Abstract
New Delhi metallo-β-lactamase-1 (NDM-1) has recently emerged as a global threat because of its ability to confer resistance to all common β-lactam antibiotics. Understanding the molecular basis of β-lactam hydrolysis by NDM is crucial for designing NDM inhibitors or β-lactams resistant to their hydrolysis. In this study, for the first time, NMR was used to study the influence of Zn(II) ions on the dynamic behavior of NDM-1. Our results highlighted that the binding of Zn(II) in the NDM-1 active site induced several structural and dynamic changes on active site loop 2 (ASL2) and L9 loops and on helix α2. We subsequently studied the interaction of several flavonols: morin, quercetin, and myricetin were identified as natural and specific inhibitors of NDM-1. Quercetin conjugates were also synthesized in an attempt to increase the solubility and bioavailability. Our NMR investigations on NDM-1/flavonol interactions highlighted that both Zn(II) ions and the residues of the NDM-1 ASL1, ASL2, and ASL4 loops are involved in the binding of flavonols. This is the first NMR interaction study of NDM-1/inhibitors, and the models generated using HADDOCK will be useful for the rational design of more active inhibitors, directed against NDM-1.
Introduction
Gram-negative bacteria (GNB), particularly Enterobacterales, Pseudomonas aeruginosa, and Acinetobacter baumannii, have reemerged as major actors in antimicrobial resistance worldwide.1,2 Pandrug resistance (PDR) and multidrug resistance (MDR) have been observed among them.3 Currently, β-lactamase-mediated resistance does not spare even the newest and most potent β-lactams (carbapenems), whose activity is challenged by the class B metallo-β-lactamases (MBLs) and by the serine-carbapenemases (classes A and D).4−6 MBLs are by far the β-lactamases of greatest concern because there are currently not clinically useful inhibitors for this class.6 The spread of MBL genes among Enterobacterales is of great clinical concern, given the importance of these pathogens as causes of nosocomial and community-acquired infections and the fact that MBLs inactivate expanded-spectrum cephalosporins and carbapenems, the major antibiotics used to treat infections due to Enterobacterales.
β-Lactamases catalyze the opening of the β-lactam ring, thereby inactivating β-lactam antibiotics.7 Based on their amino acid sequence, the β-lactamases have been grouped into four major classes. Classes A, C, and D use an active site serine to catalyze hydrolysis, while class B (MBLs) requires one or two zinc ions (Zn(II)) for their activity.8 Based on the structural diversity of the active sites, such as differences in metal content and residues involved in metal coordination, MBLs have been divided into three subclasses: B1, B2, and B3.8
The pandemic New Delhi metallo-β-lactamase-1 (NDM-1) disseminating worldwide in Gram-negative organisms threatens to take medicine back into the preantibiotic era since mortality associated with infections caused by these “superbugs” is very high and the choices of treatment are very limited. NDM-1, a member of subclass B1 MBLs, has rapidly spread worldwide on several MDR plasmids.9−12 Numerous NDM-1 structures, complexed or not with metal ions and a variety of substrates, are available in the Protein Data Bank (PDB). NDM-1 adopts the general αβ/βα fold of MBLs,13−18 consisting of two central β-sheets and five solvent-exposed α-helices. Numerous experimental and theoretical studies have been conducted to define the catalytic mechanism of NDM-1.14,18−22 The active site is a hydrophobic cavity, which consists of two Zn(II) (Zn1 and Zn2) surrounded by five mobile labeled loops called active site loops (ASL1–5) responsible for substrate binding and specificity.14 Zn1 and Zn2 are coordinated, respectively, by H120, H122, H189 and D124, C208, H250, with different affinities,23,24 and by a water molecule located between these two Zn(II) ions, which acts as a nucleophile during β-lactam hydrolysis.
Like all MBLs, NDM-1 is not sensitive to any commercially available serine-β-lactamase inhibitors. The development of chemically active inhibitors for the NDM-1 enzyme has been extensively undertaken.17,25−46 The metal chelator edetate disodium (EDTA) has also been shown to inactivate the enzymatic activity of NDM-1 by removing the catalytically required Zn(II) ions,17,47 but its toxicity impedes its clinical use. Potent inhibitors of NDM-1 have also been identified through virtual screening of natural compounds, such as the natural product aspergillomaramine, analogous to the metal-ion chelator EDTA,48,49 baicalin,50 and magnolol, which has a significant impact on NDM-1 enzyme activity in vitro,51 and hesperidin, which acts directly on key residues near the active site of NDM-1.52
To better understand the impact of Zn(II) on the structural and dynamic behavior of NDM-1, we analyzed this protein in the presence and absence of Zn(II) by NMR. We showed that not only loops carrying the zinc ligands undergo a significant change in their dynamic behavior but also other regions far from the active site. In addition, we identified three flavonols: morin, quercetin, and myricetin as specific inhibitors of NDM-1 activity. Flavonols are naturally occurring polyphenolic compounds that are ubiquitous in a wide range of vascular plants.53 They are a class of flavonoids that are recognized to have a variety of biological activities and important therapeutic applications.54−62 We used NMR spectroscopy to map the site of interaction on NDM-1 during complexation to the three flavonols. Our results show that the NDM-1 loops, ASL1, ASL2, and ASL4 are perturbated by flavonol binding and that Zn(II) ions are involved in their binding. Our NMR data provide support for the NDM-1/flavonol complex models, generated using the HADDOCK program, which will be useful for the rational design of ligands with increased activity.
Results and Discussion
NDM-1 Is Stabilized in the Presence of Zn(II)
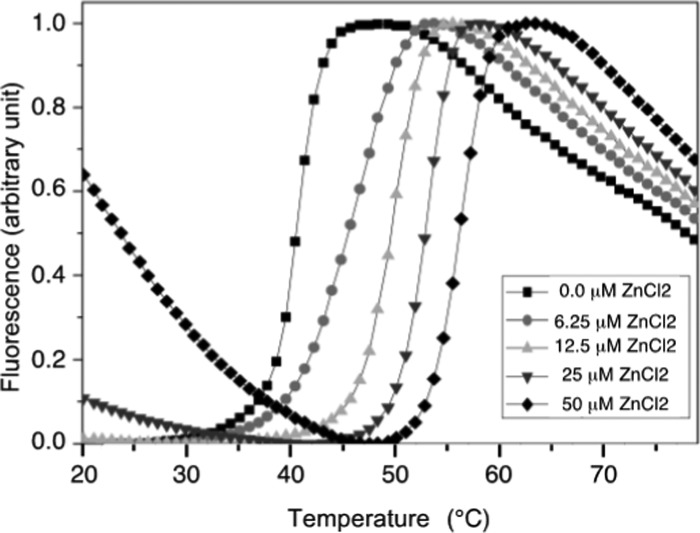
We verified by NMR spectroscopy and thermal shift experiments that NDM-1 was produced without Zn(II). The addition of EDTA did not induce any chemical shift variation in the heteronuclear single quantum correlation (HSQC) spectra and did not modify the thermostability profiles of NDM-1 (data not shown). The addition of increasing amounts of Zn(II) to the metal-free NDM-1, followed by thermal shift assay resulted in an increase in the denaturation temperature of the protein (Figure 1 and Supporting Information Table S1), indicating that Zn(II) strongly stabilizes NDM-1. However, at a high concentration of Zn(II), we observed a notable increase in fluorescence background prior to denaturation of NDM-1, suggesting the appearance of an exposed hydrophobic region or the formation of oligomers. This effect occurred at a Zn(II) concentration greater than 2 molar equiv relative to the protein concentration and increased sharply with increasing Zn(II) concentration. It could be explained by intermolecular interactions between several NDM-1 proteins, mediated by His-tag/Zn(II) complexes.
Figure 1.
Effect of Zn(II) ions on the denaturation of NDM-1 followed by thermal shift assay. Thermal denaturation profiles of NDM-1 were studied in the presence of various concentrations of ZnCl2 (0–50 μM) and 9 μM of NDM-1.
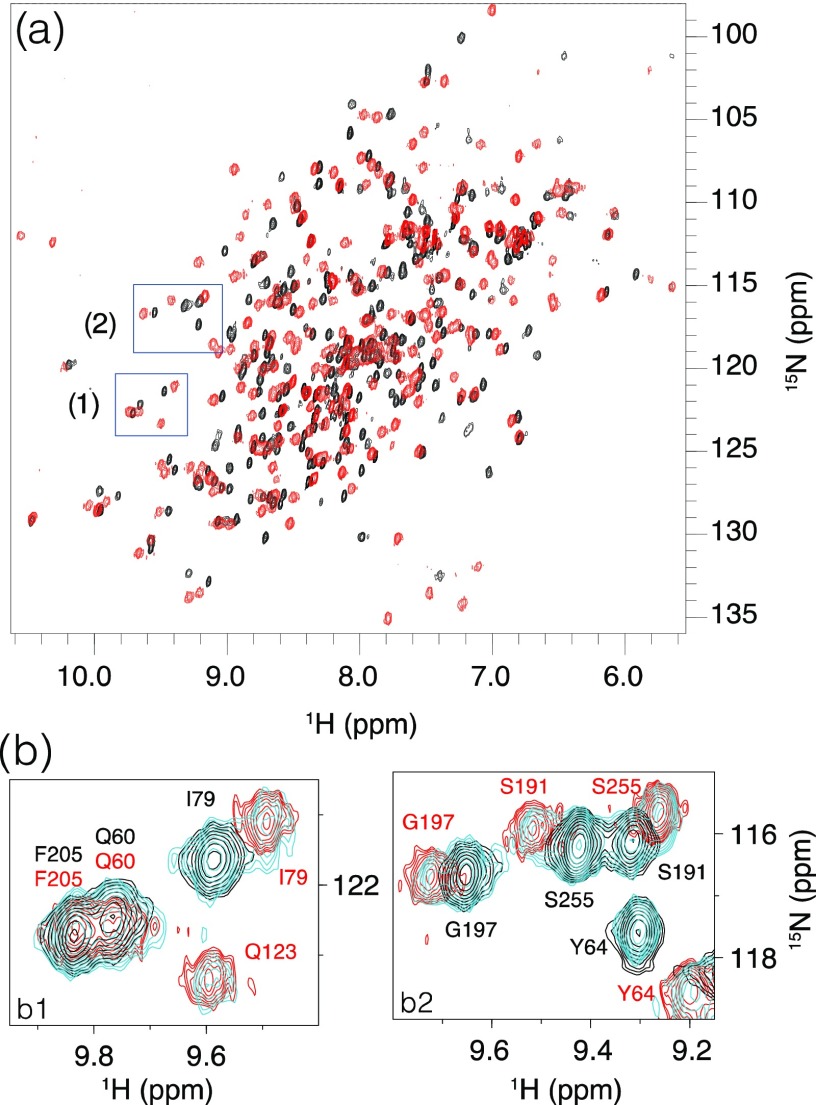
The effect of Zn(II) binding on the structural and dynamic properties of NDM-1 was investigated by NMR. The 1H–15N HSQC spectra of 15N-labeled NDM-1, recorded in the absence and presence of 2 molar equiv of Zn(II), showed well-dispersed cross-peaks characteristic of a well-folded protein (Figure 2a).
Figure 2.
(a) Superimposition of the 1H–15N HSQC spectra recorded in the absence of Zn(II) (black) and in the presence of 2 molar equiv of Zn(II) (red). (b) Superimposition of the 1H–15N HSQC spectra recorded on NDM-1 in the absence of Zn(II) (dark blue) and in the presence of 1 (cyan) and 2 molar equiv of Zn(II) (red). Zoomed-in images corresponding to the boxes (1) and (2) highlighted in (a).
In the presence of 1 Zn(II) equivalent, we observed a slow exchange, on the NMR chemical shift timescale, between the metal-free and the di-Zn(II) states of NDM-1 (Figure 2b), in agreement with the high affinity of NDM-1 for Zn(II).23,24 Upon the addition of 2 molar equiv of Zn(II), we only observed resonances of the NDM-1 bound state, demonstrating that NDM-1 binds two Zn(II) ions, in accordance with the crystal structures of NDM-1. The 1H–15N HSQC spectrum underwent a drastic change that affected the majority of cross-peaks (Figure 2a), as already mentioned by Zheng et al.63 Above 2 Zn(II) equivalents, a dramatic decrease of all of the 1H–15N cross-peak intensities was observed (Supporting Information Figure S1) as well as heavy precipitation, suggesting the formation of insoluble NDM-1 oligomers. These NMR data are in agreement with thermal shift experiments (Figure 1), which show that a slight excess of Zn(II) induces NDM-1 oligomerization. Therefore, to limit protein self-association for the rest of this study, we decided to never exceed the 2 molar equiv of Zn(II).
NMR Backbone Resonance Assignment of NDM-1
To determine the NDM-1 residues most heavily influenced by Zn(II) binding, we assigned the backbone NMR resonances of NDM-1 alone and complexed with Zn(II), using the classical triple resonance and nuclear Overhauser effect spectroscopy (NOESY) correlation experiments. The optimal quality of the spectra, used for assignment, was observed at 20 °C in the absence of Zn(II) (Supporting Information Figures S2 and S3) and at 27 °C in the presence of 150 mM NaCl and 2 molar equiv of Zn(II) relative to the NDM-1 concentration (Supporting Information Figures S4 and S5). A total of 226 of the 253 metal-free NDM-1 backbone amide protons were assigned. Missing assignments are those of unstructured N-terminus residues (M28-E30) and residues in flexible loops (e.g., V118-D130, Q151-V155, G207, and C208) (Supporting Information Figure S6a). From the di-Zn(II) NDM-1 sample, 94% of the 1H–15N cross-peaks were assigned. Only backbone proton assignments of 14 residues (residues M28-E30, N57, K125, M126, G186, G207, K216, N220, L221, A257, L271, and E272) were missing, probably due to the conformational exchange or fast exchange with the solvent, which resulted in a significant broadening for these amino acids. Many of these residues play an important role in the enzymatic catalysis. Indeed, the residues N220 and L221 are located in the ASL4 loop, which plays a direct role in substrate recognition.64 Additional 10% backbone resonances have been assigned over the available assignment of NDM-1 in the presence of Zn(II)65 (Supporting Information Figure S6b).
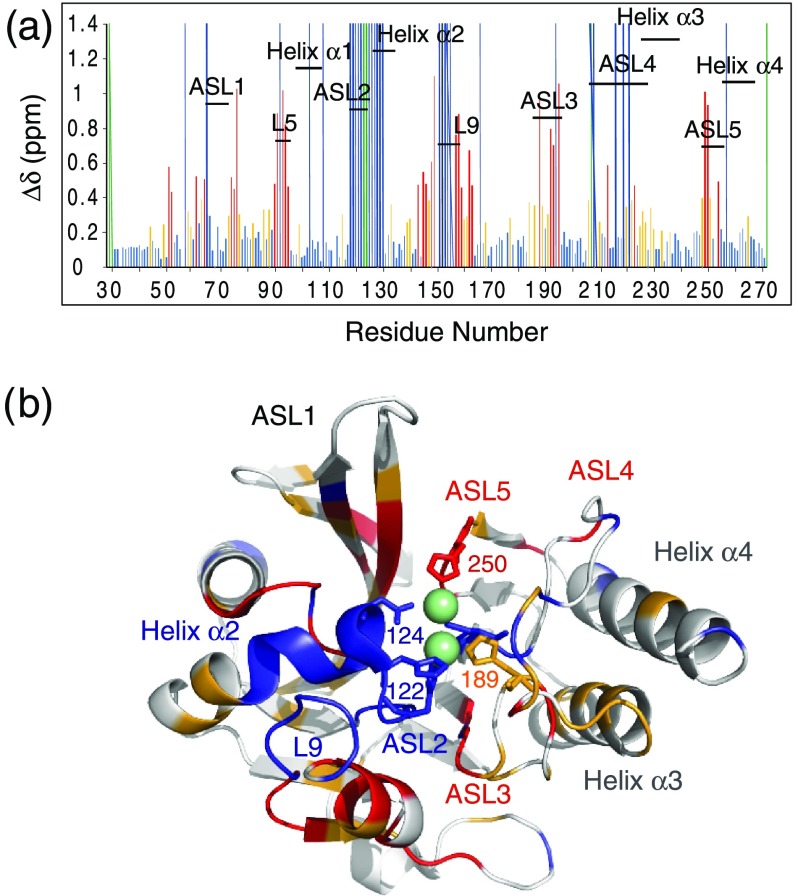
Substantial differences were observed between the 1H–15N HSQC spectra recorded on metal-free NDM-1 and di-Zn(II) NDM-1 (Supporting Information Figures S2 and S4). Analysis of the 15N–1H chemical shift differences after the addition of increasing concentration of Zn(II) to NDM-1 suggests that the protein undergoes local structural changes upon binding to Zn(II) (Figure 3a). Since the resonances of the ASL2 and L9 loops and of the helix α2 could not be observed in the absence of Zn(II), no information could be obtained on the chemical shift variations for these three domains in the presence of Zn(II). The largest observable variations are in the ASL3 and ASL5 loops, which include the H189, C208, and H250 residues involved in the coordination of the two Zn(II). It is interesting to note that domains remote from the Zn(II) binding site, such as the β-sheet, which flanks the ASL1 loop and the L5 and L9 loops (Figure 3b), are also affected by the Zn(II) binding. On the contrary, the ASL1 and ASL4 loops show only a minor chemical shift variation upon Zn(II) binding, with the exception of the first residues of the ASL4 loop (residues 206–209), probably due to the coordination of C208 with Zn(II).
Figure 3.
Specific interactions between NDM-1 and Zn(II). (a) Chemical shift variations of the nitrogen and amide proton nuclei between metal-free NDM-1 and di-Zn(II) NDM-1. The 1H and 15N chemical shifts were extracted from NDM-1 spectra recorded at 25 °C and 600 MHz 1H frequency in the absence of Zn(II), on a sample containing 50 mM phosphate buffer (pH 7.0 and 150 mM NaCl), in the presence of 2 molar equiv of Zn(II) on a solution sample containing 0.1 mM bis tris (pH 7.0, 150 mM NaCl) (Δδ (1H, 15N) = [((ΔδH)2 + (ΔδN)2 × 0.14) × 0.5]1/2). In red and orange are indicated the residues with Δδ > 0.4 ppm and 0.2 > Δδ > 0.4 ppm, respectively. The unassigned residues of NDM-1 in the absence and presence of Zn(II) are represented by dark blue and green lines, respectively. (b) Location of the residues whose amide proton and nitrogen resonances are significantly disturbed during Zn(II) binding, mapped on the NDM-1 crystal structure (PDB: 3SPU) with the same color code. Zn(II) are represented by green spheres.
The location of the secondary structure elements in the metal-free NDM-1 and di-Zn(II) NDM-1 complex derived from TALOS–N software66 was in very good agreement with that of the NDM-1 crystal structures in the absence and presence of Zn(II)13,67 (Supporting Information Figure S6). The results also show that the binding of Zn(II) ions does not significantly modify the NDM-1 secondary structures.
Analysis of NDM-1 Dynamics by NMR
To better understand the impact of Zn(II) binding on the dynamics of NDM-1, we collected 15N R1 and R2 relaxation rates and 1H–15N heteronuclear NOE (hetNOE) values that provide information on the local and global dynamics in the protein. The results show that the overall structure of NDM-1 is rigid in the presence or absence of Zn(II), with the exception of the N-terminal extremity, which has shorter transversal relaxation rates and lower hetNOE values, indicating greater flexibility in the 100 ps timescale (Supporting Information Figure S7). The 15N relaxation rates are homogeneous over the folded domains with mean values of R1 and R2 of 0.73 ± 0.08 and 24.22 ± 1.69 s–1, respectively, in the absence of Zn(II), and 0.64 ± 0.05 and 31.19 ± 1.27 s–1, respectively, in the presence of Zn(II) (Supporting Information Table S2). The lower mean value of R1 and the higher mean value of R2 calculated in the presence of Zn(II) could be explained by the NDM-1 intermolecular interactions induced by a slight excess of Zn(II), as already seen in the thermal shift experiments. In addition, the relaxation data show that only the ASL1 loop (residues 65–74) is slightly more flexible than the rigid zones of the protein in both the absence and presence of Zn(II) (for additional information, see Table S2 and Figure S7).
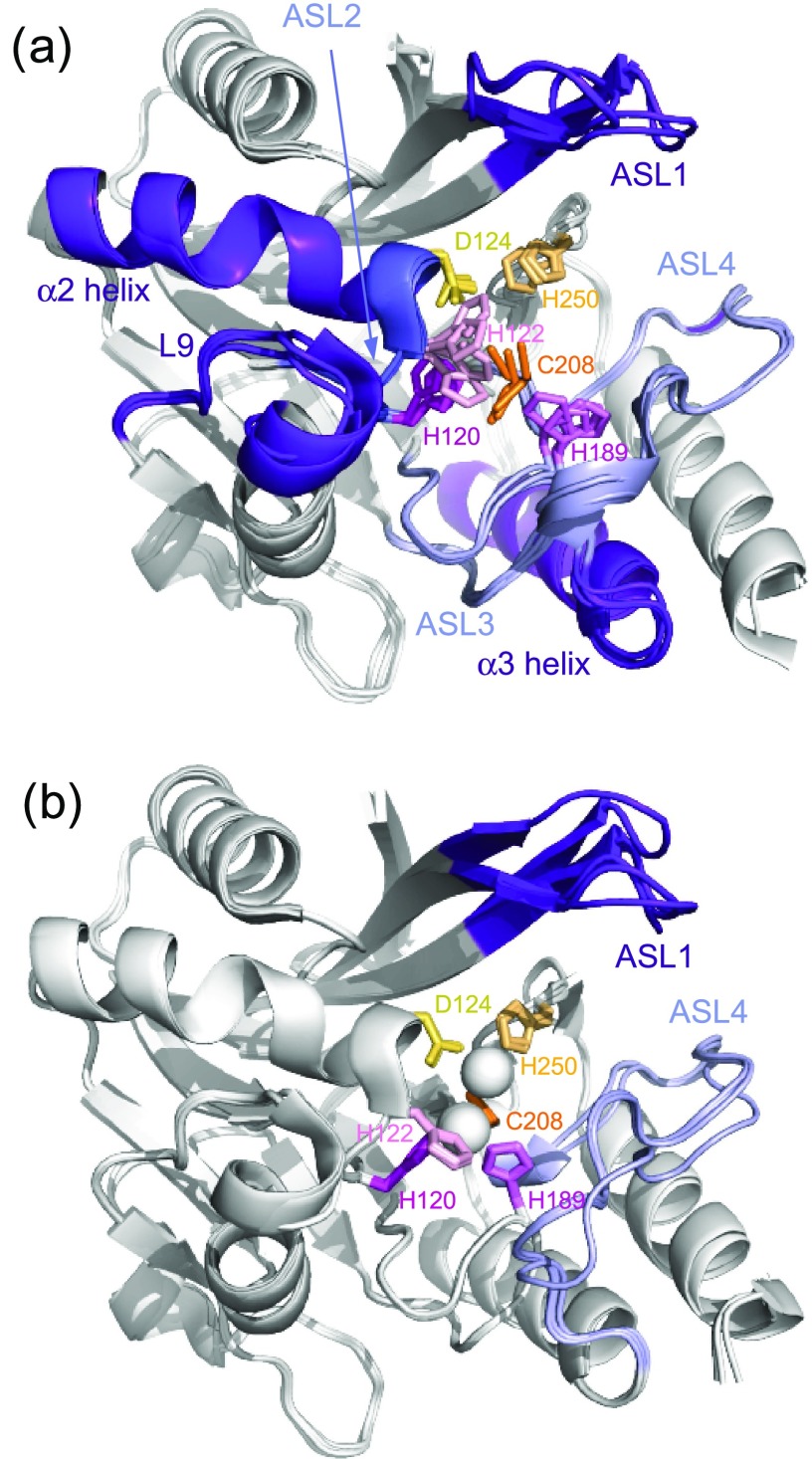
The main effects of the binding of the two Zn(II) occurred at the loops ASL2 and L9, and at α2 helix. In the absence of zinc, the residues belonging to these domains showed disappearance of their signals, consistent with an intermediate exchange at the NMR chemical shift timescale, whereas in the presence of zinc, the signals of these residues are perfectly visible and the measured relaxation parameters for these three domains indicate their significant stiffening (Supporting Information Table S2). Indeed, in the absence of Zn(II), the three Zn(II) ligands (H120, H122, and D124) belonging to the ASL2 loop undergo fluctuations that propagate through the ASL2 loop and α2 helix. The ASL2 and L9 loops could influence each other due to their spatial proximities in the three-dimensional (3D) structure (Figure 4a). The same effects of Zn(II) were observed by Chen et al. using molecular dynamics (MD) simulations.22 In addition, the authors also showed that the di-Zn(II) coordination exerts stronger restrictions on the movements of ASL1 and ASL4 loops (denoted L3 and L10, respectively, in Chen et al.22). Our relaxation data did not indicate a decrease in the internal flexibility of ASL1 and ASL4 upon Zn(II) binding contrary to that observed in their MD simulations.22
Figure 4.
(a) Superimposition on the backbone atoms for the domain 45–270 of four metal-free apo-NDM-1 (PDB: 3RKK, 3SBL, 3RKJ, 3PG4) and (b) four apo-NDM-1 (PDB: 3SPU, 4TYF, 4TZE, 4TZF) crystallographic structures. Highly flexible domains, far from the active site, are colored purple, and flexible domains bearing Zn(II) ligands are colored light blue and dark blue. The residues interacting directly with Zn(II) are represented by stick, and the residues interacting with Zn1 and in Zn2 are in pink and orange, respectively. Both Zn(II) are represented by white spheres.
Our results are consistent with crystallographic structures that show that in the absence of zinc, the variabilities in the orientation of ligands that can bind to Zn1 and Zn2 are higher for Zn1 (H120, H122, and H189) than for Zn2 (D124, C208, and H250) (Figure 4a). These observations are validated by comparing the B-factors of several NDM-1 structures crystallized in the absence and presence of Zn(II). B-factors can be used to identify and interpret the dynamic characteristics of proteins.68 Furthermore, the flexibilities of ASL1 observed by NMR for di-Zn(II) NDM-1 (Figure 4b) are in agreement with that observed by comparison of several X-ray crystal structures of di-Zn(II) NDM-1, which show only significant differences in the orientations of the ASL1 loop (Figure 4b). These observations are also consistent with the results showing that the loop ASL1 is a key loop capable of correctly orienting the substrate in the active site and should therefore, due to its flexibility, be able to adapt to many types of ligands.13,14,67,69,70
Flavonol Molecules Inhibit NDM-1 Activity
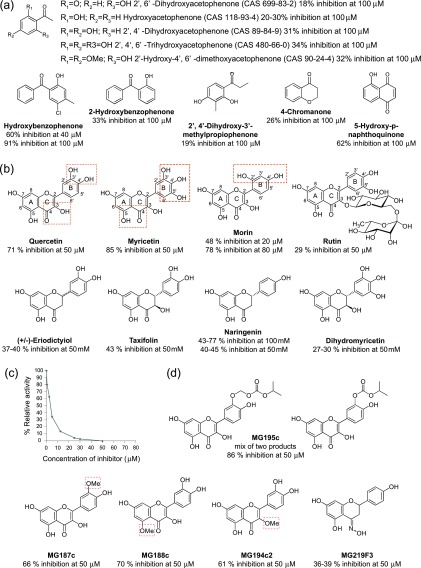
NDM-1 uses two Zn(II) ions and a water molecule coordinated in between in its active site, for the hydrolysis of the antibiotics β-lactam ring.8 Based on this mechanism, we sought to determine the inhibitory activity of simplified structures, providing an initial structure–activity relationship (SAR) that highlighted a common fragment, which was a phenol, substituted with a keto moiety in the position 2 (Figure 5a). The second ketone substituent may be aromatic, enolic, or aliphatic, the best inhibitory activities being obtained in the first two cases. On the basis of this minimal structure, a chemoinformatic analysis identified 222 molecules of this type available within the LabEx LERMIT chemical library (Figure 5). Biological screening of this library using the miniaturized automated assay allowed us to identify three flavonols (quercetin, myricetin, and morin) as the best inhibitors of NDM-1 (Figure 5b). Overall, the best results have been obtained for myricetin (85% of inhibition at 50 μM). The IC50 for myricetin was determined as 3.3 μM (Figure 5c).
Figure 5.
(a) Summary of the inhibitory activity of commercially available hydroxyketone derivatives. (b) All of these molecules have the general structure of the flavonoids, a 15-carbon skeleton, which consists of the two phenyl rings A and B and the heterocyclic ring C. Quercetin, myricetin, morin, and rutin differ from each other in the number and position of the hydroxy groups: quercetin, myricetin, and morin. In rutin, the hydroxy group at position C-3 is substituted by glucose and rhamnose sugar groups. The potential chelation sites of Zn(II) are surrounded by red rectangles. Eriodictyiol, taxifolin, naringenin, and dihydromyricetin have two stereocenters on the C-ring, unlike the previous ones which have none. They also differ from each other in the number and position of hydroxy groups. (c) IC50 determination for myricetin on purified NDM-1. Representation of the IC50 curve for myricetin with the residual activity of NDM-1 monitored by hydrolysis of different concentrations of inhibitor with a constant concentration of imipenem (100 μM). (d) Quercetin derivatives have been synthesized and tested as inhibitors of NDM-1, with methylation on the 3′ (MG187c), 3 (MG194c2), or 5 (MG188c) hydroxy group, oxime formation on the 4-carbonyl group of naringenin (MG219F3), and the addition of isopropyl carbonate or methyl isopropyl carbonate on the 3′ hydroxy group (MG195c).
Due to solubility problems, IC50 values for quercetin could only be estimated between 5 and 10 μM. To solve this solubility problem, derivatives of quercetin were synthesized (Figure 5d, Supporting Information Figures S8–S10, and Supporting Information Experimental Section on the chemical synthesis of the compounds) that were expected to have improved solubility or bioavailability. Unfortunately, their inhibitory activities were not very different from that observed for quercetin (Figure 5d). Only the mixture of quercetin derivatives with the addition of isopropyl carbonate or methyl isopropyl carbonate to the 3′ hydroxy group (MG195c) has a higher inhibitory activity than quercetin and also higher than that observed with myricetin (Figure 5d). On the other hand, oxime formation on the 4-carbonyl group of naringenin (MG219F3) induced a slight decrease in the inhibitory activity of this compound (Figure 5b,d). On the basis of these results, it appears that: the 4-carbonyl group is important for the activities of the molecules; the possible steric gene induced by OMe in position 3′, 5, or 3 has little effect on their activities; and the addition of polar compounds such as isopropyl carbonate or methyl isopropyl carbonate to hydroxy group 3 increases the activity of the quercetin derivative. None of the quercetin derivatives produced better result than myricetin. Myricetin was also tested at a concentration of 50 μM against other carbapenemases: OXA-48 and KPC-2, with inhibitions of 90 and 78%, respectively.
In Vivo Activities of Flavonol Molecules on Clinical NDM-1 Producing Escherichia coli and Klebsiella pneumoniae Isolates
Minimum inhibitory concentrations (MICs) of imipenem for K. pneumoniae CAG andE. coli GUE expressing NDM-1 were determined in the presence of increasing amounts of myricetin (Table 1) to assess in vivo activity on bacteria. Height-fold drop in MICs was observed with 500 μM myricetin (decreasing from 128 to 16 μg/mL) for K. pneumoniae CAG, and a 16-fold drop in MICs for E coli GUE with 500 μM myricetin (from 128 to 8 μg/mL), illustrating that these molecules have an in vivo activity. A concentration of 500 μM myricetin has no effect on the growth of the bacteria in the absence of imipenem. The values for quercetin could not be determined accurately, due to solubility problems. Nevertheless, at 100 and 200 μM, two and three dilution drops of MICs were observed with the E. coli GUE NDM-1 strain, respectively. No effect was observed with the strain K. pneumoniae CAG (Table 1). From our inhibitor activity assay data, we identified the flavonols as candidate inhibitors of the NDM-1 activity. The flavonols are abundant in various fruits and vegetables and are known to interact with proteins involved in cancer, as well as cardiovascular and brain diseases.54−62,71,72
Table 1. Minimum Inhibitory Concentrations of Imipenem in the Presence of Myricetin and Quercetin for E. coli GUE NDM-1 and K. pneumoniae CAG NDM-1.
| myricetin |
quercetin |
|||||
|---|---|---|---|---|---|---|
| concentration (μM) | 0 | 50 | 100 | 0 | 100 | 200 |
| MICs (μg/mL) | ||||||
| E. coli NDM-1 GUE | 128 | 32 | 8 | 128 | 32 | 16 |
| K. pneumonia NDM-1 CAG | 128 | 64 | 16 | 128 | 128 | 128 |
NMR Characterization of the Binding Interface between NDM-1 and the Three Flavonols: Morin, Myricetin, and Quercetin
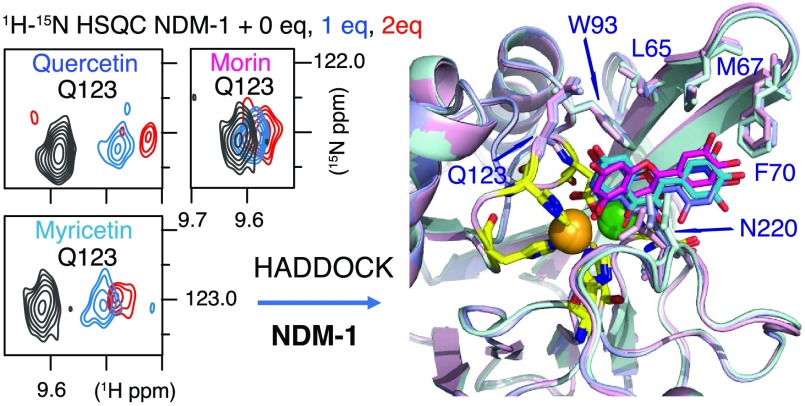
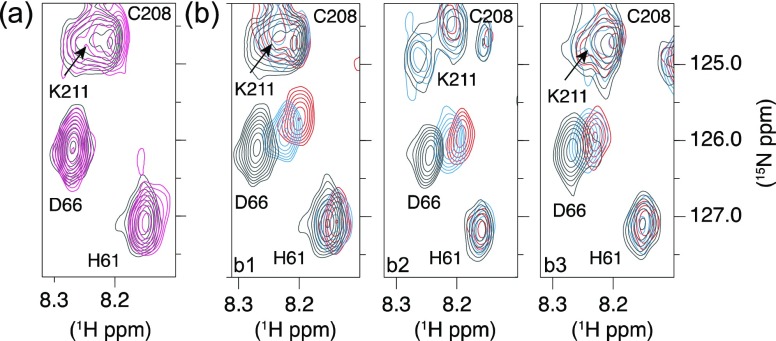
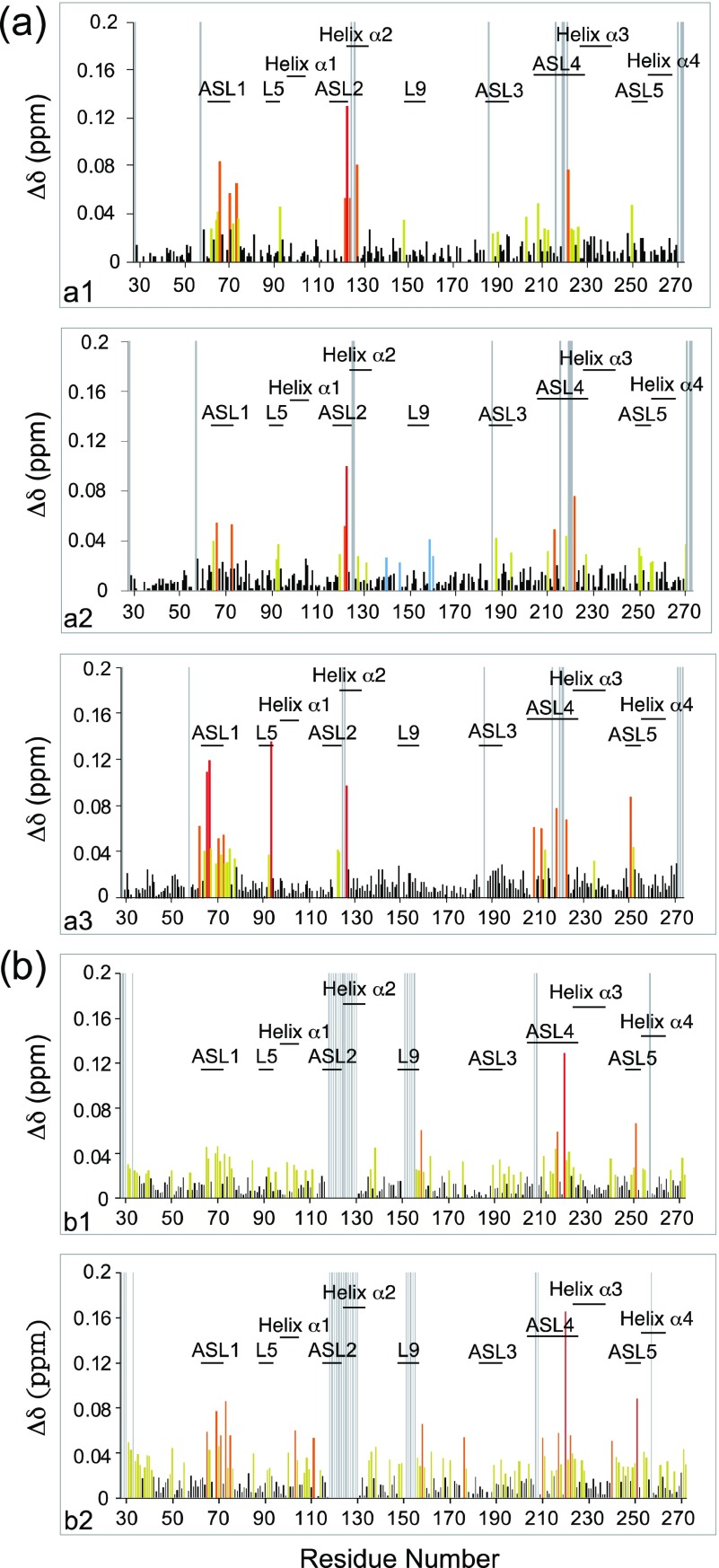
The NDM-1/flavonol interactions were investigated by NMR in the presence of Zn(II) (Figure 6). As the ligands (morin, myricetin, and quercetin) were poorly soluble in aqueous solution (these molecules are hydrophobic, and the Log P estimate indicated that myricetin had the best solubility in aqueous buffer (Supporting Information Table S3)), they were introduced into the NDM-1 samples from a concentrated stock of a deuterated dimethyl sulfoxide (DMSO). The addition of small amounts of DMSO did not cause precipitation of the protein or chemical shift variations (Figure 6a). The interaction of NDM-1 with the three flavonols resulted in significant chemical shift perturbations (CSPs) of many backbone amide protons and nitrogens. Representative extracts of 1H–15N HSQC spectra along the titration are shown in Figure 6b, and the 1H–15N chemical shift perturbations after the addition of 2 molar equiv of inhibitors are shown in Figure 7. The analysis highlighted a specific site for interaction of the three flavonols with NDM-1. The NDM-1 residues involved in the interaction with the three flavonols belong mainly to the flexible loops ASL1, ASL2, and ASL4, and to a lesser extent to ASL3 and L5 (Figure 7) with subtle differences between the three flavonols. The residues in the ASL1 and ASL4 loops have the highest CSPs in the presence of morin, and W93 and H250 have much higher CSP in the presence of morin (Figure 7a3) than in the presence of quercetin (Figure 7a1) or myricetin (Figure 7a2).
Figure 6.
Specific interaction between NDM-1 and flavonols. Selected regions of the 1H–15N HSQC spectra probing the NDM-1–flavonol interactions. (a) Addition of DMSO (black in the absence of DMSO and pink in the presence of the amount of DMSO corresponding to that added to have 2 molar equiv of flavonol relative to the concentration of NDM-1: 5.36 μL of DMSO) does not induce significant chemical shift variations of the NDM-1 resonances. (b) Titrations of NDM-1 (black in the absence of flavonol) by 1 molar equiv (in blue) and 2 molar equiv (in red) of morin (b1), quercetin (b2), and myricetin (b3) showing the influence of flavonols on several NDM-1 resonances. The spectra were recorded in the presence of 2 molar equiv of Zn(II).
Figure 7.
(a) Plots of the measured chemical shift perturbations of the amide protons and nitrogens of NDM-1, in the presence of 2 molar equiv of Zn(II), after addition of 2 molar equiv of flavonols: (a1) quercetin, (a2) myricetin, and (a3) morin. (b) Plots of the measured chemical shift perturbations of the amide protons and nitrogens of NDM-1, in the absence of Zn(II) and 2 molar equiv of morin (b1) and 4 molar equiv of morin (b2). In red, orange, and yellow are represented the residues with Δδ > 0.09 ppm, 0.09 > Δδ > 0.05 ppm, and Δδ > 0.02 ppm, respectively. The unassigned residues of NDM-1 are represented by gray lines.
On the contrary, the highest CSPs were observed in the ASL2 loop in the presence of quercetin compared to those observed after the addition of morin. Although specific, the flavonol binding induced CSPs over relatively large regions of the flexible domains such as 62–80, 208–223, and 250–254 (Figure 7a). CSPs were used to estimate the dissociation constant, Kd, of morin and myricetin despite the uncertainty in the actual concentration of the ligands available in solution due to their precipitation. As the exchange is rapid on the chemical shift timescale, an estimated Kd value of around 300 μM for the two ligands (Supporting Information Table S4 and Figure S11) could be calculated using the residues with the highest CSPs (L65 and D66). To determine the influence of Zn(II) on flavonol interactions, the CSPs of the NDM-1 backbone resonances were followed using the 1H–15N HSQC spectra recorded at each flavonol increment in the absence of Zn(II) (Figure 7b and Supporting Information Figure S12). As for the flavonol titrations performed in the presence of Zn(II) (Figure 6a), we verified that the addition of DMSO did not induce significant variations in the chemical shift of the NDM-1 resonances in the absence of zinc (Supporting Information Figure S12).
The analysis shows that flavonols also interact with NDM-1, but the CSPs are lower at equal ligand concentration and more dispersed along the protein sequence compared to what happens in the presence of 2 molar equiv of Zn(II). In the absence of Zn(II) and in the presence of 2 (Figure 7b1) and 4 molar equiv (Figure 7b2) of morin, for example, the ASL1 and ASL4 loops are the domains of NDM-1 that undergo the major CSPs. However, in the absence of Zn(II), several other regions are also targeted by the flavonols, including residues 100–111, 135–138, and 156–177, compared to that happened in the presence of Zn(II) (Figure 7a3). It is interesting to note that the two tryptophan residues (W104 and W168) and the many localized hydrophobic residues in these domains could interact with the highly hydrophobic flavonols. Thus, the two Zn(II) ions contributed to increase the selectivity of the interaction of NDM-1 for flavonols.
Our results indicate that morin, quercetin, and myricetin interact with NDM-1 residues at the active site, without ejection of Zn(II). Despite some chemical shift variations upon ligand binding, the 1H–15N HSQC spectra retain their initial appearance, namely, the appearance of the NDM-1 spectrum in the presence of 2 molar equiv of Zn(II), and not the NDM-1 spectrum recorded in the absence of Zn(II). In addition, the hydrophobic residues around the active site L65, F70, and W93 also appear to play an important role in substrate recognition.
Altogether, the residues L65, M67, F70, W93, H122, Q123, D124, N220, and H250 and the two Zn(II) are involved in the binding of flavonols. Our NMR data demonstrated the specific interaction of morin, quercetin, and myricetin at the NDM-1 active site.
These results are in agreement with the previous in silico modeling and docking with the generated NDM-1 models, from an alignment with two templates VIM-2 and VIM-4 by Ganugapati et al.73 and the crystal structure of NDM-1 (PDB: 3SPU) by Padmavathi et al.74 The results of these two studies indicated that quercetin may be the best inhibitor among the flavonoids present in green tea (flavan-3-ols, flavones, and flavonols) tested during dockings, in terms of energy values for the first study,73 and calculated binding affinity after docking for the second study.74
Reconstruction of the NDM-1/Flavonol Binding Interfaces by Molecular Docking Simulations
As no intermolecular NOE was observed in the 15N–13C filtered 1H–1H NOESYs recorded on 13C–15N-labeled NDM-1/flavonol complexes, the structural models of the NDM-1/morin, NDM-1/quercetin, and NDM-1/myricetin complexes were constructed using molecular docking simulations in the NMR data-driven program HADDOCK (High Ambiguity Driven biomolecular DOCKing)75 on the basis of experimental CSPs. To propose relevant models, several series of calculation were undertaken using 19 crystallographic structures of NDM-1 as the docking results may be influenced by the starting protein structure,76 whether obtained in the presence or absence of ligand. Ten of them were complexed with a ligand and were used after removal of the complexed ligand, and we called them “artificial” NDM-1 crystal structures (art-apo NDM-1). Five structures were apo NDM-1 crystal structures (crystallized in the absence of ligand), and four were metal-free apo forms of NDM-1 (crystallized in the absence of ligand and in the absence of Zn(II)). The superimposition on the backbone atoms (domain 45–270) of the 10 art-apo NDM-1 structures with each other and with the five apo NDM-1 structures highlights that the main structural differences between all of these structures are located in the loops ASL1 and ASL4 (Supporting Information Figure S13). Furthermore, the distances between the two Zn(II) (Zn1 and Zn2) fluctuate from one structure to the other depending on the presence or absence of a ligand and also of the type of ligand, with the Zn1–Zn2 distances varying from 3.1 to 3.8 Å in the five apo NDM-1 crystal structures, and from 3.6 to 4.6 Å in the 10 art-apo NDM-1 structures used for our docking. In fact, it has been shown by Zhang et al.77 that Zn1–Zn2 distance in the active site of the native protein is affected by crystal packing, pH, and buffer components. Although the Zn1–Zn2 distances in the enzyme/product complexes are mostly at 4.6 Å,77 this distance is smaller in several other NDM-1 complexes (Supporting Information Table S5).
Based on our experimental NMR CSP values, the residues L65, M67, F70, W93, H122, Q123, D124, N220, and H250 (Supporting Information Figure S13d,e) were determined to belong to the interface and used as ambiguous interaction restraints in docking calculation with morin, which induced the largest CSPs (Figure 7) in the NDM-1 spectra. As the NDM-1 CSPs could be the result of direct interactions with flavonol or/and the result of conformation changes of the flexible loops, NDM-1 L65, M67, F70, W93, H122, Q123, D124, N220, and H250 residues were defined as “passive” residues and flavonol as an “active” residue (as defined in the HADDOCK software) in a first step. The best generated models, selected on the basis of the HADDOCK score (Supporting Information Table S6), using the four metal-free apo (Supporting Information Figure S14 and Table S6), the five apo (Supporting Information Figure S15 and Table S6), or the 10 art-apo NDM-1 structures (Supporting Information Figure S16 and Table S6), show that the morin is located in the substrate-binding pocket and is able to contact the Zn(II) ions via polar contacts.
The flavonols are recognized as metal-ion chelators;78−82 they bind to Zn(II) ions82 with a lower affinity than NDM-1 does.23,24 A binding association constant of 3.57 ± 0.1 × 104 M–1 was found between quercetin and Zn(II) in protein-free solution by Bhuiya et al.82 A flavonol molecule has three potential sites for cation chelation: (i) the 3′,4′ or 2′,4′-dihydroxy group or 3′,4′,5′-trihydroxy group located on the B-ring (depending on the type of flavonol); (ii) between the 3-hydroxy and the 4-carbonyl groups in the C-ring; (iii) and via the 5-hydroxy and the 4-carbonyl group (Figure 5b). In addition, we determined by NMR that the Zn(II) ions direct flavonols binding to the active site of NDM-1 (Figure 7). All of these data showed that both Zn(II) ions probably play a role in the coordination of flavonols at the NDM-1 binding site. Therefore, in the second step of docking simulations, both Zn(II) were defined as active residues in addition to the L65, M67, F70, W93, H122, Q123, D124, N220, H250, and NDM-1 residues defined as passive residues and morin as an active residue using again the five apo (Supporting Information Figure S17 and Table S7) and the 10 art-apo NDM-1 crystallographic structures (Supporting Information Figure S18 and Table S7).
Analysis of the four best structures of each top clusters shows that, as expected, the morin targets the Zn(II) ions through polar contacts. The orientation of the morin in the active site of NDM-1 was strongly dependent on the crystallographic structure used for docking (Supporting Information Figures S17–S18). Either the morin adopts only a single orientation in the NDM-1 active site for the four structures in each top cluster or the orientations are very different from each other. In the first case, the rmsd values calculated on the morin atoms after superimposition on the (45–271) backbone atoms of the NDM-1 structures (art-apo PDB: 4EY2, 4EYB, 4HL2, 4RL0, 4EYF) are between 0.68 and 1.43 Å, and in the second case, (art-apo PDB: 4EXS, 4EYL, 4RAM, 4RBS, 4RL2) between 2.58 and 5.40 Å (Supporting Information Table S8). It is interesting to note that this difference in behavior seems to be related to the Zn1–Zn2 distances in the starting structures of NDM-1 used for the docking since they vary from 4.5 to 4.6 Å in the first case and from 3.1 to 4.1 Å in the second case (Supporting Information Table S5) and also to the orientation of the ASL1 loop toward the active site (Supporting Information Figure S13c). A 62.5% of the structures analyzed adopt orientation 1 (Figure 8a,c) with an average rmsd of 0.91 ± 0.32 Å for 52.5% of them and 3.24 ± 0.59 Å for the remaining. A 17.5% of the structures are in orientation 2 (Figure 8b,d) with a main rmsd value of 3.35 ± 1.09 Å between them. We observed a loss of Zn2 coordination in the remaining structures (20%), so we did not associate them to any group. It is interesting to note that the loss of Zn2 coordination is only observed for the NDM-1 starting structures with the shortest Zn1–Zn2 distances. More precisely, the Zn1–Zn2 distance is the factor that most strongly affected the positioning of the ligand in the active site during molecular docking simulations. As already described by Zhang et al.,77 although the Zn1–Zn2 distances observed in different structures of NDM-1 complexed with hydrolyzed antibiotics are longer than those observed for NDM-1 in the native form, they fluctuate after all between 3.8 and 4.6 Å and are also highly dependent on the type of ligand. Therefore, the choice of NDM-1 starting structures for docking with ligands capable of binding Zn(II) strongly influences the final results. Our docking results show that the PDB crystal structures 4EY2, 4EYB, 4HL2, and 4EYF are the most suitable for docking calculations with the morin structure. Indeed, the smallest deviations of Zn1 and Zn2 after docking from their positions in the original crystal structures were observed with these four crystal structures (Supporting Information Figure S19). These structures have the largest distances between Zn1 and Zn2, 4.6 Å (Supporting Information Table S5). In addition, the rmsd values calculated on the morin atoms are also the lowest (Supporting Information Figure S18 and Table S8).
Figure 8.
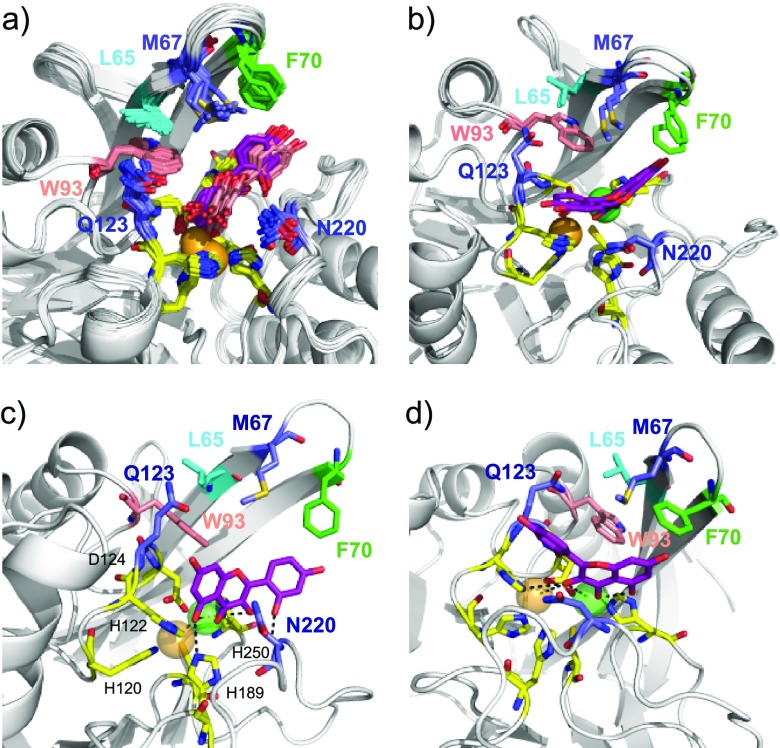
Models of the NDM-1/morin complexes generated using HADDOCK. (a) Superimposition of 21 structures over the 40 structures clustered in the top clusters, generated using the 10 art-apo NDM-1 crystallographic structures (PDB: 4EXS, 4EY2, 4EYB, 4EYF, 4EYL, 4HL2, 4RAM, 4RBS, 4RL2, 4RL0) showing the predominant orientation of morin at the active site of NDM-1 (orientation 1). These 21 structures adopt two slightly different orientations in the active site, 11 are in dark pink, and 10 in light pink. (b). A minority orientation is also observed in the docking generated structures (orientation 2). Orientation 2 differs from orientation 1 by a rotation of about 180°. (c) Zoom on one of the 21 structures in which morin adopts orientation 1 showing the polar contacts between the ligand and Zn(II) ions (PDB: 4HL2) (cluster 1: structure 2). (d) Zoom on one of the structures in which morin adopts orientation 2 showing the polar contacts between the ligand and Zn(II) ions (PDB: 4RBS (cluster 1: structure 1)). The Zn(II) ligands are represented by stick and colored yellow, Zn1 is orange, and Zn2 is green and represented by spheres. L65, M67, F70, W93, Q123, and N220, defined as active residues (in addition to H122, D124, and H250 in yellow) upon docking are represented by stick and colored differently for better identification.
In orientation 1 and orientation 2, electrostatic interactions are observed between Zn2 and O-3 (cycle C), O-4 (cycle C), and O-5 (cycle A). Zn1 is either targeted by 5-O (cycle A) in orientation 1 or by O-3 (cycle C) in orientation 2 (Figure 8c,d). In addition, hydrogen-bond interactions are also observed between O-7 (cycle A) and the amide proton of Q123 and between O-2′ (cycle B) and the amide proton of N220 in orientation 1 in several complexes (Figure 8c). Morin also showed a H-bonding with H189 in orientation 1 (Figure 8c). In addition, the three-ring system forms extensive nonpolar interactions with L65, M67, F70, and W93 (Figure 8). Moreover, the CSPs observed for several Zn(II) ligands such as H122, D124, and H250, and to a lesser extent C208, could be explained by a slight movement of Zn1 and Zn2 and their ligands during the interaction between NDM-1 and flavonols, as well as a direct interaction with flavonols (Figure 8).
To investigate whether the number and position of hydroxy groups on the B-ring could influence the orientations of flavonol in the active site of NDM-1, docking simulations were also undertaken using quercetin (Supporting Information Table S9) and myricetin (Supporting Information Table S10) using the protocol already described in the previous paragraph. The molecular docking results show that morin, quercetin, and myricetin are located in the same binding site (Supporting Information Figure S20). The variability of ligand orientations in the active site of NDM-1 is greater for quercetin and myricetin than for morin and the number of structures rejected (usually due to loss of Zn2 coordination (Supporting Information Figure S18)) is also greater for quercetin and myricetin than for morin. Quercetin and myricetin also predominantly adopt orientation 1. In orientation 1 and orientation 2, the Zn(II) ions/morin, Zn(II) ions/quercetin, and Zn(II) ions/myricetin interactions are quite similar regardless of flavonol.
It is interesting to note that the addition of methoxy groups at the 3′, 5, or 3 positions slightly decrease the inhibition of native quercetin (Figure 5d). We verified using molecular docking simulation that the three quercetin derivatives are still able to interact with Zn(II) (Supporting Information Figure S21) with approximately the same structural statistics (Table S11) as quercetin wild type. As for quercetin, orientation 1 is mainly adopted by quercetin derivatives. In all cases, their C4 carbonyl group interacts with Zn2 in the active site of NDM-1. Moreover, despite the possible steric hindrance generated by the replacement of the hydroxyl group by a methoxy at positions 3 (MG194c2) or 5 (MG188c) (Figure 5d), the structures resulting from docking show that these two quercetin derivatives are still capable of complexing Zn1 by their 5-O (cycle A). This is also true for the derivative in which the hydroxyl group has been replaced by a methoxy at the 3′ position (MG187c). The slight decreases in inhibition are explained by the steric hindrance generated by the methoxy groups.
There are very few flavonol structures that interact with metal ions within a protein. Interestingly, quercetin has been shown to have strong inhibitory activities against yeast alcohol dehydrogenase, which has been explained on the basis of the formation of a complex between quercetin and the Zn(II) ion that maintains the tertiary structure of the metallo-enzyme.7,82 In addition, the crystal structures of quercetin 2,3-dioxygenase, a mononuclear copper-dependent dioxygenase (PDB: 1H1I, 1H1M),83 and quercetin 2,4-dioxygenases, which uses nickel as an active site cofactor to catalyze oxidative cleavage, complexed with the quercetin substrate (PDB: 5FLI),84 show that the coordination of the metal ions involved in both cases, the 3-hydroxy and the 4-carbonyl groups in the C-ring of quercetin with distances around 2.3 and 3.5 Å, respectively. We also observed in orientation 1 and orientation 2 of our NDM-1/flavonol models the electrostatic interactions between Zn2 and the 3-hydroxy and the 4-carbonyl groups of quercetin.
NDM-1 hydrolyzed antibiotics such as methicillin (PDB: 4EY2), oxacillin (PDB: 4EYB), meropenems (PDB: 4RBS, 4EYL), benzylpenicillin (PDB: 4EYF), cephalosporins (PDB: 4RL2, 4RL0), penicillin G (PDB: 4RAM), and ampicillin (PDB: 4HL2) bind the two Zn(II) through polar interaction with the carbapenem C3 and C6 carboxylate oxygens (Supporting Information Figure S22). Most of MBL inhibitors belong to one of two categories: either they act by sequestration of Zn(II), such as EDTA17,47,85,86 and aspergillomarasmine A,87 resulting in inactivation of the enzyme, or they have several electron donor groups, such as carboxylate, carboxyl, thiol, and/or nitrogen, which are capable of binding one or both Zn(II) in the active site17,27−31,33−44,50,88−91 to act as competitive inhibitors. We show that flavonols belong here to the second category and that they are able to bind to both Zn(II) in the active site of NDM-1, without expelling the Zn(II) ions from the protein.
Conclusions
Our analysis of NDM-1 by NMR in the absence and presence of Zn(II) shows that specific regions (ASL2, ASL3, ASL5, and L9 loops and Helix α2) of the protein were affected by the binding of two Zn(II). Ion binding led to the rigidification of the protein with respect to the metal-free NDM-1, but the ASL1 loop retained its slight flexibility even after Zn(II) binding. Our assay of inhibitor activities and structural characterization of NDM-1/flavonols complexes in the presence of Zn(II) show that morin, myricetin, and quercetin interact in the same binding site as antibiotics and that these interactions are largely driven by the interaction with both Zn(II) as is the case for substrate recognition in MBLs, which is largely determined by the interaction with metal ions.92,93
Experimental Section
Protein Production and Purification
The gene sequence encoding the NDM-1 mature enzyme from G29 to R270 was subcloned into a pET41b vector with a C-terminal His6 tag to facilitate protein purification. For the overexpression of the double 15N/13C/-labeled NDM-1 sample, the fusion protein was expressed in BL21 E. coli cells. A 100 mL Lysogeny broth (LB) culture supplemented with 40 μg/mL kanamycin was inoculated with 10 mL of a fresh overnight LB preculture and grown at 37 °C until OD600 = 1.5. After a 10 min centrifugation at 5000 rpm, the bacterial pellet was resuspended in 1 L of M9 minimal medium (Na2HPO4, 6 g/L; KH2PO4, 3 g/L; NaCl, 0.5 g/L; MgSO4, 1 mM; CaCl2, 1.10–4 M) containing 2.0 g/L [13C]glucose, 1 g/L [15N]NH4Cl, and 40 μg/mL kanamycin preincubated at 37 °C. The culture was maintained in this medium at 37 °C until OD600 ∼ 0.6. Subsequently, induction with 0.4 mM isopropyl-β-d-thiogalacto-pyranoside took place for 20 h at 28 °C.
The bacteria were spun down at 5000 rpm for 10 min, and the pellet (3.5 g) was washed in phosphate-buffered saline (PBS); sonicated 15 times with 10 s pulses at 4 °C in 30 mL of buffer A (20 mM sodium phosphate pH 7.5, 0.5 M NaCl, 10 mM imidazole, 1 mM β-ME) containing 0.5 mg/mL lysosyme, 100 μg/mL DNase, and one tablet of complete mini EDTA-free protease inhibitor cocktail (Roche Diagnostics); and centrifuged (100 000g for 30 min). The supernatant was applied two times on a 1 mL His GraviTrap affinity column (GE Healthcare) equilibrated with buffer A, and the column was washed with 20 mL of buffer A containing 20 mM imidazole. Then, elution of the protein was carried out in buffer A containing 0.3 M imidazole. The eluted fraction containing pure NDM-1 (about 50 mg) was brought to a concentration of 500 μM by ultrafiltration and dialysis against 0.1 M Bis-Tris-HCl pH 7.0.
Thermal Shift Assay
The thermal shift assay was performed using the ABI7900HT Real-Time PCR system (Applied Biosystems). The assay quantifies the binding of the Sypro Orange probe to exposed hydrophobic regions of proteins challenged to a temperature gradient.94 The dye becomes highly fluorescent when bound to protein hydrophobic sites. NDM-1 samples (9 μM) were incubated in buffer 100 mM Bis-Tris pH 7 containing different ZnCl2 concentrations and Sypro Orange (650-fold diluted from stock solution; Invitrogen). Reactions were carried out in duplicate in a 96-well fast PCR plate at a final volume of 20 μL. The samples were submitted to denaturation from 15 to 95 °C at a rate of 3 °C/min, and fluorescence of Sypro Orange dye was recorded in real time (excitation spectra, 488 nm; emission spectra, 500–650 nm). The protein denaturation temperature was calculated using the software SDS2.4.1 as the maximum of the derivative of the resulting fluorescence curves.
Enzyme Activity Assay Screening Library
All of the compounds tested as potential inhibitors of NDM-1 were either purchased from Sigma-Aldrich Corporation or available from LabEx LERMIT. The CAS numbers are as follows: hydroxybenzophenone (CAS 68751-90-6), 2-hydroxybenzophenone (CAS 117-99-7), 2′,4′-dihydroxy-3′-methylpropiophenone (CAS 63876-46-0), 5-hydroxy-p-naphthoquinone (CAS 481-39-0), 4-chromanone (CAS 491-37-2), quercetin (CAS 117-39-5), myricetin (CAS 529-44-2) and morin (CAS 654055-01-3), rutin (CAS 207671-50-9), eriodictyiol (CAS 4049-38-1), taxifolin (CAS 480-18-2), naringenin (CAS 67604-48-2), and dihydromyricetin (CAS 27200-12-0). They were dissolved in 100% DMSO as 5 or 10 mM stock solutions. The imipenem antibiotic was purchased from Ranbaxy and dissolved in Milli-Q water as a 10 mM stock solution. To determine the percentage of inhibition, a miniaturized automated assay was set up based on a 96-well plate format with a concentration of 0.5 μg/mL NDM-1. The screening assay conditions were as follows: 100 mM phosphate buffer, pH 7.0 (Na2HPO4/NaH2PO4) added with 50 μM ZnSO4; 100 μM imipenem, and 50 μM of compound (dissolved in DMSO) monitored by a Varian Cary 300 Bio UV–Visible spectrophotometer at 297 nm; time course, 600 s; 25 °C. Imipenem was added into the reaction after 5 min of incubation. In parallel, two controls were performed: positive control was performed with DMSO instead of compound and the negative control with 1 mM EDTA. The equation for the calculation of the percentage of inhibition is: Percentage inhibition = (1 – (I0 – IEDTA)/(IDMSO – IEDTA)) x 100%, where I0 is the intensity of the product signal in each well, IEDTA is the intensity in the presence of EDTA, and IDMSO is the intensity in the presence of DMSO.
IC50 Determination
IC50 of the inhibitors was determined in 100 mM phosphate buffer, pH 7.0 (Na2HPO4/NaH2PO4), added with 50 μM ZnSO4 and 100 μM imipenem, and the mixture was incubated for 3 min. The assay was performed with an ULTROSPEC 2000 UV spectrophotometer and SWIFT II software (GE Healthcare, Velizy-Villacoublay, France). IC50 values were obtained using the equation IC50 = ((1/0.5 × v0) – m)/q, where v0 is the rate of hydrolysis of the reporter substrate (v0 being the rate measured in the absence of inhibitor), q is the y axis intercept, and m is the slope of the resulting linear regression.
Minimum Inhibitory Concentration (MIC) Determination
MIC values were determined by broth microdilution, in triplicate, in cation-adjusted Mueller Hinton broth according to the Clinical Laboratory Standards Institute (CLSI) guidelines. Two enterobacterial clinical strains expressing the carbapenemase NDM-1 were used: E. coli NDM-1 GUE and K. pneumoniae NDM-1 CAG. Experiments were performed in microtiter plates containing the medium with imipenem and inhibitors (myricetin and quercetin) dissolved (DMSO) at different concentrations (0.5, 5, 50, 500, and 1000 μM). Plates were incubated overnight at 37 °C for 18–24 h.
NMR Backbone Resonance Assignment
Samples for NMR spectroscopy are composed either of 0.1 M Bis-Tris-HCl pH 7.0 buffer, supplemented by 150 mM NaCl or 50 mM phosphate sodium pH 6.5 depending, respectively, on the presence or absence or of ZnCl2 in the solutions, and 95%/5% H2O/D2O. Protein concentration was set from 100 to 996 μM. The NMR spectra were recorded on 600 and 800 MHz Bruker Avance III spectrometers at different temperatures ranging from 20 to 35 °C depending on the presence or absence of ZnCl2. The two spectrometers are equipped with a 5 mm z-gradient TCI (H/C/N) cryoprobe.
The backbone resonance assignments were performed in the absence or presence of ZnCl2 using the standard triple resonance experiments: HNCA, HN(CO)CA, HNCO, HN(CA)CO, HNCACB, and HN(CO)CACB. 3D NOESY-15N edited spectra (mixing times 100 and 200 ms) were also collected to help the assignment. NMR data were processed using TOPSPIN 3.5 software (Bruker) and analyzed using CcpNmr 2.4.2 software (http://www.ccpn.ac.uk).95 NDM-1 resonance assignments in the absence and presence of 2 molar equiv of Zn(II) were deposited in BMRB accession numbers 26950 and 26952, respectively.
15N Relaxation Measurements
The 15N R1 and R2 relaxation rates and {1H}–15N heteronuclear NOE were measured at 25 °C and 600 MHz 1H frequency on the metal-free NDM-1 and di-Zn(II) NDM-1 complex. The experiments were based on the TROSY-based relaxation experiments96 and recorded in an interleaved pseudo-3D method. For the determination of R1 relaxation constants, 13 total data sets were collected at relaxation delay times of 20 (duplicated), 100, 200, 300, 500, 600, 750, 900, 1000, 1200, 1400, and 1700 ms. For the determination of R2 relaxation constants, 13 data sets were collected at delay times of 16.96 (duplicated), 33.92, 50.88, 67.84, 101.76, 118.72, 152.64, 169.60, 203.52, 237.44, 271.36, and 305.28 ms.
Two two-dimensional (2D) 1H–15N planes were collected in the presence or absence of a 3 s 1H saturation sequence (120° 1H pulses train) during relaxation delay to extract the {1H}–15N heteronuclear NOE. Spectral widths were 1.630 and 4.208 kHz along the 15N and 1H dimensions, respectively, and the interscan delay was set to 5 s. The acquisition times were 121.65 and 41.5 ms in the 1H and 15N dimensions, respectively, with 16 scans per increment for the determination of R1 and R2 relaxation constants. The duration of the experiments was around 2 days each. For {1H}–15N heteronuclear NOE experiments, the acquisition times were 121.65 and 62.3 ms in the 1H and 15N dimensions, respectively, with 32 scans per increment and a duration of experience of 14 h.
NMR HSQC Titration Experiments
The NDM-1–Zn(II) interaction was characterized by means of chemical shift perturbations (CSPs), using a series of 1H–15N HSQC experiments recorded upon the addition of increasing concentration of ZnCl2. The protein, concentrated to 320 μM, was solubilized in 0.1 mM Bis-Tris solution, pH 7, supplemented by 150 mM NaCl. ZnCl2 was dissolved in 0.1 mM Bis-tris buffer, pH 7, to a concentration of 20 mM in the stock solution.
The flavonols (morin, quercetin, and myricetin) interaction surfaces on NDM-1 were characterized in the presence of 2 molar equiv of Zn(II). The flavonols were dissolved in 100% DMSO at a concentration of 20 mM. We titrated 320 μM of NDM-1 with the flavonols (0–4 molar equiv for morin, 0–3 molar equiv for myricetin, and 0–2 molar equiv for quercetin). Approximately 2.7 μL of DMSO was added by an equivalent of flavonol (the final concentration of DMSO-d6 did not exceed 5% in the NMR tube), and at each stage of the titration, one-dimensional (1D) 1H and 2D 1H–15N HSQC spectra were recorded. In all cases, we observed a slight precipitation of the ligands probably due to their hydrophobic properties. Each HSQC spectrum was collected with eight scans per increment with spectral widths of 9.6 and 3.2 kHz for 1H and 15N, respectively, as well as 256 data points in the indirect dimension. The experiments used the 3-9-19 watergate sequence97 for water suppression. The weighted chemical shift change of each NH signal was calculated according to:
Δδ (1H, 15N) = [((ΔδH)2 + (ΔδN)2 × 0.14) × 0.5]1/2, where δH and δN are the modifications of the chemical shift in the 1H and 15N dimensions, respectively.98
NDM-1/Flavonol Dockings Using HADDOCK
Ambiguous distance restraints based on chemical shift perturbations were used to drive the docking between NDM-1 and the three flavonols: morin, quercetin, and myricetin. Nineteen crystallographic structures of NDM-1 were used: (i) 10 of them were in complex with a ligand: hydrolyzed methicillin (PDB: 4EY2), hydrolyzed oxacillin (PDB: 4EYB), l-captopril (PDB: 4EXS); hydrolyzed meropenems (PDB: 4EYL, 4RBS); hydrolyzed benzylpenicillin (PDB: 4EYF); hydrolyzed cephalosporins (PDB: 4RL0, 4RL2), hydrolyzed penicillin G (PDB: 4RAM), hydrolyzed ampicillin (PDB: 4HL2), these structures were used after removing of the ligand localized in the active site of the NDM-1 crystallographic structures, and named “artificial apo” (art-apo) NDM-1 structures; (ii) five of them were apo NDM-1 crystal structures (PDB: 4TYF, 3S0Z, 4TZF, 4TZE, 3SPU); (iii) four were metal-free apo form of NDM-1 (PDB: 3RKJ, 3RKK, 3SBL, 3PG4). The three flavonol structures were extracted from crystal structures: morin PDB: 6AE3, 5AUY; quercetin PDB: 2UXH; myricetin PDB: 2O63 where they were in complex with proteins. The L65, M67, F70, W93, H122, Q123, D124, N220, and H250 residues were defined as belonging to the NDM-1 interaction surface with the flavonols. In a first series of calculations, the two Zn(II) were defined as passive residues and in a second series as active residues. In addition, a restraint file (unambiguous restraints) that imposed the distances between Zn1 and Zn2, on the one hand, and between Zn1 and Zn2, and their respective ligands His120, His122, His189 and Asp124, Cys208, His250, was supplied to HADDOCK75 to maintain the geometry around each Zn(II). For the two series of calculations, L65, M67, F70, W93, Q123, N220, D124, and H250 were defined as passive residues. In all of the steps, the residues belonging to the segments 65, 92–94, 123, and 218–222 were defined as fully flexible segments, and those belonging to 66–73 as semiflexible segments. During the rigid-body docking, 2000 structures were calculated, and 400 or 200 (depending on the calculation) during both simulated annealing and water refinement. All 400 or 200 water-refined structures were analyzed, and the cutoff for clustering was 7.5, or 0.6 Å for interface RMSD or Fraction of Common Contacts (FCC) respectively, with four structures per cluster. The ranking of the clusters is based on the average score of the top four members of each cluster. The HADDOCK score is calculated as a function of the intermolecular van der Waals energy, intermolecular electrostatic energy, empirical desolvation energy term, and ambiguous interaction restraints (AIRs) energy. The cluster numbering reflects the size of the cluster, with cluster 1 being the most populated cluster. After successful docking, the best complex models were selected on the basis of the HADDOCK score (Tables S6, S7, S9, and S10). The same protocol was used for the docking of NDM-1 (PDB: 4EY2) and three quercetin conjugates: quercetin-CH3A (quercetin with OMe in position 5); quercetin-CH3B (quercetin with OMe in position 3′); and quercetin-CH3C (quercetin with OMe in position 3) (Table S11).
Glossary
Abbreviations
- GNB
Gram-negative bacteria
- MBLs
metallo-β-lactamases
- MDR
multidrug resistance
- NDM-1
New Delhi metallo-β-lactamase-1
- MICs
minimum inhibitory concentrations
- CSPs
chemical shift perturbations
- ASL
active site loop
- PDB
Protein Data Bank
- HADDOCK
High Ambiguity Driven biomolecular DOCKing
- art-apo
artificial apo
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c00590.
Influence of zinc on the NDM-1 1H–15N HSQC spectra (Figures S1–S5); comparison of the NDM-1 secondary structures derived from NMR chemical shifts by TALOS+ with those observed in the X-ray structures and the Yao et al. results (Figure S6); schemes of the synthetic route of compounds MG188c, MG187c, MG195c, MG194c2, and MG219F3 (Figures S7–S9); experimental NMR titration curves (Figure S10); selected regions of the 1H–15N HSQC recorded in the absence of Zn(II) and in the presence of morin (Figure S11); superimposition of the 10 art-apo NDM-1 and apo-NDM-1 crystallographic structures (Figure S12); models of the NDM-1/morin complexes generated using the HADDOCK webserver using the conditions described in the Experimental Section (Figures S13–S17); relative deviations of Zn1 and Zn2 during docking (Figure S18); models of the NDM-1/quercetin and NDM-1/myricetin complexes (Figure S19); models of the NDM-1/quercetin conjugate complexes (Figure S20); comparison of the NDM-1/morin model structure and the PDB structures of NDM-1 in complex with hydrolyzed antibiotics (Figure S21); effect of Zn(II) on NDM-1 thermal stability (Table S1); average of the relaxation parameters of the flexible and rigid domains of NDM-1 (Table S2); log P of the flavonols (Table S3); estimated Kd values of morin and myricetin in complex with NDM-1 (Table S4); Zn1–Zn2 distances in PDB structures of NDM-1 (Table S5); NDM-1/morin docking results using HADDOCK (Tables S6 and S7); Rmsd calculated on the morin atoms in complex with NDM-1 (Table S8); NDM-1/quercetin docking results using HADDOCK (Table S9); NDM-1/myricetin docking results using HADDOCK (Table S10); NDM-1/quercetin derivatives docking results using HADDOCK (Table S11); Experimental Section: chemical synthesis of the compounds: MG188c, MG187c, MG195c, MG194c2, and MG219F3 (PDF)
Accession Codes
Atomic coordinates of the HADDOCK models of NDM-1/morin, NDM-1/quercetin, and NDM-1/myricetin have been deposited to the PDB under accession code PDB ID: 6TT8, 6TTA, and 6TTC, respectively. NDM-1 resonance assignments in the absence and presence of 2 molar equiv of Zn(II) were deposited in BMRB accession numbers 26950 and 26952, respectively.
Author Present Address
○ Sorbonne Université—Campus Pierre et Marie Curie, Laboratoire SU-UR6 “Enzymology of RNA” (Bât. B); Case Courrier 60-4, Place Jussieu, F-75252 Paris Cedex 05, France.
Author Present Address
∇ Personal address: 16 rue Sarrette, 75014 Paris.
Author Present Address
# German Center for Neurodegenerative Diseases (DZNE), Von-Siebold-Str. 3a, 37075 Göttingen, Germany.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
This work was supported by the Assistance Publique—Les Hôpitaux de Paris (AP-HP), the University Paris-Sud, the Laboratory of Excellence in Research on Medication and Innovative Therapeutics (LERMIT) supported by a grant from the French National Research Agency [ANR-10-LABX-33] and by the Joint Programming Initiative on Antimicrobial Resistance (JPIAMR) DesInMBL [ANR-14-JAMR-002], and the French National Centre for Scientific Research-CNRS. Financial support from the IR-RMN-THC Fr3050 CNRS for conducting the research is gratefully acknowledged.
The authors declare no competing financial interest.
Supplementary Material
References
- Oliveira J.; Reygaert W. C.. Gram Negative Bacteria. StatPearls; Treasure Island: FL, 2019. [PubMed] [Google Scholar]
- Ruppé E.; Woerther P. L.; Barbier F. Mechanisms of antimicrobial resistance in Gram-negative bacilli. Ann. Intensive Care 2015, 5, 61 10.1186/s13613-015-0061-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasoo S.; Barreto J. N.; Tosh P. K. Emerging issues in gram-negative bacterial resistance: an update for the practicing clinician. Mayo Clin. Proc. 2015, 90, 395–403. 10.1016/j.mayocp.2014.12.002. [DOI] [PubMed] [Google Scholar]
- Ambler R. P. The structure of beta-lactamases. Philos. Trans. R. Soc., B 1980, 289, 321–331. 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- Naas T.; Oueslati S.; Bonnin R. A.; Dabos M. L.; Zavala A.; Dortet L.; Retailleau P.; Iorga B. I. Beta-lactamase database (BLDB) - structure and function. J. Enzyme Inhib. Med. Chem. 2017, 32, 917–919. 10.1080/14756366.2017.1344235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordmann P.; Naas T.; Poirel L. Global spread of Carbapenemase-producing Enterobacteriaceae. Emerging Infect. Dis. 2011, 17, 1791–1798. 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page M. I. The reactivity of beta-lactams, the mechanism of catalysis and the inhibition of beta-lactamases. Curr. Pharm. Des. 1999, 5, 895–913. [PubMed] [Google Scholar]
- Bebrone C. Metallo-beta-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochem. Pharmacol. 2007, 74, 1686–1701. 10.1016/j.bcp.2007.05.021. [DOI] [PubMed] [Google Scholar]
- Bonomo R. A. New Delhi Metallo-beta-Lactamase and Multidrug Resistance: A Global SOS?. Clin. Infect. Dis. 2011, 52, 485–487. 10.1093/cid/ciq179. [DOI] [PubMed] [Google Scholar]
- Kumarasamy K. K.; Toleman M. A.; Walsh T. R.; Bagaria J.; Butt F.; Balakrishnan R.; Chaudhary U.; Doumith M.; Giske C. G.; Irfan S.; Krishnan P.; Kumar A. V.; Maharjan S.; Mushtaq S.; Noorie T.; Paterson D. L.; Pearson A.; Perry C.; Pike R.; Rao B.; Ray U.; Sarma J. B.; Sharma M.; Sheridan E.; Thirunarayan M. A.; Turton J.; Upadhyay S.; Warner M.; Welfare W.; Livermore D. M.; Woodford N. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 2010, 10, 597–602. 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh T. H.; Khoo C. T.; Wijaya L.; Leong H. N.; Lo Y. L.; Lim L. C.; Koh T. Y. Global spread of New Delhi metallo-beta-lactamase 1. Lancet Infect. Dis. 2010, 10, 828. 10.1016/S1473-3099(10)70274-7. [DOI] [PubMed] [Google Scholar]
- Nordmann P.; Poirel L.; Walsh T. R.; Livermore D. M. The emerging NDM carbapenemases. Trends Microbiol. 2011, 19, 588–595. 10.1016/j.tim.2011.09.005. [DOI] [PubMed] [Google Scholar]
- Kim Y.; Tesar C.; Mire J.; Jedrzejczak R.; Binkowski A.; Babnigg G.; Sacchettini J.; Joachimiak A. Structure of apo- and monometalated forms of NDM-1--a highly potent carbapenem-hydrolyzing metallo-beta-lactamase. PLoS One 2011, 6, e24621 10.1371/journal.pone.0024621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.; Hao Q. Crystal structure of NDM-1 reveals a common beta-lactam hydrolysis mechanism. FASEB J. 2011, 25, 2574–2582. 10.1096/fj.11-184036. [DOI] [PubMed] [Google Scholar]
- King D.; Strynadka N. Crystal structure of New Delhi metallo-beta-lactamase reveals molecular basis for antibiotic resistance. Protein Sci. 2011, 20, 1484–1491. 10.1002/pro.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King D. T.; Worrall L. J.; Gruninger R.; Strynadka N. C. New Delhi metallo-beta-lactamase: structural insights into beta-lactam recognition and inhibition. J. Am. Chem. Soc. 2012, 134, 11362–11365. 10.1021/ja303579d. [DOI] [PubMed] [Google Scholar]
- Guo Y.; Wang J.; Niu G.; Shui W.; Sun Y.; Zhou H.; Zhang Y.; Yang C.; Lou Z.; Rao Z. A structural view of the antibiotic degradation enzyme NDM-1 from a superbug. Protein Cell 2011, 2, 384–394. 10.1007/s13238-011-1055-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.; Cunningham M. A.; Mire J.; Tesar C.; Sacchettini J.; Joachimiak A. NDM-1, the ultimate promiscuous enzyme: substrate recognition and catalytic mechanism. FASEB J. 2013, 27, 1917–1927. 10.1096/fj.12-224014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z.; Li L.; Wang Y.; Chen L.; Kong X.; Hong Y.; Lan L.; Zheng M.; Guang-Yang C.; Liu H.; Shen X.; Luo C.; Li K. K.; Chen K.; Jiang H. Molecular basis of NDM-1, a new antibiotic resistance determinant. PLoS One 2011, 6, e23606 10.1371/journal.pone.0023606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H.; Ding J.; Zhu D.; Liu X.; Xu X.; Zhang Y.; Zang S.; Wang D. C.; Liu W. Structural and mechanistic insights into NDM-1 catalyzed hydrolysis of cephalosporins. J. Am. Chem. Soc. 2014, 136, 14694–14697. 10.1021/ja508388e. [DOI] [PubMed] [Google Scholar]
- Zhu K.; Lu J.; Ye F.; Jin L.; Kong X.; Liang Z.; Chen Y.; Yu K.; Jiang H.; Li J. Q.; Luo C. Structure-based computational study of the hydrolysis of New Delhi metallo-beta-lactmase-1. Biochem. Biophys. Res. Commun. 2013, 431, 2–7. 10.1016/j.bbrc.2012.12.141. [DOI] [PubMed] [Google Scholar]
- Chen J.; Wang J.; Zhu W. Zinc ion-induced conformational changes in new Delphi metallo-beta-lactamase 1 probed by molecular dynamics simulations and umbrella sampling. Phys. Chem. Chem. Phys. 2017, 19, 3067–3075. 10.1039/C6CP08105C. [DOI] [PubMed] [Google Scholar]
- Thomas P. W.; Zheng M.; Wu S.; Guo H.; Liu D.; Xu D.; Fast W. Characterization of purified New Delhi metallo-beta-lactamase-1. Biochemistry 2011, 50, 10102–10113. 10.1021/bi201449r. [DOI] [PubMed] [Google Scholar]
- Cheng Z.; Thomas P. W.; Ju L.; Bergstrom A.; Mason K.; Clayton D.; Miller C.; Bethel C. R.; VanPelt J.; Tierney D. L.; Page R. C.; Bonomo R. A.; Fast W.; Crowder M. W. Evolution of New Delhi metallo-beta-lactamase (NDM) in the clinic: Effects of NDM mutations on stability, zinc affinity, and mono-zinc activity. J. Biol. Chem. 2018, 293, 12606–12618. 10.1074/jbc.RA118.003835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooke C. L.; Hinchliffe P.; Bragginton E. C.; Colenso C. K.; Hirvonen V. H. A.; Takebayashi Y.; Spencer J. beta-Lactamases and beta-Lactamase Inhibitors in the 21st Century. J. Mol. Biol. 2019, 431, 3472–3500. 10.1016/j.jmb.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linciano P.; Cendron L.; Gianquinto E.; Spyrakis F.; Tondi D. Ten Years with New Delhi Metallo-beta-lactamase-1 (NDM-1): From Structural Insights to Inhibitor Design. ACS Infect. Dis. 2019, 5, 9–34. 10.1021/acsinfecdis.8b00247. [DOI] [PubMed] [Google Scholar]
- Li N.; Xu Y.; Xia Q.; Bai C.; Wang T.; Wang L.; He D.; Xie N.; Li L.; Wang J.; Zhou H. G.; Xu F.; Yang C.; Zhang Q.; Yin Z.; Guo Y.; Chen Y. Simplified captopril analogues as NDM-1 inhibitors. Bioorg. Med. Chem. Lett. 2014, 24, 386–389. 10.1016/j.bmcl.2013.10.068. [DOI] [PubMed] [Google Scholar]
- Liu S.; Jing L.; Yu Z. J.; Wu C.; Zheng Y.; Zhang E.; Chen Q.; Yu Y.; Guo L.; Wu Y.; Li G. B. ((S)-3-Mercapto-2-methylpropanamido)acetic acid derivatives as metallo-beta-lactamase inhibitors: Synthesis, kinetic and crystallographic studies. Eur. J. Med. Chem. 2018, 145, 649–660. 10.1016/j.ejmech.2018.01.032. [DOI] [PubMed] [Google Scholar]
- Klingler F. M.; Wichelhaus T. A.; Frank D.; Cuesta-Bernal J.; El-Delik J.; Muller H. F.; Sjuts H.; Gottig S.; Koenigs A.; Pos K. M.; Pogoryelov D.; Proschak E. Approved Drugs Containing Thiols as Inhibitors of Metallo-beta-lactamases: Strategy To Combat Multidrug-Resistant Bacteria. J. Med. Chem. 2015, 58, 3626–3630. 10.1021/jm501844d. [DOI] [PubMed] [Google Scholar]
- Zhang Y. L.; Yang K. W.; Zhou Y. J.; LaCuran A. E.; Oelschlaeger P.; Crowder M. W. Diaryl-substituted azolylthioacetamides: Inhibitor discovery of New Delhi metallo-beta-lactamase-1 (NDM-1). ChemMedChem 2014, 9, 2445–2448. 10.1002/cmdc.201402249. [DOI] [PubMed] [Google Scholar]
- Zhai L.; Zhang Y. L.; Kang J. S.; Oelschlaeger P.; Xiao L.; Nie S. S.; Yang K. W. Triazolylthioacetamide: A Valid Scaffold for the Development of New Delhi Metallo-beta-Lactmase-1 (NDM-1) Inhibitors. ACS Med. Chem. Lett. 2016, 7, 413–417. 10.1021/acsmedchemlett.5b00495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livermore D. M.; Mushtaq S.; Morinaka A.; Ida T.; Maebashi K.; Hope R. Activity of carbapenems with ME1071 (disodium 2,3-diethylmaleate) against Enterobacteriaceae and Acinetobacter spp. with carbapenemases, including NDM enzymes. J. Antimicrob. Chemother. 2013, 68, 153–158. 10.1093/jac/dks350. [DOI] [PubMed] [Google Scholar]
- Brem J.; van Berkel S. S.; Aik W.; Rydzik A. M.; Avison M. B.; Pettinati I.; Umland K. D.; Kawamura A.; Spencer J.; Claridge T. D.; McDonough M. A.; Schofield C. J. Rhodanine hydrolysis leads to potent thioenolate mediated metallo-beta-lactamase inhibition. Nat. Chem. 2014, 6, 1084–1090. 10.1038/nchem.2110. [DOI] [PubMed] [Google Scholar]
- González M. M.; Kosmopoulou M.; Mojica M. F.; Castillo V.; Hinchliffe P.; Pettinati I.; Brem J.; Schofield C. J.; Mahler G.; Bonomo R. A.; Llarrull L. I.; Spencer J.; Vila A. J. Bisthiazolidines: A Substrate-Mimicking Scaffold as an Inhibitor of the NDM-1 Carbapenemase. ACS Infect. Dis. 2015, 1, 544–554. 10.1021/acsinfecdis.5b00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingler F. M.; Moser D.; Buttner D.; Wichelhaus T. A.; Lohr F.; Dotsch V.; Proschak E. Probing metallo-beta-lactamases with molecular fragments identified by consensus docking. Bioorg. Med. Chem. Lett. 2015, 25, 5243–5246. 10.1016/j.bmcl.2015.09.056. [DOI] [PubMed] [Google Scholar]
- Christopeit T.; Leiros H. K. Fragment-based discovery of inhibitor scaffolds targeting the metallo-beta-lactamases NDM-1 and VIM-2. Bioorg. Med. Chem. Lett. 2016, 26, 1973–1977. 10.1016/j.bmcl.2016.03.004. [DOI] [PubMed] [Google Scholar]
- Wang X.; Lu M.; Shi Y.; Ou Y.; Cheng X. Discovery of novel new Delhi metallo-beta-lactamases-1 inhibitors by multistep virtual screening. PLoS One 2015, 10, e0118290 10.1371/journal.pone.0118290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N.; Du R. L.; Zheng Y. Y.; Guo Q.; Cai S. Y.; Liu Z. H.; Fang Z. Y.; Yuan W. C.; Liu T.; Li X. M.; Lu Y. J.; Wong K. Y. Antibacterial activity of 3-methylbenzo[d]thiazol-methylquinolinium derivatives and study of their action mechanism. J. Enzyme Inhib. Med. Chem. 2018, 33, 879–889. 10.1080/14756366.2018.1465055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Hu X.; Wu Y.; Zhang W.; Chen X.; You X.; Hu L. Synthesis and structure-activity relationship of novel bisindole amidines active against MDR Gram-positive and Gram-negative bacteria. Eur. J. Med. Chem. 2018, 150, 771–782. 10.1016/j.ejmech.2018.03.031. [DOI] [PubMed] [Google Scholar]
- Yu Z. J.; Liu S.; Zhou S.; Li H.; Yang F.; Yang L. L.; Wu Y.; Guo L.; Li G. B. Virtual target screening reveals rosmarinic acid and salvianolic acid A inhibiting metallo- and serine-beta-lactamases. Bioorg. Med. Chem. Lett. 2018, 28, 1037–1042. 10.1016/j.bmcl.2018.02.025. [DOI] [PubMed] [Google Scholar]
- Zhang E.; Wang M. M.; Huang S. C.; Xu S. M.; Cui D. Y.; Bo Y. L.; Bai P. Y.; Hua Y. G.; Xiao C. L.; Qin S. NOTA analogue: A first dithiocarbamate inhibitor of metallo-beta-lactamases. Bioorg. Med. Chem. Lett. 2018, 28, 214–221. 10.1016/j.bmcl.2017.10.074. [DOI] [PubMed] [Google Scholar]
- Chu W. C.; Bai P. Y.; Yang Z. Q.; Cui D. Y.; Hua Y. G.; Yang Y.; Yang Q. Q.; Zhang E.; Qin S. Synthesis and antibacterial evaluation of novel cationic chalcone derivatives possessing broad spectrum antibacterial activity. Eur. J. Med. Chem. 2018, 143, 905–921. 10.1016/j.ejmech.2017.12.009. [DOI] [PubMed] [Google Scholar]
- Xiang Y.; Chang Y. N.; Ge Y.; Kang J. S.; Zhang Y. L.; Liu X. L.; Oelschlaeger P.; Yang K. W. Azolylthioacetamides as a potent scaffold for the development of metallo-beta-lactamase inhibitors. Bioorg. Med. Chem. Lett. 2017, 27, 5225–5229. 10.1016/j.bmcl.2017.10.038. [DOI] [PubMed] [Google Scholar]
- Chen A. Y.; Thomas P. W.; Stewart A. C.; Bergstrom A.; Cheng Z.; Miller C.; Bethel C. R.; Marshall S. H.; Credille C. V.; Riley C. L.; Page R. C.; Bonomo R. A.; Crowder M. W.; Tierney D. L.; Fast W.; Cohen S. M. Dipicolinic Acid Derivatives as Inhibitors of New Delhi Metallo-beta-lactamase-1. J. Med. Chem. 2017, 60, 7267–7283. 10.1021/acs.jmedchem.7b00407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K.; Bradford P. A. Interplay between beta-lactamases and new beta-lactamase inhibitors. Nat. Rev. Microbiol. 2019, 17, 295–306. 10.1038/s41579-019-0159-8. [DOI] [PubMed] [Google Scholar]
- Everett M.; Sprynski N.; Coelho A.; Castandet J.; Bayet M.; Bougnon J.; Lozano C.; Davies D. T.; Leiris S.; Zalacain M.; Morrissey I.; Magnet S.; Holden K.; Warn P.; De Luca F.; Docquier J. D.; Lemonnier M. Discovery of a Novel Metallo-beta-Lactamase Inhibitor That Potentiates Meropenem Activity against Carbapenem-Resistant Enterobacteriaceae. Antimicrob. Agents Chemother. 2018, 62, e00074-18 10.1128/AAC.00074-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T.; Wang Q.; Chen F.; Li X.; Luo S.; Fang H.; Wang D.; Li Z.; Hou X.; Wang H. Biochemical characteristics of New Delhi metallo-beta-lactamase-1 show unexpected difference to other MBLs. PLoS One 2013, 8, e61914 10.1371/journal.pone.0075704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King A. M.; Reid-Yu S. A.; Wang W.; King D. T.; De Pascale G.; Strynadka N. C.; Walsh T. R.; Coombes B. K.; Wright G. D. Aspergillomarasmine A overcomes metallo-beta-lactamase antibiotic resistance. Nature 2014, 510, 503–506. 10.1038/nature13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.; Wang S.; Wei Q.; Guo Q.; Bai Y.; Yang S.; Song F.; Zhang L.; Lei X. Synthesis and biological evaluation of Aspergillomarasmine A derivatives as novel NDM-1 inhibitor to overcome antibiotics resistance. Bioorg. Med. Chem. 2017, 25, 5133–5141. 10.1016/j.bmc.2017.07.025. [DOI] [PubMed] [Google Scholar]
- Shi C.; Bao J.; Sun Y.; Kang X.; Lao X.; Zheng H. Discovery of Baicalin as NDM-1 inhibitor: Virtual screening, biological evaluation and molecular simulation. Bioorg. Chem. 2019, 88, 102953 10.1016/j.bioorg.2019.102953. [DOI] [PubMed] [Google Scholar]
- Liu S.; Zhou Y.; Niu X.; Wang T.; Li J.; Liu Z.; Wang J.; Tang S.; Wang Y.; Deng X. Magnolol restores the activity of meropenem against NDM-1-producing Escherichia coli by inhibiting the activity of metallo-beta-lactamase. Cell Death Discovery 2018, 4, 28 10.1038/s41420-018-0029-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C.; Chen J.; Xiao B.; Kang X.; Lao X.; Zheng H. Discovery of NDM-1 inhibitors from natural products. J. Global Antimicrob. Resist. 2019, 18, 80–87. 10.1016/j.jgar.2019.02.003. [DOI] [PubMed] [Google Scholar]
- Cody V.; Middleton E.; Harbone J. B. In Biochemical, Pharmacological, and Structure-Activity Relationships, Proceedings on the Plant Flavonoids in Biology and Medicine, Buffalo, Vol. 1. Alan. R. Liss, Ed.; Academic Press: New York, 1986.
- Cao S.; Jiang X.; Chen J. Effect of Zinc (II) on the interactions of bovine serum albumin with flavonols bearing different number of hydroxyl substituent on B-ring. J. Inorg. Biochem. 2010, 104, 146–152. 10.1016/j.jinorgbio.2009.10.014. [DOI] [PubMed] [Google Scholar]
- Guerrero L.; Castillo J.; Quinones M.; Garcia-Vallve S.; Arola L.; Pujadas G.; Muguerza B. Inhibition of Angiotensin-Converting Enzyme Activity by Flavonoids: Structure-Activity Relationship Studies. PLoS One 2012, 7, e49493 10.1371/journal.pone.0049493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolomeo M.; Grimaudo S.; Di Cristina A.; Pipitone R. M.; Dusonchet L.; Meli M.; Crosta L.; Gebbia N.; Invidiata F. P.; Titone L.; Simoni D. Galangin increases the cytotoxic activity of imatinib mesylate in imatinib-sensitive and imatinib-resistant Bcr-Abl expressing leukemia cells. Cancer Lett. 2008, 265, 289–297. 10.1016/j.canlet.2008.02.025. [DOI] [PubMed] [Google Scholar]
- Grundmann O.; Nakajima J.-I.; Kamata K.; Seo S.; Butterweck V. Kaempferol from the leaves of Apocynum venetum possesses anxiolytic activities in the elevated plus maze test in mice. Phytomedicine 2009, 16, 295–302. 10.1016/j.phymed.2008.12.020. [DOI] [PubMed] [Google Scholar]
- Chimenti F.; Cottiglia F.; Bonsignore L.; Casu L.; Casu M.; Floris C.; Secci D.; Bolasco A.; Chimenti P.; Granese A.; Befani O.; Turini P.; Alcaro S.; Ortuso F.; Trombetta G.; Loizzo A.; Guarino I. Quercetin as the active principle of Hypericum hircinum exerts a selective inhibitory activity against MAO-A: Extraction, biological analysis, and computational study. J. Nat. Prod. 2006, 69, 945–949. 10.1021/np060015w. [DOI] [PubMed] [Google Scholar]
- Fahlman B. M.; Krol E. S. Inhibition of UVA and UVB Radiation-Induced Lipid Oxidation by Quercetin. J. Agric. Food Chem. 2009, 57, 5301–5305. 10.1021/jf900344d. [DOI] [PubMed] [Google Scholar]
- Lu J.; Papp L. V.; Fang J. G.; Rodriguez-Nieto S.; Zhivotovsky B.; Holmgren A. Inhibition of mammalian thioredoxin reductase by some flavonoids: Implications for myricetin and quercetin anticancer activity. Cancer Res. 2006, 66, 4410–4418. 10.1158/0008-5472.CAN-05-3310. [DOI] [PubMed] [Google Scholar]
- Hsu Y.-L.; Chang J.-K.; Tsai C.-H.; Chien T.-T. C.; Kuo P.-L. Myricetin induces human osteoblast differentiation through bone morphogenetic protein-2/p38 mitogen-activated protein kinase pathway. Biochem. Pharmacol. 2007, 73, 504–514. 10.1016/j.bcp.2006.10.020. [DOI] [PubMed] [Google Scholar]
- Kwon O.; Eck P.; Chen S.; Corpe C. P.; Lee J.-H.; Kruhlak M.; Levine M. Inhibition of the intestinal glucose transporter GLUT2 by flavonoids. FASEB J. 2007, 21, 366–377. 10.1096/fj.06-6620com. [DOI] [PubMed] [Google Scholar]
- Zheng B.; Tan S.; Gao J.; Han H.; Liu J.; Lu G.; Liu D.; Yi Y.; Zhu B.; Gao G. F. An unexpected similarity between antibiotic-resistant NDM-1 and beta-lactamase II from Erythrobacter litoralis. Protein Cell 2011, 2, 250–258. 10.1007/s13238-011-1027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Materon I. C.; Beharry Z.; Huang W. Z.; Perez C.; Palzkill T. Analysis of the context dependent sequence requirements of active site residues in the metallo-beta-lactamase IMP-1. J. Mol. Biol. 2004, 344, 653–663. 10.1016/j.jmb.2004.09.074. [DOI] [PubMed] [Google Scholar]
- Yao C.; Wu Q.; Xu G.; Li C. NMR backbone resonance assignment of New Delhi metallo-beta-lactamase. Biomol. NMR Assignments 2017, 11, 239–242. 10.1007/s12104-017-9756-5. [DOI] [PubMed] [Google Scholar]
- Shen Y.; Bax A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR 2013, 56, 227–241. 10.1007/s10858-013-9741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King D.; Strynadka N. Crystal structure of New Delhi metallo-beta-lactamase reveals molecular basis for antibiotic resistance. Protein Sci. 2011, 20, 1484–1491. 10.1002/pro.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z.; Liu Q.; Qu G.; Feng Y.; Reetz M. T. Utility of B-Factors in Protein Science: Interpreting Rigidity, Flexibility, and Internal Motion and Engineering Thermostability. Chem. Rev. 2019, 119, 1626–1665. 10.1021/acs.chemrev.8b00290. [DOI] [PubMed] [Google Scholar]
- Green V. L.; Verma A.; Owens R. J.; Phillips S. E.; Carr S. B. Structure of New Delhi metallo-β-lactamase 1 (NDM-1). Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun. 2011, 67, 1160–1164. 10.1107/S1744309111029654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.; Chen H.; Shi Y.; Hu F.; Lao X.; Gao X.; Zheng H.; Yao W. Probing the effect of the non-active-site mutation Y229W in New Delhi metallo-beta-lactamase-1 by site-directed mutagenesis, kinetic studies, and molecular dynamics simulations. PLoS One 2013, 8, e82080 10.1371/journal.pone.0082080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dajas F.; Andres Abin-Carriquiry J.; Arredondo F.; Blasina F.; Echeverry C.; Martinez M.; Rivera F.; Vaamonde L. Quercetin in brain diseases: Potential and limits. Neurochem. Int. 2015, 89, 140–148. 10.1016/j.neuint.2015.07.002. [DOI] [PubMed] [Google Scholar]
- Brito A. F.; Ribeiro M.; Abrantes A. M.; Pires A. S.; Teixo R. J.; Tralhao J. G.; Botelho M. F. Quercetin in Cancer Treatment, Alone or in Combination with Conventional Therapeutics?. Curr. Med. Chem. 2015, 22, 3025–3039. 10.2174/0929867322666150812145435. [DOI] [PubMed] [Google Scholar]
- Ganugapati M.; Sirisha V.; Mukkavalli S.; Atimamula S.; Sai S. Insilico modeling and docking studies of new delhi metallo Beta lactamase-1 (super bug). Int. J. Eng. Sci. Res. Technol. 2011, 3, 2427–2434. [Google Scholar]
- Padmavathi M.; Prasanth Reddy V.; Rao R. Inhibition of NDM-1 in superbugs by flavonoids- an insilico approach. J. Adv. Bioinf. Appl. Res. 2012, 3, 328–332. [Google Scholar]
- van Zundert G. C. P.; Rodrigues J.; Trellet M.; Schmitz C.; Kastritis P. L.; Karaca E.; Melquiond A. S. J.; van Dijk M.; de Vries S. J.; Bonvin A. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J. Mol. Biol. 2016, 428, 720–725. 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- Erickson J. A.; Jalaie M.; Robertson D. H.; Lewis R. A.; Vieth M. Lessons in molecular recognition: the effects of ligand and protein flexibility on molecular docking accuracy. J. Med. Chem. 2004, 47, 45–55. 10.1021/jm030209y. [DOI] [PubMed] [Google Scholar]
- Zhang H.; Ma G.; Zhu Y.; Zeng L.; Ahmad A.; Wang C.; Pang B.; Fang H.; Zhao L.; Hao Q. Active-Site Conformational Fluctuations Promote the Enzymatic Activity of NDM-1. Antimicrob. Agents Chemother. 2018, 62, e01579-18 10.1128/AAC.01579-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira L.; Fernandez M. T.; Santos M.; Rocha R.; Florencio M. H.; Jennings K. R. Interactions of flavonoids with iron and copper ions: A mechanism for their antioxidant activity. Free Radical Res. 2002, 36, 1199–1208. 10.1080/1071576021000016463. [DOI] [PubMed] [Google Scholar]
- Grazul M.; Budzisz E. Biological activity of metal ions complexes of chromones, coumarins and flavones. Coord. Chem. Rev. 2009, 253, 2588–2598. 10.1016/j.ccr.2009.06.015. [DOI] [Google Scholar]
- Liu Y.; Guo M. Studies on transition metal-quercetin complexes using electrospray ionization tandem mass spectrometry. Molecules 2015, 20, 8583–8594. 10.3390/molecules20058583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolatabadi J. E. Molecular aspects on the interaction of quercetin and its metal complexes with DNA. Int. J. Biol. Macromol. 2011, 48, 227–233. 10.1016/j.ijbiomac.2010.11.012. [DOI] [PubMed] [Google Scholar]
- Bhuiya S.; Haque L.; Pradhan A. B.; Das S. Inhibitory effects of the dietary flavonoid quercetin on the enzyme activity of zinc(II)-dependent yeast alcohol dehydrogenase: Spectroscopic and molecular docking studies. Int. J. Biol. Macromol. 2017, 95, 177–184. 10.1016/j.ijbiomac.2016.11.047. [DOI] [PubMed] [Google Scholar]
- Steiner R. A.; Kalk K. H.; Dijkstra B. W. Anaerobic enzyme.substrate structures provide insight into the reaction mechanism of the copper-dependent quercetin 2,3-dioxygenase. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 16625–16630. 10.1073/pnas.262506299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeoung J. H.; Nianios D.; Fetzner S.; Dobbek H. Quercetin 2,4-Dioxygenase Activates Dioxygen in a Side-On O2-Ni Complex. Angew. Chem., Int. Ed. 2016, 55, 3281–3284. 10.1002/anie.201510741. [DOI] [PubMed] [Google Scholar]
- Aoki N.; Ishii Y.; Tateda K.; Saga T.; Kimura S.; Kikuchi Y.; Kobayashi T.; Tanabe Y.; Tsukada H.; Gejyo F.; Yamaguchi K. Efficacy of calcium-EDTA as an inhibitor for metallo-beta-lactamase in a mouse model of Pseudomonas aeruginosa pneumonia. Antimicrob. Agents Chemother. 2010, 54, 4582–4588. 10.1128/AAC.00511-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J.; McLeod S.; MacCormack K.; Sriram S.; Gao N.; Breeze A. L.; Hu J. Real-time monitoring of New Delhi metallo-beta-lactamase activity in living bacterial cells by 1H NMR spectroscopy. Angew. Chem., Int. Ed. 2014, 53, 2130–2133. 10.1002/anie.201308636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom A.; Katko A.; Adkins Z.; Hill J.; Cheng Z.; Burnett M.; Yang H.; Aitha M.; Mehaffey M. R.; Brodbelt J. S.; Tehrani K.; Martin N. I.; Bonomo R. A.; Page R. C.; Tierney D. L.; Fast W.; Wright G. D.; Crowder M. W. Probing the Interaction of Aspergillomarasmine A with Metallo-beta-lactamases NDM-1, VIM-2, and IMP-7. ACS Infect. Dis. 2018, 4, 135–145. 10.1021/acsinfecdis.7b00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchliffe P.; Gonzalez M. M.; Mojica M. F.; Gonzalez J. M.; Castillo V.; Saiz C.; Kosmopoulou M.; Tooke C. L.; Llarrull L. I.; Mahler G.; Bonomo R. A.; Vila A. J.; Spencer J. Cross-class metallo-beta-lactamase inhibition by bisthiazolidines reveals multiple binding modes. Proc. Natl. Acad. Sci. U.S.A. 2016, 113, E3745–E3754. 10.1073/pnas.1601368113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spyrakis F.; Celenza G.; Marcoccia F.; Santucci M.; Cross S.; Bellio P.; Cendron L.; Perilli M.; Tondi D. Structure-Based Virtual Screening for the Discovery of Novel Inhibitors of New Delhi Metallo-beta-lactamase-1. ACS Med. Chem. Lett. 2018, 9, 45–50. 10.1021/acsmedchemlett.7b00428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A. U.; Ali A.; Danishuddin; Srivastava G.; Sharma A. Potential inhibitors designed against NDM-1 type metallo-beta-lactamases: an attempt to enhance efficacies of antibiotics against multi-drug-resistant bacteria. Sci. Rep. 2017, 7, 9207 10.1038/s41598-017-09588-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou J.; Wan S.; Chan K. F.; So P. K.; He D.; Chan E. W.; Chan T. H.; Wong K. Y.; Tao J.; Chen S. Ebselen as a potent covalent inhibitor of New Delhi metallo-beta-lactamase (NDM-1). Chem. Commun. 2015, 51, 9543–9546. 10.1039/C5CC02594J. [DOI] [PubMed] [Google Scholar]
- Rasia R. M.; Vila A. J. Structural determinants of substrate binding to Bacillus cereus metallo-beta-lactamase. J. Biol. Chem. 2004, 279, 26046–26051. 10.1074/jbc.M311373200. [DOI] [PubMed] [Google Scholar]
- Meini M. R.; Gonzalez L. J.; Vila A. J. Antibiotic resistance in Zn(II)-deficient environments: metallo-beta-lactamase activation in the periplasm. Future Microbiol. 2013, 8, 947–979. 10.2217/fmb.13.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo M. C.; Aulabaugh A.; Jin G.; Cowling R.; Bard J.; Malamas M.; Ellestad G. Evaluation of fluorescence-based thermal shift assays for hit identification in drug discovery. Anal. Biochem. 2004, 332, 153–159. 10.1016/j.ab.2004.04.031. [DOI] [PubMed] [Google Scholar]
- Vranken W. F.; Boucher W.; Stevens T. J.; Fogh R. H.; Pajon A.; Llinas M.; et al. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 2005, 59, 687–696. 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- Zhu G.; Xia Y. L.; Nicholson L. K.; Sze K. H. Protein dynamics measurements by TROSY-based NMR experiments. J. Magn. Reson. 2000, 143, 423–426. 10.1006/jmre.2000.2022. [DOI] [PubMed] [Google Scholar]
- Piotto M.; Saudek V.; Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR 1992, 2, 661–665. 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- Williamson M. P. Using chemical shift perturbation to characterise ligand binding. Prog. Nucl. Magn. Reson. Spectrosc. 2013, 73, 1–16. 10.1016/j.pnmrs.2013.02.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.