Mastermind proteins are required for transcription of Notch target genes, yet the molecular basis for mastermind function remains incompletely understood. Previous work has shown that Notch can induce transcriptional responses by binding to promoters but more often by binding to enhancers, with HES4 and DTX1 as representative mammalian examples of promoter and enhancer responsiveness, respectively. Here, we show that mastermind dependence of the Notch response at these loci is differentially encoded in Jurkat T-cell acute lymphoblastic leukemia (T-ALL) cells.
KEYWORDS: Notch signaling, transcriptional activation
ABSTRACT
Mastermind proteins are required for transcription of Notch target genes, yet the molecular basis for mastermind function remains incompletely understood. Previous work has shown that Notch can induce transcriptional responses by binding to promoters but more often by binding to enhancers, with HES4 and DTX1 as representative mammalian examples of promoter and enhancer responsiveness, respectively. Here, we show that mastermind dependence of the Notch response at these loci is differentially encoded in Jurkat T-cell acute lymphoblastic leukemia (T-ALL) cells. Knockout of Mastermind-like 1 (MAML1) eliminates Notch-responsive activation of both these genes, and reduced target gene expression is accompanied by a decrease in H3K27 acetylation, consistent with the importance of MAML1 for p300 activity. Add-back of MAML1 variants in knockout cells identifies residues 151 to 350 of MAML1 as essential for expression of either Notch-responsive gene. Fusion of the Notch-binding region of MAML1 to the histone acetyltransferase (HAT) domain of p300 rescues expression of HES4 but not DTX1, suggesting that an additional activity of MAML1 is needed for gene induction at a distance. Together, these studies establish the functional importance of the MAML1 region from residues 151 to 350 for Notch-dependent transcriptional induction and reveal differential requirements for MAML1-dependent recruitment activities at different Notch-responsive loci, highlighting the molecular complexity of Notch-stimulated transcription.
INTRODUCTION
Notch proteins are receptors in a juxtacrine signaling pathway that influences numerous developmental patterning decisions (1, 2). Dysregulation of Notch signaling contributes to the pathogenesis of many human cancers, including T-cell acute lymphoblastic leukemia (T-ALL), a disease in which activating mutations of human Notch1 are well established oncogenic drivers (3–5).
Notch signaling is initiated by ligand binding, which induces proteolytic liberation of the Notch intracellular domain (NICD) into the cell, where it enters the nucleus to activate transcription. In the nucleus, it forms a Notch transcription complex (NTC) with the sequence-specific DNA binding protein RBPJ (also known as CSL) and a transcriptional coactivator of the Mastermind-like (MAML) family (6–8). The mechanisms by which NTC genomic binding activates transcription are an active area of study.
MAML proteins are required for transcription of mammalian Notch target genes, highlighting the importance of studying MAML proteins to understand the transcriptional response to Notch activation (8). In human MAML1, residues 13 to 74 directly interact with RBPJ and NICD1, but the remainder of the 1,016 amino-acid-long protein is poorly characterized (9). C-terminal truncations of MAML1 have dominant negative activity, presumably because these fragments bind the NTC but cannot interact with the other coregulators that it is responsible for recruiting (8, 10). These findings have motivated further characterization of the function of the remaining C-terminal portion of MAML1, in order to identify what cofactors it recruits and how these partners contribute to transcriptional activation.
Prior studies suggest that MAML1 has two transcriptional activation domains (TADs). A glutathione S-transferase (GST) fusion protein with the first region (TAD1), which includes residues 75 to 301, binds to p300 recovered from nuclear extracts, and a fragment from residues 1 to 301 [MAML1(1–301)], which includes both the region required for NTC formation and TAD1, is sufficient to stimulate transcription from an in vitro chromatinized template (11). However, it is not clear that MAML1 is directly responsible for recruiting p300 to the genome, as chromatin immunoprecipitation followed by PCR (ChIP-PCR) of the HES1 locus in C33A cells showed p300 at the promoter in the absence of Notch activation and before NTC binding (12). In addition, a similar fragment, MAML1(1–300), acts as a dominant negative protein in reporter assays, further complicating interpretation of how p300 binding is related to transcriptional induction by NTCs (10). The remaining C-terminal portion of MAML1 (TAD2) is not required for transcription in vitro but is required in cells (11). One line of investigation has suggested that this part of MAML1 is required for recruitment of cyclin C:CDK8 complexes (and, thus, the kinase module of the mediator complex), thereby promoting phosphorylation of NICD1, but how these activities are coordinated and how they contribute to target gene expression remain incompletely understood (12).
Such mechanistic studies looking at regulation of Notch-responsive genes have centered on canonical targets such as HES1, which is regulated by promoter proximal NTC binding (12). Most cell-type-specific Notch-responsive genes, however, are regulated by binding of the NTC to distal enhancers (13–16). The conflicting requirements reported for different MAML1 activities in vitro, in cells, and in organisms highlight the importance of studying Notch function in physiologically relevant cellular systems and suggest that more expansive approaches could uncover different roles for MAML binding at other genomic loci.
In this study, we have interrogated the function of MAML1 fragments in regulating Notch-dependent gene expression in Jurkat T-ALL cells, examining established target genes regulated by either promoter proximal Notch binding (HES4) or distal enhancer binding (DTX1). We greatly extended prior work by creating MAML1 knockout cells and by using them to show that a form of MAML1 lacking residues 151 to 350, a region that overlaps TAD1, is defective in rescuing Notch target gene expression in MAML1 knockout cells. Furthermore, we established that reduced target gene expression is accompanied by a decrease in H3K27 acetylation, consistent with the importance of the excised region from residues 151 to 350 for p300 recruitment. However, when the Notch binding region of MAML1 is fused to the histone acetyltransferase (HAT) domain of p300, the fusion protein is able to rescue expression of HES4, the proximal-promoter-regulated gene, but not DTX1, the enhancer-regulated Notch target gene, suggesting that an additional activity of MAML1 is needed for gene induction at this locus. Together, these studies establish the functional importance of the MAML1(151–350) region for Notch-dependent transcriptional induction and reveal differential requirements for MAML1-dependent recruitment activities at different Notch-responsive loci, highlighting the molecular complexity of NTC-stimulated transcription.
RESULTS
Jurkat cells exhibit dynamic Notch responsiveness dependent on MAML1.
To investigate MAML structure-function relationships, we turned to the Jurkat T-ALL cell line because Jurkat cells have a juxtamembrane insertion in Notch1 that renders the protein constitutively active, but they do not depend on Notch activity for survival (5, 17). Furthermore, the constitutive activity of the mutated Notch1 protein in these cells relies on gamma secretase cleavage for release from the membrane, allowing us to use a well-established gamma secretase inhibitor (GSI) washout assay (18) to toggle between Notch-off (GSI) and Notch-on (washout) states.
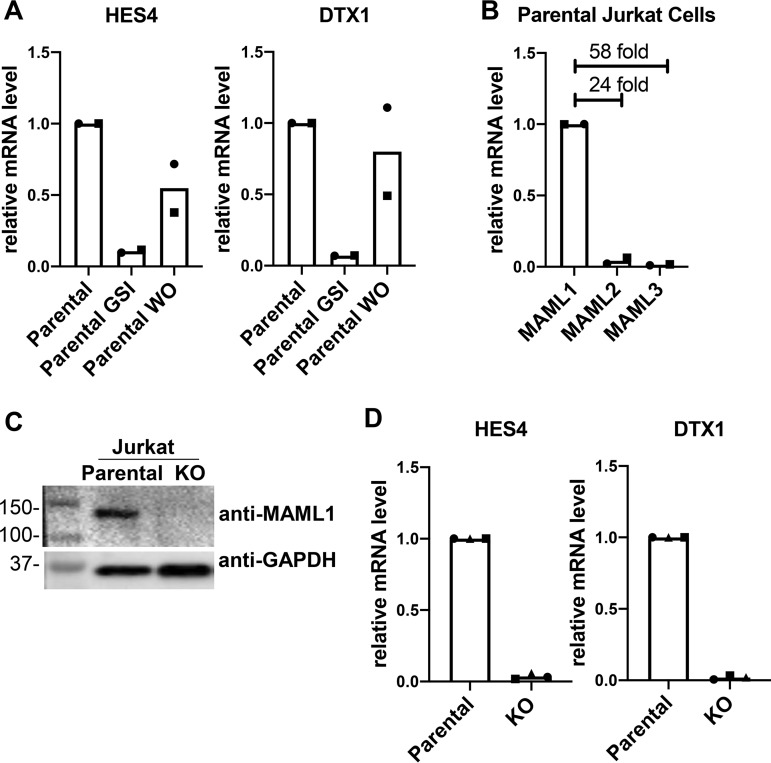
With this approach, we confirmed that HES4 and DTX1 are direct Notch1 target genes in these cells by quantitative reverse transcription-PCR (RT-qPCR) (Fig. 1A). Expression of each of these genes is suppressed by GSI treatment and restored upon inhibitor washout, a characteristic feature of Notch target genes. Because HES4 and DTX1 exhibited robust responsiveness to Notch1 in these cells, and because they have previously been shown to be Notch-responsive genes in other T-ALL lines, they were chosen as representative proximally and distally regulated genes, respectively, for this study (13, 16).
FIG 1.
Jurkat cells are dependent on MAML1 for Notch-stimulated transcription. (A) HES4 and DTX1 are Notch-responsive genes in Jurkat cells. RT-qPCR results from untreated Jurkat cells (parental), Jurkat cells treated with gamma secretase inhibitor (parental GSI), and Jurkat cells after GSI washout (parental WO) are shown. Data points show the average for technical replicates from each biological replicate, with each biological replicate indicated using a different point shape. (B) Comparison of MAML1, MAML2, and MAML3 transcript abundances in parental Jurkat cells, as determined by RT-qPCR. Data points show the average for technical replicates from each biological replicate, with each biological replicate indicated using a different point shape. (C) Western blot for MAML1 protein in parental and MAML1 KO Jurkat cells. Numbers to the left of the blot denote molecular masses of protein standards in kilodaltons. (D) Expression of Notch target genes HES4 and DTX1 in parental and MAML1 KO Jurkat cells, as determined by RT-qPCR. Data points show the average for technical replicates from each biological replicate, with each biological replicate indicated using a different point shape.
Next, we determined the transcript abundance of the three human MAML proteins in Jurkat cells by RT-qPCR. MAML1 transcripts are much more abundant in these cells than are MAML2 and MAML3 (Fig. 1B), consistent with prior studies implicating MAML1 as the most important member of this family in lymphocyte development in vivo (19). Therefore, we created a clonal MAML1 knockout (KO) cell line using CRISPR-Cas9 genome editing (Fig. 1C) to enable subsequent add-back studies for structure-function analysis. Consistent with the conclusion that HES4 and DTX1 are indeed Notch target genes in this cell context, their expression was greatly suppressed in the KO line (Fig. 1D).
Amino acids 151 to 350 of MAML1 are essential for Notch activation.
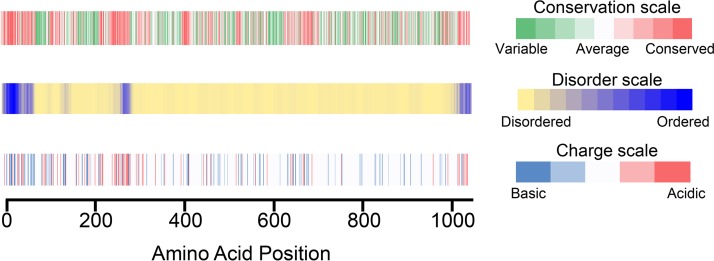
In order to elaborate structure-function relationships, we created a series of truncation and internal deletion variants of MAML1. First, we assessed the conservation, predicted disorder, and charge distribution in MAML1. Residues 13 to 74 of MAML1 interact directly with Notch1 and RBPJ, forming the core of the NTC (9, 20, 21). This N-terminal region is also more highly conserved and predicted to be more ordered than the remainder of the protein (Fig. 2). There are two other segments that are more acidic, more conserved, and more likely to be ordered, spanning residues 263 to 276 and 990 to 1016. The C-terminal segment has been reported to associate with the human papillomavirus E6 protein, but precise functions for these regions of MAML1 have otherwise not been assigned (22). The remainder of the MAML1 sequence is predicted to be disordered (Fig. 2, middle).
FIG 2.
MAML1 sequence features, including conservation, predicted disorder, and charge distribution. (Top) Conservation of MAML1 sequence, measured by the program Conseq (27–30); (middle) native disorder in the MAML1 sequence, predicted using the program Disopred3 (31, 32); (bottom) charge distribution of the MAML1 amino acid sequence.
We created several protein deletion variants to assess the function of different C-terminal regions of MAML1 (Fig. 3A). All molecules retained the N-terminal 150 amino acids in order to span the nuclear localization signal (NLS) and the Notch-interacting region and included a C-terminal green fluorescent protein (GFP) tag and a mutated protospacer adjacent motif (PAM) sequence to avoid recognition by the Cas9 enzyme in add-back cell lines.
FIG 3.
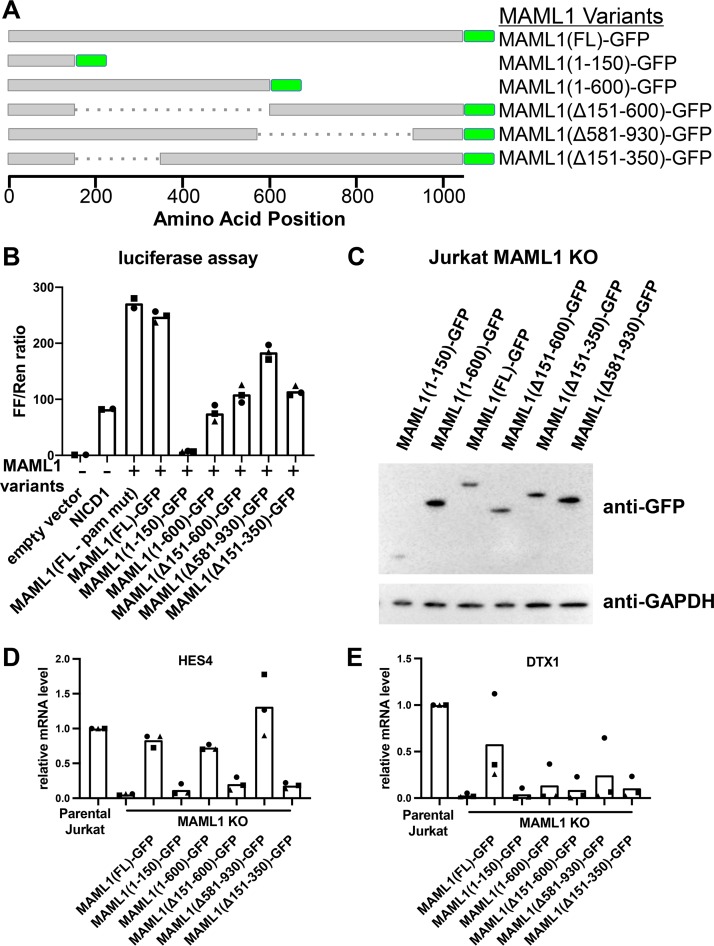
Removal of residues 151 to 350 from MAML1 impairs the Notch1-dependent transcriptional response of HES4 and DTX1. (A) MAML1 variants assayed in add-back experiments. All proteins contain the MAML1 nuclear localization sequence and a C-terminal GFP tag. (B) Luciferase assay performed in U2OS cells, showing the effect of different MAML1 variants upon Notch1-mediated transcription from a Notch-responsive reporter. Data points show the average for technical replicates from each biological replicate, with each biological replicate indicated using a different point shape. (C) Stable cell lines expressing MAML1 variants as GFP fusion proteins. Expression of the MAML1 proteins was detected by Western blotting with an anti-GFP antibody. (D and E) RT-qPCR of HES4 (D) and DTX1 (E), measuring transcript abundance in Jurkat parental, KO, and various MAML1 add-back cell lines. Data points show the average for technical replicates from each biological replicate, with each biological replicate indicated using a different point shape.
These MAML1 variants were first tested in a well-characterized luciferase reporter assay for Notch1 signaling in U2OS cells (8, 23). Previous work has shown that addition of full-length MAML1 enhances the transcriptional response to NICD1 in this assay (8). Addition of the C-terminal GFP tag to MAML1 did not reduce activity compared to untagged full-length protein (Fig. 3B). In contrast, MAML1(1–150) acts as a dominant negative protein, as has been reported for other N-terminal MAML1 fragments, which bind Notch and RBPJ but are unable to stimulate transcription (10). All other MAML1 variants reduced activity to various extents: MAML1(1–600) by 70%, MAML1(Δ151–600) and MAML1(Δ151–350) by over 50%, and MAML1(Δ581–930) by 25%. In particular, the activity difference between MAML1(1–600) and MAML1(Δ581–930) suggests an important role for the C-terminal acidic region, which is present in the more active MAML1(Δ581–930) protein but absent from the MAML1(1–600) protein. The fact that MAML1(Δ151-600) and MAML1(Δ151–350) exhibit similar degrees of reduced activity in the assay suggests that the removal of residues 151 to 350 is primarily responsible for this effect.
We then created stable cell lines expressing these MAML1 variants (Fig. 3C) in MAML1 KO cells and measured expression of the Notch target genes HES4 and DTX1 using RT-qPCR (Fig. 3D and E). All proteins that lack residues 151 to 350 were unable to rescue either HES4 or DTX1 expression. MAML1(1–600) and MAML1(Δ581-930) rescued expression of HES4 but did not rescue DTX1 to the same extent, suggesting different requirements for the C-terminal portion of MAML1 for different genes.
Defective activation by MAML1(Δ151–350) is not due to a reduction in activated Notch1 abundance.
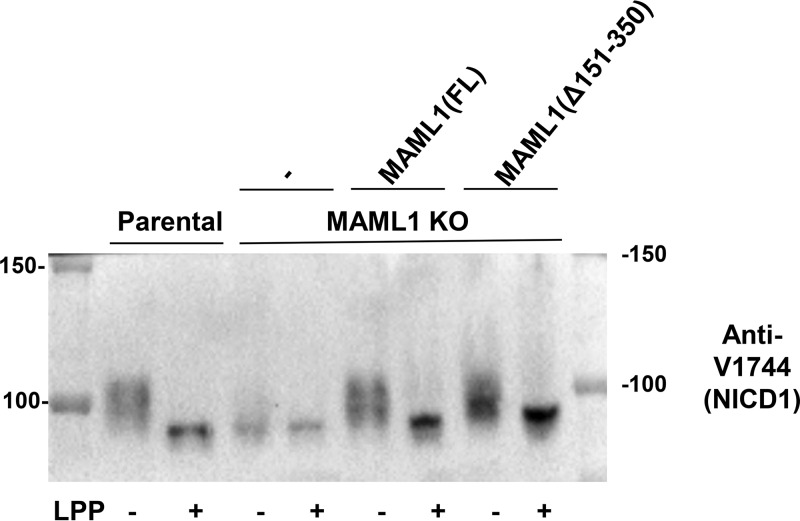
Because all variants of MAML1 missing amino acids 151 to 350 were functionally impaired in induction of both HES4 and DTX1, we focused on how this segment contributes to Notch1-dependent gene expression. First, we assessed the amount of active, intracellular Notch1 in parental, KO, full-length add-back, and MAML1(Δ151–350) add-back cell lines (Fig. 4). In the parental line, NICD1 underwent substantial phosphorylation, as judged by the ability of lambda phosphatase to collapse the NICD1 protein band on the Western blot to a more focused, faster-migrating species. In the KO cells, the total amount of NICD1 protein appeared to be reduced, and the majority of the NICD1 protein in the MAML1 KO cells migrated similarly to the lambda phosphatase-treated protein, suggesting that MAML1 is also required for most NICD1 phosphorylation in these cells. Phosphorylation of NICD1 was rescued by both the full-length and MAML1(Δ151–350) proteins, although the MAML1(Δ151–350) protein appeared to show differences in the proportions of various NICD1 phosphorylated species. Examination of the NICD1 abundance after phosphatase treatment showed, however, that the overall abundance of Notch protein was not lower in the MAML1(Δ151–350) line than in the full-length line, indicating that this form of MAML1 is not deficient in transcription because of effects on NICD1 overall abundance or stability.
FIG 4.
KO cells expressing full-length MAML1 (FL) or MAML1(Δ151–350) do not have reduced amounts of NICD1 compared to parental cells. Shown is a Western blot for activated Notch1 (NICD1) in parental Jurkat, MAML1 KO, MAML1(FL) add-back, and MAML1(Δ151–350) add-back cell lines. Lanes marked “+” were treated with lambda phosphatase (LPP). Numbers to the left and right of the blot denote molecular masses of protein standards in kilodaltons.
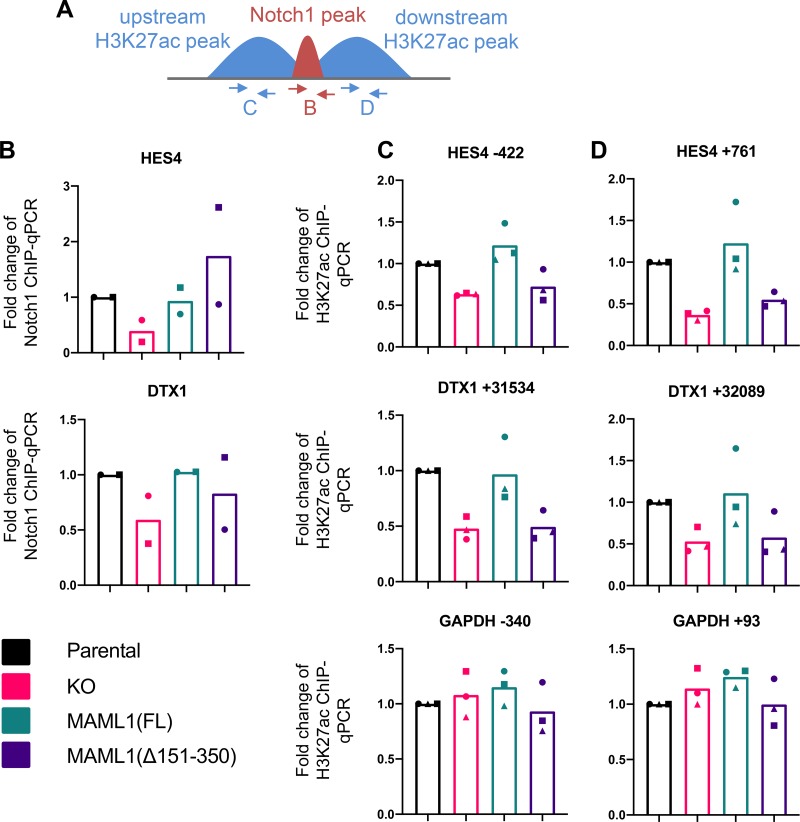
MAML1(Δ151–350) does not change the amount of chromatin-bound Notch1 but does reduce H3K27 acetylation around Notch target genes.
We then assessed chromatin-bound Notch protein amounts at the HES4 and DTX1 Notch target genes in our four cell lines [parental, KO, full-length add-back, and MAML1(Δ151–350) add-back] by ChIP-qPCR (Fig. 5A and B). We selected Notch1 binding sites for analysis based on published chromatin immunoprecipitation-sequencing (ChIP-seq) data sets from cells of the CUTLL1 line, another T-ALL line characterized by constitutively active Notch signaling (13). Both RBPJ and Notch1 bind to consensus RBPJ sites present at these loci. RBPJ/Notch1 binding at these sites also cooccurs with p300 binding and is flanked by H3K27ac marks, indicating that the selected loci are functional Notch response elements. As anticipated based on the reduced amount of total Notch1 protein in the MAML1 KO cells (Fig. 4), MAML1 KO reduced Notch1 abundance at both the HES4 and DTX1 binding sites. In contrast, both full-length and MAML1(Δ151–350) add-back restored bound Notch1 to amounts comparable to those of the parental cells at these loci, indicating that residues 151 to 350 of MAML1 are not required for stabilization of Notch1 bound to these response elements on DNA. We next measured H3K27ac amounts at flanking regions adjacent to these Notch binding sites (Fig. 5A, C, and D). MAML1 KO cells have reduced H3K27ac at these Notch targets, which is rescued by full-length MAML1 but not by MAML1(Δ151–350), indicating that this segment is required to promote histone acetylation surrounding Notch genomic responsive elements.
FIG 5.
MAML1(Δ151–350) cannot rescue H3K27ac around Notch binding sites. (A) Scheme showing primer positions for Notch1 and H3K27ac ChIP-qPCR experiments. (B) Notch1 binding at regulatory regions for HES4 and DTX1, as measured by Notch1 ChIP-qPCR. Data points show the average of technical replicates from each biological replicate, with each biological replicate indicated using a different point shape. (C and D) H3K27ac signal upstream (C) and downstream (D) of the Notch1 binding sites is shown, as measured by ChIP-qPCR. H3K27ac flanking the GAPDH promoter is shown as a non-Notch responsive control. Data points show the average of technical replicates from each biological replicate, with each of the three biological replicates indicated using a different point shape.
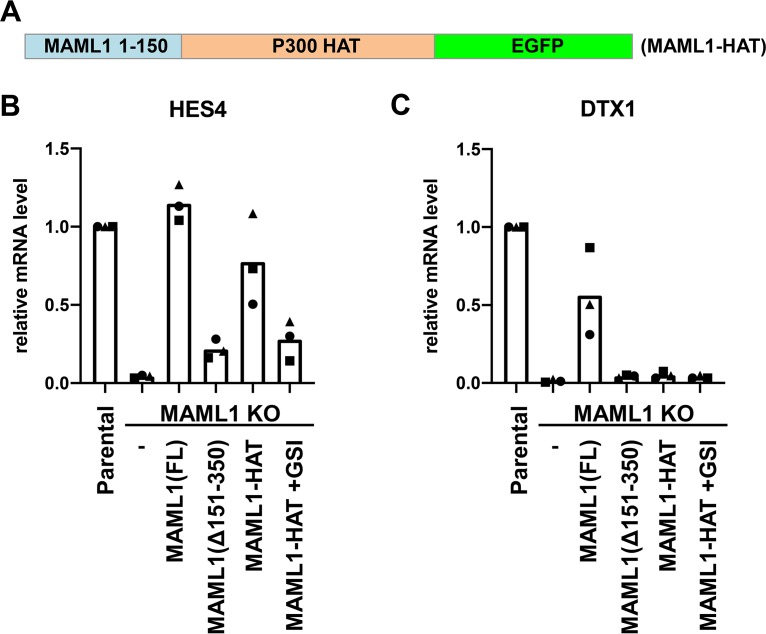
MAML1(1–150)-p300 HAT fusion partially rescues target gene expression.
To determine if gene expression could be restored by artificial recruitment of histone acetyltransferase activity, we created a fusion of MAML1 to the p300 histone acetyltransferase (HAT) domain. We linked the N-terminal 150 amino acids of MAML1, containing the NLS and Notch binding region, to the p300 HAT domain, which we designated MAML1-HAT (Fig. 6A). We created a stable cell line in which this fusion was expressed in MAML1 KO cells and measured Notch target gene expression. This fusion rescued HES4 expression, and importantly, this rescue of target gene expression was suppressed in the presence of GSI, indicating that rescue by the MAML1-HAT fusion is dependent on the presence of intracellular Notch1 (Fig. 6B). The MAML1-HAT fusion, however, did not measurably rescue expression of DTX1 (Fig. 6C).
FIG 6.
A MAML1-p300 fusion protein can rescue expression of HES4 but not DTX1. (A) Scheme of the designed MAML1-HAT fusion protein. (B and C) RT-qPCR of Notch1-responsive genes HES4 (B) and DTX1 (C) in MAML1 KO Jurkat cells expressing FL, MAML1(Δ151–350), MAML1-HAT, or MAML1-HAT in the presence of a gamma secretase inhibitor (GSI). Data points show the average for technical replicates from each biological replicate, with the three biological replicates indicated using a different point shape.
DISCUSSION
In this work, we have shown the requirement of amino acids 151 to 350 for MAML1 function both in reporter assays and at endogenous loci in Jurkat cells. This finding is consistent with previous work identifying a requirement for TAD1 (residues 75 to 301) in Notch1-dependent transcription. Interestingly, previous work has shown that a p300 interaction region of MAML1 resides in a proline-rich motif within amino acids 81 to 87, which is retained in our MAML1(Δ151–350) construct. Because MAML1(Δ151–350) cannot support histone acetylation at Notch-responsive binding sites, our data suggest that binding of MAML1 to p300 through the previously defined site (i.e., residues 81 to 87) is not sufficient for it to stimulate histone acetylation (24) and that additional segments of MAML1 beyond the previously reported p300 binding site are required for recruiting p300 acetyltransferase activity. Along these lines, others have reported a binding site for the KIX domain of p300 on the related family member MAML2, which lies in the region of MAML2 analogous to the region of MAML1 from residues 150 to 350 identified in this study (25). In addition, lysine residues in the region of MAML1 from residues 151 to 350 can be acetylated by p300, suggesting the existence of a more complex interplay between MAML1-p300 binding and enzymatic activity than previously appreciated (24).
We also showed that direct fusion of the MAML1 N terminus to the p300 HAT domain could rescue expression of HES4 but not DTX1. HES4 is regulated by promoter-proximal NTC binding, while DTX1 is regulated by an intronic enhancer approximately 31 kb downstream of the transcription start site. Previous studies tethering the p300 HAT domain to dCas9 have also found that gene activation through recruitment of p300 to enhancers is less robust than activation from promoters (26). It is intriguing to consider how different Notch-responsive genes have encoded different requirements for transcriptional cofactors. DTX1 is more dependent on the C-terminal regions of MAML1 than is HES4, highlighting that while promoter-proximally regulated genes may be primarily dependent on p300 recruitment and activation, distally regulated genes may depend on recruitment of other coactivators by the C-terminal end of MAML1. Future work will be required to identify additional MAML1-dependent cofactors, define how they interact with MAML1, and establish how they contribute to expression of Notch target genes.
MATERIALS AND METHODS
Cell culture.
Jurkat cells were maintained in RPMI medium with l-glutamine medium (Corning) plus 10% fetal bovine serum (FBS; Gemini Bio-Sciences) and penicillin-streptomycin (Gibco). U2OS and 293T cells were maintained in Dulbecco modified Eagle medium (DMEM; Corning) plus 10% FBS (Gemini Bio-Sciences) and penicillin-streptomycin (Gibco).
GSI treatment and washout.
Gamma secretase inhibitor compound E (Millipore Sigma) was dissolved in dimethyl sulfoxide (DMSO) and diluted 1:1,000 in RPMI medium to a final concentration of 1 μM. Cells were treated with compound E or an equal quantity of DMSO as a control for 8 or 16 h. Cells from compound E-treated and DMSO control-treated conditions were then recovered by centrifugation (300 × g for 5 min). Cells were then subjected to three cycles of resuspension in RPMI medium followed by recentrifugation to remove the GSI or DMSO. Cells were harvested for analysis 4 h after GSI or DMSO washout.
Cloning.
A guide sequence (GCGGTCATGGAGCGCCTTCGC) for CRISPR targeting of MAML1 was cloned into the LentiV2 vector using the Esp3I restriction enzyme. The various MAML1 truncation constructs were subcloned into the MigR1 vector using restriction sites for XhoI and NcoI. The resulting cDNAs eliminated the internal ribosome entry site (IRES) for expression of GFP and instead encoded MAML1-GFP fusion proteins directly.
The MAML1-HAT fusion protein was cloned into the MigR1 vector using the same sites, also generating a C-terminal GFP fusion protein.
Cell lines.
The MAML1 knockout cell line was generated by infecting Jurkat cells with the lentiviral CRISPR-Cas9 system to target MAML1. Infected cells expressing GFP underwent single-cell sorting and clonal expansion before Western blot validation. MAML1 rescue cDNA constructs had their PAM sequences mutated to prevent Cas9 cleavage.
Viral particles were made by cotransfecting MigR1-MAML1 plasmid, vesicular stomatitis virus G protein (VSV-G), and Gag-Pol vectors into 293T cells. Viruses were harvested after 48 h and filtered through a 0.45-μm membrane. Jurkat cells were then infected with virus by mixing 0.5 million cells with virus and 4 μg/ml of Polybrene. GFP-positive cells were sorted 3 to 4 days after infection using a FACS Aria sorter.
Reporter assays.
A total of 50,000 U2OS cells were plated per well in a 24-well cell culture plate. The next day, cells were transfected with 10 ng of ICN1 in pcDNA, 50 ng of MAML1 construct in MigR1 vector, 100 ng of reporter mix (98 ng of TP1/2 ng of PRLTK), and empty pcDNA vector to 250 ng, using polyethylenimine (PEI) in a 1:3 DNA/PEI ratio. Three replicate transfections were performed per condition. Cells were harvested the next day, and luciferase assays were performed using a dual-luciferase reporter assay kit (Promega), read out on a Turner Biosystems Modulus microplate luminometer, with 2 or 3 technical replicates per transfection. Firefly-to-Renilla luminescence ratios were normalized to the mean of the empty vector control, assigned a value of 1.
Western blots.
For Western blots, cells were pelleted, resuspended in 1× SDS running buffer at 106 cells per 100 μl, boiled, and run on a 4 to 20% gradient gel (Bio-Rad). MAML1 was detected with CST antibody D3K7B for Fig. 1C. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was detected with CST antibody 14C10, and GFP was detected with CST antibody 2956.
A total of 750,000 cells were harvested and resuspended in 40 μl of lambda protein phosphatase reaction buffer (1× NEBuffer pack for protein metallophosphatases from New England BioLabs [NEB], 1 mM MnCl2 [NEB], 0.1% Igepal NP-40, protease inhibitor tablet). One microliter of lambda protein phosphatase (NEB) was added to samples according to the Fig. 4 legend. Samples were incubated at 30°C for 30 min at 700 rpm, and then 10 μl of 5× SDS loading buffer was added. Samples were run on 8% Tris-glycine Novex gels (Invitrogen) at 150 V for 2 h, until the 75-kDa marker was at the bottom of the gel. Samples were Ponceau stained to verify equal loading and blotted with CST anti-V1744 activated Notch1 (NICD1) antibody (4147).
RT-qPCR.
RNA was harvested from 1,000,000 cells using the RNEasy kit (Qiagen). One microgram of total RNA was used as input for cDNA synthesis, using the iScript kit (Bio-Rad). qPCR was performed using PowerUp SYBR green master mix (Thermo Fisher) in a 10-μl total reaction volume with 0.25 μM forward and reverse primers, with 2 technical qPCR replicates per condition. Primer sequences are listed in Table S1 in the supplemental material. The standard thermocycler parameters suggested by the manufacturer were used, with a default melt curve step right after the PCR run to confirm primer specificity. Expression was normalized to the average GAPDH expression for each condition. In order to show biological replicates on the same scale, within each replicate expression was normalized to wild-type (WT) Jurkat cell expression, assigned a value of 1.
ChIP-qPCR.
Chromatin immunoprecipitation followed by qPCR (ChIP-qPCR) was performed with specific antibodies targeting NOTCH1 (custom-made antiserum from Covance [16]) or H3K27ac (abcam; ab4729). Briefly, cells were harvested by cross-linking with 1% formaldehyde for 5 (for H3K27ac) or 15 (for NOTCH1) min and subsequently quenched with glycine. Chromatin shearing was done in bulk with 50 to 100 million cells. Sonication was performed with a Bioruptor Pico sonicator, enriching for fragments between 200 and 500 bp. Aliquots were taken so that each NOTCH1 and H3K27ac ChIP experiment used 7.5 and 2.5 million cells, respectively. Sheared chromatin was preincubated with anti-rabbit protein A– agarose beads to reduce nonspecific binding. Precleared chromatin samples were then incubated with specific antibodies at 4°C overnight with gentle rotation. The next day, immunoprecipitation was performed by adding protein A-agarose beads to the chromatin-antibody solution and rotating for 2 h at 4°C. Beads were pelleted and subjected to low-salt (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris [pH 8], 150 mM NaCl), high-salt (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris [pH 8], 500 mM NaCl), LiCl, and Tris-EDTA (TE) washes and then eluted in SDS-NaHCO3 elution buffer. Reverse cross-linking and proteinase K (NEB) treatment were performed to release the bound DNA, which was recovered by ethanol precipitation. qPCR was performed as described in “RT-qPCR” above. Primers for qPCR are listed in Table S1. Quantification cycle (Cq) values were converted into percent input by comparison to a 1% input sample. In order to show biological replicates on the same scale, percent input was normalized to the value for WT Jurkat cells within each replicate, assigned a value of 1.
Sequence analysis.
Protein sequence conservation was determined using the ConSurf server with default settings (27–30). Predicted disorder was determined using DISOPRED3 on the Psipred server (31, 32). Amino acid charge scale was determined by assigning colors to the pI values of the amino acids.
Supplementary Material
ACKNOWLEDGMENTS
J.M.R. and B.G. were funded by LLS CDP postdoctoral fellowships, and J.M.R. was also supported by NIH training grant 2T32GM007748-40. This work was supported by NCI R35CA220340 to S.C.B.
We thank members of the Blacklow and Adelman labs for helpful discussion.
S.C.B. is a consultant for AYALA Pharmaceutical and IFM Therapeutics and is on the scientific advisory board of Erasca, Inc. He also receives funding from Erasca and from the Dana Farber-Novartis drug development translational research program. J.C.A. is a consultant for AYALA Pharmaceutical.
B.G., J.C.A., K.A., and S.C.B. designed experiments, J.M.R., B.G., and E.D.E. performed experiments, J.M.R., B.G., and S.C.B. drafted the manuscript, and all authors edited the manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Henrique D, Schweisguth F. 2019. Mechanisms of Notch signaling: a simple logic deployed in time and space. Development 146:dev172148. doi: 10.1242/dev.172148. [DOI] [PubMed] [Google Scholar]
- 2.Bray SJ. 2016. Notch signalling in context. Nat Rev Mol Cell Biol 17:722–735. doi: 10.1038/nrm.2016.94. [DOI] [PubMed] [Google Scholar]
- 3.Ellisen LW, Bird J, West DC, Soreng AL, Reynolds TC, Smith SD, Sklar J. 1991. TAN-1, the human homolog of the Drosophila Notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell 66:649–661. doi: 10.1016/0092-8674(91)90111-B. [DOI] [PubMed] [Google Scholar]
- 4.Pear WS, Aster JC, Scott ML, Hasserjian RP, Soffer B, Sklar J, Baltimore D. 1996. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J Exp Med 183:2283–2291. doi: 10.1084/jem.183.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weng AP, Ferrando AA, Lee W, Morris JP, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. 2004. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 6.Petcherski AG, Kimble J. 2000. Mastermind is a putative activator for Notch. Curr Biol 10:471–473. doi: 10.1016/S0960-9822(00)00577-7. [DOI] [PubMed] [Google Scholar]
- 7.Petcherski AG, Kimble J. 2000. LAG-3 is a putative transcriptional activator in the C. elegans Notch pathway. Nature 405:364–368. doi: 10.1038/35012645. [DOI] [PubMed] [Google Scholar]
- 8.Wu L, Aster JC, Blacklow SC, Lake R, Artavanis-Tsakonas S, Griffin JD. 2000. MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat Genet 26:484–489. doi: 10.1038/82644. [DOI] [PubMed] [Google Scholar]
- 9.Nam Y, Sliz P, Song L, Aster JC, Blacklow SC. 2006. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell 124:973–983. doi: 10.1016/j.cell.2005.12.037. [DOI] [PubMed] [Google Scholar]
- 10.Weng AP, Nam Y, Wolfe MS, Warren S, Griffin JD, Blacklow SC, Jon C, Pear WS, Aster JC. 2003. Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of Notch signaling. Mol Cell Biol 23:655–664. doi: 10.1128/mcb.23.2.655-664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fryer CJ, Lamar E, Turbachova I, Kintner C, Jones KA. 2002. Mastermind mediates chromatin-specific transcription and turnover of the notch enhancer complex. Genes Dev 16:1397–1411. doi: 10.1101/gad.991602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fryer CJ, White JB, Jones KA. 2004. Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol Cell 16:509–520. doi: 10.1016/j.molcel.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 13.Wang H, Zang C, Taing L, Arnett KL, Wong YJ, Pear WS, Blacklow SC, Liu XS, Aster JC. 2014. NOTCH1-RBPJ complexes drive target gene expression through dynamic interactions with superenhancers. Proc Natl Acad Sci U S A 111:705–710. doi: 10.1073/pnas.1315023111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castel D, Mourikis P, Bartels SJJ, Brinkman AB, Tajbakhsh S, Stunnenberg HG. 2013. Dynamic binding of RBPJ is determined by notch signaling status. Genes Dev 27:1059–1071. doi: 10.1101/gad.211912.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krejčí A, Bernard F, Housden B, Collins S, Bray SJ. 2009. Direct response to notch activation: signaling crosstalk and incoherent logic. Sci Signal 2:1–15. doi: 10.1126/scisignal.2000140. [DOI] [PubMed] [Google Scholar]
- 16.Wang H, Zou J, Zhao B, Johannsen E, Ashworth T, Wong H, Pear WS, Schug J, Blacklow SC, Arnett KL, Bernstein BE, Kieff E, Aster JC. 2011. Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells. Proc Natl Acad Sci U S A 108:14908–14913. doi: 10.1073/pnas.1109023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneider U, Schwenk HU, Bornkamm G. 1977. Characterization of EBV-genome negative “NULL” and “T” cell lines derived from children with acute lymphoblastic leukemia and leukemic transformed non-Hodgkin lymphoma. Int J Cancer 19:621–626. doi: 10.1002/ijc.2910190505. [DOI] [PubMed] [Google Scholar]
- 18.Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, Del Bianco C, Rodriguez CG, Sai H, Tobias J, Li Y, Wolfe MS, Shachaf C, Felsher D, Blacklow SC, Pear WS, Aster JC. 2006. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev 20:2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oyama T, Harigaya K, Muradil A, Hozumi K, Habu S, Oguro H, Iwama A, Matsuno K, Sakamoto R, Sato M, Yoshida N, Kitagawa M. 2007. Mastermind-1 is required for Notch signal-dependent steps in lymphocyte development in vivo. Proc Natl Acad Sci U S A 104:9764–9769. doi: 10.1073/pnas.0700240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson JJ, Kovall RA. 2006. Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA. Cell 124:985–996. doi: 10.1016/j.cell.2006.01.035. [DOI] [PubMed] [Google Scholar]
- 21.Choi SH, Wales TE, Nam Y, O’Donovan DJ, Sliz P, Engen JR, Blacklow SC. 2012. Conformational locking upon cooperative assembly of notch transcription complexes. Structure 20:340–349. doi: 10.1016/j.str.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan MJA, White EA, Sowa ME, Harper JW, Aster JC, Howley PM. 2012. Cutaneous β-human papillomavirus E6 proteins bind Mastermind-like coactivators and repress Notch signaling. Proc Natl Acad Sci U S A 109:1473–1480. doi: 10.1073/pnas.1205991109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aster JC, Xu L, Karnell FG, Patriub V, Pui JC, Pear WS. 2000. Essential roles for ankyrin repeat and transactivation domains in induction of T-cell leukemia by Notch1. Mol Cell Biol 20:7505–7515. doi: 10.1128/mcb.20.20.7505-7515.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saint Just Ribeiro M, Hansson ML, Wallberg AE. 2007. A proline repeat domain in the Notch co-activator MAML1 is important for the p300-mediated acetylation of MAML1. Biochem J 404:289–298. doi: 10.1042/BJ20061900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clark MD, Kumar GS, Marcum R, Luo Q, Zhang Y, Radhakrishnan I. 2015. Molecular basis for the mechanism of constitutive CBP/p300 coactivator recruitment by CRTC1-MAML2 and its implications in cAMP signaling. Biochemistry 54:5439–5446. doi: 10.1021/acs.biochem.5b00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hilton IB, Ippolito AMD, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA. 2015. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol 33:510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. 2010. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res 38:529–533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, Ben-Tal N. 2016. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res 44:W344–W350. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Celniker G, Nimrod G, Ashkenazy H, Glaser F, Martz E, Mayrose I, Pupko T, Ben-Tal N. 2013. ConSurf: using evolutionary data to raise testable hypotheses about protein function. Isr J Chem 53:199–206. doi: 10.1002/ijch.201200096. [DOI] [Google Scholar]
- 30.Berezin C, Glaser F, Rosenberg J, Paz I, Pupko T, Fariselli P, Casadio R, Ben-Tal N. 2004. ConSeq: the identification of functionally and structurally important residues in protein sequences. Bioinformatics 20:1322–1324. doi: 10.1093/bioinformatics/bth070. [DOI] [PubMed] [Google Scholar]
- 31.Buchan DWA, Jones DT. 2019. The PSIPRED Protein Analysis Workbench: 20 years on. Nucleic Acids Res 47:W402–W407. doi: 10.1093/nar/gkz297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones DT, Cozzetto D. 2015. DISOPRED3: precise disordered region predictions with annotated protein-binding activity. Bioinformatics 31:857–863. doi: 10.1093/bioinformatics/btu744. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.