Abstract
Fast identification of pathogens directly from positive blood cultures is of highest importance to supply an adequate therapy of bloodstream infections (BSI). There are several platforms providing molecular-based identification, detection of antimicrobial resistance genes, or even a full antimicrobial susceptibility testing (AST). Two of such test systems allowing rapid diagnostics were assessed in this study: The Biofire FilmArray® and the Genmark ePlex®, both fully automated test system with a minimum of hands-on time. Overall 137 BSI episodes were included in our study and compared to conventional culture–based reference methods. The FilmArray® is using one catridge including a panel for the most common bacterial and fungal BSI pathogens as well as selected resistance markers. The ePlex® offers three different cartridges for detection of Gram-positives, Gram-negatives, and fungi resulting in a broader panel including also rare pathogens, putative contaminants, and more genetic resistance markers. The FilmArray® and ePlex® were evaluated for all 137 BSI episodes with FilmArray® detecting 119 and ePlex® detecting 128 of these. For targets on the respective panel of the system, the FilmArray® generated a sensitivity of 98.9% with 100% specificity on Gram-positive isolates. The ePlex® system generated a sensitivity of 94.7% and a specificity of 90.7% on Gram-positive isolates. In each case, the two systems performed with 100% sensitivity and specificity for the detection of Gram-negative specimens covered by each panel. In summary, both evaluated test systems showed a satisfying overall performance for fast pathogen identification and are beneficial tools for accelerating blood culture diagnostics of sepsis patients.
Electronic supplementary material
The online version of this article (10.1007/s10096-020-03828-5) contains supplementary material, which is available to authorized users.
Keywords: Bloodstream infection, Molecular identification, Antibiotic resistance, Rapid identification system, GenMark ePlex®, Biofire FilmArray®
Introduction
As bacteremia and sepsis are still leading causes of morbidity and mortality [1, 2], fast diagnosis of the causative organism and appropriate antimicrobial therapy are essential for rapid treatment decisions [3]. Worldwide, the incidence of sepsis is about 19 million per year [4] with a lethality of 20–30% [5, 6]. This emphasizes the need for rapid identification (ID) and detection of resistance genes. Conventional microbiological, agar-based methods used for blood culture diagnostics include culture-dependent identification by MALDI-TOF and AST by phenotypic methods. As these techniques are time-consuming, automated molecular ID methods with lower hands-on and turnaround time were developed allowing for identification and the genotypic detection of resistance genes [7–12]. Therefore, using quick molecular-based ID tests directly from positive blood culture bottles can help with providing an adequate therapy in a timely manner by de-escalating broad-spectrum antimicrobial therapy or escalating insufficient treatment, respectively [13–15].
To speed up the identification and AST of Gram-negative bacteria in positive blood cultures, the Accelerate Pheno® system (Accelerate® Diagnostics, USA) was evaluated for fast ID and AST compared to culture-based diagnostics [12]. The system is implemented in our routine laboratory accelerating reports for ID and AST especially for high-risk patients having BSI with Gram-negative bacteria. To establish a workflow for faster identification of all pathogens causing bloodstream infections, two different blood culture identification panels were compared in this study.
The BioFire FilmArray® Blood Culture Identification Panel (Biomerieux, Nürtingen, Germany) is a CE-marked multiplex PCR system, which allows identification of 24 targets at once including Gram-positives, Gram-negatives, and fungi, as well as three antibiotic resistance genes. DNA extraction, multiplex-PCR, and detection are fully automated with 2 min of hands-on time. A results report is generated approximately after 75 min [16].
The ePlex® system (Genmark Diagnostics, Carlsbad, USA) is endowed with three variable cartridges for pathogen identification. Deploying this test allows the detection of Gram-positives, Gram-negatives, and fungi. Besides the coverage of a large variety of organisms (55 bacterial/fungal targets) the test is designed to detect ten antibiotic resistance genes in addition. Nucleic acid extraction, amplification, and detection are completed in about 90 min using the microfluidic eSensor technology and require only 2 min of hands-on time.
The aim of the present prospective study was the comparative analysis of two molecular based Blood Culture Identification panels in comparison to conventional culture-dependent ID methods in terms of (i) correct detection and identification of the pathogen, (ii) correct prediction of resistance genes and genotypes, and (iii) suitability for implementation in a diagnostic routine workflow.
Material and methods
Study design
The study was performed from November 2017 to January 2018 at a tertiary university hospital in Southern Germany in accordance with the local ethics committee (no. 667/2014BO1 and 139/2016BO2).
The first positive blood culture of each patient with a positive initial Gram-stain (Gram-positive or Gram-negative bacteria as well as fungi) was included in the study until 100 cultures with Gram-positive isolates were reached. Samples having more than one obvious organism according to the Gram-stain were not included in the study.
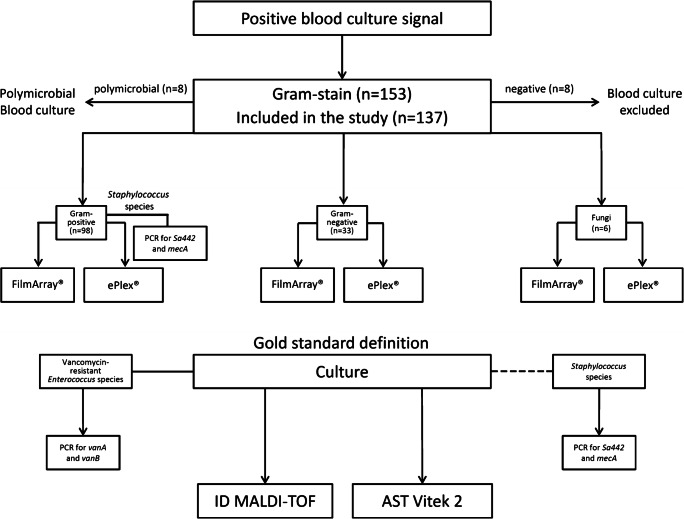
Positive blood cultures with Gram-positive (n = 98) or Gram-negative bacteria (n = 33) as well as fungi (n = 6) were analyzed by two different blood culture identification instruments: FilmArray® Blood Culture Identification panel (BioFire Diagnostics, Salt Lake City, UT, USA) and ePlex® Research Use Only (RUO) Blood Culture Identification panels (Genmark Diagnostics, Carlsbad, CA, USA). A schematic overview of the study design is shown in Fig. 1.
Fig. 1.
Flow chart of the study design. Each patient having a first positive blood culture signal (n = 153) with a positive initial Gram-stain (Gram-positive [n = 98] or Gram-negative bacteria [n = 33] as well as fungi [n = 6]) was included in the study (in total n = 137). Samples having more than one obvious organism according to the Gram-stain (n = 1) and Gram-stainings showing no organism (n = 8) were excluded from the study. Blood cultures positive for Gram-positive, Gram-negative organisms or fungi were analyzed with the FilmArray® and the ePlex® blood culture panels. On weekdays, PCR for Staphylococcus aureus and the mecA gene was performed directly from positive blood culture bottles, based on the Gram-stain result indicating the presence of staphylococci. Results obtained from the two evaluated test systems were compared to culture- (MALDI-TOF identification and Vitek-AST) or molecular-based (in-house PCR for Sa442 and mecA or vanA/vanB respectively) reference method
Conventional diagnostics
Laboratory operation hours are weekdays from 7:30 AM to 5:30 PM and weekends from 7:30 AM to 4 PM. Blood cultures are processed every morning starting at 7:30 AM and during the operating hours as soon as they are flagged positive.
Incubation of Blood culture bottles was performed in the Bactec FX blood culture instrument (BD Diagnostic Systems, Franklin Lakes, USA). The routine diagnostic workflow included subcultures of positive blood culture bottles on CNA- (Biomerieux, Nürtingen, Germany) and Brain Heart agar (Oxoid, Munich, Germany) in addition to Gram-stain. Identification to the species level was achieved by MALDI-TOF mass spectrometry (Microflex LT, Bruker Daltonics, Germany) and supplemented, if necessary, by the VITEK® 2 identification system (bioMérieux SA, France).
The appropriate testing card of VITEK® 2 (bioMérieux, SA, France) was used for Antimicrobial Susceptibility Testing (AST). All bacterial isolates were stored at − 80 °C for further analysis. The guidelines set of the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_8.1_Breakpoint_Tables.pdf) were consulted for interpretation of susceptibility results.
If necessary, resistance genes blaVIM, blaIMP, blaKPC, blaOXA-48, and blaNDM for carbapenemases were confirmed by molecular diagnostics (Light Mix Modular Realtime-PCR, Roche, Mannheim). In house-PCR for mecA [17] was performed on a T3 thermal cycler (Biometra, Göttingen) and analyzed by agarose gel electrophoresis directly from positive blood culture bottles during laboratory operating hours, but not during weekends. For these cases, the mecA-PCR was repeated from culture isolates. Enterococcus species were tested for the presence of vancomycin resistance genes vanA and vanB by in-house PCRs [18] using T3 thermal cycler (Biometra, Göttingen) followed by gel electrophoresis. Resistance of selected Enterobacterales against 3rd generation cephalosporins was further analyzed phenotypically for the presence of extended-spectrum beta lactamases (blaCTX-M) using ESBL detection disk set (Mast Diagnostica GmbH, Reinfeld, Germany). 16S rRNA-PCR directly from positive blood culture bottles [17] using T3 thermal cycler (Biometra, Göttingen) was performed in case of lacking cultural growth.
Fungi were cultivated on yeast agar supplemented with gentamicin (in-house preparation). Single colonies were identified with MALDI-TOF mass spectrometry (Microflex LT, Bruker Daltonics, Germany) and BBL CHROMagar for Candida (BD Diagnostic Systems, Franklin Lakes, USA). Resistance profiles for fungi were not assessed for the study.
Biofire FilmArray® blood culture identification panel
The FilmArray® instrument was run according to manufacturer’s instructions. In brief, the reagents in the pouch were adjusted with approximately 300 μl rehydration solution. Next, 100 μl medium from the positive blood culture bottle was mixed with sample buffer and loaded into the pouch, which requires only 2 min hands-on time. Once inserted in the FilmArray® instrument, nucleic acid extraction was performed followed by nested multiplex PCR giving results in about 1 h. The BioFire FilmArray® is using one reagent pouch for identification of Gram-positives, Gram-negatives, and fungi. The organisms covered by the assay are shown in Table S1. The software of the FilmArray® generates an automated report. One system was available during the study period to analyze one sample at a time.
Genmark ePlex® BC panels
The GenMark ePlex® system is using the eSensor technology deploying competitive DNA hybridization followed by electrochemical detection. Three different blood culture identification panels are available: one cartridge each for the identification of Gram-positives (BCID-GP), Gram-negatives (BCID-GN), and fungi (BCID-FP). The decision of which cartridge to use is based on the Gram-stain result. To run the assay in accordance with the manufacturer specifications, 50 μl of positive blood culture are transferred into the sample port of the respective cartridge followed by loading into the system. One tower with six test bays was available during the study period, offering the possibility to analyze six blood culture samples simultaneously. Result reports were generated automatically after about one and half hours from starting the test. Organisms included in the panel are illustrated in Table S1. The cartridges for Gram-negative and Gram-positive bacteria include a PAN target, that detects the presence of Gram-positives organisms in the Gram-negative (BCID-GN) cartridge and vice versa helping to identify polymicrobial mixed BSI that may have been missed by Gram-staining initially. In addition, a PAN-target for Candida is included on the bacterial detection cartridges.
Data analysis
Identification of blood culture isolates by MALDI-TOF was defined as reference method. In case of lacking growth, 16S rRNA-PCR was performed directly out of positive blood culture bottles. Dependent on the fast ID system tested, some species are only identified to the genus level. Exact genus ID—as identified by the reference method—was rated as correctly identified in case a fast ID system did not provide species level identification. If one of the two systems produced an invalid or failed run, it was repeated with a new identification cartridge. Due to operating hours of the laboratory during the study period, it was not possible to repeat every invalid or failed run generated by one of the two test systems. Therefore two samples (n = 2) were excluded from the study.
Detection of genotypic resistance markers by the test systems were compared to the results of in-house PCRs in relation to the phenotypic resistance obtained by Vitek 2 or agar diffusion (ESBL screening) respectively.
An identification was called ‘true positive’ for any organism covered by the FilmArray® or the ePlex® panel respectively and identified by the reference method as well as by FilmArray® or the ePlex®. ‘True negative’ was defined as any species not detected by the panels of the two systems and different from the reference method result. Organisms are called ‘false positive’ if identified by the FilmArray® or ePlex® system and not by the reference method. Any organism missed or misidentified by the two ID systems, but detected by the reference method, were marked as “false negative.”
The calculated specificity and the number of true negatives refer to positive blood cultures, which were negative for the respective target organism.
Results and discussion
During the study period, 153 positive blood cultures gave a positive signal and were assessed for inclusion in our analysis. Overall, eight positive blood cultures were excluded from the study, due to a negative initial Gram-stain result. In total, 137 positive blood cultures were included in our study (Fig. 1). Among those 98 episodes were monomicrobial BSI with Gram-positive organisms, 33 were monomicrobial with Gram-negative organisms and 6 episodes were caused by yeast strains (Tab.1). In addition, in eight samples, more than one organism was detected by culture. One example was not included in the study as the presence of multiple organisms was already suspected by the Gram stain, the results of the remaining seven samples are summarized in Table S2.
Table 1.
Identification results for bacteria and yeast (monomicrobial BSI)
| No. of true/false positives no. of true/false negatives | No. of true/false positives no. of true/false negatives | ||||
|---|---|---|---|---|---|
| Species (n = reference method ID) | FilmArray® | ePlex® | FilmArray® | ePlex® | Comments |
| ID of Gram-positive organisms | |||||
| S. aureus (n = 18) | 18/0 | 18/0 | 119/0 | 119/0 | |
| S. epidermidis (n = 35) | 35/0a | 35/2 | 102/0 | 100/0 | FilmArray® ID only on Staphylococcus spp. level, false ePlex® ID of S. hominis as S. epidermidis, false ePlex® ID of S. warneri as S. epidermidis |
| S. haemolyticus (n = 8) | 8/0a | 7/0 | 129/0 | 129/1 | FilmArray® ID only on Staphylococcus species level, no ePlex® ID for one S. haemolyticus isolate |
| S. hominis (n = 6) | 6/0a | 5/0 | 131/0 | 131/1 | FilmArray® and ePlex® ID only on Staphylococcus spp. level, false ePlex® ID of S. hominis as S. epidermidis |
| S. warneri (n = 2) | 1/0a | 1/0 | 135/1 | 135/1 | FilmArray® and ePlex® ID only on Staphylococcus spp. level, no FilmArray® ID for 1/2 isolates, false ePlex® ID of one S. warneri isolate as S. epidermidis |
| S. capitis (n = 2) | 2/0a | 2/0 | 135/0 | 135/0 | FilmArray® and ePlex® ID only on Staphylococcus spp. level |
| E. faecium (n = 4) | 4/0a | 4/2 | 133/0 | 131/0 | FilmArray® ID only on Enterococcus spp. level, false ePlex® ID for two E. faecalis isolates as E. faecium |
| E. faecalis (n = 5) | 5/0a | 3/0 | 132/0 | 132/2 | FilmArray® ID only on Enterococcus spp. level, false ePlex® ID for two E. faecalis isolates as E. faecium |
| S. pneumoniae (n = 1) | 1/0 | 1/0 | 136/0 | 136/0 | |
| S. gallolyticus (n = 2) | 2/0 | 2/0 | 135/0 | 135/0 | FilmArray® and ePlex® ID only on Streptococcus spp. level |
| S. mitis group (n = 1) | 1/0 | 1/0 | 136/0 | 136/0 | FilmArray® and ePlex® ID only on Streptococcus spp. level |
| S. oralis (n = 1) | 1/0 | 1/0 | 136/0 | 136/0 | FilmArray® and ePlex® ID only on Streptococcus spp. level |
| S. anginosus (n = 1) | 1/0 | 1/0 | 136/0 | 136/0 | FilmArray® ID only on Streptococcus spp. level |
| Streptococcus sp. (group C) (n = 1) | 1/0 | 1/0 | 136/0 | 136/0 | FilmArray® and ePlex® ID only on Streptococcus spp. level |
| C. acnes (n = 5) | n/a | 5/0 | n/a | 132/0 | Not included in the FilmArray® panel |
| A. meyeri (n = 1) | n/a | n/a | n/a | n/a | Not included in the FilmArray® and ePlex® panel |
| Actinomyces sp. (n = 1) | n/a | n/a | n/a | n/a | Not included in the FilmArray® and ePlex® panel |
| P. faecalis (n = 1) | n/a | n/a | n/a | n/a | Not included in the FilmArray® and ePlex® panel |
| Lactobacillus sp. (n = 1) | n/a | 1/0 | n/a | 136/0 | Not included in the FilmArray® panel, ePlex® ID only on Lactobacillus spp. level |
| Corynebacterium sp. (n = 1) | n/a | 1/0 | n/a | 136/0 | Not included in the FilmArray® panel |
| A. parvulum (n = 1) | n/a | n/a | n/a | n/a | Not included in the FilmArray® and ePlex® panel |
| Total number of organisms (n = 98) | 86/0 | 89/4 | 50/1 | 39/5 | |
| ID of Gram-negative organisms | |||||
| E. coli (n = 15) | 15/0 | 15/0 | 122/0 | 122/0 | |
| K. pneumoniae/variicola (n = 5) | 5/0 | 5/0 | 132/0 | 132/0 | Two K. variicola identified as K. pneumoniae by FilmArray® and ePlex® |
| K. oxytoca (n = 1) | 1/0 | 1/0 | 136/0 | 136/0 | |
| S. marcescens (n = 2) | 2/0 | 2/0 | 135/0 | 135/0 | |
| E. cloacae Komplex (n = 1) | 1/0 | 1/0 | 136/0 | 136/0 | |
| E. ludwigii (n = 1) | 1/0 | 1/0 | 136/0 | 136/0 | FilmArray® and ePlex® ID as E. cloacae complex |
| P. aeruginosa (n = 1) | 1/0 | 1/0 | 136/0 | 136/0 | |
| H. influenzae (n = 1) | 1/0 | 1/0 | 136/0 | 136/0 | |
| M. liquefaciens (n = 1) | n/a | n/a | n/a | n/a | Not included in the FilmArray® and ePlex®panel |
| N. polysaccharea (n = 1) | n/a | n/a | n/a | n/a | Not included in the FilmArray® and ePlex® panel |
| C. canimorsus (n = 1)a | n/a | n/a | n/a | n/a | Not included in the FilmArray® and ePlex®panel; aonly identified by 16S-rRNA-PCR |
| B. fragilis (n = 2) | n/a | 2/0 | n/a | 135/0 | Not included in the FilmArray® panel |
| B. vulgatus/dorei (n = 1) | n/a | n/a | n/a | n/a | Not included in the FilmArray® and ePlex® panel |
| Total number of organisms (n = 33) | 27/0 | 29/0 | 110/0 | 108/0 | |
| ID of yeasts | |||||
| Candida albicans (n = 2) | 2/0 | 2/0 | 135/0 | 135/0 | |
| Candida glabrata (n = 2) | 2/0 | 2/0 | 135/0 | 135/0 | |
| Candida parapsilosis (n = 1) | 1/0 | 1/0 | 136/0 | 136/0 | |
| Candida tropicalis (n = 1) | 1/0 | 1/0 | 136/0 | 136/0 | |
| Total number of organisms (n = 6) | 6 | 6 | 131/0 | 131/0 | |
aCoagulase-negative Staphylococcus spp. as well as Enterococcus spp. were detected on genus level only by the FilmArray®
Identification of Gram-positive BSI pathogens
In summary 21 different Gram-positive organisms (98 isolates in total) were determined by routine-diagnostic methods. Identification by MALDI-TOF was compared to identification results created by the BioFire FilmArray® and the GenMark ePlex® system.
Correct identification by the FilmArray® was achieved for 86/98 (87.8%) isolates on genus and species level, as defined in the system panel for the respective target organism. The system scored 12 samples negative (12.2%). These 11 samples contained organisms that are not included in the system panel, namely C. acnes (n = 5), Actinomyces meyeri (n = 1), Actinomyces species (n = 1), P. faecalis (n = 1), Lactobacillus species (n = 1), Corynebacterium species (n = 1) and Atopobium parvulum (n = 1). One Staphylococcus warneri isolate was not detected as Staphyloccus species by the system. ID failure of S. warneri has been reported before and was explained by a reduced sensitivity of the FilmArray® system for some CoNS including S. warneri [19], which is also mentioned in the package insert of the panel. The FilmArray® therefore identified 86/87 (98.9%) correctly taking only the organisms included in the panel into account. The results are shown in detail in Table 1. For the Gram-positives, the FilmArray® showed a sensitivity of 98.9% and a 100% specificity (Table 4) for organisms covered by the panel.
Table 4.
Overview of sensitivity, specificity, positive predictive values (PPV), and negative predictive values (NPV) for in-panel organisms only
| FilmArray® | ePlex® | |
|---|---|---|
| Sensitivity (%) | ||
| Gram-positives | 98.9 | 94.7 |
| Gram-negatives | 100 | 100 |
| Specificity (%) | ||
| Gram-positives | 100 | 90.7 |
| Gram-negatives | 100 | 100 |
| Positive predictive value (PPV) (%) | ||
| Gram-positives | 100 | 95.7 |
| Gram-negatives | 100 | 100 |
| Negative predictive value (NPV) (%) | ||
| Gram-positives | 98 | 88.6 |
| Gram-negatives | 100 | 100 |
In contrast to the FilmArray® system, the ePlex® system aims at identifying enterococci and coagulase negative staphylococci to the species level. In total identification to the species level was achieved in 89/98 (90.8%) of the Gram-positive bacterial isolates. Two Staphyloccus species were misidentified by the system: S. hominis as well as S. warneri were identified as S. epidermidis. A larger sample size is required to assess if this assay is more prone to misidentification of CoNS. Furthermore, it is difficult to rule-out that one or more of these specimens contained multiple Staphylococus species. In addition, as all these three CoNS species belong to the S. epidermidis group as part of the normal skin flora with similar pathogenicity [20] it is not mandatory to identify these CoNS on the species level. Worth mentioning is the inclusion of S. lugdunensis in the Genmark ePlex® panel as it is a common pathogen with species specific virulence factors [21] associated with aggressive causes of infective endocarditis [22–24].
In addition, two E. faecalis isolates were misidentified as E. faecium. The correct identity of these two E. faecalis isolates was verified by MALDI-TOF and confirmed by next generation sequencing (NGS) of the whole genome on an Illumina Nextseq platform followed by calculating the average nucleotide identity (ANI) [25]. The ePlex® cartridges used in this study were for Research Use Only and according to the package insert of the test assay in a subsequent clinical study evaluating the performance of the CE-IVD and U.S. FDA cleared assays, the ePlex® demonstrated high sensitivity and specificity for identification of Enterococcus faecalis. Enterococcus species cause approximately 10% of nosocomial blood stream infections [26] caused by a variety of different entries of the pathogen [27]. Treatment of Enterococcus related BSI is strongly dependent on correct species identification. Whereas E. faecalis is mostly susceptible to the small-spectrum antibiotic ampicillin, E. faecium is resistant [28].
Classifying CoNS and enterococci only to the genus level, correct identification could be achieved for 93/98 (94.9%) of the Gram-positive bacterial isolates. Based on the broader panel C. acnes (n = 5), Lactobacillus species (n = 1) and Corynebacterium species (n = 1) were detected in contrast to the FilmArray® system. Four bacterial isolates were not identified as they were not part of the panel: Actinomyces meyeri (n = 1), Actinomyces species (n = 1), P. faecalis (n = 1) and Atopobium parvulum (n = 1). Moreover one S. haemolyticus strain was not detected by the system. Regarding only organisms covered by the panel, the ePlex® correctly identified 93/94 (98.9%) strains to genus level and 89/94 (94.7%) to the species level. For two blood cultures, the ePlex® detected a second Gram-positive organism (Micrococcus in addition to S. aureus and accordingly Staphylococcus spp. in addition to E. faecium). None of these additional organisms could be confirmed by culture (Table 1). The ePlex® panel allows in contrast to the FilmArray® identification of potential contaminants namely Corynebacterium spp., Micrococcus spp., and C. acnes usually resulting in no need for treatment unless there is direct evidence for infection by the organism. Fast identification of such contaminants can help in quick de-escalation of the ongoing therapy and prevent redundant administration of anti-infectious therapies [29–31].
The ePlex® system showed a sensitivity of 94.7% and a specificity of 90.7% for Gram-positive pathogens included in the panel (Table 4).
Percentages of detected organisms covered by the respective blood culture identification panel as well as for all microorganism detected by the reference method are shown in Table 2.
Table 2.
Resistance genes identified for Gram-positive bacteria by FilmArray® and ePlex®
| Genotypic detection of methicillin resistance (mecA/mecC ) | ||||||
|---|---|---|---|---|---|---|
| Staphylococcus species (total n=66) | mecA in house PCR | mecC in house PCR | mecA FilmArray® | mecA ePlex® | mecC ePlex® | comment |
| S. aureus (n=18) | negative (n=18)a | n/a | negative (n=18) | negative (n=18) | negative (n=18) | |
| Coagulase-negative Staphylococcus spp. (n=48) | negative (n=15)b | n/a | negative (n=16) | negative (n=15) | negative (n=48) | False-negative mecA result by FilmArray® (n=1) |
| positive (n=33)c | positive (n=31) | positive (n=33) | No target detected by FilmArray® (n=1) | |||
| Genotypic detection of vancomycin resistance (vanA/vanB) | ||||||
| Enterococcus species (total n=9) | vanA/vanB in-house PCR | vanA/vanB FilmArray® | vanA/vanB ePlex® | comment | ||
| Enterococcus vancomycin sensitive (n= 8) | negative (n=8) | negative (n=8) | negative (n=8) | E. faecalis (n=5), E. faecium (n=3) | ||
| Enterococcus vancomycin resistant (n= 1) | positive van B (n=1) | positive vanA /vanB (n=1) | positive vanB (n=1) | vanB positive E. faecium isolate | ||
atested from blood culture (n=13), tested from culture (n=5)
btested from blood culture (n=11), tested from culture (n=4)
ctested from blood culture (n=18), tested from culture (n=15)
Identification of Gram-negative BSI pathogens
We further assessed identification of Gram-negative organisms for the BioFire FilmArray® and the Genmark ePlex® system. Routine-diagnostic analysis provided evidence for 14 different species within 33 samples in total. Due to the lack of bacterial growth, one sample with Capnocytophaga canimorsus was not verified by MALDI-TOF, but by 16S rRNA PCR out of the positive blood culture bottle. Two Klebsiella variicola isolates were reported as Klebsiella pneumoniae by the FilmArray® and GenMark ePlex®, but were not assessed as misidentification, as MALDI-TOF based differentiation of these two species has only recently become available [32]. Klebsiella variicola was described as a new species genetically isolated from K. pneumoniae and can be phenotypically distinguished by the inability of adonitol fermentation. The pathogenicity potential and analysis of virulence factors of K. variicola are still under investigation and high-risk antibiotic resistance genes are already described for K. variicola [33–35]. Thus, identification of K. variicola on species level and dissociation from K. pneumoniae could be of relevance for a future panel update.
In contrast to the FilmArray® panel, the ePlex® system can identify the Bacteroides fragilis, which is of clinical importance since the B. fragilis group belongs to the most prevalent anaerobic pathogens causing BSI [36].
In total, the FilmArray® identified 27/33 isolates (81.8%). Six samples included organisms outside the panel (Moraxella liquefaciens (n = 1), Neisseria polysaccharea (n = 1), Capnocytophaga canimorsus (n = 1), B. fragilis (n = 2) and B. vulgatus/dorei (n = 1)). This resulted in a correct identification by the FilmArray® instrument for all 27/27 (100%) organisms included in the panel.
The ePlex® system identified 29/33 isolates correctly (87.9%), whereas M. liquefaciens (n = 1), N. polysaccharea (n = 1), C. canimorsus (n = 1) and B. vulgatus/dorei (n = 1) were not included in the panel. Thus, the remaining 29/29 specimens (100%) were correctly identified.
Identification of yeast strains as BSI pathogens
During the study period, six BSI episodes with different yeast strains occurred. Both Biofire FilmArray® and GenMark ePlex® identified all 6/6 strains (100%) correctly, namely C. albicans (n = 2), C. glabrata (n = 2), C. parapsilosis (n = 1) and C. tropicalis (n = 1), which were all confirmed by MALDI-TOF analysis.
Detection of polymicrobial BSI pathogens
In total, for seven polymicrobial BSI episode samples, both test systems were performed (summarized in Table S2). The FilmArray® and ePlex® detected all pathogens covered by the panels at least on genus level with only one exception: the FilmArray® missed detecting a Streptococcus constellatus isolate although covered by the panel, which was found in addition to Proteus mirabilis in one blood culture bottle. In the initial Gram-stain, only Gram-negative rods were observed. Furthermore, culture-based diagnostic showed only a few colonies of Streptococcus on CNA agar plates, indicating a low burden of Gram-positives in this blood culture.
For the ePlex® the cartridge for Gram-negatives was used for the analysis of that blood culture resulting in the detection of the Gram-positive PAN target indicating the precence of the Streptococcus constellatus isolate. The Gram-positive PAN target was also detected in a polymicrobial BSI episode containing S. epidermidis in addition to K. pneumoniae.
In another polymicrobial blood culture, Fusobacterium nucleatum and Dialister pneumosintes were identified by culture. In the initial Gram-stain, the Gram-negative rods appeared Gram-positive, whereupon the ePlex® cartridge for Gram-positives was used. Both species are not covered by the FilmArray® panel and were not detected accordingly. However, no PAN target for Gram-negatives was detected by the ePlex®.
Detection of genotypic resistance markers
In total, 71 Staphylococcus species were identified in the study. Five Staphylococcus isolates could not be verified for the presence of mecA by in-house PCR as the isolates were not stored by the reason of routine laboratory workflow. Nevertheless, the susceptibility results of these five Staphylococcus isolates correlate with the detection results for mecA generated by the FilmArray® and the ePlex®. For the residual 66 Staphylococcus isolates in-house mecA-PCR results were available. All S. aureus isolates (n = 18) were methicillin-susceptible and negative for mecA as reported by the FilmArray®, the ePlex® and the in-house PCR (Table 3). Moreover, no mecC was detected by the ePlex® system. In the 48 BSI episodes with CoNS, 15 isolates were methicillin-susceptible and 33 methicillin-resistant according to VITEK 2 results and mecA in-house PCR. This is in accordance with the mecA-PCR results of the ePlex® system. The FilmArray® produced one false-negative mecA result for an oxacillin-resistant S. epidermidis isolate positive for mecA in the in-house PCR. For the S. warneri isolate not identified by the system no PCR result was produced. Other, recently evaluated option for accelerated AST results include the culture-based EUCAST rapid antimicrobial susceptibility testing (RAST) allowing interpreting inhibition zones after 4, 6, or 8 h of growth (The European Committee on Antimicrobial Susceptibility Testing. Zone diameter breakpoints for rapid antimicrobial susceptibility testing (RAST) directly from blood culture bottles. Version 1.0, 2018. http://www.eucast.org.).
Table 3.
Resistance genes identified for Gram-positive bacteria by FilmArray® and ePlex®
| Genotypic detection of methicillin resistance (mecA/mecC) | ||||||
| Staphylococcus species (total n = 66) | mecA in house PCR | mecC in house PCR | mecA FilmArray® | mecA ePlex® | mecC ePlex® | Comment |
| S. aureus (n = 18) | Negative (n = 18)a | n/a | Negative (n = 18) | Negative (n = 18) | Negative (n = 18) | |
| Coagulase-negative Staphylococcus spp. (n = 48) | Negative (n = 15)b | n/a | Negative (n = 16) | Negative (n = 15) | Negative (n = 48) | False-negative mecA result by FilmArray® (n = 1) |
| Positive (n = 33)c | Positive (n = 31) | Positive (n = 33) | No target detected by FilmArray® (n = 1) | |||
| Genotypic detection of vancomycin resistance (vanA/vanB) | ||||||
| Enterococcus species (total n = 9) | vanA/vanB in-house PCR | vanA/vanB FilmArray® | vanA/vanB ePlex® | Comment | ||
| Enterococcus vancomycin sensitive (n = 8) | negative (n = 8) | negative (n = 8) | negative (n = 8) | E. faecalis (n = 5), E. faecium (n = 3) | ||
| Enterococcus vancomycin resistant (n = 1) | positive van B (n = 1) | positive vanA /vanB (n = 1) | positive vanB (n = 1) | vanB positive E. faecium isolate | ||
aTested from blood culture (n = 13), tested from culture (n = 5)
bTested from blood culture (n = 11), tested from culture (n = 4)
cTested from blood culture (n = 18), tested from culture (n = 15)
During the study period, nine Enterococcus species (E. faecalis [n = 5] and E. faecium [n = 4]) were isolated. Eight isolates were negatively tested for the presence of vanA and vanB by routine-diagnostic in-house PCR as well as by the FilmArray® and ePlex® system. One vancomycin resistant isolate (positive for vanB) was identified by FilmArray® and ePlex® (Tab. 3). None of the 33 isolated Gram-negative microorganisms were carbapenem-resistant, being in line with negative PCR results from the evaluated systems and reflecting the low prevalence of BSI with carbapenem-resistant bacteria in our hospital [37]. However, due to increasing bacterial resistance, rapid detection of genes encoding carbapenemases is a desirable characteristic of molecular-based blood culture identification panels. In this regard the carbapenemase resistance genes covered by the ePlex® panel for Gram-negatives can be highlighted as the most common genes [38]. The ePlex® detected the presence of blaCTX-M for three study isolates (E. coli (n = 2) and K. pneumoniae (n = 1)), phenotypically confirmed by the presence of an ESBL phenotype in the cultured strains.
A summary of the overall performance of the two test systems is illustrated in Table 4. Concerning the detection of Gram-positive bacteria included in the panels of the FilmArray® and the ePlex®, the best performance with slightly higher sensitivity, specificity, PPV and NPV values was shown by the FilmArray® (Table 4). However, due to the broader identification panel of the ePlex®, putative contaminants or frequent anaerobes can be identified, resulting in on overall higher number of identified Gram-positive organisms (93/98) as compared to the FilmArray® with identification of 86 of 93 specimens. Regarding the identification of Gram-negative organisms and yeast strains, no major differences were found between the two systems.
In conclusion and in view of our study aims, both blood culture identification systems showed good results for fast pathogen recognition directly from positive blood cultures as well as for resistance gene detection. The broad coverage three panel approach of the ePlex® system implicate a drawback as it necessitates a Gram-staining prior to decide which cartridge to use for identification. However, the intended use of the other system also requires a Gram-stain as part of their approved instructions for use. Both systems require only short hands-on time, can easily be implemented in a routine microbiological diagnostic workflow and cause similar costs.
The Accelerate Pheno® system, which is already implemented for high risk patients in our blood culture workflow [12], allows accelerating reports for ID and especially full AST for hard to predict resistance profiles for Gram-negatives. The panels of the FilmArray® and ePlex® systems with their ability for detecting common antimicrobial resistance genes are suitable for fast identification and rough genotypic resistance characterization of Gram-positive organisms, specifically for Staphylococcus species demarcating S. aureus and MRSAs and Enterococcus species defining VREs. Specific performance characteristics of the assays, cost-benefit ratio, laboratory operating hours, manning and state of knowledge of the laboratory personnel, patient population as well as effect on patient care from individual identification panels affect the advantages of rapid PCR-based blood culture diagnostics implicating the need of a considered selection and implementation of a rapid molecular ID system [39].
Electronic supplementary material
(DOCX 32 kb)
Acknowledgments
Both companies were given the opportunity to comment on the manuscript. They had no role in the study design, experimental procedure, and data analysis.
We thank Matthias Willmann for critical discussion of the manuscript and the team of diagnostic technicians for their excellent technical assistance.
Funding information
Open Access funding provided by Projekt DEAL. The study was funded in part by the Institute of Medical Microbiology and Hygiene, Medical Faculty, University of Tübingen, Germany. During the study the FilmArray® (Biomerieux, Nürtingen, Germany) and ePlex® (GenMark Diagnostics, Carlsbad, USA) systems were provided by the companies. In addition, the reagents for the ePlex® system were provided, while for the FilmArray® reagent a discount was received during the study period.
Compliance with ethical standards
Conflict of interest
SP received a speakers’ honorarium from Biomerieux (Nürtingen, Germany) and is a consultant for IDbyDNA (San Francisco, USA).
Ethical approval
The study was approved by the local ethics committee (no. 667/2014BO1 and 139/2016BO2).
Informed consent
Not applicable to this study.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
9/1/2020
Table 2 in the originally published article is not correct and is a duplicate of Table 3. The error happened during typesetting. The correct Table 2 is shown below.
References
- 1.Annane D, Bellissant E, Cavaillon JM. Septic shock. Lancet. 2005;365(9453):63–78. doi: 10.1016/S0140-6736(04)17667-8. [DOI] [PubMed] [Google Scholar]
- 2.Mayr FB, Yende S, Angus DC. Epidemiology of severe sepsis. Virulence. 2014;5(1):4–11. doi: 10.4161/viru.27372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leibovici L, Shraga I, Drucker M, Konigsberger H, Samra Z, Pitlik SD. The benefit of appropriate empirical antibiotic treatment in patients with bloodstream infection. J Intern Med. 1998;244(5):379–386. doi: 10.1046/j.1365-2796.1998.00379.x. [DOI] [PubMed] [Google Scholar]
- 4.Adhikari NK, Fowler RA, Bhagwanjee S, Rubenfeld GD. Critical care and the global burden of critical illness in adults. Lancet. 2010;376(9749):1339–1346. doi: 10.1016/S0140-6736(10)60446-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fleischmann C, Scherag A, Adhikari NK, Hartog CS, Tsaganos T, Schlattmann P, Angus DC, Reinhart K, International Forum of Acute Care T Assessment of global incidence and mortality of hospital-treated sepsis. current estimates and limitations. Am J Respir Crit Care Med. 2016;193(3):259–272. doi: 10.1164/rccm.201504-0781OC. [DOI] [PubMed] [Google Scholar]
- 6.Stevenson EK, Rubenstein AR, Radin GT, Wiener RS, Walkey AJ. Two decades of mortality trends among patients with severe sepsis: a comparative meta-analysis*. Crit Care Med. 2014;42(3):625–631. doi: 10.1097/CCM.0000000000000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peters RP, van Agtmael MA, Danner SA, Savelkoul PH, Vandenbroucke-Grauls CM. New developments in the diagnosis of bloodstream infections. Lancet Infect Dis. 2004;4(12):751–760. doi: 10.1016/S1473-3099(04)01205-8. [DOI] [PubMed] [Google Scholar]
- 8.Chang SS, Hsieh WH, Liu TS, Lee SH, Wang CH, Chou HC, Yeo YH, Tseng CP, Lee CC. Multiplex PCR system for rapid detection of pathogens in patients with presumed sepsis - a systemic review and meta-analysis. PLoS One. 2013;8(5):e62323. doi: 10.1371/journal.pone.0062323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wojewoda CM, Sercia L, Navas M, Tuohy M, Wilson D, Hall GS, Procop GW, Richter SS. Evaluation of the Verigene gram-positive blood culture nucleic acid test for rapid detection of bacteria and resistance determinants. J Clin Microbiol. 2013;51(7):2072–2076. doi: 10.1128/JCM.00831-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mancini N, Infurnari L, Ghidoli N, Valzano G, Clementi N, Burioni R, Clementi M. Potential impact of a microarray-based nucleic acid assay for rapid detection of gram-negative bacteria and resistance markers in positive blood cultures. J Clin Microbiol. 2014;52(4):1242–1245. doi: 10.1128/JCM.00142-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altun O, Almuhayawi M, Ullberg M, Ozenci V. Clinical evaluation of the FilmArray blood culture identification panel in identification of bacteria and yeasts from positive blood culture bottles. J Clin Microbiol. 2013;51(12):4130–4136. doi: 10.1128/JCM.01835-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marschal M, Bachmaier J, Autenrieth I, Oberhettinger P, Willmann M, Peter S. Evaluation of the accelerate pheno system for fast identification and antimicrobial susceptibility testing from positive blood cultures in bloodstream infections caused by Gram-negative pathogens. J Clin Microbiol. 2017;55(7):2116–2126. doi: 10.1128/JCM.00181-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barlam TF, Cosgrove SE, Abbo LM, MacDougall C, Schuetz AN, Septimus EJ, Srinivasan A, Dellit TH, Falck-Ytter YT, Fishman NO, Hamilton CW, Jenkins TC, Lipsett PA, Malani PN, May LS, Moran GJ, Neuhauser MM, Newland JG, Ohl CA, Samore MH, Seo SK, Trivedi KK. Executive summary: implementing an antibiotic stewardship program: guidelines by the Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America. Clin Infect Dis. 2016;62(10):1197–1202. doi: 10.1093/cid/ciw217. [DOI] [PubMed] [Google Scholar]
- 14.Bauer KA, Perez KK, Forrest GN, Goff DA. Review of rapid diagnostic tests used by antimicrobial stewardship programs. Clin Infect Dis. 2014;59(Suppl 3):S134–S145. doi: 10.1093/cid/ciu547. [DOI] [PubMed] [Google Scholar]
- 15.Fournier PE, Drancourt M, Colson P, Rolain JM, La Scola B, Raoult D. Modern clinical microbiology: new challenges and solutions. Nat Rev Microbiol. 2013;11(8):574–585. doi: 10.1038/nrmicro3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blaschke AJ, Heyrend C, Byington CL, Fisher MA, Barker E, Garrone NF, Thatcher SA, Pavia AT, Barney T, Alger GD, Daly JA, Ririe KM, Ota I, Poritz MA. Rapid identification of pathogens from positive blood cultures by multiplex polymerase chain reaction using the FilmArray system. Diagn Microbiol Infect Dis. 2012;74(4):349–355. doi: 10.1016/j.diagmicrobio.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldenberger D, Kunzli A, Vogt P, Zbinden R, Altwegg M. Molecular diagnosis of bacterial endocarditis by broad-range PCR amplification and direct sequencing. J Clin Microbiol. 1997;35(11):2733–2739. doi: 10.1128/JCM.35.11.2733-2739.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bell JM, Paton JC, Turnidge J. Emergence of vancomycin-resistant enterococci in Australia: phenotypic and genotypic characteristics of isolates. J Clin Microbiol. 1998;36(8):2187–2190. doi: 10.1128/JCM.36.8.2187-2190.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Southern TR, VanSchooneveld TC, Bannister DL, Brown TL, Crismon AS, Buss SN, Iwen PC, Fey PD. Implementation and performance of the BioFire FilmArray(R) blood culture identification panel with antimicrobial treatment recommendations for bloodstream infections at a midwestern academic tertiary hospital. Diagn Microbiol Infect Dis. 2015;81(2):96–101. doi: 10.1016/j.diagmicrobio.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 20.Becker K, Heilmann C, Peters G. Coagulase-negative staphylococci. Clin Microbiol Rev. 2014;27(4):870–926. doi: 10.1128/CMR.00109-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anguera I, Del Rio A, Miro JM, Matinez-Lacasa X, Marco F, Guma JR, Quaglio G, Claramonte X, Moreno A, Mestres CA, Mauri E, Azqueta M, Benito N, Garcia-de la Maria C, Almela M, Jimenez-Exposito MJ, Sued O, De Lazzari E, Gatell JM, Hospital Clinic Endocarditis Study G. Staphylococcus lugdunensis infective endocarditis: description of 10 cases and analysis of native valve, prosthetic valve, and pacemaker lead endocarditis clinical profiles. Heart. 2005;91(2):e10. doi: 10.1136/hrt.2004.040659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petti CA, Simmon KE, Miro JM, Hoen B, Marco F, Chu VH, Athan E, Bukovski S, Bouza E, Bradley S, Fowler VG, Giannitsioti E, Gordon D, Reinbott P, Korman T, Lang S, Garcia-de-la-Maria C, Raglio A, Morris AJ, Plesiat P, Ryan S, Doco-Lecompte T, Tripodi F, Utili R, Wray D, Federspiel JJ, Boisson K, Reller LB, Murdoch DR, Woods CW, International Collaboration on Endocarditis-Microbiology I Genotypic diversity of coagulase-negative staphylococci causing endocarditis: a global perspective. J Clin Microbiol. 2008;46(5):1780–1784. doi: 10.1128/JCM.02405-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guillaume MP, Dubos F, Godart F. Staphylococcus lugdunensis endocarditis in children. Cardiol Young. 2017;27(4):784–787. doi: 10.1017/S1047951116001657. [DOI] [PubMed] [Google Scholar]
- 24.Flores Umanzor EJ, San Antonio R, Jimenez Britez G, Caldentey G (2016, 2016) Staphylococcus lugdunensis: an unusual and aggressive cause of infective endocarditis. BMJ Case Rep. 10.1136/bcr-2016-217156 [DOI] [PMC free article] [PubMed]
- 25.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57(Pt 1):81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 26.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis. 2004;39(3):309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 27.Maki DG, Agger WA. Enterococcal bacteremia: clinical features, the risk of endocarditis, and management. Medicine. 1988;67(4):248–269. doi: 10.1097/00005792-198807000-00005. [DOI] [PubMed] [Google Scholar]
- 28.Gagetti P, Bonofiglio L, Garcia Gabarrot G, Kaufman S, Mollerach M, Vigliarolo L, von Specht M, Toresani I, Lopardo HA (2018) Resistance to beta-lactams in enterococci. Revista Argentina de microbiologia. 10.1016/j.ram.2018.01.007 [DOI] [PubMed]
- 29.Dawson S. Blood culture contaminants. J Hosp Infect. 2014;87(1):1–10. doi: 10.1016/j.jhin.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 30.Bates DW, Goldman L, Lee TH. Contaminant blood cultures and resource utilization. The true consequences of false-positive results. Jama. 1991;265(3):365–369. doi: 10.1001/jama.1991.03460030071031. [DOI] [PubMed] [Google Scholar]
- 31.Gander RM, Byrd L, DeCrescenzo M, Hirany S, Bowen M, Baughman J. Impact of blood cultures drawn by phlebotomy on contamination rates and health care costs in a hospital emergency department. J Clin Microbiol. 2009;47(4):1021–1024. doi: 10.1128/JCM.02162-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dinkelacker AG, Vogt S, Oberhettinger P, Mauder N, Rau J, Kostrzewa M, Rossen JWA, Autenrieth IB, Peter S, Liese J (2018) Typing and species identification of clinical Klebsiella isolates by Fourier transform infrared spectroscopy and matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 56(11). 10.1128/JCM.00843-18 [DOI] [PMC free article] [PubMed]
- 33.Potter RF, Lainhart W, Twentyman J, Wallace MA, Wang B, Burnham CA, Rosen DA, Dantas G (2018) Population structure, antibiotic resistance, and uUropathogenicity of Klebsiella variicola. mBio 9(6). 10.1128/mBio.02481-18 [DOI] [PMC free article] [PubMed]
- 34.Andrade BG, de Veiga RN, Marin MF, Fonseca EL, Vicente AC. The genome of a clinical Klebsiella variicola strain reveals virulence-associated traits and a pl9-like plasmid. FEMS Microbiol Lett. 2014;360(1):13–16. doi: 10.1111/1574-6968.12583. [DOI] [PubMed] [Google Scholar]
- 35.Martinez-Romero E, Rodriguez-Medina N, Beltran-Rojel M, Toribio-Jimenez J, Garza-Ramos U. Klebsiella variicola and Klebsiella quasipneumoniae with capacity to adapt to clinical and plant settings. Salud publica de Mexico. 2018;60(1):29–40. doi: 10.21149/8156. [DOI] [PubMed] [Google Scholar]
- 36.Ng LS, Kwang LL, Rao S, Tan TY. Anaerobic bacteraemia revisited: species and susceptibilities. Ann Acad Med Singap. 2015;44(1):13–18. [PubMed] [Google Scholar]
- 37.Katchanov J, Asar L, Klupp EM, Both A, Rothe C, Konig C, Rohde H, Kluge S, Maurer FP. Carbapenem-resistant gram-negative pathogens in a German university medical center: prevalence, clinical implications and the role of novel beta-lactam/beta-lactamase inhibitor combinations. PLoS One. 2018;13(4):e0195757. doi: 10.1371/journal.pone.0195757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nordmann P, Naas T, Poirel L. Global spread of Carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2011;17(10):1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.She RC, Bender JM. Advances in rapid molecular blood culture diagnostics: healthcare impact, laboratory implications, and multiplex technologies. J Appl Lab Med. 2019;3(4):617–630. doi: 10.1373/jalm.2018.027409. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 32 kb)