Abstract
Poly-(ADP-ribose) polymerase 1 (PARP1) is commonly known for its vital role in DNA damage response and repair. However, its enzymatic activity has been linked to a plethora of physiological and pathophysiological transactions ranging from cellular proliferation, survival and death. For instance, malignancies with BRCA1/2 mutations heavily rely on PARP activity for survival. Thus, the use of PARP inhibitors is a well-established intervention in these types of tumors. However, recent studies indicate that the therapeutic potential of attenuating PARP1 activity in recalcitrant tumors, especially where PARP1 is aberrantly overexpressed and hyperactivated, may extend its therapeutic utility in wider cancer types beyond BRCA-deficiency. Here, we discuss treatment strategies to expand the tumor-selective therapeutic application of PARP inhibitors and novel approaches with predictive biomarkers to perturb NAD+ levels and hyperPARylation that inactivate PARP in recalcitrant tumors. We also provide an overview of genetic alterations that transform non-BRCA mutant cancers to a state of “BRCAness” as potential biomarkers for synthetic lethality with PARP inhibitors. Finally, we discuss a paradigm shift for the use of novel PARP inhibitors outside of cancer treatment, where it has the potential to rescue normal cells from severe oxidative damage during ischemia-reperfusion injury induced by surgery and radiotherapy.
Keywords: PARP Inhibitors, beta-lapachone, NQO1, PARG, NAMPT, cancer therapeutics, DNA repair, cMET
1. Introduction
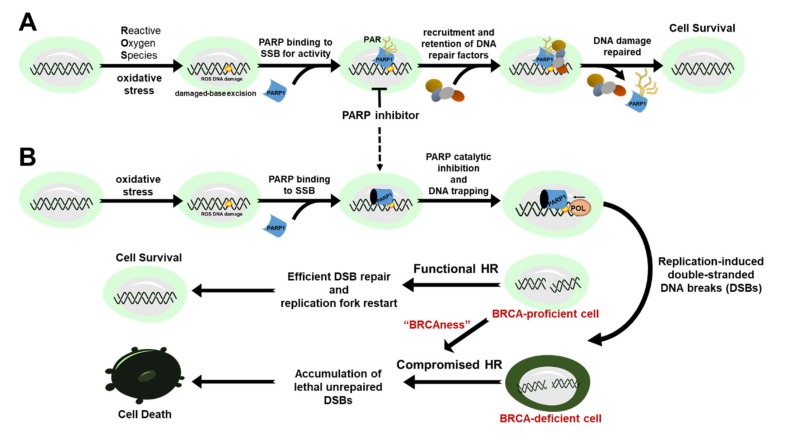
Maintenance of genomic integrity is vital to achieve normal cellular function and to prevent the development of diseases such as cancer [1]. At the heart of this intricate biological process are DNA repair factors that work harmoniously to scan, detect, and repair potentially deleterious damage to cellular genetic information. Indeed, the disruption of one or more DNA repair pathways compromises genetic stability and is a known mechanism of cancer initiation, development, and progression. Poly-(ADP-ribose) polymerase 1 (PARP1) is the classical and founding member of at least 17 human PARP enzymes that share the ability to catalyze the transfer of ADP-ribose units to target proteins to modulate chromatin structure, transcription, replication, DNA damage response and repair [2]. PARP1 is an abundant nuclear protein that acts as a DNA damage sensor and a facilitator of DNA repair pathway choice in response to cellular stress [3,4,5,6]. Specifically, it is involved in the repair of single-stranded DNA breaks (SSB) via the base-excision repair (BER) pathway. In BER, PARP1 functions to recruit other repair factors by binding to single-stranded DNA break intermediates. Additionally, it catalyzes the synthesis of poly(ADP)-ribose (PAR) chains to the acceptor proteins (e.g., histones and XRCC1), including itself, using nicotinamide adenine dinucleotide (NAD+) as the substrate and source of energy. The PARylated proteins then recruit and retain critical processing factors to the site of the lesion to facilitate the efficient repair of the SSB (Figure 1A) [7].
Figure 1.
The role of PARP1 in DNA damage response and repair and cancer therapy. (A) PARP1 binds to single-strand breaks (SSB) for activity to target, recruit and retain critical DNA repair proteins at the sites of DNA lesions. (B) PARP inhibitors convert SSBs to lethal double-stranded breaks (DSBs) that are left unrepaired in BRCA-deficient cells due to a compromised homologous recombination (HR) repair consequently leading to cell death.
In the absence of PARP1, unrepaired SSBs are converted to double-stranded breaks (DSBs) during replication or the S-phase of the cell cycle [8]. Double-stranded DNA breaks are one of the most lethal forms of DNA damage induced by exogenous DNA damaging agents (e.g., ionizing radiation (IR) and chemotherapeutic agents) or endogenous replicative stress in fast proliferating cancer cells. Thus, accurate repair of DSBs is paramount to the growth and survival of all cells. Indeed, cells have evolved a range of DSB repair mechanisms. The two major repair pathways that have been studied extensively are homologous recombination (HR) and non-homologous end-joining (NHEJ) [9]. HR is largely error-free and more prevalently activated during and after DNA duplication when an identical chromatid is accessible as a template for repair. In contrast, NHEJ is active throughout the cell cycle and promotes direct ligation of DSB ends at the cost of small insertions, deletions, substitutions at the break, and even translocations that arise if DSBs from different parts of the genome are combined [10]. Cells utilize two mechanistically distinct end-joining pathways to process DNA DSBs [10,11,12]: Classical-NHEJ (c-NHEJ) leads to a minimal sequence alteration at the repair junctions, whereas alternative-NHEJ (alt-NHEJ, also known as microhomology-mediated end-joining (MMEJ) or back-up NHEJ) causes extensive genetic changes (deletions and insertions) that scar the break sites following ligation of DSB ends.
Several studies have shown that loss-of-function mutation of canonical HR factors – such as breast cancer type 1 and 2 (BRCA1/2) susceptibility proteins that are commonly associated with breast and ovarian cancer [13,14,15] – promotes PARP1 hyperactivation in fast replicating cancer cells [16,17]. This suggests that hyperactivation of PARP1 is essential to facilitate the repair of potentially lethal DNA breaks for the survival of HR-defective (HRD) cancers. Indeed, BRCA1/2 mutant cancers are selectively killed by PARP inhibitors (Figure 1B), which has led to the approval of these agents to treat HR-deficient ovarian and breast cancers. However, several studies have reported that certain cancers without a BRCA deficiency have significant clinical benefits to PARP1 inhibitors positing that PARP inhibitors could be expanded to a target population beyond BRCA-deficiency (e.g., gBRCA mutation carriers) [18,19,20,21].
This review article highlights treatment strategies to selectively target BRCA-proficient cancers by modulating PARP activity that alters PARP binding, PAR and NAD+ levels to induce tumor-selective cell death using predictive biomarkers for therapeutic response. We summarize how aberrant alterations of PARG, NAMPT, NQO1, cMET and “BRCAness” genes that have been shown to affect PARP activity in cancers could serve as prognostic biomarkers for targeted therapy. Finally, we briefly discuss innovative approaches for the use of novel PARP inhibitors to rescue injured normal cells from severe oxidative damage during ischemia-reperfusion injury that could be induced by surgery and radiotherapy.
2. Mechanism of Action for PARP Inhibitors
PARP inhibitors are designed to mimic the substrate-protein interactions of NAD+ within the ADP-ribose transferase (ART) catalytic core of PARP1-3, which are key DNA damage response sensors and transducers. The inhibitors compete with NAD+ binding site of PARP to inhibit PAR polymerization, which then hinders the recruitment and regulation of DNA repair factors and the eventual release of PARP from DNA damage. Two mechanisms are proposed to induce the lethality of PARP inhibitors: PARP catalytic inhibition and PARP trapping [22,23]. However, the relative contributions of these two pathways in mediating the lethality of PARP inhibitors remain enigmatic. For example, there is evidence suggesting that the differences in the trapping potential of PARP1 to the DNA are more efficient at killing HR-deficient cells [22,23]. While there has been no evidence that this mechanism exists with clinically-used PARP inhibitors, a recent study has demonstrated that certain non-hydrolysable NAD+ analogs (e.g., benzamide adenine dinucleotide, BAD) binding at the catalytic site of PARP1 could greatly enhance the binding affinity of PARP1 at sites of DNA damage that prevents its release [24]. Mechanistically, this analog competes with NAD+ at the catalytic binding site that then stabilizes a conformation that induces the DNA binding domain of PARP1 to be locked on a DNA break with a significantly better binding affinity (~10-fold) – a phenomenon known as “reverse allostery.” While most PARP inhibitors (e.g., Rucaparib) bind at the catalytic site, not all of them are “created equal.” In fact, the relative contribution of DNA trapping induced by PARP inhibitors contributes significantly to the toxicity induced by these agents in cancers that are deficient in HR pathways [23,25,26,27]. Some of them have variable inhibitory effects on the PARP isoforms other than PARP1, and some may have more off-target effects on kinases than others. Regardless, PARP inhibitors are now being explored for use in several cancers, even in cancers beyond BRCA mutations [25,28,29,30]. Interestingly, a recent study has reported a correlation between therapeutic response to PARP inhibition and the patterns of ADP-ribosylation (i.e., amount of PARylation) in a panel of ovarian cancers, suggesting that ADP-ribosylation may be a useful biomarker for HR deficiency and sensitivity to PARP inhibitors regardless of BRCA status [31]. While most of the PARP inhibitors have been suggested to specifically inhibit PARP1 and/or PARP2, our limited understanding of the overlapping and non-overlapping functions and cross-talks among all of the PARP family members still raises the problem of target specificity and the risk of unintended effects and consequences promoted by the targeting of other PARP family members.
3. Approved PARP Inhibitors for Targeted Cancer Therapy
PARP inhibitors such as Olaparib [32,33], Rucaparib [34], and Niraparib [35] have been approved by the Food and Drug Administration (FDA) and the European Medicine Agency (EMA) for the treatment of adults with deleterious or suspected deleterious germline BRCA-mutated ovarian cancer in different settings [36]. Moreover, these agents have also been approved for maintenance therapy in platinum-sensitive high-grade ovarian cancer (HGOC), which has a high recurrence rate and an extremely poor prognosis following relapse [37]. Currently, PARP inhibitors are being evaluated in clinical trials in other cancer settings such as nonsmall cell lung cancer, pancreatic cancer, and gastric cancer (ClinicalTrials.gov Identifiers NCT01082549, NCT02184195, NCT03427814, respectively).
Olaparib (LYNPARZA, AstraZeneca and Merck) was the first PARP inhibitor to receive FDA approval for indications in breast cancer and pancreatic cancer, and has since been approved for metastatic breast cancer in patients with germline BRCA1 or BRCA2 (gBRCA1/2) mutations. In 2009, Olaparib was deemed safer than conventional chemotherapeutics, with mild and reversible side effects during clinical trials [26]. It was subsequently determined to have anti-tumor activity in advanced ovarian cancer patients with gBRCA1/2 mutations, particularly with primary tumors that are sensitive to platinum [18,32,38,39,40]. The phase III OlympiAD trial found a 60% reduction in metastatic BRCA1/2 mutant breast tumor size with Olaparib in comparison to 29% in patients receiving conventional chemotherapy. The FDA approved Olaparib in ovarian and breast patients with gBRCA mutations in 2014 and 2018, respectively [27,41,42,43].
Rucaparib (RUBRACA, Clovis Oncology, Inc.) was granted accelerated approval by the FDA in 2016 for the treatment of adult ovarian cancer patients with germline and/or somatic BRCA-mutations previously treated with two or more prior lines of chemotherapy. This agent is also approved as a maintenance monotherapy for adult patients with recurrent ovarian cancer who have established complete or partial response (CR/PR) to platinum-based chemotherapy. In 2018, Rucaparib gained FDA approval for the maintenance treatment of advanced or recurrent ovarian, fallopian tube, and primary peritoneal cancer in patients who have received platinum-based chemotherapies, as well as in patients with germline or somatic BRCA1/2 mutations following two or more treatments with conventional chemotherapy [44,45].
Niraparib (ZEJULA, Tesaro, Inc.) was approved by the FDA and EMA in 2017. Compared to Olaparib, this agent results in longer progression-free survival in patients with BRCA1/2-mutant tumors [35]. Niraparib also increases progression-free survival in patients with wild-type BRCA1/2 ovarian cancers, though less effectively than in patients with BRCA mutations [35]. Niraparib, however, is associated with more severe side effects than other PARP inhibitors, with patients experiencing thrombocytopenia (33.8%), anemia (25.3%), and neutropenia (19.6%) [35]. In 2019, the FDA approved Niraparib for homologous recombination-deficient (HRD) patients with advanced ovarian, fallopian tube, or primary peritoneal cancer previously treated with three or more chemotherapies [46].
Talazoparib (TALZENNA, Pfizer, Inc.) is a new active PARP inhibitor approved by the FDA in 2018 for patients with deleterious or suspected deleterious gBRCA-mutated, HER2-negative local advanced or metastatic breast cancer [47]. Compared to other PARP inhibitors, Talazoparib is still at an early stage of clinical development in gathering evidence to support its use on the treatment of epithelial ovarian cancer.
4. Harnessing the Power of PAR and PARG Inhibition for Cancer Therapy
Poly(ADP)-ribosylation (PARylation) is a covalent and reversible posttranslational modification (PTM) of acceptor proteins catalyzed by PARPs, particularly in response to DNA damage and oxidative stress [48,49]. PARylation is also a critical signaling PTM for other biological transactions such as transcription, cell cycle regulation, and genome maintenance [50,51,52]. Intracellular PAR levels are very low in unstressed cells due to low enzymatic activity of PARP. When PARP1 is activated following DNA damage, PAR formation increases and consequently depletes intracellular NAD+ and ATP levels [53]. Accumulation of PAR and loss of NAD+ and ATP can lead to severe metabolic dysfunction and eventual cell death. Thus, the balance between ADP-ribose synthesis (i.e., PAR writers) and degradation (i.e., PAR erasers) is critical for the coordination of various cellular response pathways for survival [54].
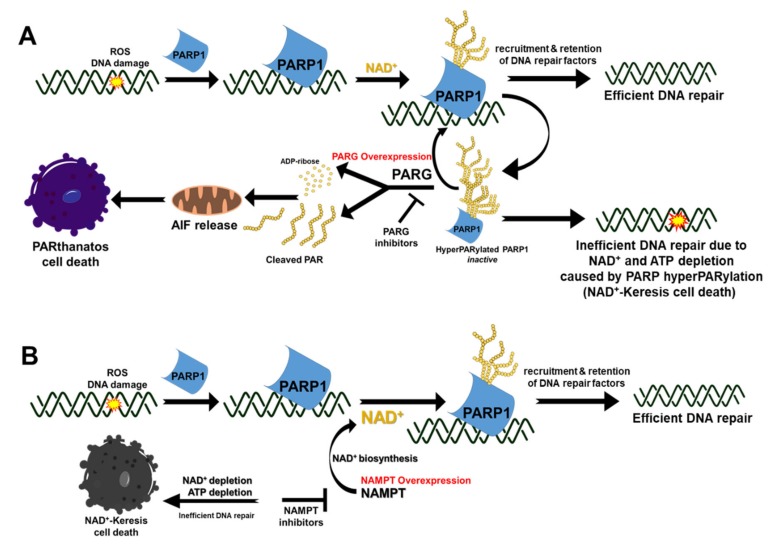
Poly(ADP-ribose) glycohydrolase (PARG) is an endo- and exo-glycohydrolase that rapidly catalyzes the degradation of PAR polymerized by PARP1 to coordinate DNA repair [48,49]. It is comprised of an N-terminal regulatory domain required for recruitment to DNA damage sites [55], a C-terminal catalytic domain, and a central mitochondrial-targeting sequence [56]. PARG hydrolyzes the glycosidic bonds between ADP-ribose units producing free ADP-ribose and PAR oligomers (Figure 2A) [57], which play distinct biological roles in cellular processes. Free ADP-ribose is predominantly involved in energy catabolism, calcium signaling, and protein glycation [57]. The long-chain of free PAR formed by PARG cleavage has been shown to interact with the mitochondrial proteins triggering a unique intrinsic PAR-mediated cell death program known as PARthanatos (Figure 2A) [58,59]. Mechanistically, a cell commits to PARthanatos when free PAR (> 60 ADP-ribose units) migrates from the nucleus to the cytosol, which then triggers the translocation of PAR-bound apoptosis-inducing factor (AIF) from the mitochondria to the nucleus to activate the cell death process (Figure 2A) [60]. Early events in PAR-mediated cell death include loss of mitochondrial membrane potential and mitochondrial permeability transition [61]. While caspase activation has been demonstrated to act as a bystander in PAR-dependent cell death, this caspase-independent process shares cytological and morphological characteristics of both necrosis and apoptosis [58,59].
Figure 2.
Pharmacological modulation of PAR ad NAD+ in cancers for therapy. (A) Mechanism of PARthanatos mediated by the translocation of cleaved PAR from the nucleus to the cytosol and mitochondria to induce the release of AIF that translocates into the nucleus to initiate death in PARG-overexpressing cancers. Alternatively, breakdown and recycling of PAR can be prevented by inhibition of PARG to enhance NAD+ depletion caused by PARP hyperactivation, ultimately starving the cell of ATP needed for various critical cellular processes (e.g., DNA repair). (B) In cancer cells that overexpress NAMPT, the use of NAMPT inhibitors interfere with generation of intracellular NAD+ levels that compromise PARP activity, consequently resulting in impaired repair of DNA damage and NAD+-Keresis.
ADP-ribose hydrolases (ARH) can catalyze PAR hydrolysis but to a lesser extent than PARG, which catalyzes the majority (~90%) of PAR catabolism [62]. PARG is aberrantly overexpressed in most human cancers, suggesting that PARG is required for tumorigenesis and cancer survival [63]. Therefore, inducing cell death via PARG inhibition and manipulation of the PAR cycle through PARP1/PARG inhibition is an attractive target for cancer therapy (Figure 2). Available PARG inhibitors consist of DNA intercalators, tannins, and substrate analogs. DNA intercalators (e.g., Tilorene, GPI116552, GPI18214, Ethacridine) indirectly inhibit PARG by forming complexes with its substrate, PAR [64,65,66]. Hydrolyzable tannins (e.g., Nobotanin B, Oenothenin B, gallotannin) inhibit PARG in vitro by competing with the PAR substrate [67,68,69]; however, poor membrane permeability, poor bioavailability, and toxicity of these compounds hinder their usage. Adenosinediphosphate (hydroxymethyl) pyrrolidinediol (ADP-HPD), a nonhydrolyzable-analogue of ADP-ribose, has been shown to be a potent competitive PARG inhibitor [66]. Other reversible PARG inhibitors under development include modified salicylanilide pharmacophore [70] and rhodanine-based small molecule analogs (RBPIs) [70]. Recently, Pillay et al. [71] demonstrated that PARG inhibitor, PDD00017273, mimics poly(ADP-ribose) polymerase (PARP) inhibitor therapy in ovarian cancer cells by exacerbating the formation of hyperPARylated-PARP1 that severely compromises PARP activity. Mechanistically, PARG inhibitors prevent the recycling of hyperPARylated-PARP1 to its more active form for efficient DNA damage response and repair. In support of this, survival studies of certain human cancer cell lines with PARG knockdown synergistically enhances the lethality to DNA-damaging agents such as alkylating agents [72,73] and cisplatin [72]. Overall, pharmacological targeting of PARG is a promising therapeutic target and future investigations are required to develop effective strategies to manipulate the dynamic PAR formation and break-down process in response to DNA damage. The relative low abundance of PARG in normal cells, compared to tumors with aberrant PARG overexpression, makes it an attractive target in cancer therapeutics for tumor-selective drug response. Moreover, unlike the human PARP family with 17 related enzymes with almost similar catalytic subunit – PARG is a unique mammalian protein without any paralogs, which may offer fewer off-target effects than PARP inhibitors.
5. Targeting NAD+ for Cancer Therapy
NAD+ is a co-enzyme that mediates redox reactions by acting as an electron carrier and a substrate in metabolic pathways including glycolysis, the tricarboxylic acid cycle (TCA), oxidative phosphorylation, and serine biosynthesis [74]. It is a cofactor for PARP and plays a key role in energy production, cell signaling, redox homeostasis, DNA repair, gene expression, and the stress response. Many of these processes are disrupted in cancer, which alters NAD+ production and consumption [75,76]. Nicotinamide phosphoribosyltransferase (NAMPT) and nicotinamide mononucleotide adenylyltransferase (NMNAT) enzymes regulate the salvage pathway critical for controlling intracellular NAD+ levels [77]. Increased NAD+ and aberrant NAMPT overexpression promotes glycolysis, which fuels the growth and survival of cancer cells [78,79]. Indeed, NAMPT inhibitors suppress cancer cell proliferation by inhibiting glycolysis [79,80,81,82,83].
Overexpression of NAMPT is observed in several types of malignant tumors, which supplies back-up NAD+ to sustain cellular proliferation and promote resistance to therapeutic agents [84,85,86,87,88,89]. NAMPT as a predictive biomarker for tumor-selective targeting of NAD+ metabolism is, therefore, a promising therapeutic strategy (Figure 2B). Mechanistically, NAMPT inhibitors induce cell death in cancer cells by depleting intracellular NAD+ and ATP levels and inhibiting glycolysis and glucose uptake [90,91]. Examples of NAMPT inhibitors include FK866, GMX-1777/8, STF-31, STF-118804, GNE-617, GNE-618, LSN3154567, KPT-9274. Vacor adenine dinucleotide (VAD) is an NAD+ analog that has been shown to inhibit both NAMPT and NMNAT, leading to necrotic cell death through NAD+ depletion (NAD+-Keresis, Figure 2B), glycolytic block, and energy failure [82,92,93,94,95,96,97,98]. Several NAMPT inhibitors have been investigated in the clinic as a monotherapy such as FK866 and GMX-1777/8. However, thrombocytopenia was demonstrated to be the dose-limiting toxicity in these clinical trials [99,100]. Though NAMPT inhibitors as a monotherapy are promising candidates for cancer therapy, drug resistance and toxicity to normal tissues when used at high concentration remain a major hurdle [99,101,102,103]. Perhaps, a combination approach with tumor-selective DNA damaging agents (e.g., NQO1-bioactivatable agents, vide infra in Section 6) may overcome this limitation to improve the tumor-selectivity of NAMPT inhibitors by severely depleting the NAD+ needed by PARPs for efficient DNA repair and survival [82,104].
6. Leveraging NQO1 as a Biomarker for Tumor-Selective Use of PARP Inhibitors with NQO1-Bioactivatable Drugs
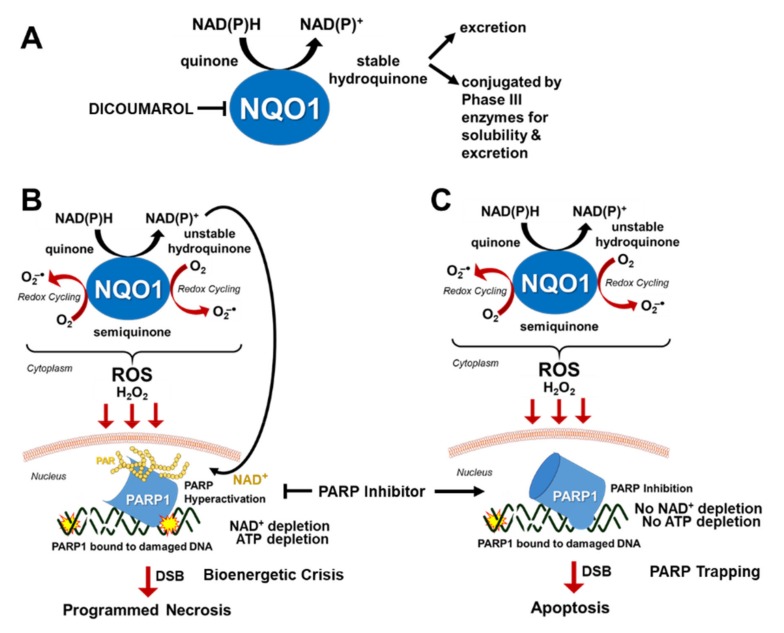
NAD(P)H:Quinone Oxidoreductase 1 (NQO1, DT-diaphorase) is a cytoplasmic Phase II detoxification enzyme that utilizes NAD(P)H to reduce certain quinones to stable hydroquinones via two-electron reductions that are readily conjugated by Phase III transporters for cellular efflux (Figure 3A) [105]. NQO1 is abnormally overexpressed in most malignant tumors, which contributes to drug resistance by metabolizing xenobiotics to their inactive forms or chemical structures that are excreted out of the cell for chemoprotection. Indeed, the addition of an NQO1-inhibitor (e.g., dicoumarol) in cancers could sensitize the anti-cancer effects of certain agents by inhibiting resistance due to drug efflux.
Figure 3.
Strategy for tumor-selective use of PARP inhibitors in solid tumors. (A) The role of NQO1 in detoxifying certain quinones for cellular efflux. (B) The role of NQO1 in bioactivating certain quinones to induce toxicity in NQO1(+) cancer cells. Note that the stability of hydroquinones determine whether bioreduction by NQO1 leads to detoxification or toxicity. (C) Mechanism of tumor-selective synergistic cell death induced by combination of PARP inhibitor and NQO1-bioactivatable agents in NQO1(+) cells.
Certain quinones are bioactivated by NQO1 to create a rapid increase in reactive oxygen species (ROS), particularly hydrogen peroxide (H2O2), which can permeate through the nucleus to induce the formation of toxic DNA damage leading to cell death (Figure 3B) [106,107,108,109,110]. For example, β-lapachone (β-lap) is a soluble ortho-naphthoquinone with potent anti-tumor and radiosensitizing activity only in the presence of high NQO1 activity [108,109,110,111,112,113,114], which is only noted in most malignant tumors but not in normal tissues [109]. Mechanistically, the two-electron reductase capacity of NQO1 catalyzes the oxidoreduction of β-lap to an inherently unstable hydroquinone, which automatically and rapidly gets reverted back to its original quinone form as it undergoes a two-step oxidation (Figure 3B) [108]. This produces what is termed as a “futile cycle” of oxidation and produces a significant level of reactive oxygen species such as superoxide radicals (O2−) that are rapidly catalyzed by superoxide dismutases to hydrogen peroxide in the cytoplasm (Figure 3B) [115]. This stable form of ROS can easily penetrate through the nucleus and is converted to genotoxic hydroxyl radicals via the Fenton reaction process, which causes massive formation of oxidative DNA base damage and SSBs (Figure 3B) [109,116]. The surge of this specific types of DNA damage in the nucleus causes hyperactivation of PARP1, which is an NAD+-dependent enzyme, to a level that consequently leads to NAD+-Keresis death due to the depletion of critical biological energy sources, NAD+ and ATP [82,107,109]. Another possibility is that PARP hyperactivation could lead to an NAD+-independent glycolytic and bioenergetics crisis that is presumably caused by PAR-dependent inhibition of glycolysis that occurs through the inhibition of hexokinase leading to a specific form of cell death termed “PARthanathos” [117]. Future studies are required to firmly establish the contribution of PARP hyperactivation, NAD+, or free PAR in specific types of cell death. Regardless, the use of NQO1-bioactivatable agents to modulate PARP activity, NAD+ and PAR to induce cell death is a viable strategy to treat cancer. As NQO1 is typically overexpressed in tumors but not at a significant level in normal healthy cells [109], the application of NQO1-bioactivatable agents is an attractive possibility for tumor-selective generation of H2O2-induced DNA damages that hyperactivate PARP and deplete NAD+ to cause eventual cell death, which is vital for developing safe and effective drugs with minimal off-target effects.
While β-lap (ARQ761 in clinical form) shows potential as a single-agent therapeutic [107,118], it has also been associated with hemolytic anemia at a dose of 450 mg/m2 [119]. Therefore, increasing its efficacy at a lower dose is therapeutically advantageous. Due to the role of PARP hyperactivation in the mechanism of action of β-lap, PARP inhibitors have been a central focus in improving the efficacy and reducing the toxicity of β-lap for use in NQO1(+) cancers. PARP inhibition followed by β-lap treatment alters the mechanism of β-lap function by blocking the repair of damaged DNA and inhibiting PARP1 hyperactivation, slowing down the process of NAD+ depletion as a result, and extending the cycling of β-lap by NQO1 to maximize hydrogen peroxide production and amplifying DNA damage in a tumor-selective manner (Figure 3C) [109]. Instead of dying by caspase-independent NAD+-Keresis, cell death from exposure to β-lap following PARP1 inhibition occurs through caspase-dependent apoptosis, presumably due to the availability of NAD+ and ATP (Figure 3C) [109]. As NQO1 will continue cycling β-lap until the drug is depleted or the required source of NAD(P)H is depleted, blocking the DNA repair process which leads to NAD+ depletion following β-lap treatment prolongs the efficacy of the drug, allowing for lower doses to be used with the synergistic effect, minimizing the potential for hemolytic anemia.
The use of NQO1-bioactivatable drug (e.g., β-lap) with PARP inhibitors have been shown to significantly expand the clinical application of PARP inhibitors outside of BRCA1/2 mutant cancers for which the majority of PARP inhibitors are currently approved. The prevalence of NQO1 overexpression in such a wide variety of malignant cancers (>90% in pancreatic cancers with KRAS mutation, >80% nonsmall cell lung cancer (NSCLC), >60% breast cancer, >60% prostate cancer, >45% head and neck, and >60% colon cancers [109]) suggests that this combination therapy may have significant future potential, especially in cancers in which prognosis is currently poor, such as head and neck cancer, pancreatic cancer and NSCLC [117,120,121,122,123,124,125,126].
7. Targeting cMET to Attenuate PARP1 Activity
Many studies have shown that most malignant tumors have increased levels of reactive oxygen species (ROS), particularly hydrogen peroxide (H2O2), compared to their normal counterparts. These genotoxic reactive species cause oxidative DNA damage and single-stranded DNA breaks that stimulate PARP activity in DNA repair [127]. Interestingly, Du and colleagues [128] have shown that cMET, which is a receptor tyrosine kinase, can further stimulate the activity of PARP1 in cancers to survive the lethal effects of ROS-induced DNA damage. In cancer, cMET is overexpressed and its abnormal activation can promote the development and progression of multiple cancers [129].
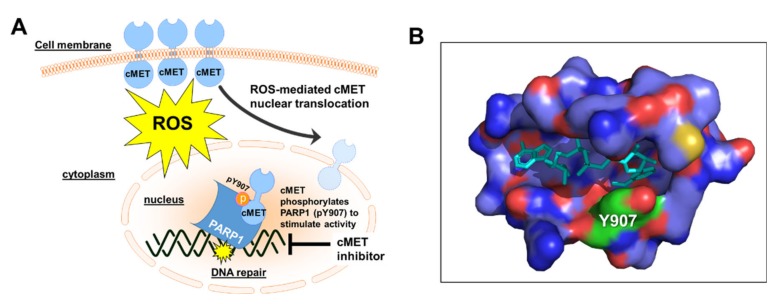
Mechanistically, the high level of ROS in cancer promotes the activation and translocation of cMET into the nucleus where it directly binds and phosphorylates the tyrosine residue 907 (Y907) of PARP1 to enhance its enzymatic activity (Figure 4A) [128,130,131]. Indeed, Du and colleagues have demonstrated that phosphorylated PARP1 (pY907) showed a higher level of PAR production than non-phosphorylated PARP1 in vitro, thereby highlighting the critical role of cMET in enhancing PARP1 activity in DNA repair [128,130,131]. Moreover, the phosphorylation of this specific residue (pY907) has been suggested to significantly decrease the binding of clinically-relevant PARP inhibitors due to a potential steric hindrance (Figure 4B) [128]. Indeed, c-MET-mediated phosphorylation of PARP1 at Y907 leads to PARP inhibitor resistance, which could be potentiated when combined with cMET inhibitors (e.g., crizotinib and foretinib) [128]. Thus, inhibition of cMET is an attractive strategy to indirectly block PARP1-mediated DNA repair and enhance the therapeutic effects of PARP inhibitors in cancer cells [128]. However, the mechanistic basis for requiring Y907 phosphorylation to promote H2O2-induced PARP1 activity remains to be firmly established. A recent report also suggests that phosphorylation of PARP1 by EGFR and cMET heterodimer contributes to PARP1 inhibitor resistance. Hence, combination treatment consisting of EGFR, cMET, and PARP1 inhibitors posits a novel therapeutic strategy in cancer treatment involving overexpression of PARP1/EGFR/cMET, which are frequent alterations in most solid tumors [132].
Figure 4.
Attenuation of PARP Activity by cMET inhibition. (A) cMET enhances PARP activity during oxidative stress. Inhibition of cMET is an excellent strategy for inhibiting PARP1-mediated DNA repair and synergy with PARP inhibitors. (B) A snapshot of non-hydrolyzable NAD+ analog (in cyan) bound to residues in PARP1 catalytic active site within 6Å from the ligand (PDB ID: 6 bhv, processed via PyMOL Molecular Graphics System). Phosphorylation of PARP1 residue Y907 (shown in green) inhibits binding of PARP inhibitors.
8. Targeting of PARP Activity in Non-Oncological Events
PARP inhibition or gene deletion has been shown to attenuate tissue injury associated with ischemia-reperfusion injury and inflammation that could arise during tumor surgical resection and radiation therapy [133,134]. During this non-oncological event, the catalytic activity of PARP1 becomes hyperactivated and consequently leads to NAD+ depletion. This process forces NAD+ replenishment through the salvage pathway, which decreases cellular ATP levels and results in bioenergetic crisis, eventually leading to necrotic cell death that is typically associated with inflammation [3,135]. Thus, inhibition of PARP activity could have protective effects by dramatically reducing NAD+ consumption and preventing energetic failure and the consequent necrotic cell death. While the trapping mechanism induced by specific PARP inhibitors might be advantageous for the treatment of certain cancers, this mechanism of action might not be optimal to rescue normal cells from severe oxidative damage during ischemia-reperfusion injury (e.g., ischemia of the lung due to ionizing radiation [136] and cerebral ischemia during surgery [137]).
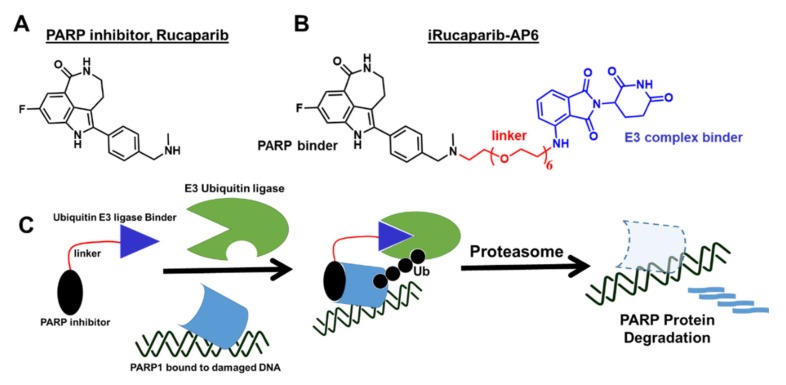
For non-oncological indications, the safety profile of most FDA-approved PARP inhibitors would be expected to be better if it had less PARP trapping activity to restore the viability of normal tissue damage from injury caused by ischemia-reperfusion. To circumvent the cytotoxic effects of PARP trapping, the Yu group [138] developed a strategy to decrease PARP activity using a novel lead compound, iRucaparib-AP6 (Figure 5), which promotes PARP degradation upon binding that mimics PARP1 genetic deletion. Mechanistically, the Rucaparib component of the small molecule specifically binds PARP at the site of damaged DNA, but it is attached to a linker with a ligand that brings the E3 ligase in close proximity to ubiquitinate PARP and subsequently gets degraded through the proteasome pathway (Figure 5C). Unlike PARP trapping, this strategy of modulating PARP activity protects cells against genotoxic stress-induced cell death [138] that could have potential clinical applications for the treatment of ischemia-reperfusion injury and neurodegeneration. However, further development is needed to firmly establish the mechanism of action and evaluate its protective effects to treat the aforementioned non-oncological indications.
Figure 5.
Development of a novel “PARP degrader” molecule as a biochemical probe and therapeutic agent. (A) Chemical structure of Rucaparib. (B) Chemical structure and elements of iRucaparib-AP6. (C) Simplified representation of the mechanism of action for a novel proteolysis targeting chimeric (PROTAC) molecule, iRucaparib-AP6.
To summarize, the enzymatic function of PARP1 – which is to catalyze the PARylation of a large number of PAR-acceptor proteins (including itself) – becomes hyperactivated upon detection of genotoxic stresses associated with various pathological conditions such as cancer, ischemia-reperfusion injury (e.g., myocardial infarction), inflammatory diseases (e.g., colitis, arthritis, asthma), vascular diseases (e.g., diabetic complications, atherosclerosis), and neurodegeneration. Thus, modulation of PARP activity using a novel class of PARP activity inhibitors (e.g., iRucaparib-AP6) with little to no “trapping” potential may be a promising strategy to ameliorate certain pathological conditions that are aimed to heal instead of killing the severely injured normal tissues (e.g., inflamed tissues). Indeed several studies have looked at the protective effects of PARP inhibitors in lung transplantation [139], neurodegeneration [140], renal injury [141], aging [142], and acute pancreatitis [143]. While these studies offer critical insights toward the potential therapeutic repurposing of PARP inhibitors in treating non-oncological indications, it is important to learn more about the possible side-effects of long-term treatment and whether PARP inhibition may increase the risk of mutagenesis and carcinogenesis.
9. Perspectives and Conclusions
Individualized treatments to patients with recalcitrant cancers in order to achieve cure are urgently needed. Currently, cancer patients have many therapeutic options for treatment; however, the lack of effective and tumor-selective treatment options still remains a major hurdle in the fight against cancer today. This is due to very limited predictive biomarkers that define individual risks of disease recurrence or sensitivity to treatments. For example, a significant number of cancer patients are over-treated in order to possibly attain an improved overall survival in early cancer stage. This “shotgun” approach, however, could have life-threatening consequences to cancer patients due to the emergence of drug resistance and secondary malignancies. Clearly, there is an urgent need for (1) a clear understanding of normal versus cancer cell biology to gain more informative decisions regarding the most effective therapeutic interventions, and (2) an arsenal of new treatment options based on predictive biomarkers for this disease to eliminate toxic effects to normal cells, drug resistance, and development of secondary cancers.
In less than two decades, studies on the use of PARP inhibitors for the treatment of gBRCA1/2 mutant cancers and beyond BRCA-deficiency have increased tremendously. Of course, these were all made possible by the collaborative effort of the scientific and medical communities to understand the functional roles of breast cancer associated genes 1 and 2 (BRCA1/2) in normal physiology and breast cancer development. Accordingly, deficiencies in BRCA1 and BRCA2 genes can compromise HR-mediated repair, conferring hypersensitivity to PARP inhibitors by chemical synthetic lethality. The rarity of breast (5–10%) and ovarian (15–20%) cancers with germline BRCA1/2 loss-of-function mutations, however, currently restricts the therapeutic utility of PARP inhibitor monotherapy. This is further complicated by several reports of potential molecular mechanisms of resistance for PARP inhibitors in BRCA-deficient breast cancers [144]. Interestingly, approximately 24% of high-grade triple negative breast cancers (TNBC) without a BRCA mutation showed great response to PARP inhibitor in Phase II clinical trials suggesting that the use of PARP inhibitors can be expanded beyond BRCA deficiency, and this has been reinforced by several studies in other cancers [18,19,20,21]. Indeed, genetic and pharmacological alterations of specific proteins (Table 1) or long non-coding RNAs (lncRNAs, e.g., PCAT-1 [145]) that can induce a state of “BRCAness” or HR-deficiency in BRCA-proficient cancers have the potential to broaden the application of PARP inhibitors beyond the current protocols approved by the FDA and EMA.
Table 1.
Representative list of genes/proteins that have been shown to cause “BRCAness” or HR defect in BRCA-proficient tumors when expression is lost or activity is inhibited. These deficiencies could be exploited as predictive biomarkers for precision treatment with PARP inhibitors.
| Protein Name | Primary Function/Activity | Association with “BRCAness” | Reference |
|---|---|---|---|
| CDK1 | Cell cycle regulation | Loss of expression or activity inhibition compromises phosphorylation of BRCA1 for proper HR function | [30] |
| CDK12/13 | Phosphorylates RNAPII CTD | Loss of expression and activity inhibition suppresses expression of specific HR proteins such as RAD51 and BRCA1 | [147] |
| AXL | A receptor tyrosine kinase associated with metastasis, invasion and migration in many cancers | Loss of expression or activity inhibition decreases expression of specific HR genes and proteins | [148] |
| Kub5-Hera, RPRD1B, CREPT | Transcription termination factor | Loss of expression compromises HR by decreasing CDK1 expression | [25] |
| WEE1 | Involved in the terminal phosphorylation and inactivation of CDK1-bound cyclin B | Activity inhibition with AZD1775 indirectly inhibits BRCA2 | [149] |
| UCHL3 | Deubiquitinase | Activity inhibition with perifosine promotes ubiquitination of RAD51 and blocks the binding of RAD51 with BRCA2 | [150] |
| BET | Transcriptional regulators | Activity inhibition with JQ1 decreases expression of RAD51 and Ku80 | [151] |
| PI3K | Kinase involve in cell growth, proliferation, differentiation, motility, survival and intracellular trafficking | Inhibition of activity impairs BRCA1/2 expression | [152] |
| Cyclin D1 | Regulator of CDKs (cyclin dependent kinases), required for cell cycle G1/S transition | Loss of expression impairs recruitment of RAD51 | [153,154] |
| AURKA | Play important role in mitosis/ regulation of cell cycle progression | Activity inhibition or loss of expression decreases expression of BRCA1 and BRCA2 | [155,156] |
| HKMT | Regulation of histone methylation | Inhibition of activity abolishes retention of BRCA1/BARD1 complexes at sites of DSB | [157,158] |
| CCDC6 | Tumor suppressor | Loss of expression compromises BRCC3 and DNA damage checkpoints in response to DNA damage. | [158,159] |
| MEK | Kinase that phosphorylates and activates MAPK | Activity inhibition or loss of expression downregulates BRCA2 | [160] |
| HDAC | Removes acetyl groups from an amino acid on a histone | Activity inhibition with SAHA reduces BRCA1 protein levels by targeting the UHRF1/BRCA1 protein complex | [161] |
| PAK1 | Regulates cytoskeleton remodeling, phenotypic signaling and gene expression | Reduced activity and loss of expression downregulates the expression of genes involved in FA/BRCA pathway | [162] |
| Androgen receptor | DNA-binding transcription factor that regulates gene expression | Activity inhibition or loss of expression suppresses the expression of HR genes, thus creating HR deficiency and BRCAness | [163] |
| TGFβ | Involved in embryonic development, cell proliferation, motility and apoptosis, extracellular matrix production, and immunomodulation | Overexpression suppresses BRCA1, ATM, and MSH2 | [164] |
In conclusion, a serious concern for the use of PARP inhibitors – as with all chemotherapeutic agents – is the development of acquired drug resistance and de novo malignancies [146] due to limited information and often unclear understanding of the full extent of the agents’ specificity and mechanism of action. A complete and clear knowledge of how different PARPs are activated to perform overlapping and non-overlapping functions or how PAR and NAD+ levels are modulated to alter specific biological events toward cellular survival or death are essential to: (1) design novel PARP inhibitors that are more specific and tumor-selective; (2) develop better mechanistic-based strategies using reliable predictive biomarkers for PARP inhibitor monotherapy as well as combination treatments; and (3) identify situations that can re-sensitize recalcitrant tumor cells to PARP activity inhibition or modulation to treat cancer patients more safely and efficiently.
Author Contributions
Conceptualization: E.A.M.; resources: E.A.M.; writing – original draft preparation: E.A.M., N.S., S.L.P., S.B.B., and U.A.; writing - review, and editing: E.A.M., N.S., and S.L.P.; visualization: E.A.M.; supervision: E.A.M.; funding acquisition: E.A.M. All authors have read and agree to the final version of the manuscript.
Funding
This work was supported by Biomedical Research Grant from IUSM, Indiana CTSI Showalter Trust and NIH/NCI R01 CA210489 to E.A.M. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funders.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Ciccia A., Elledge S.J. The DNA damage response: Making it safe to play with knives. Mol. Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hottiger M.O., Hassa P.O., Luscher B., Schuler H., Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem. Sci. 2010;35:208–219. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 3.D’Amours D., Desnoyers S., D’Silva I., Poirier G.G. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Pt 2Biochem. J. 1999;342:249–268. doi: 10.1042/bj3420249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gibson B.A., Kraus W.L. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 2012;13:411–424. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- 5.Hottiger M.O. Nuclear ADP-Ribosylation and Its Role in Chromatin Plasticity, Cell Differentiation, and Epigenetics. Annu. Rev. Biochem. 2015;84:227–263. doi: 10.1146/annurev-biochem-060614-034506. [DOI] [PubMed] [Google Scholar]
- 6.Pascal J.M. The comings and goings of PARP-1 in response to DNA damage. DNA Repair (Amst) 2018;71:177–182. doi: 10.1016/j.dnarep.2018.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei H., Yu X. Functions of PARylation in DNA Damage Repair Pathways. Genomics Proteomics Bioinforma. 2016;14:131–139. doi: 10.1016/j.gpb.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hossain M.A., Lin Y., Yan S. Single-Strand Break End Resection in Genome Integrity: Mechanism and Regulation by APE2. Int. J. Mol. Sci. 2018;19:2389. doi: 10.3390/ijms19082389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Panier S., Durocher D. Push back to respond better: Regulatory inhibition of the DNA double-strand break response. Nat. Rev. Mol. Cell Biol. 2013;14:661–672. doi: 10.1038/nrm3659. [DOI] [PubMed] [Google Scholar]
- 10.Lieber M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deriano L., Roth D.B. Modernizing the nonhomologous end-joining repertoire: Alternative and classical NHEJ share the stage. Annu. Rev. Genet. 2013;47:433–455. doi: 10.1146/annurev-genet-110711-155540. [DOI] [PubMed] [Google Scholar]
- 12.Sfeir A., Symington L.S. Microhomology-Mediated End Joining: A Back-up Survival Mechanism or Dedicated Pathway? Trends Biochem. Sci. 2015;40:701–714. doi: 10.1016/j.tibs.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chae Y.K., Anker J.F., Carneiro B.A., Chandra S., Kaplan J., Kalyan A., Santa-Maria C.A., Platanias L.C., Giles F.J. Genomic landscape of DNA repair genes in cancer. Oncotarget. 2016;7:23312–23321. doi: 10.18632/oncotarget.8196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miki Y., Swensen J., Shattuck-Eidens D., Futreal P.A., Harshman K., Tavtigian S., Liu Q., Cochran C., Bennett L.M., Ding W., et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 15.Wooster R., Bignell G., Lancaster J., Swift S., Seal S., Mangion J., Collins N., Gregory S., Gumbs C., Micklem G. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 16.Gottipati P., Vischioni B., Schultz N., Solomons J., Bryant H.E., Djureinovic T., Issaeva N., Sleeth K., Sharma R.A., Helleday T. Poly(ADP-ribose) polymerase is hyperactivated in homologous recombination-defective cells. Cancer Res. 2010;70:5389–5398. doi: 10.1158/0008-5472.CAN-09-4716. [DOI] [PubMed] [Google Scholar]
- 17.Helleday T. The underlying mechanism for the PARP and BRCA synthetic lethality: Clearing up the misunderstandings. Mol. Oncol. 2011;5:387–393. doi: 10.1016/j.molonc.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gelmon K.A., Tischkowitz M., Mackay H., Swenerton K., Robidoux A., Tonkin K., Hirte H., Huntsman D., Clemons M., Gilks B., et al. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: A phase 2, multicentre, open-label, non-randomised study. Lancet Oncol. 2011;12:852–861. doi: 10.1016/S1470-2045(11)70214-5. [DOI] [PubMed] [Google Scholar]
- 19.Evans T., Matulonis U. PARP inhibitors in ovarian cancer: Evidence, experience and clinical potential. Ther. Adv. Med. Oncol. 2017;9:253–267. doi: 10.1177/1758834016687254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ledermann J., Harter P., Gourley C., Friedlander M., Vergote I., Rustin G., Scott C.L., Meier W., Shapira-Frommer R., Safra T., et al. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: A preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol. 2014;15:852–861. doi: 10.1016/S1470-2045(14)70228-1. [DOI] [PubMed] [Google Scholar]
- 21.Scott C.L., Swisher E.M., Kaufmann S.H. Poly (ADP-ribose) polymerase inhibitors: Recent advances and future development. J. Clin. Oncol. 2015;33:1397–1406. doi: 10.1200/JCO.2014.58.8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murai J., Huang S.Y., Das B.B., Renaud A., Zhang Y., Doroshow J.H., Ji J., Takeda S., Pommier Y. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res. 2012;72:5588–5599. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pommier Y., O’Connor M.J., de Bono J. Laying a trap to kill cancer cells: PARP inhibitors and their mechanisms of action. Sci. Transl. Med. 2016;8:362ps17. doi: 10.1126/scitranslmed.aaf9246. [DOI] [PubMed] [Google Scholar]
- 24.Hopkins T.A., Shi Y., Rodriguez L.E., Solomon L.R., Donawho C.K., DiGiammarino E.L., Panchal S.C., Wilsbacher J.L., Gao W., Olson A.M., et al. Mechanistic Dissection of PARP1 Trapping and the Impact on In Vivo Tolerability and Efficacy of PARP Inhibitors. Mol. Cancer Res. 2015;13:1465–1477. doi: 10.1158/1541-7786.MCR-15-0191-T. [DOI] [PubMed] [Google Scholar]
- 25.Motea E.A., Fattah F.J., Xiao L., Girard L., Rommel A., Morales J.C., Patidar P., Zhou Y., Porter A., Xie Y., et al. Kub5-Hera (RPRD1B) Deficiency Promotes “BRCAness” and Vulnerability to PARP Inhibition in BRCA-proficient Breast Cancers. Clin. Cancer Res. 2018;24:6459–6470. doi: 10.1158/1078-0432.CCR-17-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fong P.C., Boss D.S., Yap T.A., Tutt A., Wu P., Mergui-Roelvink M., Mortimer P., Swaisland H., Lau A., O’Connor M.J., et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 27.Kaufman B., Shapira-Frommer R., Schmutzler R.K., Audeh M.W., Friedlander M., Balmana J., Mitchell G., Fried G., Stemmer S.M., Hubert A., et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J. Clin. Oncol. 2015;33:244–250. doi: 10.1200/JCO.2014.56.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang L., Zhang Y., Shan W., Hu Z., Yuan J., Pi J., Wang Y., Fan L., Tang Z., Li C., et al. Repression of BET activity sensitizes homologous recombination-proficient cancers to PARP inhibition. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aal1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mateo J., Carreira S., Sandhu S., Miranda S., Mossop H., Perez-Lopez R., Nava Rodrigues D., Robinson D., Omlin A., Tunariu N., et al. DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N. Engl. J. Med. 2015;373:1697–1708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson N., Li Y.C., Walton Z.E., Cheng K.A., Li D., Rodig S.J., Moreau L.A., Unitt C., Bronson R.T., Thomas H.D., et al. Compromised CDK1 activity sensitizes BRCA-proficient cancers to PARP inhibition. Nat. Med. 2011;17:875–882. doi: 10.1038/nm.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conrad L.B., Lin K.Y., Nandu T., Gibson B.A., Lea J.S., Kraus W.L. ADP-Ribosylation Levels and Patterns Correlate with Gene Expression and Clinical Outcomes in Ovarian Cancers. Mol. Cancer Ther. 2020;19:282–291. doi: 10.1158/1535-7163.MCT-19-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ledermann J., Harter P., Gourley C., Friedlander M., Vergote I., Rustin G., Scott C., Meier W., Shapira-Frommer R., Safra T., et al. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N. Engl. J. Med. 2012;366:1382–1392. doi: 10.1056/NEJMoa1105535. [DOI] [PubMed] [Google Scholar]
- 33.Pujade-Lauraine E., Ledermann J.A., Selle F., Gebski V., Penson R.T., Oza A.M., Korach J., Huzarski T., Poveda A., Pignata S., et al. Olaparib tablets as maintenance therapy in patients with platinum-sensitive, relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): A double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 2017;18:1274–1284. doi: 10.1016/S1470-2045(17)30469-2. [DOI] [PubMed] [Google Scholar]
- 34.Coleman R.L., Oza A.M., Lorusso D., Aghajanian C., Oaknin A., Dean A., Colombo N., Weberpals J.I., Clamp A., Scambia G., et al. Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;390:1949–1961. doi: 10.1016/S0140-6736(17)32440-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mirza M.R., Monk B.J., Herrstedt J., Oza A.M., Mahner S., Redondo A., Fabbro M., Ledermann J.A., Lorusso D., Vergote I., et al. Niraparib Maintenance Therapy in Platinum-Sensitive, Recurrent Ovarian Cancer. N. Engl. J. Med. 2016;375:2154–2164. doi: 10.1056/NEJMoa1611310. [DOI] [PubMed] [Google Scholar]
- 36.Mittica G., Ghisoni E., Giannone G., Genta S., Aglietta M., Sapino A., Valabrega G. PARP Inhibitors in Ovarian Cancer. Recent Pat. Anticancer Drug Discov. 2018;13:392–410. doi: 10.2174/1574892813666180305165256. [DOI] [PubMed] [Google Scholar]
- 37.Weng C.S., Wu C.C., Chen T.C., Chen J.R., Huang C.Y., Chang C.L. Retrospective Analysis Of Comparative Outcomes In Recurrent Platinum-Sensitive Ovarian Cancer Treated With Pegylated Liposomal Doxorubicin (Lipo-Dox) And Carboplatin Versus Paclitaxel And Carboplatin. Cancer Manag. Res. 2019;11:9899–9905. doi: 10.2147/CMAR.S217329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fong P.C., Yap T.A., Boss D.S., Carden C.P., Mergui-Roelvink M., Gourley C., De Greve J., Lubinski J., Shanley S., Messiou C., et al. Poly(ADP)-ribose polymerase inhibition: Frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J. Clin. Oncol. 2010;28:2512–2519. doi: 10.1200/JCO.2009.26.9589. [DOI] [PubMed] [Google Scholar]
- 39.Tutt A., Robson M., Garber J.E., Domchek S.M., Audeh M.W., Weitzel J.N., Friedlander M., Arun B., Loman N., Schmutzler R.K., et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: A proof-of-concept trial. Lancet. 2010;376:235–244. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- 40.Audeh M.W., Carmichael J., Penson R.T., Friedlander M., Powell B., Bell-McGuinn K.M., Scott C., Weitzel J.N., Oaknin A., Loman N., et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: A proof-of-concept trial. Lancet. 2010;376:245–251. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- 41.Olaparib Keeps Hereditary Breast Tumors in Check. Cancer Discov. 2017;7:OF10. doi: 10.1158/2159-8290.CD-NB2017-085. [DOI] [PubMed] [Google Scholar]
- 42.Robson M., Im S.A., Senkus E., Xu B., Domchek S.M., Masuda N., Delaloge S., Li W., Tung N., Armstrong A., et al. Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. N. Engl. J. Med. 2017;377:523–533. doi: 10.1056/NEJMoa1706450. [DOI] [PubMed] [Google Scholar]
- 43.Domchek S.M., Aghajanian C., Shapira-Frommer R., Schmutzler R.K., Audeh M.W., Friedlander M., Balmana J., Mitchell G., Fried G., Stemmer S.M., et al. Efficacy and safety of olaparib monotherapy in germline BRCA1/2 mutation carriers with advanced ovarian cancer and three or more lines of prior therapy. Gynecol. Oncol. 2016;140:199–203. doi: 10.1016/j.ygyno.2015.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swisher E.M., Lin K.K., Oza A.M., Scott C.L., Giordano H., Sun J., Konecny G.E., Coleman R.L., Tinker A.V., O’Malley D.M., et al. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): An international, multicentre, open-label, phase 2 trial. Lancet Oncol. 2017;18:75–87. doi: 10.1016/S1470-2045(16)30559-9. [DOI] [PubMed] [Google Scholar]
- 45.Kristeleit R., Shapiro G.I., Burris H.A., Oza A.M., LoRusso P., Patel M.R., Domchek S.M., Balmana J., Drew Y., Chen L.M., et al. A Phase I-II Study of the Oral PARP Inhibitor Rucaparib in Patients with Germline BRCA1/2-Mutated Ovarian Carcinoma or Other Solid Tumors. Clin. Cancer Res. 2017;23:4095–4106. doi: 10.1158/1078-0432.CCR-16-2796. [DOI] [PubMed] [Google Scholar]
- 46.Moore K.N., Secord A.A., Geller M.A., Miller D.S., Cloven N., Fleming G.F., Wahner Hendrickson A.E., Azodi M., DiSilvestro P., Oza A.M., et al. Niraparib monotherapy for late-line treatment of ovarian cancer (QUADRA): A multicentre, open-label, single-arm, phase 2 trial. Lancet Oncol. 2019;20:636–648. doi: 10.1016/S1470-2045(19)30029-4. [DOI] [PubMed] [Google Scholar]
- 47.Guney Eskiler G. Talazoparib to treat BRCA-positive breast cancer. Drugs Today (Barc) 2019;55:459–467. doi: 10.1358/dot.2019.55.7.3015642. [DOI] [PubMed] [Google Scholar]
- 48.Okayama H., Edson C.M., Fukushima M., Ueda K., Hayaishi O. Purification and properties of poly(adenosine diphosphate ribose) synthetase. J. Biol. Chem. 1977;252:7000–7005. [PubMed] [Google Scholar]
- 49.Brochu G., Duchaine C., Thibeault L., Lagueux J., Shah G.M., Poirier G.G. Mode of action of poly(ADP-ribose) glycohydrolase. Biochim. Biophys. Acta (BBA) Gene Struct. Expr. 1994;1219:342–350. doi: 10.1016/0167-4781(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 50.Hassa P.O., Haenni S.S., Elser M., Hottiger M.O. Nuclear ADP-ribosylation reactions in mammalian cells: Where are we today and where are we going? Microbiol. Mol. Biol. Rev. 2006;70:789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hassa P.O., Hottiger M.O. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front. Biosci. 2008;13:3046–3082. doi: 10.2741/2909. [DOI] [PubMed] [Google Scholar]
- 52.Kovacs K., Toth A., Deres P., Kalai T., Hideg K., Gallyas F., Jr., Sumegi B. Critical role of PI3-kinase/Akt activation in the PARP inhibitor induced heart function recovery during ischemia-reperfusion. Biochem. Pharmacol. 2006;71:441–452. doi: 10.1016/j.bcp.2005.05.036. [DOI] [PubMed] [Google Scholar]
- 53.Juarez-Salinas H., Sims J.L., Jacobson M.K. Poly(ADP-ribose) levels in carcinogen-treated cells. Nature. 1979;282:740–741. doi: 10.1038/282740a0. [DOI] [PubMed] [Google Scholar]
- 54.O’Sullivan J., Tedim Ferreira M., Gagne J.P., Sharma A.K., Hendzel M.J., Masson J.Y., Poirier G.G. Emerging roles of eraser enzymes in the dynamic control of protein ADP-ribosylation. Nat. Commun. 2019;10:1182. doi: 10.1038/s41467-019-08859-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Botta D., Jacobson M.K. Identification of a regulatory segment of poly(ADP-ribose) glycohydrolase. Biochemistry. 2010;49:7674–7682. doi: 10.1021/bi100973m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meyer-Ficca M.L., Meyer R.G., Coyle D.L., Jacobson E.L., Jacobson M.K. Human poly(ADP-ribose) glycohydrolase is expressed in alternative splice variants yielding isoforms that localize to different cell compartments. Exp. Cell Res. 2004;297:521–532. doi: 10.1016/j.yexcr.2004.03.050. [DOI] [PubMed] [Google Scholar]
- 57.Braun S.A., Panzeter P.L., Collinge M.A., Althaus F.R. Endoglycosidic cleavage of branched polymers by poly(ADP-ribose) glycohydrolase. Eur. J. Biochem. 1994;220:369–375. doi: 10.1111/j.1432-1033.1994.tb18633.x. [DOI] [PubMed] [Google Scholar]
- 58.David K.K., Andrabi S.A., Dawson T.M., Dawson V.L. Parthanatos, a messenger of death. Front. Biosci. (Landmark Ed) 2009;14:1116–1128. doi: 10.2741/3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Andrabi S.A., Kim N.S., Yu S.W., Wang H., Koh D.W., Sasaki M., Klaus J.A., Otsuka T., Zhang Z., Koehler R.C., et al. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc. Natl. Acad. Sci. USA. 2006;103:18308–18313. doi: 10.1073/pnas.0606526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Y., Kim N.S., Haince J.F., Kang H.C., David K.K., Andrabi S.A., Poirier G.G., Dawson V.L., Dawson T.M. Poly(ADP-ribose) (PAR) binding to apoptosis-inducing factor is critical for PAR polymerase-1-dependent cell death (parthanatos) Sci. Signal. 2011;4:ra20. doi: 10.1126/scisignal.2000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu S.W., Wang H., Poitras M.F., Coombs C., Bowers W.J., Federoff H.J., Poirier G.G., Dawson T.M., Dawson V.L. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297:259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- 62.Niere M., Mashimo M., Agledal L., Dolle C., Kasamatsu A., Kato J., Moss J., Ziegler M. ADP-ribosylhydrolase 3 (ARH3), not poly(ADP-ribose) glycohydrolase (PARG) isoforms, is responsible for degradation of mitochondrial matrix-associated poly(ADP-ribose) J. Biol. Chem. 2012;287:16088–16102. doi: 10.1074/jbc.M112.349183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marques M., Jangal M., Wang L.C., Kazanets A., da Silva S.D., Zhao T., Lovato A., Yu H., Jie S., Del Rincon S., et al. Oncogenic activity of poly (ADP-ribose) glycohydrolase. Oncogene. 2019;38:2177–2191. doi: 10.1038/s41388-018-0568-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tavassoli M., Tavassoli M.H., Shall S. Effect of DNA intercalators on poly(ADP-ribose) glycohydrolase activity. Biochim. Biophys. Acta. 1985;827:228–234. doi: 10.1016/0167-4838(85)90207-9. [DOI] [PubMed] [Google Scholar]
- 65.Li Q., Li M., Wang Y.L., Fauzee N.J., Yang Y., Pan J., Yang L., Lazar A. RNA interference of PARG could inhibit the metastatic potency of colon carcinoma cells via PI3-kinase/Akt pathway. Cell. Physiol. Biochem. 2012;29:361–372. doi: 10.1159/000338491. [DOI] [PubMed] [Google Scholar]
- 66.Putt K.S., Hergenrother P.J. A nonradiometric, high-throughput assay for poly(ADP-ribose) glycohydrolase (PARG): Application to inhibitor identification and evaluation. Anal. Biochem. 2004;333:256–264. doi: 10.1016/j.ab.2004.04.032. [DOI] [PubMed] [Google Scholar]
- 67.Aoki T., Kojima M., Adachi J., Okano M. Effect of short-term egg exclusion diet on infantile atopic dermatitis and its relation to egg allergy: A single-blind test. Acta Derm. Venereol. Suppl. (Stockh) 1992;176:99–102. [PubMed] [Google Scholar]
- 68.Tsai Y.J., Aoki T., Maruta H., Abe H., Sakagami H., Hatano T., Okuda T., Tanuma S. Mouse mammary tumor virus gene expression is suppressed by oligomeric ellagitannins, novel inhibitors of poly(ADP-ribose) glycohydrolase. J. Biol. Chem. 1992;267:14436–14442. [PubMed] [Google Scholar]
- 69.Formentini L., Arapistas P., Pittelli M., Jacomelli M., Pitozzi V., Menichetti S., Romani A., Giovannelli L., Moroni F., Chiarugi A. Mono-galloyl glucose derivatives are potent poly(ADP-ribose) glycohydrolase (PARG) inhibitors and partially reduce PARP-1-dependent cell death. Br. J. Pharmacol. 2008;155:1235–1249. doi: 10.1038/bjp.2008.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Finch K.E., Knezevic C.E., Nottbohm A.C., Partlow K.C., Hergenrother P.J. Selective Small Molecule Inhibition of Poly(ADP-Ribose) Glycohydrolase (PARG) ACS Chem. Biol. 2012;7:563–570. doi: 10.1021/cb200506t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pillay N., Tighe A., Nelson L., Littler S., Coulson-Gilmer C., Bah N., Golder A., Bakker B., Spierings D.C.J., James D.I., et al. DNA Replication Vulnerabilities Render Ovarian Cancer Cells Sensitive to Poly(ADP-Ribose) Glycohydrolase Inhibitors. Cancer Cell. 2019;35:519–533. doi: 10.1016/j.ccell.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fujihara H., Ogino H., Maeda D., Shirai H., Nozaki T., Kamada N., Jishage K., Tanuma S., Takato T., Ochiya T., et al. Poly(ADP-ribose) Glycohydrolase deficiency sensitizes mouse ES cells to DNA damaging agents. Curr. Cancer Drug Targets. 2009;9:953–962. doi: 10.2174/156800909790192419. [DOI] [PubMed] [Google Scholar]
- 73.Shirai H., Poetsch A.R., Gunji A., Maeda D., Fujimori H., Fujihara H., Yoshida T., Ogino H., Masutani M. PARG dysfunction enhances DNA double strand break formation in S-phase after alkylation DNA damage and augments different cell death pathways. Cell Death Dis. 2013;4:e656. doi: 10.1038/cddis.2013.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Murphy J.P., Giacomantonio M.A., Paulo J.A., Everley R.A., Kennedy B.E., Pathak G.P., Clements D.R., Kim Y., Dai C., Sharif T., et al. The NAD(+) Salvage Pathway Supports PHGDH-Driven Serine Biosynthesis. Cell Rep. 2018;24:2381–2391. doi: 10.1016/j.celrep.2018.07.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Verdin E. NAD(+) in aging, metabolism, and neurodegeneration. Science. 2015;350:1208–1213. doi: 10.1126/science.aac4854. [DOI] [PubMed] [Google Scholar]
- 76.Garrido A., Djouder N. NAD(+) Deficits in Age-Related Diseases and Cancer. Trends Cancer. 2017;3:593–610. doi: 10.1016/j.trecan.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 77.Yaku K., Okabe K., Nakagawa T. NAD metabolism: Implications in aging and longevity. Ageing Res. Rev. 2018;47:1–17. doi: 10.1016/j.arr.2018.05.006. [DOI] [PubMed] [Google Scholar]
- 78.Yamamoto M., Inohara H., Nakagawa T. Targeting metabolic pathways for head and neck cancers therapeutics. Cancer Metastasis Rev. 2017;36:503–514. doi: 10.1007/s10555-017-9691-z. [DOI] [PubMed] [Google Scholar]
- 79.Tan B., Young D.A., Lu Z.H., Wang T., Meier T.I., Shepard R.L., Roth K., Zhai Y., Huss K., Kuo M.S., et al. Pharmacological inhibition of nicotinamide phosphoribosyltransferase (NAMPT), an enzyme essential for NAD+ biosynthesis, in human cancer cells: Metabolic basis and potential clinical implications. J. Biol. Chem. 2013;288:3500–3511. doi: 10.1074/jbc.M112.394510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cerna D., Li H., Flaherty S., Takebe N., Coleman C.N., Yoo S.S. Inhibition of nicotinamide phosphoribosyltransferase (NAMPT) activity by small molecule GMX1778 regulates reactive oxygen species (ROS)-mediated cytotoxicity in a p53- and nicotinic acid phosphoribosyltransferase1 (NAPRT1)-dependent manner. J. Biol. Chem. 2012;287:22408–22417. doi: 10.1074/jbc.M112.357301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kato H., Ito E., Shi W., Alajez N.M., Yue S., Lee C., Chan N., Bhogal N., Coackley C.L., Vines D., et al. Efficacy of combining GMX1777 with radiation therapy for human head and neck carcinoma. Clin. Cancer Res. 2010;16:898–911. doi: 10.1158/1078-0432.CCR-09-1945. [DOI] [PubMed] [Google Scholar]
- 82.Moore Z., Chakrabarti G., Luo X., Ali A., Hu Z., Fattah F.J., Vemireddy R., DeBerardinis R.J., Brekken R.A., Boothman D.A. NAMPT inhibition sensitizes pancreatic adenocarcinoma cells to tumor-selective, PAR-independent metabolic catastrophe and cell death induced by beta-lapachone. Cell Death Dis. 2015;6:e1599. doi: 10.1038/cddis.2014.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zerp S.F., Vens C., Floot B., Verheij M., van Triest B. NAD(+) depletion by APO866 in combination with radiation in a prostate cancer model, results from an in vitro and in vivo study. Radiother. Oncol. 2014;110:348–354. doi: 10.1016/j.radonc.2013.10.039. [DOI] [PubMed] [Google Scholar]
- 84.Ju H.Q., Zhuang Z.N., Li H., Tian T., Lu Y.X., Fan X.Q., Zhou H.J., Mo H.Y., Sheng H., Chiao P.J., et al. Regulation of the Nampt-mediated NAD salvage pathway and its therapeutic implications in pancreatic cancer. Cancer Lett. 2016;379:1–11. doi: 10.1016/j.canlet.2016.05.024. [DOI] [PubMed] [Google Scholar]
- 85.Lucena-Cacace A., Otero-Albiol D., Jimenez-Garcia M.P., Munoz-Galvan S., Carnero A. NAMPT Is a Potent Oncogene in Colon Cancer Progression that Modulates Cancer Stem Cell Properties and Resistance to Therapy through Sirt1 and PARP. Clin. Cancer Res. 2018;24:1202–1215. doi: 10.1158/1078-0432.CCR-17-2575. [DOI] [PubMed] [Google Scholar]
- 86.Lucena-Cacace A., Otero-Albiol D., Jiménez-García M.P., Peinado-Serrano J., Carnero A. NAMPT overexpression induces cancer stemness and defines a novel tumor signature for glioma prognosis. Oncotarget. 2017;8:99514–99530. doi: 10.18632/oncotarget.20577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhu Y., Guo M., Zhang L., Xu T., Wang L., Xu G. Biomarker triplet NAMPT/VEGF/HER2 as a de novo detection panel for the diagnosis and prognosis of human breast cancer. Oncol. Rep. 2016;35:454–462. doi: 10.3892/or.2015.4391. [DOI] [PubMed] [Google Scholar]
- 88.Wang B., Hasan M.K., Alvarado E., Yuan H., Wu H., Chen W.Y. NAMPT overexpression in prostate cancer and its contribution to tumor cell survival and stress response. Oncogene. 2011;30:907–921. doi: 10.1038/onc.2010.468. [DOI] [PubMed] [Google Scholar]
- 89.Shackelford R.E., Bui M.M., Coppola D., Hakam A. Over-expression of nicotinamide phosphoribosyltransferase in ovarian cancers. Int. J. Clin. Exp. Pathol. 2010;3:522–527. [PMC free article] [PubMed] [Google Scholar]
- 90.Gehrke I., Bouchard E.D., Beiggi S., Poeppl A.G., Johnston J.B., Gibson S.B., Banerji V. On-target effect of FK866, a nicotinamide phosphoribosyl transferase inhibitor, by apoptosis-mediated death in chronic lymphocytic leukemia cells. Clin. Cancer Res. 2014;20:4861–4872. doi: 10.1158/1078-0432.CCR-14-0624. [DOI] [PubMed] [Google Scholar]
- 91.Del Nagro C., Xiao Y., Rangell L., Reichelt M., O’Brien T. Depletion of the central metabolite NAD leads to oncosis-mediated cell death. J. Biol. Chem. 2014;289:35182–35192. doi: 10.1074/jbc.M114.580159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hasmann M., Schemainda I. FK866, a highly specific noncompetitive inhibitor of nicotinamide phosphoribosyltransferase, represents a novel mechanism for induction of tumor cell apoptosis. Cancer Res. 2003;63:7436–7442. [PubMed] [Google Scholar]
- 93.Olesen U.H., Christensen M.K., Björkling F., Jäättelä M., Jensen P.B., Sehested M., Nielsen S.J. Anticancer agent CHS-828 inhibits cellular synthesis of NAD. Biochem. Biophys. Res. Commun. 2008;367:799–804. doi: 10.1016/j.bbrc.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 94.Watson M., Roulston A., Belec L., Billot X., Marcellus R., Bedard D., Bernier C., Branchaud S., Chan H., Dairi K., et al. The small molecule GMX1778 is a potent inhibitor of NAD+ biosynthesis: Strategy for enhanced therapy in nicotinic acid phosphoribosyltransferase 1-deficient tumors. Mol. Cell. Biol. 2009;29:5872–5888. doi: 10.1128/MCB.00112-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chan D.A., Sutphin P.D., Nguyen P., Turcotte S., Lai E.W., Banh A., Reynolds G.E., Chi J.T., Wu J., Solow-Cordero D.E., et al. Targeting GLUT1 and the Warburg effect in renal cell carcinoma by chemical synthetic lethality. Sci. Transl. Med. 2011;3:94ra70. doi: 10.1126/scitranslmed.3002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Espindola-Netto J.M., Chini C.C.S., Tarrago M., Wang E., Dutta S., Pal K., Mukhopadhyay D., Sola-Penna M., Chini E.N. Preclinical efficacy of the novel competitive NAMPT inhibitor STF-118804 in pancreatic cancer. Oncotarget. 2017;8:85054–85067. doi: 10.18632/oncotarget.18841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhao G., Green C.F., Hui Y.H., Prieto L., Shepard R., Dong S., Wang T., Tan B., Gong X., Kays L., et al. Discovery of a Highly Selective NAMPT Inhibitor That Demonstrates Robust Efficacy and Improved Retinal Toxicity with Nicotinic Acid Coadministration. Mol. Cancer Ther. 2017;16:2677–2688. doi: 10.1158/1535-7163.MCT-16-0674. [DOI] [PubMed] [Google Scholar]
- 98.Abu Aboud O., Chen C.H., Senapedis W., Baloglu E., Argueta C., Weiss R.H. Dual and Specific Inhibition of NAMPT and PAK4 By KPT-9274 Decreases Kidney Cancer Growth. Mol. Cancer Ther. 2016;15:2119–2129. doi: 10.1158/1535-7163.MCT-16-0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Von Heideman A., Berglund A., Larsson R., Nygren P. Safety and efficacy of NAD depleting cancer drugs: Results of a phase I clinical trial of CHS 828 and overview of published data. Cancer Chemother. Pharmacol. 2010;65:1165–1172. doi: 10.1007/s00280-009-1125-3. [DOI] [PubMed] [Google Scholar]
- 100.Goldinger S.M., Gobbi Bischof S., Fink-Puches R., Klemke C.D., Dreno B., Bagot M., Dummer R. Efficacy and Safety of APO866 in Patients With Refractory or Relapsed Cutaneous T-Cell Lymphoma: A Phase 2 Clinical Trial. JAMA Dermatol. 2016;152:837–839. doi: 10.1001/jamadermatol.2016.0401. [DOI] [PubMed] [Google Scholar]
- 101.Holen K., Saltz L.B., Hollywood E., Burk K., Hanauske A.R. The pharmacokinetics, toxicities, and biologic effects of FK866, a nicotinamide adenine dinucleotide biosynthesis inhibitor. Investig. New Drugs. 2008;26:45–51. doi: 10.1007/s10637-007-9083-2. [DOI] [PubMed] [Google Scholar]
- 102.Ravaud A., Cerny T., Terret C., Wanders J., Bui B.N., Hess D., Droz J.P., Fumoleau P., Twelves C. Phase I study and pharmacokinetic of CHS-828, a guanidino-containing compound, administered orally as a single dose every 3 weeks in solid tumours: An ECSG/EORTC study. Eur. J. Cancer. 2005;41:702–707. doi: 10.1016/j.ejca.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 103.Hovstadius P., Larsson R., Jonsson E., Skov T., Kissmeyer A.M., Krasilnikoff K., Bergh J., Karlsson M.O., Lonnebo A., Ahlgren J. A Phase I study of CHS 828 in patients with solid tumor malignancy. Clin. Cancer Res. 2002;8:2843–2850. [PubMed] [Google Scholar]
- 104.Liu H.Y., Li Q.R., Cheng X.F., Wang G.J., Hao H.P. NAMPT inhibition synergizes with NQO1-targeting agents in inducing apoptotic cell death in non-small cell lung cancer cells. Chin. J. Nat. Med. 2016;14:582–589. doi: 10.1016/S1875-5364(16)30068-1. [DOI] [PubMed] [Google Scholar]
- 105.Zhang K., Chen D., Ma K., Wu X., Hao H., Jiang S. NAD(P)H:Quinone Oxidoreductase 1 (NQO1) as a Therapeutic and Diagnostic Target in Cancer. J. Med. Chem. 2018;61:6983–7003. doi: 10.1021/acs.jmedchem.8b00124. [DOI] [PubMed] [Google Scholar]
- 106.Pardee A.B., Li Y.Z., Li C.J. Cancer therapy with beta-lapachone. Curr. Cancer Drug Targets. 2002;2:227–242. doi: 10.2174/1568009023333854. [DOI] [PubMed] [Google Scholar]
- 107.Beg M.S., Huang X., Silvers M.A., Gerber D.E., Bolluyt J., Sarode V., Fattah F., Deberardinis R.J., Merritt M.E., Xie X.J., et al. Using a novel NQO1 bioactivatable drug, beta-lapachone (ARQ761), to enhance chemotherapeutic effects by metabolic modulation in pancreatic cancer. J. Surg. Oncol. 2017;116:83–88. doi: 10.1002/jso.24624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bey E.A., Bentle M.S., Reinicke K.E., Dong Y., Yang C.R., Girard L., Minna J.D., Bornmann W.G., Gao J., Boothman D.A. An NQO1- and PARP-1-mediated cell death pathway induced in non-small-cell lung cancer cells by beta-lapachone. Proc. Natl. Acad. Sci. USA. 2007;104:11832–11837. doi: 10.1073/pnas.0702176104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Huang X., Motea E.A., Moore Z.R., Yao J., Dong Y., Chakrabarti G., Kilgore J.A., Silvers M.A., Patidar P.L., Cholka A., et al. Leveraging an NQO1 Bioactivatable Drug for Tumor-Selective Use of Poly(ADP-ribose) Polymerase Inhibitors. Cancer Cell. 2016;30:940–952. doi: 10.1016/j.ccell.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Motea E.A., Huang X., Singh N., Kilgore J.A., Williams N.S., Xie X.J., Gerber D.E., Beg M.S., Bey E.A., Boothman D.A. NQO1-dependent, Tumor-selective Radiosensitization of Non-small Cell Lung Cancers. Clin. Cancer Res. 2019;25:2601–2609. doi: 10.1158/1078-0432.CCR-18-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Blanco E., Bey E.A., Khemtong C., Yang S.G., Setti-Guthi J., Chen H., Kessinger C.W., Carnevale K.A., Bornmann W.G., Boothman D.A., et al. Beta-lapachone micellar nanotherapeutics for non-small cell lung cancer therapy. Cancer Res. 2010;70:3896–3904. doi: 10.1158/0008-5472.CAN-09-3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dong Y., Bey E.A., Li L.S., Kabbani W., Yan J., Xie X.J., Hsieh J.T., Gao J., Boothman D.A. Prostate cancer radiosensitization through poly(ADP-Ribose) polymerase-1 hyperactivation. Cancer Res. 2010;70:8088–8096. doi: 10.1158/0008-5472.CAN-10-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Reinicke K.E., Bey E.A., Bentle M.S., Pink J.J., Ingalls S.T., Hoppel C.L., Misico R.I., Arzac G.M., Burton G., Bornmann W.G., et al. Development of beta-lapachone prodrugs for therapy against human cancer cells with elevated NAD(P)H:quinone oxidoreductase 1 levels. Clin. Cancer Res. 2005;11:3055–3064. doi: 10.1158/1078-0432.CCR-04-2185. [DOI] [PubMed] [Google Scholar]
- 114.Silvers M.A., Deja S., Singh N., Egnatchik R.A., Sudderth J., Luo X., Beg M.S., Burgess S.C., DeBerardinis R.J., Boothman D.A., et al. The NQO1 bioactivatable drug, beta-lapachone, alters the redox state of NQO1+ pancreatic cancer cells, causing perturbation in central carbon metabolism. J. Biol. Chem. 2017;292:18203–18216. doi: 10.1074/jbc.M117.813923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pink J.J., Planchon S.M., Tagliarino C., Varnes M.E., Siegel D., Boothman D.A. NAD(P)H:Quinone oxidoreductase activity is the principal determinant of beta-lapachone cytotoxicity. J. Biol. Chem. 2000;275:5416–5424. doi: 10.1074/jbc.275.8.5416. [DOI] [PubMed] [Google Scholar]
- 116.Imlay J.A., Chin S.M., Linn S. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science. 1988;240:640–642. doi: 10.1126/science.2834821. [DOI] [PubMed] [Google Scholar]
- 117.Andrabi S.A., Umanah G.K., Chang C., Stevens D.A., Karuppagounder S.S., Gagne J.P., Poirier G.G., Dawson V.L., Dawson T.M. Poly(ADP-ribose) polymerase-dependent energy depletion occurs through inhibition of glycolysis. Proc. Natl. Acad. Sci. USA. 2014;111:10209–10214. doi: 10.1073/pnas.1405158111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kim S., Lee S., Cho J.Y., Yoon S.H., Jang I.J., Yu K.S. Pharmacokinetics and tolerability of MB12066, a beta-lapachone derivative targeting NAD(P)H: Quinone oxidoreductase 1: Two independent, double-blind, placebo-controlled, combined single and multiple ascending dose first-in-human clinical trials. Drug Des. Dev. Ther. 2017;11:3187–3195. doi: 10.2147/DDDT.S151269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hartner L.P., Rosen L., Hensley M., Mendelson D., Staddon A.P., Chow W., Kovalyov O., Ruka W., Skladowski K., Jagiello-Gruszfeld A., et al. Phase 2 dose multi-center, open-label study of ARQ 501, a checkpoint activator, in adult patients with persistent, recurrent or metastatic leiomyosarcoma (LMS) J. Clin. Oncol. 2007;25(Suppl. 18):20521. doi: 10.1200/jco.2007.25.18_suppl.20521. [DOI] [Google Scholar]
- 120.Buranrat B., Chau-in S., Prawan A., Puapairoj A., Zeekpudsa P., Kukongviriyapan V. NQO1 expression correlates with cholangiocarcinoma prognosis. Asian Pac. J. Cancer Prev. 2012;13:131–136. [PubMed] [Google Scholar]
- 121.Cui X., Jin T., Wang X., Jin G., Li Z., Lin L. NAD(P)H:quinone oxidoreductase-1 overexpression predicts poor prognosis in small cell lung cancer. Oncol. Rep. 2014;32:2589–2595. doi: 10.3892/or.2014.3494. [DOI] [PubMed] [Google Scholar]
- 122.Cui X., Li L., Yan G., Meng K., Lin Z., Nan Y., Jin G., Li C. High expression of NQO1 is associated with poor prognosis in serous ovarian carcinoma. BMC Cancer. 2015;15:244. doi: 10.1186/s12885-015-1271-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kolesar J.M., Dahlberg S.E., Marsh S., McLeod H.L., Johnson D.H., Keller S.M., Schiller J.H. The NQO1*2/*2 polymorphism is associated with poor overall survival in patients following resection of stages II and IIIa non-small cell lung cancer. Oncol. Rep. 2011;25:1765–1772. doi: 10.3892/or.2011.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li Z., Zhang Y., Jin T., Men J., Lin Z., Qi P., Piao Y., Yan G. NQO1 protein expression predicts poor prognosis of non-small cell lung cancers. BMC Cancer. 2015;15:207. doi: 10.1186/s12885-015-1227-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lin L., Qin Y., Jin T., Liu S., Zhang S., Shen X., Lin Z. Significance of NQO1 overexpression for prognostic evaluation of gastric adenocarcinoma. Exp. Mol. Pathol. 2014;96:200–205. doi: 10.1016/j.yexmp.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 126.Yang Y., Zhang Y., Wu Q., Cui X., Lin Z., Liu S., Chen L. Clinical implications of high NQO1 expression in breast cancers. J. Exp. Clin. Cancer Res. 2014;33:14. doi: 10.1186/1756-9966-33-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Morales J., Li L., Fattah F.J., Dong Y., Bey E.A., Patel M., Gao J., Boothman D.A. Review of poly (ADP-ribose) polymerase (PARP) mechanisms of action and rationale for targeting in cancer and other diseases. Crit. Rev. Eukaryot. Gene Expr. 2014;24:15–28. doi: 10.1615/CritRevEukaryotGeneExpr.2013006875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Du Y., Yamaguchi H., Wei Y., Hsu J.L., Wang H.-L., Hsu Y.-H., Lin W.-C., Yu W.-H., Leonard P.G., Lee IV G.R. Blocking c-Met–mediated PARP1 phosphorylation enhances anti-tumor effects of PARP inhibitors. Nat. Med. 2016;22:194. doi: 10.1038/nm.4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhang Y., Xia M., Jin K., Wang S., Wei H., Fan C., Wu Y., Li X., Li X., Li G. Function of the c-Met receptor tyrosine kinase in carcinogenesis and associated therapeutic opportunities. Mol. Cancer. 2018;17:45. doi: 10.1186/s12943-018-0796-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chiarugi P., Cirri P. Redox regulation of protein tyrosine phosphatases during receptor tyrosine kinase signal transduction. Trends Biochem. Sci. 2003;28:509–514. doi: 10.1016/S0968-0004(03)00174-9. [DOI] [PubMed] [Google Scholar]
- 131.Chen M.-K., Du Y., Sun L., Hsu J.L., Wang Y.-H., Gao Y., Huang J., Hung M.-C. H2O2 induces nuclear transport of the receptor tyrosine kinase c-MET in breast cancer cells via a membrane-bound retrograde trafficking mechanism. J. Biol. Chem. 2019;294:8516–8528. doi: 10.1074/jbc.RA118.005953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dong Q., Du Y., Li H., Liu C., Wei Y., Chen M.-K., Zhao X., Chu Y.-Y., Qiu Y., Qin L. EGFR and c-MET cooperate to enhance resistance to PARP inhibitors in hepatocellular carcinoma. Cancer Res. 2019;79:819–829. doi: 10.1158/0008-5472.CAN-18-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Crawford R.S., Albadawi H., Atkins M.D., Jones J.E., Yoo H.J., Conrad M.F., Austen W.G., Jr., Watkins M.T. Postischemic poly (ADP-ribose) polymerase (PARP) inhibition reduces ischemia reperfusion injury in a hind-limb ischemia model. Surgery. 2010;148:110–118. doi: 10.1016/j.surg.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zingarelli B., Hake P.W., O’Connor M., Denenberg A., Kong S., Aronow B.J. Absence of poly(ADP-ribose)polymerase-1 alters nuclear factor-kappa B activation and gene expression of apoptosis regulators after reperfusion injury. Mol. Med. 2003;9:143–153. doi: 10.1007/BF03402179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Blaser H., Dostert C., Mak T.W., Brenner D. TNF and ROS Crosstalk in Inflammation. Trends Cell Biol. 2016;26:249–261. doi: 10.1016/j.tcb.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 136.Johnson P.M., Sagerman R.H., Dombrowski C.S. Ischemia of the lung due to ionizing radiation: Quantitative studies. J. Nucl. Med. 1970;11:491–495. [PubMed] [Google Scholar]
- 137.Zhou Z.B., Meng L., Gelb A.W., Lee R., Huang W.Q. Cerebral ischemia during surgery: An overview. J. Biomed. Res. 2016;30:83–87. doi: 10.7555/JBR.30.20150126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wang S., Han L., Han J., Li P., Ding Q., Zhang Q.J., Liu Z.P., Chen C., Yu Y. Uncoupling of PARP1 trapping and inhibition using selective PARP1 degradation. Nat. Chem. Biol. 2019;15:1223–1231. doi: 10.1038/s41589-019-0379-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hatachi G., Tsuchiya T., Miyazaki T., Matsumoto K., Yamasaki N., Okita N., Nanashima A., Higami Y., Nagayasu T. The poly(adenosine diphosphate-ribose) polymerase inhibitor PJ34 reduces pulmonary ischemia-reperfusion injury in rats. Transplantation. 2014;98:618–624. doi: 10.1097/TP.0000000000000305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Xu J.C., Fan J., Wang X., Eacker S.M., Kam T.I., Chen L., Yin X., Zhu J., Chi Z., Jiang H., et al. Cultured networks of excitatory projection neurons and inhibitory interneurons for studying human cortical neurotoxicity. Sci. Transl. Med. 2016;8:333ra48. doi: 10.1126/scitranslmed.aad0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chatterjee P.K., Chatterjee B.E., Pedersen H., Sivarajah A., McDonald M.C., Mota-Filipe H., Brown P.A., Stewart K.N., Cuzzocrea S., Threadgill M.D., et al. 5-Aminoisoquinolinone reduces renal injury and dysfunction caused by experimental ischemia/reperfusion. Kidney Int. 2004;65:499–509. doi: 10.1111/j.1523-1755.2004.00415.x. [DOI] [PubMed] [Google Scholar]
- 142.Mouchiroud L., Houtkooper R.H., Moullan N., Katsyuba E., Ryu D., Canto C., Mottis A., Jo Y.S., Viswanathan M., Schoonjans K., et al. The NAD(+)/Sirtuin Pathway Modulates Longevity through Activation of Mitochondrial UPR and FOXO Signaling. Cell. 2013;154:430–441. doi: 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mota R., Sanchez-Bueno F., Berenguer-Pina J.J., Hernandez-Espinosa D., Parrilla P., Yelamos J. Therapeutic treatment with poly(ADP-ribose) polymerase inhibitors attenuates the severity of acute pancreatitis and associated liver and lung injury. Br. J. Pharmacol. 2007;151:998–1005. doi: 10.1038/sj.bjp.0707310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Montoni A., Robu M., Pouliot E., Shah G.M. Resistance to PARP-Inhibitors in Cancer Therapy. Front. Pharmacol. 2013;4:18. doi: 10.3389/fphar.2013.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Prensner J.R., Chen W., Iyer M.K., Cao Q., Ma T., Han S., Sahu A., Malik R., Wilder-Romans K., Navone N., et al. PCAT-1, a long noncoding RNA, regulates BRCA2 and controls homologous recombination in cancer. Cancer Res. 2014;74:1651–1660. doi: 10.1158/0008-5472.CAN-13-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Livraghi L., Garber J.E. PARP inhibitors in the management of breast cancer: Current data and future prospects. BMC Med. 2015;13:188. doi: 10.1186/s12916-015-0425-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Quereda V., Bayle S., Vena F., Frydman S.M., Monastyrskyi A., Roush W.R., Duckett D.R. Therapeutic targeting of CDK12/CDK13 in triple-negative breast cancer. Cancer Cell. 2019;36:545–558. doi: 10.1016/j.ccell.2019.09.004. [DOI] [PubMed] [Google Scholar]
- 148.Balaji K., Vijayaraghavan S., Diao L., Tong P., Fan Y., Carey J.P., Bui T.N., Warner S., Heymach J.V., Hunt K.K. AXL inhibition suppresses the DNA damage response and sensitizes cells to PARP inhibition in multiple cancers. Mol. Cancer Res. 2017;15:45–58. doi: 10.1158/1541-7786.MCR-16-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Garcia T.B., Snedeker J.C., Baturin D., Gardner L., Fosmire S.P., Zhou C., Jordan C.T., Venkataraman S., Vibhakar R., Porter C.C. A small-molecule inhibitor of WEE1, AZD1775, synergizes with olaparib by impairing homologous recombination and enhancing DNA damage and apoptosis in acute leukemia. Mol. Cancer Ther. 2017;16:2058–2068. doi: 10.1158/1535-7163.MCT-16-0660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Song Z., Tu X., Zhou Q., Huang J., Chen Y., Liu J., Lee S., Kim W., Nowsheen S., Luo K. A novel UCHL 3 inhibitor, perifosine, enhances PARP inhibitor cytotoxicity through inhibition of homologous recombination-mediated DNA double strand break repair. Cell Death & Disease. 2019;10:398. doi: 10.1038/s41419-019-1628-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Miller A.L., Fehling S.C., Garcia P.L., Gamblin T.L., Council L.N., van Waardenburg R.C., Yang E.S., Bradner J.E., Yoon K.J. The BET inhibitor JQ1 attenuates double-strand break repair and sensitizes models of pancreatic ductal adenocarcinoma to PARP inhibitors. EBioMedicine. 2019;44:419–430. doi: 10.1016/j.ebiom.2019.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ibrahim Y.H., García-García C., Serra V., He L., Torres-Lockhart K., Prat A., Anton P., Cozar P., Guzmán M., Grueso J. PI3K inhibition impairs BRCA1/2 expression and sensitizes BRCA-proficient triple-negative breast cancer to PARP inhibition. Cancer Discov. 2012;2:1036–1047. doi: 10.1158/2159-8290.CD-11-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhong Q., Hu Z., Li Q., Yi T., Li J., Yang H. Cyclin D1 silencing impairs DNA double strand break repair, sensitizes BRCA1 wildtype ovarian cancer cells to olaparib. Gynecol. Oncol. 2019;152:157–165. doi: 10.1016/j.ygyno.2018.10.027. [DOI] [PubMed] [Google Scholar]
- 154.Jirawatnotai S., Hu Y., Michowski W., Elias J.E., Becks L., Bienvenu F., Zagozdzon A., Goswami T., Wang Y.E., Clark A.B., et al. A function for cyclin D1 in DNA repair uncovered by protein interactome analyses in human cancers. Nature. 2011;474:230–234. doi: 10.1038/nature10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hirst J., Godwin A.K. AURKA inhibition mimics BRCAness. Aging (Albany NY) 2017;9:1945. doi: 10.18632/aging.101291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Byrum A.K., Vindigni A., Mosammaparast N. Defining and Modulating ‘BRCAness’. Trends Cell Biol. 2019 doi: 10.1016/j.tcb.2019.06.005. [DOI] [PubMed] [Google Scholar]
- 157.Wu W., Nishikawa H., Fukuda T., Vittal V., Asano M., Miyoshi Y., Klevit R.E., Ohta T. Interaction of BARD1 and HP1 is required for BRCA1 retention at sites of DNA damage. Cancer Res. 2015;75:1311–1321. doi: 10.1158/0008-5472.CAN-14-2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Criscuolo D., Morra F., Giannella R., Visconti R., Cerrato A., Celetti A. New combinatorial strategies to improve the PARP inhibitors efficacy in the urothelial bladder Cancer treatment. J. Exp. Clin. Cancer Res. 2019;38:91. doi: 10.1186/s13046-019-1089-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Cerrato A., Merolla F., Morra F., Celetti A. CCDC6: The identity of a protein known to be partner in fusion. Int. J. Cancer. 2018;142:1300–1308. doi: 10.1002/ijc.31106. [DOI] [PubMed] [Google Scholar]
- 160.Vena F., Jia R., Esfandiari A., Garcia-Gomez J.J., Rodriguez-Justo M., Ma J., Syed S., Crowley L., Elenbaas B., Goodstal S. MEK inhibition leads to BRCA2 downregulation and sensitization to DNA damaging agents in pancreas and ovarian cancer models. Oncotarget. 2018;9:11592. doi: 10.18632/oncotarget.24294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Yin L., Liu Y., Peng Y., Peng Y., Yu X., Gao Y., Yuan B., Zhu Q., Cao T., He L. PARP inhibitor veliparib and HDAC inhibitor SAHA synergistically co-target the UHRF1/BRCA1 DNA damage repair complex in prostate cancer cells. J. Exp. Clin. Cancer Res. 2018;37:153. doi: 10.1186/s13046-018-0810-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Cruz O.V., Prudnikova T.Y., Araiza-Olivera D., Perez-Plasencia C., Johnson N., Bernhardy A.J., Slifker M., Renner C., Chernoff J., Arias L.E. Reduced PAK1 activity sensitizes FA/BRCA-proficient breast cancer cells to PARP inhibition. Oncotarget. 2016;7:76590. doi: 10.18632/oncotarget.12576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Li L., Karanika S., Yang G., Wang J., Park S., Broom B., Manyam G.C., Wu W., Luo Y., Basourakos S. Enzalutamide-induced “BRCAness” and PARP inhibition is a synthetic lethal therapy for castration-resistant prostate cancer. Sci. Signal. 2017;10 doi: 10.1126/scisignal.aam7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Liu L., Zhou W., Cheng C.-T., Ren X., Somlo G., Fong M.Y., Chin A.R., Li H., Yu Y., Xu Y. TGFβ induces “BRCAness” and sensitivity to PARP inhibition in breast cancer by regulating DNA-repair genes. Mol. Cancer Res. 2014;12:1597–1609. doi: 10.1158/1541-7786.MCR-14-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]