Abstract
The ability to comprehensively profile cellular heterogeneity in functional proteome is crucial in advancing our understanding of cell behavior, organism development, and disease mechanisms. Conventional bulk measurement by averaging the biological responses across a population often loses the information of cellular variations. Single-cell proteomic technologies are becoming increasingly important to understand and discern cellular heterogeneity. The well-established methods for single-cell protein analysis based on flow cytometry and fluorescence microscopy are limited by the low multiplexing ability owing to the spectra overlap of fluorophores for labeling antibodies. Recent advances in mass-spectrometry, microchip, and reiterative staining-based techniques for single-cell proteomics have enabled the evaluation of cellular heterogeneity with high throughput, increased multiplexity, and improved sensitivity. In this review, we describe the principles, developments, advantages and limitations of these advanced technologies in analysis of single-cell proteins, along with their biological applications to study cellular heterogeneity. At last, we discuss the remaining challenges, possible strategies, and future opportunities that will facilitate the improvement and broad applications of single-cell proteomic technologies in cell biology and medical research.
Keywords: Single cell proteomics, Cellular heterogeneity, Mass cytometry, Microchip, DNA barcoded antibody
1. Introdcution
No two cells are exactly alike. Cell-to-cell heterogeneity is an intrinsic trait of any advanced organisms where cells are specialized to carry on various functions. Decades of studies confirm that cellular heterogeneity plays critical roles in disease occurrence and drug responses [1–3]. The causes for cellular heterogeneity are mainly attributed to variable biochemical processes including stochastic gene and protein expression, functional differences in cell development or cell cycle status, and tissue microenvironmental changes [1, 4]. The ever-increasing appreciation of the significance of cellular heterogeneity in tissue development and disease progression has been gradually changing the paradigm of therapeutic intervention and propelling the discovery of new targeted drugs [5]. However, the complexity of cell diversity in a system poses major challenges to the identification of key cellular subsets that may be responsible for systemic diseases, such as autoimmune diseases, Alzheimer’s disease, and various types of cancers [3, 4, 6].
Single-cell technologies for profiling cellular heterogeneity have advanced rapidly in the past decade. Varied by the measurement targets, those technologies can be grouped as genomics-, epigenomics, transcriptomics-, proteomics, or metabolomics-based approaches [5, 7]. Next-generation sequencing technologies have powered the dramatic progress of single-cell genomics, epigenomics and transcriptomics, and these analytical methods are commercially available [8–10]. On the contrary, single-cell proteomics at the genomic scale is still not attainable yet. Arguably, the proteomic level of knowledge is equally important since proteins carry most of the biological functions and participate in almost all cellular processes including signal transduction, migration, transportation and secretion, differentiation, proliferation, and apoptosis [11–13]. Both the qualitative and quantitative measurements of gene expression and mutations in single cells can only infer the possible functional consequences which have to be presented by proteins. Studies also confirm that the gene expression is not quantitatively equivalent to protein expression since there are multi-step post-translational modification in between, and protein’ life span is in general a few orders of magnitude longer than mRNA [13–15]. This is validated by Xie group that the protein and mRNA copy numbers for any given gene in single Escherichia coli cell are highly uncorrelated [15]. For these reasons, there remains an urgent need to develop single-cell proteomic tools for elucidating cellular identity and heterogeneity, and to provide another layer of knowledge other than that by single-cell transcriptomics and genomics.
Nevertheless, analysis of the entire proteome at the single-cell level is extremely challenging. The major bottleneck is the miniscule amount of protein in individual cells, which often leads to high noise background for protein assays. Different from DNAs and RNAs, proteins cannot be amplified directly to increase copy number to overcome signal-to-noise limitations [7, 16]. Particularly, some functional proteins such as secreted signaling proteins and intracellular phosphoproteins can present as low as tens of copies and exhibit a large discrepancy of protein abundance (generally spanning several orders of magnitude) [17], thereby requiring extremely sensitive assays for single-cell protein measurement. Another challenge lies in the enormous complexity of the proteome, meaning that a myriad of proteins (e.g., intracellular proteins, surface markers, and secreted proteins) can be spliced, modified, and activated on different epitopes in the same cell [18]. It is estimated that the human proteome may have over 1 million species, however only ~1,200 proteins that are functionally related have been quantitatively detected to date [19]. The level of complexity, with cellular variations at the same time, requires multiparametric and dynamic analysis of plenty of individual cells, of which information are crucial to multidimensional profiling of cell-to-cell heterogeneity and dictating the role of a cell in the entire population [18]. Therefore, a single-cell functional proteomics assay must be both high-throughput and high-multiplexity to discover biomarkers and provide quantitative correlations or inter-relationships within complex protein interacting networks.
Conventional fluorescence microscopy-based imaging approaches have been routinely used to quantify proteins in individual cells. These techniques are highly sensitive and commercially available, but the range of attainable information normally only span a few proteins per cell, which is not sufficient to cover the whole spectrum of the functional proteome. State-of-the-art multicolor flow cytometry technique can detect ~10 different types of proteins in single cells with a high-throughput manner but still limited by the low multiplexing capacity because of spectral overlap of fluorophore-labeled antibodies, and it cannot capture secreted proteins that are essential to understand cell signaling functions [20]. Besides, these techniques tend to be the end point readouts where cells are fixed and lose viability, preventing the study of protein secretion dynamics in single cells and interactions of individual cells [21].
To enable deep profiling of cellular heterogeneity, new methods for single-cell protein analysis with high detection sensitivity and multiplexing capability have recently developed, which herein be categorized as mass spectrometry-based, microchip-based, and reiterative staining methods. Mass spectrometry (MS)-based methods feature with high specificity, wide molecular coverage, semi-quantification, and structural identification capabilities [22, 23]. Microchip-based devices hold unique advantages due to their powerful single-cell handling capability, flexible custom design, and multifunctional integration within a single system [7, 24]. Reiterative staining techniques as emerging and high-content tools have a tremendous potential for single-cell protein assays with high-throughput and multiplexity [20, 25]. In this review, we will discuss these techniques that can be used for highly multiplexed single-cell proteomic assays with an emphasis on the profiling of cellular heterogeneity. Additionally, the latest advances for these assays will be presented and their distinct advantages and limitations will be compared. Finally, the future development in the field will be proposed to help us gain a deeper understanding of cellular diversity and heterogeneity.
2. Mass spectrometry-based method
2.1. Mass spectrometry
The modern mass spectrometry has shown great potential in multiplexed analysis of proteome in single cells [22]. This analysis is primarily based on liquid chromatography (LC) combined with electrospray ionization (ESI) and tandem MS, usually abbreviated as LC-MS/MS [26]. Briefly, LC-MS/MS starts with cell lysis, then purification of cell lysate proteins, followed by separation of peptides through a nano-liquid chromatographic column, and finally ionization for analysis. In the early stage, LC-MS/MS has been applied to single-cell lysates for deep and accurate quantitation of proteins from bulk samples [26–28], and it has also enabled proteomic analysis of giant cells, such as oocytes (450~800 proteins/cell) [29, 30] and muscle fibers (~2100 proteins/cell) [31, 32]. However, the detection limit is far not enough for analysis of signaling proteins that only have no more than thousands of copies. Throughput at only 10 cells/h per instrument also limits the practical use [29, 30].
To address these issues, the Slavov group developed Single-Cell ProtEomics by Mass Spectrometry (SCoPE-MS) through introducing the concept of carrier proteins barcoded with tandem-mass-tags [33, 34]. The use of these tags not only reduces the sample loss and enhances the detectability of ions, but also provides fragment ions for peptide sequence identification. By combining this concept with mechanical cell lysis, Slavov and coworkers utilized the SCoPE-MS to analyze over a thousand proteins in differentiating mouse embryonic stem cells and classify cell types based on their proteomic profiles [33]. To further improve the quantification and throughput of the LC-MS/MS method, the Slavov group proposed a next generation SCoPE-MS version, SCoPE2 (Fig. 1a) [35]. It integrates the SCoPE-MS with automated and miniaturized sample preparation, substantially lowering cost and hands-on time. Specht et al. used SCoPE2 to quantify ~2,000 proteins in 356 single monocytes and macrophages, and analyzed the emergence of cellular heterogeneity as homogeneous monocytes differentiated into macrophage-like cells in the absence of polarizing cytokines [35]. The results showed a continuous gradient of proteome states for the macrophage-like cells, indicating that the individual macrophage-like cells are highly heterogeneous even though they originate from homogeneous monocytes exposed to identical environmental conditions.
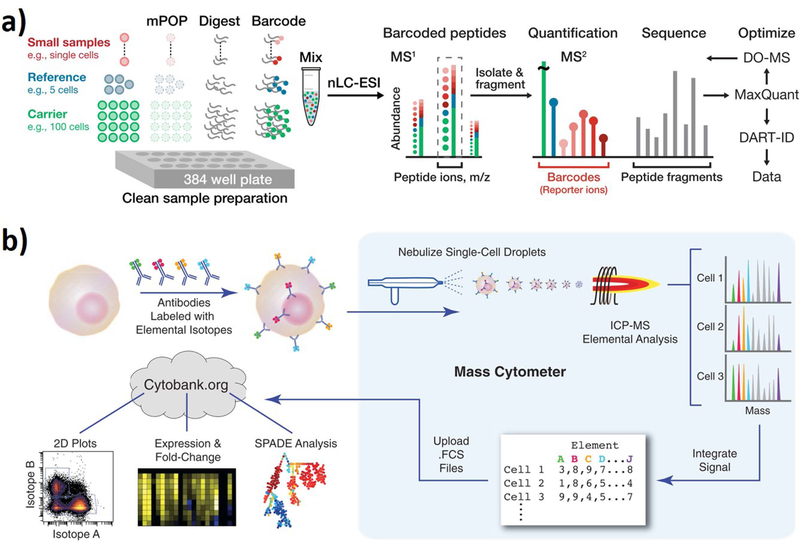
Fig. 1.
Representative mass spectrometry-based methods for single-cell proteomic analysis. a) Single-Cell ProtEomics by Mass Spectrometry (SCoPE2): single cells are isolated in individual wells, lysed, and the proteins digested to peptides. The peptides from each single cell are covalently barcoded with tandem-mass-tags. The MS instrument isolates labeled peptides with the same sequence and fragments them. The fragmentation generates reporter ions, whose abundances reflect protein abundances in the corresponding single cells. Reproduced from ref. [35] with permission from bioRxiv. b) Mass cytometry: individual cells are stained with epitope-specific antibodies conjugated to transition elemental isotope. After nebulized into single-cell droplets, the cells are measured by their m/z ratio in a time of flight mass analyzer and can then be analyzed in an elemental mass spectrum for each individual cell. Reproduced from ref. [38] with permission from the American Association for the Advancement of Science.
A challenge in the further development of LC-MS/MS technique is that without an external standard, quantitative measurements of more complex proteoform stoichiometrically is rather difficult due to ratios between specific peptides [36]. Quantification is impaired by current MS proteomics methods because of peptide associated biases. In efforts to avoid these biases, the Slavov group developed a first principles model (HIquant) for quantifying proteomics stoichiometrically [37]. Based on accurate relative quantification, the HIquant explicitly models peptide levels measured across conditions as a superposition of the levels of the proteins from which the peptides came from. The HIquant model can infer the protein levels independently from noise and make stoichiometric estimates between homologous proteins and proteoform by coupling the equations for different peptides. Although the MS methods have allowed for the measurements of thousands of proteins, the instruments are expensive and rare in ordinary laboratories, and also need professionals and skilled technician to operate, which may limit their broad applications.
2.2. Mass cytometry
Mass cytometry developed by the Nolan group is a combination of flow cytometry and inductively coupled plasma mass spectrometry (ICP-MS), which enables high-throughput, highly multiplexed, and quantitative single-cell analysis (Fig. 1b) [38]. In this approach, multiple protein targets in individual cells are labeled with antibodies coupled with transition element isotopes instead of fluorophores. Subsequently, labelled cells are sprayed as single-cell droplets, and transported into plasma by laminar flow of argon gas. The individual cells are ionized and filtered through a quadrupole, and then measured by their m/z ratio in a time of flight mass analyzer. The signal for each transition element isotope is translated to the abundances of the proteins in single cells. A distinct advantage of mass cytometry is that the use of metal isotopes tagged antibodies on targeted biomolecules overcome the spectral overlap observed in fluorescent-based cytometry, thus allowing for a greater number of parameters to be analyzed in single cells [23]. More than 36 different types of proteins can be simultaneously detected at the single-cell level with approximately 1000 cells per second throughput [39, 40]. Besides, the transition metal isotopes have low abundance in biological samples, which greatly reduces background noise [41].
Mass cytometry can measure large panels of surface markers, functional cytoplasmic proteins, and even secreted proteins from large numbers of single cells. This abundant proteomic information from single cells are useful in different types of biological researches, such as deep cellular phenotyping for discovery of heterogeneous cellular subsets and studying the relationship between disease pathology and intracellular signaling networks [38, 42–47]. Bendall et al. utilized single-cell mass cytometry to examine human bone marrow cells by simultaneously monitoring 34 cellular parameters [38]. An antibody panel targeting 13 core- and 18 subtype-specific surface markers was used for deep phenotypic profiling under a set of ex vivo stimuli and drug inhibitors. The spanning-tree progression analysis of density-normalized events (SPADE) multidimensional clustering method revealed the systematic organization and lineage relationships among the different cellular subtypes, providing a global view of immune signaling in the human hematopoietic and immune systems. Fragiadakis et al. used a mass cytometry assay to simultaneously examine the phenotype and intracellular signaling activities of all major immune cell subsets derived from fetal umbilical cord and maternal peripheral blood samples [42]. Distinct immune cell subsets can be visualized by single-cell analysis, and functional differences can be identified between fetal and maternal immune systems. The results provided a high-resolution reference map of the cellular composition and functional organization of the healthy fetal and maternal immune systems at birth. Bodenmiller et al. developed mass-tag cellular barcoding (MCB) by using n metal ion tags to multiplex up to 2n samples, significantly improving mass cytometry throughput as well as reducing antibody consumption [43]. The authors used the MCB to characterize human peripheral blood mononuclear cell (PBMC) signaling dynamics and cell-to-cell communication. Through measuring the effects of 27 inhibitors on 14 phosphorylation sites in 14 PBMC types at 96 conditions, the data set revealed 18,816 quantified phosphorylation levels from each multiplexed sample. This large number of simultaneously measured parameters enabled the high-content, high-throughput classification of inhibitors and cell types based on the cellular signaling network, which could be used as an effective tool for personalized medicine and new drug discovery.
Cancer is a heterogeneous disease with distinct genetic and phenotypic properties that can differentially promote progression, metastasis and drug resistance. Mass cytometry is also an ideal technique for highly multiplexed analysis of cancer cell and its association with tumor progression [48–50]. Giesen et al. coupled immunohistochemical and immunocytochemical methods with high-resolution laser ablation to mass cytometry, enables the simultaneous imaging of 32 proteins and protein modifications at subcellular resolution [48]. The authors applied this imaging mass cytometry to human breast cancer samples, allowing delineation of cell subpopulations and cell-cell interactions and highlighting tumor heterogeneity. However, the mass cytometry approaches usually require large amounts of input materials (>106 cells) for effective analysis [23]. Meanwhile, the sample introduction efficiency for mass cytometry is relatively low (generally ~30%−40% injected cell samples can be analyzed), which may prohibit the application to rare cells and precious clinical samples [51]. Additionally, the enriched metal isotopes typically contain 1% of isotope impurities, and some isotope ions form oxide in the plasma, causing interference to the accurate data analysis [23]. Also, as the measured cells are atomized during the assay, the cells measured by mass cytometry are hard to be retrieved for downstream analysis.
3. Microchip-based method
3.1. Microengraving
The Love group demonstrated the adaptation of a soft lithographic method, called microengraving, to parallelly detect secreted proteins from large numbers of individual live cells (>100,000 cells/assay) [52–54]. This technique uses a planar array of microwells to confine individual cells in small volumes (0.1–1 nL), and subsequently generates microarrays of proteins from each microwell (Fig. 2a). A glass slide bearing specific antibodies is compressed on the array to capture proteins secreted by confined cells. After short periods of incubation, the slide with captured proteins is removed and interrogated by sandwich-type immunoassay. Release of proteins from a cell into the enclosed small volume allows for highly sensitive detection of low abundance analytes [52]. Meanwhile, microengraving can be repeatedly performed on the same population of cells in a nondestructive manner via replacing the antibody-coated glass slides, enabling dynamic analysis of multiple parameters (e.g., cytokines, surface markers) [53]. This is a unique analytical process that are not feasible by the end-point measurement technologies like mass cytometry or immunofluorescence staining [54].
Fig. 2.
Representative microchip-based methods for single-cell proteomic analysis. a) Microengraving: individual cells are enclosed into microwells and generate microarrays of proteins from each microwells. The protein microarrays are identified by an antibodies-coated glass slide. The slide is replaced at various time points for dynamic analysis. Reproduced from ref. [53] with permission from the Proceedings of the National Academy of Sciences. b) Single-cell western blotting: single cells are captured into polyacrylamide gel microwells and go through in situ cell lysis, electrophoresis for protein separation, photo-induced protein immobilization. Through cycles of antibody stripping and protein reprobing, multiple proteins from the same cell can be detected. Reproduced from ref. [64] with permission from Nature Publishing Group. c) Droplet microfluidics: single T cells are encapsulated in agarose-gel droplets together with functionalized capture beads, and then secrete cytokines that bind to the beads confined within the droplets. The droplets are gelled and washed to break the emulsion, incubated with fluorescently labeled detection antibodies, and quantified by flow cytometry. Reproduced from ref. [71] with permission from the Royal Society of Chemistry. d) Single-cell barcode microchip: single cells are loaded and isolated into high-throughput microchambers. The secreted proteins from cells are captured by multiplex antibody barcode arrays coated on a glass substrate, and then verified by sandwich-type immunoassay. Reproduced from ref. [75] with permission from Nature Publishing Group. e) Microbeads array: each microbeads are functionalized by a specific ssDNA sequence and then converted into antibody microbeads array by cDNA-antibody conjugates for secreted proteins detection. Successive hybridization (labeling) and denaturation (quenching) are continuously performed for decoding process, exhibiting ordered fluorescent colors on every microbead. The ordered colors are corresponding to different types of antibodies carried on the microbeads. Reproduced from ref. [93] with permission from Wiley.
Embracing these advantages, the microengraving method can generate unique biological insights into various cellular systems of interest at the single-cell level, especially for immune cells [53, 55–57]. For example, Han et al. investigated the kinetics of cytokine secretion (IFN-γ, IL-2, and TNF-α) from single primary human T cells through sequential microengraving analyses [53]. Most of the T cells were found to initiate the multifunctional cytokine responses asynchronously, but the ensuing dynamic trajectories of these responses evolved programmatically in a sequential manner, which indicated that cells predominantly release one of these cytokines at a time rather than maintain active secretion of multiple cytokines simultaneously. Varadarajan et al. examined the relationship between cytolysis and the induction of secreted cytokines from thousands of individual CD8+ T cells in HIV-infected patients [55]. The cytokine secretion by CD8+ T cells isolated ex vivo from the patients, particularly IFN-γ, was found largely independent of cytolytic activity. Only a small minority of cells exhibited both functions simultaneously, while the majority of individual, functional antigen-specific cells either lysed target cells or secreted IFN-γ. The microengraving method has also been utilized to identify immune cells functional response to diseases for predicting disease progress [58, 59]. Torres et al. combined both imaging of immunological synapses formed on mobile lipid bilayers and the capture of cytokines released from individual activated T cells by microengraving [58]. This combination allowed both the physical isolation of individual cells and the parallel activation of single T cells in either pan-specific or antigen-specific manners, elucidating the biophysical and structural effects of the synapse on the activation of individual T cells and their complex functional responses. In all, the microengraving method enables facile operation and sequential detection, but it is limited by its multiplexing capacity due to the spectral overlap of organic fluorophores and fluorescent proteins.
3.2. Single-cell western blotting
Almost all the detection methods for single-cell proteomics rely on antibodies to target proteins. This antibody-dependence may generate false positive signals due to the non-specific binding from antibody cross-reactivity. The Herr group developed single-cell western blotting (scWestern) to overcome that limitation by integrating conventional protein electrophoresis and antibody recognition (Fig. 2b), and thus to add another dimension to validate the detection result [60–64]. In scWestern, the proteins from cells can be separated by their molecular mass before antibody probing, which allows for the on-target and off-target signals to be clearly differentiated [60]. This method is performed on a microchip that comprises an array of ~7670 microwells molded in a layer of polyacrylamide gel. Individual cells are first settled into the gel-coated microwells and lysed in situ with a denaturing buffer. The cell lysates run through gel electrophoresis, subsequently exposed to ultraviolet (UV) light for protein immobilization and detected by antibody immunoprobing. Through antibody stripping and target reprobing rounds, multiplexed protein detection can be achieved. The scWestern permits simultaneous assays of ~2,000 individual cells in <4 h and supports analyses of low starting cell numbers (~200), which addresses the need for high-specificity and high-sensitivity protein assays at single-cell level.
The Herr group used the scWestern method to measure cell-to-cell heterogeneity within complex populations of neural stem cells (NSC) and their differentiation responses to homogeneous in vitro stimuli [60]. After stimulation by fibroblast growth factor 2 (FGF-2), pERK5 signal (the off-target band for pERK½) was unexpectedly identified by scWestern analysis, and the signal was found uncorrelated with on-target pERK signal and contributed up to 52% of the overall pERK signal in unstimulated cells. Under mixed differentiation conditions that yielded both astrocytes and neurons, the scWesterns revealed high cell-to-cell marker expression variability, including profound increases in glial fibrillary acidic protein (GFAP) expression in the relatively rare astrocyte population. In addition, scWestern has been applied in assessing glioblastoma heterogeneity response to chemotherapy and profiling protein expression in circulating tumor cells, which provides useful information designed for personalized cancer therapy and prognosis [63, 64].
Later on, Duncombe et al. reported an advanced scWestern by using spatial and temporal control of polyacrylamide gel (PAG) [65]. The introduction of the PAG material enables thousands of pore-gradient electrophoresis microgels to be created on a standard microscope slide and also resolves a wide mass range of proteins which is essential for assessing signaling pathways. Employing the pore-gradient microgels to study HER2-related signaling in single cells dissociated from breast tumor biopsy, a five to tenfold difference in representative signaling proteins expression (mTOR, ERK, and eIF4E) was observed. Nonetheless, scWestern may have a relatively lower detection sensitivity, as up to 40% of the proteins can be lost during cell lysis, protein immobilization, and repeated antibody stripping [60].
3.3. Droplet-based microfluidics
Droplet-based microfluidic approaches for single-cell analysis permits greatly enhanced throughput of cell screening and reduce sample-handling complexity [66, 67]. They also overcome one of the major limitations of traditional flow cytometry on detection of secreted proteins [68–70]. In general, highly monodisperse aqueous droplets carrying single cells and antibody-functionalized-beads flow into an inert oil on a microchannel chip (Fig. 2c) [71]. Up to several hundred droplets can be generated per second, and each droplet serves as a miniaturized assay vessel of picoliter-nanoliter volume. The functionalized beads capture secreted proteins and then being visualized by fluorescent probes. Another distinct advantage of the droplet-based microfluidics for single cells analysis is its capability of linking cellular genotype with phenotype [24]: the same cell can be subject to cytokine detection first and subsequently be sorted and lysed for downstream genetic analysis.
This technique has greatly contributed to the quantitative understanding of cellular heterogeneity and maps subsets within cell populations with specific functions [71, 72]. Chokkalingam et al. used monodisperse agarose droplets to encapsulate single activated T-cells and detected cytokine (IL-2, IFN-γ, and TNF-α) secretion of the cells over time [71]. Analysis of 7415 cell-containing beads by flow cytometry revealed the presence of 8 different cellular subpopulations. ~85% of T cells secreted one or more cytokines upon overnight in-droplet stimulation, while ~57% of stimulated T cells secreted all 3 tested cytokines. In-depth heterogeneity analysis demonstrated that cells that secreted large amounts of IL-2 also secreted significantly higher levels of TNF-α and IFN-γ, but a subpopulation of cells that secreted intermediate levels of IL-2 had significantly higher levels of TNF-α. Debs et al. presented a droplet microfluidic platform allowing the functional screening of up to 300,000 individual hybridoma cell clones within reasonable time [72]. The authors used this platform to perform a model screen for therapeutic monoclonal antibodies (mABs) that inhibit angiotensin converting enzyme 1. The cells expressing these mABs were spiked into an unrelated hybridoma cell population in a ratio of 1∶10,000, and a 9,400-fold enrichment was observed by fluorescence activated droplet sorting.
High-throughput droplet generation and high-performance screening suggest that the droplet-based technology can perform as a potential platform for multiomics approaches, especially for single-cell proteomics. However, this technique also suffers from the low multiplexing capacity because of the lower probability of loading unique beads in droplets as a result of the Poisson distributions [73]. In addition, it cannot perform washing steps to remove excess reagents, thus resulting in nontrivial background fluorescence within the droplets.
3.4. Single-cell barcode microchip
The first generation of barcoded microfluidic device for multiplexed protein analysis, the single cell barcode chip (SCBC), was developed the Heath group [74, 75]. In SCBC platform, thousands of nanoliter-sized microchambers, which patterned as parallel antibody stripes, are utilized to isolate individual cells for simultaneous protein measurement (Fig. 2d) [75]. As the key to this platform, the antibody stripes are fabricated by a microchannel-guided flow patterning of a single-stranded DNA (ssDNA) barcode and DNA-encoded antibody library (DEAL) approach [76, 77]. Briefly, oligonucleotides are first immobilized on a poly-lysine (PLL)-coated surface as high-dense ssDNA microarrays using microfluidic flow channels. The immobilized oligonucleotides are hybridized by barcoded DNA-antibody conjugates to convert the ssDNA microarrays into antibody microarrays. Proteins from individual cells are captured by the antibody microarrays and quantified by using a sandwich-type immunoassay. The generated fluorescence signals are compared with a calibration curve (built by using protein standards) to determine the abundances of the secreted proteins.
The SCBC platform has been broadly used in single-cell proteomics for deep profiling of functional heterogeneity since its birth in 2011 [75, 78–89]. Ma et al. applied this platform for ex vivo quantification of T cell polyfunctional diversity via the simultaneous measurement of a dozen protein molecules secreted from tumor antigen-specific cytotoxic T lymphocytes (CTLs) [75]. Highly heterogeneous and functional diversity in active tumor antigen-specific CTLs, with the major functional phenotypes quantitatively identified, was observed compared to the samples from healthy donor controls. Later on, Shi et al. modified the initial SCBC design to incorporate a cell lysis module by using pneumatic valves [78]. The valves can be utilized for isolating the chambers and controlling fluid paths, enabling the introduction of lysis buffers to the microchambers for cell lysis and follow-up intracellular proteins assay. They applied this new design to uncover EGF-receptor-mediated PI3K signaling pathway in single cancer cells by quantifying 11 intracellular proteins, and they revealed the cellular heterogeneity on organ-specific markers in circulating tumor cells [78–80]. Another significant progress in SCBC platform has been reported by Wang et al [81]. The authors built a valve-less architecture by employing a set of deformable, three-state polydimethylsiloxane (PDMS) posts for controlling cell loading, on-chip cell lysis and antibody assay. This SCBC platform not only features with significantly higher density of antibody array, but each array element has a unique molecular identity for localizing an appropriately labeled capture antibody. Besides, the valve-less architecture permits 30-fold more cellular assays than the previous SCBCs that involve cell lysis. Likewise, Lu et al. developed an advanced SCBC platform by integrating a sub-nanoliter microchamber array and high-density antibody barcodes, which allows a simultaneous detection of 14 cytokines from more than a thousand single cells in parallel [82]. Analysis of the single cell secretomic signatures revealed cellular heterogeneity is related with biophysiological activities such as migration.
To further increase the multiplexing capacity of SCBC platform, Lu et al. combined the spatial and spectral barcodes on one chip [83]. Each microchamber consists of 15 isolated antibody stripes, and each stripe contains three different antibodies that are later labelled by detection antibodies with 3 different fluorescent colors. Thus, up to 45 different proteins from cells can be verified (three of them serve as positive controls), which keeps the record of the highest multiplexing capacity for a single-cell secretion assay to date. In addition, SCBC platform has also been widely applied in the study of intracellular metabolite heterogeneity, cell-to-cell communication, and cell response to drug treatments [84–88].
SCBC has many advantages compared with other single-cell proteomic technologies. First, it enables the analysis of secreted proteins from live cells, which is not possible in flow cytometry and mass cytometry where cells need to be fixed. Apart from secreted proteins, cytoplasmic and membrane proteins together with metabolites from the same single cells can be quantified simultaneously [90]. Secondly, the method can be high throughput, and a statistically significant number of single cells can be analyzed [82]. Finally, this technique can be standardized through external recombinant protein experiments, and semiquantitative fluorescence intensity can be converted to the number of molecules for each protein in single cells [73]. Despite those advantages, SCBC also has some limitations. The analytes are spatially patterned on a two-dimensional plane, on which the protein type is encoded by position [83]. There is a tradeoff between the multiplexing capacity and detection sensitivity: higher multiplexity requires expansion of microchamber size and volume, which inversely decreases assay sensitivity. Additionally, the SCBC platform suffers from complex antibody barcodes fabrication and limited high-quality capture and detection antibody pairs.
3.5. Microbeads method
Microbeads-based techniques hold a high potential for multiplexed single-cell proteomic analysis, as microbeads are not spatially limited compared to SCBC techniques and can be densely packed into a microchip [73, 91]. The microbeads are spectrally encoded with two or more fluorescent dyes and loaded into a microchip manually. Each of the microbeads are conjugated with antibodies for capturing specific proteins and then subjected to a standard ELISA procedure. All channels use the same fluorescent readout for protein quantification, and the microbeads are subsequently decoded by imaging the different barcoding channels and mapping them to their specific bead-coding regions. Diercks et al. used the similar concept for protein standard samples analysis and measured four different proteins within a microchip [91]. The microbeads in this technique do not need to be precisely positioned within the microchip, because the identity of each microbeads with captured proteins is determined by fluorescent imaging after the assay is complete. The sensitivity of this microchip is sufficient to detect thousands of copies of tumor necrosis factor in a very small volume (4.7 nL). In addition, the dynamic range of the measurement can be easily tuned for each protein by varying the number of beads in the microchip. For example, Junkin et al. developed a time-resolved system to dynamically measure key immune parameters from individual cells by using beads combined with fluorescent imaging [92]. Beads are initially loaded into a microchamber and then exposed to medium coming from individual cells through peristaltic pumping. After specific capture of proteins of interest, the beads are rinsed and moved back to the original microchamber. This process is repeated with a new set of beads for the next round, enabling multiparameter analysis of single-cell dynamics. After analysis of individual macrophage cell signal under pathogen inputs, TNF secretion from the cell was found highly heterogeneous and surprisingly uncorrelated with the dynamics of NF-κB, the transcription factor controlling TNF production.
Recently, Wang group advanced the microbeads techniques by presenting a multiplexed in situ tagging (MIST) strategy (Fig. 2e), which can potentially detect tens to hundreds of molecular targets in single cells with high throughput and high sensitivity [93, 94]. This approach is based on ssDNA encoded microbead arrays integrated with a DNA encoded antibody library. Through multicolor and multicycle successive imaging, comprehensive protein profiling can be achieved. For example, with M color type of fluorescent dye and N staining cycle or imaging round number, an overall MN protein targets can be profiled in situ in the same sample [93, 94]. After conversion of the microbeads arrays into barcoded antibody arrays and analysis of differentiated single macrophage cells upon lipopolysaccharide (LPS) stimulation, significant cellular heterogeneity was observed when the secretion of ten immune effector proteins were quantified. This technique allows easy fabrication and assay operation, as well as exhibits an unprecedented level in multiplexity, but it is also limited by high-quality antibody pairs.
4. Reiterative staining method
4.1. DNA barcoding method
Popular multiplexing methods for single-cell proteomic analysis often utilize a fluorescent antibody labeling technique, but the caveat is that there is a limit to the number of distinctive fluorescent colors [95]. The use of DNA barcode for multiplexing provides an opportunistic platform to which an unlimited number of distinctive codes can be generated based on their size or sequence [96–98]. The Weissleder group developed antibody barcoding with cleavable DNA (ABCD) platform to increase the multiplexing capacity of single-cell analysis [96]. In this method, each different antibody is tagged with a unique and short DNA barcode through a photocleavable linker. After the DNA barcoded antibody binding to cells, the cells are exposed to UV light to release the unique DNA barcode. The released DNA can be detected by gel electrophoresis or fluorescence hybridization technique, and then be translated to protein expression levels [96, 97]. This method enables characterization of the heterogeneity among different cell types with high reliability validated by flow cytometry. In addition, it allows for quantitative, high-sensitivity, and multiplexed detection of proteins in live single cells, which is very helpful for analyzing rare but biologically important cells in clinical samples. Ullal et al. used this approach to profile ~90 proteins at the single-cell level and subsequently investigated inter- and intra-tumoral heterogeneity as well as the pathways in drug response [97].
One limitation in the aforementioned platform is that short fluorophore-labeled DNA barcode is vulnerable to problematic, non-specific binding to cellular endogenous molecules (e.g., nuclei, plasmid) when applied to cells for in situ hybridization. This undesired staining is hard to quench by previously used cleavage method, thus resulting in considerable background noise during fluorescence imaging. To overcome this obstacle, the Weissleder group recently proposed to pre-hybridize fluorescent DNA imaging strands to matching DNA-antibody barcodes in vitro and use the fluorescent DNA-antibody conjugates for subsequent cellular staining (Fig. 3a) [98]. As hybridization strength is dependent on salt concentration, the fluorescent strands can be stably attached to the barcoded antibody in PBS, and can then be simply washed off with melting buffer (MB) after imaging, which can effectively reduce the background by cycle-to-cycle staining. Weissleder and coworkers demonstrated that this advanced platform can be used to interrogate drug-relevant pathways in clinical samples [98]. Using the PI3K/PTEN/CDK4/6 pathways in breast cancer as an example, considerable heterogeneity in protein expression and dose-dependent suppression were observed in given patients and between patients. Likewise, the DNA barcoding method is limited by the antibody cross-reactivity.
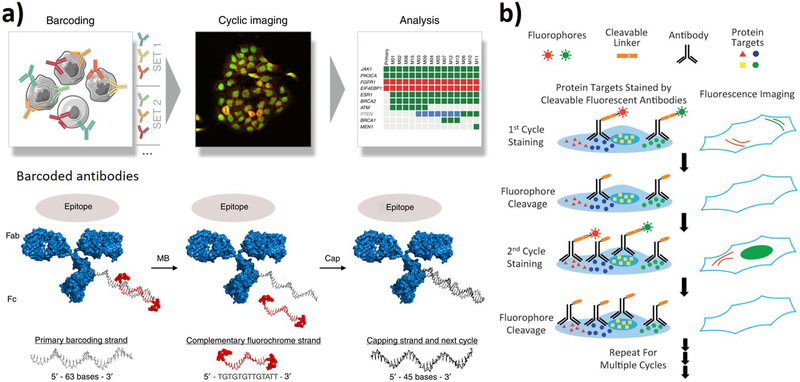
Fig. 3.
Representative reiterative staining method for single-cell proteomic analysis. a) DNA barcoding method: target proteins in individual cells are interrogated and quantitated by antibody-DNA conjugates hybridized to complementary fluorochrome strands. After imaging of each cycle, the fluorescent strands are washed off with melting buffer (MB), and the primary strands are capped with an unlabeled strand to reduce background during subsequent cellular staining. Reproduced from ref. [98] with permission from Nature Publishing Group. b) Cyclic immunofluorescence: antibodies are labeled with fluorophores for specific target proteins via a cleavable linker. After imaging, the fluorophores are removed to initiate the next fluorescent cycle. Through reiterative process of recognition, imaging, and removal, different protein in single cells can be identified. Reproduced from ref. [107] with permission from Wiley.
4.2. Cyclic immunofluorescence
One of the advantages of fluorescence staining is that ultra-high sensitivity and resolution are achievable [99]. Fluorescent hybridization probes have been utilized to quantify location and expression heterogeneity of genetically identical cells in relation to their own spatial context [100, 101]. Multiplexed single-cell in situ analysis via cyclic immunofluorescence has capabilities of quantifying over hundreds of different proteins in one tissue section or cell sample [102–106]. In principle, antibodies are labeled with distinct fluorophores for specific target proteins of interest. Once the fluorescent images are collected, the fluorescent signals are removed to initiate the next fluorescent cycle. The reiterative process of target recognition, fluorescence imaging, and fluorophore signal removal, has potential for high-content analysis of individual cells.
The ability to remove the fluorescence signals effectively in cellular environments is a key requirement for success of this technique. Both photo- and chemical-based bleaching methods have been developed to erase the fluorescence signals [100, 102, 103]. Such add-on type technologies are ready to use in many labs, but they are either suffering from long assay time and low throughput or compromising the detection sensitivity duo to the damage of antibody epitopes. Recently, Guo and coworkers demonstrated the use of an azide-based cleavable linker for the conjugation of antibodies and fluorophores (Fig. 3b) [107]. After cell staining, the linker of the antibody-fluorophore conjugates can be efficiently cleaved by the mild reducing reagent tris(2-carboxyethyl)phosphine (TCEP) within 30 minutes. Even with a 24-hour incubation, the epitopes of the conjugates remain intact without loss of protein target antigenicity. The authors labeled twelve different proteins in single HeLa cells using cleavable antibodies conjugated with three fluorescent color, and thus at least three distinct proteins can be quantified at the same time in one immunofluorescence cycle. Their protein measurement showed a combination of a basal state of low expression and a long tail of high expression, suggesting that these proteins are generated in bursts, rather than at a constant rate. The Guo group further developed the cyclic immunofluorescence probes for multiplexed proteomics and even to genetic analysis using cleavable fluorescent tyramide and biorthogonal cleavable fluorescent oligonucleotides, which both allows for the detection of cell-to-cell expression heterogeneity [108, 109]. Although this method has a large sample throughput and features with high sensitivity, it is experimentally time-consuming for the antibody-fluorophore conjugation. In addition, the relative size of cleavable linker may interfere with the binding specificity and affinity site for antibodies.
5. Conclusions and future perspectives
The enormous progress and rapid expansion of single-cell proteomic techniques offer unprecedented opportunities for the study of cellular functions and development. The new discoveries and perceptions of cellular diversity and heterogeneity from the advanced technologies have shown their great potential for numerous practical applications, including cancer diagnostics and prognosis, drug discovery, regenerative medicine, and many others. A comparison of the techniques discussed in this review is summarized in Table 1. Each of these technologies has its distinct advantages. Mass cytometry and SCBC allow the high-throughput and multiparameter analysis of single cells. Droplet microfluidics and microbeads methods enable the detection of secreted proteins from live cells. DNA barcoding method has the highest multiplexing capacity among all these techniques, and scWestern avoids the misleading background signals by cross-reactivity and nonspecific binding on antibody-based assays.
Table 1.
Comparison of single-cell proteomic analysis methods
| Description | Analyte | Multiplexity | Throughput | Standards | Cell Recovery | Ref. |
|---|---|---|---|---|---|---|
| Mass spectrometry-based method | ||||||
| Mass spectrometry | Intracellular proteins | >1000 | ~10 cells/hr | Yes | No | [29, 30] |
| Mass cytometry | Membrane proteins/ intracellular proteins | ~40 | 103 cells/s | Yes | No | [39] |
| Microchip-based method | ||||||
| Microengraving | Cytokines/membrane proteins | 3 | 103−105 cells/chip | Yes | Yes | [52, 58] |
| Single-cell western blotting | Membrane proteins/ intracellular proteins | ~10 | 103−104 cells/chip | No | No | [60, 61] |
| Droplet-based microfluidics | Cytokines | 3 | 102 droplets/s | No | Yes | [71] |
| Single-cell barcode microchip | Cytokines/intracellular proteins/metabolites | ~45 | −104 cells/chip | Yes | Yes | [83] |
| Microbeads method | Cytokines | 10 | ~500 microbeads/chip | Yes | Yes | [93] |
| Reiterative staining method | ||||||
| DNA barcoding method | Membrane proteins/ intracellular proteins | 90 | 102−104 cells | Yes | Yes | [97] |
| Cyclic immunofluorescence | Intracellular proteins | 12 | 103 cells | Yes | Yes | [107] |
Despite these remarkable achievements, the single-cell proteomics techniques are still in their infancy, and several challenges remain. The low multiplexing capacity is the major technical bottleneck for the current methodologies. Although the mass cytometry and SCBC methods can quantify dozens of proteins and the DNA barcoding platform even exhibits a multiplexing capacity of ~90 proteins, the number of proteins that can be measured only represent a tiny fraction of the whole proteome, which impairs the characterization of complex signaling network and the comprehensive information of cellular subsets. The main cause for the low multiplexing capacity is attributed to the limited antibody-encoding strategies relied on fluorophores and transition metal mass labels. In addition, not all the antibodies of desired proteins are commercially available, especially for those techniques (e.g, SCBC and microbeads method) that use antibody pairs, thereby requiring careful selection of protein panels before assays. For this concern, there is a great need to develop other high affinity probes for the recognition of proteins in cells. Some promising candidates include single-chain variable fragments, aptamers, and fluorescent biorthogonal probes [11].
In addition, to achieve multifaceted analysis of cellular types and heterogeneity, single-cell protein analysis can be coupled with other “-omics” tools and phenotypic measurements, such as genomics, transcriptomics, epigenomics, metabolomics, and cell size and morphology. For example, the biological sample can be first analyzed by genomics and transcriptomics to select the cell types of interest. The identified cells can be profiled by the following single-cell proteomics to generate the most informative footprints of proteins. After a combination of the information from these integrative multi-omics and cell behavior analysis (e.g., cell growth, proliferation and migration), a whole picture and detailed dynamic information on cellular states can be obtained.
Moreover, with the fast growth of single-cell methods for protein measurements and the increase of multiplexing capacity, data analysis and standardization should be carefully considered. For instance, new algorithms and multitask-handling software packages for statistics and bioinformatics need to be developed for the increasingly large datasets generated from multiple individual cells [2]. Similar to the development of multicolor flow cytometry technique, it is essential to standardize the experimental protocols, the reagents, and the data analysis routines. At last, the commercialization of the above single-cell proteomic techniques is far from mature. Most methods discussed herein have just recently emerged from academic laboratories and require significant skill sets in engineering, technology, biology, and computational analysis. To accelerate the widespread use of these technologies in broad clinical applications, it is highly desired to develop ease of use, trustworthy, and cost-effective systems in the future studies. Despite these technical challenges encountered by single-cell protein analysis, it promises new insight into fundamental research of cell biology and modern medicine. We expect that the ongoing developments of the single-cell proteomics will facilitate our deeper understanding of cellular heterogeneity and further contribute to the related biomedical applications including disease diagnostics, personalized precision therapy, and the discovery of novel drug targets.
Acknowledgments
This work was supported by the startup fund from State University of New York at Stony Brook, and National Institute of Health (R01GM12898401) to J.W.
References
- [1].Altschuler SJ, Wu LF, Cellular heterogeneity: Do differences make a difference? Cell 2010, 141, 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wang DJ, Bodovitz S, Single cell analysis: the new frontier in ‘omics’. Trends Biotechnol. 2010, 28, 281–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Almendro V, Marusyk A, Polyak K, Cellular heterogeneity and molecular evolution in cancer. Annu. Rev. Pathol.: Mech. Dis 2013, 8, 277–302. [DOI] [PubMed] [Google Scholar]
- [4].McClellan J, King MC, Genetic heterogeneity in human disease. Cell 2010, 141, 210–217. [DOI] [PubMed] [Google Scholar]
- [5].Heath JR, Ribas A, Mischel PS, Single-cell analysis tools for drug discovery and development. Nat. Rev. Drug Discovery 2016, 15, 204–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lawson DA, Kessenbrock K, Davis RT, Pervolarakis N, Werb Z, Tumour heterogeneity and metastasis at single-cell resolution. Nat. Cell Biol 2018, 20, 1349–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Deng YX, Finck A, Fan R, Single-cell omics analyses enabled by microchip technologies. Annu. Rev. Biomed. Eng 2019, 21, 365–393. [DOI] [PubMed] [Google Scholar]
- [8].Frei AP, Bava FA, Zunder ER, Hsieh EWY, et al. , Highly multiplexed simultaneous detection of RNAs and proteins in single cells. Nat. Methods 2016, 13, 269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Levitin HM, Yuan JZ, Sims PA, Single-cell transcriptomic analysis of tumor heterogeneity. Trends Cancer 2018, 4, 264–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Papalexi E, Satija R, Single-cell RNA sequencing to explore immune cell heterogeneity. Nat. Rev. Immunol 2018, 18, 35–45. [DOI] [PubMed] [Google Scholar]
- [11].Wu MY, Singh AK, Single-cell protein analysis. Curr. Opin. Biotechnol 2012, 23, 83–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Doerr A, Single-cell proteomics. Nat. Methods 2019, 16, 20–20. [DOI] [PubMed] [Google Scholar]
- [13].Marx V, A dream of single-cell proteomics. Nat. Methods 2019, 16, 809–812. [DOI] [PubMed] [Google Scholar]
- [14].Washburn MP, Koller A, Oshiro G, Ulaszek RR, et al. , Protein pathway and complex clustering of correlated mRNA and protein expression analyses in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2003, 100, 3107–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Taniguchi Y, Choi PJ, Li GW, Chen HY, et al. , Quantifying E-coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 2010, 329, 533–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Chappell L, Russell AJC, Voet T, Single-cell (multi)omics technologies. Annu. Rev. Genomics Hum. Genet 2018, 19, 15–41. [DOI] [PubMed] [Google Scholar]
- [17].Wang J, Yang F, Emerging single-cell technologies for functional proteomics in oncology. Expert. Rev. Proteomics 2016, 13, 805–815. [DOI] [PubMed] [Google Scholar]
- [18].Chen Z, Chen JJ, Fan R, Single-cell protein secretion detection and profiling. Annu. Rev. Anal. Chem 2019, 12, 431–449. [DOI] [PubMed] [Google Scholar]
- [19].Ponomarenko EA, Poverennaya EV, Ilgisonis EV, Pyatnitskiy MA, et al. , The size of the human proteome: The width and depth. Int. J. Anal. Chem 2016, 7436849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Mondal M, Liao RJ, Guo J, Highly multiplexed single-cell protein analysis. Chem.-Eur. J 2018, 24, 7083–7091. [DOI] [PubMed] [Google Scholar]
- [21].Spiller DG, Wood CD, Rand DA, White MRH, Measurement of single-cell dynamics. Nature 2010, 465, 736–745. [DOI] [PubMed] [Google Scholar]
- [22].Zhang LW, Vertes A, Single-cell mass spectrometry approaches to explore cellular heterogeneity. Angew. Chem., Int. Ed 2018, 57, 4466–4477. [DOI] [PubMed] [Google Scholar]
- [23].Spitzer MH, Nolan GP, Mass cytometry: single cells, many features. Cell 2016, 165, 780–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lu Y, Yang L, Wei W, Shi QH, Microchip-based single-cell functional proteomics for biomedical applications. Lab Chip 2017, 17, 1250–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gunderson KL, Kruglyak S, Graige MS, Garcia F, et al. , Decoding randomly ordered DNA arrays. Genome Res. 2004, 14, 870–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Li SY, Plouffe BD, Belov AM, Ray S, et al. , An integrated platform for isolation, processing, and mass spectrometry-based proteomic profiling of rare cells in whole blood. Mol. Cell. Proteomics 2015, 14, 1672–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Schwartz SA, Reyzer ML, Caprioli RM, Direct tissue analysis using matrix-assisted laser desorption/ionization mass spectrometry: practical aspects of sample preparation. J. Mass Spectrom 2003, 38, 699–708. [DOI] [PubMed] [Google Scholar]
- [28].Hummon AB, Amare A, Sweedler JV, Discovering new invertebrate neuropeptides using mass spectrometry. Mass Spectrom. Rev 2006, 25, 77–98. [DOI] [PubMed] [Google Scholar]
- [29].Lombard-Banek C, Moody SA, Nemes P, Single-cell mass spectrometry for discovery proteomics: Quantifying translational cell heterogeneity in the 16-cell frog (Xenopus) embryo. Angew. Chem., Int. Ed 2016, 55, 2454–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Virant-Klun I, Leicht S, Hughes C, Krijgsveld J, Identification of Maturation-Specific Proteins by Single-Cell Proteomics of Human Oocytes. Mol. Cell. Proteomics 2016, 15, 2616–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Murgia M, Nagaraj N, Deshmukh AS, Zeiler M, et al. , Single muscle fiber proteomics reveals unexpected mitochondrial specialization. Embo Rep. 2015, 16, 387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Murgia M, Toniolo L, Nagaraj N, Ciciliot S, et al. , Single muscle fiber proteomics reveals fiber-type-specific features of human muscle aging. Cell Rep. 2017, 19, 2396–2409. [DOI] [PubMed] [Google Scholar]
- [33].Budnik B, Levy E, Harmange G, Slavov N, SCoPE-MS: mass spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation. Genome Biol. 2018, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Specht H, Slavov N, Transformative opportunities for single-cell proteomics. J. Proteome Res 2018, 17, 2565–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Specht H, Emmott E, Perlman DH, Koller A, Slavov N, High-throughput single-cell proteomics quantifies the emergence of macrophage heterogeneity. bioRxiv 2019, 10.1101/665307 [DOI] [Google Scholar]
- [36].Nesvizhskii AI, Proteogenomics: concepts, applications and computational strategies. Nat. Methods 2014, 11, 1114–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Malioutov D, Chen TC, Airoldi E, Jaffe J, et al. , Quantifying homologous proteins and proteoforms. Mol. Cell. Proteomics 2019, 18, 162–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Bendall SC, Simonds EF, Qiu P, Amir EAD, et al. , Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 2011, 332, 687–696.21551058 [Google Scholar]
- [39].Bendall SC, Nolan GP, Roederer M, Chattopadhyay PK, A deep profiler’s guide to cytometry. Trends Immunol. 2012, 33, 323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Newell EW, Cheng Y, Mass cytometry: blessed with the curse of dimensionality. Nat. Immunol 2016, 17, 890–895. [DOI] [PubMed] [Google Scholar]
- [41].Han GJ, Spitzer MH, Bendall SC, Fantl WJ, Nolan GP, Metal-isotope-tagged monoclonal antibodies for high-dimensional mass cytometry. Nat. Protoc 2018, 13, 2121–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Fragiadakis GK, Baca QJ, Gherardini PF, Ganio EA, et al. , Mapping the fetomaternal peripheral immune system at term pregnancy. J. Immunol 2016, 197, 4482–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Bodenmiller B, Zunder ER, Finck R, Chen TJ, et al. , Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat. Biotechnol 2012, 30, 858–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Kaiser Y, Lakshmikanth T, Chen Y, Mikes J, et al. , Mass cytometry identifies distinct lung CD4(+) T cell patterns in lofgren’s syndrome and non-lofgren’s syndrome sarcoidosis. Front. Immunol 2017, 8, 1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].O’Gorman WE, Kong DS, Balboni IM, Rudra P, et al. , Mass cytometry identifies a distinct monocyte cytokine signature shared by clinically heterogeneous pediatric SLE patients. J. Autoimmun 2017, 81, 74–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Newell EW, Sigal N, Bendall SC, Nolan GP, Davis MM, Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8(+) T cell phenotypes. Immunity 2012, 36, 142–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Horowitz A, Strauss-Albee DM, Leipold M, Kubo J, et al. , Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci. Transl. Med 2013, 5, 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Giesen C, Wang HAO, Schapiro D, Zivanovic N, et al. , Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods 2014, 11, 417–422. [DOI] [PubMed] [Google Scholar]
- [49].Amir ED, Davis KL, Tadmor MD, Simonds EF, et al. , viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol 2013, 31, 545–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Levine JH, Simonds EF, Bendall SC, Davis KL, et al. , Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell 2015, 162, 184–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Atkuri KR, Stevens JC, Neubert H, Mass Cytometry: A highly multiplexed single-cell technology for advancing drug development. Drug Metab. Dispos 2015, 43, 227–233. [DOI] [PubMed] [Google Scholar]
- [52].Love JC, Ronan JL, Grotenbreg GM, van der Veen AG, Ploegh HL, A microengraving method for rapid selection of single cells producing antigen-specific antibodies. Nat. Biotechnol 2006, 24, 703–707. [DOI] [PubMed] [Google Scholar]
- [53].Han Q, Bagheri N, Bradshaw EM, Hafler DA, et al. , Polyfunctional responses by human T cells result from sequential release of cytokines. Proc. Natl. Acad. Sci. USA 2012, 109, 1607–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Yamanaka YJ, Szeto GL, Gierahn TM, Forcier TL, et al. , Cellular barcodes for efficiently profiling single-cell secretory responses by microengraving. Anal. Chem 2012, 84, 10531–10536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Varadarajan N, Julg B, Yamanaka YJ, Chen HB, et al. , A high-throughput single-cell analysis of human CD8(+) T cell functions reveals discordance for cytokine secretion and cytolysis. J. Clin. Invest 2011, 121, 4322–4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Han Q, Bradshaw EM, Nilsson B, Hafler DA, Love JC, Multidimensional analysis of the frequencies and rates of cytokine secretion from single cells by quantitative microengraving. Lab Chip 2010, 10, 1391–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Varadarajan N, Kwon DS, Law KM, Ogunniyi AO, et al. , Rapid, efficient functional characterization and recovery of HIV-specific human CD8(+) T cells using microengraving. Proc. Natl. Acad. Sci. USA 2012, 109, 3885–3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Torres AJ, Contento RL, Gordo S, Wucherpfennig KW, Love JC, Functional single-cell analysis of T-cell activation by supported lipid bilayer-tethered ligands on arrays of nanowells. Lab Chip 2013, 13, 90–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Bradshaw EM, Kent SC, Tripuraneni V, Orban T, et al. , Concurrent detection of secreted products from human lymphocytes by microengraving: Cytokines and antigen-reactive antibodies. Clin. Immunol 2008, 129, 10–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Hughes AJ, Spelke DP, Xu ZC, Kang CC, et al. , Single-cell western blotting. Nat. Methods 2014, 11, 749–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Kang CC, Yamauchi KA, Vlassakis J, Sinkala E, et al. , Single cell-resolution western blotting. Nat. Protoc 2016, 11, 1508–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Yamauchi KA, Herr AE, Subcellular western blotting of single cells. Microsyst. Nanoeng 2017, 3, 16079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Kang CC, Lin JMG, Xu ZC, Kumar S, Herr AE, Single-cell western blotting after whole-cell imaging to assess cancer chemotherapeutic response. Anal. Chem 2014, 86, 10429–10436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Sinkala E, Sollier-Christen E, Renier C, Rosas-Canyelles E, et al. , Profiling protein expression in circulating tumour cells using microfluidic western blotting. Nat. Commun 2017, 8, 14622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Duncombe TA, Kang CC, Maity S, Ward TM, et al. , Hydrogel pore-size modulation for enhanced single-cell western blotting. Adv. Mater 2016, 28, 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Brouzes E, Medkova M, Savenelli N, Marran D, et al. , Droplet microfluidic technology for single-cell high-throughput screening. Proc. Natl. Acad. Sci. USA 2009, 106, 14195–14200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Yin HB, Marshall D, Microfluidics for single cell analysis. Curr. Opin. Biotechnol 2012, 23, 110–119. [DOI] [PubMed] [Google Scholar]
- [68].Mazutis L, Gilbert J, Ung WL, Weitz DA, et al. , Single-cell analysis and sorting using droplet-based microfluidics. Nat. Protoc 2013, 8, 870–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Teh SY, Lin R, Hung LH, Lee AP, Droplet microfluidics. Lab Chip 2008, 8, 198–220. [DOI] [PubMed] [Google Scholar]
- [70].Guo MT, Rotem A, Heyman JA, Weitz DA, Droplet microfluidics for high-throughput biological assays. Lab Chip 2012, 12, 2146–2155. [DOI] [PubMed] [Google Scholar]
- [71].Chokkalingam V, Tel J, Wimmers F, Liu X, et al. , Probing cellular heterogeneity in cytokine-secreting immune cells using droplet-based microfluidics. Lab Chip 2013, 13, 4740–4744. [DOI] [PubMed] [Google Scholar]
- [72].El Debs B, Utharala R, Balyasnikova IV, Griffiths AD, Merten CA, Functional single-cell hybridoma screening using droplet-based microfluidics. Proc. Natl. Acad. Sci. USA 2012, 109, 11570–11575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Huang L, Michael SA, Chen YF, Wu HK, Current advances in highly multiplexed antibody-based single-cell proteomic measurements. Chem.-Asian J 2017, 12, 1680–1691. [DOI] [PubMed] [Google Scholar]
- [74].Fan R, Vermesh O, Srivastava A, Yen BKH, et al. , Integrated barcode chips for rapid, multiplexed analysis of proteins in microliter quantities of blood. Nat. Biotechnol 2008, 26, 1373–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Ma C, Fan R, Ahmad H, Shi QH, et al. , A clinical microchip for evaluation of single immune cells reveals high functional heterogeneity in phenotypically similar T cells. Nat. Med 2011, 17, 738–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Bailey RC, Kwong GA, Radu CG, Witte ON, Heath JR, DNA-encoded antibody libraries: A unified platform for multiplexed cell sorting and detection of genes and proteins. J. Am. Chem. Soc 2007, 129, 1959–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Shin YS, Ahmad H, Shi QH, Kim H, et al. , Chemistries for patterning robust dna microbarcodes enable multiplex assays of cytoplasm proteins from single cancer cells. Chemphyschem 2010, 11, 3063–3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Shi QH, Qin LD, Wei W, Geng F, et al. , Single-cell proteomic chip for profiling intracellular signaling pathways in single tumor cells. Proc. Natl. Acad. Sci. USA 2012, 109, 419–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Deng YL, Zhang Y, Sun S, Wang ZH, et al. , An integrated microfluidic chip system for single-cell secretion profiling of rare circulating tumor cells. Sci. Rep 2014, 4, 7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Yang L, Wang ZH, Deng YL, Li Y, et al. , Single-cell, multiplexed protein detection of rare tumor cells based on a beads-on-barcode antibody microarray. Anal. Chem 2016, 88, 11077–11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Wang J, Tham D, Wei W, Shin YS, et al. , Quantitating cell-cell interaction functions with applications to glioblastoma multiforme cancer cells. Nano Lett. 2012, 12, 6101–6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Lu Y, Chen JJ, Mu LY, Xue Q, et al. , High-throughput secretomic analysis of single cells to assess functional cellular heterogeneity. Anal. Chem 2013, 85, 2548–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Lu Y, Xue Q, Eisele MR, Sulistijo ES, et al. , Highly multiplexed profiling of single-cell effector functions reveals deep functional heterogeneity in response to pathogenic ligands. Proc. Natl. Acad. Sci. USA 2015, 112, E607–E615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Wei W, Shi QH, Remacle F, Qin LD, et al. , Hypoxia induces a phase transition within a kinase signaling network in cancer cells. Proc. Natl. Acad. Sci. USA 2013, 110, E1352-E1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Kravchenko-Balasha N, Wang J, Remacle F, Levine RD, Heath JR, Glioblastoma cellular architectures are predicted through the characterization of two-cell interactions. Proc. Natl. Acad. Sci. USA 2014, 111, 6521–6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Su YP, Wei W, Robert L, Xue M, et al. , Single-cell analysis resolves the cell state transition and signaling dynamics associated with melanoma drug-induced resistance. Proc. Natl. Acad. Sci. USA 2017, 114, 13679–13684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Tang Y, Wang Z, Li ZM, Kim J, et al. , High-throughput screening of rare metabolically active tumor cells in pleural effusion and peripheral blood of lung cancer patients. Proc. Natl. Acad. Sci. USA 2017, 114, 2544–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Su YP, Bintz M, Yang YZ, Robert L, et al. , Phenotypic heterogeneity and evolution of melanoma cells associated with targeted therapy resistance. Plos Comput. Biol 2019, 15, e1007034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Xue Q, Lu Y, Eisele MR, Sulistijo ES, et al. , Analysis of single-cell cytokine secretion reveals a role for paracrine signaling in coordinating macrophage responses to TLR4 stimulation. Sci. Signal 2015, 8, ra59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Yu J, Zhou J, Sutherland A, Wei W, et al. , Microfluidics-based single-cell functional proteomics for fundamental and applied biomedical applications. Annu. Rev. Anal. Chem 2014, 7, 275–295. [DOI] [PubMed] [Google Scholar]
- [91].Diercks AH, Ozinsky A, Hansen CL, Spotts JM, et al. , A microfluidic device for multiplexed protein detection in nano-liter volumes. Ana.l Biochem 2009, 386, 30–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Junkin M, Kaestli AJ, Cheng Z, Jordi C, et al. , High-content quantification of single-cell immune dynamics. Cell Rep. 2016, 15, 411–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Zhao P, Bhowmick S, Yu JC, Wang J, Highly multiplexed single-cell protein profiling with large-scale convertible DNA-antibody barcoded arrays. Adv. Sci 2018, 5, 1800672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Abdullah MAA, Wang J, Ultrasimple single-cell detection of multiple cytokines by a nanowell chip integrated with encoded microarrays. ACS sens. 2019, 4, 2296–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Armbrecht L, Dittrich PS, Recent advances in the analysis of single cells. Anal. Chem 2017, 89, 2–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Agasti SS, Liong M, Peterson VM, Lee H, Weissleder R, Photocleavable DNA barcode-antibody conjugates allow sensitive and multiplexed protein analysis in single cells. J. Am. Chem. Soc 2012, 134, 18499–18502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Ullal AV, Peterson V, Agasti SS, Tuang S, et al. , Cancer cell profiling by barcoding allows multiplexed protein analysis in fine-needle aspirates. Sci. Transl. Med 2014, 6, 219ra9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Giedt RJ, Pathania D, Carlson JCT, McFarland PJ, et al. , Single-cell barcode analysis provides a rapid readout of cellular signaling pathways in clinical specimens. Nat. Commun 2018, 9, 4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Thul PJ, Akesson L, Wiking M, Mahdessian D, et al. , A subcellular map of the human proteome. Science 2017, 356, eaal3321. [DOI] [PubMed] [Google Scholar]
- [100].Schubert W, Bonnekoh B, Pommer AJ, Philipsen L, et al. , Analyzing proteome topology and function by automated multidimensional fluorescence microscopy. Nat. Biotechnol 2006, 24, 1270–1278. [DOI] [PubMed] [Google Scholar]
- [101].Levsky JM, Singer RH, Fluorescence in situ hybridization: past, present and future. J. Cell Sci 2003, 116, 2833–2838. [DOI] [PubMed] [Google Scholar]
- [102].Gerdes MJ, Sevinsky CJ, Sood A, Adak S, et al. , Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc. Natl. Acad. Sci. USA 2013, 110, 11982–11987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Lin JR, Fallahi-Sichani M, Sorger PK, Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method. Nat. Commun 2015, 6, 8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Duose DY, Schweller RM, Hittelman WN, Diehl MR, Multiplexed and reiterative fluorescence labeling via DNA circuitry. Bioconjugate Chem. 2010, 21, 2327–2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Schweller RM, Zimak J, Duose DY, Qutub AA, et al. , Multiplexed in situ immunofluorescence using dynamic DNA complexes. Angew. Chem., Int. Ed 2012, 51, 9292–9296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Zrazhevskiy P, Gao XH, Quantum dot imaging platform for single-cell molecular profiling. Nat. Commun 2013, 4, 1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Mondal M, Liao RJ, Xiao L, Eno T, Guo J, Highly multiplexed single-cell in situ protein analysis with cleavable fluorescent antibodies. Angew. Chem., Int. Ed 2017, 56, 2636–2639. [DOI] [PubMed] [Google Scholar]
- [108].Mondal M, Liao RJ, Nazaroff CD, Samuel AD, Guo J, Highly multiplexed single-cell in situ RNA and DNA analysis with bioorthogonal cleavable fluorescent oligonucleotides. Chem. Sci 2018, 9, 2909–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Liao R, Mondal M, Nazaroff CD, Mastroeni D, et al. , Highly sensitive in situ proteomics with cleavable fluorescent tyramide reveals human neuronal heterogeneity. bioRxiv 2019, 10.1101/539106. [DOI] [PMC free article] [PubMed] [Google Scholar]