Abstract
Background
The genetic background of patients with hypertrophic cardiomyopathy (HCM) treated with alcohol septal ablation (ASA) and its relationship to the outcomes are not known. We aimed to investigate whether the outcome of genotype positive (G+) patients differs from genotype negative (G−) patients treated with ASA.
Methods
We included 129 HCM patients (mean age 54±13 years) treated with ASA in a tertiary cardiovascular center and performed next generation sequencing (NGS) based genomic testing. All patients were followed-up three months after the procedure and yearly thereafter.
Results
A total of 30 (23%) HCM patients were G+ patients. At the 3-months follow-up, both groups of patients had similar left ventricular outflow tract PG (16.9±15.7 mmHg in G+ vs. 16.3±18.8 mmHg in G−, P=0.73) and symptoms (follow-up NYHA class 1.40±0.62 vs. 1.37±0.53, P=0.99, follow-up CCS class 0.23±0.52 vs. 0.36±0.65, P=0.36). The independent predictors of all-cause mortality were baseline interventricular septum (IVS) thickness (HR 1.12, 95% CI: 1.00–1.26, P=0.049) and age at the time of ASA (HR 1.11, 95% CI: 1.06–1.17, P<0.01). The adjusted all-cause mortality rate did not differ significantly between G+ and G− patients (P=0.52). The adjusted combined mortality event rate did not differ between both groups (P=0.78).
Conclusions
Despite more severe phenotype in G+ HCM patients, ASA is an equally effective treatment for LVOTO in G+ patients as it is for treating LVOTO in G− patients. The long-term outcome after ASA is similar in G+ and G− patients.
Keywords: Cardiomyopathy, hypertrophic cardiomyopathy (HCM), hypertrophic obstructive cardiomyopathy, alcohol septal ablation (ASA), genetics
Introduction
Hypertrophic cardiomyopathy (HCM) is a heterogeneous condition in both its genetic origin and phenotypic features (1). Two-thirds of HCM patients have left ventricular (LV) outflow tract obstruction (LVOTO) (2,3). Alcohol septal ablation (ASA) is a safe and effective method of treating LVOTO (4,5). The genetic background of patients treated by ASA and its possible relationship with the outcomes of the procedure are not known. The aim of this study was to investigate whether the outcome of genotype positive (G+) patients differs from genotype negative (G−) patients in a highly symptomatic group of HCM patients treated with ASA.
Methods
Study population
We included 129 consecutive unrelated patients with a clinical diagnosis of HCM in a single tertiary cardiovascular center. The patients were treated with ASA between 1998 and 2017. An HCM diagnosis was established by experienced cardiologists based on a clinical examination, electrocardiography (ECG), and findings of LV hypertrophy ≥15 mm on echocardiography and/or magnetic resonance imaging (2,3). Secondary hypertrophy attributable to aortic valve stenosis or amyloidosis was excluded. When patients presented with mild concomitant systemic hypertension, the HCM echocardiography specialists had to claim the hypertension to be either controlled or the severity insufficient to cause the degree of LV hypertrophy. Symptomatic patients [New York Heart Association (NYHA) class III–IV or syncope on exertion] with significant LVOTO (maximal gradient at rest or during physiological provocation ≥50 mmHg), despite maximal tolerated pharmacotherapy, were offered septal reduction therapy. ASA was indicated after a careful assessment by a multidisciplinary heart team, in addition to local experience with ASA, and the patient’s preference. All ASA procedures were performed by a single operator as previously described (6-8), and all procedures were guided by myocardial contrast echocardiography. A temporary pacemaker lead was placed in the right ventricle in all patients without previous permanent pacemaker implantation. Patients were observed in the coronary care unit for at least 48 hours, and the temporary pacemaker lead was then removed if no episode of high-degree atrioventricular block occurred. All the patients remained on continuous ECG monitoring for up to seven days until discharge. Basic demographic, clinical, and echocardiographic data were collected at baseline, and the patients were followed up three months after the ASA procedure and yearly thereafter. The study protocol conforms to the ethical guidelines of the Declaration of Helsinki (2000, Fifth revision) and was approved by the institutional ethical committee. Written informed consent was obtained from all patients.
Genetic testing
The DNA samples for next generation sequencing (NGS) testing were obtained between 2005 and 2017. Genetic testing was performed in all patients treated with ASA who provided written informed consent. Genomic DNA was isolated from whole blood of all included patients. Our targeted NGS enrichment panel and methods of variant identification, prioritization, and classification were described in detail in our previous paper (9). All identified variants were classified according to the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP) guidelines (10). Patients, whose variants were classified as pathogenic or likely pathogenic (P/LP), were marked as genotype positive (G+ patients). The rest of the cohort, including patients with variants of unknown significance, was considered genotype negative (G− patients).
Statistical analysis
Data are presented as mean ± standard deviation (SD) for continuous variables and proportions for categorical variables. The Student’s t-test, Mann-Whitney test, and Fisher’s exact test were used where appropriate. Cox proportional hazards regression was used to identify predictors of mortality events. The following clinical and echocardiographic variables with potential impact on mortality events were first evaluated in a univariate model: age at the time of ASA, sex, LVOTO, interventricular septum (IVS) thickness, and left atrial diameter. Variables with P values <0.15 were then entered into a multivariable analysis, which was performed using backward stepwise Cox regression. The long-term occurrence of mortality was estimated using the Kaplan-Meier method, and differences between groups were assessed by the log-rank test. Kaplan-Meier curves of G+ and G− patients were adjusted for age at the time of ASA and baseline IVS. The level of statistical significance was set to 0.05. The Prism v.8.1.1 (GraphPad Software Inc., USA) statistical software was used for statistical analysis.
Results
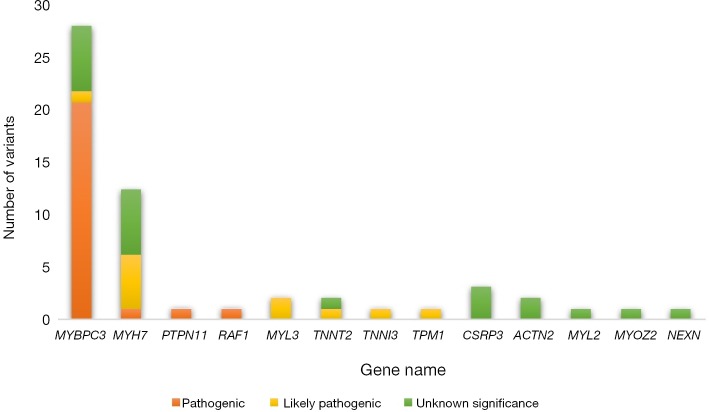
The baseline characteristics of all 129 patients are shown in Table 1. In 53 (41%) HCM patients, we identified 68 genetic variants in 25 different genes. The complete list of genes is shown in Table S1. Thirty of these variants (localized in 8 genes) identified in 30 (23%) HCM patients were classified as P/LP. The distribution of genetic variants is shown in Figure 1, and patients sorted by classified variants are shown in Table 2. Variants were identified in two major genes, MYBPC3 and MYH7, wherein the dominant role of MYBPC3 was even more apparent in the group of P/LP genetic variants (62% of identified variants).
Table 1. Clinical and echocardiographic data at baseline and during follow-up in G+ compared to G− patients.
| Genotype+ (n=30) | Genotype– (n=99) | P value | |
|---|---|---|---|
| Male sex [%] | 19 [63] | 46 [46] | 0.10 |
| Age at ASA, years | 47.3±12.3 | 58.8±11.3 | <0.01 |
| LVEF baseline, % | 78.7±5.6 | 79.4±5.7 | 0.48 |
| LVEF follow-up, % | 70.3±9.3 | 72.5±7.3 | 0.34 |
| LVEDD baseline, mm | 40.1±4.9 | 43.6±4.6 | <0.01 |
| LVEDD follow-up, mm | 45.6±4.8 | 47.2±4.7 | 0.11 |
| Angina, CCS class baseline | 1.9±1.2 | 1.3±1.0 | 0.46 |
| Angina, CCS class follow-up | 0.2±0.5 | 0.4±0.7 | 0.36 |
| Dyspnoea, NYHA class baseline | 2.8±0.6 | 2.8±0.5 | 0.81 |
| Dyspnoea, NYHA class follow-up | 1.4±0.6 | 1.4±0.5 | 0.99 |
| Episodes of syncope baseline [%] | 4 [13] | 14 [14] | 0.99 |
| Episodes of syncope follow-up [%] | 1 [3] | 13 [13] | 0.19 |
| IVS thickness baseline, mm | 23.9±5.0 | 20.3±3.6 | <0.01 |
| IVS thickness follow-up, mm | 15.5±4.0 | 13.0±4.0 | <0.01 |
| LVOTO baseline, mmHg | 63.6±31.4 | 63.5±40.2 | 0.50 |
| LVOTO follow-up, mmHg | 16.9±15.7 | 16.3±18.8 | 0.73 |
| Pacemaker implanted before ASA | 2 [7] | 4 [4] | 0.62 |
| ICD implanted before ASA | 2 [7] | 3 [3] | 0.33 |
LVEF, left ventricular ejection fraction; LVEDD, left ventricular enddiastolic diameter; NYHA, New York Heart Association; CCS, Canadian Cardiovascular Society; IVS, interventricular septum; LVOTO, left ventricular outflow tract obstruction.
Figure 1.
The distribution of genetic variants in HCM patients treated with ASA. HCM, hypertrophic cardiomyopathy; ASA, alcohol septal ablation.
Table 2. Patients sorted by identified variant class (N=129).
| Variant classification | Number of patients [%] |
|---|---|
| Pathogenic or likely pathogenic | 30 [23] |
| Unknown significance | 23 [18] |
| No significant findings* | 76 [59] |
*, includes benign and likely benign variants.
Compared to G− patients, G+ patients were treated with ASA at a younger age (47.3±12.3 vs. 58.8±11.3 years, P<0.01), had greater hypertrophy of the IVS (23.9±5.0 vs. 20.3±3.6 mm, P<0.01), and a smaller LV end-diastolic diameter (40.1±4.9 vs. 43.6±4.6 mm, P<0.01).
At baseline, both groups had similar maximal LVOT pressure gradients (PG) (63.6±31.4 vs. 63.5±40.2 mmHg, P=0.50) and symptoms (NYHA class 2.77±0.57 vs. 2.81±0.49, P=0.81; Canadian Cardiovascular Society (CCS) class 1.93±1.17 vs. 1.83±1.04, P=0.45).
At the three-month follow-up, both groups of patients had similar maximal LVOT PG (16.9±15.7 mmHg in the G+ group vs. 16.3±18.8 mmHg in the G− group, P=0.73) and symptoms (follow-up NYHA class 1.40±0.62 vs. 1.37±0.53, P=0.99, follow-up CCS class 0.23±0.52 vs. 0.36±0.65, P=0.36).
Early adverse events
None of the patients died or suffered a stroke within 30 days after the procedure. Sustained ventricular tachycardia/ventricular fibrillation requiring urgent electrical cardioversion during the hospital stay occurred in 5 patients (4%). Either transient or persistent complete heart block was documented in 22 patients (17%), and a new permanent pacemaker was implanted in 9 patients (7%) during the hospital stay. Access site complication occurred in 2 patients (2%). There were no significant differences in complication rate within 30 days between G+ and G− patients (Table 3).
Table 3. Incidence of complications within 30 days after ASA in G+ and G− patients.
| Adverse event | Genotype+ (n=30) | Genotype− (n=99) | P value |
|---|---|---|---|
| Death [%] | 0 [0] | 0 [0] | NA |
| Stroke [%] | 0 [0] | 0 [0] | NA |
| Ventricular tachycardia/fibrillation [%] | 1 [3] | 4 [4] | 0.99 |
| Complete heart block [%] | 2 [7] | 20 [20] | 0.10 |
| Permanent pacemaker implantation [%] | 1 [3] | 8 [8] | 0.68 |
| Cardiopulmonary resuscitation [%] | 1 [3] | 4 [4] | 0.99 |
| Vascular access complication [%] | [0] | 2 [2] | 0.99 |
Long-term survival
The median (interquartile range) follow-up in the survival analysis was 9.1 (6.3–12.9) years. The mean follow-up in the survival analysis was 9.9±4.8 years. A total of 21 patients (16.3%) died during 1,211 patient-years of follow-up, which resulted in an all-cause mortality rate of 1.7 deaths per 100 patient-years. In the G+ group, 2 (6.7%) of 30 patients died during follow-up. In the G− group during follow-up, 19 (19.2%) of 99 patients died. The unadjusted all-cause mortality rate did not differ significantly between G+ and G− patients (P=0.087), as shown in Figure 2. The independent predictors of all-cause mortality were baseline IVS thickness (HR 1.12, 95% CI: 1.00–1.26, P=0.049) and age at the time of ASA (HR 1.11, 95% CI: 1.06–1.17, P<0.001). The all-cause mortality rate adjusted for age at the time of ASA and baseline IVS thickness did not differ significantly between the G+ and G− patients (P=0.288), as shown in Figure 3. Combined mortality event endpoint [all-cause mortality and appropriate implantable cardioverter-defibrillator (ICD) discharge] occurred in 24 patients (18.6%) during 1,211 patient-years of follow-up, resulting in an event rate of 2.0 per 100 patient-years. The unadjusted combined mortality event rate did not differ significantly between the G+ and the G− groups (P=0.518), as shown in Figure 4. The independent predictors of combined mortality event rate were IVS thickness (HR 1.14, 95% CI: 1.03–1.26, P=0.011) and age at the time of ASA (HR 1.07, 95% CI: 1.03–1.12, P<0.001). The combined mortality event rate adjusted for age at the time of ASA and baseline IVS thickness did not differ significantly between G+ and G− patients (P=0.777), as shown in Figure 5.
Figure 2.
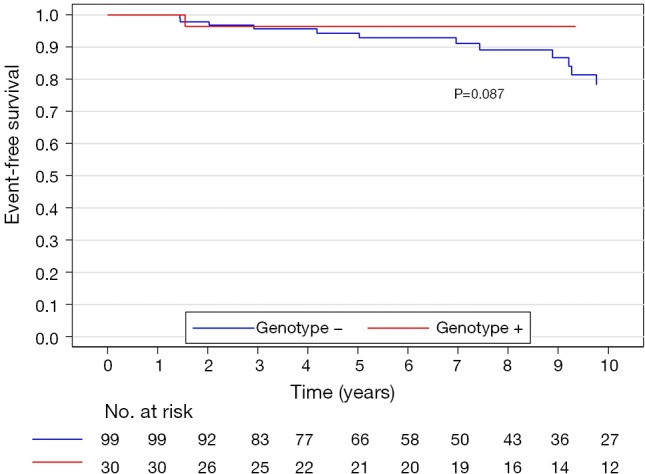
All-cause mortality rate (unadjusted) of genotype positive and genotype negative patients.
Figure 3.
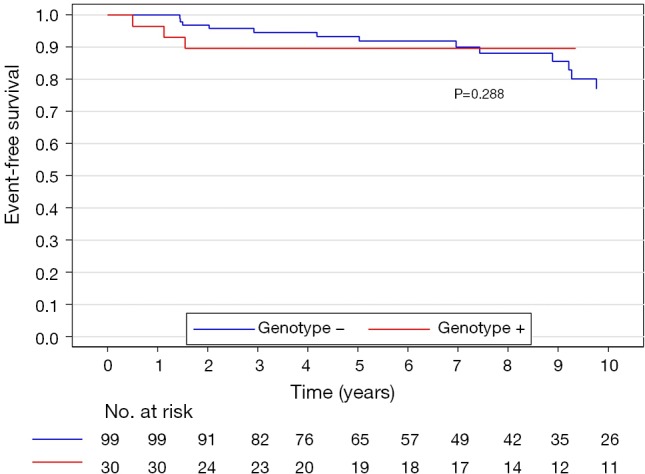
All-cause mortality rate adjusted for age at the time of ASA and baseline IVS thickness of genotype positive and genotype negative patients. ASA, alcohol septal ablation; IVS, interventricular septum.
Figure 4.
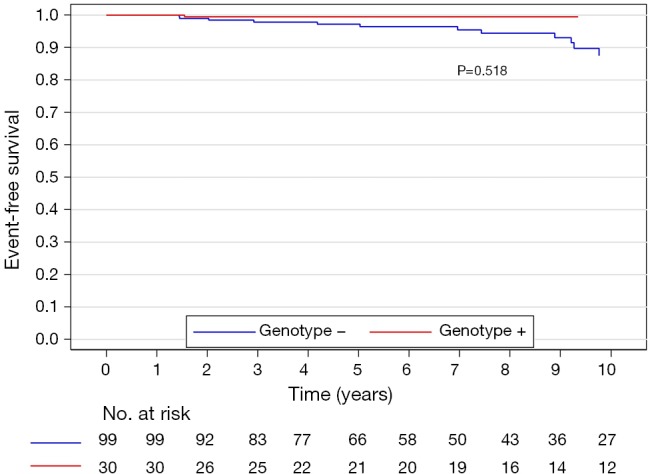
Combined mortality event rate (unadjusted) of genotype positive and genotype negative patients.
Figure 5.
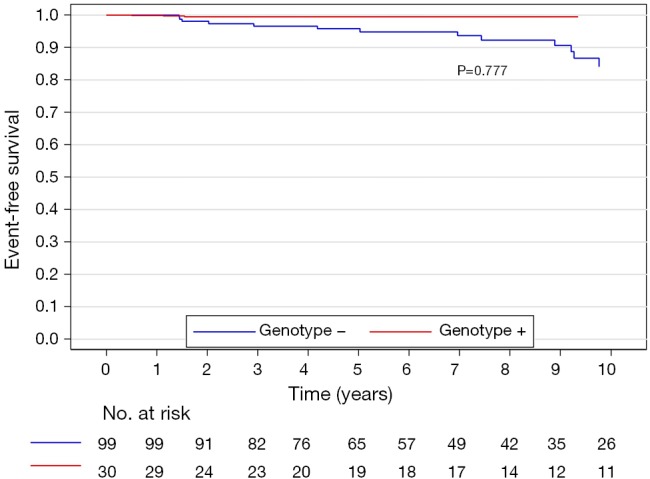
Combined mortality event rate adjusted for age at the time of ASA and baseline IVS thickness of genotype positive and genotype negative patients. ASA, alcohol septal ablation; IVS, interventricular septum.
Discussion
Our study suggests that ASA is an equally effective treatment of LVOTO in G+ HCM patients as in G− patients. This finding is important, regarding the fact, that G+ patients are considered to have more severe phenotype and worse outcome than G− patients (11,12). To date, the reported data about G+ patients included a presentation at an earlier age, greater hypertrophy of IVS and smaller LV enddiastolic diameter (13-15). Increased risk for the combined endpoints of cardiovascular death, nonfatal stroke, or progression to NYHA functional class III or IV compared to G− HCM patients was also reported (16). Recent data from a large Portuguese registry of HCM patients suggest a higher risk of sudden cardiac death in G+ patients (12). Our patients’ baseline characteristics are in line with the previously reported data; G + patients are younger at the time of diagnosis, have greater hypertrophy of IVS and smaller LV enddiastolic diameter. Surprisingly, the severity of the LVOTO was similar between both groups in our study. Furthermore, the symptoms (angina CCS class, dyspnea NYHA class, syncope) were not significantly different at baseline. In both groups, the symptoms improved after ASA procedures. This finding further supports the previously reported data from the Euro-ASA registry showing comparable outcomes among various subgroups of HCM patients treated with ASA (4). Since the residual LVOTO after ASA is known to be associated with worse prognosis (17), the favorable results in G+ patients are of great importance.
We included highly symptomatic HCM patients with significant LVOTO. After ASA, their symptoms greatly improved, as it has been already demonstrated by several studies (4,5,16,17). It is also known, that more pronounced reduction of LVOTO is associated with a lower NYHA class during the follow-up (4). This was confirmed by the current study, where both groups of patients had very low LVOT PG and only mild symptoms during the follow up. Severe LVOTO is known to be an independent predictor of adverse clinical outcome (18,19). Taking this into account, the clinical consequence of ASA is largely important. Resulting lower LVOT gradient is associated not only with better functional class, but also with better survival (18,20,21). The important finding of our current study is that HCM patients after ASA gained all these benefits irrespective of their genetic background. The possible explanation of relatively higher number of syncopes after ASA in G− patients could be the fact, that the G− patients were on average 11.5 years older than G+ patients and had higher incidence of complete heart block after ASA, as shown in Table 3. Nevertheless, most of the complete heart blocks were transient and did not require permanent pacemaker implantation.
The long-term survival was not statistically different between G+ and G− patients. Neither differed the occurrence of combined mortality endpoint (all-cause mortality and appropriate ICD discharge). Despite the long follow-up, the number of endpoints is relatively low, emphasizing the favorable prognosis of post-ASA patients. Prediction of post-ASA clinical outcome is challenging because of the marked heterogeneity of the treated HCM cohort. In our study, the independent predictors of all-cause mortality were baseline IVS thickness and age at the time of ASA. Patients in the G+ group were on average 11.5 years younger than G− patients. Despite their greater IVS thickness at baseline, their mortality rate was lower than in G− patients, that is probably attributed to lower age. The genotype positivity was not found to be independent of these two main predictors. Nevertheless, the low number of endpoints in the G+ group compromised the multivariate analysis.
Another important finding is that the genetic background of HCM patients did not influence the safety of the procedure. None of our study patients died or suffered a stroke within 30 days after ASA. Despite the greater hypertrophy of IVS, the occurrence of malignant arrhythmias and cardiopulmonary resuscitation in G+ patients were similar to G− patients. The G− patients inclined towards higher rates of a complete heart block after ASA, but the difference was not statistically significant. Neither the rates of permanent pacemaker implantation differed between our two groups.
Marked genetic heterogeneity of HCM, including incomplete penetrance, variable expressivity and existence of HCM phenocopies, makes the genotype results interpretation complicated. It is likely, that factors other than the sarcomere mutation itself influence clinical course and outcomes. These genetic, epigenetic, and environmental modifiers play an important role but are not yet fully characterized or understood. All of these aspects of heterogeneity in HCM have prevented identifying clear correlations between genotype and phenotype to date.
In our study, we present the largest ASA cohort evaluated with NGS. We identified genetic variants in 41% of patients. Only 23% of identified variants were classified as pathogenic or likely pathogenic (P/LP). This relatively low yield of genetic testing is in line with our recent findings (9). Advances in contemporary DNA-sequencing methodology make gene-based diagnosis faster and cheaper in clinical practice. Screening large numbers of genes results in the identification of many genetic variants of unknown significance (VUS) (18,19) which are not clinically actionable. In our study, we used a strict classification ACMG guidelines criteria (10), that explains the lower number of patients with P/LP variants than described in the past (13,14) but in line with the most recent works (12,20). Even with analysis of up to 229 genes, 59% of our patients do not carry any genetic variant susceptible of causing HCM. Together with a known low yield of genetic testing in patients with sigmoid septal morphology (13), these findings suggest, that some of the patients, despite having significant LVOTO treated with ASA, may not suffer from a true monogenic disorder.
While historically viewed as an autosomal dominant inherited heart condition, in patients with negative genetic tests, the inheritance pattern and utility of family screening are unclear. Recent studies have shown that expanded panel for genetic testing offers limited additional sensitivity for most patients with HCM (21-23). Also, in our study of ASA patients, almost all P/LP variants were found in sarcomeric genes, with only two exceptions. It is clear, that a large proportion of individuals with a clinical diagnosis of HCM but without sarcomere gene mutations may exhibit a distinct disease process that has a more complex, non-Mendelian inheritance pattern (24). Their phenotype fulfils the clinical criteria for the diagnosis of HCM (2,3) and they commonly suffer from typical features of the disease, including LVOTO. The results of our study revealed that both these groups of patients can be treated effectively with ASA resulting in similar clinical outcomes. Therefore, the decision about ASA procedure should not be influenced by genetic background in clinical practice.
Our study has several limitations. It is a retrospective analysis of prospectively collected data. The study is underpowered in terms of mortality, since our study groups are relatively small. We cannot be confident that these data are generalizable for non-tertiary referral centers with less experience with HCM patients and ASA. Institutional experience is a key determinant of successful outcomes and lower complication rates of ASA (25). This fact results in another selection bias—patients were carefully selected for ASA, considering the presence of septal and mitral apparatus anatomy appropriate for the procedure. Moreover, the results are only applicable to the adult HCM population, since no pediatric patients were included in our study.
Conclusions
Despite a more severe phenotype in G+ HCM patients, ASA is an equally effective treatment for LVOTO in G+ patients as it is for treating LVOTO in G− patients. The long-term outcome after ASA is similar in G+ and G− patients.
Table S1. The complete list of genes and identified variants.
| Gene | Pathogenic | Likely pathogenic | VUS |
|---|---|---|---|
| MYBPC3 | 20 | 1 | 6 |
| MYH7 | 1 | 5 | 6 |
| PTPN11 | 1 | 0 | 0 |
| RAF1 | 1 | 0 | 0 |
| MYL3 | 0 | 2 | 0 |
| TNNT2 | 0 | 1 | 1 |
| TNNI3 | 0 | 1 | 0 |
| TPM1 | 0 | 1 | 0 |
| CSRP3 | 0 | 0 | 3 |
| ACTN2 | 0 | 0 | 2 |
| DSP | 0 | 0 | 2 |
| CACNA1C | 0 | 0 | 1 |
| DES | 0 | 0 | 1 |
| DMD | 0 | 0 | 1 |
| DSG2 | 0 | 0 | 1 |
| FHL2 | 0 | 0 | 1 |
| LDB3 | 0 | 0 | 1 |
| MYH6 | 0 | 0 | 1 |
| MYL2 | 0 | 0 | 1 |
| MYOZ2 | 0 | 0 | 1 |
| NEXN | 0 | 0 | 1 |
| PKP2 | 0 | 0 | 1 |
| SCN10A | 0 | 0 | 1 |
| SOS1 | 0 | 0 | 1 |
| SCN2B | 0 | 0 | 1 |
VUS, variants of unknown significance.
Acknowledgments
The authors would like to thank Dr. Marek Maly and Eva Hansvenclova for their assistance with statistical analysis.
Funding: Supported by the Ministry of Health of the Czech Republic (grant No. 15-34904A) and Conceptual Development of Research Organization, Motol University Hospital, Prague (grant No. 00064203).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study protocol conforms to the ethical guidelines of the Declaration of Helsinki (2000, Fifth revision) and was approved by the institutional ethical committee. Written informed consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
Footnotes
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/cdt.2020.01.12). The authors have no conflicts of interest to declare.
References
- 1.Maron BJ, Maron MS, Semsarian C. Genetics of hypertrophic cardiomyopathy after 20 years: clinical perspectives. J Am Coll Cardiol 2012;60:705-15. 10.1016/j.jacc.2012.02.068 [DOI] [PubMed] [Google Scholar]
- 2.Elliott PM, Anastasakis A, Borger M, et al. 2014 ESC Guidelines on diagnosis and management of hypertrophic cardiomyopathy. Eur Heart J 2014;35:2733-79. 10.1093/eurheartj/ehu284 [DOI] [PubMed] [Google Scholar]
- 3.Gersh BJ, Maron BJ, Bonow RO, et al. 2011 ACCF/AHA guideline for the diagnosis and treatment of hypertrophic cardiomyopathy: A report of the American College of cardiology foundation/American heart association task force on practice guidelines. Circulation 2011;124:e783-831. [DOI] [PubMed] [Google Scholar]
- 4.Veselka J, Jensen MK, Liebregts M, et al. Long-term clinical outcome after alcohol septal ablation for obstructive hypertrophic cardiomyopathy: Results from the Euro-ASA registry. Eur Heart J 2016;37:1517-23. 10.1093/eurheartj/ehv693 [DOI] [PubMed] [Google Scholar]
- 5.Veselka J, Polaková E, Bonaventura J. Update on alcohol septal ablation for hypertrophic obstructive cardiomyopathy. Kardiol Pol 2019;77:160-1. 10.5603/KP.2019.0019 [DOI] [PubMed] [Google Scholar]
- 6.Veselka J. Alcohol septal ablation for hypertrophic obstructive cardiomyopathy: a review of the literature. Med Sci Monit 2007;13:RA62-8. [PubMed] [Google Scholar]
- 7.Veselka J, Zemánek D, Fiedler J, et al. Real-time myocardial contrast echocardiography for echo-guided alcohol septal ablation. Arch Med Sci 2009;5:271-2. [Google Scholar]
- 8.Veselka J, Lawrenz T, Stellbrink C, et al. Early outcomes of alcohol septal ablation for hypertrophic obstructive cardiomyopathy: A European multicenter and multinational study. Catheter Cardiovasc Interv 2014;84:101-7. 10.1002/ccd.25236 [DOI] [PubMed] [Google Scholar]
- 9.Bonaventura J, Norambuena P, Tomašov P, et al. The utility of the Mayo Score for predicting the yield of genetic testing in patients with hypertrophic cardiomyopathy. Arch Med Sci 2019;15:641-9. 10.5114/aoms.2018.78767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405-24. 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho CY, Day SM, Ashley EA, et al. Genotype and Lifetime Burden of Disease in Hypertrophic Cardiomyopathy. Circulation 2018;138:1387-98. 10.1161/CIRCULATIONAHA.117.033200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lopes LR, Brito D, Belo A, et al. Genetic characterization and genotype-phenotype associations in a large cohort of patients with hypertrophic cardiomyopathy - An ancillary study of the Portuguese registry of hypertrophic cardiomyopathy. Int J Cardiol 2019;278:173-9. 10.1016/j.ijcard.2018.12.012 [DOI] [PubMed] [Google Scholar]
- 13.Bos JM, Will ML, Gersh BJ, et al. Characterization of a phenotype-based genetic test prediction score for unrelated patients with hypertrophic cardiomyopathy. Mayo Clin Proc 2014;89:727-37. 10.1016/j.mayocp.2014.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lopes LR, Syrris P, Guttmann OP, et al. Novel genotype-phenotype associations demonstrated by high-throughput sequencing in patients with hypertrophic cardiomyopathy. Heart 2015;101:294-301. 10.1136/heartjnl-2014-306387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Binder J, Ommen SR, Gersh BJ, et al. Echocardiography-guided genetic testing in hypertrophic cardiomyopathy: Septal morphological features predict the presence of myofilament mutations. Mayo Clin Proc 2006;81:459-67. 10.4065/81.4.459 [DOI] [PubMed] [Google Scholar]
- 16.Olivotto I, Girolami F, Ackerman MJ, et al. Myofilament protein gene mutation screening and outcome of patients with hypertrophic cardiomyopathy. Mayo Clin Proc 2008;83:630-8. 10.1016/S0025-6196(11)60890-2 [DOI] [PubMed] [Google Scholar]
- 17.Veselka J, Tomašov P, Januška J, et al. Obstruction after alcohol septal ablation is associated with cardiovascular mortality events. Heart 2016;102:1793-6. 10.1136/heartjnl-2016-309699 [DOI] [PubMed] [Google Scholar]
- 18.Fokstuen S, Munoz A, Melacini P, et al. Rapid detection of genetic variants in hypertrophic cardiomyopathy by custom DNA resequencing array in clinical practice. J Med Genet 2011;48:572-6. 10.1136/jmg.2010.083345 [DOI] [PubMed] [Google Scholar]
- 19.Meder B, Haas J, Keller A, et al. Targeted Next-Generation Sequencing for the Molecular Genetic Diagnostics of Cardiomyopathies. Circ Cardiovasc Genet 2011;4:110-22. 10.1161/CIRCGENETICS.110.958322 [DOI] [PubMed] [Google Scholar]
- 20.Murphy SL, Anderson JH, Kapplinger JD, et al. Evaluation of the Mayo Clinic Phenotype-Based Genotype Predictor Score in Patients with Clinically Diagnosed Hypertrophic Cardiomyopathy. J Cardiovasc Transl Res 2016;9:153-61. 10.1007/s12265-016-9681-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walsh R, Thomson KL, Ware JS, et al. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet Med 2017;19:192-203. 10.1038/gim.2016.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ingles J, Goldstein J, Thaxton C, et al. Evaluating the Clinical Validity of Hypertrophic Cardiomyopathy Genes. Circ Genom Precis Med 2019;12:e002460. 10.1161/CIRCGEN.119.002460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alfares AA, Kelly MA, McDermott G, et al. Results of clinical genetic testing of 2,912 probands with hypertrophic cardiomyopathy: Expanded panels offer limited additional sensitivity. Genet Med 2015;17:880-8. 10.1038/gim.2014.205 [DOI] [PubMed] [Google Scholar]
- 24.Ko C, Arscott P, Concannon M, et al. Genetic testing impacts the utility of prospective familial screening in hypertrophic cardiomyopathy through identification of a nonfamilial subgroup. Genet Med 2018;20:69-75. 10.1038/gim.2017.79 [DOI] [PubMed] [Google Scholar]
- 25.Veselka J, Faber L, Jensen MK, et al. Effect of Institutional Experience on Outcomes of Alcohol Septal Ablation for Hypertrophic Obstructive Cardiomyopathy. Can J Cardiol 2018;34:16-22. 10.1016/j.cjca.2017.10.020 [DOI] [PubMed] [Google Scholar]