Abstract
Background
We investigated the efficacy of a switch to dolutegravir plus lamivudine in aviremic individuals without evidence of persistent lamivudine resistance-associated mutations in baseline proviral DNA population sequencing.
Methods
Open-label, single-arm, 48-week pilot trial. HIV-1 infected adults, naïve to integrase inhibitors, with CD4+ above 350 cell/μL and fewer than 50 HIV-1 RNA copies per mL the year prior to study entry switched to dolutegravir plus lamivudine. Participants were excluded if baseline proviral DNA population genotyping detected lamivudine resistance-associated mutations. To detect resistance minority variants, proviral DNA next-generation sequencing was retrospectively performed from baseline samples. Primary efficacy endpoint was proportion of participants with fewer than 50 HIV-1 RNA copies per mL at week 48. Safety and tolerability outcomes were incidence of adverse events and treatment discontinuations. ART-PRO is registered with ClinicalTrials.gov, NCT03539224.
Findings
41 participants switched to dolutegravir plus lamivudine, 21 with lamivudine resistance mutations in historical plasma genotypes. Baseline next-generation sequencing detected lamivudine resistance mutations (M184V/I and/or K65R/E/N) over a 5% threshold in 15/21 (71·4%) and 3/20 (15%) of participants with and without history of lamivudine resistance, respectively. At week 48, 92·7% of participants (38/41) had fewer than 50 HIV-1 RNA copies per mL. There were no cases of virologic failure. Three participants with historical lamivudine resistance were prematurely discontinued from the study (2 protocol violations, one adverse event). Ten participants (4 in the group with historical lamivudine resistance) had a transient viral rebound, all resuppressed on dolutegravir plus lamivudine. There were 28 drug-related adverse events, only one leading to discontinuation.
Interpretation
In this pilot trial, dolutegravir plus lamivudine was effective in maintaining virologic control despite past historical lamivudine resistance and presence of archived lamivudine resistance-associated mutations detected by next generation sequencing. Further studies are needed to confirm our results.
Funding
Fondo de Investigaciones Sanitarias, Instituto de Salud Carlos III PI16/00837-PI16/00678.
Research in context.
Evidence before this study
We searched PubMed for clinical trial publications, cohort studies, and review articles published between July 1, and November 30, 2019, with combinations of the search terms “lamivudine resistance”, “HIV”, “simplification”, “switch”, “proviral DNA”, “next generation sequencing” (NGS), “nucleoside reverse transcriptase inhibitor”, “integrase strand transfer inhibitor” (INSTI), “dolutegravir”, “lamivudine” and “dual therapy”, with no restrictions on language or publication date. We also searched for relevant publications from international HIV congresses (2018-2019) and we used public search engines to retrieve most updated guidelines. One large multicentric trial and other smaller-sized studies using lamivudine and dolutegravir as dual HIV-1 therapy in virologically suppressed patients have been reported. Retrospective cohort studies and one clinical trial have suggested that lamivudine might provide additional antiviral activity when combined with dolutegravir in treatment experienced patients history of lamivudine resistance-associated mutations. There is scarce data of the efficacy of lamivudine and dolutegravir as maintenance therapy in patients with historical resistance to lamivudine.
Added value of this study
ART-PRO 48-week study is the first prospective trial providing preliminary evidence that dolutegravir plus lamivudine can be efficacious in maintaining virological control in integrase inhibitor-naïve, long-term viral suppressed persons with a history of lamivudine resistance when baseline proviral DNA Sanger genotype does not detect the persistence of lamivudine resistance-associated mutations. This pilot study brings evidence against the notion that treatment with dolutegravir and lamivudine in the context of historical resistance to lamivudine is a functional dolutegravir monotherapy. We found that the presence of archived lamivudine resistance-associated mutations detected in proviral DNA by next generation sequencing, but not by population sequencing, did not affect the efficacy of the 21 patients included in this group. In ART-PRO study none of the 21 patients with either history of lamivudine resistance or detection of lamivudine resistance-associated mutations on DNA proviral sequencing experienced virological failure.
Implications of all the available evidence
Standard of care, three-drug combination antiretroviral therapy might bring toxicities leading to its interruption or to suboptimal compliance. In heavily treatment experienced persons with history of lamivudine resistance mutations, naïve to integrase-inhibitors, recycling discarded antiretrovirals such as lamivudine might open the possibility of avoiding toxicity or simplifying regimens. This is the first clinical trial specifically designed to address the efficacy of lamivudine and dolutegravir as dual therapy for maintenance of viral suppression in patients with historical lamivudine resistance as long as M184V/I and/or K65R/E/N mutations are not found on proviral DNA population sequencing prior to dual therapy initiation. Results from ART-PRO support further assessment of lamivudine and dolutegravir as dual therapy in treatment experienced HIV-1 infected persons with prior lamivudine resistance.
Alt-text: Unlabelled box
Introduction
Antiretroviral treatment (ART) has led to improved control of HIV infection which, in turn, has given the opportunity to aspire for less-toxic but equally potent reduced-drug regimens. Dolutegravir plus lamivudine is endorsed by guidelines for treatment of persons initiating HIV treatment [1,2]. In treatment-experienced individuals a large multicenter study showed that dolutegravir plus lamivudine was non-inferior for maintenance of virological suppression to a tenofovir-alafenamide based triple-drug regimen at 48 weeks [3]. Other smaller studies demonstrated that residual viremia and viral reservoir did not increase after a year switch to dolutegravir plus lamivudine compared to standard therapy [4,5]. However, these studies excluded patients with prior virologic failure and data regarding the efficacy of dolutegravir plus lamivudine in persons with history of resistance to lamivudine are currently scarce. Specifically, there are no prospective data exploring the effect that archived lamivudine resistance mutations may exert upon the efficacy of a maintenance dual regimen of lamivudine plus dolutegravir.
Studies in virologically failing patients have shown that lamivudine retains antiviral efficacy despite evidence of lamivudine resistance-associated mutations [6], [7], [8]. The MOBIDIP study was the first trial evaluating a dual regimen including lamivudine for maintenance of HIV suppression in persons with history of lamivudine resistance-associated mutations. In said study, 265 virologically suppressed participants- 96% of whom had a historical genotype detecting the M184V mutation- were randomized to either boosted protease inhibitor monotherapy or to lamivudine plus boosted protease inhibitor. The study was prematurely discontinued after 48 weeks because the proportion of virologic failures with boosted protease inhibitor monotherapy was significantly higher compared to lamivudine plus boosted protease inhibitor (difference between groups 21·8%) [9]. Considering these results and in the context of past lamivudine resistance, we hypothesized that combining lamivudine to another drug with a high genetic barrier such as dolutegravir would also succeed in maintaining virologic suppression.
At present, it is still unknown if proviral DNA genotyping in virologically suppressed patients can predict the efficacy of a given treatment, and discussion persists as to whether proviral DNA adds additional helpful information to that of a historical genotype. DHHS guidelines currently recommend to cautiously interpret the results of proviral DNA resistance testing [2], as this assay may not detect all previously existing drug-resistance mutations [10], [11], [12]. Next-generation sequencing allowed to predict virological failure of initial ART based on non-nucleoside reverse transcriptase inhibitors but, reducing the threshold to detect minority resistant variants also increased the number of persons missclassified as “at risk” of virological failure, when in fact they had good virological control- a finding that, authors argued, could lead to unnecessary changes in antiretroviral treatment [13]. The importance of detecting minority resistant populations in proviral DNA is even more questionable since a high proportion of archived HIV is non replicating [14,15]. In addition, there is currently little information on the relevance of those minority resistant populations detected by next-generation sequencing, but not through proviral DNA population sequencing.
We designed the pilot study ART-PRO to gather preliminary information on efficacy of dolutegravir plus lamivudine as maintenance therapy in virologically suppressed patients with past history of lamivudine-resistant mutations, but without present evidence of their persistence at study initiation in proviral DNA Sanger sequencing. We retrospectively performed next-generation sequencing of baseline samples to evaluate if resistance-associated mutations detected only through this technique could impact the antiviral efficacy of the regimen.
We hypothesized that long-term viral suppression could lead to clearance of historical resistance mutations, and that, if these were not detected by proviral DNA population genotyping at baseline, treatment with dolutegravir plus lamivudine would be able to maintain viral control in patients with prior history of lamivudine resistance. This would not only open the possibility to recycle discarded antiretrovirals but would also allow access to a simplified regimen endorsed by guidelines in patients who, owing to previous failures, are frequently treated with complex antiretroviral combinations that may include high pill burden or risk of toxicity. We also theorized that persisting mutations detected only through next-generation sequencing, but not in DNA bulk genotyping, would not negatively impact the efficacy of dolutegravir plus lamivudine.
The aim of this pilot study was to perform an initial test of the accuracy of our hypothesis, which, if so, would provide the grounds for a fully-powered study to confirm our findings.
2. Methods
2.1. Study design and participants
This pilot- proof of concept, single-arm, phase IIa, open label clinical trial took place in two university hospitals in Madrid (Spain): Hospital Universitario 12 de Octubre and Hospital Universitario La Paz.
Inclusion criteria were HIV-1 infected participants who were 18 years or older, on stable ART for at least 3 months, CD4 count of more 350 cell/µL, and HIV-1 RNA of less than 50 copies per mL in the year prior to study entry (allowing for one blip [HIV-1 RNA of less 500 copies/mL] in the 3 months before study entry, only if preceded and followed by HIV-1 RNA of less 50 copies/mL). For inclusion, participants had to have history of treatment with lamivudine or emtricitabine, to be naïve to integrase inhibitors and willing to switch ART owing to toxicity or for simplification reasons. Prior virologic failures were allowed and, for the purpose of the study, we included one group of participants with historical RNA genotypic test documenting a previous history of lamivudine resistance-associated mutations. Participants were excluded if lamivudine resistance-associated mutations were detected in proviral DNA by population genotyping at screening. Other exclusion criteria were pregnant, lactating or fertile women unwilling to commit to birth control and hepatitis B antigen S-positive infected persons.
All participants provided written informed consent. The study was approved by the Ethics Committee and is in accordance with applicable laws and the Declaration of Helsinki. The study protocol for ART-PRO is available online.
Although originally planned for 48 weeks, a 144-week extension of the study is currently ongoing.
2.2. Procedures
Participants were selected from our prior study that evaluated the concordance in detecting persistence of resistance-associated mutations in aviremic persons using proviral DNA population and next-generation sequencing genotyping (“GEN-PRO”) [16].
In GEN-PRO study visit, we obtained samples for peripheral blood proviral DNA genotyping (Sanger sequencing using Big Dye Terminator (Applied Biosystem), and the Stanford HIVDB algorithm to interpret resistance mutations) and peripheral blood mononuclear cells (PBMCs) to retrospectively perform next-generation sequencing. For next-generation sequencing, HIV-1 DNA was extracted from PBMCs using the QIAamp® DNA blood minikit (Qiagen, Hiden, Germany) and a 3385 bp pol gene fragment was then amplified by nested PCR as described elsewhere [17]. Next-generation sequencing was performed with Illumina in a Miseq instrument (Illumina, San Diego, CA, USA). Sequences were preprocessed with Prinseq-lite (http://prinseq.sourceforge.net/) and FLASH (http://www.cbcb.umd.edu/software/flash) programs [18,19]. The resulting aligned reads were analyzed for HIV-1 drug resistance testing with PaSeq software (https://paseq.org/). The 1% threshold was chosen for resistance interpretation. Baseline samples with lamivudine resistance-associated mutations detected through next-generation sequencing underwent hypermutation analysis with Hypermut 2.0 software (https://www.hiv.lanl.gov/content/sequence/hypermut/hypermut.html) using default settings. Individual sequencing reads from each patient were considered hypermutated when p< 0·05 was obtained in Fisher´s exact test that compared the number of APOBEC-associated G-to-A changes in each read with the G-to-A changes in the reference sequence (HXB2).
We reviewed if participants qualified for the ART-PRO study on the basis of inclusion and exclusion criteria. To be eligible, proviral DNA population genotype had to be performed at most 45 days before switching treatment without detecting lamivudine resistance-associated mutations. For the participants included in this study, we considered the GEN-PRO study visit as the screening visit.
There were two groups of participants: one group without history of lamivudine resistance or without lamivudine resistance-associated mutations in historical RNA genotype and a second group with lamivudine resistance-associated mutations in historical RNA genotype.
On the first study visit (day 1), participants were switched to dolutegravir 50 mg plus generic lamivudine 300 mg both taken orally once daily. Thereafter, scheduled visits were performed at week 4, 8,12, 24, 36 and 48. At all visits participants underwent medical evaluation including physical examination, reporting of adverse events, review of concomitant medications, HIV-1 RNA viral load, CD4+ cell count, hemogram, lipid profile, liver, urine and renal function tests. Adherence was measured using pill count.
Any viral rebound had to be followed by a re-test within a two-week window to confirm that the viral load decreased under 50 HIV-1 RNA copies per mL. If any participant had HIV-1 RNA above 200 copies per mL we performed plasma genotyping testing in the integrase and retrotranscriptase (ViroSeq HIV-1 Genotyping System, Abbott Molecular, Spain).
Adverse events were graded and classified as drug related when appropriate by the investigator. Patients could be withdrawn from the study for safety reasons, if unable to participate in study proceedings, lost to follow-up or at their own request anytime throughout the study.
2.3. Outcomes
The primary outcome was the proportion of participants with HIV RNA below 50 copies per mL at 48 weeks, in the intention-to-treat-exposed population using the US Food and Drug Administration (FDA) snapshot algorithm.
Secondary efficacy outcomes included the proportion of participants with HIV RNA below 50 copies per mL at week 24 (FDA-snapshot, intention-to-treat-exposed population), the proportion of participants with virologic failure at week 48 using FDA-snapshot algorithm, transient virological rebounds (defined as unconfirmed episodes of detectable viral load), mean change in CD4+ cells count per µL from baseline to week 48 and incidence, type and number of resistance mutations at week 48.
Safety and tolerability outcomes were incidence of adverse events and treatment discontinuation due to toxicity or intolerance.
2.4. Statistical analysis
This is a pilot, proof of concept trial and as such we performed convenience sampling and selected a small number of participants. Initially we intended to recruit 20 participants for each arm (total 40). All analysis were prespecified and described in the protocol.
Intention-to-treat-exposed population included all participants receiving at least one dose of dolutegravir plus lamivudine. We calculated the proportion of participants with HIV RNA below 50 copies per mL at 48 weeks, in the intention-to-treat-exposed population using the US Food and Drug Administration (FDA) snapshot algorithm in each of the two groups of participants separately. In the per protocol analysis we included all subjects receiving at least one dose of study treatment with no deviation to the eligibility criteria. For the primary endpoint we included the analysis of the proportion of participants with missing data following the FDA snapshot algorithm. For the secondary endpoint analysis, we included only participants with complete information.
Descriptive analysis of individuals’ characteristics was performed using frequency tables for categorical variables and median and interquartile range for continuous variables. Differences in socio-demographic and clinical characteristics according to historical mutation group were assessed with the non-parametric Kruskal-Wallis test for continuous variables and the chi-squared test for independence for categorical variables. Adjusted differences of the mean change in CD4 cell counts, lipid parameters and weight at week 48 compared to baseline and significance were assessed with a multivariable linear regression using as covariables the historical mutation group and the baseline value (analysis of covariance). We assessed the association between categorical variables using the χ2 test when samples were of sufficient size or with the Fisher exact test when they were not.
We compared the frequency of resistance mutations detected by next-generation sequencing and proviral bulk genotyping, and we analyzed the percentage of participants with M184V/I and/or K65R/E/N detected by next-generation sequencing at 1%, 5%, 10% and 20% thresholds at study entry.
We performed a planned interim analysis at week 24. To continue the study at least 80% of patients with an adherence over 90% needed to have HIV RNA below 50 copies per mL in each study group, without emergence of any new resistance mutations in case of virologic failure.
All analyses were prespecified in the protocol and were performed using Stata software (version 15.0; Stata Corporation, College Station, Texas, USA).
2.5. Role of the funding source
The funding source had no role in designing, collecting, interpreting or writing of the report. All authors had access to the data and final manuscript and vouch for the veracity of the submitted publication. The corresponding authors are responsible for the decision to submit the study for publication.
3. Results
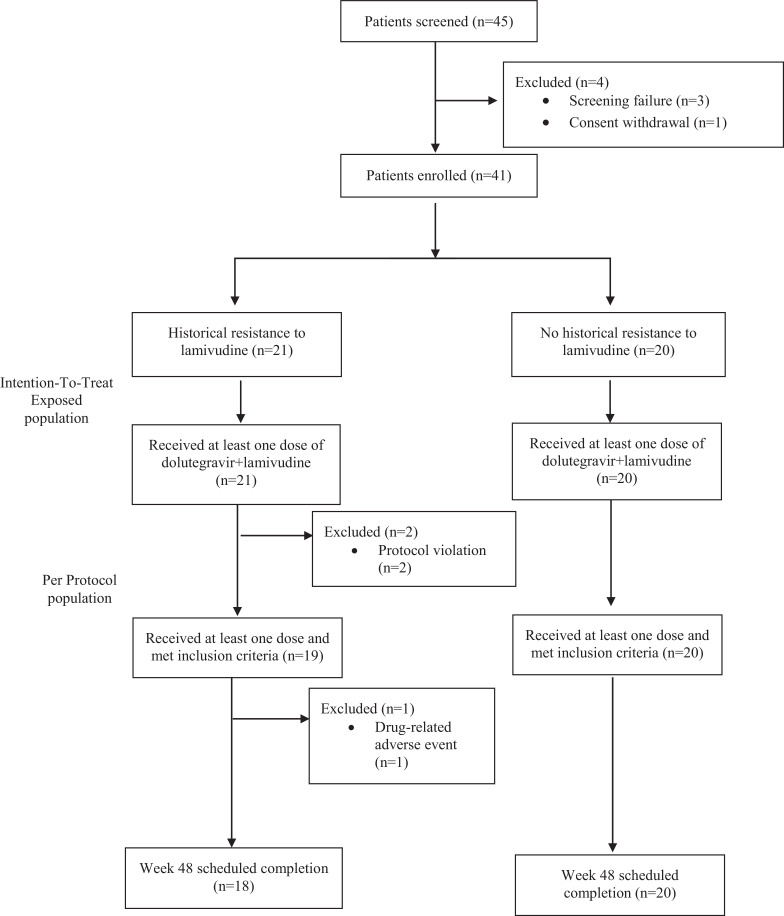
Between September 25, 2017, and April 9, 2018, we assessed the eligibility to participate in this study. Participants were selected among the 102 participating subjects of our prior study GEN-PRO, of whom 45 were offered to participate in ART-PRO to achieve our intended sample size (Figure 1). Forty-one participants on suppressive ART were included and switched to dolutegravir plus lamivudine: based on historical RNA genotype 21 had a history of resistance to lamivudine and 20 had not. Median time between the historical RNA genotype and baseline proviral DNA sequencing was 12·9 years (IQR: 6·7-14·4).
Figure 1.
Trial participant flow chart. Intention-to-treat exposed population includes every participant receiving at least one dose of dolutegravir+lamivudine. Per protocol population excludes participants who received the therapy with protocol deviations.
Baseline demographic characteristics at study entry were overall similar between groups (Table 1). Participants with previous lamivudine resistance had a non-statistically significant longer duration of HIV infection and lower CD4+ nadir count and had received a greater number of prior treatment regimens and were less likely to be receiving treatment with lamivudine or emtricitabine prior to entering the study compared to those without historical lamivudine resistance. Median duration of HIV viral suppression was 6 years (IQR 3·3-11) in both groups, which was also higher in the group with historical lamivudine resistance (7·7 years versus 5·3 years in the group without history of lamivudine resistance).
Table 1.
Demographic and Baseline characteristics for the enrolled participants (n=41).
| All participants (n=41) | Historical resistance to lamivudine | No historical lamivudine resistance | p value | |
|---|---|---|---|---|
| (n=21) | (n=20) | |||
| Male sex, n (%) | 32 (78·1) | 16 (76·2) | 16 (80) | 0·768 |
| Age (years), median (IQR) | 52·4 (45·4-56·9) | 53·4 (47·1-57·6) | 50·8 (42·9-55·3) | 0·636 |
| Time since HIV diagnosis (years), median (IQR) | 20·6 (15-25·1) | 21·5 (17·5-23·5) | 16·9 (12-27·4) | 0·342 |
| HIV mechanism of transmission, n (%) | 0·383 | |||
| Men who have sex with men | 14 (34·1) | 5 (23·8) | 9 (45) | |
| Heterosexual | 9 (22) | 6 (28·6) | 3 (15) | |
| Intravenous drug users | 13 (31·7) | 8 (38·1) | 5 (25) | |
| Other | 5 (12·2) | 2 (9·5) | 3 (15) | |
| CD4 count (cells/mm3), median (IQR) | ||||
| Nadir | 196 (93-290) | 160 (99-216) | 259 (70-314) | 0·161 |
| Baseline | 673 (531-842) | 705 (531-871) | 647 (530-800) | 0·527 |
| ART duration (years), median (IQR) | 18 (13·1-21·4) | 18·8 (17·2-21) | 13·1 (7·9-21·6) | 0·085 |
| Duration of suppressed plasma HIV RNA (years), median (IQR) | 6 (3·3-11) | 7·7 (4-12) | 5·3 (3-8.9) | 0·272 |
| Number of previous ART regimens, median (IQR) | 6 (4-10) | 7 (5-10) | 4 (2-7) | 0·427 |
| Type of ART regimen at baseline, n (%) | 0·002 | |||
| 2 NRTI + bPI | 8 (19.5%) | 7 (33·3) | 1 (5) | |
| 2 NRTI + NNRTI | 9 (22) | 2 (9·5) | 7 (35) | |
| 1 NNRTI + bPI | 5 (12·2) | 5 (23·8) | 0 (0) | |
| 1 NRTI + bPI | 14 (34·1) | 3 (14·3) | 11 (55) | |
| bPI monotheraphy | 5 (12·2) | 4 (19) | 1 (5) | |
| Baseline ART including lamivudine or emtricitabine | 28 (68·3) | 9 (42·9) | 19 (95) | <0·001 |
| Time from historical RNA genotype to baseline proviral DNA sequencing | 12·9 (11·1-14·4) | 13·4 (11·1-14·6) | 10·4 (4·3-13·6) | 0·152 |
| M184V (Proviral DNA Sanger genotype)* | 2 (4·9) | 2 (9·5) | 0 (0) | 0·488 |
| M184V/I detected by NGS in proviral DNA, n (%) | ||||
| >20% | 8 (19·5) | 7 (33) | 1 (5) | 0·046 |
| >10% | 13 (31·7) | 11 (52·4) | 2 (10) | 0·007 |
| >5% | 17 (45·5) | 14 (66·7) | 3 (15) | 0·002 |
| >1% | 27 (65·9) | 20 (95·2) | 7 (35) | <0·01 |
| K65R/E/N detected by NGS in proviral DNA, n (%)† | ||||
| >20% | 1 (2·4) | 1 (4·8) | 0 (0) | 1 |
| >10% | 2 (4·9) | 2 (9·5) | 0 (0) | 0·488 |
| >5% | 2 (4·9) | 2 (9·5) | 0 (0) | 0·488 |
| >1% | 3 (7·3) | 3 (14·3) | 0 (0) | 0·233 |
Abbreviations: 3TC: lamivudine. ART: antiretroviral treatment. bPI: boosted protease inhibitor. FTC: emtricitabine. NRTI: Nucleoside reverse transcriptase inhibitors. NNRTI: Non- Nucleoside reverse transcriptase inhibitors (NRTIs). NGS: Next-Generation Sequencing.
Protocol violation.
Of those who initiated cART prior to labour.
Proviral DNA next-generation sequencing genotypes from baseline samples obtained before switching ART were retrospectively available in all cases except for one participant belonging to the non-historical lamivudine resistance group, in whom amplification was unsuccessful. Seven participants with historical lamivudine resistance had the M184V/I and/or K65R/E/N mutations detected over the 20% threshold at baseline by next generation sequencing. When considering over a 5% threshold more than half of the participants with history of lamivudine resistance (15/21) had these mutations, and almost all participants in this group had lamivudine resistance-associated mutations when the 1% threshold was applied (20/21). Among participants without historical lamivudine resistance, none presented the K65R/E/N mutation but in three and seven cases the M184I mutation was detected with over a 5% or 1% threshold, respectively, including one participant harboring the M184I mutation with a 99% frequency. Reads with lamivudine resistance-associated mutations detected through next-generation sequencing (1% threshold) were subjected to hypermutation analysis, showing that 16/27 samples harbored retrotranscriptase defective viral genomes due to APOBEC-induced mutations. After removal of reads identified as hypermutated, lamivudine resistance-associated mutations remained present in 22 of the 27 initial samples.
Two participants in the history of lamivudine resistance group -initially misclassified as lacking lamivudine resistance-associated mutations- had the M184V mutation detected in proviral DNA Sanger sequencing at baseline (protocol violation).
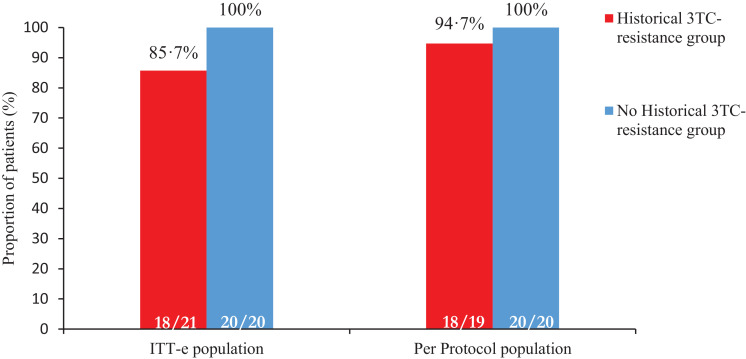
38/41 patients completed the 48 weeks of study. There were three participants that prematurely discontinued: the two protocol violations (week 12) and one withdrawal due to an adverse event (week 8), all doing so with HIV RNA below 50 copies per mL. At week 48, 92·7% of participants (38/41) remained with HIV RNA below 50 copies per mL (FDA-snapshot intention- to- treat- exposed analysis). All participants completing 48 weeks reached the primary efficacy endpoint of HIV RNA below 50 copies per mL following the FDA-snapshot algorithm (Figure 2, Table 2). The three participants that did not complete the study belonged to the historical lamivudine resistance group: primary efficacy endpoint in this group was 85·7 % in the intention- to- treat- exposed analysis (18/21 of participants with HIV RNA below 50 copies per mL at week 48) and 94·7% in the per-protocol analysis (18/19 HIV RNA below 50 copies per mL at week 48) (Table 2). The three non-completers abandoned the study before reaching week 24, hence, efficacy at week 24 does not differ from the results presented for week 48.
Figure 2.
Proportion of patients with virological suppression at W48 in Intention-to-treat-exposed (ITT-e) and Per-protocol analysis. ITT-e population is defined as all participants who received at least one dose of study treatment; PP population is defined as all participants who received at least one dose of study treatment and had no deviation to the eligibility criteria. Virological suppression was defined as an HIV RNA viral load of less than 50 copies per mL.
Abbreviations: 3TC: lamivudine.
Table 2.
FDA-snapshot at W48, Intention to treat- exposed (ITT-e) analysis population (n=41). Abbreviations: AE: adverse event.
| All participants | Historical resistance to lamivudine | No historical resistance to lamivudine | |
|---|---|---|---|
| (n=41) | (n=21) | (n=20) | |
| HIV-1 RNA ≤50 copies/mL | 38 (92·7) | 18 (85·7) | 20 (100) |
| Virologic failure | 0 (0) | 0 (0) | 0 (0) |
| HIV-1 RNA ≥50 copies/mL | 0 (0) | 0 (0) | 0 (0) |
| HIV-1 RNA ≥50 copies/mL in W48 window | 0 (0) | 0 (0) | 0 (0) |
| Discontinuation Study Drug due to Lack of Efficacy | 0 (0) | 0 (0) | 0 (0) |
| Discontinuation Study Drug due to other reasons and Last available HIV-1 RNA ≥50 copies/mL | 0 (0) | 0 (0) | 0 (0) |
| No virologic data at W48 | 3 (7·3) | 3 (14·3) | 0 (0) |
| Discontinuation Study Drug Due to AE | 1 (2·4) | 1 (4·8) | 0 (0) |
| Discontinuation Study Drug due to other reasons and Last available HIV-1 RNA <50 copies/mL | 2 (4·9) | 2 (9·5) | 0 (0) |
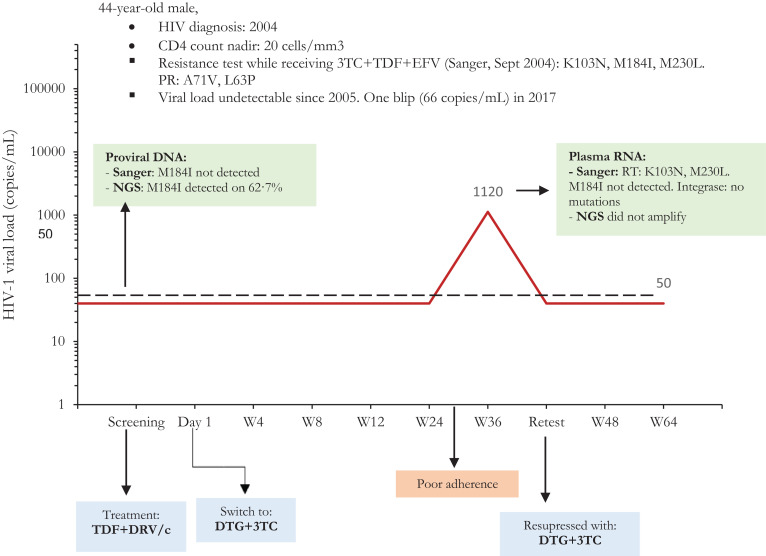
Ten participants – four from the historical lamivudine resistance and six from the non- lamivudine resistance group- had a transient low-level viral rebound followed by a re-test HIV RNA below 50 copies per mL (Table 3). Two of these occurred on day 1 of the study (before switching treatment), and in half of the cases the person admitted to temporary low adherence or was suffering a concomitant mild infection. All cases of transient viral rebound re-suppressed without changing treatment. We were able to obtain Sanger sequencing in two participants with history of M184V/I mutation who had a transient viral rebound, including one individual rebounding to 1120 copies/mL at week 36 (Figure 3): there was no re-emergence of lamivudine-resistance associated mutations nor did we identify any newly acquired integrase mutations. Three of the six participants without historical lamivudine resistance who had transient low-level viral rebound had the M184I mutation detected through next-generation sequencing in baseline proviral DNA, including the participant who had this mutation detected at a frequency above 99%. Notably, this participant's viral rebound occurred on day 1 of the study when he had not yet received the first dose of dolutegravir plus lamivudine, and remained without any blips throughout the study.
Table 3.
Therapeutic, viral and biological characteristics of patients presenting transient virological rebound during follow-up.
| Baseline | Follow-up | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| History of K65/M184VI mutation | Time suppressed before dual therapy (months) | Visit (week) | HIV-1 RNA (copies/mL) | resistance test | HIV-1 RNA after blip (copies/mL) | ART prior to study entry | Comment | Follow-up | |
| Patient A | Yes | 122 | W8 | 120 | NP | <50 | TDF/FTC+DRV/r | - | Continued on study |
| Patient B | No | 24 | W4 | 236 | Integrase NM. RT: L210W; T215D PR: 13V; 63P. | <50 | 3TC+DRV/c | Concomitant respiratory infection | Continued on study |
| Patient C | W4 | 120 | NP | <50 | TDF/FTC+EFV | W4 and W36 Concomitant respiratory infection. W24: Received intramuscular steroids | Continued on study | ||
| No | 32 | W24 | 73 | NP | <50 | ||||
| W36 | 63 | NP | <50 | ||||||
| Patient D | No | 47 | Day 1 | 108 | NP | <50 | 3TC+DRV/r | - | Continued on study |
| Patient E | No | 72 | Day 1 | 1.943 | RT: 69TN | <50 | TDF/FTC/RPV | - | Continued on study |
| Patient F | Yes | 93 | W12 | 64 | NP | <50 | ETV+DRV/r | Concomitant respiratory infection | Withdrawn (protocol violation) |
| Patient G | No | 35 | W12 | 112 | NP | <50 | 3TC+DRV/c | Continued on study | |
| Patient H | No | 92 | W36 | 202 | AU | <50 | TDF/FTC+EFV | Low adherence | Continued on study |
| Patient I | Yes | 151 | W36 | 1.120 | Integrase: NM; RT and PR: AU | <50 | TDF+DRV/c | Recent flu vaccination | Continued on study |
| Patient J | Yes | 181 | W48 | 257 | Integrase: NM; RT: M184V/I not detected. | <50 | DRV/c | - | Continued on study |
* All patents with VL> 200 copies/mL had a resistance test requested. Not performed (NP) indicates when such test was requested but not performed at laboratory discretion due to low-level viremia.
Abbreviations: AU: amplification unsuccessful. NM: no mutations NP: not performed. PR: protease. PV: protocol violation. RT: retrotranscriptase. ART: antiretroviral treatment. TDF: tenofovir disoproxil fumarate. FTC: emtricitabine. DRV/r: darunavir/ritonavir. 3TC: lamivudine. DRV/c: darunavir/cobicistat. EFV: efavirenz. RPV: rilpivirine. ETV: etravirine.
Figure 3.
Viral load evolution of the patient presenting over 1000 copies/mL at W36.
Abbreviations: 3TC: lamivudine. DRV/c: darunavir/cobicistat. DTG: dolutegravir. EFV: efavirenz. NGS: Next-generation sequencing. RT: Retrotranscriptase. TDF: tenofovir disoproxil fumarate.
Through week 48, there were no cases of withdrawal due to virologic failure and no emergence of resistance mutations. None of the 21 participants who had either history of lamivudine resistance or detection of lamivudine resistance-associated mutations above the 5% threshold in proviral DNA had virologic failure after 48 weeks of follow-up (IC95% 0-14%). At 48 weeks, there was not a significant difference in mean change of CD4 count from baseline (23·84 cells/mm3, p=0·686, IC95% -94·88-142·56).
There were 28 treatment related adverse events (Table 4). Most drug related adverse events were mild cases of flatulence or insomnia that spontaneously resolved in the first weeks after the switch. One participant had newly acquired insomnia attributed to dolutegravir leading to treatment discontinuation on week 8, after which his symptoms disappeared. The only laboratory grade 3 drug-related adverse event occurred in one participant in the group with historical lamivudine resistance (hypercholesterolemia). We found no difference in total cholesterol, HDL, LDL mean changes by group. Mean weight change from baseline was 1·36 kg (95% CI -1·11; 3·84) without significant differences by group.
Table 4.
Overview of drug-related adverse events.
| Historical resistance to lamivudine |
No historical resistance to lamivudine |
||||
|---|---|---|---|---|---|
| (n=21) |
(n=20) |
||||
| Patients | Adverse events | Patients | Adverse events | p-value | |
| Any adverse event | 10 (47·6%) | 13 | 8 (40%) | 15 | 0·624 |
| Grade 3 or 4 adverse event | 1 (4·8%) | 1 | 0 (0%) | 0 | <0·001 |
| Laboratory Grade 3 or 4 adverse event | 1 (4·8%) | 1 | 0 (0%) | 0 | <0·001 |
| Serious adverse event | 0 (0%) | 0 | 0 (0%) | 0 | |
| Discontinuation due to adverse event | 1 (4·8%) | 1 | 0 (0%) | 0 | <0·001 |
| Death | 0 (0%) | 0 | 0 (0%) | 0 | |
| Adverse events occurring in at least 5% of patients in either group | |||||
|---|---|---|---|---|---|
| Digestive | 2 (9·5%) | 2 | 4 (20%) | 5 | 0·342 |
| Neuropsychiatric | 5 (23·8%) | 5 | 3 (15%) | 3 | 0·477 |
| Dermatologic | 0 (0%) | 0 | 1 (5%) | 1 | <0·001 |
| Musculoskeletal | 2 (9·5%) | 2 | 1 (5%) | 1 | <0·001 |
| Head and neck | 0 (0%) | 0 | 2 (10%) | 2 | 0·1373 |
4. Discussion
In this pilot trial dolutegravir plus lamivudine maintained virologic suppression through 48 weeks despite history of lamivudine resistance and detection of archived lamivudine resistance-associated mutations in proviral DNA by next generation sequencing but not by population sequencing. These results provide preliminary evidence supporting the combination of dolutegravir plus lamivudine as a reduced-drug regimen for maintenance of HIV suppression in integrase naïve persons with historical lamivudine resistance, without lamivudine resistance-associated mutations detected by proviral DNA Sanger sequencing
Our results and those of others do no support the assumption that dolutegravir plus lamivudine in patients with prior history of lamivudine resistance is functional dolutegravir monotherapy. In ART-PRO none of the 21 participants reaching week 48 who had either history of lamivudine resistance or detection of lamivudine resistance mutations above the 5% threshold in proviral DNA experienced virologic failure. In two other clinical trials a total of 21 participants with history of lamivudine resistance or detection of lamivudine resistance-associated mutations in proviral DNA did not experience virologic failure 48 weeks after switching to dolutegravir plus lamivudine [20], [21], [22]. Adding the results of these three trials, none of a total of 42 participants with history and/or detection of lamivudine resistance-associated mutations have developed virologic failure after switching to dual therapy with dolutegravir plus lamivudine. In contrast, pooled data from dolutegravir monotherapy studies for maintenance treatment revealed a proportion of virologic failure of 8·9% (95% CI 4·7-16·2) at 48 weeks [23]. These differences in the rates of virological failure suggest that lamivudine might provide significant antiviral activity to dolutegravir despite the presence of archived lamivudine resistance mutations. Additional support for this hypothesis comes from two retrospective cohort studies in which a historical plasma genotype detecting the M184V mutation was not predictive of virologic failure in participants switching to lamivudine- based dual therapies with either a protease inhibitor or an integrase inhibitor [24,25].
The residual antiviral efficacy of lamivudine was demonstrated more than a decade ago in a study showing a viral load increase of 0·5 log10 after discontinuing lamivudine despite the presence of the M184V mutation [26]. Another study reported lower viral rebound in persons harboring the M184V who received lamivudine monotherapy compared to patients interrupting treatment [27]. In addition to the antiviral effect of lamivudine, the maintenance of the M184V/I mutations might contribute to the success of dolutegravir plus lamivudine by sustaining a diminished viral fitness and preventing the emergence of resistance mutations against dolutegravir [28,29].
Another factor that might have contributed to the success of the dolutegravir plus lamivudine regimen in this pilot trial is the exclusion of participants if baseline proviral DNA population genotyping detected lamivudine resistance-associated mutations. In our study GEN-PRO [16] we found that when lamivudine resistance-associated mutations were absent in proviral DNA by population genotyping, the percentage of lamivudine resistance-associated mutations detected by next-generation sequencing was much lower than when proviral DNA population sequencing did detect lamivudine resistance-associated mutations. Only 17·1% of the participants in ART-PRO had detection of lamivudine resistance-associated mutations by next generation sequencing with a threshold over 20%. This is in contrast with GEN-PRO, where 78·5% or participants with lamivudine resistance-associated mutations detected in proviral DNA by population genotyping had these mutations detected by next generation sequencing with a threshold of 20%. We believe baseline proviral DNA population sequencing renders at least some assurance that past lamivudine resistance-associated mutations are not persisting at high levels and thus, increases the probability of response to dolutegravir plus lamivudine. In our study, the hypermutation analysis showed that 18·5% of mutations associated with lamivudine resistance detected at baseline through next-generation sequencing were from hypermutated viral sequences, and therefore, not clinically relevant. However, this analysis does not completely exclude that the archived resistance mutations found in our study may be related to defective viral genomes for other reasons different to hypermutation, such as large internal deletions [15].
In our study the group with historical lamivudine resistance had longer duration of viral suppression (7·7 vs 5·3 years) compared to the group without historical lamivudine resistance. It has been postulated that the duration of virologic suppression could have some role in the dynamics of lamivudine resistance. The small sample size prevents us from inferring if this had a significant impact in our results, however, in the study by Gagliardini et al. [30] dual therapy with either boosted protease inhibitor or integrase inhibitor plus lamivudine in persons with past M184V demonstrated only a statistically higher risk of virological failure for those with a viral suppression equal or under three years, which is inferior to the median viral suppression duration in our study.
The most important limitations of our study are the small sample size-characteristic of a pilot proof-of-concept clinical trial and the relative short follow-up. We intentionally recruited a small number of participants because treatment with dolutegravir plus lamivudine in suppressed persons with prior history of resistance to lamivudine had never been prospectively tested before. A 144-week extension of our study is currently ongoing, but further studies are necessary to confirm our findings.
To our knowledge, our study is the only specifically designed to address the outcomes of dolutegravir plus lamivudine for maintenance of viral suppression in patients with historical resistance to lamivudine. Our results generate the hypothesis that a switch to dolutegravir plus lamivudine in integrase-inhibitor naïve persons with historical lamivudine resistance-associated mutations could be effective in preserving virologic suppression at 48 weeks, under the condition that, before switching to dual therapy, proviral DNA population sequencing does not detect the persistence of lamivudine resistance-associated mutations. The findings of this pilot trial call for a larger, fully-powered study to confirm our results.
Declaration of Competing Interest
RDM reports grants from Fondo de Investigaciones Sanitarias, during the conduct of the study; personal fees and non-financial support from Janssen, non-financial support from ViiV, non-financial support from Gilead, outside the submitted work.
DR reports personal fees from Gilead Sciences Inc, personal fees from Janssen Cilag, grants and personal fees from ViiV Healthcare, outside the submitted work.
LD reports payment for lectures from Gilead and Janssen, and financial support for expert courses and congress from Merck Sharp and Dome, Gilead and Abbvie, outside the submitted work.
RM reports grants from Juan Rodes 18/00039, during the conduct of the study; personal fees from ViiV Health care, personal fees from Janssen Cilag, outside the submitted work.
OB reports non-financial support from GILEAD, personal fees from GILEAD, personal fees from VIIV, grants from ViiV, non-financial support from MSD, grants from VIIV, outside the submitted work.
AE reports grants from Instituto de Salud Carlos III, during the conduct of the study.
PA reports personal fees from ViiV Healthcare, outside the submitted work.
NS reports personal fees from Janssen, personal fees from Gilead, outside the submitted work.
LB has nothing to disclose.
MG reports grants and personal fees from Hologic, grants from Roche diagnostics, grants from Beckman coulter, personal fees from ViiV Health Care, grants from Instituto de Salud Carlos III, outside the submitted work.
JC reports grants from Instituto de Salud Carlos III - Ministerio de Ciencia, Innovación y Universidades, outside the submitted work.
MS reports personal fees from Janssen Cilag, personal fees from ViiV Healthcare, outside the submitted work.
BA has nothing to disclose.
AH has nothing to disclose.
MM has nothing to disclose.
JC has nothing to disclose.
VM reports personal fees from ViiV Health Care, personal fees from Gilead Sciences, personal fees and non-financial support from Janssen Cilag, personal fees from Merck Sharp & Dohme, outside the submitted work.
LM reports personal fees from Gilead, personal fees from Viiv, personal fees from MSD, personal fees from Janssen, outside the submitted work.
RR reports personal fees from ViiV Healht Care, personal fees from Gilead Sciences, personal fees from Janssen Cilag, personal fees from Merck Sharp & Dohme, outside the submitted work.
RD has received conference fees from ViiV.
FP reports grants from Instituto de Salud Carlos III, during the conduct of the study; personal fees from Gilead Sciences, personal fees from Janssen, personal fees from MSD, personal fees from ViiV Healthcare, outside the submitted work.
JRA reports grants from Instituto de Salud Carlos III, during the conduct of the study; grants and personal fees from VIIV, Gilead, personal fees from Janssen, MSD, Alexa, TEVA, outside the submitted work.
Acknowledgments
Contributors
JRA, FP, LD, RM, NS, OB, AH, and RD participated in the conceptualization and design of the study. RDM, DR, LD, RM, OB, JC, VM, LM, RR, FP and JRA were study investigators and participated in the conduct of the study, including the recruitment and follow-up of participants. AE, PA, NS, MG, and RD performed DNA sequencing and resistance analysis. LB, AH, MS, MM and JMC curated data, project administration, and coordination. BA, RDM, DR, LD and RM were involved with formal data analysis. JRA and FP were responsible for funding acquisition and supervision of all the processes of the trial. All authors participated in the drafting and review of the manuscript.
Data Sharing
Study protocol will be available online at the time of publication. Deidentified participant data and study documents will be accessible with a signed data access agreement upon approval of a proposal.
Acknowledgments
This study was funded by Fondo de Investigaciones Sanitarias, Instituto de Salud Carlos III PI16/00837 - PI16/00678. RDM is supported by a Rio Hortega fellowship from Fondo de Investigaciones Sanitarias (CM17/00064). M.G.A is supported by “Instituto de Salud Carlos III” (JR15/00031). AE is supported by a PFIS fellowship from Fondo de Investigaciones Sanitarias (FI17/00194). JC is supported by a Rio Hortega fellowship from Fondo de Investigaciones Sanitarias (CM19/00059).
The authors thank the study participants; their families and caregivers; investigators and site staff who participated in the study.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102779.
Contributor Information
Federico Pulido, Email: federico.pulido@salud.madrid.org.
José Ramón Arribas, Email: joser.arribas@salud.madrid.org.
Appendix
ART-PRO, PI16/00837-PI16/00678 study group:
Hospital Universitario La Paz - IdiPAZ, Madrid, Spain: J.R. Arribas, R. De Miguel Buckley, R. Montejano, A. Esteban-Cantos, N. Stella-Ascariz, J. Cadiñanos, M. Mayoral, J.M. Castro, V. Moreno, L. Martin-Carbonero, E. Valencia, I. Bernardino, C. Busca, R. Micán, I. Pérez-Valero, J. González, ML. Montes, J. Rodríguez Centeno.
Hospital Universitario 12 de Octubre - Imas12, Madrid, Spain: F. Pulido, D. Rial-Crestelo, L. Dominguez-Dominguez, P. Aranguren-Rivas, O. Bisbal, L. Bermejo Plaza, M. Garcia-Alvarez, M. Santacreu-Guerrero, M. de Lagarde, M. Matarranz, J. Luzckoviak, A. Sotillo, R. Delgado, R. Rubio.
Instituto de Salud Carlos III, Madrid, Spain: B. Alejos
Universidad Europea de Madrid- Imas12, Madrid, Spain: A. Hernando
Appendix B. Supplementary materials
References
- 1.EACS Guidelines version 10.0, November 2019. 2019;(November).
- 2.Panel on Antiretroviral Guidelines for Adults and Adolescents Guidelines for the Use of Antiretroviral Agents in Adults and Adolescents with HIV. Dep Heal Hum Serv. 2018 [Google Scholar]
- 3.van Wyk J, Ajana F, Bisshop F, De Wit S, Osiyemi O, Portilla J. Efficacy and Safety of Switching to Dolutegravir/Lamivudine Fixed-Dose Two-Drug Regimen Versus Continuing a Tenofovir Alafenamide–Based Three- or Four-Drug Regimen for Maintenance of Virologic Suppression in Adults With HIV-1: Phase 3, Randomized. Non-inf. Clin Infect Dis. 2020 Jan doi: 10.1093/cid/ciz1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charpentier C, Peytavin G, Burdet C, Landman R, Lê M, Katlama C. Residual HIV-1 RNA, HIV-1 DNA, and Drug Plasma Cmin in Dual DTG+3TC. ANRS 167 Lamidol. CROI 2019. 2019 [Google Scholar]
- 5.Li JZ, Sax PE, Marconi VC, Fajnzylber J, Berzins B, Nyaku AN. No significant changes to residual viremia after switch to dolutegravir and lamivudine in a randomized trial. Open Forum Infect Dis. 2019;6(3):5–8. doi: 10.1093/ofid/ofz056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paton NI, Kityo C, Hoppe A, Boles J, Thompson J, Tumukunde D. Assessment of second-line antiretroviral regimens for HIV therapy in Africa. N Engl J Med. 2014 Jul;371(3):234–247. doi: 10.1056/NEJMoa1311274. [DOI] [PubMed] [Google Scholar]
- 7.La Rosa AM, Harrison LJ, Taiwo B, Wallis CL, Zheng L, Kim P. Raltegravir in second-line antiretroviral therapy in resource-limited settings (SELECT): a randomised, phase 3, non-inferiority study. lancet HIV. 2016 Jun;3(6):e247–e258. doi: 10.1016/S2352-3018(16)30011-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Study Group SECOND LINE, Boyd M, Kumarasamy N, Moore CL, Nwizu CA. Ritonavir-boosted lopinavir plus nucleoside or nucleotide reverse transcriptase inhibitors versus ritonavir-boosted lopinavir plus raltegravir for treatment of HIV-1 infection in adults with virological failure of a standard first-line ART regimen (SECOND. Lancet. 2013 Jun;381(9883):2091–2099. doi: 10.1016/S0140-6736(13)61164-2. [DOI] [PubMed] [Google Scholar]
- 9.Ciaffi L, Koulla-Shiro S, Sawadogo AB, Ndour CT, Eymard-Duvernay S, Mbouyap PR. Boosted protease inhibitor monotherapy versus boosted protease inhibitor plus lamivudine dual therapy as second-line maintenance treatment for HIV-1-infected patients in sub-Saharan Africa (ANRS12 286/MOBIDIP): a multicentre, randomised, parallel, open-la. Lancet HIV. 2017;4(9):e384–e392. doi: 10.1016/S2352-3018(17)30069-3. [DOI] [PubMed] [Google Scholar]
- 10.Wirden M, Soulie C, Valantin MA, Fourati S, Simon A, Lambert-Niclot S. Historical HIV-RNA resistance test results are more informative than proviral DNA genotyping in cases of suppressed or residual viraemia. J Antimicrob Chemother. 2011;66(4):709–712. doi: 10.1093/jac/dkq544. [DOI] [PubMed] [Google Scholar]
- 11.Allavena C, Rodallec A, Leplat A, Hall N, Luco C, Le Guen L. Interest of proviral HIV-1 DNA genotypic resistance testing in virologically suppressed patients candidate for maintenance therapy. J Virol Methods. 2018;251(October 2017):106–110. doi: 10.1016/j.jviromet.2017.10.016. [DOI] [PubMed] [Google Scholar]
- 12.Delaugerre C, Braun J, Charreau I, Delarue S, Nere ML, de Castro N. Comparison of resistance mutation patterns in historical plasma HIV RNA genotypes with those in current proviral HIV DNA genotypes among extensively treated patients with suppressed replication. HIV Med. 2012;13(9):517–525. doi: 10.1111/j.1468-1293.2012.01002.x. [DOI] [PubMed] [Google Scholar]
- 13.Inzaule SC, Hamers RL, Noguera-Julian M, Casadellà M, Parera M, Kityo C. Clinically relevant thresholds for ultrasensitive HIV drug resistance testing: a multi-country nested case-control study. Lancet HIV. 2018;5(11):e638–e646. doi: 10.1016/S2352-3018(18)30177-2. [DOI] [PubMed] [Google Scholar]
- 14.Dauwe K, Staelens D, Vancoillie L, Mortier V, Verhofstede C. Deep Sequencing of HIV-1 RNA and DNA in Newly Diagnosed Patients with Baseline Drug Resistance Showed No Indications for Hidden Resistance and Is Biased by Strong Interference of Hypermutation. Caliendo AM, editor. J Clin Microbiol. 2016 Jun;54(6):1605–1615. doi: 10.1128/JCM.00030-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ho YC, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DIS. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155(3):540. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Domínguez-Domínguez L, Montejano R, Esteban-cantos A, García M, Stella-ascariz N, Bisbal O. 17th European AIDS Conference. 2019. Prevalence and factors associated to the detection (population and next generation sequencing) of archived 3TC resistance mutations in aviremic HIV-infected adults (GEN- PRO) PE13/2. [Google Scholar]
- 17.Perrier M, Visseaux B, Landman R, Joly V, Todesco E, Yazdanpanah Y. No impact of HIV-1 protease minority resistant variants on the virological response to a first-line PI-based regimen containing darunavir or atazanavir. J Antimicrob Chemother. 2018;73(1):173–176. doi: 10.1093/jac/dkx366. [DOI] [PubMed] [Google Scholar]
- 18.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27(6):863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magoč T, Salzberg SL.FLASH. Fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Wyk J, F Ajana, Bisshop F, Dewit S, Osiyemi O, Portilla J. Switching To DTG-3TC Fixed-Dose Combination (Fdc) Is Non-Inferior To Continuing a Taf-Based Regimen in Maintaining Virologic Suppression Through 48 Weeks (Tango Study) IAS 2019. 2019 [Google Scholar]
- 21.Reynes J, Meftah N, Tuaillon E, Charpentier C, Montes B. Dual regimen with dolutegravir and lamivudine maintains virologic suppression even in heavily treatment experienced HIV-infected patients: 96 weeks results from maintenance DOLULAM study. IAS 2017. 2017 [Google Scholar]
- 22.Charpentier C, Montes B, Perrier M, Meftah N, Reynes J. HIV-1 DNA ultra-deep sequencing analysis at initiation of the dual therapy dolutegravir + lamivudine in the maintenance DOLULAM pilot study. J Antimicrob Chemother. 2017 Oct;72(10):2831–2836. doi: 10.1093/jac/dkx233. [DOI] [PubMed] [Google Scholar]
- 23.Wandeler G, Buzzi M, Anderegg N, Sculier D, Béguelin C, Egger M. Virologic failure and HIV drug resistance on simplified, dolutegravir-based maintenance therapy: Systematic review and meta-analysis. F1000Research. 2019;7:1359. doi: 10.12688/f1000research.15995.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gagliardini R, Ciccullo A, Borghetti A, Maggiolo F, Bartolozzi D, Borghi V. Impact of the M184V Resistance Mutation on Virological Efficacy and Durability of Lamivudine-Based Dual Antiretroviral Regimens as Maintenance Therapy in Individuals With Suppressed HIV-1 RNA: A Cohort Study. Open Forum Infect Dis. 2018 Jun;5(6):1–8. doi: 10.1093/ofid/ofy113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baldin G, Ciccullo A, Borghetti A, Di Giambenedetto S. Virological efficacy of dual therapy with lamivudine and dolutegravir in HIV-1-infected virologically suppressed patients: long-term data from clinical practice. J Antimicrob Chemother. 2019;74(5):1461–1463. doi: 10.1093/jac/dkz009. [DOI] [PubMed] [Google Scholar]
- 26.Campbell TB, Shulman NS, Johnson SC, Zolopa AR, Young RK, Bushman L. Antiviral Activity of Lamivudine in Salvage Therapy for Multidrug‐Resistant HIV‐1 Infection. Clin Infect Dis. 2005;41(2):236–242. doi: 10.1086/430709. [DOI] [PubMed] [Google Scholar]
- 27.Castagna A, Danise A, Menzo S, Galli L, Gianotti N, Carini E. Lamivudine monotherapy in HIV-1-infected patients harbouring a lamivudine-resistant virus: A randomized pilot study (E-184V study) Aids. 2006;20(6):795–803. doi: 10.1097/01.aids.0000218542.08845.b2. [DOI] [PubMed] [Google Scholar]
- 28.Wainberg MA. The impact of the M184V substitution on drug resistance and viral fitness. Expert Rev Anti Infect Ther. 2004;2(1):147–151. doi: 10.1586/14787210.2.1.147. [DOI] [PubMed] [Google Scholar]
- 29.Oliveira M, Ibanescu RI, Pham HT, Brenner B, Mesplede T, Wainberg MA. The M184I/V and K65R nucleoside resistance mutations in HIV-1 prevent the emergence of resistance mutations against dolutegravir. AIDS. 2016;30(15):2267–2273. doi: 10.1097/QAD.0000000000001191. [DOI] [PubMed] [Google Scholar]
- 30.Gagliardini R, Ciccullo A, Borghetti A, Maggiolo F, Bartolozzi D, Borghi V. Impact of the M184V Resistance Mutation on Virological Efficacy and Durability of Lamivudine-Based Dual Antiretroviral Regimens as Maintenance Therapy in Individuals With Suppressed HIV-1 RNA: A Cohort Study. Open forum Infect Dis. 2018 Jun;5(6) doi: 10.1093/ofid/ofy113. ofy113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.