The extensive array of basic helix-loop-helix (bHLH) transcription factors and their combinations as dimers underpin the diversity of molecular function required for cell type specification during embryogenesis. The bHLH factor TWIST1 plays pleiotropic roles during development. However, which combinations of TWIST1 dimers are involved and what impact each dimer imposes on the gene regulation network controlled by TWIST1 remain elusive. In this work, proteomic profiling of human TWIST1-expressing cell lines and transcriptome analysis of mouse cranial mesenchyme have revealed that TWIST1 homodimers and heterodimers with TCF3, TCF4, and TCF12 E-proteins are the predominant dimer combinations.
KEYWORDS: E-protein, TWIST1, bHLH factor, embryonic stem cells, lineage differentiation
ABSTRACT
The extensive array of basic helix-loop-helix (bHLH) transcription factors and their combinations as dimers underpin the diversity of molecular function required for cell type specification during embryogenesis. The bHLH factor TWIST1 plays pleiotropic roles during development. However, which combinations of TWIST1 dimers are involved and what impact each dimer imposes on the gene regulation network controlled by TWIST1 remain elusive. In this work, proteomic profiling of human TWIST1-expressing cell lines and transcriptome analysis of mouse cranial mesenchyme have revealed that TWIST1 homodimers and heterodimers with TCF3, TCF4, and TCF12 E-proteins are the predominant dimer combinations. Disease-causing mutations in TWIST1 can impact dimer formation or shift the balance of different types of TWIST1 dimers in the cell, which may underpin the defective differentiation of the craniofacial mesenchyme. Functional analyses of the loss and gain of TWIST1–E-protein dimer activity have revealed previously unappreciated roles in guiding lineage differentiation of embryonic stem cells: TWIST1–E-protein heterodimers activate the differentiation of mesoderm and neural crest cells, which is accompanied by the epithelial-to-mesenchymal transition. At the same time, TWIST1 homodimers maintain the stem cells in a progenitor state and block entry to the endoderm lineage.
INTRODUCTION
The helix-loop-helix (HLH) superfamily is an ancient group of transcription factors that appeared in unicellular organisms >600 million years ago and has continuously evolved and diversified since then to carry out ever-elaborating activities required for lineage specification (1). Through dimerization with different protein partners, basic HLH (bHLH) factors drive the specification of many cell types during neurogenesis, hematopoiesis, and myogenesis (2–5). Eukaryotic HLH factors can be classified based on their structural and functional attributes (6). Class I, II, and V factors are known to engage in homo- and heterodimerization. Class I proteins, comprised of all E-proteins (e.g., E12/TCF3, HEB/TCF12, and E2-2/TCF4), are broadly expressed in various tissues (7). Class II proteins are generally present in specific cell lineages, for example, MYOD in myogenic cells, NEUROD in neurogenic cells, and TWIST1 in mesenchymal tissues (e.g., craniofacial and limb mesenchyme) (8–14). Class V represents a group of non-DNA-binding HLH proteins, such as the ID proteins that compete for E-protein binding (15). It was hypothesized that the bHLH dimer composition is regulated by the expression level of each bHLH factor, their level of phosphorylation, and their proportion relative to each other (16–18). However, the diversity of dimer combinations and their functional specificity during development remain enigmatic.
The bHLH factor TWIST1 is highly expressed in cranial mesoderm- and neural crest-derived mesenchyme (12, 13, 19), where it is critical for craniofacial development. Twist1+/− mice display craniosynostosis (20, 21) that partly phenocopies skeletal defects associated with TWIST1 haploinsufficiency in human Saethre-Chotzen syndrome (SCS) (AHC) (MIM: 101400). Conditional ablation of Twist1 in the cranial mesoderm (CM) or the cranial neural crest (CNC) leads to malformations of the cranium, facial skeleton, brain, cranial nerves, and muscles (22–24). At the cellular level, Twist1 is required for maintaining the mesenchymal cell morphology and their potency for osteo-, chondro-, and adipogenesis (12, 13, 19, 25). Previous studies have highlighted the differential functions of TWIST1 dimers in the osteogenic differentiation of the cranial sutural mesenchyme (21, 26), which is mediated by their targeted action on fibroblast growth factor (FGF) signaling (25, 27, 28). For example, the TWIST1-TCF3 heterodimer promotes mesenchymal stem cell (MSC) proliferation, while the TWIST1 homodimer activates FGFR2, OCN, and BSP expression for ossification. Identifying TWIST1 dimerization partners and their transcriptional targets in the cranial mesenchyme will, therefore, allow for a better understanding of the mechanisms of development regulated by TWIST1 and bHLH factor dimers.
In this study, the diversity and expression of dimerization partners of TWIST1 were determined by mass spectrometry (MS) analysis, following immunoprecipitation of human TWIST1 (hTWIST1) from mesenchymal cells, and cross-compared with Twist1 coexpression analysis in mouse embryonic head tissues. We employed the bimolecular fluorescence complementation (BiFC) assay to elucidate the balance between hetero- and homodimerization and to assess the potential impact of pathological mutations. Finally, to dissect the specific functions of each TWIST1 dimer and their immediate downstream targets, we genetically engineered embryonic stem cells (ESCs), in which the expression of different TWIST1–E-protein dimers could be tightly controlled, and analyzed their ability to differentiate and migrate. By delineating TWIST1 molecular interactions, our work has revealed previously unappreciated layers of control in lineage determination and cellular behavior: TWIST1–E-protein heterodimers promote mesoderm and neural crest differentiation through epithelial-mesenchymal transition (EMT), while the TWIST1 homodimer maintains a progenitor-like state and blocks entry to the endoderm lineage. Using recent quantitative approaches and engineered cell models, this study has generated new insights into an ancient group of bHLH factors, the regulation of their dimerization activity, and their role in fine-tuning lineage specification and differentiation.
RESULTS
Identification of bHLH partners of TWIST1 in the embryonic head mesenchyme.
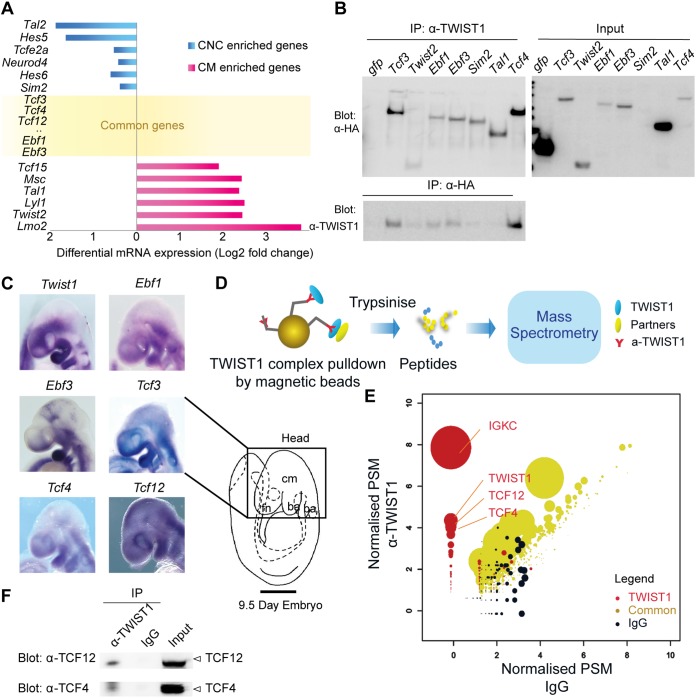
In order to identify potential candidates dimerizing with TWIST1 protein, we first focused on genes coexpressed with Twist1 in vivo by investigating tissues of the embryonic mouse head. Microarray analysis of CNC and CM cells sorted from heads of embryonic day 9.5 (E9.5) embryos of Wnt1-Cre:GFP and Mesp1-Cre:GFP transgenic mice, respectively (14, 29), revealed that 58 out of 158 known bHLH factors (30) were expressed in the head mesenchyme (see Table S1 in the supplemental material). Twelve bHLH factors were significantly enriched in CNC or CM (Fig. 1A), and 46 were expressed in both tissues (Fig. 1A and Table S1). Based on their known roles in craniofacial development, seven candidates were selected for validation, including SIM2, TCF4, EBF1, EBF3, TAL1, TWIST2, and TCF3 (an isoform of E2A, a known TWIST1 partner as the positive control). Hemagglutinin (HA)-tagged protein (including HA-tagged green fluorescent protein [GFP] as a negative control) expression constructs were transfected into Madin-Darby canine kidney (MDCK) cells that stably overexpress hTWIST1 (referred here as MDCK/hTWIST1-OE cells) and have previously been used to investigate the role of TWIST1 in inducing mesenchymal phenotypes (14, 31). These factors were coimmunoprecipitated with TWIST1. Reciprocally, TWIST1 was coimmunoprecipitated with EBF1, EBF3, and TCF4 but enriched less efficiently with TWIST2, SIM1, and TAL1 (Fig. 1B). Therefore, we further analyzed Ebf1, Ebf3, and Tcf4 by in situ hybridization and observed their overlapping expression with Twist1 in the frontonasal tissues, the cranial mesoderm, and the first branchial arch of the mouse embryo (Fig. 1C). Collectively, the microarray coexpression screen identified putative TWIST1 dimerization partners, from which we validated the physical interaction of EBF1/EBF3/TCF4 with TWIST1.
FIG 1.
Unbiased high-throughput identification of proteins interacting with TWIST1 by coexpression analysis, coimmunoprecipitation (IP), and mass spectrometry. (A) Relative expression of genes encoding known bHLH factors in the cranial neural crest (CNC) versus cranial mesoderm (CM) of the E9.5 embryonic head collated from transcriptome data previously published (14, 29). Forty-one additional genes expressed in both tissues can be found in Table S1 in the supplemental material. (B) Detection of HA-tagged proteins (left blot, α-HA) after IP of TWIST1 (IP: α-TWIST1) and reciprocal IP of HA-tagged proteins (IP: α-HA) and detection of TWIST1 (Blot: α-TWIST1) from lysates of hTWIST1-expressing MDCK cells transfected with constructs expressing the HA-tagged protein partners, with expression confirmed in the input (right blot, α-HA). (C) Expression of candidate transcripts analyzed by whole-mount in situ hybridization in regions of the E9.5 embryonic head. Lateral views with anterior to the left are shown. cm, cranial mesoderm; fn, frontonasal tissue; ba, brachial arch. Scale bar, 500 μm. (D) Experimental strategy for identifying TWIST1 interacting partners from hTWIST1-expressing MDCK cells. TWIST1-containing protein complexes are isolated on beads using an anti-TWIST1 antibody before being analyzed by tandem mass spectrometry, which determines the ion spectrum of the different peptides (one example is shown here). (E) Groups of proteins identified using the nonparametric rank product method based on a cumulative count of reporting a protein across independent experiments. Sets: red, TWIST1 IP; black, nonspecific IgG; yellow, common to TWIST1 IP and IgG control. Normalized unique peptide spectrum matches (PSM) of the detected proteins are shown on both axes for each condition. The size of the dots corresponds to –log10 P value. IGKC, immunoglobulin kappa constant. (F) Detection of TCF12 (Blot: α-TCF12) or TCF4 (Blot: α-TCF4) endogenous proteins after IP of TWIST1 (IP, α-TWIST1) from lysates of embryoid bodies overexpressing TWIST1, compared with results obtained for nonspecific IgG and the input.
Proteomic screening of MDCK/TWIST1-OE cells revealed TCF factors as the prevalent partners of TWIST1.
Complementary to the above approach, TWIST1 dimer partners were identified through unbiased proteomic screening using affinity purification of the TWIST1 complexes from MDCK/hTWIST1-OE cells, followed by mass spectrometry (Fig. 1D). We performed three biological replicates of immunoprecipitation using an anti-TWIST1 antibody in parallel with a mouse IgG control. Differential analysis using the ranked-product method identified 846 proteins and divided them into three sets (32) (Fig. 1E; see Table S2 in the supplemental material). Set 1 represents 103 identified proteins that bound specifically to TWIST1, set 2 comprises the proteins that bound only to IgG (377 proteins), and set 3 contains proteins that bound to both IgG and TWIST1 (362 proteins). As expected, TWIST1 was one of the top enriched proteins in set 1 (Fig. 1E). Only two other bHLH factors, TCF12 and TCF4, were also highly enriched. The rest of set 1 TWIST1 putative partners included RNA-binding proteins and transcription factors such as cyclin T2, TRIP6, GATA1, FHL2, and CRTC2 (Table S2). Tcf4 and Tcf12, like Tcf3, are expressed in the facial primordia and within Twist1 expression domains in the frontonasal, the branchial arches, and the mesoderm in the embryonic head (Fig. 1C). To recapitulate early development in vivo, we engineered mouse ESCs in which TWIST1 expression is inducible and differentiated these ESCs into embryoid bodies (EBs). Coimmunoprecipitation experiments performed with these EBs demonstrated that TWIST1 interacted with endogenous TCF4 and TCF12 (Fig. 1F).
Mutations of conserved residues in the bHLH domain impaired TWIST1-TCF12 dimer formation and function.
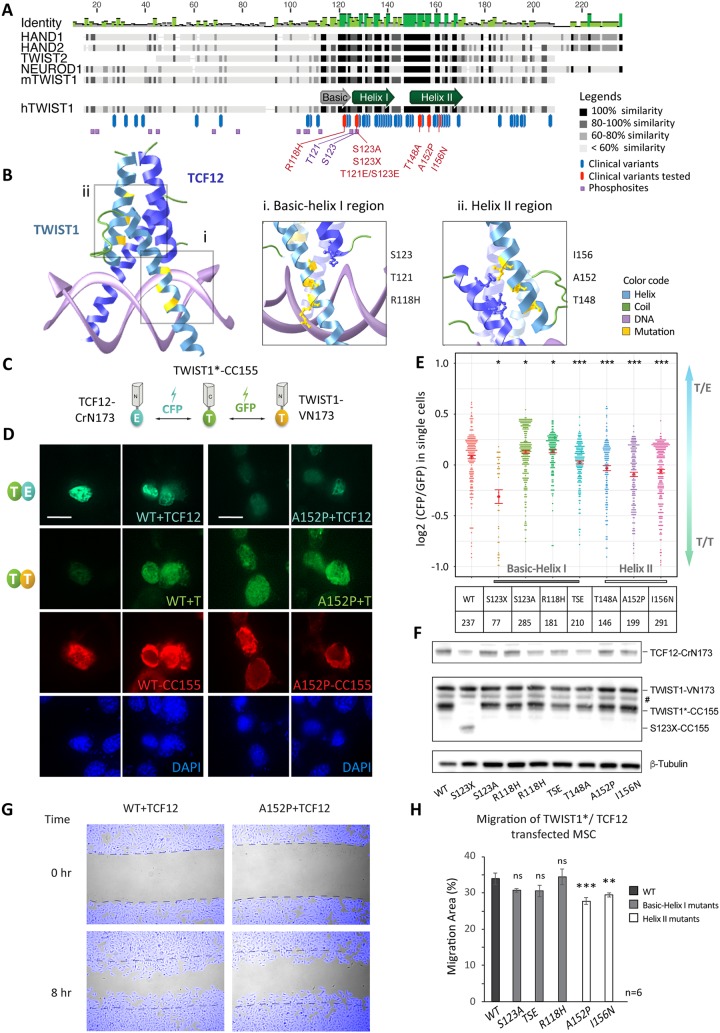
The TWIST1 bHLH dimerization domain shows 84 to 100% sequence identity among vertebrate orthologues (human, mouse, frog, and zebrafish) (Fig. 2A). It is also highly conserved (∼70% similarity) among several TWIST family proteins, i.e., TWIST2, HAND1, HAND2, TCF15 (paraxis), and scleraxis, suggesting functional importance (33, 34). Clinically significant point mutations (63 reported mostly in SCS patients) showed high incidence in the bHLH domain of TWIST1 (Fig. 2A) (human genomic mutation database) (16–18, 35, 36). Some of these mutations are located at or around the threonine and serine phosphorylation sites (Fig. 2A) and could inhibit TWIST1 phosphorylation (17, 37). It was previously shown that TWIST1 SCS mutations in the basic helix I region, such as R118H, may impair phosphorylation at T121 and S123 residues and alter the preference of dimerization of TWIST1 with TCF3 and HAND2 (17).
FIG 2.
Human disease-causing mutations disrupt the TWIST1-TCF12 dimer function. (A) Alignment of selected mouse TWIST family bHLH proteins and hTWIST1 sequences. The degree of similarity of the different regions is indicated in dark shades. Blue and red bars indicate missense pathological mutations reported in the Human Gene Mutation Database (35). The purple squares are predicted phosphosites. Mutations (red) and phosphorylation sites (purple) relevant for this study are labeled. (B) In silico 3D structure prediction of the bHLH regions of TWIST1-TCF12 dimer binding to DNA, based on the structure of the NeuroD-TCF3 dimer as determined by crystallography (65) (http://aquaria.ws). Enlarged representations of the basic helix I (i) and helix II (ii) domains of TWIST1 are shown. TWIST1 residues mutated in disease and their atomic interaction with TCF12 are highlighted in yellow. (C) Experimental setup for the BiFC assay. Three independent constructs expressing TWIST1*-CC155, TWIST1-VN173, and TCF12-CrN173 were equimolarly cotransfected in C3H10T1/2 cells. TWIST1*, wild type or SCS variants. Interactions between the different proteins and the relative proportion of TWIST1 heterodimers versus TWIST1 homodimers are then assessed by quantifying and comparing CFP and GFP fluorescence intensities, as shown in panel E. (D) Representative confocal microscopy images of BiFC assays performed for wild-type (WT) and A152P mutant TWIST1, with two replicate images each. Signals for dimers: TWIST1*-CC155/TWIST1-VN173 (T/T) or TWIST1-VN173/TCF12-CrN173 (T/E) as indicated. The red channel shows TWIST1*-CC155 detected by immunostaining with an anti-HA antibody. The cell nucleus was stained by DAPI. Scale bar, 10 μm. (E) Heterodimer/homodimer CFP/GFP ratio quantified for individual cells (each dot corresponding to one cell) in BiFC assays. The TWIST1 variant and number of cells assayed are indicated in the chart below. TSE corresponds to the T121E S123E double mutant. Red bars represent standard errors of the mean. Data were analyzed by the nonparametric Mann-Whitney U test (two-tailed): *, P ≤ 0.05; ***, P ≤ 0.001. (F) Western blot analysis of BiFC protein expression in C3H10T1/2 cells. TWIST1 BiFC constructs were detected with an anti-GFP antibody; TCF12-CRN173 was detected with an anti-TCF12 antibody. The loading control (β-tubulin) is also shown. #, a nonspecific band. (G) Example images from a scratch assay. C3H10T1/2 cells were cotransfected with TWIST1*-CC155 (WT or A152P) and TCF12-CrN173 expression vectors. Time-lapse images were taken every 15 min over an 8-h period after a scratch was made on the cell monolayer. Cells were tracked (pseudocolored in blue), and the area of the cell-free gap was quantified in ImageJ software. Dashed lines indicate original gap boundaries. (H) Quantification of migration (percentage of the total area in 8 h) of C3H10T1/2 cells cotransfected with TWIST1*-CC155 and TCF12-CrN173 expression vectors. Data represent the means ± standard errors from n = 6 independent experiments, analyzed by one-way ANOVA with Holm-Sidak’s posttest. **, P ≤ 0.01; ***, P ≤ 0.001; ns, nonsignificant.
To validate the putative interactions identified above and test the hypothesis that TWIST1 pathological mutations in the bHLH region may shift the balance of homo- versus heterodimers, we employed the BiFC system, a bimolecular complementation assay for detecting the direct physical interaction between two partners by fluorescence (38). A subset of mutations with known impact on the basic helix I region was selected (Fig. 2A, red): a truncation mutation in the bHLH domain (S123X), phosphorylation-inhibitory mutations (S123A and R118H), and phosphomimetic mutations (T121E and S123E). We also selected previously uncharacterized mutations in helix II (T148A, A152P, and I156N), which affect highly conserved residues in the TWIST family and are positioned near the dimerization interface (Fig. 2A and B-ii). When examined by three-dimensional (3D) in silico modeling of the bHLH regions of a TWIST1/TCF12 dimer in the presence of DNA, the phosphoregulatory residues appear to be in close proximity to the DNA interface rather than the site in contact with the E-protein (Fig. 2B-i), while clinical variants on helix II affect residues located at the protein interaction interface (Fig. 2B-ii).
We tested the impact of these TWIST1 mutations on the ability of TWIST1 to dimerize with TCF12, which is also associated with SCS and other craniofacial anomalies (39). MSCs were transfected with constructs for TWIST1-VN173 and TCF12-CrN173, which competitively bind TWIST1*-CC155 (* for wild type or SCS variants). Fluorescence is emitted only when one of the N-terminal fluorophores in TWIST1-VN173 (GFP fluorescence) or TCF12-CrN173 (cyan fluorescent protein [CFP] fluorescence) is brought together with their shared C-terminal domain in the TWIST1*-CC155 construct, therefore indicating an interaction between TWIST1 or TCF12 with TWIST1* (Fig. 2C). We then measured the interaction of TWIST1-CC155 with TCF12-CrN173 versus TWIST1-VN173 by the ratio of fluorescence intensity from the two different fluorophores (Fig. 2D and E). When expressed at comparable levels (Fig. 2F), TWIST1 mutants displayed significant changes in their preference to form homodimers (GFP) or heterodimers (CFP) (Fig. 2D and E). Among all mutations tested, truncation at the bHLH domain (S123X) most severely disrupted dimerization (Fig. 2E). Phosphorylation-inhibitory mutations S123A and R118H increased the preference for heterodimer formation, while the phosphomimetic mutant (TSE for T121E and S123E) of TWIST1 favored homodimerization (Fig. 2E). Finally, all three missense mutations at helix II compromised heterodimerization severely (Fig. 2E).
For mutations associated with a significant shift in dimer balance, we further tested their potential impact on cell migration, a known TWIST1-driven biological process. Cell migration was tracked and quantified after a scratch was made on a confluent monolayer of mesenchymal stem cells transiently expressing TCF12 and different variants of TWIST1 (Fig. 2G). Mutations in helix I showed no effect on the cells, but cells expressing TWIST1 with helix II mutations (A152P and I156N) showed reduced cell migration (Fig. 2G and H). Altogether, these results demonstrate that residues in helix I and helix II are essential determinants for dimerization and that disrupting TWIST1/TCF12 dimerization with mutations at the interaction site in helix II may compromise mesenchymal migration.
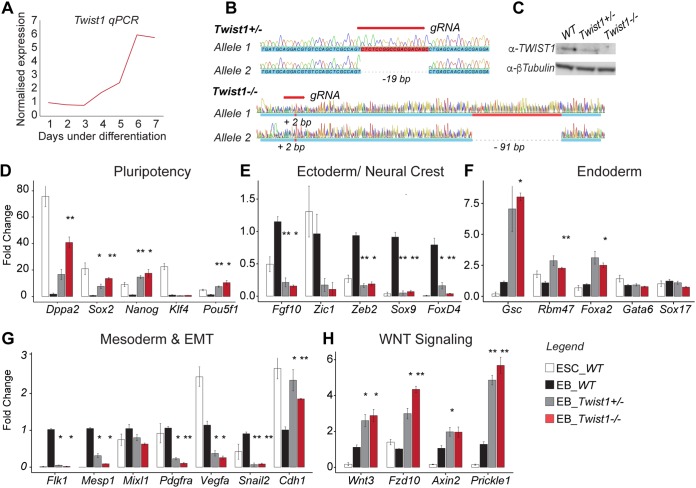
Loss of Twist1 impairs exit from pluripotency and cell fate specification.
During mouse ESC differentiation, the expression of endogenous Twist1 was first detected from day 3 and then peaked at day 6 to 7 (Fig. 3A). We first generated and studied Twist1 loss-of-function (LOF) ESCs to appreciate the overall function of the ensemble of TWIST1 dimers during differentiation. Mono- and biallelic Twist1 knockout mouse ESCs were engineered using the CRISPR-Cas9 methodology (Fig. 3B and C), and the expression of pluripotency and lineage gene markers was analyzed (Fig. 3D to H). Pairwise statistical tests revealed that both full and partial loss of Twist1 disrupted lineage specification in a gene dosage-dependent manner. On day 3 of differentiation, when Twist1 became active, genes associated with pluripotency, such as Dppa2, Nanog, Sox2, and Pou5f1, were downregulated in wild-type parental cells, whereas the expression was maintained at the predifferentiated levels in the mutant cells (Fig. 3D). Concurrently, ectodermal and mesodermal genes were not activated in the mutant cells (Fig. 3E and G). Expression of EMT genes such as Pdgfra, Snail2, and Vegfa was also reduced in the mutant cells, while the epithelial marker, Cdh1, was upregulated (Fig. 3G). In contrast, endodermal markers such as Gsc, Rbm47, Foxa2, and WNT signaling pathway components (ligand, Wnt3; receptors, Fzd10 and Prickle1; target gene, Axin2) were upregulated in the mutant cells (Fig. 3F and H). These functions of Twist1 in selectively directing lineage differentiation and maintaining the mesenchymal progenitors are likely to be orchestrated by a combination of TWIST1 dimers.
FIG 3.
TWIST1 is required for mesoderm and ectoderm differentiation. (A) RT-qPCR analysis of Twist1 expression relative to the mean value of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene expression during mouse ESC differentiation. Each time point represents the mean value from n = 3 independent experiments. (B) Twist1 alleles of two independent ESC clones (Twist1+/− and Twist1−/−) generated by CRISPR-Cas9 editing. Genomic positions targeted by the gRNA, Sanger sequencing chromatograms, and deduced gDNA sequences are shown. Mutations are frameshifts in the coding sequence of exon 1. Twist1+/− clone, allele 1 is unmodified and allele 2 has a 19-bp deletion; Twist1−/− clone, allele 1 has a 2-bp insertion and allele 2 has a 2-bp insertion and a 91-bp deletion. (C) Western blot analysis of TWIST1 proteins in the clones characterized in panel B and the parental ES wild-type (WT) cell line. The loading control (β-tubulin) is shown. (D to H) RT-qPCR analysis of the expression of selected genes involved in cell lineage, EMT, or WNT signaling in mutant, wild-type (WT), and undifferentiated ESCs at day 3 of in vitro differentiation. Expressions were normalized against the mean values of TBP and GAPDH gene expression and the wild-type sample at day 3 of differentiation. Each data point represents the mean ± standard error from n = 3 independent experiments. Data were analyzed by the nonparametric Mann-Whitney U test (two tailed). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Comparison of the target specificity that differs between different TWIST1 dimers.
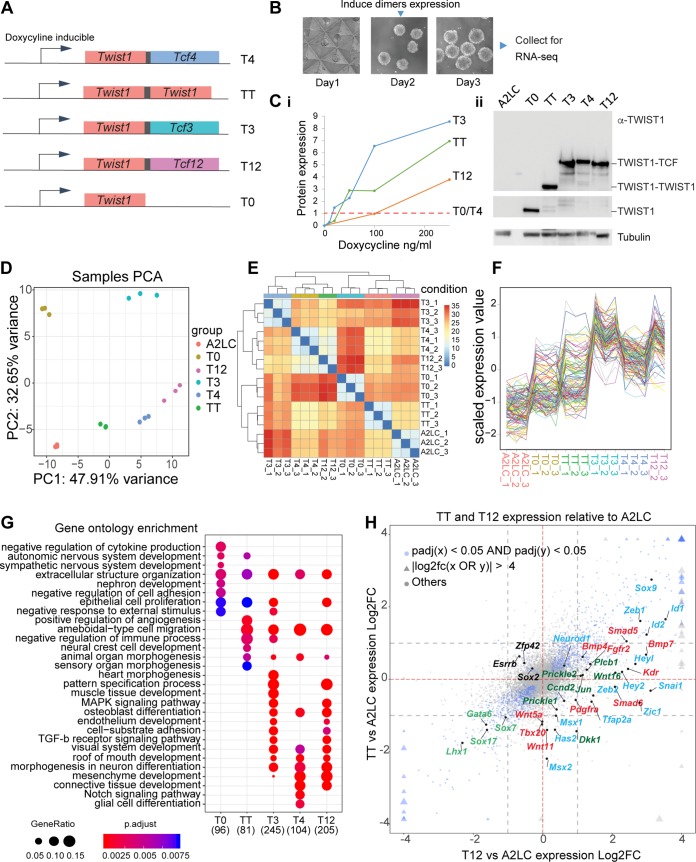
In order to deconstruct the function of each TWIST1 homodimer and heterodimers during lineage differentiation, we took a gain-of-function (GOF) approach to analyze the downstream genes controlled by individual dimers. In Drosophila, tethered Twist-Twist and Twist-Daughterless dimers could recapitulate the function of dimers formed naturally by Twist1 and Daughterless. The Twist homodimer induced Mef2 expression and muscle differentiation in vivo, whereas the Twist-Daughterless dimer antagonized this activity (40). Following the same strategy, we tethered TWIST1 and TCF proteins into dimers via flexible polyglycine linkers, i.e., TWIST1-TWIST1 (TT), TWIST1-TCF3 (T3), TWIST1-TCF4 (T4), and TWIST1-TCF12 (T12), and expressed them in ESCs (Fig. 4A to C). The expression constructs were stably integrated into the designated docking site at the Hprt locus downstream of a doxycycline-inducible promoter. In the presence of doxycycline, the rtTA protein is expressed from the Rosa26 locus and binds and activates the inducible promoter. This design ensures consistent transgene expression (41–43). ESCs were differentiated for 3 days into EBs (Fig. 4B) and assayed for the expression of developmental/lineage marker genes. Before doxycycline induction, we detected no expression of the transgene (Fig. 4C-i). Dimer expression was induced at day 2 of differentiation in synchrony with the onset of expression of endogenous Twist1 (Fig. 3A). To ensure that the effects of each dimer were not confounded by the different levels of expression, we tested different doses of doxycycline and selected the optimal concentration for each cell line to achieve comparable levels of expression for each construct (Fig. 4C-i and -ii).
FIG 4.
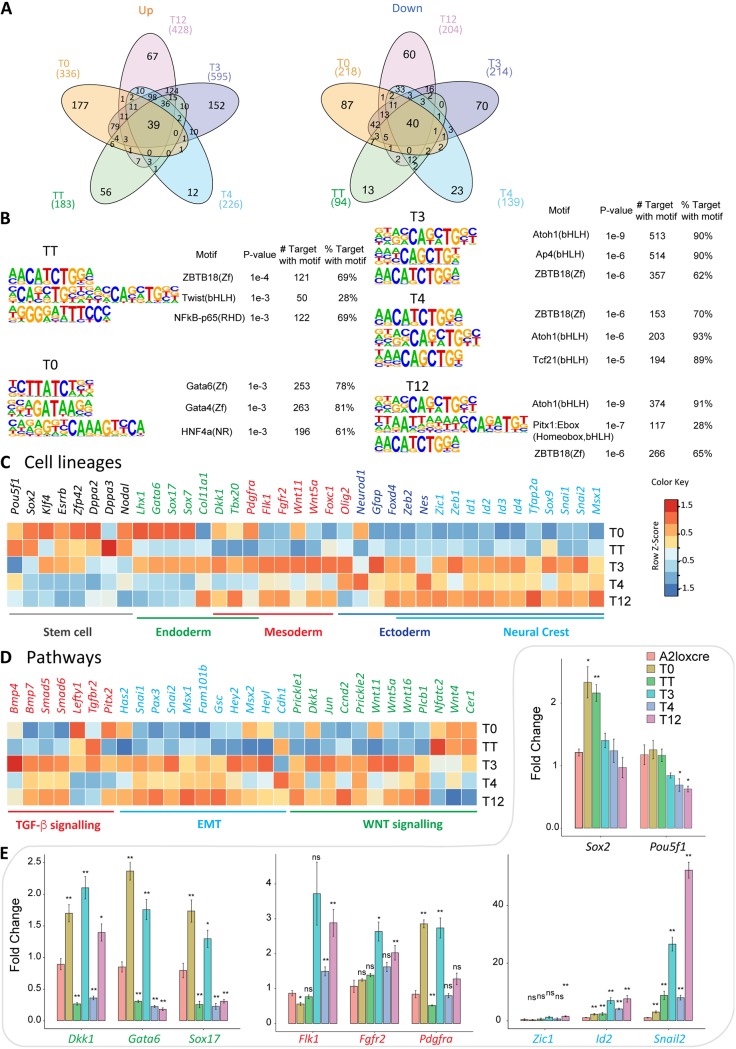
Transcriptome profiles of embryoid bodies expressing different TWIST1 dimers and monomer. (A) Schematic of doxycycline-inducible expression constructs Twist1-Tcf3 (T3), Twist1-Tcf4 (T4), Twist1-Tcf12 (T12), Twist1-Twist1 (TT), and Twist1 only (T0; TWIST1 monomer) recombined into the designated locus in A2loxCre parental ESCs (41, 42). Proteins in the dimer are joined by a flexible polyglycine linker (black box). (B) Experimental setup to obtain dimers expressing EBs. ESCs are differentiated into EBs for 3 days. Dimer expression is induced with doxycycline 16 h before collection (C) Panel i, optimization of doxycycline dosage to induce comparable levels of expression of the constructs between the different ESC lines. The maximal protein expression for cell lines expressing T0 and T4 was reached at 1,000 ng/ml doxycycline. Treatments for the resting cell lines were adjusted accordingly. The protein expression under different concentrations of doxycycline were quantified by Western blotting using an anti-TWIST1 antibody and normalized against β-tubulin expression. Panel ii, Western blot showing comparable expression of TWIST1 dimers and monomer in the induced EBs used in the final RNA-seq experiment. The expected protein location is labeled on the right. The parental A2loxCre cell line (A2LC) was used as a control. The loading control (β-tubulin) is also shown. (D) Principal-component analysis of the transcriptomes of the dimer- and monomer-expressing ESCs and A2LC EBs (n = 3 independent RNA-seqs for each cell line), showing the first two primary components. (E) Correlation map of the different RNA-seq data, showing Euclidean distance between cell lines (from a high correlation in blue to a low correlation in red). (F) Scaled expression of the 125 genes with top PC loadings across all cell lines. Each line corresponds to one gene. (G) Gene ontology (GO) analysis of upregulated genes (adjp < 0.05, log2 FC > 1) in all cell lines versus A2LC. The top 10 most significant nonredundant GO categories for each cell line were plotted. Dot sizes represent “GeneRatio” (gene upregulated/gene in GO set), and the adjusted P value is color coded. The number of identified genes in each group is shown in parentheses. (H) Four-way plot visualization of DESeq2 result. Log2 FC comparisons between TT and T12 treatments versus A2LC were performed. Blue nodes indicate genes significantly changed for both cell lines. Triangles indicate values exceeding the limit of the plot. Size change reflects the magnitude of log2 FC values. Gray dashed lines indicate |log2 FC| = 1. Key markers and pathway genes are indicated: black, pluripotency; green, endoderm; red, mesoderm; blue, ectoderm and EMT; dark green, WNT signaling pathway.
The transcriptomes of EBs differentiated for 3 days expressing comparable levels of dimers were then analyzed by transcriptome sequencing (RNA-seq) and compared to the transcriptome of the parental A2loxcre cell line (A2LC) as well as to the transcriptome of a cell line expressing TWIST1 monomer (T0) that were both also differentiated for 3 days. Unsupervised principal-component analysis (PCA) and hierarchical clustering confirmed groupings of replicates (Fig. 4D and E). Global gene expression profiles separated T3, T4, and T12 from TT, T0, and A2LC EBs (Fig. 4E and F), a result which was driven mainly by genes upregulated in the heterodimers. Among the three heterodimers, cells expressing T4 and T12 dimer exhibited more similar gene expression profiles (Fig. 4E). Differentially expressed gene (DEG) analysis was performed for T0, TT, T3, T4, and T12 against the A2LC cells. Examination of the upregulated gene groups revealed that while all dimers activated the cell migration program, the heterodimers specifically activated the expression of genes associated with pattern specification, mesenchymal tissue development (muscle, skeletal, connective tissue, etc.) and transforming growth factor β (TGF-β) signaling (Fig. 4G). A four-way comparison between TT or T12 against A2LC also demonstrated a stronger ability of the heterodimers to activate developmental markers, especially mesenchyme-related genes (Fig. 4H).
It was previously demonstrated by in vitro mobility shift assay that different dimers preferentially bind different DNA sequences (18, 44). We performed sequence motif analyses on the proximal regulatory sequences for dimer-specific DEGs. For each group, 100 to 600 genes were upregulated, and 100 to 200 were downregulated (adjusted P value [adjp] of ≤0.05 and fold change [FC] of ≥2) (Fig. 5A; see Table S3 in the supplemental material). Top ranking regulatory motifs for heterodimer-activated promoters contained a single canonical E-box CATCTG or CAGCTG (Fig. 5B), which is consistent with TWIST1 chromatin immunoprecipitation sequencing (ChIP-seq) and mobility shift assays results (18, 44). TT targets were uniquely enriched in double E-boxes with a five-nucleotide spacer, as previously reported by ChIP-seq in human mammary epithelial cells overexpressing TWIST1 (44). Additionally, the TWIST1 monomers showed a greater variety of motifs in addition to canonical E-box sequences. This observation suggests that the free TWIST1 monomer may form a mixture of alternative dimers/complexes with other proteins, potentially the GATA family members (Fig. 5B).
FIG 5.
Differential specificity analysis of TWIST1 dimers and monomer. (A) Pairwise differential gene expression analysis of the dimer- and monomer-expressing ESCs against the parental A2LoxCre ESCs performed with DESeq2 (61) (adjp < 0.05, fold change 2). The number of individual and common upregulated (Up) and downregulated (Down) genes are shown in Venn diagrams. (B) Top three predicted binding motifs enriched in the regulatory sequences (TSS ± 2 kb) of the differentially regulated target genes of TWIST1 dimers generated with HOMER (64). Names of the known motif and percentages of targets containing these motifs are listed. (C and D) Heat maps showing the relative expression (log2 fold versus A2LC) of selected genes involved in cell lineage, EMT, or cell signaling pathways for the dimer- and monomer-expressing EBs. Color-coded normalized Z-scores are shown. (E) RT-qPCR analysis of the expression of key lineage markers in different EBs. Expression was normalized against the value for the TBP gene and the wild-type sample at day 3 of differentiation. Each data point represents the mean ± standard error from n = 3 independent experiments. Data were analyzed by the nonparametric Mann-Whitney U test (two tailed). *, P ≤ 0.05; **, P ≤ 0.01; ns, nonsignificant. Black, pluripotency markers; green, endoderm markers; red, mesoderm markers; blue, neural crest markers.
To further dissect the functionality of each dimer, we performed additional enrichment analyses focusing on development-related gene sets and taking into account gene expression changes relative to the parental A2LC cells. Neural ectoderm differentiation derivatives were repressed by the monomer, while mesoderm and endoderm cell fates were repressed by TT (Table 1 and Table S3). On the other hand, heterodimers activated overlapping sets of genes associated with mesoderm and neural ectoderm development (Table 1 and Table S3). The T3 dimer activated heart developmental processes, including heart vasculature, muscle, and blood cells, while the most frequent processes induced by T4 were related to neuron differentiation (Table 1 and Table S3). T12 activity was intermediate between those of T3 and T4. A comparison down to key cell lineage and pathway genes also reflected these changes (Fig. 5C and D), which were furthermore validated by reverse transcription-quantitative PCR (RT-qPCR) experiments (Fig. 5E). The most evident changes in the heterodimer groups were the downregulation of pluripotency genes and a burst of expression of lineage markers, especially those for mesoderm and neural crest ectoderm development (Fig. 5C and E). All dimers except T3 repressed endoderm markers (Fig. 5D and E and Table S3).
TABLE 1.
Summary of gene ontology analysis of dimer-specific target genesa
| TWIST type | Gene ontologiesb |
|---|---|
| T0 | Cardiac atrium development, olfactory lobe development, endocrine pancreas development, pallium development, hippocampus development |
| TT | Cardiac atrium development, endoderm development, placenta blood vessel development, mesoderm development, embryonic skeletal system development |
| T3 | Artery development, coronary vasculature development, myeloid leukocyte differentiation, cardiac septum development, regulation of skeletal muscle tissue development |
| T4 | Dendrite development, regulation of erythrocyte differentiation, face development, neuron projection development, regulation of epidermal cell differentiation |
| T12 | Cardiac septum development, dendrite development, cerebral cortex development, positive regulation of dendrite development, cardiac chamber development |
Gene ontology enrichment analysis of genes induced by TWIST1 dimers and TWIST1 monomer against the parental A2LoxCre ESCs. Gene sets were filtered by total gene number of >20 and P value of <0.05.
The top five significant gene ontologies are indicated. Bold, upregulated gene sets; italic, downregulated gene sets.
Taken together, our results have demonstrated that TWIST1 dimerization partners drive different gene expression programs, which may be influenced by the preference for specific DNA motifs. Heterodimers appeared to drive ectoderm and mesoderm differentiation, whereas the TWIST1 homodimer repressed the expression of developmental genes while maintaining the pluripotency state.
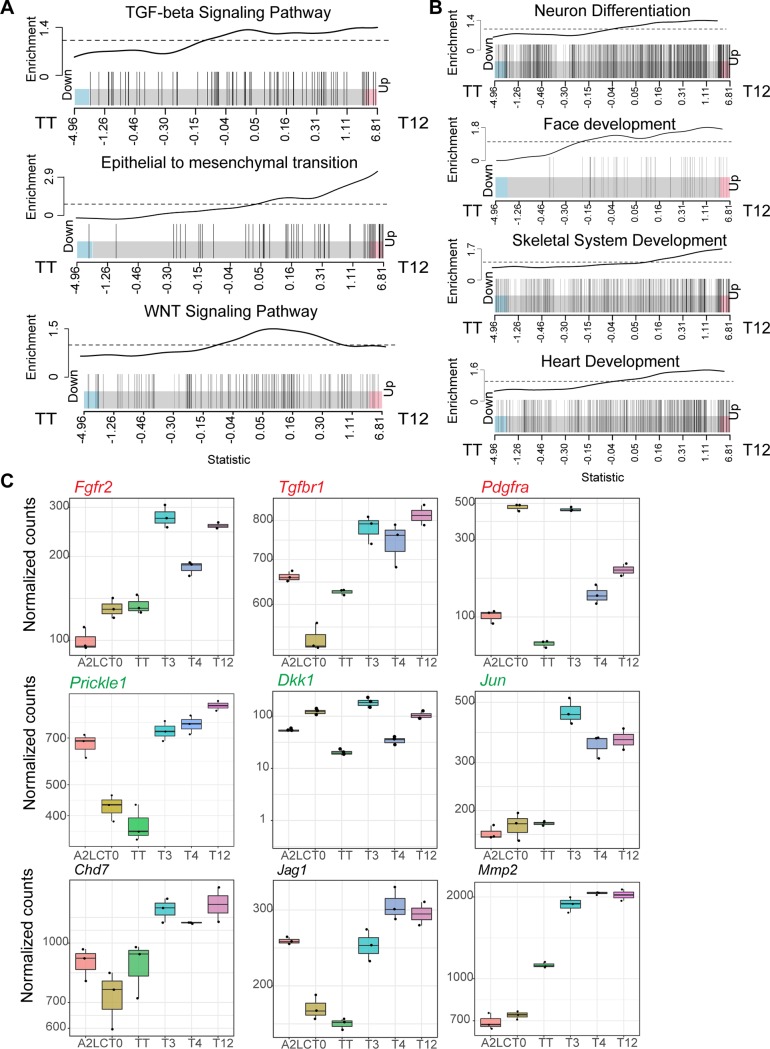
The TCF12-TWIST1 dimer activates EMT more robustly and antagonizes TWIST1 homodimer activity for critical signaling pathway genes.
Twist1 and Tcf12 have been shown to act synergistically for cranial tissue development (39). Previous studies have suggested that homodimers and TWIST1-TCF3 (E2A) heterodimers regulate the skull suture closure (21, 26). Therefore, we focused further on our analysis of the previously uncharacterized T12 dimer and its ability to induce transcription of developmental pathways in comparison with TT (see Table S4 in the supplemental material). T12-expressing cells significantly activated more genes associated with TGF-β signaling (false-discovery rate [FDR], 0.007), WNT signaling (FDR, 0.017), and EMT (FDR, 0.002) (around 50% of all EMT genes were enriched in T12 cells) (Fig. 6A). T12-expressing cells were also enriched for genes associated with processes of neuronal, skeletal, face, and cardiovascular system development (Fig. 6B and Table S4). Examining specifically genes encoding transcriptional regulators associated with craniofacial development (Fig. 6B, Face development), we noted that the homodimer and the heterodimer antagonistically regulated a subset of critical developmental regulators such as those associated with TGF-β signaling (e.g., Tgfbr1 and Pdgfrα) and WNT signaling (Prickle1 and Dkk1) (Fig. 6C). Other genes, represented by Fgfr2, Jun, Chd7, and Mmp2, were activated more effectively by T12 than by TT (Fig. 6C).
FIG 6.
The TWIST1-TCF12 heterodimer antagonizes TWIST1 homodimer. (A and B) Barcode plots of the enrichment of cell signaling- and developmental process-related genes in TWIST1-TCF12 EBs (right) versus TWIST1-TWIST1 EBs (left). Individual genes, represented by vertical bars, are ranked from left to right based on log2 fold change (x axis) for TWIST1-TCF12 EBs versus TWIST1-TWIST1 EBs. The enrichment line shows the relative local enrichment of the bars in the plot. The dashed line indicates neutral enrichment. Values above this line show enrichment, while values below the line show depletion. (C) Box plots depicting the expression of selected genes important for craniofacial development and cell signaling pathways that are enriched in TWIST1-TCF12- versus TWIST1-TWIST1-expressing EBs. Red, TGF-β signaling-related genes; green, WNT signaling-related genes. Dots represent replicate data points. The top and bottom edge of each box show first and third quantiles (Q1 and Q3) of the data values, respectively; the middle bar shows median values. Upper whisker, highest observed value within [Q3, 1.5 × Q3]; lower whisker, lowest observed value within [0.5 × Q1, Q1].
The differential expression of downstream genes between T12 and TT and the functional consequences for craniofacial development are, therefore, likely to be the outcome of the balancing activity of these two dimers for transcriptional regulation.
DISCUSSION
TCF factors are the major dimerization partner and transcriptional modulator of TWIST1.
Combining affinity purification-coupled mass spectrometry, coexpression data from cranial mesenchyme, and coimmunoprecipitation analysis, we identified TCF3, TCF4, and TCF12 E-proteins as the predominant interacting partners of TWIST1. This is also supported by the results of a TWIST1 proximity labeling screen showing that the TCF factors partner with TWIST1 consistently in neural crest cells, mesenchymal stem cells, and 3T3 cells (X. Fan et al., unpublished results). E-proteins are also major dimerization partners of several other class II bHLH factors (45, 46). For example, TCF3, TCF4, and TCF12 are top candidates for interacting with HAND2 and enhance its transcriptional activity at the promoter of target genes (45). Consistent with these observations, we found that TWIST1 induced differentiation programs more effectively when coupled with E-proteins. Also, similar to the case for HAND2, coupling TWIST1 with different partners resulted in altered DNA-binding activity as previously shown by electromobility shift assay (18, 45). Binding of in vitro-synthesized tethered TWIST1 dimers against isotope-labeled oligonucleotides has revealed variations of the E-box consensus sequence (CANNTG). By measuring the intensity of the shifted DNA, it was found that TWIST1 homodimers bind preferably to the CATATG sequence, while TWIST1-HAND2 and TWIST1-TCF3 bind more effectively to the CATCTG sequence (18). These observations suggest that E-proteins might be the primary transcriptional modulators of TWIST1, and potentially other class II bHLH factors, for the regulation of various tissue-specific programs during development.
Despite a focus on the E-protein partners, we do not exclude the possibility that TWIST1 interacts with other bHLH factors that were not detected in this study. It is likely that TWIST1 may also interact with additional TWIST family members, such as HAND2, which has been shown to interact genetically with TWIST1 in limb development (17). The interaction between TWIST1 and other bHLH factors may be selectively enhanced in certain cell populations, such as cells undergoing osteogenesis. Activation of TGF-β signaling in MSCs induces expression of ID proteins as well as phosphorylation that may disrupt TWIST1-TCF interaction (21, 26) and thus free up TWIST1 binding for alternative partners.
Pathological mutations induce bHLH factor dimerization imbalance.
For TWIST family bHLH factors, dimerization can be modulated by phosphorylation of the bHLH domain (16–18). Hypophosphorylation mutations (T121A and S123A) near or at the helix I phosphorylation sites were previously shown to favor homodimerization (17). Nevertheless, a triple phosphomutant model of TWIST1 (S123A, T148A, and A184A) in oncogenic cell lines showed a preference for dimerization with TCF3 and HAND2 (47). In line with this, we found that two individual hypophosphorylation mutations (R118H and S123A) both increased preference for TCF12 heterodimer formation, while phosphomimetic forms at T121 and S123 reduced heterodimerization. We noted that incorporation of the negatively charged phosphate group into these residues might reduce binding to DNA, rather than directly interfering with the dimerization interface. Therefore, T121/S123-dephosphorylated TWIST1 may favor heterodimerization through stabilizing DNA binding or adapting a helix conformation that enhances E-protein binding. Driven by a potentially different mechanism, TWIST1 helix II mutations at the interaction site severely compromised hetero- versus homodimerization potential, which had more significant functional consequence than helix I variants and impacted on the migration of mesenchymal cells. Altogether, these observations demonstrate that these residues constitute an essential interface for dimer balance adjustment and therefore play a critical role in cell differentiation and development.
RNA-seq analysis of differentiating ESCs revealed that the TWIST1-TCF12 heterodimer is a potent driver of EMT and mesenchymal differentiation compared to the TWIST1 homodimer. TWIST1-TCF12 also induced signaling pathways and key transcription regulators associated with head mesenchyme development. These genes were either inactive or repressed in cells expressing the TWIST1 homodimer. The Twist1 heterozygous (Twist1+/−) mouse model, which recapitulates SCS phenotypes, displays excessive FGF and TGF-β signaling at the osteogenic front, as revealed by the expansion of FGFR2, pSmad 1/5/8, and ID protein signals (21, 26). These misregulated signaling activities could, therefore, be a consequence of imbalanced TWIST1–E-protein activities.
Dimerization-dependent activities of TWIST1 in progenitor cell lineage specification.
TWIST1 regulates multiple pathways during lineage specification. Loss of Twist1 function leads to retention of pluripotency, defective mesoderm, and ectoderm differentiation, as well as disrupted EMT. EMT is tightly associated with ESC differentiation, especially the specification of mesoderm (42, 48, 49). Major EMT drivers, including Twist1, Zeb2, Snail1, and Snail2, are induced by canonical WNT signaling in mesendoderm differentiation (50), while constitutive activation of WNT further directs precursor cells into primitive endoderm (51). Both Twist1 and Snail1 are required for WNT-induced mesoderm differentiation (39; this study), but only Twist1 negatively feeds back on WNT signaling regulation and represses endoderm cell fates. Overexpression of the TWIST1 homodimer blocked WNT activity and endoderm differentiation, while the loss of TWIST1 shifted differentiation toward endoderm. On the other hand, the ablation of Snail1 promotes ectodermal instead of mesendodermal fate without affecting WNT pathway genes (39). These observations suggest a dichotomic control carried out by TWIST1 during development: (i) an EMT-dependent role in promoting mesoderm and neural crest commitment and (ii) an EMT-independent role in endodermal repression potentially by negative feedback on WNT activity.
E-proteins direct these activities undertaken by TWIST1 during lineage commitment. E-proteins display overlapping expression patterns and developmental functions in embryonic tissues (7, 45). They are expressed in the cranial mesenchyme and proliferating neuroepithelium at midgestation (E9.5 to E11.5) (45). Tcf3 and Tcf12 expression is restricted to the proliferative ventricular and subventricular zones and is downregulated during neurogenesis (52, 53). Tcf4 expression persists in the adult brain in regions that overlap regions expressing genes encoding neuronal bHLH factors such as Atoh1, Ascl1, and Neurog2 and initiates neuronal specification (46). Consistent with these findings, we have shown that tethered TWIST1-TCF4 modulates the activity of neural ectoderm genes, whereas tethered TWIST1-TCF3 most strongly activated the mesoderm genes. TWIST1–E-protein heterodimers activated the neural crest cell differentiation and TGF-β signaling pathway, which in turn upregulated ID proteins that could compete for E-protein binding, providing a potential negative feedback mechanism to modulate the activity of the heterodimers (21).
In regard to studies conducted by Connerney et al. (21, 26) on TWIST1 homodimers and TWIST1-TCF3 in cranial suture MSCs, we included two additional E-proteins and systemically analyzed the differential activities of in total four major TWIST1 dimers in early embryonic patterning. Using the tethered-dimer inducible system and quantitative transcriptome analysis, we have demonstrated the functions of designated dimers by their global downstream changes in genes and pathways. In the MSCs, the TWIST1-TCF3 heterodimer represses ossification, while in the ESCs, all three dimers (TWIST1-TCF3, -TCF4, and -TCF12) drive differentiation toward multiple cell lineages, whereas the homodimer maintains pluripotency. The distinct output of TWIST1 dimers in pluripotent versus multipotent stem cells could be related to differences in the chromatin context or in the switching between downstream activating/repressive transcriptional regulatory partners.
In conclusion, our study has uncovered the predominant dimerization partners of TWIST1 and their role in regulating lineage differentiation during early development. TWIST1 interacts with TCF proteins to promote the differentiation of neural crest and mesoderm progenitors and to repress endoderm lineage differentiation with the help of the TWIST1 homodimer. The TWIST1-TCF dimers enable the mesenchyme precursors to acquire myogenic, skeletogenic, and neurogenic fates. In the context of defective development, we showed that the mutations of the TWIST1 bHLH domain are likely to alter the balance of dimer compositions, which may lead to the mesenchymal tissue defects.
MATERIALS AND METHODS
Cell lines and cell culture conditions.
Cell lines and their treatments are summarized in Table 2. The Madin-Darby canine kidney (MDCK) cell line overexpressing human TWIST1 was received from Xue and colleagues (54). Cells were maintained in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) (Gibco), 10% fetal calf serum (FCS), and 10 mM β-mercaptoethanol at 37°C and 5% CO2. TWIST1 expression was under constant selection using 4 μg/ml of puromycin (54). The C3H10T1/2 cell line was received from ATCC. Cells were maintained in high-glucose DMEM with 10% FCS (Life Tech, Australia) and 10 mM β-mercaptoethanol at 37°C in 5% CO2.
TABLE 2.
Summary of cell lines and treatments used in this study
| Cell line(s) | Description (reference or source) | Treatment(s) | Expt(s) |
|---|---|---|---|
| MDCK (hTWIST1 O/E) | MDCK cells stably overexpressing hTWIST1 (54) | Transfection with pCMV-XX-HA constructs for partners | IP MS, co-IP validation |
| C3H10T1/2 | Mesenchymal stem cells (ATCC) | Transfection with BiFC constructs | BiFC, scratch assay |
| mESC | EB formation and doxycycline induction | qPCR analysis or RNA-seq | |
| A2loxcre | 41 | ||
| Twist1−/− and Twist1+/− | Derived from A2loxcre by CRISPR editing | ||
| T0, TT, T3, T4, and T12 | Derived from A2loxcre by induced cassette exchange; doxycycline inducible |
Mouse ESCs (A2loxCre) were a gift from the Kyba lab (Lillehei Heart Institute, MN) and were originally described by Mazzoni et al. (41). The ESCs were cultured in complete mESC medium (high-glucose DMEM [Gibco], 12.5% [vol/vol] heat-inactivated fetal bovine serum [FBS] [Fisher Biotec Australia], 10 mM β-mercaptoethanol, 0.09% [vol/vol] 1× nonessential amino acids [100×; Thermo Fisher Scientific], 1% [vol/vol] nucleosides [100×; Merck], and 10 million U/ml ESGRO mouse leukemia inhibitory factor [Merck]). All cell lines tested free of mycoplasma.
In situ hybridization.
The use of C57BL/6-Arc E9.5 mouse embryos was approved by the Children’s Medical Research Institute/Children’s Hospital at Westmead Animal Ethics Committee. Embryo collection and in situ hybridization were performed as previously described (14).
Protein immunoprecipitation.
For the validation of protein interaction, hTWIST1-expressing MDCK cells were transfected 24 h before immunostaining using Lipofectamine 3000 (Life Tech) according to the manufacturer’s instructions with one of the following plasmids: pCMV-gfp-HA, pCMV-Tcf3-HA, pCMV-Tcf4-HA, pCMV-Twist2-HA, pCMV-Ebf1-HA, pCMV-Ebf3-HA, pCMV-Sim2-HA, or pCMV-Tal1-HA.
Cell pellets were thawed in 300 μl hypotonic lysis buffer (20 mM HEPES, 1 mM MgCl2, 10% glycerol, 0.5% Triton X-100, 1 mM dithiothreitol [DTT], 1× complete protease inhibitor [Roche], 1,000 U/ml benzonuclease) and incubated at room temperature for 15 min. An equal volume of hypertonic lysis buffer (20 mM HEPES, 500 mM NaCl2, 1 mM MgCl2, 10% glycerol, 1 mM DTT, 1× complete protease inhibitor) was applied to the lysate. Cells were further broken down by passage through a 25-gauge needle 10 times and rotated at 4°C for 30 min before to be centrifuged at 14,000 × g for 15 min. The supernatant was incubated with an anti-TWIST1 or an anti-FLAG antibody (2 μg/ml) at 4°C for 2 h, and 1/10 volume of protein G-agarose beads (Roche) was then added and the sample rotated for 30 min at room temperature. Beads were finally washed with ice-cold wash buffer (a 1:1 mixture of the two lysis buffers) 6 times and transferred to a new 1.5-ml tube. Samples were eluted in 2× lithium dodecyl sulfate (LDS) loading buffer (Life Technologies) at 70°C for 10 min, and half of the eluate was loaded for SDS-PAGE in parallel with the “input” control for Western blot analysis.
Western blotting.
Proteins were extracted using radioimmunoprecipitation assay (RIPA) lysis buffer (1× phosphate-buffered saline [PBS], 1.5% Triton X-100, 1% Igepal, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM DTT, 1× complete protease inhibitor [Roche]) for 30 min at 4°C while rotating. Lysed samples were then centrifuged at 15,000 × g, and the supernatant was collected. Protein concentration was measured using the Direct Detect spectrometer (Millipore). Twenty micrograms of protein was denatured at 70°C for 10 min in 1× LDS loading buffer. Protein electrophoresis and transfer were performed using the NuPage system (Life Technologies, catalog number NP0322BOX), following the manufacturer’s instructions.
Primary antibodies used were mouse monoclonal anti-TWIST1 (1:1,000; Abcam, catalog number ab50887), mouse anti-α-tubulin (1:1,000; Sigma, catalog number T6199), rabbit anti-HA (1:1,000; Abcam, catalog number ab9110), mouse anti-FLAG M2 (1:1,000; Sigma, catalog number F1804), mouse anti-TCF4 (1:500; Santa Cruz, catalog number sc-166699), rabbit anti-TCF12 (1:400; Santa Cruz, catalog number SC-357 1), and mouse anti-GFP (1:1,000; Thermo Fisher Scientific, catalog number A-11120). Secondary antibodies used were horseradish peroxidase (HRP)-conjugated donkey anti-rabbit IgG (1:8,000; Jackson Immunoresearch, catalog number 711-035-152) and HRP-conjugated donkey anti-mouse IgG (1:8,000; Jackson Immunoresearch, catalog number 711-035-150).
Protein affinity purification and mass spectrometry.
A total of 2 × 107 MDCK hTWIST1-overexpressing cells were collected for each experiment. Cell pellets were thawed in 500 μl hypotonic lysis buffer (20 mM HEPES, 1 mM MgCl2, 10% glycerol, 0.5% Triton X-100, 1 mM DTT, 1× complete protease inhibitor [Roche], 1,000 U/ml benzonuclease) and incubated at room temperature for 15 min. An equal volume of hypertonic lysis buffer (20 mM HEPES, 500 mM NaCl2, 1 mM MgCl2, 10% glycerol, 1 mM DTT, 1× complete protease inhibitor) was applied to the lysate. Cells were further broken down by passage through a 25-gauge needle 10 times and then rotated at 4°C for 30 min. After centrifugation at 14,000 × g for 15 min, 450 μl of lysate was incubated with 5 μg of an anti-TWIST1 antibody or mouse IgG at 4°C overnight with rotation. Meanwhile, a Dynabead-protein G slurry (Invitrogen, catalog number 10003D) was prepared by washing and blocking for 1 h at room temperature in 1% BSA–0.1% Triton X-100 dissolved in 1× PBS before use. Fifty microliters of Dynabeads was then added to each sample and rotated at room temperature for 30 min. Beads were then washed with 500 μl ice-cold wash buffer (1:1 mixture of the lysis buffer, without nuclease) 6 times and transferred to a new 1.5-ml tube before the last wash. Beads were then washed quickly with 1 ml of cold 50 mM Tris-HCl (pH 7.4) and then 500 μl of triethylammonium bicarbonate (TEAB) buffer (75 mM). Lastly, beads were collected by centrifugation for 5 min at 2,000 × g before being processed for mass spectrometry analysis.
Tryptic digestion of the immunoprecipitated proteins was performed by adding trypsin at 1:20 (μg) directly to the washed beads in 50 mM TEAB buffer, vortexing, and incubating at 37°C overnight. The second digestion was performed by adding trypsin at 1:40 the next day and incubating for 4 h. The tubes were then placed on a magnet, and the supernatant was collected and directly transferred into trifluoroacetic acid (TFA) to obtain a final concentration of 0.5%.
Proteolytic peptides were desalted using Oligo R3 reversed-phase resin (Thermo Fisher Scientific) in custom-made stage tips (55). Mass spectrometry (MS) was performed using an LTQ Velos-Orbitrap MS (Thermo Fisher Scientific) coupled with an UltiMate RSLCnano-LC system (Thermo Fisher Scientific). Raw MS data files were processed using Proteome Discoverer v.1.3 (Thermo Fisher Scientific). Processed files were searched against the UniProt mammalian database (downloaded November 2016) using the Mascot search engine version 2.3.0. Searches were done with tryptic specificity allowing up to two missed cleavages and tolerance on the mass measurement of 10 ppm in MS mode and 0.3 Da for MS/MS ions. Using a reversed decoy database, a false-discovery rate (FDR) threshold of 1% was used. The lists of protein groups were filtered for first hits.
Rank product analysis for MS data.
We generated three sets of data using the nonparametric rank product method (32) and based on how many times a protein was reported across independent experiments (see Table S2 in the supplemental material). To test for cases where proteins were preferentially detected (here “preferentially” is defined as present in at least 2 samples for Twist1, i.e., set 1, and 2 samples for control, i.e., set 2, and not detected in at least 2 samples of their respective contrast), we employed a single-class test. If proteins were present in at least two Twist and two control samples, we deemed this set suitable for a two-class comparison, i.e., direct comparison of protein levels (set 3). The overall schema is represented in Fig. 1E and summarized below as follows: set 1, present preferentially in TWIST1 IPs (n ≥ 2); set 2, present preferentially in control IPs (n ≥ 2); and set 3, present in Twist and control cell IPs (n ≥ 2). P values and FDRs were then determined using the rank product method implemented in the RankProd R package (version 2.34) using 1,000 permutations (56). We used an FDR threshold of 0.20 in each set. If a protein for set 3 was present in set 1 or 2, it was removed after FDR calculation.
Plasmid construction.
Plasmids used in the BiFC experiments were a generous gift from the Hu lab and were imported via the Addgene depository. The plasmid backbone and the describing publication are listed in Table S3 in the supplemental material (57). Twist1 and Tcf12 coding sequences were cloned from cDNAs generated from E9.5 mouse head embryos and inserted into the relevant expression plasmids. Mouse homologs of S123A, R118H, T121E;S123E, T148A, A152P, and I156N variants were generated by site-directed mutagenesis as described previously (58) using primers listed in Table S5 in the supplemental material.
BiFC experiments.
Bimolecular fluorescence complementation (BiFC) experiments and quantitative analyses were performed as described previously (38). The day before transfection, C3H10T1/2 cells were seeded on 0.1% gelatin-coated coverslips at a density of 1.66 × 106 cells per 9.6 cm2 (well of a 6-well plate), and 24 h later, transient transfection was performed using Lipofectamine 3000 (Life Technologies) according to the manufacturer’s instructions. Equimolar quantities (1 μg each) of plasmids encoding TWIST1-VN173, TCF12-CrN173, and wild-type or mutant TWIST1*-CC155 were cotransfected.
Cells were imaged at 16 to 24 h following transfection. Before imaging, cells were brought to room temperature, washed twice with 1× PBS, and fixed in 4% paraformaldehyde (dissolved in 1× PBS) for 10 min. Coverslips were then washed three times with 1% Tween 20 in 1× PBS (PBT), once with 1% Triton X-100 in 1× PBS for 5 min, and three times with PBT. This was followed by incubation with 4′,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich; 1:10,000) for 10 min at room temperature. Coverslips were then washed three times with PBT for 5 min in the dark and mounted using Fluoromount-G (Invitrogen). All immunofluorescence slides were imaged using Zeiss Axio Imager A1 (Carl Zeiss, Australia). TWIST1*-CC155/TCF12-CrN173 heterodimers were visualized using a CFP filter set, while a GFP filter set was used for the detection of TWIST1*-CC155/TWIST1-VN173 homodimers. DAPI was imaged at 405 nm. For BiFC quantification, regions of the slides to be imaged were randomly chosen and pictured at a magnification of ×20. At least 1,000 cells for each treatment (based on DAPI counting) were sampled across three biological replicates. Fluorescence intensity for the nucleus (DAPI stained) was quantified using ImageJ software and analyzed in R (scripts are available upon request).
After filtering for background fluorescence, using the same intensity threshold for all treatment groups, the CFP/GFP ratio was taken for each cell. The mean value of this ratio was used to compare the CC155/CrN173 and CC155/VN173 dimer formation efficiencies between wild-type and mutant TWIST1. The nonparametric Mann-Whitney U test was used for statistical analysis. TWIST1* represents mouse homologs of the wild type or the S123A, R118H, T121E; S123E, T148A, A152P, or I156N variant.
Scratch assay.
TWIST1*-CC155 (1 μg) and TCF12-CrN173 (1 μg) were cotransfected into C3H10T1/2 cells seeded in wells of a 6-well plate. At 24 h after transfection and when cells had reached confluence, a p200 pipette tip was used to make a controlled scratch on the cell monolayer. Cells were then washed once with cell culture medium to remove floating cells detached by the scratch. Plates were then installed into the chamber (37°C, 5% CO2) of a Cell Observer wide-field microscope (Zeiss International), and bright-field images of the region scratched were taken every 15 min over 8 h. ImageJ was used to process the imaging data. Briefly, images were enhanced by subtracting the background, and binary images were created to assist the quantification of the gap closure. The total area of migration was calculated for each cell line, from the start of imaging to the time point when the first cell line closed the gap. The area of migration was represented as a percentage of the field of view. One-way analysis of variance (ANOVA), followed by a two-tailed t test, was used to determine the significance of the difference observed between the different conditions.
Generation of Twist1 mutant ESCs by CRISPR-Cas9.
ESC editing and selection by GFP were performed as described in detail previously (43). Briefly, 3 μg of pSpCas9(BB)-2A-GFP (Addgene plasmid number 48138, a gift from Feng Zhang) expressing the guide RNA (gRNA) of interest was electroporated into 1 × 106 A2loxCre ESCs using the Neon transfection system (Thermo Fisher Scientific). Colonies expressing GFP were picked 4 days after electroporation.
For genotyping, the region surrounding the mutation site (±200 bp) was amplified from cell lysates, purified, and cloned for sequencing. Clones with mono- or biallelic frameshift mutations were expanded and used for subsequent experiments.
Generation of dimer-inducible ESC lines.
The inducible TWIST1 dimer ESC lines were generated using the inducible cassette exchange method described previously (41, 42). The protein-coding sequences were cloned from mouse embryo cDNAs into the p2lox plasmid downstream of the FLAG tag-coding sequence (41, 42). Plasmids were transfected into A2loxCre cells treated with 1 μg/ml doxycycline for 24 h. The selection was performed in 300 μg/ml of G418 (Gibco) antibiotic for 1 week. Colonies were then picked and tested for construct expression following doxycycline treatment.
EB formation.
Embryoid bodies (EBs) were maintained in EB medium, consisting of high-glucose DMEM supplemented with 15% heat-inactivated FCS, 1% nonessential amino acid supplement, and 10 mM β-mercaptoethanol. Single ESCs were seeded onto Aggrewells in a 24-well plate (Stem Cell Technologies, catalog number 34411) at 1.5 × 106 cells per well in EB medium. Cell aggregates were transferred to non-tissue culture-treated 10-cm plates and EB medium 24 h later and placed onto an orbital shaker under slight rotation at 37°C with 5% CO2. Drug treatment conditions were optimized, as shown in Fig. 4C-i. Doxycycline was supplied at 50 ng/ml for T3 and T12, 100 ng/ml for TT, and 1,000 ng/ml for other cell lines after 2 days in differentiation medium. Cells were collected 16 h later, as the expression of known TWIST1 targets peaked at 12 to 16 h after induction.
RNA-seq and data analysis.
RNA library preparation (Truseq mRNA kit) and sequencing (Hiseq2500, 100bpPE) were performed by Macrogen (Seoul, South Korea). Raw Fastq sequence files (20 million reads per sample) were imported into Galaxy (https://usegalaxy.org.au/) and analyzed using a standard pipeline (59). The FastQC package was used to assess data quality, and adapter sequences and low-quality reads were trimmed from the original reads (Slidingwindow method in Trimmomatic package). A minimum of a 50-bp read length filter was applied. The alignment was performed using the Tophat2 package using the built-in mm10 genome. The resulting bam file was sorted and assessed before reads were counted using the htseq-count package and Genecode mouse basic genome assembly (GRCm38, vM10).
Read count values were imported into the R software for DEG analysis (60). Genes that were not expressed at a biologically meaningful level under any conditions were discarded. A standard counts-per-million (CPM) reads value threshold of 1 (or log CPM value of 0) was used for considering a gene to be expressed. To be kept for downstream analysis, a gene had to be expressed in at least three samples across the entire experiment. After filtering using these criteria, the number of genes was reduced to 14,165, approximately half of the original number. One replicate of the T12 sample was removed from further analysis due to low Tcf12 induction level and abnormal sample distance from other replicates in correlation map. For EdgeR and limma-voom analysis, the data were normalized by library size before further processing.
Preprocessed data were analyzed with R/Bioconductor package DESeq2 (61). The standard adjusted P value (adjp) threshold of 0.05 and log2 FC of 2 were used. Two types of gene set analysis were applied: a competitive gene set analysis using the clusterProfiler R package (62) for Fig. 4G and a self-contained gene set analysis using the ROAST package for Table S2 (summarized in Table 1) (63). In contrast to the commonly used competitive gene set analysis, the self-contained test takes gene expression into account and examines whether the genes in the set/pathway are differentially expressed as a whole for different dimers. Motif analysis on DEGs was performed using the Homer package (64).
Web resources.
The following web resources were used: OMIM (http://www.omim.org/), Galaxy Australia (https://usegalaxy.org.au/), the Human Gene Mutation Database (HGMD) (http://www.hgmd.cf.ac.uk/), and AQUARIA (http://aquaria.ws).
Data availability.
The raw RNA-seq sequence data were deposited into the NCBI GEO database and can be accessed with accession number GSE130252.
Supplementary Material
ACKNOWLEDGMENTS
Imaging analysis was performed at the ACRF Telomere Analysis Centre, and proteomics analysis was performed at the Biomedical Proteomics Facility, both supported by the Australian Cancer Research Foundation. Our work was supported by the Australian Research Council (DP 1094008 and DP 160100933) and by James Fairfax (Bridgestar Pty Ltd.). X.F. was supported by the University of Sydney International Postgraduate Research Scholarship, the Australian Postgraduate Award, and the CMRI Scholarship. P.P.L.T. is an NHMRC Senior Principal Research Fellow (grant ID 1110751).
X.F., P.P.L.T., and N.F. designed the project; X.F., M.D., P.O., and J.Q.J.S. conducted the experiments; D.A.F.L. and M.G. provided technical assistance with proteomics and transcriptome analysis; X.F., A.J.W., and P.O. performed the bioinformatics analysis; and X.F., A.J.W., P.P.L.T., and N.F. wrote the manuscript.
We declare no competing interests
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Murre C. 2019. Helix-loop-helix proteins and the advent of cellular diversity: 30 years of discovery. Genes Dev 33:6–25. doi: 10.1101/gad.320663.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dennis DJ, Han S, Schuurmans C. 2019. bHLH transcription factors in neural development, disease, and reprogramming. Brain Res 1705:48–65. doi: 10.1016/j.brainres.2018.03.013. [DOI] [PubMed] [Google Scholar]
- 3.Schlaeger TM, Schuh A, Flitter S, Fisher A, Mikkola H, Orkin SH, Vyas P, Porcher C. 2004. Decoding hematopoietic specificity in the helix-loop-helix domain of the transcription factor SCL/Tal-1. Mol Cell Biol 24:7491–7502. doi: 10.1128/MCB.24.17.7491-7502.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belle I, Zhuang Y. 2014. E proteins in lymphocyte development and lymphoid diseases. Curr Top Dev Biol 110:153–187. doi: 10.1016/B978-0-12-405943-6.00004-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Comai G, Tajbakhsh S. 2014. Molecular and cellular regulation of skeletal myogenesis. Curr Top Dev Biol 110:1–73. doi: 10.1016/B978-0-12-405943-6.00001-4. [DOI] [PubMed] [Google Scholar]
- 6.Massari ME, Murre C. 2000. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol 20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L-H, Baker NE. 2015. E proteins and ID proteins: helix-loop-helix partners in development and disease. Dev Cell 35:269–280. doi: 10.1016/j.devcel.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thisse B, el Messal M, Perrin-Schmitt F. 1987. The twist gene: isolation of a Drosophila zygotic gene necessary for the establishment of dorsoventral pattern. Nucleic Acids Res 15:3439–3453. doi: 10.1093/nar/15.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee JE, Hollenberg SM, Snider L, Turner DL, Lipnick N, Weintraub H. 1995. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science 268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- 10.Spicer DB, Rhee J, Cheung WL, Lassar AB. 1996. Inhibition of myogenic bHLH and MEF2 transcription factors by the bHLH protein Twist. Science 272:1476–1480. doi: 10.1126/science.272.5267.1476. [DOI] [PubMed] [Google Scholar]
- 11.El Ghouzzi V, Legeai-Mallet L, Aresta S, Benoist C, Munnich A, de Gunzburg J, Bonaventure J. 2000. Saethre-Chotzen mutations cause TWIST protein degradation or impaired nuclear location. Hum Mol Genet 9:813–819. doi: 10.1093/hmg/9.5.813. [DOI] [PubMed] [Google Scholar]
- 12.Bildsoe H, Loebel DA, Jones VJ, Chen Y-T, Behringer RR, Tam PP. 2009. Requirement for Twist1 in frontonasal and skull vault development in the mouse embryo. Dev Biol 331:176–188. doi: 10.1016/j.ydbio.2009.04.034. [DOI] [PubMed] [Google Scholar]
- 13.Bildsoe H, Loebel DA, Jones VJ, Hor ACC, Braithwaite AW, Chen Y-T, Behringer RR, Tam PP. 2013. The mesenchymal architecture of the cranial mesoderm of mouse embryos is disrupted by the loss of Twist1 function. Dev Biol 374:295–307. doi: 10.1016/j.ydbio.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bildsoe H, Fan X, Wilkie EE, Ashoti A, Jones VJ, Power M, Qin J, Wang J, Tam PPL, Loebel D. 2016. Transcriptional targets of TWIST1 in the cranial mesoderm regulate cell-matrix interactions and mesenchyme maintenance. Dev Biol 418:189–203. doi: 10.1016/j.ydbio.2016.08.016. [DOI] [PubMed] [Google Scholar]
- 15.Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. 1990. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell 61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 16.Centonze VE, Firulli BA, Firulli AB. 2004. Fluorescence resonance energy transfer (FRET) as a method to calculate the dimerization strength of basic helix-loop-helix (bHLH) proteins. Biol Proc Online 6:78–82. doi: 10.1251/bpo75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Firulli BA, Krawchuk D, Centonze VE, Vargesson N, Virshup DM, Conway SJ, Cserjesi P, Laufer E, Firulli AB. 2005. Altered Twist1 and Hand2 dimerization is associated with Saethre-Chotzen syndrome and limb abnormalities. Nat Genet 37:373–381. doi: 10.1038/ng1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Firulli BA, Redick BA, Conway SJ, Firulli AB. 2007. Mutations within helix I of Twist1 result in distinct limb defects and variation of DNA binding affinities. J Biol Chem 282:27536–27546. doi: 10.1074/jbc.M702613200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch DH, McMahon AP, Sommer L, Boussadia O, Kemler R. 2001. Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development (Cambridge, England) 128:1253–1264. [DOI] [PubMed] [Google Scholar]
- 20.Carver EA, Oram KF, Gridley T. 2002. Craniosynostosis in Twist heterozygous mice: a model for Saethre-Chotzen syndrome. Anat Rec 268:90–92. doi: 10.1002/ar.10124. [DOI] [PubMed] [Google Scholar]
- 21.Connerney J, Andreeva V, Leshem Y, Muentener C, Mercado MA, Spicer DB. 2006. Twist1 dimer selection regulates cranial suture patterning and fusion. Dev Dyn 235:1345–1357. doi: 10.1002/dvdy.20717. [DOI] [PubMed] [Google Scholar]
- 22.Chen ZF, Behringer RR. 1995. Twist is required in head mesenchyme for cranial neural tube morphogenesis. Genes Dev 9:686–699. doi: 10.1101/gad.9.6.686. [DOI] [PubMed] [Google Scholar]
- 23.Soo K, O'Rourke MP, Khoo P-L, Steiner KA, Wong N, Behringer RR, Tam PPL. 2002. Twist function is required for the morphogenesis of the cephalic neural tube and the differentiation of the cranial neural crest cells in the mouse embryo. Dev Biol 247:251–270. doi: 10.1006/dbio.2002.0699. [DOI] [PubMed] [Google Scholar]
- 24.Ota MS, Loebel DAF, O'Rourke MP, Wong N, Tsoi B, Tam PPL. 2004. Twist is required for patterning the cranial nerves and maintaining the viability of mesodermal cells. Dev Dyn 230:216–228. doi: 10.1002/dvdy.20047. [DOI] [PubMed] [Google Scholar]
- 25.Miraoui H, Severe N, Vaudin P, Pagès J-C, Marie PJ. 2010. Molecular silencing of Twist1 enhances osteogenic differentiation of murine mesenchymal stem cells: implication of FGFR2 signaling. J Cell Biochem 110:1147–1154. doi: 10.1002/jcb.22628. [DOI] [PubMed] [Google Scholar]
- 26.Connerney J, Andreeva V, Leshem Y, Mercado MA, Dowell K, Yang X, Lindner V, Friesel RE, Spicer DB. 2008. Twist1 homodimers enhance FGF responsiveness of the cranial sutures and promote suture closure. Dev Biol 318:323–334. doi: 10.1016/j.ydbio.2008.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rice DP. 2005. Craniofacial anomalies: from development to molecular pathogenesis. Curr Mol Med 5:699–722. doi: 10.2174/156bp652405774641043. [DOI] [PubMed] [Google Scholar]
- 28.Guenou H, Kaabeche K, Mée SL, Marie PJ. 2005. A role for fibroblast growth factor receptor-2 in the altered osteoblast phenotype induced by Twist haploinsufficiency in the Saethre-Chotzen syndrome. Hum Mol Genet 14:1429–1439. doi: 10.1093/hmg/ddi152. [DOI] [PubMed] [Google Scholar]
- 29.Fan X, Loebel DAF, Bildsoe H, Wilkie EE, Qin J, Wang J, Tam P. 2016. Tissue interactions, cell signaling and transcriptional control in the cranial mesoderm during craniofacial development. AIMS Genetics 3:74. doi: 10.3934/genet.2016.1.74. [DOI] [Google Scholar]
- 30.Skinner MK, Rawls A, Wilson-Rawls J, Roalson EH. 2010. Basic helix-loop-helix transcription factor gene family phylogenetics and nomenclature. Differentiation 80:1–8. doi: 10.1016/j.diff.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xue G, Hemmings BA. 2013. PKB/Akt-dependent regulation of cell motility. J Natl Cancer Inst 105:393–404. doi: 10.1093/jnci/djs648. [DOI] [PubMed] [Google Scholar]
- 32.Breitling R, Armengaud P, Amtmann A, Herzyk P. 2004. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett 573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 33.Castanon I, Baylies MK. 2002. A Twist in fate: evolutionary comparison of Twist structure and function. Gene 287:11–22. doi: 10.1016/s0378-1119(01)00893-9. [DOI] [PubMed] [Google Scholar]
- 34.Qin Q, Xu Y, He T, Qin C, Xu J. 2012. Normal and disease-related biological functions of Twist1 and underlying molecular mechanisms. Cell Res 22:90–106. doi: 10.1038/cr.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stenson PD, Ball EV, Mort M, Phillips AD, Shiel JA, Thomas NS, Abeysinghe S, Krawczak M, Cooper DN. 2003. Human Gene Mutation Database (HGMD): 2003 update. Hum Mutat 21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- 36.Barnes RM, Firulli AB. 2009. A twist of insight—the role of Twist-family bHLH factors in development. Int J Dev Biol 53:909–924. doi: 10.1387/ijdb.082747rb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Firulli AB, Conway SJ. 2008. Phosphoregulation of Twist1 provides a mechanism of cell fate control. Curr Med Chem 15:2641–2647. doi: 10.2174/092986708785908987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu C-D, Kerppola TK. 2003. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat Biotechnol 21:539–545. doi: 10.1038/nbt816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharma VP, Fenwick ALL, Brockop MS, McGowan SJ, Goos JA, Hoogeboom AJ, Brady AF, Jeelani NO, Lynch SA, Mulliken JB, Murray DJ, Phipps JM, Sweeney E, Tomkins SE, Wilson LC, Bennett S, Cornall RJ, Broxholme J, Kanapin A, Consortium, Johnson D, Wall SA, van der Spek PJ, Mathijssen IM, Maxson RE, Twigg SR, Wilkie AO. 2013. Mutations in TCF12, encoding a basic helix-loop-helix partner of TWIST1, are a frequent cause of coronal craniosynostosis. Nat Genet 45:304–307. doi: 10.1038/ng.2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Castanon I, Von Stetina S, Kass J, Baylies MK. 2001. Dimerization partners determine the activity of the Twist bHLH protein during Drosophila mesoderm development. Development (Cambridge, England) 128:3145–3159. [DOI] [PubMed] [Google Scholar]
- 41.Mazzoni EO, Mahony S, Iacovino M, Morrison CA, Mountoufaris G, Closser M, Whyte WA, Young RA, Kyba M, Gifford DK, Wichterle H. 2011. Embryonic stem cell-based mapping of developmental transcriptional programs. Nat Methods 8:1056–1058. doi: 10.1038/nmeth.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iacovino M, Roth ME, Kyba M. 2014. Rapid genetic modification of mouse embryonic stem cells by inducible cassette exchange recombination. Methods Mol Biol 1101:339–351. doi: 10.1007/978-1-62703-721-1_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sibbritt T, Osteil P, Fan X, Sun J, Salehin N, Knowles H, Shen J, Tam PPLPL. 2019. Gene editing of mouse embryonic and epiblast stem cells. Methods Mol Biol 1940:77–95. doi: 10.1007/978-1-4939-9086-3_6. [DOI] [PubMed] [Google Scholar]
- 44.Chang AT, Liu Y, Ayyanathan K, Benner C, Jiang Y, Prokop JW, Paz H, Wang D, Li H-R, Fu X-D, Rauscher FJ, Yang J. 2015. An evolutionarily conserved DNA architecture determines target specificity of the TWIST family bHLH transcription factors. Genes Dev 29:603–616. doi: 10.1101/gad.242842.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murakami M, Kataoka K, Tominaga J, Nakagawa O, Kurihara H. 2004. Differential cooperation between dHAND and three different E-proteins. Biochem Biophys Res Commun 323:168–174. doi: 10.1016/j.bbrc.2004.08.072. [DOI] [PubMed] [Google Scholar]
- 46.Forrest MP, Hill MJ, Quantock AJ, Martin-Rendon E, Blake DJ. 2014. The emerging roles of TCF4 in disease and development. Trends Mol Med 20:322–331. doi: 10.1016/j.molmed.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 47.Wang J, Nikhil K, Viccaro K, Chang L, Jacobsen M, Sandusky G, Shah K. 2017. The Aurora-A-Twist1 axis promotes highly aggressive phenotypes in pancreatic carcinoma. J Cell Sci 130:1078–1093. doi: 10.1242/jcs.196790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Evseenko D, Zhu Y, Schenke-Layland K, Kuo J, Latour B, Ge S, Scholes J, Dravid G, Li X, MacLellan WR, Crooks GM. 2010. Mapping the first stages of mesoderm commitment during differentiation of human embryonic stem cells. Proc Natl Acad Sci U S A 107:13742–13747. doi: 10.1073/pnas.1002077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakaya Y, Sheng G. 2008. Epithelial to mesenchymal transition during gastrulation: an embryological view. Dev Growth Differ 50:755–766. doi: 10.1111/j.1440-169X.2008.01070.x. [DOI] [PubMed] [Google Scholar]
- 50.ten Berge D, Koole W, Fuerer C, Fish M, Eroglu E, Nusse R. 2008. Wnt signaling mediates self-organization and axis formation in embryoid bodies. Cell Stem Cell 3:508–518. doi: 10.1016/j.stem.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Price FD, Yin H, Jones A, van Ijcken W, Grosveld F, Rudnicki MA. 2013. Canonical Wnt signaling induces a primitive endoderm metastable state in mouse embryonic stem cells. Stem Cells 31:752–764. doi: 10.1002/stem.1321. [DOI] [PubMed] [Google Scholar]
- 52.Uittenbogaard M, Chiaramello A. 2002. Expression of the bHLH transcription factor Tcf12 (ME1) gene is linked to the expansion of precursor cell populations during neurogenesis. Brain Res Gene Expr Patterns 1:115–121. doi: 10.1016/s1567-133x(01)00022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pfurr S, Chu Y-H, Bohrer C, Greulich F, Beattie R, Mammadzada K, Hils M, Arnold SJ, Taylor V, Schachtrup K, Uhlenhaut NH, Schachtrup C. 2017. The E2A splice variant E47 regulates the differentiation of projection neurons via p57(KIP2) during cortical development. Development 144:3917–3931. doi: 10.1242/dev.145698. [DOI] [PubMed] [Google Scholar]
- 54.Xue G, Restuccia DF, Lan Q, Hynx D, Dirnhofer S, Hess D, Ruegg C, Hemmings BA. 2012. Akt/PKB-mediated phosphorylation of Twist1 promotes tumor metastasis via mediating cross-talk between PI3K/Akt and TGF-beta signaling axes. Cancer Discov 2:248–259. doi: 10.1158/2159-8290.CD-11-0270. [DOI] [PubMed] [Google Scholar]
- 55.Rappsilber J, Mann M, Ishihama Y. 2007. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 56.Hong F, Breitling R, McEntee CW, Wittner BS, Nemhauser JL, Chory J. 2006. RankProd: a bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics 22:2825–2827. doi: 10.1093/bioinformatics/btl476. [DOI] [PubMed] [Google Scholar]
- 57.Shyu YJ, Liu H, Deng X, Hu C-D. 2006. Identification of new fluorescent protein fragments for bimolecular fluorescence complementation analysis under physiological conditions. Biotechniques 40:61–66. doi: 10.2144/000112036. [DOI] [PubMed] [Google Scholar]
- 58.Liu H, Naismith JH. 2008. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol 8:91. doi: 10.1186/1472-6750-8-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK, Huber W, Robinson MD. 2013. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc 8:1765–1786. doi: 10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
- 60.Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Čech M, Chilton J, Clements D, Coraor N, Eberhard C, Grüning B, Guerler A, Hillman-Jackson J, Von Kuster G, Rasche E, Soranzo N, Turaga N, Taylor J, Nekrutenko A, Goecks J. 2016. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res 44:W3–W10. doi: 10.1093/nar/gkw343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu G, Wang L-G, Han Y, He Q-Y. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu D, Lim E, Vaillant F, Asselin-Labat M-L, Visvader JE, Smyth GK. 2010. ROAST: rotation gene set tests for complex microarray experiments. Bioinformatics 26:2176–2182. doi: 10.1093/bioinformatics/btq401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.O'Donoghue SI, Sabir KS, Kalemanov M, Stolte C, Wellmann B, Ho V, Roos M, Perdigão N, Buske FA, Heinrich J, Rost B, Schafferhans A. 2015. Aquaria: simplifying discovery and insight from protein structures. Nat Methods 12:98–99. doi: 10.1038/nmeth.3258. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw RNA-seq sequence data were deposited into the NCBI GEO database and can be accessed with accession number GSE130252.