Dear Editor,
We read with interest the paper by Onno J. de Boer and colleagues,1 titled “Comparison of Two Different Immunohistochemical Quadruple Staining Approaches to Identify Innate Lymphoid Cells in Formalin-fixed Paraffin-embedded Human Tissue.”
We are familiar with the “virtual quadruple staining protocol” having recently published the method in detail,2 together with other authors,3,4 and we found it peculiar that almost half (four out of nine) of the antibodies used by the authors in that paper either did not survive a single cycle of antibody removal or required prioritization in the staining sequence. This is in contrast to our experience, where we found that the epitope for just 3 antibodies out of more than 300 tested was destroyed by the very first stripping cycle (CD30/BerH2, CD45RO/UCHL1, CTLA4/BSB-88); the remaining antibodies, including some of the same clones used by the authors, did survive more than 30 cycles of staining and stripping, with intensity variations less than 10% from time zero and occasional increases above that.2,5
Thus, for the 314 antibodies we tested, there was no need to prioritize the staining sequence with the exceptions named above and for phosphorylated epitopes (not shown). An updated list of validated antibodies is published and updated.6
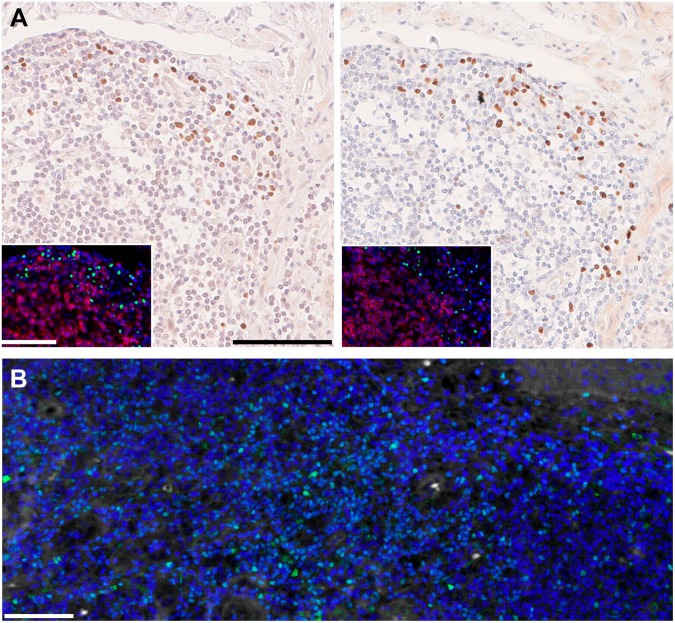
For retinoic acid receptor related orphan receptor C (RORC), we directly assessed the effect of the stripping buffer by extending the incubation time to be equivalent to the cumulative exposure during 4 or 10 staining and stripping cycles (as previously published3) (Figure 1) and we confirmed that epitopes which survive the first cycle will not be affected by numerous subsequent staining and stripping cycles.
Figure 1.
(A) Normal human intestine was stained in indirect immunohistochemistry on FFPE (formalin-fixed, paraffin embedded) serial sections for RORC, clone 6F3.1 as per Gendusa et al.3 and in indirect immunofluorescence (inset; green) together with CD3 (clone CD3-12; red) as per Bolognesi et al.2 Nuclear counterstain is hematoxylin and 4′,6-diamidino-2-phenylindole (DAPI; blue) respectively. On the left, no stripping occurred before staining. On the right, sections were stripped for 10×-equivalent (5 hr) or 4×-equivalent (2 hr; inset) before staining. and (B) normal human FFPE tonsil was stained for GATA3, goat antibody AF2605-SP (green) in indirect immunofluorescence as per Bolognesi et al.2 as the 12th cycle of staining and stripping. Nuclear counterstain DAPI (blue), no autofluorescence was digitally subtracted. Bars A and B = 100 µm
However, there are other steps in the process in which the antigens potentially are damaged.
After our first publication,3 we noticed occasional “re-masking” (loss of detection) of the epitopes, the cause of which we identified and solved by adding disaccharides throughout the process.7 Subsequently, we found that glycerol/gelatin mounting media affect epitope detection affecting reproducibility.2
Strict adherence to protocols is critical, such as avoiding washing the slides in tap water containing Ca++, as calcium is a known “re-masking agent.”8
Upon close inspection, key differences between our published work2 and the manuscript by de Boer were noted. In the de Boer et al. protocol, slides were washed in tap water and mounted in glycerol/gelatin and, moreover, no disaccharides were present in the antibody diluent and the washing buffer.
One last issue vaguely addressed by the authors is the removal of the NovaRed pigment after each cycle; we know that 2ME (β mercapto ethanol)/SDS (sodium dodecyl sulfate) can remove hematoxylin and eosin (H&E) staining, probably by the same mechanism by which it removes 4′,6-diamidino-2-phenylindole (DAPI) and ribonucleic acid (RNA)-bound FISH (fluorescent in-situ hybridization) probes, but we would be interested knowing the details by which it removes a dye reported to be insoluble or only partially in organic solvents.
In summary, we failed to confirm the decrease in antigenicity upon stripping reported by de Boer and colleagues. We believe that deviations from the published protocols2,6 result in re-masking or damaging of some antigens during washing and mounting, leading to erroneous conclusions about the staining sequence for the “virtual quadruple staining” approach.
Footnotes
Competing Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: MMB, FMB, and GC designed the study; FMB provided essential reagents; GC performed the experiments; and MMB, FMB, and GC wrote the letter.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work has been supported by Departmental University of Milano–Bicocca funds. Maddalena Maria Bolognesi is a PhD student of the DIMET course XXXV, University of Milano–Bicocca.
Contributor Information
Maddalena Maria Bolognesi, Pathology, Department of Medicine and Surgery, Università di Milano–Bicocca, Monza, Italy.
Francesca Maria Bosisio, Laboratory of Translational Cell and Tissue Research, KU Leuven, Leuven, Belgium.
Giorgio Cattoretti, Pathology, Department of Medicine and Surgery, Università di Milano–Bicocca, Monza, Italy.
Literature Cited
- 1. de Boer OJ, Krebbers G, Mackaaij C, Florquin S, de Rie MA, van der Wal AC, Teunissen MBM. Comparison of two different immunohistochemical quadruple staining approaches to identify innate lymphoid cells in formalin-fixed paraffin-embedded human tissue. J Histochem Cytochem. 2019;118(6):127–38. doi: 10.1369/0022155419897257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bolognesi MM, Manzoni M, Scalia CR, Zannella S, Bosisio FM, Faretta M, Cattoretti G. Multiplex staining by sequential immunostaining and antibody removal on routine tissue sections. J Histochem Cytochem. 2017;65(8):431–44. doi: 10.1369/0022155417719419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gendusa R, Scalia CR, Buscone S, Cattoretti G. Elution of high-affinity (>10-9 KD) antibodies from tissue sections: clues to the molecular mechanism and use in sequential immunostaining. J Histochem Cytochem. 2014;62(7):519–31. doi: 10.1369/0022155414536732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kim M, Soontornniyomkij V, Ji B, Zhou X. System-wide immunohistochemical analysis of protein co-localization. PLoS ONE. 2012;7(2):e32043. doi: 10.1371/journal.pone.0032043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Manzoni M, Bolognesi MM, Antoranz A, Mancari R, Carinelli S, Faretta M, Bosisio FM, Cattoretti G. The adaptive and innate immune cell landscape of uterine leiomyosarcomas. Sci Rep. 2020;10(1):702. doi: 10.1038/s41598-020-57627-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cattoretti G, Bosisio FM, Marcelis L, Bolognesi MM. Multiple iterative labeling by antibody neodeposition (MILAN). Nature Res. 2019. September 25 [cited 2020. February 4]. Available from: https://protocolexchange.researchsquare.com/article/nprot-7017/v5
- 7. Boi G, Scalia CR, Gendusa R, Ronchi S, Cattoretti G. Disaccharides protect antigens from drying-induced damage in routinely processed tissue sections. J Histochem Cytochem. 2016;64(1):18–31. doi: 10.1369/0022155415616162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Morgan JM, Navabi H, Schmid KW, Jasani B. Possible role of tissue-bound calcium ions in citrate-mediated high-temperature antigen retrieval. J Pathol. 1994;174(4):301–7. doi: 10.1002/path.1711740410. [DOI] [PubMed] [Google Scholar]