Abstract
Artificial intelligence (AI) has contributed substantially to the resolution of a variety of biomedical problems, including cancer, over the past decade. Deep learning, a subfield of AI that is highly flexible and supports automatic feature extraction, is increasingly being applied in various areas of both basic and clinical cancer research. In this review, we describe numerous recent examples of the application of AI in oncology, including cases in which deep learning has efficiently solved problems that were previously thought to be unsolvable, and we address obstacles that must be overcome before such application can become more widespread. We also highlight resources and datasets that can help harness the power of AI for cancer research. The development of innovative approaches to and applications of AI will yield important insights in oncology in the coming decade.
Keywords: artificial intelligence, deep learning, machine learning, oncology, personalized medicine
Artificial intelligence (AI) has contributed substantially to the resolution of a variety of biomedical problems, including cancer, over the past decade. In this review, we describe numerous recent examples of the application of AI in oncology, including cases in which deep learning has efficiently solved problems that were previously thought to be unsolvable, and we address obstacles that must be overcome before such application can become more widespread.
1. INTRODUCTION
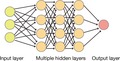
Artificial intelligence (AI) is a field of research in which computers are applied to mimic human intelligence. 1 Machine learning is a subfield of AI in which mathematical and statistical approaches are applied to improve the performance of computers. Deep learning is a subfield of machine learning characterized by the operation of multilayered artificial neural networks (Figure 1). The term “deep learning” refers to a set of new techniques that together have shown marked improvements in performance compared with existing best‐in‐class machine learning algorithms in several disciplines. For instance, these methods have revolutionized image classification and speech recognition as a result of their flexibility and high accuracy. 2 These breakthroughs have allowed deep learning to be adopted as an approach that can efficiently solve various problems in biomedicine. The application of deep learning to the diagnosis of diseases on the basis of the classification of radiological or pathological images has demonstrated a performance that equals or actually exceeds that of clinical experts. 3 , 4 Deep learning has also proved highly accurate in the detection of retinopathy from fundus photographs. 5 With high expectations for this technology, it is now being applied to the field of drug discovery. 6
Figure 1.
Artificial Intelligence (AI), machine learning and deep learning. AI refers to a broad range of computational methods that mimic human intelligence. Machine learning is a subfield of AI that relies on statistical methods to detect hidden patterns within a dataset. Deep learning is a subfield of machine learning that harnesses the power of multilayered networks
Biology and medicine are rapidly becoming data‐intensive. Stephens et al (2015) claimed that the application of AI to genomics alone will equal or exceed that to social media, online videos and other data‐intensive disciplines with regard to data generation and analysis within the next 10 years. 7 Automated algorithms that extract meaningful patterns can provide practical knowledge and change the way in which treatments are developed, patients are classified and diseases are studied. In contrast, AI may infringe on privacy because of potential access to personal information such as genomic sequences during data processing. Given that large datasets with appropriate data annotation are required for the application of deep learning technology, it is essential that both medical professionals and biological scientists possess a basic knowledge of deep learning, including its applications and potential drawbacks, when collaborating with AI researchers and using deep learning in their projects.
Cancer is the most common cause of death in developed countries, and it is estimated that the number of cases will increase further in aging populations. 8 , 9 In Japan, almost 1 million individuals are diagnosed with cancer and nearly 400 000 die of the disease every year. Cancer research will, thus, continue to be a top priority for the saving of lives in the coming decade.
In this review, we focus on the application of deep learning to cancer. For more comprehensive information on deep learning, including its mathematical aspects, we recommend several recent reviews 2 , 10 , 11 , 12 , 13 , 14 and a book. 15
2. DEEP LEARNING ESSENTIALS
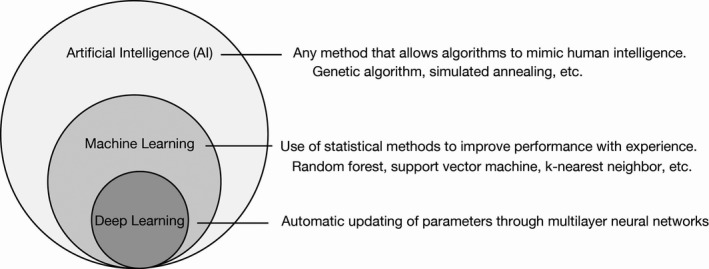
The deep learning approach evolved from the study of artificial neurons, being first proposed in 1943 as a model for the processing of information by neurons in the biological brain. 16 In a neural network, the input is provided to an input layer, which transfers its computed value to one or more hidden layers that are linked to an output layer. A layer consists of a set of nodes, known as “units” or “features,” that are connected through edges to both the previous and the next layers. Each unit transforms the data in a nonlinear manner by applying an activation function (Figure 2A). A deep neural network possesses multiple (sometimes > 100) hidden layers (Figure 2B). The training process allows deeper layers in the network to combine high‐level features coming from the previous layer and build more such features. As a result, these algorithms can automatically design features that are appropriate for solving the task at hand.
Figure 2.
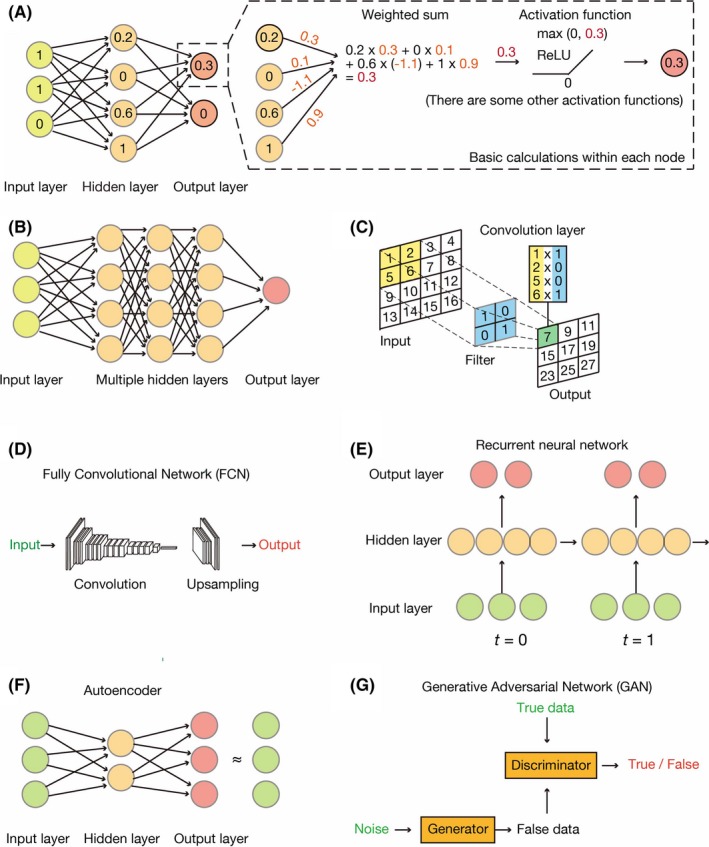
Common architectures of neural networks. A, The simplest neural network comprises three layers: an input layer, a hidden layer and an output layer. Each node has some value and transmits its signal to the next layer. It first sums all weighted inputs and then transmits the resulting value to an activation function (rectified linear unit, or ReLU, in this case). B, A typical deep neural network, also known as a dense neural network, has multiple hidden layers, the nodes of each of which calculate values in the same manner as shown in (A). C, A convolutional neural network (CNN) applies multiple convolution layers before feeding the data into a dense neural network. The convolution layers apply filters (or kernels) to grid‐based data. D, A fully convolutional network (FCN) is a variant of a CNN in that it lacks densely connected layers. E, A recurrent neural network (RNN) is a special network designed for time‐series data. Each hidden layer holds certain variables and transmits them to the next time step. F, An autoencoder resembles a dense neural network but is trained to output signals that are identical to the inputs. Such networks offer a means to encode and decode data, with encoded features being stored in the hidden layer. G, A generative adversarial network (GAN) consists of two independent neural networks: a generator and a discriminator. The generator attempts to create new data (false data) that resemble the true data. In contrast, the discriminator discriminates real data from artificial data created by the generator. Alternate training of these two networks helps to decipher the complex rules underlying the data
There are many architectures of deep neural networks. 15 A convolutional neural network (CNN) can be applied to data that have a grid topology, such as images, and uses multiple filters to detect patterns within the data (Figure 2C). A fully convolutional network (FCN) differs from a CNN in that fully connected layers of the CNN are replaced by upsampling and deconvolution layers, which are considered the inverse of pooling and convolution layers, respectively (Figure 2D). An FCN generates a score map for each class instead of one probability score. 17 This map is the same precise size as the input image and classifies the image by pixels. These new layers have been used to develop deep learning algorithms in many applications. Finally, a recurrent neural network (RNN) is useful for the analysis of sequential data, such as language and genomic sequences (Figure 2E). These neural networks correspond to supervised learning methods, which require answers to train the network.
In contrast, no labels are necessary for unsupervised learning. An autoencoder is a representative example of such an unsupervised method that attempts to provide output signals that are identical to the original input (Figure 2F). A generative adversarial network (GAN) actually consists of two networks: a generator and a discriminator. The generator neural network creates fake data to cheat the discriminator network, whereas the discriminator distinguishes the false data from true data. Training these networks alternately allows the generator to finally learn how to generate fake data indistinguishable from real data (Figure 2G).
Although we do not dive into detail here, various novel architectures, such as graphical convolution 18 , 19 and capsuleNet, are continually being proposed and applied in biomedicine. Given that each type of neural network is specialized in its specific data structure, the combination of multiple existing frameworks or the development of new architectures is expected to greatly improve the feasibility of using neural networks for interpretation of complex phenomena.
3. ARTIFICIAL INTELLIGENCE FOR CANCER IMAGES
3.1. Application to image analysis
Early detection of cancer is key to saving the lives of affected individuals. Deep learning has revolutionized image analysis since its spectacular win in the image recognition contest ILSVRC (ImageNet Large Scale Visual Recognition Challenge) in 2012, 2 with many researchers and physicians having attempted to harness the power of AI (or, more precisely, CNN) for application to clinical radiology and pathology because it obviates the need to generate detailed features by craftsmanship. One example of high impact is the successful classification of dermoscopy images. 20 , 21 AI was, thus, found to be able to annotate skin lesions (including melanoma) as precisely as were expert dermatologists (with an area under the curve [AUC] of 0.94‐0.96). Given that smartphones extend the reach of dermatologists outside of the clinic, this achievement has the potential to provide universal access to dermatologist‐level diagnoses. AI has also achieved a level of accuracy similar to that of medical specialists in the interpretation of mammograms for breast cancer screening (AUC for AI of 0.840, compared with 0.814 for physicians). 22 In addition, deep neural networks have been able to detect enlarged lymph nodes or colonic polyps in computed tomography images. 23 The multiple applications of deep learning in radiology are covered in more detail elsewhere. 24 , 25
Whole‐slide imaging is becoming routine in developed countries, which has resulted in the accumulation of digital pathology images and allowed the application of deep learning to pathological diagnosis. 26 One proposed model extracts features with a CNN and supplies them to a support vector machine, another machine learning algorithm, to train for the detection of breast mitosis. 27 A deep learning system has been used to detect areas of cancer in whole‐slide images of radical prostatectomy specimens and to automatically assign the Gleason score with an accuracy of 0.70 (which is superior to that of general pathologists). 28 A CNN has also been applied to pathological slides from The Cancer Genome Atlas (TCGA) for the automatic detection of tumor‐infiltrating lymphocytes (TIL), and the features extracted in this model (TIL maps) were found to be prognostic factors for 13 different cancer types, including breast, lung and colorectal tumors. 29 In addition, given that deep learning can also output a map of where potential cancer cells are present on a slide and determine with what likelihood the cells are cancerous, pathologists may need only to evaluate slides for which AI is not able to provide a clear answer.
Deep learning is also able to elucidate the molecular status of a tumor from pathological data. In addition to being able to assess and score the expression of tumor marker proteins such as HER2 in tissue slides, 30 a CNN model (inception v3) successfully predicted which genes in tumor tissue harbor mutations (with AUC from 0.733 to 0.856). 31 Another CNN model, designated HE2RNA, can predict transcriptome profiles from pathological images, without expert annotation. 32 Collectively, these achievements show that deep learning has already been used to perform versatile tasks, such as cancer diagnosis, at a level equal to or sometimes greater than that of expert physicians. 33
One of the remaining obstacles in automatic histopathology analysis is the establishment of standard protocols. Differences in color tone on pathology slides may occur among institutions as a result of differences in staining reagents, protocols and section thickness. It will, therefore, be necessary to standardize color tones in digital slides for the development of accurate AI algorithms. 34 Some automated CNN‐based tools, such as HistoQC and DeepFocus, 35 , 36 have been developed to standardize the quality of whole‐slide imaging. GAN‐based image generators were also recently proposed to tackle this obstacle. In this case, a neural network receives a grayscale image, instead of noise data, as input, and a normalized hematoxylin‐eosin‐stained image can be generated. 37 GAN‐based approaches have also been adopted beyond diagnosis. For example, they have already been used to correct feature segmentation 38 and to score levels of the immune‐checkpoint protein PD‐L1 in needle biopsy specimens of non–small cell lung cancer. 39 Hematoxylin‐eosin–stained specimens have also been successfully converted to immunohistologically stained images for cytokeratins 18/19. 40
These various examples indicate that AI can be of great help in reducing the burden on medical staff involved in tumor assessment. In the near future, deep learning will become an important support tool for pathologists that will improve the accuracy and efficiency of histopathologic diagnosis and thereby inform treatment selection. For further reading, we recommend two recent reviews in this field. 41 , 42
3.2. Limitations and their potential solutions in medical image analysis
In almost all cases, the number of medical images available for training is <1 million, which is many orders of magnitude smaller than the natural image collection. Researchers have developed several strategies to overcome this limitation. Data augmentation, whereby images are randomly cropped, tilted, inverted or flipped to increase their number, is one effective strategy for dealing with the small size of training sets. An example of this approach is provided by a series of papers on the analysis of mammograms. 22 , 43 A second strategy, known as transfer learning, involves reuse for the new purpose of medical imaging of features extracted by deep learning from natural image datasets such as ImageNet. The potential to diagnose diabetic retinopathy from fundus photographs has attracted many deep learning researchers since 2015, when a large, labeled dataset of images was released at a machine learning competition called Kaggle. For example, researchers reused a 48‐layer CNN architecture known as inception v3 that was pretrained with natural images and succeeded in exceeding the specificity and sensitivity of the then state‐of‐the‐art model. 5 These approaches have also been exploited in training AI to detect melanoma, the most deadly skin cancer. 20 , 21
Such findings motivate researchers to share image data and make it freely usable among communities. Some useful resources of tumor images are summarized in Table 1. The increasing rate of adoption of AI in oncology research will result in an increase in the number of medical images becoming available, thus allowing researchers to build more robust and sophisticated algorithms.
Table 1.
Examples of available image datasets related to cancer
4. ARTIFICIAL INTELLIGENCE FOR CANCER GENOMICS
With the cost of genome sequencing declining, the use of a supercomputer to analyze genomic data from cancer patients often results in the identification of between 1000 and 100 000 genomic mutations for each tumor sample. 44 However, it is necessary to clarify the association of each of these mutations with clinical phenotypes, which is currently a bottleneck for genomic medicine. 45 The clinical interpretation of genetic variants depends mainly on information in the scientific and medical literature. In other words, researchers need to find the relevant literature to link the identified genomic mutations to information on disease states, effective drugs and prognosis. With more than 200 000 new cancer‐related articles being published in 2019 alone, human resources are not sufficient for manual curation. An example of a database that summarizes the relation between genomic variation and disease is the COSMIC database provided by the Sanger Center. 46 COSMIC version 90, released in September 2019, extracted 9 733 455 mutations in gene coding regions from 26 829 papers.
The use of AI will, therefore, become increasingly indispensable. Before the application of AI to genomic data, the sequence is transformed into a binary table (one‐hot encoding) that shows the presence or absence of each of the four bases at each position. Several filters are then applied one‐dimensionally for convolution of the table (Figure 3A). There are two main reasons why deep learning is useful in cancer genomics. First, in addition to single‐task learning, it allows multitask learning, in which AI learns multiple different tasks simultaneously by sharing parts of a model (Figure 3B). User‐defined losses (differences in predicted and actual values for each task) are minimized during the training process by an appropriate optimizer algorithm. Second, it allows multimodal learning, a method for integration of different types of data (such as sequence and chromatin accessibility) and their entry as inputs. In this process, AI automatically learns how to combine these different kinds of data. Given that cancer is a complex disease, it is preferable to integrate multilayered data. The application of AI to analysis of a large amount of “omics” (exome, transcriptome, and epigenome) data as well as data on the susceptibility of patients with acute myeloid leukemia to anticancer drugs resulted in the identification of drug‐susceptibility genes. 47 Watson for Genomics also analyzed 323 patients to identify genomic alterations with potential clinical effects that were not recognized by the conventional Molecular Tumors panel. 48
Figure 3.
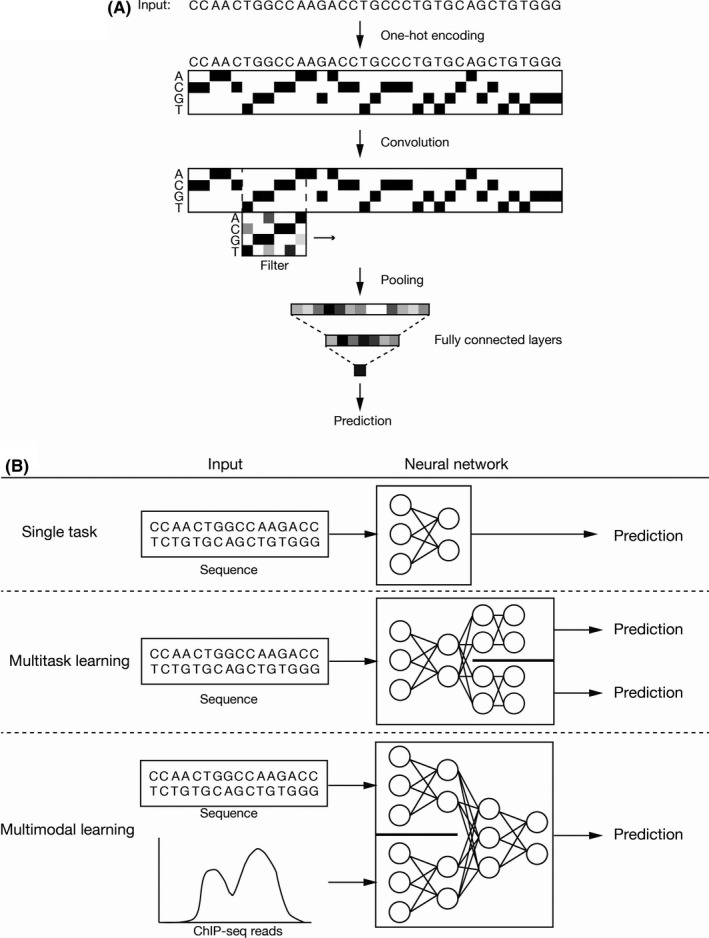
Deep learning for cancer genomics. A, Sequence data are converted to a binary map (one‐hot encoding) and several filters are applied (1D‐CNN), resulting in the transformation of genomics data into numerical vectors. The remaining procedure is the same as for common tasks (updating of weights to minimize loss). B, A typical workflow receives one type of data and outputs prediction. In multitask learning, multiple types of prediction (such as clinical impact of a mutation and biological activity of a gene) are generated by the shared network and specified networks for each task. Conversely, in multimodal learning, the network integrates different types of information (such as sequence data and chromatin accessibility) and outputs prediction
Deep learning is also applied to the variant calling process. In addition to the standard variant detection framework, Google’s DeepVariant exploits the Inception TensorFlow framework, which was initially developed for image classification. This workflow converts variant calling into an image recognition task by changing the BAM file into an image similar to the genome browser snapshot and determining variants based on likelihood. 49
Another algorithm, ExPecto, links genetic mutations with disease prediction. 50 ExPecto predicts the level of gene expression in each tissue on the basis of wide regulatory regions consisting of 40‐kb promoter‐proximal sequences. This framework was built with the use of all publicly available genome‐wide association studies and has been experimentally validated. Estimation of gene expression level by this approach might help to decipher the complex etiology of cancer on the basis of genome‐wide sequence data.
More and more clinical cancer genomic data are gradually being accumulated. In 2017, the US FDA approved several genome sequence‐based panels related to oncology, including the Oncomine Dx Target Test, the Praxis Extended RAS Panel, MSK‐IMPACT and FoundationOne CDx. In Japan, the Ministry of Health, Labor, and Welfare set a goal in May 2019 to perform genome‐wide sequencing of 100 000 individuals over the next 3 years through the full‐scale introduction of genome‐wide sequencing to the clinic. AI is, thus, set to play a more important role in the interpretation of cancer genomic findings.
Machine learning also has the potential to provide novel biological insights. As one example, a regulatory role for Fbxw7 (one of the most frequently mutated E3 ubiquitin ligases in cancer) in the oxidative metabolism of cancer cells was discovered with the use of a machine learning algorithm (kernelized Bayesian transfer learning). 51 In combination with our previous studies showing that Fbxw7 plays an essential role in the maintenance of quiescence and stemness in cancer stem cells, 52 , 53 , 54 this finding may provide important clues to uncovering the metabolic characteristics of cancer stem cells. AI can also precisely predict RNA splicing. Precursor mRNA transcripts undergo splicing to generate multiple mature mRNA isoforms, and prediction of splicing sites has been a major goal over the past decade. Early studies adopted a naive Bayesian model, 55 but the advent of deep learning allowed the development of more complex models that have provided better predictive accuracy. SpliceAI, a 32‐layer deep CNN, predicts splicing from a given pre–mRNA sequence. 56 Abnormal splicing has been frequently observed in cancer, and the splicing machinery is being targeted for therapeutic purposes. 57 Further advances in this field might allow researchers to predict tissue‐specific and exon‐specific splicing patterns from genomic sequences.
5. ARTIFICIAL INTELLIGENCE AND PERSONALIZED MEDICINE
A key challenge in medical science is the precise classification of diseases and the development of optimal therapies, which would be expected to improve the outcome of many patients. Current “gold standard” approaches to disease classification in oncology include histological examination by expert pathologists and evaluation of the expression of molecular markers such as cell surface receptors at the protein or mRNA level. One example is the PAM50 classification, in which breast cancer is classified into several subtypes on the basis of the expression of marker genes. 58 There is still considerable heterogeneity within these subtypes, however. 59 Given the increasing amount of available molecular data, it may be possible to identify disease subtypes more comprehensively to predict future disease behavior and treatment response.
We recently developed a tool, termed the molecular Prognostic Score (mPS), that is able to predict the prognosis of breast cancer patients precisely (Figure 4A). 60 We integrated almost 6000 breast cancer patients by meta‐analysis, which is mainly applied in the field of epidemiology, and comprehensively identified 184 prognosis‐related genes for breast cancer without any biological information. We built a random forest classifier combined with a neural network and trained the model to predict overall survival with half of the METABRIC cohort. 61 This scoring system stratified prognosis more accurately than existing methods in several independent test datasets. Unlike previous tools, 62 , 63 it can even be applied to estrogen receptor‐negative breast cancer patients (Figure 4B). In addition, the score provides useful information for avoidance of overtreatment (Figure 4C).
Figure 4.
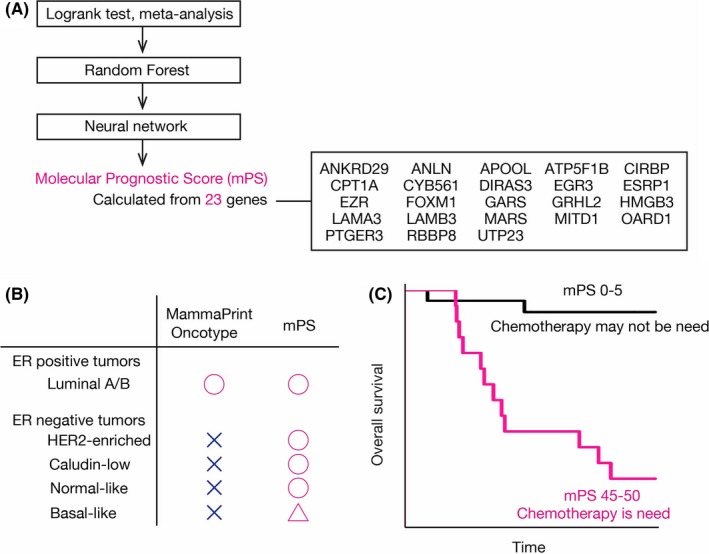
Prediction of prognosis of breast cancer patients. A, All human protein‐coding genes were evaluated using a series of methods (log‐rank test, meta‐analysis, machine learning with the random forest method and a neural network). The molecular Prognostic Score (mPS) based on 23 prognostic genes predicts the prognosis of breast cancer patients. B, mPS stratifies not only estrogen receptor (ER)‐positive but also ER‐negative patients, in contrast to existing methods such as MammaPrint and Oncotype. C, Chemotherapy may not be necessary for patients with a low mPS because their prognosis is fairly good
Attempts have also been made to predict the risk of cancer recurrence from pathological images. A digital pathology test thus predicts the risk of recurrence in breast cancer patients with the use of stored samples from a completed clinical trial, ECOG 2197. 64 This AI‐driven approach allows the stratification of patients with a hazard ratio of 2.41 (95% confidence interval, 1.21‐4.79).
Artificial intelligence is a key driver of the transformation of health care to precision medicine. Numerous international competitions are held each year to further revolutionize the role of AI in health care (Table 2) and have yielded novel algorithms to tackle complicated tasks. Together with crowdsourcing, 65 such open and innovative challenges will broaden further applications of AI to cancer research. On the basis of recent advances, the FDA has finally begun the approval of clinical medical devices based on deep learning (Table 2). In 2018, the cloud‐based Arterys imaging platform was approved by the FDA as a tool to help physicians track tumors on the basis of MRI and computed tomography scans of lung and liver cancer patients. In 2019, the digital pathology solution PAIGE.AI was designated as a Breakthrough Device by the FDA. Startups such as PAIGE.AI, Proscia, and PathAI use deep learning‐based AI algorithms to detect, diagnose and predict certain cancer types.
Table 2.
Summary of open competitions, companies with FDA‐approved artificial intelligence devices, useful tools, and educational resources for beginners
| Type | Name | URL |
|---|---|---|
| Open challenge | Dream Challenges | http://dreamchallenges.org/ |
| Open challenge | Grand Challenge | https://grand-challenge.org/challenges/ |
| Open challenge | PrecisionFDA Truth Challenge | https://precision.fda.gov/challenges/truth |
| Open challenge | CAGI | https://genomeinterpretation.org/ |
| Company | Arterys | https://www.arterys.com/ |
| Company | PAIGE.AI | https://paige.ai/ |
| Company | Proscia | https://proscia.com/ |
| Company | PathAI | https://www.pathai.com/ |
| Library | Scikit‐learn | https://scikit-learn.org/stable/ |
| Library | TensorFlow | https://www.tensorflow.org/ |
| Library | Keras | https://keras.io/ja/ |
| Library | PyTorch | https://pytorch.org/ |
| Library | DragoNN | https://kundajelab.github.io/dragonn/ |
| Library | Kipoi | https://kipoi.org/ |
| Education | DeepOncology | https://github.com/deeponcology/PyTorchMedicalAI |
| Education | fast.ai | https://www.fast.ai/ |
| Education | Medical‐ai‐course‐materials | https://japan-medical-ai.github.io/medical-ai-course-materials/ |
6. CONCLUSION
Digitization of health care is dramatically changing clinical workflow by providing both healthcare providers and patients with access to information based on big data. Experience‐based medicine is being replaced with an evidence‐based, patient‐centric approach. Rapidly evolving AI technology will continue to have a large impact on the field of cancer in the near future. Both physicians and researchers need to be ready for this coming revolutionary era. 66 Medical education must, therefore, include not only life sciences and clinical medicine, but also advanced statistical and computational skills. Indeed, some medical schools have already begun to include AI education courses in the curriculum. Implementation of AI technology is becoming easier as a result of the availability of various open‐source tools and cloud computing. Some useful resources for beginners in this field are listed in Table 2.
Deep learning has gained much attention in recent years, but several obstacles must be overcome before its application to health care and cancer research becomes more widespread. First, maximization of the power of deep learning will require the deposition of medical data with sufficient annotation in large‐scale databases. International collaborative projects (such as The Cancer Imaging Archive [http://www.cancerimagingarchive.net] and Genomic Data Commons Data Portal %5B http://broken-link/) that build large, labeled datasets should make a substantial contribution to meeting this challenge. A second obstacle is that deep learning is now a black box that does not explain the decision‐making process clearly. The large number of parameters involved makes it difficult to understand the details of how deep learning analyzes data and makes decisions. The development of a “white box” approach has become a major research topic in biomedical science. 67 Such innovative approaches in this area are also likely to provide key insights in cancer research. Given how rapidly the field is evolving and the many potential applications of AI in cancer science, AI will revolutionize oncology in the coming decade.
CONFLICT OF INTERESTS
The authors declare no potential conflicts of interest.
ACKNOWLEDGMENTS
We thank members of our laboratory for discussion and A. Ohta for help with preparation of the manuscript. This work was supported by KAKENHI grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan to HS (19K20403) and to KIN (18H05215).
Shimizu H, Nakayama KI. Artificial intelligence in oncology. Cancer Sci. 2020;111:1452–1460. 10.1111/cas.14377
REFERENCES
- 1. Rajkomar A, Dean J, Kohane I. Machine learning in medicine. N Engl J Med. 2019;380:1347‐1358. [DOI] [PubMed] [Google Scholar]
- 2. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436‐444. [DOI] [PubMed] [Google Scholar]
- 3. Teare P, Fishman M, Benzaquen O, Toledano E, Elnekave E. Malignancy detection on mammography using dual deep convolutional neural networks and genetically discovered false color input enhancement. J Digit Imaging. 2017;30:499‐505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ehteshami Bejnordi B, Veta M, Johannes van Diest P, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA. 2017;318:2199‐2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316:2402‐2410. [DOI] [PubMed] [Google Scholar]
- 6. Chen H, Engkvist O, Wang Y, Olivecrona M, Blaschke T. The rise of deep learning in drug discovery. Drug Discov Today. 2018;23:1241‐1250. [DOI] [PubMed] [Google Scholar]
- 7. Stephens ZD, Lee SY, Faghri F, et al. Big data: astronomical or genomical? PLoS Biol. 2015;13:e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7‐34. [DOI] [PubMed] [Google Scholar]
- 9. DeSantis CE, Miller KD, Dale W, et al. Cancer statistics for adults aged 85 years and older, 2019. CA Cancer J Clin. 2019;69:452‐467. [DOI] [PubMed] [Google Scholar]
- 10. Jang HJ, Cho KO. Applications of deep learning for the analysis of medical data. Arch Pharm Res. 2019;42:492‐504. [DOI] [PubMed] [Google Scholar]
- 11. Ching T, Himmelstein DS, Beaulieu‐Jones BK, et al. Opportunities and obstacles for deep learning in biology and medicine. J R Soc Interface. 2018;15:20170387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60‐88. [DOI] [PubMed] [Google Scholar]
- 13. Miotto R, Wang F, Wang S, Jiang X, Dudley JT. Deep learning for healthcare: review, opportunities and challenges. Brief Bioinform. 2018;19:1236‐1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Angermueller C, Parnamaa T, Parts L, Stegle O. Deep learning for computational biology. Mol Syst Biol. 2016;12:878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Goodfellow I, Bengio Y, Courville A. Deep Learning. Cambridge, MA: The MIT Press; 2016. [Google Scholar]
- 16. Mcculloch W, Pitts W. A logical calculus of the ideas immanent in nervous activity. Bull Math Biol. 1990;52:99‐115; discussion 173‐197. [PubMed] [Google Scholar]
- 17. Wang S, Yang DM, Rong R, Zhan X, Xiao G. Pathology image analysis using segmentation deep learning algorithms. Am J Pathol. 2019;189:1686‐1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Haehn D, Tompkin J, Pfister H. Evaluating ‘Graphical Perception’ with CNNs. IEEE Trans Vis Comput Graph. 2019;25:641‐650. [DOI] [PubMed] [Google Scholar]
- 19. Tsubaki M, Tomii K, Sese J. Compound‐protein interaction prediction with end‐to‐end learning of neural networks for graphs and sequences. Bioinformatics. 2019;35:309‐318. [DOI] [PubMed] [Google Scholar]
- 20. Esteva A, Kuprel B, Novoa RA, et al. Dermatologist‐level classification of skin cancer with deep neural networks. Nature. 2017;542:115‐118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yu L, Chen H, Dou Q, Qin J, Heng PA. Automated melanoma recognition in dermoscopy images via very deep residual networks. IEEE Trans Med Imaging. 2017;36:994‐1004. [DOI] [PubMed] [Google Scholar]
- 22. Rodriguez‐Ruiz A, Lång K, Gubern‐Merida A, et al. Stand‐alone artificial intelligence for breast cancer detection in mammography: comparison with 101 radiologists. J Natl Cancer Inst. 2019;111:916‐922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Roth HR, Lu L, Liu J, et al. Improving computer‐aided detection using convolutional neural networks and random view aggregation. IEEE Trans Med Imaging. 2016;35:1170‐1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shen D, Wu G, Suk HI. Deep learning in medical image analysis. Annu Rev Biomed Eng. 2017;19:221‐248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ueda D, Shimazaki A, Miki Y. Technical and clinical overview of deep learning in radiology. Jpn J Radiol. 2019;37:15‐33. [DOI] [PubMed] [Google Scholar]
- 26. Niazi MKK, Parwani AV, Gurcan MN. Digital pathology and artificial intelligence. Lancet Oncol. 2019;20:e253‐e261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Albayrak A, Bilgin G. Mitosis detection using convolutional neural network‐based features In Proceedings of the IEEE Seventh International Symposium on Computational Intelligence and Informatics (CINTI); 2016. 10.1109/CINTI.2016.7846429 [DOI] [Google Scholar]
- 28. Nagpal K, Foote D, Liu Y, et al. Development and validation of a deep learning algorithm for improving Gleason scoring of prostate cancer. NPJ Digit Med. 2019;2:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Saltz J, Gupta R, Hou L, et al. Spatial organization and molecular correlation of tumor‐infiltrating lymphocytes using deep learning on pathology images. Cell Rep. 2018;23:181‐193.e187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vandenberghe ME, Scott ML, Scorer PW, Soderberg M, Balcerzak D, Barker C. Relevance of deep learning to facilitate the diagnosis of HER2 status in breast cancer. Sci Rep. 2017;7:45938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Coudray N, Ocampo PS, Sakellaropoulos T, et al. Classification and mutation prediction from non‐small cell lung cancer histopathology images using deep learning. Nat Med. 2018;24:1559‐1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schmauch B, Romagnoni A, Pronier E, et al. Transcriptomic learning for digital pathology. bioRxiv. 2019. 10.1101/760173 [DOI] [Google Scholar]
- 33. Litjens G, Sanchez CI, Timofeeva N, et al. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci Rep. 2016;6:26286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shaban MT, Baur C, Navab N, Albarqouni S. StainGAN: stain style transfer for digital historical images. arXiv. 2018. https://arxiv.org/abs/1804.01601 [Google Scholar]
- 35. Janowczyk A, Zuo R, Gilmore H, Feldman M, Madabhushi A. HistoQC: an open‐source quality control tool for digital pathology slides. JCO Clin Cancer Inform. 2019;3:1‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Senaras C, Niazi MKK, Lozanski G, Gurcan MN. DeepFocus: detection of out‐of‐focus regions in whole slide digital images using deep learning. PLoS One. 2018;13:e0205387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zanjani FG, Zinger S, Bejnordi BE, et al. Stain normalization of histopathology images using generic adaptive networks 2018 IEEE 15th International Symposium on Biomedical Imaging, ISBI 2018. https://ieeexplore.ieee.org/document/8363641 [Google Scholar]
- 38. Mahmood F, Borders D, Chen R, et al. Deep adversarial training for multi‐organ nuclei segmentation in histopathology images. IEEE Trans Med Imaging. 2019. 10.1109/TMI.2019.2927182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kapil A, Meier A, Zuraw A, et al. Deep semi supervised generative learning for automated tumor proportion scoring on NSCLC tissue needle biopsies. Sci Rep. 2018;8:17343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xu Z, Moro CF, Bozóky B, Zhang Q. GAN‐based virtual re‐staining: a promising solution for whole slide image analysis. ArXiv.org. 2019. https://arxiv.org/abs/1901.04059 [Google Scholar]
- 41. Komura D, Ishikawa S. Machine learning methods for histopathological image analysis. Comput Struct Biotechnol J. 2018;16:34‐42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chang HY, Jung CK, Woo JI, et al. Artificial intelligence in pathology. J Pathol Transl Med. 2019;53:1‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dhungel N, Carneiro G, Bradley AP. A deep learning approach for the analysis of masses in mammograms with minimal user intervention. Med Image Anal. 2017;37:114‐128. [DOI] [PubMed] [Google Scholar]
- 44. Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703‐713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hoskinson DC, Dubuc AM, Mason‐Suares H. The current state of clinical interpretation of sequence variants. Curr Opin Genet Dev. 2017;42:33‐39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Forbes SA, Beare D, Boutselakis H, et al. COSMIC: somatic cancer genetics at high‐resolution. Nucleic Acids Res. 2017;45:D777‐D783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lee JS, Das A, Jerby‐Arnon L, et al. Harnessing synthetic lethality to predict the response to cancer treatment. Nat Commun. 2018;9:2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Patel NM, Michelini VV, Snell JM, et al. Enhancing next‐generation sequencing‐guided cancer care through cognitive computing. Oncologist. 2018;23:179‐185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Szegedy C, Liu W, Jia Y, et al. Going deeper with convolutions. arXiv. 2014. https://arxiv.org/abs/1409.4842 [Google Scholar]
- 50. Zhou J, Theesfeld CL, Yao K, et al. Deep learning sequence‐based ab initio prediction of variant effects on expression and disease risk. Nat Genet. 2018;50:1171‐1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Davis RJ, Gonen M, Margineantu DH, et al. Pan‐cancer transcriptional signatures predictive of oncogenic mutations reveal that Fbw7 regulates cancer cell oxidative metabolism. Proc Natl Acad Sci USA. 2018;115:5462‐5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Takeishi S, Matsumoto A, Onoyama I, Naka K, Hirao A, Nakayama KI. Ablation of Fbxw7 eliminates leukemia‐initiating cells by preventing quiescence. Cancer Cell. 2013;23:347‐361. [DOI] [PubMed] [Google Scholar]
- 53. Takeishi S, Nakayama KI. To wake up cancer stem cells, or to let them sleep, that is the question. Cancer Sci. 2016;107:875‐881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shimizu H, Takeishi S, Nakatsumi H, Nakayama KI. Prevention of cancer dormancy by Fbxw7 ablation eradicates disseminated tumor cells. JCI insight. 2019;4:125138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Xiong HY, Barash Y, Frey BJ. Bayesian prediction of tissue‐regulated splicing using RNA sequence and cellular context. Bioinformatics. 2011;27:2554‐2562. [DOI] [PubMed] [Google Scholar]
- 56. Jaganathan K, Kyriazopoulou Panagiotopoulou S, McRae JF, et al. Predicting splicing from primary sequence with deep learning. Cell. 2019;176:535‐548.e524. [DOI] [PubMed] [Google Scholar]
- 57. Lee SC, Abdel‐Wahab O. Therapeutic targeting of splicing in cancer. Nat. Med. 2016;22:976‐986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ohnstad HO, Borgen E, Falk RS, et al. Prognostic value of PAM50 and risk of recurrence score in patients with early‐stage breast cancer with long‐term follow‐up. Breast Cancer Res. 2017;19:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yeo SK, Guan JL. Breast cancer: multiple subtypes within a tumor? Trends Cancer. 2017;3:753‐760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shimizu H, Nakayama KI. A 23 gene‐based molecular prognostic score precisely predicts overall survival of breast cancer patients. EBioMedicine. 2019;46:150‐159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Curtis C, Shah SP, Chin S‐F, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346‐352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nicolini A, Ferrari P, Duffy MJ. Prognostic and predictive biomarkers in breast cancer: past, present and future. Semin Cancer Biol. 2018;52:56‐73. [DOI] [PubMed] [Google Scholar]
- 63. Nasrazadani A, Brufsky AM. Artificial intelligence‐directed prognostication of breast cancer. EBioMedicine. 2019;46:6‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Verma N, Harding D, Mohammadi A, et al. Image‐based risk score to predict recurrence of ER+ breast cancer in ECOG‐ACRIN Cancer Research Group E2197. J Clin Oncol. 2018;36:540. [Google Scholar]
- 65. Horowitz S, Koepnick B, Martin R, et al. Determining crystal structures through crowdsourcing and coursework. Nat. Commun. 2016;7:12549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mesko B, Drobni Z, Benyei E, Gergely B, Gyorffy Z. Digital health is a cultural transformation of traditional healthcare. Mhealth. 2017;3:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yang JH, Wright SN, Hamblin M, et al. A white‐box machine learning approach for revealing antibiotic mechanisms of action. Cell. 2019;177:1649‐1661.e1649. [DOI] [PMC free article] [PubMed] [Google Scholar]


