Abstract
Resistance to chemotherapy is a major challenge for the treatment of patients with colorectal cancer (CRC). Previous studies have found that microRNAs (miRNAs) play key roles in drug resistance; however, the role of miRNA‐373‐3p (miR‐375‐3p) in CRC remains unclear. The current study aimed to explore the potential function of miR‐375‐3p in 5‐fluorouracil (5‐FU) resistance. MicroRNA‐375‐3p was found to be widely downregulated in human CRC cell lines and tissues and to promote the sensitivity of CRC cells to 5‐FU by inducing colon cancer cell apoptosis and cycle arrest and by inhibiting cell growth, migration, and invasion in vitro. Thymidylate synthase (TYMS) was found to be a direct target of miR‐375‐3p, and TYMS knockdown exerted similar effects as miR‐375‐3p overexpression on the CRC cellular response to 5‐FU. Lipid‐coated calcium carbonate nanoparticles (NPs) were designed to cotransport 5‐FU and miR‐375‐3p into cells efficiently and rapidly and to release the drugs in a weakly acidic tumor microenvironment. The therapeutic effect of combined miR‐375 + 5‐FU/NPs was significantly higher than that of the individual treatments in mouse s.c. xenografts derived from HCT116 cells. Our results suggest that restoring miR‐375‐3p levels could be a future novel therapeutic strategy to enhance chemosensitivity to 5‐FU.
Keywords: 5‐fluorouracil, chemosensitivity, colorectal cancer, miR‐375‐3p, nanoparticles
Resistance to chemotherapy is a major challenge for the treatment of patients with colorectal cancer (CRC). Our results suggest that the restoration of microRNA‐375‐3p levels could be a future novel therapeutic strategy to modulate and enhance chemosensitivity to 5‐fluorouracil treatment in CRC.

Abbreviations
- 5‐FU
5‐fluorouracil
- ALT
alanine transaminase
- AST
aspartate transaminase
- BUN
blood urea nitrogen
- Cr
creatinine
- CRC
colorectal cancer
- DOTAP
1,2‐dioleoyl‐3‐ trimethylammonium‐propane
- FDR
false discovery rate
- GSEA
gene set enrichment analysis
- GSVA
gene set variation analysis
- HCC
hepatocellular carcinoma
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- miR
microRNA
- miRNA
microRNA
- NC
negative control
- NOM
nominal
- NP
nanoparticle
- PI
propidium iodide
- qRT‐PCR
quantitative real‐time PCR
- TCGA
The Cancer Genome Atlas
- TYMS
thymidylate synthase
1. INTRODUCTION
Although considerable progress has been made in the treatment of CRC in recent years, CRC accounts for approximately 13% of all tumors and is the second leading cause of tumor‐related death in developed countries.1, 2, 3 Fluorouracil‐based chemotherapy has served as the first‐line standard of care and most common regimen for CRC over the past 50 years.4, 5 However, patient resistance to 5‐FU is a major obstacle to effective therapy. Therefore, efforts to clarify the molecular mechanism underlying 5‐FU resistance and to identify new diagnostic biomarkers for 5‐FU resistance are urgently needed to facilitate the development of therapeutic strategies for CRC patients.
Thymidylate synthase is one of the key enzymes in the 5‐FU catabolic pathway and is associated with the response to 5‐FU‐based therapy.6 As the TYMS‐catalyzed enzymatic reaction provides the sole intracellular de novo source of thymidylate, an essential precursor for DNA biosynthesis, TYMS has been a major target for anticancer therapy.7 Hence, TYMS activity is the best predictor of sensitivity to 5‐FU.
MicroRNAs have been shown to play significant roles in cancer progression by regulating target gene expression at the posttranscriptional level.8 MicroRNAs act as either tumor suppressors or oncomiRs, which are broadly implicated in tumor proliferation, invasion, and resistance to therapy.9 The aberrant expression of certain miRNAs can lead to drug resistance by affecting the expression levels of genes involved in stress responses, drug transport, metabolism, cell survival, and cell death.10, 11 For example, miR‐433 enhances the chemosensitivity of HeLa cells to 5‐FU by reducing TYMS expression, as described in a report addressing the regulation of TYMS by miRNAs in the cellular response to 5‐FU.12 In addition, miR‐20313 and miR‐197 have been reported to mediate the response of CRC cells to 5‐FU by regulating TYMS expression.14
Our research team has studied and identified the aberrantly expressed miRNAs involved in HCC through the comparison of miRNA expression profiling in cancerous hepatocytes with that in normal primary human hepatocytes, and found miR‐375 was one of the greatest changes in decreased expression (GEO database: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE20077). MicroRNA‐375, as an important tumor suppressor, has considerable clinical value in early diagnosis15 and has been reported to be an effective therapeutic target. MicroRNA‐375 has been shown to inhibit tumor growth, metastasis, and epithelial‐mesenchymal transition, and thereby control tumor progression in various cancers, including gastric cancer,16 HCC,17 and esophageal squamous cell carcinoma.18 In addition, it has been reported to be an effective prognostic biomarker.19 Interestingly, emerging evidence suggests that miR‐375 can be used as a sensitizer for cancer treatment and can reverse resistance to chemotherapy drugs in various cancers, such as HCC,20 breast cancer,21 and cervical cancer.22 However, studies on the role of miR‐375 in colorectal cancer resistance and the underlying mechanisms are limited.
MicroRNA mimics cannot efficiently enter cells independently and typically require the assistance of a transport system. If 5‐FU and miR‐375 mimic are given as separate therapies, differences between them in physicochemical properties could result in differences in biodistribution and tumor accumulation between the 2 drugs. This problem can potentially be overcome by codelivering the drugs by NPs.23, 24, 25 Nanoparticles can accumulate in tumors due to their enhanced permeability and retention properties and have been widely applied in drug delivery to enhance therapeutic effects and reduce adverse reactions.26, 27 Cationic lipids, such as DOTAP, and miRNAs can form NPs by electrostatic adsorption. However, such NPs typically have limited circulation time and stability. In previous work, to enhance the antitumor effects of HCC treatment, miR‐375 was loaded onto calcium carbonate NPs along with chemotherapeutic drugs to produce miR‐375/drug coloaded lipid‐coated calcium carbonate NPs.28, 29 This design offers the advantages conferred by cationic liposomes and calcium carbonate, which include: (i) a strong proton sponge effect that permits rapid, pH‐dependent drug delivery in the cytoplasm and promotes endosomal lysis;30 (ii) the rapid release of highly stable encapsulated drugs into the tumor microenvironment;31 (iii) excellent biocompatibility and degradability;32 and (iv) improved therapeutic efficacy and prolonged circulation time as a result of escaping clearance through the reticuloendothelial system.
Herein, we first investigated miRNA expression in a panel of 3 CRC cell lines and in tumor tissues using qRT‐PCR. The present study described the downregulation of miR‐375‐3p in human CRC. MicroRNA‐375‐3p overexpression increased 5‐FU sensitivity. Further bioinformatic analysis revealed that miR‐375‐3p contained a binding site in the 3′‐UTR of the TYMS gene. TYMS was proved to be a direct target of miR‐375‐3p, and TYMS knockdown exerted similar influences on cellular response to 5‐FU as miR‐375‐3p overexpression. Next, we developed an NP formulation that was coloaded with 5‐FU and miR‐375‐3p (named miR‐375 + 5‐FU/NPs) and evaluated the therapeutic efficacy of these NPs in vivo in CRC models. Finally, we found TYMS‐related signaling pathways in colon cancer using GSEA and GSVA.
2. MATERIALS AND METHODS
2.1. Cell culture, miRNA precursors and inhibitors, and treatment
The human CRC cell lines, which included HCT116, HT29, SW480, Caco2, and NCM460, were stored by Hubei Clinical Center and Key Laboratory of Intestinal and Colorectal Diseases (Zhongnan Hospital of Wuhan University) and were thawed and cultured in DMEM or RMPI‐1640 containing 10% FCS (Invitrogen Gibco) and 1% penicillin‐streptomycin at 37°C in a humidified environment with 5% CO2. The 5‐FU‐resistant cell line HCT‐15/FU was purchased from the Chinese Academy of Sciences and subjected to short tandem repeat genotyping. The precursor and inhibitor of miR‐375‐3p and the negative control (miR‐NC) or miR‐NC inhibitor were synthesized by RiboBio.
2.2. Expression datasets
Colorectal cancer patients with at least 5 years of follow‐up from TCGA database (hereafter, TCGA cohort) were enrolled in this study for clinical analyses. Among these patients, 367 had corresponding gene expression data (read counts) and relatively complete clinical information. Cases from TCGA with gene expression in both colon adenocarcinoma cancer tissues and normal colon tissues were used to analyze the miR‐375‐3p and TYMS mRNA levels.
2.3. 5‐Fluorouracil treatment and CCK‐8 assays
Colon cancer cells that were transfected with miR‐375‐3p mimics or miR‐NC were seeded in 96‐well plates. Then 5‐FU solution (MedChem Express; CAS No. 51‐21‐8) was added for 24, 48, or 72 hours. The 5‐FU concentration varied from 0.1 to 100 µg/mL. The cytotoxicity was measured using CCK‐8 kits (Promoter) according to the manufacturer’s instructions.
2.4. Flow cytometric analysis of apoptosis assays
A total of 5 × 105 cancer cells were transfected with 100 nmol/L mimics or nonspecific controls. After 48 hours, the cells were double‐stained with PI and annexin V (Vybrant Apoptosis Assay Kit; Invitrogen). Fluorescence intensity was detected by flow cytometer (BD FACSCanto II cell sorting system, BD Biosciences) to identify apoptotic cells.
2.5. Cell migration and invasion assays
The migration and invasion assays were carried out with Transwell insert chambers (8‐mm pore size; Corning). For the migration assay, after transfection for 24 hours, 10 × 104 HCT116 cells and 20 × 104 HT29 cells were placed in serum‐free medium in the upper chamber. The lower chamber was filled with 20% serum medium. After 24‐36 hours of incubation, the cells in the upper chamber were removed with a cotton swab, and the cells in the lower chamber were fixed and dyed. For the invasion assay, cells were plated in the upper chamber, which had been coated with diluted Matrigel (ECM gel; Sigma), and then harvested after incubation for 24‐36 hours.
2.6. Quantitative real‐time PCR
Total RNA was extracted from tumor tissues or cells using TRIzol reagent (Invitrogen). A specific miR‐375‐3p primer and reverse transcriptase (Toyobo) were applied. Then qRT‐PCR was carried out using SYBR‐Green PCR Master Mix and primers on a Bio‐Rad real‐time PCR system. U6 small nuclear RNA was used to normalize miR‐375‐3p level. For miRNA quantification, Bulge‐loop miRNA qRT‐PCR Primer Sets specific for miR‐375‐3p were designed by RiboBio. The primers used for SYBR Green qRT‐PCR are listed in Table S1.
2.7. Small interfering RNA and plasmid transfection
Both TYMS and control siRNA were purchased from RiboBio. The human TYMS ORF cDNA plasmid pCMV3, which contains the full‐length TYMS coding sequence, was purchased from Sino Biological (Cat. HG17389‐UT). Cells were transiently transfected with TYMS siRNA or control siRNA and TYMS plasmid or negative control vector with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. The siRNA sequences are listed in Table S2.
2.8. Western blot analysis
Western blotting was carried out as previously described33 with Abs targeting TYMS (15047‐1‐AP; Proteintech) and β‐actin (#4970; Cell Signaling Technology). Antibodies targeting p‐Akt (#13038), p‐mTOR (#5536), and Bax (#5023T) were purchased from Cell Signaling Technology, whereas those targeting Bcl‐2 (A0208), MMP‐2 (A6247), MMP‐9 (A0289), cyclin D1 (A0310), and GAPDH (AC001) were purchased from ABclonal.
2.9. Luciferase assay
To precisely identify the target of miR‐375‐3p, the pMIR‐REPORT system (Applied Biosystems) was applied. HT29 cells were seeded in 96‐well plates and cotransfected with pMIR‐TYMS 3′‐UTR or pMIR‐TYMS 3′‐UTR‐mut plasmid and miR‐375‐3p mimic or miR‐NC. The pMIR‐REPORT luciferase vector was transfected into each group to serve as a normalized control. After 48 hours of transfection, the cells were analyzed for luciferase intensity by dual‐luciferase assay (Promega). The firefly luciferase intensity was normalized to Renilla luciferase intensity.
2.10. Preparation of miR‐375 + 5‐FU/NPs
Anionic lipid‐coated CaCO3 cores were prepared using a water‐in‐oil reverse emulsion method as described previously, with modifications.29, 34, 35 The synthesis of the 5‐FU NP cores was carried out following a previous study.29
For the synthesis of 5‐FU/NPs, DOTAP, cholesterol, methoxy‐polyethylene‐glycol‐distearoyl‐phosphatidyl‐ethanolamine, and 5‐FU NP cores were dissolved in chloroform. After chloroform removal by rotary evaporation, residual lipids were dispersed in H2O to form 5‐FU/NPs. To prepare miR‐375 + 5‐FU/NPs, 5‐FU/NPs and miR‐375‐3p were mixed at a 5‐FU/NP : miR‐375‐3p ratio of 200:1 (w/w) in RNase‐free H2O. The samples were vortexed for 30 seconds and then incubated at room temperature for 20 minutes to ensure the formation of miR‐375 + 5‐FU/NPs.
2.11. In vitro pH‐triggered drug release
The pH‐dependent release of miR‐375‐3p and 5‐FU from 375 + 5‐FU/NPs was detected at room temperature using dialysis. Acetate and PBS solutions adjusted to different pH values (pH 7.4, 6.8, and 5.5) were used as drug‐release media to simulate normal blood, tissue, and tumor environments. The experimental protocols followed previous publications.28, 29
2.12. Intracellular distribution of miR‐375 + 5‐FU/NPs
To analyze the uptake of 375 + 5‐FU/NPs, CRC cancer cells were seeded in 6‐well plates and transfected with Cy3‐labeled miR‐375‐3p for 1.5 hours. The cells were then washed and fixed. The nuclei were counterstained with DAPI. The cells were then imaged under an Olympus fluorescence microscope.
2.13. Xenograft assay
The mouse experiments were approved by the Animal Experiment Center of Zhongnan Hospital of Wuhan University and were carried out under specific pathogen‐free conditions. HCT116 cells (2.5 × 106) were suspended in PBS and s.c. injected into the flanks of 4‐week‐old BALB/c nude mice. The tumor sizes were measured every other day starting from day 10 after inoculation, at which point the tumor volume was 100‐200 mm3. The mice were randomly separated into 5 treatment groups as follows: PBS, 5‐FU/NPs, miR‐NC/NPs, miR‐375/NPs, and miR‐375 + 5‐FU/NPs. On day 12, the mice were intratumorally injected with their respective treatments. The doses of 5‐FU and miR‐375‐3p were 1.0 mg/kg and 2 nmol per mouse, respectively. The treatments were carried out every 3 days until day 24. Tumor volume was determined by measuring tumor length (L) and width (W) with calipers and was calculated as (L × W2)/2.
2.14. Histology
Tumor tissues were collected, fixed in 4% paraformaldehyde, embedded in paraffin, sectioned, and mounted on slides. The sections were then stained with H&E. Ki‐67 was used for histological evaluations of tumor tissues prepared for immunohistochemistry. The primary Abs anti‐TYMS (1:100; Proteintech), anti‐Ki‐67 (1:200; Abcam), anti‐Bax (1:100; Proteintech), and anti‐Bcl2 (1:100; Proteintech) were applied to the tissue sections, which were then incubated overnight at 4°C. The procedures were carried out as previously described.36
2.15. Liver and kidney toxicity
Blood samples from the eye were collected after the mice were killed and then centrifuged to separate the cells and plasma. The plasma was used to assess liver and kidney function by using an AST, ALT, Cr, and BUN detection kit (NJJCBio) following the manufacturer’s instructions.
2.16. Gene set enrichment analysis
To determine whether an a priori defined set of genes significantly differed between 2 biological states, a computational method called GSEA was used.37 The TYMS expression data were divided into high and low groups according to the median TYMS expression level, and gene set permutations were undertaken 1000 times for each analysis. In the whole process, the expression level of TYMS was regarded as a phenotype. The pathways enriched for each phenotype were classified by the NOM P value and normalized enrichment score. Gene sets with a normal P value less than .05 and FDR less than 0.25 were considered significantly enriched.
2.17. Gene set variation analysis with TYMS expression
The GSVA package in R was used to explore biological processes and KEGG pathways of genes showing differential enrichment between the low and high TYMS expression groups.38 Gene terms with |logFC| ≥0.1 and P less than .05 were considered statistically significant, and a heatmap was generated using R version 3.5.1.
2.18. Statistical analysis
All data are represented as the means of at least 3 samples ± SD unless otherwise indicated. Statistical differences between groups were assessed by paired or unpaired 2‐tailed Student’s t tests using GraphPad 5.0 software. Two‐tailed P values of less than .05 were considered statistically significant. Differences among more than 2 groups were assessed by one‐way ANOVA. The Wilcoxon rank‐sum test was used to analyze the differences in the distributions of the demographic characteristics of patients between the 2 different TYMS and miR‐375 expression groups. Pearson’s χ2 test was used to evaluate the correlation of clinicopathologic features between 2 groups with differential expression.
3. RESULTS
3.1. MicroRNA‐375‐3p widely downregulated in human CRC and promotes sensitivity of CRC cells to 5‐FU
To measure the expression levels of miR‐375‐3p in human CRC specimens, qRT‐PCR analysis was undertaken on 30 pairs of tumor specimens and matched adjacent normal tissues. The significant downregulation of miR‐375‐3p was observed in tumor tissues compared with adjacent normal tissues (Figure 1A). In addition, the expression level of miR‐375‐3p was downregulated in CRC cells (SW480, HCT116, HT29, and Caco2) compared with a normal colon epithelium cell line (NCM460) (Figure 1B). Next, we investigated the role of miR‐375‐3p in regulating resistance to 5‐FU in CRC. Quantitative real‐time PCR analysis indicated that a miR‐375‐3p mimic promoted miR‐375‐3p expression in both HCT116 and HT29 cells (Figure 1C). The response of HT29 and HCT116 cells to 5‐FU increased after treatment with miR‐375‐3p mimic compared with miR‐NC (Figure 1D,E). Thus, the results indicated that miR‐375‐3p can overcome 5‐FU resistance in CRC.
Figure 1.
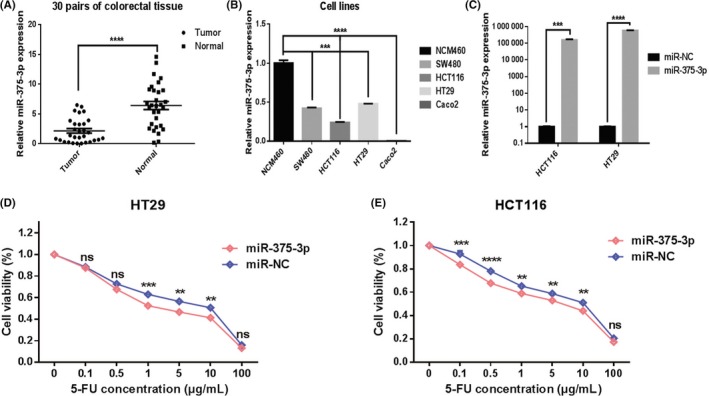
MicroRNA (miR)‐375‐3p is widely downregulated in human colorectal cancer and promotes the sensitivity of cells to 5‐fluorouracil (5‐FU). A, Expression levels of miR‐375‐3p were detected by quantitative real‐time (qRT)‐PCR in 30 pairs of tumor tissues and matched adjacent normal tissues. B, Expression levels of miR‐375‐3p in different CRC cells and in normal colon epithelium cell lines were detected by qRT‐PCR. C, Expression levels of miR‐375‐3p in HCT116 and HT29 cells treated with miR‐375‐3p mimic or miR‐NC (negative control) for 24 h were measured by qRT‐PCR analysis. D, E, HT29 (D) and HCT116 cells (E) treated with miR‐375‐3p mimic or miR‐NC were incubated with different concentrations of 5‐FU for 24 h, and 5‐FU chemosensitivity was determined by CCK‐8 assay. All data are shown as the means ± SEM of 3 independent experiments. *P < .05, **P < .01, ***P < .001, ****P < .0001
3.2. MicroRNA‐375‐3p increased 5‐FU sensitivity by inducing CRC cell apoptosis and cell cycle arrest and inhibiting cell proliferation, migration, and invasion in vitro
To explore the mechanisms by which miR‐375‐3p increased 5‐FU sensitivity, cell proliferation, migration, and invasion assays were carried out. We found that cell proliferation, migration, and invasion were attenuated in miR‐375‐3p mimic‐transfected cells relative to miR‐NC‐transfected cells (Figure 2A‐C). Moreover, the suppressive effects of combined miR‐375‐3p and 5‐FU were stronger than those of the individual treatments. We then investigated the role of miR‐375‐3p in regulating cell apoptosis by using flow cytometry. As shown in Figure 2D, compared with the control (9.28% and 13.87% in HT29 and HCT116, respectively), miR‐375‐3p (9.9% and 13.93% in HT29 and HCT116, respectively) or 5‐FU (42.22% and 18.09% in HT29 and HCT116, respectively) alone could increase the proportion of apoptotic cells in both cells. Moreover, more apoptotic cells in both cell types were found when miR‐375‐3p was combined with 5‐FU (52.71% and 22.37% in HT29 and HCT116, respectively). We next investigated the effects of miR‐375‐3p on cell cycle progression. The results showed that miR‐375‐3p can arrest cells in G0/G1 phase. (Figure S1A). The levels of functional proteins regulated by miR‐375‐3p associated with the malignant phenotype of CRC cells are shown in Figure S1B; these proteins include cell proliferation markers p‐Akt and p‐mTOR, classical proapoptotic (Bax) and antiapoptotic (Bcl‐2) markers, the migration markers MMP‐2 and MMP‐9, and the cyclic protein cyclin D1. We also used the 5‐FU‐resistant cell line HCT‐15/FU to study how miR‐375‐3p regulates tolerance to 5‐FU; the results for the malignant phenotype are presented in Figure S2.
Figure 2.
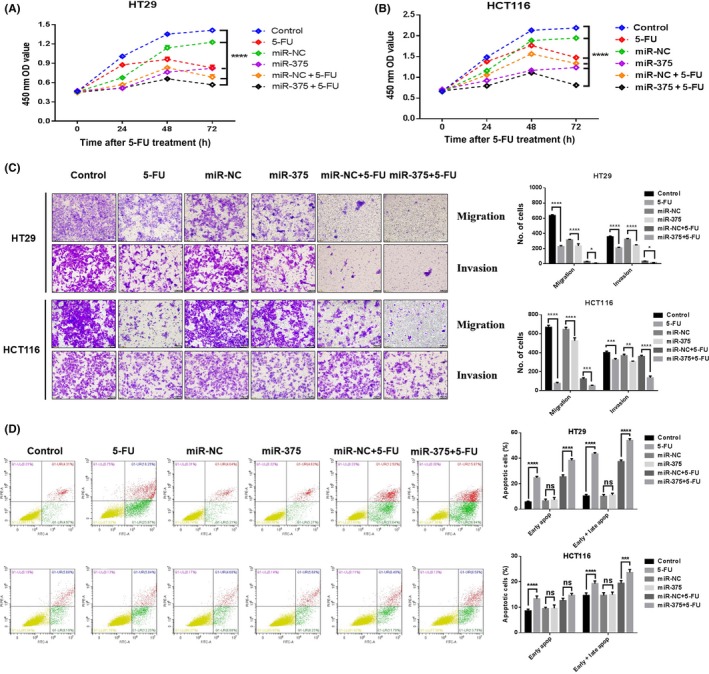
MicroRNA (miR)‐375‐3p increased the sensitivity of cells to 5‐fluorouracil (5‐FU) in vitro by regulating malignant phenotypes. A, B, Treatment with miR‐375‐3p in combination with 5‐FU resulted in significantly lower viability of (A) HT29 and (B) HCT116 cells than did any other treatment. OD, optical density. C, Transwell invasion (with Matrigel) and migrations (no Matrigel) assays with HCT116 and HT29 cells subjected to different treatments (×200) (left). Histograms indicate the number of invading and migrating cells (right). D, Apoptosis of HT29 and HCT116 cells at 48 h after transfection was detected by annexin V/propidium iodide labeling and flow cytometry. Quadrants from the lower left (counterclockwise) represent healthy, early apoptotic, late apoptotic, and necrotic cells, respectively. Evaluation of apoptosis was based on the number of apoptotic cells relative to the total cell number. Cells were treated with 100 nmol/L miRNA and 1 μg/mL 5‐FU concentration. All data are shown as the means ± SEM of 3 independent experiments. *P < .05, **P < .01, ****P < .0001
These data suggested that the induction of apoptosis and cell cycle arrest and the inhibition of cell proliferation, migration, and invasion were likely involved in the miR‐375‐3p regulation of 5‐FU sensitivity.
3.3. Potential target gene of miR‐375‐3p
Using the TargetScan and RNAhybrid algorithms, TYMS was predicted to be a direct target of miR‐375‐3p (Figure 3A). TYMS mRNA and protein levels were downregulated in miR‐375‐3p‐overexpressing cells relative to the levels in controls (Figure 3B‐E). In contrast, the overexpression of an miR‐375‐3p inhibitor upregulated TYMS protein levels (Figure 3F). Finally, the cotransfection of miR‐375‐3p mimics significantly decreased the WT TYMS luciferase reporter activity, whereas the luciferase activity was not reduced when the cells were transfected with a mutated TYMS reporter (Figure 3G).
Figure 3.
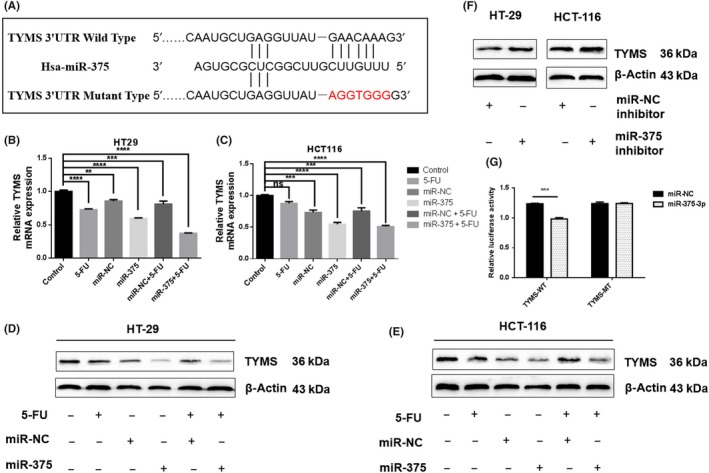
Thymidylate synthase (TYMS) is a direct target of microRNA (miR)‐375‐3p in colorectal cancer. A, Alignment of the potential miR‐375 binding site in the 3′‐UTR of TYMS mRNA. B‐E, mRNA (B, C) and protein levels (D, E) of TYMS were measured by quantitative real‐time PCR and western blotting, respectively, in HT29 and HCT116 cells transfected with miR‐375‐3p mimics. F, Protein levels of TYMS were measured by western blotting in HT29 and HCT116 cells transfected with an miR‐375‐3p inhibitor. G, Luciferase assay of HT29 cells transfected with a dual‐luciferase reporter vector containing WT or mutant 3′‐UTR of TYMS mRNA. At 24 h after transfection, cells were further transfected with miR‐375‐3p or miR‐NC (negative control), and the luciferase activity was measured. Cells were treated with 100 nmol/L miRNA and 1 μg/mL 5‐fluorouracil (5‐FU) concentration. All data are shown as the means ± SEM of 3 independent experiments. **P < .01, ***P < .001, ****P < .0001
Collectively, these results indicate that there is a negative correlation between miR‐375‐3p and TYMS, suggesting that miR‐375‐3p might exert its effects through the direct suppression of TYMS.
3.4. Silencing of TYMS reverses the suppression of miR‐375‐3p
To investigate whether TYMS plays a significant role in miR‐375‐3p‐mediated 5‐FU resistance, 3 TYMS siRNAs were selected for TYMS silencing. We found that the knockdown ability of TYMS siR‐2 was superior to that of the others (Figure S3A), and this si‐TYMS was selected for subsequent functional assays. As TYMS was a direct target of miR‐375‐3p, we first investigated the effects of TYMS on cell sensitivity to 5‐FU in colon cancer cells. Our results indicated that the responses of HT29, HCT116, and HCT‐15/FU cells to 5‐FU increased after treatment with si‐TYMS relative to their responses after transfection with si‐NC (Figures 4Aa,B and S4A). As expected, the downregulation of TYMS simulated miR‐375‐3p‐inhibited cell proliferation, migration, and invasion and miR‐375‐3p‐promoted cell apoptosis (Figures 4Ac,Ad,B,C and S4A‐C), suggesting that miR‐375‐3p suppressed human colon cancer cell proliferation, migration, and invasion as well as induced cell apoptosis by inhibiting its target gene TYMS. However, after knocking out TYMS, the cell cycle was blocked in S phase (Figures S4D and S5A), and after 5‐FU treatment, cell cycle was arrested at G0/G1 phase (HT29 and HCT116 cells) or S phase (HCT‐15/FU cells). These results differed from those of the miR‐375‐3p overexpression experiments. The levels of functional proteins following si‐TYMS treatment associated with the malignant phenotype of CRC cells are shown in Figure S5B. Additionally, we undertook similar functional experiments in which TYMS was overexpressed using special plasmids (Figure S3B), and the results are presented in Figure S6. Taken together, these results suggest that miR‐375‐3p mediates 5‐FU resistance by targeting TYMS in colon cancer.
Figure 4.
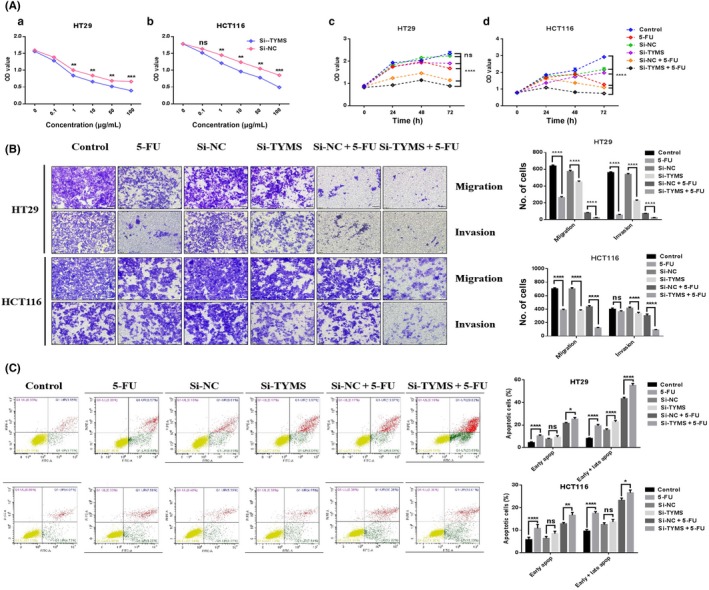
Silencing thymidylate synthase (TYMS) reverses the suppression of microRNA (miR)‐375‐3p in colorectal cancer cells. A, Effects of TYMS knockdown on cell proliferation. HT29 (a) and HCT116 (b) cells treated with si‐TYMS and si‐NC (negative control) were incubated with different concentrations of 5‐FU for 24 h, and 5‐fluorouracil (5‐FU) chemosensitivity was determined by CCK‐8. Compared with all other treatments, si‐TYMS given in combination with 5‐FU resulted in significantly lower viability of HT29 (c) and HCT116 (d) cells. OD, optical density. B, Transwell invasion (with Matrigel) and migration (without Matrigel) assays with HT29 and HCT116 cells subjected to different treatments (× 200) (left). Histograms indicate the numbers of invading and migrating cells (right). C, Apoptosis of HT29 and HCT116 cells at 48 h after transfection was detected by annexin V/propidium iodide labeling and flow cytometry. Quadrants from the lower left (counterclockwise) represent healthy, early apoptotic, late apoptotic, and necrotic cells, respectively. The evaluation of apoptosis was based on the number of apoptotic cells relative to the total cell number. Cells were treated with the 100 nmol/L miRNA and 1 μg/mL 5‐FU. All data are shown as the means ± SEM of 3 independent experiments. Comparisons were evaluated using a t test. *P < .05, **P < .01, ***P < .001, ****P < .0001
3.5. Restoration of miR‐375‐3p overcomes 5‐FU resistance in vivo
In light of our findings that 5‐FU in association with miR‐375‐3p mimic synergistically increased the anticancer effect of 5‐FU in colon cancer cells in vitro, we examined the effect of this combination in mice.
To achieve the cotransport of 5‐FU and miR‐375‐3p into cells, a cationic liposome drug‐loading system was designed, as described in the Materials and Methods section. The cationic liposomes had a calcium carbonate core and a cationic lipid shell. Transmission electron microscopy revealed that the NPs were spherical and had a light lipid shell and a dark calcium carbonate core. The average diameter of the miR‐375 + 5‐FU/NP core coated with 1,2‐dioleoyl‐sn‐glycerol‐3‐phosphate and DOTAP was approximately 50 nm.28 To study the uptake of the cationic liposome drug‐loading system, cells were treated with NPs or 5‐FU/NPs loaded with Cy3‐labeled miR‐375‐3p. As shown in Figure 5A, miR‐375‐3p was uniformly distributed throughout the cytoplasm and had rapidly and efficiently entered the nucleus 1.5 hours after transfection. The pH‐dependent release of the drug was analyzed by dialysis. Buffer solutions at weakly acidic pH values (pH 5.5) were used as drug‐release media to simulate the tumor environment.31 After 72 hours, more than 90% of the drug was released at pH 5.5, as reported by others.28
Figure 5.
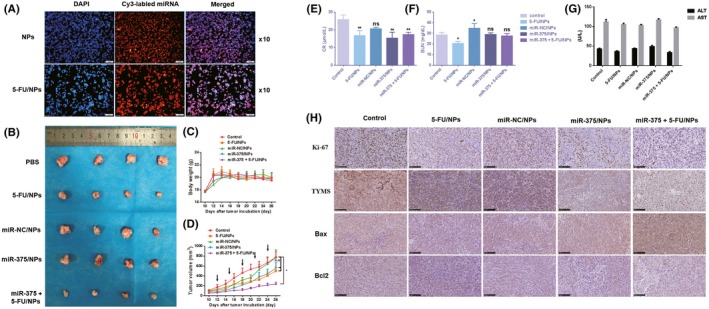
Restoration of microRNA (miR)‐375‐3p expression overcomes 5‐fluorouracil (5‐FU) resistance in vivo. A, Cellular uptake of miR‐375‐3p delivered by cationic liposomes in HT29 cells was visualized by fluorescence microscopy. Colorectal cancer cells were incubated with Cy3‐labeled miR‐375‐3p for 1.5 h, stained with DAPI, and observed under a fluorescence microscope (×100). B, Morphology of tumors excised from mice at the end of the experiment. C, Body weight of mice after they were killed. D, Growth curves of xenograft tumors. Results are presented as the means ± SEM of 4 animals per group. E‐G, Changes in renal and liver function after different drug treatments. H, Immunohistochemical analysis of the proliferation marker Ki‐67, thymidylate synthase (TYMS), and apoptotic proteins Bax and Bcl‐2 (×200). ALT, alanine transaminase; AST, aspartate transaminase; NP, nanoparticle
To determine whether the delivery of miR‐375‐3p in vivo increases 5‐FU chemosensitivity, we established a colon cancer model by s.c. injecting 2.5 × 106 HCT116 cells in BALB/c nude mice. When the tumor volume had reached 100‐200 mm3, the mice were randomly separated into 5 groups and treated with PBS, 5‐FU/NPs, miR‐NC/NPs, miR‐375/NPs, or miR‐375 + 5‐FU/NPs (on day 12). The tumor sizes were measured every other day starting from day 10, and the injections were carried out every 3 days until day 24. MicroRNA‐375 + 5‐FU/NPs inhibited the growth of tumors more efficiently than the 5‐FU/NPs or miR‐375/NPs individual treatments (Figure 5B,D), which indicated that miR‐375 + 5‐FU/NPs possessed great potential for reversing the resistance to 5‐FU treatment in colon cancer. Particularly, the body weight of 375 + 5‐FU/NPs showed no distinct decline among these groups (Figure 5C). Renal function, as measured by Cr and BUN levels, differed little among the groups (Figure 5E,F). Similarly, there were no differences in the liver function tests for ALT and AST among the different groups (Figure 5G). To verify the apoptotic effect of 375 + 5‐FU/NPs on tumor tissue, the expression of Bax and Bcl‐2 was measured by immunohistochemistry. As shown in Figure 5H, in response to the combination treatment, the expression of the proapoptotic protein Bax was significantly increased, whereas the expression of the antiapoptotic protein Bcl‐2 was markedly downregulated. The expression level of Ki‐67, a marker of cellular proliferation, was found to markedly decrease in response to the combination treatment. Additionally, consistent with the in vitro results, the immunohistochemistry of xenografts also revealed a marked reduction in TYMS expression in the miR‐375/NP‐treated group and the miR‐375 + 5‐FU/NP‐treated group (Figure 5H) relative to TYMS expression in the other groups. All of these results indicate the efficacy of 375 + 5‐FU/NPs, which was significantly higher than the efficacies of the individual treatments in vivo.
3.6. Correlation between TYMS or miR‐375 expression and clinical features of colon cancer
To investigate the correlation between TYMS or miR‐375 expression and the clinicopathological features of colon cancer, patients from TCGA database with at least 5 years of follow‐up were enrolled in this study for clinical analyses. We divided patients into 2 groups (high vs. low TYMS or miR‐375 expression) according to the media value. The associations identified between TYMS expression and the clinical characteristics of colon cancer cases are summarized in Table 1. Age (P = .035), preoperative pretreatment carcinoembryonic antigen level (P = .013), and tumor stage (P = .00072) were significantly correlated with TYMS expression, whereas only age (P = .005) was significantly correlated with miR‐375 expression.
Table 1.
Correlation of clinicopathologic features with tumor microRNA (miR)‐375 and thymidylate synthase (TYMS) mRNA levels in a cohort of colorectal cancer patients (n = 367)
| miR‐375 | TYMS | |||||
|---|---|---|---|---|---|---|
| High | Low | P value | High | Low | P value | |
| Gender | ||||||
| Female | 91 | 84 | .495 | 93 | 82 | .27100 |
| Male | 93 | 99 | 91 | 101 | ||
| Age (y) | ||||||
| <50 | 28 | 13 | .005 | 24 | 17 | .03500 |
| 50‐70 | 68 | 94 | 69 | 93 | ||
| >70 | 88 | 76 | 91 | 73 | ||
| Colon polyps present | ||||||
| Yes | 32 | 31 | .426 | 30 | 33 | .08300 |
| No | 52 | 63 | 49 | 66 | ||
| Unknown | 100 | 89 | 105 | 84 | ||
| Lymphatic invasion | ||||||
| Yes | 68 | 67 | .982 | 60 | 75 | .24100 |
| No | 98 | 97 | 105 | 90 | ||
| Unknown | 18 | 19 | 19 | 18 | ||
| Cancer status | ||||||
| With tumor | 41 | 46 | .534 | 36 | 51 | .12400 |
| Tumor‐free | 116 | 105 | 114 | 107 | ||
| Unknown | 27 | 32 | 34 | 25 | ||
| Preoperative pretreatment CEA level (ng/mL) | ||||||
| <5.9 | 85 | 79 | .661 | 79 | 85 | .01300 |
| ≥5.9 | 37 | 34 | 27 | 44 | ||
| Unknown | 62 | 70 | 78 | 54 | ||
| Tumor stage | ||||||
| Stage I + II | 105 | 92 | .336 | 119 | 77 | .00072 |
| Stage III + IV | 75 | 84 | 60 | 100 | ||
| Unknown | 4 | 7 | 5 | 6 | ||
| Venous invasion | ||||||
| Yes | 37 | 45 | .508 | 36 | 46 | .23600 |
| No | 120 | 116 | 119 | 117 | ||
| Unknown | 27 | 22 | 29 | 20 | ||
Bold indicates significant P values
Data were subjected to the chi‐Square test
4. Thymidylate synthase‐related signaling pathways in colon cancer based on GSEA and GSVA
To explore the potential functions of TYMS in colon cancer, we undertook KEGG pathway analyses with GSEA, a robust computational method to identify a priori‐defined set of genes associated with a special phenotype, and GSVA, which is designed to integrate genes that share common biological functions. The former revealed significant differences (FDR < 0.25, NOM P value < .05) in the enrichment of the MSigDB Collection; details of the top 20 enriched gene sets are provided in Table S3. In particular, gene sets related to the cell cycle, p53 signaling pathway, homologous recombination, RNA degradation, arginine and proline metabolism, and DNA replication were differentially enriched in the high TYMS expression phenotype (Figure 6). Similar conclusions were also obtained from GSVA, and representative terms are presented in the heatmap in Figure 7.
Figure 6.
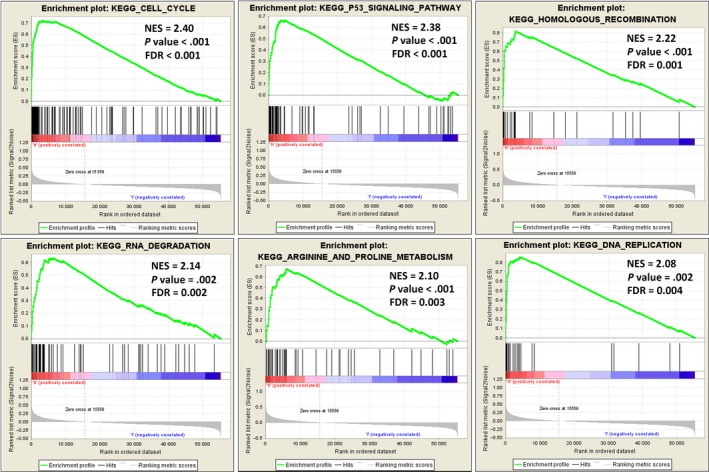
Enrichment plots from gene set enrichment analysis. Several pathways and biological processes, including the cell cycle, p53 signaling pathway, homologous recombination, RNA degradation, arginine and proline metabolism, and DNA replication, were differentially enriched in colon adenocarcinoma cases with high thymidylate synthase expression. ES, enrichment score; FDR, false discovery rate; KEGG, Kyoto Encyclopedia of Genes and Genomes; NES, normalized enrichment score; NOM P‐val, nominal P value
Figure 7.
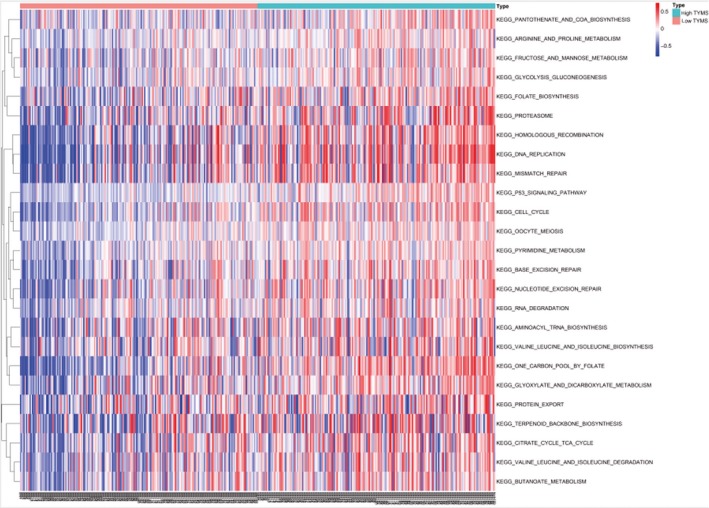
Thymidylate synthase (TYMS)‐related signaling pathways in colon cancer. TYMS‐related Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways based on the gene set variation analysis of The Cancer Genome Atlas data. Representative terms are presented in the heatmap
5. DISCUSSION
Colorectal cancer is the most frequently diagnosed malignancy of the gastrointestinal tract. Despite the rapid development of novel targeted therapies, fluoropyrimidine, either in combination or as a single agent, remains the mainstay of treatment for disseminated, metastatic CRC. However, despite the steady improvement of 5‐FU‐based treatment regimens, the patient response rate to 5‐FU‐based chemotherapy remains modest, mainly due to the development of drug resistance.39 The acquisition of drug resistance is a multifactorial process mediated by complex determinants. Further studies are needed to identify effective drivers and to elucidate the mechanisms underlying resistance to chemotherapeutic agents in cancer cells.
Evidence indicates that abnormal miRNA expression is linked to the initiation and development of diverse cancers as well as to cancer cell resistance to therapeutic drugs.40, 41, 42 Because personalized medicine is rapidly being more widely adopted, the discovery of specific biomarkers for chemotherapy response is an important step toward implementing individualized treatment.
MicroRNA‐375 was first described as a specific miRNA of the murine pancreatic islet β‐cell line MIN6, with its gene located in a region widely conserved in both humans and mice.43 Since its first description and characterization, miR‐375 has been identified as having roles in islet development, glucose homeostasis, the production and secretion of pulmonary surfactants, mucosal‐mediated immunity, and tumorigenesis.44 Studies have revealed that miR‐375 expression is highly aberrant in diverse human cancer types and could offer potential diagnostic and prognostic value.36
Thymidylate synthase is an enzyme that catalyzes the methylation of dUMP to yield dTMP and thereby impacts DNA synthesis and repair.45 Inhibition of TYMS is considered the main mechanism of 5‐FU action.46 Increasing evidence shows that patients with high expression of TYMS develop cell resistance to 5‐FU.47, 48 We measured the protein expression levels of TYMS in the normal colon epithelial‐derived cell line NCM460, the colon cancer cell lines HCT116, HT29, SW480, and Caco2, and the 5‐FU drug‐resistant cell line HCT‐15/FU (Figure S3C). We found that our experimental cell lines HCT116, HT29, and HCT‐15/FU had higher levels of TYMS expression than did the normal colon‐derived cells and other colon cancer cell lines, indicating that these cells have some degree of intrinsic drug resistance. Our results further indicated that miR‐375‐3p exerts its role by targeting TYMS, which is also a common target of 5‐FU. Functional experiments showed that either the overexpression of miR‐375‐3p or the silencing of TYMS enhanced cell sensitivity to 5‐FU by inhibiting proliferation, invasion, and migration and by inducing apoptosis and cell cycle arrest. We also analyzed patient information from TCGA database and found that miR‐375 was significantly downregulated in colon cancer tumor tissue relative to normal tissue (Figure S7A), and TYMS was significantly upregulated in cancer tissues relative to normal ones (Figure S7B). Targeting miR‐375‐3p could be an effective method for maximizing the effects of 5‐FU chemotherapy in vitro. Our results indicate that miR‐375‐3p could serve as a therapeutic target for 5‐FU‐based chemotherapy in colon cancer.
The combination of traditional chemotherapy with miRNA therapy has advantages for cancer therapy. However, the differences between the 2 pharmaceuticals in molecular weight can lead to differences in their biodistribution and tumor accumulation. Many studies have reported that the coloading of chemotherapeutic drugs and RNA interference drugs onto NPs can allow the 2 treatments to exert their maximal effects.23, 24, 25, 49, 50 In particular, lipid‐coated calcium carbonate NPs have recently been exploited for nucleic acid delivery.35, 51 In the present study, miR‐375 + 5‐FU/NPs were successfully prepared for colon cancer therapy. The NPs showed a high loading efficiency and rapid drug release in a weakly acidic tumor microenvironment. In a murine s.c. tumor model, miR‐375 + 5‐FU/NPs exerted the expected combinatory inhibitory effects in vivo, and seemed to have no obvious toxicity to the liver and kidney.
The GSEA and GSVA results indicated that the effect of TYMS on colon cancer progression involved the cell cycle, p53 signaling pathway, homologous recombination, RNA degradation, arginine and proline metabolism, and DNA replication. Unfortunately, we did not have the chance to investigate the correlation between miR‐375‐3p and TYMS levels and their prognostic values in our own clinical specimens. In addition, the expression levels of miR‐375‐3p in patients with chemical‐resistant tumors and in those who respond well to chemotherapy need to be determined. Furthermore, research on the clinical value of miR‐375‐3p in chemotherapy is needed. In summary, we showed that miR‐375‐3p plays a crucial role in reducing resistance to 5‐FU. Although additional studies are necessary to confirm the functions of miR‐375‐3p, our results suggest that the restoration of miR‐375‐3p levels could be a novel therapeutic strategy to modulate and enhance chemosensitivity to 5‐FU treatment.
DISCLOSURE
The authors have no conflicts of interest.
ETHICS STATEMENT
All patients provided informed consent and all experimental procedures were approved by the Committee on Ethical Animal Experiment at Zhongnan Hospital of Wuhan University. The methods used in this study were carried out in accordance with the approved guidelines.
Supporting information
Supinfo
ACKNOWLEDGMENTS
This work was supported by the Hubei Provincial Natural Science Foundation of China (Project 2018CFB446), the Health Commission of Hubei Province Scientific Research (Project WJ2019H025) and Zhongnan Hospital of Wuhan University Science, Technology and Innovation Seed Fund (Project cxpy2017062). We would like to thank the School of Pharmacy, Tongji Medical College, Huazhong University of Science and Technology for the synthesis of the drug loading system of cationic liposome.
Xu F, Ye M‐L, Zhang Y‐P, et al. MicroRNA‐375‐3p enhances chemosensitivity to 5‐fluorouracil by targeting thymidylate synthase in colorectal cancer. Cancer Sci. 2020;111:1528–1541. 10.1111/cas.14356
Ming‐Liang Ye, Yu‐Peng Zhang, Wen‐Jie Li, and Meng‐Ting Li contributed equally.
REFERENCES
- 1. Wang L, Qian L, Li X, Yan J. MicroRNA‐195 inhibits colorectal cancer cell proliferation, colony‐formation and invasion through targeting CARMA3. Mol Med Rep. 2014;10:473‐478. [DOI] [PubMed] [Google Scholar]
- 2. Mansour MA, Hyodo T, Ito S, et al. SATB2 suppresses the progression of colorectal cancer cells via inactivation of MEK5/ERK5 signaling. Febs J. 2015;282:1394‐1405. [DOI] [PubMed] [Google Scholar]
- 3. Balaguer F, Link A, Lozano JJ, et al. Epigenetic silencing of miR‐137 is an early event in colorectal carcinogenesis. Cancer Res. 2010;70:6609‐6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Benson AR, Bekaii‐Saab T, Chan E, et al. Localized colon cancer, version 3.2013: featured updates to the NCCN Guidelines. J Natl Compr Canc Netw. 2013;11:519‐528. [DOI] [PubMed] [Google Scholar]
- 5. Benson AR, Venook AP, Bekaii‐Saab T, et al. Version 2.2015. J Natl Compr Canc Netw. 2015;13:719‐728. [DOI] [PubMed] [Google Scholar]
- 6. Qiu LX, Tang QY, Bai JL, et al. Predictive value of thymidylate synthase expression in advanced colorectal cancer patients receiving fluoropyrimidine‐based chemotherapy: evidence from 24 studies. Int J Cancer. 2008;123:2384‐2389. [DOI] [PubMed] [Google Scholar]
- 7. Chu E, Callender MA, Farrell MP, Schmitz JC. Thymidylate synthase inhibitors as anticancer agents: from bench to bedside. Cancer Chemother Pharmacol. 2003;52(Suppl 1):S80‐S89. [DOI] [PubMed] [Google Scholar]
- 8. Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64‐71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Svoronos AA, Engelman DM, Slack FJ. OncomiR or tumor suppressor? The duplicity of MicroRNAs in cancer. Cancer Res. 2016;76:3666‐3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. An X, Sarmiento C, Tan T, Zhu H. Regulation of multidrug resistance by microRNAs in anti‐cancer therapy. Acta Pharm Sin B. 2017;7:38‐51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fojo T. Multiple paths to a drug resistance phenotype: mutations, translocations, deletions and amplification of coding genes or promoter regions, epigenetic changes and microRNAs. Drug Resist Updat. 2007;10:59‐67. [DOI] [PubMed] [Google Scholar]
- 12. Gotanda K, Hirota T, Matsumoto N, Ieiri I. MicroRNA‐433 negatively regulates the expression of thymidylate synthase (TYMS) responsible for 5‐fluorouracil sensitivity in HeLa cells. BMC Cancer. 2013;13:369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li T, Gao F, Zhang XP. miR‐203 enhances chemosensitivity to 5‐fluorouracil by targeting thymidylate synthase in colorectal cancer. Oncol Rep. 2015;33:607‐614. [DOI] [PubMed] [Google Scholar]
- 14. Sun Z, Zhou N, Han Q, et al. MicroRNA‐197 influences 5‐fluorouracil resistance via thymidylate synthase in colorectal cancer. Clin Transl Oncol. 2015;17:876‐883. [DOI] [PubMed] [Google Scholar]
- 15. Wang S, Wang L, Bayaxi N, et al. A microRNA panel to discriminate carcinomas from high‐grade intraepithelial neoplasms in colonoscopy biopsy tissue. Gut. 2013;62:280‐289. [DOI] [PubMed] [Google Scholar]
- 16. Ding L, Xu Y, Zhang W, et al. MiR‐375 frequently downregulated in gastric cancer inhibits cell proliferation by targeting JAK2. Cell Res. 2010;20:784‐793. [DOI] [PubMed] [Google Scholar]
- 17. Chang Y, Yan W, He X, et al. miR‐375 inhibits autophagy and reduces viability of hepatocellular carcinoma cells under hypoxic conditions. Gastroenterology. 2012;143:177‐187. [DOI] [PubMed] [Google Scholar]
- 18. Kong KL, Kwong DL, Chan TH, et al. MicroRNA‐375 inhibits tumour growth and metastasis in oesophageal squamous cell carcinoma through repressing insulin‐like growth factor 1 receptor. Gut. 2012;61:33‐42. [DOI] [PubMed] [Google Scholar]
- 19. Huang X, Yuan T, Liang M, et al. Exosomal miR‐1290 and miR‐375 as prognostic markers in castration‐resistant prostate cancer. Eur Urol. 2015;67:33‐41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Provvisiero DP, Negri M, de Angelis C, et al. Vitamin D reverts resistance to the mTOR inhibitor everolimus in hepatocellular carcinoma through the activation of a miR‐375/oncogenes circuit. Sci Rep. 2019;9:11695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ward A, Balwierz A, Zhang JD, et al. Re‐expression of microRNA‐375 reverses both tamoxifen resistance and accompanying EMT‐like properties in breast cancer. Oncogene. 2013;32:1173‐1182. [DOI] [PubMed] [Google Scholar]
- 22. Shen Y, Wang P, Li Y, et al. miR‐375 is upregulated in acquired paclitaxel resistance in cervical cancer. Br J Cancer. 2013;109:92‐99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sun TM, Du JZ, Yao YD, et al. Simultaneous delivery of siRNA and paclitaxel via a "two‐in‐one" micelleplex promotes synergistic tumor suppression. ACS Nano. 2011;5:1483‐1494. [DOI] [PubMed] [Google Scholar]
- 24. Xiong XB, Lavasanifar A. Traceable multifunctional micellar nanocarriers for cancer‐targeted co‐delivery of MDR‐1 siRNA and doxorubicin. ACS Nano. 2011;5:5202‐5213. [DOI] [PubMed] [Google Scholar]
- 25. Xu X, Xie K, Zhang XQ, et al. Enhancing tumor cell response to chemotherapy through nanoparticle‐mediated codelivery of siRNA and cisplatin prodrug. Proc Natl Acad Sci. 2013;110:18638‐18643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang X, Zeng X, Liang X, et al. The chemotherapeutic potential of PEG‐b‐PLGA copolymer micelles that combine chloroquine as autophagy inhibitor and docetaxel as an anti‐cancer drug. Biomaterials. 2014;35:9144‐9154. [DOI] [PubMed] [Google Scholar]
- 27. Cho K, Wang X, Nie S, Chen ZG, Shin DM. Therapeutic nanoparticles for drug delivery in cancer. Clin Cancer Res. 2008;14:1310‐1316. [DOI] [PubMed] [Google Scholar]
- 28. Zhao P, Li M, Wang Y, et al. Enhancing anti‐tumor efficiency in hepatocellular carcinoma through the autophagy inhibition by miR‐375/sorafenib in lipid‐coated calcium carbonate nanoparticles. Acta Biomater. 2018;72:248‐255. [DOI] [PubMed] [Google Scholar]
- 29. Zhao P, Wu S, Cheng Y, et al. MiR‐375 delivered by lipid‐coated doxorubicin‐calcium carbonate nanoparticles overcomes chemoresistance in hepatocellular carcinoma. Nanomedicine‐Uk. 2017;13:2507‐2516. [DOI] [PubMed] [Google Scholar]
- 30. Kim SK, Foote MB, Huang L. Targeted delivery of EV peptide to tumor cell cytoplasm using lipid coated calcium carbonate nanoparticles. Cancer Lett. 2013;334:311‐318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lim EK, Huh YM, Yang J, Lee K, Suh JS, Haam S. pH‐triggered drug‐releasing magnetic nanoparticles for cancer therapy guided by molecular imaging by MRI. Adv Mater. 2011;23:2436‐2442. [DOI] [PubMed] [Google Scholar]
- 32. Ma X, Chen H, Yang L, Wang K, Guo Y, Yuan L. Construction and potential applications of a functionalized cell with an intracellular mineral scaffold. Angew Chem Int Ed Engl. 2011;50:7414‐7417. [DOI] [PubMed] [Google Scholar]
- 33. Xu F, Liao JZ, Xiang GY, et al. MiR‐101 and doxorubicin codelivered by liposomes suppressing malignant properties of hepatocellular carcinoma. Cancer Med. 2017;6:651‐661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Poojari R, Kini S, Srivastava R, Panda D. Intracellular interactions of electrostatically mediated layer‐by‐layer assembled polyelectrolytes based sorafenib nanoparticles in oral cancer cells. Colloids Surf B Biointerfaces. 2016;143:131‐138. [DOI] [PubMed] [Google Scholar]
- 35. Li J, Yang Y, Huang L. Calcium phosphate nanoparticles with an asymmetric lipid bilayer coating for siRNA delivery to the tumor. J Control Release. 2012;158:108‐114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. He XX, Chang Y, Meng FY, et al. MicroRNA‐375 targets AEG‐1 in hepatocellular carcinoma and suppresses liver cancer cell growth in vitro and in vivo. Oncogene. 2012;31:3357‐3369. [DOI] [PubMed] [Google Scholar]
- 37. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge‐based approach for interpreting genome‐wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545‐15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA‐seq data. BMC Bioinformatics. 2013;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Allen WL, Stevenson L, Coyle VM, et al. A systems biology approach identifies SART1 as a novel determinant of both 5‐fluorouracil and SN38 drug resistance in colorectal cancer. Mol Cancer Ther. 2012;11:119‐131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fujita Y, Kojima K, Hamada N, et al. Effects of miR‐34a on cell growth and chemoresistance in prostate cancer PC3 cells. Biochem Biophys Res Commun. 2008;377:114‐119. [DOI] [PubMed] [Google Scholar]
- 41. Hwang JH, Voortman J, Giovannetti E, et al. Identification of microRNA‐21 as a biomarker for chemoresistance and clinical outcome following adjuvant therapy in resectable pancreatic cancer. PLoS ONE. 2010;5:e10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bitarte N, Bandres E, Boni V, et al. MicroRNA‐451 is involved in the self‐renewal, tumorigenicity, and chemoresistance of colorectal cancer stem cells. Stem Cells. 2011;29:1661‐1671. [DOI] [PubMed] [Google Scholar]
- 43. Baroukh NN, Van Obberghen E. Function of microRNA‐375 and microRNA‐124a in pancreas and brain. Febs J. 2009;276:6509‐6521. [DOI] [PubMed] [Google Scholar]
- 44. Yan JW, Lin JS, He XX. The emerging role of miR‐375 in cancer. Int J Cancer. 2014;135:1011‐1018. [DOI] [PubMed] [Google Scholar]
- 45. Lehman NL. Future potential of thymidylate synthase inhibitors in cancer therapy. Expert Opin Investig Drugs. 2002;11:1775‐1787. [DOI] [PubMed] [Google Scholar]
- 46. Houghton JA, Tillman DM, Harwood FG. Ratio of 2'‐deoxyadenosine‐5'‐triphosphate/thymidine‐5'‐triphosphate influences the commitment of human colon carcinoma cells to thymineless death. Clin Cancer Res. 1995;1:723‐730. [PubMed] [Google Scholar]
- 47. Cho YB, Chung HJ, Lee WY, et al. Relationship between TYMS and ERCC1 mRNA expression and in vitro chemosensitivity in colorectal cancer. Anticancer Res. 2011;31:3843‐3849. [PubMed] [Google Scholar]
- 48. Galbiatti AL, Caldas HC, Maniglia JV, Pavarino EC, Goloni‐Bertollo EM. Gene expression profile of 5‐fluorouracil metabolic enzymes in laryngeal cancer cell line: predictive parameters for response to 5‐fluorouracil‐based chemotherapy. Biomed Pharmacother. 2014;68:515‐519. [DOI] [PubMed] [Google Scholar]
- 49. Patil YB, Swaminathan SK, Sadhukha T, Ma L, Panyam J. The use of nanoparticle‐mediated targeted gene silencing and drug delivery to overcome tumor drug resistance. Biomaterials. 2010;31:358‐365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yang T, Zhao P, Rong Z, et al. Anti‐tumor efficiency of lipid‐coated cisplatin nanoparticles co‐loaded with MicroRNA‐375. Theranostics. 2016;6:142‐154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li J, Chen YC, Tseng YC, Mozumdar S, Huang L. Biodegradable calcium phosphate nanoparticle with lipid coating for systemic siRNA delivery. J Control Release. 2010;142:416‐421. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supinfo


