Abstract
Metastasis‐associated lung adenocarcinoma transcript 1 (MALAT1) is an oncogenic long noncoding RNA that has been found to promote carcinogenesis and metastasis in many tumors. However, the underlying role of MALAT1 in the progression and metastasis of hepatocellular carcinoma (HCC) remains unclear. In this study, aberrantly elevated levels of MALAT1 were detected in both HCC specimens and cell lines. We found that knockdown of MALAT1 caused retardation in proliferation, migration, and invasion both in vivo and in vitro. Mechanistic investigations showed that Snail family transcriptional repressor 1 (SNAI1) is a direct target of microRNA (miR)‐22 and that MALAT1 modulates SNAI1 expression by acting as a competing endogenous RNA for miR‐22. Inhibition of miR‐22 restored SNAI1 expression suppressed by MALAT1 knockdown. Furthermore, MALAT1 facilitated the enrichment of enhancer of zeste homolog 2 (EZH2) at the promoter region of miR‐22 and E‐cadherin, which was repressed by MALAT1 knockdown. Cooperating with EZH2, MALAT1 positively regulated SNAI1 by repressing miR‐22 and inhibiting E‐cadherin expression, playing a vital role in epithelial to mesenchymal transition. In conclusion, our results reveal a mechanism by which MALAT1 promotes HCC progression and provides a potential target for HCC therapy.
Keywords: EZH2, hepatocellular carcinoma, MALAT1, miR‐22, SNAI1
Metastasis‐associated lung adenocarcinoma transcript 1 (MALAT1) cooperating with enhancer of zeste homolog 2, positively regulated Snail family transcriptional repressor 1 (SNAI1) by repressing microRNA (miR)‐22 and inhibiting E‐cadherin expression, playing a vital role in epithelial to mesenchymal transition. These findings indicated that MALAT1 promotes hepatocellular carcinoma (HCC) progression by modulating the miR‐22/SNAI1 axis and provides a potential target for HCC therapy.

Abbreviations
- Ago2
argonaute RISC catalytic component 2
- ceRNA
competing endogenous RNA
- EMT
epithelial‐mesenchymal transition
- EZH2
enhancer of zeste homolog 2
- H3K27
histone H3 lysine 27
- HCC
hepatocellular carcinoma
- HOTAIR
HOX transcript antisense RNA
- lncRNA
long noncoding RNA
- MALAT1
metastasis‐associated lung adenocarcinoma transcript 1
- miR
microRNA
- PRC2
polycomb repressive complex 2
- qPCR
quantitative PCR
- RIP
RNA immunoprecipitation
- RISC
RNA‐induced silencing complex
- SNAI1
Snail family transcriptional repressor 1
- TCGA
The Cancer Genome Atlas
1. INTRODUCTION
Hepatocellular carcinoma is one of the most common malignancies worldwide. 1 , 2 In China, it was estimated that approximately 466 100 cases were newly diagnosed and caused approximately 422 100 deaths in 2015. 3 Despite significant recent advances in surgical techniques and chemotherapeutics, the 5‐year survival rate of HCC patients remains poor, eg, only 18% in the United States. 4 An improved understanding of the molecular pathogenesis of HCC could help us to develop new effective targeted therapies for HCC patients.
More than 90% of the human genome is actively transcribed, and the majority of transcripts are noncoding RNAs, 5 which have no protein‐coding capacity. Long noncoding RNAs are members of this family and are temporally and spatially specific in different tissue. 6 , 7 In several diseases, including cancer, the expression and function of lncRNAs are often deregulated. 8 Metastasis‐associated lung adenocarcinoma transcript 1, a widely studied lncRNA, is upregulated in several types of solid tumors and is defined as an independent prognostic factor in non‐small‐cell lung cancer. 9 , 10 , 11 , 12 , 13 , 14 However, there are only preliminary studies of MALAT1 in HCC, and the biological role and underlying mechanism of MALAT1 in HCC progression and metastasis remain largely unknown.
Enhancer of zeste homolog 2, the catalytic subunit of PRC2, functions as a H3K27 methyltransferase at the target gene promoter and has been identified as a key chromatin repressive epigenetic modification. 15 , 16 It is reported to be overexpressed in almost all human cancers. 17 However, the mechanism that confers specificity to EZH2 or polycomb targeting is unknown. 18 Long noncoding RNAs exert their functions by interacting with special proteins, including EZH2. Hirata et al reported that MALAT1 interacts with EZH2 in renal cancer, which induces EMT and Wnt/β‐catenin signaling hyperactivation. 11 It is interesting to study whether lncRNAs participate in specifying the targets of PRC2 through the enrollment of EZH2. Another well‐defined mechanism of lncRNA is that it can function as a natural miRNA sponge to regulate gene expression posttranscriptionally. 19 , 20 Our previous study confirmed that EZH2 represses tumor‐suppressive miR‐22 expression in HCC, 21 but it remains unknown whether MALAT1 participates in the regulation of miR‐22 in HCC. Accordingly, it is interesting to study the interaction between MALAT1, EZH2, and miRNA in HCC.
Collectively, in this study, we found that MALAT1 positively regulates the expression of SNAI1, a key transcription factor in EMT, by sponging miR‐22. Moreover, MALAT1 interacts with and facilitates EZH2 occupation at the promoter of miR‐22 and E‐cadherin. Together, the results of our study elucidate the role of MALAT1/miR‐22 in HCC progression and shed new light on lncRNA‐directed diagnostics and therapeutics in liver cancer.
2. MATERIALS AND METHODS
2.1. Clinical samples
Thirty pairs of well‐established primary HCC specimens and matched adjacent normal tissues were obtained from Union Hospital, Tongji Medical College (stage Ⅰ, 6 cases; stage Ⅱ, 8 cases; stage Ⅲ, 12 cases; stage Ⅳ, 4 cases). Each tumor tissue sample was snap‐frozen in liquid nitrogen and stored at −80°C for 10 minutes. The protocols used in this study were approved by the Institutional Review Board of Tongji Medical College of Huazhong University of Science and Technology.
2.2. Cell culture
HepG2, Hep3B, HuH7, and PLC/PRF5 cells were obtained from the Chinese Academy of Sciences Cell Bank. The cells were cultured in DMEM supplemented with 10% FBS (Life Technologies), penicillin (100 U/mL), and streptomycin (100 mg/mL) and were maintained in a humidified incubator at 37°C with 5% CO2.
2.3. Antibodies and reagents
E‐cadherin, EZH2, vimentin, and β‐actin Abs were purchased from Cell Signaling Technology. The SNAI1, Ago2, and HRP‐conjugated secondary Abs were purchased from Proteintech. Matrigel matrix was obtained from BD Biosciences. The primers for quantitative real‐time PCR and sequences for ChIP/RIP/RNA pull‐down were shown in Table S1.
2.4. Plasmids and transfection
Three shRNAs targeting MALAT1 and a control were synthesized by GeneChem. The miR‐22‐3p mimic and inhibitor (anti‐22‐3p) and negative controls were purchased from RiboBio. Transfection was carried out using a Lipofectamine 3000 Kit (Invitrogen) according to the manufacturer’s instructions. Stable cell lines were selected by puromycin treatment (1.5 µg/mL) for 2 weeks.
2.5. RNA extraction and qPCR assays
Total RNA, including miRNA, was isolated with an RNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol. Reverse transcription was undertaken with a PrimeScript RT Reagent Kit (Takara Biomedical Technology). The stem‐loop method was used to reverse transcribe mature miR‐22. All data analyses were undertaken using the StepOnePlus Real‐Time PCR System (Applied Biosystems). All of the results were normalized to the expression of GAPDH or U6.
2.6. Western blot analysis
Tissue or cellular proteins were extracted with 1× cell lysis buffer (Promega). Whole‐cell lysates were separated on 10% SDS‐PAGE gels and blotted on Immobilon PVDF membranes, which were hybridized with Abs overnight at 4°C before being incubated with the appropriate secondary Ab for 1 hour at room temperature. The membranes were then incubated with a chemiluminescence substrate according to the manufacturer’s protocol (Pierce) and visualized by autoradiography.
2.7. Proliferation assay
An EdU kit (Cell‐Light EdU Apollo 643 In Vitro Imaging Kit; RiboBio) was used to evaluate the cell proliferation viability according to the manufacturer’s instructions. Images were detected and analyzed with a microscope at 200× (Olympus). The ratio of EdU‐stained cells (with red fluorescence) to Hoechst‐stained cells (with blue fluorescence) was used to evaluate cell proliferation activity.
2.8. Chromatin immunoprecipitation assay
Hepatocellular carcinoma cells were seeded in 15‐cm wells until they reached approximately 80% confluence. The ChIP assay was then carried out using a SimpleChIP Enzymatic Chromatin IP Kit (Cell Signaling Technology) according to the manufacturer’s instructions. Quantitative PCR was undertaken with SYBR Green PCR Master Mix and primer sets targeting the putative EZH2 binding region in the MIR22HG promoter. The amount of immunoprecipitated DNA was calculated and normalized to the amount of input DNA.
2.9. RNA immunoprecipitation
The RIP assay was performed using the EZ‐Magna RIP Kit (Millipore) according to the manufacturer’s instructions. Specific primers were designed to detect whether MALAT1 was associated with the RISC and other proteins.
2.10. RNA pull‐down assay
Cells were transfected with biotinylated miR‐22, biotinylated mutant miR‐22, or biotinylated control (synthesized by TsingKe), and the final concentration of miRNA was 50 nmol/L. To determine whether MALAT1 was associated with the RISC, the DNA fragment covering the miRNA‐22‐3p seed region binding site of MALAT1 was PCR‐amplified and cloned into pEASY‐T5 Zero (Transgene). The resultant plasmid was linearized with the restriction enzyme Not1 or Spe1. Biotin‐labeled RNAs were in vitro transcribed with Biotin RNA Labeling Mix (Roche Diagnostics) and purified with the RNeasy Mini Kit (Qiagen). The antisense strand of the resultant vector was also transcribed as a control. Cells were harvested and lysed, and a pull‐down assay was carried out as previously described. 22
2.11. Luciferase reporter assay
psiCHECK‐2, psiCHECK2‐MALAT1 wt, or psiCHECK2‐MALAT1 mut was cotransfected with miRNA‐22 mimics or controls into HCC cells. The Renilla luciferase activity was normalized to firefly luciferase activity 48 hours after transfection.
2.12. Wound healing assay
Cells were seeded in 6‐well plates and grown under permissive conditions to 90% confluency in complete media. The wound was made in the confluent monolayer using a conventional pipette tip and then washed with PBS to remove the detached cells. Cell migration was observed by microscopy for the indicated times and analyzed objectively using Image J. Wound closure percentages were calculated using the following formula: (1 − [current wound size/initial wound size]) × 100.
2.13. Migration and invasion assay
A total of 5 × 104 cells were resuspended in 200 µL serum‐free medium and placed into the upper chamber of a Transwell insert (Corning, 8 µm). The lower chamber was filled with 15% FBS as a chemoattractant and incubated for 24 hours for the migration assay and 48 hours for the invasion assay. For the invasion assay, the inserts were previously coated with Matrigel matrix gel (No. 356234, Corning). At the end of the experiments, the cells on the upper surface of the membrane were removed, and the cells on the lower surface were fixed and stained with 0.1% crystal violet. Five visual fields of each insert were randomly chosen and counted under a light microscope.
2.14. Xenograft mouse model
HepG2 cells (1 × 106) stably expressing control shRNA or MALAT1 shRNA and the miR‐22 inhibitor or inhibitor control were s.c. injected into the flank area of 4‐week‐old female BALB/c nude mice (n = 5 mice per group). Tumor volumes were measured (0.5 × length × width2) weekly. After 4 weeks, mice were killed and tumor fluorescence was visualized using an In Vivo FX PRO system (Bruker). Tumors were excised and subjected to immunohistochemistry analysis. Animal experiments were approved by the Animal Care Committee of Tongji Medical College (IACUC: 602).
2.15. Statistical analysis
Data are expressed as the mean ± SD of at least 3 replicated experiments. Student’s t test was used to analyze differences between 2 groups, and Pearson’s coefficient correlation was used to analyze relationships between the expression levels of specific genes. A P value less than .05 indicated that a particular difference was statistically significant.
3. RESULTS
3.1. Metastasis‐associated lung adenocarcinoma transcript 1 upregulated in HCC tissues and cell lines and negatively correlated with miR‐22
Quantitative PCR was used to detect MALAT1 expression in 30 pairs of clinical samples and HCC cell lines. We found elevated MALAT1 levels in most liver cancer samples compared to adjacent noncancerous tissues (Figure 1A,B); similarly, HCC cell lines expressed higher levels of MALAT1 and lower levels of miR‐22 than normal liver tissues (Figure 1C). Additionally, higher levels of MALAT1 were detected in high‐stage patients (III+IV) than low‐stage patients (I+II) (Figure 1D). Through data mining of TCGA HCC datasets by third‐party tools, higher levels of MALAT1 were found in HCC tissues, similar to our result (Figure 1E). Additionally, overall survival was significantly shorter in the high MALAT1 expression group compared to the low MALAT1 group (Figure 1F).
Figure 1.

Metastasis‐associated lung adenocarcinoma transcript 1 (MALAT1) is upregulated in hepatocellular carcinoma (HCC) tissues and cell lines and negatively correlated with microRNA (miR)‐22. A, MALAT1 expression was determined by quantitative PCR and normalized to GAPDH in 30 paired HCC tissues and adjacent noncancerous tissues. B, MALAT1 expression levels of HCC and paired normal tissues were analyzed and expressed as log2 fold change (HCC/normal), and the log2 fold changes were presented as follows: >1, overexpression (17 cases); <−1, underexpression (3 cases); and the remainder were defined as unchanged (10 cases). C, HCC cell lines expressed higher levels of MALAT1 and lower levels of miR‐22 than normal liver tissues. D, Higher MALAT1 and lower miR‐22 levels were detected in high‐stage patients than low‐stage patients. E, Box plot representation of MALAT1 levels in normal liver (n = 10) and HCC (n = 35) samples. Analysis is based on RNA sequencing data from the Oncomine database (https://www.oncomine.org). F, G, Associations of MALAT1 (F) and miR‐22 (G) expression with overall survival were analyzed by Kaplan‐Meier plots. Survival data were based on The Cancer Genome Atlas (TCGA) datasets and analyzed by SurvExpress (http://bioinformatica.mty.itesm.mx/SurvExpress) and UCSC Xena (http://xena.ucsc.edu/), respectively. H, I, Inverse correlations were found between MALAT1 and miR‐22 in both clinical specimens (H) and TCGA datasets (I). (*P < .05, **P < .01)
Our previous study confirmed the tumor‐suppressive role of miR‐22 in HCC. 21 Lower levels of miR‐22 were detected in high‐stage patients (III+IV) than low‐stage patients (I+II) (Figure 1D). It was validated in TCGA datasets that higher levels of miR‐22 indicated longer overall survival in HCC patients (Figure 1G). Interestingly, an inverse correlation between MALAT1 and miR‐22 was found in cell lines, our clinical samples, and TCGA HCC datasets (Figure 1C, H, I).
3.2. Knockdown of MALAT1 inhibits proliferation, migration, and invasion of HCC cells
To determine whether MALAT1 is essential for HCC maintenance, we studied the effect of knocking down MALAT1 on HCC cells. Three siRNAs targeting MALAT1 were tested for their knockdown efficiency, and the 2 most efficient (si‐MALAT1‐1 and si‐MALAT1‐2) were selected for further experiments (Figure 2A). The CCK‐8 assays revealed that knockdown of MALAT1 significantly attenuated the proliferation ability of HCC cells (Figure 2B). As shown in the Edu proliferation assay (Figure 2C), lower proliferation rates were observed in the knockdown group compared to the control. The wound healing assay showed that cells transfected with siMALAT1 underwent slower closure of scratch wounds compared to the control group (Figure 2D,E). Transwell assays were also carried out. As shown in Figure 2F, knockdown of MALAT1 impeded both the migration and invasion capacities of HCC cells.
Figure 2.
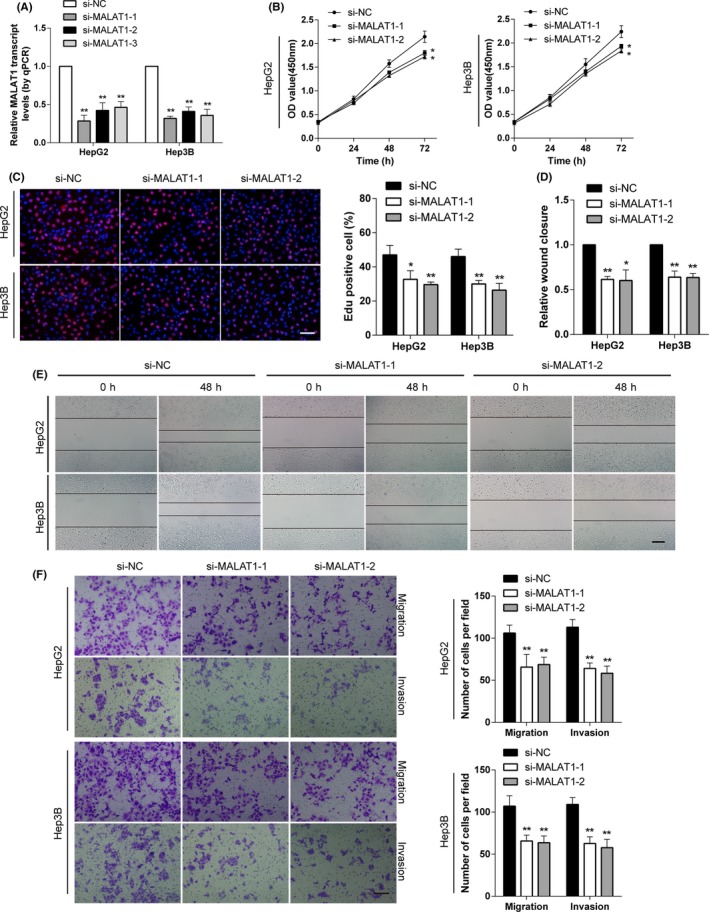
Knockdown of metastasis‐associated lung adenocarcinoma transcript 1 (MALAT1) inhibits cell proliferation, migration, and invasion of hepatocellular carcinoma (HCC) cells in vitro. A, MALAT1 levels were detected after transfection with siRNAs specific for MALAT1 in HepG2 and Hep3B cells. The most effective 2 were selected for further study. B, CCK‐8 assays showed that knockdown of MALAT1 decreased the proliferation abilities of HCC cells. C, Edu assays showed that suppression of MALAT1 attenuated the proliferation rate of HCC cells (scale bar, 50 µm). D, E, Wound healing assays showed that MALAT1 knockdown resulted in a delayed closure of scratch wounds (scale bar, 100 µm). F, Transwell migration and invasion assays showed that knockdown of MALAT1 attenuated the migration and invasion abilities of HCC cells (scale bar, 50 µm). *P < .05, **P < .01, compared with control (si‐NC)
3.3. Direct interaction between MALAT1 and miR‐22 in HCC
Because lncRNA can function as a natural sponge for miRNAs and MALAT1 is inversely correlated with miR‐22 in HCC, we investigated whether MALAT1 directly interacts with miR‐22 in HCC. Using the 2 online bioinformatics databases, starBase (http://starbase.sysu.edu.cn) and DIANA‐LncBase, we observed that miR‐22‐3p had several putative binding sites within MALAT1 (Figure 3A). A series of experiments were undertaken to verify this ceRNA interaction. First, after overexpression of miR‐22, the MALAT1 level significantly decreased, whereas inhibition of miR‐22 induced elevated levels of MALAT1 in HCC cell lines (Figure 3B). Knockdown of MALAT1 significantly increased miR‐22 expression in HCC cancer cells (Figure 3C). It has been shown that miRNAs exert gene silencing through the RISC, and Ago2 is the core component of RISC. To verify whether MALAT1 and miR‐22 were in the same RISC, RIP assays were carried out using the Ago2 Ab first. Higher levels of MALAT1 and miR‐22 were detected in Ago2 immunoprecipitates compared to IgG (Figure 3D). RNA‐induced silencing complex is a heterogeneous complex; MALAT1 and miR‐22 could still be part of different RISC complexes. Biotin‐labeled RNA pull‐down assays were utilized to explore whether miR‐22 could pull down MALAT1, and vice versa. Hepatocellular carcinoma cells were transfected with synthesized biotinylated miR‐22, a mutated form, or a control and were then lysed for the pull‐down assay. Then MALAT1 was pulled down by miR‐22 as identified by qPCR analysis, but the introduction of mutations hampered this interaction (Figure 3E). We also used an inverse pull‐down assay to test whether MALAT1 could pull down miR‐22 using a biotin‐labeled MALAT1 probe containing the predicted binding sites for miR‐22 and the biotin‐labeled antisense strand as an inner control. Higher levels of miR‐22 were precipitated in the MALAT group than in the control. To further confirm MALAT1 as a target of miR‐22, a dual‐luciferase reporter assay was undertaken. Our results showed that the miR‐22 mimic diminished the luciferase activity of the psiCHECK2 vector bearing the putative binding site of miR‐22, but had almost no effect on the mutated vector (Figure 3F). Collectively, the above results showed that MALAT1 was able to sponge miR‐22, as predicted in HCC.
Figure 3.

Metastasis‐associated lung adenocarcinoma transcript 1 (MALAT1) regulates microRNA (miR)‐22 expression by directly binding to miR‐22 in hepatocellular carcinoma (HCC) cells. A, Schematic representation of the putative binding sites of miR‐22 on the MALAT1 transcript, as predicted by LncBase Predicted version 2. B, Expression levels of MALAT1 in HCC cells transfected with miR‐22 mimic or inhibitor. C, miR‐22 levels were detected after knockdown and overexpression MALAT1, respectively. D, RNA immunoprecipitation assays were carried out using anti‐Ago2 Ab. Relative expression levels of MALAT1 and miR‐22 in HCC cells were detected by quantitative PCR (qPCR). Expressions of MALAT1 and miR‐22 were normalized by GAPDH and U6, respectively. E, HCC cell lysates were incubated with biotinylated MALAT1 probe containing the putative binding sites of miR‐22 or the biotinylated antisense strand. Lysates incubated with beads only were used as negative control (NC), input was used for normalization. MiR‐22 levels were analyzed by qPCR. F, Luciferase activities of HCC cells were detected after cotransfection with indicated vectors and miR‐22 mimic or control. *P < .05, **P < .01, compared with respective control
3.4. Enhancer of zeste homolog 2 interacts with 3′‐end of MALAT1 and knockdown of MALAT1 disrupts occupancy of EZH2 on miR‐22 promoter
Previous studies have shown the interaction of MALAT1 and EZH2 in renal cancer, 11 and thus, we undertook an RNA pull‐down assay to identify the interaction between MALAT1 and EZH2 in HCC cells. EZH2 was enriched in the sense‐strand probe compared to the antisense probe and beads‐only groups, as shown in Figure 4A. Furthermore, EZH2 was shown to bind to the 3′‐end of MALAT1 through truncated MALAT1 pull‐down assays, as confirmed by cross‐linking and sonication combined with anti‐EZH2 RIP assays and followed by qPCR (Figure 4B‐D). Next, to determine whether the interaction between MALAT1 and EZH2 was affected by miR‐22, anti‐EZH2 RIP assays were undertaken after transfection of miR‐22 mimics. Surprisingly, even the level of MALAT1 decreased after transfection of miR‐22 mimics, the relative enrichment of MALAT1 by the anti‐EZH2 Ab was not influenced by miR‐22 mimics (Figure 4E).
Figure 4.

Metastasis‐associated lung adenocarcinoma transcript 1 (MALAT1) interacts with enhancer of zeste homolog 2 (EZH2) in hepatocellular carcinoma. A, RNA pull‐down assays were carried out with biotinylated MALAT1 probe followed by western blot assay using anti‐EZH2 Ab. B, Schematic illustration of MALAT1 regions used for truncated RNA pull‐down assays. C, RNA pull‐down assays using biotinylated truncated MALAT1 confirmed that MALAT1 bound EZH2 mainly at its 3′‐end. D, Enrichment of the MALAT1 fractions in cross‐link and sonication RNA immunoprecipitation (RIP) using anti‐EZH2 Ab further validated EZH2 interacted with the 3′‐end of MALAT1. E, Anti‐EZH2 RIP was carried out after transiently overexpressing microRNA (miR)‐22 followed by detecting MALAT1 with quantitative PCR. miR‐22 did not compete with EZH2 for binding MALAT1. F, G, ChIP assays were undertaken with anti‐EZH2 and anti‐H3K27me3 Abs after knockdown of MALAT1. Knockdown of MALAT1 attenuated the enrichment of EZH2 at miR‐22 (F) and E‐cadherin (G) promoter. *P < .05, **P < .01, compared with respective control
Our previous study showed that EZH2 represses miR‐22 expression through occupancy of the promoter of the miR‐22 host gene MIR22HG. 21 In this study, after knockdown of MALAT1 in HCC cells, interestingly, the enrichment of EZH2 and H3K27me3 on the promoter region of miR‐22 was significantly decreased, as determined by ChIP assays (Figure 4F). Recent studies reported E‐cadherin expression was repressed by H3H27me3 mediated by EZH2 in HCC and renal cancer. 11 , 23 We checked the ENCODE (Encyclopedia of DNA Elements) database, based on the ChIP sequencing data of H3K27mes and EZH2 in HepG2 cells, enrichment peaks of both proteins were found near the promoter region of E‐cadherin (CHD1) and MIR22HG (Figure S1). We also found that knockdown of MALAT1 attenuated enrichment of EZH2 at E‐cadherin in HCC (Figure 4G). These results suggest that EZH2 represses miR‐22 expression in a MALAT1‐dependent manner.
3.5. SNAI1 is a direct target of miR‐22 and positively correlated with MALAT1 in HCC
To elucidate the molecular mechanism by which miR‐22 suppresses the proliferation and metastasis of HCC cells, bioinformatics tools were utilized to identify potential target genes. As a key transcription factor in EMT, SNAI1 was predicted to be a direct target of miR‐22. Dual‐luciferase reporter assays were undertaken to verify this connection (Figure 5A). After cotransfection with miR‐22 mimics, a remarkable decrease in luciferase activity was found in psiCHECK‐2 vectors harboring the 3′‐UTR sequence of SNAI1, but not the vector that contained a mutant in the seed sequence (Figure 5B). Furthermore, western blot assays showed that transfection of the miR‐22 mimic downregulated SNAI1 expression, whereas the miR‐22 inhibitor upregulated SNAI1 in HCC cells (Figure 5C). For the MALAT1/miR‐22 ceRNA network, a luciferase reporter assay and western blot were carried out after overexpression and knockdown of MALAT1. As predicted, upregulated MALAT1 caused high expression of SNAI1, whereas knockdown of MALAT1 decreased the expression of SNAI1 (Figure 5D‐F). The effects of MALAT1 were opposite to those of miR‐22.
Figure 5.

Snail family transcriptional repressor 1 (SNAI1) is a direct target of microRNA (miR)‐22 in hepatocellular carcinoma (HCC). A, Schematic representation and sequence of the intact miR‐22 binding site (WT) and its mutation form (MUT) within the luciferase reporter vector. B, HCC cells were cotransfected with indicated vector and miR‐22 mimic or control (NC), luciferase activities were determined 48 h after transfection. C, HCC cells were transfected with miR‐22 mimic (miR‐22 mmc) or inhibitor (anti‐miR‐22), followed by western blot assays to identify expression of SNAI1. D, Relative luciferase activities of HCC cells transfected with SNAI1 3′‐UTR WT vectors were determined after knockdown or upregulation of metastasis‐associated lung adenocarcinoma transcript 1 (MALAT1). E, F, SNAI1 expression levels in HCC cells were determined by western blot assays after knockdown or upregulation of MALAT1. G, A negative correlation was found between miR‐22 and SNAI1 mRNA levels in clinical specimens. H, MALAT1 was positively correlated with SNAI1 levels, confirmed by quantitative PCR in HCC tissues. *P < .05, **P < .01, compared with respective control
To exclude other miRNAs targeting both MALAT1 and SNAI1, we used TargetScan to predict miRNAs targeting SNAI1. StarBase version 2.0 and miRcode were used to screen miRNAs targeting MALAT1. The above results were intersected and several miRNAs were identified. Then SNAI1 and MALAT1 levels were examined after transferring mimics into HCC cells. As the result showed, miR‐22 was the only one that matched all 3 algorithms and downregulated both MALAT1 and SNAI1 markedly (Figure S2). To further confirm our findings, we analyzed the correlation between MALAT1/miR‐22 levels and SNAI1 expression levels in the specimens. As shown in Figure 5G, SNAI1 was negatively correlated with miR‐22, but was positively correlated with MALAT1 (Figure 5H).
3.6. Inhibition of miR‐22 partially reverses the effect of MALAT1 knockdown in HCC cells in vitro
To investigate the biological association of MALAT1 and miR‐22 in HCC cells, rescue experiments were undertaken by transfection of miR‐22 antagomir in MALAT1 knockdown cells. Western blot assays showed that the expression of E‐cadherin, vimentin, and SNAI1 were partially restored after inhibition of miR‐22 in MALAT1 knockdown cells (Figures 6A and S3). The Edu assay showed that the cell proliferation rate was decreased in MALAT1 knockdown cells, whereas the proliferation ability was partially restored after transfection of miR‐22 antagomir (Figure 6B). The colony formation assay revealed that the clonogenicity ability of HCC cells suppressed by MALAT1 knockdown was regained by miR‐22 inhibition (Figure 6C). In addition, similar effects were also observed in the migration and invasion abilities of HCC cells after transfection of miR‐22 antagomir (Figure 6D). In summary, rescue experiments proved that miR‐22 antagonizes the function of MALAT1 in HCC progression.
Figure 6.

Decreased abilities of proliferation, migration, and invasion caused by metastasis‐associated lung adenocarcinoma transcript 1 (MALAT1) knockdown were partially reversed by microRNA (miR)‐22 inhibition in vitro. Hepatocellular carcinoma (HCC) cells stably transfected with empty vector (mock) or sh‐MALAT1 were cotransfected with negative control inhibitor (anti‐ctrl, 100 nmol/L) or anti‐miR‐22 inhibitor (100 nmol/L). A, Western blot assays of epithelial‐mesenchymal transition markers (ie, E‐cadherin, vimentin, or Snail family transcriptional repressor 1 [SNAI1]) indicated that altered expressions caused by MALAT1 knockdown were reversed by miR‐22 inhibition. B, Representative images (upper) and quantification (lower) of Edu proliferation assays showed that inhibition of miR‐22 restored the proliferation abilities of HCC cells decreased by MALAT1 knockdown (scale bar, 50 µm). C, Representation (left panel) and quantification (right panel) of colony formation assays showed the clonogenicity abilities of HCC cells transfected with indicated vectors (scale bar, 1 cm). D, Knockdown of MALAT1 attenuated the migration and invasion of HCC cells, which was reversed by transfection of anti‐miR‐22 (scale bar, 50 µm). *P < .05, **P < .01, compared with respective control
3.7. Metastasis‐associated lung adenocarcinoma transcript 1 exerts its oncogenic activity partially by downregulating miR‐22 in vivo
To further identify the oncogenic potential of MALAT1 in vivo, HepG2 cells stably transfected with the desired vectors were injected s.c. into nude mice. As shown in Figure 7A‐C, the tumor volumes in the shMALAT1 group were dramatically smaller than those of the empty vector group, whereas cotransfection of the miR‐22 inhibitor partially restored the tumorigenicity ability inhibited by MALAT1 knockdown. Furthermore, the immunohistochemistry of the xenograft tumors indicated that expression of SNAI1 from the sh‐MALAT1 group was lower than that from the control group, and the miRNA‐22 inhibitor decreased the discrepancies (Figure 7D). In the experimental metastasis studies, HepG2 cells stably transfected with shMALAT1 established markedly fewer lung metastatic nodules than the mock group. Moreover, cotransfection of the miR‐22 inhibitor partially rescued the decrease in metastasis caused by MALAT1 knockdown in athymic nude mice (Figure 7E).
Figure 7.

Knockdown of metastasis‐associated lung adenocarcinoma transcript 1 (MALAT1) inhibited tumor growth and metastasis in vivo, and it could be restored by inhibition of microRNA (miR)‐22. HepG2 cells stably transfected with empty vector (mock) or sh‐MALAT1 were cotransfected with negative control inhibitor (anti‐ctrl, 100 nmol/L) or anti‐miR‐22 inhibitor (100 nmol/L). A, Representative fluorescence images of mice (n = 5 each group) injected with HepG2 cells stably transfected with indicated plasmids bearing the cherry fluorescent protein. Bar graph showing the quantification of normalized total photon counts of s.c. xenografts in mice of each group. B, In vivo growth curves of xenograft tumors after hypodermic injection of indicated HepG2 cells. C, Representation (upper panel) and quantification (lower panel) of xenograft tumors formed by hypodermic injection of HepG2 cells transfected with indicated vectors. D, Immunohistochemical staining for Snail family transcriptional repressor 1 (SNAI1) and E‐cadherin expression within tumors formed by hypodermic injections of HepG2 cells stably transfected with indicated vectors (scale bar, 100 µm). E, Representation (left panels, arrowhead; scale bar, 500 µm) and quantification (right panels; scale bar, 100 µm) of lung metastasis of nude mice injected with HepG2 cells (2 × 106) stably transfected with mock or sh‐MALAT1, and those cotransfected with anti‐miR‐22 or control through the tail vein (n = 5, each group)
4. DISCUSSION
Long noncoding RNA MALAT1, also known as nuclear‐enriched transcript 2 (NEAT2), has been reported to be overexpressed in many types of malignancies. 24 , 25 , 26 , 27 In this study, we also found higher levels of MALAT1 in HCC tissues compared to normal tissues. High levels of MALAT1 are associated with a high tumor grade and can be an independent prognosis factor for HCC patients. Knockdown of MALAT1 attenuated the proliferation, migration, and invasion capacities of HCC cells. Furthermore, we proved that MALAT1 interacts with EZH2 and exerts its oncogenic role to promote EMT by sponging miR‐22. Snail family transcriptional repressor 1, the key transcription factor of EMT, 28 , 29 , 30 , 31 is a direct target of miR‐22 and is positively correlated with MALAT1 in HCC.
Long noncoding RNAs can function as natural miRNA sponges. 20 , 32 In the present study, we identified the interaction between MALAT1 and miRNAs first. Our previous study showed that miR‐22 inhibits HCC cell proliferation and metastasis both in vitro and in vivo. 21 Interestingly, we found that MALAT1 was inversely correlated with miR‐22 in both clinical sample and TCGA datasets. Based on the bioinformatics database, we speculated that MALAT1 could interact with miR‐22 and form a ceRNA network. Anti‐Ago2 RIP assays identified MALAT and miR‐22 in the same RISC, and pull‐down assays showed that MALAT1 was able to pull down miR‐22 in HCC cells. Our results further confirmed the functional reciprocal repression between MALAT1 and miR‐22. In addition, inhibition of miR‐22 partially reversed the effect caused by MALAT1 knockdown. Taken together, these data revealed that MALAT1 could function as an effective miR‐22 sponge.
MicroRNA‐22, primitively cloned from HeLa cells, is an evolutionarily conserved miRNA. Increasingly, studies have confirmed that miR‑22 functions as an oncogenic or tumor‐suppressive miRNA in malignancies, depending on the cancer types and complex contexts. 33 In most situations, it functions as a tumor suppressor, including for HCC. Several targets of miR‐22 associated with tumor initiation and progression have been identified, including cyclin D1, MMP14, CD147, and histone deacetylase 6. 34 , 35 , 36 , 37 , 38 Our study also proved that SNAI1 is a direct target of miR‐22, similar to a recent report. 39 Snail family transcriptional repressor 1 is a key transcription factor in EMT, 31 and upregulation of SNAI1 increases binding to E‐box consensus sequences on the promoter, which promotes repression of the adhesion molecule E‐cadherin. Our study also found that MALAT1 is positively associated with SNAI1 and that knockdown of MALAT1 causes downregulation of SNAI1 at the protein level, which could be rescued by miR‐22 inhibition. These findings suggested that MALAT1/miR‐22 interacts with SNAI1 to promote the progression of HCC.
In addition to sponging miRNAs, lncRNAs also function as protein scaffold. 40 It was first discovered that HOTAIR mediates its effect by interacting with PRC2, enhancing the methylation of H3K27, leading to the silencing of tumor suppressor genes. 41 , 42 A recent study also found that HOTAIR mediates the physical interaction between SNAI1 and EZH2, which directly represses a broad repertoire of epithelial genes. 28 A previous study found that MALAT1 can interact with EZH2 in renal cancer, prostate cancer, and colorectal cancer. 25 , 43 , 44 Through data mining of TCGA datasets, we found a positive correlation between MALAT1 and EZH2 in liver cancer, and RIP and RNA pull‐down assays also confirmed that MALAT1 interacts with EZH2 through its 3′‐end in HCC cell lines. One interesting phenomenon was that the interaction between MALAT1 and EZH2 was slightly affected even the level of MALAT1 was dramatically decreased by miR‐22 mimics. We inferred that miR‐22 titrated MALAT1 located in cytoplasm, but not MALAT1 localized to nuclear speckles, which interacted with EZH2 and other RNA binding proteins.
As the catalytic subunit of PRC2, EZH2 mediates the gene silencing involved in tumor progression and metastasis through trimethylation of H3K27. 45 It plays a fundamental role in the cell cycle, proliferation, and aerobic metabolism by regulating the expression of a repertoire of genes. However, it is not known how the specificity of PRC2 is determined in various contexts or cell types. Here, in the frame of HCC, MALAT1 is pinpointed to function by directing PRC2 to various targets by enrollment of EZH2. It is also reported that H19 inhibits the adipocyte differentiation of bone marrow mesenchymal stem cells by modulating histone deacetylases. 46 We concluded that the interaction between lncRNA and epigenetic modifiers could be a mechanism to determine the specificity of epigenetic modifiers as well as a vital mechanism for lncRNAs to exert their profound biological functions.
In summary, our study shows that MALAT1 is an oncogene that interacts with EZH2 and exerts its role by sponging the tumor suppressor miR‐22 to upregulate SNAI1. At the same time, MALAT1 represses miR‐22 transcription by enhancing the enrichment of EZH2 and H3K27me3 on the miR‐22 promoter. Knockdown of MALAT1 decreased SNAI1 expression, leading to inhibition of EMT in HCC cells, which can be partially reversed by inhibition of miR‐22. These results suggest that downregulation of MALAT1 in tumors in which MALAT1 is overexpressed should be considered a new strategy for HCC therapy.
CONFLICT OF INTEREST
The authors have no conflict of interest for this article.
Supporting information
Fig S1‐S3
Table S1
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (Grant/Award Number 81772967), Natural Science Foundation of Hubei Province (Grant/Award Number 2019CFB496), Funds for training young and middle‐aged medical backbone talents in Wuhan (Grant/Award Number 02.05.18030002), Huazhong University of Science and Technology Innovation Research Fund (Grant/Award Number 2017KFYXJJ251), and National Key Basic Research Program of China (Grant/Award Number 2015CB5540007).
Chen S, Wang G, Tao K, et al. Long noncoding RNA metastasis‐associated lung adenocarcinoma transcript 1 cooperates with enhancer of zeste homolog 2 to promote hepatocellular carcinoma development by modulating the microRNA‐22/Snail family transcriptional repressor 1 axis. Cancer Sci. 2020;111:1582–1595. 10.1111/cas.14372
Shaofei Chen and Guobin Wang contributed equally to this work.
Contributor Information
Jiarui Pu, Email: 2013xh0964@hust.edu.cn.
Hang Li, Email: 2013xh0884@hust.edu.cn.
REFERENCES
- 1. El‐Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557‐2576. [DOI] [PubMed] [Google Scholar]
- 2. Li H, Wu K, Tao K, et al. Tim‐3/galectin‐9 signaling pathway mediates T‐cell dysfunction and predicts poor prognosis in patients with hepatitis B virus‐associated hepatocellular carcinoma. Hepatology. 2012;56:1342‐1351. [DOI] [PubMed] [Google Scholar]
- 3. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115‐132. [DOI] [PubMed] [Google Scholar]
- 4. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. [DOI] [PubMed] [Google Scholar]
- 5. Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489:101‐108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26‐46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci. 2008;105:716‐721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629‐641. [DOI] [PubMed] [Google Scholar]
- 9. Shi X, Sun M, Liu H, Yao Y, Song Y. Long non‐coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339:159‐166. [DOI] [PubMed] [Google Scholar]
- 10. Fang Z, Zhang S, Wang Y, et al. Long non‐coding RNA MALAT‐1 modulates metastatic potential of tongue squamous cell carcinomas partially through the regulation of small proline rich proteins. BMC Cancer. 2016;16:706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hirata H, Hinoda Y, Shahryari V, et al. Long noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR‐205. Cancer Res. 2015;75:1322‐1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Malakar P, Shilo A, Mogilevsky A, et al. Long noncoding RNA MALAT1 promotes hepatocellular carcinoma development by SRSF1 upregulation and mTOR activation. Cancer Res. 2017;77:1155‐1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Guo D, Ma J, Yan L, et al. Down‐regulation of Lncrna MALAT1 attenuates neuronal cell death through suppressing beclin1‐dependent autophagy by regulating Mir‐30a in cerebral ischemic stroke. Cell Physiol Biochem. 2017;43:182‐194. [DOI] [PubMed] [Google Scholar]
- 14. Li H, Yuan X, Yan D, et al. Long non‐coding RNA MALAT1 decreases the sensitivity of resistant glioblastoma cell lines to temozolomide. Cell Physiol Biochem. 2017;42:1192‐1201. [DOI] [PubMed] [Google Scholar]
- 15. Ezhkova E, Pasolli HA, Parker JS, et al. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue‐specific stem cells. Cell. 2009;136:1122‐1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gao SB, Zheng QF, Xu B, et al. EZH2 represses target genes through H3K27‐dependent and H3K27‐independent mechanisms in hepatocellular carcinoma. Mol Cancer Res. 2014;12:1388‐1397. [DOI] [PubMed] [Google Scholar]
- 17. Kondo Y. Targeting histone methyltransferase EZH2 as cancer treatment. J Biochem. 2014;156:249‐257. [DOI] [PubMed] [Google Scholar]
- 18. Simon JA, Kingston RE. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol Cell. 2013;49:808‐824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li Y, Wu Z, Yuan J, et al. Long non‐coding RNA MALAT1 promotes gastric cancer tumorigenicity and metastasis by regulating vasculogenic mimicry and angiogenesis. Cancer Lett. 2017;395:31‐44. [DOI] [PubMed] [Google Scholar]
- 20. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell. 2011;146(3):353‐358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen S, Pu J, Bai J, et al. EZH2 promotes hepatocellular carcinoma progression through modulating miR‐22/galectin‐9 axis. J Exp Clin Cancer Res. 2018;37:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Feng Y, Hu X, Zhang Y, Zhang D, Li C, Zhang L. Methods for the study of long noncoding RNA in cancer cell signaling. Methods Mol Biol. 2014;1165:115‐143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xu X, Gu J, Ding X, et al. LINC00978 promotes the progression of hepatocellular carcinoma by regulating EZH2‐mediated silencing of p21 and E‐cadherin expression. Cell Death Dis. 2019;10:752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yoshimoto R, Mayeda A, Yoshida M, Nakagawa S. MALAT1 long non‐coding RNA in cancer. Biochim Biophys Acta. 2016;1859:192‐199. [DOI] [PubMed] [Google Scholar]
- 25. Li P, Zhang X, Wang H, et al. MALAT1 is associated with poor response to oxaliplatin‐based chemotherapy in colorectal cancer patients and promotes chemoresistance through EZH2. Mol Cancer Ther. 2017;16:739‐751. [DOI] [PubMed] [Google Scholar]
- 26. Wang C, Mao ZP, Wang L, et al. Long non‐coding RNA MALAT1 promotes cholangiocarcinoma cell proliferation and invasion by activating PI3K/Akt pathway. Neoplasma. 2017;64:725‐731. [DOI] [PubMed] [Google Scholar]
- 27. Chen R, Liu Y, Zhuang H, et al. Quantitative proteomics reveals that long non‐coding RNA MALAT1 interacts with DBC1 to regulate p53 acetylation. Nucleic Acids Res. 2017;45:9947‐9959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Battistelli C, Cicchini C, Santangelo L, et al. The Snail repressor recruits EZH2 to specific genomic sites through the enrollment of the lncRNA HOTAIR in epithelial‐to‐mesenchymal transition. Oncogene. 2017;36:942‐955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miao L, Yang L, Li R, et al. Disrupting androgen receptor signaling induces snail‐mediated epithelial‐mesenchymal plasticity in prostate cancer. Cancer Res. 2017;77:3101‐3112. [DOI] [PubMed] [Google Scholar]
- 30. Zheng M, Jiang YP, Chen W, et al. Snail and Slug collaborate on EMT and tumor metastasis through miR‐101‐mediated EZH2 axis in oral tongue squamous cell carcinoma. Oncotarget. 2015;6:6797‐6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kaufhold S, Bonavida B. Central role of Snail1 in the regulation of EMT and resistance in cancer: a target for therapeutic intervention. J Exp Clin Cancer Res. 2014;33:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Militello G, Weirick T, John D, Doring C, Dimmeler S, Uchida S. Screening and validation of lncRNAs and circRNAs as miRNA sponges. Brief Bioinform. 2017;18:780‐788. [DOI] [PubMed] [Google Scholar]
- 33. Wang J, Li Y, Ding M, Zhang H, Xu X, Tang J. Molecular mechanisms and clinical applications of miR‐22 in regulating malignant progression in human cancer (Review). Int J Oncol. 2017;50:345‐355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ting Y, Medina DJ, Strair RK, Schaar DG. Differentiation‐associated miR‐22 represses Max expression and inhibits cell cycle progression. Biochem Biophys Res Commun. 2010;394:606‐611. [DOI] [PubMed] [Google Scholar]
- 35. Kong LM, Liao CG, Zhang Y, et al. A regulatory loop involving miR‐22, Sp1, and c‐Myc modulates CD147 expression in breast cancer invasion and metastasis. Cancer Res. 2014;74:3764‐3778. [DOI] [PubMed] [Google Scholar]
- 36. Huang SC, Wang M, Wu WB, et al. Mir‐22‐3p inhibits arterial smooth muscle cell proliferation and migration and neointimal hyperplasia by targeting HMGB1 in arteriosclerosis obliterans. Cell Physiol Biochem. 2017;42:2492‐2506. [DOI] [PubMed] [Google Scholar]
- 37. Zhang K, Li XY, Wang ZM, Han ZF, Zhao YH. MiR‐22 inhibits lung cancer cell EMT and invasion through targeting Snail. Eur Rev Med Pharmacol Sci. 2017;21:3598‐3604. [PubMed] [Google Scholar]
- 38. Xia SS, Zhang GJ, Liu ZL, et al. MicroRNA‐22 suppresses the growth, migration and invasion of colorectal cancer cells through a Sp1 negative feedback loop. Oncotarget. 2017;8:36266‐36278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xu M, Li J, Wang X, et al. MiR‐22 suppresses epithelial‐mesenchymal transition in bladder cancer by inhibiting Snail and MAPK1/Slug/vimentin feedback loop. Cell Death Dis. 2018;9:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ribeiro DM, Zanzoni A, Cipriano A, et al. Protein complex scaffolding predicted as a prevalent function of long non‐coding RNAs. Nucleic Acids Res. 2018;46:917‐928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yang F, Zhang L, Huo X‐S, et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679‐1689. [DOI] [PubMed] [Google Scholar]
- 42. Wang KC, Yang YW, Liu B, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120‐124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Huo Y, Li Q, Wang X, et al. MALAT1 predicts poor survival in osteosarcoma patients and promotes cell metastasis through associating with EZH2. Oncotarget. 2017;8:46993‐47006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang D, Ding L, Wang L, et al. LncRNA MALAT1 enhances oncogenic activities of EZH2 in castration‐resistant prostate cancer. Oncotarget. 2015;6:41045‐41055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chinaranagari S, Sharma P, Chaudhary J. EZH2 dependent H3K27me3 is involved in epigenetic silencing of ID4 in prostate cancer. Oncotarget. 2014;5:7172‐7182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Miard S, Girard MJ, Joubert P, et al. Absence of Malat1 does not prevent DEN‐induced hepatocarcinoma in mice. Oncol Rep. 2017;37:2153‐2160. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1‐S3
Table S1


