Abstract
Background
It was shown that the major part of human genome is transcribed and produces a large number of long noncoding RNAs (lncRNAs). Today there are many evidences that lncRNAs play important role in the regulation of gene expression during different cellular processes. Moreover, lncRNAs are involved in the development of various human diseases. However, the function of the major part of annotated transcripts is currently unknown, whereas different lncRNAs annotations tend to have low overlap. Recent studies revealed that some lncRNAs have small open reading frames (smORFs), that produce the functional microproteins. However, the question whether the function of such genes is determined by microprotein or RNA itself or both remains open. Thus, the study of new lncRNA genes is important to understanding the functional role of such a heterogeneous class of genes.
Results
In the present study, we used reverse transcription PCR and rapid amplification of cDNA ends (RACE) analysis to determine the structure of the LINC01420 transcript. We revealed that LINC01420 has two isoforms that differ in length of the last exon and are localized predominantly in the cytoplasm. We showed that expression of the short isoform is much higher than the long. Besides, MTT and wound-healing assays revealed that LINC01420 inhibited cell migration in human melanoma cell line A375, but does not influence on cell viability.
Conclusion
During our work, D’Lima et al. found smORF in the first exon of the LINC01420 gene. This smORF produces functional microprotein named non-annotated P-body dissociating polypeptide (NoBody). However, our results provide new facts about LINC01420 transcript and its function.
Keywords: LINC01420, NBDY, Long noncoding RNA, lncRNA, Cell migration, smORF, RACE, MTT, Wound healing
Background
After the appearance of the GENCODE project [1], the mRNA-centric paradigm for transcript annotation has dramatically changed. It was shown that the major part of the human genome (~ 62–75%) is transcribed and produces a large number of long noncoding RNAs (lncRNAs) [2, 3]. LncRNAs are transcripts with the length more than 200 nucleotides that do not possess long open reading frames (ORFs) [4]. They are actively studied relatively recently, and today there is not even a good understanding of their number. According to the current GENCODE release (version 29), 16′066 lncRNA genes were annotated in the human genome. At the same time, the FANTOM CAT project revealed 27′919 human lncRNA genes and predicted the potential functionality of ~ 69% of them [5].
Despite this, only a small number of lncRNAs has experimentally defined function [6]. It is already known, that they are involved in various gene-expression regulation processes on transcriptional [7–13] and post-transcriptional [14–16] levels. Besides, lncRNAs play a role in the development of various human diseases [17]. The lncRNADisease database contains entries about 914 lncRNAs associated with 329 diseases [18], including different cancer types, genomic imprinting disorders, neurodegenerative, cardiovascular diseases and other pathologies. In some cases, non-coding transcripts play a key role in the molecular pathogenesis of the diseases. Thus, lncRNAs have the potential to be used as biomarkers and therapeutic targets [19].
There are several lncRNA annotations, which tend to have low overlap. For example, FANTOM CAT and GENCODE have 25,7% and 57,8% common genes respectively. Moreover, the most annotations of transcripts are 5′-end and 3′-end incomplete [20]. The challenge of lncRNA gene annotation is related to their differences from mRNAs, such as weak conservation, low and tissue-specific expression, lack of information about functional elements [20, 21]. Despite this trend, there are lncRNAs with high and widespread expression (MALAT1, NEAT1, TINCR) and cross-species conservation (MALAT1, TINCR, PVT1). It suggests that some lncRNAs may play a role in essential cellular processes. Thus, the study of new lncRNA genes is intriguing and requires in the experimental validation of transcript structure.
Recent studies revealed that some lncRNAs have small open reading frames (smORFs) (< 100 amino acids), that are translated. The resulting short peptides are functional and play a role in cell physiology [22]. At the same time, some studies have shown the dual function of RNAs: coding and intrinsic RNA [23].
In the present study, we investigated widely expressed lncRNA LINC01420, the function of which was not described at the time of the beginning of the study. We determined the structure of LINC01420 transcript, its localization, and influence on cell physiology. During our work, D’Lima et al. found smORF in LINC01420. This smORF produces microprotein named non-annotated P-body dissociating polypeptide (NoBody) [24]. Authors demonstrated the potential functionality of this microprotein as a component of the mRNA decapping complex. However, our results provide new facts about LINC01420 transcript and its function.
Results
LINC01420 has conservative sequences and high expression level in human tissues and cell lines
Using nucleotide BLAST search, we revealed that LINC01420 transcript has homologs across Mammals, but not in other Vertebrates. Moreover, multiple alignment of 100 Vertebrates genomes presented in UCSC browser confirms that this gene is present only in Mammals. HMMER analysis of NoBody homologous proteins showed the same result.
Analysis of the FANTOM5 and GTEx expression data revealed that LINC01420 is highly expressed in most human cell lines and tissues. Moreover, an expression profile of LINC01420 in 975 human samples from FANTOM5 allows classifying this gene as a housekeeping gene with broad and uniform expression [25]. We validated the high widespread expression of this transcript using RT-qPCR analysis of 12 human cell lines, as well as human primary skin fibroblasts (Fig. 1b).
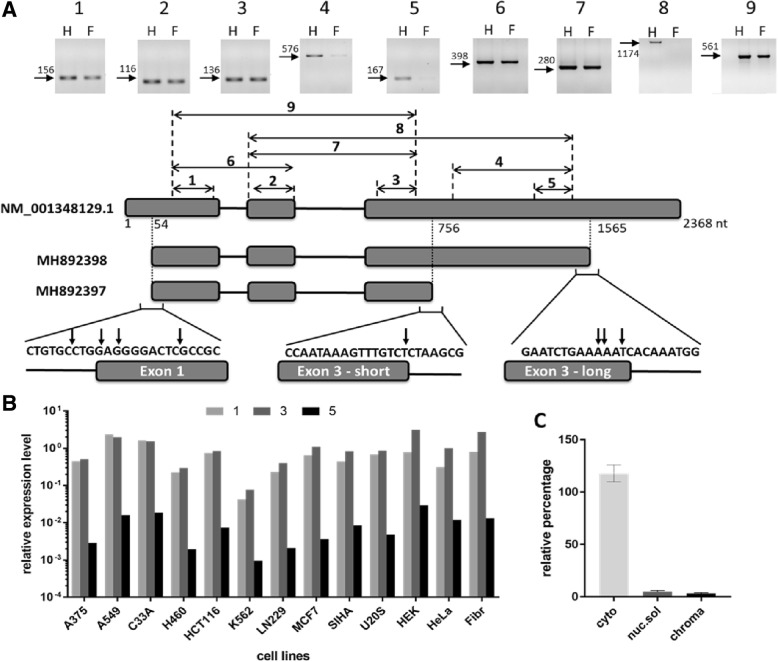
Fig. 1.
Analysis of the structure and expression of the LINC01420 transcript. a. RT-PCR analysis of nine LINC01420 loci of total RNA isolated from HeLa cells (H) and human skin fibroblasts (F) above. The bold numbers (1–9) indicate the amplified locus. The numbers above horizontal arrows show the length of observed products in base pairs (bp). Scheme of the ResSeq (NM_001348129.1) and two experimentally established (MH892397, MH892398) isoforms of the LINC01420 transcript with the amplified loci are shown below. Results of 5′- and 3′- RACE analysis is presented under the scheme. The vertical arrows represent the genomic position of exact 5′- and 3′-ends. Nucleotide numbering was based on reference sequence NM_001348129.1 b. RT-qPCR analysis revealed expression level of three LINC01420 loci relative to four reference genes (HPRT1, B2M, TFRC, TBP) in 12 human cell lines and human skin fibroblasts. Loci 1 and 3 are common for short and long isoforms, whereas locus 5 is long isoform specific. For illustrative purposes expression data are given in logarithmic coordinates. c. To identified the subcellular localization of the LINC01420 transcript total RNA was extracted from three separated cell fractions: cytoplasmic (cyto), nuclear-soluble (nuc.sol) and chromatin-bound (chroma) of HEK293T cells. The amount of transcript in the different fractions relative to whole-cell RNA was measured by RT-qPCR. The error bars represent SEM (standard error mean)
LINC01420 has two isoforms
Different lnсRNA annotations revealed various possible structures of the LINC01420 transcript. To determine a real structure of the LINC01420 isoforms, we performed reverse transcription PCR and rapid amplification of cDNA ends (RACE) analysis on HEK293T, HeLa cell lines, and human primary skin fibroblasts (Fig. 1a). We revealed that the LINC01420 RNA consists of three exons and has two polyadenylated isoforms that differ in length of the last exon. The total length of the short and long LINC01420 isoforms is 701 bp and 1510 bp respectively. Nucleotide sequences of short and long isoforms were deposited into GenBank under accession numbers MH892397 and MH892398, respectively. To determine the expression level of two different isoforms, we performed qPCR with primers common for both isoforms (pairs 1 and 3) and with primers specific for the long isoform (pair 5) (Fig. 1b). We found that expression of the long isoform is ~ 150 times lower than the short one.
Cytoplasmic localization of LINC01420
Since a large group of lncRNA functions is associated with the regulation of transcription and binding to chromatin, we investigated the subcellular localization of LINC01420 using soft lysis method. RNA was isolated from cytoplasmic, nuclear and chromatin-bound fractions of HEK293T cells. RT-qPCRs were performed to determine the level of the transcript of interest in each fraction. As control transcripts, we used U1 [25] and BIRC5 [26], which are localized predominantly in the nucleus and the cytoplasm, respectively. We revealed the LINC01420 transcript has mostly cytoplasmic localization (Fig. 1c). It suggests that the function of this transcript is not related to the transcription regulation and chromatin binding.
The influence of LINC01420 on cell physiology
To determine the potential effect of LINC01420 on cell physiology we performed knockdown experiments using RNA-interference. We designed LINC01420 siRNA and optimized transfection conditions of human melanoma cell line A375. The expression level of a gene of interest was measured by qPCR. The LINC01420 knockdown efficiency was ~ 60% (data not shown).
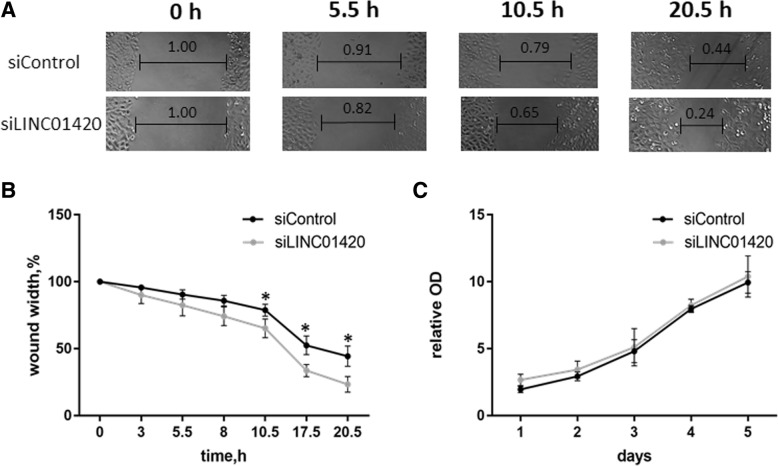
After LINC01420 knockdown, the proliferation of A375 cells was measured by the MTT assay, and the cell migration was examined by the wound healing assay. We found that the knockdown of LINC01420 does not affect A375 proliferation (Fig. 2c), while the positive control knockdown of the EIF3D gene demonstrated an adverse effect on cell viability [27–29]. Wound-healing assay revealed that LINC01420 knockdown leads to increased cell migration (Fig. 2a, b). Thus, we showed that expression of LINC01420 inhibited A375 migration.
Fig. 2.
LINC01420 knockdown revealed activation of A375 cells migration, but does not influence on cell viability. Wound-healing and MTT assays were performed on A375 melanoma cell line. Cells were transfected by siRNAs against LINC01420 (siLINC01420) or nonspecific siRNA (siControl). a. Representative images of wound-healing experiments are shown. b. The graph shows the dependence of wound width on time. Cells treated by siLINC01420 more quickly heal the wound than control cells. c. Result of MTT did not reveal the difference between cells treated by siLINC01420 and control cells. Error bars represent the mean ± SEM (standard error mean) of three independent experiments. *p < 0.01, vs. control (according to Mann–Whitney U test)
Discussion
In the present study, we experimentally determined the structure of LINC01420 transcript and revealed that it has two polyadenylated isoforms that differ in length of the last exon. We showed that the short isoform expressed at a much higher level than the long form and is localized predominantly in the cytoplasm. Besides, we found that LINC01420 inhibited cell migration in human melanoma cell line A375, but does not influence cell viability. Recently Yang L et al. have shown that expression of the LINC01420 gene in nasopharyngeal carcinoma (NPC) is higher than in normal nasopharyngeal epithelial tissues and correlates with NPC distant metastasis and poor prognosis [30]. The authors observed that knockdown of LINC01420 inhibited NPC cell migration and invasion in 5-8F cell line. These contradictory results allow us to assume that this transcript affects cell migration in cell context-specific manner.
In silico analysis of CLIP-seq data about miRNA-mRNA interactions [31–34] from starBase v2.0 database [35] revealed that the LINC01420 transcript interacts with several miRNAs. Top miRNA interactors are involved in the cell migration regulation pathways (miR-876-5p [36], miR-197-3p [37], miR-410-3p [38, 39], miR-340-5p [40], miR-873-5p [41, 42]). Moreover, expression of this miRNAs could either activate or inhibit cell migration depending on cell context. These observations confirm the contradictory results obtained in the present work and Yang L et al. However, this question requires further investigation.
Cytoplasmic localization of the LINC01420 transcript is consistent with the work of D’Lima et al. in which the authors revealed smORF that is translated from the first exon of LINC01420 [24]. This smORF produces microprotein NoBody (non-annotated P-body dissociating polypeptide). D’Lima et al. demonstrated that NoBody interacts with mRNA decapping protein EDC4 and loss of NoBody causes a decrease in the cellular levels of an NMD substrate. The influence of this microprotein on cell migration was not studied. Thus, it remains unclear whether the LINC01420 effect on cell migration is associated with the microprotein or with the RNA itself.
Using SPLASH data [43] available in starBase v2.0 [35] we found that the LINC01420 transcript interacts with ACAD11, NPHP3 and PPP1R3F RNAs. However, the possible functional role of these interactions requires further investigation. At the same time, analysis of LINC01420 RNA-binding proteins revealed that many of them are involved in regulation of mRNA nuclear export, translation and stability (e.g. EIF4A3 [44], IGF2BP1 [45], IGF2BP2 [46], IGF2BP3 [47], UPF1 [48]). It indicates the protein coding function of this transcript.
LncRNA genes became a breakthrough in our understanding of gene regulation and RNA metabolism. smORFs are a new class of genetic elements that expand the coding potential of the genome. High-throughput RNA sequencing (RNA-seq) and ribosome footprinting followed by sequencing (Ribo-seq) increase evidence of the existence of lncRNAs with smORFs [49]. Some of these microproteins are evolutionary conserved and play a functional role not related to the host lncRNA. Taking into account the common features of lncRNAs (weak conservation, low and tissue-specific expression) we speculate that most conservative and widely-expressed lncRNA genes contain smORF, as in the case of the NBDY/LINC01420 gene. However, the question of whether microprotein or RNA itself or both determine the function of such genes remains open.
Conclusions
We provide an analysis of widely expressed lncRNA LINC01420, the function of which was not described at the time of the beginning of the study. Bioinformatics analysis showed that the LINC01420 gene has conservation across mammalian and “housekeeping” broad and uniform expression. Our experimental work revealed that LINC01420 has two isoforms, that are localized predominantly in the cytoplasm. Besides, cell physiology experiments showed that LINC01420 does not affect cell viability, but inhibits melanoma cell line migration.
Recently published work of D’Lima et al. revealed functional NoBody microprotein, that is translated from the first exon of LINC01420. However, the influence of this microprotein on cell migration was not studied. Thus, it remains unclear whether LINC01420 effect on cell migration is associated with the microprotein or with the RNA itself.
Methods
Bioinformatic methods
Nucleotide sequences of the studied gene were found in following databases: RefSeq release 90 [50], Ensemble release 93 [51], GENCODE release 28. Conservation and expression level in various human cell lines and tissues were analyzed using data from the UCSC genomic browser [52]. Nucleotide sequences were analyzed using the BLAST (Basic Local Alignment Search Tool) search of the NCBI NR nucleotide database with standard parameters [53].
RNA extraction and reverse transcription-quantitative PCR (RT-qPCR)
Total RNA from cell lines was extracted using ExtractRNA reagent (Evrogen, Russia) according to the manufacturer’s instruction. RNA was treated with DNAseI (Thermo Fisher Scientific, USA) and reverse transcribed using ImProm-II™ Reverse Transcription System (Promega, USA). qPCR experiments were performed using EvaGreen® Dye (Biotium). Primers used for amplification of different LINC01420 loci are presented in Table 1. PCR amplification reactions were run in triplicates for each cDNA sample. For normalization we used expression of four reference genes (B2M, HPRT, TFRC, TBP), primers are listed in Table 1.
Table 1.
Primer sequences used for amplification of different LINC01420 loci and reference genes and siRNA sequences used for knockdown
| Primer | Sequence |
|---|---|
| F1 | 5′ - GCCCACCGGAGAAAACTGAC – 3 |
| R1 | 5′ – CCTTCCGGATAATCCCAACG - 3‘ |
| R1–1 | 5′ – GAGGCTTGGCTTCCCGTG - 3‘ |
| R1–2 | 5′ – TTCCAGGTGGGAGAGTGGA – 3 |
| F2 | 5′ – AGTGATTGCAGTATGACTCCA - 3‘ |
| R2 | 5′ – TTCCAGGTTCAGGACACCAGA - 3‘ |
| F3 | 5′ – TCAGCGCGATTTCACTTCCTG – 3’ |
| F3–1 | 5′ – CTGAATTTCGATGAATTCTAAGAC – 3’ |
| F3–2 | 5′ – ACCTCTGAGATTTAAGGCCATG – 3’ |
| R3 | 5′ – GACCATCTCACAGGCATTGTT – 3’ |
| F4 | 5′ – GTACACTTTCTTTAATTTGCTGTC – 3’ |
| F5 | 5′ – GAAAATGTCAGATAAACTTGGCT – 3’ |
| R5 | 5′ – CTTTAATTCTGATGCTAGGGACT – 3’ |
| HPRTf | 5′ – TGTAATGACCAGTCAACAGGG - 3’ |
| HPRTr | 5′ – TGCGACCTTGACCATCTTTG - 3’ |
| B2Mf | 5′ – TCTTTCAGCAAGGACTGGTC - 3’ |
| B2Mr | 5′ – GGCATCTTCAAACCTCCATG - 3’ |
| TBPf | 5′ – CGGAGAGTTCTGGGATTGTAC - 3’ |
| TBPr | 5′ – GTGGTTCGTGGCTCTCTTATC - 3’ |
| TFRCf | 5′ – TCCTTGCATATTCTGGAATCCC - 3’ |
| TFRCr | 5′ – ATCACGAACTGACCAGCG - 3’ |
| siLINC01420 |
5′-UCCGGAGAAGUAGAGAAAUdTdT-3′/ 5′-AUUUCUCUACUUCUCCGGAdTdT-3′ |
| siEIF3D#1 |
5′-GCGUCAUUGACAUCUGCAUdTdT-3′/ 5′-AUGCAGAUGUCAAUGACGCdTdT-3′ |
| siEIF3D#2 |
5′-CGACAUGGAUAAGAAUGAAdTdT-3′/ 5′-TTCATTCTTATCCATGTCGdTdT-3’ |
| siControl |
5′-AGGUAGUGUAAUCGCCUUGdTdT-3′/ 5′-CAAGGCGAUUACACUACCUdTdT-3′ |
| FAM-control |
5′-FAM-AGGUCGAACUACGGGUCAAdTdT-3′/ 5′-FAM- UUGACCCGUAGUUCGACCUdTdT-3′ |
RACE
We isolated total RNA from HEK293T, HeLa cell lines and human primary skin fibroblasts using ExtractRNA reagent (Evrogen, Russia) according to the manufacturer’s instruction. cDNAs synthesis and rapid amplification of cDNA ends were performed using Mint RACE cDNA amplification set (Evrogen, Russia) according to the manufacturer’s instruction. All primers used for RACE are presented in Table 1. Amplicons were analyzed by 1% agarose gel electrophoresis. 5′- and 3′-RACE fragments were cloned into pGEM-T Easy vector. Ten random insert clones were obtained and sequenced.
Subcellular localization (fractionation of RNA)
For RNA fractionation was used soft lysis procedure [12]. The HEK293T cells were detached by treating with 1× Trypsin, transferred into 1.5 ml tube and centrifuged at RT 168 g for 5′. The pellet was lysed with 175 μl/106 cells of cold RLN1 solution (50 mM Tris HCl pH 8, 140 mM NaCl, 1.5 mM MgCl2, 0.5% NP-40, RNasin Plus RNase Inhibitor, Promega) and incubated 5′ in ice. Next, the suspension was centrifuged at 4 °C 300 g for 2′ and the supernatant, corresponding to the cytoplasmic fraction, was transferred into a new tube and stored in ice. The pellet containing nuclei was extracted with 175 μl/106 cells of cold RLN2 solution (50 mM Tris HCl pH 8, 500 mM NaCl, 1.5 mM MgCl2, 0.5% NP-40, RNasin Plus RNase Inhibitor, Promega) and 5′ incubated in ice. The suspension was centrifuged at 4 °C 16360 g for 2′ and the supernatant, corresponding to the nuclear-soluble fraction, was transferred into a new tube and stored in ice. The remaining pellet corresponds to the chromatin-associated fraction. The ratio of fractions to total RNA was estimated using RT-qPCR. All experiments were performed in triplicate.
RNA interference
For LINC01420 knockdown we designed two siRNAs using in-house software.
The A375 cells were maintained in DMEM (PanEco, Russia) with 10% fetal bovine serum (Biosera, France) in a with 5% CO2 at 37 °C. Knockdown experiments were conducted as described in Vyakhireva et al. [54]. Briefly, 5 × 103 cells were seed in 96-well plates overnight and transfected with siRNA using METAFECTENE® (Biontex, Germany) according to the manufacturer’s instructions.
For LINC01420 knockdown we used siLINC01420; for EIF3D knockdown – mix of two siRNAs siEIF3D#1 and siEIF3D#2; for control we used nonspecific siControl; for transfection efficiency control we used FAM-labeled nonspecific siRNA (Table 1).
MTT assay
At 0, 24, 48, 72 h, 96 h and 120 h after transfection of A375 cells in 96-well plate, the 20 μl of MTT (Sigma-Aldrich, USA) solution (5 mg/ml) was added to each well with 200 μl culture media and incubated 3 h at 37 °C. After this media was removed. Formazan pellets in each well were dissolved in 200 μl DMSO, and the absorbance of formazan solutions was measured at 570 nm and 670 nm (for background signals).
All the experiments were carried out at three biological and five technical replicates. Data analyzed by a nonparametric paired Mann-Whitney U Test.
Wound healing assay
At 24 h after transfection of A375 cells in 96-well plate. Cells in monolayer culture were scraped using the pipette tips. At 0, 3, 5.5, 8, 10.5, 17.5 and 20.5 h after wounding, 1 field/well was visualized by microscopy. Then, images were analyzed using Image J program (National Institutes of Health). Changes of the remaining wound area were measured relative to total wound square at 0 h.
All the experiments were carried out at three biological and five technical replicates. Data analyzed by a nonparametric paired Mann-Whitney U Test.
Acknowledgements
Not applicable.
Funding
The publication cost was covered by The Ministry of Education and Science of the Russian Federation.
Availability of data and materials
Not applicable.
About this supplement
This article has been published as part of BMC Genomics Volume 20 Supplement 3, 2019: Selected articles from BGRS\SB-2018: genomics. The full contents of the supplement are available online at https://bmcgenomics.biomedcentral.com/articles/supplements/volume-20-supplement-3.
Abbreviations
- DMSO
Dimethyl sulfoxide
- lncRNA
Long noncoding RNA
- NPC
Nasopharyngeal carcinoma
- ORF
Open reading frame
- PCR
Polymerase chain reaction
- RACE
Rapid amplification of cDNA ends
- RT-PCR
Reverse transcription PCR
- siRNA
Small interfering RNA
- smORF
Small open reading frame
Authors’ contributions
MS and AF designed the experiments, DK and AF performed the experiments and wrote the manuscript. MS revised the manuscript. All authors reviewed, considered and approved the manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Daria O. Konina, Email: darya.konina@phystech.edu
Alexandra Yu. Filatova, Phone: +7(916)335-33-29, Email: maacc@yandex.ru
Mikhail Yu. Skoblov, Email: mskoblov@gmail.com
References
- 1.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.St Laurent G, Wahlestedt C, Kapranov P. The Landscape of long noncoding RNA classification. Trends Genet. 2015;31(5):239–251. doi: 10.1016/j.tig.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schlackow M, et al. Distinctive patterns of transcription and RNA processing for human lincRNAs. Mol Cell. 2017;65(1):25–38. doi: 10.1016/j.molcel.2016.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark BS, Blackshaw S. Long non-coding RNA-dependent transcriptional regulation in neuronal development and disease. Front Genet. 2014;5:164. doi: 10.3389/fgene.2014.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hon CC, et al. An atlas of human long non-coding RNAs with accurate 5′ ends. Nature. 2017;543(7644):199–204. doi: 10.1038/nature21374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kornienko AE, et al. Long non-coding RNAs display higher natural expression variation than protein-coding genes in healthy humans. Genome Biol. 2016;17:14. doi: 10.1186/s13059-016-0873-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang L, et al. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147(4):773–788. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pandey RR, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32(2):232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 9.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao J, et al. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322(5902):750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai MC, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cabianca DS, et al. A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell. 2012;149(4):819–831. doi: 10.1016/j.cell.2012.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leucci E, et al. Melanoma addiction to the long non-coding RNA SAMMSON. Nature. 2016;531(7595):518–522. doi: 10.1038/nature17161. [DOI] [PubMed] [Google Scholar]
- 14.Engreitz JM, et al. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent pre-mRNAs and chromatin sites. Cell. 2014;159(1):188–199. doi: 10.1016/j.cell.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karreth FA, et al. In vivo identification of tumor- suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell. 2011;147(2):382–395. doi: 10.1016/j.cell.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoon JH, et al. LincRNA-p21 suppresses target mRNA translation. Mol Cell. 2012;47(4):648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delas MJ, Hannon GJ. lncRNAs in development and disease: from functions to mechanisms. Open Biol. 2017;7(7):170121. doi: 10.1098/rsob.170121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ning S, et al. LincSNP: a database of linking disease-associated SNPs to human large intergenic non-coding RNAs. BMC Bioinformatics. 2014;15:152. doi: 10.1186/1471-2105-15-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu Filatova A, et al. Long noncoding RNAs are a promising therapeutic target in various diseases. Bulletin of RSMU. 2017;6:5–16.
- 20.Uszczynska-Ratajczak B, et al. Towards a complete map of the human long non-coding RNA transcriptome. Nat Rev Genet. 2018;19(9):535–548. doi: 10.1038/s41576-018-0017-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ransohoff JD, Wei Y, Khavari PA. The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol. 2018;19(3):143–157. doi: 10.1038/nrm.2017.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi SW, Kim HW, Nam JW. The small peptide world in long noncoding RNAs. Brief Bioinform. 2018. [DOI] [PMC free article] [PubMed]
- 23.Nam JW, Choi SW, You BH. Incredible RNA: dual functions of coding and noncoding. Mol Cells. 2016;39(5):367–374. doi: 10.14348/molcells.2016.0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.D'Lima NG, et al. A human microprotein that interacts with the mRNA decapping complex. Nat Chem Biol. 2017;13(2):174–180. doi: 10.1038/nchembio.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Consortium F, et al. A promoter-level mammalian expression atlas. Nature. 2014;507(7493):462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reid DW, Nicchitta CV. Primary role for endoplasmic reticulum-bound ribosomes in cellular translation identified by ribosome profiling. J Biol Chem. 2012;287(8):5518–5527. doi: 10.1074/jbc.M111.312280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu GZ, et al. Knockdown of eukaryotic translation initiation factor 3 subunit D (eIF3D) inhibits proliferation of acute myeloid leukemia cells. Mol Cell Biochem. 2018;438(1–2):191–198. doi: 10.1007/s11010-017-3127-5. [DOI] [PubMed] [Google Scholar]
- 28.Fan Y, Guo Y. Knockdown of eIF3D inhibits breast cancer cell proliferation and invasion through suppressing the Wnt/beta-catenin signaling pathway. Int J Clin Exp Pathol. 2015;8(9):10420–10427. [PMC free article] [PubMed] [Google Scholar]
- 29.Lee AS, et al. eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature. 2016;536(7614):96–99. doi: 10.1038/nature18954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang L, et al. High expression of LINC01420 indicates an unfavorable prognosis and modulates cell migration and invasion in nasopharyngeal carcinoma. J Cancer. 2017;8(1):97–103. doi: 10.7150/jca.16819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xue Y, et al. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell. 2013;152(1–2):82–96. doi: 10.1016/j.cell.2012.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kishore S, et al. A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat Methods. 2011;8(7):559–564. doi: 10.1038/nmeth.1608. [DOI] [PubMed] [Google Scholar]
- 33.Gottwein E, et al. Viral microRNA targetome of KSHV-infected primary effusion lymphoma cell lines. Cell Host Microbe. 2011;10(5):515–526. doi: 10.1016/j.chom.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Memczak S, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 35.Li JH, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bao L, et al. MiR-876-5p suppresses epithelial-mesenchymal transition of lung cancer by directly down-regulating bone morphogenetic protein 4. J Biosci. 2017;42(4):671–681. doi: 10.1007/s12038-017-9722-5. [DOI] [PubMed] [Google Scholar]
- 37.Yang Y, et al. miR-137 and miR-197 induce apoptosis and suppress Tumorigenicity by targeting MCL-1 in multiple myeloma. Clin Cancer Res. 2015;21(10):2399–2411. doi: 10.1158/1078-0432.CCR-14-1437. [DOI] [PubMed] [Google Scholar]
- 38.Wu H, et al. MiR-410 acts as a tumor suppressor in estrogen receptor-positive breast Cancer cells by directly targeting ERLIN2 via the ERS pathway. Cell Physiol Biochem. 2018;48(2):461–474. doi: 10.1159/000491777. [DOI] [PubMed] [Google Scholar]
- 39.Guo R, et al. MicroRNA-410 functions as a tumor suppressor by targeting angiotensin II type 1 receptor in pancreatic cancer. IUBMB Life. 2015;67(1):42–53. doi: 10.1002/iub.1342. [DOI] [PubMed] [Google Scholar]
- 40.Mohammadi-Yeganeh S, et al. MicroRNA-340 inhibits the migration, invasion, and metastasis of breast cancer cells by targeting Wnt pathway. Tumour Biol. 2016;37(7):8993–9000. doi: 10.1007/s13277-015-4513-9. [DOI] [PubMed] [Google Scholar]
- 41.Wang RJ, et al. MicroRNA-873 (miRNA-873) inhibits glioblastoma tumorigenesis and metastasis by suppressing the expression of IGF2BP1. J Biol Chem. 2015;290(14):8938–8948. doi: 10.1074/jbc.M114.624700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han G, et al. MicroRNA-873 promotes cell proliferation, migration, and invasion by directly targeting TSLC1 in hepatocellular carcinoma. Cell Physiol Biochem. 2018;46(6):2261–2270. doi: 10.1159/000489594. [DOI] [PubMed] [Google Scholar]
- 43.Aw JG, et al. In vivo mapping of eukaryotic RNA Interactomes reveals principles of higher-order organization and regulation. Mol Cell. 2016;62(4):603–617. doi: 10.1016/j.molcel.2016.04.028. [DOI] [PubMed] [Google Scholar]
- 44.Shibuya T, et al. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat Struct Mol Biol. 2004;11(4):346–351. doi: 10.1038/nsmb750. [DOI] [PubMed] [Google Scholar]
- 45.Stohr N, Huttelmaier S. IGF2BP1: a post-transcriptional "driver" of tumor cell migration. Cell Adhes Migr. 2012;6(4):312–318. doi: 10.4161/cam.20628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kessler SM, Haybaeck J, Kiemer AK. Insulin-like growth factor 2 - the oncogene and its accomplices. Curr Pharm Des. 2016;22(39):5948–5961. doi: 10.2174/1381612822666160713100235. [DOI] [PubMed] [Google Scholar]
- 47.Deforzh E, et al. IMP-3 protects the mRNAs of cyclins D1 and D3 from GW182/AGO2-dependent translational repression. Int J Oncol. 2016;49(6):2578–2588. doi: 10.3892/ijo.2016.3750. [DOI] [PubMed] [Google Scholar]
- 48.Lee SR, et al. Target discrimination in nonsense-mediated mRNA decay requires Upf1 ATPase activity. Mol Cell. 2015;59(3):413–425. doi: 10.1016/j.molcel.2015.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bazzini AA, et al. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 2014;33(9):981–993. doi: 10.1002/embj.201488411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O'Leary NA, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44(D1):D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zerbino DR, et al. Ensembl 2018. Nucleic Acids Res. 2018;46(D1):D754–D761. doi: 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miller W, et al. 28-way vertebrate alignment and conservation track in the UCSC genome browser. Genome Res. 2007;17(12):1797–1808. doi: 10.1101/gr.6761107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson M, et al. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36(Web Server issue):W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vyakhireva JV, et al. siRNA-mediated gene silencing. Bulletin of RSMU. 2017;3:17–29. doi: 10.24075/brsmu.2017-03-02. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.