Abstract
Lung cancer is the most commonly diagnosed cancer worldwide, and metastasis in lung cancer is the leading cause of cancer‐related deaths. Thus, understanding the mechanism of lung cancer metastasis will improve the diagnosis and treatment of lung cancer patients. Herein, we found that expression of cluster of differentiation 109 (CD109) was correlated with the invasive and metastatic capacities of lung adenocarcinoma cells. CD109 is upregulated in tumorous tissues, and CD109 overexpression was associated with tumor progression, distant metastasis, and a poor prognosis in patient with lung adenocarcinoma. Mechanistically, expression of CD109 regulates protein kinase B (AKT)/mammalian target of rapamycin (mTOR) signaling via its association with the epidermal growth factor receptor (EGFR). Inhibition of CD109 decreases EGFR phosphorylation, diminishes EGF‐elicited activation of AKT/mTOR, and sensitizes tumor cells to an EGFR inhibitor. Taken together, our results show that CD109 is a potential diagnostic and therapeutic target in lung cancer patients.
Keywords: CD109, EGFR, lung cancer, metastasis, mTOR
CD109 promotes lung cancer metastasis through promoting EGFR‐AKT‐mTOR signaling and CD109 is an independent prognostic marker for lung adenocarcinoma.
![]()
1. INTRODUCTION
Lung cancer is the most often‐occurring and deadliest cancer worldwide, and it has been ranked first in cancer‐associated deaths globally for over a decade. In the United States, about 220 000 people are diagnosed and over 150 000 people die due to lung cancer each year. 1 Lung cancer is classified into two major categories of small cell lung cancer (SCLC) and non‐small cell lung cancer (NSCLC). SCLC accounts for 15% of all lung carcinomas. SCLC was shown to be more responsive to chemotherapy and radiation therapy; however, the rapid proliferative rate and high recurrence cause the poorest survival among all types of lung carcinomas. NSCLC is comprised of squamous carcinomas, large cell carcinomas, and adenocarcinomas, and it accounts for nearly 85% of lung carcinomas. 2 Although chemotherapy and radiation therapy show responses during early treatment of NSCLC, molecular changes in NSCLC are a major problem that cause resistance and distant metastasis. 3 , 4 Therefore, understanding gene expression patterns and identifying underlying mechanisms will improve diagnoses and treatments for lung cancer.
Cluster of differentiation 109 (CD109) is a glycosylphosphatidylinositol (GPI)‐anchored protein which belongs to the α2‐macroglobulin/C3, C4, and C5 family. CD109 was found to be expressed by endothelial, platelet, and hematopoietic progenitor cells. 5 The physiological function of CD109 in normal cells remains unclear, but it was reported that CD109 is upregulated in a wide variety of malignancies including squamous cell carcinomas, 6 glioblastomas, 7 melanomas, 8 and breast carcinomas. 9 Overexpression of CD109 was correlated with a poor survival probability in hepatocellular carcinoma. 10 Previous studies identified that CD109 acts as a co‐receptor of the transforming growth factor (TGF)‐β receptor (TGF‐βR) which inhibits TGF‐β signaling in keratinocytes. 11 , 12 Additionally, CD109 was reported to regulate epidermal growth factor (EGF) signaling in gliomas, 13 and to participate in the Janus kinase (JAK)/signal transducer and activator of transcription (STAT) axis in lung cancer.14 However, the contribution of CD109 to lung tumorigenesis and its underlying mechanism are largely unknown.
In the present study, we found that CD109 promotes the growth and invasiveness of lung adenocarcinoma cells. CD109 expression was associated with the EGFR and regulated its downstream signaling including the protein kinase B (AKT)/mammalian target of rapamycin (mTOR) axis. Moreover, CD109 participates in the sensitivity to EGFR‐tyrosine kinase inhibitors (TKIs), and CD109 overexpression was correlated with poor survival outcomes in lung adenocarcinoma patients.
2. MATERIALS AND METHODS
2.1. Cell lines and reagents
The human lung cancer cell lines A549, H460, H441, and PC9 were purchased from the Bioresource Collection Research Center (BCRC, Hsinchu, Taiwan) or American Type Culture Collection (ATCC). Cells were maintained in RPMI medium supplemented with 7% fetal bovine serum (FBS), 1% antibiotic‐antimycotic, and 1% GlutaMAX (Gibco, Thermo Fisher Scientific). The cell lines were authenticated through short tandem repeat profiling (GenePrint 10 System). An antibody against CD109 was purchased from Santa Cruz Biotechnology. Antibodies against AKT, phospho‐AKT, EGFR, phospho‐EGFR, mTOR, phospho‐mTOR, and phospho‐70S6K were purchased from Cell Signaling Technologies. Antibodies for α‐tubulin, phospho‐glycogen synthase kinase‐3β (GSK3β), and cyclin D1 were obtained from GeneTex (San Antonio, TX, USA). Recombinant human EGF and gefitinib were obtained from Selleckchem.
2.2. Short hairpin RNA and lentiviral infection
shRNAs for human CD109 (TRCN0000073649 and TRCN0000073650) were obtained from the National RNAi Core Facility (Academia Sinica, Taipei, Taiwan). Lentiviral preparation and viral infection were performed as previously described. 15 In brief, HEK293T cells were cotransfected with pLKO.shRNA together with the pCMV‐∆R8.91 and pMDG plasmids. At 48 hours post‐transfection, virus‐containing supernatants were mixed with polybrene (8 µg/mL) and incubated with target cells for another 48 hours. Transduced cells were selected using puromycin (2 µg/mL). A control shRNA targeting red fluorescent protein (RFP) was used as a negative control.
2.3. Transwell assay
Cells at (1 ~ 4)×105 were seeded in 24‐well transwell inserts (8‐µm pore size, Corning Costar) coated with Matrigel (BD Biosciences) or left uncoated for 24 hours. Cells in the upper wells were removed with a cotton swab, and cells in the lower well were fixed, stained with crystal violet solution, and observed under a microscope.
2.4. Immunoprecipitation and western blotting
Cells were lysed in ice‐cold radioimmunoprecipitation assay (RIPA) buffer supplemented with a protease and phosphatase inhibitor cocktail (Millipore). Whole‐cell extracts were incubated overnight with the indicated antibodies and protein A/G beads (Santa Cruz Biotechnology). The beads were washed three times with RIPA buffer and then boiled in sodium dodecyl sulfate (SDS) loading buffer. For Western blots, cell extracts were separated by SDS‐polyacrylamide gel electrophoresis (PAGE) and transferred to polyvinylidene difluoride membranes. Membranes were blocked with 1% bovine serum albumin/TBST blocking buffer at room temperature for 30 minutes and then incubated overnight at 4°C with specific primary antibodies. The membranes were washed with TBST wash buffer followed by incubation with a horseradish peroxidase‐conjugated secondary antibody at room temperature for 1 hour. Bands were detected with an enhanced chemiluminescence (ECL) system (Millipore). Western blotting was performed at least three times, and representative experiments are shown. Quantification was carried out using ImageJ software.
2.5. Phosphoprotein array analysis
Cells (107) were washed with cold phosphate‐buffered saline (PBS) and incubated with lysis buffer containing protease and phosphatase inhibitor cocktails (Millipore). Protein lysates were then subjected to a Human and Mouse MAPK (mitogen‐activated protein kinase) Pathway Phosphorylation Array C1 (Ray‐Bio) according to the manufacturer's instruction. Protein expression was visualized by ECL and detected using ImageQuant LAS4000 (GE Healthcare Life Science). Quantification was carried out using ImageJ software.
2.6. Cell viability
Cells (2 × 104) were seeded in 24‐well plates overnight, and then they were refreshed with complete culture medium. Cell growth was measured with a trypan blue exclusion assay. To evaluate the effect of CD109 on EGFR‐TKI sensitivity, 2 × 105 cells were seeded in 24‐well plates overnight, and then they were refreshed with culture medium containing 3% FBS and different concentrations of gefitinib for 24 or 48 hours of incubation. Cell viability was measured by a trypan blue exclusion assay, and data are expressed as multiples relative to the vehicle (DMSO)‐treated group.
2.7. Real‐time polymerase chain reaction
Target cell total RNA was extracted with a GENzolTM TriRNA Pure kit and reverse‐transcribed with a high‐capacity cDNA conversion kit (Invitrogen). cDNA was amplified with EvaGreen Master Mix (Biotium) in a StepOne Plus Real‐Time PCR system (Applied Biosystems) with specific primers (Table S1). Results were calculated using the ΔΔCT equation and are expressed as multiples of change relative to a control sample. 16
2.8. Immunohistochemistry
A lung cancer tissue microarray (HLug‐Ade060PG‐01) was purchased from Biomax. A slide was deparaffinized, rehydrated, and heated in an antigen‐unmasking solution (BioGenex), and incubated with an anti‐CD109 primary antibody (1:100, Santa Cruz Biotechnology) overnight at 4°C. The slide was then incubated with SignalStain Boost IHC detection reagent (Cell Signaling Technology) followed by staining with 3,3'‐diaminobenzidine (DAB) peroxidase substrate (BioGenex).
2.9. Animal study
All animal studies were performed according to guidelines and with approval of the Animal Care and Use Committee of Taipei Medical University. 2 × 106 A549/shRFP or A549/shCD109 cells were intravenously injected into the tail vein of 8‐week‐old BALB/c nude mice. 17 , 18 , 19 Eight weeks later, the mice were sacrificed, all dissected lungs were embedded in paraffin, and tissue sections were stained with hematoxylin and eosin (H&E) and scanned using TissueFax (TissueGnostics). The metastatic burden was calculated as the percentage of the lung area with tumors. 16
2.10. Bioinformatic analyses
Gene expression patterns of CD109 and its downstream targets, and clinicopathological values from The Cancer Genome Atlas (TCGA) were downloaded from University of California, Santa Cruz (UCSC) Xena browser (https://xenabrowser.net/). A high‐expression group was defined as occurring in >30% of patients. Expressions of CD109 in cancerous and normal tissues were provided by Oncomine (https://www.oncomine.org/). The survival prognosis of different cancer types as a function of CD109 was determined using PrognoScan (https://www.prognoscan.org/). Associations of CD109 with oncogenic signatures in three independent cohorts (GSE31210, GSE37745, and GSE8894) 20 , 21 , 22 were analyzed using a gene set enrichment analysis (GSEA) algorithm.
2.11. Statistical analyses
Data are presented as the mean ± standard error (SE) of three independent experiments. Statistical significance was determined by an unpaired, two‐tailed Student's t‐test unless stated otherwise. *P < .05; **P < .01. A correlation coefficient was analyzed by the Pearson test. The survival probability was plotted by Kaplan‐Meier and analyzed by a log‐rank (Mantel‐Cox) statistical test. All statistical analyses were carried out using GraphPad Prism software.
3. RESULTS
3.1. CD109 is upregulated and associated with aggressiveness of lung cancer cells
In order to characterize the role of CD109 in lung tumorigenesis, we analyzed CD109 expression in a panel of lung cancer cell lines, and results showed that CD109 was expressed in A549, H460, and PC9 cells. Interestingly, CD109 expression was closely associated with the migratory capacity of lung cancer cells (Figure 1A, left panel, Figure 1B). In support of our findings that CD109 is upregulated in CL1‐5 cells, which is an invasive subline of CL1‐3 cells 23 (Figure 1A, right panel), these data suggested that CD109 may play a role in lung cancer mobility. We further knocked‐down CD109 in A549, CL1‐5, and H460 cells (Figure 1C, Figure S1A), and results showed that silencing of CD109 significantly downregulated the migratory and invasive capacities (Figure 1D, Figure S1B), and substantially decreased the growth of tumor cells (Figure 1E, Figure S1C). Conversely, ectopic expression of CD109 enhanced migration and invasion in PC9 cells (Figure S2A and B).
Figure 1.
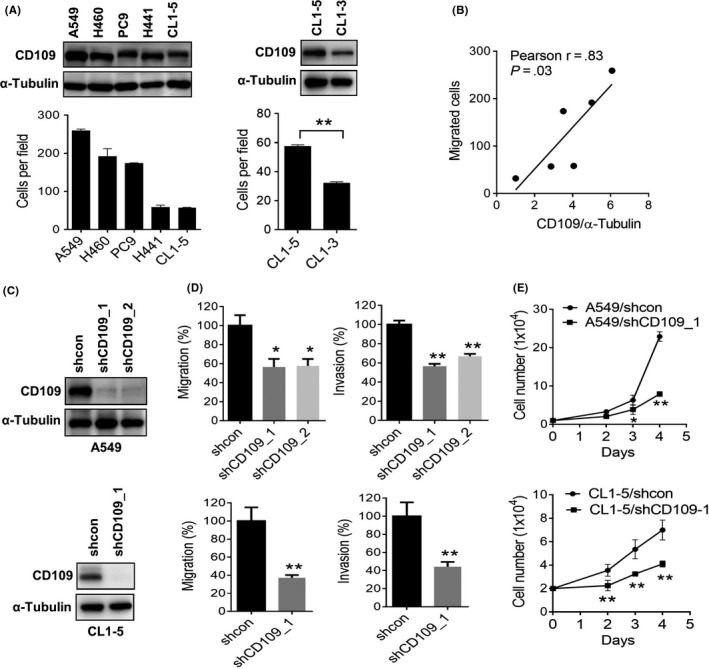
Expression of CD109 is associated with aggressiveness in lung cancer cells. A, Western blot analysis of CD109 expression in lung cancer cell lines (upper panel). The migratory capacity of lung cancer cell lines was evaluated by transwell assay (lower panel). B, Pearson correlation analysis showed a positive correlation of CD109 with the migratory capacity of lung cancer cells. C, Western blot analysis of the knockdown efficiency of CD109 shRNA‐transduced A549 and CL1‐5 cells. D, Transwell analysis of cell migration (left panel) and invasion (right panel) in CD109‐knockdown A549 and CL1‐5 cells. Data are presented as the percentage relative to the control group. E, Trypan blue exclusion analysis of cell growth in CD109‐knockdown A549 and CL1‐5 cells. *P < .05; **P < .01, as determined by an unpaired t‐test. Error bars indicate the mean ± SE from at least three independent replicates
3.2. Overexpression of CD109 is associated with poor clinicopathological and survival outcomes in lung adenocarcinoma patients
To validate that CD109 is an independent biomarker in lung cancer, we analyzed the expression of CD109 in tumorous and adjacent normal tissues by immunohistochemistry. Results showed that CD109 was overexpressed in approximately 40% of lung cancer patients (n = 30) (Figure 2A). Consistent results were identified at the transcriptional level from two other independent studies which reported that CD109 was significantly elevated in lung cancer tissues (Figure 2B). Importantly, CD109 expression was associated with aggressive clinicopathological features, including the presence of lymph node metastasis (P = .001), and more‐advanced tumor grades (P < .001) and stages (P < .001) (Figure 2C). Moreover, high CD109 expression was associated with low overall survival rates in lung adenocarcinoma patients (hazard ratio (HR) = 2.40, P < .001) but not in squamous cell carcinoma patients (Figure 2D, Figure S3). Additionally, high CD109 expression was found to be associated with worse recurrence‐free probabilities in adenocarcinoma patients (HR = 1.66, P = .023), but not in squamous cell carcinoma patients (Figure 2D). Furthermore, the association of CD109 with a poor survival prognosis was found in lung cancer, as well as in bladder and colon cancers (Figure 2E).
Figure 2.
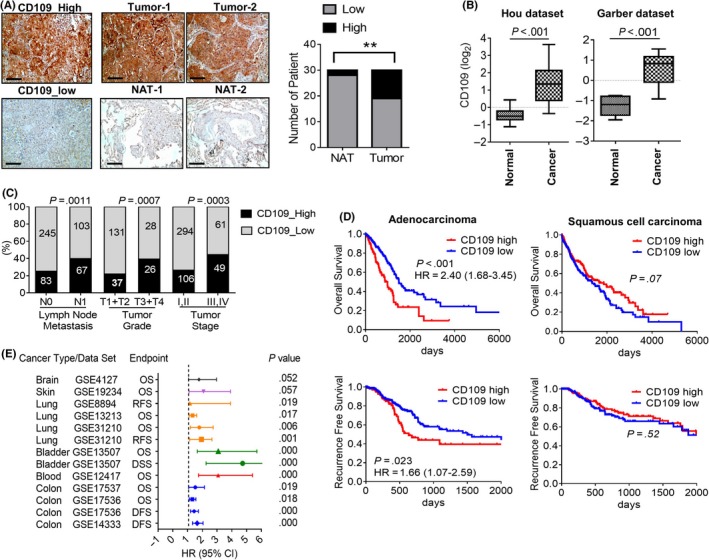
Clinicopathological associations of CD109 in lung cancer patients. A, IHC analysis of CD109 in cancerous and normal adjacent tissues (NATs) in lung cancer patients (n = 30). Representative images show high and low staining of CD109 in tumor sections (left panel). Statistical analyses were performed with a Chi‐squared test (right panel). Representative images are shown from three patients. Scale bar = 100 µm. B, Transcriptional levels of CD109 expression in normal and lung tumor tissues from the Hou and Garber datasets. C, High CD109 expression was associated with lymph node metastasis, grade, stage, and poor survival prognosis (D) in lung cancer patients using TCGA dataset. Clinicopathological correlations were examined by a Chi‐squared test, and the survival prognosis was calculated by Kaplan‐Meier analysis. E, Survival outcomes of CD109 in different cancer types. OS, overall survival; DFS, disease‐free survival; RFS, recurrence‐free survival; HR, hazard ratio
3.3. CD109 regulates the ATK/mTOR signaling cascade
To elucidate the mechanism underlying CD109‐promoted tumor invasiveness, a protein kinase array was employed in mock‐ and CD109‐silenced A549 cells. Results showed that the AKT signaling cascade, including mTOR, 70S6K, and GSK3β, was downregulated after knocking‐down CD109 (Figure 3A). Consistent results were obtained by Western blotting that CD109 silencing suppressed phosphorylation of AKT, mTOR, and 70S6K in A549, H460, and CL1‐5 cells (Figure 3B upper panel). Conversely, overexpression of CD109 increased phosphorylation of AKT and mTOR (Figure S2C). Likewise, GSK3β, an AKT substrate, and its downstream target, cyclin D1, were downregulated in CD109‐knockdown lung cancer cells (Figure 3B lower panel). Moreover, we analyzed associations of CD109 with oncogenic signatures in lung cancer patients using GSEA. The intersection of results from three independent cohorts revealed that overexpression of CD109 was particularly correlated with signatures associated with the vascular endothelial growth factor (VEGF), SRC, platelet‐derived growth factor (PDGF), and mTOR (Figure 3C and D). These data suggest that the AKT/mTOR pathway is major downstream signaling regulated by CD109.
Figure 3.
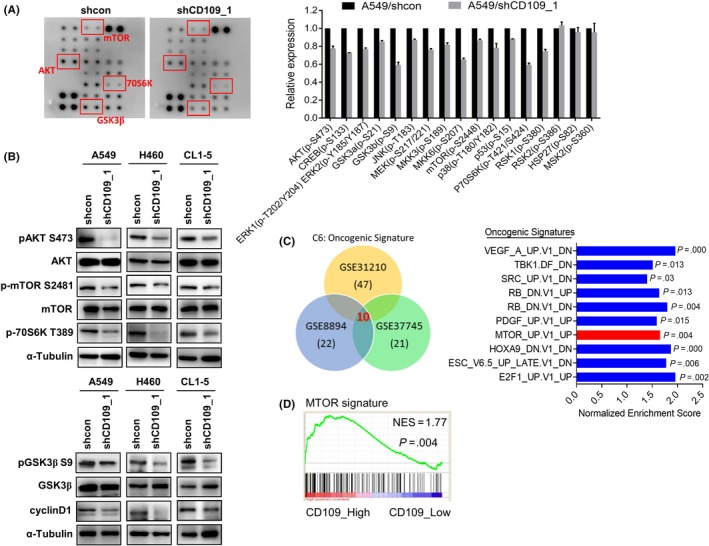
Expression of CD109 is associated with AKT/mammalian target of rapamycin (mTOR) signaling. A, Blot images from a Human Phospho‐Kinase array performed in A549/shcon and A549/shCD109 cells (left panel). Densitometry analyses were normalized to multiples relative to the sh‐control group (right). B, Western blot analysis of AKT signaling molecules in control and CD109‐knockdown lung cancer cell lines. C, Gene set enrichment analysis (GSEA) of CD109‐associated oncogenic signatures in lung cancer datasets (left panel). Intersecting results are depicted in the right panel, and mTOR signaling is shown in red. D, Representative GSEA showing a correlation of CD109 with the mTOR signature. NES, net enrichment score
3.4. CD109 is associated with the EGFR and dominant resistance to EGFR‐TKI therapy
The EGFR plays a critical role in lung cancer progression and shows targetable benefits in lung cancer patients. A previous study reported that CD109 regulates EGFR activity in gliomas, and our aforementioned data identified that suppression of CD109 decreased AKT/mTOR signaling. Because CD109 lacks an intracellular domain, we wondered whether CD109 is associated with the EGFR and cooperates in its downstream signaling. Coimmunoprecipitation assay showed an association between the EGFR and CD109 in A549 cells (Figure 4A). In addition, phosphorylation of the EGFR decreased in CD109‐knockdown A549 cells; however, the total EGFR level was not affected (Figure 4B). We hypothesized that the presence of CD109 might enhance activation of the EGFR towards its ligand, including EGF. Indeed, inhibition of CD109 diminished EGF‐induced phosphorylation of the EGFR and the downstream AKT (Figure 4C). Moreover, we also found that suppression of CD109 sensitized gefitinib‐mediated downregulation of AKT/mTOR signaling both with and without stimulation by EGF (Figure 4D and E). To test whether CD109 contributes to gefitinib resistance, A549 and CL1‐5 cells were treated with different concentrations of gefitinib for 48 hours. A549 cells harbor a KRAS mutation which confers resistance to EGFR‐TKIs; in contrast, CL1‐5 cells exhibit wild‐type KRAS and are more sensitive to gefitinib. Interestingly, CD109 knockdown increased the susceptibility towards gefitinib in both A549 and CL1‐5 cells (Figure 4F). To further evaluate clinical applications of CD109 in patients with lung cancer, we examined the role of CD109 in PC9 cells that harbor an EGFR‐activating mutation (exon 19 deletion). Results showed that inhibition of CD109 increased sensitivity of EGFR‐AKT signaling and cell viability in response to gefitinib (Figure 4G and H). These data indicate that CD109 plays a crucial role in sensitivity of lung cancer cells to EGFR‐TKIs.
Figure 4.
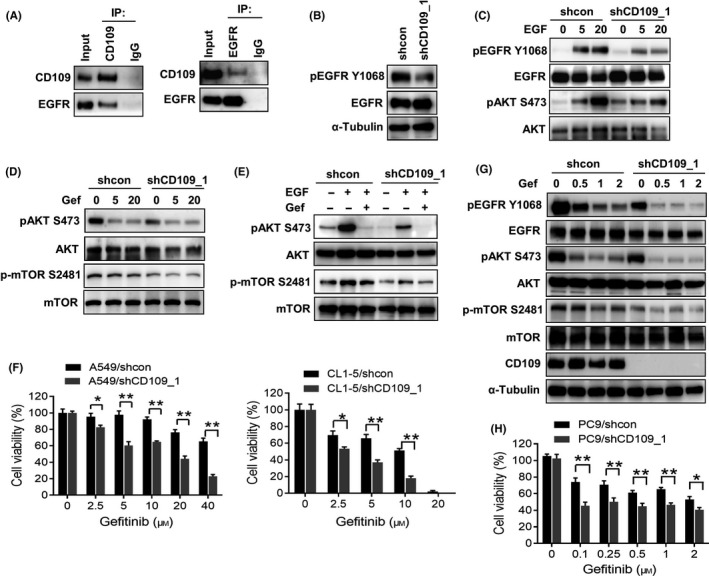
CD109 is associated with EGFR and confers resistance to EGFR‐tyrosine kinase inhibitor. A, Coimmunoprecipitation analysis of the association between CD109 and the EGFR in A549 cells. B, Western blot analysis of phosphorylated EGFR in control and CD109‐silenced A549 cells. C, Suppression of CD109 blocked the EGFR signaling cascade. A549/shcon and A549/shCD109 cells were treated with the EGF (25 ng/mL) for different time periods, and phosphorylation of the EGFR and AKT was analyzed by Western blotting. D, A549/shcon and A549/shCD109 cells were treated with different concentrations of gefitinib (0, 5, and 20 µmol/L), and phosphorylation of AKT and mTOR was analyzed by Western blotting. E, A549 cells were treated with the EGF (25 ng/mL) in the presence of gefitinib (20 µmol/L). Cell lysates were subjected to Western blotting using specific antibodies. F, Silencing of CD109 increased the sensitivity of A549 and CL1‐5 cells to gefitinib. A549 and CL1‐5 cells were treated with different concentrations of gefitinib for 48 h, and cell viability was determined by trypan blue exclusion assay. G, Western blot analysis of the EGFR signaling cascade in PC9/shcon and PC9/shCD109 cells in response to gefitinib for 2 h. H, PC9 cells were treated with different concentrations of gefitinib for 48 h, and cell viability was determined by trypan blue exclusion assay. Data are presented as the percentage relative to the control. *P < .05; **P < .01, as determined by an unpaired t‐test. Error bars indicate the mean ± SE of at least three independent replicates
3.5. Identification of CD109‐regulated genes and clinical significance in lung cancer patients
To further identify downstream genes that are regulated by CD109, we analyzed differentially expressed genes (DEGs) in mock‐ and CD109‐silenced A549 cells. Gene ontology revealed that CD109 regulated genes associated with inflammatory and wound‐healing responses (Figure 5A). Accordingly, we also found that the responses to phosphatidylinositol 3‐kinase signaling and growth factor stimulus were altered after knockdown of CD109 (Figure 5A). These data echoed findings of the downregulation of AKT/mTOR/GSK3β signaling by CD109 silencing (Figure 3). Moreover, we analyzed CD109‐regulated tumorigenic signatures by GSEA, and results revealed that genes associated with cell proliferation and invasion were downregulated by CD109 (Figure 5B). We selected genes which coexisted in the two datasets, and their downregulation by CD109 inhibition was further confirmed by quantitative PCR analysis (Figure 5C). We further identified that most of these genes, including ASPM, CDK1, CCNB2, CENPA, CENPF, DTL, MAD2L1, NEK2, RRM2, and TYMS, were positively associated with CD109 and with a poor survival prognosis in lung cancer patients (Figure 5D). Moreover, expressions of CD109‐associated gene signatures by A549 cells were confirmed to be downregulated in response to an AKT inhibitor (Figure 5E), and treatment with an AKT inhibitor blocked CD109‐promoted migration and invasion in PC9 cells (Figure S2D).
Figure 5.
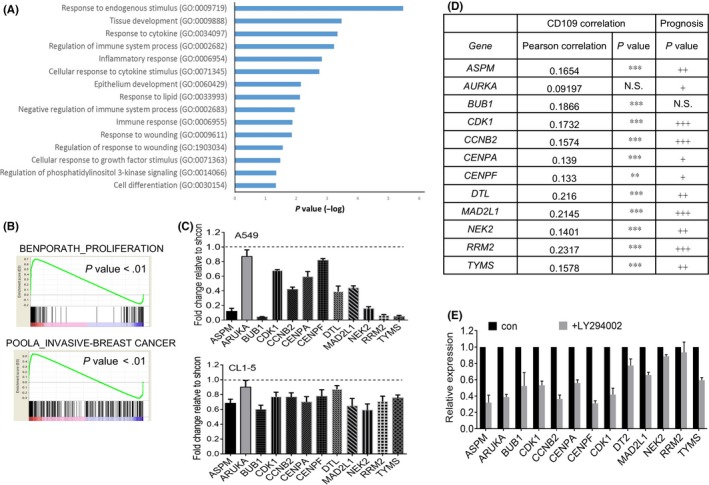
Expression of CD109 regulates the proliferation and invasion of genes. A, Gene ontology analysis of CD109‐regulated genes. B, Gene set enrichment analysis (GSEA) plots of proliferation and invasion signatures based on CD109‐downregulated genes in A549 cells. C, Real‐time PCR analyses of CD109‐regulated genes in A549 and CL1‐5 cells. Data are presented as multiples of change relative to the shcontrol group. D, Associations of CD109 and downstream genes and their prognostic values in TCGA lung cancer patients. Correlations were determined using Pearson's test. **P < .01; ***P < .001. The survival probability was analyzed by a log‐rank (Mantel‐Cox) statistical test. # P < .05; ## P < .01; ### P < .001. NS, no significant difference. E, A549 cells were treated with an AKT inhibitor (LY294002; 20 µmol/L) for 24 h, and expressions of CD109‐associated genes were measured by real‐time PCR analysis. Error bars indicate the mean ± SE for at least three independent replicates
3.6. Inhibition of CD109 suppresses tumor metastasis in mice
To validate the contribution of CD109 to lung cancer metastasis, nude mice were intravenously injected with mock‐ or CD109‐silenced A549 cells for 8 weeks. Results showed that suppression of CD109 effectively reduced the metastatic burden, compared to mock‐transduced cells (Figure 6A and Figure S4). Moreover, immunohistochemistry analysis showed that expressions of CD109 and phospho‐EGFR were concomitantly downregulated in shCD109 tumor sections (Figure 6B). These data support the notion that CD109 synergizes EGFR signaling and contributes to tumor metastasis.
Figure 6.
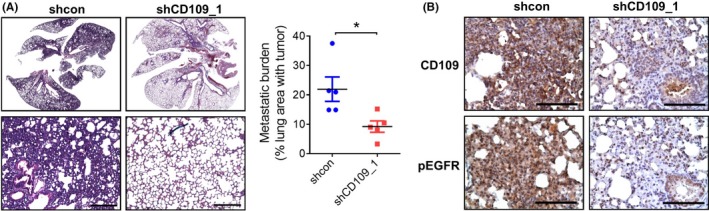
Knockdown of CD109 suppresses in vivo tumor metastasis. A, Images of lung tissues stained with H&E 8 wks after tumor inoculation in nude mice (n = 5 mice per group). High‐power magnification fields are shown in the lower panel. Scale bar = 200 µm. The metastatic burden was calculated as the percentage of the lung area with tumors. *P < .05 (right panel). B, IHC analysis of CD109 and phosphorylated EGFR in lung tumor sections. Scale bar = 50 µm
4. DISCUSSION
CD109 belongs to the α2‐macroglobulin/C3, C4, and C5 family which has the ability to bind with cytokines and growth factors. Previous studies widely reported that platelet CD109 carries the biallelic platelet‐specific alloantigen, Gova/b, which is responsible for neonatal alloimmune thrombocytopenia. 24 , 25 In addition, CD109 binds the TGF‐βR and glucose‐regulated protein 78 (GRP78) and promotes lysosomal degradation of the TGF‐βR by inducing Smad7 and Smurf2, resulting in blocking of the transduction of TGF‐β signaling. 26 , 27 Moreover, previous studies found that soluble CD109, which is cleaved by furinase, can associate with TGF‐β to negatively regulate TGF‐β signaling as well. 28 However, inhibition of TGF‐β signaling by CD109 cannot fully explain the role of CD109 in tumors, because TGF‐β is considered an inducer of the epithelial‐mesenchymal transition by tumor cells. Interestingly, Sunagawa et al identified that CD109 plays a crucial role in skin tumorigenesis that is involved in suppression of TGF‐β, as its antigrowth activity against normal cells. 29 Additionally, Chuang and colleagues found that CD109 is a metastatic driver in lung cancer via regulating JAK/signal transducer and activator of transcription 3 (STAT3) signaling. 14 In our study, we showed that CD109 was associated with the EGFR and enhanced EGFR downstream signaling including the AKT/mTOR axis. Contrarily, suppression of CD109 decreased phosphorylation of the EGFR but had no effect on the EGFR protein level.
EGFR‐TKIs show therapeutic benefits in cancer patients with activated mutation of the EGFR. However, a secondary mutation in the EGFR (T790M) or a mutation of KRAS results in acquired resistance to EGFR‐TKIs. 30 , 31 Additionally, it was also reported that lipid raft localization of the EGFR participates in sensitivity to EGFR‐TKI‐induced growth inhibition. The EGFR localizes to plasma membrane lipid rafts in EGFR‐TKI‐insensitive breast tumor cell lines. 32 In our study, we found that CD109 expression plays a crucial role in EGFR‐TKI sensitivity regardless of whether in KRAS wild‐type or mutant lung tumor cells. Additionally, suppression of CD109 increased sensitivity towards EGFR‐TKI‐sensitive tumor cells. Because CD109 is anchored in lipid rafts via glycophosphatidylinositol (GPI), and because we found that CD109 is associated with and regulates EGFR signaling, we speculated that the involvement of lipid rafts in EGFR‐TKI sensitivity may at least in part occur due to CD109, and that deserves more study and clarification. The involvement of CD109 in a secondary mutation of EGFR‐TKIs in lung cancer cells also needs to be clarified.
CD109 was found to be upregulated in squamous cell carcinoma tissues 33 ; however, we examined the CD109 expression level in a panel of lung cancer cell lines in the Cancer Cell Line Encyclopedia (CCLE) database, and there was no obvious upregulation of CD109 between cells lines of squamous cell carcinoma and adenocarcinoma origin (Figure S5). In the present study, we found that high CD109 expression was associated with a poor prognostic value in adenocarcinomas, but not in squamous cell carcinoma, indicating that CD109 expression plays a more‐important role in tumorigenesis of adenocarcinomas. Our data were consistent with previous findings that CD109 is the strongest predictor of metastasis and survival in patients with lung adenocarcinoma. 14 Interestingly, it is well cited that tobacco smoking is a major risk factor for lung squamous cell carcinoma. 34 We speculated that inflammatory factors from the tumor microenvironment may play a role in regulating CD109 in squamous cell carcinoma, and molecular function of CD109 in squamous cell carcinoma needs to be exploited. Moreover, the Ras family plays important roles in the cell cycle, cell division, and cell death. Mutations of Ras proteins including H‐Ras, K‐Ras (KRAS), and N‐Ras were identified to be closely associated with tumor progression. 35 , 36 A previous study found that CD109‐knockout mice (CD109−/−) exhibited increased incidences of H‐Ras mutations in a skin tumorigenesis model, compared to CD109+/+ mice. 29 Interestingly, our present study found that CD109 was overexpressed in A549, H460, and H441 cells which harbor the KRAS mutant, compared to PC9 and CL1‐5 cells that exhibit wild‐type KRAS. Likewise, it was reported that CD109 was a major driver of lung metastasis in a KrasLSL − G12D/+;Trp53flox/flox mice model. 14 Interestingly, a membrane glycoprotein study showed that CD109 was overexpressed in pancreatic BxPC‐3 cells, which lack the KRAS mutation. 37 Thus, more evidence from future studies is needed to clarify the involvement of KRAS in regulating CD109 expression. Taken together, our study results suggest that CD109 is an independent marker for lung adenocarcinomas. Targeting CD109 could provide therapeutic benefits against lung cancer metastasis and drug resistance.
DISCLOSURE
The authors declare that no competing financial interests exist.
Supporting information
Fig S1‐S5,Table S1
ACKNOWLEDGMENTS
This study was supported by grants from The Ministry of Science and Technology, Taiwan (MOST106‐2320‐B‐038‐040 and MOST107‐2320‐B‐038‐052‐MY3), Taipei Medical University and Shuang Ho Hospital (106SHH‐TMU‐03), and Far Eastern Memorial Hospital (FEMH‐2019‐C‐013).
Lee K‐Y, Shueng P‐W, Chou C‐M, et al. Elevation of CD109 promotes metastasis and drug resistance in lung cancer via activation of EGFR‐AKT‐mTOR signaling. Cancer Sci. 2020;111:1652–1662. 10.1111/cas.14373
Kang‐Yun Lee and Pei‐Wei Shueng contributed equally to this work.
REFERENCES
- 1. Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277‐300. [DOI] [PubMed] [Google Scholar]
- 2. Blandin Knight S, Crosbie PA, Balata H, Chudziak J, Hussell T, Dive C. Progress and prospects of early detection in lung cancer. Open Biol. 2017;7(9):170070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kastelijn EA, de Langen AJ, Peters BJM. Treatment of oncogene‐driven non‐small cell lung cancer. Curr Opin Pulm Med. 2019;25:300‐307. [DOI] [PubMed] [Google Scholar]
- 4. Ferrer I, Zugazagoitia J, Herbertz S, John W, Paz‐Ares L, Schmid‐Bindert G. KRAS‐Mutant non‐small cell lung cancer: from biology to therapy. Lung Cancer. 2018;124:53‐64. [DOI] [PubMed] [Google Scholar]
- 5. Mii S, Enomoto A, Shiraki Y, Taki T, Murakumo Y, Takahashi M. CD109: a multifunctional GPI‐anchored protein with key roles in tumor progression and physiological homeostasis. Pathol Int. 2019;69:249‐259. [DOI] [PubMed] [Google Scholar]
- 6. Hagiwara S, Murakumo Y, Sato T, et al. Up‐regulation of CD109 expression is associated with carcinogenesis of the squamous epithelium of the oral cavity. Cancer Sci. 2008;99:1916‐1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shiraki Y, Mii S, Enomoto A, et al. Significance of perivascular tumour cells defined by CD109 expression in progression of glioma. J Pathol. 2017;243:468‐480. [DOI] [PubMed] [Google Scholar]
- 8. Sole C, Tramonti D, Schramm M, et al. The circulating transcriptome as a source of biomarkers for melanoma. Cancers. 2019;11(1):70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hasegawa M, Moritani S, Murakumo Y, et al. CD109 expression in basal‐like breast carcinoma. Pathol Int. 2008;58:288‐294. [DOI] [PubMed] [Google Scholar]
- 10. Zong G, Xu Z, Zhang S, et al. CD109 mediates cell survival in hepatocellular carcinoma cells. Dig Dis Sci. 2016;61:2303‐2314. [DOI] [PubMed] [Google Scholar]
- 11. Finnson KW, Tam BY, Liu K, et al. Identification of CD109 as part of the TGF‐beta receptor system in human keratinocytes. FASEB J. 2006;20:1525‐1527. [DOI] [PubMed] [Google Scholar]
- 12. Hagiwara S, Murakumo Y, Mii S, et al. Processing of CD109 by furin and its role in the regulation of TGF‐beta signaling. Oncogene. 2010;29:2181‐2191. [DOI] [PubMed] [Google Scholar]
- 13. Zhang JM, Murakumo Y, Hagiwara S, et al. CD109 attenuates TGF‐beta1 signaling and enhances EGF signaling in SK‐MG‐1 human glioblastoma cells. Biochem Biophys Res Commun. 2015;459:252‐258. [DOI] [PubMed] [Google Scholar]
- 14. Chuang CH, Greenside PG, Rogers ZN, et al. Molecular definition of a metastatic lung cancer state reveals a targetable CD109‐Janus kinase‐Stat axis. Nat Med. 2017;23:291‐300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee WY, Chen PC, Wu WS, et al. Panobinostat sensitizes KRAS‐mutant non‐small‐cell lung cancer to gefitinib by targeting TAZ. Int J Cancer. 2017;141:1921‐1931. [DOI] [PubMed] [Google Scholar]
- 16. Kuo CC, Ling HH, Chiang MC, et al. Metastatic colorectal cancer rewrites metabolic program through a Glut3‐YAP‐dependent signaling circuit. Theranostics. 2019;9:2526‐2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zheng X, Cheng M, Fu B, et al. Targeting LUNX inhibits non‐small cell lung cancer growth and metastasis. Cancer Res. 2015;75:1080‐1090. [DOI] [PubMed] [Google Scholar]
- 18. Park MH, Yun HM, Hwang CJ, et al. Presenilin Mutation Suppresses Lung Tumorigenesis via Inhibition of Peroxiredoxin 6 Activity and Expression. Theranostics. 2017;7:3624‐3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhan P, Zhang B, Xi GM, et al. PRC1 contributes to tumorigenesis of lung adenocarcinoma in association with the Wnt/beta‐catenin signaling pathway. Mol Cancer. 2017;16:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Okayama H, Kohno T, Ishii Y, et al. Identification of genes upregulated in ALK‐positive and EGFR/KRAS/ALK‐negative lung adenocarcinomas. Cancer Res. 2012;72:100‐111. [DOI] [PubMed] [Google Scholar]
- 21. Botling J, Edlund K, Lohr M, et al. Biomarker discovery in non‐small cell lung cancer: integrating gene expression profiling, meta‐analysis, and tissue microarray validation. Clin Cancer Res. 2013;19:194‐204. [DOI] [PubMed] [Google Scholar]
- 22. Lee ES, Son DS, Kim SH, et al. Prediction of recurrence‐free survival in postoperative non‐small cell lung cancer patients by using an integrated model of clinical information and gene expression. Clin Cancer Res. 2008;14:7397‐7404. [DOI] [PubMed] [Google Scholar]
- 23. Chu YW, Yang PC, Yang SC, et al. Selection of invasive and metastatic subpopulations from a human lung adenocarcinoma cell line. Am J Respir Cell Mol Biol. 1997;17:353‐360. [DOI] [PubMed] [Google Scholar]
- 24. Kelton JG, Smith JW, Horsewood P, et al. ABH antigens on human platelets: expression on the glycosyl phosphatidylinositol‐anchored protein CD109. J Lab Clin Med. 1998;132:142‐148. [DOI] [PubMed] [Google Scholar]
- 25. Bordin JO, Kelton JG, Warner MN, et al. Maternal immunization to Gov system alloantigens on human platelets. Transfusion. 1997;37:823‐828. [DOI] [PubMed] [Google Scholar]
- 26. Tsai YL, Ha DP, Zhao H, et al. Endoplasmic reticulum stress activates SRC, relocating chaperones to the cell surface where GRP78/CD109 blocks TGF‐beta signaling. Proc Natl Acad Sci USA. 2018;115:E4245‐E4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bizet AA, Tran‐Khanh N, Saksena A, Liu K, Buschmann MD, Philip A. CD109‐mediated degradation of TGF‐beta receptors and inhibition of TGF‐beta responses involve regulation of SMAD7 and Smurf2 localization and function. J Cell Biochem. 2012;113:238‐246. [DOI] [PubMed] [Google Scholar]
- 28. Li C, Hancock MA, Sehgal P, Zhou S, Reinhardt DP, Philip A. Soluble CD109 binds TGF‐beta and antagonizes TGF‐beta signalling and responses. Biochem J. 2016;473:537‐547. [DOI] [PubMed] [Google Scholar]
- 29. Sunagawa M, Mii S, Enomoto A, et al. Suppression of skin tumorigenesis in CD109‐deficient mice. Oncotarget. 2016;7:82836‐82850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Watanabe S, Yoshida T, Kawakami H, et al. T790M‐Selective EGFR‐TKI combined with dasatinib as an optimal strategy for overcoming EGFR‐TKI resistance in T790M‐positive non‐small cell lung cancer. Mol Cancer Ther. 2017;16:2563‐2571. [DOI] [PubMed] [Google Scholar]
- 31. Gao J, Li HR, Jin C, Jiang JH, Ding JY. Strategies to overcome acquired resistance to EGFR TKI in the treatment of non‐small cell lung cancer. Clin Transl Oncol. 2019;21(10):1287‐1301. [DOI] [PubMed] [Google Scholar]
- 32. Irwin ME, Mueller KL, Bohin N, Ge Y, Boerner JL. Lipid raft localization of EGFR alters the response of cancer cells to the EGFR tyrosine kinase inhibitor gefitinib. J Cell Physiol. 2011;226:2316‐2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Qi R, Dong F, Liu Q, Murakumo Y, Liu J. CD109 and squamous cell carcinoma. J Transl Med. 2018;16:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers–a different disease. Nat Rev Cancer. 2007;7:778‐790. [DOI] [PubMed] [Google Scholar]
- 35. Prieto‐Dominguez N, Parnell C, Teng Y. Drugging the small GTPase pathways in cancer treatment: promises and challenges. Cells. 2019;8(3):255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McCormick F. Progress in targeting RAS with small molecule drugs. Biochem J. 2019;476:365‐374. [DOI] [PubMed] [Google Scholar]
- 37. Haun RS, Fan CY, Mackintosh SG, Zhao H, Tackett AJ. CD109 Overexpression in Pancreatic Cancer Identified by Cell‐Surface Glycoprotein Capture. J Proteomics Bioinform. 2014;(Suppl 10):S10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1‐S5,Table S1


