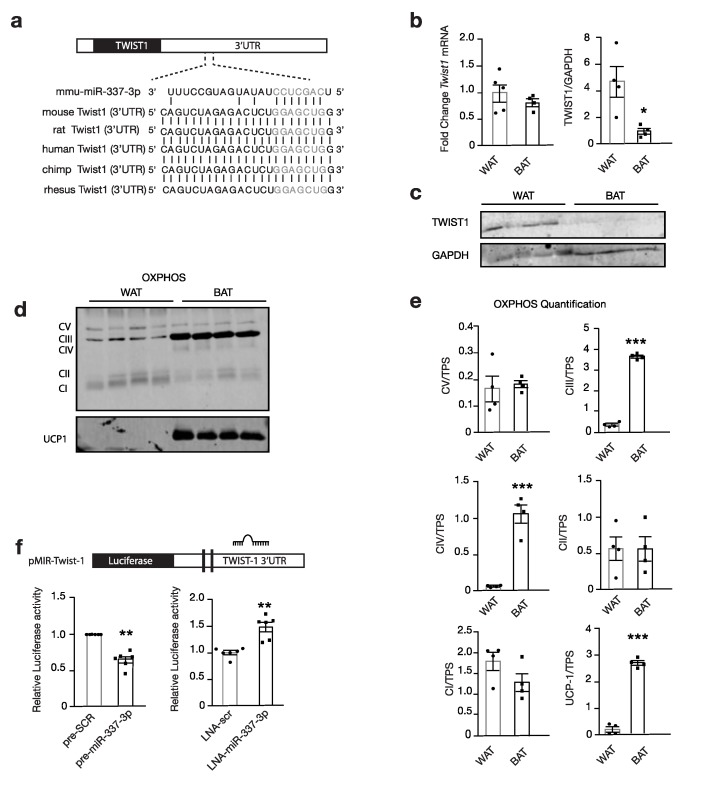
Figure 3.
miR-337-3p targets Twist1. (a) Schematic representation of predicted seed complementarity between the seed sequence of mmu-miR-337-3p, and seed region on Twist1 3′UTR and the evolutionary conservation of this region among species. (b) RT-PCR and Western blot analysis of Twist1 mRNA expression normalized to 5S (WAT n = 5, BAT n = 4) and TWIST1 protein abundance (WAT n = 4, and BAT n = 5) in mouse WAT and BAT normalized to GAPDH. (c) Representative Western blot of TWIST1 and GAPDH housekeeping protein in mouse WAT and BAT quantified in (b). (d) Western blot of mitochondrial OXPHOS, Complex V, IV, III, II, and I and thermogenic UCP1 protein. (e) Quantification of (d) OXPHOS and UCP1 protein abundance in mouse WAT and BAT normalized to TPS as the loading control. (f) Schematic of luciferase reporter plasmid containing Twist’1 3′UTR with a binding site for miR-337-3p. Relative luciferase activity quantification in Cos7 cells containing reporter plasmids, co-transfected with either mmu-miR-337-3p precursor or scrambled precursor, and co-transfected with LNA inhibitor for miR-337-3p or LNA inhibitor negative control. Data are presented as means ± SEM, and individual data points are given by the dot plot * p < 0.05; ** p < 0.01, by Independent sample t-test, equal variances not assumed. *** p < 0.001, by Independent sample t-test, equal variances assumed. n, number of biological replicas; 3′UTR, 3′ untranslated region; OXPHOS, oxidative phosphorylation; UCP1, uncoupling protein-1; GAPDH, Glyceraldehyde 3-phosphate dehydrogenase; Twist1, twist-related protein; RT-PCR, quantitative real-time polymerase chain reaction; TPS, total protein stain.