Abstract
There is increasing evidence that bone morphogenetic proteins (BMP) are involved in the proliferation and drug tolerance of kidney cancer. However, the molecular mechanism of BMP8A in renal cell proliferation and drug tolerance is not clear. Here we showed that BMP8A was highly expressed in renal cell carcinoma, which suggests a poor prognosis of ccRCC. Promotion of cell proliferation and inhibition of apoptosis were detected by CCK‐8 assay, Trypan Blue staining, flow cytometry and bioluminescence. BMP8A promoted resistance of As2O3 by regulating Nrf2 and Wnt pathways in vitro and in vivo. Mechanistically, BMP8A enhanced phosphorylation of Nrf2, which, in turn, inhibited Keap1‐mediated Nrf2 ubiquitination and, ultimately, promoted nuclear translocation and transcriptional activity of Nrf2. Nrf2 regulates the transcription of TRIM24 detected by ChIP‐qPCR. BMP8A was highly expressed in ccRCC, which suggests a poor prognosis. BMP8A was expected to be an independent prognostic molecule for ccRCC. On the one hand, activated Nrf2 regulated reactive oxygen balance, and on the other hand, by regulating the transcription level of TRIM24, it was involved in the regulation of the Wnt pathway to promote the proliferation, invasion and metastasis of ccRCC and the resistance of As2O3. Taken together, our findings describe a regulatory axis where BMP8A promotes Nrf2 phosphorylation and activates TRIM24 to promote survival and drug resistance in ccRCC.
Keywords: As2O3, BMP8A, Nrf2, TRIM24, Wnt pathway
BMP8A can promote Nrf2 phosphorylation and nuclear translocation to exert antioxidative stress and transcriptional activity. At the same time, Nrf2 acts as a transcription factor of TRIM24, promotes the expression of TRIM24, activates the Wnt pathway and increases chemotherapy tolerance.
1. INTRODUCTION
Renal cell carcinoma is a highly malignant tumor that is common in the urinary system. It has the characteristics of radiotherapy and chemotherapy tolerance. 1 , 2 Regarding the pathological types of RCC, approximately 75%‐80% of cases are clear cell renal cell carcinoma (ccRCC). Although surgical treatment is available, the recurrence rate after radical nephrectomy is still as high as 20%‐40%, and the 5‐year survival rate of ccRCC after surgical resection is only approximately 20%. 3 Therefore, revealing the molecular mechanism of renal cancer radiotherapy and chemotherapy tolerance is critical. 4
The bone morphogenetic protein (BMP) ligand family plays an important role in embryonic development, morphogenesis, differentiation, proliferation and apoptosis of various types of cells throughout the body. BMP are found to be members of the transforming growth factor‐beta (TGF‐β) family, including activin in addition to TGF‐β. Coding from 33 genes has identified members of the TGF‐β family, approximately 15 structurally‐related BMP. 5 , 6 Like TGF‐β cytokines, BMP induces the activation of the receptor’s serine/threonine kinase activity by interacting with type I and type II cell surface receptors, triggering specific cellular responses. 7 , 8
BMP8A encodes a secreted ligand of the TGF‐β superfamily of proteins. Ligands of this family bind various TGF‐beta receptors, leading to recruitment and activation of SMAD family transcription factors that regulate gene expression. The encoded preproprotein is proteolytically processed to generate each subunit of the disulfide‐linked homodimer. This protein may play a role in development of the reproductive system. BMP8A sustains spermatogenesis by activating both SMAD1/5/9 and SMAD2/3 in spermatogonia. 9
Human cells are always exposed to reactive oxygen species (ROS) produced by endogenous metabolic or environmental oxidants. 10 Maintenance of ROS homeostasis is critical for human cell survival. Therefore, ROS disorders can lead to the development of various diseases, including/even cancers. 11 However, there is still some controversy about the role of ROS in cancers. 12 , 13 It has been argued that a sustained increase in ROS promotes cell growth and promotes carcinogenesis, and once the toxicity threshold is exceeded, excessive production of ROS results in cell death. To adapt to the oxidative stress environment, cancer cells have evolved effective ROS antioxidant systems to limit excessive ROS accumulation. 14 Under oxidative stress conditions, the degradation of Nrf2 is rapidly reduced, leading to the accumulation of Nrf2 and more active nuclear translocation. In addition, Nrf2 is a conserved antioxidant response element (ARE) with a group of antioxidant and cellular defense targets (such as NADPH quinone oxidoreductase 1, heme oxygenase‐1 and ferritin heavy polypeptide 1). 15 , 16 , 17 Nrf2 promotes the transcription of target genes and exerts its antioxidant effect.
Tripartite Motif‐containing protein 24 (TRIM24), also known as transcription intermediary factor 1 alpha (TIF1a), has amino‐terminal RBCC domains. 18 , 19 TRIM24 is involved in driving different tumor types through its ability to interfere with tumor suppression and oncogenic pathways. 20 TRIM24 acts as a transcriptional coactivator that interacts with numerous nuclear receptors and coactivators and modulates the transcription of target genes. It interacts with chromatin depending on histone H3 modifications, having the highest affinity for histone H3 that is both unmodified at “Lys‐4” (H3K4me0) and acetylated at “Lys‐23” (H3K23ac), and has E3 protein‐ubiquitin ligase activity. 21 Moreover, TRIM24 upregulates ligand‐dependent transcription activation by AR, GCR/NR3C1, thyroid hormone receptor (TR) and ESR1. 22 TRIM24 modulates transcription activation by retinoic acid (RA) receptors, including RARA. TRIM24 plays a role in regulating retinoic acid‐dependent proliferation of hepatocytes. 23
In this study, we investigated the mechanism of BMP8A and attempted to elucidate its applicability as a therapeutic target by combining molecular features in ccRCC cells with analysis in renal cancer patients. We hypothesized that BMP8A promoted the activation of Nrf2, which, in turn, enhanced the transcriptional activation of TRIM24 to promote the progression of renal cell carcinoma and drug resistance.
2. MATERIALS AND METHODS
2.1. Cell culture
The human RCC cell lines (786‐O, OS‐RC‐2, Caki‐1, 769‐P, A498 and ACHN) and human renal tubular epithelial cells (HK‐2) were all purchased from the Cell Resources Center, Shanghai Academy of Life Sciences, Chinese Academy of Sciences. A498 was purchased from ATCC. ACHN cells were grown in RPMI‐DMEM (Gibco); HK‐2, 786‐O, OS‐RC‐2, A498, Caki‐1 and 769‐P cells were grown in RPMI 1640 medium (Gibco) containing 10% FBS (Biological Industries, Israel). All cells were grown at 37°C in a humidified 5% CO2 atmosphere.
2.2. Chemicals
As2O3 Sigma Cat. #202673, MG132 Selleck Cat. #S2619, NAC(N‐Acetyl‐L‐cysteine) sigma Cat. #A7250, Cycloheximide MedChemExpress Cat. #HY‐12320, DMSO Sigma Cat. #D2650 and Proteinase Inhibitor Cocktail Roche Cat. #4693116001.
2.3. Clear cell renal cell carcinoma patient samples
Tissues and their corresponding nearby non–tumor tissues were obtained from the First Affiliated Hospital of Harbin Medical University, China. Written informed consent was obtained from all patients. The study has been approved by the Research Ethics Committee of Harbin Medical University, China.
2.4. In vivo xenograft mouse study
BMP8A expression in 786‐O was suppressed by transfection with either si‐BMP8A or si‐control (786‐O/Ctl). Female 4‐week‐old nude mice were purchased from Beijing Vital River Laboratory Animal Technology. 2 × 107 paired cells were inoculated subcutaneously into each mouse. The tumor volumes were measured every week and calculated as length × width2 × 0.5. After 5 weeks, the tumors were carefully removed, photographed and weighed. For the As2O3 treatment, we used the same numbers of ACHN/Ctl or si‐BMP8A cells. When the tumor xenograft sizes reached approximately 200 mm3, the mice were randomly divided into two groups and subjected to PBS and As2O3 treatment (40 mg/kg, intraperitoneally, every other day for 4 weeks), respectively (n = 4 for each group). The tumor size and the body weights of the mice were measured weekly. All animal procedures were performed according to protocols approved by the Rules for Animal Experiments published by the Chinese Government (Beijing, China) and approved by the Research Ethics Committee of Harbin Medical University, China.
2.5. siRNA and plasmid transfection
siRNA specifically targeting BMP8A, TRIM24, Nrf2 and non–specific si‐control were synthesized by GenePharma. Full‐length TRIM24 and empty vector were purchased from Origene. All transfection experiments of siRNA and plasmids were conducted using Jetprime Transfection Reagent (Polyplus) following the manufacturer’s instructions. The corresponding siRNA sequence information is as follows:
SiRNA sequence (5′‐3′)
Si‐BMP8A #1: GGACTACCGTTCATATCCT
Si‐BMP8A #2: CGACGGACATAAGGAGACA
Si‐BMP8A #3: GCATGACTATGACGACTCA
Si‐TRIM24 #1: GAGCUCAUCAGAGGGUAAATT
Si‐TRIM24 #2: GCUGGACUCUCUAAACAAUTT
Si‐TRIM24 #3: GACUGUUCAAGUACUAUUATT
Si‐Nrf2: CCGGCAUUUCACUAAACACAA
Si‐Control: UUCUCCGAACGUGUCACGUTT
2.6. Colony formation assay
ACHN and 786‐O cells were seeded in a six‐well plate and cultured for 10 days after treatment. Colonies were then fixed with 4% paraformaldehyde for 15 minutes and stained for 10 minutes with 0.5% crystal violet.
2.7. Hoechst staining
After transfection, cells were fixed for 10 minutes with a fixation solution and washed twice with PBS for 3 minutes each time, and then the liquid was drained. Finally, 0.5 mL of Hoechst staining solution (Wanleibio) was added for 5 minutes to dye the cells. Images were taken under an Olympus microscope.
2.8. Reactive oxygen species detection
Cells were treated with 10 mmol/L 2′,7′dichlorodihydrofluorescein diacetate (H2DCFDA, Beyotime) for 30 minutes, following two washes with PBS. The reduced H2DCFDA can be oxidized and converted into fluorescent 2′,7′‐dichlorofluorescein (DCF) by intracellular ROS. The fluorescence signals were detected by flow cytometer (BD Biosciences). A total of 10 000 cells were analyzed per sample.
2.9. Western blot assay
Total protein was loaded onto a 10% SDS‐PAGE gel and transferred onto a polyvinylidene fluoride membrane (Millipore). Subsequently, the membrane was blocked with skimmed milk containing 5% TBST for 2 hours and then incubated with the corresponding primary antibody at 4°C for 24 hours. Subsequently, the membranes were incubated with an anti‐mouse or anti‐rabbit secondary antibody at room temperature for 1 hour. The blot was then labeled with HRP coupling for 1 hour. The protein bands were visualized using a chemiluminescence reagent (ECL) kit. The corresponding antibody information is as follows:
Anti–BMP8A antibody (1:1000, Abcam)
Anti–TRIM24 antibody (1:1000, Abcam and Proteintech)
Anti–Nrf2 antibody (1:1000, Abcam and Proteintech)
Anti–pNrf2 antibody (1:300, Abcam)
Anti–p62 antibody (1:3000, Proteintech)
Anti–Ubiquitin (P4D1) antibody (1:400, Santa Cruz)
Anti–Lamin B1 antibody (1:1000, Proteintech)
Anti–beta‐Actin antibody (1:5000, Proteintech)
Anti–GADPH antibody (1:1000, Proteintech)
Anti–Bax antibody (1:750, Abclonal)
Anti–Bcl2 antibody (1:750, Abclonal)
Anti–Cleaved caspase‐3 antibody (1:1000, Cell Signaling Technology)
Anti–p‐Smad1/5/9 antibody (1:1000, Cell Signaling Technology)
Anti–MMP9 antibody (1:1000, Proteintech)
Anti–MMP7 antibody (1:1000, Proteintech)
Anti–E‐cadherin antibody (1:3000, Proteintech)
Anti–N‐cadherin antibody (1:2000, Proteintech)
Anti–Snail antibody (1:500, Proteintech)
Anti–DKK1 antibody (1:500, Proteintech)
Anti–β‐catenin antibody (1:4000, Proteintech)
Anti–AKT antibody (1:1000, Proteintech)
Anti–GSK3β antibody (1:1000, Proteintech)
Anti–p‐GSK3β antibody (1:500, Proteintech)
Goat Anti–Mouse IgG H&L (1:4000, Abcam)
Goat Anti–Rabbit IgG H&L (1:4000, Abcam)
2.10. ChIP assay
An EZ‐ChIP kit (Millipore) was used. Cells were harvested, fixed and incubated with the indicated antibodies. Then DNA was sonicated and purified with a spin column and analyzed with PCR.
2.11. ELISA
BMP8A level in the culture media was measured using an ELISA Kit (R&D Systems). The indicated values represent the average of four separate measurements.
2.12. Luciferase reporter assay
Three copies of the antioxidant response element (ARE) (5′‐GTGACAAAGCAATCCCGTGA‐CAAAGCAATCCCGTGACAAAGCAATA‐3′) were cloned into pGL3‐basic luciferase reporter plasmid. The same amount of ARE‐luciferase reporter together with either BMP8A or si‐BMP8A were transfected into indicated cells. Each transfection included the same amount of Renilla, which was used to standardize transfection efficiency. At 48‐72 hours after transfection, the luciferase activities in cell lysates were measured with the Luciferase Assay System (Promega) and presented as the increase in activation over reporter alone.
2.13. Total RNA extraction and quantitative real‐time PCR
Total RNA was extracted from the RCC tissue samples and cell lines using TRIzol Reagent (Ambion, Life Technologies) according to the manufacturer’s instructions. Total RNA was reverse transcribed into cDNA using a First Strand cDNA Synthesis Kit (TOYOBO Life Science) for the detection of target genes. The relative expression of target genes was detected using FastStart Universal SYBR Green Master Mix (ROX) and normalized to that of GAPDH. The primer sequences were as follows. All the results are expressed as the mean ± SD of three independent experiments.
Real‐time RT‐PCR primer (5′‐3′)
BMP8A: forward: CACCCTTCTCATCTGGATCG.
Reverse: CAGGAAGTAGGCACCGAGAG.
TRIM24: forward: TGTGAAGGACACTACTGAGGTT
Reverse: GCTCTGATACACGTCTTGCAG
Nrf2: forward: CACATCCAGTCAGAAACCAGTGG
Reverse: GGAATGTCTGCGCCAAAAGCTG
HMOX1: forward: CCAGGCAGAGAATGCTGAGTTC
Reverse: AAGACTGGGCTCTCCTTGTTGC
FTH1: forward: TGAAGCTGCAGAACCAACGAGG
Reverse: GCACACTCCATTGCATTCAGCC
NQO1: forward: CCTGCCATTCTGAAAGGCTGGT
Reverse: GTGGTGATGGAAAGCACTGCCT
GAPDH: forward: CGACCACTTTGTCAAGCTCA
Reverse: ACTGAGTGTGGCAGGGACTC
2.14. Immunoprecipitation
Cells were lysed in IP Lysis/Wash Buffer, with Proteinase Inhibitor Cocktail (MedChemExpress) added before use. The resulting lysate was then pre–cleaned using 25‐µL protein G sepharose beads at 4°C. Immunoprecipitation mixtures, including protein lysates, blocked protein G sepharose and the indicated antibodies or IgG control derived from the same species as the indicated antibody were incubated on a rotating wheel at 4°C overnight. Sepharose beads with the bound immunoprecipitates were collected and subjected to four washes with the cold IP Lysis/Wash Buffer and then analyzed by western blot assay.
2.15. Caspase 3/7 activity assay
Cells were transfected with the indicated plasmids or siRNA oligos in the presence or absence of 10 mmol/L As2O3 for 24 hours. Following such treatments, cells were subjected to the caspase 3/7 activity assay by Caspase‐Glo 3/7 Assay Systems (Promega) according to the manufacturer’s instructions.
2.16. Trypan blue staining
Both survival and dead cells were collected and subjected to Trypan Blue staining and counted by hemocytometer 48 hours after TRIM24 expression or 72 hours after siRNA‐mediated TRIM24 or BMP8A, as indicated in the figures. The experiments were repeated independently three times.
2.17. Statistical analysis
Statistical analysis was done by GraphPad software, version 7. Data are presented as the means ± standard error of the means (SEM) or standard deviation (SD). A t test was applied to assess the statistical significance. A P‐value < 0.05 was considered significant.
3. RESULTS
3.1. Comprehensive analysis of the BMP8A in clear cell renal cell carcinoma
We downloaded 611 cases of ccRCC samples in TCGA to analyze the distribution of BMP family expression in ccRCC (Figure S1A). Combined with survival analysis, BMP8A was finally selected as the target molecule. First, we analyzed the expression of BMP8A in ccRCC and normal kidney tissues. The results showed that BMP8A expression levels were significantly increased in ccRCC cell lines and tissues (Figure 1A, B). In the renal clear cell carcinoma (KIRC) group, high BMP8A expression indicates a poor prognosis, and the difference is statistically significant (P < 0.001; Figure 1C). However, 322 kidney papillary cell carcinoma (KIRP) and 92 kidney chromophobe (KICH) did not indicate prognosis (Figure S1B and C). To assess whether BMP8A could be used as an indicator for the diagnosis of ccRCC, an ROC curve was built. The sensitivity and specificity were 0.657 and 0.889, respectively (Figure 1D). In addition, we analyzed the association between BMP8A expression and clinical characteristics, and the results were correlated with the clinical and pathological stages of the tumor (Figure 1E, F). Then we included the variables age, gender, clinical stage, tumor stage, lymph node, metastasis and BMP8A level into a multivariate Cox regression model and found that the mRNA level of BPM8A, age and grade were the independent predictors for overall status of ccRCC patients (Table 1). The results indicated that BMP8A is highly expressed in ccRCC, which suggests a poor prognosis.
FIGURE 1.
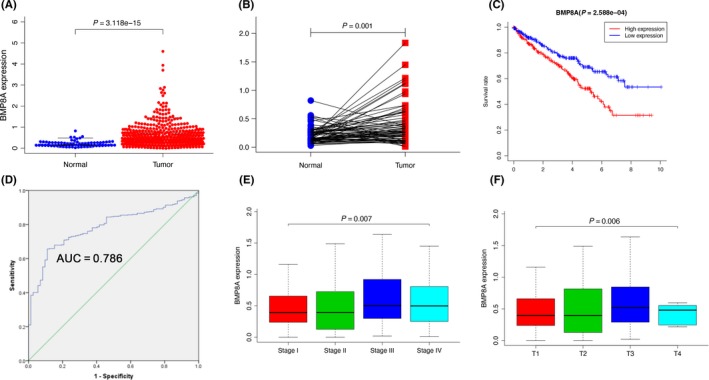
Comprehensive analysis of the BMP8A in clear cell renal cell carcinoma (ccRCC). A, the relative expression of BMP8A in ccRCC tissue (n = 539) compared with normal tissue (n = 72) was examined by qRT‐PCR and normalized to GAPDH expression and data from the TCGA database. B, The relative expression of BMP8A in ccRCC tissue (n = 72) compared with corresponding adjacent normal tissue (n = 72) is shown. C, Kaplan‐Meier plots of the KIRC cohorts. D, The ROC curve was constructed by SPSS P < 0.05. E and F, Correlation between BMP8A expression and clinical stage or pathological stage. The data represented the mean ± SD of three replicates
TABLE 1.
Clinical‐pathologic characteristic of 246 patients with clear cell renal cell carcinoma and association between BMP8A expression and these variables
| Parameter | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| Age | 1.02 | 1.0‐1.04 | 0.012 | 1.03 | 1.01‐1.05 | 0.001 |
| Gender | 1.01 | 0.66‐1.54 | 0.951 | 1.11 | 0.70‐1.76 | 0.651 |
| Grade | 2.24 | 1.68‐2.99 | 3.61E‐08 | 1.49 | 1.07‐2.09 | 0.02 |
| Stage | 1.86 | 1.54‐2.25 | 1.26E‐10 | 1.37 | 0.82‐2.28 | 0.227 |
| T classification | 1.94 | 1.54‐2.46 | 2.69E‐08 | 1.01 | 0.63‐1.65 | 0.953 |
| N classification | 2.93 | 1.52‐5.67 | 0.001 | 1.56 | 0.76‐3.22 | 0.227 |
| M classification | 4.07 | 2.63‐6.30 | 2.76E‐10 | 1.84 | 0.83‐4.10 | 0.136 |
| BMP8A | 1.48 | 1.15‐1.90 | 0.002 | 1.34 | 1.03‐1.74 | 0.03 |
The bold P‐values reflect significant difference.
3.2. BMP8A promoted clear cell renal cell carcinoma proliferation and chemoresistance inhibited apoptosis
First, we determined the mRNA and protein levels of BMP8A in renal cell carcinoma cell lines and normal renal tubular epithelial cell HK‐2. The results revealed that the mRNA and protein levels of BMP8A were significantly elevated in ccRCC cell lines (Figure 2A, B). Next, we continued to examine the expression of BMP8A in clinical samples by immunohistochemistry. It was found that the BMP8A and pSmad1/5/9 of ccRCC tissues were significantly higher than those of adjacent normal tissues (Figure 2C). Next, we investigated the role of BMP8A in the development of ccRCC. After verifying the knockdown efficiency of si‐BMP8A (BMP8A KD), the effect of si‐BMP8A on cell proliferation was examined (Figure S2A). The results showed that BMP8A KD inhibited cell proliferation (Figure 2D and S2B). We stimulated 786‐O and ACHN with the bioactive BMP8A recombinant protein according to the corresponding concentration, and then detected the proliferation level of the cells using CCK‐8 assays. The results showed that BMP8A promoted cell proliferation in a concentration‐dependent manner (Figure 2E). Cloning formation experiments also confirmed that BMP8A KD inhibits the cloning ability of ccRCC (Figure 2F). In addition, we performed Hoechst staining assays on BMP8A KD cells to detect apoptosis. The results showed that apoptosis was aggravated by BMP8A KD (Figure 2G).
FIGURE 2.
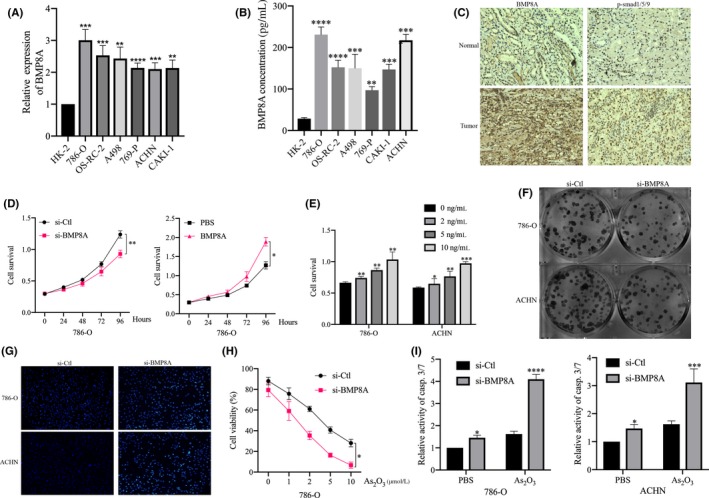
BMP8A promoted clear cell renal cell carcinoma (ccRCC) proliferation and chemoresistance and inhibited apoptosis. A, BMP8A expression levels in the RCC cell lines compared with that in the human renal tubular epithelial cell line. B, Detection of BMP8A expression in 7 cell lines by ELISA. C, The tumor sections were subjected to IHC staining using antibodies against BMP8A and p‐SMAD1/5/9. D, Using CCK‐8 assays to detect cell proliferation after different treatments. E, CCK‐8 assays to detect the effects of different concentrations of BMP8A on cell proliferation. F, Detection of si‐BMP8A/si‐Ctl clone formation assays. G, Hoechst staining assay performed in ACHN and 786‐O cells with si‐BMP8A/si‐Ctl. H, The IC50 values of As2O3 were determined by CCK‐8 assay in 786‐O cells. I, Apoptosis levels revealed by casp.3/7 activity assay followed with the indicated treatments in 786‐O and ACHN cells. The data represent the mean ± SD of three replicates. *P < 0.05, **P < 0.01, *** P < 0.001
Interestingly, we found that BMP8A KD made 786‐O and ACHN more sensitive to As2O3 treatment (Figure 2H and S2D). It was worth noting that casp.3/7 activity increased significantly after treatment with As2O3 in BMP8A KD cells (Figure 2I).
3.3. BMP8A promoted the activation of Nrf2 in clear cell renal cell carcinoma
Given that BMP8A promoted the resistance of As2O3, we hypothesize that BMP8A affected the oxidative stress balance in ccRCC cells. To confirm that the Nrf2 pathway was activated upon BMP8A treatment, we used the antioxidant response element (ARE) (Nrf2‐binding elements) reporter assay and found that BMP8A treatment promoted Nrf2‐mediated transcriptional activation, as revealed by the ARE reporter assay (Figure 3A). Conversely, BMP8A KD inhibited the transcriptional activity of ARE. In addition, we analyzed the correlation between BMP8A and Nrf2 expression based on TCGA. The data showed that there was no significant correlation (r = 0.01; Figure 3B). To determine whether BMP8A promoted upregulation of Nrf2 at the protein level. First, we detected the mRNA level of Nrf2 with BMP8A KD/stimulation. The results showed that BMP8A KD/stimulation did not significantly change the mRNA level of Nrf2 (Figure 3C, D). However, the proteins were positively correlated in the above clinical samples (Figure 3E).
FIGURE 3.
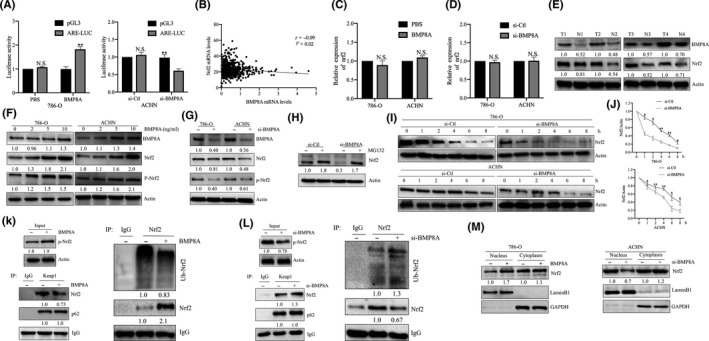
BMP8A promoted the activation of Nrf2 in clear cell renal cell carcinoma (ccRCC). A, Effect of BMP8A on ARE transcription activity detected by ARE reporter assay. B, Correlation of expression between BMP8A and Nrf2 in 540 ccRCC samples from The Cancer Genome Atlas (TCGA) dataset. C and D, Nrf2 mRNA levels were analyzed after BMP8A stimulation/KD. E‐I, Western blot analysis of the treated cell samples. J, In the si‐Ctl and si‐BMP8A cells in the presence of 100 mg/mL CHX for the indicated time periods. Nrf2 levels in the si‐Ctl cells were normalized to 1. K and L, the interaction between P62, Keap1 and Nrf2/(Ub‐Nrf2) was determined by a co–immunoprecipitation (co‐IP) assay. M, Relative amounts of Nrf2 in the nuclear or cytoplasmic fractions were determined by a cell fraction assay after BMP8A or si‐BMP8A. The data represent the mean ± SD of three replicates. n.s. not significant
The identified association between BMP8A and Nrf2 inspired us to ask whether these two proteins were correlated with each other in ccRCC. As we know, Nrf2 is mainly regulated by protein levels. Next, we examined Nrf2 protein levels, and the results showed that Nrf2 and p‐Nrf2 increased significantly after BMP8A stimulation (Figure 3F). In addition, BMP8A KD also significantly downregulated the protein levels of Nrf2 and p‐Nrf2 (Figure 3G). It is worth noting that ubiquitination and subsequent proteasome‐mediated protein degradation are the central mechanisms controlling Nrf2 in cells. Indeed, the universal proteasome inhibitor MG132 (10 μmol/L) restored Nrf2 expression. Interestingly, MG132 also completely rescued BMP8A KD‐induced Nrf2 reduction (Figure 3H). These data pointed to the concept that BMP8A promoted Nrf2 expression by preventing the degradation of proteasome‐mediated Nrf2.
To confirm this hypothesis, we used the protein synthesis inhibitor cycloheximide (100 μg/mL) to treat cells. As expected, the protein level of Nrf2 showed a sharper decline in BMP8A KD cells (Figure 3I). By analyzing the quantitative curve, we found that the half‐life of Nrf2 in the si‐Ctl group was approximately 4 hours, and in the BMP8A KD group it was probably down to 1 hour in 786‐O (Figure 3J). Next, we used immunoprecipitation to detect the effect of BMP8A on P62/Keap1/Nrf2 complex. We found that after BMP8A treatment, the ubiquitination level of Nrf2 was weakened and the total Nrf2 level was increased (Figure 3K‐L). Interestingly, the binding of p62 to Keap1 was not affected. In contrast, the ubiquitination of Nrf2 was enhanced after BMP8A KD, and the overall Nrf2 decreased, while the level of p62 did not change significantly (Figure 3K‐L). In addition, we investigated the effect of BMP8A‐stimulated/KD on nuclear translocation of Nrf2. We found that BMP8A promoted the nuclear translocation of Nrf2, and the nuclear distribution of Nrf2 decreased after BMP8A KD (Figure 3M). The above data indicated that BMP8A was an important factor in maintaining Nrf2 stability. BMP8A promoted phosphorylation of Nrf2 and nuclear translocation and enhanced antioxidative function.
3.4. Nrf2 regulated transcription of TRIM24 in clear cell renal cell carcinoma
We applied ROS scavenger NAC, which was able to partially restore the activity of casp.3/7 in BMP8A KD cells (Figure S2E). This suggests that in addition to the mechanism of apoptosis caused by Nrf2‐mediated ROS imbalance, there are other regulatory pathways. Our previous study found that TRIM24 plays a role in cancer promotion in ccRCC. We investigated possible pathways that led to the impact of TRIM24 on RCC. GSEA’s analysis of 72 RCC cases in the GSE53757 dataset of the GEO database confirmed that the TGF‐beta signaling pathway has changed significantly in RCC. Analysis of functional enrichment analysis revealed that TRIM24 was positively correlated with BMP signaling pathway (Figure 4A). Next, we analyzed the correlation between BMP8A and TRIM24 in TCGA and found that they showed a significant positive correlation (r = 0.38; Figure S2F).
FIGURE 4.
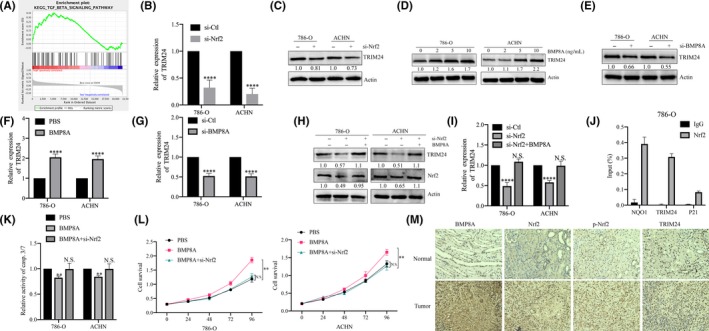
Nrf2 regulated transcription of TRIM24 in clear cell renal cell carcinoma (ccRCC). A, GSEA analysis revealed that the bone morphogenetic proteins (BMP) signaling pathway was significantly altered in ccRCC. B, TRIM24 mRNA levels were analyzed after Nrf2 KD. C‐E, H, Western blot analysis was performed to assess the expression of TRIM24 with the indicated treatments. F‐G, I, TRIM24 mRNA levels were analyzed with the indicated treatments. J, 786‐O cells were subjected to ChIP assay using Nrf2 antibody, followed by qRT‐PCR analysis using primers targeting the promoter regions of TRIM24 (NQO1 was used as a positive control and p21 was used as a negative control). K, Detecting casp.3/7 activity with the indicated treatments. L, CCK‐8 assays performed with the indicated treatments. M, Tumor sections were subjected to IHC staining using antibodies against BMP8A TRIM24, p‐Nrf2 and Nrf2. *P < 0.05, **P < 0.01; ***P < 0.001; ****P < 0.0001
It is well known that Nrf2 plays an important biological role as a transcription factor. We speculate whether Nrf2 acts as a transcription factor to regulate the expression of TRIM24. Next, we examined TRIM24 in Nrf2 KD cells. The results showed that the mRNA and protein levels of TRIM24 were significantly downregulated (Figure 4B, C). The mRNA and protein levels of TRIM24 were detected after BMP8A stimulation/KD, and we found that TRIM24 was positively correlated with BMP8A (Figure 4D‐G). In addition, Nrf2 KD was able to restore the positive regulation of BMP8A on TRIM24 and Nrf2 (Figure 4H, I). ChIP‐qPCR assays demonstrated that Nrf2 bind to the TRIM24 promoter (Figure 4J). The above data indicated that Nrf2 acts as a transcription factor for TRIM24 to regulate the transcriptional level of TRIM24 in 786‐O and ACHN.
Next, we tested the casp.3/7 activity in BMP8A stimulated cells/Nrf2 KD. The results showed that BMP8A inhibited casp.3/7 activity, while Nrf2 KD restored the inhibitory effect of BMP8A on apoptosis (Figure 4K). In addition, we analyzed the proliferation of cells with the above treatment and found that BMP8A stimulation also accelerated the proliferation of 786‐O and ACHN, while the knockdown of Nrf2 restored this change (Figure 4L). We used immunohistochemistry (IHC) to analyze the distribution of BMP8A, pNrf2, Nrf2 and TRIM24 in ccRCC clinical samples, and found that they also showed high expression in cancer tissues (Figure 4M). These data together suggest that Nrf2 promoted the transcriptional activation of TRIM24.
3.5. TRIM24 promoted clear cell renal cell carcinoma progression through the Wnt/β‐catenin pathway
TRIM24 has been studied in other tumors, mostly as a cancer‐promoting gene. Therefore, we were curious about the role of TRIM24 in ccRCC. We used GSEA to analyze ccRCC expression data in TCGA. Functional enrichment analysis revealed that TRIM24 has the best positive correlation with Wnt signaling pathway (Figure 5A). We investigated the effect of TRIM24 overexpression/KD on cell proliferation and found that TRIM24 KD inhibited cell proliferation and TRIM24 overexpression restored this change (Figure 5B, C). Next, we examined the casp.3/7 activity on the same treated cells, and found that TRIM24 KD enhanced the activity of casp.3/7 (Figure 5D). In addition, Hoechst staining also demonstrated that TRIM24 KD induced apoptosis in 786‐O and ACHN (Figure 5E). Trypan Blue Staining confirmed that TRIM24 KD promoted apoptosis and chemosensitivity of As2O3 (Figure 5F). In parallel, TRIM24 KD enhanced Bax but cleaved caspase‐3 and inhibited Bcl2, which was more significant after treated with As2O3 (Figure 5G).
FIGURE 5.
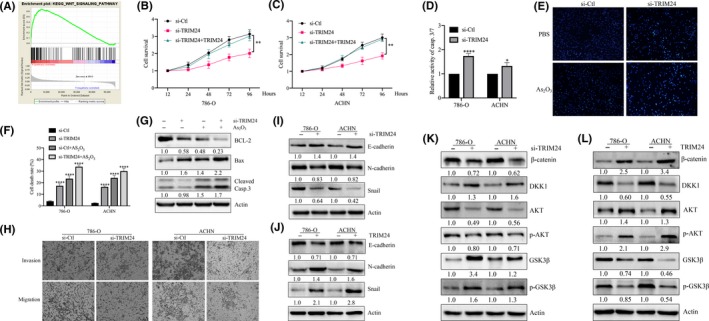
TRIM24 promoted clear cell renal cell carcinoma (ccRCC) progression through the Wnt/β‐catenin pathway. A, GSEA analyzed signal pathways related to TRIM24. B and C, The CCK‐8 assays measure cell proliferation with the indicated treatments. D, Detecting Casp.3/7 activity assay with the indicated treatments. E, Hoechst staining assay performed in 786‐O with indicated treatments. F, The average cell death rates are performed with the indicated treatments. G, Bax, Bcl2 and cleaved caspase‐3 were determined by western blotting. H, Detection of invasion and migration of cells with the TRIM24 KD. I‐L, The effect of TRIM24 on EMT‐related and Wnt pathway proteins was analyzed by western blotting. *P < 0.05, **P < 0.01; ***P < 0.001; ****P < 0.0001
The effect of TRIM24 on the invasion and migration of ccRCC was evaluated and we found that TRIM24 KD inhibited the invasion and migration of 786‐O and ACHN (Figure 5H). Next, we verified the expression levels of key molecules in EMT. The results showed that TRIM24 KD inhibited epithelial‐mesenchymal transition. E‐cadherin was highly expressed, and N‐cadherin and Snail were downregulated (Figure 5I). Conversely, TRIM24 overexpression reversed this process (Figure 5J). Accordingly, we speculate that TRIM24 exhibits such biological effects through the Wnt pathway. Western blot was used to detect Wnt pathway‐related proteins, and it was found that TRIM24 activated key factors in the Wnt pathway and inhibited the Wnt pathway inhibitor DKK1 (Figure 5K, L). To sum up, we determined that TRIM24 promoted proliferation, invasion, metastasis and As2O3 resistance through the Wnt pathway and ROS balance in ccRCC.
3.6. BMP8A/TRIM24 promoted the survival of clear cell renal cell carcinoma both in vivo and in vitro
First, we examined the effects of TRIM24 and BMP8A on ccRCC cell proliferation. The results showed that both BMP8A and TRIM24 KD inhibited cell proliferation, and BMP8A stimulation reversed this effect in 786‐O and ACHN (Figures 2D, 6A and S2G). Next, we examined the effects of BMP8A and TRIM24 on casp.3/7 activity and found that BMP8A/TRIM24 KD increased casp.3/7 activity; conversely, BMP8A/TRIM24 restored the above change (Figure 6B, C). Western blot analysis showed that BMP8A stimulation inhibited Nrf2 and Bax and increased Bcl2 (Figures 3F and 6D). Interestingly, TRIM24 restored the effects of BMP8A KD on apoptosis (Figure 6E). The above data indicated that BMP8A promoted the survival of ccRCC by promoting expression of TRIM24 in vitro.
FIGURE 6.
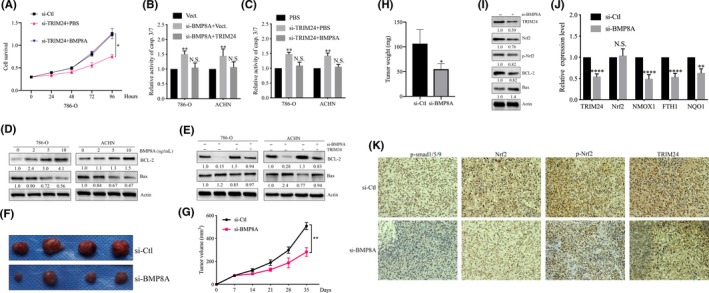
BMP8A/TRIM24 promoted the survival of clear cell renal cell carcinoma (ccRCC) cells both in vivo and in vitro. A, CCK‐8 cell proliferation assays performed with the indicated treatments. B and C, Detecting Casp.3/7 activity in 786‐O and ACHN cells following the indicated treatments. D and E, Bax, Bcl2 and cleaved caspase‐3 were determined by western blotting. F‐H, Tumor volumes at the indicated dates and images as well as tumor weight at day 35. The average values are present in the bar graphs (means ± SD). I and J, TRIM24, Nrf2, p‐Nrf2, Bax, Bcl2 and cleaved caspase‐3 were determined by western blotting and antioxidative Nrf2 targets were measured by real‐time RT‐PCR in samples derived from 786‐O xenografts. K, The tumor sections were subjected to IHC staining using antibodies against p‐Smad1/5/9, TRIM24, p‐Nrf2 and Nrf2. *P < 0.05, **P < 0.01; ***P < 0.001; ****P < 0.0001
In vivo experiments showed that BMP8A promoted ccRCC tumor growth. Si‐Ctl xenografts grew more rapidly than those with BMP8A KD, and the difference in average tumor volume continued to increase to approximately 2.5‐fold at the experimental endpoint (Figure 6F, G). Similarly, the size and weight of the transplanted tumors in the BMP8A KD group were significantly lower than those in the si‐Ctl group (Figure 6H). Western blot analysis showed that the protein level of Nrf2, p‐Nrf2 and Bcl2 decreased; Bax increased after BMP8A KD (Figure 6I). In addition, in xenograft tumors, the mRNA level of Nrf2 did not change with BMP8A KD, but the targeted antioxidative stress factors (FTH1, HMOX1 and NQO1) of Nrf2 and TRIM24 were reduced (Figure 6J). Immunohistochemical analysis of subcutaneous xenografts revealed that p‐smad1/5/9, Nrf2, p‐Nrf2 and TRIM24 decreased with BMP8A KD (Figure 6K). These data indicated that BMP8A KD also inhibited Nrf2‐TRIM24 activity to induce in vivo apoptosis.
3.7. BMP8A/Nrf2/TRIM24‐induced reactive oxygen species imbalance and Wnt pathway activation correlated with As2O3 resistance
Previously, it was thought that the ROS level in cells is crucial for the response to chemotherapy. In view of the above‐identified antioxidant activity of BMP8A, we further examined whether the ROS can increase the chemosensitivity of ccRCC to As2O3. The results proved that BMP8A KD promoted the accumulation of ROS, and As2O3 promoted the accumulation of ROS in BMP8A KD cells (Figure 7A). Similarly, As2O3 upregulated the dead cell ratio in BMP8A KD cells by Trypan Blue Staining (Figure 7B). In addition, treating with PBS/As2O3 in BMP8A KD cells revealed that Bax and cleaved caspase‐3 were upregulated and Nrf2, TRIM24 and Bcl2 were downregulated (Figure 7C). These data indicated that BMP8A KD promoted ccRCC apoptosis and chemosensitivity of As2O3.
FIGURE 7.
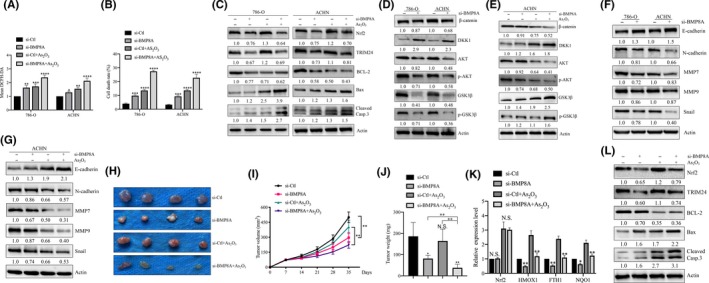
BMP8A/Nrf2/TRIM24‐induced reactive oxygen species (ROS) inhibition and Wnt pathway activation correlated with As2O3 resistance. A, ROS levels as indicated by DCFH‐DA fluorescence. B, The average cell death rates in 786‐O and ACHN. C‐G, TRIM24, Nrf2, Bax, Bcl2, cleaved caspase‐3, Wnt pathway and EMT‐related proteins were determined by western blotting. H‐J, Images as well as tumor weight at day 35 (H) and tumor volumes at the indicated dates (I). J, Represents the weight of the tumor. K, L, TRIM24, Nrf2, p‐Nrf2, Bax, Bcl2 and cleaved caspase‐3 were determined by western blotting and antioxidative Nrf2 targets were measured by real‐time RT‐PCR in samples derived from ACHN xenografts. *P < 0.05, **P < 0.01; ***P < 0.001; ****P < 0.0001
Previous studies have shown that As2O3 has an inhibitory effect on the Wnt pathway in blood tumors. In view of this, it provided a theoretical basis for the treatment of As2O3 combined with BMP8A KD. When BMP8A stimulated cells, the level of DKK1 (Wnt pathway inhibitor) and GSK3β and p‐GSK3β were inhibited, while the expression of β‐catenin, AKT p‐AKT were increased (Figure S2H). Conversely, BMP8A KD reversed this process and As2O3 can inhibit the activity of Wnt pathway to a greater extent (Figure 7D, E). In addition, EMT‐related proteins N‐cadherin, Snail, MMP2 and MMP9 were also upregulated and E‐cadherin was decreased by BMP8A treatment (Figure S2I). In contrast, BMP8A KD inhibits the EMT process, and As2O3 further strengthens this therapeutic effect (Figure 7F, G). The above data indicated that BMP8A stabilized the ROS balance and activated the Wnt pathway by regulating Nrf2‐TRIM24, and As2O3 can break the ROS balance and inhibit Wnt pathway activity. Therefore, BMP8A KD with As2O3 has a better therapeutic effect at the in vitro level of ccRCC.
Substantially consistent with the results of in vitro experiments, the subcutaneous tumor model further showed that As2O3 had a mild inhibitory effect on tumor growth derived from ACHN/si‐Ctl, but completely delayed the growth of BMP8A KD tumors (Figure 7H, I). As2O3 had no significant effect on body weight, suggesting that BMP8A KD cells became more sensitive to As2O3 treatment and had good biological safety (Figures S2J and 7J). Consistent with the experimental results in the cell lines, Nrf2 and TRIM24 protein and the transcriptional level of Nrf2‐related targets were inhibited in the subcutaneous tumors of BMP8A KD cells (Figure 7K). In BMP8A KD xenografts, As2O3 obviously induced the expression of proapoptotic makers. In contrast, anti–apoptotic Bcl2 was significantly suppressed in As2O3 treated si‐BMP8A xenografts (Figure 7L). These results show that BMP8A inhibition enhances the As2O3 efficacy of elevated ROS levels and inhibits the Wnt pathway in chemotherapy for ccRCC by inhibiting Nrf2.
4. DISCUSSION
Each step of the BMP signal pathway is controlled by a positive and negative modulator, allowing the signal to be integrated with other signal pathways. Because of the increased expression of BMP in many tumors, we speculate that BMP8A may be oncogenic. Consistent with this hypothesis, we observed that overexpression of BMP8A has a pro–tumorigenic effect in both ccRCC cell lines. At the same time, this carcinogenic activity of BMP8A was also observed in vivo. In the present study, we hypothesized that BMP8A promotes transcription of TRIM24 by activating Nrf2.
In addition, we found that BMP8A can aggravate the resistance of ccRCC cells to As2O3 by regulating ROS balance and the Wnt pathway. BMP8A stimulates phosphorylation of Nrf2 and stabilizes ROS balance, and this level of ROS promotes cell proliferation. However, when BMP8A KD, the activation of Nrf2 is decreased, resulting in the continuous accumulation of ROS in the cells, inhibiting the proliferation of ccRCC and promoting apoptosis. In addition, BMP8A KD also downregulated the expression of TRIM24 and inhibited the activity of the Wnt pathway, which enhanced the chemosensitivity of ccRCC to As2O3, both in vivo and in vitro. It is noteworthy that we have only studied the promotion of TRMP24 by BMP8A, and other BMP4 and BMP7 related to type I receptor have not been verified. More comprehensive and in‐depth research is needed.
Drug resistance remains a major obstacle in the treatment of kidney cancer. TRIM24 is a potential target for treatment in a variety of tumors. 24 TRIM24 is well known as a co–regulator of the NR signaling pathway. Recent studies have revealed new functions of this multidomain protein in regulating p53 stability, chromatin regulation, cellular metabolism and growth signaling. 25 Given the high tolerance of RCC to chemotherapy, an important association between TRIM24 and As2O3 chemotherapy tolerance is particularly important. In addition to drug resistance, TRIM24 has been reported to promote tumor cell growth in multiple cancers. However, it is still unknown whether it is the same in ccRCC. Our data suggest that TRIM24 acts as an oncogene to promote RCC growth, both in vivo and in vitro. More importantly, we have confirmed that the increased expression of TRIM24 in human specimens is positively correlated with BMP8A and Nrf2. Here, we demonstrate that this effect of TRIM24 on drug resistance can also be achieved in vitro and in vivo through the Wnt pathway.
In addition, we have demonstrated that BMP8A can promote Nrf2 phosphorylation and nuclear transfer to exert antioxidative stress and transcriptional activity (Figure 8). At the same time, Nrf2 acts as a transcription factor of TRIM24, promotes the expression of TRIM24 and activates the Wnt pathway.
FIGURE 8.
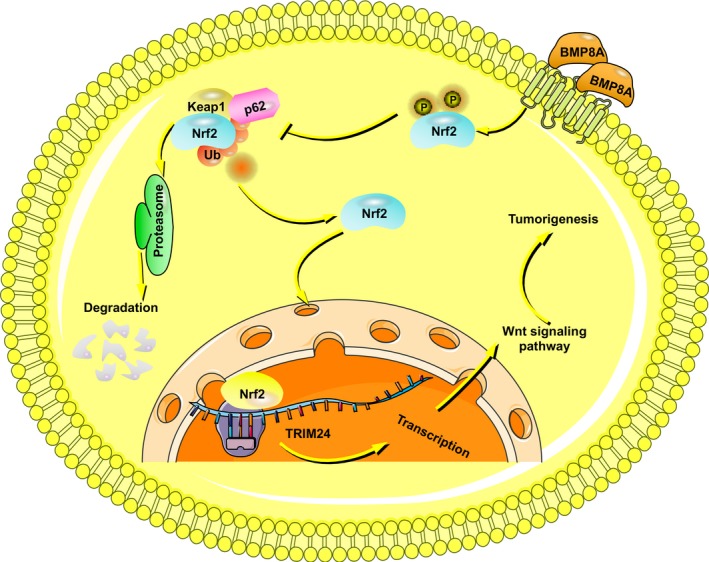
Schematic diagram of the article
DISCLOSURE
The authors have no conflict of interest.
Supporting information
Fig S1
Fig S2
ACKNOWLEDGMENTS
This study was supported in part by the National Natural Science Foundation of China (grant No. 81872084) and Harbin Medical University Graduate Innovation Research Project (grant No. YJSKYCX2018‐42HYD).
Yu Y‐P, Cai L‐C, Wang X‐Y, et al. BMP8A promotes survival and drug resistance via Nrf2/TRIM24 signaling pathway in clear cell renal cell carcinoma. Cancer Sci. 2020;111:1555–1566. 10.1111/cas.14376
Contributor Information
Kong‐bin Yang, Email: ykbneurosurgery@sina.com.
Cheng Zhang, Email: doctorcheng77@163.com.
REFERENCES
- 1. Buti S, Bersanelli M, Sikokis A, et al. Chemotherapy in metastatic renal cell carcinoma today? A systematic review. Anticancer Drugs. 2013;24:535‐554. [DOI] [PubMed] [Google Scholar]
- 2. Coppin C, Kollmannsberger C, Le L, Porzsolt F, Wilt TJ. Targeted therapy for advanced renal cell cancer (RCC): a cochrane systematic review of published randomised trials. BJU Int. 2011;108:1556‐1563. [DOI] [PubMed] [Google Scholar]
- 3. Ricketts CJ, Crooks DR, Sourbier C, Schmidt LS, Srinivasan R, Linehan WM. SnapShot: renal cell carcinoma. Cancer Cell. 2016;29:610‐610.e1. [DOI] [PubMed] [Google Scholar]
- 4. Turajlic S, Xu H, Litchfield K, et al. Tracking cancer evolution reveals constrained routes to metastases: TRACERx renal. Cell. 2018;173:581‐594.e512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Krishnan V, Chong YL, Tan TZ, et al. TGFbeta promotes genomic instability after loss of RUNX3. Cancer Res. 2018;78:88‐102. [DOI] [PubMed] [Google Scholar]
- 6. Sagorny K, Chapellier M, Laperrousaz B, Maguersatta V. BMP and cancer: the Yin and Yang of stem cells. Medecine Sci M/s. 2012;28:416‐422. [DOI] [PubMed] [Google Scholar]
- 7. Bragdon B, Moseychuk O, Saldanha S, King D, Julian J, Nohe A. Bone morphogenetic proteins: a critical review. Cell Signal. 2011;23:609‐620. [DOI] [PubMed] [Google Scholar]
- 8. Xu R‐H, Peck RM, Li DS, Feng X, Ludwig T, Thomson JA. Basic FGF and suppression of BMP signaling sustain undifferentiated proliferation of human ES cells. Nature Methods. 2005;2:185. [DOI] [PubMed] [Google Scholar]
- 9. Wu FJ, Lin TY, Sung LY, Chang WF, Wu PC, Luo CW. BMP8A sustains spermatogenesis by activating both SMAD1/5/8 and SMAD2/3 in spermatogonia. Sci Signal. 2017;10:eaal1910. [DOI] [PubMed] [Google Scholar]
- 10. Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239‐247. [DOI] [PubMed] [Google Scholar]
- 11. Galadari S, Rahman A, Pallichankandy S, Thayyullathil F. Reactive oxygen species and cancer paradox: To promote or to suppress? Free Radic Biol Med. 2017;104:144‐164. [DOI] [PubMed] [Google Scholar]
- 12. Prasad S, Gupta SC, Tyagi AK. Reactive oxygen species (ROS) and cancer: role of antioxidative nutraceuticals. Cancer Lett. 2017;387:95‐105. [DOI] [PubMed] [Google Scholar]
- 13. Kardeh S, Ashkani‐Esfahani S, Alizadeh AM. Paradoxical action of reactive oxygen species in creation and therapy of cancer. Eur J Pharmacol. 2014;735:150‐168. [DOI] [PubMed] [Google Scholar]
- 14. Ray PD, Huang BW, Tsuji Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell Signal. 2012;24:981‐990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. DeNicola GM, Karreth FA, Humpton TJ, et al. Oncogene‐induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106‐109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Duong H‐Q, Yi YW, Kang HJ, et al. Inhibition of NRF2 by PIK‐75 augments sensitivity of pancreatic cancer cells to gemcitabine. Int J Oncol. 2014;44:959‐969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Keum YS, Choi BY. Molecular and chemical regulation of the Keap1‐Nrf2 signaling pathway. Molecules. 2014;19:10074‐10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shigetsugu H. TRIM proteins and cancer. Nature Rev Cancer. 2011;770:792‐804. [DOI] [PubMed] [Google Scholar]
- 19. Groner AC, Cato L, de Tribolet‐Hardy J, et al. TRIM24 Is an oncogenic transcriptional activator in prostate cancer. Cancer Cell. 2016;29:846‐858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Appikonda S, Thakkar KN, Barton MC. Regulation of gene expression in human cancers by TRIM24. Drug Discov Today Technol. 2016;19:57‐63. [DOI] [PubMed] [Google Scholar]
- 21. Tsai W‐W, Wang Z, Yiu TT, et al. TRIM24 links a noncanonical histone signature to breast cancer. Nature. 2010;468:927‐932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lv D, Li Y, Zhang W, et al. TRIM24 is an oncogenic transcriptional co‐activator of STAT3 in glioblastoma. Nature Commun. 2017;8:1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang LH, Yin AA, Cheng JX, et al. TRIM24 promotes glioma progression and enhances chemoresistance through activation of the PI3K|[sol]|Akt signaling pathway. Oncogene. 2015;34:600‐610. [DOI] [PubMed] [Google Scholar]
- 24. Herquel B, Ouararhni K, Khetchoumian K, et al. Transcription cofactors TRIM24, TRIM28, and TRIM33 associate to form regulatory complexes that suppress murine hepatocellular carcinoma. Proc Natl Acad Sci U S A. 2011;108:8212‐8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jain AK, Allton K, Duncan AD, Barton MC. TRIM24 is a p53‐induced E3‐ubiquitin ligase that undergoes ATM‐mediated phosphorylation and autodegradation during DNA damage. Mol Cell Biol. 2014;34:2695‐2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2


