Abstract

Biofilm formation is most commonly combatted with antibiotics or biocides. However, proven toxicity and increasing resistance of bacteria increase the need for alternative strategies to prevent adhesion of bacteria to surfaces. Chemical modification of the surfaces by tethering of functional polymer brushes or films provides a route toward antifouling coatings. Furthermore, nanorough or superhydrophobic surfaces can delay biofilm formation. Here we show that submicrometer-sized roughness can outweigh surface chemistry by testing the adhesion of E. coli to surfaces of different topography and wettability over long exposure times (>7 days). Gram-negative and positive bacterial strains are tested for comparison. We show that an irregular three-dimensional layer of silicone nanofilaments suppresses bacterial adhesion, both in the presence and absence of an air cushion. We hypothesize that a 3D topography can delay biofilm formation (i) if bacteria do not fit into the pores of the coating or (ii) if bending of the bacteria is required to adhere. Thus, such a 3D topography offers an underestimated possibility to design antibacterial surfaces that do not require biocides or antibiotics.
Keywords: roughness, silicone nanofilaments, antifouling, biofouling, bacterial size
Introduction
Pathogenic bacteria cause millions of infections annually. Billions of dollars are spent on decontamination of equipment and cleaning of tubes, pipes, or ship hulls.1−5 However, it is still unclear how to reliably prevent the irreversible attachment of bacterial cells to surfaces.6 An improved understanding of cell–substratum interactions is required to tackle this question.7−10 Research focuses on the investigation of both the chemical and physical factors finding a major division into two main groups: biocidal (killing) strategies,11,12 and antifouling, which can be either chemical or physical-based (see the Supporting Information for bacterial adhesion details). We demonstrate that irregular 3D structures with submicrometer spacings can also greatly suppress bacterial adhesion.
For a long time, killing bacteria in close distance to surfaces13 by releasing antibiotics or biocides was considered the all-encompassing remedy. However, this approach suffers from limitations such as increasing bacterial resistances14−16 and toxicity of the substances.17−19 Among the antibiofouling (not biocide-releasing) chemical strategies, different nontoxic polymers have been tested, including polyethylene glycol, polyethylenimine, or dextran.20−24 Various surface-tethered polymers with antimicrobial groups have been exploited in biomaterials and biomedical devices.6,25 Notwithstanding, this route can be affected by insufficient durability of the coating or reduced efficiency due to the accumulation of contaminants and/or oxidative degradation.26
Physical strategies to delay bacterial adhesion based on properties such as electrostatic interactions,27−29 roughness,30−33 superhydrophobicity,34−37 and lubricant-impregnated surfaces21,38 have been investigated. Hasan et al. summarized representative investigations on the use of micrometer- and submicrometer-sized patterned surfaces to minimize bacterial adhesion.11 For example, a reduced bacterial adhesion was reported for a surface coated with micrometer-sized colloids. It was related to unfavorable cell bending on the curved colloids.26,39 The results reported for micro- and nanometer-sized rough coatings are still controversial. Many works on the enhanced bacterial adhesion of rough surfaces can be found, which is explained by a larger anchoring area.30,40−42
Surface physical modifications have been developed, resulting in surface features analogous to those found in nature. For example, the surfaces of cicada wings are not only antiadhesive but even bacteria-killing. Ivanova et al. attributed this behavior to deformation stresses within the bacteria caused by the topography of the surface with a nano- or microstructure capable of piercing the membrane, thereby destroying the cell wall and killing the bacteria. However, dead bacteria can serve as anchoring points for living ones.43
Another attractive route in the development of antiadhesive and self-cleaning materials focuses on nature-inspired liquid-repellent surfaces. Several groups investigated whether superhydrophobicity inhibits34,44 or promotes biofilm formation.37,45,46 Superhydrophobic surfaces are characterized by an entrapped air layer. It causes the bacterial suspension to partially rest on an air cushion and partially on low-energy protrusions. Lately, a few reports convincingly showed that the loss of an air cushion could cause a significant increase in bacterial adhesion.47−49 Often, superhydrophobic model surfaces were designed using micropillar arrays, i.e., coatings showing characteristic length scales exceeding the size of the bacterium. Indeed, on micropillar arrays, bacteria can adhere well, as they can lay flat on the top faces or align parallel to the side walls.50
In summary, the relationship between topography, wettability, and functional coatings with antimicrobial groups is still under discussion.51,52 How can bacterial adhesion be further delayed by well-tuned roughness? In contrast to most strategies focusing on roughness, we aim to investigate the influence of an irregular 3D roughness where the characteristic length scales fall just below the size of bacterial cells. Here, we demonstrate that for preventing biofilm formation and growth, surface structures with submicrometer length scale and 3D topography can greatly suppress bacterial adhesion over the tested long exposure times (up to 168 h). Indeed, surface roughness can outweigh the wetting properties of the surface.
Methods
Materials
The following chemicals were used to fabricate the solid substrates: trichloromethylsilane (TCMS, 99%, Sigma-Aldrich), 1H,1H,2H,2H-perfluorodecyltrichlorosilane (PFDTS, 96%, Alfa Aesar), n-hexane (99.99%, Fisher Chemical), toluene (99.99%, Fisher Chemical), ethanol (absolute, 99.96%, VWR Chemicals), SU-8 305 photoresist (Microchem), mr-Dev 600 developer (micro resist technology). Reagents were used as received. Thin glass slides of 24 × 60 mm2 and 170 ± 5 μm thickness were obtained from Menzel-Gläser or Carl Roth GmbH (thickness: 170 ± 5 μm) and a flow cell from Nunc Lab-Tek (Thermo-Fisher Scientific, Germany). Water with a typical resistivity of 18.2 MΩ cm was obtained from a Sartorius Arium 661 VF Water Purification System.
Stellar Competent bacteria (an E. coli HST08 strain, Cat. Nr. 636763) were purchased from Clontech (USA). The 5429 bp long plasmid EGFP-pBAD (Cat. Nr. 54762) was obtained from Addgene (USA). The Plasmid Plus Maxi Kit (Cat. Nr. 12963) was purchased from Qiagen (NL). The culture medium, LB (lysogeny broth) medium, supplemented with ampicillin (Cat. fas-am-b) as well as LB-based agar supplemented with ampicillin (Cat. fas-am-s) were purchased from InvivoGen (USA). Phosphatebuffered saline (PBS) was obtained from Gibco (USA). Glutaraldehyde 25% was purchased from Sigma-Aldrich (USA). A BacLight Live/Dead Bacterial Viability Kit (L-7007, Molecular Probes) comprising propidium iodide 18.3 mM and SYTO9 1.67 mM nucleic acid fluorophores in DMSO solution was obtained from Invitrogen (USA). Super optimal broth with catabolite repression (SOC) medium was purchased from Sigma-Aldrich (USA). Gram-positive Micrococcus luteus (DSM 20030) and Gram-negative Pseudomonas fluorescens Migula 1895 (DSM 4358) were purchased from the Leibniz Institute DSMZ-German Collection of Microorganisms and Cells Collection.
Surface Preparation and Characterization
Glass Slides
To fluorinate the glass slides, we mixed 50 μL of PFDTS with 100 mL of n-hexane. Glass slides were cleaned with acetone and were subsequently activated by oxygen plasma under 300 W for 5 min. The plasma-activated glass slides were immersed in the solution for 30 min. Subsequently, the glass slides were rinsed with n-hexane. The cleaned glass slides were dried under a nitrogen stream.
Silicone Nanofilaments
To coat surfaces with silicone nanofilaments, we immersed plasma-activated glass slides into a mixture of toluene (100 mL) with trace amounts of water (∼180 ppm) and trichloromethylsilane (0.5 mL). A spontaneous hydrolysis reaction of trichloromethylsilane with the hydroxyl groups on the glass surface induced the formation of silicone nanofilaments. After a reaction time of 5 h, the glass slides were covered by a 1–2 μm thick layer of nanofilaments. To render them superhydrophilic (OH-NF), we activated the as-prepared superhydrophobic methyl-terminated nanofilaments (Me-NF) by oxygen plasma (2 min, 120 W). Some of the hydrophilized nanofilament-coated glass slides were fluorinated. Therefore, the coated slides were immersed into 100 mL of hexane containing 50 μL of 1H,1H,2H,2H-perfluorodecyltrichlorosilane for 30 min (F-NF).
SU-8 Pillars
SU-8 micropillar arrays were prepared on thin glass slides by photolithography, as previously reported.53,54 The round pillars were designed to be 5 μm high with 314 μm2 top areas. The pillar–pillar distance between the centers of two adjacent pillars in a row was 10 μm. The fabrication process consisted of the following steps. First, glass slides were cleaned by acetone and were subsequently activated by oxygen plasma under 300 W for 5 min. SU-8 photoresist was then spin-coated (500 rpm for 5 s followed by 3000 rpm for 30 s, SÜSS MicroTec) on the glass slides. The coated glass slides were heated at 65 °C for 3 min, 95 °C for 10 min, and then at 65 °C for 30 min, respectively. Subsequently, the samples were slowly cooled down within 2 h and exposed to UV light (mercury lamp, 350 W) under a photolithography mask for 14 s (masker aligner SÜSS MicroTec MJB3 UV400). To cross-link the photoresist, the samples were heated at 65 °C for 1 min, 95 °C for 3 min and 65 °C for 30 min, and then cooled down slowly. Next, the samples were immersed in the SU-8 developer solution for 6 min, washed with isopropanol and deionized water, and then dried in air. The dried samples were immersed in a 0.1 M NaOH solution overnight to hydrolyze the surfaces of the SU-8 pillars. After rinsing the hydrolyzed surfaces with water and ethanol, the samples were immersed in a solution containing 50 μL of PFDTS dispersed in 100 mL of n-hexane for 30 min to lower the surface energy. Finally, the fluorinated SU-8 micropillar surfaces were rinsed with n-hexane and dried under a nitrogen stream.
Establishing of eGFP-Expressing E. coli
Stellar Competent bacteria were stored at −80 °C until usage. The bacterial stock (in stab culture format) contained the 5429 bp long plasmid EGFP-pBAD. Plasmids were isolated using the Qiagen Plasmid Plus Maxi Kit according to the manufacturer’s protocol. For transformation, Stellar Competent bacteria were thawed in an ice bath for 30 min. All of the following steps were carried out in 1.5 mL microcentrifuge tubes. Five ng of EGFP-pBAD plasmid DNA was added directly into the bacterial suspension and mixed gently. The vials were again incubated for 30 min on ice. The cells were then heat-shocked for exactly 45 s at 42 °C. The tubes were placed on ice for another 2 min. Afterward, SOC medium was added to a final volume of 500 μL, and the tubes were incubated for 1 h at 37 °C and 300 rpm. Finally, 100 μL of bacterial suspension was plated on agar plates containing ampicillin (100 μg/mL) and incubated overnight at 37 °C. The next day, a single bacterial colony was picked and transferred to the LB medium containing 100 μg/mL ampicillin and incubated for 2 h at 37 °C and 300 rpm. To induce the PBAD promotor for eGFP expression, l-arabinose was added to a final concentration of 0.1% in the LB medium. After another incubation of 1 h at 37 °C and 300 rpm, cells exhibited a bright GFP signal. Bacterial cells were subsequently analyzed via confocal laser scanning microscopy or SEM. The value of OD600 for eGFP-expressing E. coli was controlled within 0.13 (1.0 × 108 cells/mL) to 0.15 (1.2 × 108 cells/mL).
Culture Preparation of Freeze-Dried Bacteria (Pseudomonas fluorescens) and Active Culture
Gram-negative P. fluorescens Migula 1895 freeze-dried lyophilized cells isolated from raw milk for cheese production were rehydrated in special trypticase soy broth (TSB) medium for fluorescence (15 g of trypticase soy broth, 500 mL if distilled water, pH 7.3, autoclaved for 15 min at 121 °C) for 30 min and inoculated on agar plates. This allowed active colonies to grow under specified conditions. For surface incubation experiments, single colonies were extracted, transferred to liquid medium, and incubated overnight at 300 rpm under 30 or 28 °C. Gram-positive M. luteus was obtained as an active culture on agar plates. Upon receiving, culture was transferred to fresh medium 1 (2.5 g of peptone, 1.5 g of meat extract, 500 mL of distilled water, pH 7) and incubated at 30 °C for 24 h at 300 rpm.
Agar Plate Test
The bacterial suspensions used in this part of the work were prepared as follows. The E. coli (Stellar Component) bacteria suspended in LB (lysogeny broth) medium mother broths were harvested with sterilized micropipette tips and resuspended in 5 mL of LB medium contained in sterile 15 mL Falcon tubes. These suspensions were subsequently incubated in an orbital shaker (Eppendorf Thermomixer Comfort Type 5355, Eppendorf AG, Hamburg, Germany) at 37 °C and 300 rpm for a period of 24 h to a bacterial concentration of 1.0–1.2 × 108 cells/mL, corresponding to an optical density (OD600) of 0.13–0.15. The bacterial concentrations were checked with the help of turbidimetry using a standard photometer (Eppendorf BioPhotometer, Eppendorf AG, Hamburg, Germany). An amount of 1 mL of the bacterial suspension has been gently applied to the sample surfaces contained in the chambered glass and then incubated in an incubating cupboard (Memmert UM 200, Memmert GmbH + Co. KG, Schwabach, Germany) at 37 °C for 1 week (168 h). Half of the amount of growth medium has been removed and replaced by an equal amount of fresh LB medium daily. Following incubation, the bacterial suspension was removed, and planktonic/nonadherent bacteria were removed by three cycles of gently rinsing the sample surfaces with sterile phosphate-buffered saline (PBS 1×).
Subsequently, the sample surfaces were manually removed (using sterilized tweezers) from the chambered glass and placed into sterile 15 mL Falcon tubes containing 5 mL of sterile PBS 1×. Subsequently, they were subjected to 5 min of sonication at 35 kHz and 120 W (SONOREX RK31, BANDELIN electronic GmbH & Co. KG, Berlin, Germany), followed by 20 s of vortexing to remove the adhered bacteria and to achieve a homogeneous distribution of bacteria. Sonication and vortexing have been repeatedly applied in other works and inherently constitutes a trade-off between detaching/declumping of bacterial aggregates and killing of the bacteria considered. To ensure a countable number of colonies, rows of serial dilutions containing sterile PBS 1× of the recovered liquid were prepared with a dilution factor of 10 for each dilution step (dilutions up to a dilution factor of 104 were prepared). Aliquots of a volume of 20 μL (in duplicates) were spread onto LB agar plates with the help of flame-bent Pasteur pipets and were then incubated upside-down for 24 h at 37 °C (Memmert UM 200, Memmert GmbH + Co. KG, Schwabach, Germany). Following the incubation of the agar plates, the number of colonies was counted by visual inspection or with the help of the open-source image processing software ImageJ/Fiji.55 The number of counted colonies (CFUs = colony-forming units) was then converted to the number of colony-forming units (CFU) per milliliter (CFU):
| 1 |
Scanning Electron Microscopy (SEM)
Surfaces exposed to bacterial media were imaged by SEM (LEO 1530 Gemini, Zeiss, 3 kV) to measure the covered surface area. To prepare the samples for SEM investigations, after 72 h or 168 h long incubation periods, we removed the culture medium, and the samples were washed with a phosphate buffer solution (PBS, 1 mL, 3 times). Adhered bacteria were then fixated, adding a 2.5% (v/v) glutaraldehyde with PBS-based solution reagent for 30 min at room temperature. The fixative was afterward removed, and remaining material was washed by thorough rinsing with subsequent volumes of buffer solution. The fixated bacteria were dehydrated by successive ethanol soaking (i.e., soaking in water–ethanol mixtures, 25, 50, 60, 75, 80, 90, and 100% (v/v), 15 min each, last step twice). To increase the imaging contrast, the dried surfaces were sputter-coated with 5 nm of Pt (BalTec MED 020 Modular High Vacuum Coating System, argon at 2 × 10–2 mbar and 45 mA).
Laser Scanning Confocal Microscopy (LSCM)
Coatings were prepared on coverslips and mounted on flow cells. Attachment of bacteria over time was imaged with an inverted laser scanning confocal microscope (LSCM Leica TCS SP8 SMD) which allows recording images with a lateral resolution of approximately 500 nm and an axial resolution of 1 μm for an HCX PL APO 40 × /1.1 (water immersion objective). E. coli were treated with l-arabinose to express GFP, which can be excited using the argon line at 488 nm The contact angles were determined from the LSCM images. Therefore, a tangent was aligned to the drop shape, and the angle at the intersection of the tangent with the surface was measured.
Data Evaluation of Bacteria Coverage Area
For high-resolution imaging of individual bacteria or colonies, we used SEM. For comparison, LSCM images were investigated. The mean area coverage and the standard deviation of this value were calculated. In Figure S3, original LSCM data (left, green) and recognized bacteria areas (right, cyan) are shown. Areas consisting of bacteria were semiautomatically detected by manually setting a threshold and subsequently using the AnalyzeParticles Plugin in ImageJ/Fiji.55 This procedure allows defining a threshold for bacteria recognition directly. In Figure S5–S7, examples of recognized bacteria areas for SEM images are shown. To prevent noise from contributing to the detected area, we applied background subtraction before thresholding to correct for an inhomogeneous background.
Data Statistical Analysis
Experimental data are plotted, including a mean value and standard deviation (±), using a one-way ANOVA analysis of variance as the statistical method to calculate the significance of the difference. Statistical significant differences are expressed in the figures as follows: * for p < 0.05, ** for p < 0.01, and *** for p < 0.001.
Results and Discussion
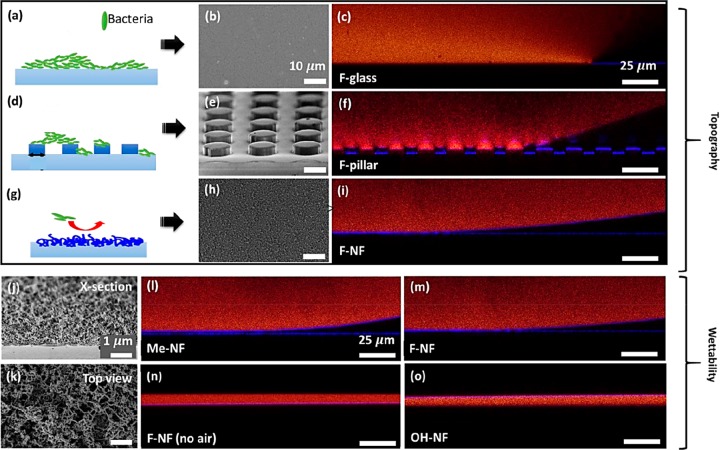
To investigate the influence of surface topography and surface wettability (chemistry and presence of an air cushion), three types of structured surfaces were incubated with bacterial suspensions (Figure 1, Methods): (I) Flat perfluorinated glass (F-glass) served as hydrophobic control surface (Figure 1a–c). (II) Superhydrophobic perfluorinated micropillar arrays (F-pillar, Figure 1d–f) were used as reference surfaces, possessing microscale roughness. (III) Glass substrates coated with silicone nanofilaments56−58 (NF) show submicroscale roughness (Figure 1g–o). Surfaces coated with nanofilaments enabled the investigation of the surface chemistry and surface wettability under otherwise identical conditions. First, superhydrophobic methyl-terminated nanofilaments (Me-NF) and perfluorinated nanofilaments (F-NF) were prepared. The role of the air cushion was investigated by enforcing wetting of the nanofilaments prior to the deposition of the E. coli solution (F-NF, no air). The fully wetted Wenzel state can also be obtained by activating the methyl-terminated filaments by oxygen plasma (OH-NF).
Figure 1.
(a, d, g) Schemes of bacteria (green) attaching on (a) flat surface, (d) SU-8 micropillar arrays, and (g) surface coated with silicone nanofilaments. For simplicity, the bacteria are drawn straight. (b, e, h) Corresponding scanning electron microscope (SEM) micrographs of the surfaces are displayed in gray (scale bar, 10 μm). (c, f, i, l–o) Wetting properties were investigated by laser scanning confocal microscopy (LSCM) using an inverted microscope (Leica TCS SP8 SMD) and a 40× water immersion objective. Red, dyed medium for bacteria cultivation; blue reflection of light from the glass, culture medium; and air, culture medium interface; black, air, glass substrate, or coating (scale bar, 25 μm). (j, k) Cross-section SEM image and top view of a nanofilament-coated sample (dcale bar: 1 μm).
The cylindrical micropillars were 5 μm in height, 13 μm in diameter, and pillar–pillar distance of 20 μm.54,59 For comparison, pillars having an identical height but a diameter of 5 μm and a pillar–pillar distance of 10 μm were investigated. The micropillars were arranged on a 170 μm thick glass slide and successively fluorinated to render them superhydrophobic (Figure 1d–f.)
Glass slides were coated with silicone nanofilaments (see Methods)58 rendering filaments of approximately 50 ± 11 nm diameter. In a few places, approximately 100 nm thick nanofilaments had formed. The reaction time determines the length of the filaments and, thus, the thickness of the coating. However, so far we are not able to fine-tune the characteristic length scales by varying the reaction parameters. The flexibility of the nanofilaments is likely the cause of the high mechanical robustness of surfaces coated with nanofilaments.35 Even under outdoor conditions the nanofilaments did not lose their 3D irregular topography or liquid repellency.60 Without further treatment, these methyl-terminated silicone-nanofilaments (Me-NF) are superhydrophobic. They become superhydrophilic (OH-NF) after oxygen plasma activation. To investigate the influence of the air cushion, we compared the adhesion of bacteria on fluorinated nanofilaments (F-NF) surrounded by air with the adhesion of bacteria on fluorinated nanofilaments that were in the fully wetted Wenzel state. The latter can be achieved by first prewetting the filaments with ethanol. Because of its low surface tension γ = 0.022 N/m, ethanol wets the fluorinated nanofilaments.61 To avoid residues of ethanol on the prewetted surfaces, the prewetted surfaces were rinsed with the LB medium (growth medium) three times to remove residuals of ethanol. Care was taken that the surface remained fully wetted during these and the following steps to ensure that the solution remained in the fully wetted Wenzel state.
The wetting properties of the surfaces were quantified using the culture medium (Methods for details on the preparation). Because of the higher accuracy, we measured receding contact angles (θrec) of a 6 μL drop of the culture medium on the different surfaces using an inverted laser scanning confocal microscope (LSCM), Table S1. The dyed culture medium appears red, whereas the air and the substrate appear black. A 6 μL sized droplet of the culture medium showed receding contact angles of θrec = 91° ± 4 on the fluorinated glass, θrec = 155° ± 10° on the fluorinated SU-8 micropillar arrays, θrec = 168° ± 2° on the methyl terminated nanofilaments, θrec = 176° ± 3° on the fluorinated nanofilaments, and θrec < 10° on the plasma-activated nanofilaments. On all superhydrophobic surfaces, droplets of 6 μL volume rolled off when tilting the surface by less than 10°. The change of the surface chemistry after oxygen plasma treatment was confirmed by XPS (Figure S1).
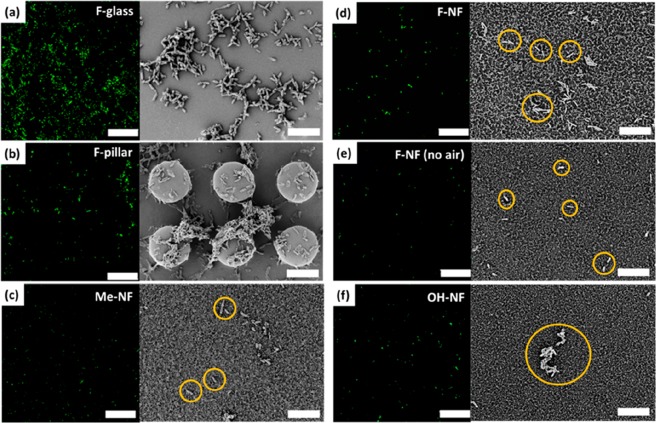
To investigate the adhesion of bacteria on the different surfaces, we proceeded as follows: The surfaces (1 cm × 1 cm) were fixed by double-side tapes onto a sterile chambered glass. Each surface was covered with a 4–5 mm thick layer of bacterial suspension, a mixture of the nutritionally rich cultivation medium and the green fluorescence protein (eGFP) expressing E. coli. The bacteria concentration varied between 1.0 × 108 cells/mL and 1.2 × 108 cells/mL. After 72 h of static incubation at 37 °C, the suspension was replaced by phosphate-buffered saline (PBS, 1 mL) for observation under a laser scanning confocal microscope under wet conditions (Figure 2). To enhance contrast, the E. coli bacteria (green) were labeled by exposure to l-Arabinose for expression of the green fluorescent protein. For the investigation by scanning electron microscopy (SEM), the samples were washed with PBS (phosphate-buffered saline) after incubation of 72 or 168 h. To ensure that proliferation and adhesion were not affected by nutrient depletion, we exchanged the culture medium daily (Methods). After that, the adhered bacteria were fixated by adding a 2.5% (v/v) glutaraldehyde with PBS-based solution reagent for 30 min at room temperature. The fixative was removed by subsequently rinsing with phosphate buffer (PBS) and dried for SEM characterization to quantify the coverage of the surfaces with bacterial cells (see Methods for a detailed discussion of incubation and characterization of the samples and analysis of the images, Figures S2, S5, and S6). After 72 h of stationary incubation, the perfluorinated glass has been covered with several three-dimensional microcolonies revealed in the confocal microscopy and SEM images (Figure 2a).
Figure 2.
Biofilm formation investigated by LSCM and SEM after 72 h of incubation at 37 °C. The bacterial suspension was incubated on the following surfaces: (a) fluorinated flat glass, (b) fluorinated micropillar array with 5 μm height and 7 μm of spacing and 13 μm of diameter (image shows the coverage with bacteria (green spots) close to the pillars’ top surface (see Figures S2 and S3 for enlarged versions of the confocal images), (c) methyl-terminated nanofilaments (d) fluorinated nanofilaments, (e) fluorinated nanofilament coating where the air cushion was removed prior to incubation, and (f) plasma-activated nanofilament coating (some of the bacterial cells on the silicone nanofilaments in c–f are highlighted in yellow circles to enhance the contrast with the surface background) The E. coli were exposed to l-arabinose for expression of the green fluorescent protein, which can be excited at 488 nm. Scale bar of LSCM, 50 μm; scale bar of SEM, 10 μm.
The fluorinated glass slides showed a large number of bacteria colonies separated by regions almost free of bacteria (Figure 2a, Figure S4a). On the superhydrophobic SU-8 micropillar arrays, the space between the pillars, their top faces, and the flat bottom surface are covered with individual bacteria and several microcolonies (Figure 2b shows the focal plane corresponding to the top faces of the pillars, Figure S5). The presence of bacteria at the sidewalls and bottom surface demonstrates that the solution passed the Cassie-to-Wenzel transition. Notably, only sparse and isolated bacteria were observed on all surfaces coated with nanofilaments (Figure 2c–f, Figure S4b–e).
Bacterial adhesion was consistently found to be reduced by approximately 2 orders of magnitude on confocal and electron microscopy images. The confocal images have the advantage of a larger imaged area. However, it turned out to be difficult to work at a constant brightness, which depends on the roughness of the surface and the details of the experimental protocol. Small changes in the brightness greatly influenced the number of detected bacteria. Therefore, for the calculation of the percentage area covered by bacteria, typically, 12 SEM images per surface and incubation time were evaluated (Methods). Areas consisting of bacteria were semiautomatically detected by manually setting a threshold, and subsequently, using the AnalyzeParticles Plugin in ImageJ/Fiji.55 Bacteria recognition on nanofilament surfaces is more challenging compared to flat surfaces due to the inhomogeneous background. The threshold was chosen to yield the best consistency of recognized bacteria with visual inspection (Figures S5 and S6). In the case of pillar substrates, the regions on top of the pillars as well as in-between pillars were evaluated, Figure 3.
Figure 3.
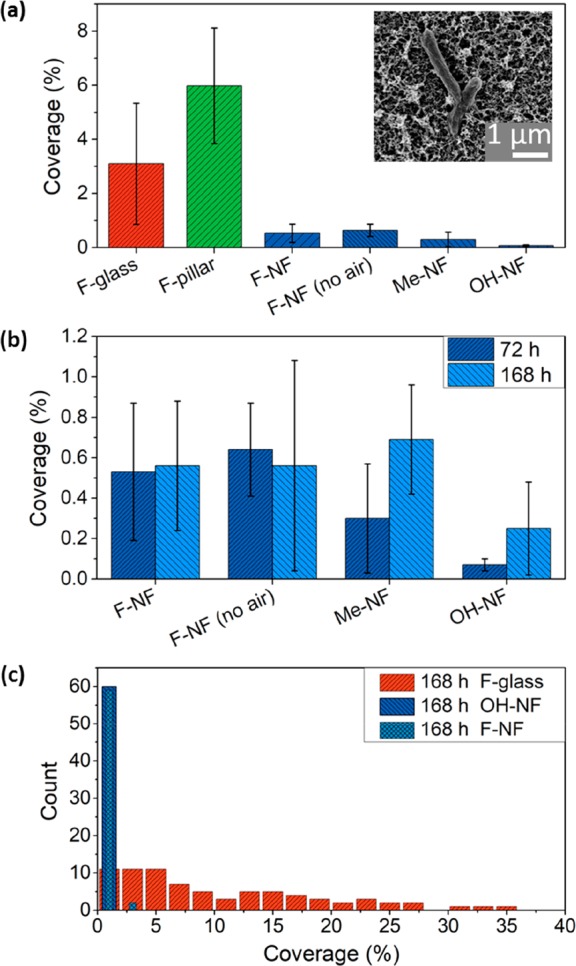
(a) Coverage area based on SEM images (Table S2, Figures S4 and S8a) of different surfaces after 72 h of incubation with E. coli. F-glass, fluorinated glass surfaces, F-pillar, superhydrophobic micropillar arrays, F-NF, fluorinated nanofilaments in the presence of an air cushion; F-NF (no-air), fully wetted fluorinated filaments; Me-NF, methyl-terminated nanofilaments; OH-NF, plasma-activated nanofilaments. The inset SEM image shows E. coli bacterial cells attached to a surface coated with nanofilaments (Figure S8b). (b) Comparison of coverage of adhered bacteria on incubation duration for surfaces coated with nanofilaments showing different surface functionalities in the presence and absence of an air cushion. (c) Histogram based on the number of evaluated SEM images, showing the coverage on three surfaces: fluorinated glass, fully wetted fluorinated nanofilament-coated surface, and superhydrophilic nanofilament-coated surface (see also Figure S9). The y-axis stands for the number of bacteria aggregates of a certain size.
To gain insight into the long-term antibacterial effect of nanofilament-coated surfaces, the duration of static incubation was extended to 168 h (Figure 3a, b). Even after more than doubled incubation times, the surface coverage on the fluorinated nanofilament-coated surfaces remained below 0.7% regardless of the existence of the air plastron. On OH-terminated nanofilament-coated surfaces, the bacterial coverage was even as low as 0.25%. In contrast, the surface coverage obtained on fluorinated glass (F-glass) reached 10.5 ± 8.6% after 168h of incubation, which is 2 orders of magnitude larger (Table S1). These differences could be attributed to effects such as surface charge interactions with the bacterial cell membrane and surface free energy barriers. However, elucidating the mechanism is beyond the scope and the aim of this work.62,63
Not only the average coverage but also the spatial distribution of the surface area covered with bacteria differ (Figure 3c). After 168 h of incubation, the coverage on the flat fluorinated glass surfaces varied between 1.4% and up to 40%. Thus, many areas were covered with large three-dimensional bacterial biofilms, Figure S4a. In contrast, on the nanofilament-coated surfaces—even after wetting by the bacterial solution—the coverage remained low, varying between 0 and 1%, Figure 3c. Only isolated bacteria and no colonies are observed (Figure S4b–e and Figure S7). The variation in the coverage between different positions gives rise to the error bars in Figure 3a, b.
SEM images provided detailed information on how bacteria adhere to the surface. It suffers, however, from the fact that the investigated areas are small and provide only local information. Therefore, we quantified bacterial adhesion using the Plate Count Agar (PCA) protocol (Figure 4, Methods). Following incubation (168 h), after nonadherent (planktonic) bacteria were removed by rinsing in sterile phosphate-buffered saline (PBS 1X), adherent bacteria were removed by sonication followed by vortexing of the sample surface in 5 mL of sterile PBS. The recovered suspension was then serially diluted (dilution factor up to 1 × 104), and aliquot volumes of 20 μL were spread onto LB agar plates in duplicates and subsequently incubated for 24 h at 37 °C. Image processing (Image/Fiji) and conversion yield the number of colony-forming units (Methods). The results obtained at a dilution factor of 1 × 102 are presented in Figure 4.64−66
Figure 4.
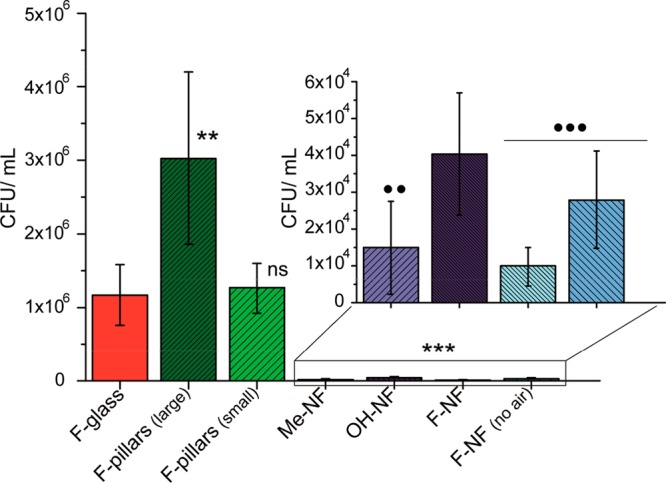
CFU/mL of E. coli for differently treated surfaces. The sample surfaces were incubated for 168 h at 37 °C. The inset shows the CFU/mL for the surfaces coated with silicone nanofilaments at enlarged magnification. F-pillar: diameter = 13 μm, height = 5 μm, pillar–pillar distance = 20 μm. F-pillar 2: diameter = 5 μm, height = 5 μm, pillar–pillar distance = 10 μm. Six independent samples were prepared. Results, standard deviations, and statistical paired t test (95, 99, and 99.9% confidence levels) are calculated from independent experiments, using as calculated probabilities (p): *p < 0.05; **p < 0.01, and ***p < 0.001 as significant differences; ns corresponds to no difference. Asterisks denote comparison between F-glass, pillars, and all the nanofilament samples (main plot), whereas circles mark the comparison between OH-NF and the rest of the nanofilaments (inset).
Analogous to our results obtained with SEM and LSCM (Figures 2 and 3), the largest number of adhered colonies was found on fluorinated pillars, followed by the flat fluorinated glass (Figure 4). Again, the difference in the number of adhered colonies on all nanofilament-based coatings is 2 orders of magnitude lower compared to the flat fluorinated glass and the fluorinated pillar substrate. This also holds for pillars of smaller diameter (5 μm instead of 13 μm) and spacing (10 μm instead of 20 μm). At least three independent experiments were performed for each sample surface type (Table S3).
The small differences between the average coverage determined by SEM and CFU might be caused by the different protocols. The average coverage measured by SEM does not resolve the number of bacteria contributing to a biofilm. As soon as a biofilm formed, bacteria can lie on top of each other. The measurement of the colony-forming units takes each adhered bacterium into account. This led to the SEM images possibly slightly underestimating the ratio between the flat surfaces and the surfaces coated with nanofilaments.
To explore whether the nanofilaments are effective in delaying or preventing adhesion of other bacterial strains, we investigated spherical Gram-positive Micrococcus luteus (Figure 5) and rod-shaped Gram-negative Pseudomonas fluorescens (Figure S10). After 72 h, all nanofilament-coated surfaces with fluorine, methyl, and hydroxyl groups showed low bacterial coverage, independent of the presence (F-NF, Me-NF) or absence (OH-NF) of an air cushion.
Figure 5.
SEM images after incubating with Micrococcus luteus a for 3 days: (a) fluorinated glass (F-glass), (b) fluorinated nanofilament coating (F-NF), (c) methyl-terminated nanofilament (Me-NF), and (d) plasma-activated nanofilament (OH-NF). Scale bar, 10 μm; scale bar of inset SEM image, 1 μm.
Conclusion
The design of nanofilament coatings with different surface wettability and modification supports the hypothesis of the importance of the 3D topography and the length scales of the coating. The spacing between the irregularly arranged nanofilaments (approximately 0.2–1 μm) falls just below the size of bacteria cells (approximately 1 to 5 μm in length). This has several advantages: first, the spacing is sufficiently small that the stiff bacteria cannot fit in-between the filaments. Second, the spacing is sufficiently large to greatly reduce the number of possible adhesion points for the bacteria. The effective anchoring area is further reduced by the local curvature of the filaments. In that respect, our surfaces differ from surfaces possessing a nanoroughness, which often leads to an increase of the effective anchoring area. The presented strategy suppresses the adhesion of bacteria but does not release biocides. Superhydrophobic surfaces can delay bacteria adhesion if their 3D topography and the characteristic length scales fit the above criteria. Notably, the presence of an air plastron is of minor importance for the tested surfaces. Once the surface structure has been designed based on the above principles, the antibiofouling properties of the coating can be further optimized by tethering polymer films or brushes possessing fouling-resistant, fouling release or antimicrobial properties. We envision applications in coating water tubes, medical tubing such as catheters, or materials used in hospitals where no biocides shall be released.
Acknowledgments
This work was supported by the Marie Skłodowska-Curie fellowship 660523-NoBios-ESR (N.E.), the DAAD-MoST sandwich scholarship program (C.-Y.Y.), the ERC advanced grant 340391 – SUPRO (H.-J.B), the Collaborative Research Center 1194 (H.-J.B., F.G.) and the European Union’s Horizon 2020 research and innovation program LubISS No 722497 (D.V., P.B.). G. Schäfer and G. Glasser are acknowledged for technical support.
Glossary
Abbreviations
- SU-8
epoxy-based photoresist
- F-glass
fluorinated flat glass
- F-pillar
fluorinated micropillar structure
- F-NF
perfluorinated nanofilaments
- Me-NF
superhydrophobic methyl-terminated nanofilaments
- OH-NF
superhydrophilic oxygen plasma activated methyl-terminated nanofilaments
- LSCM
laser scanning confocal microscopy
- SEM
scanning electron microscopy
- PBS
phosphate-buffered saline solution
- CFU
colony forming unit
- PCA
plate count agar
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsami.9b22621.
Bacterial adhesion information, XPS spectra, enlarged versions of confocal images, LSCM images, SEM images, histograms, table of bacterial cell coverage analysis, table of number of elevated SEM images, and table of PCA protocol results (PDF)
Author Contributions
† N.E. and C.-Y.Y. contributed equally to this work. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Klevens R. M.; Edwards J. R.; Richards C. L.; Horan T. C.; Gaynes R. P.; Pollock D. A.; Cardo D. M. Estimating Health Care-Associated Infections and Deaths in U.S. Hospitals, 2002. Public Health Rep. 2007, 122, 160–166. 10.1177/003335490712200205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmielewski R. A. N.; Frank J. F. Biofilm Formation and Control in Food Processing Facilities. Compr. Rev. Food Sci. Food Saf. 2003, 2, 22–32. 10.1111/j.1541-4337.2003.tb00012.x. [DOI] [PubMed] [Google Scholar]
- Dinh H. T.; Kuever J.; Mußmann M.; Hassel A. W.; Stratmann M.; Widdel F. Iron Corrosion by Novel Anaerobic Microorganisms. Nature 2004, 427, 829–832. 10.1038/nature02321. [DOI] [PubMed] [Google Scholar]
- Enning D.; Venzlaff H.; Garrelfs J.; Dinh H. T.; Meyer V.; Mayrhofer K.; Hassel A. W.; Stratmann M.; Widdel F. Marine Sulfate-reducing Bacteria Cause Serious Corrosion of Iron under Electroconductive Biogenic Mineral Crust. Environ. Microbiol. 2012, 14, 1772–1787. 10.1111/j.1462-2920.2012.02778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Stoodley L.; Costerton J. W.; Stoodley P. Bacterial Biofilms: from the Natural Environment to Infectious Diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- Yang W. J.; Neoh K. G.; Kang E. T.; Teo S. L. M.; Rittschof D. Polymer Brush Coatings for Combating Marine Biofouling. Prog. Polym. Sci. 2014, 39, 1017–1042. 10.1016/j.progpolymsci.2014.02.002. [DOI] [Google Scholar]
- Renner L. D.; Weibel D. B. Physicochemical Regulation of Biofilm Formation. MRS Bull. 2011, 36, 347–355. 10.1557/mrs.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zobell C. E. The Effect of Solid Surfaces upon Bacterial Activity. J. Bacteriol. 1943, 46, 39–56. 10.1128/JB.46.1.39-56.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan J.; Crawford R. J.; Ivanova E. P. Antibacterial Surfaces: the Quest for a New Generation of Biomaterials. Trends Biotechnol. 2013, 31, 295–304. 10.1016/j.tibtech.2013.01.017. [DOI] [PubMed] [Google Scholar]
- Bixler G. D.; Bhushan B. Biofouling: Lessons from Nature. Philos. Trans. R. Soc., A 2012, 370, 2381–2417. 10.1098/rsta.2011.0502. [DOI] [PubMed] [Google Scholar]
- Hasan J.; Chatterjee K. Recent Advances in Engineering Topography Mediated Antibacterial Surfaces. Nanoscale 2015, 7, 15568–15575. 10.1039/C5NR04156B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adlhart C.; Verran J.; Azevedo N. F.; Olmez H.; Keinanen-Toivola M. M.; Gouveia I.; Melo L. F.; Crijns F. Surface Modifications for Antimicrobial Effects in the Healthcare Setting: a Critical Overview. J. Hosp. Infect. 2018, 99, 239–249. 10.1016/j.jhin.2018.01.018. [DOI] [PubMed] [Google Scholar]
- Sharma V. K.; Yngard R. A.; Lin Y. Silver Nanoparticles: Green Synthesis and their Antimicrobial Activities. Adv. Colloid Interface Sci. 2009, 145, 83–96. 10.1016/j.cis.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Stewart P. S.; William Costerton J. Antibiotic Resistance of Bacteria in Biofilms. Lancet 2001, 358, 135–138. 10.1016/S0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- Neu H. C. The Crisis in Antibiotic Resistance. Science 1992, 257, 1064–1073. 10.1126/science.257.5073.1064. [DOI] [PubMed] [Google Scholar]
- Spellberg B.; Guidos R.; Gilbert D.; Bradley J.; Boucher H. W.; Scheld W. M.; Bartlett J. G.; Edwards J. The Epidemic of Antibiotic-Resistant Infections: A Call to Action for the Medical Community from the Infectious Diseases Society of America. Clin. Infect. Dis. 2008, 46, 155–164. 10.1086/524891. [DOI] [PubMed] [Google Scholar]
- Marambio-Jones C.; Hoek E. M. V. A Review of the Antibacterial Effects of Silver Nanomaterials and Potential Implications for Human Health and the Environment. J. Nanopart. Res. 2010, 12, 1531–1551. 10.1007/s11051-010-9900-y. [DOI] [Google Scholar]
- Bondarenko O.; Juganson K.; Ivask A.; Kasemets K.; Mortimer M.; Kahru A. Toxicity of Ag, CuO and ZnO Nanoparticles to Selected Environmentally Relevant Test Organisms and Mammalian Cells in Vitro: a Critical Review. Arch. Toxicol. 2013, 87, 1181–1200. 10.1007/s00204-013-1079-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Alba A. R.; Hernando M. D.; Piedra L.; Chisti Y. Toxicity Evaluation of Single and Mixed Antifouling Biocides Measured with Acute Toxicity Bioassays. Anal. Chim. Acta 2002, 456, 303–312. 10.1016/S0003-2670(02)00037-5. [DOI] [Google Scholar]
- Salwiczek M.; Qu Y.; Gardiner J.; Strugnell R. A.; Lithgow T.; McLean K. M.; Thissen H. Emerging Rules for Effective Antimicrobial Coatings. Trends Biotechnol. 2014, 32, 82–90. 10.1016/j.tibtech.2013.09.008. [DOI] [PubMed] [Google Scholar]
- Epstein A. K.; Wong T.-S.; Belisle R. A.; Boggs E. M.; Aizenberg J. Liquid-infused Structured Surfaces with Exceptional Anti-biofouling Performance. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 13182–13187. 10.1073/pnas.1201973109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova E. P.; Hasan J.; Webb H. K.; Gervinskas G.; Juodkazis S.; Truong V. K.; Wu A. H. F.; Lamb R. N.; Baulin V. A.; Watson G. S.; Watson J. A.; Mainwaring D. E.; Crawford R. J. Bactericidal Activity of Black Silicon. Nat. Commun. 2013, 4, 2838. 10.1038/ncomms3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z.; Mi L.; Mendiola J.; Ella-Menye J.-R.; Zhang L.; Xue H.; Jiang S. Reversibly Switching the Function of a Surface between Attacking and Defending against Bacteria. Angew. Chem., Int. Ed. 2012, 51, 2602–2605. 10.1002/anie.201106466. [DOI] [PubMed] [Google Scholar]
- Chung K. K.; Schumacher J. F.; Sampson E. M.; Burne R. A.; Antonelli P. J.; Brennan A. B. Impact of Engineered Surface Microtopography on Biofilm Formation of Staphylococcus Aureus. Biointerphases 2007, 2, 89–94. 10.1116/1.2751405. [DOI] [PubMed] [Google Scholar]
- Wu J.-G.; Chen J.-H.; Liu K.-T.; Luo S.-C. Engineering Antifouling Conducting Polymers for Modern Biomedical Applications. ACS Appl. Mater. Interfaces 2019, 11 (24), 21294–21307. 10.1021/acsami.9b04924. [DOI] [PubMed] [Google Scholar]
- Lagree K.; Mon H. H.; Mitchell A. P.; Ducker W. A. Impact of surface topography on biofilm formation by Candida albicans. PLoS One 2018, 13, e0197925 10.1371/journal.pone.0197925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruins M. R.; Kapil S.; Oehme F. W. Microbial Resistance to Metals in the Environment. Ecotoxicol. Environ. Saf. 2000, 45, 198–207. 10.1006/eesa.1999.1860. [DOI] [PubMed] [Google Scholar]
- Liu H.; Du Y.; Wang X.; Sun L. Chitosan kills bacteria through cell membrane damage. Int. J. Food Microbiol. 2004, 95, 147–155. 10.1016/j.ijfoodmicro.2004.01.022. [DOI] [PubMed] [Google Scholar]
- Li P.; Poon Y. F.; Li W.; Zhu H.-Y.; Yeap S. H.; Cao Y.; Qi X.; Zhou C.; Lamrani M.; Beuerman R. W.; Kang E.-T.; Mu Y.; Li C. M.; Chang M. W.; Jan Leong S. S.; Chan-Park M. B. A Polycationic Antimicrobial and Biocompatible Hydrogel with Microbe Membrane Suctioning Ability. Nat. Mater. 2011, 10, 149–156. 10.1038/nmat2915. [DOI] [PubMed] [Google Scholar]
- Truong V. K.; Lapovok R.; Estrin Y. S.; Rundell S.; Wang J. Y.; Fluke C. J.; Crawford R. J.; Ivanova E. P. The Influence of Nano-scale Surface Roughness on Bacterial Adhesion to Ultrafine-grained Titanium. Biomaterials 2010, 31, 3674–3683. 10.1016/j.biomaterials.2010.01.071. [DOI] [PubMed] [Google Scholar]
- Subramani A.; Huang X.; Hoek E. M. V. Direct Observation of Bacterial Deposition onto Clean and Organic-fouled Polyamide Membranes. J. Colloid Interface Sci. 2009, 336, 13–20. 10.1016/j.jcis.2009.03.063. [DOI] [PubMed] [Google Scholar]
- Liu L.; Ercan B.; Sun L.; Ziemer K. S.; Webster T. J. Understanding the Role of Polymer Surface Nanoscale Topography on Inhibiting Bacteria Adhesion and Growth. ACS Biomater. Sci. Eng. 2016, 2, 122–130. 10.1021/acsbiomaterials.5b00431. [DOI] [PubMed] [Google Scholar]
- Cao X.; Pettitt M. E.; Wode F.; Arpa Sancet M. P.; Fu J.; Ji J.; Callow M. E.; Callow J. A.; Rosenhahn A.; Grunze M. Interaction of Zoospores of the Green Alga Ulva with Bioinspired Micro- and Nanostructured Surfaces Prepared by Polyelectrolyte Layer-by-Layer Self-Assembly. Adv. Funct. Mater. 2010, 20, 1984–1993. 10.1002/adfm.201000242. [DOI] [Google Scholar]
- Zhang X.; Wang L.; Levänen E. Superhydrophobic Surfaces for the Reduction of Bacterial Adhesion. RSC Adv. 2013, 3, 12003–12020. 10.1039/c3ra40497h. [DOI] [Google Scholar]
- Geyer F.; D’Acunzi M.; Yang C.-Y.; Müller M.; Baumli P.; Kaltbeitzel A.; Mailänder V.; Encinas N.; Vollmer D.; Butt H.-J. How to Coat the Inside of Narrow and Long Tubes with a Super-Liquid-Repellent Layer—A Promising Candidate for Antibacterial Catheters. Adv. Mater. 2019, 31, 1801324. 10.1002/adma.201801324. [DOI] [PubMed] [Google Scholar]
- Meier M.; Dubois V.; Seeger S. Reduced Bacterial Colonisation on Surfaces Coated with Silicone Nanostructures. Appl. Surf. Sci. 2018, 459, 505–511. 10.1016/j.apsusc.2018.08.003. [DOI] [Google Scholar]
- Hwang G. B.; Page K.; Patir A.; Nair S. P.; Allan E.; Parkin I. P. The Anti-Biofouling Properties of Superhydrophobic Surfaces are Short-Lived. ACS Nano 2018, 12, 6050–6058. 10.1021/acsnano.8b02293. [DOI] [PubMed] [Google Scholar]
- Wong T.-S.; Kang S. H.; Tang S. K. Y.; Smythe E. J.; Hatton B. D.; Grinthal A.; Aizenberg J. Bioinspired Self-repairing Slippery Surfaces with Pressure-stable Omniphobicity. Nature 2011, 477, 443–447. 10.1038/nature10447. [DOI] [PubMed] [Google Scholar]
- Kargar M.; Chang Y.-R.; Khalili Hoseinabad H.; Pruden A.; Ducker W. A. Colloidal Crystals Delay Formation of Early Stage Bacterial Biofilms. ACS Biomater. Sci. Eng. 2016, 2, 1039–1048. 10.1021/acsbiomaterials.6b00163. [DOI] [PubMed] [Google Scholar]
- Puckett S. D.; Taylor E.; Raimondo T.; Webster T. J. The Relationship between the Nanostructure of Titanium Surfaces and Bacterial Attachment. Biomaterials 2010, 31, 706–713. 10.1016/j.biomaterials.2009.09.081. [DOI] [PubMed] [Google Scholar]
- Singh A. V.; Vyas V.; Patil R.; Sharma V.; Scopelliti P. E.; Bongiorno G.; Podestà A.; Lenardi C.; Gade W. N.; Milani P. Quantitative Characterization of the Influence of the Nanoscale Morphology of Nanostructured Surfaces on Bacterial Adhesion and Biofilm Formation. PLoS One 2011, 6, e25029 10.1371/journal.pone.0025029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D. H. K.; Pham V. T. H.; Truong V. K.; Sbarski I.; Wang J.; Balčytis A.; Juodkazis S.; Mainwaring D. E.; Crawford R. J.; Ivanova E. P. Role of Topological Scale in the Differential Fouling of Pseudomonas Aeruginosa and Staphylococcus Aureus Bacterial Cells on Wrinkled Gold-coated Polystyrene Surfaces. Nanoscale 2018, 10, 5089–5096. 10.1039/C7NR08178B. [DOI] [PubMed] [Google Scholar]
- Ivanova E. P.; Hasan J.; Webb H. K.; Truong V. K.; Watson G. S.; Watson J. A.; Baulin V. A.; Pogodin S.; Wang J. Y.; Tobin M. J.; Lobbe C.; Crawford R. J. Natural Bactericidal Surfaces: Mechanical Rupture of Pseudomonas Aeruginosa Cells by Cicada Wings. Small 2012, 8, 2489–2494. 10.1002/smll.201200528. [DOI] [PubMed] [Google Scholar]
- Ye W.; Shi Q.; Hou J.; Jin J.; Fan Q.; Wong S.-C.; Xu X.; Yin J. Superhydrophobic Coating of Elastomer on Different Substrates Using a Liquid Template to Construct a Biocompatible and Antibacterial Surface. J. Mater. Chem. B 2014, 2, 7186–7191. 10.1039/C4TB01126K. [DOI] [PubMed] [Google Scholar]
- Sousa C.; Rodrigues D.; Oliveira R.; Song W.; Mano J. F.; Azeredo J. Superhydrophobic Poly(L-lactic acid) Surface as Potential Bacterial Colonization Substrate. AMB Express 2011, 1, 34. 10.1186/2191-0855-1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrov I.; Polishchuk I.; Shemesh M.; Pokroy B. Superhydrophobic Wax Coatings for Prevention of Biofilm Establishment in Dairy Food. ACS Appl. Bio Mater. 2019, 2, 4932–4940. 10.1021/acsabm.9b00674. [DOI] [PubMed] [Google Scholar]
- Fadeeva E.; Truong V. K.; Stiesch M.; Chichkov B. N.; Crawford R. J.; Wang J.; Ivanova E. P. Bacterial Retention on Superhydrophobic Titanium Surfaces Fabricated by Femtosecond Laser Ablation. Langmuir 2011, 27, 3012–3019. 10.1021/la104607g. [DOI] [PubMed] [Google Scholar]
- Loo C.-Y.; Young P. M.; Lee W.-H.; Cavaliere R.; Whitchurch C. B.; Rohanizadeh R. Superhydrophobic, Nanotextured Polyvinyl Chloride Films for Delaying Pseudomonas Aeruginosa Attachment to Intubation Tubes and Medical Plastics. Acta Biomater. 2012, 8, 1881–1890. 10.1016/j.actbio.2012.01.015. [DOI] [PubMed] [Google Scholar]
- Dou X.-Q.; Zhang D.; Feng C.; Jiang L. Bioinspired Hierarchical Surface Structures with Tunable Wettability for Regulating Bacteria Adhesion. ACS Nano 2015, 9, 10664–10672. 10.1021/acsnano.5b04231. [DOI] [PubMed] [Google Scholar]
- Hochbaum A. I.; Aizenberg J. Bacteria Pattern Spontaneously on Periodic Nanostructure Arrays. Nano Lett. 2010, 10, 3717–3721. 10.1021/nl102290k. [DOI] [PubMed] [Google Scholar]
- Graham V. M.; Cady C. N. Nano and Microscale Topographies for the Prevention of Bacterial Surface Fouling. Coatings 2014, 4, 37–59. 10.3390/coatings4010037. [DOI] [Google Scholar]
- Serrano C.; García-Fernández L.; Fernández-Blázquez J. P.; Barbeck M.; Ghanaati S.; Unger R.; Kirkpatrick J.; Arzt E.; Funk L.; Turón P.; del Campo A. Nanostructured Medical Sutures with Antibacterial Properties. Biomaterials 2015, 52, 291–300. 10.1016/j.biomaterials.2015.02.039. [DOI] [PubMed] [Google Scholar]
- Gao N.; Geyer F.; Pilat D. W.; Wooh S.; Vollmer D.; Butt H. J.; Berger R. How Drops Start Sliding Over Solid Surfaces. Nat. Phys. 2018, 14, 191. 10.1038/nphys4305. [DOI] [Google Scholar]
- Mammen L.; Bley K.; Papadopoulos P.; Schellenberger F.; Encinas N.; Butt H. J.; Weiss C. K.; Vollmer D. Functional Superhydrophobic Surfaces Made of Janus Micropillars. Soft Matter 2015, 11, 506–515. 10.1039/C4SM02216E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J.; Arganda-Carreras I.; Frise E.; Kaynig V.; Longair M.; Pietzsch T.; Preibisch S.; Rueden C.; Saalfeld S.; Schmid B.; Tinevez J.-Y.; White D. J.; Hartenstein V.; Eliceiri K.; Tomancak P.; Cardona A. Fiji: an Open-source Platform for Biological-image Analysis. Nat. Methods 2012, 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artus G. R. J.; Jung S.; Zimmermann J.; Gautschi H. P.; Marquardt K.; Seeger S. Silicone Nanofilaments and Their Application as Superhydrophobic Coatings. Adv. Mater. 2006, 18, 2758–2762. 10.1002/adma.200502030. [DOI] [Google Scholar]
- Zhang J.; Seeger S. Superoleophobic Coatings with Ultralow Sliding Angles Based on Silicone Nanofilaments. Angew. Chem., Int. Ed. 2011, 50, 6652–6656. 10.1002/anie.201101008. [DOI] [PubMed] [Google Scholar]
- Geyer F.; Schönecker C.; Butt H.-J.; Vollmer D.. CO2 Capture: Enhancing CO2 Capture using Robust Superomniphobic Membranes Adv. Mater. 2017, 29, 10.1002/adma.201770035. [DOI] [PubMed] [Google Scholar]
- Papadopoulos P.; Mammen L.; Deng X.; Vollmer D.; Butt H.-J. How Superhydrophobicity Breaks down. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 3254. 10.1073/pnas.1218673110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer F.; D’Acunzi M.; Sharifi-Aghili A.; Saal A.; Gao N.; Kaltbeitzel A.; Sloot T.-F.; Berger R.; Butt H.-J.; Vollmer D. When and How Self-cleaning of Superhydrophobic Surfaces Works. Sci. Adv. 2020, 6, eaaw9727. 10.1126/sciadv.aaw9727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez G.; Alvarez E.; Navaza J. M. Surface Tension of Alcohol Water + Water from 20 to 50 °C. J. Chem. Eng. Data 1995, 40, 611–614. 10.1021/je00019a016. [DOI] [Google Scholar]
- Hori K.; Matsumoto S. Bacterial Adhesion: From Mechanism to Control. Biochem. Eng. J. 2010, 48, 424–434. 10.1016/j.bej.2009.11.014. [DOI] [Google Scholar]
- Bos R.; Mei H.C.; Gold J.; Busscher H.J. Retention of Bacteria on a Substratum Surface with Micro-patterned Hydrophobicity. FEMS Microbiol. Lett. 2000, 189, 311–315. 10.1111/j.1574-6968.2000.tb09249.x. [DOI] [PubMed] [Google Scholar]
- Bjerkan G.; Witsø E.; Bergh K. Sonication is Superior to Scraping for Retrieval of Bacteria in Biofilm on Titanium and Steel Surfaces in Vitro. Acta Orthop. 2009, 80, 245–250. 10.3109/17453670902947457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell C.; Vu T. L.; Lin J. J.; Kolle S.; Juthani N.; Watson E.; Weaver J. C.; Alvarenga J.; Aizenberg J. Self-Replenishing Vascularized Fouling-Release Surfaces. ACS Appl. Mater. Interfaces 2014, 6, 13299–13307. 10.1021/am503150y. [DOI] [PubMed] [Google Scholar]
- Kobayashi H.; Oethinger M.; Tuohy M. J.; Procop G. W.; Bauer T. W. Improved Detection of Biofilm-formative Bacteria by Vortexing and Sonication: a Pilot Study. Clin. Orthop. Relat. Res. 2009, 467, 1360–1364. 10.1007/s11999-008-0609-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.