Sir,
Surveillance studies of ESBL-producing Escherichia coli have identified a globally disseminated high-risk clone named ST131, with strains belonging to three clades (A, B and C) and three different subclades (C1, C1-M27 and C2). While C2 is associated with CTX-M-15, clade C1-M27 has been associated with CTX-M-27.1 Nowadays, the MDR and CTX-M-27-producing ST131-C1 cluster has been considered a novel epidemic clone.1–3 In South America, neither blaCTX-M-27 nor E. coli ST131 C1-M27 have been reported so far.
During a local surveillance study conducted to monitor the presence of WHO critical priority pathogens in impacted marine ecosystems, brown mussels (Perna perna) and oysters (Crassostrea spp.) were collected from 14 near-shore sites located at different distances from the port of Santos (the largest port of Latin America). Mussel (n = 10) and oyster (n = 10) samples, collected from each site, were placed into sterile plastic bags. The samples were kept refrigerated and processed within 3 h after collection. Following standard methods for the examination, 25 g of bivalves were distributed in sterile plastic bags containing 225 mL of Brain Heart Infusion broth and incubated at 37°C for 24 h. Subsequently, the samples were streaked onto MacConkey agar plates supplemented with ceftriaxone (2 mg/L), meropenem (2 mg/L) or colistin (2 mg/L), following incubation at 37°C for 24 h.
Two ceftriaxone-resistant E. coli isolates were recovered from mussel (E. coli 6M) and oyster (E. coli MO) samples collected from two different sites (23.987125S, 46.308609 W and 23.976040S, 46.372580 W) close to the port. Antimicrobial susceptibility testing, performed by disc diffusion and/or Etest methods,4,5 revealed that both strains were resistant to amoxicillin/clavulanic acid, aztreonam, trimethoprim/sulfamethoxazole, ceftiofur (>32 mg/L), ceftazidime (>32 mg/L), cefotaxime (>32 mg/L) and tetracycline. Additionally, E. coli MO was resistant to nalidixic acid (>32 mg/L) and ciprofloxacin (>4 mg/L). PCR screening and Sanger sequencing revealed that these isolates were positive for the blaCTX-M-27 ESBL gene.
E. coli strains were subjected to WGS using the Illumina NextSeq (2 × 150 bp) platform (Illumina, USA). De novo assemblies were performed using Spades v. 3.11. WGS data were analysed using bioinformatics tools available from the Center for Genomic Epidemiology (www.cge.dtu.dk).
E. coli 6M (accession number: NCWA00000000.1) belonged to serotype O86:H18 and sequence type ST38/CC38, whereas E. coli MO (accession number: NCVZ00000000.1) belonged to serotype O25b:H4 and ST131/CC131. These STs have been globally disseminated among humans, animals and aquatic environments, being commonly associated with CTX-M variants.1,2,6–10 Both strains belonged to the high-virulence phylogenetic group B2. In this regard, virulome analysis of E. coli 6M revealed the presence of iss (increased serum survival), astA (EAST-1 toxin), eatA (enterotoxigenic autotransporter A), capU (hexosyltransferase homologue), nfaE (diffuse adherence fibrillar adhesin) and eilA (Salmonella HilA homologue) genes, whereas iha (adherence protein), sat (secreted autotransporter toxin), gad (glutamate decarboxylase), senB (enterotoxin) and iss genes were found in E. coli MO. Moreover, E. coli MO carried fimH30 (associated with ST131) and the C1 subclade-specific prophage-like region (M27PP1).11 In this regard, the virulome content (i.e. iha, sat, gad, iss and senB genes) of E. coli MO was identical to that of other E. coli strains of ST131 and C1-M27 clade.2,3 On the other hand, both strains displayed an identical resistome for aminoglycosides (strA, strB and aadA5), β-lactams (blaCTX-M-27), sulphonamides (sul1 and sul2), trimethoprim (dfrA17) and tetracycline [tet(A)], as previously observed in E. coli of ST131 and C1-M27;2,3 whereas mutations in the quinolone resistance-determining regions of gyrA (Ser83Leu, Asp87Asn), parC (Ser80Ile, Glu84Val) and parE (Ile529Leu) genes were only identified in the E. coli MO strain. FIB and FII, and Col156, FIA, FIB and FII replicon types were identified in E. coli 6M and MO strains, respectively.
Mobilization of plasmids ∼130 kb in size (named pMO and p6M), bearing blaCTX-M-27 genes, was achieved by bacterial transformation using E. coli TOP10. FIB and FII replicons were identified in p6M (FAB formula F2:A−:B10), whereas FIA, FIB and FII replicon types were confirmed in pMO (FAB formula F1:A2:B20). The complete sequence of the pMO plasmid (GenBank accession no. MG886288) was obtained using de novo assembly, followed by gap closure by PCR and Sanger sequencing.
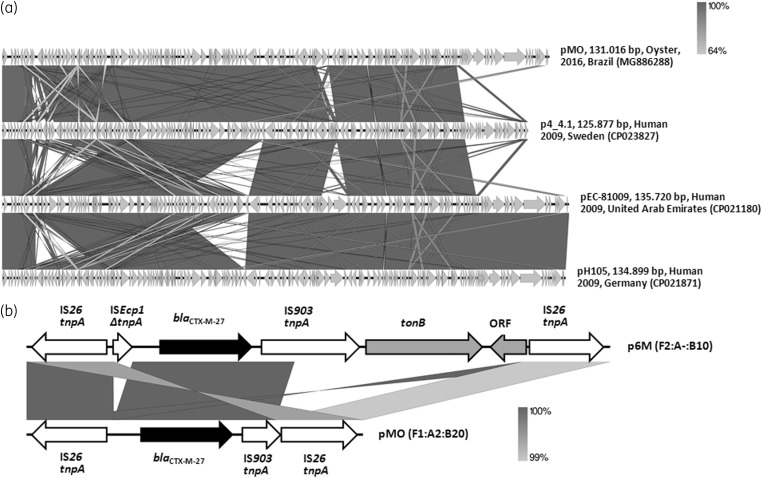
The pMO plasmid was 131 016 bp in length, containing 52.1% GC and 171 coding regions (CDS), of which 129 CDS encoded proteins with known functions (i.e. proteins related to plasmid replication, partition, maintenance, conjugation, toxin–antitoxin systems and antimicrobial resistance). Besides blaCTX-M-27, the pMO plasmid harboured aadA5, sul1, dfrA17, tet(A) and mphA resistance genes, similarly to F1:A2:B20 plasmids harboured by the C1-M27 clade (Figure 1a). In fact, pMO showed a high nucleotide identity (>95%) to other F1:A2:B20 plasmids harboured by CTX-M-27-producing E. coli strains of ST131 and clade C1-M27, identified in European, Asian and North American countries (Figure 1a), which could support intercontinental dissemination of this sort of plasmid.
Figure 1.
(a) Full-length alignment of four F1:A2:B20 plasmids harboured by CTX-M-27-producing E. coli strains of ST131 and clade C1-M27, identified in Brazil (pMO, GenBank accession number: MG886288), Sweden (p4_4.1, 125.877, GenBank accession number: CP023827), the United Arab Emirates (pEC-81009, GenBank accession number: CP021180) and Germany (pH105, 134.899, GenBank accession number: CP021871). Arrows indicate the direction of transcription, whereas the grey bands denote regions of identity. Overall, the identity was more than 95%. (b) Schematic representation of the genetic environment of blaCTX-M-27 genes carried by the pMO and p6M plasmids identified in CTX-M-27-producing E. coli strains, isolated from areas impacted by intensive maritime traffic and transoceanic shipping activities, in Brazil. Arrows indicate the direction of transcription of blaCTX-M-27 genes (black arrow), genes related to mobile elements (white arrows) and tonB (related to energy transduction functions) and ORF genes (grey arrows).
Although analysis of the genetic environment of blaCTX-M-27 genes, carried by both E. coli strains, revealed the presence of IS26 and IS903 mobile elements, E. coli MO presented a truncated IS903 upstream of the blaCTX-M-27 gene, whereas E. coli 6M presented a truncated ISEcp1 downstream of the blaCTX-M-27 gene, and tonB and ORF genes (Figure 1b).
In summary, to our knowledge, we report the first identification of CTX-M-27-producing E. coli strains, of ST131 and clade C1-M27, in Brazil. In this regard, since CTX-M-27-positive E. coli strains were recovered from areas impacted by intensive maritime traffic and transoceanic shipping activities, a possible introduction of international clones via commercial shipping routes could be speculated.12 Another option could be polluted effluents with previously unnoticed presence of CTX-M-27-positive strains. In fact, in Brazil, aquatic environments receiving large quantities of urban wastewater, animal waste and hospital effluents have been recognized as potential sources for the dissemination of CTX-M- and carbapenemase-producing Enterobacterales.13 Therefore, continued monitoring of ESBL-producing E. coli in South American countries remains necessary to elucidate the local epidemiology and dynamics of the transmission of high-risk clades with pandemic potential.
Acknowledgements
We thank Cefar Diagnóstica Ltda. (Brazil) for kindly supplying antibiotic discs for susceptibility testing.
Funding
This work was funded by research grants from Bill & Melinda Gates Foundation (Grand Challenges Explorations Brazil – New approaches to characterize the global burden of antimicrobial resistance, grant OPP1193112); Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) (grant 2016/08593-9; as well as 2015/13527-2 and 2014/11523-7 to M.R.F. and M.P.V.C.); and by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (grants 433128/2018-6 and 443819/2018-1; as well as 312249/2017-9 to N.L.).
Transparency declarations
None to declare.
References
- 1. Peirano G, Pitout J.. Extended-spectrum β-lactamase-producing Enterobacteriaceae: update on molecular epidemiology and treatment options. Drugs 2019; 79: 1529–41. [DOI] [PubMed] [Google Scholar]
- 2. Matsumura Y, Johnson JR, Yamamoto M. et al. CTX-M-27- and CTX-M-14-producing, ciprofloxacin-resistant Escherichia coli of the H30 subclonal group within ST131 drive a Japanese regional ESBL epidemic. J Antimicrob Chemother 2015; 70: 1639–49. [DOI] [PubMed] [Google Scholar]
- 3. Ghosh H, Doijad S, Falgenhauer L. et al. bla CTX-M-27-encoding Escherichia coli sequence type 131 lineage C1-M27 clone in clinical isolates. Emerg Infect Dis 2017; 23: 1754–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.CLSI. Performance Standards for Antimicrobial Susceptibility Testing—Twenty-Ninth Edition: M100 2019.
- 5.CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals—Fifth Edition: VET01 2018.
- 6. Hu YY, Cai JC, Zhou HW. et al. Molecular typing of CTX-M-producing Escherichia coli isolates from environmental water, swine feces, specimens from healthy humans, and human patients. Appl Environ Microbiol 2013; 79: 5988–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guenther S, Semmler T, Stubbe A. et al. Chromosomally encoded ESBL genes in Escherichia coli of ST38 from Mongolian wild birds. J Antimicrob Chemother 2017; 72: 1310–3. [DOI] [PubMed] [Google Scholar]
- 8. Riley LW. Pandemic lineages of extraintestinal pathogenic Escherichia coli. Clin Microbiol Infect 2014; 20: 380–90. [DOI] [PubMed] [Google Scholar]
- 9. Bevan ER, Jones AM, Hawkey PM.. Global epidemiology of CTX-M β-lactamases: temporal and geographical shifts in genotype. J Antimicrob Chemother 2017; 72: 2145–55. [DOI] [PubMed] [Google Scholar]
- 10. Ghosh H, Bunk B, Doijad S. et al. Complete genome sequence of blaCTX-M-27-encoding Escherichia coli strain H105 of sequence type 131 lineage C1/H30R. Genome Announc 2017; 5: e00736–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Birgy A, Bidet P, Levy C. et al. CTX-M-27-producing Escherichia coli of sequence type 131 and clade C1-M27, France. Emerg Infect Dis 2017; 23: 885.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ruiz GM, Rawlings TK, Dobbs FC. et al. Global spread of microorganisms by ships. Nature 2000; 408: 49–50. [DOI] [PubMed] [Google Scholar]
- 13. Conte D, Palmeiro JK, da Silva Nogueira K. et al. Characterization of CTX-M enzymes, quinolone resistance determinants, and antimicrobial residues from hospital sewage, wastewater treatment plant, and river water. Ecotoxicol Environ Saf 2017; 136: 62–9. [DOI] [PubMed] [Google Scholar]