Abstract
The study explores the role of neddylation in early trophoblast development and its alteration during the pathogenesis of recurrent spontaneous abortion (RSA). Immunofluorescence and western blot were conducted to evaluate the expression pattern of NEDD8 protein in the first-trimester placentas of healthy control and RSA patients. Neddylated-cullins, especially neddylated-cullin1, were downregulated and their substrate, p21, was accumulated in RSA samples. NEDD8 cytoplasmic recruitment was observed in extravillous trophoblast (EVT) progenitors of RSA placentas. Consistent with the results of clinical samples, neddylation inhibition using MLN4924 in trophoblast cell lines caused obvious p21 accumulation and free NEDD8 cytoplasmic recruitment. Further in vitro study demonstrated neddylation inhibition attenuated proliferation of Jeg-3 cells via p21 accumulation. Moreover, when trophoblast stem (TS) cells derived from first-trimester placentas were cultured for differentiation analyses. MLN4924 impaired the differentiation of TS cells towards EVTs by downregulating HLA-G and GATA3. p21 knockdown could partly rescue MLN4924-suppressed HLA-G and GATA3 expression. In conclusion, cullin1 neddylation-mediated p21 degradation is required for trophoblast proliferation and can affect trophoblast plasticity by affecting HLA-G and GATA3 expression. The results provide insights into the pathological mechanism of RSA and the biological regulation of trophoblast development.
Keywords: neddylation, NEDD8, recurrent spontaneous abortion, trophoblast stem cell, p21
Introduction
Recurrent miscarriage (RM), also termed as recurrent spontaneous abortion (RSA), is defined as two or more consecutive pregnancy losses of a clinically established intrauterine pregnancy (Practice Committee of American Society for Reproductive Medicine 2013). It is one of the most complicated pregnancy-related diseases affecting 2–5% of couples (El Hachem et al., 2017). The etiology includes chromosomal abnormality, genital malformation, infection, endocrine disorder, immune dysregulation and hemostatic dysfunction (Carrington et al., 2005; Garrido-Gimenez and Alijotas-Reig 2015; Grimstad and Krieg 2016). However, almost half of cases are diagnosed without an exact etiology and most cases occur during the first trimester of pregnancy (El Hachem et al., 2017). A successful pregnancy requires a normal functioning placenta that connects the fetus and uterus. Trophoblasts are placenta-specialized cells arising from the trophectoderm, which are divided into subtypes including cytotrophoblasts (CTBs), syncytiotrophoblast (ST) and extravillous trophoblasts (EVTs) (Red-Horse et al., 2004). CTBs are the bipotential trophoblast progenitor that form a mitotically active epithelial layer and can proliferate as well as differentiate into EVTs and STs (Bischof and Irminger-Finger 2005). CTBs fuse to form multinuclear ST which covers the outer layer of the chorionic villus performing gas/nutrient exchange and produce hormones (James et al., 2012). CTBs on the anchoring villi aggregate to form cell column trophoblasts (CCTs) and further differentiate into EVTs. The differentiated EVTs invade into the decidua to participate in spiral artery remodeling and establishment of maternal-fetal immune tolerance (Rouas-Freiss et al., 1997; Cierna et al., 2016; Ferreira et al., 2017). Although accumulating evidence has demonstrated that abnormal trophoblast development is involved in the pathogenesis of RSA, the underlying mechanism has not been elucidated (Choi et al., 2003; Baek et al., 2007; Li et al., 2019; Lv et al., 2019).
Several signaling pathways have been reported to be involved in trophoblast development, including Wnt, Notch, Hippo and transforming growth factor β (Nishioka et al., 2009; Knofler 2010; Knofler and Pollheimer 2013; Haider et al., 2016). Recently, several studies have demonstrated that cullins, the scaffold protein of the Cullin RING ligase (CRL) complex, the largest E3 ubiquitin ligase family for protein ubiquitylation and degradation (Zhao and Sun 2013), are essential for proper trophoblast development. Cullin7 has been reported to trigger epithelial–mesenchymal transition of choriocarcinoma JEG-3 cells (Fu et al., 2010). Cullin1 (CUL1) is reported to be significantly downregulated in placentas from preeclampsia patients, and knockdown of CUL1 affects the invasion of trophoblasts (Zhang et al., 2013). Placenta-specific deletion of Cullin3 in mice results in less invasive trophoblasts in the maternal decidua (Zhang et al., 2015). Generally, CRLs consist of four components: cullins, RINGs, adaptor proteins and substrate recognition receptors. These complexes are responsible for the ubiquitylation of ~ 20% of cellular protein (Sarikas et al., 2011; Zhao and Sun 2013). The assembly of an active CRL complex requires reversible modification of cullins by a NEDD8 molecule, a process called neddylation. The neddylation process is similar to protein ubiquitination, in that the NEDD8 is activated by NEDD8-activating enzyme E1, loaded by NEDD8-conjugating enzyme E2 (UBE2M or UBE2F), and conjugated to a substrate by an E3 ligase via covalent attachment (Hori et al., 1999; Zhou et al., 2019). The reverse process in which conjugated NEDD8 is removed from a neddylated cullin by the action of an NEDD8 isopeptidase, COP9 signalosome complex (CSN), is called deneddylation (Pan et al., 2004; Duda et al., 2008; Zhao and Sun 2013; Zhao et al., 2014; Lan et al., 2016). However, unlike ubiquitination, this process is not directed to substrate degradation but rather to modulate protein activity and function (Zhao et al., 2014). The activation of CRLs further triggers the ubiquitin-mediated degradation of its substrate protein and thereby maintains cellular homeostasis (Zhao et al., 2014). MLN4924, an inhibitor of NEDD8-activating enzyme E1, can result in decreased NEDD8-Cullins conjugates and impair CRL complex assembly.
Neddylation has been reported to affect human endometrial stromal cell (HESC) proliferation and decidualization (Liao et al., 2015). However, limited studies have been performed to explore the role of neddylation in trophoblast development and its relationship with RSA. In the present study, we first demonstrated that the cullin neddylation level was significantly lower in RSA first-trimester placenta compared with the healthy control. In vitro cell assays confirmed that neddylation inhibition-related p21 accumulation inhibited trophoblast cell proliferation and affected the differentiation of TS cells toward EVTs.
Materials and Methods
Patient recruitment and tissue collection
This study was approved by the Sir Run Run Shaw Hospital Research and Ethics Committee (Number of approval: 20140224-9; February 2014). Written informed consent was obtained from all participants before enrollment. First-trimester human placental tissues of RSA patients and healthy controls (HCs) were collected from women undergoing dilatation and curettage at the Department of Obstetrics and Gynecology of the Sir Run Run Shaw Hospital affiliated to the School of Medicine, Zhejiang University.
RSA is regarded as two or more consecutive early pregnancy losses of a clinically established intrauterine pregnancy with the same spouse including the present pregnancy. Ultrasound examination indicated an empty sac or an embryo with no fetal heartbeat. Patients with the following conditions were excluded: (i) genital malformation; (ii) abnormal karyotype of the parents and abortuses; (iii) endocrine or metabolic disorders; (iv) autoimmune diseases; (v) other major disease; and (vi) improper drug treatment, and exposure to chemicals or radiation. The HCs were women with at least one live birth and no history of any pregnancy-related diseases such as miscarriage, preterm labor and preeclampsia. Ultrasound examination indicated normal gestation with a fetal heartbeat. A total of 34 HCs and 25 RSA first-trimester placentas were collected. The clinical characteristics of recruited patients are provided in Table I.
Table I. Clinical characteristics of the recruited patients.
| Healthy controls | Recurrent spontaneous abortion | P value | |
|---|---|---|---|
| Sample size | 34 | 25 | |
| Maternal age (years) | 31.2 ± 0.7 | 33.0 ± 1.1 | ns |
| Gestational age at D&C (days) | 53 ± 1.3 | 53 ± 1.9 | ns |
| Number of spontaneous abortions | n/a | 2.4 ± 0.18 | n/a |
Data are mean ± SEM. D&C, dilatation and curettage; ns, not statistically significant; n/a, not applicable.
The placental tissues were washed with PBS to remove blood. Tissues for immunofluorescence (IF) and immunohistochemistry (IHC) analysis were fixed in 4% (w/v) paraformaldehyde. For protein and RNA extraction, the villi from placentas were scraped from the chorionic membrane to avoid maternal contamination (Liu et al., 2018) and cut into small pieces of 3–5 mm. All the pieces were mixed, aliquoted and subsequently snap frozen in liquid nitrogen. Three HC placentas were used for TS cell derivation.
First-trimester villous explant culture
Matrigel (8 mg/mL) (Coring, USA) was added to a 24-well plate and incubated at 37°C to form a drop of gel. MLN4924 was added to Matrigel to a final concentration of 1 μM before gel formation; the same amount of DMSO was added to the control group. The placenta villus (6th–8th weeks of gestation) tissues were dissected into small pieces (2–3 mm) from villus tips. Each piece of villus was carefully placed on the top of the gel, covered with 50 μL of medium and incubated at 37°C with 5% CO2 in air for 4–6 h to allow anchorage. Subsequently, explants were cultured in 500 μL of medium with the indicated treatment. The outgrowth of explants was recorded and photographed at Days 1 and 4 under a microscope (Carl Zeiss Primovert, Oberkochen, Germany).
Trophoblast cell culture and siRNA transfection
Jeg-3 cells were cultured in Dulbecco’s modified Eagle’s medium/Nutrient Mixture F-12 medium (DMEM/F12) (Gibco, Carlsbad, CA, USA) with 15% (v/v) fetal bovine serum (FBS) and 100 IU/mL penicillin and streptomycin each. HTR8/Sveno (HTR8) cells were cultured in RPMI-1640 medium with 10% (v/v) FBS and 100 IU/mL penicillin and streptomycin each. Both cell lines were obtained from American Type Culture Collection. All cell lines were incubated with at 37°C with 5% CO2. Jeg-3 cells were transfected with siRNA oligonucleotides using Lipofectamine 3000 (Invitrogen USA), according to the manufacturer’s instructions. MLN4924 was added after siRNA transfection. The sequences of the siRNAs were as follows: si-p21: 5′-GAUGGAACUUCGACUUUGUTT-3′; si-cullin1: 5′-GGUCGCUUCAUAAACAACAdTdT-3′.
Cell proliferation assay
We used the Cell Counting Kit 8 (CCK-8) (Tojindo, Shanghai, China) according to the manufacturer’s instructions to measure cell proliferation. The kit utilizes WST-8 [2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt] to produce a water-soluble formazan dye upon reduction in the presence of an electron carrier. Jeg-3 cells were seeded in 96-well plates at a density of 2 × 103 cells per well. After 24 h of incubation, the cells were transfected with p21 siRNAs with or without MLN4924 treatment. The cell growth rate was measured using a cell counting kit-8 (CCK8) every 24 h.
TS cell culture and EVT differentiation
The isolation and culture of TS cells was conducted following a previously published protocol (Okae et al., 2018). In brief, the placental villus of first-trimester placentas was dissected into small pieces and enzymatically digested three times in a TrypLE (Thermo Fisher Scientific, USA) and Accumax (Innovative Cell Tech, USA) mixture (1:1) for 30 min at 37°C. Cell suspensions were filtered through a 70-μm mesh filter. CTBs were immunomagnetically purified using an EasySep phycoerythrin (PE)-positive selection kit (Stemcell Technologies, Vancouver, Canada) and PE-conjugated anti-ITGA6 antibody (BioLegend, CA, USA). The selected cells were seeded in a collagen IV (Col IV) (Coring, USA)-coated plate and cultured in TS medium (Supplementary Table SI). CTBs cells proliferated and were passaged. Cells at passages 10–30 were used for analyses and differentiation assays.
For EVT differentiation, TS cells were seeded in a plate precoated with Col IV and cultured in EVT medium (Supplementary Table SI). Matrigel was added at a final concentration of 2% (v/v) shortly after suspending the cells in the medium. At Day 2, the medium was replaced with EVT medium without NRG1, and Matrigel was added at a final concentration of 0.5%. The cells were cultured for an additional 3 days and passaged to a new Col IV-coated plate and cultured with EVT medium without NRG1 and KSR for another 2 days. Differentiated cells were collected at Days 5 and 8. EVT cells treated with MLN4924 were collected at Day 5. MLN4924 was added at 72 h before the cells were collected.
For ST differentiation, TS cells were seeded in a plate precoated with Col IV and cultured in ST medium (Supplementary Table SI) for 3 days and fixed in 4% (w/v) paraformaldehyde for IF or collected for RNA extraction.
Immunofluorescence
Paraffin-embedded tissues were deparaffinized, rehydrated, subjected to antigen retrieval, blocked and incubated with primary antibodies (Supplementary Table SII). Cultured cells on a glass coverslip were fixed in 4% (w/v) paraformaldehyde for 30 min. After permeabilization with cold methanol at −20°C or 10 min, the cells were blocked with 3% (w/v) bovine serum albumin (BSA) for 1 h and then incubated with primary antibodies at 4°C overnight. Then, the tissues or cells were washed three times and incubated for 1 h with goat anti-mouse or anti-rabbit IgG conjugated to Alexa Fluor 488 or Alexa Fluor 549 (Muti-Science, Hangzhou, China) at room temperature. 4′,6-Diamidino-2-phenylindole (DAPI) was used to stain nuclei. For the IF of HLA-G, cultured cells were blocked with 10% (v/v) goat serum and 3% (w/v) BSA/phosphate-buffered saline (PBS) for 20 min at 4°C and then stained with HLA-G primary antibody for 1 h. Subsequently, cells were fixed with 1% (w/v) paraformaldehyde (PFA) for 10 min and stained with goat anti-mouse IgG conjugated to Alexa Fluor 488 for 30 min. DAPI was used to stain nuclei. A normal rabbit IgG was used as negative control. Tissues and cells were digitally photographed by a fluorescence microscope (Olympus BX53, Cell Sens DI software) or a confocal laser scanning microscope (Zeiss LSM800, Carl Zeiss, Germany).
Immunohistochemistry
Tissues fixed in 4% (w/v) paraformaldehyde were embedded in paraffin, sectioned at 3-μm thicknesses onto glass slides and analyzed by immunohistochemistry (IHC) according to a standard protocol. Primary antibodies used in this study are listed in Supplementary Table SII. A GTvisonII detection kit was used for staining. The sections were counterstained with hematoxylin, dehydrated and mounted in neutral balsam. Images were captured under a light microscope (80i; Nikon, Tokyo, Japan).
Western blotting
Whole cell lysates were prepared with RIPA buffer using standard protocols. First-trimester placenta tissue was cut into small pieces as described previously before adding an appropriate volume of RIPA buffer. Subsequently, the mixtures were homogenized (Servicebio, China). Nuclear and cytoplasmic extracts were obtained using a NE-PER nuclear and cytoplasmic protein extraction kit (Thermo Scientific; USA). Western blotting (WB) was conducted using a standard protocol. Primary antibodies are listed in Supplementary Table SII. Signals were detected using an ECL Detection Kit (Millipore, Boston, MA, USA) and visualized with a ChemiDoc™ Touch Imaging System (Bio-Rad, Hercules, CA, USA). Quantification was performed using ImageJ software.
RNA extraction and quantitative real-time transcription-polymerase chain reaction
Total RNA was extracted using TRIzol reagent (Invitrogen, USA), according to the manufacturer’s instructions. RNA concentrations were quantified by a NanoDrop 2000 (NanoDrop, Wilmington, DE, USA). Reverse transcription of 1 μg total RNA was performed using a Quantscript RT kit (Tiangen Biotechnology, Beijing, China). mRNA expression levels were determined by quantitative real time polymerase chain reaction (PCR) using a SYBR Green Master Mix Kit (DBI Bioscience, Ludwigshafen, Germany) and Roche 480 Real-Time PCR System (Roche, Basel, Switzerland). Primers are listed in Supplementary Table SIII. Melting curve analysis was performed to verify the specificity of the PCR after each run.
Flow cytometry
For flow cytometric analysis of cell cycle, cells under the indicated treatments were harvested and fixed in 70% (v/v) ethanol at 4°C overnight. The cells were washed twice with ice-cold PBS and then stained with propidium iodide (PI, 50 μg/mL; Sigma-Aldrich) for 30 min in the dark, and RNA-aseA was added to destroy RNA. The samples were then analyzed for cell cycle distributions using a BD LSRFortessa flow cytometer (BD, NJ, USA).
A PE Annexin V Apoptosis Detection Kit (BD, NJ, USA) was used for flow cytometric analysis of apoptosis according to the manufacturer’s instruction. In brief, cells were harvested, washed twice with cold PBS, resuspended in 100 μL 1× binding buffer and stained with PE Annexin V and 5 μL 7-AAD for 15 min at room temperature in the dark. After incubation, 400 μL of 1× Binding Buffer was added to each tube. The samples were analyzed for cell cycle distributions using a BD LSRFortessa flow cytometer (BD, NJ, USA).
For flow cytometric analysis of ITGA6, TS cells were disassociated with TrypLE and incubated with PE-conjugated anti-ITGA6 antibody for 30 min at room temperature. PE-conjugated mouse IgG1 was used as isotype control. For flow cytometric analysis of HLA-G, TS and EVT cells were disassociated with TrypLE and incubated with HLA-G antibody for 30 min at room temperature and stained with Alexa Fluor 488-conjugated anti-rabbit IgG (Muti-Science, Hangzhou, China) for 30 min. For flow cytometric analysis of CK7, disassociated TS cells were permeabilized using eBioscience™ Fixation/Permeabilization Concentrate and Diluent(Invitrogen USA) and then incubated with anti-CK7 antibody for 30 min at room temperature and stained with Alexa Fluor 549-conjugated anti-rabbit IgG (Muti-Science, Hangzhou, China) for 30 min. PE-conjugated mouse IgG1 and FITC-conjugated mouse IgG1 were used as isotype controls. The samples were then analyzed for using BD LSRFortessa flow cytometer (BD, NJ, USA).
SA-β-galactosidase staining
The expression of senescence-associated β-galactosidase was determined by a SA-β-galactosidase (SA-β-Gal) staining kit (Beyotime, Shanghai, China), according to the manufacturer’s specifications. Blue dye-positive cells were recorded as senescent cells. In situ β-galactosidase activity assay was performed following a recently published protocol (Velicky et al., 2018): whole mount placentas were fixed with the fixative solution at 4°C overnight, washed with PBS and incubated over night with ß-galactosidase staining working solution at 37°C without CO2 (pH 6). Placentas were subsequently dehydrated and perfused with paraplast (Hualing, China), then embedded in paraffin. Serial sections of the embedded tissue were counterstained with antibodies against HLA-G and DAPI. The ratio of SA-β-GAL-positive area in CCT was analyzed using ImageJ software.
Statistical analysis
All statistical analysis tests were calculated using GraphPad Prism 7 software (GraphPad Software, San Diego, California, USA). The Gaussian distribution and equality of variances were examined by Shapiro–Wilk normality test. Normally distributed datasets were analyzed using unpaired t test; those datasets that did not pass the normality test were analyzed using Wilcoxon–Mann–Whitney U tests. A P value of < 0.05 was considered as statistically significant. Unless otherwise noted, all experiments were conducted in duplicates and repeated at least three times.
Results
NEDD8 is highly activated in proliferating trophoblast subsets
The expression pattern of NEDD8 protein was examined by IF in HC first trimester villus. In the floating villus, NEDD8 was prominently expressed in the single layer of CTB cells and significantly downregulated in STB labeled for hCGβ (Fig. 1A). Human leukocyte antigen G (HLA-G) was used to label the anchoring villus and its direction towards decidual. In the anchoring villus, the most intensive expression of NEDD8 was observed in the proximal cell column trophoblasts (pCCTs) generated by aggregating CTBs. Because CTBs and pCCTs are highly proliferating cells, this result demonstrated that NEDD8 expression was more intensive in cells with a higher proliferation potential.
Figure 1. Expression pattern of NEDD8 in first trimester placentas and change in subcellular localization of NEDD8 protein in recurrent spontaneous abortion (RSA) placentas. (A–B) Nedd8 immunofluorescence (IF) in first trimester placentas: sections of healthy control (HC) (n = 4) and RSA (n = 4) early pregnancy placentas from 6–8-week pregnancies were stained with antibodies against NEDD8 (red), HGCβ (green) or HLA-G (green). Nuclei were stained with DAPI. For negative controls, primary antibodies were replaced with rabbit monoclonal isotype IgG (mAb IgG). VS, villus stroma; CTB, cytotrophoblast; STB, syncytiotrophoblast; pCCT: proximal cell column trophoblast; dCCT: distal cell column trophoblast. Scale bars: 50 μm. (A) Top: representative images of NEDD8 staining in the first-trimester floating villus. HCGβ (green) was costained to indicate STB. Bottom: representative images of NEDD8 staining in the first trimester anchoring villus. HLA-G (green) was costained to indicate dCCTs. (B) Representative images of NEDD8 subcellular localization of the anchoring villus of HC and RSA first trimester placentas. (C) Scatter plot of nuclear/cytoplasmic ratio of NEDD8 measured by ImageJ. Sixty single nuclear HLA-G (+) cells from four first trimester villus were analyzed for each group. Means ± interquartile range (IQR) are shown. ****P < 0.0001.
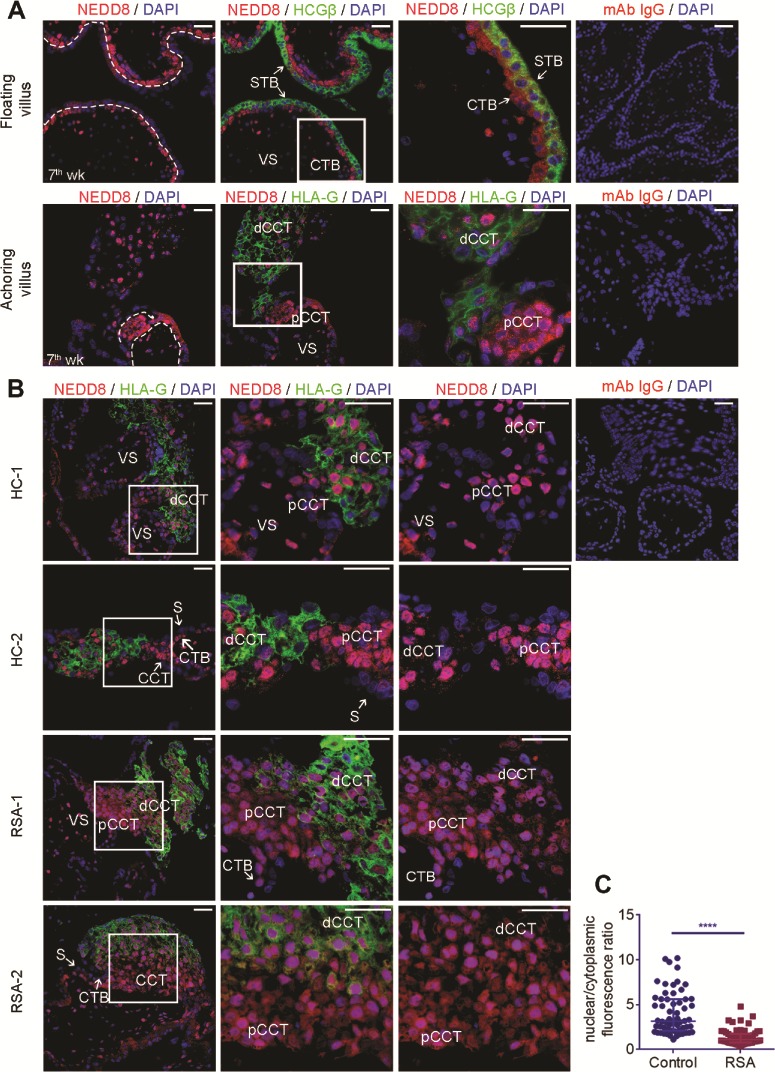
NEDD8 subcellular localization and activity is altered in the RSA anchoring villus
IF was performed to explore the NEDD8 subcellular distribution in HC and RSA first trimester villus. We found the distribution of NEDD8 in HLA-G (+) CCTs, which differentiate towards EVTs, were different between HCs and RSA patients. In the RSA group, the NEDD8 cytoplasmic distribution was largely increased in HLA-G (+) CCTs (Fig. 1B and C), indicating that the activity of NEDD8 might be altered in RSA placentas. For confirmation, we conducted WB of first-trimester chorionic villus tissues from RSA patients and HCs. Figure 2A shows that the expression of neddylated cullins (NEDD8-cullin) was significantly downregulated in the RSA villus. However, the total NEDD8 mRNA level was not significantly different between the two groups (Supplementary Fig. S1A), indicating that the decrease in the neddylated cullin protein level was largely due to neddylation inhibition. These results inferred that in the RSA anchoring villus, more free NEDD8 was localized in the cytoplasm and neddylation activity was lower.
Figure 2. Neddylation inhibition causes cytoplasmic recruitment of NEDD8 in trophoblasts. (A) Representative western blotting (WB) of neddylated cullins (NEDD8-Cullins), Cullin1 (CUL1) and p21 in the recurrent spontaneous abortion (RSA) early pregnancy villus (n = 6) compared with healthy controls (HCs) (n = 5). Bar graph on the left is quantification of Nedd8 and p21. Means ± SD normalized to β-actin are shown. **P = 0.0013; *P = 0.0225. (B) Representative WB of Jeg-3 cell line treated with MLN4924 at increasing concentrations. (C–F) Jeg-3 cells were treated with 1 μM MLN4924 for 48 h. (C) Representative WB of NEDD8-Cullins (90 kD) and free NEDD8 (12 kD). WCL: whole cell lysis. (D) RT-PCR analysis of NEDD8 mRNA expression in Jeg-3 cells after MLN4924 treatment. Means ± SD normalized to 18 s rRNA are shown. (E) Representative WB of Jeg-3 cells separated into cytoplasmic and nuclear fractions. (F) Representative immunofluorescence (IF) images and nuclear/cytoplasmic ratio analysis showing the subcellular localization change of NEDD8 after MLN4924 treatment. Scale bars: 50 μm. Nuclear/cytoplasm ratio was calculated from 30 single cells for each group of three independent assays. Means ± IQR are shown. ****P < 0.0001.

Neddylation inhibition mediates downstream p21 accumulation observed in RSA first-trimester placentas or induced by MLN4924 treatment
Because NEDD8 binds to all cullins and regulates their activity, we analyzed various cullins and NEDD8. WB of several members of the cullin family indicated that the inhibition of CUL1 neddylation was the most significant (Fig. 2A, Fig. S1B). Moreover, a well-known substrate of CUL1, p21, was accumulated in the RSA villus (Fig. 2A, Supplementary Fig. S1C). IHC staining demonstrated that the p21 accumulation in RSA placentas was mostly observed in trophoblast cells instead of villus stroma (Fig. S1C). Thus, in this study, we mainly focused on CUL1 neddylation related p21 degradation and its role in trophoblast development.
MLN4924 is a selective inhibitor of NAE, a NEDD8-activating enzyme. It efficiently blocks cullin neddylation and inactivates CRL/SCF E3 ligase (Soucy et al., 2009; Brownell et al., 2010). To further investigate the effect of neddylation inhibition on trophoblasts, we treated Jeg-3 cells with MLN4924 and found dose-dependent inhibition of CUL1 neddylation and accumulation of p21 (Fig. 2B). Accordingly, the protein level of the NEDD8-Cullins complex was highly downregulated and the free NEDD8 upregulated (Fig. 2C) without change in the NEDD8 mRNA expression level (Fig. 2D). Moreover, MLN4924 treatment of Jeg-3 cells resulted in free NEDD8 accumulation in cytoplasm and correspondingly reduced the Cul1-NEDD8 level in nuclei (Fig. 2E). The subcellular distribution change of NEDD8 was also confirmed by IF: an obvious increase in NEDD8 fluorescence signal was observed in the cytoplasm (Fig. 2F). Similar results were also observed in HTR8 cells (Supplementary Fig. S2). These results suggested that MLN4924 causes cytoplasmic recruitment of free NEDD8 protein. Together with the alteration in subcellular localization of NEDD8 in the RSA anchoring villus and inhibited neddylation state, we considered that abnormal neddylation might participate in the regulation of normal development of the invasive trophoblast lineage and might contribute to the pathogenesis of RSA.
Neddylation inhibition restricts cell proliferation via p21 accumulation
The IHC staining intensity of Ki67 in the RSA villus was significantly lower than that in HCs (Fig. 3A), indicating attenuated proliferation of RSA trophoblasts. Next, we explored the influence of neddylation inhibition by MLN4924 on growth of primary villus explants form HC early pregnancy placentas: Upon MLN4924 treatment, the outgrowth of villus explants was significantly suppressed (Fig. 3B). In vitro study revealed that MLN4924 treatment suppressed proliferation and caused G2 arrest and apoptosis in the trophoblast cell line Jeg-3 cells (Fig. 3D–G; Fig. S3). To further elucidate the mechanism of inhibited cell proliferation under neddylation inhibition, p21 expression of Jeg-3 cells was knocked down using siRNA (Fig. 3C and D). IF staining of proliferating cell markers, p-H3 and ki67 as well as CCK8 assays showed that the MLN4924-induced inhibition of Jeg-3 proliferation was rescued by p21 silencing (Fig. 3E and F). Moreover, p21 has been widely reported to be related to cell cycle arrest (Jia et al., 2011; Lan et al., 2016; Zhang et al., 2019). In the present study, MLN4924 causes G2 arrest in Jeg-3 cells, and this effect was partly rescued by p21 knockdown (Fig. 3G).
Figure 3. Proliferation restriction in recurrent spontaneous abortion (RSA) placentas is related to p21 accumulation caused by neddylation inhibition. (A) Representative immunohistochemical staining of Ki67 in healthy control (HC) and RSA floating and anchoring villus. n = 4 for each group. Scale bars: 50 μm. Bar graph shows the means ± SD of the HSCORE. ****P < 0.0001. (B) Representative images of HC villus explants culture (4 days) after MLN4924 (1 μM) or DMSO treatment for 48 h. Scale bars: 500 μm. Bar graph on the left is quantification of the villus outgrowth distance. Fifteen explants for each condition derived from four different HC placentas were measured. Means ± SD are shown. ***P = 0.006. (C–F) Jeg-3 cells were treated with 1 μM MLN4924 or DMSO for 48 h following p21 silencing: (C) western blot (WB) and (D) RT-PCR results showing p21 knockdown efficiency after siRNA transfection. Bar graph of the mRNA level shows means ± SD normalized to 18 s rRNA. *P = 0.0266; ***P = 0.001. (E) CCK8 assay measuring the proliferation rate of Jeg-3 cells. (F) IF staining of Ki67 (red) and p-H3 (red). Nuclei were stained with DAPI. Scale bars: 50 μm. (G) Jeg-3 cells were synchronized overnight before treated with 1 μM MLN4924 or DMSO for 48 h following p21 silencing. The cell cycle distributions were analyzed by flow cytometry. Bar graph on the left indicates the percentage of cells in G1, S or G2/M phase. Means ± SD are shown, t test was conducted to compare the G2 phase ratio between each group. **P = 0.0023; *P = 0.00187.

Neddylation activation is required for the differentiation of human TS cells toward EVTs
A normal functioning placenta requires not only cell proliferation, but also fine-tuned differentiations of TS cells and progenitors. To further investigate the effect of neddylation during trophoblast differentiation, we derived TS cells referring to a newly established system (Okae et al., 2018) from the early pregnancy placenta (Fig. 4A). Immunostaining of the derived cell lines confirmed positive expression of CK7 (a pan-trophoblast marker) and CDH1 (E-cadherin, a CTB marker) and negative expression of vimentin (a stromal marker) (Fig. 4B). Flow cytometry of CK7 and IGTA6 confirmed the purity of selected and cultured cells (Fig. 4C). Similar to the results in Jeg-3 cells, MLN4924 treatment inhibited TS cells proliferation (Supplementary Fig. S4). TS cells have been reported to have the ability to differentiate into EVT- and STB-like cells under certain conditions. In the present study, we directed the TS cells to differentiate into EVT-like cells as well as ST-like cells according to the method introduced in the original article (Supplementary Fig. S5) (Okae et al., 2018). Time-dependent upregulation of HLA-G and downregulation of p63 and CDH1 were observed (Supplementary Fig. S5A and B). Because significant HLA-G upregulation had been achieved at Day 5 of differentiation culture, we treated EVTs at the beginning of differentiation induction with MLN4924 and collected the cells at Day 5. WB, Q-PCR and IF analysis revealed the upregulation of HLA-G and downregulation of CDH1 after EVT differentiation was hindered by MLN4924 in both the protein and mRNA level (Fig. 4D–F; Supplementary Fig. S6). Moreover, the mRNA level of two more genes upregulated in EVT, MMP2 (Sagi et al., 2016) and ITGA1 (Haider et al., 2016) was downregulated under MLN4924 treatment (Fig. 4D), indicating impaired differentiation of TS towards EVT under neddylation inhibition.
Figure 4. Neddylation inhibition hinders trophoblast stem cell differentiation toward extravillous trophoblasts (EVTs). (A) Phase-contrast image of trophoblast stem (TS) cells. Scale bar: 100 μm. (B) Representative immunofluorescence (IF) images showing CK7 (trophoblast marker), CDH1 (CTB marker) and VIM (vimentin, a stromal marker) in trophoblast stem cells (TS). Scale bars: 50 μm. (C) Flow cytometry histogram of CK7 and IGTA6 expression in TS cells. (D–F) TS cells, TS cells treated with MLN4924, differentiating EVT-Day 5 (EVT-D5) and EVT-D5 treated with MLN4924 (EVT-D5 + MLN4924) were processed for further analysis: (D) RT-PCR analysis of HLA-G (***P = 0.001; ****P < 0.0001), MMP2 (***P = 0.001; ****P < 0.0001), ITGA1 (****P < 0.0001; *P = 0.0419), GATA3 (****P < 0.0001; ***P = 0.0003; *P = 0.0194) and CDH1 (****P < 0.0001; ***P = 0.0006) mRNA expression. Means ± SD normalized to 18 s rRNA are shown. (E) Representative western blot (WB) results and quantification (n = 3) showing the protein expression levels of HLA-G (***P = 0.0002; *P = 0.0211), CHD1 (***P = 0.0008; *P = 0.0102; **P = 0.0049) and GATA3 (****P < 0.0001; *P = 0.0211). NEDD8-Cullins were regarded as an indicator of Neddylation inhibition. Mean values ± SD normalized to β-actin are shown. (F) Representative IF images showing expression of HLA-G, GATA3 and CDH1. Scale bars: 20 μm.

Neddylation activation regulates the expression of critical factors that affect the plasticity of human trophoblast progenitors
As a well-known marker of EVTs and EVT progenitors (Kovats et al., 1990; Ferreira et al., 2017), HLA-G is crucial for embryo implantation and maternal–fetal immune tolerance establishment (Moffett and Loke 2006; Tilburgs et al., 2015). Obvious time and dose-dependent downregulation of HLA-G after MLN4924 treatment was also observed in Jeg-3 cells (Fig. 5A and B). A similar effect was achieved by single CUL1 silencing (Fig. 5C), indicating that the effect of HLA-G downregulation might be due to CUL1 downstream substrate accumulation. Intriguingly, GATA3, an important transcription factor that regulates gene expression in trophoblast progenitors (Home et al., 2009), was downregulated by MLN4924 treatment in both EVTs and Jeg-3 cells (Figs 4D–F and 5A and B). This transcription factor has been reported to directly affect expression of genes at multiple stages of trophoblast development, including HLA-G (Ferreira et al., 2016; Home et al., 2017). In addition to the total and nuclear downregulation of GATA3, cytoplasmic recruitment of the protein was observed after MLN4924 treatment (Fig. 5E), which indicates that the function of the transcription factor was dysregulated by neddylation inhibition.
Figure 5. HLA-G and GATA3 downregulation caused by MLN4924 is related to p21 accumulation. (A) Representative western blot (WB) results of HLA-G and GATA3 proteins in Jeg-3 cells after treatment with various concentration of MLN4924 for 48 h. (B) Representative WB results of HLA-G, GATA3 and p21 expression in Jeg-3 cells after MLN4924 treatment for indicated hours. (C) Representative WB results of Jeg-3 cells after CUL1 silencing. (D) Representative phase contrast image of SA-β-GAL staining of senescent Jeg-3 cells. Jeg-3 cells were treated with 1 μM MLN4924 or DMSO following p21 silencing. Scale bars: 50 μm. (E) Representative WB of Jeg-3 cells separated into cytoplasmic and nuclear fractions after MLN4924 treatment for 48 h. (F) Representative WB results of Jeg-3 cells after p21 silencing followed by MLN4924 treatment for 24 h.

In situ SA-β-GAL staining of whole-mount first-trimester placentas reveals that the CCTs of RSA placentas exert higher level of SaβG activity compare to HC placentas (Supplementary Fig. S7), indicating early onset of cell senescence in CCTs, because p21 has been widely reported to induce cell senescence (Jia et al., 2011; Lan et al., 2016), its accumulation might also affect plasticity of trophoblast progenitors. The cell senescence caused by MLN4924 was largely alleviated by p21 silencing (Fig. 5D). Moreover, p21 accumulation happened prior to the downregulation of HLA-G and GATA3 (Fig. 5B): obvious p21 accumulation was observed within 12 h of MLN4924 treatment, while HLA-G and GATA3 downregulation was observed only after 24 h. A partial rescue of MLN4924 inhibited HLA-G and GATA3 expression were observed after silencing p21 (Fig. 5F).
Discussion
The pathology of RSA is complicated and multifactorial and has not been clearly elucidated yet. In this study, we first found neddylation inhibition in the RSA villus, and downstream p21 accumulation inhibited trophoblast proliferation and affected their plasticity.
Development of trophoblast lineages is initiated at the very early stage of pregnancy (Zdravkovic et al., 2015). It is a multistep process and relies on proper spatiotemporal gene expression (Paul et al., 2017). The neddylation pathway has been frequently reported to be overactivated in several human cancers and is related to an unfavorable prognosis (Soucy et al., 2009; Li et al., 2014; Chen et al., 2016; Zhou et al., 2018; Zhang et al., 2019). Dynamic neddylation and deneddylation of cullins require a subtle regulation of numerous regulators in the pathway. Including E1 NEDD8-activating enzyme (NAE), E2 NEDD8-conjugating enzyme, E3 NEDD8 ligase and CSN. The fine-tuned regulation of neddylation and deneddylation allows targeted ubiquitin-dependent degradation of numerous proteins in order to maintain the cellular homoeostasis (Petroski and Deshaies 2005; Sarikas et al., 2011). The results of our study showed that NEDD8 was highly expressed in the nucleus in proliferating subsets of trophoblasts of the human first-trimester placenta. We observed obvious neddylation inhibition and subcellular localization changes of NEDD8 in the RSA first trimester placenta. The direct consequence of neddylation inhibition is accumulation of the CRL complex substrate. In the present study, we observed obvious neddylation inhibition and accumulation of the well-known CUL1 substrate p21 in the RSA placenta. Both siRNA downregulation of CUL1 (Zhang et al., 2013) and MLN4924 treatment inhibited the outgrowth of HC primary villus explant cultures. Consistent to a previous study(Wu et al., 2017), moderate p21 expression can be observed in trophoblast cells and villus stroma in HC placenta, while in RSA sample a global increasing of p21 expression in CTBs, syncytiotrophoblast and stroma cells can be observed (Supplementary Fig. S1C). As the majority of p21-positive cells are trophoblast cells, we consider its accumulation would more prominently affect the function of trophoblast cells. By silencing p21, the effect of MLN4924 on the proliferation rate of Jeg-3 cells was prominently rescued, and the G2 arrest state was largely alleviated, suggesting that activation of NEDD8-CUL1-mediated p21 degradation is essential for proper trophoblast growth.
Because the most obvious subcellular localization change of NEDD8 was in EVT progenitors of RSA placental samples, moreover, impaired EVT functioning has been reported to be associated with RSA (Staun-Ram and Shalev 2005; Tian et al., 2016; Li et al., 2019; Lv et al., 2019). We suspected that the dysregulated neddylation in RSA might affect the plasticity of EVT progenitors by affecting the process of TS cell differentiation toward EVT (Okae et al., 2018). The TS cell-derived EVTs in the present study were considered to correspond to the EVT subtype of dCCTs, as they had very similar transcriptome profiles to the primary EVTs isolated from villus tissues (Okae et al., 2018). This subtype of EVTs undergoes endoreduplication and further differentiates into subsets of invasive interstitial and endovascular EVTs (Tilburgs et al., 2015; Velicky et al., 2016; Velicky et al., 2018). Our results demonstrate that neddylation inhibition significantly hindered EVT differentiation by downregulating EVT markers HLA-G MMP2 and ITGA1 and upregulating the CTB marker CDH1 during the differentiation process.
As a well-known marker of EVT, the soluble HLA-G level and HLA-G gene polymorphisms have been reported to be associated with unfavorable pregnancy outcomes such as RSA and recurrent implantation failure (Pfeiffer et al., 2000; Jassem et al., 2012; Wang et al., 2013; Fan et al., 2017). HLA-G (+) EVTs interact with decidual natural killer (dNK) cells, and the latter physically acquire HLA-G by trogocytosis promoting a tolerogenic NK cell signature while maintaining the potential for antiviral immunity at the maternal-fetal interface (Chumbley et al., 1994; Rajagopalan and Long 2012; Tilburgs et al., 2015). Besides maternal–fetal immunotolerance establishment, HLA-G has been reported to regulates the invasive properties of trophoblast cells (Liu et al., 2013). The expression of HLA-G requires activation of a cis-regulatory element, enhancer L that is 12 kb upstream of the HLA-G locus, and motifs for transcription factors of GATA families have been identified in this region (Ferreira et al., 2017). The GATA family are transcription factors implicated in cell fate establishment during mammalian development (Paul et al., 2017). GATA3 along with GATA2 has been proved to be essential to ensure trophoblast development during mammalian placental development (Home et al., 2009; Home et al., 2017). One previous study reported downregulated GATA3 mRNA expression in the fetal–maternal interface of RSA patients (Qiu and Lin 2007). In the present study, we observed consistent alterations in expression of GATA3 and HLA-G in both differentiated EVT and Jeg-3 cells. These results indicated that the plasticity of EVT progenitors was impaired under neddylation inhibition condition.
We explored further the possible mechanism of HLA-G and GATA3 downregulation. Silencing of p21 partially restored HLA-G and GATA3 levels upon MLN4925 treatment. Together with the result that cell senescence can be alleviated by p21 si-RNA, we consider the impaired HLA-G and GATA3 expression was most probably due to the excessive cell senescence caused by p21 rather than any direct regulation. Moreover, according to Velicky et al., (Velicky et al., 2018) and in situ SA-β-GAL staining of HC placenta in the present study, EVT cell senescence occurs upon invading into the decidual, while the EVT progenitor dCCT cells undergo endoreduplicative cycle and only exert mild SA-β-GAL activity. In contrast, excessive SA-β-GAL activity in CCTs as well as p21 accumulation were observed in RSA placenta samples, we speculated neddylation inhibition related p21 accumulation contribute to the early onset of CCT senescence and further give rise to the dysregulation of EVT differentiation. However, the specific mechanism of neddylation regulating trophoblast progenitor plasticity requires further exploration.
In conclusion, through the combined study of clinical samples, primary derived TS cells, and a trophoblast cell line we demonstrated that CUL1 neddylation activation-mediated p21 degradation is required for trophoblast proliferation and is implicated in the regulation of trophoblast plasticity by affecting GATA3 and HLA-G expression (Fig. 6). Together with the inhibition of neddylation observed in the RSA first-trimester placenta, abnormal neddylation might be a possible pathological mechanism of RSA.
Figure 6. Schematic of the present study. Neddylation inhibition in recurrent spontaneous abortion (RSA) trophoblasts causes cytoplasmic retention of free NEDD8 and p21 accumulation, thereby causing proliferation restriction and plasticity impairment.

Supplementary data
Supplementary data are available at Molecular Human Reproduction online.
Supplementary Material
Acknowledgements
We are most grateful to all the patients that participated in our study. The team of assisted reproductive technology department of Sir Run Run Shaw Hospital is sincerely appreciated.
Authors’ roles
X.H.S. and X.M.T. conceived and designed the experiments; S.Y.Z. designed and supervised the project. Y.Q.H., C.L. and T.Z. collected the clinical samples, X.H.S. carried out the experiments and drafted the manuscript, and X.M.T. finalized the manuscript. Y.L.Z., Y.B.P. and L.L. analyzed data and revised manuscript. All authors read and approved the final revised manuscript.
Funding
Zhejiang Provincial Natural Science Foundation of China (LY19H040013); Natural Science Foundation of Zhejiang Province (LGF18H040005); Chinese Medical Association clinical doctors scientific research fund (18010280757); National Natural Science Foundation of China (81401264; 81601308; 81701514; 81871135; 81971358).
Conflict of interest
The authors report no conflicts of interest in this work.
Data availability
All data included in this study are available upon request by contact with the corresponding author.
References
- Baek KH, Lee EJ, Kim YS. Recurrent pregnancy loss: the key potential mechanisms. Trends Mol Med 2007;13:310–317. [DOI] [PubMed] [Google Scholar]
- Bischof P, Irminger-Finger I. The human cytotrophoblastic cell, a mononuclear chameleon. Int J Biochem Cell Biol 2005;37:1–16. [DOI] [PubMed] [Google Scholar]
- Brownell JE, Sintchak MD, Gavin JM, Liao H, Bruzzese FJ, Bump NJ, Soucy TA, Milhollen MA, Yang X, Burkhardt AL et al. Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Mol Cell 2010;37:102–111. [DOI] [PubMed] [Google Scholar]
- Carrington B, Sacks G, Regan L. Recurrent miscarriage: pathophysiology and outcome. Curr Opin Obstet Gynecol 2005;17:591–597. [DOI] [PubMed] [Google Scholar]
- Chen P, Hu T, Liang Y, Li P, Chen X, Zhang J, Ma Y, Hao Q, Wang J, Zhang P et al. Neddylation inhibition activates the extrinsic apoptosis pathway through ATF4-CHOP-DR5 Axis in human esophageal cancer cells. Clin Cancer Res 2016;22:4145–4157. [DOI] [PubMed] [Google Scholar]
- Choi HK, Choi BC, Lee SH, Kim JW, Cha KY, Baek KH. Expression of angiogenesis- and apoptosis-related genes in chorionic villi derived from recurrent pregnancy loss patients. Mol Reprod Dev 2003;66:24–31. [DOI] [PubMed] [Google Scholar]
- Chumbley G, King A, Robertson K, Holmes N, Loke YW. Resistance of HLA-G and HLA-A2 transfectants to lysis by decidual NK cells. Cell Immunol 1994;155:312–322. [DOI] [PubMed] [Google Scholar]
- Cierna Z, Varga I, Danihel L Jr, Kuracinova K, Janegova A, Danihel L. Intermediate trophoblast--a distinctive, unique and often unrecognized population of trophoblastic cells. Ann Anat 2016;204:45–50. [DOI] [PubMed] [Google Scholar]
- Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell 2008;134:995–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hachem H, Crepaux V, May-Panloup P, Descamps P, Legendre G, Bouet PE. Recurrent pregnancy loss: current perspectives. Int J Womens Health 2017;9:331–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Huang Z, Li S, Xiao Z. The HLA-G 14-bp polymorphism and recurrent implantation failure: a meta-analysis. J Assist Reprod Genet 2017;34:1559–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira LM, Meissner TB, Mikkelsen TS, Mallard W, O'Donnell CW, Tilburgs T, Gomes HA, Camahort R, Sherwood RI, Gifford DK et al. A distant trophoblast-specific enhancer controls HLA-G expression at the maternal-fetal interface. Proc Natl Acad Sci U S A 2016;113:5364–5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira LMR, Meissner TB, Tilburgs T, Strominger JL. HLA-G: at the interface of maternal-fetal tolerance. Trends Immunol 2017;38:272–286. [DOI] [PubMed] [Google Scholar]
- Fu J, Lv X, Lin H, Wu L, Wang R, Zhou Z, Zhang B, Wang YL, Tsang BK, Zhu C et al. Ubiquitin ligase cullin 7 induces epithelial-mesenchymal transition in human choriocarcinoma cells. J Biol Chem 2010;285:10870–10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido-Gimenez C, Alijotas-Reig J. Recurrent miscarriage: causes, evaluation and management. Postgrad Med J 2015;91:151–162. [DOI] [PubMed] [Google Scholar]
- Grimstad F, Krieg S. Immunogenetic contributions to recurrent pregnancy loss. J Assist Reprod Genet 2016;33:833–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider S, Meinhardt G, Saleh L, Fiala C, Pollheimer J, Knofler M. Notch1 controls development of the extravillous trophoblast lineage in the human placenta. Proc Natl Acad Sci U S A 2016;113:E7710–E7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Home P, Kumar RP, Ganguly A, Saha B, Milano-Foster J, Bhattacharya B, Ray S, Gunewardena S, Paul A, Camper SA et al. Genetic redundancy of GATA factors in the extraembryonic trophoblast lineage ensures the progression of preimplantation and postimplantation mammalian development. Development 2017;144:876–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Home P, Ray S, Dutta D, Bronshteyn I, Larson M, Paul S. GATA3 is selectively expressed in the trophectoderm of peri-implantation embryo and directly regulates Cdx2 gene expression. J Biol Chem 2009;284:28729–28737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T, Osaka F, Chiba T, Miyamoto C, Okabayashi K, Shimbara N, Kato S, Tanaka K. Covalent modification of all members of human cullin family proteins by NEDD8. Oncogene 1999;18:6829–6834. [DOI] [PubMed] [Google Scholar]
- James JL, Carter AM, Chamley LW. Human placentation from nidation to 5 weeks of gestation. Part I: what do we know about formative placental development following implantation? Placenta 2012;33:327–334. [DOI] [PubMed] [Google Scholar]
- Jassem RM, Shani WS, Loisel DA, Sharief M, Billstrand C, Ober C. HLA-G polymorphisms and soluble HLA-G protein levels in women with recurrent pregnancy loss from Basrah province in Iraq. Hum Immunol 2012;73:811–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L, Li H, Sun Y. Induction of p21-dependent senescence by an NAE inhibitor, MLN4924, as a mechanism of growth suppression. Neoplasia 2011;13:561–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knofler M. Critical growth factors and signalling pathways controlling human trophoblast invasion. Int J Dev Biol 2010;54:269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knofler M, Pollheimer J. Human placental trophoblast invasion and differentiation: a particular focus on Wnt signaling. Front Genet 2013;4:190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovats S, Main EK, Librach C, Stubblebine M, Fisher SJ, DeMars R. A class I antigen, HLA-G, expressed in human trophoblasts. Science 1990;248:220–223. [DOI] [PubMed] [Google Scholar]
- Lan H, Tang Z, Jin H, Sun Y. Neddylation inhibitor MLN4924 suppresses growth and migration of human gastric cancer cells. Sci Rep 2016;6:24218. doi: 10.1038/srep24218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Wang M, Yu G, Chen P, Li H, Wei D, Zhu J, Xie L, Jia H, Shi J et al. Overactivated neddylation pathway as a therapeutic target in lung cancer. J Natl Cancer Inst 2014;106: dju083. doi: 10.1093/jnci/dju083 [DOI] [PubMed] [Google Scholar]
- Li X, Ma XL, Tian FJ, Wu F, Zhang J, Zeng WH, Lin Y, Zhang Y. Downregulation of CCNA2 disturbs trophoblast migration, proliferation, and apoptosis during the pathogenesis of recurrent miscarriage. Am J Reprod Immunol 2019;82:e13144. doi: 10.1111/aji.13144 [DOI] [PubMed] [Google Scholar]
- Liao Y, Jiang Y, He H, Ni H, Tu Z, Zhang S, Wang B, Lou J, Quan S, Wang H. NEDD8-mediated neddylation is required for human endometrial stromal proliferation and decidualization. Hum Reprod 2015;30:1665–1676. [DOI] [PubMed] [Google Scholar]
- Liu X, Gu W, Li X. HLA-G regulates the invasive properties of JEG-3 choriocarcinoma cells by controlling STAT3 activation. Placenta 2013;34:1044–1052. [DOI] [PubMed] [Google Scholar]
- Liu Y, Fan X, Wang R, Lu X, Dang YL, Wang H, Lin HY, Zhu C, Ge H, Cross JC et al. Single-cell RNA-seq reveals the diversity of trophoblast subtypes and patterns of differentiation in the human placenta. Cell Res 2018;28:819–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv S, Wang N, Lv H, Yang J, Liu J, Li WP, Zhang C, Chen ZJ. The attenuation of trophoblast invasion caused by the downregulation of EZH2 is involved in the pathogenesis of human recurrent miscarriage. Mol Ther Nucleic Acids 2019;14:377–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffett A, Loke C. Immunology of placentation in eutherian mammals. Nat Rev Immunol 2006;6:584–594. [DOI] [PubMed] [Google Scholar]
- Nishioka N, Inoue K, Adachi K, Kiyonari H, Ota M, Ralston A, Yabuta N, Hirahara S, Stephenson RO, Ogonuki N et al. The hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev Cell 2009;16:398–410. [DOI] [PubMed] [Google Scholar]
- Okae H, Toh H, Sato T, Hiura H, Takahashi S, Shirane K, Kabayama Y, Suyama M, Sasaki H, Arima T. Derivation of human trophoblast stem cells. Cell Stem Cell 2018;22:50–63e56. [DOI] [PubMed] [Google Scholar]
- Pan ZQ, Kentsis A, Dias DC, Yamoah K, Wu K. Nedd8 on cullin: building an expressway to protein destruction. Oncogene 2004;23:1985–1997. [DOI] [PubMed] [Google Scholar]
- Paul S, Home P, Bhattacharya B, Ray S. GATA factors: master regulators of gene expression in trophoblast progenitors. Placenta 2017;60:S61–S66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 2005;6:9–20. [DOI] [PubMed] [Google Scholar]
- Pfeiffer KA, Rebmann V, Passler M, van der Ven K, van der Ven H, Krebs D, Grosse-Wilde H. Soluble HLA levels in early pregnancy after in vitro fertilization. Hum Immunol 2000;61:559–564. [DOI] [PubMed] [Google Scholar]
- Practice Committee of American Society for Reproductive M Definitions of infertility and recurrent pregnancy loss: a committee opinion. Fertil Steril 2013;99:63. [DOI] [PubMed] [Google Scholar]
- Qiu LH, Lin QD. Study on the expression of transcription factor GATA-3 and T-bet mRNA in decidua of women with unexplained recurrent spontaneous abortion. Zhonghua Fu Chan Ke Za Zhi 2007;42:96–98. [PubMed] [Google Scholar]
- Rajagopalan S, Long EO. Cellular senescence induced by CD158d reprograms natural killer cells to promote vascular remodeling. Proc Natl Acad Sci U S A 2012;109:20596–20601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Red-Horse K, Zhou Y, Genbacev O, Prakobphol A, Foulk R, McMaster M, Fisher SJ. Trophoblast differentiation during embryo implantation and formation of the maternal-fetal interface. J Clin Invest 2004;114:744–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouas-Freiss N, Goncalves RM, Menier C, Dausset J, Carosella ED. Direct evidence to support the role of HLA-G in protecting the fetus from maternal uterine natural killer cytolysis. Proc Natl Acad Sci U S A 1997;94:11520–11525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagi I, Chia G, Golan-Lev T, Peretz M, Weissbein U, Sui L, Sauer MV, Yanuka O, Egli D, Benvenisty N. Derivation and differentiation of haploid human embryonic stem cells. Nature 2016;532:107–111. [DOI] [PubMed] [Google Scholar]
- Sarikas A, Hartmann T, Pan ZQ. The cullin protein family. Genome Biol 2011;12:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 2009;458:732–736. [DOI] [PubMed] [Google Scholar]
- Staun-Ram E, Shalev E. Human trophoblast function during the implantation process. Reprod Biol Endocrinol 2005;3:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian FJ, Cheng YX, Li XC, Wang F, Qin CM, Ma XL, Yang J, Lin Y. The YY1/MMP2 axis promotes trophoblast invasion at the maternal-fetal interface. J Pathol 2016;239:36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilburgs T, Crespo AC, Zwan A, Rybalov B, Raj T, Stranger B, Gardner L, Moffett A, Strominger JL. Human HLA-G+ extravillous trophoblasts: immune-activating cells that interact with decidual leukocytes. Proc Natl Acad Sci U S A 2015;112:7219–7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velicky P, Knofler M, Pollheimer J. Function and control of human invasive trophoblast subtypes: intrinsic vs. maternal control. Cell Adh Migr 2016;10:154–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velicky P, Meinhardt G, Plessl K, Vondra S, Weiss T, Haslinger P, Lendl T, Aumayr K, Mairhofer M, Zhu X et al. Genome amplification and cellular senescence are hallmarks of human placenta development. PLoS Genet 2018;14:e1007698. doi: 10.1371/journal.pgen.1007698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Jiang W, Zhang D. Association of 14-bp insertion/deletion polymorphism of HLA-G gene with unexplained recurrent spontaneous abortion: a meta-analysis. Tissue Antigens 2013;81:108–115. [DOI] [PubMed] [Google Scholar]
- Wu F, Tian F, Zeng W, Liu X, Fan J, Lin Y, Zhang Y. Role of peroxiredoxin2 downregulation in recurrent miscarriage through regulation of trophoblast proliferation and apoptosis. Cell Death Dis 2017;8:e2908. doi: 10.1038/cddis.2017.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdravkovic T, Nazor KL, Larocque N, Gormley M, Donne M, Hunkapillar N, Giritharan G, Bernstein HS, Wei G, Hebrok M et al. Human stem cells from single blastomeres reveal pathways of embryonic or trophoblast fate specification. Development 2015;142:4010–4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Chen Q, Lu X, Zhou Z, Zhang H, Lin HY, Duan E, Zhu C, Tan Y, Wang H. CUL1 promotes trophoblast cell invasion at the maternal-fetal interface. Cell Death Dis 2013;4:e502. doi: 10.1038/cddis.2013.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Yu S, Huang X, Tan Y, Zhu C, Wang YL, Wang H, Lin HY, Fu J, Wang H. New insights into the function of Cullin 3 in trophoblast invasion and migration. Reproduction 2015;150:139–149. [DOI] [PubMed] [Google Scholar]
- Zhang W, Liang Y, Li L, Wang X, Yan Z, Dong C, Zeng MS, Zhong Q, Liu XK, Yu J et al. The Nedd8-activating enzyme inhibitor MLN4924 (TAK-924/Pevonedistat) induces apoptosis via c-Myc-Noxa axis in head and neck squamous cell carcinoma. Cell Prolif 2019;52:e12536. doi: 10.1111/cpr.12536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Morgan MA, Sun Y. Targeting Neddylation pathways to inactivate cullin-RING ligases for anticancer therapy. Antioxid Redox Signal 2014;21:2383–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Sun Y. Cullin-RING ligases as attractive anti-cancer targets. Curr Pharm Des 2013;19:3215–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Jiang Y, Luo Q, Li L, Jia L. Neddylation: a novel modulator of the tumor microenvironment. Mol Cancer 2019;18:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Zhang W, Sun Y, Jia L. Protein neddylation and its alterations in human cancers for targeted therapy. Cell Signal 2018;44:92–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data included in this study are available upon request by contact with the corresponding author.


