Abstract
Objective.
Microfabricated multielectrode arrays are widely used for high throughput recording of extracellular neural activity, which is transforming our understanding of brain function in health and disease. Currently there is a plethora of electrode-based tools being developed at higher education and research institutions. However, taking such tools from the initial research and development phase to widespread adoption by the neuroscience community is often hindered by several obstacles. The objective of this work is to describe the development, application, and open dissemination of silicon microprobes for recording neural activity in vivo.
Approach.
We propose an open source dissemination platform as an alternative to commercialization. This framework promotes recording tools that are openly and inexpensively available to the community. The silicon microprobes are designed in house, but the fabrication and assembly processes are carried out by third party companies. This enables mass production, a key requirement for large-scale dissemination.
Main results.
We demonstrate the operation of silicon microprobes containing up to 256 electrodes in conjunction with optical fibers for optogenetic manipulations or fiber photometry. These data provide new insights about the relationship between calcium activity and neural spiking activity. We also describe the current state of dissemination of these tools. A file repository of resources related to designing, using, and sharing these tools is maintained online.
Significance.
This paper is likely to be a valuable resource for both current and prospective users, as well as developers of silicon microprobes. Based on their extensive usage by a number of labs including ours, these tools present a promising alternative to other types of electrode-based technologies aimed at high throughput recording in head-fixed animals. This work also demonstrates the importance of validating fiber photometry measurements with simultaneous electrophysiological recordings.
1. Introduction
Device miniaturization, enabled by micro- and nano-fabrication techniques, has transformed how we interrogate brain function [1]. For decades, microelectromechanical systems (MEMS) with high-density electrode arrays have been developed for monitoring extracellular brain activity [2–7]. The sophistication of these tools has grown steadily [8], with current state-of-the-art tools providing simultaneous access to over one thousand neurons [9]. Today there is a wealth of emerging electrode-based technologies offering improvements in features such as electrode number [9–14], long-term stability [15–17], integrated electronics for signal processing [9, 18], integrated photonics or transparent materials for optogenetics or imaging [19–24], and integrated microfluidics for drug delivery [25, 26]. While it is clearly the hope that every new tool can be successfully adopted, there are major challenges in transitioning neural recording devices from the initial research and development phase into the basic science laboratory. A universal challenge is providing a strong incentive for neuroscientists to use a specific type of device over alternative products [27]. This incentive could be based on favorable technical capabilities (e.g., structural dimensions, number or arrangement of electrodes), as well as less quantifiable but nonetheless important considerations such as availability and convenience [28].
One of the prerequisites for creating widely accessible recording tools is the ability to fabricate devices en masse. Here, we describe a high electrode count microprobe which leverages the mass production capabilities of a commercial MEMS foundry. In order to streamline the development process, these devices are designed in house, but components are fabricated and assembled by third party manufacturers. These silicon microprobes contain up to 256 channels (ch) for recording single-unit and local field potential signals, which are compatible with commercially available recording instrumentation, and enable acute recordings from tens to hundreds of neurons in head-fixed mice. We also show a 32 ch packaging option developed for recording from freely moving animals. Furthermore, by combining microprobes with optical fibers to create opto-microprobes, we demonstrate simultaneous electrophysiological and optogenetic or photometric capabilities. To promote broad accessibility, we adopt an open source dissemination framework [29, 30]. We provide detailed information related to the development of the microprobes or opto-microprobes (e.g., photolithography mask and printed circuit board (PCB) design files, schematics for head fixation parts, and instructional videos). This allows any user to either replicate or modify our existing designs to create high electrode count neural probes using previously validated mass production methods. Finally, we describe efforts to share pre-fabricated probes under this open source framework.
2. Materials and methods
2.1. Overview of silicon microprobe development
The preparation of silicon microprobes for recording entails six steps illustrated in figure 1: 1) photolithography mask design, 2) silicon microfabrication, 3) device release and sorting, 4) PCB design and manufacture, 5) probe assembly, and 6) encapsulation, electroplating, and optional attachment of optical fibers. We describe how to navigate these steps and subsequently use the probes to record from head-fixed behaving mice. It is important to acknowledge that the majority of potential neuroscience users lack the resources, or interest, to perform all of these steps on their own. Therefore, under the open source dissemination framework, users can circumvent the first four steps which are the most labor and resource-intensive, by relying on pre-fabricated probes and PCBs. The fifth step, assembly, is straightforward as it can be performed by third-party companies. Additionally, the final step involving encapsulation, electroplating and optional fiber attachment can be performed using commercially available hardware and supplies. Finally, to record with these devices users can leverage commercially available data acquisition systems with matching electrical connectors, as well as open source spike sorting programs developed by other groups [31, 32]. Up-to-date information on microprobe designs, as well as resources for obtaining and using these tools is provided online at https://github.com/sotmasman/Silicon-microprobes, with the currently available resources listed in table 1.
Figure 1. Overview of the main steps involved in silicon microprobe development.
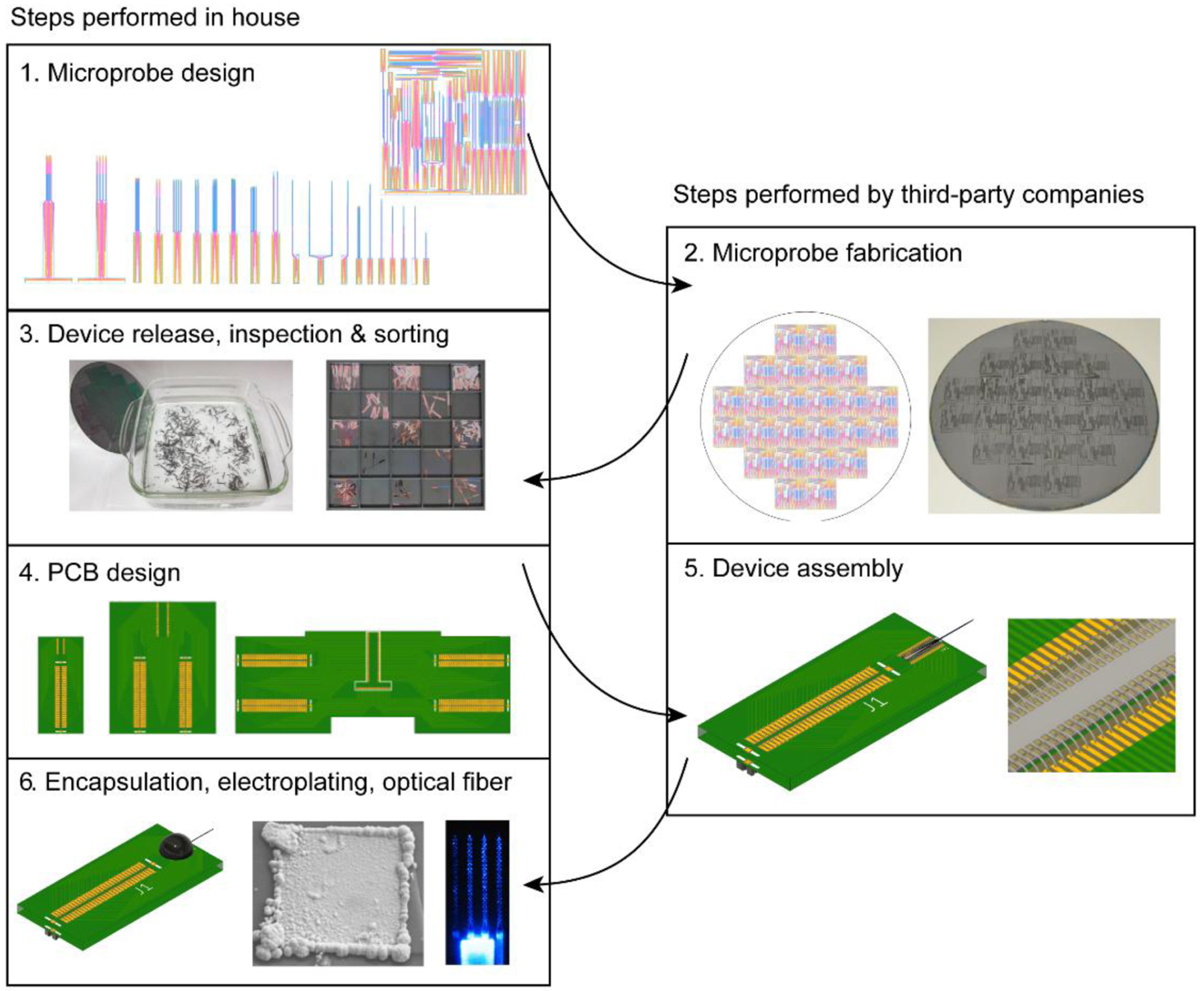
The numbered steps on the left side are performed in house or by the end user, while the steps on the right side are performed by third-party companies. Step 1 shows CAD drawings of the individual probes and reticle. Step 2 shows the reticle copied multiple times across a wafer, and a photograph of a processed wafer. Step 3 shows hot water release of the probes from the carrier wafer, and sorting the devices by design into trays for storage. Step 4 shows the three standard PCB designs. Step 5 shows a probe assembled onto a PCB with wire bonding. Step 6 shows an assembled probe with epoxy encapsulation, a gold-plated electrode, and an opto-microprobe.
Table 1.
List of information found on the online file repository (https://github.com/sotmasman/Silicon-microprobes) related to the silicon microprobes described in this paper.
| Product or procedure | File format |
|---|---|
| Silicon microprobe & reticle designs | AutoCAD DWG & GDS files |
| PCB designs | AutoCAD DWG & Gerber files |
| Encapsulation procedure | Video |
| Optical fiber attachment procedure | Video |
| Designs for device holders, head bars, head bar holders for head-fixed recordings | Fusion 360 STEP files |
| Plots of electrode maps | Matlab scripts |
| Dissemination of pre-fabricated microprobes as open source tools | Readme file |
2.2. Silicon microprobe design and fabrication (Steps 1 & 2)
Microfabrication is often the most resource-intensive process in neural probe development. This step is commonly carried out in academic clean room facilities which are well suited for prototype development and refinement, but may be less suitable for mass production. Thus, to enable scalable production of silicon microprobes we rely on a 6 inch (150 mm) wafer process at commercial MEMS foundry (Innovative Micro Technology). The structural properties of the probes (e.g., number and arrangement of electrodes, number, length, and spacing of shafts) are determined by masks, which are used to transfer patterns onto wafers via UV photolithography, followed by depositing or etching a variety of materials. The masks are designed in house using CAD software (AutoCAD, Autodesk) before being sent to the foundry for production. The masks are confined to a square area of 22 mm × 22 mm referred to as the reticle (figure 1). The reticle is populated with an assortment of different microprobe designs. During production the patterns on the reticle are transferred onto multiple different sections of the wafer, eventually leading to several copies of the devices contained in the reticle.
There are three stages to designing the reticle. First, the implantable shaft section of each microprobe is specified (figure 2) in terms of total electrode number (maximum: 256), electrode area and layout (standard area: 10 μm × 10 μm), shaft spacing, and shaft length and tip angle (default length: 7 mm, maximum length: 18 mm). The shaft width is mainly constrained by the wiring bottleneck at the base of the electrode array; the minimum interconnecting wire pitch is 0.8 μm (0.4 μm wire width and 0.4 μm spacing to the nearest feature). The second stage is combining the shafts with the base section of the probe, which contains wire bond contact pads for PCB assembly (figure 3A). To facilitate standardization of these devices, we have developed standard size bases for 64, 128, and 256 ch probes (figure 3B; 32 ch probes also use the 64 ch base). Each microprobe requires a set of three masks which are used for: exposing the electrode recording sites and wire bond contact pads from a silicon nitride insulating layer; patterning the electrodes, wire bond contact pads and interconnecting wires; and defining the 2D profile of the probes as seen from the top (figure 3C). Each device also contains an identifying label such as 64D sharp, 128K, 256AS on the base section. After designing a variety of individual microprobes, the third stage is arranging these devices without overlap in a 22 mm × 22 mm space defining the reticle (figure 3D). To increase the quantity of devices, and provide some redundancy against small defects introduced during fabrication, devices are arranged such that unused space in the reticle is reduced as much as possible (minimum device spacing: ~ 50 μm), and each type of probe is copied multiple times into the reticle. There is a tradeoff between the number of unique designs, and the number of copies per design, and we recommend between 10 – 20 unique designs per reticle to ensure sufficiently large quantities for widespread distribution. AutoCAD designs of individual microprobes and the reticle are provided in the online file repository.
Figure 2. Common design variables for the implantable shaft section.
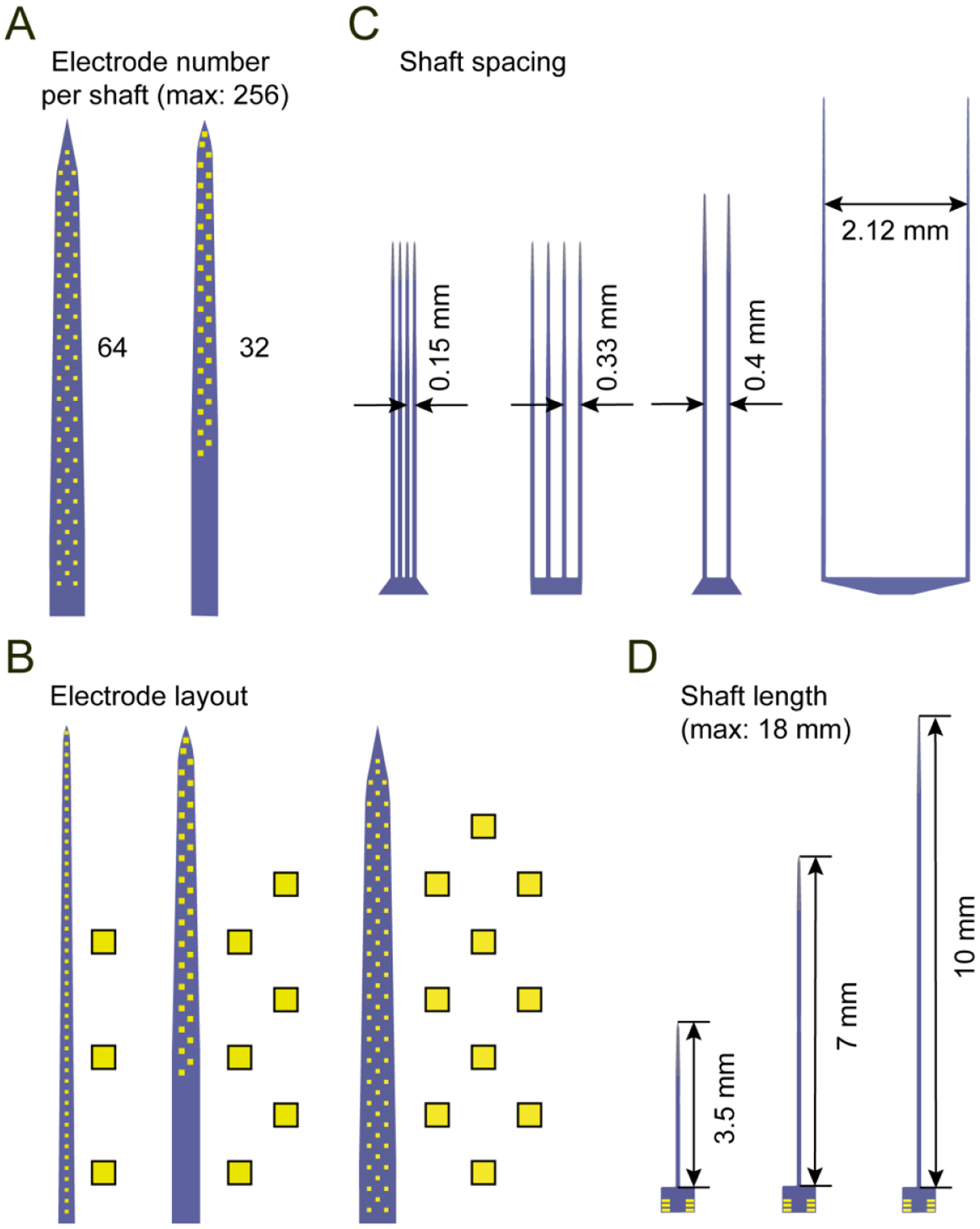
(A) Two possible shaft designs containing 64 and 32 electrodes. The maximum number of total electrodes is 256.
(B) Three possible electrode layouts. Left: linear, middle: staggered, right: honeycomb.
(C) Four possible shaft spacings.
(D) Three possible shaft lengths. The default length is 7 mm. The maximum possible length is ~ 18 mm imposed by the reticle size.
Figure 3. Design of individual microprobes and the reticle.
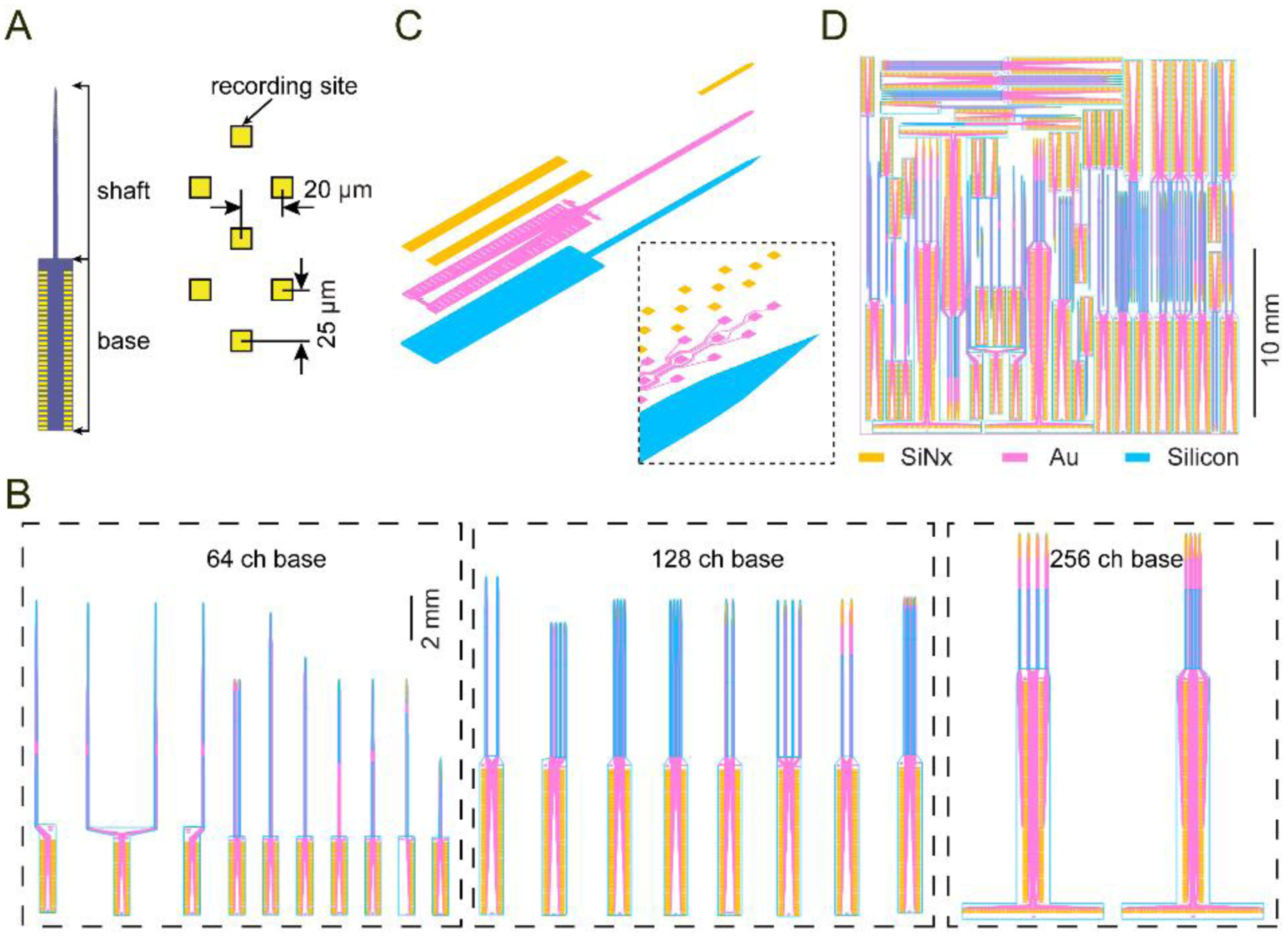
(A) A 64 ch microprobe (64D short) illustrating the shaft and base sections, and the honeycomb electrode layout.
(B) Assortment of different microprobe designs grouped by base section (64, 128, or 256 ch).
(C) Expanded view of the three structural layers used for microfabrication (orange: silicon nitride window for exposing the electrodes and wire bond contact pads; magenta: metal layer comprising the electrodes, interconnecting wires, and wire bond contact pads; blue: silicon microprobe 2D profile). The inset shows a closeup of the probe tip.
(D) Reticle layout containing an assortment of individual devices arranged closely together to minimize empty space.
Fabrication was performed on silicon-on-oxide (SOI) wafers containing a 21 μm thick Si device layer. Low stress silicon nitride (SiNx) electrical insulation layers (~ 400 nm thick) were deposited between a metallization layer consisting of 10 nm Ti, 100 nm Au, 10 nm Ti. Nitride and metal layers were patterned via plasma etching, and silicon was patterned with a deep reactive ion etching (DRIE) process, which produced vertical device side walls and chisel-shaped tips (figure 4). The next step involved using a water soluble polymer to mount the SOI wafer upside down on a mechanical carrier wafer DRIE was then used to etch the SOI wafer down to the buried oxide (BOX) stop layer. Finally, the BOX layer was removed with a reactive ion etch, completing the fabrication process.
Figure 4. High electrode density silicon microprobes.
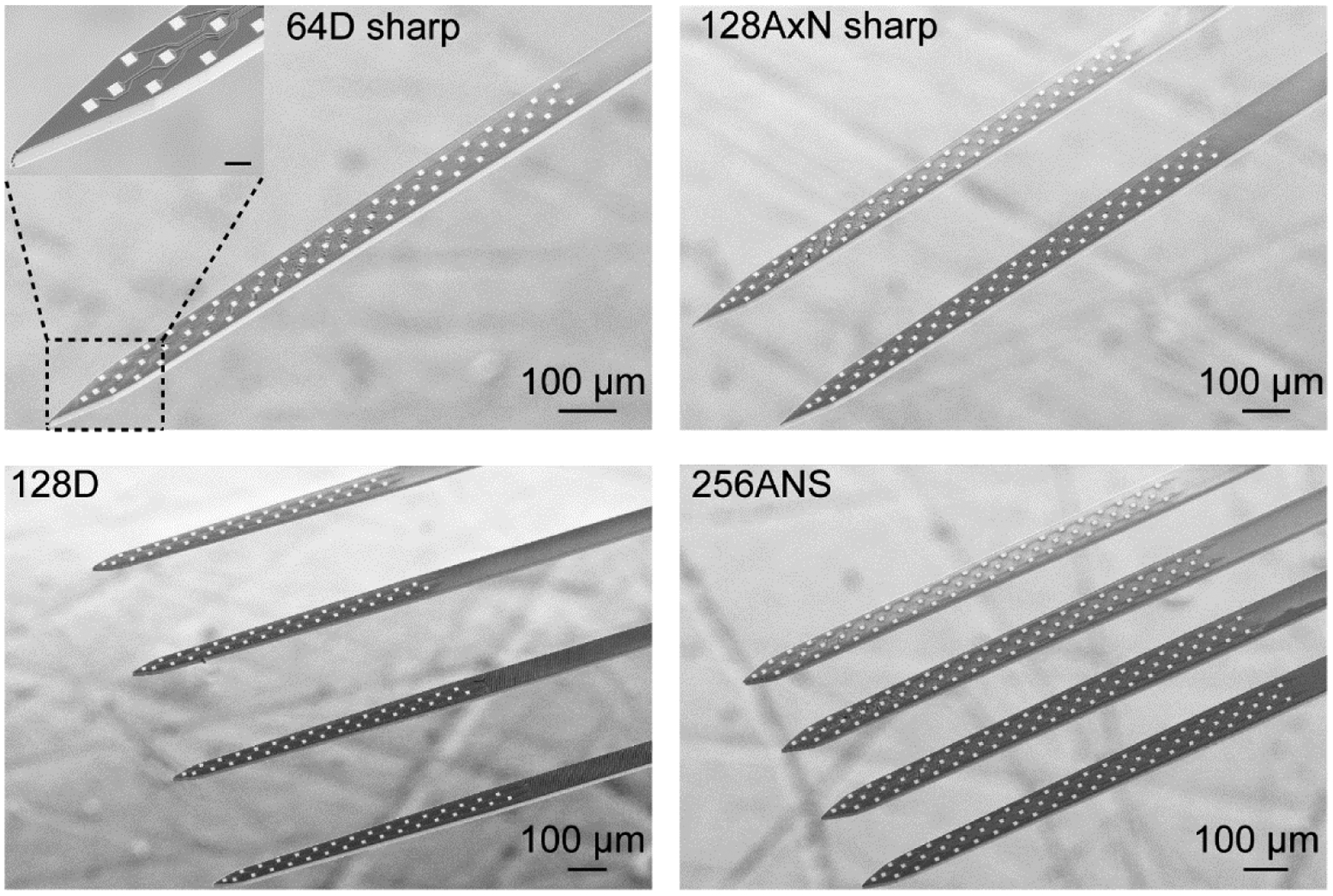
Scanning electron microscope images of four popular silicon microprobes (64D sharp, 128AxN sharp, 128D, 256ANS). Note that three of the probes use the same honeycomb electrode layout. The scale bar in the inset represents 20 μm.
2.3. Device release, inspection and sorting (Step 3)
Devices were delivered still attached to the carrier wafer, and a multi-step process was used to retrieve them. First, probes were released from the carrier by dissolving the polymer in hot DI water (80 °C). The probes were cleaned three times in a fresh bath of hot water, followed by acetone and finally ethanol. While in the acetone bath probes were briefly cleaned by sonication (10 s, Model 1510, Branson). Probes were poured out of the ethanol bath and air dried on coffee filter paper. From this point forward, each device was handled individually with a vacuum pick-up system (tip: 520–9, Ted Pella). Devices were checked for visible defects (e.g., broken of chipped shafts, or excessive residues on top of the electrodes) under a 5 – 50 × optical microscope (ME320T, AmScope). Some devices were also inspected under higher magnification using scanning electron microscopy (figure 4). Finally, devices which appeared nondefective were sorted by their design and stored under ambient conditions. To package the devices for assembly, their base section was taped onto glass slides (tape: KPT-1/4, Kapton; slides: M1002, Denville Scientific; approx. 5 – 10 devices per slide), and shipped to the assembly company in standard plastic petri dishes (351029, Falcon). Historically the yield per wafer has ranged from high (over 90 % devices with fewer than five defective channels) to moderate (over 90 % devices with 10 – 20 defective channels). Thus, a highly conservative estimate of the average yield is 50 % devices with fewer than five defective channels, but this still corresponds to hundreds of probes. The most common yield-lowering factor has been electrical shorts between adjacent interconnecting wires. However, this issue appears to be absent from the most recent batch of wafers produced by the foundry, suggesting yields in significant excess of 50 %.
2.4. PCB design and device assembly (Steps 4 & 5)
We developed three standard PCBs for 64, 128, and 256 ch probes (figure 5A), with designs provided in the online file repository. PCB manufacturing and device assembly are carried out by third party companies (PCB: Hughes Circuits, assembly: IDAX Microelectronics). PCBs are made from 0.46 mm thick FR4 material, and finished with electroless nickel immersion gold. Assembly involves soldering compact electrical connectors onto the PCB (connectors: 5024306410, Molex Slimstack, purchased from Digikey or Mouser Electronics), epoxying the silicon microprobe onto the PCB, and finally wire bonding. The connectors are matched to a commercially available 128 ch head stage for data acquisition (C3316 128 ch amplifier board, Intan Technologies). The PCBs are designed to accommodate two possible connector assembly modes: the ‘top’ mode involves placing the connectors on the same side as the silicon microprobe, while the ‘bottom’ mode involves placing the connectors on the opposite side (figure 5B). The bottom mode is recommended as it allows attachment of optical fibers without interfering with the use of the electrical connectors (see next section). Finally, for recording from freely moving rodents, we have developed a 32 ch PCB with an Omnetics connector (A79022–001, Omnetics Corporation), which has the advantage of being shorter than the three standard PCBs with Molex connectors (weight of PCB with connector: 0.55 g, figure 5D). We also created an Omnetics-to-Molex electrical adapter to facilitate electroplating probes wire bonded to the 32 ch PCB (figure 5E).
Figure 5. PCB design, device assembly, and micromanipulator holders.
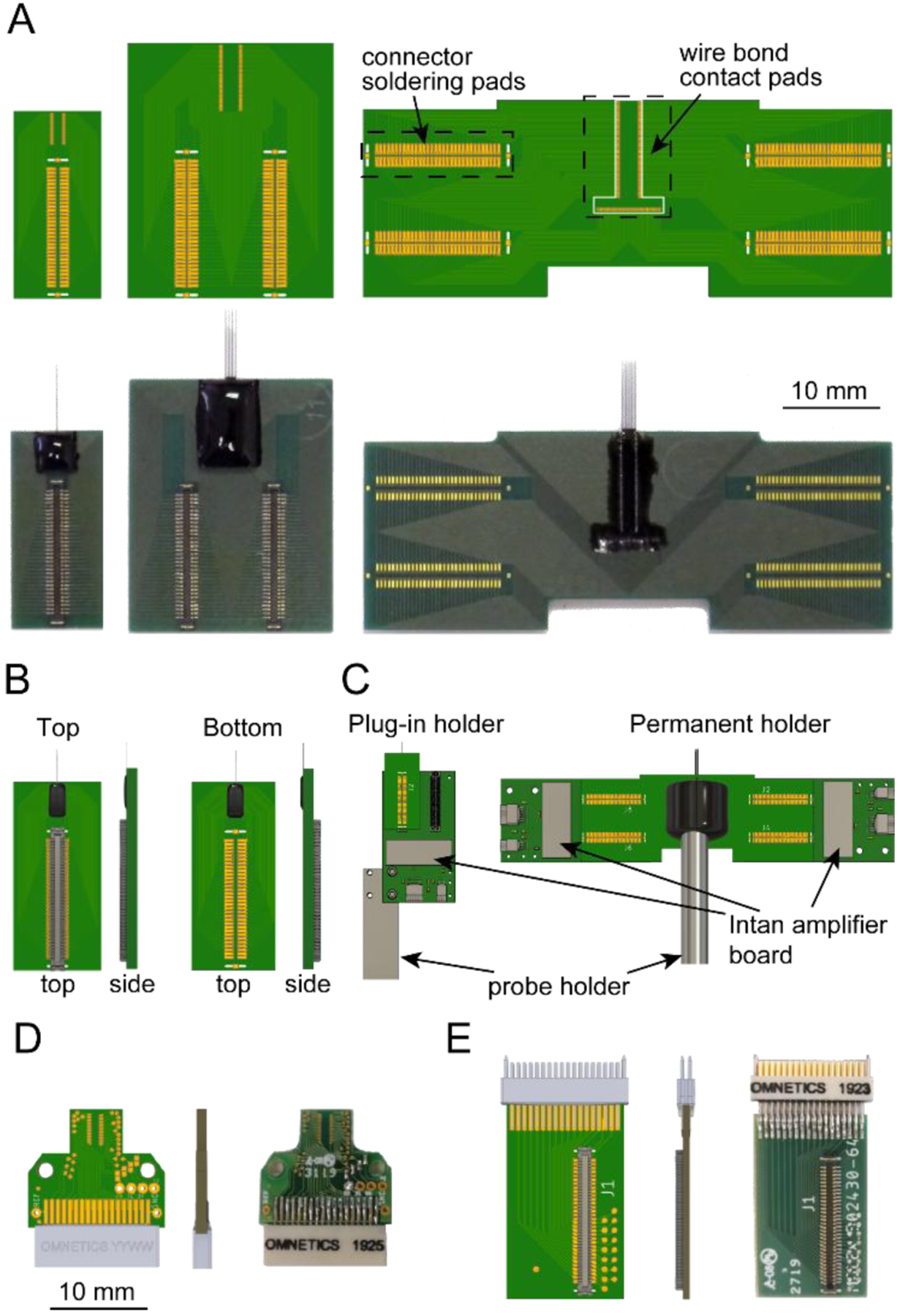
(A) Top: three standard PCB designs. The connector solder pads connect to a Molex Slimstack connector (part # 5024306410). Bottom: images of the corresponding PCBs after assembly and epoxy encapsulation. Note that the connectors on the lower right device are assembled in the bottom mode.
(B) Illustration of the top and bottom connector assembly modes.
(C) Illustration of the plug-in and permanent holders. The drawing of the 128 ch amplifier board was downloaded from www.intantech.com.
(D) 32 ch PCB with Omnetics connector for freely moving animal recordings.
(E) Omnetics-to-Molex connector adapter for the 32 ch PCB in (D).
2.5. Encapsulation, electroplating, and optional optical fiber attachment (Step 6)
The wire bonding area was encapsulated with epoxy (EP965, ResinLab) to prevent damage and isolate the electrical conduits from the recording environment (see video in online file repository). An epoxy curing time of at least 6 hrs was used under room temperature conditions. Prior to recording it is necessary to apply metal electrodeposition to reduce the electrode impedance, and hence noise. Here, we applied gold plating via a commercial electroplating and impedance measurement system (C3180 electroplating board, Intan Technologies) and gold solution (80535500 gold non-cyanide 31 gr/L, Sifco; also available from Neuralynx). The electrodes were immersed in a small bath of gold solution, and we applied 1 s pulses of constant voltage (−2.2 V) with respect to a platinum reference wire (Pt wire: 267201, Sigma). Pulses were applied repeatedly until the target impedance of less than 0.5 MΩ was reached. To improve the plating process, the electrodes were occasionally retracted out of the gold solution and re-immersed. Electrodes that could not reach the target impedance after repeated attempts were considered defective. Figure 6 shows the change in electrode impedance and noise after completing the gold plating process. The initial and final impedance values were measured at 4.8 ± 0.3 MΩ and 0.2 ± 0.1 MΩ, respectively (mean ± SD). The corresponding root mean squared noise was 4.5 ± 0.4 μV and 3.5 ± 0.6 μV before and after gold plating, respectively. In addition to gold, the devices can be electroplated with other electrode materials, such as PEDOT [33] and platinum-iridium alloys (Platinum Group Coatings) [34].
Figure 6. Device electroplating and noise characterization.
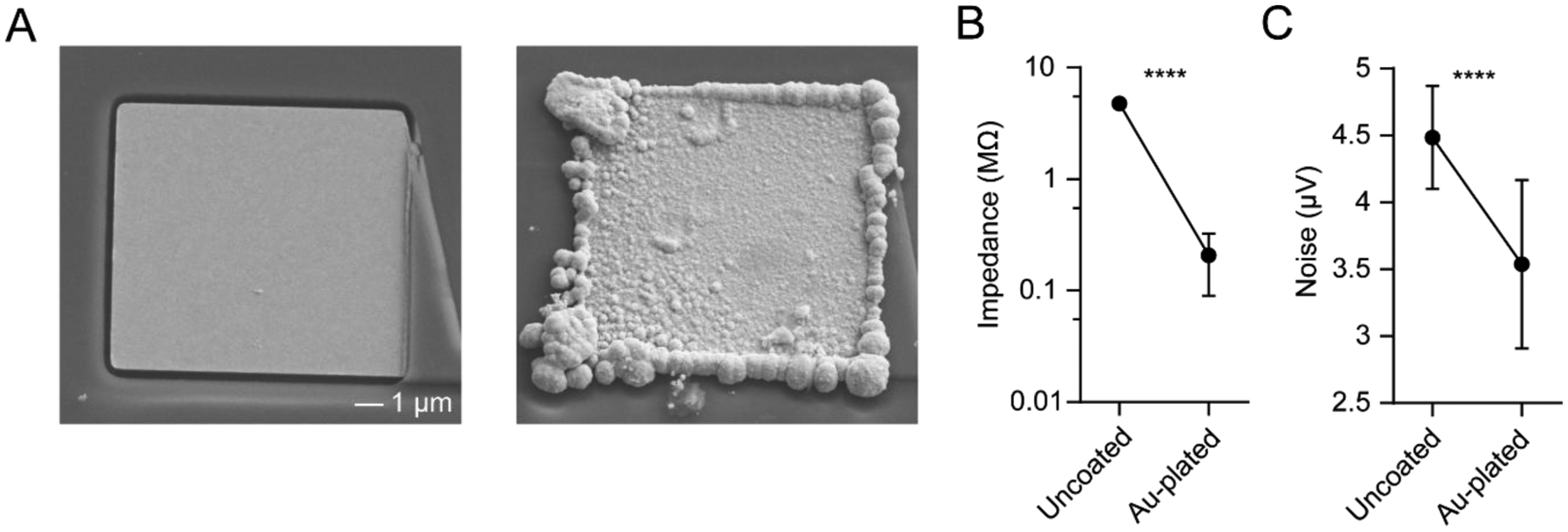
(A) Scanning electron microscope image of an uncoated (left) and gold-plated (right) electrode.
(B) Impedance of 128 electrodes before and after electroplating, measured at 1 kHz in saline (paired t-test, p < 0.0001).
(C) Root mean square noise of 128 electrodes before and after electroplating, measured in saline after applying a 0.3 – 7 kHz bandpass filter (paired t-test, p < 0.0001). Values in B and C represent mean ± SD.
Prior to the encapsulation step users have the option of attaching one or more optical fibers in proximity to the electrodes. These hybrid structures, referred to as opto-microprobes [35], are intended for electrophysiology in tandem with optogenetic manipulations (figure 8) or fiber photometry (figure 9). The opto-microprobe assembly process is shown in a video in the online file repository. The video shows a 200 μm diameter fiber (UM22–200, 0.22 NA, Thor Labs) being attached to a 64 ch silicon microprobe (64D sharp), which was used to demonstrate simultaneous electrophysiology and optogenetic manipulations (figure 8A). The fiber is cut to a length of 35 mm above a ferrule (UM22–200, Thor Labs), and attached to a rod with a fitting sleeve. The rod is mounted on a holder, and the PCB with the silicon microprobe is connected to an L-bar. The L-bar is attached to a three-axis goniometer allowing control of probe pitch, roll, and yaw (GN2, Thor Labs), which is attached to a three-axis motorized micromanipulator allowing control of probe position in the x, y, and z directions (MPC-200, Sutter Instruments). Thus, the alignment system has a total of six degrees of freedom. Assembly took place under a stereo microscope (SM-3BZ-80S, AmScope). After aligning the optical fiber parallel to the silicon microprobe shaft, the tip of the fiber is placed around 200 μm above the nearest electrode. Finally, the fiber is permanently attached to the silicon shaft with epoxy (EP965, ResinLab), and allowed to cure for at least 6 hrs at room temperature. For simultaneous electrophysiology and photometry (figure 9A, top), we attached a low autofluorescence optical fiber (430 μm outer diameter, MFC_400/430–0.48_23MM_MF2.5_FLT, Doric Lenses) to a 256 ch silicon microprobe (256ANS) using identical procedures.
Figure 8. Simultaneous electrophysiology and optogenetics with opto-microprobes.
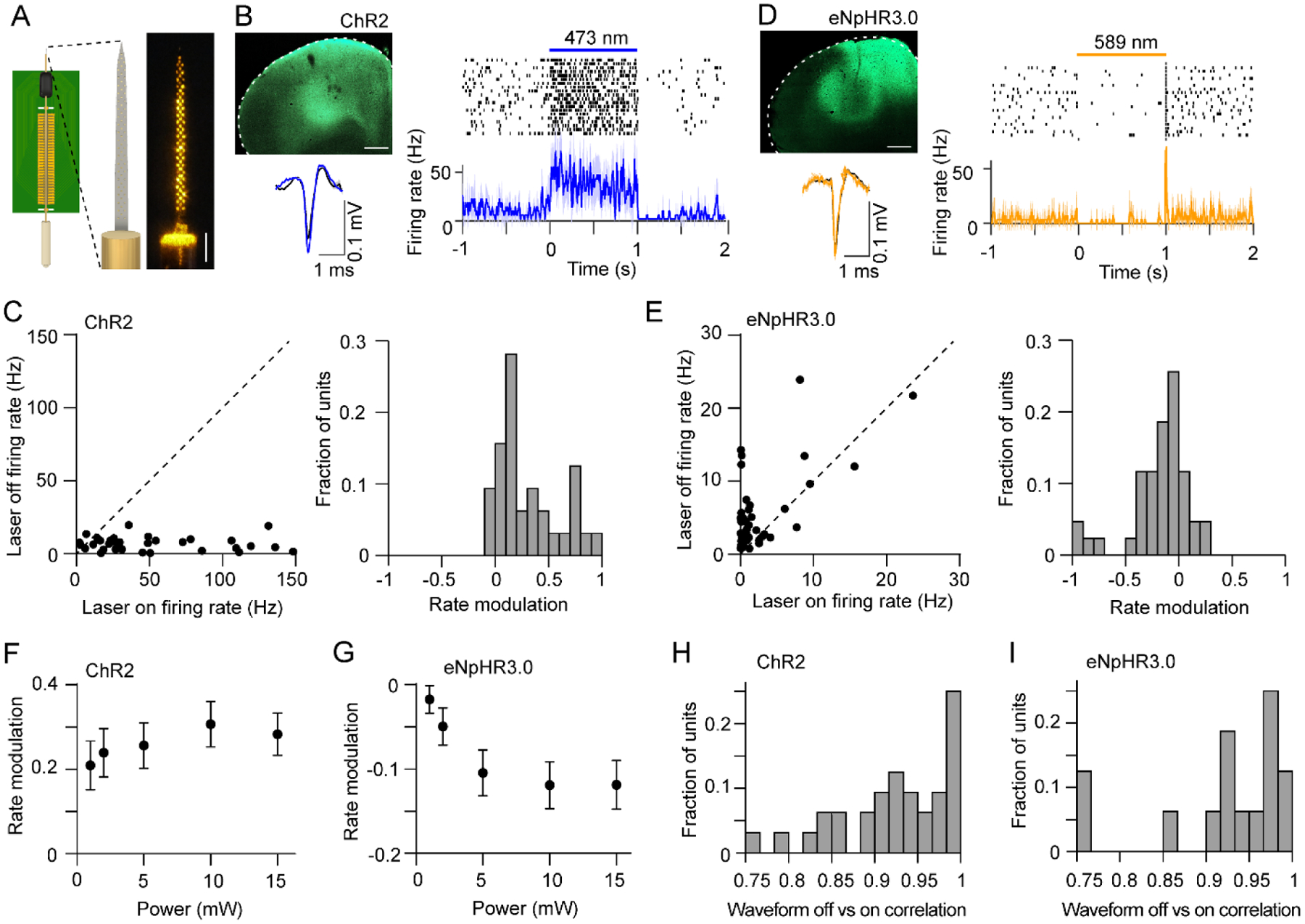
(A) Left: Illustration of opto-microprobe consisting of a 64 electrode silicon probe (64D sharp) combined with a 200 μm diameter optical fiber. Right: Photograph of the electrodes under 589 nm illumination from the fiber.
(B) Top left: Expression of ChR2-eYFP in M2 (scale bar: 0.5 mm). Right: spike raster and mean firing rate versus time of a cortical neuron from a ChR2-expressing mouse during application of 1 s continuous light pulses (10 mW, 473 nm). Bottom left: average spike waveform of the same unit under light off (black) and light on (blue) conditions.
(C) Left: Mean firing rate during laser off versus laser on of 32 cortical neurons recorded from ChR2-expressing mice. The firing rate was significantly higher during laser on (paired t-test, p < 0.0001). The light intensity was 10 mW. Right: Rate modulation index distribution of 32 cortical neurons recorded from ChR2-expressing mice. Dashed line represents the diagonal.
(D) Top left: Expression of eNpHR3.0-eYFP in M2 (scale bar: 0.5 mm). Right: spike raster and mean firing rate versus time of a cortical neuron from a eNpHR3.0-expressing mouse during application of 1 s continuous light pulses (10 mW, 589 nm). Bottom left: average spike waveform of the same unit under light off (black) and light on (orange) conditions.
(E) Left: Mean firing rate during laser off versus laser on of 43 cortical neurons recorded from eNpHR3.0-expressing mice. The firing rate was significantly lower during laser on (paired t-test, p = 0.001). The light intensity was 10 mW. Right: Rate modulation index distribution of 43 cortical neurons recorded from eNpHR3.0-expressing mice.
(F) There was no significant effect of optical power on the rate modulation in ChR2-expressing mice (one-way ANOVA, F4,149 = 0.51, p = 0.73).
(G) There was a significant effect of optical power on the rate modulation in eNpHR3.0-expressing mice (one-way ANOVA, F4,210 = 3.41, p = 0.01). Data in F and G represent mean ± SEM.
(H) Pearson correlation coefficient between each unit’s spike waveform during laser off and on conditions (mean correlation coefficient = 0.93). The light intensity was 10 mW.
(I) Pearson correlation coefficient between each unit’s spike waveform during laser off and on conditions (mean correlation coefficient = 0.92). Note that 28 out of 43 units had zero spikes during the laser on period, and thus could not be included in this analysis. The light intensity was 10 mW.
Figure 9. Simultaneous electrophysiology and fiber photometry with opto-microprobes.
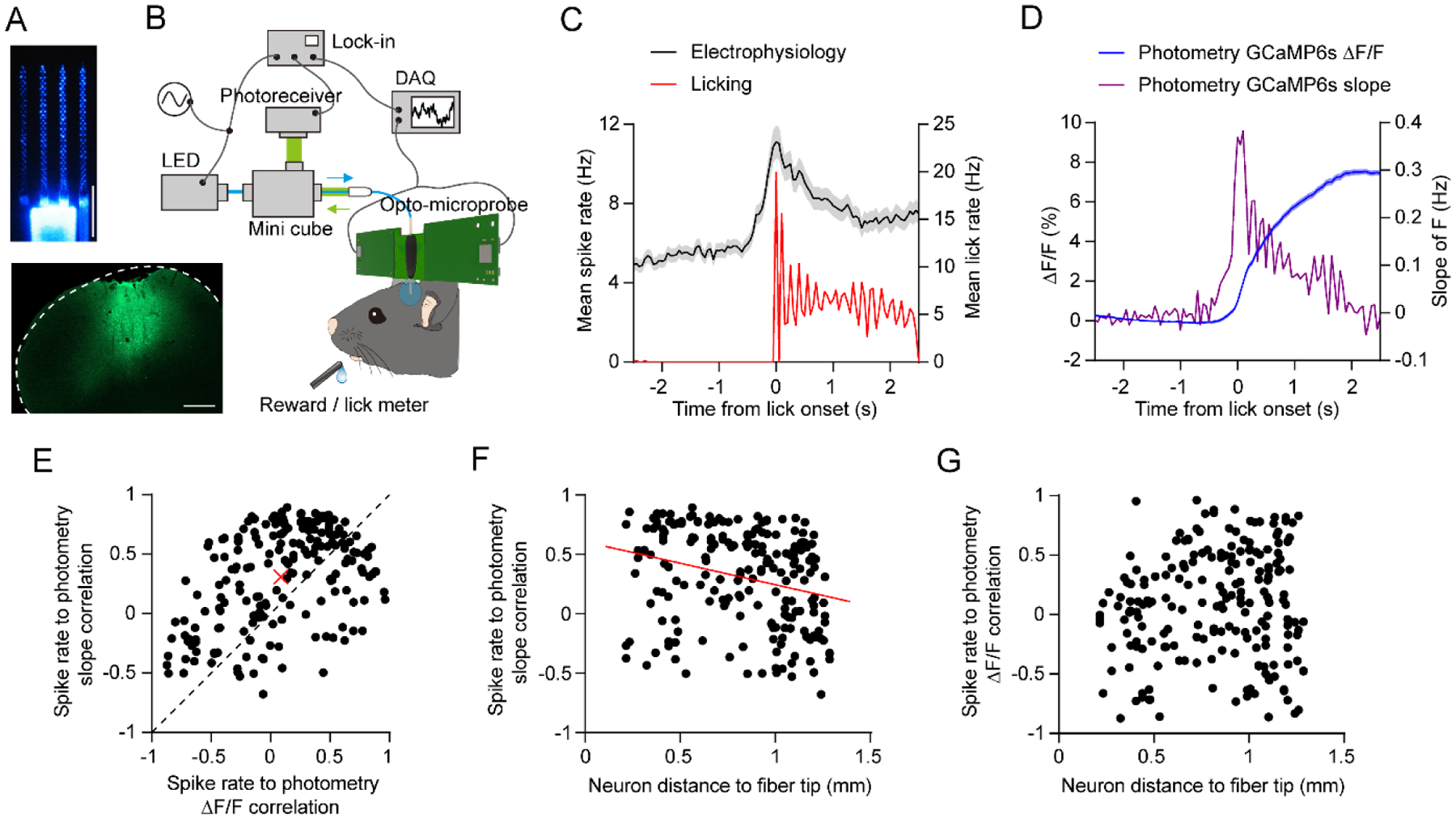
(A) Top: Image of the opto-microprobe used for simultaneous electrophysiology with a 256 ch probe and photometry with a 430 μm outer diameter optical fiber. The image was taken under 465 nm LED illumination from the fiber. Bottom: GCaMP6s expression in M2. Scale bars: 0.5 mm.
(B) Setup for performing recordings in mice trained on a self-initiated operant licking task.
(C) Black curve represents the mean firing rate versus time of 204 cortical neurons recorded from one mouse using the opto-microprobe, aligned to lick onset on rewarded trials. Red curve represents the mean licking rate during the same time period.
(D) Blue curve represents the fractional change in photometry fluorescence signal (ΔF/F) versus time. Violet curve represents the slope, or derivative, of the photometry fluorescence signal. Electrophysiological, licking, and photometry data in C and D were measured simultaneously from the same animal.
(E) Pearson correlation coefficient of each neuron’s firing rate to the photometry slope signal, versus each neuron’s firing rate to the photometry ΔF/F signal. Each black circle represents one neuron (n = 204). The correlation is significantly higher for the slope (paired t-test, p < 0.0001). Red cross represents the mean value. Dashed line represents the diagonal.
(F) Pearson correlation coefficient of each neuron’s firing rate to the photometry slope signal, versus the neuron’s diagonal distance to the optical fiber tip. There is a significant negative correlation with distance (n = 204, r = −0.27, p = 0.0001). Red line represents the best line fit.
(G) Pearson correlation coefficient of each neuron’s firing rate to the photometry ΔF/F signal, versus the neuron’s diagonal distance to the optical fiber tip. There is no significant correlation with distance (n = 204, r = 0.09, p = 0.18).
2.6. Micromanipulator device holders for in vivo recording in head-fixed mice
For acute head-fixed animal recordings, devices are inserted in the brain with a motorized micromanipulator (MPC-200, Sutter Instruments). As shown in figure 5C, devices can be attached to the micromanipulator either with a plug-in or permanent holder. We have designed a plug-in holder optimized for 64 and 128 ch probes (online file repository, manufactured at UCLA machine shop). This holder attaches to the Intan 128 ch amplifier board via two holes for size 0–80 screws, allowing straightforward assembly and disassembly. However, this holder is incompatible with 256 ch probes, which is operated with two horizontally oriented amplifier boards. Therefore, 256 ch probes are attached with epoxy (EP965, ResinLab) to a permanent holder (online file repository) consisting of an aluminium rod, 6.3 mm diameter, 155 mm length (OnlineMetals). To ensure the silicon shafts are aligned parallel to the insertion direction, the permanent holder is attached using the six degree of freedom alignment system described in the previous section.
2.7. Animal surgeries for head-fixed mouse recordings
Recording from head-fixed mice involves two survival surgical procedures: first, to attach head fixation bars and optionally inject virus for optogenetics or fiber photometry; second, to prepare a craniotomy for inserting the microprobe. Surgeries were performed under a stereo microscope (Seiler iQ). All procedures were approved by the University of California, Los Angeles Chancellor’s Animal Research Committee. Male C57BL/6J, 8 – 12 week old mice were obtained from The Jackson Laboratory (stock no. 000664). Animals were maintained on a 12 hr light cycle, and group housed until the first surgery. Animals underwent the first aseptic surgery under isoflurane anesthesia on a stereotaxic instrument (Model 1900, Kopf Instruments). A stainless steel head fixation bar (online file repository, laser cut at Mainstay Manufacturing), was attached to each side of the skull with dental cement (Metabond, Parkell). Using two separate pieces helps reduce weight and improves access to the top of the skull for microprobe insertion (figure 7A). For experiments involving optogenetics or photometry, adeno-associated virus (AAV) was obtained from the University of North Carolina Vector Core. Craniotomies were made with a surgical drill, 0.3 mm drill bit diameter (X000QVJVF9, Daewon Industries). For optogenetics, AAV for the either the excitatory opsin AAV5/CaMKIIa-hChR2-eYFP or inhibitory opsin AAV5/CaMKIIa-eNpHR3.0-eYFP was injected in the secondary motor cortex (M2, coordinates relative to bregma: 2.5 mm anterior, 1.5 mm lateral, 1.3 mm ventral) at a volume of 300 nL by pulled glass pipettes (Nanoject II, Drummond Scientific). For photometry, 300 nL of AAVDJ/hsyn-GCaMP6s was injected in the same coordinates. The exposed skull was protected by covering with silicone elastomer (Kwik-Cast, World Precision Instruments) and then dental cement (Metabond, Parkell). A daily carprofen injection (5 mg/kg, s.c.) was administered for the first three days post-operatively, and analgesics (ibuprofen) and antibiotics (amoxicillin) were administered in the drinking water for the first week post-operatively. All mice were individually housed after surgery, and were allowed to recover for at least 2 weeks (3 weeks for animals injected with virus). The second surgery was also performed on stereotaxic equipment under isoflurane anesthesia. This procedure involved using a surgical drill to remove the protective dental cement, then tweezers to remove the Kwik-Cast layer to expose the skull, making a rectangular craniotomy above the recording site, and gently removing the dura to facilitate device insertion. An additional craniotomy was made over the posterior cerebellum to accommodate an electrical reference wire. The craniotomies were temporarily sealed with Kwik-Cast for the duration of the recovery period. Mice recovered for 6 – 10 hrs in their cage with unrestricted water access before being attached to a head bar holder (online file repository, manufactured at the UCLA Physics Machine Shop), and transferred to the recording setup.
Figure 7. High electrode density neural recording in head-fixed mice.
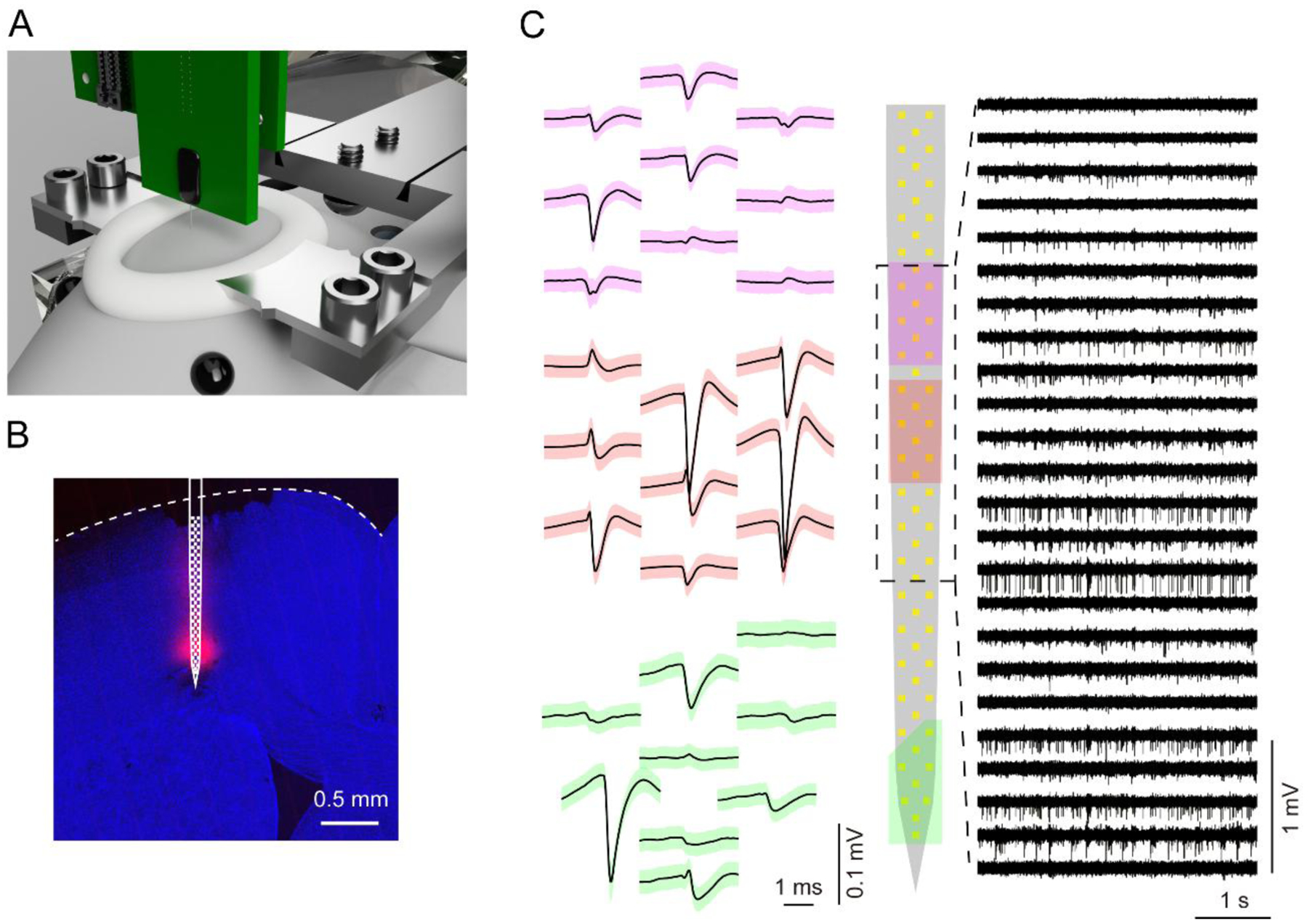
(A) Illustration of a recording setup for head-fixed mice.
(B) Confocal microscope image of the microprobe insertion site in M2. The red signal is DiD fluorescence marking the shaft position. The approximate electrode positions are superimposed in white. The slice is labeled with DAPI (blue).
(C) Left: Spike waveforms corresponding to three putative single units (mean ± SD). Each unit is color-coded according to its position on the electrode array (middle). Right: Voltage versus time traces from 24 electrodes filtered from 0.3 – 7 kHz.
2.8. In vivo data acquisition with microprobes and opto-microprobes
Before each recording session, probes were sometimes coated in fluorescent dye (DiD, Invitrogen) to assist with determining their position in the brain. The silicon microprobe connected to one or more 128 ch amplifier boards (C3316, Intan Technologies) was attached to a motorized micromanipulator with a plug-in or permanent holder (figure 5C). After transferring animals to the recording setup, the protective Kwik-Cast layer was removed to expose the recording site and electrical reference craniotomies. A silver/silver-chloride reference wire was inserted in the cerebellum and connected to the reference pin on the 128 ch amplifier board (Intan Technologies, ground and reference pins were shorted together). To improve electrical contact the reference craniotomy was capped with an absorbable gelatin sponge (Gel Foam, Pfizer) soaked in artificial cerebrospinal fluid (ACSF, 59–7316, Harvard Apparatus), and subsequently held in place with Kwik-Cast. Silicon microprobe insertion was performed under a stereo microscope (Zeiss OPMI Pico). Before insertion, the probe’s position relative to bregma was callibrated using landmarks on the skull or head fixation bars. The device was then lowered to the target coordinates in M2 (2.5 mm anterior, 1.5 mm lateral, 2.2 mm ventral to bregma). After reaching the target region, mineral oil (M8410, Sigma) was placed over the craniotomy to protect the exposed brain surface. This was followed by a 30 – 45 minute settling time before commencing data acquisition. Data were acquired with open source software (Intan Technologies) at a rate of 25 kHz per ch.
For simultaneous electrophysiology and optogenetics, optical stimulation was provided by coupling the optical fiber to a laser (for ChR2: 473 nm, MBL-FN-473–100mW, Opto Engine; for eNpHR3.0: 589 nm, MGL-F-589–100mW, Changchun New Industries Optoelectronics). The optical power output from the fiber was calibrated at the beginning of each experiment. Recordings were carried out in M2 during quiet wakefulness. Optical stimulation was applied in continuous 1 s pulses of varying intensity (1, 2, 5, 10, 15 mW, 20 trials per power level).
For simultaneous electrophysiology and fiber photometry, after recovering from the first surgery mice were food restricted to maintain their weight at around 90 % of their baseline level (unrestricted water access), and trained on a self-initiated operant licking task. During the task, mice had to lick twice within a 1 s period in order to receive a fluid reward (5 μL, 10 % sweetened condensed milk). Licking at the reward port was monitored with a lickometer based on an infrared light beam detector (UCLA Physics Machine Shop). After receiving reward mice had to withhold licking for at least 2 s until the next opportunity for reward became available. Mice received around 120 rewards per daily training session, and were trained for 7 days before recording. Recordings were carried out in M2 using a 256 ch opto-microprobe (figure 9A, top) and photometry lock-in measurement setup illustrated in figure 9B. The photometry fiber was coupled to a fluorescence mini cube (FMC4_AE(405)_E(460–490)_F(500–550)_S, Doric Lenses). Optical excitation was provided by a 465 nm LED (211 Hz sinusoidal power modulation from 10 – 100 μW). The light emitted from the GCaMP6s calcium indicator was detected by a femtowatt photoreceiver (Model 2151, Newport). The photoreceiver output was connected to a lock-in amplifier (SR810, Stanford Research). The demodulated photometry lock-in signal was sampled synchronously with the electrophysiology signals at 25 kHz by a DAQ (C3004, Intan Technologies).
2.9. Probe cleaning and histology
After each recording, probes were cleaned by soaking in undiluted trypsin solution (15400054, Gibco), rinsed in DI water and ethanol, and stored for possible reuse under ambient conditions. For histology, animals were perfused with ice cold phosphate buffered saline (PBS, pH 7.3, 10010023, Gibco) and formalin solution (neutral buffered 10 %, HT501128, Sigma) in sequence. After overnight fixation in formalin, the brain was sectioned into 100 μm sections on a vibratome (VT1000S, Leica). The section shown in figure 7B was stained with DAPI (D1306, Invitrogen). The sections shown in figures 8B, 8D, 9A were blocked using normal serum, then incubated overnight at 4 °C with chicken anti-GFP (ab13970, Abcam) as a primary antibody (1:1000 dilution). After washing three times with PBS, the sections were incubated at 4 °C with Alexa Fluor 488–conjugated donkey antibody to chicken IgG (703-545-155, Jackson ImmunoResearch) as a secondary antibody (1:200 dilution) for 4 hr. Finally, all sections were mounted using tissue mounting medium (C9368, Sigma), and imaged under laser confocal microscopy (LSM 880, Zeiss).
2.10. In vivo data analysis
Offline single-unit analysis was carried out using Kilosort spike sorting software [31], with data band-pass filtered between either 0.3 – 7 kHz, or 0.6 – 6.5 kHz for optogenetics experiments. Matlab scripts specifying each probe’s electrode maps (both top and bottom PCB assembly modes) with respect to the Intan Technologies 128 ch amplifier output are provided in the online file repository. The rate modulation index (RMI) describing the change in firing rate during optogenetic excitation or inhibition is defined by:
Where Ron and Roff represent the mean number of spikes in the 1 s laser and the preceding 1 s baseline period, respectively. Photometry signals were analyzed by downsampling first to 1,000 Hz followed by 20 Hz. Photometry signals and corresponding neural activity were aligned with respect to the onset of rewarded licking trials. The fractional change in fluorescence was calculated with respect to the average signal in a baseline period from −2 to −1 s relative to the onset of licking. The slope was calculated by applying the Matlab diff function to the downsampled data and dividing by the time bin size (0.05 s), with the resulting values having units of inverse time (Hz).
2.11. Silicon microprobe dissemination methods
Since the wafer-scale fabrication process yields hundreds of functional devices (up to approximately 500 per wafer), these tools are well-suited for dissemination to the research community. We have developed a pipeline for openly sharing (non-commercially), pre-fabricated microprobes with other users. This resource is publicized online (see online file repository readme file for up-to-date information on dissemination). The dissemination process begins with users sending a request to the corresponding author specifying the type and quantity of probes. Probes are assembled with PCBs on-demand (IDAX Microlectronics). Users are responsible for the cost of assembly, which has ranged from approximately $250 per 64 ch probe to $400 (US dollars) per 256 ch probe. This provides an inexpensive alternative to commercially available multichannel silicon probes. To quantify the number of probes recently shared using this dissemination method (figure 10), we tallied how many 64, 128, and 256 ch probes were requested and shipped to users by the third-party assembly company (18 month period between July 1 2018 and October 1 2019). To minimize any bias the tally excluded probes shipped to our home institution (UCLA). In figure 10, probes 64D sharp and 64D short are referred to as 64D, and 128AxN sharp is referred to as 128AxN.
Figure 10. Open source silicon microprobe dissemination.
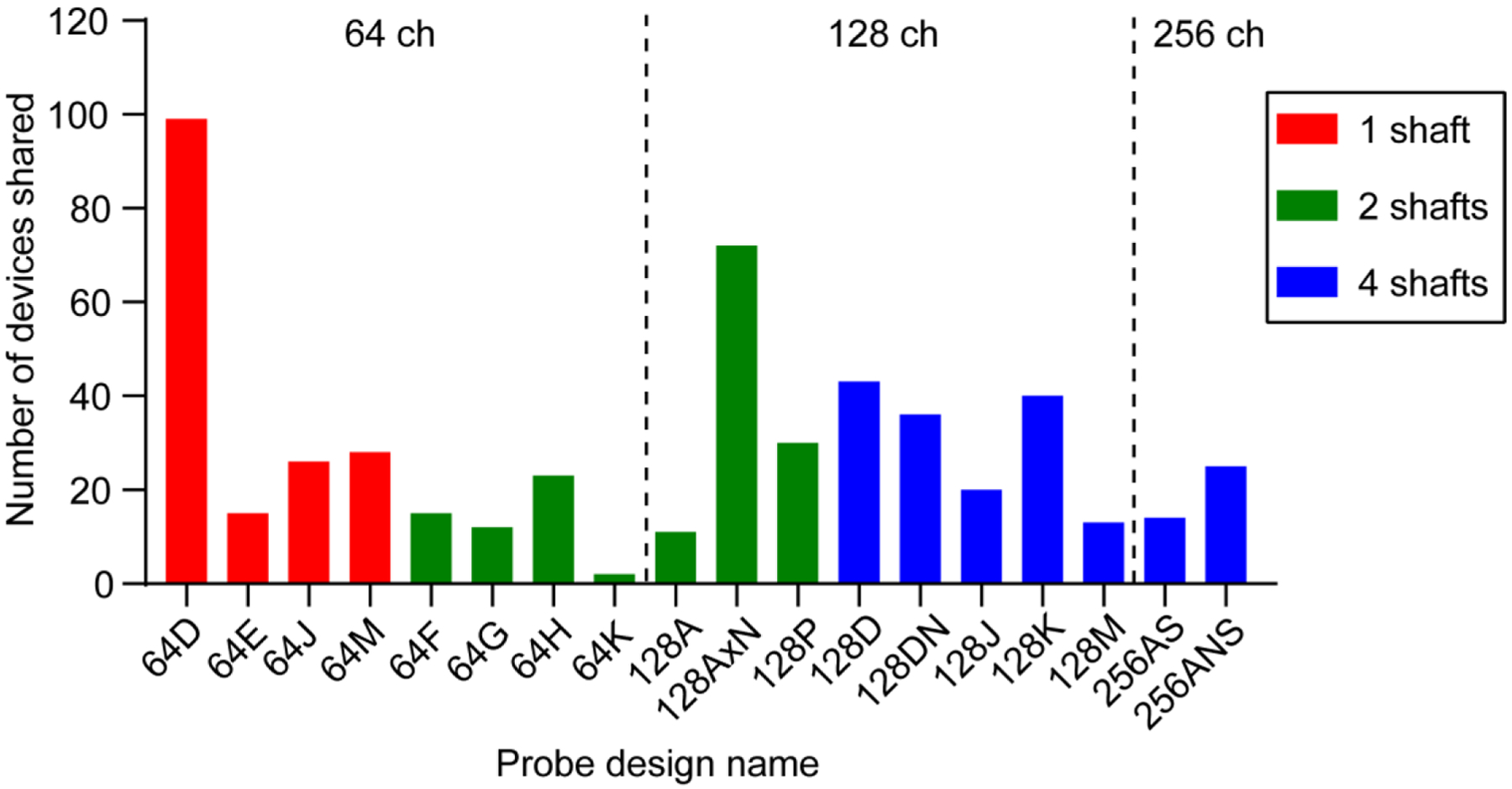
Quantity of each probe design shared with external users (n = 524 total probes). The tally is over an 18 month period from July 1 2018 to October 1 2019. The probes are grouped by electrode number and color coded by the number of shafts.
3. Results
3.1. High electrode density recording in head-fixed mice
We demonstrated recordings with a gold-plated silicon microprobe in a head-fixed mouse preparation (figure 7A). A 64 ch device (64D sharp) comprised of one shaft was inserted in the M2 region of cortex, and recordings were carried out during quiet wakefulness. The shaft was coated with fluorescent DiD to facilitate reconstruction of its position (figure 7B). Spiking activity was observed on several electrodes, and frequently, the same putative unit was detected on multiple electrodes (figure 7C). Consistent with previous measurements [11], spike sorting yielded multiple single units (median yield was 0.4 units per electrode measured across 5 recording sessions in M2). After being properly cleaned, probes can be reused multiple (between 5 and 20) times without requiring new gold plating between recording sessions.
3.2. Simultaneous electrophysiology and optogenetics with opto-microprobes
Optogenetics is a method for optically controlling the activity of genetically defined brain circuits [36]. In neuroscience, optogenetics is frequently used to establish causal relationships between the activity of certain circuits and animal behavior, or to assess interactions between circuits. It is therefore often useful or necessary to combine this method with neural recordings [37–39]. To demonstrate simultaneous electrophysiological recordings and optogenetic manipulations with the opto-microprobe (figure 8A), we virally expressed the excitatory opsin ChR2 or inhibitory opsin eNpHR3.0 in glutamatergic neurons in M2. Application of light led to spiking activity increases in ChR2-expressing animals (figures 8B, 8C), and decreases in eNpHR3.0-expressing animals (figures 8D, 8E). We found that ChR2-mediated excitatory effects were not significantly affected by light intensity (figure 8F), suggesting a saturated response at those light intensities, whereas the eNpH3.0-mediated inhibitory effects were intensity-dependent (figure 8G). We also found a high correlation between each unit’s mean spike waveform in the light off and on conditions (figures 8H, 8I), suggesting that the optical stimulus does not adversely affect the recording quality.
3.3. Simultaneous electrophysiology and fiber photometry with opto-microprobes
Fiber photometry is a widely used method for measuring population-level changes in fluorescence from genetically encoded calcium indicators, reflecting changes in neural activity [40]. While this method has unquestionably provided important insights about brain function, surprisingly, there have only been few published studies comparing photometric signals to electrophysiological activity [41]. To better understand the relationship between these two types of signals, we constructed an opto-microprobe consisting of a 256 ch electrode array and a low autofluorescence fiber (figure 9A). We built a setup allowing simultaneous acquisition of electrophysiological and GCaMP6s fluorescence data with the same DAQ (figure 9B). To examine behaviorally evoked activity, we trained head-fixed mice to perform a self-initiated operant licking task, rewarded with a drop of sweetened condensed milk. Recordings were carried out in well-trained mice with the device inserted in M2, a region that is known to be involved in licking [42]. Electrophysiological measurements yielded a total of 204 putative neurons, which on average showed an increase in firing around the time of licking (figure 9C). The concurrently measured photometry signal (F) was plotted either as the fractional change in fluorescence (ΔF/F), or, as suggested by one study [41], the slope of F. Both of these values increased around the time of licking (figure 9D); however, the slope appeared more time-locked to the mean pattern of spiking activity seen in figure 9C. To quantify the relationship of individual neuron spiking to photometric signals, we calculated the correlation coefficient between each neuron’s firing rate and either ΔF/F or slope of F. We found that, on average, individual neurons were more strongly correlated to the slope of F (figure 9E). Next, we examined whether neurons that were closer to the optical fiber were also more correlated to the photometry signal. We estimated the geometric distance between each neuron and the center of the fiber tip. We found a negative correlation between distance and the similarity of neural firing to the slope of F (figure 9F), but not ΔF/F (figure 9G). Taken together, the results suggest that under the experimental conditions used here, the slope of photometry fluorescence signals is more closely related to electrophysiological spiking activity than the more widely used measure of fractional change in fluorescence. A study performing simultaneous multi-unit recordings and photometry in the striatum reported similar observations [41]. However, since many neurobiological studies report ΔF/F but not the slope of photometry signals, our data appear to show that further characterization of photometric signals via simultaneous electrophysiological recordings is worthwhile.
3.4. Open source silicon microprobe dissemination
Under the open source framework our group has been sharing pre-fabricated microprobes with the research community. To minimize the cost to the end user, we supply the individual components needed for assembly (microprobes, PCBs, and connectors), and ask users to pay for the assembly step. Using this non-commercial dissemination strategy, over an 18 month period, we have shared over 500 microprobes with around 35 different research groups (average rate of 29 probes shared per month). Figure 10 shows the total shared quantity of each type of probe during this time period.
4. Discussion
We have described an open source paradigm for developing mass-produced silicon microprobes. These devices contain as many as 256 electrodes, and provide access to tens to hundreds of neurons in behaving head-fixed mice. We also have described a straightforward process for enhancing their functionality through the addition of optical fibers (“opto-microprobes”), enabling simultaneous electrophysiology and either optogenetics or photometry. We have made all device designs openly available on an online file repository. In a previous publication, we also described a process for assembling these devices into 3D structures targeting multiple brain areas with as many as 1,024 electrodes [11]. Following the tradition of early pioneers of silicon-based neural probes [1], we have made these tools openly accessible through non-commercial dissemination methods. This effort currently hinges on the support of funding which specifically promotes neurotechnology dissemination (see Acknowledgments). We envision that in the long term, this effort could be sustained without commercialization by pooling resources from several user groups to fund SOI wafer fabrication and PCB production.
Through our own work, as well as dissemination to other users, our silicon microprobes have contributed to at least 18 neuroscience studies related to the visual [43–48] and auditory [49] system, cortical plasticity [50], temporal processing [51], reward conditioning [52, 53], neural circuit connectivity [54, 55], interneuron function [35], astrocyte-neuron interactions [56, 57], and models of addiction [58], autism [59], and epilepsy [60]. Thus, we anticipate this work will provide a valuable resource or reference guide for both current and prospective users of silicon microprobes.
In this study, the evaluation of silicon microprobe performance focused on acute head-fixed animal recordings, which are widely used in neuroscience [61]. However, many applications require long-term recordings in freely moving animals, which present additional challenges for electrode performance [62, 63]. Other silicon-based neural probes have been successfully used chronically [9, 64–67], although it is thought that softer polymer-based devices [17] or smaller and less invasive implants [16, 68, 69] may provide superior long-term recording performance compared to silicon. The lack of extensively characterized chronic recording capabilities is a potential drawback of our technology over other electrode-based tools. On the other hand, some users are already evaluating probes attached to the 32 ch PCB (figure 5D) in freely moving animals.
A further potential pitfall of our devices is that the light source on the opto-microprobes is not monolithically integrated with the multielectrode array, as found on some other probes [19]. Nevertheless, our opto-microprobes are still useful for applications which do not require precise spatial control of light [35]. Another limitation is that the microprobes, at least as currently manufactured, are unlikely to be suitable for electrical stimulation requiring application of large currents through the electrodes.
One of the most significant advances in silicon-based neural probe technology in recent years has been the development of CMOS-based devices, which can dramatically increase the number of recording channels and reduce the wiring requirements relative to passive probes [18]. Examples include the Neuropixels probe with 384 configurable channels from a total of 960 recording sites on a single shaft [9] and Neuroseeker probes with 1,356 channels [70, 71]. Furthermore, the Neuropixels probes are already available to the community at a cost of ~ $1,100 [28]. By comparison, our silicon microprobes, which have passive wiring, offer a smaller number of electrodes, but at a lower cost to the end user (~ $400 for a 256 ch probe). The acute recording performance per individual channel on our silicon microprobes and Neuropixels probes appears comparable in head-fixed animals. With our 256 ch microprobes we reported an average yield of 115 units in the mouse striatum (0.45 units per ch) [52], which is similar to the yield reported with Neuropixels probes in this brain structure [9]. Moreover, based on our experiences with disseminating probes, some users do not appear to consider the channel count as the main selection criterion. In fact, as seen in figure 10, many users actually preferred our 64 ch over 128 or 256 ch designs, despite a relatively small difference in assembly cost between those probes. A further insight from figure 10 is that despite identifying some popular designs (64D sharp, 128AxN sharp, and 128D), no single probe could meet the needs of all users, as researchers chose among all of the ~ 18 available probe designs. This suggests that, rather than the entire community converging toward a single all-purpose neural probe, having a diverse ecosystem of devices varying in channel count, array geometry, length, mechanical stiffness, cost, and additional functionalities, remains crucial for advancing our understanding of brain function in health and disease.
Acknowledgements
We are grateful to Suresh Sampath at IMT for device fabrication, Will Lee at IDAX for assembly, and Reid Harrison at Intan for developing a compatible amplifier and electroplating board. We thank Konstantin Bakhurin, Leslie Claar, and Justin Shobe for their invaluable role in the initial development of these probes. We thank Tim Blanche, Anatol Bragin, Stephen David, Peyman Golshani, Nick Lesica, Michael Long, Markus Meister, Mu Qiao, and Michael Stryker for contributing microprobe design ideas. We also thank the dozens of users who have provided valuable feedback on these devices, and Tom Davidson for providing technical assistance on photometry. This work was supported by NIH grants NS100050, NS096994, DA042739, DA005010, and NSF NeuroNex Award 1707408. The open source dissemination of these silicon microprobes is currently funded by NSF NeuroNex Award 1707408.
Footnotes
Data availability statement
Design files and other resources related to silicon microprobe development, use, and sharing of pre-fabricated devices are available online at https://github.com/sotmasman/Silicon-microprobes. Recorded electrophysiological datasets are available from the corresponding author upon request.
Disclosures
The authors declare the absence of any conflict of interest.
References
- 1.Wise KD, Sodagar AM, Yao Y, Gulari MN, Perlin GE, and Najafi K, Microelectrodes, microelectronics, and implantable neural microsystems. Proceedings of the Ieee, 2008. 96(7): p. 1184–1202. [Google Scholar]
- 2.Wise KD and Angell JB, Low-Capacitance Multielectrode Probe for Use in Extracellular Neurophysiology. Ieee Transactions on Biomedical Engineering, 1975. Bm22(3): p. 212–219. [DOI] [PubMed] [Google Scholar]
- 3.Seymour JP and Kipke DR, Neural probe design for reduced tissue encapsulation in CNS. Biomaterials, 2007. 28(25): p. 3594–607. [DOI] [PubMed] [Google Scholar]
- 4.Blanche TJ, Spacek MA, Hetke JF, and Swindale NV, Polytrodes: high-density silicon electrode arrays for large-scale multiunit recording. J Neurophysiol, 2005. 93(5): p. 2987–3000. [DOI] [PubMed] [Google Scholar]
- 5.Du J, Blanche TJ, Harrison RR, Lester HA, and Masmanidis SC, Multiplexed, high density electrophysiology with nanofabricated neural probes. PLoS One, 2011. 6(10): p. e26204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buzsaki G, Large-scale recording of neuronal ensembles. Nature Neuroscience, 2004. 7(5): p. 446–451. [DOI] [PubMed] [Google Scholar]
- 7.Cheung KC, Djupsund K, Dan Y, and Lee LP, Implantable multichannel electrode array based on SOI technology. Journal of Microelectromechanical Systems, 2003. 12(2): p. 179–184. [Google Scholar]
- 8.Stevenson IH and Kording KP, How advances in neural recording affect data analysis. Nature Neuroscience, 2011. 14(2): p. 139–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jun JJ, Steinmetz NA, Siegle JH, Denman DJ, Bauza M, Barbarits B, Lee AK, Anastassiou CA, Andrei A, Aydin C, Barbic M, Blanche TJ, Bonin V, Couto J, Dutta B, Gratiy SL, Gutnisky DA, Hausser M, Karsh B, Ledochowitsch P, Lopez CM, Mitelut C, Musa S, Okun M, Pachitariu M, Putzeys J, Rich PD, Rossant C, Sun WL, Svoboda K, Carandini M, Harris KD, Koch C, O’Keefe J, and Harris TD, Fully integrated silicon probes for high-density recording of neural activity. Nature, 2017. 551(7679): p. 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berenyi A, Somogyvari Z, Nagy AJ, Roux L, Long JD, Fujisawa S, Stark E, Leonardo A, Harris TD, and Buzsaki G, Large-scale, high-density (up to 512 channels) recording of local circuits in behaving animals. J Neurophysiol, 2014. 111(5): p. 1132–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shobe JL, Claar LD, Parhami S, Bakhurin KI, and Masmanidis SC, Brain activity mapping at multiple scales with silicon microprobes containing 1,024 electrodes. Journal of Neurophysiology, 2015. 114(3): p. 2043–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rios G, Lubenov EV, Chi D, Roukes ML, and Siapas AG, Nanofabricated Neural Probes for Dense 3-D Recordings of Brain Activity. Nano Lett, 2016. 16(11): p. 6857–6862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scholvin J, Kinney JP, Bernstein JG, Moore-Kochlacs C, Kopell N, Fonstad CG, and Boyden ES, Close-Packed Silicon Microelectrodes for Scalable Spatially Oversampled Neural Recording. IEEE Trans Biomed Eng, 2016. 63(1): p. 120–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seidl K, Torfs T, De Maziere PA, Van Dijck G, Csercsa R, Dombovari B, Nurcahyo Y, Ramirez H, Van Hulle MM, Orban GA, Paul O, Ulbert I, Neves H, and Ruther P, Control and data acquisition software for high-density CMOS-based microprobe arrays implementing electronic depth control. Biomed Tech (Berl), 2010. 55(3): p. 183–91. [DOI] [PubMed] [Google Scholar]
- 15.Kozai TDY, Marzullo TC, Hooi F, Langhals NB, Majewska AK, Brown EB, and Kipke DR, Reduction of neurovascular damage resulting from microelectrode insertion into the cerebral cortex using in vivo two-photon mapping. Journal of Neural Engineering, 2010. 7(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel PR, Zhang H, Robbins MT, Nofar JB, Marshall SP, Kobylarek MJ, Kozai TD, Kotov NA, and Chestek CA, Chronic in vivo stability assessment of carbon fiber microelectrode arrays. J Neural Eng, 2016. 13(6): p. 066002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung JE, Joo HR, Fan JL, Liu DF, Barnett AH, Chen S, Geaghan-Breiner C, Karlsson MP, Karlsson M, Lee KY, Liang H, Magland JF, Pebbles JA, Tooker AC, Greengard LF, Tolosa VM, and Frank LM, High-Density, Long-Lasting, and Multi-region Electrophysiological Recordings Using Polymer Electrode Arrays. Neuron, 2019. 101(1): p. 21–31 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruther P and Paul O, New approaches for CMOS-based devices for large-scale neural recording. Curr Opin Neurobiol, 2015. 32: p. 31–7. [DOI] [PubMed] [Google Scholar]
- 19.Wu F, Stark E, Ku PC, Wise KD, Buzsaki G, and Yoon E, Monolithically Integrated muLEDs on Silicon Neural Probes for High-Resolution Optogenetic Studies in Behaving Animals. Neuron, 2015. 88(6): p. 1136–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scharf R, Tsunematsu T, McAlinden N, Dawson MD, Sakata S, and Mathieson K, Depth-specific optogenetic control in vivo with a scalable, high-density muLED neural probe. Sci Rep, 2016. 6: p. 28381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuzum D, Takano H, Shim E, Reed JC, Juul H, Richardson AG, de Vries J, Bink H, Dichter MA, Lucas TH, Coulter DA, Cubukcu E, and Litt B, Transparent and flexible low noise graphene electrodes for simultaneous electrophysiology and neuroimaging. Nat Commun, 2014. 5: p. 5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Segev E, Reimer J, Moreaux LC, Fowler TM, Chi D, Sacher WD, Lo M, Deisseroth K, Tolias AS, Faraon A, and Roukes ML, Patterned photostimulation via visible-wavelength photonic probes for deep brain optogenetics. Neurophotonics, 2017. 4(1): p. 011002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li B, Lee K, Masmanidis SC, and Li M, A nanofabricated optoelectronic probe for manipulating and recording neural dynamics. J Neural Eng, 2018. 15(4): p. 046008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reddy JW, Kimukin I, Stewart LT, Ahmed Z, Barth AL, Towe E, and Chamanzar M, High Density, Double-Sided, Flexible Optoelectronic Neural Probes With Embedded muLEDs. Front Neurosci, 2019. 13: p. 745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seidl K, Spieth S, Herwik S, Steigert J, Zengerle R, Paul O, and Ruther P, In-plane silicon probes for simultaneous neural recording and drug delivery. Journal of Micromechanics and Microengineering, 2010. 20(10): p. –. [Google Scholar]
- 26.Chen J, Wise KD, Hetke JF, and Bledsoe SC Jr., A multichannel neural probe for selective chemical delivery at the cellular level. IEEE Trans Biomed Eng, 1997. 44(8): p. 760–9. [DOI] [PubMed] [Google Scholar]
- 27.Bentler C and Stieglitz T, Building wireless implantable neural interfaces within weeks for neuroscientists. Conf Proc IEEE Eng Med Biol Soc, 2017. 2017: p. 1078–1081. [DOI] [PubMed] [Google Scholar]
- 28.Steinmetz NA, Koch C, Harris KD, and Carandini M, Challenges and opportunities for large-scale electrophysiology with Neuropixels probes. Curr Opin Neurobiol, 2018. 50: p. 92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siegle JH, Hale GJ, Newman JP, and Voigts J, Neural ensemble communities: open-source approaches to hardware for large-scale electrophysiology. Curr Opin Neurobiol, 2014. 32C: p. 53–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siegle JH, Lopez AC, Patel YA, Abramov K, Ohayon S, and Voigts J, Open Ephys: an open-source, plugin-based platform for multichannel electrophysiology. J Neural Eng, 2017. 14(4): p. 045003. [DOI] [PubMed] [Google Scholar]
- 31.Pachitariu M, Steinmetz N, Kadir S, Carandini M, and Harris KD, Kilosort: realtime spike-sorting for extracellular electrophysiology with hundreds of channels. bioRxiv, 2016. [Google Scholar]
- 32.Chung JE, Magland JF, Barnett AH, Tolosa VM, Tooker AC, Lee KY, Shah KG, Felix SH, Frank LM, and Greengard LF, A Fully Automated Approach to Spike Sorting. Neuron, 2017. 95(6): p. 1381–1394 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ludwig KA, Langhals NB, Joseph MD, Richardson-Burns SM, Hendricks JL, and Kipke DR, Poly(3,4-ethylenedioxythiophene) (PEDOT) polymer coatings facilitate smaller neural recording electrodes. Journal of Neural Engineering, 2011. 8(1): p. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cassar IR, Yu C, Sambangi J, Lee CD, Whalen JJ 3rd, Petrossians A, and Grill WM, Electrodeposited platinum-iridium coating improves in vivo recording performance of chronically implanted microelectrode arrays. Biomaterials, 2019. 205: p. 120–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee K, Holley SM, Shobe JL, Chong NC, Cepeda C, Levine MS, and Masmanidis SC, Parvalbumin Interneurons Modulate Striatal Output and Enhance Performance during Associative Learning. Neuron, 2017. 93(6): p. 1451–1463 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boyden ES, Zhang F, Bamberg E, Nagel G, and Deisseroth K, Millisecond-timescale, genetically targeted optical control of neural activity. Nature Neuroscience, 2005. 8(9): p. 1263–1268. [DOI] [PubMed] [Google Scholar]
- 37.Buzsaki G, Stark E, Berenyi A, Khodagholy D, Kipke DR, Yoon E, and Wise KD, Tools for probing local circuits: high-density silicon probes combined with optogenetics. Neuron, 2015. 86(1): p. 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rudmann L, Alt MT, Ashouri Vajari D, and Stieglitz T, Integrated optoelectronic microprobes. Curr Opin Neurobiol, 2018. 50: p. 72–82. [DOI] [PubMed] [Google Scholar]
- 39.Sileo L, Bitzenhofer SH, Spagnolo B, Popplau JA, Holzhammer T, Pisanello M, Pisano F, Bellistri E, Maglie E, De Vittorio M, Ruther P, Hanganu-Opatz IL, and Pisanello F, Tapered Fibers Combined With a Multi-Electrode Array for Optogenetics in Mouse Medial Prefrontal Cortex. Front Neurosci, 2018. 12: p. 771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gunaydin LA, Grosenick L, Finkelstein JC, Kauvar IV, Fenno LE, Adhikari A, Lammel S, Mirzabekov JJ, Airan RD, Zalocusky KA, Tye KM, Anikeeva P, Malenka RC, and Deisseroth K, Natural neural projection dynamics underlying social behavior. Cell, 2014. 157(7): p. 1535–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Markowitz JE, Gillis WF, Beron CC, Neufeld SQ, Robertson K, Bhagat ND, Peterson RE, Peterson E, Hyun M, Linderman SW, Sabatini BL, and Datta SR, The Striatum Organizes 3D Behavior via Moment-to-Moment Action Selection. Cell, 2018. 174(1): p. 44–58 e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Komiyama T, Sato TR, O’Connor DH, Zhang YX, Huber D, Hooks BM, Gabitto M, and Svoboda K, Learning-related fine-scale specificity imaged in motor cortex circuits of behaving mice. Nature, 2010. 464(7292): p. 1182–6. [DOI] [PubMed] [Google Scholar]
- 43.Shanks JA, Ito S, Schaevitz L, Yamada J, Chen B, Litke A, and Feldheim DA, Corticothalamic Axons Are Essential for Retinal Ganglion Cell Axon Targeting to the Mouse Dorsal Lateral Geniculate Nucleus. J Neurosci, 2016. 36(19): p. 5252–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ito S, Feldheim DA, and Litke AM, Segregation of Visual Response Properties in the Mouse Superior Colliculus and Their Modulation during Locomotion. J Neurosci, 2017. 37(35): p. 8428–8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dadarlat MC and Stryker MP, Locomotion Enhances Neural Encoding of Visual Stimuli in Mouse V1. J Neurosci, 2017. 37(14): p. 3764–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dyballa L, Hoseini MS, Dadarlat MC, Zucker SW, and Stryker MP, Flow stimuli reveal ecologically appropriate responses in mouse visual cortex. Proc Natl Acad Sci U S A, 2018. 115(44): p. 11304–11309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kissinger ST, Pak A, Tang Y, Masmanidis SC, and Chubykin AA, Oscillatory Encoding of Visual Stimulus Familiarity. J Neurosci, 2018. 38(27): p. 6223–6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McBride EG, Lee SJ, and Callaway EM, Local and Global Influences of Visual Spatial Selection and Locomotion in Mouse Primary Visual Cortex. Curr Biol, 2019. 29(10): p. 1592–1605 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ito S, Si Y, Feldheim DA, and Litke AM, Spectral cues are necessary for encoding the azimuthal map of auditory space in the mouse superior colliculus. bioRxiv, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoseini MS, Rakela B, Flores-Ramirez Q, Hasenstaub AR, Alvarez-Buylla A, and Stryker MP, Transplanted Cells Are Essential for the Induction But Not the Expression of Cortical Plasticity. J Neurosci, 2019. 39(38): p. 7529–7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bakhurin KI, Goudar V, Shobe JL, Claar LD, Buonomano DV, and Masmanidis SC, Differential Encoding of Time by Prefrontal and Striatal Network Dynamics. J Neurosci, 2017. 37(4): p. 854–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bakhurin KI, Mac V, Golshani P, and Masmanidis SC, Temporal correlations among functionally specialized striatal neural ensembles in reward-conditioned mice. J Neurophysiol, 2016. 115(3): p. 1521–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shobe JL, Bakhurin KI, Claar LD, and Masmanidis SC, Selective Modulation of Orbitofrontal Network Activity during Negative Occasion Setting. J Neurosci, 2017. 37(39): p. 9415–9423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nigam S, Shimono M, Ito S, Yeh F-C, Timme N, Myroshnychenko M, Lapish CC, Tosi Z, Hottowy P, Smith WC, Masmanidis SC, Litke AM, Sporns O, and Beggs JM, Rich-club organization in effective connectivity among cortical neurons. Journal of Neuroscience, 2016. 36(3): p. 670–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Choe KY, Sanchez CF, Harris NG, Otis TS, and Mathews PJ, Optogenetic fMRI and electrophysiological identification of region-specific connectivity between the cerebellar cortex and forebrain. Neuroimage, 2018. 173: p. 370–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nagai J, Rajbhandari AK, Gangwani MR, Hachisuka A, Coppola G, Masmanidis SC, Fanselow MS, and Khakh BS, Hyperactivity with Disrupted Attention by Activation of an Astrocyte Synaptogenic Cue. Cell, 2019. 177(5): p. 1280–1292 e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Octeau JC, Gangwani MR, Allam SL, Tran D, Huang S, Hoang-Trong TM, Golshani P, Rumbell TH, Kozloski JR, and Khakh BS, Transient, Consequential Increases in Extracellular Potassium Ions Accompany Channelrhodopsin2 Excitation. Cell Rep, 2019. 27(8): p. 2249–2261 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith WC, Rosenberg MH, Claar LD, Chang V, Shah SN, Walwyn WM, Evans CJ, and Masmanidis SC, Frontostriatal Circuit Dynamics Correlate with Cocaine Cue-Evoked Behavioral Arousal during Early Abstinence. Eneuro, 2016. 3(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lazaro MT, Taxidis J, Shuman T, Bachmutsky I, Ikrar T, Santos R, Marcello GM, Mylavarapu A, Chandra S, Foreman A, Goli R, Tran D, Sharma N, Azhdam M, Dong H, Choe KY, Penagarikano O, Masmanidis SC, Racz B, Xu X, Geschwind DH, and Golshani P, Reduced Prefrontal Synaptic Connectivity and Disturbed Oscillatory Population Dynamics in the CNTNAP2 Model of Autism. Cell Rep, 2019. 27(9): p. 2567–2578 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shuman T, Aharoni D, Cai DJ, Lee CR, Chavlis S, Taxidis J, Flores SE, Cheng K, Javaherian M, Kaba CC, Shtrahman M, Bakhurin KI, Masmanidis S, Khakh BS, Poirazi P, Silva AJ, and Golshani P, Breakdown of spatial coding and neural synchronization in epilepsy. bioRxiv, 2018. [Google Scholar]
- 61.Guo ZV, Hires SA, Li N, O’Connor DH, Komiyama T, Ophir E, Huber D, Bonardi C, Morandell K, Gutnisky D, Peron S, Xu NL, Cox J, and Svoboda K, Procedures for behavioral experiments in head-fixed mice. PLoS One, 2014. 9(2): p. e88678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ward MP, Rajdev P, Ellison C, and Irazoqui PP, Toward a comparison of microelectrodes for acute and chronic recordings. Brain Research, 2009. 1282: p. 183–200. [DOI] [PubMed] [Google Scholar]
- 63.Schwartz AB, Cortical neural prosthetics. Annu Rev Neurosci, 2004. 27: p. 487–507. [DOI] [PubMed] [Google Scholar]
- 64.Vetter RJ, Williams JC, Hetke JF, Nunamaker EA, and Kipke DR, Chronic neural recording using silicon-substrate microelectrode arrays implanted in cerebral cortex. IEEE Trans Biomed Eng, 2004. 51(6): p. 896–904. [DOI] [PubMed] [Google Scholar]
- 65.Bragin A, Hetke J, Wilson CL, Anderson DJ, Engel J Jr., and Buzsaki G, Multiple site silicon-based probes for chronic recordings in freely moving rats: implantation, recording and histological verification. J Neurosci Methods, 2000. 98(1): p. 77–82. [DOI] [PubMed] [Google Scholar]
- 66.Grand L, Wittner L, Herwik S, Gothelid E, Ruther P, Oscarsson S, Neves H, Dombovari B, Csercsa R, Karmos G, and Ulbert I, Short and long term biocompatibility of NeuroProbes silicon probes. Journal of Neuroscience Methods, 2010. 189(2): p. 216–229. [DOI] [PubMed] [Google Scholar]
- 67.Chauviere L, Pothof F, Gansel KS, Klon-Lipok J, Aarts AAA, Holzhammer T, Paul O, Singer WJ, and Ruther P, In vivo Recording Quality of Mechanically Decoupled Floating Versus Skull-Fixed Silicon-Based Neural Probes. Front Neurosci, 2019. 13: p. 464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Patel PR, Na K, Zhang H, Kozai TD, Kotov NA, Yoon E, and Chestek CA, Insertion of linear 8.4 mum diameter 16 channel carbon fiber electrode arrays for single unit recordings. J Neural Eng, 2015. 12(4): p. 046009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kozai TD, Langhals NB, Patel PR, Deng X, Zhang H, Smith KL, Lahann J, Kotov NA, and Kipke DR, Ultrasmall implantable composite microelectrodes with bioactive surfaces for chronic neural interfaces. Nat Mater, 2012. 11(12): p. 1065–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Raducanu BC, Yazicioglu RF, Lopez CM, Ballini M, Putzeys J, Wang S, Andrei A, Rochus V, Welkenhuysen M, Helleputte NV, Musa S, Puers R, Kloosterman F, Hoof CV, Fiath R, Ulbert I, and Mitra S, Time Multiplexed Active Neural Probe with 1356 Parallel Recording Sites. Sensors (Basel), 2017. 17(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dimitriadis G, Neto JP, Aarts A, Alexandru A, Ballini M, Battaglia F, Calcaterra L, David F, Fiath R, Frazao J, Geerts JP, Gentet LJ, Helleputte NV, Holzhammer T, Hoof C.v., Horvath D, Lopes G, Lopez CM, Maris E, Marques-Smith A, Marton G, McNaughton BL, Meszena D, Mitra S, Musa S, Neves H, Nogueira J, Orban GA, Pothof F, Putzeys J, Raducanu B, Ruther P, Schroeder T, Singer W, Tiesinga P, Ulbert I, Wang S, Welkenhuysen M, and Kampff AR, Why not record from every channel with a CMOS scanning probe? bioRxiv, 2019. [Google Scholar]


