Abstract
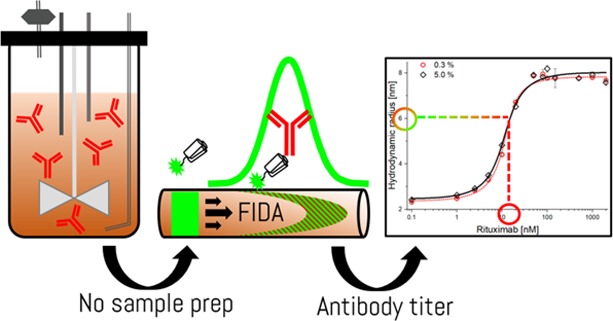
Biopharmaceuticals such as protein and peptide-based drugs are often produced by fermentation processes where it is necessary to monitor the amount and quality of the product expressed during fermentation and for release testing of the final drug product. Standard procedures involve surface-based ligand binding technologies such as enzyme-linked immunosorbent assay and biolayer interferometry, or extensive purification using, e.g., preparative chromatography followed by spectrophotometric protein quantification. The multistep nature of these methodologies leads to lengthy protocols and renders real-time process control impractical. Recently, flow-induced dispersion analysis (FIDA) was introduced as a novel in-solution ligand binding technology, requiring only nano/microliter sample volumes. FIDA is based on Taylor dispersion analysis in narrow fused silica capillaries and provides the hydrodynamic radius of the binding ligand and complex in addition to the detailed binding characterization. Here, we demonstrate the use of FIDA for quantification of monoclonal IgG antibodies (rituximab) directly in mammalian cell fermentation broth with only 4 min of analysis time. The FIDA assay utilizes a small anti-IgG affibody, conjugated to a fluorophore, as a selective rituximab binder. The apparent change in the hydrodynamic radius of the affibody, as it interacts with known concentrations of rituximab, is used for generating a binding curve in a blank fermentation medium, and hence determining the dissociation constant and complex size. Finally, the binding curve is utilized for quantifying the rituximab titer concentration in clarified fermentation broth samples.
Introduction
The vast majority of biopharmaceuticals are produced by fermentation processes via genetically modified cell lines based, primarily, on mammalian, bacterial, or yeast cells.1−3 These cells are exploited for their inherent protein synthesis apparatus, which is harnessed for assembling, folding, and expressing protein-based drugs. The cells require complex media for sustainable growth and satisfactory protein expression. Furthermore, the cells express host cell proteins (HCPs), as part of their metabolism, and may release cellular debris components (DNA, cell wall, and HCPs) into the growth medium upon apoptosis. Consequently, the growth medium, i.e., fermentation broth, is, at the end of a fermentation process, a complex solution. This presents a substantial challenge to most analytical assays; thus, purification steps are typically implemented prior to analysis,4−7 which impede fast and inexpensive selection of clones and further cell line optimization. Currently, surface-based methodologies such as biolayer interferometry (BLI) and enzyme-linked immunosorbent assay (ELISA) are the preferred methods for assessing IgG titers via immobilization of selective ligands (e.g., protein A).8,9 However, these methods rely on surface-based interactions and thus may not report the actual in-solution titer due to nonspecific surface adsorption.
Flow-induced dispersion analysis (FIDA) has emerged as a new immobilization-free ligand binding technology with abilities for in-solution characterization of protein size, stability, and binding affinity.10−14 Briefly, FIDA utilizes Taylor dispersion analysis (TDA) in narrow capillaries under a laminar flow for measuring the change in the apparent hydrodynamic radius of a selective ligand (termed the indicator) as it interacts with the analyte of interest. The apparent hydrodynamic radius (i.e., size) of the indicator is measured at different analyte concentrations and thereby forms the basis for generation of a binding curve and determination of the dissociation constant (Kd) and complex size. Furthermore, the binding curve serves as a standard curve for selectively quantifying analyte concentrations in unknown samples based on the apparent indicator size measured.
Here, we present a FIDA-based approach for quantifying the rituximab titer directly in clarified Chinese hamster ovary (CHO) cell fermentation broth using an anti-IgG affibody as the indicator. The FIDA assay was developed in an uninoculated fermentation medium using known quantities of rituximab and subsequently utilized for quantifying the rituximab titer in CHO cell fermentation broth samples. The titers measured by the FIDA assay are in good agreement with traditional measurements performed using UV–vis absorption spectroscopy, following a protein A-based purification procedure.
Experimental Section
Equipment
The FIDA experiments were conducted on a FIDA 1 instrument employing light-emitting diode (LED)-induced fluorescence detection (FIDA Biosystems ApS, Copenhagen, Denmark) with an excitation wavelength of 480 nm and a high-pass emission filter (515 nm cutoff).
TDA experiments were performed on a PrinCE NEXT 870 instrument (PrinCE Technologies, Emmen, The Netherlands) employing an ActiPix D100 UV imaging system (Paraytec Ltd., York, United Kingdom) for detection at 214 nm.
PEG-coated fused silica capillaries (inner diameter: 75 μm, outer diameter: 375 μm, length total: 100 cm, length to detection window: 84 cm) from FIDA Biosystems were used.
Materials and Chemicals
Rituximab (lot H0156B04, 144,544 kDa) from Roche (MabThera, Basel, Switzerland), clarified (without cells) CHEF1 CHO cell fermentation broths from two parallel runs (TF262, PM01, and PM02 respectively), and uninoculated fermentation media were obtained from AGC Biologics (Soeborg, Denmark). The anti-IgG affibody (cat. no. ab31900) was purchased from Abcam (Cambridge, UK). Alexa Fluor 488 C5-maleimide was purchased from Thermo Fisher (Waltham, Massachusetts, USA). Dithiothreitol (DTT) and glycine were purchased from Sigma Aldrich (St Louis, Missouri, USA). Sodium dihydrogen phosphate monohydrate and disodium hydrogen phosphate dodecahydrate were purchased from Merck (Darmstadt, Germany). Dimethyl sulfoxide (DMSO) was obtained from EMP Biotech (Berlin, Germany). Tris ultrapure was obtained from ICN Biomedical (Aurora, Ohio, USA). Ultrapure water (18.2 MΩ-cm at 25 °C) was tapped from a water purification system (SG Ultraclear, SG Water, Barsbüttel, Germany).
Assay Buffer
A 67 mM phosphate buffer, pH 7.4, was prepared with ultrapure water, filtered through a Q-max 0.45 μm nylon syringe filter (Frisenette, Knebel, Denmark), and used as the assay buffer.
Fluorophore Labelling of the Anti-IgG Affibody
The anti-IgG affibody (200 μg at 1 mg/mL) was reduced in 0.02 M DTT for 2 h at room temperature. Subsequently, the DTT was removed by buffer exchange into the assay buffer utilizing a pre-equilibrated PD SpinTrap G-25 column (GE Healthcare, Marlborough, Massachusetts, USA). Alexa Fluor 488 C5-maleimide was dissolved in DMSO to a stock concentration of 10 mM and then added to the affibody solution with a molar ratio of 19:1 (Alexa Fluor/affibody) immediately after the buffer exchange. The labelling reaction was incubated for 2 h at room temperature and subsequently purified in the assay buffer utilizing a pre-equilibrated SpinTrap G-25 column.
The concentration of the affibody–alexa488 conjugate was determined using a microvolume spectrophotometer (NanoDrop 2000C, Thermo Fisher Scientific, Wilmington, Delaware, USA), using the molar absorption coefficient of alexa488 (ε = 72,000 cm–1 M–1) at 493 nm15 since the anti-IgG affibody does not contain any tyrosine or tryptophan residues.
Protein A Purification Buffers
The equilibration and binding buffers were a 20 mM phosphate buffer, pH 7.0; the elution buffer was 0.1 M glycine–HCl, pH 2.7; and the neutralizing buffer was 1 M Tris–HCl, pH 9.0. All buffers were prepared with ultrapure water.
FIDA and TDA Procedures
The PEG-coated capillary was flushed and equilibrated prior to sample analysis with the 67 mM phosphate buffer, pH 7.4, at 1500 mbar for 5 min. The analyte sample was injected at 1500 mbar for 45 s followed by injection of the indicator sample at 50 mbar for 10 s (39 nL, corresponding to 1% of the capillary volume). Subsequently, the injected indicator sample was mobilized toward the LED detector with the analyte sample at 400 mbar for 240 s. The capillary, samples, and buffer vials were kept in a temperature-controlled environment at 25 °C inside the instrument. All samples were analyzed in triplicate.
Development of the FIDA Assay
The hydrodynamic radius of the unconjugated anti-IgG affibody was measured using TDA at a concentration of 1 mg/mL in a neat phosphate buffer.
Subsequently, the indicator, anti-IgG affibody–alexa488, was diluted to a fixed indicator concentration of 20 nM in varying concentrations (0–10% v/v) of CHO cell fermentation broth (TF262, PM02). The fermentation broth was diluted with the assay buffer. All samples were preincubated for >10 min to attain equilibrium prior to the FIDA measurements.
Standard Curves for Rituximab Quantification in the Fermentation Broth
Standard curves were established in 0.3 and 5.0% v/v uninoculated fermentation media. The indicator, anti-IgG affibody–alexa488, was added to provide a fixed indicator concentration of 20 nM in the medium with varying and known concentrations of rituximab (0–2000 nM). All samples were preincubated for >10 min to attain equilibrium prior to the FIDA measurements.
Determination of the Rituximab Titer by FIDA
Quantification of rituximab in two parallel CHO cell fermentation broths (TF262 PM01 and TF262 PM02) was performed for three different concentrations of fermentation broth (0.05, 0.075, and 0.10% v/v). The indicator, anti-IgG affibody–alexa488, was added to provide a fixed indicator concentration of 20 nM, and an uninoculated fermentation medium was added to reach a final medium concentration of 5% v/v (i.e., 4.95, 4.925, and 4.90% v/v, respectively). All samples were preincubated for >10 min to attain equilibrium prior to the FIDA measurements.
Determination of the Rituximab Titer by Protein A Purification and UV Spectrophotometry
The rituximab content was also determined using disposable protein A HP SpinTrap columns from GE Healthcare, following the supplier instructions (#28-9031-32). Three parallel purification runs were performed for each of the two fermentations (i.e., TF262 PM01 and TF262 PM02), and every column was loaded with 300 μL of fermentation broth. After purification, the concentration of rituximab was determined utilizing a microvolume spectrophotometer (NanoDrop 2000C) and the extinction coefficient of rituximab (ε = 1.7 cm–1 (mg/mL)−1) at 280 nm.16
Data Analysis
The Taylorgrams were processed using the FIDA software, version 1.03 (FIDA Biosystems ApS, Copenhagen, Denmark), for calculating the apparent hydrodynamic radius of the indicator (anti-IgG affibody–alexa-488) at each analyte concentration. The Taylorgram fraction setting was consistently fixed to 75% for all data points, and minor fluctuations in sample viscosity were corrected according to a reference measurement of the indicator in a neat phosphate buffer, as previously described.10
Typically, the following binding isotherm is used for FIDA assays, assuming an indicator concentration below the dissociation constant, 1:1 binding stoichiometry, and equilibrium10
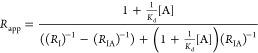 |
1 |
where Rapp, RI , RIA are the apparent, indicator, and complex hydrodynamic radii, respectively; Kd is the dissociation constant; and [A] is the formal analyte concentration.
However, in the cases where the binding affinity between the indicator and analyte is very high (low nM – pM range), the indicator concentration cannot be assumed to be less than Kd. Therefore, the excess indicator binding isotherm applies
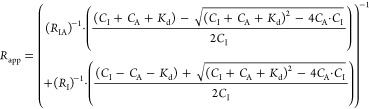 |
2 |
where CI and CA are the formal concentrations of the indicator and analyte, respectively. The details of the excess indicator binding isotherm are described in the Supporting Information.
Results and Discussion
Development of the FIDA Assay
Protein quantification using FIDA is based on an indicator molecule (anti-IgG affibody) interacting selectively with the analyte protein (rituximab, IgG) in the sample matrix. An apparent change in the indicator size due to the interaction with the analyte present is used for the quantification. An anti-IgG affibody was selected as the indicator due to an expected selectivity towards the Fc region of rituximab and, hence, not interacting with other components of the sample matrix. The affibody was fluorescently labelled at the dedicated C-terminal cysteine for single-point modification,17 ensuring selective and high sensitivity measurements in the complex samples. Furthermore, the affibody was considered a promising indicator due to the small size (7 kDa) relative to rituximab (145 kDa), thus potentially providing high sensitivity and a wide dynamic range. Other anti-IgG specific ligands, e.g., protein A (similar specificity as the affibody used in the present work), protein G, aptamers, or nanobodies,18 might be suitable for FIDA-based rituximab quantification as well. The specificity of the methods is directly linked to the specificity of the interaction between the affibody and the constant region of the antibody. As the affibody recognizes the constant region of the antibody, the same methodology is applicable to the detection of any human IgG antibody.
The hydrodynamic radii of the unconjugated dimeric affibody (14 kDa) and monomeric affibody–alexa488 conjugate (7 kDa) were determined in a neat phosphate buffer by TDA and FIDA to 3.27 ± 0.05 and 2.24 ± 0.10 nm, respectively. These values were slightly higher than expected for globular proteins,19 thereby suggesting a relatively loose conformation.
Subsequently, a feasibility study was performed to assess whether the anti-IgG affibody could be measured directly in a clarified fermentation broth containing rituximab (TF262 PM02). The apparent hydrodynamic radius of the anti-IgG affibody was measured in increasing fermentation broth concentrations using FIDA (Figure 1). The apparent hydrodynamic radius of the anti-IgG affibody was found to increase from 2.2 to a plateau at ∼8 nm (Figure 1), thereby indicating affibody binding to rituximab.
Figure 1.
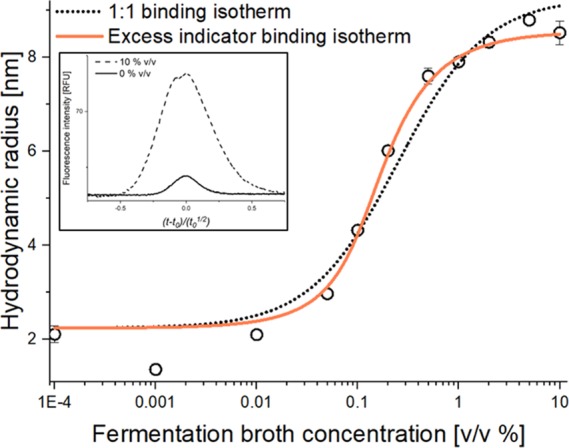
Apparent hydrodynamic radius of the 20 nM anti-IgG affibody–alexa488 as a function of fermentation broth concentration (PM02 (containing rituximab), 0–10% v/v) determined by FIDA at 25 °C (black open circles, n = 3, error bars represent standard deviation). The dotted black line and solid orange line represents fitting to 1:1 (R2 = 0.980) and excess indicator (R2 = 0.990) binding isotherms, respectively. (Insert) visual comparison of viscosity-corrected Taylorgrams at 0 and 10% v/v fermentation broth (solid and dashed line, respectively).
It was attempted to fit the data in Figure 1 to the binding isotherm assuming a 1:1 binding stoichiometry (eq 1). However, the binding curve appeared steeper than anticipated, which could point to the affibody concentration being higher than the Kd at low rituximab concentrations. Therefore, the data was also fitted to an excess indicator binding isotherm (eq 2), resulting in a significantly better fit (Figure 1, orange line). This was further investigated by a second set of experiments in which the affibody concentration was reduced from 20 to 5 nM. Here, a shift in the binding isotherm was observed (Figure S1 in the Supporting Information), indicating that the affibody was indeed in excess at low rituximab concentrations. This scenario has previously been described theoretically for FIDA experiments.13
The Taylorgram overlay at the 0 and 10% v/v fermentation broth in Figure 1 (insert, corrected for viscosity-related differences20) revealed a noteworthy change in the peak area, thereby indicating adsorption of the indicator (i.e., affibody) to labware surfaces (e.g., vials and capillary) at low fermentation broth concentrations. Here, the fermentation broth was simply added to prevent indicator (and potentially rituximab) adsorption to the labware surfaces. Taylorgrams of indicator peak areas depicted as a function of fermentation broth concentration (Figure S2) showed increased peak areas and minimized adsorption at broth concentrations above 0.05% v/v. This initial set of experiments demonstrated that the FIDA assay was capable of characterizing interactions directly in a clarified fermentation broth. We did not observe any nonspecific binding to matrix components.
Standard Curves for Rituximab in Fermentation Media
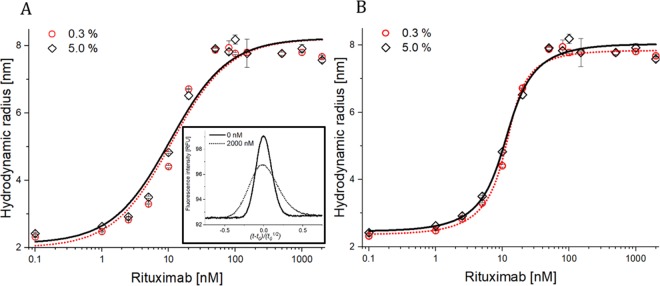
Rituximab standard curves (0–2000 nM) were established in 0.3 and 5% v/v uninoculated (i.e., blank) fermentation media in order to define optimal conditions for quantification. The apparent hydrodynamic radius of the anti-IgG affibody increased with an increasing rituximab concentration, as expected (Figure 2), and a plateau was reached around 8 nm (Figure 2), indicating full binding, in line with the initial experiments (Figure 1).
Figure 2.
Apparent hydrodynamic radius of the 20 nM anti-IgG affibody–alexa488 as a function of rituximab concentration (0–2000 nM) determined by FIDA at 25 °C. The black open diamonds and red open circles represent measurements in 0.3 and 5.0% v/v uninoculated fermentation media, respectively (n = 3, error bars represent standard deviation). The solid black line and dotted red line represent fitting to (A) 1:1 and (B) excess indicator binding isotherms, respectively. Insert: visual comparison of viscosity-corrected Taylorgrams in a 5.0% v/v uninoculated fermentation medium at 0 and 2000 nM rituximab (solid and dashed line, respectively).
As expected from the assay development experiment (Figure 1), the excess indicator isotherm (eq 2) was most suitable for the binding curves (Figure 2). This was further verified by the binding parameters summarized in Table 1. Here, the R2-values for the excess ligand binding isotherm were superior to the 1:1 in terms of describing the data. The affinity was notably high, and using the 1:1 binding model (eq 1), the apparent dissociation constants (Kd) were in the low nM range, indicating that the indicator concentration is not lower than the Kd. While eq 1 provides an apparent Kd for the interaction, it would, however, require lower indicator concentrations to quantify the true Kd using this model. In this case, it is therefore more accurate to apply the excess indicator model (eq 2). For quantification, the binding curve is used as a standard curve linking the apparent size to antibody concentration. When the limit of quantification (LOQ) is defined as 10 times the relative standard deviation of the blank sample, an LOQ of 0.9 nM or 135 ng/mL is determined, which is about two orders of magnitude lower than HPLC procedures.21 In the FIDA methodology, there is a direct link between the apparent size, fraction unbound, and ultimately concentration. There is no requirement for linearity for connecting the apparent size, fraction bound, and concentration.
Table 1. Fitted Binding Parameters (Standard Curves) in 0.3 and 5% Uninoculated Fermentation Medium, Representing Both 1:1 Binding Stoichiometry and Excess Indicator Models.
| medium concentration | 0.3% v/v | 5% v/v | 0.3% v/v | 5% v/v |
|---|---|---|---|---|
| binding isotherm model | 1:1 (eq 1) | 1:1 (eq 1) | excess (eq 2) | excess (eq 2) |
| indicator size (RI) (nm) | 2.00 | 2.12 | 2.37 | 2.46 |
| indicator-complex size (RIA) (nm) | 8.22 | 8.23 | 7.84 | 8.03 |
| dissociation constant, Kd (nM) | 2.66 | 2.70 | 0.66 | 1.12 |
| coefficient of determination (R2) | 0.965 | 0.970 | 0.999 | 0.995 |
The standard curves in 0.3 and 5% fermentation media were highly similar (Figure 2). However, it was decided to perform the rituximab quantification in the 5% v/v fermentation medium since this concentration was superior in terms of eliminating nonspecific surface adsorption (Figure S2). The standard curve covers rituximab quantification in the 5% v/v fermentation medium, thus the samples (PM01 and PM02) were diluted with the uninoculated fermentation medium in order to be within the dynamic range of the binding curve since the interaction was fully saturated at 5% broth as seen in Figure 1. The excess indicator fitting model was chosen since it fitted the data with the highest accuracy (Table 1).
Quantification of Rituximab in CHO Cell Fermentation Broth with the Anti-IgG Affibody Using FIDA
The samples (PM01 and PM02) were diluted to fermentation broth concentrations of 0.05, 0.075, and 0.10% v/v with a blank fermentation medium and assay buffer to a final medium concentration of 5% v/v. This approach was applied in order to be within the dynamic range of the binding curve (Figure 2) and eliminate nonspecific adsorption. The selected dilutions were also chosen for verifying the concentration measurements in the dynamic range of the standard curve. The apparent hydrodynamic radius of the 20 nM anti-IgG affibody–alexa488 was measured in each dilution and related to an apparent rituximab concentration using the standard curve (5% v/v, Figure 2), which led to the results in Table 2 upon correction for dilution.
Table 2. Quantification of Rituximab in CHO Cell Fermentation Broth Samples TF262 PM01 and TF262 PM02a.
| TF262
PM01 |
TF262 PM02 |
|||||
|---|---|---|---|---|---|---|
| fermentation broth concentration [% v/v] | 0.05 | 0.075 | 0.10 | 0.05 | 0.075 | 0.10 |
| rituximab concentration by FIDA [mg/mL] | 1.34 ± 0.14 | 1.28 ± 0.07 | 1.38 ± 0.06 | 1.46 ± 0.02 | 1.34 ± 0.06 | 1.28 ± 0.04 |
| rituximab concentration by UV spectrophotometry following protein A purification | 1.22 ± 0.08 | 1.41 ± 0.04 | ||||
The rituximab concentration was found to be within the expected titer for a typical CHO cell fermentation process.1
Quantification of Rituximab in CHO Cell Fermentation Broth by Protein A Purification and UV Spectrophotometry
The content of rituximab in the fermentation broth samples (PM01 and PM02) was quantified using a conventional method involving protein A-based solid phase purification followed by quantification using UV absorbance at 280 nm.21,22 Although we employed spin columns (see the Experimental Section), HPLC methodologies are also applicable.21 Whereas an HPLC-based procedure often requires sample preparation, a recent study reported a fast (less than 5 min) HPLC-based procedure but with a detection limit two orders of magnitude higher than the methodology reported in the present work.21 The results are depicted in Table 2. The rituximab titers determined by this approach are in good agreement with the FIDA measurements. This demonstrates that the developed FIDA method is compatible with specific measurements in a clarified fermentation broth. Furthermore, FIDA can measure selectively in a clarified fermentation broth, thus eliminating any requirement for purification prior to sample analysis, which, in combination with a fully automated instrument, allows for a considerably high throughput of samples with few procedural steps. The simple FIDA methodology enables assessment of the in-solution IgG titer in less than 4 min, thereby allowing nearly real-time measurements of IgG titer during fermentation processes.
Conclusions
The present work shows that FIDA is a viable technology for direct (no sample pretreatment) quantification of the antibody titer in a CHO cell fermentation broth (nanomolar sensitivity) and for characterization of in-solution binding to monoclonal antibodies. The developed FIDA assay utilized a selective anti-IgG affibody as indicator and thereby accurately determining the rituximab concentration without any preceding purification process. This demonstrates that FIDA is characterized by being highly tolerant to the sample matrix, hence, little or no sample pretreatment is required. Furthermore, the developed method characterized the affibody–rituximab interaction with regards to determining the dissociation constant (Kd), complex size (Rh), and binding stoichiometry.
This protocol may be extended to include a full functional assessment of the antibody by assessment of specific target binding. We thus envision that the FIDA methodology will find applications in development and production of therapeutic proteins.
Acknowledgments
AGC Biologics is acknowledged for providing samples and reference materials. Financial support from the Innovation Fund Denmark (grant no. 9065-00009B), the Market Development Fund (grant no. 2016-10649), and Carlsberg Foundation (grant no. 2013_01_0544) are gratefully acknowledged.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c00791.
Additional experimental details (PDF)
Author Contributions
All authors have given approval to the final version of the manuscript.
The authors declare the following competing financial interest(s): HJ, JO, and MEP have commercial interests in FIDA Biosystems ApS. MEP and HJ are employees of FIDA Biosystems ApS.
Supplementary Material
References
- Li F.; Vijayasankaran N.; Shen A.; Kiss R.; Amanullah A. Cell Culture Processes for Monoclonal Antibody Production. mAbs 2010, 2, 466–479. 10.4161/mabs.2.5.12720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantardjieff A.; Zhou W. Mammalian Cell Cultures for Biologics Manufacturing. Adv. Biochem. Eng./Biotechnol. 2014, 10.1007/10_2013_255. [DOI] [PubMed] [Google Scholar]
- dos Santos N. V.; de Carvalho Santos-Ebinuma V.; Pessoa Junior A.; Pereira J. F. B. Liquid–Liquid Extraction of Biopharmaceuticals from Fermented Broth: Trends and Future Prospects. J. Chem. Technol. Biotechnol. 2018, 93, 1845–1863. 10.1002/jctb.5476. [DOI] [Google Scholar]
- Reusch D.; Haberger M.; Selman M. H. J.; Bulau P.; Deelder A. M.; Wuhrer M.; Engler N. High-Throughput Work Flow for IgG Fc-Glycosylation Analysis of Biotechnological Samples. Anal. Biochem. 2013, 432, 82–89. 10.1016/j.ab.2012.09.032. [DOI] [PubMed] [Google Scholar]
- Estes S.; Melville M.. Mammalian Cell Line Developments in Speed and Efficiency. In Mammalian Cell Cultures for Biologics Manufacturing. Advances in biochemical engineering/biotechnology; Springer, 2014; pp 11–33. 10.1007/10_2013_260. [DOI] [PubMed] [Google Scholar]
- Dong J.; Migliore N.; Mehrman S. J.; Cunningham J.; Lewis M. J.; Hu P. High-Throughput, Automated Protein A Purification Platform with Multiattribute LC-MS Analysis for Advanced Cell Culture Process Monitoring. Anal. Chem. 2016, 88, 8673–8679. 10.1021/acs.analchem.6b01956. [DOI] [PubMed] [Google Scholar]
- Jiang M.; Severson K. A.; Love J. C.; Madden H.; Swann P.; Zang L.; Braatz R. D. Opportunities and Challenges of Real-Time Release Testing in Biopharmaceutical Manufacturing. Biotechnol. Bioeng. 2017, 114, 2445–2456. 10.1002/bit.26383. [DOI] [PubMed] [Google Scholar]
- Barnard G. C.; Kull A. R.; Sharkey N. S.; Shaikh S. S.; Rittenhour A. M.; Burnina I.; Jiang Y.; Li F.; Lynaugh H.; Mitchell T.; et al. High-Throughput Screening and Selection of Yeast Cell Lines Expressing Monoclonal Antibodies. J. Ind. Microbiol. Biotechnol. 2010, 37, 961–971. 10.1007/s10295-010-0746-1. [DOI] [PubMed] [Google Scholar]
- Yu Y.; Mitchell S.; Lynaugh H.; Brown M.; Paul Nobrega R.; Zhi X.; Sun T.; Caffry I.; Cao Y.; Yang R.; et al. Understanding Fortebio’s Sensors for High-Throughput Kinetic and Epitope Screening for Purified Antibodies and Yeast Culture Supernatant. J. Biomol. Screening 2016, 21, 88–95. 10.1177/1087057115609564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen M. E.; Gad S. I.; Østergaard J.; Jensen H. Protein Characterization in 3D: Size, Folding, and Functional Assessment in a Unified Approach. Anal. Chem. 2019, 91, 4975–4979. 10.1021/acs.analchem.9b00537. [DOI] [PubMed] [Google Scholar]
- Jensen H.; Østergaard J. Flow Induced Dispersion Analysis Quantifies Noncovalent Interactions in Nanoliter Samples. J. Am. Chem. Soc. 2010, 132, 4070–4071. 10.1021/ja100484d. [DOI] [PubMed] [Google Scholar]
- Poulsen N. N.; Pedersen M. E.; Østergaard J.; Petersen N. J.; Nielsen C. T.; Heegaard N. H. H.; Jensen H. Flow-Induced Dispersion Analysis for Probing Anti-DsDNA Antibody Binding Heterogeneity in Systemic Lupus Erythematosus Patients: Toward a New Approach for Diagnosis and Patient Stratification. Anal. Chem. 2016, 88, 9056–9061. 10.1021/acs.analchem.6b01741. [DOI] [PubMed] [Google Scholar]
- Poulsen N. N.; Andersen N. Z.; Østergaard J.; Zhuang G.; Petersen N. J.; Jensen H. Flow Induced Dispersion Analysis Rapidly Quantifies Proteins in Human Plasma Samples. Analyst 2015, 140, 4365–4369. 10.1039/C5AN00697J. [DOI] [PubMed] [Google Scholar]
- Pedersen M. E.; Østergaard J.; Jensen H.. Flow-Induced Dispersion Analysis (FIDA) for Protein Quantification and Characterization. In Methods in Molecular Biology; Humana Press, 2019. 10.1007/978-1-4939-9213-3_8. [DOI] [PubMed] [Google Scholar]
- To A. G.; Probes F.; Technologies L.. Thiol-Reactive Probes Thiol-Reactive Probes. In The Molecular Probes Handbook: A Guide to Fluorescent Probes and Labeling Technologies; 2010; pp 96–121.
- Nupur N.; Chhabra N.; Dash R.; Rathore A. S. Assessment of Structural and Functional Similarity of Biosimilar Products: Rituximab as a Case Study. mAbs 2018, 10, 143–158. 10.1080/19420862.2017.1402996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justino C. I. L.; Duarte A. C.; Rocha-Santos T. A. P. Analytical Applications of Affibodies. TrAC Trends Anal. Chem. 2015, 65, 73–82. 10.1016/j.trac.2014.10.014. [DOI] [Google Scholar]
- Kruljec N.; Bratkovič T. Alternative Affinity Ligands for Immunoglobulins. Bioconjugate Chem. 2017, 28, 2009–2030. 10.1021/acs.bioconjchem.7b00335. [DOI] [PubMed] [Google Scholar]
- Erickson H. P. Size and Shape of Protein Molecules at the Nanometer Level Determined by Sedimentation, Gel Filtration, and Electron Microscopy. Biol. Proced. Online 2009, 11, 32–51. 10.1007/s12575-009-9008-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamieh J.; Merdassi H.; Rossi J. C.; Jannin V.; Demarne F.; Cottet H. Size Characterization of Lipid-Based Self-Emulsifying Pharmaceutical Excipients during Lipolysis Using Taylor Dispersion Analysis with Fluorescence Detection. Int. J. Pharm. 2018, 537, 94–101. 10.1016/j.ijpharm.2017.12.032. [DOI] [PubMed] [Google Scholar]
- Satzer P.; Jungbauer A. High-Capacity Protein A Affinity Chromatography for the Fast Quantification of Antibodies: Two-Wavelength Detection Expands Linear Range. J. Sep. Sci. 2018, 41, 1791–1797. 10.1002/jssc.201701481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willoughby N.; Martin P.; Titchener-Hooker N. Extreme Scale-down of Expanded Bed Adsorption: Purification of an Antibody Fragment Directly from Recombinant E Coli Culture. Biotechnol. Bioeng. 2004, 87, 641–647. 10.1002/bit.20173. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.