Berberoglu et al., 2017; Pipalia et al., 2016; Seger et al., 2011; Siegel et al., 2013) and pax7 function is required for proper adult muscle repair (Berberoglu et al., 2017). Our previous work demonstrated that Pax7-positive SLCs are sparse in uninjured zebrafish adult muscle and, when present, these rare Pax7-positive SLCs are concentrated in slow muscle; after injury, they increase dramatically at the injury site (Berberoglu et al., 2017). We utilized two methods to identify Pax7-positive SLCs, a pax7a:GFP transgene (Seger et al., 2011) and a Pax7 antibody. In mymk mutant adult muscle, pax7a:GFP expression is widespread (Fig. 6 D, E), suggesting extensive SLC activation. Correspondingly, the number of Pax7-positive cells is higher in mymk mutant muscle than in wild-type muscle, with a marked increase both at the horizontal myoseptum (a slow muscle domain) and in fast muscle (Fig. 6 F). Because pax7a-driven GFP perdures longer than Pax7 protein, we infer that GFP-positive cells with strong Pax7-positive nuclei are SLCs, whereas GFP-positive cells with little Pax7 label are likely former SLCs that have begun to differentiate (Berberoglu et al., 2017). Alongside pax7a:GFP and Pax7, we examined expression of two muscle differentiation markers: Rbfoxll, which marks myoblast and myofiber nuclei (Berberoglu et al., 2017; Gallagher et al., 2011) and myog:H2B-mRFP which marks differentiating myonuclei (Berberoglu et al., 2017; Tang et al., 2016) (Fig. 6 L). In mymk mutant adults, myoblast number is increased and myog:H2B-mRFP-positive myonuclei number is decreased in both fast and slow muscle compared to wild-type adults (Fig. 6 G–M). These data suggest that adult mymk mutant muscle has an abundance of newly formed myofibers, because proliferation, SLCs, and myoblast density are significantly increased.
Figure 6: Cell proliferation and Pax7-positive satellite-like cell number are dramatically increased in adult mymk mutants.
(A, B) Transverse sections near the horizontal myoseptum (HM) of wild-type (WT) (A) and mymk mutant (B) adult skeletal muscle showing Pcna expression (grey) which marks proliferating nuclei. (A’ and B’) Magnified images of boxed regions in A and B. (C) Graph showing the average number of Pcna-positive cells per mm2 near the HM in WT and mymk mutant adults. (D, E) Transverse sections of pax7:GFP (green); myog:H2B-mRFP (magenta) transgenic WT (D) and mymk mutant (E) adult skeletal muscle colabeled for Pax7 (red), Rbfoxll (blue), and pax7a:GFP (green). pax7a:GFP marks satellite-like cells (SLCs) as well as cells that have begun to differentiate; SLCs can be unambiguously identified by their intensely Pax7-positive nuclei. myog:H2B-mRFP (magenta) marks myonuclei; expression of myog:H2B-mRFP and pax7:GFP transgenes rarely overlap in adult WT muscle (Berberoglu et al., 2017). (F) The average number of Pax7-positive cells per mm2 near the HM and in dorsal fast muscle of WT and mymk mutant adults. (G-K’) Magnified views of boxed regions in D and E show the merged (G, G’) and individual confocal channels for Pax7 (H, H’), pax7a:GFP (I, I’), myog:H2B-mRFP (J, J’) and Rbfoxll (K, K’) in adult WT (G-K) and mymk mutant (G’-K’) muscle. Arrowheads indicate SLCs (yellow), myoblasts (teal), and myofibers (magenta). (L) Summary of cell types based on expression overlap. (M) Number of SLCs (strongly Pax7-positive and GFP-positive), myoblasts (GFP-and Rbfoxll-positive), and myofibers (Rbfoxll-and RFP-positive) in WT and mymk mutant adult slow and fast muscle domains. Student’s f-test (p*** < 0.001, p** < 0.01). Scale bar in B (for A and B) and E (for D, E) is 100 μm, in B’ (for A’ and B’) is 15 μm, and in K’ (for G-K’) is 20 μm.
2.8. Myofiber volume scales proportionally with fusion capacity
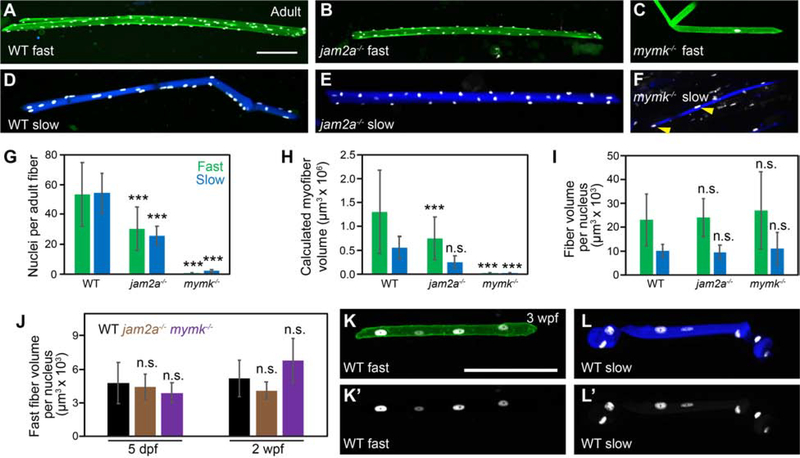
To examine myofiber size and nuclei number in adults, we isolated individual myofibers from 3-month old wild-type, jam2a mutant, and mymk mutant adults carrying muscle-specific transgenes and quantified myonuclei number per fiber for each genotype (Fig. 7 A–G). We find that adult mymk mutant fast myofibers (carrying the mylfpa:lyn-cyan fast muscle-specific transgene) are invariably mononucleate, consistent with a permanent block to fusion (Fig. 7 C). We also examined jam2a mutant fast fibers, which showed partial recovery of fusion by 2 weeks post fertilization (Figure 3 F); in adults, these fibers contain about half the myonuclei per fiber compared to wild-type sibling fast myofibers (Fig. 7 A, B, G). To assess myofiber volume, we measured length and area of each fiber and calculated volume using the formula for a cylindrical object. As expected, fast fiber volume is larger in wild-type than jam2a or mymk mutant adults (Fig. 7 H). Correspondingly, the average fast fiber volume per nucleus, a proxy for myonuclear domain, is the same in fast muscle fibers isolated from wild-type, jam2a mutant, and mymk mutant adults (Fig. 7 I). We observe the same phenomena during larval (5 dpf) and late-larval (2 wpf) stages (Fig. 7 J). These findings show that jam2a mutant fast myofiber fusion does not fully recover by adulthood and there is no recovery in mymk mutants. However, because fiber size scales with the number of nuclei per fiber, we find that the fast myonuclear domain (fiber volume per nucleus) is remarkably similar across all genotypes examined.
Figure 7: Fast and slow myofiber size is tightly correlated to myonuclei number in both wild-type and fusion-impaired mutant fish.
(A-F) Individual myofibers were isolated from wild-type (WT) (A, D), jam2ahu3319 mutant (B, E) and mymkoz17 mutant (C, F) adults. Myonuclei are marked by expression of the myog:H2B-mRFP transgene (white), fast myofibers by the mylfpa:lyn-cyan transgene (A-C, green), and slow myofibers by the smyhc1:EGFP transgene (D-F, blue). Yellow arrowheads indicate two of the nuclei in a mymkoz17 mutant slow fiber (F). (G) Graph showing the average nuclei per fast (green) and slow (blue) myofiber isolated from WT, jam2a mutant, and mymk mutant adults. (H) Myofiber volume was calculated for the same isolated myofibers by measuring length, area, and then applying the formula for volume of a cylinder. (I) The average myofiber volume per nucleus, an approximation of myonuclear domain size, is similar for all fast myofibers and for all slow myofibers, regardless of genotype. (J) At earlier times, we also found no difference in fast muscle myonuclear domain size among WT (black), jam2ahu3319 mutant (brown), and mymkoz17 mutant (purple) larvae (myofiber volume per nucleus was calculated using 5 dpf and 2 wpf datasets from Figure 3). (K-L’) Individual fast (K) and slow (L) myofibers were isolated from 3 wpf WT fish, and shown as merged images, color-coded as above (K, L), and as myog:H2B-mRFP single channel images (K’, L’). Scale bars in A (for A-F) and K (for K-L’) are 100 μm. Significance determined by ANOVA and Tukey-Kramer post-hoc analysis (p*** < 0.001 compared to WT; n.s. indicates not significantly different from WT at P of 0.05).
2.9. Slow muscle fibers become multinucleate during post-embryonic stages
To compare fusion capacity of the two major muscle cell types, we compared myonuclei number in isolated adult myofibers, using slow (smyhc1:EGFP) and fast (mylpfa:lyn-Cyan) muscle transgenes to identify fiber type (Fig. 7). Because slow myofibers are strictly mononucleate in zebrafish embryos, we expected to observe striking differences in myonuclei number between slow and fast muscle types. Surprisingly, isolated adult slow myofibers are multinucleate, containing a similar number of myonuclei per fiber as fast myofibers (Fig. 7 D, E, G). In jam2a mutants, slow myofibers, like fast myofibers, are multinucleate and contain about half the myonuclei per fiber compared to wild-type sibling myofibers (Fig. 7 D, E, G). Whereas adult mymk mutant fast myofibers are strictly mononucleate, some adult mymk mutant slow myofibers are multinucleate (Fig. 7 D–G). However, myonuclei number per slow fiber is >10-fold lower than in wild-type and jam2a mutant adults and slow myofiber size is equivalently smaller (Fig. 7 G, H). The average myonuclear domain size in slow myofibers is similar among wild-t ype, jam2a mutant, and mymk mutant fish but is smaller in slow myofibers than in fast (Fig. 7 I). To determine when slow muscle becomes multinucleate, we performed live imaging of 3MuscleGlow transgenic fish at late-larval stages. Slow myofibers are mononucleate at 2 wpf, are multinucleate by 3 wpf and have around 5 nuclei by 4 wpf (data not shown). We then isolated individual myofibers from 3-week-old larvae and found around 3 nuclei per fiber (3 wpf: 2.9±0.8 nuclei per slow myofiber, N=9; 4.3±1.3 nuclei per fast myofiber, N=10; ± indicates standard deviation) (Fig. 6 K–L’). Between late-larval and adult stages, myonuclei number increases for both fiber types (compare Fig. 7 A, D to K, L). Collectively, these data reveal that slow muscle fibers become multinucleate much later in development than fast muscle fibers, and that efficient slow muscle multinucleation requires mymk.
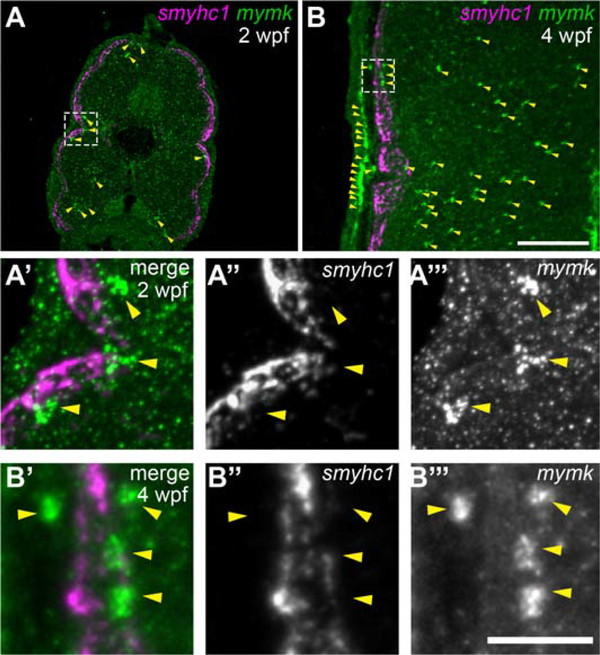
2.10. During multinucleation, slow muscle fibers do not express mymk
To investigate whether slow muscle fibers express mymk when they become fusion-competent, we examined mymk expression at 2 and 4 wpf (Fig. 8 A–B”’), which spans the time interval when slow myofibers become multinucleate. At these stages, mymk is expressed in cells throughout the myotome (Fig. 8 A, B). Although mymk does not appear to be expressed in slow muscle fibers, it is expressed in nearby cells (Fig. 8 A’–B”’). These data suggest that slow myofiber multinucleation occurs as neighboring mymk-expressing cells fuse into differentiated mymk-negative slow muscle fibers. Thus, the addition of new nuclei in late-larval zebrafish may be similar to what has been described in adult mammals undergoing injury-induced muscle growth, in which Mymk is expressed in muscle progenitors but not muscle fibers (Quinn et al., 2017). Altogether, these data support our proposal that zebrafish fusion mechanisms are context-specific and differ between embryonic and post-embryonic development.
Figure 8: As slow muscle fusion ensues, mymk is expressed in cells near myofibers, but not in myofibers themselves.
(A-B) Transverse sections of wild-type larvae at 2 weeks post fertilization (wpf) (A) and 4 wpf (B), processed by fluorescent in situ hybridization (FISH) to detect smyhc1 (magenta) and mymk (green) transcripts; arrowheads (yellow) indicate a subset of the mymk-positive cells. Magnified images (A’ and B’) of boxed regions in A and B show that smyhc1 (A” and B”) and mymk (A”’ and B”’) are expressed in distinct cell types at both time points. Scale bar in B (for A, B) is 100 μm and in B”’ (for A’-B”’) is 25 μm.
Discussion
Our findings indicate that muscle fusion mechanisms differ between embryonic and late-larval stages. In embryos, both jam2a and mymk are essential for muscle fusion, only fast myoblasts fuse, and mymk is expressed throughout the somite fast muscle domain (Fig. 9 A–C), consistent with previous reports (Cortés et al., 2003; Devoto et al., 1996; Di Gioia et al., 2017; Landemaine et al., 2014; Powell and Wright, 2011; Shi et al., 2018; Si et al., 2019; Zhang and Roy, 2016). We find that zebrafish myofibers at all stages examined proportionally scale volume with nuclei number, regardless of fusion capacity, suggesting that the myonuclear domain size is not influenced by fusion. Our live imaging experiments offer new insights into embryonic fusion by elucidating further how the dynamic movements of slow and fast muscle cells influence fusion and by revealing that muscle membranes are internalized at the fusion interface (Fig. 9 B). By late-larval stages, we show that slow muscle becomes multinucleate, jam2a is no longer required for fast muscle fusion, and mymk expression is more restricted (Fig. 9 D–F). Our work sheds light on how muscle fusion mechanisms change over developmental time and how fusion impacts muscle health and function.
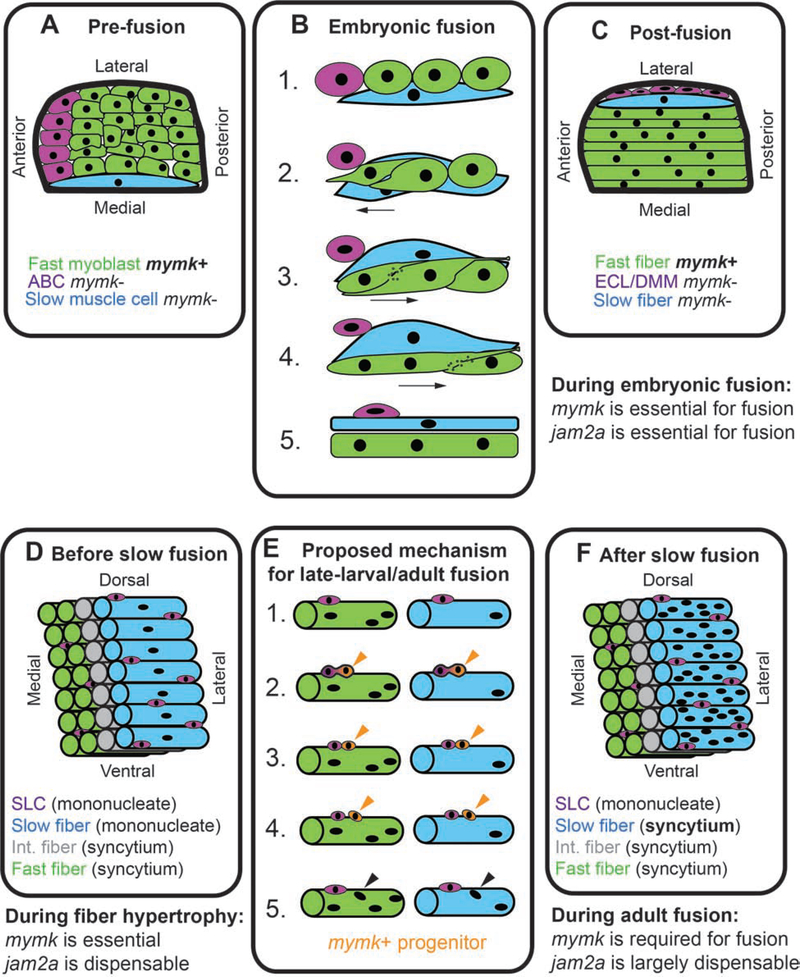
Figure 9: Muscle fusion mechanisms are developmentally regulated.
Model describing developmentally-regulated muscle fusion in zebrafish. (A) Diagram of cell positions in a newly formed somite, viewed from dorsal, showing anterior border cells (ABCs; purple), fast myoblasts (green), and an elongated slow muscle cell (blue). (B) Cell dynamics during embryonic cell fusion. (B1, B2) As a slow muscle cell moves laterally, ABCs also shift position and are replaced at the anterior border by fast myoblasts. (B3-B5) Fast myoblasts then sequentially fuse toward the posterior somite border. (C) Diagram of a somite after slow muscle cell migration. At this stage, slow muscle cells are mononucleate and fast muscle cells are multinucleate. After shifting position, ABCs are called the external cell layer (ECL) in some studies and dermomyotome (DMM) in others. See reviews for further details on embryonic muscle development in zebrafish (Goody et al., 2017; Jackson and Ingham, 2013; Li et al., 2017; Sampath et al., 2018; Stellabotte and Devoto, 2007). (D) Muscle arrangement in late larval stages, prior to slow muscle fusion. By this stage there are three fiber types: fast (green), intermediate (int., gray), and slow (blue), and the slow muscle region is enriched for satellite-like cells (SLCs, purple) (Berberoglu et al., 2017). (E) In late-larval stages, we hypothesize that SLCs produce mymk-positive muscle progenitors (orange). These progenitors could potentially fuse into both fast and slow muscle fibers (orange arrowheads), resulting in the addition of new myonuclei (black arrowheads). (F) By late-larval stages, all muscle fiber types are multinucleate.
3.1. Multinucleate fibers are rare in the mymkoz17 mutant
The mymkoz17 mutants show a dramatic reduction in multinucleation, consistent with previous findings in fish and mouse (Di Gioia et al., 2017; Landemaine et al., 2014; Millay et al., 2016, 2014, 2013; Zhang and Roy, 2016). Our studies also revealed that larval fast myofibers and adult slow myofibers are occasionally binucleate in mymk°z17 mutants. We did not observe binucleate cells in adult mymkoz17 mutant fast fibers, either because they are no longer present or because the sample size was not large enough to detect rare events. Except for these rare multinucleate fibers, the mymkoz17 phenotype we describe is consistent with phenotypes of two recently published frameshifting alleles (Di Gioia et al., 2017; Shi et al., 2018) and potentially stronger than phenotypes of a deletion allele, mymksq36, which swam normally at one month (Zhang and Roy, 2017). We speculate that the milder phenotypes of mymksq36 mutants may be due to the presence of fusion modifier genes. While the frameshifting alleles could possibly encode neomorphic or dominant-negative proteins, we find this explanation unlikely for four reasons. First, our findings in zebrafish are consistent with mouse studies showing that Mymk is key to adult muscle fusion and growth (Goh and Millay, 2017; Millay et al., 2014). Second, we do not detect any phenotype in mymkoz17 heterozygotes. Third, we do not detect any mymk transcript in mymkoz17 mutant embryos, suggesting that mutant transcripts are rapidly degraded by nonsense-mediated decay. Fourth, it is unlikely that translation of functional protein could begin 3’ to the mymkoz17 lesion because several Mymk transmembrane domains are encoded by sequences 5’ to this mutation. Together, these findings indicate that the mymkoz17 lesion causes strong loss of gene function. We offer two alternative explanations for rare multinucleation in mymkoz17 myofibers. Genetic compensation by other fusogens could potentially enable mymk-independent fusion. Alternatively, zebrafish myonuclei may occasionally divide within myofibers at rates too low to be easily detected. Further work using fusion reporters (Sawamiphak et al., 2017) may reveal whether these rare multinucleate mymkoz17 myofibers arise via fusion-dependent or independent processes.
3.2. mymkoz17 mutant skeletal muscle is a model for Carey-Fineman-Ziter syndrome and chronic muscle wasting diseases
Recently, gene variants that reduce human MYMK function have been shown to cause Carey-Fineman-Ziter syndrome, a muscle weakness disease (Di Gioia et al., 2017; Hedberg-Oldfors et al., 2018). Consistent with these symptoms, we show that adult mymk mutant zebrafish have smaller muscle fibers and reduced muscle strength. Others have shown that mymk mutant muscle displays fatty infiltration, a characteristic of Carey-Fineman-Ziter syndrome (Di Gioia et al., 2017; Shi et al., 2018). Although there are parallels between zebrafish mymk mutant phenotypes and Carey-Fineman-Ziter syndrome, the overall muscle defects in the mymkoz17 mutant is much more severe than in the human syndrome. In zebrafish, mymk mutant adult muscle contains an abundance of cells that appear to be immature, fusion-incompetent myoblasts. Additionally, we observe more proliferating cells and SLCs and fiber-type organizational defects in mymk mutant muscle, which suggest that the tissue is continually regenerating. We propose that muscle fusion is thoroughly compromised in the zebrafish mymkozri mutant, whereas fusion is only partially blocked in individuals with Carey-Fineman-Ziter syndrome.
The mymkoz17 mutant displays hallmarks of skeletal muscle wasting diseases (Tabebordbar et al., 2013; Tisdale, 2002; Wallace and McNally, 2009), such as reduced Laminin, an indicator of muscle damage, and dramatic increases in Pax7-positive and Pcna-positive cells, indicators of satellite cell activation. Our findings suggest that activated SLCs in mymk mutants contribute to muscle fibers because pax7a-driven GFP expression perdures in immature fibers that also express muscle differentiation markers Rbfox1l or myog:H2B-mRFP (Berberoglu et al., 2017). Continuous regeneration could also contribute to other adult mymkoz17 mutant muscle defects, such as the presence of fast and slow fiber types outside of their usual spatial domains. It is challenging to pinpoint when and how the mymkoz17 wasting phenotype begins because of potential interplay between muscle fiber growth defects and muscle damage. Mononucleate fibers may become damaged simply because they are not strong enough to withstand normal contractions; alternatively, the tissue may be in a constant state of regeneration because fusion is essential to complete the repair process for normally-occurring damage. Further investigation will parse out which component of the phenotype is the primary cause of tissue damage.
3.3. Zebrafish slow muscle migration triggers timely embryonic skeletal muscle fusion
We began our work by examining muscle fusion mechanisms in zebrafish embryos, where fast myoblast fusion is closely associated with slow muscle migration and requires slow muscle cells for timely onset (Cortés et al., 2003; Devoto et al., 1996; Henry and Amacher, 2004; Yin et al., 2018). Fast muscle precursors extend lengthy cytoplasmic processes before fusion (Snow et al., 2008) that typically elongate the cells anteriorly (Yin et al., 2018). In some cells, fusion begins only after these cytoplasmic extensions reach both the anterior and posterior somite boundaries (Snow et al., 2008). Our results suggest that other fast myoblasts begin to fuse before they span the somite and that other events could also trigger fusion (Fig. 9 B). We show that fast muscle precursors often extend processes anteriorly when they are in contact with the slow muscle cells; these movements appear to displace the anterior-most cells in the somite, the anterior border cells (Fig. 9 B #1–2 and (Yin et al., 2018)). We show that anterior border cells lack mymk expression, so replacement of anterior border cells by mymk-expressing fast muscle precursors at the somite border could potentially facilitate cell-cell fusion. After a fast muscle precursor contacts the anterior somite border, it begins fusing with more posterior myoblasts, suggesting that border contact may provide an activating signal (Fig. 9 B #3). Then, cell-cell fusion adds new myoblasts until reaching the posterior somite boundary (Fig. 9 B #4–5), where the extracellular matrix is thought to block further fiber elongation (Henry et al., 2005; Snow et al., 2008). We speculate that migrating slow muscle cells can trigger cell-cell fusion by activating myoblast elongation and by displacing mymk-positive fast muscle precursors to the anterior somite border where they initiate fusion with more posterior precursors to generate a multinucleate muscle fiber. In future work, time-lapse analysis of mutants that lack anterior-posterior identity within somites could reveal how cell positioning affects fusion.
3.4. Muscle precursor fusion uses plasma membrane breakdown and dispersal
Our live imaging studies offer insights into how cell membranes merge during fusion. Membrane merging has been viewed at highest resolution under an electron microscope (Doberstein et al., 1997; Fumagalli et al., 1981; Lipton and Konigsberg, 1972; Raamsdonk et al., 1974; Rash and Fambrough, 1973; Shimada, 1971). In these studies, membranes were proposed to fuse by fusion pore formation and subsequent pore widening (Lipton and Konigsberg, 1972), and/or by plasma membrane breakdown and dispersal (Doberstein et al., 1997; Fumagalli et al., 1981; Raamsdonk et al., 1974; Rash and Fambrough, 1973; Shimada, 1971). During Drosophila fusion, a single large pore appears to initiate myoblast fusion (Sens et al., 2010); if multiple pores form along a broader interface, the membrane at the interface would need to be disposed of, rather than contributing to the joint surface of both cells. Our time-lapse images of living embryos suggest that at the fusion interface membrane particles disperse into the newly formed syncytium, indicating that fusing cells internalize their surface membrane to the cytoplasm. Consistent with this model, recent work shows Myomerger-dependent membrane pores also form in cultured mammalian muscle cell lines (Leikina et al., 2018). Similarly, EM studies have shown multiple cytoplasmic bridges between fusing cells along with membrane fractions within the cytoplasm of myotubes (Fumagalli et al., 1981; Rash and Fambrough, 1973). Thus, our live imaging studies indicate that membranes at the fusion interface are internalized and degraded after pore formation (Fig. 9 B #3–4).
3.5. Zebrafish myofibers may operate near the optimum myonuclear domain size
Our findings reveal that the average myonuclear domain size for each fiber type and developmental stage is the same in wild-type, jam2a mutant (fusion-reduced), and mymk mutant (fusion-impaired) fish. Previous work has suggested that each muscle nucleus can support a certain maximal volume of cytoplasm, known as the myonuclear domain (Abmayr and Pavlath, 2012; Gundersen and Bruusgaard, 2008; Hall and Ralston, 1989; Van der Meer et al., 2011). Myonuclear domain size can be influenced by species, fiber type, age, exercise, inactivity, position within fiber, and overall muscle size (Bruusgaard and Gundersen, 2008; Conceição et al., 2018; Gundersen and Bruusgaard, 2008; Liu et al., 2009; Mantilla et al., 2008; Ohira et al., 1999; Rosser et al., 2002; Schwartz et al., 2016; Van der Meer et al., 2011; Windner et al., 2019). Similarly, we note that in zebrafish, the myonuclear domain appears smaller in slow than fast fibers and in larvae than adults. Within fibers of a given type and age, we find that myonuclear number reliably predicts fiber size. Consistent with this finding, a recent meta-analysis revealed that human myofibers can only increase volume by 15–26% during induced fiber hypertrophy before new myonuclei are added, suggesting that fibers normally operate close to their near-maximal myonuclear domain size (Conceição et al., 2018). We find that zebrafish jam2a and mymk mutant larvae are mononucleate but have normal myonuclear domain size and normal muscle force, supporting a correlation between myonuclear domain size and force. When the myonuclear domain size of Drosophila larvae was experimentally altered, embryonic motility was dramatically reduced, revealing that myonuclear domain size is carefully regulated to ensure muscle function (Windner et al., 2019). Together, these findings suggest that healthy myofibers scale their myonuclear domain for optimal muscle function for each fiber type and developmental stage.
3.6. jam2a is essential to muscle fusion in embryonic, but not in late-larval or adult zebrafish
Although mymk is required throughout life in zebrafish, jam2a becomes partially dispensable for fusion by 2 wpf. In this way, larval and adult zebrafish are similar to the mouse, which requires Mymk but not Jam2 for muscle fusion (Goh and Millay, 2017; Millay et al., 2013; Powell and Wright, 2011). To our knowledge, mammalian Jam function has not yet been examined in early embryonic muscle, nor have muscle nuclei been carefully quantified post-embryonically. Although recent work indicated that zebrafish jam2a mutant adults have a normal number of nuclei per fiber (Si et al., 2019), we find that adult jam2a mutant fibers have half the normal nuclei number suggesting that muscle multinucleation partially requires jam2a. Future work will clarify whether the jam2a mutant myonuclei are reduced due to delayed fusion or a lower rate of secondary fusion.
3.7. Slow muscle fibers become multinucleate by late-larval stages
Fish continually add new muscle fibers and expand fiber size (Koumans and Akster, 1995; Nguyen et al., 2017; Weatherley et al., 1988). As body size increases during larval growth, fish begin to rely primarily on slow muscle for casual swimming (Voesenek et al., 2018), and this shift to slow muscle function may necessitate changes in slow muscle fiber growth and/or physiology. We show that slow muscle fibers become multinucleate by late-larval stages during a period of pronounced muscle fiber growth. Because slow muscle fibers are mononucleate in embryos but become multinucleate between 2 and 3 wpf, we propose that late-larval slow muscle cells become fusion competent. Our findings suggest that slow muscle becomes multinucleate primarily via cell-cell fusion, because slow muscle has few nuclei per fiber in the fusion-deficient mymk mutant. In principle, multinucleation could arise via other mechanisms like endoreplication, however we think this is at most a minor source of new nuclei since we observe little or no Pcna-label in muscle fibers and mature myofibers are considered post-mitotic (Bryson-Richardson and Currie, 2008; Moss and Leblond, 1971, 1970; Simionescu and Pavlath, 2011). These lines of evidence suggest mymk-dependent cell-cell fusion is the main source of new nuclei in slow muscle fibers (Fig. 9 D–F).
3.8. Muscle fusion may become asymmetric in late-larval zebrafish
Studies of mosaicjam2a and jam3b mutants suggested that zebrafish embryonic muscle cell fusion occurs symmetrically, without defined founder cells and fusion-competent myoblasts (Powell and Wright, 2012, 2011). We find that jam2a is no longer essential for fusion by 2 wpf, leading us to focus on mymk expression in late-larval fish. In zebrafish embryos, expression of fast muscle-specific genes, including mymk, is repressed in slow muscle precursors (Jackson et al., 2015; von Hofsten et al., 2008; Yao et al., 2013; this study); by late-larval stages, neither slow nor fast muscle fibers express mymk, but small cells adjacent to muscle fibers do express mymk. These mymk-positive cells may be satellite-like cells, activated myoblasts, or other cell types that contribute to muscle (Berberoglu et al., 2017; Liu et al., 2017; Mitchell et al., 2010; Qu-Petersen et al., 2002). In the mouse, Mymk is also expressed in small cells adjacent to adult myofibers and lineage tracing shows that Mymk-expressing cells contribute to new myofibers (Goh and Millay, 2017; Millay et al., 2014). These findings are consistent with the idea that muscle cell fusion becomes asymmetric in late-larval zebrafish and adult mouse (Millay et al., 2014; Sampath et al., 2018), suggesting a possible similarity between fusogenic cells in adult vertebrates with the founder cell/fusion competent cell pair in Drosophila. The shift in symmetry corresponds with changes in cell behaviors; during embryogenesis, cell-cell fusion generates muscle fibers de novo, but at late-larval and adult stages, fiber addition is slower and cell-cell fusion is thought to primarily add nuclei to existing fibers (Gurevich et al., 2015; Nguyen et al., 2017; Patterson et al., 2008; Rowlerson and Veggetti, 2001). Such asymmetric fusion could explain why slow muscle fibers become multinucleate, since neither slow nor fast fibers express mymk in late-larval fish. A shift in fusogenic cell type could also explain why jam2a is dispensable for larval muscle fusion, since jam2a function has only been described in symmetric embryonic fusion events. Together our findings suggest that by late-larval stages, zebrafish muscle cell fusion mechanisms are similar to the adult mouse in terms of gene requirements, gene expression, and fusion symmetry; future work may further clarify how fusion mechanisms change through development in both species.
Conclusions
Recent work has highlighted key differences between Drosophila and vertebrate muscle fusion mechanisms. Our work in zebrafish highlights how muscle fusion mechanisms can vary over time within a single organism and that, similar to the findings in Drosophila larvae, adult muscle fusion may proceed asymmetrically in zebrafish. Furthermore, similar to findings in Drosophila and mammals, we find that nuclear number is a predictor of myofiber size. In zebrafish embryos, slow muscle cell migration helps to orchestrate fast myoblast fusion, which at this stage is both jam2a-and mymk-dependent. In late-larval and adult zebrafish, both fast and slow muscle fibers are fusion-competent; at these later stages, fusion is mymk-dependent, but does not rely on jam2a. Furthermore, whereas embryonic fusion is largely symmetric, it appears to become asymmetric by the late-larval stage, with mymk-expressing cells fusing with mymk-negative muscle fibers. Although mononucleate embryonic muscles function normally, fusion becomes essential for muscle function, fiber size, and integrity by adulthood. Adult zebrafish mymk mutants are weak, with small muscle fibers that appear to be continuously regenerating. Together, these findings highlight that muscle cell fusion is an essential and dynamically-regulated process.
Experimental Procedures
4.1. Animal Stocks and husbandry
We raised and housed wild-type (AB strain), mutant, and transgenic zebrafish at 28.5°C on a 14 hour light and 10 hour dark cycle (Westerfield, 2007). We collected embryos by natural spawning of adult fish and staged as described (Kimmel et al., 1995; McMenamin et al., 2016; Parichy et al., 2009; Singleman and Holtzman, 2014). Zebrafish lines were maintained according to The Ohio State University Institutional Animal Care and Use Committee (IACUC). We intercrossed transgenic fish carrying Tg(myog:Has.HIST1H2BJ-mRFP)fb121 (Tang et al., 2016) (called myog:H2B-mRFP for simplicity), Tg(smyhc1:EGFP)i104 (Elworthy et al., 2008), and Tg(mylpfa:lyn-Cyan)fb122 (Ignatius et al., 2012) (called mylpfa:lyn-Cyan for simplicity) to generate the 3MuscleGlow line, which carries all three transgenes. The Tg(−10kb smyhc1:lyn-tdTomato) line was generated from a plasmid gifted by the Ingham Lab that had been used to generate the Tg(−10kb smyhc1:lyn-tdTomato)i261 transgenic line (Wang et al., 2011); our new derivative transgenic line is designated Tg(−10kb smyhc1:lyn-tdTomato)oz29 (or, smyhc1:lyn-tdTomato for simplicity). The BAC Tg(six1b:lyn-GFP)oz5 transgenic line (six1b:lyn-GFP for simplicity) was generated using BAC CH73–301I9, which contains six1b, six4b, and over 100 kb of surrounding sequence; construction of this line is described elsewhere (Talbot et al., In press). To inactivate six4b and to replace six1b coding sequence with sequence encoding lyn-GFP, we modified this BAC using previously described protocols (Suster et al., 2011) and then used this modified BAC to generate a stable transgenic line. The jam2ahu3319 line was generously provided by Gareth Powell and Gavin Wright. Two new mymk alleles, mymkoz17 and mymkoz25, were generated in this study and are described below.
4.2. mymk mutant construction and identification
To generate new knockout alleles of mymk, we performed CRISPR mutagenesis as previously described (Talbot and Amacher, 2014). The mymk CRISPR target site used was 5’-GGCATTTACTCCGGCCCCAT-3’. A 10 bp deletion allele that creates a premature stop codon at amino acid 125 of 220 was recovered and designated mymkoz17. We also recovered a 5 bp deletion allele, mymkoz25, that creates a premature stop codon at amino acid 170 of 220. Both alleles can be identified using the primers (Forward) 5’CGCAGCTGTGAGGATCTACC-3’ and (Reverse) 5’-GACGTGTCTCAAACTCACCCA-3’, followed by analysis on a 3% gel or by using high-resolution melt analysis. mymkoz25 can also be identified by digesting the 109 bp PCR product with Haelll which cuts the wild-type amplicon, but not the mutant amplicon, into 51 and 58 bp fragments. mymk mutants are also readily identified after 24 hpf by centrally localized myonuclei, which can be visualized using the Tg(myog:H2B-mRFP) transgenic line.
4.3. Live imaging of skeletal muscle fusion
To label all cell nuclei, H2B-CFP mRNA in 0.2 M KCl with 0.05% Phenol Red (5 ng/pl dose) was microinjected at the one-cell stage into six1b:lyn-GFP;smyhc1:lyn-tdTomato embryos. At 18–20 hpf, embryos were mounted in 0.1% agarose containing 5% Tricaine anesthetic. During time-lapse, fish were held in a 28.5°C Okagawa-heated chamber and imaged for 4–5 hours. MetaMorph software (Molecular Devices) was used for image and video processing and the MTRackJ tool in ImageJ (Meijering et al., 2012) was used for cell tracking.
4.4. In situ hybridization and fluorescent in situ hybridization (FISH)
In situ hybridization and FISH were performed as previously described (Jowett, 1999; Talbot et al., 2016). The smyhc1 probe has been previously described (von Hofsten et al., 2008). We synthesized a template for mymk in situ by PCR-amplifying a 617 bp region of 3’ UTR using (Forward) 5’-TGGCAGACTTCACAACCTCAGA-3’ and (Reverse) 5’-taatacgactcactatagggCAAGGGAGCTAATAATTCAGGGGGGCTAAT-3’ primers. For antisense probe synthesis, we included the T7 promoter sequence (lowercase) in the reverse primer. We amplified and purified template for the probe using the Qiagen MinElute PCR Purification Kit (Cat. No. 28006) and transcribed DIG-labeled antisense riboprobe using T7 RNA polymerase.
4.5. Sectioning and Immunohistochemistry
Embryos, larva, and adult fish > 3 months of age were processed for cryostat sectioning and antibody stained as previously described (Berberoglu et al., 2017; Bird et al., 2012). The following primary antibodies were used at the noted dilutions: chicken anti-GFP (ab13970, Abcam; 1:2000), mouse anti-Pax7 (DSHB; 1:20), rabbit anti-Laminin (L9393, Sigma; 1:200), rabbit anti-Rbfox1l (1:1000) (Berberoglu et al., 2017), and anti-Pcna (GTX124496, GeneTex; 1:500). Pcna labeling was performed as previously described (Katz et al., 2016). Fiber diameter was measured in ImageJ and ≥ 80 fibers were measured from 2–3 sections for 2–4 fish per genotype for each time point.
4.6. Myofiber Preps
Myofibers were prepared from adult zebrafish essentially as previously described (Horstick et al., 2013). Adult transgenic fish were anesthetized in Tricaine, then sacrificed by decapitation, de-finned, skinned, and internal organs were removed. The resulting muscle fillets were placed in 3.125mg/mL Collagenase IV in L15 media for 2–2.5 hours and washed twice in L15 media before trituration with a Pasteur pipet. Single fibers were selected for immediate confocal imaging in L15 media.
4.7. Nuclei, myofiber volume, cross-sectional area, and cell density quantification
Myonuclei counts (Fig. 2 H) were performed using Velocity software (PerkinElmer) on 1–3 dpf zebrafish embryos and larvae containing the myog:H2B-mRFP transgene. Somites 12 and 15 were imaged and nuclei were manually counted through the entire z-stack (step size: 1.0 μm) for each somite imaged. The values reported are the average of the averages for combined total nuclei from both somites 12 and 15. For fast muscle cell density on cross sections (Figure 4 J), mylpfa:lyn-Cyan positive cells were counted in fast muscle, but not slow or intermediate domains, and counts were divided by the area of quantification. Two sections were quantified per fish and 2–4 fish per genotype were examined at each developmental stage. Using ImageJ, we determined cross-sectional area (CSA) (Figure 4 K) for over 50 myofibers per section and 3 non-consecutive sections per fish in two wild-type (WT) and two mymk mutant fish. CSA was averaged and compared per genotype using student’s T-test. For total myofiber counts (Figure 4 L), we counted fast myofibers using the fast muscle label mylpfa:lyn-cyan and slow muscle label smyhc1:GFP at 5 dpf and 2 wpf, stages where whole sections fit into a single image; average counts per genotype were compared using student’s T-test. To quantify the density of labeled cell types in adult fish (Fig. 6 C, F, M), cells were counted throughout the entire slow or fast muscle domain within each image and then divided by the area of quantification. Two sections per fish were quantified, and the average cell density per fish was compared for three WT and three mymk mutant fish using students T-test. For nuclei counts in isolated myofibers (Fig. 7 G and Section 2.8 results text), counts are determined per myofiber then compared among genotypes and fiber types using ANOVA and Tukey-Kramer post-hoc analysis. We calculated the volume of isolated myofibers (Figure 7 H) by measuring the length and area of each fiber on confocal projection, then dividing area by length to find the average width, then applied the formula for area of a cylinder (Volume = Pi x [1/2 width] 2 x Length) on at least 10 fibers per fiber type for each genotype. Fiber volume per nucleus (Figure 7 I) was calculated and averaged over at least 10 fibers per genotype per fiber type. For larval myofibers (Figure 7 J), we measured cross-sectional area and length on whole mount images and used these measurement to calculate cylindrical volumes. We calculated larval volume per nuclei ratios for 10 fibers per fish, and at least 8 fish per genotype at 5 dpf and 2 wpf.
4.8. Swim Tunnel Assay
Adult zebrafish (4–5 months) were tested in a swim tunnel adapted from Gilbert et al. (2014). The swim tunnel is a 2 cm by 30 cm long tube connected to an EHEIM aquarium pump with King Instrument flowmeter that is adjusted by the use of a water valve. Each fish was individually acclimated to the swim tunnel for 15 minutes at the lowest flow rate of 6 cm/s. After acclimation, the flow was increased by 6 cm/s every 10 minutes for the endurance assay and every 1 minute for the sprint assay until the subject fatigued, which was defined as the time when the fish was pushed to the back of the tunnel for more than 5 seconds. After the initial endurance and sprint assays, we let the fish rest for 45 minutes and then performed a second endurance assay on each fish. Maximum endurance (Ucrit) and maximum sprint speed (Umax) values were calculated as described and reported as a ratio (Ucrit/Umax) (Gilbert et al., 2014).
4.9. Muscle Contractile Analysis
Contractile analysis of 3 dpf larvae was performed as described previously (Martin et al., 2015) and summarized briefly here. To measure contraction, live 3 dpf larvae were anesthetized in 0.02% weight/volume tricaine buffered with TrisHCl in Krebs-Henseleit solution and then mounted on a custom-built set up between a force transducer and a hook. To induce contraction, larvae were stimulated at increasing frequencies, and contractile strength was measured at each frequency. The maximal contractile force reached during each contraction was recorded, analyzed, and reported per larva. Measurements were compared at each contraction frequency using student’s T-test. At 180 Hz we observe fused tetanic contractions, wherein muscle tension is sustained at a plateau level.
4.10. RT-PCR
Twenty embryos at 4–8 cell, 6 somite, 20 somite, and 48 hpf stages were solubilized in TRIzol (ThermoFisher) for RNA extraction. At larval stages (5 dpf, N=20; 2 wpf, N=12), fish were anesthetized in Tricaine and decapitated, then the rest of the fish was immediately placed into Ringer’s solution before TRIzol solubilization. Total RNA (2.5 pg) was purified and reverse transcribed into cDNA using Superscript VILO master mix (ThermoFisher) following manufacturer’s instructions. Products were amplified using a primer in the first (5’-ATGGGAGCGTTTATCGCCAAG-3’) and sixth (5’-TCATACACAGCAGCAGAGGGT-3’) exons.
4.11. Confocal Imaging
Confocal imaging, except for the images shown in Figure 3, was performed on an inverted Nikon TiE microscope equipped with an Andor Revolution WD spinning disk. For Figure 3, imaging used an inverted Olympus IX-81 scanning microscope. Lasers used for all figures were 405 nm, 445 nm, 488 nm, 561 nm, and 640nm using 10x, 20x, 40x, and 60x objectives.
Supplementary Material
Highlights.
Live imaging reveals membrane fusion and dispersal during muscle fiber formation.
Myonuclear domain size is established independently of fusion capacity
The embryonic fusogen jam2a is not vital for fusion by late-larval stages.
During late-larval stages, zebrafish slow-twitch muscle fibers become multinucleate.
By adulthood, fusion is critical for muscle fiber growth, structure, and performance
Acknowledgments
We thank the Amacher lab fish facility staff for excellent fish husbandry, the OSU Neuroscience Imaging Core for equipment and advice, and Geremy Lerma, Joseph Beljan, and Danielle Pvirre for assistance. We thank Shaojun Du and Yufeng Si for exchanging ideas in advance of publication. We thank David Langenau for sharing the mylpfa:lyn-cyan and myog:H2B-mRFP transgenic lines, Phil Ingham for sharing the smyhc1:GFP transgenic line and the plasmid used to construct the Tg(−10kb smyhc1:lyn-tdTomato)oz29 line, and Gavin Wright for the jam2ahu3319 mutant line. We thank Clarissa Henry and Teresa Easterbrooks for comments on the manuscript.
Funding: Work was supported by NIH grants GM061952, GM088041, and GM117964 (to S.L.A.), by an NIH T32 training grant NS077984 (to K.J.H and J.C.T) by an NIH loan repayment program contract and Pelotonia Postdoctoral Fellowship (to J.C.T.). The OSU Neuroscience Imaging Core is supported by NIH grants P30-NS045758, P30-NS104177, and S10-OD010383.
Footnotes
Declaration of interests: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abmayr SM, Pavlath GK, 2012. Myoblast fusion: lessons from flies and mice. Development 139, 641–656. 10.1242/dev.068353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharyya S, Butchbach MER, Sahenk Z, Wang H, Saji M, Carathers M, Ringel MD, Skipworth RJE, Fearon KCH, Hollingsworth MA, Muscarella P, Burghes AHM, Rafael-Fortney JA, Guttridge DC, 2005. Dystrophin glycoprotein complex dysfunction: A regulatory link between muscular dystrophy and cancer cachexia. Cancer Cell 8, 421–432. 10.1016/j.ccr.2005.10.004 [DOI] [PubMed] [Google Scholar]
- Aguilar PS, Baylies MK, Fleissner A, Helming L, Inoue N, Podbilewicz B, Wang H, Wong M, 2013. Genetic basis of cell-cell fusion mechanisms. Trends in Genetics 29, 427–437. 10.1016/j.tig.2013.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcangeli M-L, Bardin F, Frontera V, Bidaut G, Obrados E, Adams RH, Chabannon C, Aurrand-Lions M, 2014. Function of Jam-B/Jam-C Interaction in Homing and Mobilization of Human and Mouse Hematopoietic Stem and Progenitor Cells. Stem Cells 32, 1043–1054. 10.1002/stem.1624 [DOI] [PubMed] [Google Scholar]
- Baas D, Caussanel-Boude S, Guiraud A, Calhabeu F, Delaune E, Pilot F, Chopin E, Machuca-Gayet I, Vernay A, Bertrand S, others, 2012. CKIP-1 regulates mammalian and zebrafish myoblast fusion. Journal of cell science 125, 3790–3800. [DOI] [PubMed] [Google Scholar]
- Barresi MJ, D’Angelo JA, Hernández LP, Devoto SH, 2001. Distinct mechanisms regulate slow-muscle development. Current Biology 11, 1432–1438. [DOI] [PubMed] [Google Scholar]
- Berberoglu MA, Gallagher TL, Morrow ZT, Talbot JC, Hromowyk KJ, Tenente IM, Langenau DM, Amacher SL, 2017. Satellite-like cells contribute to pax7 -dependent skeletal muscle repair in adult zebrafish. Developmental Biology 424, 162–180. 10.1016/j.ydbio.2017.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi P, Ramirez-Martinez A, Li H, Cannavino J, McAnally JR, Shelton JM, Sánchez-Ortiz E, Bassel-Duby R, Olson EN, 2017. Control of muscle formation by the fusogenic micropeptide myomixer. Science 356, 323–327. 10.1126/science.aam9361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird NC, Windner SE, Devoto SH, 2012. Immunocytochemistry to Study Myogenesis in Zebrafish. Myogenesis 798, 153–169. [DOI] [PubMed] [Google Scholar]
- Bruusgaard JC, Gundersen K, 2008. In vivo time-lapse microscopy reveals no loss of murine myonuclei during weeks of muscle atrophy. J. Clin. Invest. 118, 1450–1457. 10.1172/JCI34022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryson-Richardson RJ, Currie PD, 2008. The genetics of vertebrate myogenesis. Nature Reviews Genetics 9, 632–646. 10.1038/nrg2369 [DOI] [PubMed] [Google Scholar]
- Conceição MS, Vechin FC, Lixandráo M, Damas F, Libardi CA, Tricoli V, Roschel H, Camera D, Ugrinowitsch C, 2018. Muscle Fiber Hypertrophy and Myonuclei Addition: A Systematic Review and Meta-analysis. Medicine & Science in Sports & Exercise 50, 1385–1393. 10.1249/MSS.0000000000001593 [DOI] [PubMed] [Google Scholar]
- Cortés F, Daggett D, Bryson-Richardson RJ, Neyt C, Maule J, Gautier P, Hollway GE, Keenan D, Currie PD, 2003. Cadherin-mediated differential cell adhesion controls slow muscle cell migration in the developing zebrafish myotome. Developmental cell 5, 865–876. [DOI] [PubMed] [Google Scholar]
- Devoto SH, Melangon E, Eisen JS, Westerfield M, 1996. Identification of separate slow and fast muscle precursor cells in vivo, prior to somite formation. Development 122, 3371–3380. [DOI] [PubMed] [Google Scholar]
- Di Gioia SA, Connors S, Matsunami N, Cannavino J, Rose MF, Gilette NM, Artoni P, de Macena Sobreira NL, Chan W-M, Webb BD, Robson CD, Cheng L, Van Ryzin C, Ramirez-Martinez A, Mohassel P, Leppert M, Scholand MB, Grunseich C, Ferreira CR, Hartman T, Hayes IM, Morgan T, Markie DM, Fagiolini M, Swift A, Chines PS, Speck-Martins CE, Collins FS, Jabs EW, Bönnemann CG, Olson EN, Andrews CV, Barry BJ, Hunter DG, Mackinnon SE, Shaaban M, Frempong T, Hao K, Naidich TP, Rucker JC, Zhang Z, Biesecker BB, Bonnycastle LL, Brewer CC, Brooks BP, Butman JA, Chien WW, Farrell K, FitzGibbon EJ, Gropman AL, Hutchinson EB, Jain MS, King KA, Lehky TJ, Lee J, Liberton DK, Narisu N, Paul SM, Sadeghi N, Snow J, Solomon B, Summers A, Toro C, Thurm A, Zalewski CK, Carey JC, Robertson SP, Manoli Engle, E.C., 2017. A defect in myoblast fusion underlies Carey-Fineman-Ziter syndrome. Nature Communications 8, 16077 10.1038/ncomms16077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doberstein SK, Fetter RD, Mehta AY, Goodman CS, 1997. Genetic Analysis of Myoblast Fusion: blown fuse Is Required for Progression Beyond the Prefusion Complex. The Journal of Cell Biology 136, 1249–1261. 10.1083/jcb.136.6.1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dray N, Bedu S, Vuillemin N, Alunni A, Coolen M, Krecsmarik M, Supatto W, Beaurepaire E, Bally-Cuif L, 2015. Large-scale live imaging of adult neural stem cells in their endogenous niche. Development 142, 3592–3600. 10.1242/dev.123018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elworthy S, Hargrave M, Knight R, Mebus K, Ingham PW, 2008. Expression of multiple slow myosin heavy chain genes reveals a diversity of zebrafish slow twitch muscle fibres with differing requirements for Hedgehog and Prdm1 activity. Development 135, 2115–2126. 10.1242/dev.015719 [DOI] [PubMed] [Google Scholar]
- Fiorotto M, 2012. The making of a muscle. Biochem (Lond) 34, 4–11. [PMC free article] [PubMed] [Google Scholar]
- Fumagalli G, Brigonzi A, Tachikawa T, Clementi F, 1981. Rat myoblast fusion: morphological study of membrane apposition, fusion, and fission during controlled myogenesis in vitro. J. Ultrastruct. Res. 75, 112–125. [DOI] [PubMed] [Google Scholar]
- Gallagher TL, Arribere JA, Geurts PA, Exner CRT, McDonald KL, Dill KK, Marr HL, Adkar SS, Garnett AT, Amacher SL, Conboy JG, 2011. Rbfox-regulated alternative splicing is critical for zebrafish cardiac and skeletal muscle functions. Developmental Biology 359, 251–261. 10.1016/j.ydbio.2011.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganassi M, Badodi S, Ortuste Quiroga HP, Zammit PS, Hinits Y, Hughes SM, 2018. Myogenin promotes myocyte fusion to balance fibre number and size. Nature Communications 9 10.1038/s41467-018-06583-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert MJH, Zerulla TC, Tierney KB, 2014. Zebrafish (Danio rerio) as a model for the study of aging and exercise: Physical ability and trainability decrease with age. Experimental Gerontology 50, 106–113. 10.1016/j.exger.2013.11.013 [DOI] [PubMed] [Google Scholar]
- Gliki G, Ebnet K, Aurrand-Lions M, Imhof BA, Adams RH, 2004. Spermatid differentiation requires the assembly of a cell polarity complex downstream of junctional adhesion molecule-C. Nature 431, 320–324. 10.1038/nature02877 [DOI] [PubMed] [Google Scholar]
- Goh Q, Millay DP, 2017. Requirement of myomaker-mediated stem cell fusion for skeletal muscle hypertrophy. eLife 6 10.7554/eLife.20007 [DOI] [Google Scholar]
- Gundersen K, Bruusgaard JC, 2008. Nuclear domains during muscle atrophy: nuclei lost or paradigm lost?: Nuclear domains during muscle atrophy. The Journal of Physiology 586, 2675–2681. 10.1113/jphysiol.2008.154369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günther S, Kim J, Kostin S, Lepper C, Fan C-M, Braun T, 2013. Myf5-Positive Satellite Cells Contribute to Pax7-Dependent Long-Term Maintenance of Adult Muscle Stem Cells. Cell Stem Cell 13, 590–601. 10.1016/j.stem.2013.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich D, Siegel A, Currie PD, 2015. Skeletal Myogenesis in the Zebrafish and Its Implications for Muscle Disease Modelling, in: Brand-Saberi B (Ed.), Vertebrate Myogenesis. Springer Berlin Heidelberg, Berlin, Heidelberg, pp. 49–76. [DOI] [PubMed] [Google Scholar]
- Gurevich DB, Nguyen PD, Siegel AL, Ehrlich OV, Sonntag C, Phan JMN, Berger S, Ratnayake D, Hersey L, Berger J, Verkade H, Hall TE, Currie PD, 2016. Asymmetric division of clonal muscle stem cells coordinates muscle regeneration in vivo. Science 353, aad9969–aad9969. 10.1126/science.aad9969 [DOI] [PubMed] [Google Scholar]
- Hall ZW, Ralston E, 1989. Nuclear domains in muscle cells. Cell 59, 771–772. 10.1016/0092-8674(89)90597-7 [DOI] [PubMed] [Google Scholar]
- Haralalka S, Shelton C, Cartwright HN, Guo F, Trimble R, Kumar RP, Abmayr SM, 2014. Live Imaging Provides New Insights on Dynamic F-Actin Filopodia and Differential Endocytosis during Myoblast Fusion in Drosophila. PLoS ONE 9, e114126 10.1371/journal.pone.0114126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He WA, Berardi E, Cardillo VM, Acharyya S, Aulino P, Thomas-Ahner J, Wang J, Bloomston M, Muscarella P, Nau P, Shah N, Butchbach MER, Ladner K, Adamo S, Rudnicki MA, Keller C, Coletti D, Montanaro F, Guttridge DC, 2013. NF-KappaB-mediated Pax7 dysregulation in the muscle microenvironment promotes cancer cachexia. Journal of Clinical Investigation 123, 4821–4835. 10.1172/JCI68523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedberg-Oldfors C, Lindberg C, Oldfors A, 2018. Carey-Fineman-Ziter syndrome with mutations in the myomaker gene and muscle fiber hypertrophy. Neurol Genet 4 10.1212/NXG.0000000000000254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry CA, Amacher SL, 2004. Zebrafish slow muscle cell migration induces a wave of fast muscle morphogenesis. Developmental cell 7, 917–923. [DOI] [PubMed] [Google Scholar]
- Henry CA, McNulty IM, Durst WA, Munchel SE, Amacher SL, 2005. Interactions between muscle fibers and segment boundaries in zebrafish. Developmental Biology 287, 346–360. 10.1016/j.ydbio.2005.08.049 [DOI] [PubMed] [Google Scholar]
- Horstick EJ, Gibbs EM, Li X, Davidson AE, Dowling JJ, 2013. Analysis of Embryonic and Larval Zebrafish Skeletal Myofibers from Dissociated Preparations. Journal of Visualized Experiments 81, e50259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatius MS, Chen E, Elpek NM, Fuller AZ, Tenente IM, Clagg R, Liu S, Blackburn JS, Linardic CM, Rosenberg AE, Nielsen PG, Mempel TR, Langenau DM, 2012. In vivo imaging of tumor-propagating cells, regional tumor heterogeneity, and dynamic cell movements in embryonal rhabdomyosarcoma. Cancer Cell 21,680–693. 10.1016/j.ccr.2012.03.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson HE, Ono Y, Wang X, Elworthy S, Cunliffe VT, Ingham PW, 2015. The role of Sox6 in zebrafish muscle fiber type specification. Skeletal Muscle 5, 2 10.1186/s13395-014-0026-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jowett T, 1999. Analysis of protein and gene expression. Methods in cell biology 59, 63–85. [DOI] [PubMed] [Google Scholar]
- Katz S, Cussigh D, Urban N, Blomfield I, Guillemot F, Bally-Cuif L, Coolen M, 2016. A Nuclear Role for miR-9 and Argonaute Proteins in Balancing Quiescent and Activated Neural Stem Cell States. Cell Reports 17, 1383–1398. https://doi.Org/10.1016/j.celrep.2016.09.088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Jin P, Duan R, Chen EH, 2015. Mechanisms of myoblast fusion during muscle development. Current Opinion in Genetics & Development, Developmental mechanisms, patterning and organogenesis 32, 162–170. 10.1016/j.gde.2015.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF, 1995. Stages of embryonic development of the zebrafish. Developmental dynamics 203, 253–310. [DOI] [PubMed] [Google Scholar]
- Koumans JTM, Akster HA, 1995. Myogenic cells in development and growth of fish. Comparative Biochemistry and Physiology Part A: Physiology 110, 3–20. 10.1016/0300-9629(94)00150-R [DOI] [Google Scholar]
- Landemaine A, Rescan P-Y, Gabillard J-C, 2014. Myomaker mediates fusion of fast myocytes in zebrafish embryos. Biochemical and Biophysical Research Communications 451,480–484. 10.1016/j.bbrc.2014.07.093 [DOI] [PubMed] [Google Scholar]
- Leikina E, Gamage DG, Prasad V, Goykhberg J, Crowe M, Diao J, Kozlov MM, Chernomordik LV, Millay DP, 2018. Myomaker and Myomerger Work Independently to Control Distinct Steps of Membrane Remodeling during Myoblast Fusion. Developmental Cell 46, 767–780.e7. 10.1016/j.devcel.2018.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipton BH, Konigsberg IR, 1972. A Fine-Structural Analysis of the Fusion of Myogenic Cells. The Journal of Cell Biology 53, 348–364. 10.1083/jcb.53.2.348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J-X, Hoglund A-S, Karlsson P, Lindblad J, Qaisar R, Aare S, Bengtsson E, Larsson L, 2009. Myonuclear domain size and myosin isoform expression in muscle fibres from mammals representing a 100 000-fold difference in body size: Myonuclear domain size. Experimental Physiology 94, 117–129. 10.1113/expphysiol.2008.043877 [DOI] [PubMed] [Google Scholar]
- Liu N, Garry GA, Li S, Bezprozvannaya S, Sanchez-Ortiz E, Chen B, Shelton JM, Jaichander P, Bassel-Duby R, Olson EN, 2017. A Twist2-dependent progenitor cell contributes to adult skeletal muscle. Nature Cell Biology 19, 202–213. 10.1038/ncb3477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Li E, Nie Q, Zhang X, Luo W, Li E, Nie Q, Zhang X, 2015. Myomaker, Regulated by MYOD, MYOG and miR-140–3p, Promotes Chicken Myoblast Fusion. International Journal of Molecular Sciences 16, 26186–26201. 10.3390/ijms161125946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltzahn J. von, Jones AE, Parks RJ, Rudnicki MA, 2013. Pax7 is critical for the normal function of satellite cells in adult skeletal muscle. PNAS 110, 16474–16479. 10.1073/pnas.1307680110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantilla CB, Sill RV, Aravamudan B, Zhan W-Z, Sieck GC, 2008. Developmental effects on myonuclear domain size of rat diaphragm fibers. Journal of Applied Physiology 104, 787–794. 10.1152/japplphysiol.00347.2007 [DOI] [PubMed] [Google Scholar]
- Martin BL, Gallagher TL, Rastogi N, Davis JP, Beattie CE, Amacher SL, Janssen PML, 2015. In vivo assessment of contractile strength distinguishes differential gene function in skeletal muscle of zebrafish larvae. Journal of Applied Physiology 119, 799–806. 10.1152/japplphysiol.00447.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsakas A, Otto A, Elashry MI, Brown SC, Patel K, 2010. Altered Primary and Secondary Myogenesis in the Myostatin-Null Mouse. Rejuvenation Research 13, 717–727. 10.1089/rej.2010.1065 [DOI] [PubMed] [Google Scholar]
- McMenamin SK, Chandless MN, Parichy DM, 2016. Working with zebrafish at postembryonic stages, in: Methods in Cell Biology. Elsevier, pp. 587–607. 10.1016/bs.mcb.2015.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijering E, Dzyubachyk O, Smal I, 2012. Methods for Cell and Particle Tracking, in: Methods in Enzymology. Elsevier, pp. 183–200. 10.1016/B978-0-12-391857-4.00009-4 [DOI] [PubMed] [Google Scholar]
- Millay DP, Gamage DG, Quinn ME, Min Y-L, Mitani Y, Bassel-Duby R, Olson EN, 2016. Structure-function analysis of myomaker domains required for myoblast fusion. PNAS 113, 2116–2121. 10.1073/pnas.1600101113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millay DP, O’Rourke JR, Sutherland LB, Bezprozvannaya S, Shelton JM, Bassel-Duby R, Olson EN, 2013. Myomaker is a membrane activator of myoblast fusion and muscle formation. Nature 499, 301–305. 10.1038/nature12343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millay DP, Sutherland LB, Bassel-Duby R, Olson EN, 2014. Myomaker is essential for muscle regeneration. Genes Dev. 28, 1641–1646. 10.1101/gad.247205.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell KJ, Pannérec A, Cadot B, Parlakian A, Besson V, Gomes ER, Marazzi G, Sassoon DA, 2010. Identification and characterization of a non-satellite cell muscle resident progenitor during postnatal development. Nature Cell Biology 12, 257–266. 10.1038/ncb2025 [DOI] [PubMed] [Google Scholar]
- Moore CA, Parkin CA, Bidet Y, Ingham PW, 2007. A role for the Myoblast city homologues Dock1 and Dock5 and the adaptor proteins Crk and Crk-like in zebrafish myoblast fusion. Development 134, 3145–3153. 10.1242/dev.001214 [DOI] [PubMed] [Google Scholar]
- Moss FP, Leblond CP, 1971. Satellite cells as the source of nuclei in muscles of growing rats. Anat. Rec. 170, 421–435. 10.1002/ar.1091700405 [DOI] [PubMed] [Google Scholar]
- Moss FP, Leblond CP, 1970. Nature of dividing nuclei in skeletal muscle of growing rats. J. Cell Biol. 44, 459–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen PD, Gurevich DB, Sonntag C, Hersey L, Alaei S, Nim HT, Siegel A, Hall TE, Rossello FJ, Boyd SE, Polo JM, Currie PD, 2017. Muscle Stem Cells Undergo Extensive Clonal Drift during Tissue Growth via Meox1-Mediated Induction of G2 Cell-Cycle Arrest. Cell Stem Cell 21, 107–119.e6. 10.1016/j.stem.2017.06.003 [DOI] [PubMed] [Google Scholar]
- Ohira Y, Yoshinaga T, Ohara M, Nonaka I, Yoshioka T, Yamashita-Goto K, Shenkman BS, Kozlovskaya IB, Roy RR, Edgerton VR, 1999. Myonuclear domain and myosin phenotype in human soleus after bed rest with or without loading. Journal of Applied Physiology 87, 1776–1785. 10.1152/jappl.1999.87.5.1776 [DOI] [PubMed] [Google Scholar]
- Pajcini KV, Pomerantz JH, Alkan O, Doyonnas R, Blau HM, 2008. Myoblasts and macrophages share molecular components that contribute to cell-cell fusion. The Journal of Cell Biology 180, 1005–1019. 10.1083/jcb.200707191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panza P, Maier J, Schmees C, Rothbauer U, Sollner C, 2015. Live imaging of endogenous protein dynamics in zebrafish using chromobodies. Development 142, 1879–1884. 10.1242/dev.118943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parichy DM, Elizondo MR, Mills MG, Gordon TN, Engeszer RE, 2009. Normal table of postembryonic zebrafish development: Staging by externally visible anatomy of the living fish. Developmental Dynamics 238, 2975–3015. 10.1002/dvdy.22113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson SE, Mook LB, Devoto SH, 2008. Growth in the larval zebrafish pectoral fin and trunk musculature. Developmental Dynamics 237, 307–315. 10.1002/dvdy.21400 [DOI] [PubMed] [Google Scholar]
- Pavlath GK, Rich K, Webster SG, Blau HM, 1989. Localization of muscle gene products in nuclear domains. Nature 337, 570–573. 10.1038/337570a0 [DOI] [PubMed] [Google Scholar]
- Pipalia TG, Koth J, Roy SD, Hammond CL, Kawakami K, Hughes SM, 2016. Cellular dynamics of regeneration reveals role of two distinct Pax7 stem cell populations in larval zebrafish muscle repair. Disease Models & Mechanisms 9, 671–684. 10.1242/dmm.022251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell GT, Wright GJ, 2012. Do muscle founder cells exist in vertebrates? Trends in Cell Biology 22, 391–396. https://doi.org/10.1016Zj.tcb.2012.05.003 [DOI] [PubMed] [Google Scholar]
- Powell GT, Wright GJ, 2011. Jamb and Jamc Are Essential for Vertebrate Myocyte Fusion. PLoS Biology 9, e1001216 10.1371/journal.pbio.1001216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn ME, Goh Q, Kurosaka M, Gamage DG, Petrany MJ, Prasad V, Millay DP, 2017. Myomerger induces fusion of non-fusogenic cells and is required for skeletal muscle development. Nature Communications 8, 15665 10.1038/ncomms15665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu-Petersen Z, Deasy B, Jankowski R, Ikezawa M, Cummins J, Pruchnic R, Mytinger J, Cao B, Gates C, Wernig A, Huard J, 2002. Identification of a novel population of muscle stem cells in mice: potential for muscle regeneration. The Journal of Cell Biology 157, 851–864. 10.1083/jcb.200108150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raamsdonk W, van der Stelt A, Diegenbach PC, van de Berg W, Bruyn H, van Dijk J, Mijzen P, 1974. Differentiation of the musculature of the teleost Brachydanio rerio. I. Myotome shape and movements in the embryo. Anat. Embryol. 145, 321–342. [DOI] [PubMed] [Google Scholar]
- Rash JE, Fambrough D, 1973. Ultrastructural and electrophysiological correlates of cell coupling and cytoplasmic fusion during myogenesis in vitro. Developmental Biology 30, 166–186. 10.1016/0012-1606(73)90055-9 [DOI] [PubMed] [Google Scholar]
- Rochlin K, Yu S, Roy S, Baylies MK, 2010. Myoblast fusion: When it takes more to make one. Developmental Biology, Special Section: Morphogenesis 341, 66–83. 10.1016/j.ydbio.2009.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosser BWC, Dean MS, Bandman E, 2002. Myonuclear domain size varies along the lengths of maturing skeletal muscle fibers. Int. J. Dev. Biol. 46, 747–754. [PubMed] [Google Scholar]
- Rowlerson A, Veggetti A, 2001. Cellular mechanisms of post-embryonic muscle growth in aquaculture species, in: Johnston IA (Ed.), Muscle Development and Growth, Fish Physiology; pp. 103–140. [Google Scholar]
- Roy S, Wolff C, Ingham PW, 2001. The u-boot mutation identifies a Hedgehog-regulated myogenic switch for fiber-type diversification in the zebrafish embryo. Genes & development 15, 1563–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SD, Williams VC, Pipalia TG, Li K, Hammond CL, Knappe S, Knight RD, Hughes SM, 2017. Myotome adaptability confers developmental robustness to somitic myogenesis in response to fibre number alteration. Developmental Biology 431, 321–335. 10.1016/j.ydbio.2017.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Gómez M, Coutts N, Price A, Taylor MV, Bate M, 2000. Drosophila dumbfounded: a myoblast attractant essential for fusion. Cell 102, 189–198. [DOI] [PubMed] [Google Scholar]
- Russell B, Motlagh D, Ashley WW, 2000. Form follows function: how muscle shape is regulated by work. Journal of Applied Physiology 88, 1127–1132. 10.1152/jappl.2000.88.3.1127 [DOI] [PubMed] [Google Scholar]
- Saera-Vila A, Kasprick DS, Junttila TL, Grzegorski SJ, Louie KW, Chiari EF, Kish PE, Kahana A, 2015. Myocyte Dedifferentiation Drives Extraocular Muscle Regeneration in Adult Zebrafish. Invest. Ophthalmol. Vis. Sci. 56, 4977–4993. 10.1167/iovs.14-16103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saera-Vila A, Kish PE, Kahana A, 2016. Fgf regulates dedifferentiation during skeletal muscle regeneration in adult zebrafish. Cellular Signalling 28, 1196–1204. 10.1016/j.cellsig.2016.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath Srihari C., Sampath Srinath C., Millay DP, 2018. Myoblast fusion confusion: the resolution begins. Skeletal Muscle 8 10.1186/s13395-017-0149-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawamiphak S, Kontarakis Z, Filosa A, Reischauer S, Stainier DYR, 2017. Transient cardiomyocyte fusion regulates cardiac development in zebrafish. Nature Communications 8, 1525 10.1038/s41467-017-01555-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz LM, 2019. Skeletal Muscles Do Not Undergo Apoptosis During Either Atrophy or Programmed Cell Death-Revisiting the Myonuclear Domain Hypothesis. Front. Physiol. 9, 1887 10.3389/fphys.2018.01887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz LM, Brown C, McLaughlin K, Smith W, Bigelow C, 2016. The myonuclear domain is not maintained in skeletal muscle during either atrophy or programmed cell death. American Journal of Physiology-Cell Physiology 311, C607–C615. 10.1152/ajpcell.00176.2016 [DOI] [PubMed] [Google Scholar]
- Seale P, Sabourin LA, Girgis-Gabardo A, Mansouri A, Gruss P, Rudnicki MA, 2000. Pax7 Is Required for the Specification of Myogenic Satellite Cells. Cell 102, 777–786. 10.1016/S0092-8674(00)00066-0 [DOI] [PubMed] [Google Scholar]
- Seger C, Hargrave M, Wang X, Chai RJ, Elworthy S, Ingham PW, 2011. Analysis of Pax7 expressing myogenic cells in zebrafish muscle development, injury, and models of disease. Developmental Dynamics 240, 2440–2451. 10.1002/dvdy.22745 [DOI] [PubMed] [Google Scholar]
- Sens KL, Zhang S, Jin P, Duan R, Zhang G, Luo F, Parachini L, Chen EH, 2010. An invasive podosome-like structure promotes fusion pore formation during myoblast fusion. The Journal of Cell Biology 191, 1013–1027. 10.1083/jcb.201006006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Bi P, Pei J, Li H, Grishin NV, Bassel-Duby R, Chen EH, Olson EN, 2017. Requirement of the fusogenic micropeptide myomixer for muscle formation in zebrafish. Proceedings of the National Academy of Sciences 114, 11950–11955. 10.1073/pnas.1715229114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Cai M, Si Y, Zhang J, Du S, 2018. Knockout of myomaker results in defective myoblast fusion, reduced muscle growth and increased adipocyte infiltration in zebrafish skeletal muscle. Human Molecular Genetics. 10.1093/hmg/ddy268 [DOI] [PubMed] [Google Scholar]
- Shimada Y, 1971. ELECTRON MICROSCOPE OBSERVATIONS ON THE FUSION OF CHICK MYOBLASTS IN VITRO. J Cell Biol 48, 128–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si Y, Wen H, Du S, 2019. Genetic Mutations in jamb, jamc, and myomaker Revealed Different Roles on Myoblast Fusion and Muscle Growth. Marine Biotechnology 21, 111–123. 10.1007/s10126-018-9865-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel AL, Gurevich DB, Currie PD, 2013. A myogenic precursor cell that could contribute to regeneration in zebrafish and its similarity to the satellite cell. FEBS Journal 280, 4074–4088. 10.1111/febs.12300 [DOI] [PubMed] [Google Scholar]
- Simionescu A, Pavlath GK, 2011. Molecular Mechanisms of Myoblast Fusion Across Species, in: Dittmar T, Zänker KS (Eds.), Cell Fusion in Health and Disease. Springer Netherlands, Dordrecht, pp. 113–135. [DOI] [PubMed] [Google Scholar]
- Singleman C, Holtzman NG, 2014. Growth and Maturation in the Zebrafish, Danio Rerio: A Staging Tool for Teaching and Research. Zebrafish 11, 396–406. 10.1089/zeb.2014.0976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow CJ, Goody M, Kelly MW, Oster EC, Jones R, Khalil A, Henry CA, 2008. Time-Lapse Analysis and Mathematical Characterization Elucidate Novel Mechanisms Underlying Muscle Morphogenesis. PLoS Genetics 4, e1000219 10.1371/journal.pgen.1000219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn RL, Huang P, Kawahara G, Mitchell M, Guyon J, Kalluri R, Kunkel LM, Gussoni E, 2009. A role for nephrin, a renal protein, in vertebrate skeletal muscle cell fusion. Proceedings of the National Academy of Sciences 106, 9274–9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas BP, Woo J, Leong WY, Roy S, 2007. A conserved molecular pathway mediates myoblast fusion in insects and vertebrates. Nature Genetics 39, 781–786. 10.1038/ng2055 [DOI] [PubMed] [Google Scholar]
- Suster ML, Abe G, Schouw A, Kawakami K, 2011. Transposon-mediated BAC transgenesis in zebrafish. Nature Protocols 6, 1998–2021. 10.1038/nprot.2011.416 [DOI] [PubMed] [Google Scholar]
- Tabebordbar M, Wang ET, Wagers AJ, 2013. Skeletal Muscle Degenerative Diseases and Strategies for Therapeutic Muscle Repair. Annual Review of Pathology: Mechanisms of Disease 8, 441–475. 10.1146/annurev-pathol-011811-132450 [DOI] [PubMed] [Google Scholar]
- Talbot JC, Amacher SL, 2014. A streamlined CRISPR pipeline to reliably generate zebrafish frameshifting alleles. Zebrafish 11, 583–585. 10.1089/zeb.2014.1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot JC, Nichols JT, Yan Y-L, Leonard IF, BreMiller RA, Amacher SL, Postlethwait JH, Kimmel CB, 2016. Pharyngeal morphogenesis requires fras1-itga8-dependent epithelial-mesenchymal interaction. Dev. Biol. 416, 136–148. https://doi.Org/10.1016/j.ydbio.2016.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot JC, Teets EM, Ratnayake D, Duy PQ, Currie PD, Amacher SL, In press. Muscle precursor cell movements in zebrafish are dynamic and require six-family genes. Development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamir-Livne Y, Mubariki R, Bengal E, 2017. Adhesion molecule Kirrel3/Neph2 is required for the elongated shape of myocytes during skeletal muscle differentiation. Int. J. Dev. Biol. 61, 337–345. 10.1387/ijdb.170005eb [DOI] [PubMed] [Google Scholar]
- Tang Q, Moore JC, Ignatius MS, Tenente IM, Hayes MN, Garcia EG, Torres Yordan N, Bourque C, He S, Blackburn JS, Look AT, Houvras Y, Langenau DM, 2016. Imaging tumour cell heterogeneity following cell transplantation into optically clear immune-deficient zebrafish. Nature Communications 7, 10358 10.1038/ncomms10358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee J-M, Sartori da Silva MA, Rygiel AM, Muncan V, Bink R, van den Brink GR, van Tijn P, Zivkovic D, Kodach LL, Guardavaccaro D, Diks SH, Peppelenbosch MP, 2012. asb11 Is a Regulator of Embryonic and Adult Regenerative Myogenesis. Stem Cells and Development 21, 3091–3103. 10.1089/scd.2012.0123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisdale MJ, 2002. Cachexia in cancer patients. Nature Reviews Cancer 2, 862–871. 10.1038/nrc927 [DOI] [PubMed] [Google Scholar]
- Van der Meer SFT, Jaspers RT, Degens H, 2011. Is the myonuclear domain size fixed? J Musculoskelet Neuronal Interact 11,286–297. [PubMed] [Google Scholar]
- Voesenek CJ, Muijres FT, Leeuwen JL van, 2018. Biomechanics of swimming in developing larval fish. Journal of Experimental Biology 221, jeb149583 10.1242/jeb.149583 [DOI] [PubMed] [Google Scholar]
- von Hofsten J, Elworthy S, Gilchrist MJ, Smith JC, Wardle FC, Ingham PW, 2008Prdml-and Sox6-mediated transcriptional repression specifies muscle fibre type in the zebrafish embryo. EMBO reports 9, 683–689. 10.1038/embor.2008.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace GQ, McNally EM, 2009. Mechanisms of Muscle Degeneration, Regeneration, and Repair in the Muscular Dystrophies. Annual Review of Physiology 71, 37–57. 10.1146/annurev.physiol.010908.163216 [DOI] [PubMed] [Google Scholar]
- Wang X, Ono Y, Tan SC, Chai RJ, Parkin C, Ingham PW, 2011. Prdm1a and miR-499 act sequentially to restrict Sox6 activity to the fast-twitch muscle lineage in the zebrafish embryo. Development 138, 4399–4404. 10.1242/dev.070516 [DOI] [PubMed] [Google Scholar]
- Weatherley AH, Gill HS, Lobo AF, 1988. Recruitment and maximal diameter of axial muscle fibres in teleosts and their relationship to somatic growth and ultimate size. Journal of Fish Biology 33, 851–859. 10.1111/j.1095-8649.1988.tb05532.x [DOI] [Google Scholar]
- Westerfield M, 2007. The Zebrafish Book: A guide for the laboratory use of zebrafish (Danio rerio). University of Oregon Press, Eugene. [Google Scholar]
- White RB, Biérinx A-S, Gnocchi VF, Zammit PS, 2010. Dynamics of muscle fibre growth during postnatal mouse development. BMC Developmental Biology 10, 21 10.1186/1471-213X-10-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windner SE, Manhart A, Brown A, Mogilner A, Baylies MK, 2019. Nuclear Scaling Is Coordinated among Individual Nuclei in Multinucleated Muscle Fibers. Developmental Cell 49, 48–62.e3. 10.1016/j.devcel.2019.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Farr GH, Tapscott SJ, Maves L, 2013. Pbx and Prdm1a transcription factors differentially regulate subsets of the fast skeletal muscle program in zebrafish. Biology Open 2, 546–555. 10.1242/bio.20133921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Lee R, Ono Y, Ingham PW, Saunders TE, 2018. Spatiotemporal Coordination of FGF and Shh Signaling Underlies the Specification of Myoblasts in the Zebrafish Embryo. Developmental Cell 46, 735–750.e4. https://doi.Org/10.1016/j.devcel.2018.08.024 [DOI] [PubMed] [Google Scholar]
- Zhang Q, Vashisht AA, O’Rourke J, Corbel SY, Moran R, Romero A, Miraglia L,Zhang J, Durrant E, Schmedt C, Sampath Srinath C., Sampath Srihari C., 2017. The microprotein Minion controls cell fusion and muscle formation. Nature Communications 8, 15664 10.1038/ncomms15664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Roy S, 2017. Myomaker is required for the fusion of fast-twitch myocytes in the zebrafish embryo. Developmental Biology 423, 24–33. 10.1016/j.ydbio.2017.01.019 [DOI] [PubMed] [Google Scholar]
- Zhang W, Roy S, 2016. The zebrafish fast myosin light chain mylpfa:H2B-GFP transgene is a useful tool for in vivo imaging of myocyte fusion in the vertebrate embryo. Gene Expression Patterns 20, 106–110. https://doi.org/10.1016Zj.gep.2016.02.001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.