Abstract
To assess the accuracy and physiological relevance of circulating microRNA (miRNA) as a biomarker of pediatric concussion, we compared changes in salivary miRNA and cerebrospinal fluid (CSF) miRNA concentrations after childhood traumatic brain injury (TBI). A case-cohort design was used to compare longitudinal miRNA concentrations in CSF of seven children with severe TBI against three controls without TBI. The miRNAs “altered” in CSF were interrogated in saliva of 60 children with mild TBI and compared with 18 age- and sex-matched controls. The miRNAs with parallel changes (Wilcoxon rank sum test) in CSF and saliva were interrogated for predictive accuracy of TBI status using a multivariate regression technique. Spearman rank correlation identified relationships between miRNAs of interest and clinical features. Functional analysis with DIANA mirPath identified related mRNA pathways. There were 214 miRNAs detected in CSF, and 135 (63%) were also present in saliva. Six miRNAs had parallel changes in both CSF and saliva (miR-182-5p, miR-221-3p, mir-26b-5p, miR-320c, miR-29c-3p, miR-30e-5p). These miRNAs demonstrated an area under the curve of 0.852 for identifying mild TBI status. Three of the miRNAs exhibited longitudinal trends in CSF and/or saliva after TBI, and all three targeted mRNAs related to neuronal development. Concentrations of miR-320c were directly correlated with child and parent reports of attention difficulty. Salivary miRNA represents an easily measured, physiologically relevant, and accurate potential biomarker for TBI. Further studies assessing the influence of orthopedic injury and exercise on peripheral miRNA patterns are needed.
Keywords: biomarker, concussion, microRNA, pediatrics, saliva
Introduction
Three million concussions occur in the United States each year. Two-thirds occur in children and adolescents,1 and more than 80% result from mild traumatic brain injury (mTBI).2 Many pediatricians report that clinical tools for patient treatment and family education are lacking.3 Clinicians currently have no standard method for pediatric concussion diagnosis or prognosis.
Despite important biomechanical differences in the developing brain (including skull geometry, brain water content, and myelination),4 current concussion guidelines rely heavily on adult studies.2,5 The 2012 Consensus Statement on Concussion in Sport recognized that children report concussion symptoms differently than adults5 and recommended that age-appropriate symptom checklists be administered to children, parents, teachers, and caregivers. Unfortunately, the feasibility of administering and scoring numerous age-specific questionnaires within the time of a typical clinical encounter makes adopting these recommendations difficult.3 As a result, researchers have begun to explore biomarker approaches to concussion diagnosis.
An emerging class of biomarkers includes micro-ribonucleic acids (miRNAs), noncoding fragments of RNA that inhibit protein translation through targeted degradation of mRNA transcripts.6 The abundance, stability, and essential regulatory role of miRNAs make them ideal biomarker candidates.7 Studies of rat models8 and human adults9,10 have reported changes in peripheral miRNA expression after TBI. These peripheral changes mirror miRNA patterns in the central nervous system (CNS).11,12 In 18 adults with severe TBI (sTBI), serum levels of miR-16, miR-92a, and miR-765 were predictive of injury and demonstrated 100% accuracy for differentiating control and TBI participants.10 Another study involving nine adults with mTBI and comorbid post-traumatic stress disorder looked at miRNA expression in peripheral blood mononuclear cells.9 The authors identified four miRNAs significantly down-regulated in the mTBI group (miR-671-5p, miR-1285, miR-455-3p, and miR-US4) and three noncoding RNAs with 89% classification accuracy for differentiating mTBI participants from controls.
Still, no study has specifically examined miRNA expression patterns in pediatric subjects with concussion. Further, no consensus has been reached on the ideal bio-fluid for miRNA concussion assessment. Given the robust, stabile expression of miRNA in saliva,13,14 its ease of collection in pediatric patients, and our previous experience demonstrating its biomarker utility in disorders of the CNS,15 we sought to explore the utility of salivary miRNA as a diagnostic tool in children and adolescents with TBI. This study tests the hypotheses that salivary miRNA profiles of children and adolescents with mTBI: (1) reflect cerebrospinal fluid (CSF) profiles in children and adolescents with sTBI; (2) accurately identify the presence of mTBI; and (3) differ from adult miRNA biomarkers of mTBI.
Methods
This study was approved by the Independent Review Board at the Penn State College of Medicine. Written informed consent was obtained from the parents or guardians of all children who served as subjects of the investigation and, when appropriate, assent from the subjects themselves was obtained.
sTBI recruitment and sample collection
The CSF samples collected previously for a study of F2-isoprostane levels in children and adolescents with sTBI16 were utilized for a longitudinal characterization of CSF miRNA. Briefly, ventricular CSF samples collected from eight children with sTBI were selected at random for the current study. To remove sample selection bias, researchers were blind to participant characteristics before sample selection. The selected cohort included children ages 4–17 years with a Glasgow Coma Score (GCS) <8 and a clinically indicated extraventricular drain (EVD) for increased intracranial pressure after sTBI. Mechanisms of injury included fall and motor vehicle crash. CSF was extracted passively from each subject's EVD in a sterile fashion at three times after injury: day 1, day 4–7, and day 8–17. Age, sex, mechanism of injury, and times of collection were recorded for each subject (Supplementary Table 1; see online supplementary material at ftp.liebertpub.com). Control CSF included 12 samples taken at one time point from three subjects (ages 1–8 years) undergoing clinically indicated spinal tap for epilepsy, or as part of a rule-out-sepsis protocol. All CSF was freshly aliquoted and frozen immediately at −80C. Samples did not undergo any freeze/thaw cycles and therefore remained frozen continuously from collection time until time of the present analysis.
mTBI recruitment and sample collection
Salivary miRNA profiles were investigated in subjects (age 5–21 years) with or without a clinical diagnosis of mTBI. The mTBI cohort included 61 children and adolescents presenting to the Penn State Hershey Medical Center for evaluation of mTBI within 14 days of initial injury. The 14-day cutoff was chosen based on previous investigations that suggested most clinical symptoms and biomarker profiles return to baseline within two weeks of concussion.17 Exclusion criteria for the mTBI group included GCS <12, clinical diagnosis of sTBI, penetrating head injury, skull fracture, intracranial bleed, or symptoms attributable to underlying psychological disorder (e.g., depression or anxiety). The control cohort included 19 children and adolescents presenting to the Penn State Hershey Medical Center Primary Pediatrics Clinic for a regularly scheduled well child visit. Exclusion criteria for this group included a history of concussion, ongoing rheumatological condition, or recent orthopedic injury. Subjects with periodontal disease, upper respiratory infection, seizure disorder, intellectual disability, history of migraine headaches, or drug/alcohol use disorder were excluded from both groups.
Saliva samples were collected from each participant at the time of enrollment in a nonfasting state after an oral tap-water rinse through expectoration into an Oragene RE-100 saliva collection kit (DNA Genotek; Ottawa, Canada). Samples were shaken by hand 5–10 times and stored at room temperature for up to 10 days before transfer into a 4°C refrigerator. Because of circadian variations in miRNA levels, time of day was recorded for all saliva collections. There was no difference in average collection time between control and mTBI groups.
Medical and demographic characteristics
Medical and demographic information was collected from both mTBI and control participants, including: age, sex, race/ethnicity, height, weight, medical history, and medications (Supplementary Table 2; see online supplementary material at ftp.liebertpub.com). Given the potential impact of underlying psychological conditions on mTBI symptom reports, selective serotonin reuptake inhibitor use was recorded for all participants. Food/medical allergies and dietary restrictions were also recorded to assess potential influence of oropharyngeal status on the salivary miRNA profile. The mTBI cohort reported history of concussions, details of current concussion (days since injury, mechanism, associated emesis, weakness, amnesia, fractures, or loss of consciousness), and time of last analgesic use (nonsteroidal anti-inflammatory or acetaminophen). Finally, mTBI subjects and their parent/guardian completed an inventory of concussive symptoms using the child sport concussion assessment tool (SCAT-3).
RNA processing and quantification
The CSF samples were stored at −80°C and saliva samples were stored at −20°C before RNA extraction. RNA was extracted from saliva and CSF samples using a Norgen Circulating and Exosomal RNA Purification Kit (Norgen Biotek, Ontario, Canada) as we have reported previously.18 Samples of saliva or CSF were combined with lysis buffer, Norgen Slurry C2 solution, and β-mercaptoethanol and vortexed for 15 sec per manufacturer instructions. The solution was incubated at 60°C for 10 min, before adding 3 mL of 96% ethanol. Next, each sample was centrifuged for 30 sec at 1000 rotations per minute, followed by an additional 60°C lysis step and ethanol incubation. Approximately 650 μL of this mixture was centrifuged for 1 min at 14,000 RPM. The minifilter spin column then underwent three washes for 1 min at 14,000 RPM before a final two-step elution procedure: 2 min at 2000 RPM, followed by 3 min at 14,000 RPM. Final RNA concentrations were quantified with a Nanodrop Spectrophotometer, and extracted RNA was stored at −80°C before sequencing. The RNA yield and quality were assessed with the Agilent 2100 Bioanalyzer before library construction.
Sequencing of salivary RNA occurred at the Penn State Genomics Core Facility using a NEXTflex Small RNA-Seq Kit v3 (Bioo Scientific; Austin, TX), an Illumina HiSeq 2500 Instrument, and a targeted depth of three million reads per sample. The CSF RNA samples were sequenced at the SUNY Molecular Analysis Core at Upstate Medical University using an Illumina TruSeq Small RNA Sample Prep protocol (Illumina; San Diego, CA), an Illumina MiSeq instrument, and a targeted depth of three million reads per sample. Reads for all samples were aligned to the hg38 build of the human genome in Partek Flow (Partek; St. Louis, MO) using the SHRiMP2 aligner. Local alignment was performed with a match window length of 100% and gap open scores of −255. Single best mapping was completed with a maximum of 10 hits per read and ungapped alignment. Total miRNA counts within each sample were quantified with miRBase precursor and mature-microRNA v21. Saliva samples with less than 5 × 103 total counts were excluded from the final analysis, resulting in 60 mTBI and 18 control saliva samples.
Only miRNAs with raw read counts greater than 10 in at least 25% of samples were evaluated in the differential expression analysis for CSF and saliva, respectively. The miRNAs present in 25% of sTBI CSF samples and absent from all control CSF samples were also investigated as “up-regulated” miRNAs. Before statistical analysis, read counts were sum-normalized, mean-centered, and divided by the standard deviation of each variable. The data set supporting the results of this article will be made available in the NCBI Sequence Read Archive.
Statistical analysis
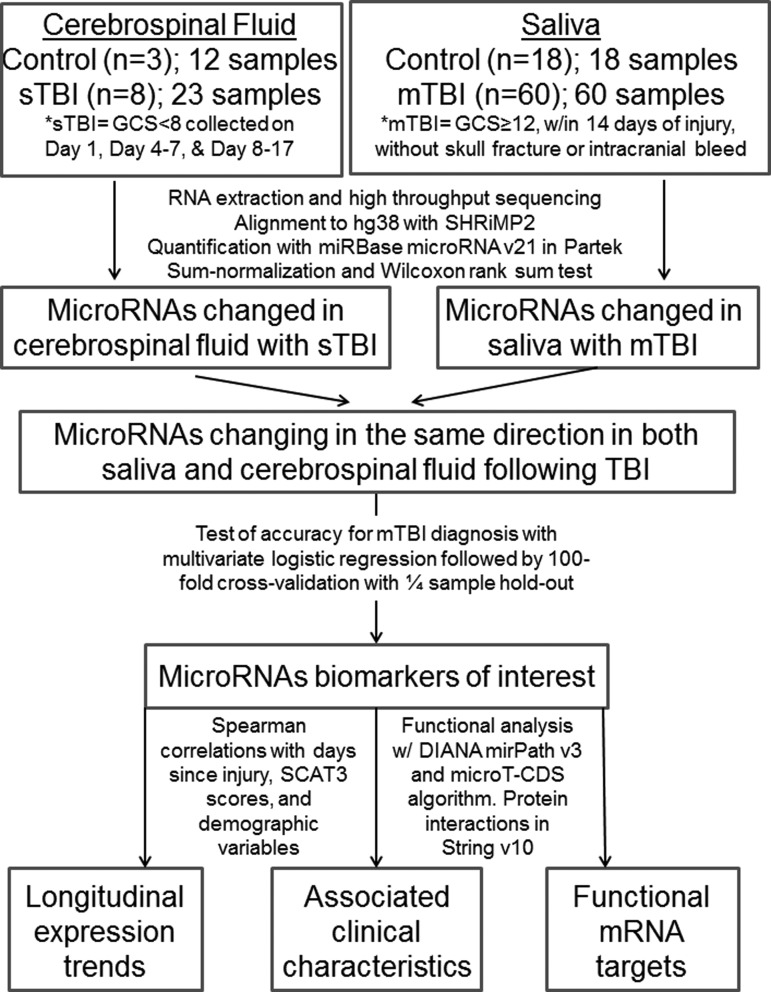
The miRNAs with the greatest physiological relevance as concussion biomarkers were identified using a three-step procedure: (1) The miRNAs present only in sTBI CSF samples (absent in control CSF samples) or miRNAs with “altered” concentrations in sTBI CSF (measured as reads per million; RPM) were identified with a nonparametric Wilcoxon rank sum test with Benjamini Hochberg false detection rate (FDR) correction; (2) concentrations (RPM) of these miRNA targets were investigated in mTBI saliva samples (compared with control saliva) using a Wilcoxon rank sum test; (3) the miRNAs altered in both CSF and saliva TBI samples were examined for parallel up- or down-regulation relative to controls (Fig. 1). For each step, “altered” miRNAs were those with raw p value <0.05. The miRNAs of interest were inspected for longitudinal trends in both CSF and saliva concussion samples using a Spearman rank correlation metric (correlating miRNA concentrations with days since injury). The diagnostic accuracy of these biomarker prospects was assessed with a multivariate logistic regression analysis, and results were visualized with a receiver operating characteristic curve. To avoid “overmodeling” of the dataset and to ensure that the miRNA biomarkers accurately differentiated control and mTBI subjects, we employed a secondary approach involving a 100-fold Monte-Carlo Cross Validation technique alongside a random 1/4 sample hold-out procedure in Metaboanalyst software.18 Relationships between medical/demographic characteristics and salivary miRNAs of interest were examined with Spearman rank correlations. Analysis of medical and demographic data across mTBI and control groups was accomplished with a two-tailed Student t test.
FIG. 1.
Methodological pipeline for identifying accurate and physiologically relevant microribonucleic acid markers of concussion. sTBI, severe traumatic brain injury; GCS, Glasgow Coma Scale; mTBI, mild traumatic brain injury; RNA, ribonucleic acid;
Functional analysis
The miRNA biomarkers of mTBI underwent functional annotation analysis in DIANA mirPath v3 online software (http://snf-515788.vm.okeanos.grnet.gr/) using the microT-CDS algorithm to identify species-specific mRNA targets.19 DIANA mirPath identified gene ontology (GO) categories with significant (FDR <0.05) target enrichment using a Fisher exact test. A list of high confidence mRNA targets (microT-CDS score ≥0.99) was interrogated for protein-protein interaction networks using moderate stringency settings (interaction score >0.40) in String v10 software (http://string-db.org).20
Results
Medical and demographic characteristics
There was no difference in participant age (p = 0.48), sex (p = 0.27), or race/ethnicity (% white; p = 0.70) between the mTBI and control groups (Supplementary Table 2; see online supplementary material at ftp.liebertpub.com). There was no difference in the percentage of participants with food/medicine allergies (p = 0.63), dietary restrictions (p = 0.79), or anti-depressant medications (p = 0.88). The mTBI group was significantly taller (p = 0.002) and had utilized nonsteroidal anti-inflammatory medications (p = 0.001), and acetaminophen (p = 0.003) with a higher frequency in the 6 h before saliva collection. Salivary collection for mTBI participants occurred, on average, 6.5 days post-concussion.
The most common mechanisms of injury for this group included sport-related injury (59%), motor vehicle crash (18%), and fall (16%). Post-concussive symptoms within the mTBI group included loss of consciousness (25%), emesis (21%), weakness (31%), and memory loss (44%). The mean SCAT-3 score for mTBI participants was 23.7 on child report and 21.8 on parental report, with an average of 11 symptoms per participant. Symptoms lasted beyond four weeks in 66% of mTBI participants, and 43% reported a history of concussion.
CSF miRNA in sTBI
There was more robust miRNA expression in CSF after sTBI (mean aligned miRNA reads per sample = 565,805) than in control CSF (22,885 aligned reads per sample). The known 2813 mature human miRNAs were interrogated, and 214 (7.6%) were present in CSF samples (Supplementary Table 3; see online supplementary material at ftp.liebertpub.com). One-hundred and fourteen of those miRNAs had nominal differences in expression (p < 0.05), and 86 had significant changes (FDR <0.05) between sTBI and control groups. Seventy-two were down-regulated and 42 were up-regulated in sTBI.
Salivary miRNA in mTBI
There were 214 salivary miRNAs with robust expression across both control and mTBI samples (Supplementary Table 4; see online supplementary material at ftp.liebertpub.com). Forty of the miRNAs measured in saliva had nominal differences in normalized read counts (p < 0.05), and 10 had significant differences between control and mTBI groups (FDR <0.05). Nine of the miRNAs were down-regulated in mTBI saliva, and 31 were up-regulated.
Combined analysis of CSF and salivary miRNAs
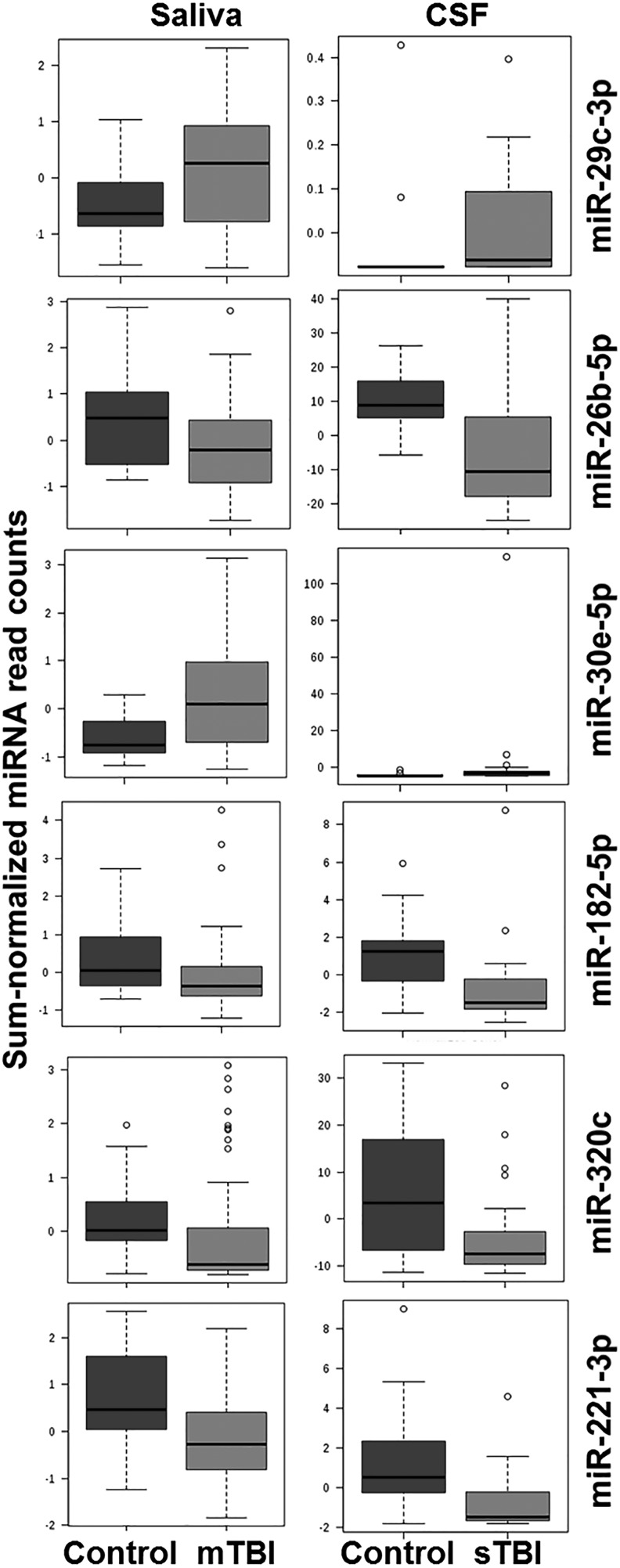
Of the 214 miRNAs detected in CSF, 135 (63%) were also present in saliva. Of the 114 miRNAs with nominal changes in the CSF of sTBI subjects, 64 (56%) were present in saliva, and 10 (8.7%) demonstrated nominal differences in the mTBI group. Six of these 10 miRNAs have been reported in previous serum studies of adults with TBI.10,12,21 None of the miRNAs have overlapping seed sequences. Six miRNAs were altered in the same direction in both saliva and CSF TBI samples (Table 1). Four were down-regulated (miR-182-5p, miR-221-3p, mir-26b-5p, miR-320c) and two (miR-29c-3p, miR-30e-5p) were up-regulated (Fig. 2).
Table 1.
miRibonucleic Acids Altered in Both Cerebrospinal Fluid and Saliva after Traumatic Brain Injury
| MicroRNA | Seed sequence | CSF | Saliva | Previous study |
|---|---|---|---|---|
| hsa-let-7f-5p | GAGGUAG | ↑ | ↓ | Mitra et al., 2017 |
| hsa-miR-151a-5p | CGAGGAG | ↑ | ↓ | |
| hsa-miR-182-5p | UUGGCAA | ↓ | ↓ | Mitra et al., 2017 |
| hsa-miR-221-3p | GCUACAU | ↓ | ↓ | Redell et al., 2010 |
| hsa-miR-26b-5p | UCAAGUA | ↓ | ↓ | Redell et al., 2010 |
| hsa-miR-29c-3p | AGCACCA | ↑ | ↑ | Bhomia et al., 2016 |
| hsa-miR-30e-5p | GUAAACA | ↑ | ↑ | |
| hsa-miR-320c | AAAGCUG | ↓ | ↓ | Redell et al., 2010 |
| hsa-miR-532-5p | AUGCCUU | ↑ | ↓ | |
| hsa-miR-744-5p | GCGGGGC | ↑ | ↓ |
microRNA, microribonucleic acid; CSF, cerebrospinal fluid.
Arrows indicate direction of change in traumatic brain injury samples.
FIG. 2.
Whisker box plots depicting mean concentrations in cerebrospinal fluid (CSF) and saliva for the six microribonucleic acids (miRNAs) of interest across concussion and control groups. Nominally significant changes were detected for miR-29c-3p (CSF p = 0.032; saliva p = 0.008), miR-26b-5p (CSF p = 0.003; saliva p = 0.016), miR-30e-5p (CSF p = 0.045; saliva p = 0.009), miR-182-5p (CSF p = 0.009; saliva p = 0.013), miR-320c (CSF p = 0.037; saliva p = 0.016), and miR-221-3p (CSF p = 0.014; saliva p = 0.005) with Wilcoxon rank sum testing. False detection rate correction was ≤0.15 for all six miRNAs. mTBI, mild traumatic brain injury; sTBI, severe traumatic brain injury.
Predictive accuracy of miRNA biomarker panel
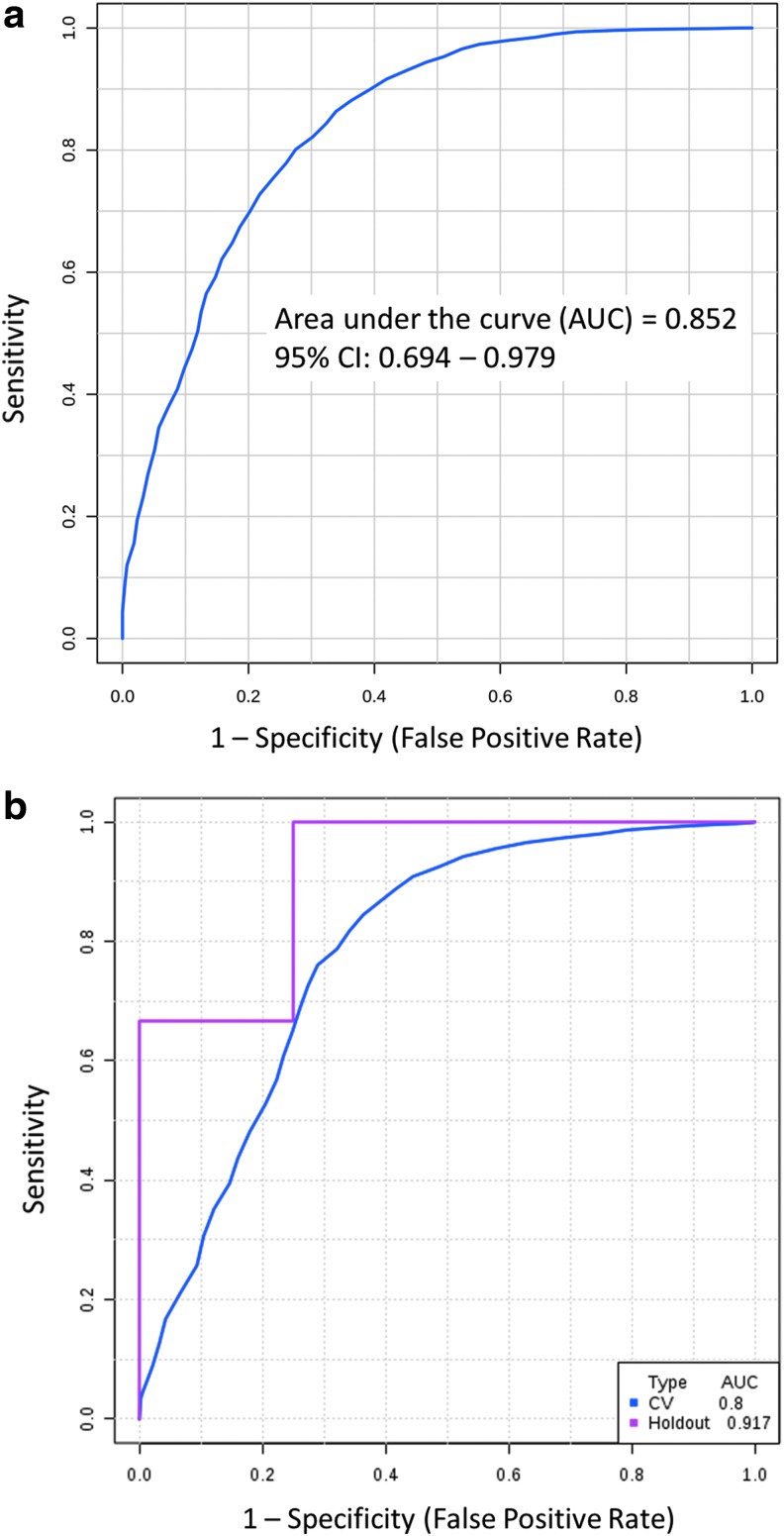
When used in a random forest multivariate regression analysis differentiating mTBI and control saliva samples, the six miRNAs had a combined area under the curve (AUC) of 0.852 (Fig. 3a). The algorithm misclassified 2/18 control subjects and 15/60 mTBI subjects, yielding a sensitivity of 75% and a specificity of 89% with 78% accuracy. A 100-fold cross-validation procedure holding out 25% of samples at random validated this model with an AUC of 0.800 in the cross-validation set and an AUC of 0.917 in the hold-out set (Fig. 3b).
FIG. 3.
The six microribonucleic acids (miRNAs) of interest accurately identify mild traumatic brain injury (mTBI) status in a multivariate regression analysis. A receiver operator characteristics curve utilizing salivary concentrations of six miRNAs (miR-29c-3p, miR-26b-5p, miR-30e-5p, miR-182-5p, miR-320c, and miR-221-3p) demonstrated an area under the curve (AUC) of 0.852 on random forest testing of mTBI status (a). The established algorithm misclassified two control subjects and 15 mTBI subjects. The 100-fold cross-validation of this tool holding out one-fourth of control and mTBI subjects at random exhibited similar accuracy (b). CI, confidence interval.
Longitudinal changes in concussion-related miRNAs
The six miRNAs with parallel changes in CSF and saliva samples were interrogated for longitudinal trends after concussion. Spearman rank correlation between miRNA concentration and time since injury (in days) was determined for both CSF and saliva samples (Table 2). Of the six miRNAs, three showed parallel correlations in CSF and saliva. Relative concentrations (RPM) of miR-29c-3p and miR-182-5p trended down over time in both CSF and saliva. Relative concentrations of miR-320c trended up over time in both biofluids. This trend was significant (FDR <0.05) for miR-320c in both CSF and saliva, and for miR-29c-3p in saliva.
Table 2.
Spearman Correlations between microRibonucleic Acid Concentration and Days since Injury in Saliva and Cerebrospinal Fluid
| Saliva | CSF | |||||||
|---|---|---|---|---|---|---|---|---|
| MicroRNA | R | t-stat | p value | FDR | R | t-stat | p value | FDR |
| hsa-miR-30e-5p | −0.5545 | 23598 | 0.0001 | 0.0008 | 0.2470 | 1524 | 0.2546 | 0.3855 |
| hsa-miR-29c-3p | −0.5196 | 23068 | 0.0003 | 0.0016 | −0.1751 | 2378 | 0.4242 | 0.5561 |
| hsa-miR-320c | 0.4566 | 8249 | 0.0016 | 0.0058 | 0.7164 | 574 | 0.0002 | 0.0039 |
| hsa-miR-221-3p | −0.2833 | 19480 | 0.0594 | 0.1010 | 0.1345 | 1752 | 0.5406 | 0.6683 |
| hsa-miR-182-5p | −0.0519 | 15968 | 0.7348 | 0.7937 | −0.0751 | 2176 | 0.7330 | 0.8201 |
| hsa-miR-26b-5p | −0.4002 | 21256 | 0.0064 | 0.0162 | 0.7065 | 594 | 0.0002 | 0.0041 |
CSF, cerebrospinal fluid; microRNA, microribonucleic acid; FDR, false detection rate.
Functional analysis
The six miRNAs with predictive utility for mTBI status had 700 predicted high-confidence mRNA targets, 354 of which had been experimentally validated (Supplementary Table 5; see online supplementary material at ftp.liebertpub.com). There were 34 mRNAs targeted by more than one miRNA. The 700 mRNA targets had significant associations with 30 GO categories (Supplementary Table 6; see online supplementary material at ftp.liebertpub.com). Notably, there was significant enrichment for mRNA targets associated with nervous system development (p = 2.67E-07), a pathway including 37 genes targeted by four miRNAs (miR-182-5p, miR-29c-3p, miR-30e-5p, and miR-320c). Of note, one of these genes is S100-PBP, whose protein product is affiliated with concussion biomarkers. Protein-protein interaction networks were defined for the 280 highest confidence mRNA targets (microT-CDS score ≥0.999) in String v10. This analysis identified a significant protein-protein interaction network (p < 0.0001) containing 269 nodes and 247 edges with a clustering coefficient of 0.775 (Supplementary Fig. 1; see online supplementary material at ftp.liebertpub.com). Analysis of this network identified 67 biologic processes with significant enrichment (Supplementary Table 7; see online supplementary material at ftp.liebertpub.com), including nervous system development (61 genes; p = 8.56E-09), neuron development (29 genes; p = 8.45E-05), and axon development (21 genes; p = 4.89E-04).
Relationships between medical characteristics and salivary miRNAs
Correlations of the six salivary miRNAs of interest with child SCAT-3 scores, parental SCAT-3 scores, and medical/demographic factors were explored. There were significant correlations (FDR <0.05) between child-reported measures on SCAT-3 and salivary concentrations of miR-26b-5p and miR-320c (Supplementary Table 8a; see online supplementary material at ftp.liebertpub.com). Levels of miR-26b-5p were inversely correlated with reports of “I get tired a lot” and “I get tired easily,” while levels of miR-320c were directly correlated with reports of “I daydream too much” and “I get confused.” There were also significant direct correlations between miR-320c and parent-reported SCAT-3 measures, including “has trouble sustaining attention” and “is easily distracted” (Supplementary Table 8b; see online supplementary material at ftp.liebertpub.com). There were nominal correlations between female sex and salivary concentrations of miR-182-5p and miR-221-3p (Supplementary Table 8c; see online supplementary material at ftp.liebertpub.com). No significant correlations were found, however, between the six miRNAs of interest and other medical/demographic characteristics, including participant age, ethnicity, weight, height, anti-depressant medication use, or dietary restrictions. There was also no correlation between concentrations of the six miRNAs and presence of broken bones or concussions obtained during sport (exercise).
Discussion
More than 50% of the miRNAs found in CSF are also found in saliva, and nearly 10% undergo parallel changes after concussive head trauma. Salivary concentrations of six of these miRNAs are predictive of concussion status, and five have been described in previous studies of adult human serum. Importantly, these six miRNAs have no correlation with bony injury, sports participation, or participant demographic characteristics. They also display striking enrichment for mRNA targets related to neuronal development. These factors, coupled with ease of collection and quantification, make salivary miRNAs an ideal substrate for concussion assessment with potential utility for initiating personalized medical interventions, tracking therapy response, and making return-to-play decisions.
Potential mechanisms for salivary transfer of brain-related miRNAs
In a medical community dominated by blood-based assays, the idea that salivary sampling provides a window into the brain might be difficult to fathom. This is because the vast majority of medical tests rely on measurements of proteins that are easily degraded in the enzymatic milieu of the mouth. In comparison, the short, single-stranded structure of miRNAs renders them relatively resistant to enzymatic degradation.7 They are also commonly protected by microvesicle or protein-bound mechanisms during extracellular transport.22
These factors account for the stability and reproducibility of salivary miRNA signatures in healthy subjects over time.13 They also help explain how brain-related miRNAs may travel to saliva. Exosomal transport of miRNAs could occur directly from cranial nerves that innervate the oropharynx (glossopharyngeal, facial, vagus, and trigeminal nerves)23 or indirectly through extraction from the blood by specialized cells in salivary glands.13 This latter mechanism demonstrates, in part, why many of the peptides and lipids found in blood are also present in saliva24 and why the current study finds such high overlap between serum-based miRNA biomarkers of concussion and those detected in saliva. The glymphatic system, which helps regulate CSF turnover via peri-arterial tissue within the myelin sheath of cranial nerves and the olfactory bulb, represents a primary route by which brain-related molecules enter the peripheral circulation.25 Given the proximity of these structures to the oropharynx, it seems likely that the glymphatic system could also play a role in the transfer of brain-related miRNA to saliva.
The role of miRNAs in the physiological response to TBI
The six miRNAs identified in the current investigation are not merely correlated with the presence or absence of concussion. They also have neurobiological implications in the physiological response to TBI. For example, miR-320c is down-regulated in CSF of sTBI subjects and saliva of mTBI subjects. In both biofluids, concentrations of miR-320c are directly correlated with time since injury (i.e., they increase toward baseline over time). MiR-320c is implicated in several pathways critical to nervous system function, including plasticity, mood, and circadian rhythm.
One mRNA target of miR-320c is phospholipid phosphatase related 1, a member of the plasticity-related gene family that is dynamically expressed during neuronal excitation and regulates neuronal plasticity.26 Plasticity-related genes are implicated in attentional deficits, and in the current investigation, concentrations of miR-320c were directly correlated with child report of increased daydreaming and parental report of child distraction. Initial suppression of miR-320c levels may mitigate these symptoms. On the other hand, unfettered increases in miR-320c could lead to mood dysregulation commonly reported in post-concussive syndrome. This idea is supported by a study of miRNA expression in the adult forebrain after successful suicide completion that found significant increases in miR-320c.27
Implications for concussion management
The salivary miRNAs identified in this exploratory investigation may one day be applied to the diagnosis and management of pediatric concussion. Benefits of such an approach would include ease of collection (relative to serum), speed of assessment (relative to multiple office-based surveys), and cost (relative to standard imaging methods). Because miRNA signatures remain altered nearly two weeks beyond injury and trend toward baseline during that time, they have potential clinical application at time of initial presentation to an acute clinic or emergency department setting, as well as at follow-up encounters with concussion specialists. Because miRNA can be measured with polymerase chain reaction techniques already employed in most medical facilities, results can be returned within 12–24 h. Thus, longitudinal trends in miRNA concentrations could be used potentially to triage specialist referrals, initiate personalized medical therapies, track responses to therapy, or make return-to-play decisions. Pediatricians often struggle to determine when educational modifications ought to be initiated (and discontinued) in patients with concussion. Levels of miR-320c provide a potential subjective biomarker of attention difficulty that deserves further study.
The panel of miRNAs identified in this investigation misclassified 17 of 78 subjects. The misclassified controls included one subject with food allergies and type 1 diabetes mellitus who was taking antidepressant medication and a nonsteroidal anti-inflammatory medicine, as well as one subject with no identifiable medical conditions. The 15 misclassified mTBI subjects were characterized by a history of previous concussion (n = 5), weakness (n = 3), emesis (n = 3), myopia (n = 3), and anti-inflammatory medication use (n = 6). Thus, future investigations will be needed to examine the relationship of these factors to salivary miRNA levels. Controlling for such factors may help improve the sensitivity of salivary miRNA testing in mTBI. Improved sensitivity would be critical if this miRNA approach were to be applied in clinical settings of occult TBI, such as that seen in abusive heat trauma of infants.
Limitations
There are several limitations to the design of this investigation that must be considered when interpreting the results. Retrospective analysis of CSF collected over a decade before carries the potential for RNA degradation. Assessment of RNA yield and quality with an Agilent Bioanalyzer, however, demonstrated remarkable preservation of ∼18–25 nucleotide species in the miRNA range (Supplementary Fig. 2; see online supplementary material at ftp.liebertpub.com). In addition, we applied rigorous quality control parameters to alignment and quantification steps and still identified a diverse population of miRNA species consistent with previous CSF reports.12 Finally, control and sTBI CSF samples were stored and thawed in an identical manner, limiting the possibility that miRNA degradation occurred preferentially in one group.
In interpreting these findings, one must also consider that the CSF and saliva samples analyzed in this study arose from different sets of individuals. The CSF collection in children with mTBI is not methodologically possible, so CNS miRNA changes must be modeled from sTBI subjects (even though the latter population is likely to experience different intracranial pathophysiology after injury). This approach, comparing peripheral miRNA expression in mTBI subjects with CSF miRNA expression in sTBI subjects, has been utilized in previous studies.12 Because the current study used previously collected CSF, paired saliva samples from sTBI subjects were not available. A comparison of individual saliva and CSF expression patterns should be pursued in future investigations, to validate the hypothesis that patterns of CSF miRNAs are reflected in the saliva after sTBI.
Obtaining CSF from a group of healthy control subjects is not methodologically possible. Thus, the comparison of TBI and control CSF employed in the present study highlights miRNAs that are present in the CNS and “altered” in TBI, but do not represent differences from a healthy control state. In addition, there are differences in age between the control and TBI CSF groups that may impact miRNA expression. Cross-referencing the 114 miRNAs altered in sTBI CSF against a list of 62 miRNAs with differential expression across the developing human brain,28 reveals that only five (miR-3615, miR-99a-5p, miR-96-5p, miR-486-5p, and miR-338-5p) show longitudinal expression differences, and none of these was included in the final biomarker set.
The control arm of this study was relatively underpowered, providing 80% power to detect a two-fold difference between groups for 500 miRNAs with a per-gene alpha of 0.002. For this reason, we focused on miRNAs with nominal differences between control and mTBI groups and used filters of physiological relevance based on CSF findings. The sample size of the sTBI cohort is also relatively small. Thus, miRNA changes observed in the CSF may be influenced by variation existing between individuals, rather than sTBI itself. This error would likely reduce overlap between CSF miRNA alterations and those observed in saliva. It may explain (in-part) why only six miRNAs were identified in this study. Another possible explanation for this small number of overlapping miRNAs is the broad timing of saliva collection in mTBI subjects. A collective average of miRNA expression from 0–14 days after mTBI could have washed out changes in miRNAs that were acutely altered but returned to baseline within one week.
Finally, important differences between control and mTBI subject characteristics must be taken into account when interpreting the present data. The two groups were well matched for sex, age, ethnicity, and antidepressant medication use. There was significantly higher utilization of nonsteroidal anti-inflammatory medication and acetaminophen in the mTBI group, however. MiRNAs are important mediators of inflammation and may be affected by these medications, but we found no correlation between anti-inflammatory utilization and concentrations of the six miRNAs of interest. Previous studies have also noted influences of exercise and orthopedic injury on concussion biomarkers. This study had a low proportion of mTBI participants with orthopedic injuries (8%), but a relatively high number of sports-related concussions (59%). There were 10 samples (of 60) obtained the day of injury and only five of these (8%) were sports-related concussions. Thus, the influence of exercise on the changes observed in the mTBI group as a whole is likely minimal. Nonetheless, because of the prevalence of these features in pediatric concussion, it will be important to investigate the influence of bony injury and exercise on miRNA expression in future studies.
Conclusion
This investigation identified six salivary miRNAs (miR-182-5p, miR-221-3p, mir-26b-5p, miR-320c, miR-29c-3p, and miR-30e-5p) altered in mTBI that reflect CSF patterns in sTBI and demonstrate diagnostic accuracy for mTBI status. These six miRNAs are functionally related to neuronal development and demonstrate intriguing correlations with concussion symptom reports. Although several have been identified in previous serum studies of adult concussion, here we show that they are easily measured in saliva and exhibit sustained dysregulation for up to two weeks after injury.
Supplementary Material
Acknowledgments
The authors thank Matthew Silvis, MD and Alyssa Kate Weller, MD for their contributions to the design of this study. In addition, we thank Jennifer Stokes, Andrij Tarasiuk, and Debbie Spear for assistance with sample/data collection as well as Patricia Silveyra, PhD, Karen Gentile, Susan DiAngelo, and Yuka Imamura, PhD for assistance with RNA processing. We thank Frank Middleton, PhD for aid with statistical analysis.
Funding Sources: Supported by the Children's Miracle Network (Grant# 417-51HY-4BFB to SDH) and Quadrant Biosciences Inc. (Research agreement with AL and SDH).
Author Disclosure Statement
Dr. Hicks is a co-inventor of preliminary patents for microRNA biomarkers in disorders of the central nervous system that is assigned to the SUNY Upstate and Penn State Research Foundations and licensed to Quadrant Biosciences (formerly Motion Intelligence). Dr. Hicks is also a consultant for Quadrant Biosciences. These conflicts of interest are currently managed by the Penn State College of Medicine. For the remaining authors, no competing financial interests exist.
References
- 1.McCarthy M.T. and Kosofsky B.E. (2015). Clinical features and biomarkers of concussion and mild traumatic brain injury in pediatric patients. Ann. N.Y. Acad. Sci. 1345, 89–98 [DOI] [PubMed] [Google Scholar]
- 2.Kirkwood M.W., Yeates K.O., and Wilson PE. (2006). Pediatric sport-related concussion: a review of the clinical management of an oft-neglected population. Pediatrics 117,1359–1371 [DOI] [PubMed] [Google Scholar]
- 3.Zonfrillo M.R., Master C.L., Grady M.F., Winston F.K., Callahan J.M., and Arbogast K.B. (2012). Pediatric providers' self-reported knowledge, practices, and attitudes about concussion. Pediatrics 130, 1120–1125 [DOI] [PubMed] [Google Scholar]
- 4.Bauer R. and Fritz H. (2004). Pathophysiology of traumatic injury in the developing brain: an introduction and short update. Exp. Toxicol. Pathol. 56, 65–73 [DOI] [PubMed] [Google Scholar]
- 5.McCrory P., Meeuwisse W.H., Aubry M., Cantu B., Dvorak J., Echemendia R.J., Engebretsen L., Johnston K., Kutcher J.S., Raftery M., Sills A., Benson B.W., Davis G.A., Ellenbogen R., Guskiewicz K.M., Herring S.A., Iverson G.L., Jordan B.D., Kissick J., McCrea M., McIntosh A.S., Maddocks D., Makdissi M., Purcell L., Putukian M., Schneider K., Tator C.H., and Turner M. (2013). Consensus statement on concussion in sport: the 4th International Conference on Concussion in Sport, Zurich, November 2012. J. Athl. Train. 48, 554–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rogelj B. and Giese K.P. (2004). Expression and function of brain-specific small RNAs. Rev. Neurosci. 15, 185–198 [DOI] [PubMed] [Google Scholar]
- 7.Gilad S., Meiri E., Yogev Y., Benjamin S., Lebanony D., Yerushalmi N., Benjamin H., Kushnir M., Cholakh H., Melamed N., Bentwich Z., Hod M., Goren Y., and Chajut A. (2008). Serum microRNAs are promising novel biomarkers. PLoS One 3, e3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Redell J.B., Zhao J., and Dash P.K. (2011). Altered expression of miRNA-21 and its targets in the hippocampus after traumatic brain injury. J. Neurosci. Res. 89, 212–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pasinetti G.M., Ho L., Dooley C., Abbi B., and Lange G. (2012). Select non-coding RNA in blood components provide novel clinically accessible biological surrogates for improved identification of traumatic brain injury in OEF/OIF Veterans. Am. J. Neurodegen. Dis. 1, 88–98 [PMC free article] [PubMed] [Google Scholar]
- 10.Redell J.B., Moore A.N., Ward III N.H., Hergenroeder G.W., and Dash P.K. (2010). Human traumatic brain injury alters plasma microRNA levels J. Neurotrauma 27, 2147–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zetterberg H., Smith D.H., and Blennow K. (2013). Biomarkers of mild traumatic brain injury in cerebrospinal fluid and blood. Nat. Rev. Neurology 9, 201–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhomia M., Balakathiresan N.S., Wang K.K., Papa L., and Maheshwari R.K. (2016). A panel of serum miRNA biomarkers for the diagnosis of severe to mild traumatic brain injury in humans. Sci. Rep. 6, 28148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bahn J.H., Zhang Q., Li F., Chan T.M., Lin X., Kim Y., Wong D.T., and Xiao X. (2015). The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin. Chem. 61, 221–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weber J.A., Baxter D.H., Zhang S., Huang D.Y., Huang K.H., Lee M.J., Galas D.J., and Wang K. (2010). The microRNA spectrum in 12 body fluids. Clin. Chem. 56, 1733–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hicks S.D., Ignacio C., Gentile K., and Middleton F.A. (2016). Salivary miRNA profiles identify children with autism spectrum disorder, correlate with adaptive behavior, and implicate ASD candidate genes involved in neurodevelopment. BMC Pediatr 16, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Varma S., Janesko K.L., Wisniewski S.R., Bayir H., Adelson P.D., Thomas N.J., and Kochanek P.M. (2003). F2-isoprostane and neuron-specific enolase in cerebrospinal fluid after severe traumatic brain injury in infants and children. J. Neurotrauma 20, 781–786 [DOI] [PubMed] [Google Scholar]
- 17.Yokobori S., Hosein K., Burks S., Sharma I., Gajavelli S., and Bullock R. (2013). Biomarkers for the clinical differential diagnosis in traumatic brain injury—a systematic review. CNS Neurosci Ther 19, 556–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia J., and Wishart D.S. (2016). Using MetaboAnalyst 3.0 for comprehensive metabolomics data analysis. Curr. Protoc. Bioinformatics 55,1014–1019 [DOI] [PubMed] [Google Scholar]
- 19.Vlachos I.S., Zagganas K., Paraskevopoulou M.D., Georgakilas G., Karagkouni D., Vergoulis T., Dalamagas T., and Hatzigeorgiou A.G. (2015). DIANA-miRPath v3. 0: deciphering microRNA function with experimental support. Nucleic Acids Res. 43, 460–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P., Kuhn M., Bork P., Jensen L.J., and von Mering C. (2015). STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, 447–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitra B., Rau T.F., Surendran N., Brennan J.H., Thaveenthiran P., Sorich E., Fitzgerald M.C., Rosenfeld J.V., and Patel S.A. (2017). Plasma micro-RNA biomarkers for diagnosis and prognosis after traumatic brain injury: A pilot study. J. Clin. Neurosci. 38, 37–42 [DOI] [PubMed] [Google Scholar]
- 22.Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J.J., and Lötvall J.O. (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659 [DOI] [PubMed] [Google Scholar]
- 23.Majem B., Rigau M., Reventós J., and Wong D.T. (2015). Non-coding RNAs in saliva: emerging biomarkers for molecular diagnostics. Intt J Mol Sci 16, 8676–8698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan W., Apweiler R., Balgley B.M., Boontheung P., Bundy J.L., Cargile B.J., Cole S., Fang X., Gonzalez‐Begne M., Griffin T.J., Hagen F., Hu S., Wolinsky L.E., Lee C.S., Malamud D., Melvin J.E., Menon R., Mueller M., Qiao R., Rhodus N.L., Sevinsky J.R., States D., Stephenson J.L., Than S., Yates J.R., Yu W., Xie H., Xie Y., Omenn G.S., Loo J.A., and Wong D.T. (2009). Systematic comparison of the human saliva and plasma proteomes. Proteomics Clin. Appl. 3, 116–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plog B.A., Dashnaw M.L., Hitomi E., Peng W., Liao Y., Lou N., Deane R., and Nedergaard M. (2015). Biomarkers of traumatic injury are transported from brain to blood via the glymphatic system. J. Neurosci. 35, 518–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Savaskan N.E., Bräuer A.U., and Nitsch R. (2004). Molecular cloning and expression regulation of PRG‐3, a new member of the plasticity‐related gene family. Eur. J. Neurosci. 19, 212–220 [DOI] [PubMed] [Google Scholar]
- 27.Lopez J.P., Fiori L.M., Gross J.A., Labonte B., Yerko V., Mechawar N., and Turecki G. (2014). Regulatory role of miRNAs in polyamine gene expression in the prefrontal cortex of depressed suicide completers. Int. J. Neuropsychopharmacol. 17, 23–32 [DOI] [PubMed] [Google Scholar]
- 28.Ziats M.N. and Rennert O.M. (2014) Identification of differentially expressed microRNAs across the developing human brain. Mol. Psychiatry 19, 848–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.