Abstract
Current studies have shown that the clock gene Period 1 (Per1) is downregulated in various tumors and plays an important role in promoting tumor progression. However, the biological functions and mechanism of Per1 in tumors remain largely unknown. In this study, 86 specimens of oral squamous cell carcinoma (OSCC) tissues and adjacent noncancerous tissues were collected to determine the Per1 expression level and the clinical significance of Per1 expression. Per1 was stably inhibited or overexpressed in OSCC cells to investigate its function and mechanism in vitro and in vivo. We found that Per1 was remarkably downregulated in OSCC and that low Per1 expression was significantly associated with TNM clinical stage and poor prognosis of OSCC patients. Per1 overexpression in SCC15 OSCC cells (Per1‐OE SCC15 cells) significantly promoted autophagy and apoptosis while inhibiting proliferation and the AKT/mTOR pathway. However, the results obtained in Per1‐silenced TSCCA OSCC cells were opposite those obtained in Per1‐OE SCC15 cells. After addition of the AKT activator SC79 to Per1‐OE SCC15 cells, the increased autophagy and apoptosis as well as decreased proliferation were remarkably rescued. Furthermore, increased apoptosis was significantly rescued in Per1‐OE SCC15 cells treated with the autophagy inhibitor autophinib. In vivo tumorigenicity assays also confirmed that Per1 overexpression suppressed tumor growth. Taken together, our findings demonstrate for the first time that Per1 promotes OSCC progression by inhibiting autophagy‐mediated cell apoptosis and enhancing cell proliferation in an AKT/mTOR pathway‐dependent manner, and Per1 could be used as a valuable therapeutic target for OSCC.
Keywords: autophagy, carcinogenesis, oral cancer, period 1, prognosis
Low Per1 expression was significantly associated with poor prognosis of oral squamous cell carcinoma (OSCC) patients. Per1 can regulate autophagy‐mediated cell apoptosis and cell proliferation in OSCC cells in an AKT/mTOR pathway‐dependent manner. Per1 overexpression suppressed OSCC growth in vivo.
1. INTRODUCTION
Head and neck cancer is the sixth most common cancer diagnosed globally.1 Oral squamous cell carcinoma (OSCC) is the most common head and neck cancer.2 There are more than 350 000 new cases of OSCC every year in the world, and approximately 170 000 people die from OSCC annually.3 OSCC has a poor prognosis. Currently, the main methods of treatment for OSCC are surgical resection, adjuvant radiotherapy and chemotherapy. Although surgical techniques, radiotherapy and chemotherapy have made great progress over the past 30 years, the 5‐year survival rate of OSCC patients has remained at approximately 50% and has not increased significantly.4, 5, 6 Therefore, in‐depth study of the molecular mechanism of OSCC development is the key to developing new therapeutic strategies and increasing the survival rate of OSCC patients.
Current research shows the presence of circadian clock genes in almost all cells of the human body.7, 8 The abnormal expression of circadian clock genes is closely related to the occurrence and development of tumors.9, 10 Per1 is an important core clock gene.11, 12 Current studies have shown that Per1 expression is reduced in many cancers, such as gastric cancer and non–small cell lung cancer, and that Per1 has an important cancer‐promoting effect,13, 14, 15 but the mechanism of this effect is unclear. We previously found that Per1 expression is decreased in OSCC tissues compared to that in normal tissues, and Per1 expression was significantly associated with TNM clinical stage.16 However, whether low Per1 expression affects the survival time of OSCC patients and the mechanism of how Per1 promotes the development of OSCC are unclear.
Autophagy is a lysosome‐mediated process that degrades abnormal proteins or damaged organelles in the cytoplasm.17 Autophagy plays an essential role in the development of tumors,18, 19 and autophagy and apoptosis often interact.20 Recently, in a study on the etiology of cerebral ischemia, Wiebking et al21 found that autophagy was significantly decreased, and apoptosis was significantly increased in the hippocampal neuronal cells of Per1 knockout mice compared to wild‐type mice, which suggested that Per1 regulates autophagy and apoptosis in hippocampal neurons. However, whether Per1 can regulate tumor cell autophagy is unclear. The AKT/mTOR signaling pathway is frequently activated in the development of many cancers, including OSCC,22, 23, 24 and is a crucial pathway that regulates cell autophagy, apoptosis and proliferation.25, 26, 27 However, it is unclear whether Per1 can regulate autophagy and apoptosis in tumor cells and tumor cell proliferation through the AKT/mTOR pathway.
In this study, we found that Per1 expression is decreased in OSCC tissues and cells and that the mean overall survival time of OSCC patients with low Per1 expression is shorter than that of OSCC patients with high Per1 expression. In addition, OSCC cells in which Per1 was stably inhibited and overexpressed were established and used to demonstrate that Per1 can regulate autophagy and apoptosis in OSCC cells and OSCC cell proliferation in an AKT/mTOR pathway‐dependent manner in vitro and in vivo. Furthermore, we found that autophagy can mediate apoptosis.
2. MATERIAL AND METHODS
2.1. Human tissue samples
Tissue sections obtained from 86 OSCC patients hospitalized in the Oral and Maxillofacial Surgery Department of the First Affiliated Hospital of Chongqing Medical University from January 2009 to December 2011 were embedded in paraffin. Clinicopathological data from the patients are shown in Table 1. Thirty‐two specimens with adjacent noncancerous tissues were selected from the 86 paraffinized sections as a control group. All patients in this study were diagnosed by pathology, and no other treatments, such as radiotherapy or chemotherapy, were performed before surgery. This study was approved by the Biomedical Ethics Committee of the First Affiliated Hospital of Chongqing Medical University (approval number: 2016‐124), and all patients provided informed consent.
Table 1.
The expression of Per1 and its relationship with clinicopathological features of patients with OSCC
| Parameters | Total | Per1 expression | χ 2 | P‐value | |
|---|---|---|---|---|---|
| Low | High | ||||
| Tissue type | |||||
| OSCC | 86 | 54 | 32 | 9.348 | .002* |
| ANT | 32 | 10 | 22 | ||
| Age | |||||
| ≥60 | 39 | 27 | 12 | 2.151 | .341 |
| 40‐60 | 36 | 22 | 14 | ||
| <40 | 11 | 5 | 6 | ||
| Gender | |||||
| Male | 48 | 29 | 19 | 0.262 | .609 |
| Female | 38 | 25 | 13 | ||
| Tumor differentiation | |||||
| Well | 30 | 21 | 9 | 2.616 | .270 |
| Moderate | 31 | 16 | 15 | ||
| Poor | 25 | 17 | 8 | ||
| T staging | |||||
| T1 | 15 | 5 | 10 | 10.926 | .012* |
| T2 | 21 | 11 | 10 | ||
| T3 | 32 | 23 | 9 | ||
| T4 | 18 | 15 | 3 | ||
| Lymph node metastasis | |||||
| No | 50 | 24 | 26 | 11.184 | .001* |
| Yes | 36 | 30 | 6 | ||
| Clinical stage | |||||
| I | 13 | 4 | 9 | 15.932 | .001* |
| II | 14 | 5 | 9 | ||
| III | 38 | 27 | 11 | ||
| IV | 21 | 18 | 3 | ||
| Site | |||||
| Gingiva | 13 | 10 | 3 | 6.020 | .198 |
| Tongue | 33 | 24 | 9 | ||
| Buccal | 26 | 14 | 12 | ||
| The floor of the oral | 8 | 3 | 5 | ||
| Palate | 6 | 3 | 3 | ||
P‐values reflect the relationship between Per1 expression and clinicopathological parameters with χ2 test. P < 0.05 was considered statistically significant.
Abbreviations: ANT, adjacent noncancerous tissues; OSCC, oral squamous cell carcinoma.
P < 0.05. High (score 6‐12), low (score 0‐4).
2.2. Cell culture and reagents
Normal oral mucosal HOMEC cells were purchased from Shanghai Bioleaf Biotechnology (Shanghai, China); TSCCA, SCC15 and CAL27 OSCC cells were purchased from Shanghai Zhong Qiao Xin Zhou Biotechnology. The cells were cultured in DMEM (Gibco) containing 10% FBS in an incubator containing 5% CO2 at 37°C.
2.3. Construction and transfection of Per1 overexpression vectors
According to the full‐length open reading frame of Per1 (NM_002616), the lentivirus‐expressing Per1 were designed and synthesized by Shanghai GeneChem. Lentivirus‐expressing Per1 was used to infect SCC15 cells to construct SCC15 cells stably overexpressing Per1 (Per1‐OE SCC15 cells). Scramble plasmid was used to construct lentivirus that was used to infect SCC15 cells as a negative control (Per1‐NC SCC15 cells), and SCC15 cells that did not undergo treatment were used as a blank control (Blank).
2.4. Construction of Per1 interference vectors and cell transfection
Lentivirus‐expressing Per1‐shRNA was provided by Shanghai GeneChem. To construct Per1 stable knockdown TSCCA cells (Per1‐shRNA TSCCA cells), Per1‐shRNA lentivirus was used to infect TSCCA cells; scramble plasmid was used to construct lentivirus that was used to infect TSCCA cells as a negative control (Per1‐NC TSCCA cells), and TSCCA cells that did not undergo treatment were used as a blank control (Blank).
2.5. Immunohistochemistry
Immunohistochemistry was carried out according to the operation instructions of an immunohistochemical detection kit (Beijing, Zhongshan Jinqiao, SP‐9000). Four‐micrometer‐thick paraffinized sections were routinely treated and incubated with primary antibody against Per1 at 4°C overnight. The biotin‐labeled goat anti–rabbit IgG secondary antibody was incubated with the slices for 10 minutes at room temperature. Then, the slices were incubated with streptavidin‐peroxidase for 15 minutes at room temperature, followed by development with DAB. Mayer’s hematoxylin was used to counterstain the sections for 2 minutes. Subsequently, the slices were dehydrated and sealed with neutral gum. For the negative control, PBS was used instead of the primary antibody. The obtained immunohistochemical results were reviewed by double grading and semiquantitative grading.28
2.6. Quantitative real‐time PCR
Total RNA from cells and tissues was extracted according to the instructions of RNAiso Plus (9180, TaKaRa). The total RNA concentration and purity were measured with a Nanodrop ND 2000 (Thermo Scientific). cDNA was generated with a PrimeScript RT Reagent Kit with gDNA Eraser (RR047A, TaKaRa). RT‐PCR was performed using TB Green Premix Ex Taq II (Tli RNaseH Plus) (RR820A, TaKaRa) on an AC‐1000 Thermal Cycler (Bio‐Rad). PCR primers for Per1, AKT, mTOR, LC3B, Beclin1, BAX, Ki67 and the housekeeping gene GAPDH were designed with Oligo 7.0 software, and the sequences of the primers are shown in Table S1. The relative expression level of each gene was determined using the 2−ΔΔ C t analysis method.
2.7. Western blotting
Total protein was isolated using RIPA lysis buffer (P0013B; Beyotime) containing 1% PMSF and 2% phosphatase inhibitor. The protein concentration was quantified using a BCA Protein Quantification Kit (P0010; Beyotime), 40 µg of total protein was separated on an 8%‐15% SDS‐PAGE gel and transferred onto a 0.45‐µm PVDF membrane. After blocking for 1 hour at 37°C, the membrane was incubated with primary antibodies against Per1, AKT, p‐AKT, mTOR, p‐mTOR, LC3B, P62, Beclin1, BAX, Ki67 and GAPDH overnight at 4°C. Details of the antibodies are listed in Table S2. Subsequently, the membrane was incubated with HRP‐conjugated secondary antibody at 37°C for 1 hour. The protein bands were detected using an ECL Advance Western blot detection system (Bio‐Rad) with an enhanced chemiluminescent substrate (34577, Thermo Scientific). ImageJ 5.0 software (Windows, 64‐bit Java 1.8.0_112) was used to analyze the gray values of the bands.
2.8. In vivo tumorigenicity assay
The use of 10 SPF‐grade BALB/c nu/nu female nude mice (18‐22 g, 4‐6 weeks old) without a specific pathogen in this study was approved by the Chongqing Medical University Laboratory Animal Research Institute. After 1 week of adaptive feeding, the nude mice were randomly divided into the Per1‐OE group and the Per1‐NC group, with five mice in each group. Then, 0.2 mL of Per1‐OE and Per1‐NC SCC15 cell suspensions at a concentration of 5 × 106 cells/mL were subcutaneously injected into the left back of each mouse. Tumor size was measured every 4 days after injection. After 4 weeks, tumor formation was obvious, and the nude mice were killed by cervical dislocation. Then, the tumors were weighed with an electronic balance (A250; Denver Instrument), and the tumor volume (V) was calculated using the following formula: V = 0.5 × a × b 2 (where a is the maximum long diameter and b is the minimum short diameter of the tumor). RT‐qPCR was used to detect the mRNA expression levels of Per1, LC3B, Beclin1, BAX and Ki67 in the tumor tissues. The protein expression levels of Per1, AKT, p‐AKT, mTOR, p‐mTOR, LC3B, P62, Beclin1, BAX and Ki67 in the tumor tissues were detected by western blotting. All animal experimental procedures were approved by the Laboratory Animal Use Management Committee of the Experimental Animal Institute of Chongqing Medical University (approval number: 2018‐102).
2.9. Statistical analysis
GraphPad Prism 7.0 (GraphPad Software) and SPSS 23 (IBM, SPSS) were used for data processing and statistical analysis. The relationships between Per1 expression level and clinicopathological parameters were analyzed using the χ2 test. Multivariate analysis with the Cox regression model was used to analyze the statistical significance of survival‐related factors. The Kaplan‐Meier method was used to plot survival curves, and the log‐rank test was used to analyze the difference in overall survival time between the two groups. Statistical comparisons between two independent groups were analyzed using the two‐tailed Student’s t‐test, and comparisons between three or more means were carried out using one‐way ANOVA. The results are shown as the means ± standard deviations (SD) from at least three independent experiments. A value of P < 0.05 indicated statistical significance.
Other methods are shown in Figure S1.
3. RESULTS
3.1. Low expression of Per1 is related to poor prognosis in oral squamous cell carcinoma patients
Immunohistochemistry showed that Per1 expression in OSCC tissues was significantly lower than that in adjacent noncancerous tissues (P < 0.01) (Figure 1A and Table 1). Per1 expression was significantly correlated with tumor size, cervical lymph node metastasis and TNM clinical stage (P < 0.05) (Table 1). Kaplan–Meier survival analysis showed that the mean overall survival times of OSCC patients with low and high Per1 expression levels were 48.4 ± 4.2 months and 64.5 ± 6.5 months, respectively, and that the mean overall survival time of OSCC patients with low Per1 expression was significantly shorter than that of OSCC patients with high Per1 expression (P < 0.05). The 5‐year survival rates of OSCC patients with low and high Per1 expression levels were 40.7% and 59.4%, respectively. That is, the 5‐year survival rate of patients with low Per1 expression was significantly lower than that of patients with high Per1 expression (P < 0.05) (Figure 1B). Multivariate Cox regression analysis showed that the Per1 expression level is an independent prognostic factor in OSCC patients (Table 2). These results suggest that Per1 plays an essential role in the development of OSCC.
Figure 1.
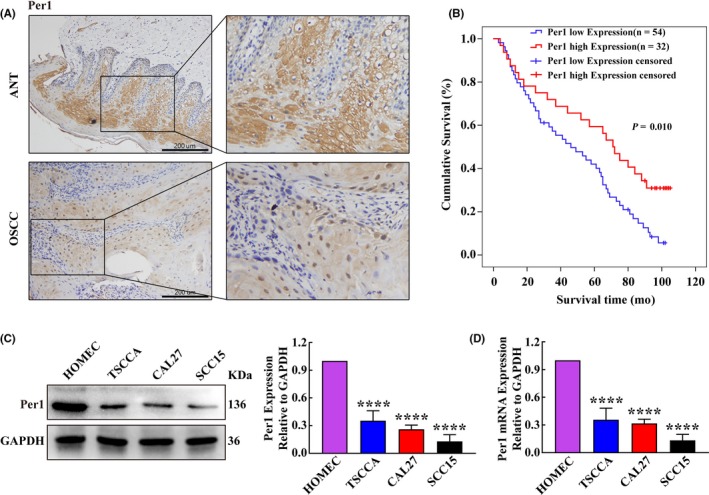
Per1 expression is decreased in oral squamous cell carcinoma (OSCC) tissues and cell lines. A, Immunohistochemistry results showed that Per1 expression in OSCC tissues was significantly lower than that in adjacent noncancerous tissues (n = 86; scale bars = 200 μm). B, The mean overall survival time of OSCC patients with low Per1 expression was significantly shorter than that of patients with high Per1 expression. C, D, Western blotting (C) and RT‐qPCR (D) showed that Per1 expression was significantly decreased in TSCCA, CAL27 and SCC15 OSCC cells compared with that in normal oral mucosal HOMEC cells. All data represent three independent experiments. The results are shown as the mean ± SD (n ≥ 3). *P < .05; **P < .01; ***P < .001; ****P < .0001
Table 2.
Univariate analysis and multivariate analysis of various progression in patients with OSCC Cox regression analysis
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| P‐value | HR | 95% CI | P‐value | HR | 95% CI | |
| Per1 | .012* | 0.519 | 0.312‐0.864 | .040* | 2.119 | 1.036‐4.338 |
| Age | .578 | 0.915 | 0.669‐1.251 | .985 | 0.996 | 0.669‐1.483 |
| Gender | .652 | 0.897 | 0.561‐1.437 | .669 | 0.891 | 0.525‐1.513 |
| Tumor differentiation | .357 | 1.149 | 0.855‐1.543 | .343 | 1.180 | 0.838‐1.660 |
| T classification | <.001* | 6.513 | 4.341‐9.771 | <.001* | 3.795 | 2.129‐6.766 |
| Lymph node metastasis | <.001* | 8.133 | 4.448‐14.869 | .007* | 2.694 | 1.319‐5.504 |
| Clinical stage | <.001* | 5.227 | 3.540‐7.720 | .011* | 2.378 | 1.224‐4.620 |
| Site | .716 | 0.958 | 0.759‐1.208 | .899 | 1.017 | 0.788‐1.311 |
Abbreviations: CI, confidence interval; HR, hazard ratio; OSCC, oral squamous cell carcinoma.
P < 0.05
3.2. Per1 regulates oral squamous cell carcinoma cell proliferation and apoptosis
To explore the effect of changes in Per1 expression on the proliferation and apoptosis of OSCC cells, we first detected Per1 expression in normal oral mucosal HOMEC cells and three OSCC cell lines (TSCCA, CAL27 and SCC15 cells) by RT‐qPCR and western blotting. Per1 expression in the three OSCC cells was significantly lower than that in normal oral mucosal cells (P < 0.0001) (Figure 1C,D). We selected SCC15 cells, which had the lowest Per1 expression, to stably overexpress Per1 (Per1‐OE SCC15 cells) and TSCCA cells, which had the highest expression of Per1, to stably inhibit Per1 (Per1‐shRNA TSCCA cells). The Per1 overexpression efficiency in Per1‐OE SCC15 cells was 2.14‐fold, and the Per1 knockdown efficiency in Per1‐shRNA TSCCA cells was 61.7% (Figure 2A,B).
Figure 2.

Changes in the proliferation and apoptosis of oral squamous cell carcinoma (OSCC) cells after the overexpression and silencing of Per1. A, B, Verification of Per1 overexpression and knockdown efficiency in OSCC cells showed that the mRNA and protein expression of Per1 was significantly higher in Per1‐OE SCC15 cells and lower in TSCCA cells compared to that in control cells. C, Flow cytometry analysis showed that the apoptotic index of Per1‐OE SCC15 cells was significantly increased, while that of Per1‐shRNA TSCCA cells was significantly decreased. D, TUNEL assay showed significantly more TUNEL‐positive Per1‐OE SCC15 cells and significantly fewer TUNEL‐positive Per1‐shRNA TSCCA cells compared to control cells. E, F, CCK8 and MTT assays revealed that the proliferation of Per1‐OE SCC15 cells was significantly decreased, while that of Per1‐shRNA TSCCA cells was significantly increased. All data represent three independent experiments. The results are shown as the mean ± SD (n ≥ 3). *P < .05; **P < .01; ***P < .001; ****P < .0001
Using flow cytometry and TUNEL, CCK8 and MTT assays, we detected the effect of changes in Per1 expression on the apoptosis and proliferation of OSCC cells. Detection of apoptosis by flow cytometry showed that the apoptotic index of Per1‐OE SCC15 cells was significantly increased (P < 0.05), while that of Per1‐shRNA TSCCA cells was significantly decreased (P < 0.05) (Figure 2C). TUNEL experiments showed that the apoptosis rate in Per1‐OE SCC15 cells was significantly increased (P < 0.05), while that in Per1‐shRNA TSCCA cells was significantly decreased (P < 0.05) (Figure 2D). The CCK8 and MTT assays showed that the proliferation of Per1‐OE cells was significantly decreased compared with that of the control group (P < 0.05), while the proliferation of Per1‐shRNA TSCCA cells was significantly increased (P < 0.05) (Figure 2E,F). In addition, Per1‐shRNA lentivirus was used to infect Per1‐OE SCC15 cells to knock down Per1. It was found that the decreased proliferation and increased apoptosis were significantly rescued (P < 0.05) (Figure S1A–E). All the above results demonstrate that Per1 knockdown in OSCC cells inhibits apoptosis and promotes cell proliferation, whereas the overexpression of Per1 has the opposite effects.
3.3. Per1 regulates autophagy in oral squamous cell carcinoma cells
We further explored the effect of Per1 expression changes on autophagy in OSCC cells. As detected by TEM, the density of autophagosomes in Per1‐OE SCC15 cells increased significantly (P < 0.05), while that in Per1‐shRNA TSCCA cells decreased significantly (P < 0.05) compared to the density of autophagosomes in the control group (Figure 3A). Western blotting showed that the LC3BII/LC3BI ratio and Beclin1 expression in Per1‐OE SCC15 cells were significantly increased (P < 0.05), while P62 expression was significantly decreased (P < 0.05). However, the LC3BII/LC3BI ratio and Beclin1 expression in Per1‐shRNA TSCCA cells were significantly decreased (P < 0.05), while P62 expression was significantly increased (P < 0.05) (Figure 3B). RT‐qPCR showed that the mRNA expression of LC3B and Beclin1 in Per1‐OE SCC15 cells was significantly increased (P < 0.05). The mRNA expression of LC3B and Beclin1 in Per1‐shRNA TSCCA cells was significantly decreased (P < 0.05) (Figure 3C). To further detect autophagy flux, the lysosomal inhibitor chloroquine (CQ) was added to the Per1‐OE cells group at a final concentration of 20 µmol/L. After 16 hours, it was found that the expression of LC3BII and P62 was increased significantly (P < 0.05) (Figure 3D). The above results demonstrate that autophagy in OSCC cells is significantly attenuated by Per1 knockdown, while Per1 overexpression had the opposite effect.
Figure 3.
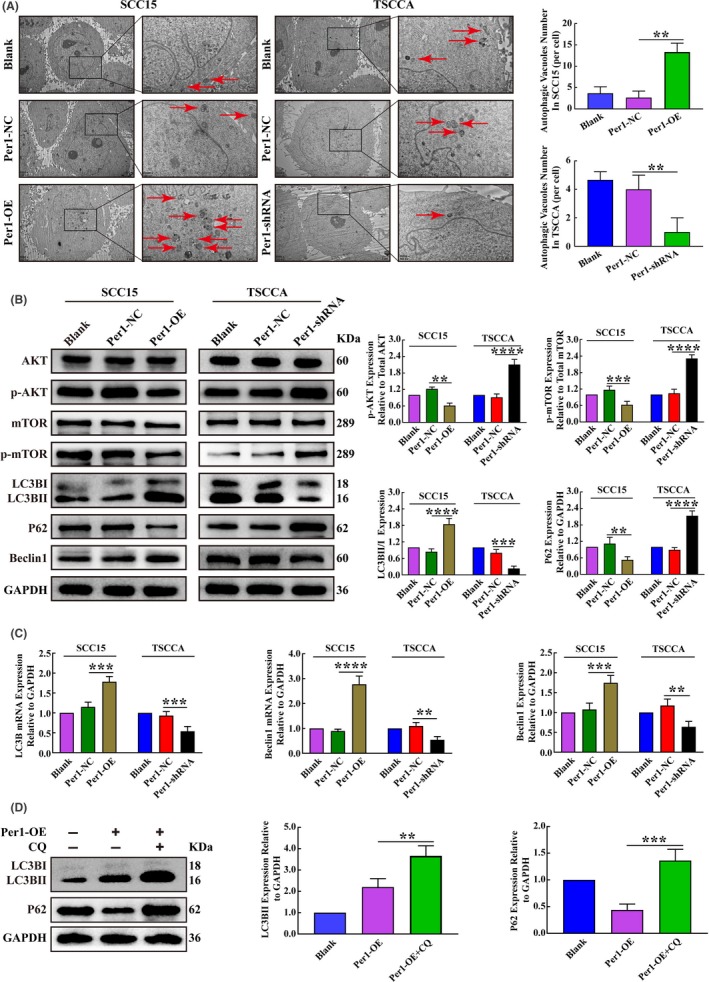
Changes in autophagy and AKT/mTOR pathway activity in oral squamous cell carcinoma (OSCC) cells after the overexpression and silencing of Per1. A, Transmission electron microscopy assay showed that the autophagosome density in Per1‐OE SCC15 cells was significantly increased, while that in Per1‐shRNA TSCCA cells was significantly decreased (low magnification scale bars = 2 μm; high magnification scale bars = 1 μm). B, Western blotting showed no significant changes in the total protein levels of AKT and mTOR in Per1‐OE SCC15 cells and Per1‐shRNA TSCCA cells. In Per1‐OE SCC15 cells, the expression of p‐AKT, p‐mTOR and P62 was significantly decreased, while the LC3BII/LC3BI ratio and Beclin1 expression were significantly increased. However, in Per1‐shRNA TSCCA cells, the expression of p‐AKT, p‐mTOR and P62 was significantly increased, while the LC3BII/LC3BI ratio and Beclin1 expression were significantly decreased. C, RT‐qPCR analysis revealed that the mRNA expression of LC3B and Beclin1 was significantly increased in Per1‐OE SCC15 cells; however, the mRNA expression of LC3B and Beclin1 was significantly decreased in Per1‐shRNA TSCCA cells. D, Western blotting showed that the LC3BII and P62 expression in Per1‐OE SCC15 cells were significantly increased after addition of the lysosomal inhibitor CQ. All data represent three independent experiments. The results are shown as the mean ± SD (n ≥ 3). *P < .05; **P < .01; ***P < .001; ****P < .0001
3.4. Per1 regulates the AKT/mTOR signaling pathway in oral squamous cell carcinoma cells
To determine whether Per1 regulates the AKT/mTOR pathway in OSCC, we examined the expression of AKT, p‐AKT, mTOR and p‐mTOR, which are key molecules in the AKT/mTOR pathway, in Per1‐OE SCC15 and Per1‐shRNA TSCCA cells. Western blotting showed that in Per1‐OE SCC15 cells, the expression of total AKT and mTOR proteins was not significantly changed (P > 0.05), but the expression of p‐AKT and p‐mTOR was significantly decreased (P < 0.05); in Per1‐shRNA TSCCA cells, there was no significant change in total AKT and mTOR protein expression (P > 0.05), but the expression of p‐AKT and p‐mTOR was significantly increased (P < 0.05) (Figure 3B). These results demonstrate that Per1 regulates the AKT/mTOR signaling pathway in OSCC cells.
3.5. Per1 regulates autophagy, proliferation and apoptosis in an AKT/mTOR pathway‐dependent manner
To determine whether the AKT/mTOR signaling pathway is a key pathway for Per1‐mediated regulation of autophagy, apoptosis and proliferation in OSCC, the AKT activator SC79 (HY‐18749; MCE) was added to the Per1‐OE SCC15 cells at a final concentration of 8 µg/mL. After 16 hours, the changes in autophagy, apoptosis and proliferation were detected. After the addition of the AKT activator SC79 to Per1‐OE SCC15 cells, the increased autophagosome density, apoptotic index, apoptotic cell rate, LC3BII/LC3BI ratio and Beclin1 expression due to Per1 overexpression were significantly rescued (P < 0.05) (Figure 4A‐D), and decreased proliferation and P62 expression were remarkably rescued by SC79 treatment (P < 0.05) (Figure 4D,E). To further detect autophagy flux, SC79 was added to Per1‐OE cells with CQ. After 16 hours, it was found that the increased expression of LC3BII was remarkably rescued (P < 0.05), and the expression of P62 increased significantly (P < 0.05) (Figure 4F ). These results demonstrate that Per1 regulates autophagy, apoptosis and proliferation in OSCC cells in an AKT/mTOR signaling pathway‐dependent manner.
Figure 4.
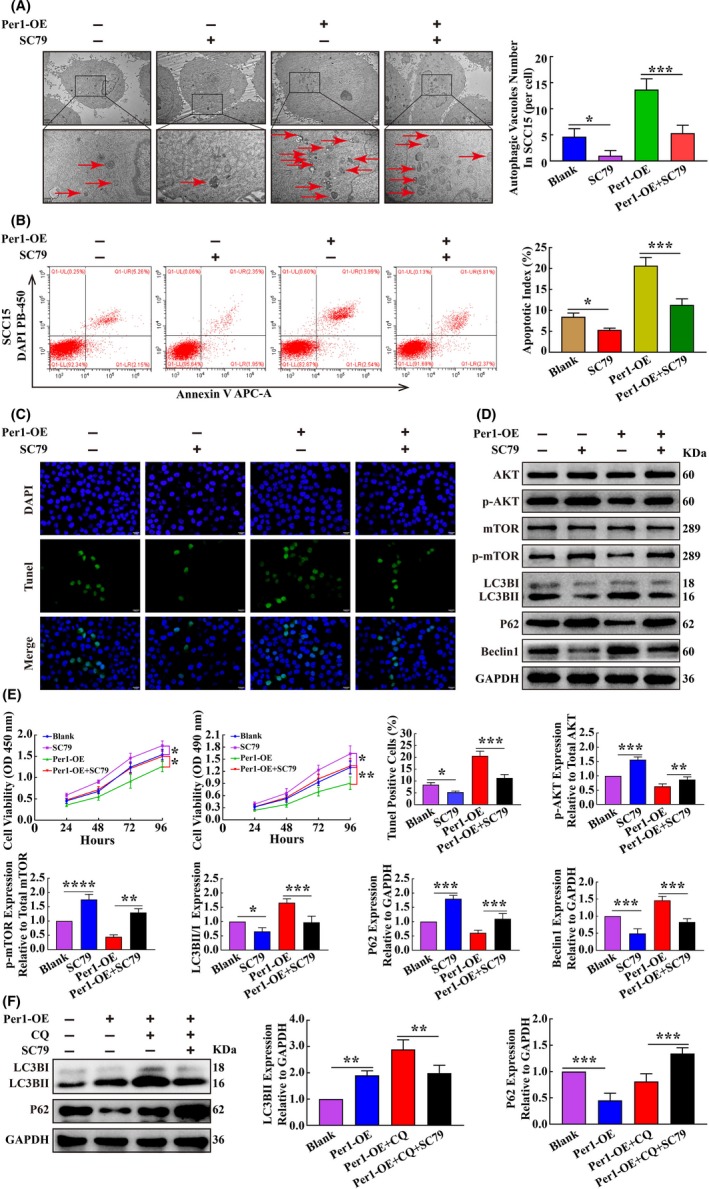
Per1 inhibits autophagy in oral squamous cell carcinoma (OSCC) cells and the apoptosis and proliferation of OSCC cells by regulating the AKT/mTOR pathway. A, Transmission electron microscopy experiments revealed that after addition of the AKT activator SC79 to Per1‐OE SCC15 cells, the autophagosome density was significantly reduced (low magnification scale bars = 2 μm; high magnification scale bars = 1 μm). B, Flow cytometry showed that the apoptotic index of Per1‐OE SCC15 cells was significantly reduced after addition of the AKT activator SC79. C, TUNEL assay indicated that the increased TUNEL‐positive rate in Per1‐OE SCC15 cells was remarkably rescued after addition of the AKT activator SC79. D, Western blotting showed that the decreased protein expression of p‐AKT, p‐mTOR and P62 in Per1‐OE SCC15 cells was remarkably increased after addition of the AKT activator SC79, while the increased protein expression of Beclin1 and LC3BII/LC3BIratio were remarkably decreased. E, CCK8 and MTT assays revealed that the proliferation of Per1‐OE SCC15 cells was significantly increased after addition of the AKT activator SC79. F, Western blotting showed that the increased expression of LC3BII in Per1‐OE SCC15 cells with CQ was remarkably decreased after addition of the AKT activator SC79, while the expression of P62 was remarkably increased. All data represent three independent experiments. The results are shown as the mean ± SD (n ≥ 3). *P < .05; **P < .01; ***P < .001; ****P < .0001
3.6. Increased expression of Per1 promotes autophagy‐mediated cell apoptosis
To further explore the relationship between autophagy and apoptosis in OSCC cells, the autophagy inhibitor autophinib (HY‐101920; MCE) was added to the Per1‐OE SCC15 cells at a final concentration of 80 nmol/L. After 16 hours, the changes in autophagy and apoptosis were detected. Compared with those in Per1‐OE SCC15 cells, in Per1‐OE SCC15 cells to which autophinib was added to inhibit autophagy, the autophagosome density, apoptotic index and apoptotic cell rate were significantly lower (P < 0.05) (Figure 5A‐C). Western blotting showed that the LC3BII/LC3BI ratio and Beclin1 expression were significantly decreased and that P62 expression was significantly increased (P < 0.05) with the addition of autophinib (Figure 5D). These results indicated that the increased apoptosis in Per1‐OE SCC15 cells was significantly rescued by the addition of the autophagy inhibitor autophinib and demonstrate that Per1‐mediated regulation of apoptosis in OSCC cells is dependent on autophagy regulation.
Figure 5.
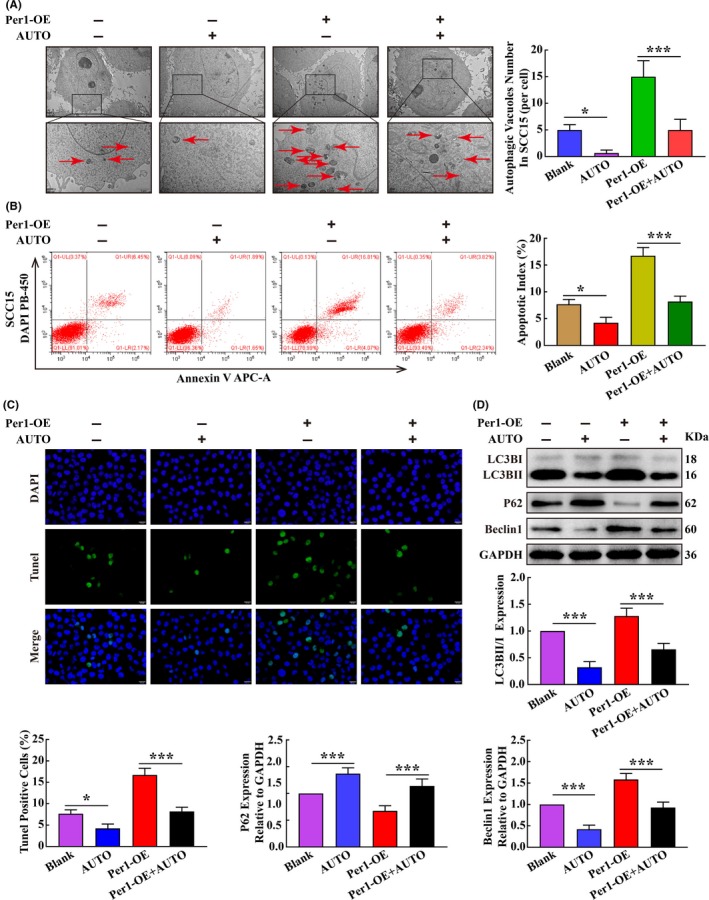
Per1 regulates apoptosis depending on autophagy in oral squamous cell carcinoma (OSCC) cells. A, Transmission electron microscopy experiments revealed that after addition of the autophagy inhibitor autophinib to Per1‐OE SCC15 cells, the autophagosome density was significantly reduced (low magnification scale bars = 2 μm; high magnification scale bars = 1 μm). B, Flow cytometry showed that the apoptotic index of Per1‐OE SCC15 cells was significantly reduced after the addition of autophinib. C, TUNEL assay indicated that the increased TUNEL‐positive rate in Per1‐OE SCC15 cells was remarkably decreased after the addition of autophinib. D, Western blotting showed that the decreased protein expression of P62 in Per1‐OE SCC15 cells was remarkably increased after the addition of autophinib, while the increased LC3BII/LC3BI ratio and Beclin1 expression were remarkably decreased. All data represent three independent experiments. The results are shown as the mean ± SD (n ≥ 3). *P < .05; **P < .01; ***P < .001; ****P < .0001
3.7. Overexpression of Per1 suppressed tumor growth in vivo
An in vivo tumorigenicity assay in nude mice revealed that the weights and volumes of tumors in the Per1‐OE cell group were significantly lower than those in the Per1‐NC cell group (P < 0.05) (Figure 6A). The growth of tumors in the Per1‐OE cell group was slower than that in the Per1‐NC cell group (P < 0.05) (Figure 6B). RT‐qPCR showed that Per1, LC3B and BAX mRNA expression was significantly increased in the Per1‐OE cell group compared with that in the Per1‐NC cell group, while Ki67 mRNA expression was significantly decreased (P < 0.05) (Figure 6C). Western blotting showed no significant differences in AKT and mTOR protein expression between the Per1‐OE cell group and the Per1‐NC cell group (P > 0.05), whereas p‐AKT, p‐mTOR, P62 and Ki67 protein expression was significantly reduced, and the LC3BII/LC3BI ratio and BAX protein expression were significantly increased in the Per1‐OE cell group compared to those in the Per1‐NC cell group (P < 0.05) (Figure 6D). These findings suggest that AKT/mTOR pathway activity and cell proliferation were inhibited in the Per1‐OE cell group, while autophagy and apoptosis were elevated compared to those in the Per1‐NC cell group. The above results indicate that Per1 overexpression can significantly inhibit the growth of tumors in vivo and that its regulatory mechanism is consistent with the mechanism suggested by the results obtained in vitro.
Figure 6.
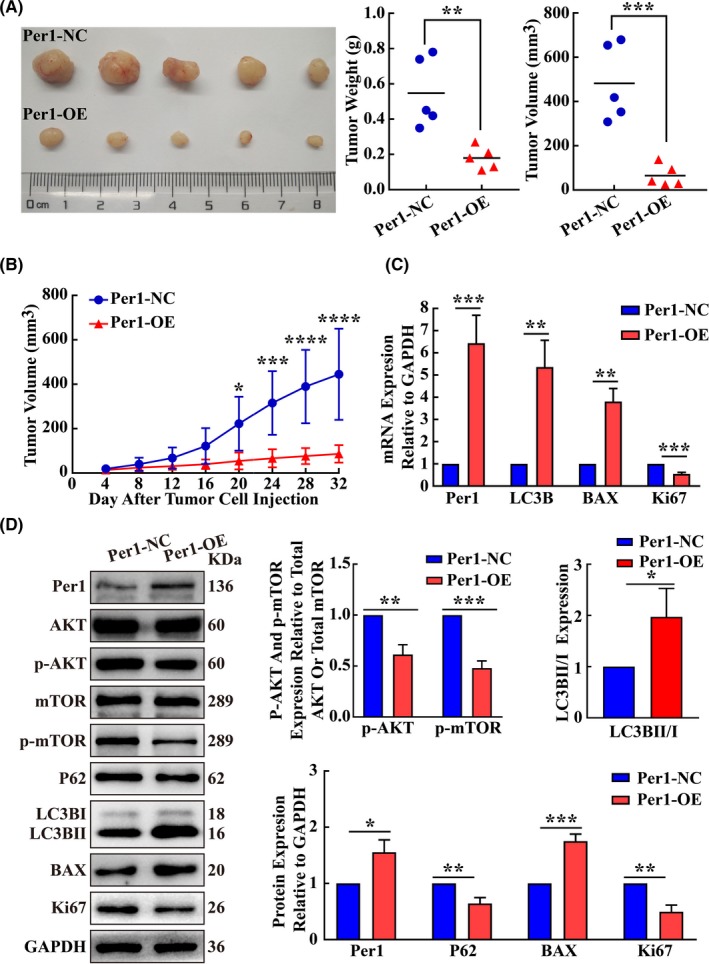
Increased expression of Per1 inhibits the tumorigenesis of oral squamous cell carcinoma (OSCC) cells in vivo. A, The weights and volumes of tumors in the Per1‐OE group were significantly lower than those in the Per1‐NC group (N = 5). B, The growth curve from an in vivo tumorigenicity assay showed that subcutaneous tumor growth in the Per1‐OE group was significantly slower than that in the Per1‐NC group. C, Tumor‐forming tissues in vivo were assessed by RT‐qPCR. The mRNA expression levels of Per1, LC3B and BAX in the Per1‐OE group were significantly increased, while the mRNA expression levels of Ki67 were significantly decreased. D, Western blotting was used to detect protein expression in tumor‐forming tissues in vivo. The LC3BII/LC3BI ratio and protein expression of Per1 and BAX were significantly increased in the Per1‐OE group, while the protein expression of p‐AKT, p‐mTOR, P62 and Ki67 was significantly decreased. All data represent three independent experiments. The results are shown as the mean ± SD (n ≥ 3). *P < .05; **P < .01; ***P < .001; ****P < .0001
4. DISCUSSION
Current studies have shown that the abnormal expression of circadian clock genes is closely related to the occurrence and development of various diseases, such as tumors.9, 10, 29 Fourteen circadian clock genes have been found: Period1, Period2, Period3, Bmal1, Clock, Tim, CK1ε, NPAS2, REV‐ERBs, Dec1/2, Cry1/2 and the ROR. These clock genes regulate approximately 43% of the protein‐coding genes in the human genome. Thus, they have essential regulatory effects on many physiological processes in the body.30, 31, 32 Per1 is among the core circadian clock genes.11, 12 In this study, we analyzed 86 OSCC tissues and showed that the mean overall survival time of OSCC patients with low Per1 expression was significantly shorter than that of OSCC patients with high Per1 expression. This result further suggests that Per1 plays an essential role in OSCC development.
Changes in autophagy, apoptosis and proliferation are crucial causes of cancer development.26, 33 Recent studies have demonstrated that low Per1 expression can inhibit tumor cell apoptosis and promote proliferation.14, 15 The AKT/mTOR signaling pathway is a crucial pathway that regulates apoptosis, proliferation and autophagy,25, 27 but whether Per1 can regulate the AKT/mTOR pathway and autophagy in tumor cells remains unclear. In a study of circadian clock gene function in hypothalamic suprachiasmatic nucleus cells, Cao et al34 observed a significant correlation between Per1 expression and mTOR activity in hypothalamic suprachiasmatic nucleus cells from transgenic Per1 reporter gene mice. Rami et al35 added rapamycin to primary cultured Per1−/− and wild‐type hippocampal cells and found that autophagy was significantly increased in wild‐type hippocampal cells, while Per1−/− hippocampal neurons were resistant to autophagy induced by rapamycin, suggesting that Per1−/− has an inhibitory effect on autophagy in hippocampal neurons. In this study, Per1‐shRNA TSCCA cells and Per1‐OE SCC15 cells were established by silencing and overexpressing Per1, respectively, in OSCC cells. p‐AKT and p‐mTOR expression levels and cell proliferation were found to be significantly increased, while autophagy and apoptosis were significantly reduced in Per1‐shRNA TSCCA cells compared to those in control cells. The opposite changes were found in Per1‐OE SCC15 cells, and these effects of Per1 overexpression were significant. After addition of an AKT activator to Per1‐OE SCC15 cells, the changes in p‐mTOR expression levels, autophagy, apoptosis and proliferation were all significantly rescued. For in vivo verification, an animal xenograft model was further established using Per1‐OE SCC15 cells, and the effects of Per1 overexpression were the same as those obtained through in vitro experiments. Taken together, these results demonstrate for the first time that Per1 regulates the AKT/mTOR pathway and autophagy in cancer cells and that the AKT/mTOR pathway plays an essential role in Per1‐mediated regulation of autophagy, apoptosis and proliferation.
Changes in autophagy and apoptosis play an essential role in the development of cancer,33 and autophagy and apoptosis often interact in a complex manner.36, 37 Autophagy can inhibit or promote cell apoptosis due to different cell types, environments and stimuli.38, 39 In this study, we found that autophagy and apoptosis were significantly increased in Per1‐overexpressing SCC15 cells, but increased apoptosis was significantly rescued after the addition of the autophagy inhibitor autophinib. These results prove that in this study, autophagy had a positive regulatory effect on apoptosis, but the specific regulatory mechanism of this effect remains to be further studied.
In summary, the results of this study demonstrate for the first time that the mean overall survival time of OSCC patients with low Per1 expression was significantly shorter than that of patients with high Per1 expression. Per1 can regulate autophagy‐mediated cell apoptosis and cell proliferation in OSCC cells in an AKT/mTOR pathway‐dependent manner. These findings suggest that Per1 can potentially be used as a valuable therapeutic target for OSCC patients.
DISCLOSUREs
Authors declare no conflicts of interest for this article.
Supporting information
Fig S1
Table S1
Table S2
Document S1
ACKNOWLEDGMENTS
This study was supported by Natural Science Foundation of Chongqing, China (cstc2018jcyjA0481).
Yang G, Yang Y, Tang H, Yang K. Loss of the clock gene Per1 promotes oral squamous cell carcinoma progression via the AKT/mTOR pathway. Cancer Sci. 2020;111:1542–1554. 10.1111/cas.14362
REFERENCES
- 1. Parfenov M, Pedamallu CS, Gehlenborg N, et al. Characterization of HPV and host genome interactions in primary head and neck cancers. Proc Natl Acad Sci USA. 2014;111:15544‐15549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Angela C, Chi AC, Day TA, Neville BW. Oral cavity and oropharyngeal squamous cell carcinoma–an update. CA Cancer J Clin. 2015;65:401‐421. [DOI] [PubMed] [Google Scholar]
- 3. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394‐424. [DOI] [PubMed] [Google Scholar]
- 4. Gonzalez‐Ramirez I, Soto‐Reyes E, Sanchez‐Perez Y, Herrera LA, Garcia‐Cuellar C. Histones and long non–coding RNAs: the new insights of epigenetic deregulation involved in oral cancer. Oral Oncol. 2014;50:691‐695. [DOI] [PubMed] [Google Scholar]
- 5. Chien HT, Cheng SD, Liao CT, Wang HM, Huang SF. Amplification of the EGFR and CCND1 are coordinated and play important roles in the progression of oral squamous cell carcinomas. Cancers. 2019;11:760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kim JW, Park Y, Roh JL, et al. Prognostic value of glucosylceramide synthase and P‐glycoprotein expression in oral cavity cancer. Int J Clin Oncol. 2016;21:883‐889. [DOI] [PubMed] [Google Scholar]
- 7. Turek FW. Circadian clocks: not your grandfather’s clock. Science. 2016;354:992‐993. [DOI] [PubMed] [Google Scholar]
- 8. Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annu Rev Neurosci. 2012;35:445‐462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shostak A. Circadian clock, cell division, and cancer: from molecules to organism. Int J Mol Sci. 2017;18:873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Filipski E, King VM, Li X, et al. Host circadian clock as a control point in tumor progression. J Natl Cancer Inst. 2002;94:690‐697. [DOI] [PubMed] [Google Scholar]
- 11. Siepka SM, Yoo SH, Park J, et al. Circadian mutant overtime reveals F‐box protein FBXL3 regulation of cryptochrome and period gene expression. Cell. 2007;129:1011‐1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shearman LP, Sriram S, Weaver DR, et al. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013‐1019. [DOI] [PubMed] [Google Scholar]
- 13. Zhao H, Zeng ZL, Yang J, et al. Prognostic relevance of period1 (Per1) and period2 (per2) expression in human gastric cancer. Int J Clin Exp Pathol. 2014;7:619‐630. [PMC free article] [PubMed] [Google Scholar]
- 14. Liu B, Xu K, Jiang Y, Li X. Aberrant expression of Per1, Per2 and Per3 and their prognostic relevance in non–small cell lung cancer. Int J Clin Exp Pathol. 2014;7:7863‐7871. [PMC free article] [PubMed] [Google Scholar]
- 15. Cao Q, Gery S, Dashti A, et al. A role for the clock gene Per1 in prostate cancer. Cancer Research. 2009;69:7619‐7625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen R, Yang K, Zhao NB, et al. Abnormal expression of Per1 circadian‐clock gene in oral squamous cell carcinoma. Onco Targets Ther. 2012;5:403‐407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Levine B, Klionsky DJ. Development by self‐digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463‐477. [DOI] [PubMed] [Google Scholar]
- 18. Shintani T, Klionsky DJ. Autophagy in health and disease: a double‐edged sword. Science. 2004;306:990‐995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. White E, DiPaola RS. The double‐edged sword of autophagy modulation in cancer. Clin Cancer Res. 2009;15:5308‐5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Levine B. Cell biology: autophagy and cancer. Nature. 2007;446:745‐747. [DOI] [PubMed] [Google Scholar]
- 21. Wiebking N, Maronde E, Rami A. Increased neuronal injury in clock gene Per‐1 deficient‐mice after cerebral ischemia. Curr Neurovasc Res. 2013;10:112‐125. [DOI] [PubMed] [Google Scholar]
- 22. Katayama K, Fujita N, Tsuruo T. Akt/protein kinase B‐dependent phosphorylation and inactivation of WEE1Hu promote cell cycle progression at G2/M transition. Mol Cell Biol. 2005;25:5725‐5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sussman MA, Völkers M, Fischer K, et al. Myocardial AKT: the omnipresent nexus. Physiol Rev. 2011;91:1023‐1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Watanabe S, Sato K, Okazaki Y, Tonogi M, Tanaka Y, Yamane GY. Activation of PI3K‐AKT pathway in oral epithelial dysplasia and early cancer of tongue. Bull Tokyo Dent Coll. 2009;50:125‐133. [DOI] [PubMed] [Google Scholar]
- 25. Kim YC, Guan KL. mTOR: a pharmacologic target for autophagy regulation. J Clin Invest. 2015;125:25‐32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chang F, Lee JT, Navolanic PM, et al. Involvement of PI3K/Akt pathway in cell cycle progression, apoptosis, and neoplastic transformation: a target for cancer chemotherapy. Leukemia. 2003;17:590‐603. [DOI] [PubMed] [Google Scholar]
- 27. Martini M, De Santis MC, Braccini L, Gulluni F, Hirsch E. PI3K/AKT signaling pathway and cancer: an updated review. Ann Med. 2014;46:372‐383. [DOI] [PubMed] [Google Scholar]
- 28. Zhang H, Pan Y, Cheung M, et al. LAMB3 mediates apoptotic, proliferative, invasive, and meta static behaviors in pancreatic cancer by regulating the PI3K/Akt signaling pathway. Cell Death Dis. 2019;10:019‐1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Morris CJ, Purvis TE, Hu K, Scheer FA. Circadian misalignment increases cardiovascular disease risk factors in humans. Proc Natl Acad Sci USA. 2016;113:E1402‐E1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rana S, Mahmood S. Circadian rhythm and its role in malignancy. J Circadian Rhythms. 2010;8:1‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhao Q, Zheng G, Yang K, et al. The clock gene per1 plays an important role in regulating the clock gene network in human oral squamous cell carcinoma cells. Oncotarget. 2016;7:70290‐70302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang R, Lahens NF, Ballance HI, Hughes ME. Hogenesch JB. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci USA. 2014;111:16219‐16224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ravegnini G, Sammarini G, Nannini M, et al. Gastrointestinal stromal tumors (GIST): facing cell death between autophagy and apoptosis. Autophagy. 2017;13:452‐463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cao R, Anderson FE, Jung YJ, Dziema H, Obrietan K. Circadian regulation of mammalian target of rapamycin signaling in the mouse suprachiasmatic nucleus. Neuroscience. 2011;181:79‐88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rami A, Fekadu J, Rawashdeh O. The hippocampal autophagic machinery is depressed in the absence of the circadian clock protein PER1 that may lead to vulnerability during cerebral ischemia. Curr Neurovasc Res. 2017;14:207‐214. [DOI] [PubMed] [Google Scholar]
- 36. Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self‐eating and self‐killing: crosstalk between autophagy and apoptosis. Nature Rev Mol Cell Biol. 2007;8:741‐752. [DOI] [PubMed] [Google Scholar]
- 37. Wirawan E, Vande Walle L, Kersse K, et al. Caspase‐mediated cleavage of Beclin‐1 inactivates Beclin‐1‐induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis. 2010;1:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mariño G, Niso‐Santano M, Baehrecke EH, Kroemer G. Self‐consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol. 2014;15:81‐94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Song S, Tan J, Miao Y, Li M, Zhang Q. Crosstalk of autophagy and apoptosis: involvement of the dual role of autophagy. J Cell sPhysiol. 2017;232:2977‐2984. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Table S1
Table S2
Document S1


