Abstract
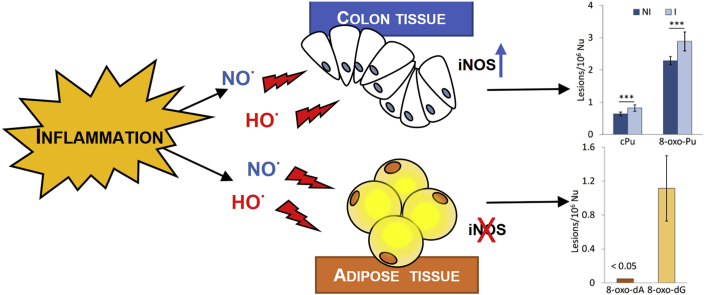
Chronic inflammation is estimated to be a causative factor in a variety of diseases. Under inflammatory conditions reactive oxygen species (ROS) and nitrogen species (RNS) are released leading to DNA damage accumulation and genomic instability. Purine 5′,8-cyclo-2′-deoxynucleosides (cPu) are oxidative DNA lesions, exclusively derived from the attack of HO• radicals, which are known to have cytotoxic and mutagenic properties. Herein, we have analyzed the presence of cPu in genomic DNA isolated from fresh colon and visceral adipose tissue biopsies collected from inflammatory bowel diseases (IBD)-affected patients and severely obese subjects, respectively, versus what observed in the control specimens. In colon biopsies, characterized by a higher gene expression level of inducible nitric oxide synthase (iNOS), a significant increase of 8-oxo-7,8-dihydro-2′-deoxyadenosine (8-oxo-dA) and 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxo-dG) lesions and an accumulation of both diastereomeric cPu have been detected. In contrast, the 8-oxo-dA and 8-oxo-dG levels were extremely lower compared to the colon tissues values and no accumulation of cPu, in the inflamed visceral adipose tissue biopsies isolated from bariatric patients versus the lean counterpart was reported. In addition, in adipose tissue undetectable levels of iNOS have been found. These data suggest a potential involvement of cPu in the colon cancer susceptibility observed in IBD patients.
Keywords: Reactive oxygen species, Free radicals, DNA damage, Cyclopurines, Inflammatory bowel diseases, Obesity
Graphical abstract
1. Introduction
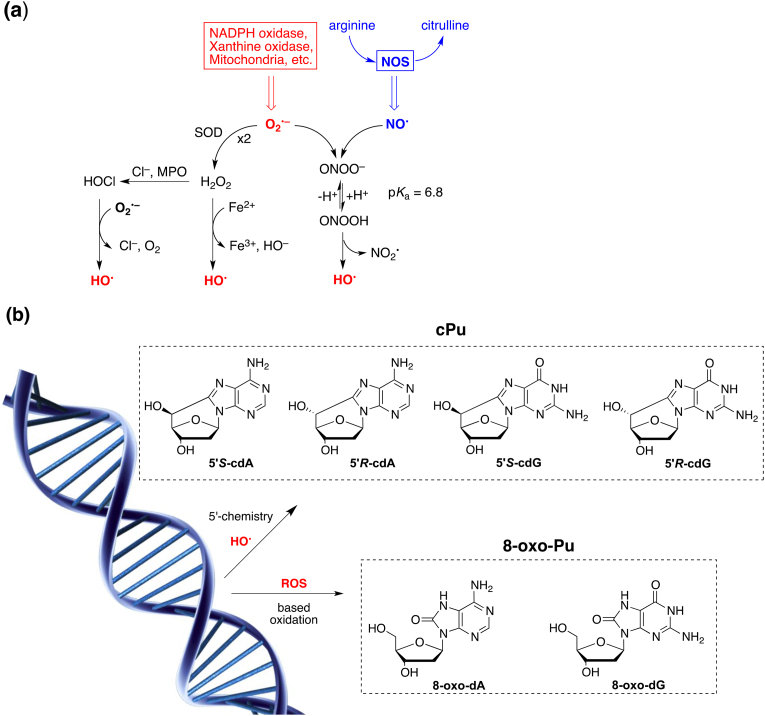
Superoxide dismutase (SOD) and nitric oxide synthase (NOS) are key enzymes known to prevent deleterious effects due to oxidative stress, by regulating the level of superoxide radical anion (O2•–) and inducing the formation of nitric oxide (NO•), respectively (Fig. 1a) [1,2]. These two radicals are the precursors of endogenous reactive oxygen/nitrogen species (ROS/RNS) network that includes molecules, such as hydrogen peroxide (H2O2), hypochlorous acid (HOCl) and peroxynitrite (ONOO–), as well as radicals such as hydroxyl radical (HO•), nitrogen dioxide (•NO2) and carbonate radical anion (CO3•–). The ROS/RNS are well known for playing a dual role as both deleterious and beneficial species. In healthy tissue, the reactive species are present at very low concentrations and perform important functions as an efficient cellular defense mechanism eliminating viral and microbial infections. Whereas, the overproduction of ROS/RNS under conditions of acute or chronic inflammation has been linked with the etiology of various diseases though the reactivity of these species towards DNA, proteins and lipids and the induced regulation of several signaling pathways affecting a variety of cellular processes. Among the reactive species, the HO• radical is a strong oxidizing agent and can directly react with the DNA, either by hydrogen abstraction from the sugar or by addition/hydrogen abstraction involving the base moieties. The main processes that generate HO• radicals are reported in Fig. 1a [1,2]. Peroxynitrite (ONOO−/ONOOH) associated with a pKa value of 6.8 and the protonated form decomposes through homolysis of the weak O–O bond forming •OH and •NO2 radicals. The Fenton reaction, i.e., H2O2 reacts with the reduced-state transition metal ions, like Fe2+ or Cu1+, to give HO• radicals. H2O2 is used by peroxidase for the synthesis of HOCl, which then interacts with O2•– yielding HO• radicals.
Fig. 1.
(a) Endogenous ROS/RNS network and molecular pathways of hydroxyl radical (HO•) generation. (b) Structures of cPu lesions generated by H-abstraction from H5’ position by HO• radical and 8-oxo-Pu lesions generated by based oxidation from HO• radical and other ROS species.
The reaction of HO• radicals with isolated DNA has been studied in some details, the site of attack being both the base moieties (85–90%) and the 2-deoxyribose (10-15%) [3,4]. Among the DNA modifications the purine 5′,8-cyclo-2′-deoxynucleosides (cPu) are peculiar of the chemistry of the C5′ position and they are formed exclusively by reaction of the hydroxyl radical (•OH) with 2′-deoxyguanosine (dG) or 2′-deoxyadenosine (dA) moieties, resulting in the formation of an additional C5′−C8 covalent bond. The cPu can exist in a 5′R and 5′S configuration (Fig. 1b) [[4], [5], [6]]. The presence of C5’-C8 bond renders the cPu lesions attractive substrates for nucleotide excision repair (NER) which is the only repair mechanism able to process these lesions. A different repair efficiency of the R and S forms has been observed [7,8].
Unrepaired cPu lesions may lead to harmful biological consequences. Both 5′,8-cyclo-2′-deoxyadenosine (cdA) and 5′,8-cyclo-2′-deoxyguanosine (cdG), are able to block DNA and RNA polymerases leading to cell death. In vitro studies showed that these lesions can be bypassed, during DNA replication, giving rise to a specific mutation pattern [9]. These lesions are known to accumulate with aging in a tissue specific manner, in: liver > kidney > brain [10,11], are highly cytotoxic and mutagenic [12,13] while their extended presence in cellular DNA is also associated with cancer [14,15] and neurodegeneration [[16], [17], [18], [19]]. On the other hand, the addition of HO• radicals to the guanine and adenine moieties leads to a variety of products including the well-known 8-oxo-7,8-dihydro-2′-deoxyadenosine (8-oxo-dA) and 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxo-dG) lesions (Fig. 1b) [20]. Like HO• radicals, other oxidizing species such as H2O2, singlet oxygen or ONOO— are able to generate 8-oxo-Pu lesions that are removed by the base excision repair (BER) system [21].
Leading theories suggest that oxidative stress can have a crucial role in the development of different pathologies characterized by chronic inflammation, such as inflammatory bowel disorders (IBD) [[22], [23], [24]] and the several dysfunctions frequently associated with severe obesity, such as insulin resistance, cardiovascular diseases and diabetes [25]. It is known that IBD patients have an increased risk of colorectal cancer (CRC) and that severe obesity can influence cancer risk as well as cancer outcomes [[26], [27], [28]]. Elevated ROS production and high proliferation rate of epithelial cells can concur to the early development of genetic and genomic instability typically observed, at early stages, during IBD-associated colon cancer progression [29]. In IBD patients, gene expression level of inducible nitric oxide synthase (iNOS) was increased in inflamed colon biopsies versus the normal counterpart [30], and a positive correlation between the degree of inflammation, the iNOS expression levels and the subsequent p53 activation in IBD tissues has been found [31]. In addition, both enzymatic and non-enzymatic endogenous antioxidant defense, such as catalase, superoxide dismutase and the reduced form of glutathione, was lowered in colon biopsies [32,33]. The increase of both oxidative DNA damage and lipid peroxidation in tissue biopsies and body fluids also testifies an imbalance between antioxidant defence and ROS/RNS production in IBD [[34], [35], [36]].
The adipose tissue, held for long time solely responsible for the thermogenesis and the energy storage, is an endocrine organ with high plasticity, able to secrete anti-inflammatory and pro-inflammatory cytokines, e.g. adipokine and leptin, respectively [37]. It is well known that obesity is a risk factor for several co-morbidities, such as cardiovascular diseases, diabetes and cancer. Obese patients are characterized by a low grade of chronic and sterile inflammation, a condition that suggests a potential impact on genome stability. Consistently, recent lines of evidence testify that the oxidative stress and the inflammation, associated with obesity, lead to DNA damage accumulation: increased levels of 8-oxoguanine, in urine, blood and plasma, as well as single and double DNA strand breaks in human lymphocytes, have been found in obese humans when compared to what observed in the normal weight counterpart [38,39].
The aim of this study was to investigate the presence of cPu in genomic DNA isolated from tissue biopsies collected from patients affected by chronic inflammatory diseases, such as IBD and severe obesity, assuming that these highly mutagenic adducts could contribute to genomic abnormalities as well as the increased cancer susceptibility associated with these pathologies.
2. Materials and methods
2.1. Materials
Nuclease P1 from Penicilliumcitrinum, phosphodieasterase I and II, alkaline phosphatase from bovine intestinal mucosa, DNase I and DNase II, benzonase 99%, BHT, deferoxaminemesylate and pentostatin were purchased from Sigma-Aldrich (Steinheim, Germany). RNase T1 was from Thermo Fisher Scientific (Waltham, MA, USA) and RNase A from Roche Diagnostic GmbH, (Mannheim, Germany). 2′-Deoxyadenosine monohydrate and 2′-Deoxyguanosine monohydrate were purchased from Berry & Associates Inc. (Dexter, NY, USA). Isotopically labelled internal standards of 5′R-cdA, 5′S-cdA, 5′R-cdG, 5′S-cdG, 8-oxo-dA and 8-oxo-dG were prepared according to the previously reported procedures [40]. Solvents (HPLC-grade) were purchased from Fisher Scientific (Waltham, MA, USA). The 3 kDa cut-off filters were obtained from Millipore (Bedford, OH, USA). Written consent was obtained from all participants. The approval by the ethics committee was obtained for IBD and obese patients (Istituto Superiore di Sanità, PROT. PRE 173/16; University of Rome Tor Vergata Register 169/15).
2.2. DNA and RNA isolation and gene expression analysis
Colon biopsies were collected from macroscopic inflamed and normal areas of patients undergoing routine colonoscopy for a suspected or already diagnosed IBD and kept in RNA later solution to preserve the integrity of the nucleic acids during transport and storage. Before DNA and RNA extractions, both colon and adipose tissue biopsies were homogenized. Total DNA was extracted by genomic DNA isolation kit (Norgenbiotek corp. 24700) after homogenization by mixer mill (MM 400, Retsch) of tissue samples (approximately 200 mg). Total RNA extraction from colon biopsies was carried out by using RNeasy mini kit (Qiagen) and then converted to single stranded cDNA by using a reverse transcriptase with the High-Capacity cDNA Archive Kit (Life technologies). Conversely, total RNA from adipose tissue biopsies was performed by Invitrogen TRIzol reagent. Gene expression levels of genes of interest were analyzed by TaqMan gene expression assays (7500 fast real time PCR system, Life Technologies).
2.3. Enzymatic digestion protocol
DNA samples (10 μg) were hydrolyzed to single nucleosides using an 100 μL of Ar flushed enzymatic cocktail containing10 mMTris-HCl (pH 7.9), 10 mM MgCl2, 50 mM NaCl, 0.2 mM pentostatin, 5 μM butylated hydroxytoluene (BHT) and 3 mM deferoxamine and the internal standards ([15N5]-5′S-cdA, [15N5]-5′R-cdA, [15N5]-5′S-cdG, [15N5]-5′R-cdG, [15N5]-8-oxo-dG and [15N5]-8-oxo-dA) as previously described (see Fig. S1) [14,41]. Benzonase (3 U in 20 mMTris-HCl pH 8.0, 2 mM MgCl2 and 20 mM NaCl), 4 mU phosphodiesterase I, 3 U DNase I, 2 mU of phosphodiesterase II and 2 U of alkaline phosphatase were added and the mixture was incubated at 37 °C. After 21 h, 35 μL of Ar flushed buffer containing 0.3 M NaOAc (pH 5.6) and 10 mM ZnCl2 were added along with 0.5 U of Nuclease P1 (in 30 mM NaOAc pH 5.3, 5 mM ZnCl2 and 50 mM NaCl), 4 mU PDE II and 125 mU of DNase II and the mixture was further incubated at 37 °C for extra 21 h. A step-quenching with 1% formic acid solution (final pH~7) was followed, the digestion mixture was placed in a microspin filter (3 kDa) and the enzymes were filtered off by centrifugation at 14,000×g (4 °C) for 20 min. Subsequently, the filtrate was freeze-dried before HPLC analysis, clean-up and enrichment.
2.4. Measurement of modified nucleosides by LC-MS/MS
The samples were analyzed by an HPLC-UV system coupled with a sample collector, while the fractions containing the lesions were collected, freeze-dried, pooled, freeze-dried again, redissolved in Milli-Q water and subsequently injected to the LC-MS/MS system [4,5,14,[40], [41], [42]]. A triple-stage quadrupole mass spectrometer equipped with electrospray ionization (ESI) source in positive mode was employed for the detection and quantification of the lesions in the enzymatically digested DNA samples. The gradient elution program used for the chromatographic separation of the DNA lesions initiated with 99% of 2 mM ammonium formate (solvent A) and 1% acetonitrile (solvent B) (held for 1 min), increasing solvent B from 1% to 9.8% within 20 min and then immediately to 15% solvent B (held for 5 min), closing with initial conditions for 10 min re-equilibration. The flow rate remained constant at 0.2 mL/min, the injection volume was 30 μL and column temperature was set at 30 °C. Detection was performed in multiple reaction monitoring mode (MRM) using the two most intense and characteristic precursor/product ion transitions for each DNA lesion (Supplementary Fig. S2 and Table S1) [40,43].
2.5. Statistical analysis
The data were expressed as mean ± standard deviation (SD) of the nine samples measurements. The unpaired t-test was used for statistical analysis and a two-tailed p-value < 0.05, p-value < 0.01and p-value <0.001 were considered to indicate a statistical significant difference.
3. Results
3.1. Analysis of oxidative DNA damage in colon biopsies isolated from IBD patients
Genomic DNA was isolated from fresh tissue biopsies collected from inflamed and normal areas during routine colonoscopy and then hydrolyzed to single nucleosides by using a nuclease-containing enzymatic cocktail and analyzed by liquid chromatography with tandem mass spectrometry for the determination of the six modified purine nucleosides (see in accordance to a recently optimized protocol) [4,42]. The quantification of the lesions was executed in two separate steps, increasing the overall sensitivity of the method. Initially, the hydrolyzed DNA sample was analyzed by an HPLC-UV system coupled with a sample collector, where the quantification of the unmodified nucleosides took place, based on their absorbance at 260 nm. The collected samples, which contain the modified lesions, were concentrated and injected subsequently to LC-ESI-MS/MS to be analyzed independently [4,42].
In Table 1 the levels of the oxidatively-induced purine lesions in DNA isolated from nine patients affected by IBD are summarized, in both non-inflamed (NI) and inflamed (I) samples. As shown in Fig. 2a, the 5′S-cdG is the most predominant lesion with levels of 0.27-0.48/106 nucleobases (Nu) followed by 5′S-cdA with 0.16-0.31/106 Nu, 5′R-cdA with 0.06-0.15/106 Nu and 5′R-cdG with 0.04-0.08/106 Nu. In general, the I samples exhibited substantially increased levels of cPu lesions compared to NI samples (Supplementary Table S2 and Table S3). Moreover, significant higher levels of both 8-oxo-dA and 8-oxo-dG were also observed in I samples, as Fig. 2b depicts (p = 0.0012 and p = 0.0002, respectively, cf. Supplementary Table S3). Overall, NI and I samples accumulate approximately 5-6 times elevated levels of 8-oxo-dG (1.81-2.67/106 Nu) comparing with the corresponding 8-oxo-dA ones (0.27-0.60/106 Nu).
Table 1.
The levels (lesions/106 Nu) of 5′S-cdA, 5′R-cdA, 5′S-cdG, 5′R-cdG, 8-oxo-dA and 8-oxo-dG in genomic DNA samples isolated from non-inflamed (NI) and inflamed (I) tissues. The numbers represent the values of DNA lesions levels from the measurement of each sample.
| Sample | 5′S-cdA | 5′R-cdA | 5′S-cdG | 5′R-cdG | 8-oxo-dA | 8-oxo-dG |
|---|---|---|---|---|---|---|
| 1 NI | 0.16 | 0.06 | 0.32 | 0.06 | 0.32 | 1.89 |
| 1 I | 0.29 | 0.13 | 0.42 | 0.07 | 0.60 | 2.52 |
| 2 NI | 0.22 | 0.09 | 0.33 | 0.06 | 0.35 | 2.03 |
| 2 I | 0.31 | 0.14 | 0.48 | 0.08 | 0.55 | 2.43 |
| 3 NI | 0.22 | 0.07 | 0.32 | 0.04 | 0.27 | 2.11 |
| 3 I | 0.27 | 0.10 | 0.39 | 0.06 | 0.37 | 2.67 |
| 4 NI | 0.20 | 0.08 | 0.34 | 0.04 | 0.33 | 2.06 |
| 4 I | 0.25 | 0.10 | 0.41 | 0.08 | 0.48 | 2.63 |
| 5 NI | 0.22 | 0.09 | 0.30 | 0.04 | 0.28 | 1.81 |
| 5 I | 0.29 | 0.11 | 0.36 | 0.06 | 0.42 | 2.50 |
| 6 NI | 0.21 | 0.09 | 0.31 | 0.04 | 0.38 | 2.07 |
| 6 I | 0.27 | 0.15 | 0.39 | 0.07 | 0.53 | 2.67 |
| 7 NI | 0.25 | 0.11 | 0.32 | 0.05 | 0.41 | 1.93 |
| 7 I | 0.25 | 0.11 | 0.32 | 0.06 | 0.42 | 2.00 |
| 8 NI | 0.19 | 0.06 | 0.27 | 0.04 | 0.32 | 1.82 |
| 8 I | 0.26 | 0.08 | 0.32 | 0.05 | 0.42 | 2.39 |
| 9 NI | 0.20 | 0.07 | 0.28 | 0.04 | 0.30 | 1.90 |
| 9 I | 0.24 | 0.08 | 0.33 | 0.05 | 0.34 | 2.06 |
Fig. 2.
The levels of lesions/106Nu in genomic DNA samples measured by LC-MS/MS. (a) The levels of 5′S-cdA 5′R-cdA, 5′S-cdG and 5′R-cdG in NI (dark blue) and I (light blue) colon tissues samples from IBD patients; (b) The levels of 8-oxo-dA and 8-oxo-dG in NI and I samples from IBD patients; (c) Diastereomeric ratios (5′R/5′S) of cdA and cdG in NI and I from IBD patients; (d) The levels of cPu and 8-oxo-Pu, in NI and I samples from IBD patients; (e) The levels of 8-oxo-dA and 8-oxo-dG in adipose tissues samples from severely obese patients. The values represent the mean ± SD of n = 9 independent samples of each group,* denotes a statistically significant difference (p < 0.05) between NI and I groups, ** denotes a statistically significant difference (p < 0.01) between NI and I group sand *** denotes a statistically significant difference (p < 0.001) between NI and I groups. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In Fig. 2c, the ratios of 5′R/5′S for cdG and cdA, measured in NI and I colon biopsies are presented. In general, the ratios of I groups for both cdA and cdG were found higher compared with NI (0.32-0.46 and 0.12-0.19, respectively), while a statistically significant enhancement was revealed for cdA (p = 0.045) in I samples (Supplementary Table S4, Table S5 and Table S6). Total 8-oxo-Pu levels were found approximately 3.5-fold more elevated compared with total cPu in NI and I samples as depicted in Fig. 2d (Supplementary Table S7). Comparison between NI and I samples highlighted a significant increase for both cPu (p = 0.0003) and 8-oxo-Pu (p = 0.0002) in I samples (Supplementary Table S8).
3.2. Analysis of oxidative DNA damage in adipose tissue biopsies isolated from severely obese patients
The levels of oxidized bases were also determined, similarly to what executed with IBD specimens, in biopsies of visceral adipose tissue collected from nine severely obese patients during a sleeve gastrectomy intervention. A strong reduction of DNA base oxidation was observed in adipose tissue versus colon biopsies. In particular, in adipose tissue, the cPu levels were below the detection limits; the 8-oxo-dG levels show a high variability (0.49-1.57/106 Nu) and the 8-oxo-dG levels were extensively higher compared to 8-oxo-dA (from 27 to 150 times) (Table 2 and Fig. 2e). It was also worth mentioning that four out of nine samples the 8-oxo-dA levels were below the detection limits.
Table 2.
The levels (lesions/106 Nu) of 8-oxo-Pu in genomic DNA of adipose tissue samples. The numbers represent the values of DNA lesions levels from the measurement of each sample.
| Sample | 8-oxo-dA | 8-oxo-dG |
|---|---|---|
| 1 OB | 0.04 | 1.57 |
| 2 OB | – | 0.65 |
| 3 OB | 0.05 | 1.35 |
| 4 OB | – | 0.49 |
| 5 OB | 0.01 | 1.39 |
| 6 OB | 0.02 | 0.90 |
| 7 OB | – | 0.97 |
| 8 OB | 0.01 | 1.52 |
| 9 OB | 0.01 | 1.21 |
3.3. Gene expression analysis in colon and adipose tissue biopsies
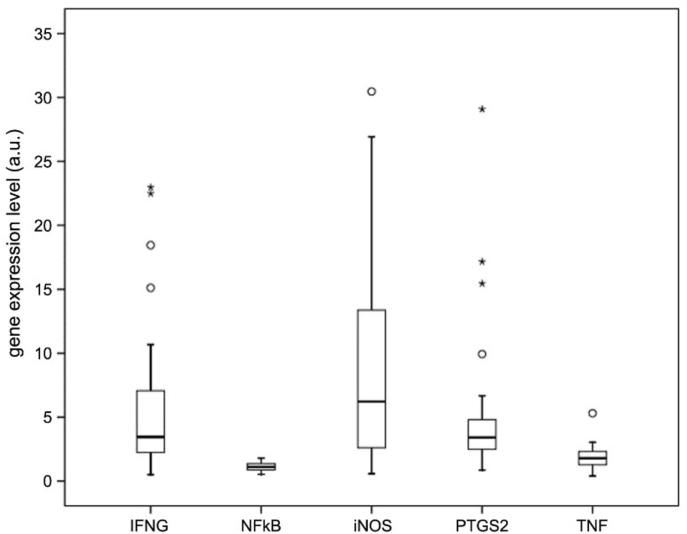
Gene expression levels of interferon-γ(IFNγ), nuclear factor-k (NF-kB), inducible nitric oxide synthase (iNOS), prostaglandin-endoperoxide synthase 2 (PTGS2) and tumor necrosis factor (TNF) have been analyzed by quantitative PCR to monitor the inflammation status. A higher gene expression level of INFγ, PTGS2 and iNOS has been observed in the macroscopically inflamed colon biopsies versus the non-inflamed counterpart testifying a clear discrimination between inflamed and normal tissue at the molecular level too (Fig. 3) [36].
Fig. 3.
Analysis of the mRNA expression levels of inflammation-related genes in inflamed colon biopsies isolated from IBD patients normalized versus non-inflamed mucosa.
Several studies have demonstrated that high levels of expression of iNOS are correlated with increased levels of intracellular RNS and ROS, resulting in DNA damage [30,31,44]. Since it is well documented that one of the main pathways to generating HO• radicals can be from homolysis of protonated peroxynitrite [1,2,45], the analysis of iNOS expression, together with the measurement of 8-oxodG and cPu, represents the determining parameter for measuring the tissue inflammation state. Therefore, the iNOS gene expression level has also been analyzed in adipose tissue biopsies isolated from severely obese patients (7 patients) who have undergone bariatric surgery (sleeve gastrectomy). These results were compared with what observed in adipose tissue biopsies isolated from normal weight subjects, i.e. patients (5 patients) who, preventively, undergo cholecystectomies for gallstones without ongoing inflammation process. In the majority of samples, the levels of iNOS were not measurable (below the detection limits) in both obese and normal biopsies. These data are in agreement with what was previously observed in human subcutaneous adipose tissue [46] and with what is reported in the Protein Atlas online immunohistochemistry re-source (http://www.proteinatlas.org).
4. Discussion
The majority of DNA lesions induced by oxidative stress is recognized by DNA glycosylases, which catalyze the first step of the base excision repair (BER), an error-free stepwise repair mechanism that leads to a complete sequence restoration. The cPu, oxidative DNA lesions specifically generated by HO• radicals, are bulky adducts formed by abstraction of the H5′ atom of the 2-deoxyribose moiety, followed by intramolecular cyclization and subsequent oxidation of the resulting N7-radical [4,41]. The peculiar structural changes caused by cPu, i.e. the displacement of the purine base, the unusual sugar pucker and the deformation of the sugar-phosphate backbone, prevent the recognition by DNA glycosylases. Therefore, the cPu can be recognized and repaired by nucleotide excision repair (NER) and no other repair pathways can process these adducts. Unrepaired cPu lead to deleterious biological consequences since these lesions are able to block both DNA and RNA polymerases and can be bypassed by error-prone Y family DNA polymerases, then causing cell death and mutagenesis [16]. In details, in vitro steady-state kinetic assays, carried out in human cells, showed that both 5′S-cdA and 5′S-cdG can be bypassed by a concerted action of pol η/ι and pol ζ, which are able to execute the insertion and extension step, respectively [9]. The replicative bypass of 5′S-cdG was mutagenic, at a frequency of 50%, leading to both G to T and G to A mutations. Conversely, the replicative bypass across 5′S-cdA is less mutagenic leading to approximately 10% of A to T transvertions.
Recently, translesion activity similarly to what observed for the Y family DNA polymerases was shown for DNA polymerase β by Liu and coworkers [47]. In this study was found that Pol β can efficiently bypass a 5′R-cdA incorporating dT opposite the lesion and still to perform strand displacement synthesis, but can also misincorporate dC and both purine nucleosides (dG and dA). Moreover, in case of 5′S-cdA, pol β inserts dT opposite the lesion, but fails to continue the synthesis resulting in accumulation of single-strand DNA breaks. Thus, pol β bypass of an R-cdA can lead to nucleotide misincorporation causing mutations, whereas its bypass of an S-cdA can cause the accumulation of DNA strand break intermediates that in turn results in recombination and genome instability [47].
Based on these lines of evidence we wondered whether these lesions could be accumulated in vivo, at target tissue level, in human specimens derived from patients affected by diseases characterized by chronic inflammation, such as IBD. The results presented in this study about the abundance of 5′S on 5′R, both for cdA than cdG, showed a significant accumulation of cPu in inflamed IBD-colon biopsies versus the normal counterpart. Additionally, the 5′S diastereomers of cdA and cdG were found at higher levels than the 5′R counterparts, supporting the more efficient repair of the 5′R diastereomers of the 5′,8-cyclopurine reported in human cell extracts [8]. Previous studies carried out in calf-thymus DNA [48] by Fenton-type reagents in vitro showed that cPu are induced at much lower levels than 8-oxo-dG, and R diastereomers of the cPu lesions are produced more efficiently than the S counterparts. The relatively small differences in the levels of cPu vs. 8-oxo-Pu, and of the two diastereomers in tissue DNA (compared to results obtained from in vitro Fenton reaction) suggest that cPu lesions are poorly repaired compared to 8-oxo-Pu. The abundance of cPu lesions in our study suggests their potential role in the known CRC susceptibility of IBD patients. This hypothesis is also supported by the recent whole-exome sequencing analyses of IBD-associated colon cancers (IBD-CRC) and sporadic-CRC (S-CRC) [49]. Although no sharp differences have been found in the mutational landscape observed in IBD-CRC versus S-CRC, it is interesting to note a predominance of G to A transitions and an excess of A to C transversions. The majority (48%) of G to A transitions were located at CpG sites and these are mutations commonly found in other cancers and derived from the spontaneous deamination of methylated cytosine. For the remaining (14%) G to T randomly located it should be interesting to remind that this kind of mutations is correlated to the expression of iNOS, which is known to be highly expressed in IBD tissues [50,51]. In addition, G to A mutations can also rise by translesion synthesis across 5′S-cdG [9].
Similarly, to what previously found in esophageal adenocarcinomas, another tumor characterized by an environment of inflammation, the majority of AT>CG transversions are located in a context of AAG trinucleotides. These transversions can also derive from the misincorporation of the 8-oxo-dG triphosphate, as oxidatively damaged DNA precursor [52]. Of note, no significant contribution of GC>TA transversions, the canonical signature of oxidative DNA damage, has been observed in the IBD-CRC either in nuclear or mitochondrial genome [49,53], although a clear increase of 8-oxo-dG has been observed. An increased DNA repair capacity of oxidative DNA damage could explain this apparent discrepancy. Actually, it has been recently reported that OGG1 and MTH1, the 8-oxoguanine-DNA glycosylase and the nucleotide pool-sanitizing enzyme, respectively, were upregulated in IBD, through an inflammatory condition that progresses to CRC, suggesting that an adaptive response to oxidative stress can concur to the pathogenesis of IBD-CRC [54].
The cPu are expected to persist in the mitochondrial genome since the well-known lack of NER in these organelles. Interestingly, in a recent study, higher levels of circulating mitochondrial DNA have been found in both IBD-patients and mouse colitis induced by dextran sulfate-sodium compared to the controls, suggesting that mtDNA fragments, released during the active phase, might be a key pro-inflammatory factor in IBD [55]. In addition, the analysis of the mitochondrial DNA mutations in biopsies from IBD patients with and without cancer, as well as, at different stages of dysplastic progression, showed a dual pattern: an accumulation of mitochondrial mutations, from control to pre-neoplastic dysplasia and a negative selection in cancer [53]. All these data suggest that the accumulation of cPu in inflamed colon biopsies might be a relevant predictive marker for IBD-CRC susceptibility.
Conversely, in adipose tissue biopsies a decrease of oxidative DNA damage, testified by a strong reduction of 8-oxo-dA (Fig. 2e and Table 2) and the lack of cPu, together with the absence of iNOS expression has been detected.
In literature the results of investigations of DNA damage in obesity patients are conflicting [56], and the data provide only limited evidence for an association between overweight/obesity and genomic instability. The current discrepancy in the studies concerning the DNA damage and DNA repair capacity in obesity might be due to potential confounding factors such as: uncontrolled intake of vitamins and minerals and consumption of various nutrition as well as the different methodologies applied to quantify DNA lesions and assess DNA integrity. The dietary heterogeneity is thought to be, at least partially, responsible for the poor reproducibility among the distinct human studies. Consistently, the standard diets and the controlled housing conditions, applied in the animal studies, lead to reproducible results that show a link between obesity and genomic instability [[57], [58], [59]].
These studies were mainly conducted with C57BL/6j mice and these animals develop symptoms which are characteristic for obese humans namely hyperinsulinemia, hyperglycemia, and hypertension [60]. The bariatric patients enrolled in this study, although affected by severe obesity (average BMI = 43.2 ± 9.9), do not have any obesity-associated comorbidity and show a low grade of inflammation (our unpublished data) consistently with the reduced level of oxidative DNA lesions.
The methodologies used for the quantification of the oxidative DNA lesions, may be another important factor that might explain the conflicting results found in literature. It should be noticed that data obtained in different laboratories gave rise to different results, mainly due to deviations among experimental conditions used in the various laboratories or inconsistent quantification of lesions because of oxidation artefacts [61,62]. Also 8-oxo-dG measurements in humans do not allow to draw firm conclusions (positive, as well as negative correlations were detected); notably, the study with the highest quality score found even a significant inverse association [63].
In this study, we followed a rigorous protocol, developed in our laboratory, to analyse and quantify, simultaneously, the occurrence of the four cPu and the two 8-oxo-Pu lesions [14,40,41,43]. The lesions were analyzed by LC-MS/MS, the use isotopically labelled internal standards spiked in the DNA samples prior to the treatment with enzymes, provide robust information regarding the analyte stability and the overall method accuracy, which is critical for quantification.
5. Conclusions
In this study we tested the role of oxidative stress in two different pathologies characterized by a chronic inflammation process, such as inflammatory bowel disorders (IBD) and severe obesity. We show that the inflamed colon biopsies, collected from IBD patients, are characterized by a significant increase of cPu in parallel to the overexpression of iNOS, whose activity leads to the formation of HO• which is known to be the radical species of election for the cPu formation. These findings suggest that the accumulation of these mutagenic adducts might be involved in the CRC susceptibility observed in people affected by these chronic diseases. In contrast, in adipose tissue biopsies, collected from severely obese patients, the undetectable levels of cPu and the lack of iNOS induction, lead to exclude the potential involvement of these adducts in the higher obesity-related cancer susceptibility. The potential causal role of the cPu in IBD-related tumorigenesis suggests their usefulness as early predictive biomarkers for IBD-associated colon cancer while ongoing experiments with biopsies of different stage of the disorder received from the inflamed areas and compared with healthy counterpart will elucidate lesions accumulation and their direct correlation to HO•-induced DNA damage.
Funding
This work was partially supported by RF2013-02357791 of Italian Ministry of Public Health. This work was also supported by a grant from the Ministero dell'Istruzione, dell'Università e della Ricerca (prot. 2017L8Z2EM).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101562.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Winterbourn C.C. Biological chemistry of reactive oxygen species. In: Chatgilialoglu C., Studer A., editors. Encyclopedia of Radicals in Chemistry, Biology and Materials. Wiley; Chichester: 2012. p. 1259. [Google Scholar]
- 2.Geacintov N.E., Shafirovich V. Reactions of small reactive species with DNA. In: Chatgilialoglu C., Studer A., editors. Encyclopedia of Radicals in Chemistry, Biology and Materials. Wiley; Chichester: 2012. p. 1284. [Google Scholar]
- 3.Chatgilialoglu C., Ferreri C., Terzidis M.A. Purine 5’,8-cyclonucleoside lesions: chemistry and biology. Chem. Soc. Rev. 2011;40:1368–1382. doi: 10.1039/c0cs00061b. [DOI] [PubMed] [Google Scholar]
- 4.Chatgilialoglu C., Ferreri C., Geacintov N.E., Krokidis M.G., Liu Y., Masi A., Shafirovich V., Terzidis M.A., Tsegay P.S. 5’,8-Cyclopurine lesions in DNA damage: chemical, analytical, biological and diagnostic significance. Cells. 2019;8:513. doi: 10.3390/cells8060513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chatgilialoglu C., Krokidis M.G., Masi A., Barata-Vallejo S., Ferreri C., Terzidis M.A., Szreder T., Bobrowski K. New insights into the reaction paths of hydroxyl radicals with purine moieties in DNA and double-stranded oligonucleotides. Molecules. 2019;24:3860. doi: 10.3390/molecules24213860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaruga P., Dizdaroglu M. 8,5'-Cyclopurine-2'-deoxynucleosides in DNA: mechanisms of formation, measurement, repair and biological effects. DNA Repair. 2008;7:1413–1425. doi: 10.1016/j.dnarep.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Kuraoka I., Bender C., Romieu A., Cadet J., Wood R.D., Lindahl T. Removal of oxygen free-radical-induced 5',8-purine cyclodeoxynucleosides from DNA by the nucleotide excision-repair pathway in human cells. Proc. Natl. Acad. Sci. U.S.A. 2000;97:3832–3837. doi: 10.1073/pnas.070471597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kropachev K., Ding S., Terzidis M.A., Masi A., Liu Z., Cai Y., Kolbanovskiy M., Chatgilialoglu C., Broyde S., Geacintov N.E., Shafirovich V. Structural basis for the recognition of diastereomeric 5',8-cyclo-2'-deoxypurine lesions by the human nucleotide excision repair system. Nucleic Acids Res. 2014;42:5020–5032. doi: 10.1093/nar/gku162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.You C., Swanson A.L., Dai X., Yuan B., Wang J., Wang Y. Translesion synthesis of 8,5'-cyclopurine-2'-deoxynucleosides by DNA polymerases eta, iota, and zeta. J. Biol. Chem. 2013;288:28548–28556. doi: 10.1074/jbc.M113.480459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J., Clauson C.L., Robbins P.D., Niedernhofer L.J., Wang Y. The oxidative DNA lesions 8,5’-cyclopurines accumulate with aging in a tissue-specific manner. Aging Cell. 2012;11:714–716. doi: 10.1111/j.1474-9726.2012.00828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson A.R., Yousefzadeh M.J., Rozgaja T.A., Wang J., Li X., Tilstra J.S., Feldman C.H., Gregg S.Q., Johnson C.H., Skoda E.M., Frantz M.-C., Bell-Temin H., Pope-Varsalona H., Gurkar A.U., Nasto L.A., Robinson R.A.S., Fuhrmann-Stroissnigg H., Czerwinska J., McGowan S.J., Cantu-Medellin N., Harris J.B., Maniar S., Ross M.A., Trussoni C.E., LaRusso F.N., Cifuentes-Pagano E., Pagano P.J., Tudek B., Vo N.V., Rigatti L.H., Opresko P.L., Stolz D.B., Watkins S.C., Burd C.E., Croix C.M.St, Siuzdak G., Yates N.A., Robbins P.D., Wang Y., Wipf P., Kelley E.E., Niedernhofer L.J. Spontaneous DNA damage to the nuclear genome promotes senescence, redox imbalance and aging. Redox Biol. 2018;17:259–273. doi: 10.1016/j.redox.2018.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan B., Wang J., Cao H., Sun R., Wang Y. High throughput analysis of the mutagenic and cytotoxic properties of DNA lesions by next-generation sequencing. Nucleic Acids Res. 2011;39:5945–5954. doi: 10.1093/nar/gkr159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.You C., Dai X., Yuan B., Wang J., Wang J., Brooks P.J., Niedernhofer L.J., Wang Y. A quantitative assay for assessing the effects of DNA lesions on transcription. Nat. Chem. Biol. 2012;8:817–822. doi: 10.1038/nchembio.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krokidis M.G., Louka M., Efthimiadou E., Zervou S.-K., Papadopoulos K., Hiskia A., Ferreri C., Chatgilialoglu C. Membrane lipidome reorganization and accumulation of tissue DNA lesions in tumor-bearing mice: an exploratory study. Cancers. 2019;11:480. doi: 10.3390/cancers11040480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nyaga S.G., Jaruga P., Lohani A., Dizdaroglu M., Evans M.K. Accumulation of oxidatively induced DNA damage in human breast cancer cell lines following treatment with hydrogen peroxide. Cell Cycle. 2007;6:1472–1478. [PubMed] [Google Scholar]
- 16.Brooks P.J. The cyclopurinedeoxynucleosides: DNA repair, biological effects, mechanistic insights, and unanswered questions. Free Radic. Biol. Med. 2017;107:90–100. doi: 10.1016/j.freeradbiomed.2016.12.028. [DOI] [PubMed] [Google Scholar]
- 17.Kirkali G., de Souza-Pinto N.C., Jaruga P., Bohr V.A., Dizdaroglu M. Accumulation of (5’S)-8,5’-cyclo-2’-deoxyadenosine in organs of Cockayne syndrome complementation group B gene knockout mice. DNA Repair. 2009;8:274–278. doi: 10.1016/j.dnarep.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mori T., Nakane H., Iwamoto T., Krokidis M.G., Chatgilialoglu C., Tanaka K., Kaidoh T., Hasegawa M., Sugiura S. High levels of oxidatively generated DNA damage 8,5’-cyclo-2’-deoxyadenosine accumulate in the brain tissues of xerodermapigmentosum group A gene-knockout mice. DNA Repair. 2019;80:52–58. doi: 10.1016/j.dnarep.2019.04.004. [DOI] [PubMed] [Google Scholar]
- 19.Krokidis M.G., Parlanti E., D'Errico M.R., Pascucci B., Pino A., Alimonti A., Pietraforte D., Masi A., Ferreri C., Chatgilialoglu C. Purine DNA lesions varying oxygen concentration in DNA repair-impaired human embryonic epithelial cells (EUE-siXPA) Cells. 2019;8:1377. doi: 10.3390/cells8111377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu Y., Wang P., Cui Y., Wang Y. Chemical analysis of DNA damage. Anal. Chem. 2018;90:556–576. doi: 10.1021/acs.analchem.7b04247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cui L., Ye W., Prestwich E.G., Wishnok J.S., Taghizadeh K., Dedon P.C., Tannenbaum S.R. Comparative analysis of four oxidized guanine lesions from reactions of DNA with peroxynitrite, single oxygen, and γ-radiation. Chem. Res. Toxicol. 2013;26:195–202. doi: 10.1021/tx300294d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kruidenier L., Verspaget H.W. Oxidative stress as a pathogenic factor in inflammatory bowel disease—radicals or ridiculous? Aliment. Pharmacol. Ther. 2002;16:1997–2015. doi: 10.1046/j.1365-2036.2002.01378.x. [DOI] [PubMed] [Google Scholar]
- 23.Pravda J. Radical induction theory of ulcerative colitis. World J. Gastroenterol. 2005;11:2371–2384. doi: 10.3748/wjg.v11.i16.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graham D.B., Xavier R.J. Pathway paradigms revealed from the genetics of inflammatory bowel disease. Nature. 2020;578:527–539. doi: 10.1038/s41586-020-2025-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Mello A.H., Costa A.B., Della Giustina Engel J., Rezin G.T. Mitochondrial dysfunction in obesity. Life Sci. 2018;192:26–32. doi: 10.1016/j.lfs.2017.11.019. [DOI] [PubMed] [Google Scholar]
- 26.Arnold M., Sierra M.S., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683–691. doi: 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- 27.Terzić J., Grivennikov S., Karin E., Karin M. Inflammation and colon cancer. Gastroenterology. 2010;138:2101–2114.e5. doi: 10.1053/j.gastro.2010.01.058. [DOI] [PubMed] [Google Scholar]
- 28.Donohoe C.L., Lysaght J., O'Sullivan J., Reynolds J.V. Emerging concepts linking obesity with the hallmarks of cancer trends in endocrinology & metabolism. Cell. 2017;28:46–62. doi: 10.1016/j.tem.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 29.Seril D.N., Liao J., Yang G.-Y., Yang C.S. Oxidative stress and ulcerative colitis-associated carcinogenesis: studies in humans and animal models. Carcinogenesis. 2003;24:353–362. doi: 10.1093/carcin/24.3.353. [DOI] [PubMed] [Google Scholar]
- 30.Kolios G., Valatas V., Ward S.G. Nitric oxide in inflammatory bowel disease: a universal messenger in an unsolved puzzle. Immunology. 2004;113:427–437. doi: 10.1111/j.1365-2567.2004.01984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goodman J.E., Hofseth L.J., Hussain S.P., Harris C.C. Nitric oxide and p53 in cancer-prone chronic inflammation and oxyradical over load disease. Environ. Mol. Mutagen. 2004;44:3–9. doi: 10.1002/em.20024. [DOI] [PubMed] [Google Scholar]
- 32.Grisham M.B., Gaginella T.S., von Ritter C., Tamai H., Be R.M., Granger D.N. Effects of neutrophil-derived oxidants on intestinal permeability, electrolyte transport, and epithelial cell viability. Inflammation. 1990;14:531–542. doi: 10.1007/BF00914274. [DOI] [PubMed] [Google Scholar]
- 33.Pereira C., Grácio D., Teixeira J.P., Magro F. Oxidative stress and DNA damage: implications in inflammatory bowel disease. Inflamm. Bowel Dis. 2015;21:2403–2417. doi: 10.1097/MIB.0000000000000506. [DOI] [PubMed] [Google Scholar]
- 34.Nair J., Gansauge F., Beger H., Dolara P., Winde G., Bartsch H. Increased etheno-DNA adducts in affected tissues of patients suffering from Crohn's disease, ulcerative colitis, and chronic pancreatitis. Antioxidants Redox Signal. 2006;8:1003–1010. doi: 10.1089/ars.2006.8.1003. [DOI] [PubMed] [Google Scholar]
- 35.Bartsch H., Nair J. Chronic inflammation and oxidative stress in the genesis and perpetuation of cancer: role of lipid peroxidation, DNA damage, and repair. Langenbeck's Arch. Surg. 2006;391:499–510. doi: 10.1007/s00423-006-0073-1. [DOI] [PubMed] [Google Scholar]
- 36.Grisham M.B., MacDermott R.P., Deitch E.A. Oxidant defense mechanisms in the human colon. Inflammation. 1990;14:669–680. doi: 10.1007/BF00916370. [DOI] [PubMed] [Google Scholar]
- 37.Cinti S. The adipose organ at a glance. Dis. Model Mech. 2012;5:588–594. doi: 10.1242/dmm.009662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Setayesh T., Nersesyan A., Misik M., Ferk F., Langie S., Andrade V.M., Haslberger A., Knasmuller S. Impact of obesity and overweight on DNA stability: few facts and many hypotheses. Mutat. Res. 2018;777:64–91. doi: 10.1016/j.mrrev.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 39.Wlodarczyk M., Nowicha G. Obesity, DNA damage, and development of obesity-related diseases. Int. J. Mol. Sci. 2019;20:1146. doi: 10.3390/ijms20051146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terzidis M.A., Chatgilialoglu C. An ameliorative protocol for the quantification of purine 5′,8-cyclo-2′-deoxynucleosides in oxidized DNA. Front. Chem. 2015;3:47. doi: 10.3389/fchem.2015.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chatgilialoglu C., Eriksson L.A., Krokidis M.G., Masi A., Wang S., Zhang R. Oxygen Dependent Purine lesions in double-stranded oligodeoxynucleotides: kinetic and computational studies highlight the mechanism for 5',8-cyclopurine formation. J. Am. Chem. Soc. 2020;142:5825–5833. doi: 10.1021/jacs.0c00945. [DOI] [PubMed] [Google Scholar]
- 42.Chatgilialoglu C. Cyclopurine (cPu) lesions: what, how and why? Free Radic. Res. 2019;53:941–943. doi: 10.1080/10715762.2019.1643017. [DOI] [PubMed] [Google Scholar]
- 43.Krokidis M.G., Terzidis M.A., Efthimiadou E., Zervou S.-K., Kordas G., Papadopoulos K., Hiskia A., Kletsas D., Chatgilialoglu C. Purine 5',8-cyclo-2'-deoxynucleoside lesions: formation by radicalstress and repair in human breast epithelial cancer cells. Free Radic. Res. 2017;51:470–482. doi: 10.1080/10715762.2017.1325485. [DOI] [PubMed] [Google Scholar]
- 44.Tamir S., Tannenbaum S.R. The role of nitric oxide (NO) in the carcinogenic process. Biochim. Biophys. Acta. 1996;1288:F31–F36. doi: 10.1016/0304-419x(96)00021-2. [DOI] [PubMed] [Google Scholar]
- 45.Valko M., Leibfritz D., Moncol J., Cronin M.T., Mazur M., Telser J. Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol. 2007;39:44–84. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 46.Elizalde M., Rydén M., van Harmelen V., Eneroth P., Gyllenhammar H., Holm C., Ramel S., Olund A., Arner P., Andersson K. Expression of nitric oxide synthases in subcutaneous adipose tissue of nonobese and obese humans. J. Lipid Res. 2000;41:1244–1251. [PubMed] [Google Scholar]
- 47.Jiang Z., Xu M., Lai Y., Laverde E., Terzidis M.A., Masi A., Chatgilialoglu C., Liu Y. Bypass of a 5',8-cyclopurine-2'-deoxynucleoside by DNA polymerase β during DNA replication and base excision repair leads to nucleotide misinsertions and DNA strand breaks. DNA Repair. 2015;33:24–34. doi: 10.1016/j.dnarep.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guerrero C.R., Wang J., Wang Y. Induction of 8,5'-cyclo-2'-deoxyadenosine and 8,5'-cyclo-2'-deoxyguanosine in isolated DNA by Fenton-type reagents. Chem. Res. Toxicol. 2013;26:1361–1366. doi: 10.1021/tx400221w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robles A.I., Traverso G., Zhang M., Roberts N.J., Khan M.A., Joseph C., Lauwers G.Y., Selaru F.M., Popoli M., Pittman M.E., Ke X., Hruban R.H., Meltzer S.J., Kinzler K.W., Vogelstein B., Harris C.C., Papadopoulos N. Whole-exome sequencing analyses of inflammatory bowel disease-associated colorectal cancers. Gastroenterology. 2016;150:931–943. doi: 10.1053/j.gastro.2015.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ambs S., Bennett W.P., Merriam W.G., Ogunfusika M.O., Oser S.M., Harrington A.M., Shields P.G., Felley-Bosco E., Hussain S.P., Harris C.C. Relationship between p53 mutations and inducible nitric oxide synthase expression in human colorectal cancer. J. Natl. Cancer Inst. 1999;91:86–88. doi: 10.1093/jnci/91.1.86. [DOI] [PubMed] [Google Scholar]
- 51.Hussain S.P., Amstad P., Raja K., Ambs S., Nagashima M., Bennett W.P., Shields P.G., Ham A.J., Swenberg J.A., Marrogi A.J., Harris C.C. Increased p53 mutation load in noncancerous colon tissue from ulcerative colitis: a cancer-prone chronic inflammatory disease. Canc. Res. 2000;60:3333–3337. [PubMed] [Google Scholar]
- 52.Rudd S.G., Valerie N.C.K., Helleday T. Pathways controlling dNTP pools to maintain genome stability. DNA Repair. 2016;44:193–204. doi: 10.1016/j.dnarep.2016.05.032. [DOI] [PubMed] [Google Scholar]
- 53.Baker K.T., Nachmanson D., Kumar S., Emond M.J., Ussakli C., Brentnall T.A., Kennedy S.R., Risques R.A. Mitochondrial DNA mutations are associated with ulcerative colitis preneoplasia but tend to be negatively selected in cancer. Mol. Canc. Res. 2019;17:488–498. doi: 10.1158/1541-7786.MCR-18-0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kumagae Y., Hirahashi M., Takizawa K., Yamamoto H., Gushima M., Esaki M., Matsumoto T., Nakamura M., Kitazono T., Oda Y. Overexpression of MTH1 and OGG1 proteins in ulcerative colitis-associated carcinogenesis. Oncol. Lett. 2018;16:1765–1776. doi: 10.3892/ol.2018.8812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boyapati R.K., Dorward D.A., Tamborska A., Kalla R., Ventham N.T., Doherty M.K., Whitfield P.D., Gray M., Loane J., Rossi A.G., Satsangi J., Ho G.T. Mitochondrial DNA is a pro-inflammatory damage-associated molecular pattern released during active IBD. Inflamm. Bowel Dis. 2018;24:2113–2122. doi: 10.1093/ibd/izy095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Setayesha T., Nersesyana A., Mišíka M., Ferka F., Langieb S., Andradec V.M., Haslbergerd A., Knasmüllera S. Impact of obesity and overweight on DNA stability: few facts and many hypotheses. Mutat. Res. 2018;777:64–91. doi: 10.1016/j.mrrev.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 57.Sinha J.K., Ghosh S., Swain U., Giridharan N.V., Raghunath M. Increased macromolecular damage due to oxidative stress in the neocortex and hippocampus of WNIN/Ob, a novel rat model of premature aging. Neuroscience. 2014;269:256–264. doi: 10.1016/j.neuroscience.2014.03.040. [DOI] [PubMed] [Google Scholar]
- 58.Remely M., Ferk F., Sterneder S., Setayesh T., Kepcija T., Roth S., Noorizadeh R., Greunz M., Rebhan I., Wagner K.H., Knasmüller S., Haslberger A. Vitamin E modifies high-fat diet-induced increase of DNA strand breaks, and changes in expression and DNA methylation of Dnmt1 and MLH1 inC57BL/6J male mice. Nutrients. 2017;9:607. doi: 10.3390/nu9060607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leffa D.D., da Silva J., Daumann F., Dajori A.L., Longaretti L.M., Damiani A.P., de Lira F., Campos F., Ferraz Ade B., Côrrea D.S., de Andrade V.M. Corrective effects of acerola (Malpighia emarginata DC.) juice intake on biochemical and genotoxical parameters in mice fed on a high-fat diet. Mutat. Res. 2014;770:144–152. doi: 10.1016/j.mrfmmm.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 60.Collins S., Martin T.L., Surwit R.S., Robidoux J. Genetic vulnerability to diet-induced obesity in the C57BL/6J mouse: physiological and molecular characteristics. Physiol. Behav. 2004;81(2):243–248. doi: 10.1016/j.physbeh.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 61.Yu Y., Cui Y., Niedernhofer L.J., Wang Y. Occurrence, biological consequences, and human health relevance of oxidative stress-induced DNA damage. Chem. Res. Toxicol. 2016;29:2008–2039. doi: 10.1021/acs.chemrestox.6b00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gedik C.M., Collins A. Establishing the background level of base oxidation in human lymphocyte DNA: results of an interlaboratory validation study. Faseb. J. 2005;19:82–84. doi: 10.1096/fj.04-1767fje. [DOI] [PubMed] [Google Scholar]
- 63.Loft S., Vistisen K., Ewertz M., Tjønneland A., Overvad K., Poulsen H.E. Oxidative DNA damage estimated by 8-hydroxydeoxyguanosine excretion in humans: influence of smoking, gender and body mass index. Carcinogenesis. 1992;13:2241–2247. doi: 10.1093/carcin/13.12.2241. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.