Abstract
Chronic infection with Helicobacter pylori cagA‐positive strains is causally associated with the development of gastric diseases, most notably gastric cancer. The cagA‐encoded CagA protein, which is injected into gastric epithelial cells by bacterial type IV secretion, undergoes tyrosine phosphorylation at the Glu‐Pro‐Ile‐Tyr‐Ala (EPIYA) segments (EPIYA‐A, EPIYA‐B, EPIYA‐C, and EPIYA‐D), which are present in various numbers and combinations in its C‐terminal polymorphic region, thereby enabling CagA to promiscuously interact with SH2 domain‐containing host cell proteins, including the prooncogenic SH2 domain‐containing protein tyrosine phosphatase 2 (SHP2). Perturbation of host protein functions by aberrant complex formation with CagA has been considered to contribute to the development of gastric cancer. Here we show that SHIP2, an SH2 domain‐containing phosphatidylinositol 5′‐phosphatase, is a hitherto undiscovered CagA‐binding host protein. Similar to SHP2, SHIP2 binds to the Western CagA‐specific EPIYA‐C segment or East Asian CagA‐specific EPIYA‐D segment through the SH2 domain in a tyrosine phosphorylation‐dependent manner. In contrast to the case of SHP2, however, SHIP2 binds more strongly to EPIYA‐C than to EPIYA‐D. Interaction with CagA tethers SHIP2 to the plasma membrane, where it mediates production of phosphatidylinositol 3,4‐diphosphate [PI(3,4)P2]. The CagA‐SHIP2 interaction also potentiates the morphogenetic activity of CagA, which is caused by CagA‐deregulated SHP2. This study indicates that initially delivered CagA interacts with SHIP2 and thereby strengthens H. pylori‐host cell attachment by altering membrane phosphatidylinositol compositions, which potentiates subsequent delivery of CagA that binds to and thereby deregulates the prooncogenic phosphatase SHP2.
Keywords: CagA; gastric cancer; Helicobacter pylori; PI(3,4)P2; SHIP2
The SH2 domain‐containing phosphatidylinositol 5′‐phosphatase, SHIP2, is a hitherto undiscovered CagA‐binding host protein. The CAgA‐SHIP2 interaction potentiates Helicobacter pylori‐mediated CagA delivery into gastric epithelial cells, which then promotes the formation of the oncogenic CagA‐SHP2 complex.
Abbreviations
- CRISPR
clustered regularly interspaced short palindromic repeats
- EPIYA
Glu‐Pro‐Ile‐Tyr‐Ala
- ITIM
immunoreceptor tyrosine‐based inhibition motif
- PI
phosphoinositide
- PI(3,4)P2
phosphatidylinositol 3,4‐diphosphate
- PI(3,4,5)P3
phosphatidylinositol 3,4,5‐trisphosphate
- PLA
proximity ligation assay
- PR
phosphorylation resistant
- pY
phosphotyrosine
- sgRNA
single guide RNA
- SHIP
SH2 domain‐containing phosphatidylinositol 5′‐phosphatase
- SHP
SH2 domain‐containing protein tyrosine phosphatase
- T4SS
type IV secretion system
- Tir
translocated intimin receptor
1. INTRODUCTION
Chronic infection with cagA‐positive Helicobacter pylori is strongly associated with the development of gastric cancer. 1 , 2 The H. pylori CagA protein, encoded by the cagA gene, is directly injected into gastric epithelial cells through the T4SS. 3 CagA possesses a unique tyrosine phosphorylation sequence motif termed the EPIYA motif, comprising the 5‐amino‐acid sequence Glu‐Pro‐Ile‐Tyr‐Ala, in variable numbers in its C‐terminal polymorphic region. 4 , 5 The EPIYA motif‐containing region of individual CagA consists of various combinations of 4 distinct segments (termed EPIYA‐A, EPIYA‐B, EPIYA‐C, and EPIYA‐D), each of which is defined by differences in the amino acid sequences spanning the EPIYA motif. 5 , 6 The EPIYA‐containing region of H. pylori CagA isolated in East Asian countries (East Asian CagA), where the incidence of gastric cancer is among the highest in the world, comprises EPIYA‐A, EPIYA‐B, and EPIYA‐D segments. In contrast, H. pylori strains isolated all over the world except East Asian countries comprise EPIYA‐A, EPIYA‐B, and variable numbers of tandem‐repeated EPIYA‐C segments (in most cases, 1‐3 times) (Western CagA). CagA injected into gastric epithelial cells undergoes tyrosine phosphorylation at the EPIYA motifs by host kinases such as Src family kinases and c‐Abl, 7 which enables pathological interaction of CagA with host SH2 domain‐containing proteins. Most notably, CagA forms a complex with the SHP2 tyrosine phosphatase, which contains 2 SH2 domains in its N‐terminal region, through tyrosine‐phosphorylated EPIYA‐C or EPIYA‐D segments. 6 Importantly, SHP2 binds to EPIYA‐D with 2 orders of magnitude stronger affinity than to EPIYA‐C. 8 The CagA‐SHP2 interaction leads to deregulation of the SHP2 phosphatase activity, which is essential for full activation of the RAS‐ERK signaling pathway. 4 , 9 As SHP2 is a prooncogenic phosphatase, 10 CagA has been considered to promote gastric carcinogenesis at least partly by aberrant activation of SHP2.
Although PIs are minor components of plasma membrane lipids, they are crucial for fundamental cellular processes, including cell signaling, membrane trafficking, and cytoskeletal rearrangements. 11 , 12 Metabolic abnormalities of PIs cause various diseases, especially cancer. For example, gene amplification or gain‐of‐function mutation of PIK3CA and loss‐of‐function mutation of PTEN have been shown to be associated with a diverse array of cancers. 13 , 14 Both SHIP1 and its homologue SHIP2 are phosphatidylinositol 5′‐phosphatases that contain a single SH2 domain in their N‐terminal regions. 15 , 16 , 17 , 18 The major role of these lipid phosphatases is to dephosphorylate PI(3,4,5)P3 at the 5′‐position and convert it to PI(3,4)P2. Although the expression of SHIP1 is restricted to hematopoietic cells, SHIP2 is more ubiquitously expressed. 17 , 18 SHIP2 is diffusely distributed to the cytoplasm and is translocalized to the plasma membrane upon growth factor stimuli. 16 SHIP2 plays a critical role in local cytoskeletal rearrangement that mediates focal adhesion turnover, generation of podosomes, or lamellipodia formation in response to a growth factor by increasing the local concentration of membranous PI(3,4)P2, which interacts with several PH domain‐containing proteins, including lamellipodin and TAPP1/2, and thereby causes actin cytoskeletal rearrangements. 19 , 20 Independently of its catalytic function, SHIP2 also acts as a protein scaffold that binds to actin‐related proteins such as p130Cas and filamin, which regulate cell adhesion and membrane ruffling. 21 , 22 These interactions also play substantial roles in cell adhesion and migration.
The SH2 domain of SHIP2 has been shown to interact with ITIMs. 23 Interestingly, 1 of the SH2 domain‐containing proteins that are also known to interact with ITIM motifs is SHP2, 24 suggesting that the SH2 domains of SHP2 and SHIP2 share binding targets in common. Indeed, in this study, we found that SHIP2 binds to the tyrosine‐phosphorylated EPIYA‐C or EPIYA‐D segments of CagA through the SH2 domain. Following complex formation, CagA tethers SHIP2 to the plasma membrane and thereby increases the level of membranous PI(3,4)P2, which could strengthen the attachment of H. pylori to gastric epithelial cells and thereby enhance the delivery of CagA into the host cells, which would enhance the formation of the oncogenic CagA‐SHP2 complex.
2. MATERIALS AND METHODS
2.1. Cells and transfection
All cell lines have been reported previously 4 , 25 and were tested for mycoplasma contamination by PCR prior to use. Human gastric cancer‐derived gastric epithelial AGS cells and nontransformed human gastric epithelial GES‐1 cells were cultured in RPMI‐1640 medium supplemented with 10% FBS. Monkey kidney COS‐7 cells were cultured in DMEM with 10% FBS. Cells were transfected with expression vectors using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions. SHIP2 gene KO cell lines, KO#1 and KO#2, were independently established from AGS cells using the CRISPR‐Cas9 system using 2 different sgRNAs.
2.2. Expression vectors
The cDNA fragments encoding Western ABCCC‐CagA [hereafter referred as wild‐type (WT)‐CagA], abccc‐CagA [hereafter referred as phospho‐resistant (PR)‐CagA], ABC‐CagA, ABCC‐CagA, and East Asian ABD‐CagA have been described previously. 4 , 6 , 26 , 27 A cDNA fragment encoding AB‐CagA was constructed by deletion of the sequence for the EPIYA‐C segment from a cDNA fragment encoding ABC‐CagA. These fragments with a C‐terminal Flag‐tag sequence were cloned into the pSP65SRα mammalian expression vectors. 4 A cDNA fragment encoding human SHIP2 was amplified using cDNA obtained from AGS cells as the PCR template, and the fragment with a C‐terminal Myc‐tag sequence was cloned into the pSP65SRα vector. A cDNA fragment encoding a SHIP2 mutant lacking the SH2 domain (ΔSH2‐SHIP2) was constructed by deletion of sequence residues 1‐117 from a cDNA fragment encoding SHIP2. Plasmids harboring sgRNA for the CRISPR/Cas9 system were constructed by inserting sgRNA#1 or sgRNA#2 into the pX330 vector. The target sequences for each sgRNA are as follows: sgRNA#1, 5ʹ‐GCTGGGACTTAATGAGCTGGCGG‐3ʹ; sgRNA#2, 5ʹ‐GAGGCATCCCGGTCATCCGGTGG‐3ʹ.
2.3. Antibodies
Monoclonal anti‐Flag (clone M2; Sigma), monoclonal anti‐SHIP2 (clone C76A7; Cell Signaling Technology), monoclonal anti‐PI(3,4)P2 (clone Z‐P034 and clone Z‐P034b; Echelon Biosciences), monoclonal anti‐phosphotyrosine (clone 4G10; Merck Millipore), monoclonal anti‐Myc (clone 9E10; Santa Cruz Biotechnology), monoclonal anti‐SH‐PTP2(SHP2) (clone B‐1; Santa Cruz Biotechnology), polyclonal anti‐phospho‐Src family (Tyr416) (Cell Signaling Technology), and polyclonal anti‐β‐actin (Cell Signaling Technology) Abs were used as a primary Ab for immunoprecipitation, immunoblotting, immunofluorescence staining, and PLA.
2.4. Helicobacter pylori infection
The H. pylori NCTC11637 standard strain, which contains the cagA gene encoding the Western ABCCC‐CagA protein, has been reported previously. 4 ΔcagA and ΔvirD4 are the H. pylori NCTC11637 strain‐derived isogenic mutants, which lack the cagA or virD4 gene, respectively. Cells were infected with H. pylori at a MOI of 100 in RPMI‐1640 supplemented with 10% FBS. After infection with H. pylori for the indicated time periods, cells were stained by immunofluorescence or lysed for immunoblotting.
2.5. Immunoprecipitation and immunoblotting
Cells were harvested and lysed in lysis buffer (50 mmol/L Tris‐HCl [pH7.5], 100 mmol/L NaCl, 5 mmol/L EDTA, 0.2% Triton X‐100, 10% glycerol, 2 mmol/L Na3VO4, 10 mmol/L NaF, 10 mmol/L β‐glycerophosphate, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 10 μg/ml trypsin inhibitor, and 2 mmol/L PMSF). The obtained lysates were immunoprecipitated with the indicated Ab. The immunoprecipitates and total cell lysates were resolved by SDS‐PAGE and subjected to immunoblotting, as described previously. 25
2.6. Immunofluorescence staining
Cells were seeded into 8‐well chamber slides (Lab‐Tek, Thermo Fisher Scientific), and cultured for the indicated time periods after transfection with respective vectors or infection with H. pylori. After fixing with 4% paraformaldehyde for 15 minutes and permeabilizing with 0.05% Triton X‐100 for 10 minutes, samples were subjected to immunofluorescence staining analysis as previously described. 25 Cellular nuclei and H. pylori were stained with DAPI.
2.7. Phosphatidylinositol 3,4‐bisphosphate staining
Immunofluorescence staining with anti‐PI(3,4)P2 Ab was carried out according to the manufacturer’s instructions. Briefly, cells were fixed with 4% paraformaldehyde for 20 minutes and permeabilized with 0.5% saponin (Sigma) for 15 minutes. After washing with TBS, cells were blocked with 10% normal goat serum (Wako) for 1 hour. The samples were treated with primary Abs and visualized with Alexa Fluor‐conjugated secondary Ab and DAPI. Images were acquired using the FV1200 confocal microscope system (Olympus).
2.8. Proximity ligation assay
Cells were seeded into 8‐well chamber slides. After transfection with respective vectors for 24 hours, cells were fixed with 4% paraformaldehyde for 20 minutes and permeabilized with 0.25% Triton X‐100 for 10 minutes. The PLA was carried out with a Duolink in situ PLA Kit (Sigma) according to the manufacturer’s instructions. Before or after polymerase reaction, samples were treated with Alexa Fluor 488‐conjugated secondary Ab to visualize CagA‐expressing cells. Images were acquired using the FV1200 confocal microscope system.
2.9. Cell morphological analysis
Cell morphology was analyzed using images of cells infected with H. pylori NCTC11637 strain. Elongated cell morphology with the longest diameter more than 2‐fold the shortest diameter was defined as the hummingbird phenotype. The diameters of cells were measured using ImageJ software (https://imagej.nih.gov/ij/).
2.10. Statistical analysis
Statistical analyses were carried out by Student’s t test or the Mann‐Whitney U test. P values less than .05 were considered to be statistically significant.
3. RESULTS
3.1. CagA interacts with SHIP2 through its tyrosine‐phosphorylated EPIYA motif
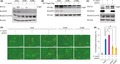
To test whether CagA binds to SHIP2, human gastric cancer‐derived gastric epithelial AGS cells or human nontransformed gastric epithelial GES‐1 cells were transiently transfected with a series of Flag‐tagged CagA vectors (Figure 1A), and the cell lysates were immunoprecipitated with an anti‐Flag Ab. The results of immunoblotting showed that endogenous SHIP2 was coprecipitated with WT‐CagA (ABCCC‐CagA) (Figure 1B). To examine whether the CagA‐SHIP2 interaction is mediated through a tyrosine‐phosphorylated EPIYA segment(s) of CagA, we made use of a PR‐CagA, in which all tyrosine residues in the 5 EPIYA segments of ABCCC‐CagA were substituted by phenylalanine (Figure 1A). In contrast to WT‐CagA, PR‐CagA did not coprecipitate SHIP2 (Figure 1C). The results of the experiment revealed that CagA forms a complex with SHIP2 in a tyrosine phosphorylation‐dependent manner.
Figure 1.
CagA interacts with SH2 domain‐containing phosphatidylinositol 5′‐phosphatase 2 (SHIP2) in a tyrosine phosphorylation‐dependent manner. A, Schematic diagram of Flag‐tagged CagA constructs. B‐E, AGS cells (B, C) or GES‐1 cells (B) were transiently transfected with a Flag‐tagged WT‐CagA or phosphorylation‐resistant (PR)‐CagA vector. COS‐7 cells (D, E) were transiently transfected with the indicated Flag‐tagged CagA vector together with a Myc‐tagged SHIP2 or control empty vector. Total cell lysates (TCLs) were immunoprecipitated (IP) with an anti‐Flag Ab and were subjected to immunoblotting (IB) with the respective Abs. EPIYA, Glu‐Pro‐Ile‐Tyr‐Ala motif
The EPIYA segments have been classified into 4 distinct types, 5 and Western CagA and East Asian CagA are characterized by the presence of EPIYA‐C and EPIYA‐D, respectively. To determine the tyrosine‐phosphorylated EPIYA segment(s) to which SHIP2 binds among these 4 segments, we next examined binding of SHIP2 with AB‐CagA, ABC‐CagA, or ABD‐CagA (Figure 1A). To this end, COS‐7 cells were transfected with the respective Flag‐tagged CagA vector together with a Myc‐tagged SHIP2 vector, and the cell lysates were immunoprecipitated with an anti‐Flag Ab. The results of anti‐Myc immunoblotting revealed that SHIP2 was most abundantly coprecipitated with ABC‐CagA among the 3 CagA species (Figure 1D), indicating that SHIP2 shows the strongest binding with the tyrosine‐phosphorylated EPIYA‐C segment. A comparison of SHIP2 binding with AB‐CagA and that with ABD‐CagA showed that SHIP2 also binds to the EPIYA‐D segment, although much less strongly than its binding to the EPIYA‐C segment (Figure 1D). As Western CagA contains varying numbers of EPIYA‐C segments, 5 we also compared the degrees of SHIP2 binding with Western CagA carrying 0‐3 repeats of the EPIYA‐C segment. We found that the degree of SHIP2 binding of Western CagA was proportional to the repeat number of EPIYA‐C segments (Figure 1A,E).
3.2. SH2 domain of SHP2 is responsible for CagA binding
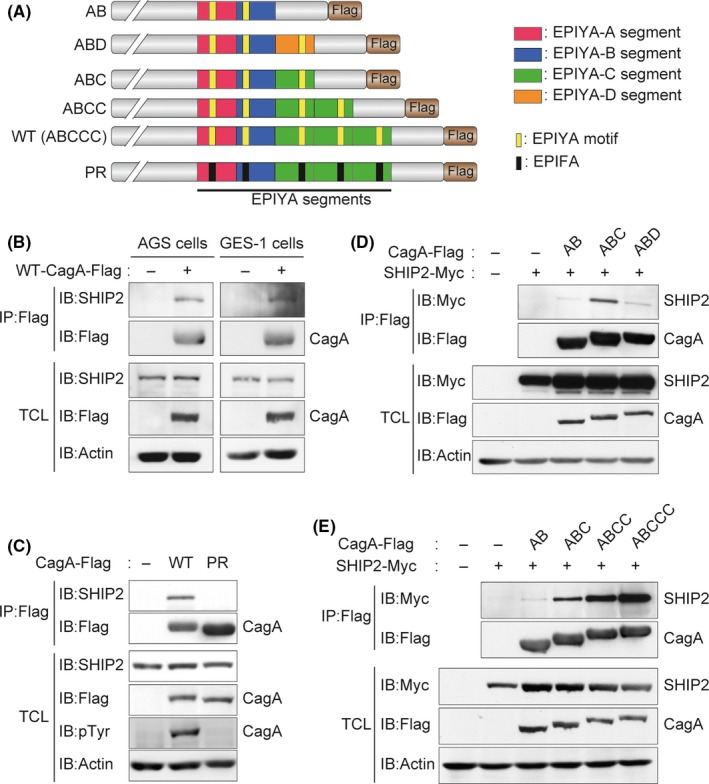
As CagA tyrosine phosphorylation was found to be indispensable for binding with SHIP2, we next examined whether the SH2 domain of SHIP2 is imperative for CagA interaction. The results of a coimmunoprecipitation experiment using lysates prepared from COS‐7 cells transiently transfected with a Flag‐tagged ABCCC‐CagA vector together with a vector for WT‐SHIP2 or a mutant SHIP2 lacking the SH2 domain (ΔSH2‐SHIP2) revealed that the SH2 domain was indispensable for binding of SHIP2 with CagA (Figure 2A,B). Thus, CagA binds to SHIP2 through the tyrosine‐phosphorylated EPIYA‐C/EPIYA‐D segment and the SH2 domain, respectively. Incidentally, SHP2 also interacts with the EPIYA‐C and EPIYA‐D segments. 6 Consistently, CagA‐SHP2 complex formation was reduced in cells overexpressing WT‐SHIP2 but not in cells overexpressing ΔSH2‐SHIP2 (Figure 2B), suggesting that SHIP2 competes with SHP2 for binding with the EPIYA‐C segment. Unexpectedly, however, SHIP2 binding to EPIYA‐C was substantially stronger than that to EPIYA‐D (Figure 1C), which was in sharp contrast to the case of CagA‐SHP2 interaction, in which SHP2‐binding affinity of EPIYA‐C was less than 1/100 of that of EPIYA‐D. 8
Figure 2.
SH2 domain of SH2 domain‐containing phosphatidylinositol 5′‐phosphatase 2 (SHIP2) is essential for binding to CagA. A, Schematic diagram of Myc‐tagged SHIP2 constructs. B, COS‐7 cells were transiently transfected with a Myc‐tagged WT‐SHIP2 or mutant SHPI2 lacking the SH2 domain (ΔSH2‐SHIP2) vector together with a Flag‐tagged WT‐CagA or control empty vector. Total cell lysates (TCLs) were immunoprecipitated (IP) with an anti‐Flag Ab and subjected to immunoblotting (IB) with the respective Abs
3.3. Fraction of SHIP2 translocalized to the plasma membrane by CagA expression
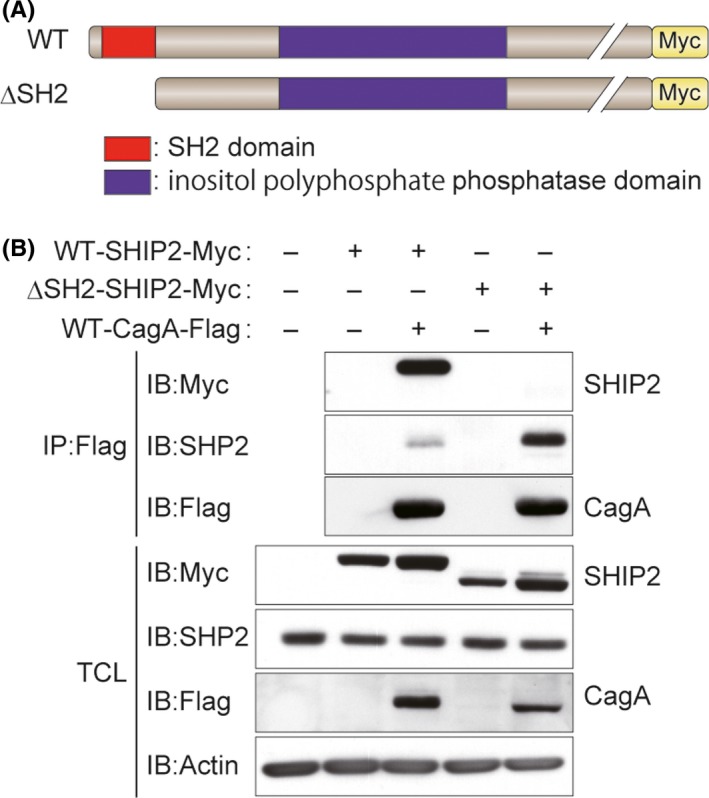
CagA is tethered to the plasma membrane after delivery into host cells. 4 In contrast, SHIP2 is distributed broadly in the cytoplasm, although it accumulates in the plasma membrane periphery following stimulation of cells with growth factors. 16 To determine the intracellular localization of the CagA‐SHIP2 complex, we undertook a PLA with an anti‐Flag Ab and an anti‐SHIP2 Ab in AGS or GES‐1 cells transiently transfected with a Flag‐tagged WT or a PR‐CagA vector and observed the PLA spots, which represent the CagA‐SHIP2 complexes in the cells. The results of the experiment confirmed specific complex formation of endogenous SHIP2 with tyrosine‐phosphorylated CagA (Figure 3A,B). Furthermore, most of the PLA spots were localized in the vicinity of the plasma membrane (Figure 3A).
Figure 3.
CagA induces translocalization of SH2 domain‐containing phosphatidylinositol 5′‐phosphatase 2 (SHIP2) to the plasma membrane. A, B, AGS (upper) or GES‐1 (lower) cells were transiently transfected with a Flag‐tagged WT‐ or phosphorylation‐resistant (PR)‐CagA vector. Physical interactions between endogenous SHIP2 and CagA in cells were analyzed by the proximity ligation assay (PLA) with an anti‐Flag Ab and an anti‐SHIP2 Ab. A, Red PLA spots indicate CagA‐SHIP2 interaction. Cellular nuclei and CagA‐expressing cells were visualized in blue and green, respectively. Upper images of AGS cells show maximum intensity projections (2.5 μm thick) and others are confocal images. Scale bar, 20 μm. B, Dots show the number of PLA spots per CagA‐expressing cell in (A). Bars indicate median. n = 50 cells. ***P < .001 (Mann‐Whitney U test). C, AGS cells were infected with the Helicobacter pylori NCTC11637 strain or its isogenic mutants (ΔcagA or ΔvirD4) at an MOI of 100 for 6 h, and were then immunofluorescence stained with an anti‐SHIP2 Ab. SHIP2 was visualized in gray. Cellular nuclei and H. pylori were visualized in blue by DAPI staining. Arrowheads indicate H. pylori dense area. Scale bar, 10 μm. Enlarged images of red boxes indicated at the third row panels are shown in the bottom row of panels (panels 2, 4, and 6 from left). Vertical xz‐sections (upper) and their scanning fluorescence intensities (lower) along the yellow lines indicated at the third row panels are also shown in the bottom row of panels (panels 1, 3, 5, and 7 from left). In the scanning data, intensities of H. pylori and SHIP2 are shown in blue and red lines, respectively
We also examined the subcellular localization of SHIP2 in AGS cells infected with the H. pylori NCTC11637 standard strain that produces ABCCC‐CagA. The CagA‐injected cells display a characteristically elongated cell shape, termed the hummingbird phenotype, 28 which is caused by CagA‐deregulated SHP2. 4 Immunofluorescence staining for endogenous SHIP2 revealed that a fraction of SHIP2 was localized to the plasma membrane of elongated cells infected with the H. pylori NCTC11637 strain (Figure 3C). In contrast, infection of AGS cells with an isogenic strain lacking the cagA gene (ΔcagA) or an isogenic strain incapable of injecting CagA due to the lack of the functional T4SS (ΔvirD4) 29 failed to induce translocalization of SHIP2 to the plasma membrane (Figure 3C). These results indicated that tyrosine‐phosphorylated CagA recruited SHIP2 to the plasma membrane following complex formation. Furthermore, SHIP2 was abundantly accumulated in the area of the plasma membrane where H. pylori was densely adhered (Figure 3C, lower panels).
3.4. Accumulation of PI(3,4)P2 in the plasma membrane of CagA‐injected cells
The plasma membrane contains several different types of PIs, each of which exerts various cellular functions by specific binding with respective partner proteins. 11 , 12 The function of SHIP2 as a lipid phosphatase is to dephosphorylate PI(3,4,5)P3 to PI(3,4)P2. As SHIP2 was accumulated to the plasma membrane to which cagA‐positive H. pylori adhered, we investigated the existence of PI(3,4)P2, the SHIP2 product, in the plasma membrane of AGS cells infected with the H. pylori NCTC11637 strain by immunostaining with an anti‐PI(3,4)P2 Ab. The results of the experiment revealed that PI(3,4)P2 was detectable at the plasma membrane of CagA‐injected cells with the hummingbird phenotype (Figure 4A). In contrast, such a membrane accumulation of PI(3,4)P2 was not observed in AGS cells infected with the isogenic ΔcagA strain (Figure 4A). Similarly, infection of SHIP2‐KO AGS cells (clones #1 and #2), which were made by using the CRISPR/Cas9 system, did not induce PI(3,4)P2 accumulation at the plasma membrane (Figure 4B). Staining of PI(3,4)P2 at the plasma membrane was also observed in AGS cells or GES‐1 cells expressing WT‐CagA but was not observed in cells expressing PR‐CagA (Figure 4C). From these observations, we concluded that injected CagA enhanced the lipid phosphatase activity of SHIP2 at the plasma membrane by tethering SHIP2 to the plasma membrane through complex formation.
Figure 4.
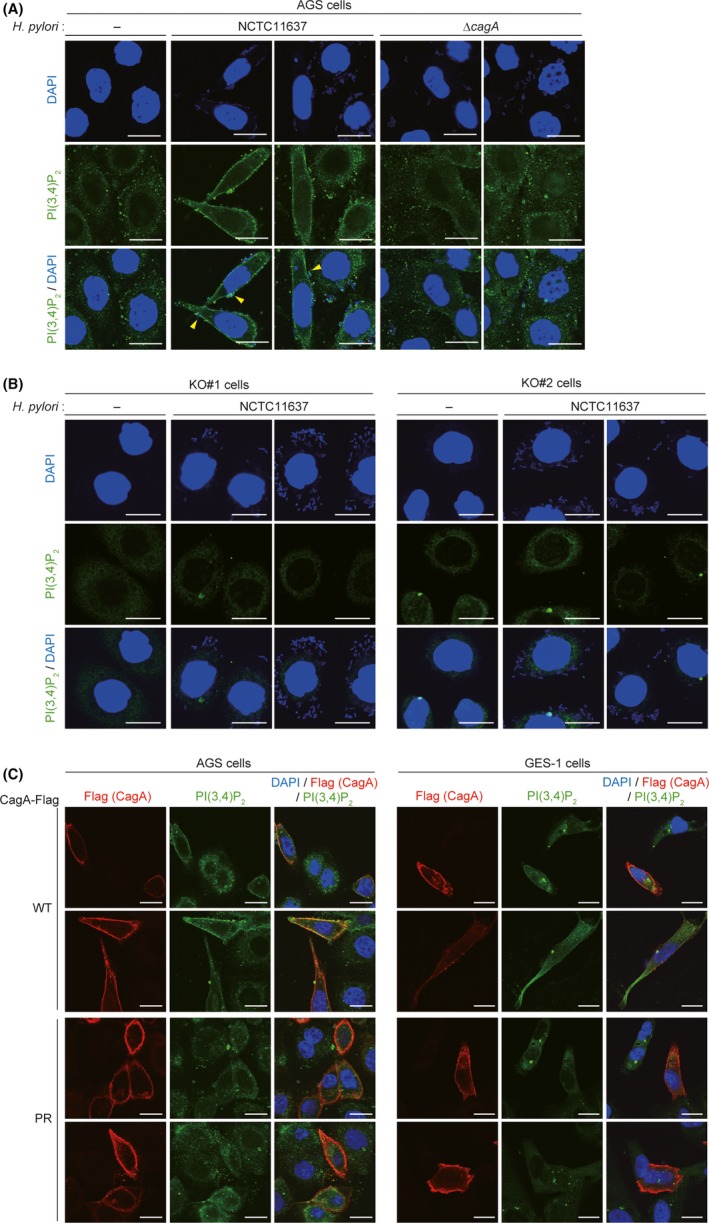
Phosphatidylinositol 3,4‐diphosphate (PI(3,4)P2) accumulates in the plasma membrane of CagA‐injected cells. A, B, AGS cells (A) or AGS‐derived SHIP2‐KO KO#1 and KO#2 cells (B) were infected with the Helicobacter pylori NCTC11637 strain or its isogenic ΔcagA strain at an MOI of 100 for 6 h, and subjected to PI(3,4)P2 staining. PI(3,4)P2 was visualized in green. Cellular nuclei and H. pylori were visualized in blue by DAPI staining. Arrowheads indicate CagA‐injected cells. Scale bar, 20 μm. C, AGS or GES‐1 cells were transiently transfected with a Flag‐tagged WT‐ or PR‐CagA vector, and subjected to PI(3,4)P2 staining. PI(3,4)P2, CagA‐Flag, and cellular nuclei were visualized in green, red, and blue, respectively. Scale bar, 20 μm
3.5. Role of CagA‐SHIP2 interaction in prooncogenic action of CagA
Enteropathogenic Escherichia coli produces an effector protein known as the Tir. The Tir is injected into intestinal epithelial cells through the bacterial type III secretion system, where it strengthens the attachment of the bacterium to the host cells by binding and thereby activating SHIP2. 30 We therefore wished to know the pathobiological role of the CagA‐SHIP2 interaction in the prooncogenic action of CagA. To this end, parental AGS cells or the SHIP2‐KO AGS cells (KO#1 and KO#2) were infected with an H. pylori cagA‐positive strain, and the amounts of CagA delivered into cells were determined. As a result, the levels of tyrosine‐phosphorylated CagA were substantially diminished in SHIP2‐KO AGS cells compared to the levels in WT AGS cells (Figure 5A). The results of an anti‐phospho‐Src (Tyr416) immunoblotting analysis showed that the level of activated Src, which phosphorylates CagA, was not decreased by knocking out SHIP2 (Figure 5A). Furthermore, the levels of CagA tyrosine phosphorylation were comparable in parental and SHIP2‐KO AGS cells expressing CagA (Figure 5B). Conversely, overexpression of SHIP2 increased the amount of H. pylori‐delivered CagA in parental AGS cells (Figure 5C). From these observations, we concluded that CagA‐SHIP2 interaction potentiated H. pylori‐mediated delivery of CagA into gastric epithelial cells.
Figure 5.
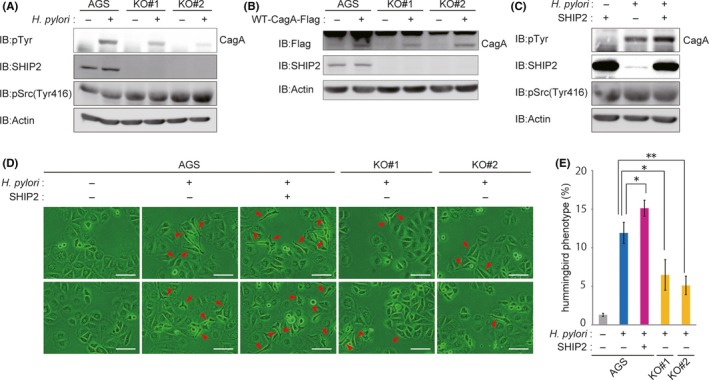
CagA‐SH2 domain‐containing phosphatidylinositol 5′‐phosphatase 2 (SHIP2) interaction potentiated delivery of CagA from Helicobacter pylori to gastric epithelial cells. A, AGS, AGS‐derived SHIP2‐KO KO#1, or AGS‐derived SHIP2‐KO KO#2 cells were infected with the H. pylori NCTC11637 strain at an MOI of 100 for 9 h. Total cell lysates were then immunoblotted (IB) with the respective Abs. B, AGS, AGS‐derived SHIP2‐KO KO#1, or AGS‐derived SHIP2‐KO KO#2 cells were transiently transfected with a WT‐CagA vector or a control empty vector for 24 h. Total cell lysates were IB with the respective Abs. C, AGS cells were transfected with a SHIP2 vector or control empty vector. At 24 h after transfection, cells were infected with the H. pylori NCTC11637 strain at an MOI of 100 for an additional 9 h. Total cell lysates were subjected to IB with the respective Abs. D, AGS, AGS‐derived SHIP2‐KO KO#1, or AGS‐derived SHIP2‐KO KO#2 cells were transfected with a SHIP2 vector or control empty vector. At 24 h after transfection, cells were infected with the H. pylori NCTC11637 strain at an MOI of 100 for an additional 9 h before microscopic analysis. Arrowheads indicate the hummingbird phenotype. Scale bar, 100 μm. E, Percentage of cells with the hummingbird phenotype shown in (D). Error bars, ±SD; n = 3. *P < .05, **P < .01 (Student’s t test)
Because induction of the hummingbird phenotype by in vitro infection with H. pylori requires CagA‐SHP2 interaction, 4 , 9 CagA‐SHIP2 interaction was thought to strengthen H. pylori‐mediated hummingbird phenotype induction by increasing CagA delivery into the host cells. To test this idea, parental and SHIP2‐overexpressing AGS cells were infected with an H. pylori cagA‐positive strain and the degree of hummingbird phenotype induction was evaluated. The results of the experiment revealed that the number of cells displaying the hummingbird phenotype was increased in SHIP2‐overexpressing AGS cells (Figure 5D,E). Conversely, induction of the hummingbird phenotype was significantly diminished in SHIP2‐KO AGS cells (KO#1 and KO#2) compared to that in WT AGS cells (Figure 5D,E). These observations indicated that CagA‐SHIP2 interaction promoted CagA delivery into host gastric epithelial cells, which in turn potentiated CagA‐SHP2 interaction that induced the epithelial‐mesenchymal transition‐like hummingbird phenotype, which is considered to reflect enhanced cell motility and cell invasion potential.
4. DISCUSSION
In the host gastric epithelial cells, to which it has been delivered by bacterial T4SS, H. pylori CagA undergoes tyrosine phosphorylation by host kinases such as Src family kinases (SFKs) and c‐Abl. Tyrosine‐phosphorylated CagA promiscuously interacts with SH2 domain‐containing host proteins, including SHP2, PI3K, and Crk, and thereby perturbs their functions to promote neoplastic transformation of the cells. 31 , 32 The results of the present study add SHIP2 to the list of CagA‐binding SH2‐containing proteins. The CagA‐SHIP2 interaction is mediated between the tyrosine‐phosphorylated EPIYA‐C (in the case of Western CagA) or EPIYA‐D segment (in the case of East Asian CagA) and the SH2 domain of SHIP2. Hence, binding specificity of SHIP2 to the CagA EPIYA segment is the same as that of CagA with SHP2. 4 , 5 , 6 However, the interaction mode of SHIP2 with CagA could be substantially different from that of SHP2 with CagA, based on the observation that SHIP2 binds EPIYA‐C more strongly than EPIYA‐D, whereas SHP2 binds EPIYA‐D more strongly than EPIYA‐C does. 8
In general, binding of SH2 domains with phosphotyrosyl peptides is mediated by a stretch of 3‐6 amino acid residues that flanks the pY on its C‐terminal side. 33 The comparison of the sequences spanning pY between the EPIYA‐C and EPIYA‐D segments of CagA shows that they are identical between pY‐3 and pY + 4, indicating that sequence variations after pY + 5 might underlie inverse binding preferences between SHIP2 and SHP2 toward Western CagA and East Asian CagA. In this regard, the pY + 5 residue of EPIYA‐C is Asp, and that of EPIYA‐D is Phe. The large aromatic side chain of Phe at pY + 5 on EPIYA‐D fills the hollow structure comprising Leu65, Gly67, Gly68, and Tyr81 on the N‐SH2 domain of SHP2, conferring strong SHP2 binding on EPIYA‐D. 8 We speculate that the SH2 domain of SHIP2 does not have such a hollow and thus the large aromatic side chain of Phe at pY + 5 sterically interferes with binding of EPIYA‐D to the SH2 domain of SHIP2. Alternatively, the presence of a positively charged residue such as aspartic acid at pY + 5 might mediate electrostatic interaction of EPIYA‐C with the SHIP2 SH2 domain.
The results of the present study revealed that a reduced level of SHIP2 subverted translocalization of CagA into the host cell infected with H. pylori, indicating a role of SHIP2 in facilitating CagA delivery by H. pylori. An intriguing possibility is that PI(3,4)P2, which is generated at the plasma membrane through activation of SHIP2 by delivered CagA during initial phase of H. pylori infection, strengthens H. pylori adherens to the gastric epithelial cells and thereby facilitates subsequent delivery of CagA through T4SS. Notably, SHIP2 plays an important role in the formation of focal adhesion spots, 21 , 22 , 34 , 35 where PI(3,4)P2 also accumulates. In contrast, in CagA‐expressing cells, SHIP2 as well as PI(3,4)P2 is accumulated to the plasma membrane periphery (Figures 3C and 4A), most probably due to impaired focal adhesions by CagA. 36 It is possible that interaction of SHIP2 with CagA is involved in the perturbation of focal adhesion spots. Given that interaction of T4SS components with integrins, the major components of focal adhesions, is crucial for CagA injection by H. pylori, 37 CagA‐SHIP2 complex formation could be facilitated by disruption of focal adhesions, which diffuses SHIP2 and integrins through the plasma membrane and thereby increases the chance of interacting with CagA. Elevated CagA‐SHIP2 interaction in turn facilitates CagA delivery and subsequent disruption of adhesion spots, making a feedforward regulation between CagA‐SHIP2 interaction and CagA delivery.
Enhanced CagA delivery by SHIP2 is of particular interest in light of the fact that Western EPIYA‐C binds SHIP2 more strongly than does East Asian EPIYA‐D. This could indicate that SHIP2 is more important in the regulation and/or function of Western CagA than East Asian CagA. Of note, a fraction (20%‐30%) of Western CagA contains tandemly duplicated EPIYA‐C segments (2‐3 times), which bind and deregulate SHP2 more strongly than Western CagA with a single EPIYA‐C segment does. 6 , 8 , 26 The results of the present study indicate that EPIYA‐C duplication potentiates the ability of Western CagA to deregulate SHP2 not only by qualitatively increasing SHP2‐binding affinity but also quantitatively facilitating CagA delivery by H. pylori T4SS through CagA‐SHIP2 interaction, further supporting the clinical observations that Western CagA with multiple EPIYA‐C is a distinct risk factor of gastric cancer development in Western countries. 38
The results of proteome analysis suggest that the amount of SHP2 is relatively in excess to that of SHIP2 in the cells. 39 We therefore assume that the EPIYA‐C segment of Western CagA binds SHIP2 more strongly than SHP2 so that CagA delivered during early H. pylori infection preferentially interacts with SHIP2. CagA‐activated SHIP2 then modifies PI compositions of plasma membrane to strengthen the H. pylori attachment to the host cell, which in turn facilitates T4SS‐mediated CagA delivery. As substantial amounts of SHIP2 interact with CagA, SHP2 becomes a primary interactor of delivered CagA through the EPIYA‐C segment. In any case, the present study indicates that CagA‐SHIP2 interaction plays an important role in quantitatively increasing the pathobiological activity of Western CagA, which is qualitatively less bioactive than East Asian CagA, by facilitating CagA delivery.
CONFLICT OF INTEREST
The authors have no conflict of interest to declare.
ACKNOWLEDGMENT
This work was supported by Grants‐in‐Aid for Scientific Research “S” (#16H06373) and “C” (#19K08436), and by the Max‐Planck Society, Germany.
Fujii Y, Murata-Kamiya N, Hatakeyama M. Helicobacter pylori CagA oncoprotein interacts with SHIP2 to increase its delivery into gastric epithelial cells. Cancer Sci. 2020;111:1596–1606. 10.1111/cas.14391
REFERENCES
- 1. Blaser MJ, Perez‐Perez GI, Kleanthous H, et al. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 1995;55:2111‐2115. [PubMed] [Google Scholar]
- 2. Parsonnet J, Friedman GD, Orentreich N, Vogelman H. Risk for gastric cancer in people with CagA positive or CagA negative Helicobacter pylori infection. Gut. 1997;40:297‐301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Covacci A, Rappuoli R. Tyrosine‐phosphorylated bacterial proteins: Trojan horses for the host cell. J Exp Med. 2000;191:587‐592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Higashi H, Tsutsumi R, Muto S, et al. SHP‐2 tyrosine phosphatase as an intracellular target of Helicobacter pylori CagA protein. Science. 2002;295:683‐686. [DOI] [PubMed] [Google Scholar]
- 5. Hatakeyama M. Oncogenic mechanisms of the Helicobacter pylori CagA protein. Nat Rev Cancer. 2004;4:688‐694. [DOI] [PubMed] [Google Scholar]
- 6. Higashi H, Tsutsumi R, Fujita A, et al. Biological activity of the Helicobacter pylori virulence factor CagA is determined by variation in the tyrosine phosphorylation sites. Proc Natl Acad Sci USA. 2002;99:14428‐14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mueller D, Tegtmeyer N, Brandt S, et al. c‐Src and c‐Abl kinases control hierarchic phosphorylation and function of the CagA effector protein in Western and East Asian Helicobacter pylori strains. J Clin Invest. 2012;122:1553‐1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hayashi T, Senda M, Suzuki N, et al. Differential Mechanisms for SHP2 binding and activation are exploited by geographically distinct Helicobacter pylori CagA Oncoproteins. Cell Rep. 2017;20:2876‐2890. [DOI] [PubMed] [Google Scholar]
- 9. Higashi H, Nakaya A, Tsutsumi R, et al. Helicobacter pylori CagA induces Ras‐independent morphogenetic response through SHP‐2 recruitment and activation. J Biol Chem. 2004;279:17205‐17216. [DOI] [PubMed] [Google Scholar]
- 10. Chan G, Kalaitzidis D, Neel BG. The tyrosine phosphatase Shp2 (PTPN11) in cancer. Cancer Metastasis Rev. 2008;27:179‐192. [DOI] [PubMed] [Google Scholar]
- 11. Schink KO, Tan KW, Stenmark H. Phosphoinositides in control of membrane dynamics. Annu Rev Cell Dev Biol. 2016;32:143‐171. [DOI] [PubMed] [Google Scholar]
- 12. Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651‐657. [DOI] [PubMed] [Google Scholar]
- 13. Thorpe LM, Yuzugullu H, Zhao JJ. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat Rev Cancer. 2015;15:7‐24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hollander MC, Blumenthal GM, Dennis PA. PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nat Rev Cancer. 2011;11:289‐301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Suwa A, Kurama T, Shimokawa T. SHIP2 and its involvement in various diseases. Expert Opin Ther Targets. 2010;14:727‐737. [DOI] [PubMed] [Google Scholar]
- 16. Erneux C, Edimo WE, Deneubourg L, Pirson I. SHIP2 multiple functions: a balance between a negative control of PtdIns(3,4,5)P₃ level, a positive control of PtdIns(3,4)P₂ production, and intrinsic docking properties. J Cell Biochem. 2011;112:2203‐2209. [DOI] [PubMed] [Google Scholar]
- 17. Geier SJ, Algate PA, Carlberg K, et al. The human SHIP gene is differentially expressed in cell lineages of the bone marrow and blood. Blood. 1997;89:1876‐1885. [PubMed] [Google Scholar]
- 18. Schurmans S, Carrió R, Behrends J, Pouillon V, Merino J, Clément S. The mouse SHIP2 (Inppl1) gene: complementary DNA, genomic structure, promoter analysis, and gene expression in the embryo and adult mouse. Genomics. 1999;62:260‐271. [DOI] [PubMed] [Google Scholar]
- 19. Krause M, Leslie JD, Stewart M, et al. Lamellipodin, an Ena/VASP ligand, is implicated in the regulation of lamellipodial dynamics. Dev Cell. 2004;7:571‐583. [DOI] [PubMed] [Google Scholar]
- 20. Dowler S, Currie RA, Campbell DG, et al. Identification of pleckstrin‐homology‐domain‐containing proteins with novel phosphoinositide‐binding specificities. Biochem J. 2000;351:19‐31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Prasad N, Topping RS, Decker SJ. SH2‐containing inositol 5'‐phosphatase SHIP2 associates with the p130(Cas) adapter protein and regulates cellular adhesion and spreading. Mol Cell Biol. 2001;21:1416‐1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dyson JM, O'Malley CJ, Becanovic J, et al. The SH2‐containing inositol polyphosphate 5‐phosphatase, SHIP‐2, binds filamin and regulates submembraneous actin. J Cell Biol. 2001;155:1065‐1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Muraille E, Bruhns P, Pesesse X, Daëron M, Erneux C. The SH2 domain containing inositol 5‐phosphatase SHIP2 associates to the immunoreceptor tyrosine‐based inhibition motif of FcγRIIB in B cells under negative signaling. Immunol Lett. 2000;72:7‐15. [DOI] [PubMed] [Google Scholar]
- 24. Unkeless JC, Jin J. Inhibitory receptors, ITIM sequences and phosphatases. Curr Opin Immunol. 1997;9:338‐343. [DOI] [PubMed] [Google Scholar]
- 25. Fujii Y, Yoshihashi K, Suzuki H, et al. CDX1 confers intestinal phenotype on gastric epithelial cells via induction of stemness‐associated reprogramming factors SALL4 and KLF5. Proc Natl Acad Sci USA. 2012;109:20584‐20589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nagase L, Hayashi T, Senda T, Hatakeyama M. Dramatic increase in SHP2 binding activity of Helicobacter pylori Western CagA by EPIYA‐C duplication: its implications in gastric carcinogenesis. Sci Rep. 2015;5:15749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Naito M, Yamazaki T, Tsutsumi R, et al. Influence of EPIYA‐repeat polymorphism on the phosphorylation‐dependent biological activity of Helicobacter pylori CagA. Gastroenterology. 2006;130:1181‐1190. [DOI] [PubMed] [Google Scholar]
- 28. Segal ED, Cha J, Lo J, Falkow S, Tompkins LS. Altered states: involvement of phosphorylated CagA in the induction of host cellular growth changes by Helicobacter pylori . Proc Natl Acad Sci USA. 1999;96:14559‐14564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Selbach M, Moese S, Meyer TF, Backert S. Functional analysis of the Helicobacter pylori cag pathogenicity island reveals both VirD4‐CagA‐dependent and VirD4‐CagA‐independent mechanisms. Infect Immun. 2002;70:665‐671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Smith K, Humphreys D, Hume PJ, Koronakis V. Enteropathogenic Escherichia coli recruits the cellular inositol phosphatase SHIP2 to regulate actin‐pedestal formation. Cell Host Microbe. 2010;7:13‐24. [DOI] [PubMed] [Google Scholar]
- 31. Hatakeyama M. Anthropological and clinical implications for the structural diversity of the Helicobacter pylori CagA oncoprotein. Cancer Sci. 2011;102:36‐43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hatakeyama M Helicobacter pylori CagA and gastric cancer: a paradigm for hit‐and‐run carcinogenesis. Cell Host Microbe. 2014;15:306‐316. [DOI] [PubMed] [Google Scholar]
- 33. Liu BA, Engelmann BW, Nash PD. The language of SH2 domain interactions defines phosphotyrosine‐mediated signal transduction. FEBS Lett. 2012;586:2597‐2605. [DOI] [PubMed] [Google Scholar]
- 34. Elong Edimo W, Vanderwinden JM, Erneux C. SHIP2 signalling at the plasma membrane, in the nucleus and at focal contacts. Adv Biol Regul. 2013;53:28‐37. [DOI] [PubMed] [Google Scholar]
- 35. Fukumoto M, Ijuin T, Takenawa T. PI(3,4)P2 plays critical roles in the regulation of focal adhesion dynamics of MDA‐MB‐231 breast cancer cells. Cancer Sci. 2017;108:941‐951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tsutsumi R, Takahashi A, Azuma T, Higashi H, Hatakeyama M. Focal adhesion kinase is a substrate and downstream effector of SHP‐2 complexed with Helicobacter pylori CagA. Mol Cell Biol. 2006;26:261‐276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kwok T, Zabler D, Urman S, et al. Helicobacter exploits integrin for type IV secretion and kinase activation. Nature. 2007;449:862–866. [DOI] [PubMed] [Google Scholar]
- 38. Li Q, Liu J, Gong Y, Yuan Y. Association of CagA EPIYA‐D or EPIYA‐C phosphorylation sites with peptic ulcer and gastric cancer risks: a meta‐analysis. Medicine. 2017;96:e6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wilhelm M, Schlegl J, Hahne H, et al. Mass‐spectrometry‐based draft of the human proteome. Nature. 2014;509:582‐587. [DOI] [PubMed] [Google Scholar]


