Abstract
Background
Breastfeeding contributes to gastrointestinal microbiota colonization in early life, but its long-term impact is inconclusive. We aimed to evaluate whether the type of feeding during the first six months of life was associated with oral microbiota in adolescence.
Methods
This is a cross-sectional sub-study using baseline information of 423 adolescents from the Finnish Health in Teens (Fin-HIT) cohort. Type of feeding was recalled by parents and dichotomized as (i) No infant formula; (ii) Infant formula (breastmilk + formula or only formula). Saliva microbiota was analysed using 16S rRNA (V3–V4) sequencing. Alpha diversity and beta diversity were compared between feeding type groups using ANCOVA and PERMANOVA, respectively. Differential bacteria abundance was tested using appropriate general linear models.
Results
Mean age and body mass index were 11.7 years and 18.0 kg/m2, respectively. The No formula group contained 41% of the participants. Firmicutes (51.0%), Bacteroidetes (19.1%), and Proteobacteria (16.3%) were the most abundant phyla among all participants. Alpha and beta diversity indices did not differ between the two feeding groups. Three Operational Taxonomic Units (OTUs) belonging to Eubacteria and Veillonella genera (phylum Firmicutes) were more abundant in the No formula than in the Infant formula group (log2fold changes/ p - values − 0.920/ < 0.001, − 0.328/ 0.001, − 0.577/ 0.004).
Conclusion
Differences exist in abundances of some OTUs in adolescence according to feeding type during the first six months of life, but our findings do not support diversity and overall oral microbiota composition in adolescents being affected by early feeding type.
Keywords: Microbiota, Saliva, Breastfeeding, Adolescent
Background
The Developmental Origins of Health and Disease (DOHaD) theory has been a target of many studies to explain the global epidemic of non-communicable diseases. DOHaD theory proposes that exposures during critical development periods, such as the first 1000 days of life, could unleash metabolic programming that is able to modify structure and function of organs and systems, impacting health status later in life [1–3].
Recently, the role of microbiota in the context of DOHaD has been evaluated [4]. Gut microbiota is known to be established during the first two years of life, reaching stability by the third year [5, 6]. Perturbations of microbiota during early life have been associated with later inflammatory or immune-mediated diseases, such as allergy, asthma, and obesity [7–10], indicating that microbiota also has a critical period of development.
Evidence of possible protection of breastfeeding against obesity and related outcomes in adulthood supports the DOHaD theory [11, 12]. An explanation for the protective effect is its role during the gut microbiota establishment period [7, 13]. Relative to formula-fed infants, breastfed infants seem to have a lower abundance of Clostridium and a predominance of Bifidobacteria and Lactobacilli in the gut [6, 8, 14–18].
Studies have suggested that also the infant oral microbiota is influenced by the type of feeding [19–21]. Proteobacteria and Actinobacteria phyla were more abundant in the oral cheek of breastfed neonates, while formula-fed infants had a higher predominance of Bacteriodetes [20]. In examining saliva culture data of three-month-old infants, vital Lactobacillus species were found in breastfed but not formula-fed ones [19]. A longer term effect, assessed at four and 12 months after birth, reinforced differences in oral microbiota composition between breast and formula-fed infants [21].
Although differences exist between gut and oral microbiota compositions, some similarities have been described [22–24]. Community types present in these sites seem to be associated, i.e. types of bacteria detected in one site are able to predict those found in the other [22, 23]. It has been suggested that oral microbiota seeds gut microbiota in infancy. Observations that both mother’s milk and infant’s faeces are colonized by some identical bacteria support the hypothesis that human milk is an important source of bacteria, contributing to define gut microbiota composition [22].
Considering that oral microbiota is relatively stable over time in healthy individuals, that perturbations could favour non-oral diseases such as type 2 diabetes [24, 25], and that saliva is easily collected, the use of saliva samples represents a unique opportunity to investigate factors associated with microbial composition. As far as we know, no study has evaluated whether the association between breastfeeding and oral microbiota composition is maintained until adolescence, which is a critical period in the life course for the prevention of chronic diseases. We aimed to evaluate whether the type of feeding during the first six months of life is associated with oral microbiota diversity and composition in Finnish adolescents. Our hypothesis is that those who received infant formula at early life (combined or not with breastmilk) had a lower exposure to breastmilk (in quantitative terms) compared to those who were fed only with breastmilk, which could influence the microbiota development.
Methods
Design and study population
This is a cross-sectional sub-study using baseline data from the Finnish Health in Teens (Fin-HIT) cohort [26]. Briefly, Fin-HIT is a representative study of the most populated areas of Finland, for which approximately 11,400 adolescents and 9900 parents (one per adolescent, mostly mothers) were recruited between 2011 and 2014. The Fin-HIT study protocol was approved by the Ethics Committee of the Hospital District of Helsinki and Uusimaa (169/13/03/00/10), and all participants provided informed consent.
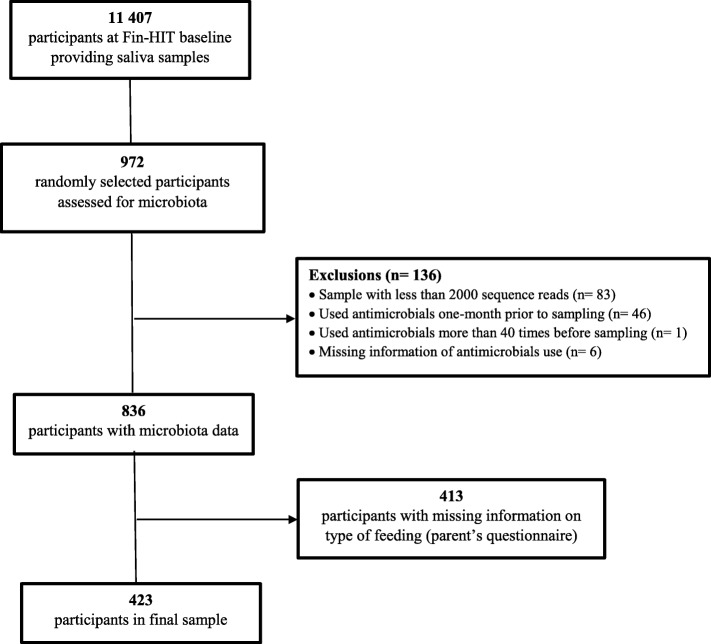
Saliva samples from all adolescents were collected and a random subsample (n = 972) had the oral microbiota analysed, maintaining representativeness of the cohort population. Participants whose samples generated less than 2000 sequence reads (n = 83) were excluded. Based on register data from the Social Insurance Institution of Finland, those who had used antimicrobials in the last month (n = 46) or had used them for more than 40 times throughout life (taken as a proxy of chronic disease) (n = 1) or without this information were also excluded (n = 6). Thus, 836 adolescents had saliva microbiota data, 423 (aged 10–14 years) of whom also had information on feeding type during the first six months of life, comprising the final number of participants in this study (Fig. 1).
Fig. 1.
Flow chart of Fin-HIT participants included in this study
Variables
The type of feeding during the first six months of life was the main exposure of this analysis. At baseline, these data were retrospectively collected using a web questionnaire answered by parents. This variable was dichotomized as follows: (i) No infant formula and (ii) Infant formula (referring to those who received formula, combined or not with breastmilk).
Other variables of interest were adolescent’s gender (male/female) and age (years); and parent’s gender (male/female), age (years), language (Finnish, Swedish/others) and education level (high school or technical level/university degree). Adolescent’s height (nearest 0.1 cm) and weight (nearest 0.01 kg) were measured by trained fieldworkers using portable stadiometers (Seca model 217) and portable digital scales (CAS model PB). Body mass index (BMI) was calculated as weight (kg)/height2 (m). Type of delivery (vaginal/ C-section) was obtained from the national health register managed by the National Institute for Health and Welfare [https://www.thl.fi/en/web/thlfi-en/statistics/ information-for-researchers].
Saliva collection and oral microbiome analysis
Adolescents’ unstimulated saliva samples were collected using the Oragene® DNA (OG-500) Self-Collection Kit (DNA Genotek Inc., Ottawa, ON, Canada), mixed with stabilizing reagent and stored at ambient temperature, according to the manufacturer’s instructions. A protocol with an intensive lysis step and mechanical disruption of microbial cells was employed [27]. Afterwards, total DNA was extracted; sample amplification and sequencing were prepared according to a simplified in-house 16S rRNA gene-based PCR amplification protocol. Amplification was performed using 16S primers targeting V3-V4 region and the Truseq (TS)-tailed1-step amplification protocol. The Illumina HiSeq1500 instrument (Illumina Inc., San Diego, CA, USA) was used for PCR amplicons sequencing. MiSeq SOP in the mothur pipeline (version V.1.35.1) was used to process sequences [28]. The SILVA 16S rRNA database (version V119) and taxonomy were used for alignment and classification of the high-quality sequence reads, which were clustered into operational taxonomic units (OTUs) at a cut-off value > 98% sequence similarity. Detailed procedures of saliva collection and microbial analysis were previously described [29].
Alpha diversity (Shannon and Inverse Simpson Indices) was calculated per sample and beta diversity between the samples using Bray Curtis dissimilarity indices. Sequencing depths were categorized as (i) low ≤10,000; (ii) medium > 10,000 and ≤ 100,000; (iii) high > 100,000 sequences.
Statistical analysis
Kolmogorov-Smirnov test was used to verify normal distribution of variables, which were described with means (standard deviation) or frequency (%). Student t-test and Chi-squared test were used to compare continuous and categorical variables according to feeding type groups. ANCOVA was used to compare alpha diversity between groups. Pearson correlation was employed to test correlations between alpha diversity indices and continuous variables, while Student t-test and ANOVA were used to compare these indices between categories of the other variables. These analyses were performed using Stata Statistical Software (release 12, 2011, StataCorp LP, College Station, TX, USA), and statistical significance was set at the level of 5 %. Permutational analysis of variance (PERMANOVA) was used to test difference in beta diversity between feeding type groups. Principal coordinate analysis based on Bray Curtis distances was used to illustrate beta diversity between groups.
General linear models (GLMs) with negative binomial distribution were employed for comparisons of bacteria abundance between feeding type groups as OTU and considering phylum and genus levels. All OTUs with low counts (< 20) were excluded. The P - values were corrected by false discovery rate. PERMANOVA and GLM were carried out using DESeq2 [30], Vegan, and phyloseq in R (version 3.4.3). Gender, age, BMI, type of delivery, parent’s education, and sequence reads were considered confounders in all adjusted models.
Multiple imputation for missing values of BMI (n = 10), type of delivery (n = 22), and parent’s education (n = 15) was performed with “mi impute chained” procedure in Stata 12.0. Imputed values were considered in the multivariate analysis.
Results
Of the 423 adolescents at baseline of Fin-HIT, 52% were female; their mean age and BMI were 11.7 years and 18.0 kg/m2 (73% were normal weight), respectively. Most participants were born by vaginal delivery (81.1%) and received infant formula (solely or combined with breastmilk) (58.6%) during their first six months of life. The majority of participating parents were female (87.4%), Finnish speakers (90.3%), and had a university degree (59.6%) (Table 1).
Table 1.
Main data of participants at Fin-HIT baseline and comparison according to type of feeding
| All n = 423 | No infant formula n = 175 | Infant formula n = 248 | P - value | |
|---|---|---|---|---|
| Continuous variables | Mean (SD) | |||
| Adolescent’s age (y) | 11.7 (0.3) | 11.7 (0.3) | 11.7 (0.3) | 0.074 |
| Parent’s age (y) | 44.0 (5.7) | 44.3 (5.4) | 43.8 (5.8) | 0.389 |
| Adolescent’s body mass index (kg/m2) a | 18.0 (2.9) | 17.7 (2.8) | 18.2 (2.9) | 0.077 |
| Alpha diversity b | ||||
| Shannon Index | 2.9 (0.3) | 2.9 (0.3) | 2.9 (0.3) | 0.877 |
| Inverse Simpson | 10.1 (3.1) | 10.1 (3.2) | 10.1 (2.3) | 0.949 |
| Categorical variables | Frequency (%) | |||
| Adolescent’s gender | ||||
| Male | 204 (48.2) | 82 (46.9) | 122 (49.2) | 0.636 |
| Female | 219 (51.8) | 93 (53.1) | 126 (50.8) | |
| Type of delivery c | ||||
| Vaginal | 325 (81.1) | 144 (88.3) | 181 (76.1) | 0.002 |
| C-section | 76 (18.9) | 19 (11.7) | 57 (23.9) | |
| Parent’s language | ||||
| Finnish | 382 (90.3) | 156 (89.1) | 226 (91.1) | 0.638 |
| Swedish | 36 (8.5) | 16 (9.2) | 20 (8.1) | |
| Other | 5 (1.2) | 3 (1.7) | 2 (0.8) | |
| Parent’s education d | ||||
| High school/ technical level | 165 (40.4) | 72 (43.4) | 93 (38.4) | 0.318 |
| University degree | 243 (59.6) | 94 (56.6) | 149 (61.6) | |
an = 413 due to missing data
b Adjusted for adolescent’s gender, age, and body mass index, type of delivery, parent’s education, and sequence reads
cn = 401 due to missing data
dn = 408 due to missing data
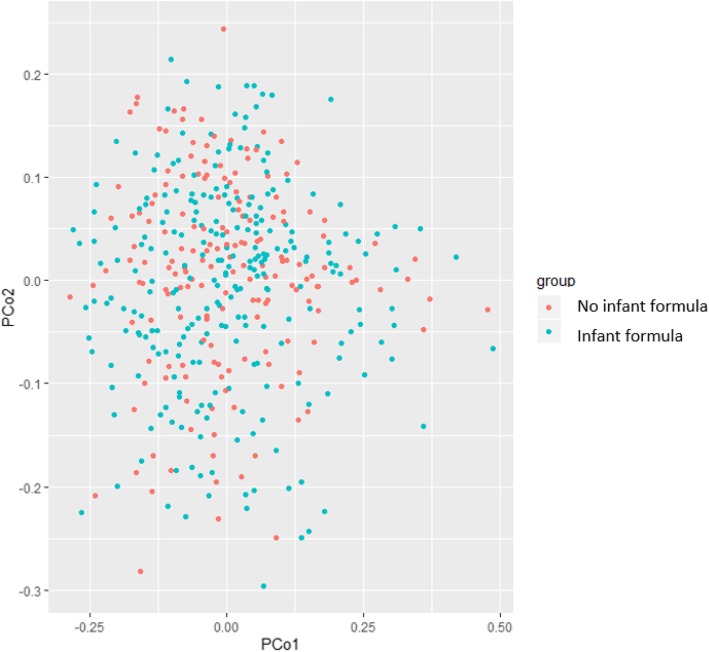
Mean (standard deviation) values of alpha diversity indices were 2.9 (0.3) and 10.1 (3.1) for Shannon and Inverse Simpson, respectively, and these did not differ between the No formula and Infant formula groups (Table 1). Beta diversity was also similar between groups (P - value = 0.881) (Fig. 2). In the No formula group, vaginal delivery was more frequent than in the Formula group (88.3% vs. 76.1%, P - value = 0.002) (Table 1). Alpha diversity was not associated with adolescents’ or parents’ characteristics (Table 2).
Fig. 2.
Principal coordinate analysis (beta-diversity) for the saliva microbiota according to type of feeding (P – value = 0.881)
Table 2.
Correlations and comparison of mean values of diversity indices by participants’ characteristics
| Shannon | Inverse Simpson | |||
|---|---|---|---|---|
| Continuous variables | r | P- value | r | P- value |
| Adolescent’s age | 0.015 | 0.763 | −0.003 | 0.949 |
| Adolescent’s body mass index | −0.047 | 0.344 | −0.037 | 0.456 |
| Categorical variables | Mean (SD) | P- value | Mean (SD) | P- value |
| Adolescent’s gender | ||||
| Male | 2.9 (0.3) | 0.338 | 10.2 (3.0) | 0.412 |
| Female | 2.9 (0.3) | 9.9 (3.2) | ||
| Type of delivery | ||||
| Vaginal | 2.9 (0.3) | 0.579 | 10.1 (3.1) | 0.579 |
| C-section | 2.9 (0.3) | 9.9 (3.0) | ||
| Parent’s language | ||||
| Finnish | 2.9 (0.3) | 0.776 | 10.0 (3.1) | 0.802 |
| Swedish | 2.9 (0.2) | 10.2 (3.0) | ||
| Other | 3.0 (0.4) | 10.9 (4.1) | ||
| Parent’s education | ||||
| High school/ technical level | 2.9 (0.3) | 0.544 | 9.8 (3.0) | 0.190 |
| University degree | 2.9 (0.3) | 10.2 (3.2) | ||
| Sequence reads | ||||
| Low | 2.8 (0.2) | 9.3 (2.1) | ||
| Medium | 2.9 (0.3) | 0.274 | 10.0 (2.9) | 0.360 |
| High | 2.9 (0.4) | 10.3 (3.4) | ||
In 423 samples of adolescents’ saliva, 22,924,455 raw sequences reads were obtained and 1049 OTUs were evaluated. In total, 11 phyla, 16 classes, 24 orders, 44 families, and 76 genera were identified. As depicted in Fig. 3, a great predominance of phylum Firmicutes was found, followed by Bacteroidetes, Proteobacteria, and Actinobacteria (panel A), while the predominant genera were Veillonella, Prevotella, and Streptococcus (panel B).
Fig. 3.
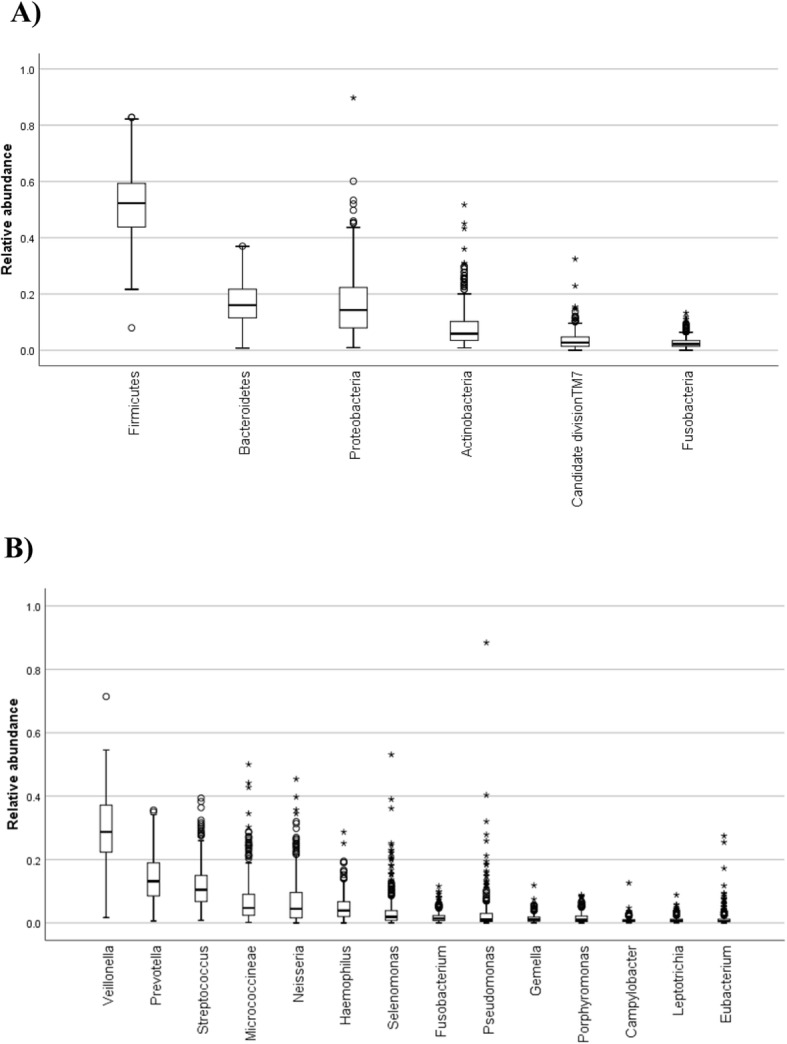
Relative abundances of phyla (a) and genera (b) in saliva microbiota
Comparing groups of feeding type according to abundance of bacteria, no difference was observed at phylum level. Three OTUs (#019, #232, and #158) were more abundant in No formula than in Infant formula group (log2fold changes/ adjusted p-values: − 0.920/ < 0.001, − 0.328/ 0.001, and − 0.577/ 0.004, respectively) (Fig. 4; and see table at Additional file 1 for detailed information). These three OTUs belonged to phylum Firmicutes, class Clostridia, and order Clostridiales; OTU #019 belonged to genus Eubacteria and #232 and #158 to genus Veillonella. Considering the entire genera, genus Eubacteria (log2fold change − 0.742, adjusted p - value < 0.0003), but not genus Veillonella, was more abundant in the No formula group.
Fig. 4.
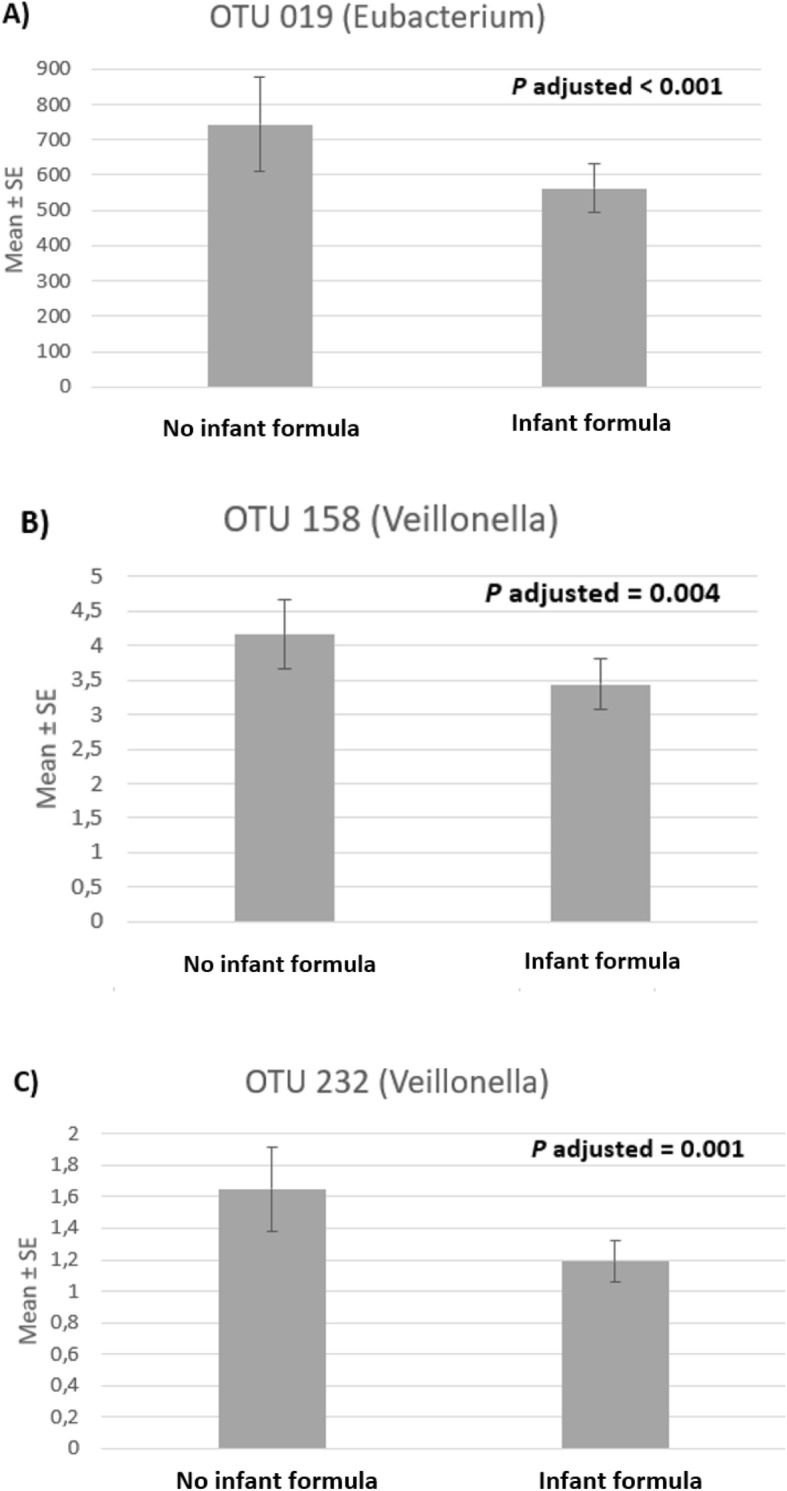
Means and standard errors (SEs) of abundance for significantly different OTUs between feeding type groups
Discussion
We investigated the long-term association of early life feeding type with oral microbiota composition in adolescence. A few differences emerged in abundances of some specific genera according to early feeding type, but our data do not suggest a significant impact on the diversity of microbiota. Abundances of commensal OTUs belonging to genera Veillonella and Eubacteria were higher in adolescents who had not been fed with formula than in others who had received infant formula, combined or not with breastmilk.
To the best of our knowledge this is the first study to explore associations of early feeding type with oral microbiota diversity and composition at this stage of life. Such relationships have been investigated mainly during the first year of life [19–21]. Despite higher abundance of certain OTUs, the overall composition (beta diversity) or the alpha diversity of oral microbiota did not differ between adolescents with different early life feeding type. In general, high phylogenetic diversity in gut microbiota has been associated with several health outcomes [31], but long-term health implications of this picture are unclear yet. We speculate that our negative results regarding overall microbiota diversity and composition could result from a stronger impact of other exposures during childhood such as weaning age, time of introduction of solid foods, Westernized eating pattern and use of antimicrobials [18, 31–33].
As far as the gut is concerned, it was reported that microbiota composition in breastfed babies tended to be more stable and has less diverse bacterial community compared with formula-fed babies [34, 35]. From three years of age, their composition converge to resemble that of an adult gut [5]. A multicentre cohort of 903 infants (including Finns) followed from three to 46 months of age showed that breastfeeding explained the greatest part of microbiome establishment during the first year of life. Also, breastfed infants had lower diversity than those weaned at an earlier stage. As age increased and breastmilk exposure decreased, diversity became more similar between groups, supporting the hypothesis that weaning is an important event affecting maturation of the gut microbiome [6]. Some studies have evaluated the long-term effect of breastfeeding on gut microbiota [6, 18]. A prospective birth cohort study of 281 Dutch children reported that breastfeeding duration was associated with gut microbiota composition in children aged 6–9 years [18].
Little is known how these changes occur in saliva, driven by diet, physical activity and antibiotics administration, which could alter the host health status. Although gut and oral microbiota had distinct compositions, community types of these sites seem to be associated [22, 24]. A study conducted with neonates 20 days old, found a positive correlation of subdominant family Lactobacillaceae in saliva and faeces samples of the same infant, and showed that Staphylococcus spp. and Streptococcus spp. were shared between the two ecosystems and also with human milk [22]. Comparable results have been reported in studies assessing gut and oral microbiota; i.e. early feeding seems to impact on microbiota profile [6, 14, 19, 20].
Comparisons with our findings are limited because we were the first to evaluate the long-term effect of early feeding on oral microbiota. Furthermore, exposures occurring during infancy, particularly related to antibiotics use, may affect microbiota composition mainly at the gut level [36, 37]. The protective role of breastfeeding against increased body adiposity and antibiotic use in childhood, especially by promoting beneficial microbiota, was eliminated by the use of antibiotics in early life [38]. Additionally, differences in microbiota composition from one population to another are recognized [39–41].
According to unpublished data of the Fin-HIT, most of the participants used antimicrobial drugs during their first years of life, which could contribute to a lasting impact on oral microbiota diversity and composition at adolescence. Regarding diet, it is suggested that especially the high and frequent intake of sucrose and other fermentable carbohydrates result in the accumulation of acidogenic and aciduric microorganisms, driving a pathogenic biofilm community formation. Particularly, high frequency of sugar intake could disrupt the homeostasis between commensals and pathogens resulting in dysbiosis, which could increase chances of caries, inflammation and periodontitis in more susceptible individuals [42]. Thus, we suppose that sugar intake during childhood could be an important factor affecting diversity and composition of oral microbiota. A previous study, conducted in public child health service in Finland, showed that sugar introduction occurs early in life. Almost half of six month old children were receiving sugar-sweetened beverages and more than 90% of those older than 16 months were receiving sweets [43]. Further, another Finnish study showed that 95% of one year-old babies consume mass-produced baby foods, of which some are sweetened with juice-concentrates (70–80% of sugar) [44].
Our finding of a major predominance of phylum Firmicutes in the human oral cavity, followed by Bacteroidetes, Proteobacteria, and Actinobacteria is consistent with previous studies in one to two month-old [20] and 12 month-old children [21]. A study conducted in 38 healthy, full-term, vaginally delivered neonates from Australia found similar alpha diversity and proportion of Firmicutes between breastfed and formula-fed groups (96.3 vs. 95.3%) [20], in accord with our findings in an older age group. A higher relative abundance of Bacteroidetes and a lower abundance of Actinobacteria and Proteobacteria were also observed in formula-fed infants than in breastfed infant.
Holgerson et al. [19] showed that three month old exclusively breastfed infants clustered separately from formula-fed infants, according to their oral microbiota. In total, 14 probes differed significantly between these two feeding types. Among these probes, cultivable Lactobacilli and Eubacterium yurii were more abundant in breastfed than exclusively formula-fed infants, in whom the mean number of species per child was higher. Lactobacillus, but not Eubacterium, has been consistently reported in breastmilk, and thus, the presence of the former in infants’ oral microflora was expected. We do not know whether the higher abundance of Eubacteria observed in our breastfed adolescents could be detected earlier, during their infancy. Another study reported higher species richness in 4-month-old formula-fed infants and marked differences in saliva microbiota composition between feeding type subgroups [21]. However, at 12 months of age these differences were no longer significant.
We found that Eubacteria and Veillonella, both genera of the normal oral microbiota, were more abundant in adolescents of No formula group compared with those who received infant formula in early life. Eubacteria is an atypical genus in samples of breastmilk, but Veillonella, Gemella, Rothia, Lactobacillus, Streptococcus, and Staphylococcus have been previously described as the most abundant genera in breastmilk [45]. The association of No formula with higher Veillonella abundance seems favourable, considering that this genus has a beneficial effect on dental plaques by metabolizing lactate to weaker acids such as acetate and propionate [46]. Since lactate is a cariogenic factor produced by Streptococcus mutans, increased abundance of Veillonella could be interpreted as a compensatory effect to protect against dental injuries. Higher abundance of the species Veillonella parvula was previously described in subgingival biofilm samples from healthy subjects compared with those with chronic periodontitis, and its presence was inversely correlated with inflammatory biomarkers from gingival crevicular fluid [47]. Periodontal disease is considered a risk factor for cardiovascular disease, and the proposed underlying mechanism was based on increased circulating pro-inflammatory cytokines [48].
Strict anaerobic bacteria from the genus Eubacteria are chemoheterotrophs, i.e. are unable to synthesize their own organic molecules, requiring mixed organic acids from the host. Thus, Eubacteria energy is obtained from carbohydrates or protein metabolism, resulting in end-products such as butyrate and acetate [49]. These short-chain fatty acids have been associated with beneficial effects, such as cardiovascular and colonic disease prevention [50]. The presence of Eubacteria in our sample of healthy adolescents was expected since this a member of normal saliva microflora and plaques. Our methods were unable to identify species of Eubacteria that would be desirable since periodontal pathogenic Eubacteria species were previously described [47, 51]. A study of patients undergoing haemodialysis identified higher taxa of Eubacterium nucleatum in sulcular fluid from the periodontal pocket of non-diabetics than diabetics. Despite being a pathogen, the prevalence of periodontitis was similar between the groups [51].
Although evidence has suggested beneficial roles of Veillonella and Eubacteria, it was not possible to extrapolate these in our sample since the study design does not allow comparing health outcomes according to abundances of specific bacteria. Prospective studies are needed to investigate whether individuals with distinct abundances of specific bacteria have different chances of developing cardiovascular disease or other outcomes.
A strength of this study was the large sample size in which next-generation sequencing technology was used to analyse microbiota. This is a sub-study of a well-designed prospective cohort, including reliable information obtained from standardized questionnaires and national health registers [26]. Statistical analyses were duly adjusted, minimizing confounding bias. The main limitations of our study comprised a design impeding establishment of causality and retrospective data collection (feeding type answered by parents), which could generate recall bias. Considering that we evaluated associations of an event occurred long time ago (type of feeding) and another occurring in adolescence, there is a possibility of recall error, especially among parents who had multiple children. However, it has been reported that information about breastfeeding recalled by mothers after 20 years still shows reasonable accuracy [52]. We could not consider “exclusive breastfeeding” since no information regarding water, tea, and food intake in early life was available, or “exclusive formula-fed” (not combined with breastmilk) since its prevalence in our study was relatively low (2.4%). Previously, these categories of “exclusive” feeding type were clearly separated by microbiota composition, but partially breastfed infants were interspersed between these groups [19]. Despite this limitation that could have attenuated differences, we still detected some differences according to feeding type. We speculate that differences identified are explained by a higher exposure to breastmilk at early life by participants of the No formula group compared to those of the Infant formula group.
Finally, the lack of oral health status could be regarded as a limitation. It is known that diet, especially in combination with poor oral health, salivary dysfunction, scarce fluoride exposure and poor oral hygiene, can modulate oral microbiota resulting in dysbiosis [42]. Since Finland public oral health care services are freely available for individuals younger than 18 years, most children and adolescents have relatively good oral health, although differences exist depending on maternal educational level and region. Not having information on oral health could contribute to confounding bias and thus is a limitation in this study.
Conclusion
The type of feeding in early life is not associated with overall changes in composition or diversity but with some alterations in adolescents’ oral microbiota (higher abundance of Veillonella and Eubacteria). The clinical relevance of these findings and their health implications need to explored in future studies. Other exposures during childhood (e.g. e.g. antimicrobial use, sugar consumption and oral health) could play a stronger role in adolescents’ oral microbiota diversity and composition than the early feeding. Further prospective studies, considering the type of feeding in early life, age of weaning, and other factors during childhood, are needed to confirm our findings.
Supplementary information
Additional file 1. Differentially abundant bacteria at OTU-level by type of feeding (No infant formula vs. Infant formula) during the first six months of life.
Acknowledgements
The authors acknowledge the Fin-HIT researchers and participants.
Abbreviations
- DOHaD
Developmental Origins of Health and Disease
- Fin-HIT
Finnish Health in Teens cohort
- BMI
Body Mass Index
- OTU
Operational Taxonomic Unit
- GLM
General Linear Model
Authors’ contributions
IE designed the study, carried out the initial analyses, drafted the initial manuscript, and reviewed and revised the manuscript. HTV and SRG designed the study and critically reviewed and revised the manuscript. SCR handled the bioinformatics data and reviewed and revised the manuscript. EW conceptualized and designed the Fin-HIT cohort, coordinated data collection, designed the study, and reviewed and revised the manuscript. RAOF designed the study, carried out the statistical analyses, and critically reviewed and revised the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by Samfundet Folkhälsan, Medicinska Understödsföreningen Liv och Hälsa rf, Swedish Cultural Foundation, the Päivikki and Sakari Sohlberg Foundation, and the São Paulo Research Foundation (FAPESP).
Availability of data and materials
The data that support the findings of this study are available from Heli T. Viljakainen but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are however available from the authors upon reasonable request and with permission of Heli T. Viljakainen.
Ethics approval and consent to participate
The Fin-HIT study was approved by the Ethics Committee of the Hospital District of Helsinki and Uusimaa (169/13/03/00/10), and all participants provided informed consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests. Where authors are identified as personnel of the International Agency for Research on Cancer / World Health Organization, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy, or views of the International Agency for Research on Cancer / World Health Organization.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13006-020-00285-w.
References
- 1.Lucas A, Fewtrell MS, Cole TJ. Fetal origins of adult disease-the hypothesis revisited. BMJ. 1999;319(7204):245–249. doi: 10.1136/bmj.319.7204.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gluckman PD, Cutfield W, Hofman P, Hanson MA. The fetal, neonatal, and infant environments-the long-term consequences for disease risk. Early Hum Dev. 2005;81(1):51–59. doi: 10.1016/j.earlhumdev.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Barnes MD, Heaton TL, Goates MC, Packer JM. Intersystem implications of the developmental origins of health and disease: advancing health promotion in the 21st century. Healthcare (Basel) 2016;4(3):E45. doi: 10.3390/healthcare4030045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stiemsma LT, Michels KB. The role of the microbiome in the developmental origins of health and disease. Pediatrics. 2018;141(4):e2017243. doi: 10.1542/peds.2017-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stewart CJ, Ajami NJ, O'Brien JL, Hutchinson DS, Smith DP, Wong MC, et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature. 2018;562(7728):583–588. doi: 10.1038/s41586-018-0617-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ottman N, Smidt H, de Vos WM, Belzer C. The function of our microbiota: who is out there and what do they do? Front Cell Infect Microbiol. 2012;2:104. doi: 10.3389/fcimb.2012.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moreno-Indias I, Cardona F, Tinahones FJ, Queipo-Ortuño MI. Impact of the gut microbiota on the development of obesity and type 2 diabetes mellitus. Front Microbiol. 2014;5:190. doi: 10.3389/fmicb.2014.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collado MC, Rautava S, Isolauri E, Salminen S. Gut microbiota: a source of novel tools to reduce the risk of human disease? Pediatr Res. 2015;77(1–2):182–188. doi: 10.1038/pr.2014.173. [DOI] [PubMed] [Google Scholar]
- 10.Tamburini S, Shen N, Wu HC, Clemente JC. The microbiome in early life: implications for health outcomes. Nat Med. 2016;22(7):713–722. doi: 10.1038/nm.4142. [DOI] [PubMed] [Google Scholar]
- 11.Kelishadi R, Farajian S. The protective effects of breastfeeding on chronic non-communicable diseases in adulthood: a review of evidence. Adv Biomed Res. 2014;3:3. doi: 10.4103/2277-9175.124629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Victora CG, Bahl R, Barros AJ, França GV, Horton S, Krasevec J, et al. Breastfeeding in the 21st century: epidemiology, mechanisms, and lifelong effect. Lancet. 2016;387(10017):475–490. doi: 10.1016/S0140-6736(15)01024-7. [DOI] [PubMed] [Google Scholar]
- 13.Ho NT, Li F, Lee-Sarwar KA, Tun HM, Brown BP, Pannaraj PS, et al. Meta-analysis of effects of exclusive breastfeeding on infant gut microbiota across populations. Nat Commun. 2018;9(1):4169. doi: 10.1038/s41467-018-06473-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bergström A, Skov TH, Bahl MI, Roager HM, Christensen LB, Ejlerskov KT, et al. Establishment of intestinal microbiota during early life: a longitudinal, explorative study of a large cohort of Danish infants. Appl Environ Microbiol. 2014;80(9):2889–2900. doi: 10.1128/AEM.00342-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Isolauri E, Salminen S, Rautava S. Early microbe contact and obesity risk: evidence of causality? J Pediatr Gastroenterol Nutr. 2016;63(Suppl 1):S3–S5. doi: 10.1097/MPG.0000000000001220. [DOI] [PubMed] [Google Scholar]
- 16.Wagner CL, Taylor SN, Johnson D. Host factors in amniotic fluid and breast milk that contribute to gut maturation. Clin Rev Allergy Immunol. 2008;34(2):191–204. doi: 10.1007/s12016-007-8032-3. [DOI] [PubMed] [Google Scholar]
- 17.Adlerberth I, Wold AE. Establishment of the gut microbiota in Western infants. Acta Paediatr. 2009;98(2):229–238. doi: 10.1111/j.1651-2227.2008.01060.x. [DOI] [PubMed] [Google Scholar]
- 18.Zhong H, Penders J, Shi Z, Ren H, Cai K, Fang C, et al. Impact of early events and lifestyle on the gut microbiota and metabolic phenotypes in young school-age children. Microbiome. 2019;7(1):2. doi: 10.1186/s40168-018-0608-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holgerson PL, Vestman NR, Claesson R, Ohman C, Domellöf M, Tanner AC, et al. Oral microbial profile discriminates breast-fed from formula-fed infants. J Pediatr Gastroenterol Nutr. 2013;56(2):127–136. doi: 10.1097/MPG.0b013e31826f2bc6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Al-Shehri SS, Sweeney EL, Cowley DM, Liley HG, Ranasinghe PD, Charles BG, et al. Deep sequencing of the 16S ribosomal RNA of the neonatal oral microbiome: a comparison of breast-fed and formula-fed infants. Sci Rep. 2016;6:38309. doi: 10.1038/srep38309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Timby N, Domellöf M, Holgerson PL, West CE, Lönnerdal B, Hernell O, et al. Oral microbiota in infants fed a formula supplemented with bovine milk fat globule membranes - A randomized controlled trial. PLoS One:122017. 2017;12(1):e0169831. 10.1371/journal.pone.0169831. [DOI] [PMC free article] [PubMed]
- 22.Biagi E, Quercia S, Aceti A, Beghetti I, Rampelli S, Turroni S, et al. The bacterial ecosystem of mother's milk and infant's mouth and gut. Front Microbiol. 2017;8:1214. doi: 10.3389/fmicb.2017.01214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding T, Schloss PD. Dynamics and associations of microbial community types across the human body. Nature. 2014;509(7500):357–360. doi: 10.1038/nature13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kodukula K, Faller DV, Harpp DN, Kanara I, Pernokas J, Pernokas M, et al. Gut microbiota and salivary diagnostics: the mouth is salivating to tell us something. Biores Open Access. 2017;6(1):123–132. doi: 10.1089/biores.2017.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zaura E, Nicu EA, Krom BP, Keijser BJ. Acquiring and maintaining a normal oral microbiome: current perspective. Front Cell Infect Microbiol. 2014;4:85. doi: 10.3389/fcimb.2014.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Figueiredo RAO, Simola-Strom S, Rounge TB, Viljakainen H, Eriksson JG, Roos E, et al. Cohort profile: the Finnish health in teens (fin-HIT) study: a population-based study. Int J Epidemiol. 2019;48(1):23–24h. 10.1093/ije/dyy189. [DOI] [PMC free article] [PubMed]
- 27.Yuan S, Cohen DB, Ravel J, Abdo Z, Forney LJ. Evaluation of methods for the extraction and purification of DNA from the human microbiome. PLoS One. 2012;7(3):e33865. doi: 10.1371/journal.pone.0033865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75(23):7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raju SC, Lagström S, Ellonen P, de Vos WM, Eriksson JG, Weiderpass E, et al. Gender-specific associations between saliva microbiota and body size. Front Microbiol. 2019;10:767. 10.3389/fmicb.2019.00767. [DOI] [PMC free article] [PubMed]
- 30.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rinninella E, Raoul P, Cintoni M, Franceschi F, Miggiano GAD, Gasbarrini A, et al. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms. 2019;7(1). 10.3390/microorganisms7010014. [DOI] [PMC free article] [PubMed]
- 32.Houghteling PD, Walker WA. Why is initial bacterial colonization of the intestine important to infants' and children's health? J Pediatr Gastroenterol Nutr. 2015;60(3):294–307. doi: 10.1097/MPG.0000000000000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edwards CA. Determinants and duration of impact of early gut bacterial colonization. Ann Nutr Metab. 2017;70(3):246–250. doi: 10.1159/000466711. [DOI] [PubMed] [Google Scholar]
- 34.Fallani M, Young D, Scott J, Norin E, Amarri S, Adam R, et al. Intestinal microbiota of 6-week-old infants across Europe: geographic influence beyond delivery mode, breast-feeding, and antibiotics. J Pediatr Gastroenterol Nutr. 2010;51(1):77–84. doi: 10.1097/MPG.0b013e3181d1b11e. [DOI] [PubMed] [Google Scholar]
- 35.Harmsen HJ, Wildeboer-Veloo AC, Raangs GC, Wagendorp AA, Klijn N, Bindels JG, et al. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J Pediatr Gastroenterol Nutr. 2000;30(1):61–67. doi: 10.1097/00005176-200001000-00019. [DOI] [PubMed] [Google Scholar]
- 36.Parker EPK, Praharaj I, John J, Kaliappan SP, Kampmann B, Kang G, et al. Changes in the intestinal microbiota following the administration of azithromycin in a randomised placebo-controlled trial among infants in South India. Sci Rep. 2017;7(1):9168. doi: 10.1038/s41598-017-06862-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Korpela K, Salonen A, Virta LJ, Kekkonen RA, Forslund K, Bork P, et al. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat Commun. 2016;7:10410. doi: 10.1038/ncomms10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korpela K, Salonen A, Virta LJ, Kekkonen RA, de Vos WM. Association of early-life antibiotic use and protective effects of breastfeeding: role of the intestinal microbiota. JAMA Pediatr. 2016;170(8):750–757. doi: 10.1001/jamapediatrics.2016.0585. [DOI] [PubMed] [Google Scholar]
- 39.Haffajee AD, Bogren A, Hasturk H, Feres M, Lopez NJ, Socransky SS. Subgingival microbiota of chronic periodontitis subjects from different geographic locations. J Clin Periodontol. 2004;31(11):996–1002. doi: 10.1111/j.1600-051X.2004.00597.x. [DOI] [PubMed] [Google Scholar]
- 40.Li J, Quinque D, Horz HP, Li M, Rzhetskaya M, Raff JA, et al. Comparative analysis of the human saliva microbiome from different climate zones: Alaska, Germany, and Africa. BMC Microbiol. 2014;14:316. doi: 10.1186/s12866-014-0316-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kemppainen KM, Ardissone AN, Davis-Richardson AG, Fagen JR, Gano KA, León-Novelo LG, et al. Early childhood gut microbiomes show strong geographic differences among subjects at high risk for type 1 diabetes. Diabetes Care. 2015;38(2):329–332. doi: 10.2337/dc14-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lamont RJ, Koo H, Hajishengallis G. The oral microbiota: dynamic communities and host interactions. Nat Rev Microbiol. 2018;16(12):745–759. doi: 10.1038/s41579-018-0089-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laitala ML, Vehkalahti MM, Virtanen JI. Frequent consumption of sugar-sweetened beverages and sweets starts at early age. Acta Odontol Scand. 2018;76(2):105–110. doi: 10.1080/00016357.2017.1387929. [DOI] [PubMed] [Google Scholar]
- 44.Hauta-Alus HH, Korkalo L, Holmlund-Suila EM, Rosendahl J, Valkama SM, Enlund-Cerullo M, et al. Food and nutrient intake and nutrient sources in 1-year-old infants in Finland: A cross-sectional analysis. Nutrients. 2017;9(12). 10.3390/nu9121309. [DOI] [PMC free article] [PubMed]
- 45.Williams JE, Carrothers JM, Lackey KA, Beatty NF, Brooker SL, Peterson HK, et al. Strong multivariate relations exist among milk, oral, and fecal microbiomes in mother-infant dyads during the first six months postpartum. J Nutr. 2019;149(6):902–914. doi: 10.1093/jn/nxy299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luppens SB, Kara D, Bandounas L, Jonker MJ, Wittink FR, Bruning O, et al. Effect of Veillonella parvula on the antimicrobial resistance and gene expression of Streptococcus mutans grown in a dual-species biofilm. Oral Microbiol Immunol. 2008;23(3):183–189. doi: 10.1111/j.1399-302X.2007.00409.x. [DOI] [PubMed] [Google Scholar]
- 47.Teles R, Sakellari D, Teles F, Konstantinidis A, Kent R, Socransky S, et al. Relationships among gingival crevicular fluid biomarkers, clinical parameters of periodontal disease, and the subgingival microbiota. J Periodontol. 2010;81(1):89–98. doi: 10.1902/jop.2009.090397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Teles R, Wang CY. Mechanisms involved in the association between periodontal diseases and cardiovascular disease. Oral Dis. 2011;17(5):450–461. doi: 10.1111/j.1601-0825.2010.01784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng L, Cheng X, Deng M, Deng X, Du Q, Ge Y, et al. Subgingival microbes. In: Atlas of oral microbiology: Elsevier; 2015. p. 67–93. 10.1016/C2013-0-19154-4.
- 50.Wong JM, de Souza R, Kendall CW, Emam A, Jenkins DJ. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol. 2006;40(3):235–243. doi: 10.1097/00004836-200603000-00015. [DOI] [PubMed] [Google Scholar]
- 51.Schmalz G, Schiffers N, Schwabe S, Vasko R, Müller GA, Haak R, et al. Dental and periodontal health, and microbiological and salivary conditions in patients with or without diabetes undergoing haemodialysis. Int Dent J. 2017;67(3):186–193. doi: 10.1111/idj.12282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Natland ST, Andersen LF, Nilsen TI, Forsmo S, Jacobsen GW. Maternal recall of breastfeeding duration twenty years after delivery. BMC Med Res Methodol. 2012;12:179. doi: 10.1186/1471-2288-12-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Differentially abundant bacteria at OTU-level by type of feeding (No infant formula vs. Infant formula) during the first six months of life.
Data Availability Statement
The data that support the findings of this study are available from Heli T. Viljakainen but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are however available from the authors upon reasonable request and with permission of Heli T. Viljakainen.