Abstract
In recent years, the genomics community has witnessed the growth of large research biobanks, which collect DNA samples for research purposes. Depending on how and where the samples are genotyped, biobanks also offer the potential opportunity to return actionable genomic results to the clinical setting. We developed a preemptive clinical pharmacogenomic implementation initiative via a health system-wide research biobank at the University of Colorado. Here, we describe how preemptive return of clinical pharmacogenomic results via a research biobank is feasible, particularly when coupled with strong institutional support to maximize the impact and efficiency of biobank resources, a multidisciplinary implementation team, automated clinical decision support tools, and proactive strategies to engage stakeholders early in the clinical decision support tool development process.
Keywords: : biobank, implementation, personalized medicine, pharmacogenetics, pharmacogenomics, precision medicine
Pharmacogenomics, a component of personalized and precision medicine, uses an individual’s genetic information, along with other clinical factors, to inform decisions about drug selection and dosing [1]. The goal of pharmacogenomics is to minimize trial and error associated with drug prescribing in order to maximize efficacy and reduce adverse effects. Over the last decade, implementation of pharmacogenomics in the clinical setting has gained momentum, largely due to the availability of evidence-based clinical practice guidelines that provide recommendations for translating pharmacogenomic test results into actionable prescribing decisions for select medications [2–4]. The implementation of pharmacogenomics spans a variety of different facilities (e.g., academic health centers, community health systems) and settings (e.g., inpatient, outpatient) [5–7].
Currently, two types of testing models, reactive or preemptive, are used in clinical pharmacogenomic implementation initiatives. Reactive testing occurs at the time a drug is prescribed and usually involves genotyping a few variants within a gene of interest. In contrast, some institutions are moving toward a preemptive testing model, whereby genotyping occurs before it may be needed, and the results are stored in the electronic health record (EHR) for future use [8]. Preemptive testing typically occurs using a panel that interrogates many pharmacovariants spanning many genes. For preemptive testing to be maximally useful, structured results should be entered into the EHR and coupled with automated clinical decision support (CDS) tools, such that pharmacogenomic test results and clinical recommendations are available to the provider at the point of care when a medication is prescribed at any time following testing.
Most institutions conduct pharmacogenomic implementation either as a clinical initiative or under the auspices of a research study [9–22]. In recent years, the genomics community has witnessed the growth of large biobanks, which collect DNA for research purposes [23–25]. Depending on how and where the samples are genotyped, biobanks offer the unique opportunity to systematically return actionable genomic results to the clinical setting over time. This ‘hybrid’ approach is advantageous because it simultaneously facilitates genomic research and clinical implementation initiatives, thus promoting the efficient and cost-effective use of institutional resources. Here, we describe a preemptive clinical pharmacogenomic implementation initiative via a health system-wide research biobank at the University of Colorado. Some questions that we sought to answer during this initiative were:
How do we facilitate the use of pharmacogenomic results in direct patient care, when these results are generated as part of a biobank research study?
How can the results of preemptive genotyping be returned to the EHR to support future timely clinical decision-making based on evolving pharmacogenomic knowledge?
What pharmacogenomic results and medications should be targeted first in a heterogeneous biobank population?
What is the best way to educate clinicians and patients about this unique pharmacogenomic implementation approach?
What resources are required to design and deploy scalable EHR-based pharmacogenomic CDS tools across a large healthcare system for this initiative?
What are the challenges and lessons learned from delivering clinical pharmacogenomic results to clinicians and patients via this hybrid model?
The University of Colorado Anschutz Medical Campus
The University of Colorado Anschutz Medical Campus, located in Aurora, Colorado, is the largest academic health center in the Rocky Mountain region. The campus includes six health professional schools, multiple centers and institutes, and two nationally ranked hospitals, University of Colorado Hospital (part of the UCHealth system) and Children’s Hospital Colorado. UCHealth is divided into three regions (Metro Denver, Northern Colorado and Southern Colorado) and spans the state of Colorado and portions of Wyoming and Nebraska. UCHealth includes 13 hospitals, approximately 600 clinics, covers over 5 million patient lives, and operates on a single instance of the Epic EHR (Epic Systems, WI, USA). UCHealth has grown through mergers, acquisitions and new construction, resulting in a variety of clinical workflows, both new and historically well established.
The Colorado Center for Personalized Medicine
The Colorado Center for Personalized Medicine (CCPM) was established in 2014 as a collaboration between the University of Colorado, UCHealth, CU Medicine, and Children’s Hospital Colorado. The mission of CCPM is to apply personalized medicine research, education, and clinical care across diseases to accelerate the development and application of personally tailored prevention, diagnosis, and treatment strategies. The academic home for faculty working within CCPM is the Division of Biomedical Informatics and Personalized Medicine, housed within the University of Colorado School of Medicine, with faculty contributing from multiple divisions and departments across the Schools of Medicine, Pharmacy, and Public Health.
In 2015, the CCPM, in partnership with UCHealth, established the CCPM Biobank Research Study to facilitate large-scale genomics and other ‘omics’-based research to advance personalized medicine for UCHealth patients across the Rocky Mountain region (www.cobiobank.org/). As of January 2020, over 120,000 participants have enrolled in the CCPM Biobank Research Study and approximately 53,000 of those individuals have had a blood sample collected for genomic analysis. Key operational units within CCPM include: the Clinical Laboratory Improvement Amendments (CLIA)-certified Biobank Laboratory (described below); the enterprise data warehouse, Health Data Compass; and the computing infrastructure, the Translational Informatics and Computational Resource (TICR). Health Data Compass is a multi-institutional data warehouse that integrates, harmonizes, and links a variety of large-scale datasets, including clinical data from the EHRs of two health systems (greater than 6.8 million patient records) and genomic data from the Biobank Research Study (www.healthdatacompass.org/). TICR manages a high-performance computing cluster and oversees the clinical workflow between the Biobank Laboratory and a third-party commercial genomic data management platform, BC Platforms (www1.ucdenver.edu/offices/office-of-information-technology/ticr-high-performance-computing).
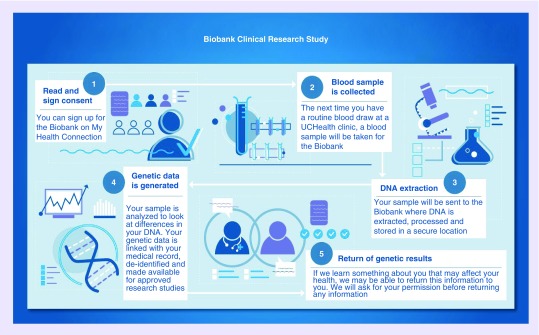
A schematic of the Biobank Research Study process is shown in Figure 1. Briefly, UCHealth patients aged 18 years or older sign an electronic research consent form through their online UCHealth patient portal (‘My Health Connection’), which triggers an order to collect a dedicated biospecimen for future research. The blood sample is obtained at the patient’s next routine blood test at a UCHealth-affiliated site. The use of a robust, high-traffic EHR for the ‘opt-in’ process allows for rapid recruitment, low overhead cost, and the ability to leverage existing health system resources to collect research specimens in a clinical setting. Subsequently, Biobank Research Study blood samples are sent to the Biobank Laboratory, located on the University of Colorado Anschutz Medical Campus, where DNA is extracted and genotyped. The Biobank Laboratory uses a customized version of the Illumina® Infinium® Expanded Multi-Ethnic Genotyping Array (MEGAEX, hereafter referred to as ‘MEGA’). The array tests for the presence of nearly 2.1 million genetic variants, primarily single nucleotide variants and some small indels [26–28]. The MEGAEX contains over 17,000 variants chosen to be relevant to clinical and pharmacogenomic studies and has the capacity to add 300,000 custom markers to the array [26]. As of December 2019, approximately 30,000 Biobank Research Study samples have been genotyped on the MEGA.
Figure 1. . Schematic of the Colorado Center for Personalized Medicine Biobank Research Study.
This schematic illustrates the components of the Colorado Center for Personalized Medicine Biobank Research Study, as explained to potential participants. Participants sign an electronic research consent form through ‘My Health Connection,’ the online UCHealth patient portal. This triggers an order to collect a dedicated biospecimen for future research at the patient's next routine blood draw. Samples are sent to the CLIA-certified Biobank Laboratory, where DNA is extracted and genotyped. When clinical genetic test results are available, participants are sent an electronic secondary clinical consent form through their online patient portal asking permission to return these results to the EHR. Pharmacogenomic results are the first type of clinical genetic test results being preemptively returned to the EHR for this initiative.
Permission for use of this figure granted by Kathleen Barnes.
CLIA: Clinical Laboratory Improvement Amendments; EHR: Electronic health record.
Unlike other biobanks across the country, the CCPM Biobank Laboratory is CLIA-certified for high-complexity testing, thus providing the ability to return clinical genetic test results back to Biobank Research Study participants and the EHR. When clinical genetic test results are available, Biobank Research Study participants are sent an electronic secondary clinical consent form through their online patient portal asking permission to preemptively return these results to the EHR and their patient portal. If a patient declines permission, their results are used for research purposes only. Given the challenges associated with coordinating and maintaining an electronic two-consent process for a large biobank, CCPM moved to a unified (single) consent process, which includes permissions for both research and the return of clinical results, for new participants as of October 2019. There are several types of clinical genetic test results that may be returned to participants and the EHR, including pharmacogenomic information, genetic diagnoses, carrier status, and predictors of disease risk (e.g., secondary or incidental findings). Pharmacogenomic results were the first type of clinical genetic test developed for preemptive return of results to the EHR.
The Pharmacogenomics Implementation Committee Colorado
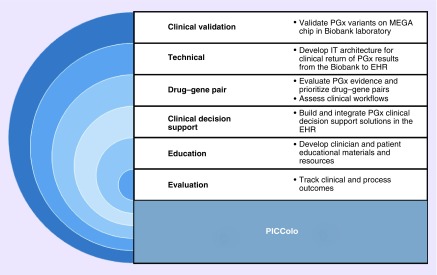
The CCPM Pharmacogenomics Implementation Committee Colorado (PICColo) was formed in August 2016 with the goal of making selected pharmacogenomic test results for CCPM Biobank Research Study participants available for clinical use in UCHealth's EHR. PICColo is a multidisciplinary committee, comprised of approximately 40 members from UCHealth, University of Colorado School of Medicine, University of Colorado Skaggs School of Pharmacy and Pharmaceutical Sciences, University of Colorado College of Nursing, and the Colorado School of Public Health. PICColo is co-led by two faculty members from the Schools of Pharmacy and Medicine. The multidisciplinary nature of the committee has proven to be advantageous, particularly during the early stages of the initiative, when diverse expertise was required to navigate the challenges associated with preemptively returning clinical results from a research biobank. As PICColo has grown and matured, so have its development processes; it has transitioned from brainstorming to collaborative problem solving to strategic design and implementation. Currently, PICColo activities are carried out by six working groups, as outlined in Figure 2. In addition, PICColo leaders meet regularly with executive-level leaders at UCHealth, which has been an invaluable means to garner institutional support and obtain feedback throughout the process.
Figure 2. . Pharmacogenomics Implementation Committee Colorado working groups and associated activities.
This schematic describes the six working groups, which carry out the activities of PICColo.
EHR: Electronic health record; IT: Information technology; MEGA: Multi-ethnic genotyping array; PGx: Pharmacogenomics; PICColo: Pharmacogenomics Implementation Committee Colorado.
The majority of PICColo’s efforts during its first two years were spent building the end-to-end information technology (IT) infrastructure needed to send structured genetic results from the Biobank Laboratory to the EHR. At our institution, genetic results have historically been stored in the EHR as a mixture of structured results and free text or scanned reports, many of which are unsuitable for search and triggering functions for CDS applications. Consequently, the prior system would not be able to deliver pharmacogenomic information to clinicians at the point of care. Therefore, we invested substantial effort upfront to return genetic results as structured data elements within the EHR so we could leverage state-of-the-art CDS tools to engage as many clinicians as possible, particularly nongeneticists.
The genomic results pipeline involves several key components: the Biobank Laboratory, which generates raw genomic results from the MEGA, including validated pharmacogene results; a Research Electronic Data Capture (REDCap) report from Health Data Compass that matches samples to UCHealth patient identifiers; ©BC Platforms (Helsinki, Finland), a third-party commercial genomic data management platform that generates variant allele calls; and UCHealth’s EHR. Each interface requires dedicated effort to develop and test the return of results, maintain linkage with patient identifiers, and implement security protocols to comply with the Health Insurance Portability and Accountability Act (HIPAA). Results are transferred from the genomic data management platform to the EHR via EHR Application Programming Interfaces and inserted into custom-built lab result records with identifiers unique to the gene in question. Results are visible in the same interfaces as other lab results because they are structured to be used in pharmacogenomic CDS rules. Clinical decisions generated as a result of pharmacogenomic CDS tools are captured within the EHR and returned to Health Data Compass, the local data warehouse via previously constructed protocols, thereby completing a true learning healthcare system.
Once a structured pharmacogenomic result is available in the EHR, it can be used in four primary types of electronic CDS tools: hyperlinks to external resources such as clinical tip sheets and guidelines; passive advisories (text embedded in lists of patient-specific suggestions in the navigation sections of the EHR that require clinicians to seek out the information); inline medication order warnings (passive suggestions that are visible within the routine clinician workflow but are not disruptive); and interruptive advisories (pop-up alerts that must be acknowledged to continue workflow). All applications utilize native EHR functionality and both the passive and interruptive advisories include the capacity to offer alternative orders to that originally placed. We have found that a multimodal CDS approach is sometimes necessary to accommodate a variety of clinicians’ workflows (e.g., family practice vs psychiatrists). These IT infrastructures and CDS technologies form the basis of our drug–gene pair development process, as described below.
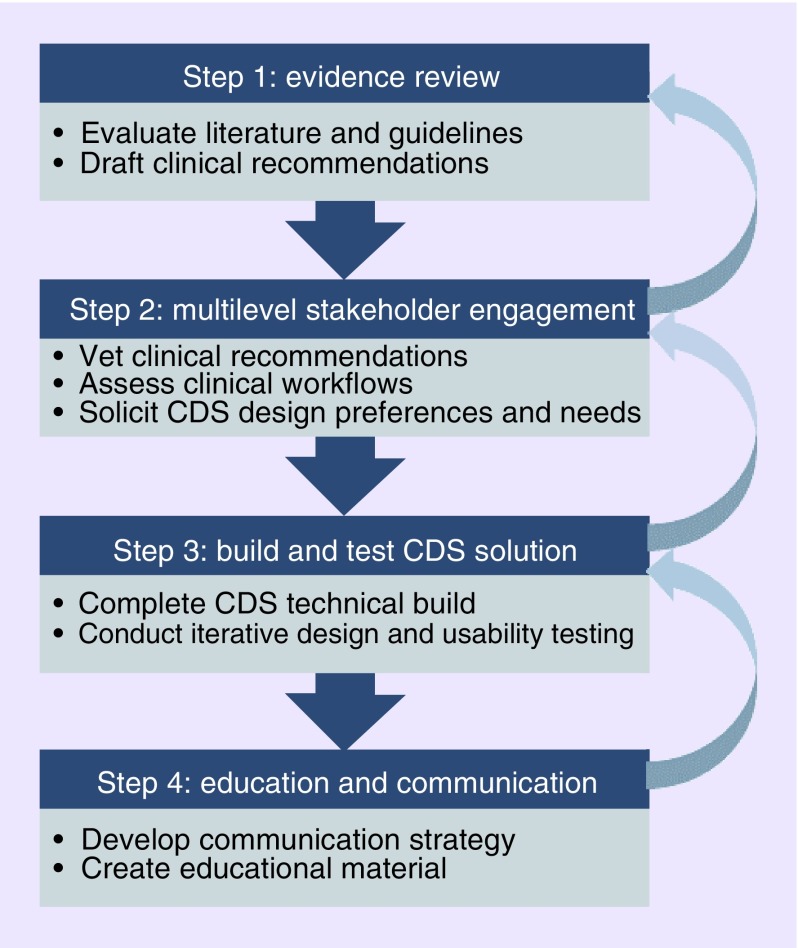
The CCPM drug–gene pair development process follows a systematic approach that is grounded in the Practical, Robust Implementation and Sustainability Model (PRISM) and best practices of CDS design [29–33]. As depicted in Figure 3, this is an iterative approach which includes: an in-depth review of the evidence and formulation of clinical recommendations; multilevel stakeholder engagement to endorse clinical recommendations, assess clinical workflows, and solicit CDS preferences; a user-centered process involving technical, design, and usability testing; and education and dissemination. Multilevel stakeholder engagement includes input from leadership at all levels, management, subject matter experts, and end users. Leadership and management are asked about internal and external drivers that could influence the implementation. Clinical subject matter experts are asked to weigh in on the evidence and associated clinical recommendations (e.g., Clinical Pharmacogenetics Implementation Consortium [CPIC] Guidelines). End users are asked how pharmacogenomic CDS tools could be best integrated into their current workflows.
Figure 3. . Pharmacogenomics Implementation Committee Colorado systematic drug–gene pair development process.
This figure describes the four steps that PICColo uses to develop new drug–gene pairs for implementation. The arrows on the right highlight the iterative nature of this process.
CDS: Clinical decision support; PICColo: Pharmacogenomics Implementation Committee Colorado.
The stakeholder engagement process helps define the general scope of construction of the CDS tools, including how the information is displayed in the user interface and how the CDS tools interface with clinicians within their workflow. Prior to deployment, the CDS solution undergoes significant design and usability testing to ensure that the content and format reflect stakeholders’ needs and preferences and to mitigate features that cause confusion or limit ease of use. Working in concert with multiple stakeholder groups is not without its challenges and requires continual awareness of the need to balance ‘need to have’ and ‘nice to have’ elements in clinical recommendations and CDS tools. Geographic, specialty-based, and clinician-specific implementation strategies have proven useful for early testing and deployment.
First pharmacogenomic drug–gene pair use case: clopidogrel & CYP2C19 variants
We selected the CYP2C19*2, *3 and *17 variants as the first pharmacogenomic results to return to the EHR. This decision was based on the known functional significance of these variants (i.e., no function or increased function alleles), their performance and validation on the MEGA, and the likelihood of detecting the variant alleles in our Biobank Research Study population. Clopidogrel was the first CYP2C19-affected drug selected for implementation. Clopidogrel is an antiplatelet agent used to treat patients with acute coronary syndromes (ACS), particularly those undergoing percutaneous coronary intervention (PCI) [34]. As a prodrug, clopidogrel is metabolized, in part by CYP2C19, to an active metabolite [34]. Studies have shown that certain variants in the CYP2C19 gene can cause enzyme abnormalities that interfere with the metabolism of clopidogrel, which puts affected patients at risk for thrombotic events following ACS or PCI [34,35]. At our institution, clopidogrel was an ideal medication to start with based on its prescribing frequency, the strength of the pharmacogenomic evidence, the availability of CPIC guidelines, a boxed warning on the US FDA approved label regarding CYP2C19 variants, a clear clinical scenario, and a well-defined provider group. As such, we set out to develop a CDS tool for clopidogrel in relation to CYP2C19 genotype in the setting of ACS and PCI.
Our initial target population was Biobank Research Study participants undergoing elective PCI at the UCHealth metro-area cardiac catheterization laboratory. We chose to start with a narrow indication and a focused setting because this was the first pharmacogenomic implementation of its kind at our institution. Subsequently, we expanded the geographic catchment area to include UCHealth cardiac catheterization laboratories in the Northern and Southern Colorado regions. We then expanded the indication to include patients with acute ACS and PCI at all three major UCHealth inpatient settings (i.e., Metro Denver, Northern Colorado and Southern Colorado regions). This staggered approach, both in terms of geographic location and indication (i.e., elective vs acute ACS and PCI), was advantageous because it allowed us to intensively engage a small group of clinicians early in the drug–gene pair development process, with progressive stakeholder engagement and expansion across the health system. This multilevel stakeholder engagement process also served a dual purpose, providing an efficient means to educate and disseminate knowledge about our initiative.
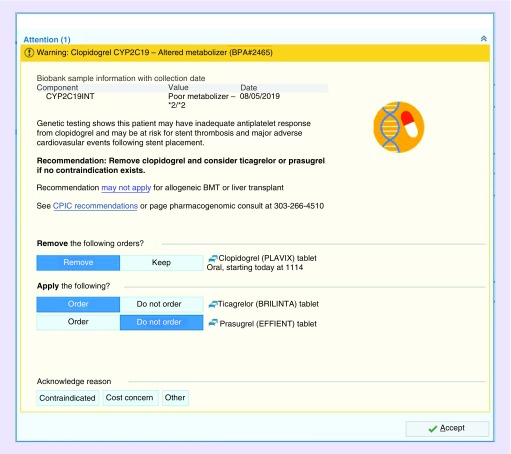
In the case of clopidogrel and CYP2C19, the CDS solution is an interruptive alert that notifies a prescribing clinician when they order clopidogrel in the inpatient setting for a patient who is a genetically mediated CYP2C19 intermediate or poor metabolizer and has had a documented PCI within the past year. The CDS alert identifies the problem, provides an actionable recommendation to change the medication order, displays pertinent information to endorse the recommendation (e.g., genotype, phenotype, and link to references), and provides the clinician with options to explain their reason(s) for not following the recommendation (Figure 4). The actionable recommendation includes the option to cancel the order for clopidogrel and choose one of two suggested alternatives (ticagrelor or prasugrel), thus making it easier for clinicians to follow the recommendation. Given the acuity and risks of the clinical situation, users cannot dismiss the alert and instead are required to either change their choice of an antiplatelet agent or provide a reason for their decision not to make a change. Although our institution generally prefers not to use interruptive CDS alerts, this type of alert was deemed necessary in this instance based on the patient care situation and a collective request from the clinical stakeholders.
Figure 4. . Representative interruptive clinical decision support alert for clopidogrel-CYP2C19 at UCHealth.
The clinical decision support alert notifies a prescribing clinician when they order clopidogrel in the inpatient setting for a patient who is a genetically mediated CYP2C19 intermediate or poor metabolizer and has had a documented percutaneous coronary intervention within the past year. In this example, the patient is a CYP2C19 poor metabolizer (*2/*2 genotype). The clinical decision support alert provides an actionable recommendation to change the medication order, displays pertinent information to endorse the recommendation (e.g., link to references), and provides acknowledge reason options if the clinician chooses not to follow the recommendation.
BMT: Bone marrow transplant; CPIC: Clinical Pharmacogenetics Implementation Consortium.
To increase the potential impact of the clopidogrel-CYP2C19 pharmacogenomic initiative, clinical cardiovascular characteristics of Biobank Research Study participants were extracted from Health Data Compass with the goal of identifying those at highest risk for future cardiovascular events. A cohort of 26,879 patients were identified from a 2-year period, between 2016 and 2018 (mean age, 50 ± 16 years; female, 64.5%; European–American, 78.6%; African–American, 4.7%; Asian, 2%; Hispanic, 7.7%). Approximately 75% of the cohort were active users of the electronic patient portal and 95.4% had a recent encounter at UCHealth. Clinical cardiovascular characteristics are shown in Table 1. CYP2C19 phenotype frequencies in a representative subset (n = 1141) of the population were: ultrarapid (4%), rapid (24%), normal (43%), intermediate (26%) and poor (3%) metabolizers. Based on this information, the first 900 clinical consents were released to participants with a history of at least one of the following: PCI in the last year at UCHealth, coronary artery bypass grafting, cardiac stent thrombosis, myocardial infarction, or coronary heart disease.
Table 1. . Clinical cardiovascular characteristics of Biobank Research Study participants (N = 26,879).
| Cardiovascular or cardiovascular risk conditions | n (%) |
|---|---|
| Coronary heart disease | 2771 (10.3%) |
| Cerebrovascular disease | 1549 (5.8%) |
| Peripheral vascular disease | 899 (3.3%) |
| Myocardial infarction | 726 (2.7%) |
| Hypertension | 9709 (36.1%) |
| Type 1 or type 2 diabetes | 5378 (20.0%) |
| Percutaneous coronary intervention | 658 (2.5%) |
| Stent thrombosis | 37 (0.14%) |
Participants could have one or more of these conditions.
In terms of benefits, the clopidogrel-CYP2C19 use case has positively affected socialization of the Biobank Research Study return of results initiative across our health system. Specifically, the implementation of a drug–gene pair with strong evidence in a high-impact patient population (ACS/PCI) garnered substantial provider- and executive-level support for the concept of personalized medicine. In implementing this use case, we established a foundational development framework for stakeholder engagement, user-centered application design, and predeployment technical testing that we now use for building all of our pharmacogenomic applications. Since the initial gene we selected also affects several other clinically actionable medications, we were able to rapidly develop additional pharmacogenomic CDS tools, thus increasing the potential reach of this initiative, as described below.
Current pharmacogenomic initiatives & future plans
Following the clopidogrel-CYP2C19 pharmacogenomic initiative, institutional leaders directed CCPM to develop and deploy CDS tools for all clinically actionable (CPIC Level A) medications affected by CYP2C19 genetics, across the UCHealth system over roughly 6 months (Figure 5). This ‘one gene, many drugs’ approach within a finite period of time allowed for rapid CDS prototyping and deployment, given that structured results for this gene already existed in the EHR. The trade-off of this tactic was the need for increased resources upfront for stakeholder engagement, because the affected medications cut across many therapeutic areas and settings. However, this method proved advantageous in our academic setting because it prevented the ‘siloing’ of implementation efforts in one therapeutic area (e.g., cardiology), thus socializing the pharmacogenomic initiative broadly across the health system in a relatively short period of time.
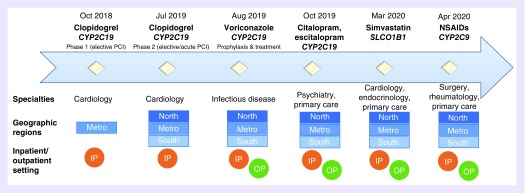
Figure 5. . Timeline of Pharmacogenomics Implementation Committee Colorado clinical decision support implementation.
Over approximately 18 months, the PICColo team designed and deployed pharmacogenomic CDS tools for nine drug–gene pairs (some in progress) across multiple specialties, geographic regions, and care settings in the UCHealth system.
CDS: Clinical decision support; PCI: Percutaneous coronary intervention; IP: Inpatient; NSAID: Nonsteroidal anti-inflammatory drugs (includes celecoxib, meloxicam, ibuprofen and piroxicam); OP: Outpatient; PICColo: Pharmacogenomics Implementation Committee Colorado.
Reprinted with permission, © 2019 Epic Systems Corporation.
Our drug–gene pair work-up and CDS development processes incorporated principles of Agile software development, such as iterative design, simplicity, user-centric focus, self-organizing teams, and collaboration [36,37]. Using this approach, we built and deployed interruptive CDS alerts for the antifungal, voriconazole (prophylaxis and treatment indications), and CYP2C19 in outpatient and inpatient settings across the UCHealth system. Next, we built and deployed a multimodal CDS solution for the selective serotonin reuptake inhibitors, citalopram and escitalopram, and CYP2C19 in outpatient and inpatient settings across the system. Sertraline, another selective serotonin reuptake inhibitor, was not included in this implementation, based on feedback from stakeholders that existing evidence was not strong enough to warrant clinical action.
The citalopram/escitalopram multimodal CDS solution consisted of a passive (noninterruptive) advisory that captured participants already prescribed these medications and a passive (noninterruptive) inline medication order warning that is displayed for new or renewed citalopram or escitalopram prescriptions. This multimodal approach was favored by stakeholder groups and addressed different steps in the continuum of prescribing and monitoring these drugs. We also initiated the process of stakeholder engagement for proton pump inhibitors and CYP2C19 but are awaiting publication of the CPIC guideline before embarking on the CDS design process. We have begun the initial stages of stakeholder outreach for medications that have CYP2C19 genetic information in their FDA-approved labeling but do not have CPIC guidelines (e.g., clobazam and brivaracetam).
The next pharmacogenes and variants we selected for integration into the EHR were SLCO1B1 (rs4149056 variant) and CYP2C9 (*2 and *3). This decision was based on the functional significance of these variants, their performance on the MEGA (validation currently underway), and the likelihood of detecting the variant alleles in our Biobank Research Study population. We are in the process of CDS usability testing for a passive (noninterruptive) inline medication order warning for simvastatin, a cholesterol-lowering medication, and SLCO1B1. We are also in the stakeholder engagement phase for selected nonsteroidal anti-inflammatory drugs (celecoxib, meloxicam, ibuprofen and piroxicam) and CYP2C9, which will likely also result in a passive (noninterruptive) inline medication order warning. Based on our work thus far, we are in the process of creating an evaluation plan using the RE-AIM framework, which is part of PRISM [32,38].
Innovations, challenges & lessons learned
Throughout this pharmacogenomic initiative, several innovations were fostered at the UCHealth system. We established a multidisciplinary team, comprised of clinical, pharmacogenomic, and technical experts from the University of Colorado and UCHealth, which allowed for rapid cycle innovation and the ability to change course in response to operational and technical barriers. Aligned with the user-centered design process and PRISM, our team led nearly 100 multilevel stakeholder meetings to introduce pharmacogenomics to practicing clinicians and inform them of the CDS tools they would see in the EHR. This tactic proved to be an efficient way to simultaneously obtain stakeholder buy-in and educate clinicians about the initiative. When coupled with widespread dissemination via our EHR provider newsletter and an internal provider education website, we were able to adopt a streamlined educational approach for clinicians that was consistent with other large-scale EHR-based implementation initiatives at our institution. We developed creative CDS solutions that moved beyond interruptive alerts and that could be deployed across the UCHealth system to accommodate heterogeneous clinical workflows, thus streamlining future CDS management efforts. In addition, we delivered pharmacogenomic results directly back to patients via the online patient portal with links to additional information about the meaning of these results and implications for their care.
We also met our share of challenges and lessons learned during this process [7]. Some stakeholders questioned whether the ‘one gene, many drugs’ approach was the most efficient way to construct the program. Our rationale for doing this was largely to prioritize resources and develop a solid implementation framework during the early stages of the initiative. We first identified genes that were adequately covered by the MEGA platform, second determined the clinical impact (e.g., number of patients, number of prescriptions, and potential for clinical benefit), and then obtained stakeholder input. By establishing this systematic developmental process up front, the resulting pipeline will allow us to expand to multiple genes and drug–gene pairs simultaneously in the future. The applicability of this approach at other institutions will ultimately depend on the technical and human resources that can be devoted to pharmacogenomics at any given institution.
A major lesson learned was that, across a large health system, clinical workflows vary extensively. For a given drug–gene pair, our ultimate goal was to build a single CDS solution that worked seamlessly across the system; however, in some cases technical builds differed because of upstream differences in clinical workflows or formularies. For example, in delivering clopidogrel CDS alerts for post-PCI patients with at-risk CYP2C19 genotypes, clinicians in different regions and provider groups place orders or order sets at different times within the clinical encounter. In addition, the format and content of order sets vary across the health system. Accordingly, we had to take this significant variation into account when designing the CDS (e.g., different ‘rules’ or ‘triggers’), and in some cases, we had to change the technical build to accommodate these differences.
We also learned that choosing the right scope and message for the project can make all the difference in its success. For example, at numerous times, initial conversations about the pharmacogenomic initiative were sidetracked by concerns about genetic testing, which were largely a result of stakeholders' experiences with genetic testing for diseases. Conflating these two categories of results (pharmacogenomics and disease risk) sometimes stopped the discussion about pharmacogenomics during governance meetings. As we moved forward with the initiative, we explicitly delineated the risks associated with different types of clinical genetic testing and developed consistent messaging to describe the scope of the initiative, which helped to address pharmacogenomic exceptionalism.
Another important lesson learned during this effort was that obtaining buy-in and approval from subject matter experts and end users is a dynamic and interactive process. Buy-in from subject matter experts is essential to validating clinical need, while buy-in from end users is necessary to ensure the acceptance and practicality of the CDS tool and associated clinical actions. One challenge we faced when engaging these two stakeholder groups was ensuring that the CDS solution promoted autonomy of decision-making while preserving accuracy and patient safety. At times, subject matter experts and end users had differing opinions, which was difficult to navigate. We worked to strike a balance between autonomy and patient safety. To preserve end user autonomy, a CDS solution that solely recommended consultation of or referral to a subject matter expert was avoided, when possible. In the future, engaging subject matter experts and end users collaboratively rather than separately may help mitigate some of these challenges.
A major challenge identified in this process is the sustainability of preemptive genotyping at scale. While it is anticipated that eventually reimbursement processes and the full engagement of payers will be widely implemented across healthcare systems, to date healthcare systems that have successfully implemented genotype-guided drug therapy have relied on industry-led partnerships and federal/private funding for preemptive genotyping initiatives. Another limitation of our program is the reliance on a genotyping platform for which a substantial number of clinically actionable pharmacogenes (e.g., CYP2D6) are not available. Improvements in genotyping platforms that incorporate structural variant calling and integrating next-generation sequencing platforms with array-based platforms have the potential to broaden the deliverable of drug–gene pairs.
Finally, clinical return of pharmacogenomic results from the Biobank Research Study is unsolicited. This means that neither the patient nor the treating clinician requested the result, nor are they expecting to see it. We frequently grappled with the question of how a healthcare organization should effectively deliver a result that is not requested. Along these lines, the most common ethical concern we encountered was the potential return of CYP2C19 genetic results to the EHR without having CDS tools in place for all actionable CYP2C19 medications. Specifically, return of these results without clinician awareness raised questions regarding the clinical and legal liability of not using these data in decision-making. To obviate these concerns, we developed and turned on point-of-prescribing CDS tools for CPIC Level A medications affected by CYP2C19 genetics well before a substantial number of genetic results were returned to the EHR. We also created a website to educate clinicians and patients about the Biobank Research Study, pharmacogenomic results, and their potential impact on patient care (www.cobiobank.org).
Conclusion
In summary, we developed a preemptive clinical pharmacogenomic implementation initiative via a health system-wide research biobank at the University of Colorado. As research biobanking initiatives continue to grow, investigators should proactively consider the utility of a hybrid implementation model. Preemptive implementation of clinical pharmacogenomic results via a research biobank is feasible, particularly when coupled with strong institutional support to maximize the impact and efficiency of biobank resources, a multidisciplinary implementation team, and proactive strategies to engage stakeholders early in the CDS development process.
Acknowledgments
Portions of this work were presented, in part, as an abstract at the Clinical Pharmacogenetics Implementation Consortium (CPIC) Open Meeting in Memphis, Tennessee, June 2019. The authors would like to thank members of the Pharmacogenomics Implementation Committee Colorado (past and present); N da Silva, J Eväsoja, and B Jackson from BC Platforms; and T Deppe, J Grabau, and E Giraud from Illumina, Inc. for their contributions to this initiative. The authors acknowledge the editorial services of EL Langmack.
Footnotes
Financial & competing interests disclosure
This work was supported, in part, by NIH grants R01HL104608-01 (KCB), K08HL125725 (DPK), and R35GM124939 (AAM). Contents are the authors' sole responsibility and do not necessarily represent official NIH views. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Open access
This work is licensed under the Attribution-NonCommercial-NoDerivatives 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
References
- 1.Relling MV, Evans WE. Pharmacogenomics in the clinic. Nature 526(7573), 343–350 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Relling MV, Klein TE, Gammal RS, Whirl-Carrillo M, Hoffman JM, Caudle KE. The Clinical Pharmacogenetics Implementation Consortium: 10 years later. Clin. Pharmacol. Ther. 107(1), 171–175 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caudle KE, Klein TE, Hoffman JM. et al. Incorporation of pharmacogenomics into routine clinical practice: the Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline development process. Curr. Drug Metab. 15(2), 209–217 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the pharmacogenomics research network. Clin. Pharmacol. Ther. 89(3), 464–467 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weitzel KW, Elsey AR, Langaee TY. et al. Clinical pharmacogenetics implementation: approaches, successes, and challenges. Am. J. Med. Genet. C Semin. Med. Genet. 166C(1), 56–67 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klein ME, Parvez MM, Shin JG. Clinical implementation of pharmacogenomics for personalized precision medicine: barriers and solutions. J. Pharm. Sci. 106(9), 2368–2379 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Krebs K, Milani L. Translating pharmacogenomics into clinical decisions: do not let the perfect be the enemy of the good. Hum. Genomics 13(1), 39 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunnenberger HM, Crews KR, Hoffman JM. et al. Preemptive clinical pharmacogenetics implementation: current programs in five US medical centers. Annu. Rev. Pharmacol. Toxicol. 55, 89–106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pulley JM, Denny JC, Peterson JF. et al. Operational implementation of prospective genotyping for personalized medicine: the design of the Vanderbilt PREDICT project. Clin. Pharmacol. Ther. 92(1), 87–95 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'donnell PH, Bush A, Spitz J. et al. The 1200 patients project: creating a new medical model system for clinical implementation of pharmacogenomics. Clin. Pharmacol. Ther. 92(4), 446–449 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bielinski SJ, Olson JE, Pathak J. et al. Preemptive genotyping for personalized medicine: design of the right drug, right dose, right time-using genomic data to individualize treatment protocol. Mayo Clin. Proc. 89(1), 25–33 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rasmussen-Torvik LJ, Stallings SC, Gordon AS. et al. Design and anticipated outcomes of the eMERGE-PGx project: a multicenter pilot for preemptive pharmacogenomics in electronic health record systems. Clin. Pharmacol. Ther. 96(4), 482–489 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shuldiner AR, Palmer K, Pakyz RE. et al. Implementation of pharmacogenetics: the University of Maryland Personalized Anti-platelet Pharmacogenetics Program. Am. J. Med. Genet. C Semin. Med. Genet. 166C(1), 76–84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffman JM, Haidar CE, Wilkinson MR. et al. PG4KDS: a model for the clinical implementation of pre-emptive pharmacogenetics. Am. J. Med. Genet. C Semin. Med. Genet. 166C(1), 45–55 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weitzel KW, Alexander M, Bernhardt BA. et al. The IGNITE network: a model for genomic medicine implementation and research. BMC Med. Genomics 9, 1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hicks JK, Stowe D, Willner MA. et al. Implementation of Clinical Pharmacogenomics within a large health system: from electronic health record decision support to consultation services. Pharmacotherapy 36(8), 940–948 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Scott SA, Owusu Obeng A, Botton MR. et al. Institutional profile: translational pharmacogenomics at the Icahn School of Medicine at Mount Sinai. Pharmacogenomics 18(15), 1381–1386 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cavallari LH, Weitzel KW, Elsey AR. et al. Institutional profile: University of Florida Health Personalized Medicine Program. Pharmacogenomics 18(5), 421–426 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Wouden CH, Cambon-Thomsen A, Cecchin E. et al. Implementing pharmacogenomics in Europe: design and implementation strategy of the Ubiquitous Pharmacogenomics Consortium. Clin. Pharmacol. Ther. 101(3), 341–358 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Dressler LG, Bell GC, Ruch KD, Retamal JD, Krug PB, Paulus RA. Implementing a personalized medicine program in a community health system. Pharmacogenomics 19(17), 1345–1356 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Volpi S, Bult CJ, Chisholm RL. et al. Research directions in the clinical implementation of pharmacogenomics: an overview of US programs and projects. Clin. Pharmacol. Ther. 103(5), 778–786 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramsey LB, Prows CA, Zhang K. et al. Implementation of pharmacogenetics at Cincinnati Children's Hospital Medical Center: lessons learned over 14 years of personalizing medicine. Clin. Pharmacol. Ther. 105(1), 49–52 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reisberg S, Krebs K, Lepamets M. et al. Translating genotype data of 44,000 biobank participants into clinical pharmacogenetic recommendations: challenges and solutions. Genet. Med. 21(6), 1345–1354 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gottesman O, Kuivaniemi H, Tromp G. et al. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet. Med. 15(10), 761–771 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collins FS, Varmus H. A new initiative on precision medicine. N. Engl. J. Med. 372(9), 793–795 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Illumina, Inc. Infinium® Expanded Multi-Ethnic GenotypingArray (MEGAEX) (2020). www.illumina.com/content/dam/illumina-marketing/documents/products/datasheets/mega-ex-data-sheet-370-2015-004.pdf
- 27.Johnston HR, Hu YJ, Gao J. et al. Identifying tagging SNPs for African specific genetic variation from the African Diaspora Genome. Sci. Rep. 7, 46398 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wojcik GL, Graff M, Nishimura KK. et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature 570(7762), 514–518 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bates DW, Kuperman GJ, Wang S. et al. Ten commandments for effective clinical decision support: making the practice of evidence-based medicine a reality. J. Am. Med. Inform. Assoc. 10(6), 523–530 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horsky J, Schiff GD, Johnston D, Mercincavage L, Bell D, Middleton B. Interface design principles for usable decision support: a targeted review of best practices for clinical prescribing interventions. J. Biomed. Inform. 45(6), 1202–1216 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Marcilly R, Ammenwerth E, Roehrer E, Nies J, Beuscart-Zephir MC. Evidence-based usability design principles for medication alerting systems. BMC Med. Inform. Decis. Mak. 18(1), 69 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feldstein AC, Glasgow RE. A practical, robust implementation and sustainability model (PRISM) for integrating research findings into practice. Jt. Comm. J. Qual. Patient Saf. 34(4), 228–243 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Osheroff JA. Healthcare Information and Management Systems Society. Improving Outcomes with Clinical Decision Support: An Implementer's Guide (2nd Edition). HIMSS, IL, USA: (2012). [Google Scholar]
- 34.Scott SA, Sangkuhl K, Stein CM. et al. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin. Pharmacol. Ther. 94(3), 317–323 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klein MD, Williams AK, Lee CR, Stouffer GA. Clinical utility of CYP2C19 genotyping to guide antiplatelet therapy in patients with an acute coronary syndrome or undergoing percutaneous coronary intervention. Arterioscler. Thromb. Vasc. Biol. 39(4), 647–652 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Kane DW, Hohman MM, Cerami EG, Mccormick MW, Kuhlmman KF, Byrd JA. Agile methods in biomedical software development: a multi-site experience report. BMC Bioinformatics 7, 273 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beck K, Beedle M, Bennekum VA . et al. (2019). www.agilemanifesto.org
- 38.Glasgow RE, Vogt TM, Boles SM. Evaluating the public health impact of health promotion interventions: the RE-AIM framework. Am. J. Public Health 89(9), 1322–1327 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]