Abstract
Objectives
Glucose-stimulated insulin secretion is a critical function in the regulation of glucose homeostasis, and its deregulation is associated with the development of type 2 diabetes. Here, we performed a genetic screen using islets isolated from the BXD panel of advanced recombinant inbred (RI) lines of mice to search for novel regulators of insulin production and secretion.
Methods
Pancreatic islets were isolated from 36 RI BXD lines and insulin secretion was measured following exposure to 2.8 or 16.7 mM glucose with or without exendin-4. Islets from the same RI lines were used for RNA extraction and transcript profiling. Quantitative trait loci (QTL) mapping was performed for each secretion condition and combined with transcriptome data to prioritize candidate regulatory genes within the identified QTL regions. Functional studies were performed by mRNA silencing or overexpression in MIN6B1 cells and by studying mice and islets with beta-cell-specific gene inactivation.
Results
Insulin secretion under the 16.7 mM glucose plus exendin-4 condition was mapped significantly to a chromosome 2 QTL. Within this QTL, RNA-Seq data prioritized Crat (carnitine O-acetyl transferase) as a strong candidate regulator of the insulin secretion trait. Silencing Crat expression in MIN6B1 cells reduced insulin content and insulin secretion by ∼30%. Conversely, Crat overexpression enhanced insulin content and secretion by ∼30%. When islets from mice with beta-cell-specific Crat inactivation were exposed to high glucose, they displayed a 30% reduction of insulin content as compared to control islets. We further showed that decreased Crat expression in both MIN6B1 cells and pancreatic islets reduced the oxygen consumption rate in a glucose concentration-dependent manner.
Conclusions
We identified Crat as a regulator of insulin secretion whose action is mediated by an effect on total cellular insulin content; this effect also depends on the genetic background of the RI mouse lines. These data also show that in the presence of the stimulatory conditions used the insulin secretion rate is directly related to the insulin content.
Keywords: Insulin secretion, Recombinant inbred mice, Carnitine acetyltransferase, Genetics, Diabetes, Beta-cells
Abbreviations: RI, recombinant inbred; QTL, quantitative trait loci; Crat, carnitine O-acetyl transferase; GRP, genetic reference population; LRS, likelihood ratio statistics; GSIS, glucose-stimulated insulin secretion; R, Pearson's correlation coefficient; eQTL, expression quantitative trait loci; RC, regular chow; HFHS, high fat high sucrose; OCR, oxygen consumption rate.
HIGHLIGHTS
-
•
A QTL analysis in BXD mice identifies Crat as a regulator of insulin secretion.
-
•
Crat regulates insulin content in MIN6B1 cells and pancreatic islets.
-
•
Crat regulates glucose oxidation in MIN6B1 cells and pancreatic islets.
-
•
Crat links glucose metabolism to the control of beta-cell insulin content.
-
•
Insulin content limits insulin secretion in response to high glucose and exendin-4 level.
1. Introduction
Glucose-stimulated insulin secretion by pancreatic beta-cells is a finely tuned process that allows for the timely delivery of the exact quantity of insulin needed to maintain normoglycemia. Insulin achieves this by stimulating glucose uptake and utilization by muscle, fat, and liver and by suppressing hepatic glucose production. In the pathogenesis of type 2 diabetes, progressive reduction in the sensitivity to insulin of target tissues combined with reduced glucose-stimulated insulin secretion leads to impaired glycemic control. The failure of beta-cells to secrete enough insulin to compensate for the prevailing insulin resistance is a critical event in triggering diabetic hyperglycemia [1,2]. Therefore, identifying the molecular pathways and genes that control beta-cell insulin secretion capacity is of high current interest as insight in these mechanisms could yield new therapeutic targets for the treatment of diabetes.
To identify novel regulators of beta-cell function in an unbiased way, genetic analysis of genetic reference populations (GRPs) of recombinant inbred (RI) mice is a powerful method. The most used and best-characterized mouse GRP is the BXD panel of advanced RI mice derived from crosses of C57Bl/6 and DBA2 fully sequenced mice [3]. The BXD panel can be used to map quantitative trait loci (QTL) controlling almost any heritable physiological function that can be measured in mice [[4], [5], [6], [7], [8]]. Insulin secretion is a complex trait. In humans, genome-wide association studies have now identified over 250 type 2 diabetes susceptibility loci [9,10]. Many of the associated genes are expressed in pancreatic beta-cells suggesting their potential role in controlling beta-cell function in diabetes. However, many of these genes are also expressed in other tissues involved in the control of glucose homeostasis and whether their primary implication in the pathogenesis of diabetes is linked to their role in beta-cells or in these other tissues is not known and difficult to assess in humans.
Here, we used islets isolated from a panel of 36 BXD mouse lines to identify new candidate regulators of insulin secretion. This genetic study combining QTL mapping for insulin secretion with islet gene expression analysis ensures that the identified candidate genes control beta-cell function; furthermore, the role of these genes can be tested in insulin cell lines and primary islets. Our study identified Crat as a new regulator of glucose-stimulated insulin secretion through its capacity to control insulin content. This effect depends on the capacity of Crat to regulate glucose oxidation rates and on the individual RI mouse genetic architecture. Our data also indicate that insulin content is a limiting factor in 16.7 mM glucose plus exendin-4 stimulated insulin secretion.
2. Material and methods
2.1. Animals
BXD mice were purchased from the Jackson Laboratory (Bar Harbor, ME, USA). Animals were housed 3–5 per cage at a constant temperature of 23 °C and under 12 h light/dark cycle, and they had ad libitum access to regular chow (RC) diet (# 3436, Provimi Kliba, Kaiseraugst, Switzerland) and water. Experiments were performed with 13-week-old male mice. All animal experiments were approved by the Veterinary Office of Canton de Vaud.
2.2. Islet isolation and insulin secretion
Pancreatic islets were isolated by handpicking following the Liberase (Roche) digestion of the pancreas, as described by Basco et al. [11]. They were cultured overnight in RPMI containing 11 mM glucose and supplemented with 10% fetal bovine serum (FBS), 1 mM l-glutamine, and 1% penicillin and streptomycin. The following day, pancreatic islets were washed in Krebs–Ringer bicarbonate HEPES buffer (120 mM NaCl, 4 mM KH2P04, 20 mM HEPES, 1 mM MgCl2, 1 mM CaCl2, 5 mM NaHCO3, and 0.5% BSA, pH 7.4; KRBH-BSA), supplemented with 2.8 mM glucose. Then, ten size-matched islets were distributed in nonadherent 12-well plates (Thermo Fischer, Lausanne, Switzerland) and incubated for 2 h in KRBH-BSA with 2.8 mM glucose. The medium was then replaced with fresh KRBH-BSA containing 2.8 mM glucose, 16.7 mM glucose, or 16.7 mM glucose plus 100 nM exendin-4 (Bachem, Bubendorf, Switzerland) for 1 h at 37 °C. Every condition was performed in duplicate using three independent islet preparations for each mouse line. The media were then collected and the islets were lysed in ethanol acid (75% ethanol, 23.5% water, and 1.5% HCl 37%) and sonicated 2 times for 25 s at 4 °C using a Bioruptor UCD-200 (Diagénode, NJ, USA). Insulin levels were determined by radioimmunoassay (RIA) (Millipore, MA, USA)
2.3. Quantitative trait loci (QTL) mapping
QTL mapping was performed using the R package R/qtl [12] with an extended genotype map from the BXD panel composed of GeneNetwork (www.genenetwork.org) genotypes merged with variants detected from RNA-Seq variant calling analysis, as described in Picard et al. [4] and available on figshare [13]. QTL interval mapping was calculated using the expected-maximization algorithm, a 5% genotyping error rate, and pseudomarkers were generated every cM. QTL location was obtained by 6.915 likelihood ratio statistics (LRS) support intervals. Significant QTLs were determined for each trait using a 5% false discovery rate threshold estimated from 1000 permutations.
2.4. RNA extraction and sequencing
Islets were isolated as described above and kept overnight in a cell culture medium. The next morning, they were collected in 1.5 mL Eppendorf tubes, washed once with Phosphate Buffered Saline (PBS), and then lysed in RLT plus buffer (RNeasy Plus Micro Kit, QIAGEN, Hombrechtikon, Switzerland) supplemented with 40 mM dithiothreitol using QIAshredder homogenizer tube (QIAGEN). The total RNA was purified using RNAeasy Plus Micro Kit (QIAGEN) according to the manufacturer's instructions. The quality of the extracted RNA was determined using a fragment analyzer (Agilent Technologies, CA, USA). The RNA quality number (RQN) for all the preparations was between 7.7 and 9.4. RNA-Seq libraries were prepared using 300 ng of RNAs pooled in equal amounts from three independent islet preparations per strain. RNA-Seq libraries were prepared using the Illumina TruSeq Stranded mRNA reagents (Illumina, CA, USA) on a Sciclone liquid handling robot with a PerkinElmer-developed automated script (PerkinElmer, MA, USA). Cluster generation was performed with the resulting libraries using the Illumina HiSeq SR Cluster Kit v4 reagents. The sequencing was performed on the Illumina HiSeq 2500 using HiSeq SBS Kit v4 reagents, to achieve a depth of 40 million reads using 100 base pair (bp) single-end reads. Sequencing data were demultiplexed through the bcl2fastq Conversion Software (v. 2.20, Illumina). Read counts were aligned to Mus musculus GRCm38.86 genome with the STAR software (v. 2.5.2b), and the quality of alignment was assessed using RSeQC script (v. 2.3.7). The number of read counts per gene was calculated with htseq-count script (v. 0.6.1) using Mus musculus GRCm38.86 gene annotation to generate a count matrix. The latter was filtered to exclude transcripts with an average number of reads lower than 1 over the replicate measurements before being normalized using a trimmed mean (TMM) normalization method with the R package “edgeR”.
2.5. Association between QTL genes and insulin secretion
Pearson's correlation coefficients between all expressed pancreatic islet mRNAs and the insulin secretion trait were calculated using the R software. The P values were calculated by the Fisher transformation and corrected for multiple testing using the Benjamini-Hochberg procedure. The associations between single nucleotide polymorphisms (SNPs) and gene expression of the QTL-constituent genes were calculated using the R/qtl package, with the same BXD panel genotype map and settings described for the QTL mapping. A mediation analysis was conducted to assess which of the genes identified in Figure 2B could explain the relationship between the genotype and the insulin secretion trait. This was performed using the mediation/r package as described in [14] using the genotype of the QTL SNP (rs27222258) as the “exposure”, the gene expression of the targeted genes as the “mediator”, and phenotype (secreted insulin) as the “outcome”. The significant average causal mediation effects were selected at P < 0.05, based on 10.000 permutations.
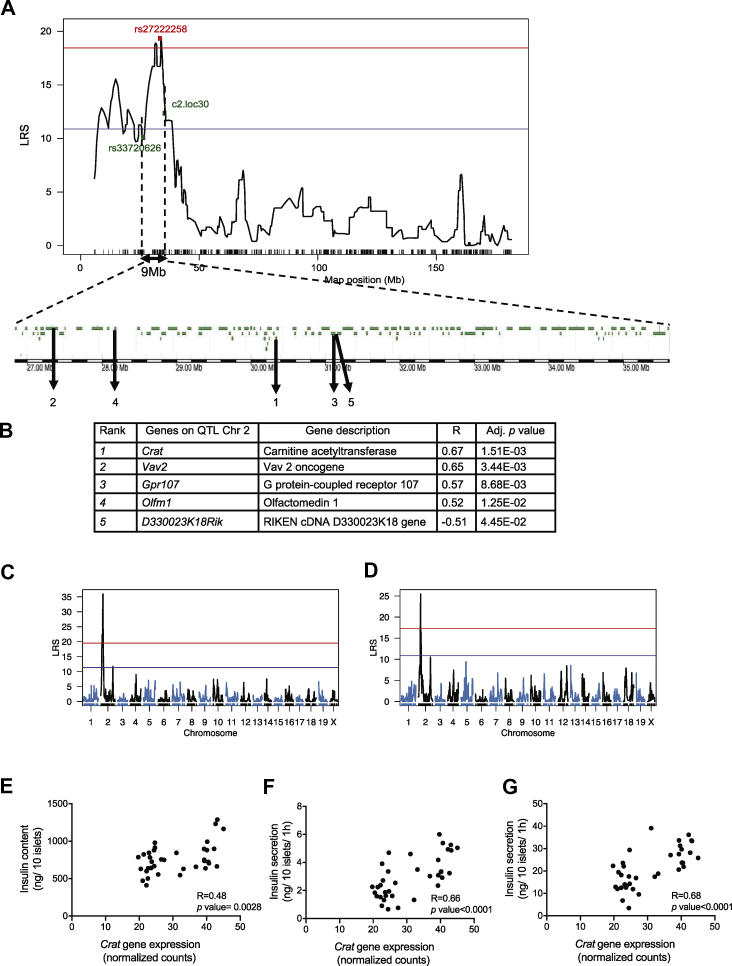
Figure 2.
Identification of QTL genes controlling the insulin secretion trait. (A) The QTL on Chr 2 spans 9 Mb and contains 173 genes; 147 of these genes are expressed in pancreatic islets (green). (B) Pearson's coefficients (R) with adjusted p values for the five genes that showed best correlation with the insulin trait. (C,D) eQTL mapping identifies a cis eQTL for Crat gene expression on Chr 2 at 21 Mb with a LRS = 35.9 (C) and for D330023K18Rik on Chr 2 at 22.1 Mb with a LRS = 25.4 (D). (E–G) Pearson correlation between Crat mRNA expression and insulin content (E), 16.7 mM glucose (F) and 16.7 mM plus exendin-4 (G) stimulated insulin secretion across all BXD islet preparations.
2.6. Cell culture and Crat silencing
The mouse MIN6B1 beta-cells were grown as described in [15]. Cells were used between passages 15 and 30. Control and Crat-specific siRNAs were designed and synthesized by Microsynth (Balgash, Switzerland). In total, three different siCrat were tested in MIN6B1 cells. Cells were reverse-transfected as follows. First, we prepared siRNA-lipid complexes inside 24-well plates by mixing 1.5 μL of siRNA from a 40 uM stock in 100 μL of Opti-MEM media (#31985-047, Thermo Fisher) and 3 μL of lipofectamine RNAiMax (#13778150, Thermo Fisher). After 15–20 min at room temperature, 3.5.105 MIN6B1 cells in 400 μL of culture medium were added to each well. The cells were then incubated for 48 h before performing the experiments. The selected CTL and Crat-specific siRNAs had, respectively, the following sequences: 5′-AGGUAGUGUAAUCGCCUUG-3′ and 5′-AGAGCCUGUUGGCAUCCUA-3′.
2.7. RNA extraction and quantitative real-time polymerase reaction chain
MIN6B1 cells were collected either by scrapping with a policeman or by trypsinization (Trypsin 0.02 mM, EDTA 0.48 mM, diluted 1/2 in PBS, #25300, Thermo Fischer). RNA was extracted as described for pancreatic islets. RNA concentrations were determined by the Nanodrop ND8000 spectrophotometer. Complementary DNAs (cDNA) were synthesized by reverse transcription (RT) of 1 μg of RNA primed with 50 pmol of random hexamers (Promega, Dubendorf, Switzerland) and 1 mM of dNTPs and M-MLV reverse transcriptase (Promega) following the manufacturer's instructions. For real-time quantitative polymerase chain reaction (qPCR), 2 μL of diluted cDNA (diluted 1/8 in Milli-Q water) was analyzed in a 10 μL final reaction volume, using 2x Power SYBR Green PCR master mix (#4368702, Thermo Fischer, MA, USA), in the presence of 10 pmol of forward and reverse primers. Amplification was performed using a 7500 Fast Real-Time PCR system (Applied Biosystems, Zug, Switzerland). Primer sequences for Crat were as follows: forward, 5′-GGAAGAATGGGCTCACACCA-3′, and reverse, 5′-CGGACAGCCAGTTCTCCATT-3′. Gene expression was normalized to the GusB mRNA (forward: 5′-GGCATTGTGCTACCTCAGAGT-3′, and reverse: 5′-GATTTTTGTCCCGGCGAACC-3′). Negative controls for the RT procedure and DNA contamination were performed by omitting reverse transcriptase and replacing cDNA by water, respectively. Gene expression was analyzed using Log 2 ˆ -ΔΔ Ct. All the primers were designed using the NCBI BLAST program and synthesized by Microsynth.
2.8. Establishment of stable MIN6B1 cell lines
Crat-specific shRNAs (forward: 5′-CCGGGACAGACTGTGTCATGTTCTTCTCG AGAAGAACATGACACAGTCTGTCTTTTTG-3′, and reverse: 5′- AATTCAAAAAGACAGACTGTGTCATGTTCTTCTCGAGAAGAACATGACACAGTCTGTC-3′) were selected from MISSION shRNA (Sigma) and synthesized by Microsynth. They were inserted into the lentiviral plasmid pLKO.1 TRC Cloning vector (#10878, Addgene, MA, USA) following the manufacturer's instructions. A Crat cDNA containing a sequence encoding a mycDDK tag was subcloned in the pRRLSIN.cPPT.PGK-GFP.WPRE vector (#12252, Addgene). Lentiviruses were produced as described by [16]. Viral titers were evaluated as described in [17] by measuring the number of integrated lentiviral DNA copies per cell. Fifty thousand HEK293T cells were infected with four different dilutions of the lentivirus stock. After 48–72 h, genomic DNA was extracted and qPCR was performed to quantitate the lentivirus WPRE sequence and the endogenous human albumin gene. Primers for WPRE were forward: 5′-GGCACTGACAATTCCGTGGT-3′ and reverse: 5′-AGGGACGTAGCAGAAGGACG-3′, and for human albumin, forward: 5′ GCTGTCATCTCTTGTGGGCTG-3′, and reverse: 5′-ACTCATGGGAGCTGCTGGTTC-3′. Viral titers ranged between 108 and 1011 transducing unit/mL (TU/mL). The infection of MIN6B1 cells was performed on cells seeded for 24 h on 6-well plates using a multiplicity of infection of 20–50.72 h after infection, transduced cells were selected in the presence of 1 μg/mL puromycin (Invivogen, California, USA) for 8 days.
2.9. Western blot analysis
MIN6B1 cells were washed twice with PBS and lysed on ice in 200 μL of lysis buffer containing 5% Sodium Dodecyl Sulfate (SDS), 5 mM EDTA, 80 mM Tris, pH = 6.8, and 2m M phenylmethylsulphonyl fluoride. The lysates were sonicated twice for 15 s at 4 °C using a Bioruptor. The protein concentration was determined by bicinchoninic acid (BCA) assay (ThermoScientific). 40 μg proteins were heated for 5 min at 95 °C in SDS sample buffer (5% SDS, 30% glycerol, 25% β-mercaptoethanol, 250 mM Tris, pH = 6.8, and 0.01% of Bromophenol blue), separated by gel electrophoresis on a 7.5% SDS-polyacrylamide gel, and transferred onto nitrocellulose membranes. The membranes were incubated with mouse anti-DDK (#F-3165, Sigma Aldrich) or mouse anti β-actin antibody (# A2066, Sigma Aldrich). The signals were revealed using the Sirius ECL chemiluminescence detection reagents (#K12043, Advansta CA, USA). Digital images were taken with the Fusion FX system (Vilber Lourmat, Collégien, France) and quantified with the Bio1D software (Vilber Lourmat).
2.10. In vitro insulin secretion assays
MIN6B1 cells were seeded for 48 or 72 h before the experiment to reach 70%–80% confluence. Incubations were as for the islet secretion experiments. The cells were then scrapped and lysed on ice in 500 μL of TETG buffer (20 mM Tris HCl, pH 8.0, 1% Triton X-100, 10% Glycerol, 137 mM NaCl, 2 mM EGTA, and protease inhibitor cocktail tablet (Roche)). After centrifugation at 4 °C for 10 min at 3000 g, the supernatants were kept aside for insulin measurement. Secreted insulin and insulin content were normalized to total protein content determined by BCA assay or to the cell number counted in wells seeded in parallel. Insulin was measured by enzyme-linked immunosorbent antigen (ELISA) (kit #1247, Mercodia, Uppsala, Sweden).
2.11. Beta-cell-specific Crat deficient mice
To generate Cratlox/lox;Ins1Cre/+mice, we crossed Ins1cre/+ mice [18] with Cratlox/lox mice [19], kindly provided by Dr. Mynatt (Pennington Biomedical Research Institute, USA). These mice were in the C57Bl/6L background. Genomic DNA was extracted with the Quick-gDNA MiniPrep Kit according to the manufacturer's instructions (#D3024, Zymo Research) and genotyped by PCR using the following primers: P1, 5′-TTGCATGAGTGGACGGAGAC-3′, and PCrat 5′-GCTGCAGTCCTGTAGTGTTCT-3′. Mice were fed with an RC or a high-fat, high-sucrose diet (#235 HF, Safe diets, Augy, France), composed of 18.0% proteins, 22.9% fat, and 46.0% carbohydrates. Studies were randomly assigned to experimental groups to ensure an unbiased distribution of animals. Studies were conducted in animals of 8–25 weeks of age and included age-matched and sex-matched littermate control mice. Mice from each cohort were weighed weekly. Glycemia and insulin levels were measured in three different metabolic states: random fed, after overnight fasting, and after overnight fasting and 6 h refeeding. Glycemia was measured in tail vein blood, using a glucometer (Ascensia Breeze 2, Bayer, Zurich, Switzerland). Plasma insulin concentrations were measured by ELISA.
2.12. Glucose tolerance tests
Following an overnight fast, glucose was administrated intraperitoneally to male mice (2 g/kg body weight). Glycemia was measured at −30, 0, 15, 30, 60, and 120 min after glucose administration from tail vein blood using the glucometer.
2.13. Oxygen consumption rates
Oxygen consumption rates (OCRs) were determined with the Seahorse XF24 Extracellular Flux Analyzer (Seahorse Biosciences). OCR is expressed as pmol of O2 per minute and was normalized by protein content (determined by BCA assay). Pancreatic islets incubated overnight in RPMI culture media were seeded at 50 islets/well in the XF 24-well islet capture microplates in 300 μL RPMI culture media. Before the assay, islets were incubated for 60–90 min in KRH-0.2% BSA supplemented with 2.5 mM glucose, without CO2. For OCR measurements with MIN6B1, cells were grown to 80% confluency on XF 24-well microplates in 500 μL of culture media for 48 h before the measurements.
2.14. Statistics
Statistical analyses and graphics were performed using GraphPad Prism 6.0 by Student's t-test, one-way ANOVA, or two-way ANOVA followed by Bonferroni's post hocc test.All the data were presented as means ± SEM.
3. Results
3.1. QTL analysis of glucose-induced insulin secretion
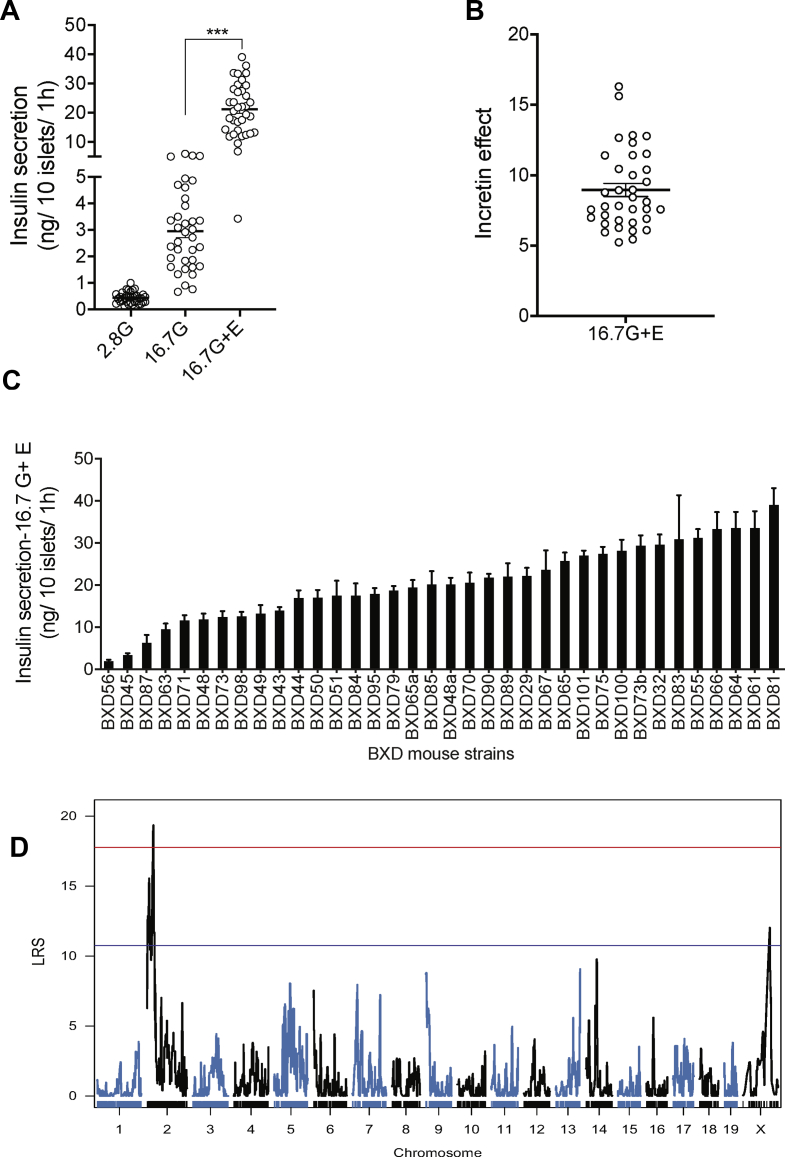
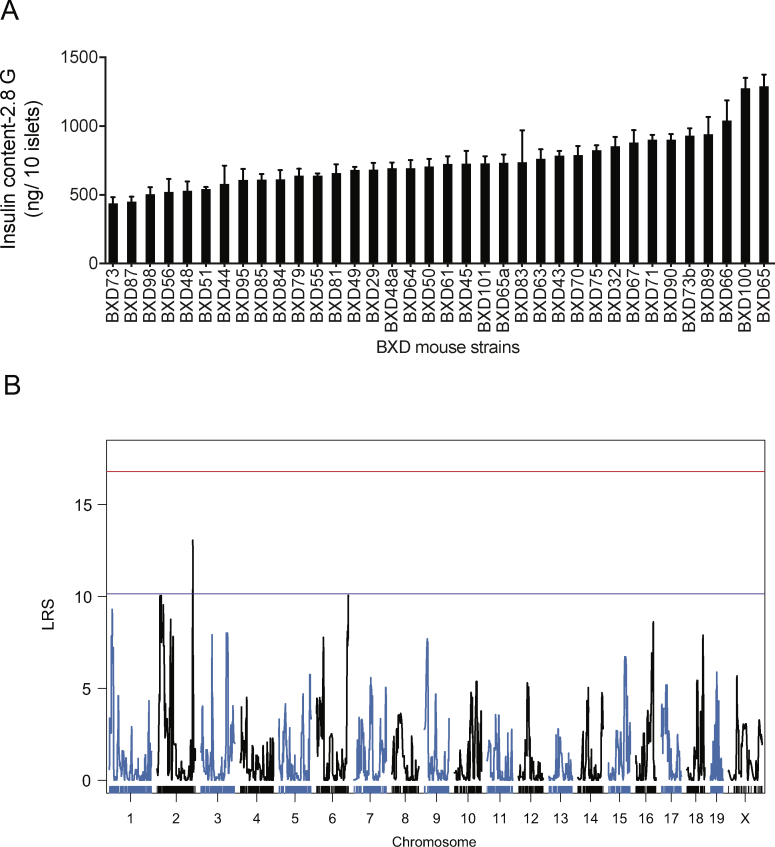
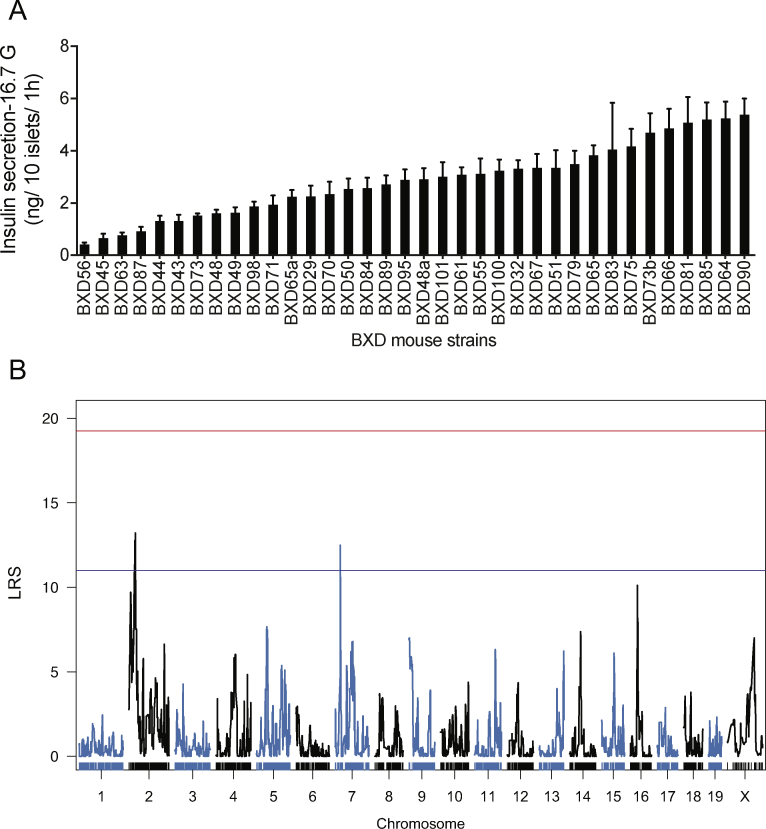
To discover unknown regulators of glucose-stimulated insulin secretion (GSIS), we isolated pancreatic islets from 36 different BXD mouse lines. This number of BXD lines is sufficient to identify QTLs accounting for approximately 40 percent of the variance of the trait with a power of ∼0.8 [20]. For each line, independent islet preparations were obtained from three mice and separately kept in culture overnight. Then, from each preparation, six batches of ten size-matched islets were hand-picked and incubated for 2 h in a KRBH-BSA solution containing 2.8 mM glucose. Duplicate islet batches were then incubated for 1 h in the presence of 2.8 mM glucose, 16.7 mM glucose, or 16.7 mM glucose plus 100 nM exendin-4. Secreted insulin and intracellular insulin were then measured by RIA. In the presence of 2.8 mM glucose, the secretion rates were low and similar for all islet preparations (Figure 1A). When islets were exposed to 16.7 mM glucose, the insulin secretion rates were markedly increased for all islet preparations but varied by ∼13-fold across mouse lines (Figure 1A). The addition of exendin-4 to 16.7 mM glucose further increased GSIS rates (Figure 1A), and this effect varied by 5- to 16-fold between islet preparations (Figure 1B). Figure 1C shows the 16.7 mM glucose plus exendin-4 insulin secretion rates for the islets prepared from each BXD mouse line with the individual mean ± SEM for each data set. The insulin content of each islet preparation is presented in Supplemental Figure 1A.
Figure 1.
A QTL on Chr 2 controls insulin secretion in response to high glucose and exendin-4. (A) Insulin secretion from islets isolated from 36 BXD mouse strains in the presence of 2.8 mM (2.8G), 16.7 mM (16.7G), or 16.7 mM glucose plus exendin-4 (16.7G + E). Each data point represents the average secretion from three islet preparations, each performed in duplicates. ∗∗∗p < 0.001. (B) Exendin-4 potentiation of 16.7 mM glucose-stimulated insulin secretion (incretin effect). (C) Insulin secretion in response to 16.7 mM plus exendin-4 for each BXD line islet preparation. Data are expressed as mean ± SEM. (D) A QTL for insulin secretion in response to 16.7 mM glucose and exendin-4 was identified on Chr 2. The red line indicates the genome-wide significant threshold and the blue line indicates the genome-wide suggestive threshold. QTL: quantitative trait loci, LRS: likelihood ratio statistics.
QTLs for each of the measured parameters were then searched. For islet insulin content, a suggestive QTL was identified on the distal part of Chr2 (between 167.77 and 172.56 Mb; LRS = 13.6) (Supplemental Figure 1B). For the 16.7 mM GSIS trait (Supplemental Figure 2A), two suggestive QTLs were identified, one on Chr2 (between 25.73 and 39.67 Mb; LRS = 13.20) and the other on Chr7 (between 25.31 and 28.31 Mb; LRS = 12.51) (Supplemental Figure 2B). A genome-wide significant QTL for the 16.7 mM glucose plus exendin-4 insulin secretion data was mapped on Chr2 (Figure 1D). This QTL has an LRS of 19.36 and was located between single nucleotide polymorphism markers rs337220626 and c2.loc30, which defines a 9 Mb segment between positions 26.78 and 35.77 Mb (Figure 2A). It explains 41 percent of the variance of the trait and contains 173 genes. It maps at the same position as the suggestive QTL for the 16.7 mM GSIS trait.
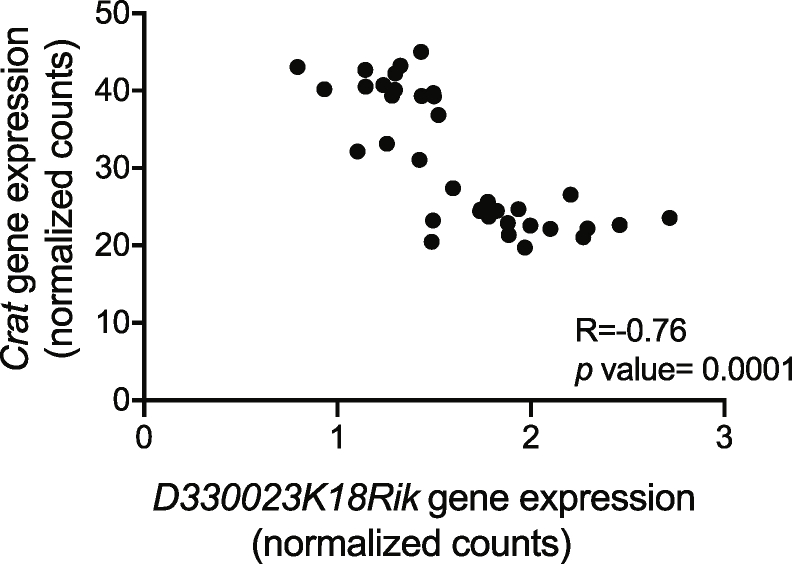
We next searched for QTL-associated genes that were expressed in pancreatic islets and whose expression was correlated with the insulin secretion trait. To this end, we performed RNA-Seq analysis of RNAs extracted and pooled from three islet preparations for each of the 36 BXD mouse strains. We found that among the 173 genes located in the QTL, 147 were expressed in pancreatic islets (Figure 2A). Pearson's correlation coefficients between the expressions of each of the 147 genes and insulin secretion were then calculated. Five transcripts with |R|≥0.5 and adjusted P value <0.05 were identified (Figure 2A,B): Crat, Vav2, Gpr107, and Olfm1, which showed a positive correlation with insulin secretion, and D330023K18Rik which was negatively correlated with the insulin secretion trait. Among those genes, Crat showed the best correlation to the trait (R = 0.67 and adjusted P = 0.0015). We then calculated the correlations between all the 28′455 islet expressed mRNAs and the insulin trait. In this genome-wide analysis, Crat showed the second highest correlation with the trait. The first gene in this list was Zmynd19 (R = −0.70, adjusted P = 0.016), a zinc transcription factor gene that is also located on Chr2 but outside of the 16.7 mM glucose plus exendin-4 QTL. We next searched for expression QTL (eQTL) for the expression of each of the five identified mRNAs. Significant eQTLs were found for Crat (LRS 35.00, P = 5e-04) and D330023K18Rik (LRS 25.50, P = 0.0025) at the same locus as the secretion QTL (Figure 2D). This indicates that the expressions of these two genes are controlled by the same QTL that controls the insulin secretion trait. It is noteworthy that D330023K18Rik is a lncRNA whose expression was strongly and negatively correlated with the expression of Crat (R = −0.76; P = 4.5e-08, Supplemental Figure 3A) suggesting that it may control Crat expression. No eQTL was found for the other 3 mRNAs. We next performed a mediation analysis, i.e., a statistical method that allows us to assess whether Crat expression, directed by the genetic background of the BXD mice, can explain (mediate) the relationship between the BXD genotype and the insulin secretion trait. A clear absolute average causal mediation effect (ACME) was observed for Crat (ACME = 3.73, P = 0.029).
Finally, we found that the expression of Crat mRNA across all the BXD islet preparations was significantly correlated with islet insulin content and insulin secretion stimulated by 16.7 mM glucose or by 16.7 mM glucose plus exendin-4 (Figure 2E–G). The Crat genes of the C57Bl/6J and DBA/2J parental strains have an identical protein-coding sequence, excluding that mutations affecting Crat activity could explain the observed phenotype.
Together, this genetic analysis suggested that Crat was the most likely regulator of the glucose plus exendin-4 secretion trait. Crat (carnitine O-acetyltransferase) is a mitochondrial enzyme that catalyzes the interconversion of acetyl-CoA into acetyl-carnitine. One of the important regulatory functions of this enzyme is to increase glucose utilization by reducing the intramitochondrial levels of acetyl-CoA [21,22], an allosteric inhibitor of the pyruvate dehydrogenase complex [23].
3.2. Silencing Crat in MIN6B1 cells reduces insulin content
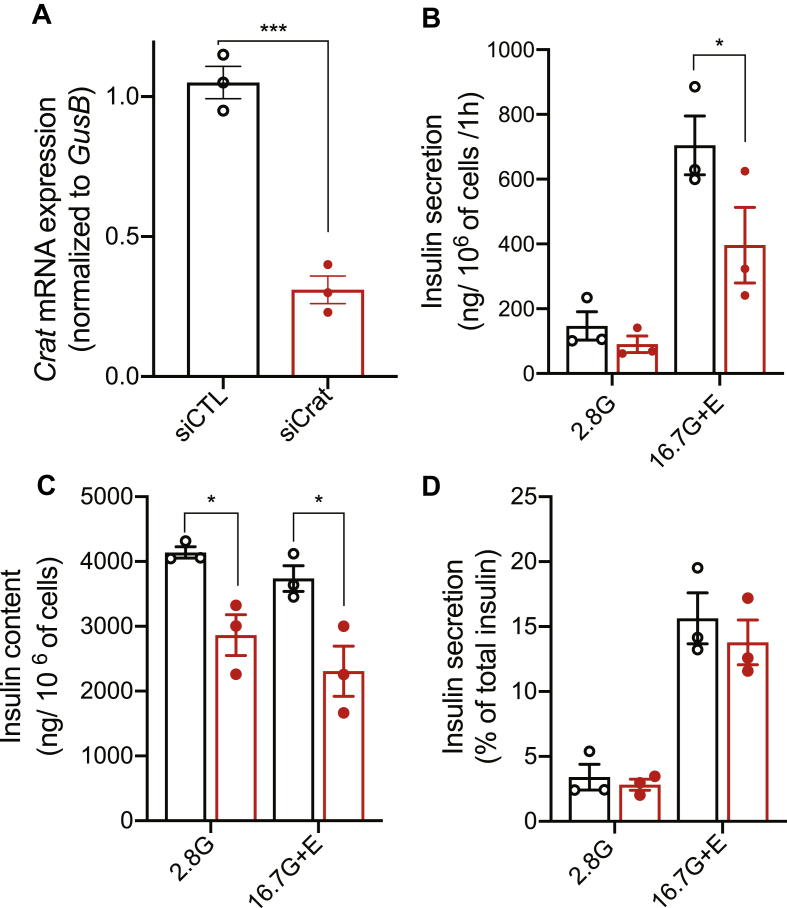
To assess whether Crat plays a role in insulin secretion, we first transfected the insulin-secreting cell line MIN6B1 with control (CTL) or Crat-specific siRNA. Insulin secretion in response to 2.8 mM glucose or 16.7 mM glucose plus exendin-4 was then measured. As shown in Figure 3A, Crat siRNA transfection reduced Crat mRNA expression by ∼60% as compared to CTL siRNA-transfected cells. This led to a ∼30% decrease in insulin secretion in response to high glucose plus exendin-4 (Figure 3B). This was associated with a similar ∼30 percent reduction of insulin content (Figure 3C). Thus, insulin secretion expressed as a percentage of insulin content was unchanged between the two transfection conditions (Figure 3D).
Figure 3.
Crat transient knockdown in MIN6B1 cells reduced insulin content. (A) Crat mRNA levels in MIN6B1 cells transfected with either siCTL (black) or siCrat (red). Data are expressed as mean ± SEM of 3 independent experiments, each performed in triplicates. Unpaired t-test, ∗∗∗p < 0.001. (B–D) Insulin secretion in response to 2.8 mM glucose (2.8G) or 16.7 mM glucose plus exendin-4 (16.7G + E). (B) Insulin secretion. (C) Insulin content. (D) Insulin secretion expressed as percent of total insulin. Data are expressed as mean ± SEM of 3 independent experiments, each performed in triplicates. Two-way ANOVA and Bonferroni's post-hoc tests; ∗p < 0.05.
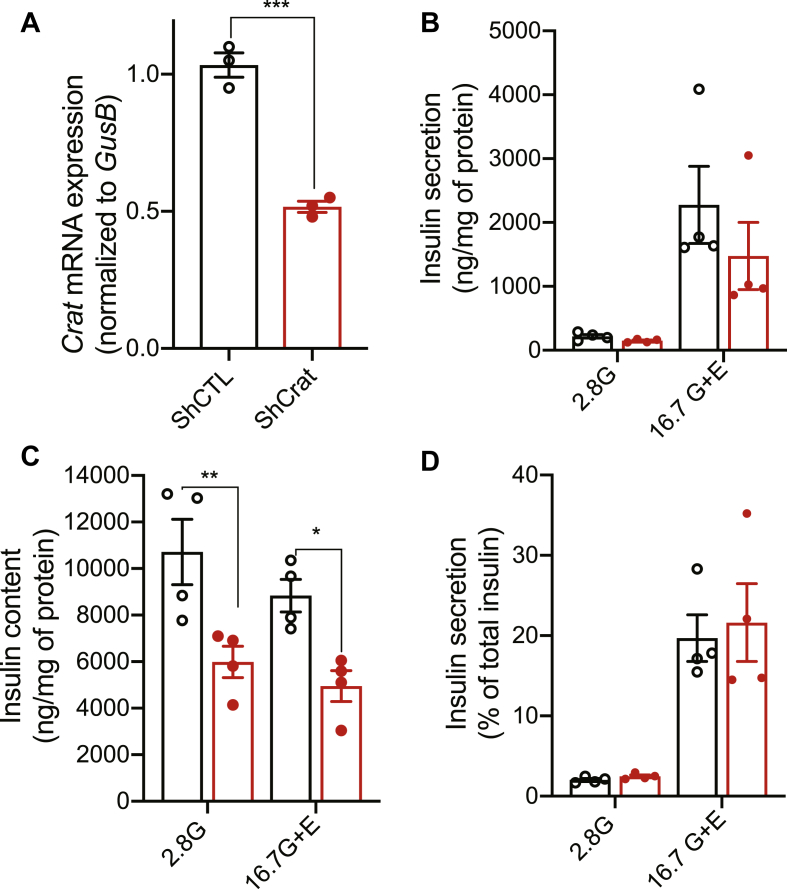
To find further support for this observation, we transduced MIN6B1 cells with recombinant lentiviruses encoding a Crat-specific short hairpin RNA (shRNA, targeting a different sequence than the Crat siRNA) or a CTL shRNA. The established MIN6B1 cell line stably expressing the Crat shRNA had a 50 percent reduction in Crat expression (Figure 4A). This caused a ∼50% decrease in both glucose plus exendin-4-stimulated insulin secretion (Figure 4B) and insulin content (Figure 4C). Again, insulin secretion expressed as a percentage of total insulin was unchanged (Figure 4D).
Figure 4.
Crat stable silencing decreases insulin content in MIN6B1 cells. Insulin secretion experiments using MIN6B1 cell lines expressing shCTL (black) or shCrat (red). (A) Crat mRNA levels. Data are expressed as mean ± SEM of 3 independent experiments, each performed in triplicates. Unpaired t-test, ∗∗∗p < 0.001. (B–D) Insulin secretion in response to 2.8 mM glucose (2.8G) or 16.7 mM of glucose plus exendin-4 (16.7G + E). (B) Insulin secretion. (C) Insulin content. (D) Insulin secretion expressed as percent of total insulin. Data are expressed as mean ± SEM of 4 independent experiments, each performed in triplicates. Two-way ANOVA and Bonferroni's post-hoc test. ∗p < 0.05; ∗∗p < 0.001
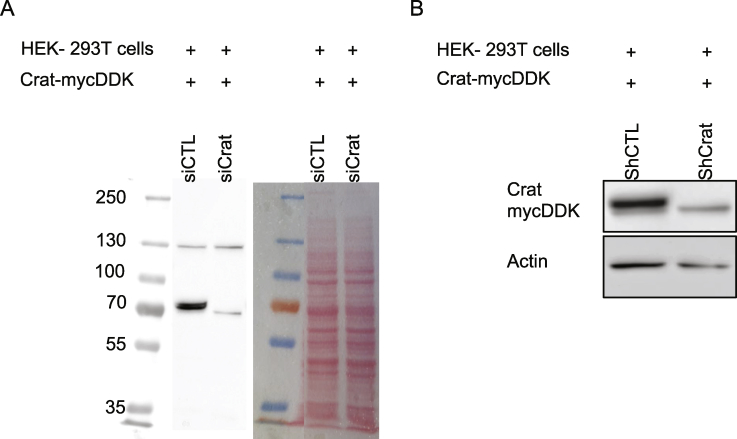
To verify that the siRNA and shRNA used above were also reducing Crat protein expression, we performed western blot analysis. However, because we could not find suitable antibodies to detect Crat, we tested whether the overexpression of a Crat-mycDDK in HEK293 cells could be efficiently silenced using an anti-DDK antibody. Supplemental Figure 4 showed that overexpressed Crat-mycDDK was strongly decreased by the selected siRNA and shRNA.
Thus, reduced Crat expression in MIN6B1 cells led to a reduced insulin content and a reduced quantity of insulin secreted; secretion expressed as a percentage of insulin content was, however, not modified, suggesting that the process of insulin secretion was not altered but that the amount of secreted insulin was determined by the insulin content.
3.3. Crat overexpression increases insulin content in MIN6B1 cells
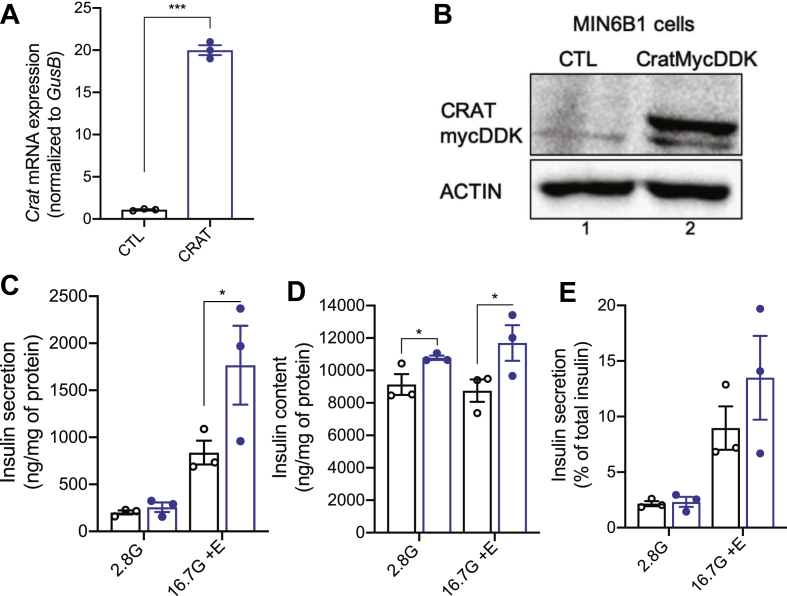
To further explore the link between Crat expression and insulin content, we transduced MIN6B1 cells with a recombinant lentivirus encoding a green fluorescent protein (GFP) or a cDNA encoding a Crat-mycDDK sequence. Figure 5A,B show Crat mRNA and protein overexpression in stably transduced MIN6B1 cells. Secretion experiments performed with these cells showed a 30 percent increase in insulin secretion as compared to the control cells in the presence of 16.7 mM glucose plus exendin-4 (Figure 5C) and a 25 percent increase in insulin content (Figure 5D). Again, insulin secretion was increased because of the higher insulin content but was unchanged between cell lines when expressed as a percentage of insulin content (Figure 5E).
Figure 5.
Crat overexpression in MIN6B1 cells increases insulin content. (A) Crat mRNA expression in MIN6B1 cells transduced with a GFP (CTL, black) or a Crat-mycDDK (CRAT, blue) expression plasmid. Data are expressed as mean + SEM of 3 independent experiments, each performed in triplicates. Unpaired t-test, ∗∗∗p < 0.001. (B) Western blot analysis of Crat-mycDDK expression using an anti-DDK antibody. (C–E) Insulin secretion in response to 2.8 mM glucose (2.8G) or 16.7 mM of glucose plus exendin-4 (16.7G + E). (C) Insulin secretion. (D) Insulin content. (E) Insulin secretion expressed as percent of total insulin. Data are expressed as mean ± SEM of 3 independent experiments, each performed in triplicates. Two-way ANOVA and Bonferroni's post-hoc test. ∗p < 0.05.
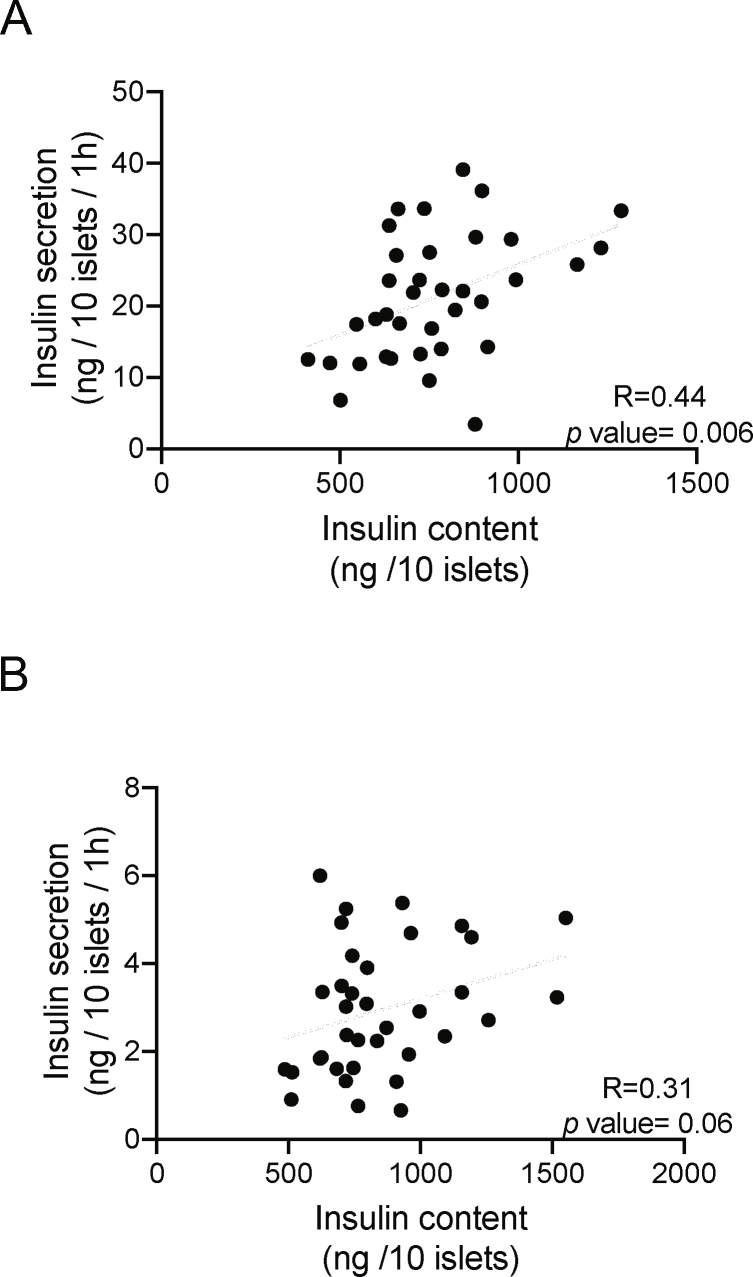
Together with the data of Figure 2E–G (which showed positive correlations between Crat expression, insulin content, and stimulated insulin secretion), the results of the Crat silencing or overexpression experiments lend further support for a primary role of Crat on insulin content and indicate that insulin content is an important determinant of the amount of secreted insulin. This is also in agreement with the finding that insulin content and 16.7 mM glucose plus exendin-4-stimulated insulin secretion were highly correlated across all BXD islet preparations (R = 0.44, P = 0.006), a correlation that was also suggestive for the 16.7 mM GSIS (R = 0.31, P = 0.06) (Supplemental Figures 6A and 6B).
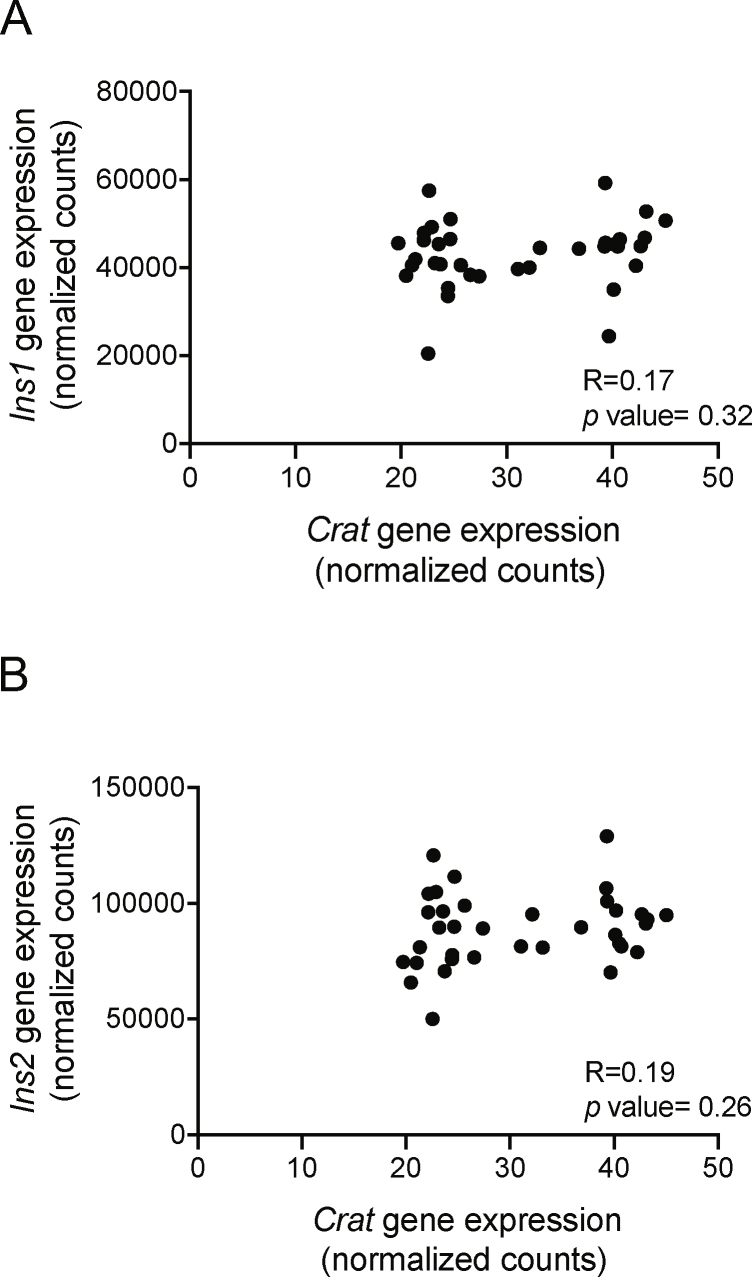
Finally, the analysis of the correlation between Crat expression and the expression of the insulin mRNAs Ins1 and Ins2 in the BXD islet RNA-Seq data revealed no relationship between these gene expressions (Crat versus Ins1: R = 0.17; P = 0.32 and Crat versus Ins2: R = 0.19; P = 0.26; Supplemental Figures 7A and 7B). This suggests that Crat regulates insulin content at a post-transcriptional level.
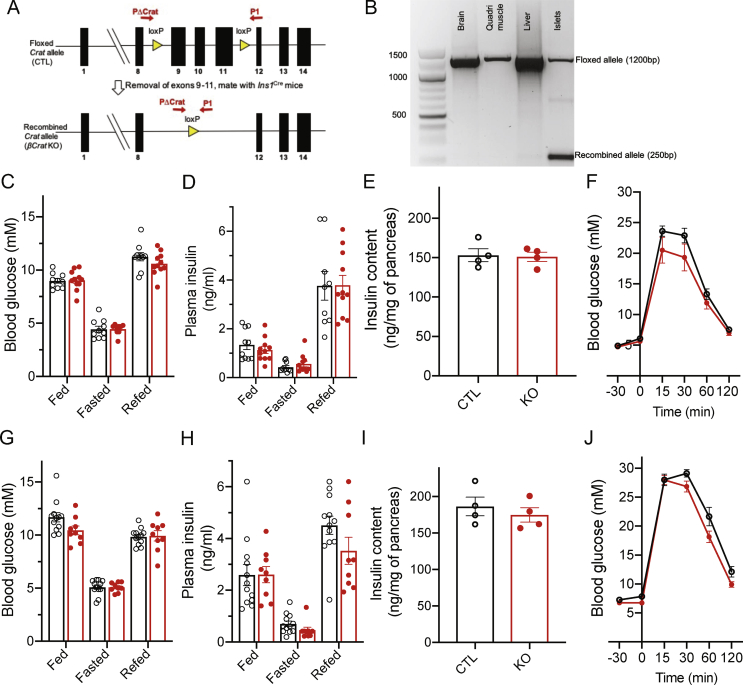
3.4. Phenotyping of ß-cell-specific Crat KO mice
We, then, investigated the role of Crat in primary islets isolated from mice with ß-cell-specific Crat inactivation. These mice were obtained by crossing Cratlox/lox mice (Supplemental Figure 5A) with Ins1cre/+ mice to obtain Cratlox/lox;Ins1cre/+ mice (ßCratKO) and Cratlox/lox;Ins1+/+ (CTL) mice. Supplemental Figure 5B shows that the recombined Crat allele was found only in the islet from ßCratKO mice and not in the other tissues tested.
We first measured blood glucose and plasma insulin levels in random fed, overnight fasting, and fasting and 6 h refed CTL and ßCratKO mice (Supplemental Figures 5C and 5D). These measurements were identical for the two mouse groups. The total pancreatic insulin contents and glucose tolerance tests were also similar in CTL and ßCratKO mice (Supplemental Figures 5E and 5F).
Mice were then fed with a high-fat high-sucrose (HFHS) diet for 20 weeks starting at 6 weeks of age. Random fed, overnight fasting, and 6 h refed mice of both genotypes showed the same blood glucose, plasma insulin levels, pancreatic insulin content, and glucose tolerance tests (Supplemental Figures 5G–5J).
Thus, Crat inactivation in β-cells does not impact insulin secretion or insulin content when mice are fed with an RC of an HFHS diet.
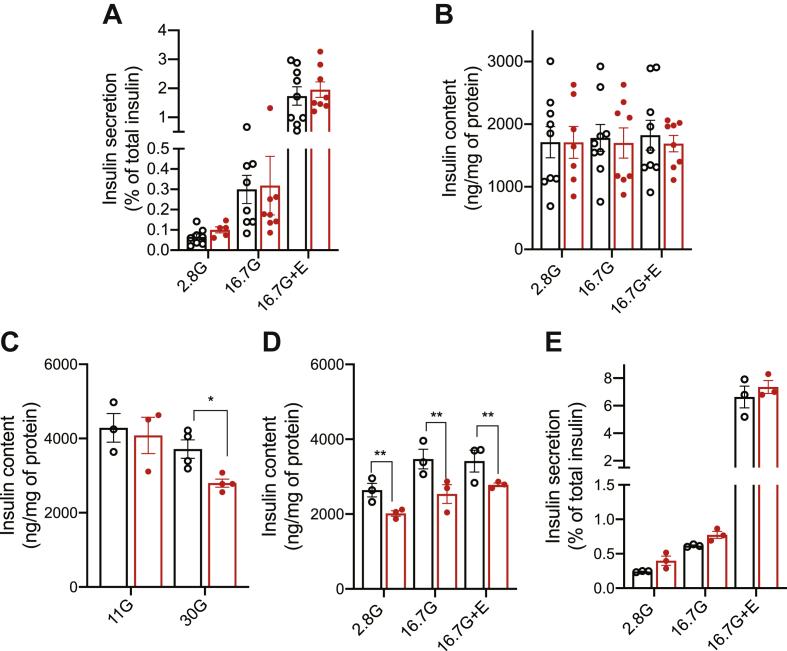
3.5. Prolonged exposure to high glucose impairs insulin content in ßCrat KO islets
We next isolated islets from CTL and ßCratKO mice and performed GSIS experiment in the presence of 2.8 mM glucose and 16.7 mM glucose with or without exendin-4. Insulin secretion expressed as a percentage of total insulin and insulin contents was identical for islets isolated from CTL and ßCratKO mice (Figure 6A,B). Similar data were obtained when using islets from female or male mice.
Figure 6.
Insulin secretion from βCratKO mouse islets. Insulin secretion from islets isolated from CTL (black) or βCratKO mice (red). (A) Insulin secretion as percent of total insulin. (B) Insulin content. Data are expressed mean ± SEM of 3 independent experiments. (C) Insulin content of CTL or βCratKO mouse islets pre-incubated for 24 h with 11 mM (11G) or 30 mM glucose (30G). (D–E) Insulin secretion from islets isolated from CTL or βCratKO mice pre-incubated for 24 h with 30 mM glucose. (D) Insulin content. (E) Insulin secretion as percent of total insulin. Data are expressed as mean ± SEM of 3 independent experiments, each performed using 3–5 animals per genotype. Unpaired t-test in C and two-way ANOVA in D; ∗p < 0.05 and ∗∗p < 0.01
Crat increases glucose oxidation rates and, consequently, OCR by increasing pyruvate dehydrogenase activity [21]. As insulin production is strongly regulated by glucose metabolism and Krebs cycle-derived intermediates have been reported to control insulin biosynthesis [[24], [25], [26]], we wondered whether an impact of Crat inactivation on insulin content would be observed when the cells are exposed to high glucose concentrations. To test this hypothesis, we incubated islets from CTL mice and ßCratKO mice for 24 h in culture medium containing 11 mM glucose (the standard glucose concentration for maintenance of mouse islets) or 30 mM glucose and measured their insulin content. When incubated in the presence of 11 mM glucose, the insulin content of both types of islets was identical (Figure 6C). However, when incubated in the presence of 30 mM glucose, ßCratKO islets displayed a 25 percent lower insulin content as compared to CTL islets (Figure 6C). To test whether this affected GSIS, we next performed secretion experiments after the preincubation of CTL and ßCratKO islets in the presence of 30 mM glucose for 24 h. Figure 6D shows that the insulin content of ßCratKO islets was decreased as compared to those of CTL islets. However, insulin secretion, expressed as a percentage of total content, was identical between the two types of islets (Figure 6E).
3.6. Reduced oxygen consumption rate in the absence of Crat
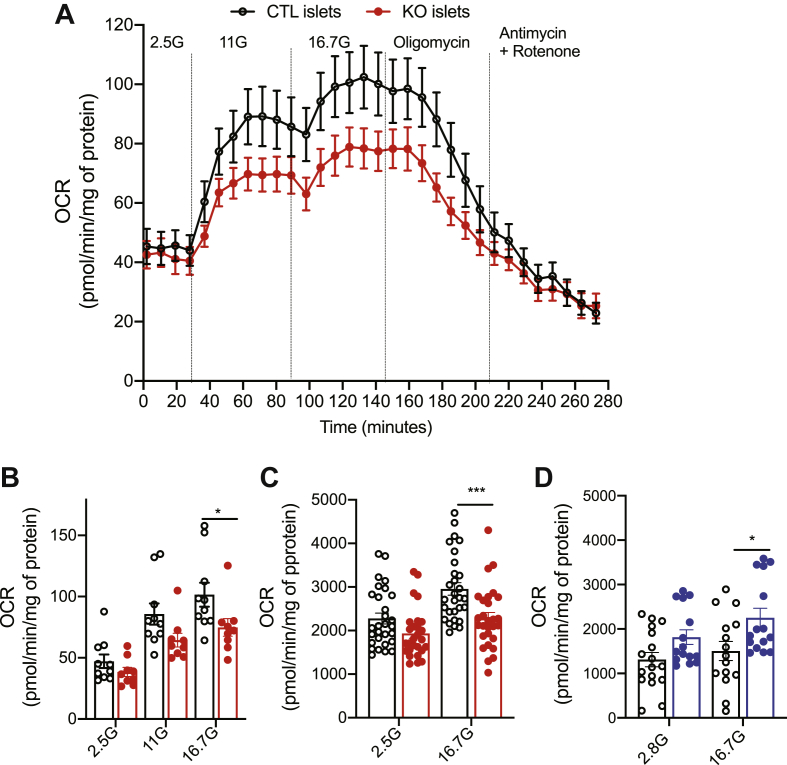
Because the above observations suggested that glucose utilization was regulated by Crat when the cells were exposed to high glucose concentrations, we measured OCR of pancreatic islets isolated from CTL or ßCratKO mice (Figure 7A,B). The OCR at 2.5 mM glucose was similar in the two groups of islets. Increasing glucose concentrations to 11 and 16.7 mM increased OCR in both types of islets but the increase was significantly less in the ßCratKO islets than that in CTL islets. When oligomycin, an ATP synthase inhibitor, was added to assess the ATP-linked respiration, the OCR reached the same low level in islets from both genotypes. The addition of rotenone and antimycin A, which block complexes I and III of the electron transport chain, respectively, enabled the measurement of nonmitochondrial respiration, which was the same in both islet groups.
Figure 7.
Reduced oxygen consumption rates in βCratKO mouse islets. (A–B) Oxygen consumption rates (OCR) of islets from CTL mice (black) or βCratKO mice (red) in the presence of the indicated concentrations of glucose and in the presence of oligomycin, and antimycin A and rotenone. (B) Quantification of the OCRs measured in A. Data are expressed as mean ± SEM of 2 independent experiments (n = 9–10 replicates, 8 mice per genotype). (C) OCR of MIN6B1 cells transfected with siCTL (black) or siCrat (red) at the indicated glucose concentrations. Data are expressed as mean ± SEM of 3 independent experiments (n = 27–29 replicates). (D) OCR of MIN6B1 cells overexpressing GFP (CTL, black) or Crat (blue) at the indicated glucose concentrations. Data are expressed as mean ± SEM of 3 independent experiments (n = 15–16 replicates). Statistical analyses for B-D were performed using rwo-way ANOVA and Bonferroni's post-hoc test, ∗p < 0.05, ∗∗∗p < 0.001.
Finally, we tested whether Crat silencing in MIN6B1 cells would lead to a similar reduction in OCR at high glucose as seen in primary islets from ßCratKO mice. We, thus, performed Seahorse experiments using MIN6B1 cells transfected with Crat-specific siRNA (Figure 7C) and observed that reduced Crat expression led to a 25 percent lower OCR than that observed in CTL-transfected cells in the presence of 16.7 mM glucose. Conversely, Crat overexpression (Figure 7D) led to a 35 percent increase in OCR. Together the above experiments demonstrated that Crat regulates glucose metabolism at high glucose concentrations both in pancreatic islets and in MIN6B1 cells.
4. Discussion
In the present work, we performed a genetic screen in the BXD genetic reference population to search for unknown regulators of GSIS. Our screen yielded a genome-wide significant QTL on Chr2 for the 16.7 mM glucose plus exendin-4 condition. Combining this information with islet transcript profiling led to the identification of Crat as the most likely regulator of the insulin secretion trait. Our further studies in insulin-secreting cell lines and primary mouse islets showed that Crat regulates insulin content rather than the process of insulin secretion itself. Thus, in the stimulatory conditions used, insulin content determines the amount of secreted insulin.
The analysis of genetic reference populations of RI mice is a powerful approach at identifying chromosomal regions controlling physiological functions when these can be assayed by precise functional tests [[4], [5], [6], [7], [8]]. Here, we identified a QTL on Chr2 for the 16.7 mM glucose plus exendin-4 condition. The same chromosomal region was identified as a suggestive QTL for the 16.7 mM GSIS further supporting the role of this locus in the control of insulin secretion.
It is interesting to compare our data with previous mouse genetic studies aimed at identifying genes controlling insulin secretion. In studies of (C57Bl/6 x DBA2) x C57Bl/6 backcross mice, a QTL for insulin secretion in response to intravenous glucose administration was found on Chr13 and the Nnt gene (nicotinamide nucleotide transhydrogenase) was identified as the gene controlling insulin hypersecretion in DBA2 mice [27]. Subsequent studies from the same group, using in vivo insulin secretion data and genetic screening of 20 BXD lines, identified Herpud1 (endoplasmic reticulum stress-induced protein) as a further regulator of insulin secretion [28]. More recently, a genetic analysis of insulin secretion by islets isolated from Diversity Outbred mice identified five genome-wide significant QTLs for insulin secretion stimulated by different glucose concentrations or by glucose plus GLP-1 or amino acids [25]. A QTL for the fold change of insulin secretion induced by GLP-1 was located on Chr2 at ∼79 Mb, a region distinct from that containing the 16.7 mM glucose plus exendin-4 QTL identified in our study. None of the other QTLs identified in the Diversity Outbred study were present in Chr2. The fact that our screens and these previous screens yielded different QTLs and causal genes further indicates that GSIS is a quantitative trait controlled by many genes. It also reflects the difference in the physiological measurements performed (in vivo or in vitro GSIS) and in the mouse populations studied, BXD mice issued from the cross of C57Bl/6 and DBA2 mice [27,28], and Diversity Outbred mice generated by crossing eight different mouse strains [25], which do not include the DBA2 mice.
The Chr2 QTL for the 16.7 mM glucose plus exendin-4 trait contains 173 genes and 147 are expressed in islets. Of those, only five show a significant correlation with the secretion trait. These genes include Vav2, which encodes a guanine nucleotide exchange factor for Rho GTPase; this has been described as a positive regulator of insulin secretion [29,30]. Gpr107 encodes a receptor for neuronostatin, a hormone produced from the somatostatin preprohormone in pancreatic delta cells and which participates in the regulation of glucagon secretion [31,32]. Olfm1 codes for a secreted glycoprotein, described in neuronal development and axon elongation [33]. D330023K18Rik is a long noncoding RNA of unknown function but the observation that its expression is strongly anticorrelated with that of Crat suggests that it may have a regulatory role in Crat expression. Although Vav2 was previously linked to insulin secretion, we selected to focus our attention on Crat because (i) it displayed the highest correlation with the trait with the lowest adjusted P value of the five genes, (ii) it came second when the trait was correlated with all islet expressed genes, (iii) an eQTL was located in the same region as the insulin secretion trait QTL, and (iv) mediation analysis identified Crat and none of the other 4 mRNAs as a significant contributor to the insulin secretion trait QTL.
Crat is a mitochondrial enzyme that catalyzes the conversion of acetyl-CoA into acetyl-carnitine. In the presence of high extracellular glucose and glycolytic activity, pyruvate production is increased. Pyruvate enters into the mitochondria through the mitochondrial pyruvate carrier and is converted to acetyl-CoA by the pyruvate dehydrogenase complex. This enzyme complex is subject to strong allosteric inhibition, in particular, by acetyl-CoA. By converting acetyl-CoA into acetyl-carnitine, Crat reduces the intramitochondrial concentration of acetyl-CoA, thereby alleviating pyruvate dehydrogenase inhibition leading to increased glucose oxidation. In muscles, Crat has been shown to have an important role in the control of metabolic flexibility, regulating the balance between glucose and lipid usage for energy generation. Indeed, mice with muscle-specific Crat inactivation showed reduced muscle pyruvate metabolism and increased fatty acid oxidation [19]. Inactivation of Crat in hypothalamic AgRP orexigenic neurons induces, through a change in sympathetic outflow, a decrease in metabolic flexibility in peripheral tissues and a change in nutrient partitioning during refeeding after a period of fast [34].
We were somewhat surprised that the inactivation of Crat in islet beta-cells did not produce any measurable effect on the whole-body glucose homeostasis nor on the insulin secretion and insulin content of isolated islets. The only observed phenotype was a reduced insulin content in islets cultured in high glucose concentrations, which is in line with their reduced OCR and with observations made in MIN6B1 cells. The low impact of Crat inactivation in the ßCratKO mice and islets can, nevertheless, be explained by the fact that this gene was identified in a screen performed with a panel of RI mice. In such a population genetic approach, the impact of a gene on a phenotype is strongly dependent on the genetic background of each mouse line tested. Indeed, similar levels of Crat expression in different islet preparations are associated with very different insulin contents, as shown in the correlation analysis of Figure 2E, which also shows that there is a correlation between Crat expression levels and insulin content. As the ßCratKO mice were in a C57Bl/6J background, they represent only one genetic architecture, which may not be conducive to the development of a strong phenotype. Introducing the Crat inactivating mutation in other genetic backgrounds may reveal stronger, genetic background-dependent phenotypes. Such a situation has been observed, for instance, when testing the impact of double heterozygous mutations of the insulin receptor and insulin receptor substrate genes in C57Bl/6J, DBA, and SV129J mice. Whereas the mutations induced major glucose homeostasis deregulation in C57Bl/6J and DBA mice, the SV129J mice were totally resistant to diabetes development [35]. Thus, Crat is likely part of a metabolic regulatory network influenced by epistatic interactions that define the coupling between glucose metabolism and insulin content.
Whether Crat expression is modified in the beta-cells of type 2 diabetic patients is not known. It may, however, be speculated that variations in Crat expression levels, as seen across the BXD panel, may contribute to the reduction in insulin secretion and beta-cell mass observed during the progression of the disease [1,2].
In the MIN6B1 cell lines, the effect of Crat expression levels on insulin content was clearly established. Indeed, silencing Crat in these cells reduces OCR and insulin content, an effect that is more prominent in the presence of high glucose concentrations. On the other hand, the overexpression of Crat leads to the opposite phenotype, with increased OCR and insulin content in the presence of 16.7 mM glucose. How does Crat regulate insulin content? Insulin stores are regulated by the balance between numerous processes, including gene transcription, mRNA stability, biosynthetic rates, and the processes of insulin granule exocytosis or degradation by autophagy (crinophagy) [25,36,37]. We found that Crat silencing or genetic inactivation was not associated with changes in insulin mRNA levels (not shown) and there was no correlation between Crat expression and that of Ins1 or Ins2 mRNAs in the BXD islet RNA-Seq data. Therefore, the reduction in insulin content must be controlled at a post-transcriptional level by the control of either insulin biosynthesis or insulin degradation. Although we have not tested either hypothesis here, it is tempting to favor the regulation of the translation rate. Indeed, glucose strongly regulates insulin biosynthesis [37], by a metabolic coupling mechanism that is distinct from that inducing insulin secretion [38]. It has been shown that pyruvate entry into the mitochondria and production of Krebs cycle intermediates, in particular succinate, are required for the stimulation of insulin mRNA translation by glucose [26].
Another interesting observation made in our study is the positive correlation between insulin secretion and insulin content across all islet preparations. This correlation is stronger when secretion is stimulated by 16.7 mM glucose plus exendin-4 than with 16.7 mM glucose only. This is somewhat surprising since the amount of insulin secreted in our experiments represents only a small fraction of the total insulin content and would, thus, not be expected to limit the amount of insulin secreted. However, insulin granules are heterogeneous in age, protein composition, and cellular locations [[39], [40], [41]]. Therefore, the amount of insulin secreted may depend on a subpool of granules, possibly predocked to the plasma membrane and/or located close to the plasma membrane and ready to be translocated to the plasma membrane for exocytosis [42,43]. The size of this subpool of granules may, thus, be determined by the total intracellular insulin content. If this could be confirmed in humans, this would suggest that insulin secretion measured under maximal stimulatory conditions, such as those described by Stumvoll et al. [44] that include high glucose and GLP-1, may actually reflect the total islet insulin content.
In summary, our work identified a novel regulator of insulin secretion, the mitochondrial enzyme Crat. Its effect is mostly mediated by a regulation of total insulin content, which is related to its control of glucose oxidation rates. The magnitude of the effect of Crat on insulin content also depends on epistatic interactions specific to each BXD mouse line. Our data also show that insulin content is a determinant of insulin secretion rates, an effect that could depend on the size of insulin granule subpopulations ready to be secreted. Whether such a link between insulin content and maximal stimulation of insulin secretion also exists in humans is of interest as this could represent an indirect way of measuring total insulin content in humans, which could be of great interest in following type 2 diabetes development and response to various drug treatments.
Authors' contributions
B. T. and B. D. conceived the experimental design, analyzed the data, and wrote the manuscript. X. B and B. D. performed the islet isolations, the generation of BXD data, and the in vivo experiments. B. D. performed all the experiments in vitro and ex vivo. M. J. performed the QTL mapping and calculated the QTL intervals. A. R. performed the eQTL mapping, RNA-Seq analysis, and correlation analysis. C. R. helped with the analysis. IC. L-M. helped with Seahorse's analysis.
Acknowledgments
We thank Professor Paul Franken (UNIL-CIG) for his careful reading of the manuscript and his many suggestions to improve its presentation. This project has received funding from the Innovative Medicines Initiative 2 Joint Undertaking under grant agreements no. 115797 (INNODIA). This Joint Undertaking receives support from the European Union’s Horizon 2020 research and innovation program, EFPIA, “JDRF”, and “The Leona M. and Harry B. Helmsley Charitable Trust”. This work was also supported by the Swiss State Secretariat for Education‘ Research and Innovation (SERI) under contract number 16.0097, and by a grant from the Swiss National Science Foundation (310030_182496) to B. T.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2020.100993.
Contributor Information
Dassine Berdous, Email: dassine.berdous@unil.ch.
Xavier Berney, Email: xavierpascal.berney@unil.ch.
Ana Rodriguez Sanchez-Archidona, Email: ana.rodriguez.1@unil.ch.
Maxime Jan, Email: maxime.jan@unil.ch.
Clara Roujeau, Email: clara.roujeau@unil.ch.
Isabel C. Lopez-Mejia, Email: isabel.lopezmejia@unil.ch.
Randall Mynatt, Email: Randall.Mynatt@pbrc.edu.
Bernard Thorens, Email: Bernard.thorens@unil.ch.
Conflict of interest
The authors declare that they have no conflicts of interest.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Figs1.
Figs2.
Figs3.
Figs4.
Figs5.
Figs6.
Figs7.
References
- 1.Prentki M., Nolan C.J. Islet beta cell failure in type 2 diabetes. Journal of Clinical Investigation. 2006;116(7):1802–1812. doi: 10.1172/JCI29103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kahn S.E. The importance of the beta-cell in the pathogenesis of type 2 diabetes mellitus. The American Journal of Medicine. 2000;108(Suppl 6a):2S–8S. doi: 10.1016/s0002-9343(00)00336-3. [DOI] [PubMed] [Google Scholar]
- 3.Peirce J.L., Lu L., Gu J., Silver L.M., Williams R.W. A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC Genetics. 2004;5:7. doi: 10.1186/1471-2156-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Picard A., Soyer J., Berney X., Tarussio D., Quenneville S., Jan M. A genetic screen identifies hypothalamic Fgf15 as a regulator of glucagon secretion. Cell Reports. 2016;17(7):1795–1806. doi: 10.1016/j.celrep.2016.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diessler S., Jan M., Emmenegger Y., Guex N., Middleton B., Skene D.J. A systems genetics resource and analysis of sleep regulation in the mouse. PLoS Biology. 2018;16(8) doi: 10.1371/journal.pbio.2005750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franken P., Chollet D., Tafti M. The homeostatic regulation of sleep need is under genetic control. Journal of Neuroscience. 2001;21(8):2610–2621. doi: 10.1523/JNEUROSCI.21-08-02610.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keller M.P., Rabaglia M.E., Schueler K.L., Stapleton D.S., Gatti D.M., Vincent M. Gene loci associated with insulin secretion in islets from non-diabetic mice. Journal of Clinical Investigation. 2019;130 doi: 10.1172/JCI129143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu Y., Williams E.G., Dubuis S., Mottis A., Jovaisaite V., Houten S.M. Multilayered genetic and omics dissection of mitochondrial activity in a mouse reference population. Cell. 2014;158(6):1415–1430. doi: 10.1016/j.cell.2014.07.039. 1410.1016/j.cell.2014.1407.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonnefond A., Froguel P. Rare and common genetic events in type 2 diabetes: what should biologists know? Cell Metabolism. 2015;21(3):357–368. doi: 10.1016/j.cmet.2014.12.020. [DOI] [PubMed] [Google Scholar]
- 10.Stancakova A., Laakso M. Genetics of type 2 diabetes. Endocrine Development. 2016;31:203–220. doi: 10.1159/000439418. 10.1159/000439418. Epub 000432016 Jan 000439419. [DOI] [PubMed] [Google Scholar]
- 11.Basco D., Zhang Q., Salehi A., Tarasov A., Dolci W., Herrera P. alpha-cell glucokinase suppresses glucose-regulated glucagon secretion. Nature Communications. 2018;9(1):546. doi: 10.1038/s41467-018-03034-0. 510.1038/s41467-41018-03034-41460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Broman K.W., Wu H., Sen S., Churchill G.A. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19(7):889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- 13.Jan M., Gobet N., Diessler S., Franken P., Xenarios I. A multi-omics digital research object for the genetics of sleep regulation. Scientific Data. 2019;6(1):258. doi: 10.1038/s41597-019-0171-x. 210.1038/s41597-41019-40171-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tingley D., Yamamoto T., Hirose K., Keele L., Imai K. mediation:RPackage for causal mediation analysis. Journal of Statistical Software. 2014;59(5) [Google Scholar]
- 15.Lilla V., Webb G., Rickenbach K., Maturana A., Steiner D.F., Halban P.A. Differential gene expression in well-regulated and dysregulated pancreatic beta-cell (MIN6) sublines. Endocrinology. 2003;144(4):1368–1379. doi: 10.1210/en.2002-220916. [DOI] [PubMed] [Google Scholar]
- 16.Salmon P., Trono D. Production and titration of lentiviral vectors. Current Protocol Neuroscience. 2006;4 doi: 10.1002/0471142301.ns0421s37. Unit 4 21. [DOI] [PubMed] [Google Scholar]
- 17.Barczak W., Suchorska W., Rubis B., Kulcenty K. Universal real-time PCR-based assay for lentiviral titration. Molecular Biotechnology. 2015;57(2):195–200. doi: 10.1007/s12033-014-9815-4. 110.1007/s12033-12014-19815-12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thorens B., Tarussio D., Maestro M.A., Rovira M., Heikkila E., Ferrer J. Ins1(Cre) knock-in mice for beta cell-specific gene recombination. Diabetologia. 2015;58(3):558–565. doi: 10.1007/s00125-014-3468-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muoio D.M., Noland R.C., Kovalik J.P., Seiler S.E., Davies M.N., DeBalsi K.L. Muscle-specific deletion of carnitine acetyltransferase compromises glucose tolerance and metabolic flexibility. Cell Metabolism. 2012;15(5):764–777. doi: 10.1016/j.cmet.2012.04.005. 710.1016/j.cmet.2012.1004.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andreux P.A., Williams E.G., Koutnikova H., Houtkooper R.H., Champy M.F., Henry H. Systems genetics of metabolism: the use of the BXD murine reference panel for multiscalar integration of traits. Cell. 2012;150(6):1287–1299. doi: 10.1016/j.cell.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noland R.C., Koves T.R., Seiler S.E., Lum H., Lust R.M., Ilkayeva O. Carnitine insufficiency caused by aging and overnutrition compromises mitochondrial performance and metabolic control. Journal of Biological Chemistry. 2009;284(34):22840–22852. doi: 10.1074/jbc.M109.032888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seiler S.E., Martin O.J., Noland R.C., Slentz D.H., DeBalsi K.L., Ilkayeva O.R. Obesity and lipid stress inhibit carnitine acetyltransferase activity. The Journal of Lipid Research. 2014;55(4):635–644. doi: 10.1194/jlr.M043448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sugden M.C., Holness M.J. Mechanisms underlying regulation of the expression and activities of the mammalian pyruvate dehydrogenase kinases. Archives of Physiology and Biochemistry. 2006;112(3):139–149. doi: 10.1080/13813450600935263. [DOI] [PubMed] [Google Scholar]
- 24.Boland B.B., Rhodes C.J., Grimsby J.S. The dynamic plasticity of insulin production in beta-cells. Molecular Metabol. 2017;6(9):958–973. doi: 10.1016/j.molmet.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rhodes C.J., Lucas C.A., Halban P.A. Glucose stimulates the biosynthesis of rat I and II insulin to an equal extent in isolated pancreatic islets. FEBS Letters. 1987;215(1):179–182. doi: 10.1016/0014-5793(87)80137-0. [DOI] [PubMed] [Google Scholar]
- 26.Alarcon C., Wicksteed B., Prentki M., Corkey B.E., Rhodes C.J. Succinate is a preferential metabolic stimulus-coupling signal for glucose-induced proinsulin biosynthesis translation. Diabetes. 2002;51(8):2496–2504. doi: 10.2337/diabetes.51.8.2496. [DOI] [PubMed] [Google Scholar]
- 27.Wong N., Morahan G., Stathopoulos M., Proietto J., Andrikopoulos S. A novel mechanism regulating insulin secretion involving Herpud1 in mice. Diabetologia. 2013;56(7):1569–1576. doi: 10.1007/s00125-013-2908-y. [DOI] [PubMed] [Google Scholar]
- 28.Aston-Mourney K., Wong N., Kebede M., Zraika S., Balmer L., McMahon J.M. Increased nicotinamide nucleotide transhydrogenase levels predispose to insulin hypersecretion in a mouse strain susceptible to diabetes. Diabetologia. 2007;50(12):2476–2485. doi: 10.1007/s00125-007-0814-x. 2410.1007/s00125-00007-00814-x. Epub 02007 Oct 00126. [DOI] [PubMed] [Google Scholar]
- 29.Kowluru A. Tiam1/Vav2-Rac1 axis: a tug-of-war between islet function and dysfunction. Biochemical Pharmacology. 2017;132:9–17. doi: 10.1016/j.bcp.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Veluthakal R., Tunduguru R., Arora D.K., Sidarala V., Syeda K., Vlaar C.P. VAV2, a guanine nucleotide exchange factor for Rac1, regulates glucose-stimulated insulin secretion in pancreatic beta cells. Diabetologia. 2015;58(11):2573–2581. doi: 10.1007/s00125-015-3707-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elrick M.M., Samson W.K., Corbett J.A., Salvatori A.S., Stein L.M., Kolar G.R. Neuronostatin acts via GPR107 to increase cAMP-independent PKA phosphorylation and proglucagon mRNA accumulation in pancreatic alpha-cells. American Journal of Physiology - Regulatory, Integrative and Comparative Physiology. 2016;310(2):R143–R155. doi: 10.1152/ajpregu.00369.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samson W.K., Zhang J.V., Avsian-Kretchmer O., Cui K., Yosten G.L., Klein C. Neuronostatin encoded by the somatostatin gene regulates neuronal, cardiovascular, and metabolic functions. Journal of Biological Chemistry. 2008;283(46):31949–31959. doi: 10.1074/jbc.M804784200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakaya N., Sultana A., Munasinghe J., Cheng A., Mattson M.P., Tomarev S.I. Deletion in the N-terminal half of olfactomedin 1 modifies its interaction with synaptic proteins and causes brain dystrophy and abnormal behavior in mice. Experimental Neurology. 2013;250:205–218. doi: 10.1016/j.expneurol.2013.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reichenbach A., Stark R., Mequinion M., Denis R.R.G., Goularte J.F., Clarke R.E. AgRP neurons require carnitine acetyltransferase to regulate metabolic flexibility and peripheral nutrient partitioning. Cell Reports. 2018;22(7):1745–1759. doi: 10.1016/j.celrep.2018.01.067. 1710.1016/j.celrep.2018.1701.1067. [DOI] [PubMed] [Google Scholar]
- 35.Kulkarni R.N., Almind K., Goren H.J., Winnay J., Ueki K., Okada T. Impact of genetic background on development of hyperinsulinemia and diabetes in insulin receptor/insulin receptor substrate-1 double heterozygous mice. Diabetes. 2003;52(6):1528–1534. doi: 10.2337/diabetes.52.6.1528. [DOI] [PubMed] [Google Scholar]
- 36.Marsh B.J., Soden C., Alarcon C., Wicksteed B.L., Yaekura K., Costin A.J. Regulated autophagy controls hormone content in secretory-deficient pancreatic endocrine beta-cells. Molecular Endocrinology. 2007;21(9):2255–2269. doi: 10.1210/me.2007-0077. [DOI] [PubMed] [Google Scholar]
- 37.Schuit F.C., In't Veld P.A., Pipeleers D.G. Glucose stimulates proinsulin biosynthesis by a dose-dependent recruitment of pancreatic beta cells. Proceeding of the National Academy of Sciences USA. 1988;85:3865–3869. doi: 10.1073/pnas.85.11.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skelly R.H., Bollheimer L.C., Wicksteed B.L., Corkey B.E., Rhodes C.J. A distinct difference in the metabolic stimulus-response coupling pathways for regulating proinsulin biosynthesis and insulin secretion that lies at the level of a requirement for fatty acyl moieties. Biochemical Journal. 1998;331(Pt 2):553–561. doi: 10.1042/bj3310553. 510.1042/bj3310553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seino S., Shibasaki T., Minami K. Dynamics of insulin secretion and the clinical implications for obesity and diabetes. Journal of Clinical Investigation. 2011;121(6):2118–2125. doi: 10.1172/JCI45680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoboth P., Muller A., Ivanova A., Mziaut H., Dehghany J., Sonmez A. Aged insulin granules display reduced microtubule-dependent mobility and are disposed within actin-positive multigranular bodies. Proceedings of the National Academy of Sciences of the USA. 2015;112(7):E667–E676. doi: 10.1073/pnas.1409542112. 610.1073/pnas.1409542112. Epub 1409542015 Feb 1409542112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muller A., Mziaut H., Neukam M., Knoch K.P., Solimena M. A 4D view on insulin secretory granule turnover in the beta-cell. Diabetes, Obesity and Metabolism. 2017;19(Suppl 1):107–114. doi: 10.1111/dom.13015. 110.1111/dom.13015. [DOI] [PubMed] [Google Scholar]
- 42.Zhu D., Xie L., Karimian N., Liang T., Kang Y., Huang Y.C. Munc18c mediates exocytosis of pre-docked and newcomer insulin granules underlying biphasic glucose stimulated insulin secretion in human pancreatic beta-cells. Molecular Metabol. 2015;4(5):418–426. doi: 10.1016/j.molmet.2015.02.004. 410.1016/j.molmet.2015.1002.1004. eCollection 2015 May. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gaisano H.Y. Here come the newcomer granules, better late than never. Trends in Endocrinology and Metabolism. 2014 doi: 10.1016/j.tem.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 44.Fritsche A., Stefan N., Hardt E., Schutzenauer S., Haring H., Stumvoll M. A novel hyperglycaemic clamp for characterization of islet function in humans: assessment of three different secretagogues, maximal insulin response and reproducibility. European Journal of Clinical Investigation. 2000;30(5):411–418. doi: 10.1046/j.1365-2362.2000.00649.x. 410.1046/j.1365-2362.2000.00649.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.