ABSTRACT
Mitochondrial DNA of Trypanosoma brucei and related parasites is a catenated network containing thousands of minicircles and tens of maxicircles, called kinetoplast DNA (kDNA). Replication of a single nucleoid requires at least three DNA polymerase I-like proteins (i.e. POLIB, POLIC and POLID), each showing discrete localizations near the kDNA during S phase. POLIB and POLID have roles in minicircle replication but the specific role of POLIC in kDNA maintenance is less clear. Here, we use an RNA interference (RNAi)-complementation system to dissect the functions of two distinct POLIC regions, i.e. the conserved family A DNA polymerase (POLA) domain and the uncharacterized N-terminal region (UCR). While RNAi complementation with wild-type POLIC restored kDNA content and cell cycle localization of kDNA, active site point mutations in the POLA domain impaired minicircle replication similar to that of POLIB and POLID depletions. Complementation with POLA domain alone abolished the formation of POLIC foci and partially rescued the RNAi phenotype. Furthermore, we provide evidence that the UCR is crucial in cell cycle-dependent protein localization and facilitates proper distribution of progeny networks. This is the first report of a DNA polymerase that impacts on mitochondrial nucleoid distribution.
This article has an associated First Person interview with the first author of the paper.
KEY WORDS: Mitochondrial DNA replication, DNA polymerase, Trypanosoma brucei, Cell cycle-dependent protein localization, Kinetoplast DNA
Highlighted Article: A dual functioning DNA polymerase in African trypanosomes supports DNA replication and distribution of mitochondrial DNA progeny.
INTRODUCTION
Maintenance of mitochondrial DNA (mtDNA) is vital for most eukaryotic cells. Accumulation of mutations or the failure to maintain copy numbers of genes can result in loss of mtDNA-encoded proteins essential for energy generation, and also in mitochondrial dysfunction associated with aging, metabolic disease, neurodegenerative disorders and cancer (Viscomi and Zeviani, 2017). As a result, maintenance of mtDNA is a focal point of biomedical research. Despite decades of investigation, the mechanisms that regulate replication, copy number maintenance and segregation of mammalian mtDNA still remain unclear.
Although many studies on mitochondrial biology have focused on a few eukaryotic lineages (i.e. yeast and mammals), great diversity exists in mitochondrial genomes and proteomes across eukaryotes (Chen and Butow, 2005; Gray, 2015; Zíková et al., 2016). Eukaryotic microbes constitute much of this evolutionary diversity, and the parasitic protists of medical importance are by far the most studied. One of the most intriguing and structurally complex mitochondrial genomes is found in trypanosomatid protists, such as Trypanosoma brucei, the parasite responsible for African sleeping sickness. Trypanosomatids harbor a single – often branched – mitochondrion. Its mtDNA consists of two genetic elements, the minicircles and maxicircles, that are catenated into a network, the so-called kinetoplast DNA (kDNA) (Jensen and Englund, 2012; Verner et al., 2015). In vivo, kDNA is condensed into a single disk-shaped nucleoid that is always present near the flagellar basal body and is replicated once in every cell cycle in near synchrony with other single-unit organelles, including the nucleus and flagellum (Woodward and Gull, 1990). The topological complexity of kDNA provides a striking reporter system, in which lesions within the network and disruption of replication intermediates help to define mechanistic defects. kDNA, therefore, represents a good model to study mtDNA dynamics.
Variations on kDNA structure, and the sizes of minicircles and maxicircles exist (for review see Lukeš et al., 2002). In T. brucei, the 23-kb maxicircles (∼25 copies) are considered to be homologs of mammalian mtDNA, as they contain similar genes encoding subunits of the respiratory complex and also rRNAs. However, a majority of the maxicircle genes are cryptic and undergo an extensive post-transcriptional RNA editing process of uridine insertion and deletion to generate translatable mRNAs. This process is mediated by minicircle-encoded guide RNAs (gRNAs) (for reviews see Read et al., 2016; Stuart et al., 2005). Therefore, both genetic elements are essential for fitness and completion of the digenetic life cycle of the parasite (Dewar et al., 2018). However, the ∼5000 minicircles (1 kb) constitute the majority of the network, with each minicircle connected through Hopf-links to three others (Chen et al., 1995). Maxicircles are linked to the minicircles and are also likely to be interlocked with other maxicircles, thereby forming a network within a network (Shapiro, 1993). How this complex structure is replicated and segregated is an area of focused study (for reviews see Jensen and Englund, 2012; Povelones, 2014; Schneider and Ochsenreiter, 2018).
One key feature is the topoisomerase II-mediated release of minicircle monomers, to enable replication that is independent of the network. Briefly, nonreplicated covalently closed (CC) minicircles are released into a region between the kDNA disk and the mitochondrial membrane nearest to the flagellar basal body, i.e. the kinetoflagellar zone (KFZ), where initiation and synthesis occur through unidirectional theta DNA replication (Abu-Elneel et al., 2001; Melendy et al., 1988). Later stages, such as Okazaki fragment processing and topoisomerase-mediated attachment of minicircle progeny occur in two protein assemblies located at opposite poles of the network periphery called antipodal sites (Hines et al., 2001; Melendy et al., 1988). Minicircle progeny are attached to the network while still containing at least one nick or multiply gapped (N/G) DNA; a presumed counting mechanism that distinguishes between replicated versus non-replicated minicircles (Ntambi et al., 1986). The process results in a spatial and temporal separation of replication events. Maxicircles replicate through theta replication within the network and progeny also contain N/G DNA (Carpenter and Englund, 1995; Liu et al., 2009). During the later stages of network replication the remaining nicks and gaps are repaired, and the network undergoes topological remodeling prior to network scission, distribution of equal progeny networks and, finally, segregation into daughter cells.
A second key feature is the tripartite attachment complex (TAC) that physically links the kDNA network to the basal body of the flagellum and mediates segregation of the progeny kDNA networks (Ogbadoyi et al., 2003). The TAC comprises three distinct subunit components: (i) exclusion zone filaments that extend from the basal body to the mitochondrial outer membrane, (ii) differentiated mitochondrial membranes that are located between the basal body and the kDNA disk, and (iii) unilateral filaments (ULFs) that extend from the mitochondrial inner membrane to the kDNA network in the KFZ. An elegant electron microscopy cytochemistry study distinguished between inner and outer types of ULF (Gluenz et al., 2007), which might represent two subdomains – one involved in replication and another involved in structural processes or segregation. Moreover, several components of each region have been identified and genetic studies indicate a hierarchical organization for TAC biogenesis starting with components that are most proximal to the basal body (Hoffmann et al., 2018; Schneider and Ochsenreiter, 2018). One core TAC component, TAC102, localizes to the ULF and is essential for proper kDNA segregation; although it does not directly interact with the kDNA network it is the most proximal ULF component (Jakob et al., 2016; Trikin et al., 2016).
A third feature is the multiplicity of proteins that have similar activities but non-redundant roles in kDNA maintenance. Among the many additional factors not present in mammalian nucleoids are two topoisomerases, two origin-binding proteins, two primases, six helicases, six DNA polymerases, two ligases and several other factors (Abu-Elneel et al., 2001; Downey et al., 2005; Lindsay et al., 2008; Liu et al., 2006; Saxowsky et al., 2003; Scocca and Shapiro, 2008). For example, in T. brucei, PIF2 helicase and PRI1 primase have defined roles in maxicircle replication (Hines and Ray, 2010; Liu et al., 2009), while PIF1 and PRI2 have roles in minicircle replication (Hines and Ray, 2011; Liu et al., 2010). In the two life cycle stages of T. brucei – the procyclic and the bloodstream form (PCF and BSF, respectively) – three members of DNA polymerase family A are required for growth and kDNA maintenance. These are the DNA polymerase I-like proteins B, C and D (POLIB, POLIC and POLID, respectively) (Bruhn et al., 2010, 2011; Chandler et al., 2008; Klingbeil et al., 2002). Whereas POLIB and POLID were demonstrated to have roles in minicircle replication, the precise role for POLIC in kDNA maintenance has not been determined.
Approximately 30 kDNA replication proteins have been characterized at single protein level and were found to localize to specific sites surrounding the kDNA disk, including the KFZ, the antipodal sites flanking the disk or within the disk. Given the unique spatiotemporal replication mechanism of the network, several studies have recently uncovered a complex choreography of kDNA replication proteins that are likely to coordinate subsets of proteins to specific sites during cell cycle progression (Abu-Elneel et al., 2001; Concepción-Acevedo et al., 2012, 2018; Johnson and Englund, 1998; Peña-Diaz et al., 2017). POLID is the most dramatic example of a kDNA replication protein that undergoes cell cycle-dependent localization which colocalized with replicating minicircles at the antipodal sites only during kDNA S phase (1N1K*), and localized to the mitochondrial matrix during all other cell cycle stages (Concepción-Acevedo et al., 2012). Cell cycle stages are identified on the basis of single-unit structures of nuclear (N) and kinetoplast DNA (K). Thus, we distinguish between 1N1K cells in G1 (1N1K) and those that undergo kDNA S phase (1N1K*), those that have two segregated progeny networks and are completing nuclear S phase and mitosis (1N2K) or those that have not yet undergone cytokinesis (2N2K). Another kDNA polymerase, POLIC, has initially been detected in the KFZ/ULF and accumulates at the antipodal sites only during kDNA S phase colocalizing with a fraction of POLID, thus, supporting the idea of highly transient kDNA interactions (Concepción-Acevedo et al., 2018). The majority of POLIC signals do not overlap with the TAC102 signal and, additionally, when both proteins localized to the KFZ/ULF region, POLIC is more proximal to the kDNA than TAC102 (Concepción-Acevedo et al., 2018). What regulates the timing and recruitment associated with the protein choreography remains unknown.
The minicircle kDNA polymerases POLIB and POLID both contain a predicted C-terminal DNA polymerase family A (POLA) domain and a 3′-5′ exonuclease domain. POLIC, however, is a 1649 aa protein (180 kDa) that comprises only a conserved C-terminal POLA domain (315 aa). The remainder of this large protein is an uncharacterized N-terminal region that we termed UCR. Gene silencing of POLIC results in loss of fitness, a decrease in both minicircle and maxicircle copy number, and progressive loss of the kDNA network; hallmarks of a kDNA replication defect (Bruhn et al., 2010; Chandler et al., 2008; Liu et al., 2010). However, POLIC silencing also results in cells that predominantly display ‘small’ kDNAs rather than their complete loss, an increased population of free minicircles as well as the appearance of ancillary kDNA − i.e. extra-kDNA material determined through fluorescence in situ hybridization (FISH), which is abnormally positioned anterior to the nucleus in addition to the normally positioned kDNA network (Miyahira and Dvorak, 1994). Ancillary kDNA has been detected when other mitochondrial proteins were silenced and are presumed to be a secondary defect (Archer et al., 2009; Clayton et al., 2011; Grewal et al., 2016; Trikin et al., 2016; Týč et al., 2015). The POLIC phenotype is unusual by not displaying as a classic replication or segregation defect that typically result in one cell containing a double-sized network and one that lacks kDNA. In addition to decreased kDNA content, POLIC depletion appears to exhibit a distribution defect in which the kDNA network is not correctly positioned for the scission event, thereby leading to asymmetric division of the incompletely replicated networks.
Therefore, the aim of this study was to apply a genetic complementation approach to dissect the pleiotropic RNA interference (RNAi) phenotype and to further define the function of POLIC in kDNA maintenance. We present data of POLIC having two functional domains that contribute to kDNA maintenance – one for nucleotidyl incorporation and another that facilitates localization of the protein during the cell cycle as well as distribution of the kDNA network. An inactive POLA domain causes a dominant-negative effect and exacerbates the kDNA replication defect of POLIC RNAi. Interestingly the UCR domain alone can restore localization to the antipodal sites during kDNA S phase. Both domains appear to contribute to proper segregation of kDNA. To our knowledge this is the first time a DNA polymerase is implicated in the correct distribution of mitochondrial nucleoid progeny, thus far, providing the strongest evidence for interactions between the kDNA replication and segregation machineries.
RESULTS
POLIC 3′UTR RNAi and rescue with epitope-tagged wild-type protein
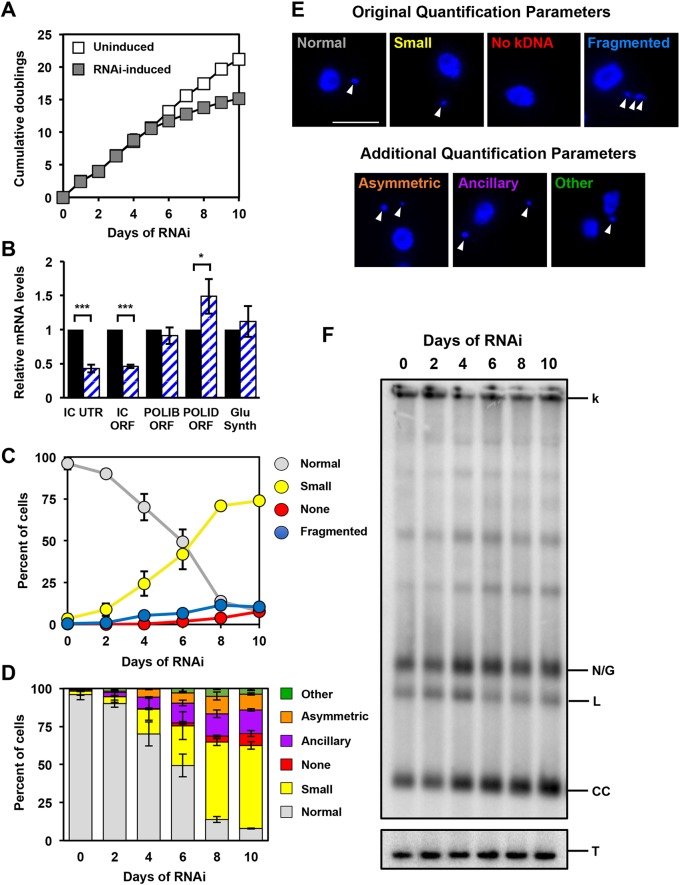
Previously, POLIC had been silenced by using an intermolecular double-stranded RNA (dsRNA) trigger targeting the open reading frame, which led to the distinct pleiotropic small, fragmented (more than two DAPI-stained spots positioned posterior to the nucleus) and ancillary kDNA phenotypes (Klingbeil et al., 2002). To examine whether the POLA and UCR domains are necessary for either kDNA replication or distribution, we coupled RNAi targeting the POLIC 3′ untranslated region (UTR) with concurrent inducible ectopic expression of epitope-tagged variants that contained an unrelated 3′-UTR to avoid RNAi targeting for a structure-function analysis. We first tested whether targeting POLIC 3′-UTR (IC-UTR) is sufficient to deplete cells of POLIC mRNA. To generate the IC-UTR cell line, a 517 bp fragment immediately downstream of the POLIC coding sequence was cloned into a stem-loop vector and subsequently introduced into 29-13 cells (Wang et al., 2000). Induction of dsRNA synthesis in response to tetracycline resulted in loss of fitness following ten rounds of proliferation (doublings) compared to uninduced cells (Fig. 1A). RT-qPCR revealed a 65% reduction of POLIC mRNA in response to induction for 48 h, but no effects on the two paralogs POLIB and POLID or the gene encoding glutathione synthetase, which is downstream of POLIC (Fig. 1B). DAPI-staining of uninduced and induced cells was used to assess the effects silencing of POLIC has on kDNA networks. Parasites per time point were classified according to kDNA size and distribution within the cell, by using the previously reported categories (i.e. normal, small, none and fragmented) according to Klingbeil et al. (2002). Ten days of IC-UTR silencing resulted in cells that displayed small kDNA (74%), fragmented/asymmetric networks (10.4%), normal kDNA (7.9%), and no kDNA (7.8%) (Fig. 1C,E), percentages that closely resembled those after POLIC ORF silencing (Klingbeil et al., 2002).
Fig. 1.
Phenotype of POLIC 3′UTR-targeted depletion. (A) Growth curves of uninduced and RNAi-induced procyclic T. brucei cells. Error bars represent the ±s.d. of the mean from three biological replicates. Error bars are too small to be displayed with respect to the size of the icon. (B) RT-qPCR analysis of uninduced (black) and tetracycline-induced (blue/white hashed) IC-UTR cells after 48 h of growth. Data are normalized to TERT and error bars represent the ±s.d. of the mean from three biological replicates. IC UTR, POLIC 3′ untranslated region; IC ORF, POLIC open reading frame; IB ORF, POLIB open reading frame; ID ORF, POLID open reading frame; Glu Synth, gene encoding glutathione synthetase. Asterisks indicate statistical significance based on pairwise Student's t-test. *P<0.1; **P<0.01; ***P<0.001. (C) Quantification of kDNA morphology by applying categories according to Klingbeil et al., 2002; i.e. Normal, Small, None and Fragmented. Over 300 DAPI-stained cells were scored at each time point and error bars represent the ±s.d. of the mean from three biological replicates. (D) Quantification of kDNA morphology utilizing new categories. Over 300 DAPI-stained cells were scored at each time point and error bars represent the ±s.d. of the mean from three biological replicates. (E) Representative images for the various kDNA phenotypes. Scale bar, 5 µm. (F) Representative Southern blot showing the increase in free minicircle species during IC 3′-UTR silencing. k, kDNA network; N/G, nicked/gapped; L, linearized; CC, covalently closed; T, tubulin.
To further dissect the multiple kDNA-associated defects, cells that contained ancillary or asymmetrically divided kDNA (progeny kDNA in which one is larger than the other) were also quantified, as ancillary kDNA had not previously been counted. Ancillary kDNA and asymmetric network division can be classified as distribution defects, and by specifically quantifying them independently would help to identify potential non-replication defects (Grewal et al., 2016; Trikin et al., 2016; Wang et al., 2002; Zhao et al., 2008). Owing to the low frequency of fragmented networks, we quantified them as part of the ‘other’ category that included any cell with an abnormal number of nuclei or kDNA.
By further subdividing the categories, IC-UTR RNAi still produced cells with a majority of small kDNA (54.7%) versus normal kDNA (7.9%), no kDNA (7.5%) and other (3.7%), but also highlighted the distribution defect signatures of asymmetric division (10.4%) and ancillary kDNA (15.6%) (Fig. 1D,E). On the basis of the proximity of POLIC and TAC102 in the KFZ and/or ULF, we wanted to test whether TAC102 localization has an impact during POLIC depletion. After 4 days of POLIC depletion, the majority of TAC102 localized properly in close proximity to the basal body, whereas some cells showed TAC102 colocalization with ancillary kDNA, as previously reported (Trikin et al., 2016). TAC102 protein appeared stable throughout the induction (Fig. S1). Moreover, by analyzing the free minicircle population by using Southern blotting and a minicircle-specific probe, both nonreplicated CC monomers and newly replicated N/G molecules increased compared to the uninduced control when POLIC was depleted (Fig. 1F) – another indication of a defect not strictly related to minicircle replication. Taken together, IC-UTR silencing phenocopied the previously reported loss of fitness and kDNA replication defect but also displayed clear signatures of a distribution defect.
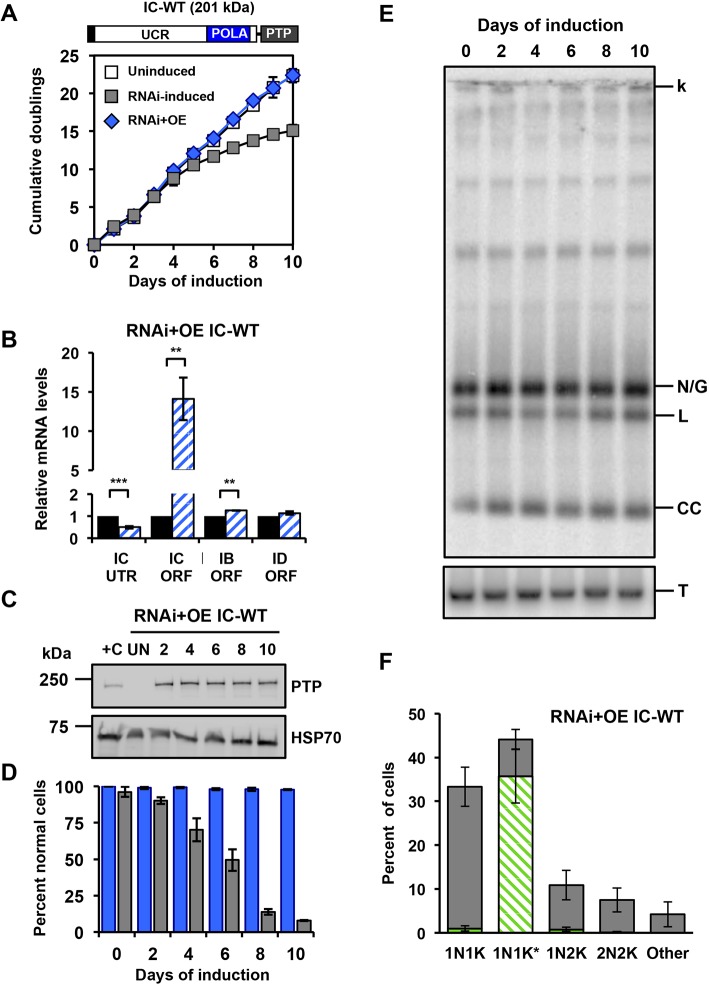
To determine whether the deficiencies caused by IC-UTR RNAi can be rescued by ectopic overexpression (OE) of PTP-tagged wild-type POLIC (IC-WT, 201 kDa), we transfected IC-UTR cells with pLewIC-WT-PTPPuro (RNAi+OE). In this genetic complementation system, the tagged POLIC variant utilizes an unrelated 3′UTR to avoid targeting by RNAi. IC-WT complementation alleviated the RNAi-mediated defects by consistently expressing IC-WT throughout the induction (Fig. 2A-E). IC-WT levels were 4-fold above levels of endogenous POLIC protein detected in a cell line engineered to endogenously express epitope-tagged POLIC from a single allele (+C, Fig. 2C) (Concepción-Acevedo et al., 2018). Additionally, we analyzed the localization of IC-WT at specific cell cycle stages and found discrete antipodal POLIC-PTP foci formation in 35% of an unsynchronized population almost exclusively in 1N1K* stage, which is similar to the 26% of endogenous POLIC-PTP antipodal site foci that were detected in the single-allele expressing control cell line (Fig. 2F) (Concepción-Acevedo et al., 2018).
Fig. 2.
Rescue with ectopically expressed IC-WT in the IC-UTR RNAi T. brucei cell line. (A) Schematic representation of full-length POLIC with the C-terminal PTP tag. Growth curves of uninduced, RNAi complementation with IC-WT, parental IC-UTR RNAi procyclic cells. Error bars represent the ±s.d. of the mean from three biological replicates. (B) RT-qPCR analysis of uninduced (black) and induced (blue/white hashed) cells following 48 h of tetracycline induction. Data are normalized to TERT and error bars represent the ±s.d. of the mean from three biological replicates. **P<0.01;***P<0.001. IC UTR, POLIC 3′ untranslated region; IC ORF, POLIC open reading frame; IB ORF, POLIB open reading frame; ID ORF, POLID open reading frame. (C) Western blot detection of the PTP tag and HSP70 over 10 days of induced complementation. At each time point, the equivalent of 1×106 cells was loaded into each well, except for +C where the equivalent of 5×106 cells was loaded. +C, single-allele-expressing control cell line endogenously expressing epitope-tagged POLIC. (D) Quantification of kDNA phenotypes. Over 300 DAPI-stained cells were scored at each time point, error bars represent the ±s.d. of the mean from three biological replicates. Blue bars, IC-WT complementation; gray bars, POLIC RNAi. Specific kDNA phenotype information can be found in Fig. 1E. (E) Representative Southern blot showing no change in the free minicircle population. k, kDNA network; N/G, nicked/gapped; L, linearized; CC, covalently closed; T, tubulin. (F) Quantification of PTP foci formation at each cell cycle stage. Percentages refer to the fraction of total cells in the population. Green/white hashed, foci positive; gray, foci negative.
By transfecting pLewIC-WT-PTPPuro into parental 29-13 cells, we generated the IC-WT cell line, to study the effects of increased IC protein in the absence of POLIC 3′UTR RNAi. Overexpression of IC-WT (8-fold) for 10 days did not interfere with fitness or kDNA maintenance, and localization of IC-WT was similar to endogenous POLIC; however, IC-WT-overexpressing (OE) cells displayed additional matrix localization (Fig. S2). The localization patterns from IC-WT overexpression and complementation were nearly identical (Fig. 2F and Fig. S2C). Therefore, IC-UTR RNAi was specific when based on the complete rescue using IC-WT and the unique pleiotropic phenotype could be attributed solely to silencing of POLIC.
POLIC DNA polymerase activity is essential for kDNA replication and cell cycle-dependent protein localization
Mutational and structural analyses of DNA polymerase family A members, have revealed two invariant Asp residues that are essential for catalytic activity in order to coordinate two divalent metal ions for transition state stabilization, which is necessary for all nucleotidyl transferases (Sousa, 1996; Steitz, 1998). POLIC and the other T. brucei mitochondrial DNA polymerases contain family A-conserved motifs A, B and C, and the non-variant Asp residues (Klingbeil et al., 2002). It is possible that the polymerase function is not the essential function of POLIC. In yeast, the DNA polymerase and 3′-5′ exonuclease activity of DNA polymerase epsilon are nonessential for cell growth, whereas the non-catalytic C-terminal domain of the protein is required (Dua et al., 1999; Kesti et al., 1999).
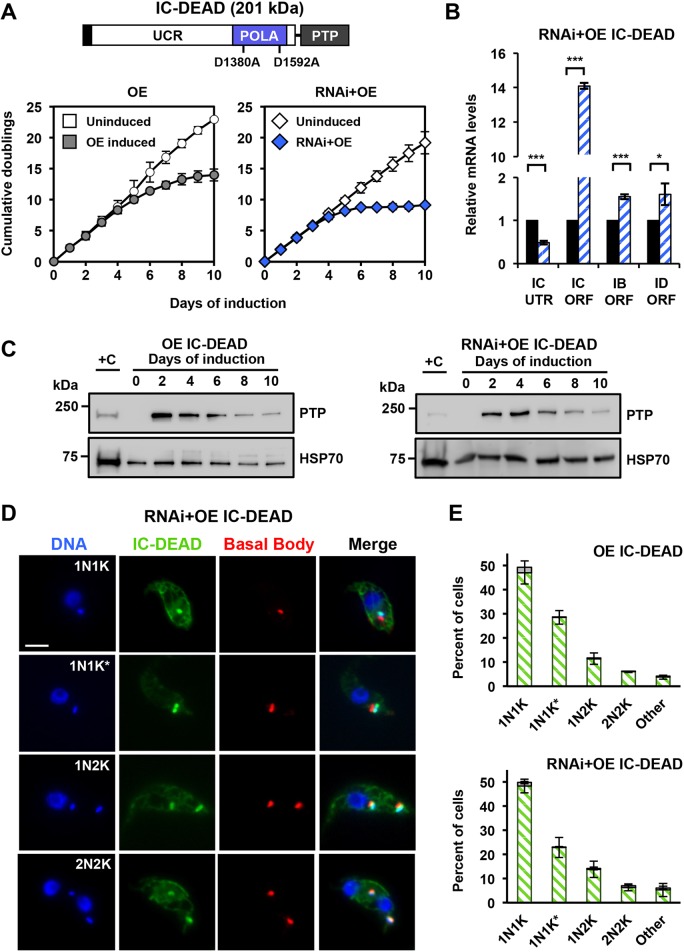
To address whether the nucleotidyl transferase activity is essential for POLIC functions in vivo, we generated a full-length POLIC PTP-tagged variant with the catalytic Asp-residue mutated to Ala (D1380A and D1592A) to inactivate polymerase activity in POLIC (IC-DEAD), which was then overexpressed (OE) and used together with RNA interference (RNAi) in the IC-UTR backgrounds (RNAi+OE). To confirm the mutated Asp residues successfully inactivated POLIC polymerase activity, IC-WT and IC-DEAD were overexpressed for 48 h, immunoprecipitated and analyzed by western blot and an in vitro primer extension assay. The POLIC variants were detected according their predicted sizes (IC-DEAD, 201 kDa; IC-POLA, 75 kDa; IC-UCR, 160 kDa) but only IC-WT demonstrated nucleotidyl transferase activity (Fig. S3).
Interestingly, IC-DEAD overexpression led to a dominant-negative loss of fitness following just 4 days of induction. The number of cumulative cell doublings after 10 days of IC-DEAD overexpression was 13.7 (Fig. 3A, left graph) compared to the 15.1 cumulative doublings following IC-UTR RNAi (Fig. 1A). However, complementation with the IC-DEAD variant exacerbated the loss of fitness and reduced the cumulative doublings to 9.1 after 10 days (Fig. 3A, right graph). RT-qPCR confirmed the depletion of endogenous POLIC transcripts and the increase of the variant transcript (Fig. 3B). The levels of the paralog transcripts were also elevated compared to those in uninduced cells. IC-DEAD protein levels were initially high compared to those of endogenous POLIC in both backgrounds (OE, 16.6-fold; RNAi+OE, 12.5-fold) but were reduced to 3-fold by the end of the 10 day induction based on ImageJ analyses from western blot data (Fig. 3C). The IC-DEAD-PTP signal concentrated on or near the kDNA during all cell cycle stages in OE (98%) and RNAi+OE (99%) cell lines (Fig. 3D,E). This is in striking contrast to endogenous POLIC-PTP and IC-WT-PTP foci that were found exclusively during 1N1K* stage (compare Fig. 3E and Fig. S2D). IC-DEAD complementation also occasionally displayed ancillary kDNA that was colocalized with TAC102 (Fig. S1).
Fig. 3.
Overexpression of IC-DEAD results in a dominant-negative fitness phenotype while complementation exacerbates loss of fitness. (A) POLIC-DEAD domain structure with the asp acid residues mutated to alanine indicated. Growth curves of uninduced and tetracycline-induced POLIC-DEAD (OE, RNAi+OE) procyclic cells. Error bars represent the ±s.d. of the mean from three biological replicates. Error bars are too small to be displayed with respect to the size of the icon. (B) RT-qPCR analysis of dsRNA in uninduced (black) cells and of cells induced with tetracycline for 48 h (blue-white hashed). Data were normalized to TERT and error bars represent the ±s.d. of the mean from three biological replicates. IC UTR, POLIC 3′ untranslated region; IC ORF, POLIC open reading frame; IB ORF, POLIB open reading frame; ID ORF, POLID open reading frame. Asterisks represent statistical significance based on pairwise Student's t-test. *P<0.1; ***P<0.001. (C) Western blot detection of the PTP tag and HSP70 protein levels following 10 days of induction. 1×106 cell equivalents was loaded into each well, except for +C where 5×106 cell equivalents was loaded. +C, single-allele-expressing control cell line. (D) Representative images of IC-DEAD at each cell cycle stage. DAPI staining (blue); anti-protein A (green); YL1/2 (red). Scale bar, 5 μm. (E) Quantification of PTP foci at each cell cycle stage in the overexpression (OE, top) and complementation (RNAi+OE, bottom) cell lines after 48 h of induction. Foci positive (green/white hashed); foci negative (gray). Error bars represent the ±s.d. of the mean from three biological replicates.
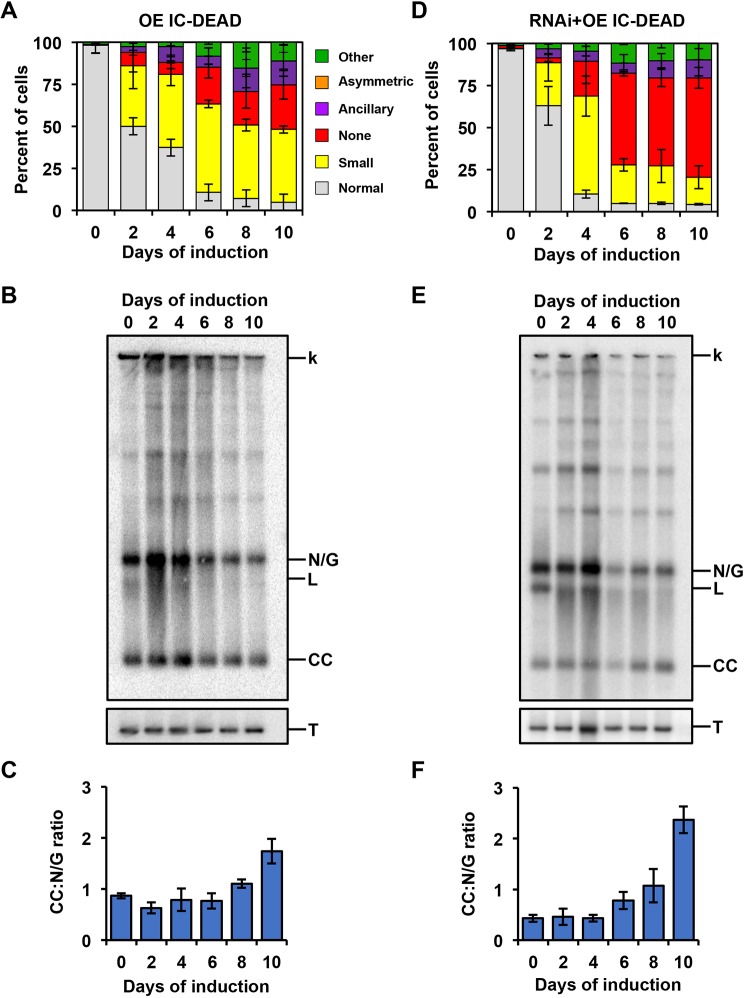
Next, we examined the effects of IC-DEAD variant on kDNA maintenance in both backgrounds (OE and RNAi+OE). Ten days of IC-DEAD overexpression in the absence of POLIC RNAi resulted in a phenotype predominantly showing small kDNA (43.5%), as well as phenotypes without (26.5%), ancillary (14.3%), other (11%) and normal (4.7%) kDNA (Fig. 4A). In striking contrast to both OE and the parental RNAi phenotypes, IC-DEAD complementation displayed a majority of cells without kDNA (59.1%), which correlates with the exacerbated loss of fitness. IC-DEAD also displayed small (16.1%), ancillary (10.8%), other (9.6%) and normal (4.4%) kDNA phenotypes, but did not display any asymmetric kDNA networks (Fig. 3D). The IC-DEAD variant caused a more rapid onset of aberrant kDNA (after 2 days) in both genetic backgrounds and a shift from a predominantly small kDNA to no kDNA phenotype for IC-DEAD complementation (Figs 1D and 3A,D).
Fig. 4.
Analyses of the kDNA replication phenotype in response to IC-DEAD overexpression and complementation. (A) Quantification of kDNA morphology for OE IC-DEAD over the course of the induction demonstrating progressive loss of kDNA. Over 300 DAPI-stained cells were scored at each time point and error bars represent the ±s.d. of the mean from three biological replicates. Gray, normal kDNA; yellow, small kDNA; red, no kDNA; purple, ancillary kDNA; orange, asymmetric size; green, other. (B) Representative Southern blot showing the effect on free minicircle population. k, kDNA network; N/G, nicked/gapped; L, linearized; CC, covalently closed; T, tubulin. (C) Ratio of CC:N/G over the course of the induction. Phosphoimager quantification was used to plot the relative abundance of nonreplicated CC and newly replicated N/G intermediates. Error bars represent the ±s.d. of the mean from three biological replicates. (D–F) Same as A–C but for the RNAi+IC-DEAD cell line.
In addition to progressive loss of the kDNA network, hallmarks of perturbing kDNA replication include a decrease in minicircle and maxicircle copy number as well as an increased ratio of nonreplicated CC to N/G progeny (Bruhn et al., 2010; Liu et al., 2010; Scocca and Shapiro, 2008). Both IC-DEAD overexpression and complementation led to a progressive decrease in free minicircle species (Fig. 3B,E). In both genetic backgrounds, the free minicircles initially increased (days 2 and 4) when the IC-DEAD variant was present. At later days during induction, the abundance of CC and N/G minicircles quickly declined, which resembled the defect caused by RNAi of the paralog POLID (Chandler et al., 2008). When comparing nonreplicated CC to replicated N/G species, overexpression of IC-DEAD resulted in a 1.7-fold increase of the CC:N/G ratio and complementation resulted in a 5.5-fold increase at day 10 when compared to uninduced cells (Fig. 3C,F).
These data indicate that IC-DEAD cannot rescue any of the pleiotropic effects from POLIC RNAi. Additionally, the variant resulted in disruption of cell cycle-dependent protein localization and displayed an overt kDNA replication defect with a decrease in N/G progeny, suggesting that nucleotidyl incorporation is essential for POLIC function.
The POLA domain partially rescues minicircle replication defects
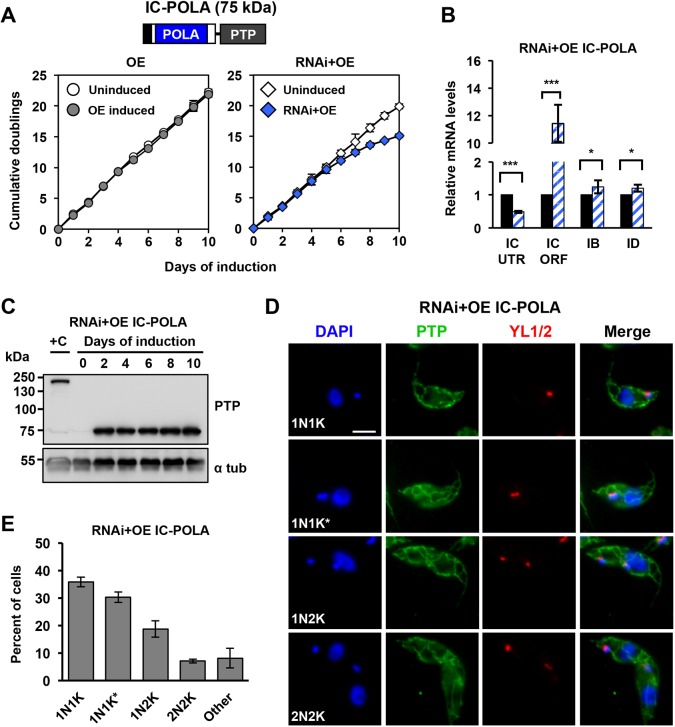
Next, we determined whether the conserved POLA domain alone is sufficient to restore POLIC function in vivo. We generated N-terminally truncated PTP-tagged POLIC variant (IC-POLA, 437 aa), and transfected it into overexpression (OE) and IC-UTR RNAi backgrounds (RNAi+OE). Consistent IC-POLA overexpression (5.4-fold increase compared to endogenous expression) did not affect parasite fitness (Fig. 4A, Fig. S4A) or kDNA maintenance, which was determined by DAPI staining (>90% normal cells at day 10) and by Southern blotting (no perturbation of free minicircle levels), respectively (Fig. S4B,C). Surprisingly, immunoprecipitated IC-POLA did not display nucleotidyl incorporation activity in the primer extension assay (Fig. S3).
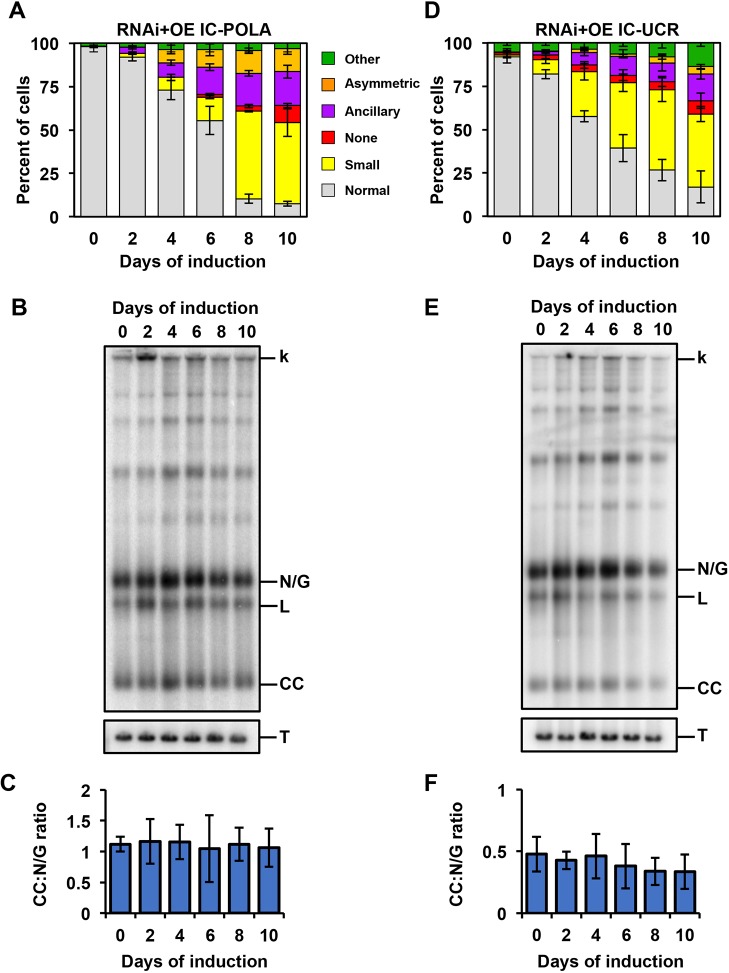
IC-POLA complementation closely resembled the parental IC-UTR loss of fitness (15.1 cumulative doublings, day 10) with a robust knockdown of endogenous POLIC mRNA (Fig. 5A,B). Western blot data showed consistent protein expression for 10 days (4.1-fold higher than endogenous) (Fig. 5C). IC-POLA showed mitochondrial matrix localization and a weak signal near the kDNA, regardless of the cell cycle stage; no antipodal PTP foci were ever detected (Fig. 5D,E). IC-POLA complementation resulted in cells with small (46.7%), no (9.8%), normal (7.5%) and other (3.1%) kDNA (Fig. 6A). Interestingly, IC-POLA cells with ancillary (19.5%) or asymmetric (13.3%) kDNA were elevated compared to 15.5% or 10.4%, respectively, for the parental IC-UTR RNAi defect, suggesting that the polymerase domain alone does not rescue the distribution defect. TAC102 was detected to colocalize with a portion of the ancillary kDNA (Fig. S1). Furthermore, in the presence of the IC-POLA variant, we neither observed detectable accumulation of free minicircles over the course of the induction (Fig. 6B,C) nor statistically significant changes in the CC:N/G ratio during the 10 day induction period, which demonstrates that the POLA domain can rescue the minicircle replication defect.
Fig. 5.
IC-POLA complementation is not sufficient to rescue the loss of fitness RNAi phenotype. (A) Growth curves of uninduced and tetracycline-induced IC-POLA (OE, RNAi+OE) procyclic T. brucei cells. Error bars represent the ±s.d. of the mean from three biological replicates. Error bars are too small to be displayed with respect to the size of the icon. (B) RT-qPCR analysis of uninduced (black) and induced (blue/white hashed) cells following 48 h of tetracycline induction. Data are normalized to TERT and error bars represent the ±s.d. of the mean from three biological replicates. IC UTR, POLIC 3′ untranslated region; IC ORF, POLIC open reading frame; IB, POLIB; ID, POLID. Asterisks represent statistical significance based on pairwise Student's t-test. *P<0.1; ***P<0.001. (C) Western blot detection of the PTP tag and alpha tubulin protein levels following 10 days of induction. (D) Representative images of IC-POLA at each cell cycle stage. Scale bar, 5 μm. (E) Quantification of foci formation at each cell cycle stage. Green, foci positive; gray, foci negative.
Fig. 6.
Analyses of the kDNA replication phenotype in response to IC-POLA and IC-UCR complementation. (A) Quantification of RNAi+IC-POLA kDNA morphology over the course of the induction. Over 300 cells were scored at each time point and error bars represent the ±s.d. of the mean from three biological replicates. Gray, normal kDNA; yellow, small kDNA; red, no kDNA; purple, ancillary kDNA; orange, asymmetric size; green, other. (B) Representative Southern blot showing the effect on free minicircles of RNAi+IC-POLA. k, kDNA network; L, linearized; N/G, nicked/gapped; CC, covalently closed; T, tubulin. (C) Quantification of the CC:N/G ratio over the course of the induction. Phosphoimager quantification was used to plot the relative abundance of nonreplicated CC and newly replicated N/G intermediates. Error bars represent the ±s.d. of the mean from three biological replicates. (D-F) Same as A–C but for the RNAi+IC-UCR cell line.
Collectively, these data show that the C-terminal polymerase domain alone can partially restore the free minicircle population in vivo, even though it did not display strict cell cycle-dependent protein localization to antipodal sites, possibly due to its ability to still localize near the kDNA disk. Additionally, the increased distribution defect suggested that the UCR is necessary for kDNA distribution and cell cycle-dependent localization.
The UCR domain partially rescues IC-UTR RNAi defects
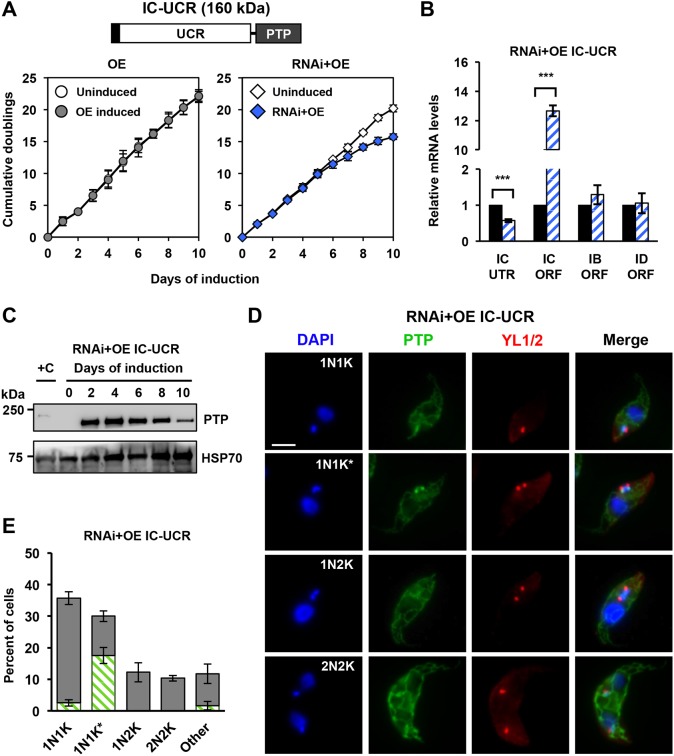
The N-terminal UCR (1291 amino acids) of POLIC has no conserved domains or motifs but does contain a predicted hydrophobic patch and Arg methylation sites (R420, R1250, R1260) that were detected in the T. brucei mitochondrial Arg methylproteome (Fisk and Read, 2011). To evaluate the role of POLIC in kDNA distribution, we generated a POLIC variant that contains only the UCR region (IC-UCR), which was transfected into the overexpression (OE) and the IC-UTR RNAi backgrounds (RNAi+OE). IC-UCR overexpression was consistent during the induction period (5.3-fold increase compared to endogenous levels), did not affect parasite fitness, and no kDNA replication defects were detected (Fig.S4D-F).
IC-UCR complementation resulted in loss of fitness, with 15.7 cumulative doublings after 10 days of induction (Fig. 7A). RT-qPCR displayed robust POLIC knockdown and western blot data revealed initially high protein levels (18-fold increase over endogenous) that continually decreased over 10 days (4-fold) (Fig. 7B,C). There was clear formation of IC-UCR-PTP foci at antipodal sites in 17.6% of 1N1K* cells compared to IC-WT complementation (30%, gray) and endogenous localization (26%); IC-UCR also displayed weak signals near the kDNA at other cell cycle stages (Figs 2F and 7D,E). IC-UCR complementation displayed mostly cells with small (42%), normal (17%), ancillary (15.3%), other (13.7%), no (7.7%), and asymmetric (4.3%) kDNA (Fig. 6D). Interestingly, the IC-UCR variant resulted in more cells with normal kDNA (17% vs 7.8%) and fewer cells with distribution defects when compared to IC-UTR RNAi alone (19.6% versus 25.5%), indicating a partial rescue of the distribution phenotype. Despite the reduction in cells displaying asymmetric networks, the percentage of cells displaying ancillary kDNA did not significantly improve and TAC102 still colocalized with ancillary kDNA (Fig. S1). In the presence of the IC-UCR variant, there were no statistically significant changes in the CC:N/G ratio, even though there was a decrease in free minicircles after 8 days of complementation (Fig. 6E,F). The POLIC variant effects are summarized in Fig. 8. Collectively, these data revealed distinct functions for POLIC in both kDNA replication and distribution, and that the UCR mediates antipodal site localization and distribution function.
Fig. 7.
IC-UCR complementation is insufficient to rescue the loss of fitness RNAi phenotype but restores cell cycle-dependent localization. (A) Schematic representation of IC-UCR variant. Growth curves of uninduced and tetracycline-induced IC-UCR (OE, RNAi+OE) procyclic cells. Error bars represent the ±s.d. of the mean from three biological replicates. (B) RT-qPCR analysis of uninduced (black) and induced (blue/white hashed) cells following 48 h of tetracycline induction. Data are normalized to TERT and error bars represent the ±s.d. of the mean from three biological replicates. IC UTR, POLIC 3′ untranslated region; IC ORF, POLIC open reading frame; IB ORF, POLIB; ID ORF, POLID. **P<0.001. (C) Western blot detection of the PTP tag and HSP70 protein levels following 10 days of induction. (D) Representative images of IC-UCR at each cell cycle stage. Scale bar, 5 μm. (E) Quantification of PTP foci formation at each cell cycle stage. Green/white hashed, foci positive; gray, foci negative. Error bars represent the ±s.d. of the mean from three biological replicates.
Fig. 8.
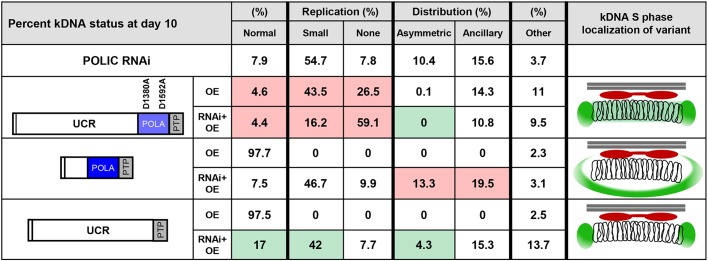
Summary of POLIC variant phenotypes and effect on kDNA network. kDNA status is shown as the percentage of total cells 10 days post induction (OE or RNAi+OE). Analyzed were two types of observed defect, i.e. replication and distribution. Replication defects are divided into 'Small' and 'None' defects; distribution defects are divided into 'Asymmetric' and 'Ancillary' defects. 'Other' refers to cells displaying any other non-normal nucleus and kDNA karyotype. Colored boxes indicate the number of cells suggesting biologically significant improved (green), worsened (red) or unchanged (white) conditions compared to the parental POLIC RNAi phenotype. The respective localization pattern during kDNA S phase is displayed on the right. In each image, the mitochondrial membrane is depicted as two parallel horizontal lines (gray), TAC102 is depicted in red, the POLIC variant is depicted in green and the catenated ring structures represent the kDNA network.
DISCUSSION
Trypanosome kDNA replication presents an interesting model system in which replication of a single nucleoid (the kDNA network) occurs once every cell cycle and segregation of daughter networks is mediated by a structure linked to the flagellar basal body called the TAC. The current kDNA replication model indicates spatial and temporal separation for early events, occurring in the KFZ and later Okazaki processing at the antipodal sites. Additionally, three DNA polymerase family A members are essential for kDNA replication. POLIB and POLID have demonstrated roles in minicircle replication but the precise role for POLIC in kDNA maintenance was unclear. Although the precise functions performed by the paralogs on minicircles and maxicircles have not yet been defined, the trio of polymerases superficially resembles the replicative polymerases α, δ and ɛ required for nuclear DNA replication. Understanding catalytic and non-catalytic roles may provide insight as to why trypanosomes utilize three distinct polymerases for kDNA replication.
POLIC shares no similarity with other proteins outside of the conserved POLA domain at the C-terminus. Consistent with an essential function in kDNA maintenance, POLIC and the overall protein structure is conserved in all of the 41 currently sequenced trypanosomatid organisms and some free-living relatives available at TriTrypDB.org (Aslett et al., 2010).
Interestingly, three Arg residues on POLIC are known to be methylated (Fisk and Read, 2011). Arg methylation is a reversible post-translational modification with regulatory roles in many biological processes, including signal transduction, transcription, RNA splicing and transport, and facilitation of protein–protein interactions (Raposo and Piller, 2018). However, at this time, it is unknown whether POLIC from other trypanosomatids retain the Arg methylation sites.
To further understand the role of POLIC in kDNA replication, we investigated the structure–function relationship by ectopically expressing several POLIC variants in trypanosome cells deficient in endogenous POLIC. Tetracycline-inducible expression of a dsRNA targeting the 3′-UTR of a gene and coexpression of an ectopic copy of the RNAi target is a validated approach for in vivo complementation studies on flagellar motility, cytokinesis and mitochondrial protein import, TAC function, and transferrin receptor trafficking in T. brucei (Käser et al., 2016, 2017; Ralston et al., 2011; Trikin et al., 2016; Weems et al., 2015; Yu et al., 2012). By using this approach, we demonstrated that IC-WT rescues the replication and distribution defects, and defined two functional domains: one for nucleotidyl incorporation, and one non-catalytic domain for localization of antipodal sites and distribution of kDNA. Two other dual-functioning proteins with roles in kDNA maintenance have been described. The putative universal minicircle sequence binding protein (UMSBP) is a minicircle protein that initiates replication (Abu-Elneel et al., 1999). Interestingly, RNAi of USMBP indicated impaired kDNA segregation, possibly, through a cell cycle checkpoint mechanism and not the – more typical – mechanism of direct TAC inhibition (Milman et al., 2007). The tricarboxylic acid (TCA) cycle enzyme, α-ketoglutarate dehydrogenase (α-KDE2, GenBank: EAN80119.1), has been reported to be an antipodal site protein with a role in the distribution of progeny networks but it displayed no role in kDNA replication (Sykes and Hajduk, 2013). Unlike UMSBP and α-KDE2, POLIC represents the clearest example of a protein bridging the replication and segregation processes.
How the POLIC polymerase domain participates at replication forks remains an open question. Although the immunoprecipitated POLA domain did not have detectable activity in a primer extension assay (Fig. S3), IC-POLA complementation indicated that the polymerase domain alone is sufficient to partially support minicircle replication, even though IC-POLA localized mainly to the mitochondrial matrix (Figs 5D and 6B,C). IC-POLA includes all the motifs conserved in DNA polymerase family A, but this truncated fragment might lack important upstream residues required for binding the primer-template in the in vitro assay. Despite IC-POLA failing to exhibit cell cycle-dependent localization during complementation, abundance around the kDNA was detected and could support the maintenance of the free minicircle population directly or indirectly. It is also possible that, in vivo, IC-POLA has access to accessory factors that allow it to maintain the free minicircle population. This truncated version of POLIC still contains two of the three Arg residues that were detected in a methylarginine proteome analysis to be able to promote protein interactions.
It is compelling that the IC-DEAD variant that is defective in nucletotidyl incorporation generated the most-striking kDNA replication defects in both overexpression and complementation (Fig. 8). IC-DEAD overexpression caused a dominant-negative loss of fitness with a defect of kDNA replication (Fig. 3A) and abolished the spatiotemporal localization of POLIC during the cell cycle. Unlike IC-POLA mitochondrial matrix localization, IC-DEAD always localized with or around the kDNA disk (Fig. 3D,E). Interestingly, there are now 250 pathogenic mutations in human mitochondrial DNA polymerase γ that interfere with deoxyribonucleotide triphosphate (dNTP) selectivity, processivity or accessory subunit binding, with many of the dominant mutations located in the polymerase domain that result in loss of mtDNA (Stumpf et al., 2013). It is possible that IC-DEAD was bound to minicircles and/or maxicircles outcompeting endogenous POLIC for substrate, inhibited access of other replication proteins or sequestered replication factors. IC-DEAD complementation results support the idea that access of other replication proteins was impacted upon. The dramatic loss of kDNA (Fig. 4D) and increase in nonreplicated free minicircles (Fig. 4E,F) are hallmarks of a kDNA replication defect that closely resembles depletion of POLID, a known minicircle replication factor that localizes to antipodal sites only during kDNA S phase (Chandler et al., 2008; Concepción-Acevedo et al., 2012). We have previously shown that POLID depletion disrupts localization of POLIC to antipodal sites. It is possible that overexpression of IC-DEAD has a similar effect on POLID or other replication proteins by inhibiting access to the kDNA (Concepción-Acevedo et al., 2018). However, the mechanisms(s) governing antipodal site localization and paralog interactions have not yet been determined.
Despite the lack of any recognizable catalytic domain, the UCR appears to contain a determinant for the localization to antipodal sites. IC-UCR complementation resulted in PTP foci formation that closely resembles the localization pattern of endogenous POLIC; there, a signal was detected within the KFZ in cells during 1N1K, IC-UCR-PTP foci at antipodal sites were detected in cells during 1N1K* and the signal was dispersed during the other cell cycle stages (Fig. 7D,E). Although two different methods were used to evaluate the percentage of cells in kDNA S phase, the proportion of PTP foci-positive cells in this stage was equivalent (59%) (Concepción-Acevedo et al., 2018). Our current model suggests that the localization of POLIC during the cell cycle is a mechanism that discriminates between the two functional domains of this protein. For example, nucleotidyl incorporation activity may be utilized when localized in the KFZ, while the distribution function may be more important at antipodal sites. The IC-UCR variant contains all three methylated Arg residues (Fisk and Read, 2011), which may contribute to POLIC localization at antipodal sites through protein-protein interactions. The methylation status of the full-length protein may also promote one of the dual functions over the other. For example, in the demethylated state, the protein may favor interactions with other replication proteins, whereas, in the fully methylated state, it may interact with proteins at antipodal sites and distribution factors or vice versa. Future studies will be focused on understanding the impact of methylation on POLIC function. IC-UCR is the first sequence from a kDNA protein shown to promote antipodal site localization, and further experimentation is necessary to refine the specific sequence determinants involved.
Another non-catalytic role for the UCR is associated with kDNA distribution. IC-UCR complementation partially rescued the RNAi response by reducing the number of cells with asymmetrically sized networks and increased the number of cells with normal kDNA (Fig. 6D). In contrast, complementation with IC-POLA (lacking the UCR) worsened the distribution defect (Figs 6A and 8), perhaps due to the elimination of the spatiotemporal localization pattern, where POLIC starts in the KFZ and then is detected at the antipodal sites at later stages of kDNA replication (Concepción-Acevedo et al., 2018). At this time, we do not understand the mechanism on how the UCR domain alone can partially rescue kDNA distribution defects, since IC-DEAD complementation displayed an apparent rescue of asymmetric networks (Figs 4D and 8). It is possible that asymmetric networks were not observed solely because the replication defect developed too rapidly, thereby preventing a distribution defect from being detected at the time points analyzed. The decline in IC-UCR abundance over the course of complementation provides one explanation for the incomplete rescue. Alternatively, the POLA domain might also have a role in distribution through protein interactions. Surprisingly, no POLIC variant complementation was able to reduce the appearance of ancillary kDNA (Figs 4D, 6A,D and 8), suggesting that a fully functional protein is crucial to suppress the ancillary kDNA phenotype. POLIC is likely not to have a direct role in distribution of the kDNA network but to impact on other proteins that are essential for the distribution.
On the basis of the criteria outlined by Schneider and Ochsenreiter (2018), POLIC is not a typical TAC component. IC-UTR RNAi does not result in one cell comprising large kDNA and another without any kDNA. Instead, there is unequal partitioning of a reduced-size network due to the POLIC replication defect. Additionally, ancillary kDNA is an unusual aberrant phenotype that appears when elements related to the mitochondrial membrane or the TAC are perturbed, as seen with functional disruptions of PUF9, ACP, PNT1, HSP70/HSP40 and TAC102 (Archer et al., 2009; Clayton et al., 2011; Grewal et al., 2016; Trikin et al., 2016; Týč et al., 2015). The TAC component most proximal to the kDNA is TAC102. When the C-terminus of this protein is obstructed, TAC102 cannot properly connect to the upstream TAC components (and, subsequently, to the basal body), resulting in mislocalization with ancillary kDNA (Trikin et al., 2016). It is interesting that complementation with all POLIC variants produced ancillary kDNA with mislocalization of TAC102 independently of their rescue status (Fig. S1). POLIC is more proximal to the kDNA than TAC102 (Concepción-Acevedo et al., 2018) and our data suggest that a fully functional POLIC is important for TAC102 to connect to upstream TAC components. At this time, we do not know whether TAC is required for proper localization of POLIC or how POLIC might interact with TAC. The recently described minicircle reattachment protein MiRF172 (GenBank: AAZ10240.1) might provide a bridge between POLIC and TAC102, since MiRF172 remains associated with isolated flagella following biochemical fractionations and its localization is more proximal to the kDNA than TAC102 (Amodeo et al., 2018). The proximity of the replication and segregation machinery in T. brucei could suggest a physical interaction of the components involved in the two processes. Thus far, the dual-functioning DNA polymerase POLIC is the clearest example of a protein able to bridge these two essential processes.
MATERIALS AND METHODS
For Primer sequences refer to Table S1.
DNA constructs
RNAi
A pStL (stem-loop) plasmid for inducible gene silencing of T. brucei DNA polymerase I-like protein C (POLIC) was constructed as previously described by Wang et al. (2000). Briefly, 517 bp of POLIC 3′-UTR sequence was PCR-amplified from TREU927 genomic DNA by using primers listed in Table S1 to generate the two fragments for subsequent cloning steps; the resulting vector, pStLIC3′UTR, was EcoRV-linearized for genome integration.
Site-directed mutagenesis
Full-length wild-type POLIC (Tb927.7.3990, TriTrypDB.org) was PCR amplified from TREU927 guide DNA by using Phusion DNA polymerase and subcloned into the pCR®-Blunt II-TOPO vector (Invitrogen). Asp-to-Ala mutations (D1380A and D1592A) were performed by site-directed mutagenesis using the QuikChange Lightning Multi Site-Directed Mutagenesis Kit (Agilent) generating pTOPO-IC-DEAD.
Inducible expression of POLIC variants
To create pLew100-PTPPuro for inducible expression of PTP (ProtC-TEV-ProtA)-tagged POLIC variants, the MYC tag of pLew100-MycPuro (gift from Dr Laurie Read, SUNY Buffalo) was replaced with the PTP tag. Briefly, the PTP tag was PCR amplified from pC-PTP-NEO (Schimanski et al., 2005) and cloned into pCR®-Blunt II-TOPO vector. Site-directed mutagenesis was used to introduce a silent mutation and to eliminate an internal BamHI site generating pTOPO-PTP-ΔBamHI. The MYC tag was replaced with PTP-ΔBamHI to generate pLew100-PTPPuro. The full-length open reading frames of wild-type POLIC or of POLIC comprising the point mutations were cloned into pLew100-PTPPuro (pLewIC-WT-PTPPuro). Gibson assembly was used to generate truncated versions of POLIC that retained only the C-terminal POLA domain or the N-terminal uncharacterized region (UCR). Briefly, PCR fragments containing the predicted mitochondrial targeting sequence (nucleotides 1–174), the POLA fragment (nucleotides 3637-4947), and the UCR fragment (nucleotides 1-3816) were generated. Gibson Assembly (NEB) was then performed using HindIII/XbaI-digested pLew100-PTPPURO with the mitochondrial targeting sequence and POLA PCR fragments or with the UCR fragment. All DNA constructs and point mutations were verified by direct sequencing. Owing to the lack of sequence conservation beyond the POLA domain, PSIpred prediction software (http://bioinf.cs.ucl.ac.uk/psipred/) was used to locate a region upstream of the POLA domain that would be least likely to affect protein folding based on the secondary structure prediction (Jones, 1999).
Trypanosome cell culture and transfection
The procyclic form of the T. brucei strain 29-13 (Wirtz et al., 1999; Wang et al., 2000) was cultured at 27°C in SDM-79 medium supplemented with FBS (15%), G418 (15 µg/ml) and hygromycin (50 µg/ml). To generate the parental RNAi cell line IC-UTR, 15 µg of pStLIC3′UTR was transfected by electroporation into T. brucei 29-13 cells, selected with 2.5 µg/ml phleomycin and cloned by limiting dilution as previously described (Chandler et al., 2008). Four individual clones were analyzed, and clone P6E2 was chosen based on growth rate, POLIC knockdown efficiency and kDNA defects. All POLIC variant expression constructs (15 µg) were transfected by nucleofection using the Amaxa Nucleofection Parasite Kit (Lonza) into T. brucei 29-13 cells to generate inducible overexpression in POLIC-PTP variant cell lines (OE) and into the IC-UTR RNAi cell line to generate complementation cell lines (RNAi+OE). Following selection with 1 µg/ml puromycin, cell lines were cloned by limiting dilution. A single clone of each cell line was selected based on variant expression for OE and confirmed POLIC knockdown and variant expression for RNAi+OE cell lines. All cell lines were maintained at a density of 1×105–1×107, and were not continuously cultured for more than three weeks. Cell density was determined using a Beckman Coulter Z2 particle counter. RNAi and/or overexpression were induced by addition of tetracycline (1 µg/ml). Cultures were supplemented with 0.5 µg/ml tetracycline on non-dilution days to maintain RNAi and/or the expression of POLIC variants (Rusconi et al., 2005).
RNA isolation and RT-qPCR analysis
Total RNA was isolated from 5×107 cells by using Trizol (Thermo Fisher) and applying standard procedures. RNA concentration was determined using a NanoDrop spectrophotometer (Thermo Fisher Scientific) and 100 ng of total RNA was reverse transcribed to cDNA using random primers and RNase inhibitor (Applied Biosystems). qPCR was performed using QuantiNova SYBR Green PCR (Qiagen) with 1 µg of cDNA and 0.2 µM of primer per reaction (see Table S1 for qPCR primers; the corresponding gene sequences were downloaded from TriTrypDB) using a Stratagene MxPro 3000x thermocycler (Aslett et al., 2010). All reactions were performed in triplicate and qPCR results were determined using three biological replicates. Gene knockdown was normalized by using TERT (Brenndörfer and Boshart, 2010).
SDS-PAGE and western blot analysis
Cells were harvested and washed once using PBS supplemented with protease inhibitor cocktail. Samples were fractionated by SDS-PAGE and transferred overnight onto a PVDF membrane in 1× transfer buffer containing 1% methanol. Membranes were blocked in Tris-buffered saline (TBS)+5% non-fat dry milk for at least 1 h. Peroxidase–anti-peroxidase soluble complex (PAP) antibody (1:2000; Sigma, catalog no. P1291) was used for 30 min for PTP tag detection. For subsequent detections, where stripping was required because proteins of interest were similarly sized to the loading control, membranes were stripped with 0.1 M glycine (pH 2.5) for 5 min at 37°C, washed in TBST (0.1% Tween-20), blocked and re-probed with specific Crithidia fasciculata anti-Hsp70 antibody (1:5000) for 1 h (Johnson and Englund, 1998) and secondary chicken anti-rabbit-HRP (1:2000; Santa Cruz, catalog no. sc-516087) for 1 h. Signals were detected using Clarity™ ECL blotting substrate (NEB) on a GE Imagequant LAS 4000 mini. Quantification of western blot intensities was carried out using ImageJ software (http://imagej.nih.gov/ij/).
DNA isolation and Southern blot analysis
Total DNA was isolated from 1×108 cells by using the Puregene Core Kit A (Qiagen). Free minicircles and total kDNA content were analyzed as previously described (Bruhn et al., 2010; Rusconi et al., 2005). Quantification was performed using a Typhoon 9210 Molecular Dynamics PhosphoImager (GE Healthcare) with background subtraction; signal intensites were normalized against the tubulin signal by using ImageQuant 5.2 software.
Immunoprecipitation and DNA polymerase activity assay
Immunoprecipitation of PTP-tagged POLIC variants was conducted according to Brandenburg et al. (2007) and Rusconi et al. (2005) with modifications. Briefly, 3×108 mid-log phase cells were harvested (9×108 IC-UCR cells were used due to low expression), washed with cold PBS and lysed in 970 μl of 150 mM KCl, 20 mM Tris-HCl pH 8, 3 mM MgCl2, 0.5% NP-40, 0.5 mM DTT supplemented with 1× cOmplete™ EDTA-free protease inhibitor (Roche). Cell lysate was bound to IgG-Sepharose beads (Roche) and washed with 100 mM NaCl, 20 mM Tris-HCl pH 8, 3 mM MgCl2, 0.1% Tween 20 buffer 5× for 5 min each by centrifugation at 1000 g. All steps were performed at 4°C. Samples bound to beads were used in a primer extension assay as follows. Reactions contained 50 mM Tris-HCl pH 8, 5 mM MgCl2, 1 mM DTT, 0.1 mg/ml BSA, 20 μg/ml activated calf thymus DNA as template, 10 μM dTTP, dGTP and dCTP, 5 μl [α-32P] dATP (Perkin Elmer) mixed with 10 μM cold dATP, and either the specific units of Klenow fragment (NEB) or the specified volumes of immunoprecipitated POLIC variants. Reactions were incubated at 37°C for 30 min, stopped by addition of 5 μl of 500 mM EDTA, were spotted onto Whatman DE81 disks, dried and then washed 4× times with 0.5 M NaPO4 (15 min each). Disks were washed with 80% EtOH for 15 min and dried before analysis using an LS2 Beckman Coulter scintillation counter.
Immunofluorescence microscopy
Procyclic cells were pelleted by centrifugation at 975 g, washed and resuspended in PBS to yield ∼5×106 cells/ml, and then adhered to poly-L-lysine-coated slides (5 min). Cells were then fixed in 4% paraformaldehyde (5 min) and washed 3× (5 min) in PBS/0.1 M glycine pH 7.4. Cells were permeabilized with 0.1% Triton X-100 (5 min) and washed 3× in PBS (5 min). PTP-tagged proteins were detected following incubation for 1 h with anti-protein A serum (1:3000; Sigma, catalog no. P3775) followed by incubation with Alexa Fluor® 594 goat anti-rabbit (1:250; Invitrogen, catalog no. A-11012) for 1 h. Detection of basal bodies (YL1/2), TAC102 and DNA (DAPI) was carried out as previously described (Concepción-Acevedo et al., 2018; Rusconi et al., 2005). Slides were then washed (3× for 5 min) in PBS prior to mounting in Vectashield (Vector Laboratories). Images were captured by using the same exposure time for each channel within the same experiment with a Nikon Eclipse E600 microscope equipped with a cooled CCD Spot-RT digital camera (Diagnostic Instruments) and a 100× Plan Fluor 1.30 (oil) objective. Quantification of kDNA morphology was performed as described previously (Bruhn et al., 2010; Chandler et al., 2008). Small networks were defined as those that appeared to have at least 50% less surface area than that found in uninduced cells; asymmetric networks were defined as those of one cell with two detectable kDNA networks; ‘other’ networks were those cells with a karyotype that neither matched a normal karyotype (i.e. 1N1K, 1N2K, 2N2K) nor aberrant kDNA morphology (small, none, ancillary or asymmetric).
Statistics
Error bars shown in figures represent the ±standard deviation (±s.d.) of the mean. Statistical values were calculated using pairwise Student's t-test, with significance values as *P<0.1; **P<0.01; ***P<0.001.
Supplementary Material
Acknowledgements
We thank Amy Springer for critical review of the manuscript, Paul Englund for the mtHsp70 antibody and Torsten Ochsenreiter for the TAC102 antibody.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: J.C.M., J.C., M.M.K.; Methodology: J.C.M., S.B.D., J.C., M.J.B.; Formal analysis: J.C.M., M.M.K.; Investigation: J.C.M., S.B.D.; Resources: J.C., M.J.B., M.M.K.; Data curation: M.M.K.; Writing - original draft: J.C.M.; Writing - review & editing: J.C.M., S.B.D., J.C., M.J.B., M.M.K.; Visualization: J.C.M.; Supervision: M.M.K.; Project administration: M.M.K.; Funding acquisition: M.M.K.
Funding
This work was partially supported by the National Institutes of Health (NIH, grant no. AI066279 and USDA NC-1173 to M.M.K.). This work was also supported by a University of Massachusetts Graduate School Dissertation Research Grant to J.C.M. and a faculty research grant to M.M.K. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.233072.supplemental
Peer review history
The peer review history is available online at https://jcs.biologists.org/lookup/doi/10.1242/jcs.233072.reviewer-comments.pdf
References
- Abu-Elneel K., Kapeller I. and Shlomai J. (1999). Universal minicircle sequence-binding protein, a sequence-specific DNA-binding protein that recognizes the two replication origins of the kinetoplast DNA minicircle. J. Biol. Chem. 274, 13419-13426. 10.1074/jbc.274.19.13419 [DOI] [PubMed] [Google Scholar]
- Abu-Elneel K., Robinson D., Drew M., Englund P. and Shlomai J. (2001). Intramitochondrial localization of universal minicircle sequence-binding protein, a trypanosomatid protein that binds kinetoplast minicircle replication origins. J. Cell Biol. 153, 725-734. 10.1083/jcb.153.4.725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amodeo S., Jakob M. and Ochsenreiter T. (2018). Characterization of the novel mitochondrial genome replication factor MiRF172 in trypanosoma brucei. J. Cell Sci. 131, jcs.211730 10.1242/jcs.211730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer S. K., Luu V.-D., de Queiroz R. A., Brems S. and Clayton C. (2009). Trypanosoma brucei PUF9 regulates mRNAs for proteins involved in replicative processes over the cell cycle. PLoS Pathog. 5, e1000565 10.1371/journal.ppat.1000565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslett M., Aurrecoechea C., Berriman M., Brestelli J., Brunk B. P., Carrington M., Depledge D. P., Fischer S., Gajria B., Gao X. et al. (2010). TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 38, D457-D462. 10.1093/nar/gkp851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandenburg J., Schimanski B., Nogoceke E., Nguyen T. N., Padovan J. C., Chait B. T., Cross G. A. and Günzl A. (2007). Multifunctional class I transcription in trypanosoma brucei depends on a novel protein complex. EMBO J. 26, 4856-4866. 10.1038/sj.emboj.7601905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenndörfer M. and Boshart M. (2010). Selection of reference genes for mRNA quantification in trypanosoma brucei. Mol. Biochem. Parasitol. 172, 52-55. 10.1016/j.molbiopara.2010.03.007 [DOI] [PubMed] [Google Scholar]
- Bruhn D. F., Mozeleski B., Falkin L. and Klingbeil M. M. (2010). Mitochondrial DNA polymerase POLIB is essential for minicircle DNA replication in African trypanosomes. Mol. Microbiol. 75, 1414-1425. 10.1111/j.1365-2958.2010.07061.x [DOI] [PubMed] [Google Scholar]
- Bruhn D. F., Sammartino M. P. and Klingbeil M. M. (2011). Three mitochondrial DNA polymerases are essential for kinetoplast DNA replication and survival of bloodstream form trypanosoma brucei. Eukaryot. Cell 10, 734-743. 10.1128/EC.05008-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter L. R. and Englund P. T. (1995). Kinetoplast maxicircle DNA replication in Crithidia fasciculata and trypanosoma brucei. Mol. Cell. Biol. 15, 6794-6803. 10.1128/MCB.15.12.6794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler J., Vandoros A. V., Mozeleski B. and Klingbeil M. M. (2008). Stem-loop silencing reveals that a third mitochondrial DNA polymerase, POLID, is required for kinetoplast DNA replication in trypanosomes. Eukaryot. Cell 7, 2141-2146. 10.1128/EC.00199-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. and Butow R. A. (2005). The organization and inheritance of the mitochondrial genome. Nat. Rev. Genet. 6, 815-825. 10.1038/nrg1708 [DOI] [PubMed] [Google Scholar]
- Chen J., Rauch C. A., White J. H., Englund P. T. and Cozzarelli N. R. (1995). The topology of the kinetoplast DNA network. Cell 80, 61-69. 10.1016/0092-8674(95)90451-4 [DOI] [PubMed] [Google Scholar]
- Clayton A. M., Guler J. L., Povelones M. L., Gluenz E., Gull K., Smith T. K., Jensen R. E. and Englund P. T. (2011). Depletion of mitochondrial Acyl carrier protein in bloodstream-form trypanosoma brucei causes a kinetoplast segregation defect. Eukaryot. Cell 10, 286-292. 10.1128/EC.00290-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concepción-Acevedo J., Luo J. and Klingbeil M. M. (2012). Dynamic localization of trypanosoma brucei mitochondrial DNA polymerase ID. Eukaryot. Cell 11, 844-855. 10.1128/EC.05291-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concepción-Acevedo J., Miller J. C., Boucher M. J. and Klingbeil M. M. (2018). Cell cycle localization dynamics of mitochondrial DNA polymerase IC in African Trypanosomes. Mol. Biol. Cell 29, mbc.E18-02-0127 10.1091/mbc.E18-02-0127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar C. E., MacGregor P., Cooper S., Gould M. K., Matthews K. R., Savill N. J. and Schnaufer A. (2018). Mitochondrial DNA is critical for longevity and metabolism of transmission stage trypanosoma brucei. PLoS Pathog. 14, e1007195 10.1371/journal.ppat.1007195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downey N., Hines J. C., Sinha K. and Ray D. S. (2005). Mitochondrial DNA ligases of trypanosoma brucei. Eukaryot. Cell 4, 765-774. 10.1128/EC.4.4.765-774.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dua R., Levy D. L. and Campbell J. L. (1999). Analysis of the essential functions of the C-terminal protein/protein interaction domain of saccharomyces cerevisiae pol ε and its unexpected ability to support growth in the absence of the DNA polymerase domain. J. Biol. Chem. 274, 22283-22288. 10.1074/jbc.274.32.22283 [DOI] [PubMed] [Google Scholar]
- Fisk J. C. and Read L. K. (2011). Protein arginine methylation in parasitic protozoa. Eukaryot. Cell 10, 1013-1022. 10.1128/EC.05103-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluenz E., Shaw M. K. and Gull K. (2007). Structural asymmetry and discrete nucleic acid subdomains in the trypanosoma brucei kinetoplast. Mol. Microbiol. 64, 1529-1539. 10.1111/j.1365-2958.2007.05749.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M. W. (2015). Mosaic nature of the mitochondrial proteome: implications for the origin and evolution of mitochondria. Proc. Natl. Acad. Sci. USA 112, 10133-10138. 10.1073/pnas.1421379112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal J. S., McLuskey K., Das D., Myburgh E., Wilkes J., Brown E., Lemgruber L., Gould M. K., Burchmore R. J., Coombs G. H., et al. (2016). PNT1 is a C11 cysteine peptidase essential for replication of the trypanosome kinetoplast. J. Biol. Chem. 291, 9492-9500. 10.1074/jbc.M116.714972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines J. C. and Ray D. S. (2010). A mitochondrial DNA primase is essential for cell growth and kinetoplast DNA replication in trypanosoma brucei▿ †. Mol. Cell. Biol. 30, 1319-1328. 10.1128/MCB.01231-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines J. C. and Ray D. S. (2011). A second mitochondrial DNA primase is essential for cell growth and kinetoplast minicircle DNA replication in Trypanosoma brucei. Eukaryot. Cell 10, 445-454. 10.1128/EC.00308-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines J. C., Engel M. L., Zhao H. and Ray D. S. (2001). RNA primer removal and gap filling on a model minicircle replication intermediate. Mol. Biochem. Parasitol. 115, 63-67. 10.1016/S0166-6851(01)00272-9 [DOI] [PubMed] [Google Scholar]
- Hoffmann A., Käser S., Jakob M., Amodeo S., Peitsch C., Týč J., Vaughan S., Zuber B., Schneider A. and Ochsenreiter T. (2018). Molecular model of the mitochondrial genome segregation machinery in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 115, 201716582 10.1073/pnas.1721248115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob M., Hoffmann A., Amodeo S., Peitsch C., Zuber B. and Ochsenreiter T. (2016). Mitochondrial growth during the cell cycle of Trypanosoma brucei bloodstream forms. Sci. Rep. 6, 36565 10.1038/srep36565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R. E. and Englund P. T. (2012). Network news: the replication of Kinetoplast DNA. Annu. Rev. Microbiol. 66, 473-491. 10.1146/annurev-micro-092611-150057 [DOI] [PubMed] [Google Scholar]
- Johnson C. E. and Englund P. T. (1998). Changes in organization of crithidia fasciculata kinetoplast DNA replication proteins during the cell cycle. J. Cell Biol. 143, 911-919. 10.1083/jcb.143.4.911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D. T. (1999). Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195-202. 10.1006/jmbi.1999.3091 [DOI] [PubMed] [Google Scholar]
- Käser S., Oeljeklaus S., Týč J., Vaughan S., Warscheid B. and Schneider A. (2016). Outer membrane protein functions as integrator of protein import and DNA inheritance in mitochondria. Proc. Natl. Acad. Sci. USA 113, E4467-E4475. 10.1073/pnas.1605497113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käser S., Willemin M., Schnarwiler F., Schimanski B., Poveda-Huertes D., Oeljeklaus S., Haenni B., Zuber B., Warscheid B., Meisinger C., et al. (2017). Biogenesis of the mitochondrial DNA inheritance machinery in the mitochondrial outer membrane of Trypanosoma brucei. PLoS Pathog. 13, e1006808 10.1371/journal.ppat.1006808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesti T., Flick K., Keränen S., Syväoja J. E. and Wittenberg C. (1999). DNA polymerase ε catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol. Cell 3, 679-685. 10.1016/S1097-2765(00)80361-5 [DOI] [PubMed] [Google Scholar]
- Klingbeil M. M., Motyka S. A. and Englund P. T. (2002). Multiple mitochondrial DNA polymerases in trypanosoma brucei. Mol. Cell 10, 175-186. 10.1016/S1097-2765(02)00571-3 [DOI] [PubMed] [Google Scholar]
- Lindsay M. E., Gluenz E., Gull K. and Englund P. T. (2008). A new function of Trypanosoma brucei mitochondrial topoisomerase II is to maintain kinetoplast DNA network topology. Mol. Microbiol. 70, 1465-1476. 10.1111/j.1365-2958.2008.06493.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Molina H., Kalume D., Pandey A., Griffith J. D. and Englund P. T. (2006). Role of p38 in replication of trypanosoma brucei kinetoplast DNA. Mol. Cell. Biol. 26, 5382-5393. 10.1128/MCB.00369-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Wang J., Yaffe N., Lindsay M. E., Zhao Z., Zick A., Shlomai J. and Englund P. T. (2009). Trypanosomes have six mitochondrial DNA helicases with one controlling Kinetoplast maxicircle replication. Mol. Cell 35, 490-501. 10.1016/j.molcel.2009.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Yildirir G., Wang J., Tolun G., Griffith J. D. and Englund P. T. (2010). TbPIF1, a trypanosoma brucei mitochondrial DNA helicase, is essential for kinetoplast minicircle replication. J. Biol. Chem. 285, 7056-7066. 10.1074/jbc.M109.084038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukeš J., Guilbride L. D., Votýpka J., Zíková A., Benne R. and Englund P. T. (2002). Kinetoplast DNA network: evolution of an improbable structure. Eukaryot. Cell 1, 495-502. 10.1128/EC.1.4.495-502.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melendy T., Sheline C. and Ray D. S. (1988). Localization of a type II DNA topoisomerase to two sites at the periphery of the kinetoplast DNA of Crithidia fasciculata. Cell 55, 1083-1088. 10.1016/0092-8674(88)90252-8 [DOI] [PubMed] [Google Scholar]
- Milman N., Motyka S. A., Englund P. T., Robinson D. and Shlomai J. (2007). Mitochondrial origin-binding protein UMSBP mediates DNA replication and segregation in trypanosomes. Proc. Natl. Acad. Sci. USA 104, 19250-19255. 10.1073/pnas.0706858104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyahira Y. and Dvorak J. A. (1994). Kinetoplastidae display naturally occurring ancillary DNA-containing structures. Mol. Biochem. Parasitol. 65, 339-349. 10.1016/0166-6851(94)90084-1 [DOI] [PubMed] [Google Scholar]
- Ntambi J., Shapiro T., Ryan K. and Englund P. (1986). Ribonucleotides associated with a gap in newly replicated kinetoplast DNA minicircles from Trypanosoma equiperdum. J. Biol. Chem. 261, 11890-11895. [PubMed] [Google Scholar]
- Ogbadoyi E. O., Robinson D. R. and Gull K. (2003). A high-order trans-membrane structural linkage is responsible for mitochondrial genome positioning and segregation by flagellar basal bodies in trypanosomes. Mol. Biol. Cell 14, 1769-1779. 10.1091/mbc.e02-08-0525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña-Diaz P., Vancová M., Resl C., Field M. C. and Lukeš J. (2017). A leucine aminopeptidase is involved in kinetoplast DNA segregation in trypanosoma brucei. PLoS Pathog. 13, e1006310 10.1371/journal.ppat.1006310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povelones M. L. (2014). Beyond replication: division and segregation of mitochondrial DNA in kinetoplastids. Mol. Biochem. Parasitol. 196, 53-60. 10.1016/j.molbiopara.2014.03.008 [DOI] [PubMed] [Google Scholar]
- Ralston K. S., Kisalu N. K. and Hill K. L. (2011). Structure-function analysis of dynein light chain 1 identifies viable motility mutants in bloodstream-form trypanosoma brucei. Eukaryot. Cell 10, 884-894. 10.1128/EC.00298-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo A. E. and Piller S. C. (2018). Protein arginine methylation: an emerging regulator of the cell cycle. Cell Div 13, 10.1186/s13008-018-0036-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read L. K., Lukeš J. and Hashimi H. (2016). Trypanosome RNA editing: the complexity of getting U in and taking U out. Wiley Interdiscip Rev Rna 7, 33-51. 10.1002/wrna.1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusconi F., Durand-Dubief M. and Bastin P. (2005). Functional complementation of RNA interference mutants in trypanosomes. BMC Biotechnol. 5, 6 10.1186/1472-6750-5-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxowsky T. T., Choudhary G., Klingbeil M. M. and Englund P. T. (2003). Trypanosoma brucei has two distinct mitochondrial DNA polymerase β Enzymes. J. Biol. Chem. 278, 49095-49101. 10.1074/jbc.M308565200 [DOI] [PubMed] [Google Scholar]
- Schimanski B., Nguyen T. N. and Günzl A. (2005). Highly efficient tandem affinity purification of trypanosome protein complexes based on a novel epitope combination. Eukaryot. Cell 4, 1942-1950. 10.1128/EC.4.11.1942-1950.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A. and Ochsenreiter T. (2018). Failure is not an option – mitochondrial genome segregation in trypanosomes. J. Cell Sci. 131, jcs221820 10.1242/jcs.221820 [DOI] [PubMed] [Google Scholar]
- Scocca J. R. and Shapiro T. A. (2008). A mitochondrial topoisomerase IA essential for late theta structure resolution in African trypanosomes. Mol. Microbiol. 67, 820-829. 10.1111/j.1365-2958.2007.06087.x [DOI] [PubMed] [Google Scholar]
- Shapiro T. (1993). Kinetoplast DNA maxicircles: networks within networks. Proc. Natl. Acad. Sci. USA 90, 7809-7813. 10.1073/pnas.90.16.7809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa R. (1996). Structural and mechanistic relationships between nucleic acid polymerases. Trends Biochem. Sci. 21, 186-190. 10.1016/S0968-0004(96)10023-2 [DOI] [PubMed] [Google Scholar]
- Steitz T. A. (1998). A mechanism for all polymerases. Nature 391, 231-232. 10.1038/34542 [DOI] [PubMed] [Google Scholar]
- Stuart K. D., Schnaufer A., Ernst N. and Panigrahi A. K. (2005). Complex management: RNA editing in trypanosomes. Trends Biochem. Sci. 30, 97-105. 10.1016/j.tibs.2004.12.006 [DOI] [PubMed] [Google Scholar]
- Stumpf J. D., Saneto R. P. and Copeland W. C. (2013). Clinical and molecular features of POLG-related mitochondrial disease. Cold Spring Harb. Perspect. Biol. 5, a011395 10.1101/cshperspect.a011395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes S. E. and Hajduk S. L. (2013). Dual functions of α-ketoglutarate dehydrogenase E2 in the krebs cycle and mitochondrial DNA inheritance in trypanosoma brucei. Eukaryot. Cell 12, 78-90. 10.1128/EC.00269-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trikin R., Doiron N., Hoffmann A., Haenni B., Jakob M., Schnaufer A., Schimanski B., Zuber B. and Ochsenreiter T. (2016). TAC102 is a novel component of the mitochondrial genome segregation machinery in trypanosomes. PLoS Pathog. 12, e1005586 10.1371/journal.ppat.1005586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Týč J., Klingbeil M. M. and Lukeš J. (2015). Mitochondrial heat shock protein machinery Hsp70/Hsp40 is indispensable for proper mitochondrial DNA maintenance and replication. Mbio 6, e02425-e02414. 10.1128/mBio.02425-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verner Z., Basu S., Benz C., Dixit S., Dobáková E., Faktorová D., Hashimi H., Horáková E., Huang Z., Paris Z., et al. (2015). Chapter three malleable mitochondrion of trypanosoma brucei. Int. Rev. Cell Mol. Biol. 315, 73-151. 10.1016/bs.ircmb.2014.11.001 [DOI] [PubMed] [Google Scholar]
- Viscomi C. and Zeviani M. (2017). MtDNA-maintenance defects: syndromes and genes. J. Inherit. Metab. Dis. 40, 587-599. 10.1007/s10545-017-0027-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Morris J. C., Drew M. E. and Englund P. T. (2000). Inhibition of trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 275, 40174-40179. 10.1074/jbc.M008405200 [DOI] [PubMed] [Google Scholar]
- Wang Z., Drew M. E., Morris J. C. and Englund P. T. (2002). Asymmetrical division of the kinetoplast DNA network of the trypanosome. EMBO J. 21, 4998-5005. 10.1093/emboj/cdf482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weems E., Singha U. K., Hamilton V., Smith J. T., Waegemann K., Mokranjac D. and Chaudhuri M. (2015). Functional complementation analyses reveal that the single PRAT family protein of trypanosoma brucei is a divergent homolog of Tim17 in saccharomyces cerevisiae. Eukaryot. Cell 14, 286-296. 10.1128/EC.00203-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz E., Leal S., Ochatt C. and Cross G. M. (1999). A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99, 89-101. 10.1016/S0166-6851(99)00002-X [DOI] [PubMed] [Google Scholar]
- Woodward R. and Gull K. (1990). Timing of nuclear and kinetoplast DNA replication and early morphological events in the cell cycle of Trypanosoma brucei. J. Cell Sci. 95, 49-57. [DOI] [PubMed] [Google Scholar]
- Yu Z., Liu Y. and Li Z. (2012). Structure–function relationship of the Polo-like kinase in Trypanosoma brucei. J. Cell Sci. 125, 1519-1530. 10.1242/jcs.094243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Lindsay M. E., Chowdhury A., Robinson D. R. and Englund P. T. (2008). p166, a link between the trypanosome mitochondrial DNA and flagellum, mediates genome segregation. EMBO J. 27, 143-154. 10.1038/sj.emboj.7601956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zíková A., Hampl V., Paris Z., Týč J. and Lukes J. (2016). Aerobic mitochondria of parasitic protists: diverse genomes and complex functions. Mol. Biochem. Parasitol. 209, 46-57. 10.1016/j.molbiopara.2016.02.007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.