Abstract
BK polyomavirus–associated nephropathy (BKpyVAN) remains a cause of graft loss in kidney transplant recipients on immunosuppressive therapy. Its diagnosis relies on the identification of BK virus (BKV) in the renal allograft biopsy by positive immunohistochemical (IHC) stain for the viral SV40 large T antigen, although in situ hybridization (ISH) for viral DNA is used in some centers. We examined tissue detection of BKV RNA by RNAscope, a novel, automated ISH test, in 61 allograft biopsies from 56 patients with BKpyVAN. We found good correlation between the estimate of BKV tissue load by RNAscope ISH and SV40 IHC (R2 = 0.65, p<0.0001). RNAscope ISH showed 88% sensitivity and 79% specificity and, as an alternative test, could confirm the presence of BKV tissue in presumed BKpyVAN and rule out BKV as the causative agent in JC virus nephropathy. We also used tissue BK viral load estimates by both RNAscope ISH and SV40 IHC to examine the relation between tissue and plasma BK levels and found significant correlation only between BK viremia and tissue BK measured by RNAscope ISH. Our findings suggest that the RNAscope ISH assay could be a reliable test for BKV detection in allograft biopsies.
Keywords: infection, nucleic acid, pathology, transplantation
Introduction
BK polyomavirus–associated nephropathy (BKpyVAN) is a challenging infectious complication of immunosuppression in recipients of renal allografts and can lead to graft loss. BK virus (BKV) infection contracted in childhood by respiratory route is highly prevalent, and antibodies against BKV can be detected in 80% to 90% of adults. After infection, the virus remains in a latent state in urothelial cells, but can be reactivated and replicate during immunosuppression, with detectable viremia developing in up to 30% of transplant recipients, and it can cause BKpyVAN in approximately 5% to 10%.1 Early detection of BKpyVAN is important to allow for prompt reduction of immunosuppression toward successful resolution of this condition. Monitoring the BK viral load in the serum by quantitative PCR (qPCR) is recommended during the first year after transplant when the risk of BKpyVAN is highest. Persistent BK viremia of >10,000 BKV DNA copies/ml for >3 weeks can indicate BKpyVAN (presumptive BKpyVAN).2 BKV can also be detected in the urine by PCR. The presence of infected “decoy” cells in the urine, especially aggregates of polyomavirus particles (“Haufen”), is also a manifestation of BKpyVAN.3 For a definite diagnosis of BKpyVAN, it is necessary to identify the presence of BKV in the renal parenchyma. Histologically, BKV infection can produce characteristic basophilic nuclear inclusions in the renal tubular and/or glomerular parietal cells, most often detected in the medullary tubules, and accompanied by variable tubulointerstitial inflammation. A more reliable and reproducible diagnostic criterion is the identification of BKV in the renal allograft biopsy by positive immunohistochemical (IHC) stain for the viral SV40 large T antigen in the nuclei of the renal epithelial cells. However, given the focality of replicating BKV in the kidney, SV40 stain can be negative in the allograft biopsy in 10% to 30% of patients with sustained viremia. Other techniques to detect BKV have been described, including the use of qPCR on RNA and/or DNA extracted from formalin-fixed, paraffin-embedded (FFPE) renal biopsy tissue4,5 and in situ hybridization (ISH) for BKV DNA.6–8 Although not widely used for routine diagnostic use, these nucleic acid–based tests may be valuable as back-up assays to identify BKV in tissue when a diagnosis of BKpyVAN is suspected. Newer techniques for ISH have been developed, which use probes designed to increase the signal-to-noise ratio to visualize RNA transcripts (RNAscope) and can be adapted for use in clinical-grade automated stainers.
In this study, we examine a single-center cohort of patients with biopsy-proven BKpyVAN and describe detection of BK polyomavirus RNA by ISH using RNAscope in paraffin-embedded tissue sections of allograft biopsies.
Materials and Methods
Patients
From 2010 to 2018, a diagnosis of BKpyVAN was made on 77 kidney allograft biopsies from 59 transplant recipients followed at our Transplant Center. Sufficient archival tissue was available in 61 biopsies from 56 patients for performing this study. Biopsies from 15 patients with negative SV40 stain were included as negative controls. Three biopsies with “presumptive” BKpyVAN and one case with JC polyomavirus infection were also analyzed. The following clinical variables were collected: age, gender, race, time posttransplant for each biopsy, serum creatinine, BK viremia, and serum creatinine at last follow-up. The BKV serum viral load was determined by qPCR, with a limit of detection of 2.3 log10 (200) BKV genome copies/ml of plasma. Immunosuppression for most patients consisted of thymoglobulin induction, tacrolimus, mycophenolate, and prednisone. Compatibility testing, desensitization protocols, and donor-specific antibody monitoring were as previously described.9 The study was approved by our institutional review board (protocol NA_00001141).
Biopsies
Allograft biopsies were performed using an 18-gauge spring-loaded biopsy needle under real-time ultrasound guidance. Biopsies were evaluated and scored according to updated Banff criteria.10–12 Diagnosis of BKpyVAN on allograft biopsies was based on the detection of one or more SV40-positive nuclei in tubular cells.
Staining Procedures
SV40 stain was performed on sections from FFPE tissue. Slides were baked for 30 min at 60C and loaded onto a Ventana Ultra Autostainer (Ventana Medical Systems; Tucson, AZ). Automated heat-induced antigen retrieval was performed at 95C with ULTRA CC1 (EDTA pH9; Ventana Medical Systems) for 30 min. Before October 2017 a rabbit polyclonal antibody (1:100 dilution, cat. no. sc-20800; Santa Cruz Biotechnology, Dallas, TX) was used with iView DAB detection kit (Ventana Medical Systems). Subsequently, primary monoclonal antibodies were used: mouse monoclonal antibody (cat. no. sc-147; Santa Cruz Biotechnology) at 1:100 dilution and prediluted mouse monoclonal antibody SV40 (MRQ-4; Cell Marque/Sigma, St Louis, MO) with ultraView DAB detection kit (Ventana Medical Systems, Tucson, AZ). Slides were counterstained with hematoxylin and coverslipped. These procedures were tested and found to be equivalent for SV40 detection.
RNA ISH was performed on sections from FFPE tissue with the RNAscope assay (Advanced Cell Diagnostics; Newark, CA). The chromogenic ISH assay was conducted on a Leica Bond Autostainer (Leica Biosystems; Wetzlar, Germany) according to the manufacturer’s protocol. In brief, 4-μm tissue sections were prepared using standard procedures. Slides were deparaffinized, rehydrated, and pretreated with RNAscope Target Retrieval at 95C, followed by treatment with RNAscope Enzyme. Slides were hybridized and detected using the RNAscope 2.5 HD Brown Kit (Advanced Cell Diagnostics) and then counterstained with hematoxylin, mounted, and coverslipped. In addition to hematoxylin, the test was also performed with manual periodic acid–Schiff (PAS) counterstain in 50 biopsies (compared with hematoxylin, the PAS counterstain did not affect the polyomavirus result on the same biopsy). The probe used for detection was a commercially available prediluted RNAscope 2.5 LS Probe—V-BKPyV-largeT (cat. no. 503528; Advanced Cell Diagnostics) targeting a consensus sequence corresponding to the BKPyVgp5 locus (large T antigen) of the BK polyomavirus complete genome (NCBI Reference Sequence: NC_001538.1).
Slides with unstained tissue serial sections were inspected and selected for SV40 and RNAscope ISH to ensure that the same amount of tissue would be present for scoring, excluding those biopsies which did not meet such requisite.
Histological Analysis and Scoring
For each biopsy stained with SV40 IHC, we counted the number of tubules showing positive nuclear staining. For each biopsy stained with the RNAscope probe, we counted the number of tubules showing positive stain for RNA ISH. Positive tubules in each biopsy were counted independently by two pathologists and a third pathologist in some of the cases (Supplemental Table 1). The tissue level of BKV (tissue viral load) was estimated by the number of positive tubules identified by SV40 or by RNA ISH per millimeter of biopsy core length.
Statistical Analysis
All values are expressed as mean ± SD, median, odds ratio (OR), 95% confidence interval (CI), and p values as appropriate. Correlations were analyzed with Pearson’s test. Categorical variables were analyzed with Chi-square statistics and Fisher’s test. A value of p<0.05 (two-tailed) was considered indicative of statistical significance, and statistical analysis was performed with GraphPad version 8 (GraphPad Software; San Diego, CA).
Results
Study Cohort
The study was performed on 61 allograft biopsies, performed between 2010 and 2018, from 56 patients, 54 with a biopsy-proven diagnosis of BKpyVAN and 2 with “presumptive” BKpyVAN. Negative control biopsies included nine allograft biopsies from transplant recipients with no evidence of BK viremia and negative SV40 stain, and six native kidney biopsies with decreased renal function and tubular injury.
Demographic information of the transplant recipients with BKpyVAN and of the transplant patients in negative control group is summarized in Table 1. For the BKpyVAN patients, the median time of diagnosis was 366 days after transplant, ranging from as early as 21 days to as late as 4.3 years after transplantation, including four patients in whom the first diagnosis occurred after 3 years of transplant. The median serum creatinine at diagnosis was 1.75 mg/dl (range, 0.7–6.6 mg/dl), and the average serum BK viral load at the time of biopsy was 205,103 copies/ml (median, 11,129 copies/ml; unknown in four patients). The median follow-up after diagnosis was 4.3 years (range, 7.1 months to 8.9 years), with one patient progressing to graft failure and six patients dying with functioning graft. The median creatinine value at last follow-up was 2.1 mg/dl (range, 0.82–8.7 mg/dl).
Table 1.
Transplant Patients.
| BKpyVAN (n=56) | Controls (n=9) | |
|---|---|---|
| Male/Female | 35/21 | 7/2 |
| African American/Caucasian/Other | 14/40/2 | 6/2/1 |
| Age at diagnosis, years (mean ± SD) | 53 ± 14 | 46 ± 20 |
| Days posttransplant at diagnosis (mean ± SD) | 453 ± 384 | 480 ± 653 |
| BK viremia at diagnosis, log value (median, range) | 4.31, 2–6.59 | Undetectable |
| Creatinine at diagnosis, mg/dl (mean ± SD) | 1.88 ± 0.64 | 5.24 ± 6.21 |
Abbreviation: BKpyVAN, BK polyomavirus–associated nephropathy.
Biopsy Features
Evaluation of the allograft biopsies showed identifiable basophilic nuclear inclusions only in 49% of cases with a definite diagnosis of BKpyVAN. In the majority of biopsies (75%), BK detection by IHC and/or ISH involved the cortex, with 24 biopsies showing positive tubules in both the cortex and the medulla, and the remaining cases showing BK localization limited to the medulla. Biopsies with the highest BKV tissue load showed SV40-positive tubules in both the cortex and the medulla, whereas significantly lower BKV tissue loads were noted in biopsies where SV40 tubules were detected in either the cortex or the medulla, which showed similar numbers of SV40-positive tubules/mm tissue (p=0.581). Interstitial inflammation and tubulitis were present in most BKpyVAN biopsies, and in 15 cases the tubulointerstitial inflammation was of sufficient degree to meet the Banff diagnostic criteria for T cell–mediated rejection. Most biopsies showed mild degree of tubular atrophy and interstitial fibrosis. A summary of the biopsy Banff scores is shown in Table 2.
Table 2.
Pathological Features and Banff Score of BKpyVAN Biopsies (Average ± SD, n=61).
| Glomerulosclerosis (%) | 6.0 ± 9.0 |
|---|---|
| g | 0.2 ± 0.5 |
| i | 1.7 ± 1.0 |
| t | 1.6 ± 1.0 |
| v | 0.0 ± 0.1 |
| ptc | 0.5 ± 1.0 |
| C4d | 0.4 ± 0.9 |
| cg | 0.0 ± 0.2 |
| ci | 1.5 ± 1.0 |
| ct | 1.5 ± 1.0 |
| cv | 0.9 ± 0.9 |
| ti | 1.6 ± 1.0 |
| ah | 0.4 ± 0.6 |
| mm | 0.2 ± 0.4 |
Glomerulosclerosis refers to globally sclerosed glomeruli. Abbreviation: BKpyVAN, BK polyomavirus–associated nephropathy.
Detection of BKV by RNAscope on Allograft Biopsies
The staining of positive cells by RNAscope ISH was generally strong, although less well defined than the nuclear stain observed by SV40 (Figs. 1A, B and 2A–C). The difference in the average number of tubules positive for SV40 (3/mm) and those positive for RNAscope (2/mm) was not significant (p=0.2064). There was a good correlation between the number of positive tubules per mm detected by the two assays in each case (R2 = 0.65, p<0.0001) (Fig. 3). Based on the examination of 76 biopsies, we estimate a sensitivity of 88% and a specificity of 79% for the detection of BKV by RNAscope in biopsy FFPE tissue, with 93% positive predictive value and 68% negative predictive value.
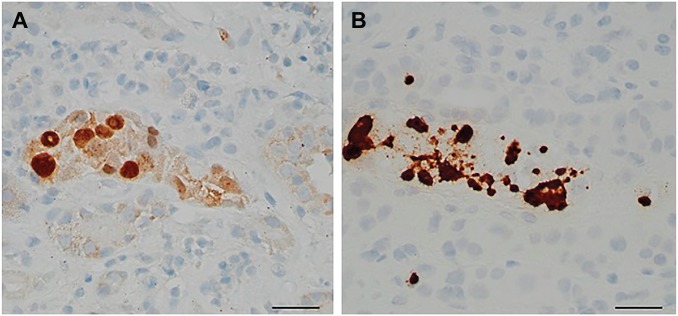
Figure 1.
Positive nuclear stain with SV40 in one affected tubule (A) and positive tubular stain by RNAscope in the same tubule (B) (scale bar, 50 µm).
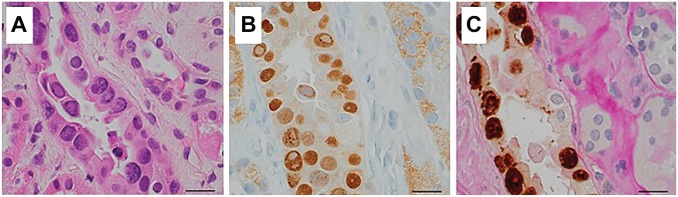
Figure 2.
Viral basophilic inclusions in one affected tubule with H&E stain (A); the same tubule shows positive nuclear stain with SV40 (B) and positive stain with RNAscope counterstained with PAS (C) (scale bar, 50 µm). Abbreviation: PAS, periodic acid–Schiff.
Figure 3.
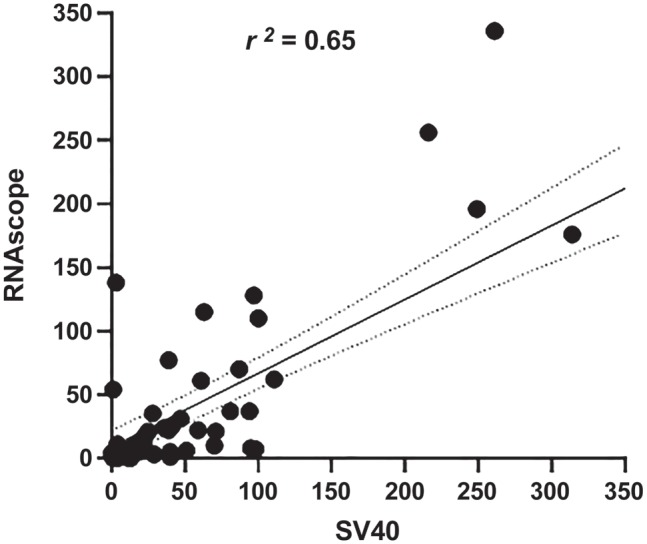
Correlation between the number of SV40-positive and RNAscope-positive tubules in individual biopsies.
The correlation between the density of BK-positive tubules (number of positive tubules per millimeter of biopsy core length) detected in the biopsy and the degree of BK viremia expressed as log of the plasma viral load at the time of biopsy was examined and shown to be significant for RNAscope ISH BK tissue levels (R2 = 0.1512, p=0.0040), but no significant correlation was seen between viremia and SV40 IHC BK levels (R2 = 0.0533, p=0.0960).
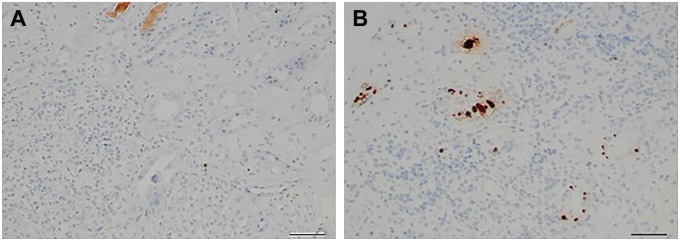
Using RNAscope ISH, we were able to detect the presence of BKV in three allograft biopsies from two patients with a presumptive diagnosis of BKpyVAN, based on BK viremia >5 log 10 DNA copies/ml at the time of biopsy, but negative SV40 IHC (appropriate staining was seen in the positive control slide). In these cases, RNAscope ISH was positive in each biopsy, allowing confirmation of a diagnosis of BKpyVAN (Fig. 4A and B).
Figure 4.
SV40-negative biopsy in a patient with BK viremia (A) showing focal positive tubular cell staining with RNAscope (B) (scale bar, 50 µm).
In nine biopsies, where SV40 IHC showed only very few tubules with positive nuclear stain (range, 1–15) in the whole tissue sample and no basophilic nuclear inclusions, RNAscope ISH was negative. We speculate that in these cases the BK infections may have been resolving because at the time of biopsy plasma BK DNA was detectable at very low levels in four patients (<201 copies/ml in three patients and 278 copies/ml in one patient) and had markedly decreased to about 7% of an earlier peak in five patients.
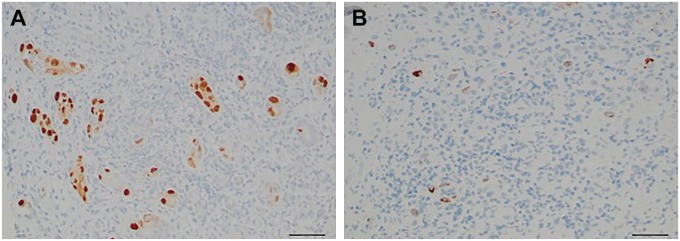
RNAscope ISH was also performed on one biopsy showing multiple SV40-positive tubules from a transplant recipient with undetectable BK viremia but evidence of JC virus viremia at the time of biopsy. In this case, positivity for RNAscope ISH was seen in only 10% of the SV40-positive tubules (Fig. 5A and B), indicating that most of the SV40 positivity was due to JC virus.
Figure 5.
Several SV40-positive tubules (A) but only sparse RNAscope-positive tubules are seen in the biopsy of a patient with JC virus infection but negative BK viremia (B) (scale bar 50 µm).
Discussion
In this study, we examine the characteristics of renal allograft biopsies in 56 transplant recipients with biopsy-proven diagnosis of BKpyVAN, and we describe detection of BK polyomavirus RNA with a novel, commercially available chromogenic RNA ISH assay, RNAscope, on FFPE sections of kidney allograft biopsies involved by BKpyVAN. The RNAscope BKV RNA ISH uses short branched oligonucleotide probes, can be automated, and has shown higher sensitivity compared with conventional ISH assays for the detection of other viruses in tissue samples.13 This assay has not been tested before for the identification of BKV RNA in renal tissue FFPE or for diagnosis of BKpyVAN, and, in previous studies, estimates of BKV RNA expression on allograft biopsies had been based on qPCR.4,5
We compared the performance of RNAscope ISH and SV40 IHC and found them comparable, with good concordance between the number of RNAscope ISH–positive tubules and SV40 IHC–positive tubules in each biopsy. We were able to detect the presence of BKV by RNAscope ISH in the allograft biopsies of two patients with repeatedly negative SV40 IHC, thus allowing a definitive diagnosis of BKpyVAN, and negative RNAscope confirmed JC virus as the infectious agent causing allograft nephropathy in a patient with negative BK viremia but positive SV40 in the biopsy. These findings underscore the importance of an alternative assay in unusual cases like these.
Nine patients in our cohort had biopsies with only rare SV40-positive nuclear stain but negative RNAscope ISH. These patients had very low or markedly decreased BK viremia from an earlier peak at the time of biopsy, likely representing a very mild infection or a resolving phase of the BKV infection, and RNAscope ISH negativity may be due to a sampling effect coupled with an extreme focality in the localization of the BKV. In these instances, detection and quantitative determination of urinary polyomavirus Haufen may be helpful as a non-invasive test, which has shown good correlation with the intrarenal polyomavirus replication in cases with rare SV40-positive tubules.3
Regarding the potential use of RNAscope ISH in a clinical setting, our data suggest that this assay could be reliable for the diagnosis of BKpyVAN in allograft recipients in selected cases, although its higher cost compared with SV40 IHC is likely to discourage routine implementation in clinical pathology labs.
When considering the time of diagnosis posttransplant, diagnosis occurred 3 years after transplant in 8% of patients with BKpyVAN, supporting suggestions made by others that screening for BKpyVAN may be indicated beyond the first 2 years.14
Analysis of the distribution of BKV in the biopsy tissue showed that, not unexpectedly, in biopsies with the highest tissue viral load, measured as the number of SV40-positive tubules/mm biopsy tissue, both the cortex and the medulla were involved. The presence of BKV was restricted to the medulla in only 20% of the biopsies similar to the regional distribution observed in a recent study.14
Finally, we also examined the relationship between the level of BK DNA in the serum and the abundance of BK in the tissue, which has been reported only in few previous studies, with somewhat conflicting results. No correlation between BK viremia and presence of BK in the tissue based on SV40 IHC was reported by Nankivell et al.,15 but a significant correlation was described by Menter et al.,16 who estimated the tissue viral load as the number of SV40-positive tubules per millimeter of biopsy length, and by Kable et al.,17 who measured the tissue viral load with a semiquantitative score including both SV40 and cytopathic changes. In our BKpyVAN cohort, the tissue level of BK at the time of biopsy based on RNAscope assay and expressed as the number of positive tubules per millimeter of biopsy core length did show significant correlation with BK viremia. However, no significant correlation was seen with the BK tissue load estimated by SV40 IHC. The reason for the discrepancy among ours and others’ findings in regard to SV40 IHC is unclear. Whether measures of the BK viral tissue levels by nucleic acid–based tests may more closely correlate with BK viremia remains to be determined and suggests the need for further study.
Supplemental Material
Supplemental material, 2020-00025R1_Production_Supplemental_Table_online_supp for BK Virus RNA in Renal Allograft Biopsies by Francesca Costigliolo, Kara Lombardo, Lois J. Arend, Avi Z. Rosenberg, Andres Matoso, Naima Carter-Monroe and Serena M. Bagnasco in Journal of Histochemistry & Cytochemistry
Footnotes
Competing Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: FC identified cases and collected clinical information, coordinated case processing for studying special stains, and reviewed biopsies. KL optimized and performed the RNA ISH assay. LJA reviewed biopsies. AZR contributed cases and helped in writing the manuscript. AM consulted on optimization and performance of RNA ISH and SV40. NC contributed cases with clinical information and reviewed biopsies. SMB designed the study, reviewed biopsies, and wrote the manuscript.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The study was supported with funds from the Johns Hopkins Department of Pathology.
Contributor Information
Francesca Costigliolo, Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, Maryland.
Kara Lombardo, Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, Maryland.
Lois J. Arend, Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, Maryland
Avi Z. Rosenberg, Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, Maryland
Andres Matoso, Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, Maryland.
Naima Carter-Monroe, Pathology and Laboratory Medicine, Veterans Administration Hospital, Baltimore, Maryland.
Serena M. Bagnasco, Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, Maryland.
Literature Cited
- 1. Hirsch H, Brennan D, Drachenberg C, Ginevri F, Gordon J, Limaye A, Mihatsch MJ, Nickeleit V, Ramos E, Randhawa P, Shapiro R, Steiger J, Suthanthiran M, Trofe J. Polyomavirus-associated nephropathy in renal transplantation: interdisciplinary analyses and recommendation. Transplantation. 2005;79:1277–1286. [DOI] [PubMed] [Google Scholar]
- 2. Hirsch HH, Randhawa PS, AST Infectious Diseases Community of Practice. BK polyomavirus in solid organ transplantation-guidelines from the American Society of Transplantation Infectious Diseases Community of Practice. Clin Transplant. 2019;33(9):e13528. [DOI] [PubMed] [Google Scholar]
- 3. Singh HK, Reisner H, Derebail VK, Kozlowski T, Nickeleit V. Polyomavirus nephropathy: quantitative urinary polyomavirus-Haufen testing accurately predicts the degree of intrarenal viral disease. Transplantation. 2015;99(3):609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schmid H, Nitschko H, Gerth J, Kliem V, Henger H, Cohen CD, Schlöndorff D, Gröne HJ, Kretzler M. Polyomavirus DNA and RNA detection in renal allograft biopsies: results from a European multicenter study. Transplantation. 2005;80:600–604. [DOI] [PubMed] [Google Scholar]
- 5. Adam B, Randhawa P, Chan S, Zeng G, Regele H, Kushner YB, Colvin RB, Reeve J, Mengel M. Banff Initiative for Quality Assurance in Transplantation (BIFQUIT): reproducibility of polyomavirus immunohistochemistry in kidney allografts. Am J Transplant. 2014;14(9):2137–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fritzsche F, Pianca S, Gaspert A, Varga Z, Wang L, Farrell MP, Chen XB, Hirsch HH, Springer E, Fehr T, Myles J, Tubbs R, Moch H. Silver-enhanced in situ hybridization for detection of polyomavirus DNA in patients with BK virus nephropathy. Diagn Mol Pathol. 2011;20(2):105–110. [DOI] [PubMed] [Google Scholar]
- 7. Cajaiba MM, Parks WT, Fuhrer K, Randhawa PS. Evaluation of human polyomavirus BK as a potential cause of villitis of unknown etiology and spontaneous abortion. J Med Virol. 2011;83(6):1031–1033. [DOI] [PubMed] [Google Scholar]
- 8. Kumari K, Pradeep I, Kakkar A, Dinda AK, Seth A, Nayak B, Singh G. BK polyomavirus and urothelial carcinoma: experience at a tertiary care centre in India with review of literature. Ann Diagn Pathol. 2019;40:77–80. [DOI] [PubMed] [Google Scholar]
- 9. Bagnasco S, Zachary A, Racusen L, Arend L, Carter-Monroe N, Alachkar N, Nazarian SM, Lonze BE, Montgomery RA, Kraus ES. Time course of pathologic changes in kidney allografts of positive crossmatch HLA-incompatible transplant recipients. Transplantation. 2014;97:440–445. [DOI] [PubMed] [Google Scholar]
- 10. Haas M, Sis B, Racusen LC, Solez K, Glotz D, Colvin RB, Castro MC, David DS, David-Neto E, Bagnasco SM, Cendales LC, Cornell LD, Demetris AJ, Drachenberg CB, Farver CF, Farris AB, 3rd, Gibson IW, Kraus E, Liapis H, Loupy A, Nickeleit V, Randhawa P, Rodriguez ER, Rush D, Smith RN, Tan CD, Wallace WD, Mengel M; Banff meeting report writing committee. Banff 2013 meeting report: inclusion of C4d-negative antibody-mediated rejection and antibody-associated arterial lesions. Am J Transplant. 2014;14(2):272–283. [DOI] [PubMed] [Google Scholar]
- 11. Loupy A, Haas M, Solez K, Racusen L, Glotz D, Seron D, Nankivell BJ, Colvin RB, Afrouzian M, Akalin E, Alachkar N, Bagnasco S, Becker JU, Cornell L, Drachenberg C, Dragun D, de Kort H, Gibson IW, Kraus ES, Lefaucheur C, Legendre C, Liapis H, Muthukumar T, Nickeleit V, Orandi B, Park W, Rabant M, Randhawa P, Reed EF, Roufosse C, Seshan SV, Sis B, Singh HK, Schinstock C, Tambur A, Zeevi A, Mengel M. The Banff 2015 kidney meeting report: current challenges in rejection classification and prospects for adopting molecular pathology. Am J Transplant. 2017;17(1):28–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Haas M, Loupy A, Lefaucheur C, Roufosse C, Glotz D, Seron D, Nankivell BJ, Halloran PF, Colvin RB, Akalin E, Alachkar N, Bagnasco S, Bouatou Y, Becker JU, Cornell LD, van Huyen JPD, Gibson IW, Kraus ES, Mannon RB, Naesens M, Nickeleit V, Nickerson P, Segev DL, Singh HK, Stegall M, Randhawa P, Racusen L, Solez K, Mengel M. The Banff 2017 kidney meeting report: revised diagnostic criteria for chronic active T cell–mediated rejection, antibody-mediated rejection, and prospects for integrative endpoints for next-generation clinical trials. Am J Transplant. 2018;18(2):293–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carossino M, Loynachan AT, James MacLachlan N, Drew C, Shuck KM, Timoney PJ, Del Piero F, Balasuriya UB. Detection of equine arteritis virus by two chromogenic RNA in situ hybridization assays (conventional and RNAscope®) and assessment of their performance in tissues from aborted equine fetuses. Arch Virol. 2016;161(11):3125–3136. [DOI] [PubMed] [Google Scholar]
- 14. Drachenberg CB, Papadimitriou JC, Chaudhry MR, Ugarte R, Mavanur M, Thomas B, Cangro C, Costa N, Ramos E, Weir MR, Haririan A. Histological evolution of BK virus–associated nephropathy: importance of integrating clinical and pathological findings. Am J Transplant. 2017;17(8):2078–2091. [DOI] [PubMed] [Google Scholar]
- 15. Nankivell BJ, Renthawa J, Sharma RN, Kable K, O’Connell PJ, Chapman JR. BK virus nephropathy: histological evolution by sequential pathology. Am J Transplant. 2017;17(8):2065–2077. [DOI] [PubMed] [Google Scholar]
- 16. Menter T, Mayr M, Schaub S, Mihatsch MJ, Hirsch HH, Hopfer H. Pathology of resolving polyomavirus-associated nephropathy. Am J Transplant. 2013;13(6):1474–1483. [DOI] [PubMed] [Google Scholar]
- 17. Kable K, Davies CD, O’Connell PJ, Chapman JR, Nankivell BJ. Clearance of BK virus nephropathy by combination antiviral therapy with intravenous immunoglobulin. Transplant Direct. 2017;3(4):e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, 2020-00025R1_Production_Supplemental_Table_online_supp for BK Virus RNA in Renal Allograft Biopsies by Francesca Costigliolo, Kara Lombardo, Lois J. Arend, Avi Z. Rosenberg, Andres Matoso, Naima Carter-Monroe and Serena M. Bagnasco in Journal of Histochemistry & Cytochemistry