Abstract
Breast cancer (BC) is a common clinical disease and the second leading cause of cancer death in women. Long noncoding RNA (lncRNA) and microRNA (miRNA) are reported to be involved in the development of BC. The present study aimed to investigate whether LncRNA ZFAS1 could regulate the proliferation, invasion and migration of breast cancer cells by targeting miR-589 through PTEN/PI3K/AKT signal pathway. The expression of ZFAS1 and miR-589 in BC cells and transfection effects were determined by RT-qPCR analysis. The abilities of proliferation, colony formation, invasion and migration of breast cancer cells were analyzed by CCK-8 assay, colony formation assay, transwell assay and wound healing assay respectively. The expression of MMP2, MMP9, Bcl2, Bax, cleaved caspase3, PTEN, p-PI3K, p-AKT, PI3K and AKT was detected by Western blot. The flow cytometry analysis was used to detect cell apoptosis. As a result, ZFAS1 expression was increased and miR-589 expression was decreased in BC cells. And, miR-589 was demonstrated to be a target of ZFAS1. ZFAS1 overexpression could inhibit the proliferation, colony formation, invasion and migration of BC cells while miR-589 overexpression could reverse the changes. In addition, ZFAS1 overexpression suppressed the expression of PI3K/AKT signal pathway by activating the PTEN expression while miR-589 overexpression could reverse the changes. Moreover, PTEN is one of the gene targets of miR-589. In conclusion, this study indicated that ZFAS1 inhibited the proliferation, invasion and migration of breast cancer cells by targeting miR-589 through regulating the PTEN/PI3K/AKT signal pathway.
Keywords: LncRNA ZFAS1/miR-589, Proliferation, Invasion, Migration, Breast cancer cells
Introduction
Breast cancer (BC) is a serious threat to women’s health all over the world. According to the relevant data of 188 countries in the global burden of disease (GBD), about 1.8 million are BC patients among the nearly 15 million newly diagnosed cancer patients. These patients accounts for about a quarter of the total number of newly diagnosed cancers in women, ranking the first among women in most countries. In the same year, a total of 464,000 patients died of BC, which is the leading cause of cancer death among women (Marmot et al. 2013; Mokdad et al. 2018). The above epidemiological evidence shows that BC has a high morbidity and mortality rate and brings a great burden to the family and society. Therefore, it is of great significance to search for targets for the treatment of BC.
Long noncoding RNA (lncRNA) usually refers to a class of non-coding RNA with molecular weight greater than 200 nucleotide sequences. In recent years, there have been more and more reports on the correlation between lncRNA and tumor, suggesting that lncRNA is an important factor in the development of tumor (Liu et al. 2015; Wang et al. 2014, 2015). LncRNA can affect the proliferation, apoptosis, migration and invasion of BC tumor cells and participate in the progress of BC by regulating epigenetic modification and key pathways of cell signal transduction (Liu et al. 2015; Askarian-Amiri et al. 2011). The literature on ZFAS1 was first reported in 2011. Askalian-amiri et al. (Askarian-Amiri et al. 2011) found that ZFAS1 was highly expressed in breast tissues, but low expressed in BC tissues. Therefore, it can be speculated that ZFAS1 may be an inhibitor gene of BC.
MicroRNA (miRNA) are small endogenous non-coding RNAs (20–25 nucleotide long). Study has shown that ZFAS1 inhibits the inhibition of its target gene ZEBl, MMPl4 and MMPl6 by binding to miR-150, thereby promoting tumor metastasis (Zhecheng et al. 2015). Bioinformatics analysis was performed and the results of starBase predicted that ZFAS1 could be combined with miR-589. The miR-589 expression was decreased in lung and liver cancer but the expression of miR-589 was abnormally increased in gastric cancer (Liu et al. 2017; Xu et al. 2018; Zhang et al. 2018). However, miR-589 has not been studied in BC cell lines.
The aim of the present study was to explore whether ZFAS1 could regulate the proliferation, invasion and migration of breast cancer cells by targeting miR-589 trough PTEN/PI3K/AKT signal pathway.
Materials and methods
Cell culture
MCF-10A, T47D, MCF-7, MDA-MB-435 and BT-549 cells were bought from American Type Culture Collection (Rockville, MD, USA). These cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco; Thermo Fisher Scientifc, Inc., Waltham, MA, USA) containing 10% fetal bovine serum (FBS) (PAA Laboratories, Inc., Pasching, Austria) which was put into constant temperature oven (37 °C with 5% CO2).
RT-qPCR analysis
The expression of ZFAS1 and miR-589 in breast cancer cells was detected by RT-qPCR analysis. Logarithmic growth phase cells were collected and total RNA was extracted from these cells with Trizol Reagent RNA extraction kit (Invitrogen; Thermo Fisher Scientific, Inc.). cDNA was synthesized with Prime Script RT reagent Kit (Takara Biotechnology Co., Ltd., Beijing, China). 20 µl reaction system for amplification was prepared according to the manufacturer’s protocol of SYBR Premix Ex Taq II (Takara Biotechnology Co., Ltd., Dalian, China). ZFAS1 forward, 5′-ACGTGCAGACATCTACAA-3′ and reverse, 5′-TACTTCCAACACCCGCAT-3′. miR-589 forward, 5′-CGAGGTCAGCGTGATTTCATGG-3′ and reverse, 5′-TGTGTCCAAGTCCCAGCCAGAG-3′. U6 forward, 5′-CTCGCTTCGGCAGCA-CATATACT-3′ and reverse, 5′-ACGCTTCACGAATTTGCGTGTC-3′. GAPDH forward, 5′-GAAGGTGAAGGTCGGAGT-3′ and reverse, 5’-GAAGATGGTGATGGGATT-3′. The PCR reaction conditions were 95 °C for 10 min, followed by 38 cycles of 95 °C for 10 s and 58˚C for 60 s. The RNA expression was quantitatively analyzed by the method of 2−ΔΔCq.
Cell transfection
After MCF-7 cells in 24-well-plate grew into the confluence of 70%–80%, 50 µl serum free medium diluting 3 µl Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) was added to each well to perform the cell transfection. In the control group, cells received no treatment. After transfection for 48 h, RT-qPCR analysis was used to evaluate the transfection effects through the expression changes of ZFAS1 and miR-589.
Dual-luciferase reporter assay
The bioinformatics software of starBase predicted that ZFAS1 could bind specifically to miR-589 and PTEN may be a potential target of miR-589. Dual-luciferase reporter assay was conducted to verify these predictions. Cells were co-transfected with ZFAS1 3′UTR plasmids and miR-589 mimic or mimic control (NC) vector. And, cells were co-transfected with PTEN 3′UTR plasmids and miR-589 mimic or mimic control (NC) vector. Based on Dual Luciferase Assay Kit (Promega), luciferase activity was detected by a dual-luciferase reporter assay system (Promega Corporation, Madison, WI, USA).
CCK-8 assay
After transfection at 24 h, 48 and 72 h, cell proliferation ability was detected by CCK-8 assay. Cells were incubated into the 96 well-plates and 10 µl CCK8 solution (Beyotime, Shanghai, China) was added into each well. After incubation for 1 h, the absorbance value of each well was measured at 450 nm with a Model 680 Microplate Reader (Bio-Rad Laboratories, Inc., Hercules, ca.).
Colony formation assay
After transfection at 24 h, 48 and 72 h, cell colony formation ability was detected by colony formation assay. The cells were inoculated in 6-well plates (1,000 cells/well) and cultured in incubators. When the formation of white colonies was visible to the naked eye, the culture was terminated. The cells were fixed with glacial methanol (Sigma-Aldrich, USA) for 15 min at room temperature and stained with Giemsa (Sigma-Aldrich) for 10 min. The number of colonies was photographed and counted, so as to show the colony formation ability of the cells.
Wound healing assay
Before the experiment, a marker was used to draw a number of parallel straight lines on the back of the 6 well-plate to insure at least 5 parallel straight lines passing through the wells. The cells were then inoculated into a 6 well plate and the groups were control group, pcDNA-NC group, pcDNA-ZFAS1 group, pcDNA-ZFAS1 + miR-NC group and pcDNA-ZFAS1 + miR-589 group. After 24 h inoculation, 10 µl pipette tip scratched perpendicular to previous straight lines. The normal saline was used to rinse the 6 well plate three times and it was replaced by the DMEM medium containing 10% FBS. Cell migration was observed and photographed after scratch at 0 and 24 h.
Transwell assay
Cells of each group were collected after 24 h transfection and digested with trypsin to obtain a single cell suspension which the cell density was 1 × 106/ml. The upper and lower chambers were separated by a polycarbonate microporous membrane (pore size of 8 µm) coated with artificial base gel. 100 µl cell suspension was added to the upper chamber and 500 µl culture medium containing 10% FBS was added to the lower chamber. Then, transwell chambers were incubated in a humid atmosphere with 5% CO2 at 37 °C for 8 h. The polycarbonate microporous membrane was taken down. Cells on the upper surface were gently wiped with a cotton swab, fixed with neutral formaldehyde and stained with hematoxylin. The number of transmembrane cells was recorded under the microscope. The number of transmembrane cells represents the cell invasion ability.
Western blot analysis
Cells were collected after transfection for 48 h and 400 µl of RIPA lysate was added. Cells were beaten repeatedly with pipette tip. The cells were placed at room temperature for 10 min and centrifuged at 12,000 r/min for 5 min. The extracted proteins were determined using BCA Protein Assay Kit (Pierce; Thermo Fisher Scientific, Inc.). Then 25 µg proteins per lane were resolved via 10% SDS-PAGE and transferred to polyvinylidene fluoride membranes (PVDF, Millipore, USA). 5% nonfat milk in TBST (Tris-buffered saline plus 0.1% Tween 20) was blocked the nonspecific binding sites. The membrane was incubated overnight at 4 ˚C with primary antibody against MMP2 (#40994), MMP9 (#13667), Bcl2 (#4223), Bax (#5023), cleaved caspase3 (#9664), PTEN (#9552), p-PI3K (#17366), PI3K (#4257), p-AKT (#4060), AKT (#4691) and GAPDH (#5174) (dilution, 1:1000; Cell Signaling Technology, Inc. Danvers, Massachusetts, USA). After washing with TBST, membrane was incubated with rabbit horseradish peroxidase-linked IgG second antibody for 30 min at room temperature. The gray values of protein bands were analyzed by Quantity one 4.0 software.
Flow cytometry analysis
After transfection for 48 h, cells were washed twice with PBS. 250 µl binding buffer was used to suspend cells to adjust cell concentration to 5 × 105/ml. 200 µl cell suspension was added with 10 µl Annexin-FITC and 10 µl 20 µg/ml propidium iodide (PI) (BD Biosciences, Franklin Lakes, NJ, USA) and incubated at room temperature in dark for 10 min. Then, harvested cells were resuspended in 500 µl binding buffer. The cell apoptosis was analyzed by flow cytometry.
Statistical analysis
In this study, all the experiments were independently repeated for 3 times. SPSS 20.0 software was used for statistical analysis of the study result data. The measurement data were expressed as mean ± standard deviation. Single factor analysis of variance (ANOVA) was used for data comparison between multiple groups, and LSD-t test was used for pairwise comparison within groups. P < 0.05 was considered statistically significant.
Results
Expression of ZFAS1 and miR-589 in breast cancer cells
The expression of ZFAS1 and miR-589 was analyzed by RT-qPCR. Figure 1a, b showed that the ZFAS1 expression was decreased and miR-589 expression was increased in T47D, MCF-7, MDA-MB-435 and BT-549 cells compared with that in MCF-10A cells. The lowest ZFAS1 expression and highest miR-589 expression were showed in MCF-7 cells. The results from starBase predicted that ZFAS1 could directly target miR-589 (Fig. 1c). The dual-luciferase reporter assay showed that the relative luciferase activity was obviously decreased in MCF-7 cells co-transfected with ZFAS1 WT and miR-589 mimic (Fig. 1d).
Fig. 1.
Expression of ZFAS1 and miR-589 in breast cancer cells. a The expression of ZFAS1 in breast cancer cells was analyzed by RT-qPCR. ***P < 0.001 vs. MCF-10A group. b The expression of miR-589 in breast cancer cells was analyzed by RT-qPCR. *P < 0.05 and ***P < 0.001 vs. MCF-10A group. c The binding sites of ZFAS1 and miR-589 were predicted by starBase. d Luciferase activity was analyzed in cells co-transfected with miR-589 mimic or miR-NC with pcDNA-ZFAS1-WT or pcDNA-ZFAS1-MUT. ***P < 0.001 vs. control group and miR-NC group
ZFAS1 inhibits the proliferation of breast cancer cells and miR-589 reverses the ZFAS1 effects
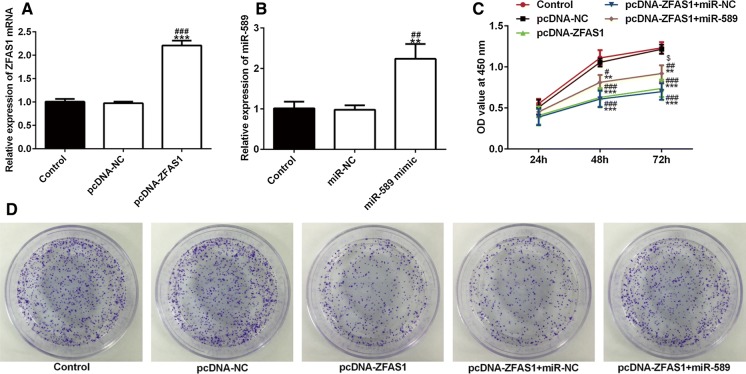
After MCF-7 cells were transfected with pcDNA-ZFAS1 or miR-589 mimic, the expression of ZFAS1 and miR-589 was all significantly up-regulated (Fig. 2a, b). After transfection at 24 h, 48 and 72 h, ZFAS1 overexpression suppressed the cell proliferation and miR-589 overexpression reversed the effects of ZFAS1 overexpression (Fig. 2c). The cell colony forming ability was obviously decreased after cells were transfected with pcDNA-ZFAS1 and pcDNA-ZFAS1 + miR-NC and the cell colony forming ability was up-regulated when cells were transfected with pcDNA-ZFAS1 + miR-589 (Fig. 2d). Above all, ZFAS1 inhibits cell proliferation and colony forming ability, which was reversed by miR-589 overexpression.
Fig. 2.
ZFAS1 inhibits the proliferation of breast cancer cells and miR-589 reverses the ZFAS1 effects. a The expression of ZFAS1 was analyzed by RT-qPCR after cell transfection. ***P < 0.001 vs. control group. ###P < 0.001 vs. pcDNA-NC group. b The expression of miR-589 was analyzed by RT-qPCR after cell transfection. **P < 0.01 vs. control group. ##P < 0.01 vs. pcDNA-NC group. c The viability of transfected cells was detected by CCK-8 assay. **P < 0.01 and ***P < 0.001 vs. control group. #P < 0.05, ##P < 0.01 and ###P < 0.001 vs. pcDNA-NC group. $P < 0.05 vs. pcDNA-ZFAS1 + miR-NC group. d The cell colony forming ability of transfected cells was detected by clone formation assay
ZFAS1 inhibits the invasion and migration of breast cancer cells and miR-589 reverses the ZFAS1 effects
To observe the cell invasion and migration after transfection, wound healing assay and transwell assay were conducted respectively(Fig. 3a, d). The decreased invasion and migration of breast cancer cells occurred in cells transfected with pcDNA-ZFAS1 and pcDNA-ZFAS1 + miR-NC while cells transfected with pcDNA-ZFAS1 + miR-589 showed increased invasion and migration. However, the cell invasion and migration in pcDNA-ZFAS1 + miR-589 group was still lower than that in control group and pcDNA-NC group. The western blot analysis presented that the expression of MMP2 and MMP9 was down-regulated in cells transfected with pcDNA-ZFAS1 and pcDNA-ZFAS1 + miR-NC while obviously up-regulated in cells transfected with pcDNA-ZFAS1 + miR-589 (Fig. 3e). These data indicated that ZFAS1 overexpression could suppress the cell invasion and migration and miR-589 overexpression could alleviate the role of ZFAS1 overexpression.
Fig. 3.
ZFAS1 inhibits the invasion and migration of breast cancer cells and miR-589 reverses the ZFAS1 effects. a, b The cell migration was observed by wound healing assay. **P < 0.01 and ***P < 0.001 vs. control group. ##P < 0.01 and ###P < 0.001 vs. pcDNA-NC group. ∆P < 0.05 vs. pcDNA-ZFAS1 group. $P < 0.05 vs. pcDNA-ZFAS1 + miR-NC group. c, d The cell invasion was observed by transwell assay. **P < 0.01 and ***P < 0.001 vs. control group. ##P < 0.01 and ###P < 0.001 vs. pcDNA-NC group. ∆∆P < 0.01 vs. pcDNA-ZFAS1 group. $$P < 0.01 vs. pcDNA-ZFAS1 + miR-NC group. e The expression of MMP2 and MMP9 was detected by western blot analysis. *P < 0.05 and ***P < 0.001 vs. control group. #P < 0.05 and ###P < 0.001 vs. pcDNA-NC group. ∆∆P < 0.01 vs. pcDNA-ZFAS1 group. $$P < 0.01 vs. pcDNA-ZFAS1 + miR-NC group
ZFAS1 promotes apoptosis of breast cancer cells and miR-589 reverses the ZFAS1 effects
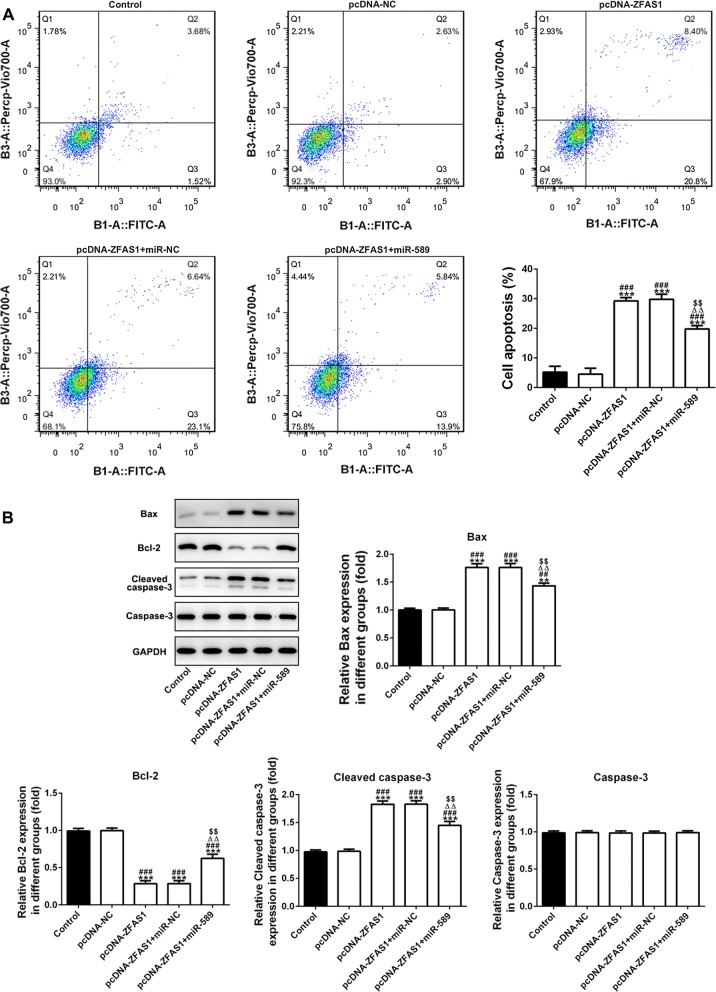
The cell apoptosis was analyzed by the flow cytometry analysis and the data showed that cell apoptosis was higher in pcDNA-ZFAS1 and pcDNA-ZFAS1 + miR-NC group than that in control group and pcDNA-NC group. The cell apoptosis was decreased in cells co-transfected with pcDNA-ZFAS1 and miR-589 compared with cells in pcDNA-ZFAS1 and pcDNA-ZFAS1 + miR-NC group (Fig. 4a). The expression of Bcl2 was decreased and the expression of Bax and cleaved caspase3 was increased in pcDNA-ZFAS1 and pcDNA-ZFAS1 + miR-NC group compared with that in control group and pcDNA-NC group. However, the expression of Bcl2 was increased and the expression of Bax and cleaved caspase3 was decreased in pcDNA-ZFAS1 + miR-589 group compared with that in pcDNA-ZFAS1 and pcDNA-ZFAS1 + miR-NC group (Fig. 4b).
Fig. 4.
ZFAS1 promotes apoptosis of breast cancer cells and miR-589 reverses the ZFAS1 effects. a The cell apoptosis was analyzed by flow cytometry analysis. ***P < 0.001 vs. control group. ###P < 0.001 vs. pcDNA-NC group. ∆∆P < 0.01 vs. pcDNA-ZFAS1 group. $$P < 0.01 vs. pcDNA-ZFAS1 + miR-NC group. b The apoptosis related expression was detected by western blot analysis. **P < 0.01 and ***P < 0.001 vs. control group. ##P < 0.01 and ###P < 0.001 vs. pcDNA-NC group. ∆∆P < 0.01 vs. pcDNA-ZFAS1 group. $$P < 0.01 vs. pcDNA-ZFAS1 + miR-NC group
ZFAS1 activates PTEN to inhibit the expression of PI3K/AKT and miR-589 reverses the ZFAS1 effects
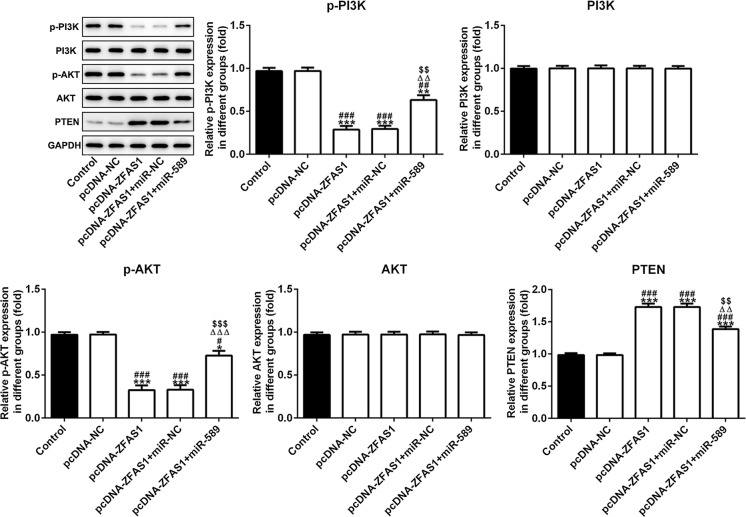
After cells were transfected with pcDNA-ZFAS1 or pcDNA-ZFAS1 + miR-NC, the PTEN was activated, and the expression of p-PI3K and p-AKT was increased. However, miR-589 overexpression inhibited the PTEN expression to up-regulate the expression of p-PI3K and p-AKT in pcDNA-ZFAS1 + miR-589 mimic group (Fig. 5).
Fig. 5.
ZFAS1 activates PTEN to inhibit the expression of PI3K/AKT and miR-589 reverses the ZFAS1 effects. The expression of PTEN, p-PI3K, p-AKT, PI3K and AKT was detected by western blot analysis. *P < 0.05, **P < 0.01 and ***P < 0.001 vs. control group. #P < 0.05, ##P < 0.01 and ###P < 0.001 vs. pcDNA-NC group. ∆∆∆P < 0.001 vs. pcDNA-ZFAS1 group. $$$P < 0.001 vs. pcDNA-ZFAS1 + miR-NC group
MiR-589 directly targets PTEN
Bioinformatics analysis was performed and the results of miRDB predicted that miR-589 could be combined with PTEN (Fig. 6a). The dual-luciferase reporter assay indicated that the relative luciferase activity was not obviously changed when cells were co-transfected with miR-NC and WT-PTEN or miR-NC and Mut-PTEN. The relative luciferase activity was significantly decreased when cells were co-transfected with miR-589 mimic and WT- PTEN, while the activity did not obviously change when cells were co-transfected with miR-589 mimic and MUT-PTEN (Fig. 6b). Above all, miR-589 directly targets PTEN.
Fig. 6.

MiR-589 directly targets PTEN. a The binding sites of ZFAS1 and miR-589 were predicted by miRDB. b Luciferase activity was analyzed in cells co-transfected with miR-589 mimic or miR-NC with PTEN-WT or PTEN-MUT. ***P < 0.001 vs. control group and miR-Control group
Discussion
Here, we explored whether ZFAS1 could regulate the proliferation, invasion and migration of breast cancer cells by targeting miR-589 trough PTEN/PI3K/AKT signal pathway. It was shown that ZFAS1 inhibited the proliferation, invasion and migration of breast cancer cells by targeting miR-589 through suppressing the PI3K/AKT signal pathway.
Literature has Mo et al. (2019) found that ZFAS1 inhibited the cell proliferation, invasion and migration by the knockdown of miR-7-5p in colorectal cancer. Dong et al. (2019) demonstrated that silencing of ZFAS1 obviously suppressed the cell proliferation, migration and invasion of clear cell renal cell carcinoma (ccRCC). Ye et al. (2018) indicated that ZFAS1 promoted cell migration and invasion by targeting the miR-27a in rheumatoid arthritis. This shows that the effects of ZFAS1 on the cell proliferation, invasion and migration are different in various diseases. In this study, ZFAS1 was studied to be decreased in breast cancer cells and ZFAS1 overexpression showed the inhibition effect on the cell proliferation, invasion and migration.
The dual-luciferase reporter assay demonstrated that ZFAS1 directly targeted miR-589. Zhu et al. (2018) presented that miR-589-5p knockdown promoted cell proliferation and migration by increasing the expression of ZMYND19 in hepatocellular carcinoma. miR-589-3p also functioned in the regulation of glioblastoma cell migration and invasion (Cesarini et al. 2018). miR-589 was found up-regulated in breast cancer cells and miR-589 overexpression could attenuate the effects of ZFAS1 overexpression on the cell proliferation, invasion and migration in breast cancer. PTEN was a potential target of miR-589. PTEN, a tumor suppressor gene, can make PIP3 dephosphorylate to maintain the low level of PIP3, thereby downregulating the PI3K/AKT pathway. The inactivation of PTEN must lead to the activation of PI3K/AKT pathway (Colakoglu et al. 2008). Activated AKT can promote tumor cell growth and proliferation, inhibit apoptosis, promote invasion and metastasis, and regulate endothelial growth and angiogenesis through catalyzing the catalytic phosphorylation of a series of proteins (Cheng et al. 2008; Kim et al. 2001; Manning and Cantley 2007; Mayo and Donner 2002). The present results indicated that ZFAS1 inhibited the cell proliferation, invasion and migration and promoted cell apoptosis by targeting miR-589 by activating PTEN to suppress the PI3K/AKT pathway.
At present, PTEN/PI3K/AKT pathway has been reported to regulate the cell proliferation, invasion, migration and apoptosis in several cancers. Cryptotanshinone inhibited bladder cancer cell proliferation, migration, and invasion and promoted apoptosis partly due to activated PTEN suppressing the PI3K/AKT/mTOR pathway (Yadong et al. 2020). PCAT18 suppressed the cell viability by increasing the ratio of p-PI3K to PI3K and p-AKT to AKT (Chen et al. 2019). MiR-203a-3p decreased the PTEN expression to activate the PI3K/AKT signal pathway for the promotion of cell proliferation (Zhang et al. 2019). Tetrandrine suppressed the proliferation of colon cancer cells (SW620 cells) partly by increasing BMP9 expression, followed by inhibiting PI3K/AKT signal pathway mediated by activated PTEN (Zhou et al. 2019). In this study, PTEN/PI3K/AKT signal pathway was also associated with the biological behaviors of breast cancer cells.
In conclusion, a novel function of ZFAS1 and miR-589 has been verified in breast cancer. Furthermore, ZFAS1 inhibits the cell proliferation, invasion and migration and promotes cell apoptosis by targeting miR-589, and miR-589 overexpression reversed these effects. ZFAS1 also can activate PTEN to suppress the PI3K/AKT pathway, thereby regulating the cell biological behavior. And, PTEN is a potential target of miR-589. Therefore, ZFAS1/miR-589/PTEN axis is important in the mechanism of breast cancer cells and it provides a new sight to treat the breast cancer. However, there still exists a limitation of one cell lines used for this experiment. This experiment is a preliminary basic experiment and we will use two more cell lines to perform the deep further study.
Acknowledgements
Not applicable.
Funding
Science and Technology Development Fund Project of Bengbu Medical College (BYKF1866). National Natural Science Foundation Item (81572992).
Complicance with ethical standards
Conflict of interest
The authors declare they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Askarian-Amiri M, Crawford J, French J, Smart C, Smith M, Clark B, Ru M, Mercer KR, Thompson TR, et al. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA. 2011;17:878–891. doi: 10.1261/rna.2528811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesarini V, Silvestris DA, Tassinari V, Tomaselli S, Alon S, Eisenberg E, Locatelli F, Gallo A. ADAR2/miR-589-3p axis controls glioblastoma cell migration/invasion. Nucleic Acids Res. 2018;46:2045–2059. doi: 10.1093/nar/gkx1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Zhao X, Wang H, Zheng M, Wang Q, Chang W. The down-regulation of lncRNA PCAT18 promotes the progression of gastric cancer via MiR-107/PTEN/PI3K/AKT signaling pathway. Onco Targets Ther. 2019;12:11017–11031. doi: 10.2147/OTT.S225235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng G, Park S, He S, Kong L, Zhang W, Yuan W, Wang Z, Cheng L. Advances of AKT pathway in human oncogenesis and as a target for anti-cancer drug discovery. Curr Cancer Drug Targets. 2008;8:1. doi: 10.2174/156800908783497104. [DOI] [PubMed] [Google Scholar]
- Colakoglu T, Yildirim S, Kayaselcuk F, Nursal TZ, Ezer A, Noyan T, Karakayali H, Haberal M. Clinicopathological significance of PTEN loss and the phosphoinositide 3-kinase/Akt pathway in sporadic colorectal neoplasms: is PTEN loss predictor of local recurrence? Am J Surg. 2008;195:719–725. doi: 10.1016/j.amjsurg.2007.05.061. [DOI] [PubMed] [Google Scholar]
- Dong D, Mu ZY, Wei N, Sun ML, Wang W, Xin N, Shao Y, Zhao CH. Long non-coding RNA ZFAS1 promotes proliferation and metastasis of clear cell renal cell carcinoma via targeting miR-10a/SKA1 pathway. Biomed Pharmacother. 2019;111:917–925. doi: 10.1016/j.biopha.2018.12.143. [DOI] [PubMed] [Google Scholar]
- Kim D, Kim S, Yoon H, Chung SO, Cho AS, Chung KS. Akt/PKB promotes cancer cell invasion via increased motility and metalloproteinase production. Faseb J Off Publi Feder Am Soc Exp Biol. 2001;15:1953. doi: 10.1096/fj.01-0198com. [DOI] [PubMed] [Google Scholar]
- Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z, Deng X, Chen H, Shen B, Peng C, Li H. Amplification of long noncoding RNA ZFAS1 promotes metastasis in hepatocellular carcinoma. Cancer Res. 2015;75:3181–91. doi: 10.1158/0008-5472.CAN-14-3721. [DOI] [PubMed] [Google Scholar]
- Liu Y, Sharma S, Watabe K. Roles of lncRNA in breast cancer. Front Biosci. 2015;7:94–108. doi: 10.2741/s427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CH, Lv DS, Li M, Zhang XF, Sun G, Bai Y, Chang DM. Hypermethylation of miRNA-589 promoter leads to upregulation of HDAC5 which promotes malignancy in non-small cell lung cancer. Int J Oncol. 2017;50:2079–2090. doi: 10.3892/ijo.2017.3967. [DOI] [PubMed] [Google Scholar]
- Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmot MG, Altman DG, Cameron DA, Dewar JA, Thompson SG, Wilcox M. The benefits and harms of breast cancer screening: an independent review. British Journal Of Cancer. 2013;108:2205. doi: 10.1038/bjc.2013.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayo LD, Donner DB. The PTEN, Mdm2, p53 tumor suppressor–oncoprotein network. Trends Biochem Sci. 2002;27:462–467. doi: 10.1016/S0968-0004(02)02166-7. [DOI] [PubMed] [Google Scholar]
- Mo DJ, Liu WW, Li YQ, Cui WB. Long non-coding RNA zinc finger antisense 1 (ZFAS1) regulates proliferation, migration, invasion, and apoptosis by targeting MiR-7-5p in colorectal cancer. Med Sci Monit. 2019;25:5150–5158. doi: 10.12659/MSM.916619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokdad A, Ballestros K, Echko M, Glenn S, Olsen E, Mullany H, Lee E, Khan A, Ahmadi A, Ferrari AA, et al. The State of US Health, 1990–2016: burden of diseases, injuries, and risk factors among US states. JAMA. 2018;19:1444–1472. doi: 10.1001/jama.2018.0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G-Y, Zhu Y-Y, Zhang Y-Q. The functional role of long non-coding RNA in digestive system carcinomas. Bull Cancer. 2014;101:E27–31. doi: 10.1684/bdc.2014.2023. [DOI] [PubMed] [Google Scholar]
- Wang J, Lei Z-J, Guo Y, Wang T, Qin Z-Y, Xiao H-L, Fan L-L, Chen D-F, Bian X. miRNA-regulated delivery of lincRNA-p21 suppresses β-catenin signaling and tumorigenicity of colorectal cancer stem cells. Oncotarget. 2015;6:37852. doi: 10.18632/oncotarget.5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Wang Y, He HT, Yang Q. MiR-589-5p is a potential prognostic marker of hepatocellular carcinoma and regulates tumor cell growth by targeting MIG-6. Neoplasma. 2018;65:753–761. doi: 10.4149/neo_2018_171125N762. [DOI] [PubMed] [Google Scholar]
- Yadong L, Fanlu L, Yaodong C, Rui W, Jiannan L, Yinshan J, Ruihua A. Cryptotanshinone inhibites bladder cancer cell proliferation and promotes apoptosis via the PTEN/PI3K/AKT pathway. J Cancer. 2020;11:488–499. doi: 10.7150/jca.31422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Gao XM, Yang NP. LncRNA ZFAS1 promotes cell migration and invasion of fibroblast-like synoviocytes by suppression of miR-27a in rheumatoid arthritis. Hum Cell. 2018;31:14–21. doi: 10.1007/s13577-017-0179-5. [DOI] [PubMed] [Google Scholar]
- Zhang F, Li K, Pan M, Li W, Wu J, Li M, Zhao L, Wang H. miR-589 promotes gastric cancer aggressiveness by a LIFR-PI3K/AKT-c-Jun regulatory feedback loop. J Exp Clin Cancer Res. 2018;37:152. doi: 10.1186/s13046-018-0821-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zheng Y, Zhu G. MiR-203a-3p targets PTEN to promote hepatocyte proliferation by regulating PI3K/Akt pathway in BRL-3A cells. Biosci Biotechnol Biochem. 2019;1:1–9. doi: 10.1080/09168451.2019.1694860. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Mu L, Liu X-L, Li Q, Ding L-X, Chen H-C, Hu Y, Li F-S, Sun W-J, He B-C, et al. Tetrandrine inhibits proliferation of colon cancer cells by BMP9/ PTEN/ PI3K/AKT signaling. Genes Dis. 2019 doi: 10.1016/j.gendis.2019.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Luo ZH, Lu GY, Gui FF, Wu JH, Li FM, Ni Y. LncRNA FABP5P3/miR-589-5p/ZMYND19 axis contributes to hepatocellular carcinoma cell proliferation, migration and invasion. Biochem Biophys Res Commun. 2018;498:551–558. doi: 10.1016/j.bbrc.2018.03.017. [DOI] [PubMed] [Google Scholar]