SUMMARY
The fan-shaped body (FB) neuropil in the Drosophila brain central complex (CX) controls a variety of adult behaviors, including navigation and sleep. How neuronal processes are organized into precise layers and columns in the FB and how alterations in FB neural-circuit wiring affect animal behaviors are unknown. We report here that secreted semaphorin 2b (Sema-2b) acts through its transmembrane receptor Plexin B (PlexB) to locally attract neural processes to specific FB laminae. Aberrant Sema-2b/ PlexB signaling leads to select disruptions in neural lamination, and these disruptions result in the formation of ectopic inhibitory connections between of FB neurons. These structural alternations and connectivity defects are associated with changes in fly sleep and arousal, emphasizing the importance of lamination-mediated neural wiring in a central brain region critical for normal sleep behavior.
In Brief
How and why do neurons form layered structures (neural lamination) in the central nervous system? The authors describe a molecular mechanism regulating lamination formation in the fruit fly central brain and demonstrate the functional importance of lamination in sleep behaviors.
INTRODUCTION
In the central nervous systems (CNSs) of both invertebrates and vertebrates, axons, dendrites, and synaptic connections are often spaced in precise three-dimensional registration. Examples include the laminar organization of the mammalian retina, neuropils in the insect visual system, the vertebrate cerebral cortex, and the dorsal horn of the spinal cord (Kolodkin and Hiesinger, 2017; Baier, 2013; Sanes and Yamagata, 2009). Though previous work suggests that murine cortical lamination is dispensable for processing somatosensory information (Guy and Staiger, 2017) and that zebrafish tectal lamination is not required for certain visual system responses (Nikolaou and Meyer, 2015), laminar organization is critical for precise synaptic connectivity and direction-selective visual system tuning in the mouse retina (Duan et al., 2014; Duan et al., 2018; Peng et al., 2017). Additional investigation, however, is required if researchers are to more fully understand the relationship between lamination and neural circuit elaboration and function.
In the insect brain, a phylogenetically conserved neuropil structure called the central complex (CX) integrates neural circuits that underlie a wide range of behaviors (Hanesch, Fischbach, Heisenberg,1989). The CX is composed of four neuropil substructures: the ellipsoid body (EB), the fan-shaped body (FB), the noduli (NO), and the protocerebral bridge (PB) (Figure 7A). In each CX substructure, axons and dendrites are precisely organized in multiple laminae (layers) along the dorsal-ventral (D-V) axis, in columns that are aligned adjacently in the horizontal plane, and in shells along the anterior-posterior (A-P) axis. At least 50 different types of small-field and large-field neurons innervate the CX (Hanesch, Fischbach, Heisenberg, 1989; Young and Armstrong, 2010). In specific CX substructures, each small-field neuron restricts its processes to one or two columns but contacts one, or multiple, laminae, whereas each large-field neuron extends its processes to cover an entire single lamina across all columns. This organization allows information to flow in precisely defined patterns between small- and large-field neurons within the CX (Lin et al., 2013; Wolff et al., 2015; Yang et al., 2013).
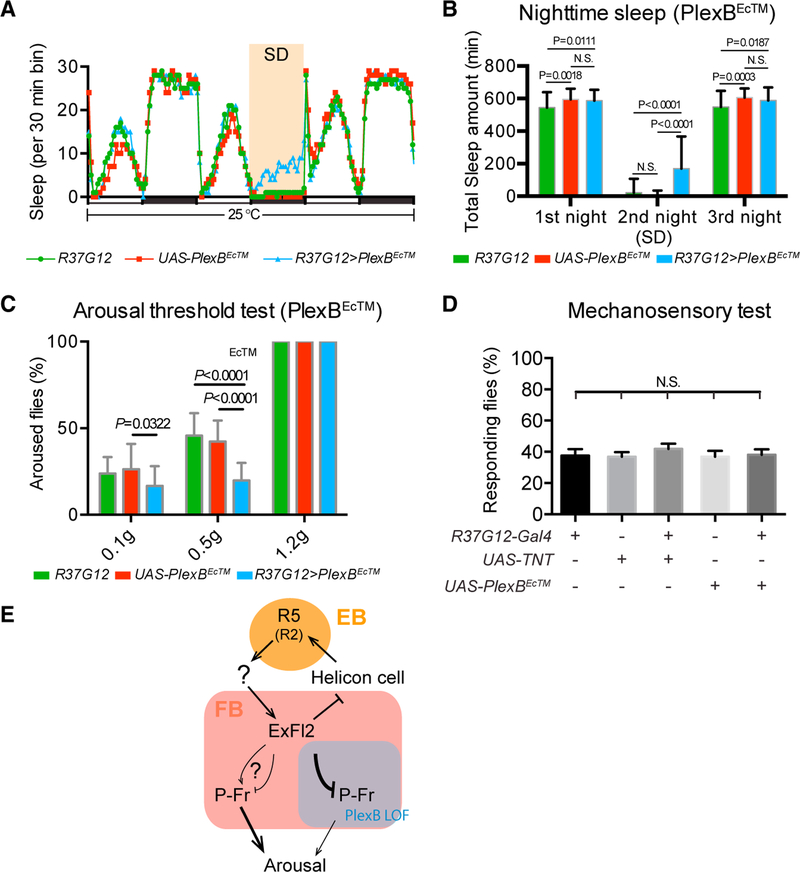
Figure 7. PlexB Functions in P-Fr Neurons to Modulate Arousal during Sleep.
(A and B) Fly sleep recording and total nighttime sleep measurements; these panels are similar to Figures 4A and 4B, except that R37G12-GAL4/+ (green line), UAS-PlexBEcTM/+ (red line), and R37G12>PlexBEcTM (blue line) flies are compared here. R37G12-GAL4/+ n = 121 flies; UAS-PlexBEcTM/+ n = 62 flies; R37G12>PlexBEcTM n = 60 flies.
(C) Sleep arousal threshold tests of flies; this panelis similar to Figure 4G, except that P-Fr neuron inactivation was accomplished through the use of R37G12-GAL4 to drive PlexBEcTM expression. A total of 13–14 flies was tested for each genotype. Fewer R37G12>PlexBEcTM flies were aroused than were flies in control groups after mild and moderate stimuli.
(D) Mechanosensory tests of a grooming response. Leg movements were examined after deflection of scutellar bristles of decapitated flies (see Star Methods for details). The results show that the grooming response is not significantly altered when P-Fr neurons are inactivated by TNT or by PlexBEcTM expression. A total of 32 flies was tested for each genotype. Two-way ANOVA with multiple comparisons was conducted for all panels.
(E) Model illustrating identified sleep circuits within the central complex (CX) and connectivity changes between ExFl2 axons and P-Fr processes within the FB when PlexB signaling was perturbed by PlexBEcTM expression in P-Fr neurons (see text for details).
The CX is required for many insect behaviors (Pfeiffer and Homberg, 2014). Several types of EB and FB neurons play important roles in sleep regulation (Liu et al., 2012; Ueno et al., 2012; Liu et al., 2016; Kottler et al., 2013; Kayser et al., 2014; Donlea et al., 2014; Donlea et al., 2018; Qian et al., 2017; Donlea et al., 2011). A group of large-field neurons, called “ExFl2,” project their axons to dorsal FB layer 6 and promote sleep (Figure 7B, red). The activity of ExFl2 neurons is locally modulated by dopamine and serotonin (Liu et al., 2012; Ueno et al., 2012; Qian et al., 2017; Donlea et al., 2011). A pair of helicon cells (Figure 7B, green) also elaborate their dendrites in dorsal FB layer 6 and are locally inhibited by the ExFl2-derived neuropeptide allatostatin A (AstA), suppressing sleep and promoting locomotion (Donlea et al., 2018). The helicon cells project their axons into the anterior EB to excite R5 axons (Omoto et al., 2017), previously named R2, that encode sleep drive in Drosophila (Figure 7B, blue) (Liu et al., 2016). Thus, FB lamination might serve to control synaptic transmission and information flow among FB-projecting neurons. However, little is known about other types of FB neurons required for regulating sleep behavior or how FB neurons across different layers interact with one another.
Here, we investigate the function of the secreted semaphorin Sema-2b and its receptor PlexB in the development of FB lamination and CX neuron function. We determine the consequences of disrupting neuronal process segregation between certain FB-projecting large- and small-field CX neurons with respect to neural connectivity between laminae. We also demonstrate a role for these small-field neurons in fly sleep behavior. These results define mechanisms that regulate neural-process lamination in the fly central brain and suggest that laminar organization of neural processes is necessary for regulating both anatomical and functional connectivity critical for normal adult behavior.
RESULTS
Neuronal Processes Are Organized in Laminae and Columns in the FB
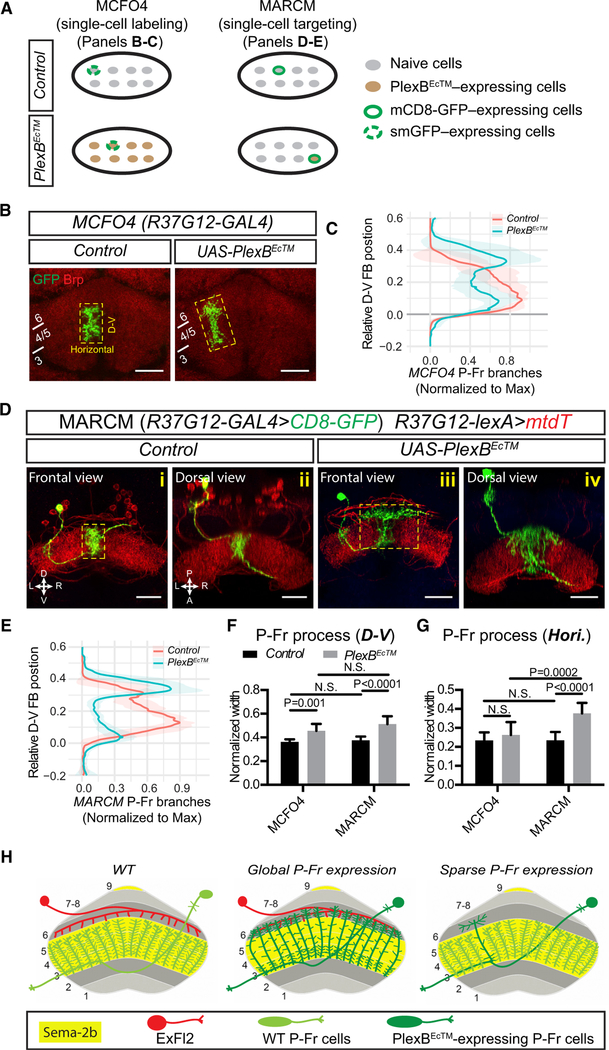
Similarly to other CX substructures, the FB is innervated by both large-field and small-field neurons. For example, R84C10-GAL4 is expressed in ~ 10–15 ExFl2 neurons per brain hemisphere, large-field neurons that project their axons to the dorsal FB (Figures 7D [red], 1Ei [green], and S1A [green]) (Young and Armstrong, 2010). On the other hand, R37G12-GAL4 is expressed in ~25–30 small-field PBG1–8.s-FBℓ3,4,5.s.b-rub.b (called “P-Fr”) neurons that elaborate their processes in the PB, medial FB (mFB), and a brain region that lies outside of the CX and is called the rubus (Figures 7C and 7D (green) and 1Eii (green)) (Wolff et al., 2015) (see Table 1 for general description of Gal4 lines used in this study).
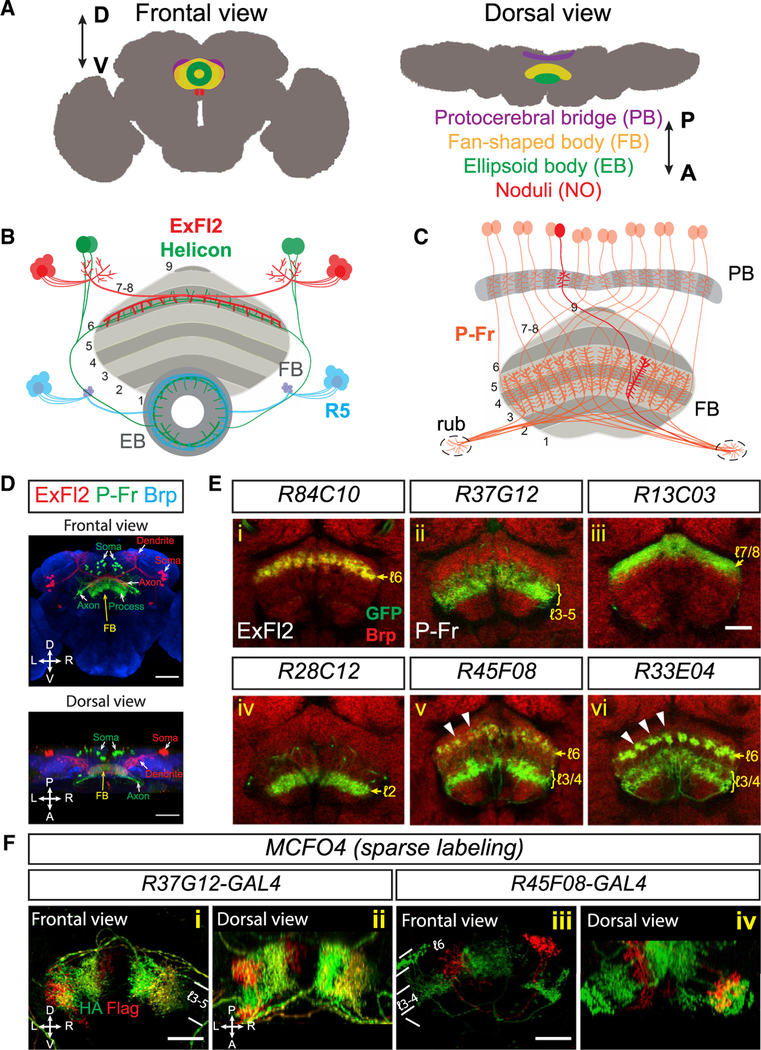
Figure 1. Neuronal Processes Are Organized in Laminae and Columns in the Fan-Shaped Body.
(A) Schematics showing frontal and dorsal views ofthe central complex (CX) in an adult fly brain. The CX is composed of four substructures: the ellipsoid body (EB), the fan-shaped body (FB), a pair of noduli (NO), and the protocerebral bridge (PB). D = dorsal; V = ventral; A = anterior; B = posterior.
B) Schematic showing local sleep circuitry that includes three different types of FB- and EB-projecting neurons. In the FB, ExFl2 neuron axons (red) and Helicon cell dendrites (green) co-stratify in dorsal FB layer 6. In the EB, Helicon cell axons and R5 axons (blue) locally interact with each other in an anterior ring.
(C) Schematic showing small-field P-Fr neuronsthat project processes to the PB, medial FB (mFB) layers 3–5, and an additional brain region called the rubus (rub). Within the PB and the FB, individual P-Fr processes precisely innervate specific dorsal-ventrl columns; P-Fr axons also target the rub. For simplification, the trajectories of P-Fr axons in the diagram do not depict their complete in vivo trajectories.
(D) Two types of FB-projecting neurons are colabeled by using (1) R84C10-lexA to express myristoylated tdTomato (mtdT, red) in about 20 large-field ExFl2 neurons and (2) R37G12-GAL4 to express CD8-GFP (green) in about 30 small-field PBG1–8.s-FℓB[3,4,5.s.b-rub.b (“P-Fr,” green) neurons in an adult fly brain (“s” spine; “b” bouton; “l” layer). Neuropils are revealed here by anti-Brp staining (blue). A frontal view (upper panel) and a dorsal view (lower panel) show that ExFl2 and P-Fr neuron soma are located at posterior-lateral and posterior-medial regions of the brain, respectively. Axons and dendrites project from these soma to the FB and other brain regions.
(E) Multiple types of neurons that target the FB are labeled by different fly lines harboring GAL4 driving expression of mCD8-GFP or lexA driving expression of mtdT (both shown in green in [i]–[vi]). AntiBrp staining (red) labels neuropil structures in adult fly brains. Single optical sections show examples of ExFl2, P-Fr and other types of small-field FB neurons that serve as markers for labeling single, or multiple, FB layers. Small-field FB neurons project their process to confined regions within each layer and form vertical columns orthogonal to FB laminae. These columns are not easily seen via Brp staining, but they can be observed in certain fly lines that label different types of small-field neurons, including R45F08 and R33E04, which label axons in layer 6 (arrowheads).
(F) Subsets of P-Fr neurons (R37G12, [i]–[ii]) and another group of small-field neurons (R45F08, [iii]–[iv]) are sparsely labeled in red and/or green by the multi-color flip-out (MCFO) technique, which illuminates restricted localization of small-field neuron processes in certain FB laminae and columns.
Scale bars represent 50 μm in (D) and 20 μm in (E) and (F).
Table 1.
GAL4 and lexA Drivers Used in This Study1
| Driver | Expression within CX in Adult Fly Brains | Expression outside of CX in Adult Fly Brains |
|---|---|---|
| R37G12-GAL4 | 25–30 small-field P-Fr neurons | A small subset of interneurons in the brain and VNC |
| R37G12-lexA | similar to R37G12-GAL4 | N/A |
| VT036267-GAL4 | small-field P-Fr and PB-EB-gal neurons | N/A |
| P-Fr-GAL4 (R37G12-p65AD; VT036267-GAL4DBD) | 25–30 small-field P-Fr neurons | 1–2 interneurons in the VNC |
| R84C10-GAL4 | 20–30 large-field ExFl2 neurons | A few interneurons in the brain |
| R84C10-lexA | Similar to R84C10-GAL4 | Weak expression in a large subset of interneurons in the brain |
| R13C03-GAL4 | Small-field neurons densely innervating the FB layer 7/8 and sparsely projecting to the FB layer 1 |
A large subset of interneurons in the brain |
| R13C03-lexA | Similar to R13C03-GAL4 | |
| R28C12-lexA | Small-field neurons innervating the FB layer 2 | N/A |
| R45F08-GAL4 | Small-field neurons innervating the FB layer 3/4/6 and the EB | A large subset of interneurons |
| R45F08-lexA | Similar to R45F08-GAL4 | |
| R33E04-GAL4 | Small-field neurons that innervating the FB layer 3/4/6 and the EB. | None |
| R33E04-lexA | Similar to R33E04-GAL4 | Two clusters of interneurons with cell bodies residing below the antennal lobes |
| R92D04-GAL4 | Small-field neurons innervating the FB layer 3–5; expression decreases in older flies | A large subset of interneurons |
| R83H12-GAL4 | Small-field neurons that innervate PB, FB layer 1, NO and EB | None |
| R40E08-lexA | A pair of ExFl2 neurons | A small subset of interneurons in the brain |
| VT036267-GAL4 | Small-field PB-EB-gall and P-Fr neurons | A small subset of interneurons in the brain |
Please see Table S1 for detailed genotypes of all fly lines used in this study and for the main text for additional descriptions of these lines and their use in this study.
The FB can be divided into at least nine laminae: from ventral to dorsal, laminae 1 through 9 (Figures 7B and 7C) (Wolff et al., 2015). Each lamina is innervated by specific types of small and large-field neurons, several of which are shown in Figure 7E utilizing fly lines that express GFP selectively in these distinct neuronal types (Table 1). For example, large-field ExFl2 neurons project their axons within FB layer 6 (Figure 7Ei), and small-field P-Fr neurons extend their processes in layers 3–5 (Figure 7Eii). The additional GAL4 drivers shown in Figure 7E ( [iii]– [vi]) label small-field neurons that serve as markers for different FB layers. Orthogonal to these FB laminae are columns formed by small-field neuron processes. FB columns, unlike FB laminae, are not often easy to recognize when they are labeled with bruchpilot (Brp) staining or when neuronal processes from particular small-field neuronal types are illuminated. However, here FB columns are revealed by the projections of axons from R45F08 and R33E04 neurons in layer 6 (Figures 7Ev and 1Evi, arrowheads) or more directly by sparse labeling of individual small-field neurons (Figure 7F). Although the FB neuropil can be observed at the beginning of pupae formation, we observed that most FB laminae are gradually formed during early pupal stages and that FB neurons display heterogeneous innervation dynamics (Figures S1A–S1D). This heterogeneity of FB neuron innervation during the formation of individual laminae suggests that multiple mechanisms underlie elaboration of complex FB connectivity.
Semaphorin 2b Is an Attractive Cue for Medial FB Neuron Lamination
To investigate mechanisms that govern FB innervation, we examined the distribution of several guidance cues in the pupal central brain. Of particular interest, expression of the secreted semaphorin Sema-2b is highly enriched in the mFB from early pupal stages (Figure S2A, arrowheads), and it is highly expressed in FB layers 3–5 and layer 9 throughout mid-pupal stages and in the adult (Figures 7A and S2A). This is in contrast to the transmembrane semaphorin Sema-1a and the other secreted semaphorin in Drosophila, Sema-2a, which are only weakly detected by immunostaining and are broadly distributed throughout the FB during pupariation (data not shown). Sema-2b expression declines after eclosion and is apparently low in the adult brain (Figure S2A, arrow). Sema-2b expression is nearly completely absent from the EB (Figure 7A).
Sema-2b-null mutant flies exhibit aberrant overall CX morphology and FB neuron lamination defects (Figure S3A and data not shown). Therefore, we used RNA interference (RNAi) to selectively knock down Sema-2b expression in the FB in order to examine Sema-2b function in FB development. The R92D04GAL4 fly line is expressed in small-field FB neurons that innervate the mFB. The identities of these neurons are not known. However, their processes co-localize with Sema-2b protein in the mFB during early pupal stages (Figure 7B, arrowheads). Sema-2b-RNAi driven by R92D04-GAL4 successfully knocked down Sema-2b expression in R92D04 neurons (but not in other Sema-2b-expressing neurons that are not targeted by R92D04-GAl4) within the mFB (Figures 7B, blue arrows, and 2C). We used several lexA driver fly lines to label different groups of FB neurons in combination with R92D04-GAL4 driving UAS-Sema-2b-RNAi. We found that R13C03 and R28C12 neurons, which project their processes to layers 7 and 8 and to layer 2, respectively, were not affected by selective Sema-2b FB knock down (Figures S2Bi and S2Bii). The processes from R40E08 and R45F08 neurons, which target layer 6 and target layers 3 and 6, respectively, showed very mild lamination defects after Sema-2b-RNAi knock down (Figures S2Biii and S2Biv, arrows and arrowheads). However, loss of Sema-2b in R92D04 neurons altered the projections of P-Fr neurons, which we labeled here by using the R37G12-lexA line to drive expression of myristoylated tdTomato (mtdT) in these neurons (Figure 7D). In the absence of Sema-2b in FB layers 3–5, P-Fr neuron processes extend dorsally into layer 6 (Figure 7Diii, arrows and dashed lines, and Figure 7E) and also mis-project outside the FB into the EB and ventral FB (Figure 7Div, arrowheads). These results show that Sema-2b regulates FB neuron development in a layer-specific manner: neurons that project to the mFB are affected by loss of Sema-2b, but dorsal FB (dFB)- or ventral FB (vFB)-projecting neurons are not. Further, these FB targeting defects strongly suggest that Sema-2b acts as an attractive guidance cue to facilitate P-Fr neuron targeting in the mFB.
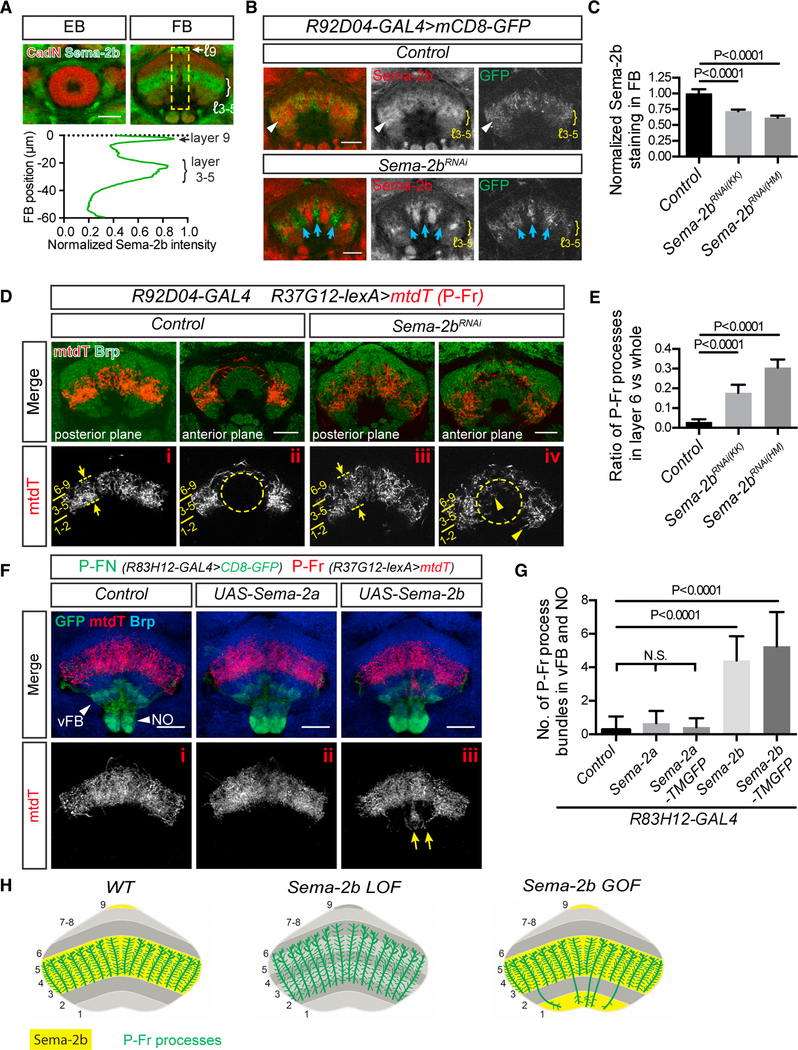
Figure 2. Semaphorin 2b (Sema-2b) Is an Attractive Cue that Regulates Medial FB Neuron Lamination.
(A) sAnti-Sema-2b staining (green) shows enrichment in the medial FB (mFB) (layers 3–5) and to a lesser extent in the dorsal FB (layer 9) during early pupal stages.Sema-2b is not expressed in the ellipsoid body (EB). Anti-N-Cadherin (CadN) staining (red) reveals pupal brain structures at 48 h after pupal formation (APF) (see Figure S2A for additional developmental stages). Quantification of the average Sema-2b immunolabeling intensity along the ventral-dorsal axis (yellow box; bottom panel) (n = 8 brains).
(B) R92D04-GAL4 is expressed in a group of small-field FB neurons (labeled by mCD8-GFP, green) that innervate the mFB (control, upper panels) and that colocalize with Sema-2b (red). When UAS-Sema-2b-RNAi is expressed in R92D04 neurons (lower panels), their processes reorganize and innervate subsets of FB columns that are now devoid of Sema-2b (blue arrows). Sema-2b is still detected in some FB columns and is expressed by other neurons that are not targeted by R92D04-GAL4.
(C) Quantification of Sema-2b immunolabeling across all mFB columns in Control and Sema-2b-RNAi-expressing animals. Two different UAS-Sema-2b-RNAi transgenic lines (KK104842 and HM05143) were used. All data points were normalized to the average value of the control group (n = 8 brains for each group). These expression data are an average of Sema-2b downregulation over a defined FB region. It is important to note that this region includes neuronal processes that extend from neurons in which Sema-2b is not knocked down, owing to the lack of Sema-2b-RNAi expression in all neurons that contribute neuronal processes to FB columns when Sema-2b-RNAi expression is driven by R92D04-GAL4
D) A group of P-Fr neurons, labeled by R37G12-lexA driving mtdT (red), elaborates their processes in mFB layers 3–5 (i) but not in the EB (ii) in control animals. Brp staining (green) reveals EB and FB morphologies and defines FB layers. Layers 2 and 6 are most recognizable because they exhibit stronger Brp staining than do other FB layers. When Sema-2b is knocked down in R92D04 neurons in the mFB, P-Fr neurons expand their projections into the dorsal FB layer 6, seen here in more posterior regions (panel iii, dashed lines and arrows), and mis-project their processes into the EB and also into ventral FB layers 1 and 2 in more anterior regions ([iv], arrowheads).>
(E) Quantification of the P-Fr process dorsal overshooting phenotype shown in (i) and (iii), shown as the ratio of P-Fr process measurement within layer 6 (revealed by higher anit-Brp immunolabeling) to all P-Fr processes across all FB layers (see Star Methods for details) (n = 10 brains for each genotype).
(F) The R83H12-GAL4 fly line was used for expressing Sema-2a or Sema-2b in P-FN neurons (green), which innervate the PB, the ventral FB (layer 1), and the NO (arrowheads). P-Fr neurons were labeled with R37G12-lexA driving mtdT (red), and anti-Brp staining (blue) labels the FB. Note that mtdT-labeled P-Fr neurons ectopically innervate the NO and vFB when Sema-2b, but not Sema2a, is expressed in the R83H12 neurons (yellow arrows).
(G) Quantification of P-Fr process innervation of the ventral FB and NO in control animals and when secreted (or membrane-tethered) Sema-2a or Sema2b were expressed in P-FN neurons using the R83H12-GAL4 driver (n = 15 brains for control; 15 for Sema-2a; 7 for Sema-2a-TMGFP; 15 for Sema-2b; and 16 for Sema-2b-TMGFP). TMGFP = transmembrane-GFP.
(H) Schematics showing co-localization of P-Fr processes with Sema-2b in FB layers 3–5 and how P-Fr processes respond to Sema-2b loss- and gain-of-function in the FB. P-Fr cell bodies and their axons and dendrites outside of the FB are not included in these diagrams. One-way ANOVA with Tukey’s multiple comparisons test was used for (C), (E), and (G). Bar graphs in this and all subsequent figures are shown as mean ± SD.
Scale bars represent 20 μm.
We next used different GAL4 drivers to overexpress Sema-2b, a secreted protein, and a membrane-tethered form of Sema-2b (Sema-2b-TMGFP) (Wu et al., 2011) in dorsal and ventral FB layers, where Sema-2b is not normally expressed; we used R37G12-lexA driving mtdT to label P-Fr processes in these gain-of-function (GoF) experiments. We found that P-Fr processes were attracted to the ventral FB and dorsal NO when R83H12-GAL4 was used to overexpress Sema-2b (Figure 7Fiii) or Sema-2b-TMGFP (Figure S2Cii) in a group of PB-FB-NO (P-FN) neurons that normally target FB layer 1 and the NO. Sema-2a and Sema-2a-TMGFP were also overexpressed in P-FN neurons, and we observed that expression of Sema-2a (Figure 7Fii) or Sema-2a-TMGFP (Figure S2Ci) in P-FN neurons had little effect on P-Fr process targeting (Figure 7G). Taken together, these results show that Sema-2b, but not Sema-2a, functions as a local attractive cue in the mFB to non-cell-autonomously regulate lamination of small-field P-Fr neurons (Figure 7H).
Plexin B Is Cell-Autonomously Required for P-Fr Neuron-Process Lamination
The transmembrane protein Plexin B (PlexB) is a Sema-2b receptor (Ayoob et al., 2006), and we next asked whether Sema-2b regulates P-Fr process development through PlexB signaling. PlexB mutant flies showed defects in CX morphology and P-Fr innervation; this is similar to what was observed in Sema-2b mutant flies (Figure S3A) and suggests that PlexB and Sema-2b function in the same pathway to regulate the CX development.
In order to examine PlexB expression, we used a MiMIC15559 insertion located within the PlexB locus (Venken et al., 2011) and employed the “Trojan” technique (Diao et al., 2015) to generate a PlexBGAL4 allele (Figure S3B). PlexBGAL4 failed to complement the PlexBKG00878 (PlexBKG) null allele (Ayoob et al., 2006), as indicated by the observation that PlexBGAL4/KG flies showed increased lethality prior to and after fly eclosion (Figures S3C and S3D). When UAS-PlexB or UAS-Myc-PlexB was expressed under the control of PlexBGAL4 in PlexBGAL4/KG flies, early lethality was almost completely rescued (Figure S3C), and late lethality was partially rescued (Figure S3D). These results suggest that PlexBGAL4 is a null, or very strongly hypomorphic, PlexB allele and that PlexBGAL4 driving PlexB expression functionally recapitulates endogenous PlexB.
We also used PlexBGAL4 to drive PlexB expression from a UAS-Myc-PlexB allele to examine PlexB expression in the pupal brain. Interestingly, Myc-PlexB was detected at relatively high levels in the ventral and dorsal FB, an expression pattern that is complementary to that of Sema-2b, which is expressed in the medial FB (Figure S3E). To determine whether P-Fr neurons express PlexB, we expresed a nuclear-localized GFP (Stinger-GFP; Vef et al., 2006) under the control of PlexBGAL4. Stinger-GFP was observed in a large proportion, but not in all, of DAPI-labeled cell nuclei in fly brains (data not shown). When P-Fr neurons were labeled using R37G12-lexA to drive mtdT expression, Stinger-GFP was detected in all pupal P-Fr cell bodies (Figure S3F); this confirms that P-Fr neurons express PlexB.
We next asked whether PlexB is cell-autonomously required for P-Fr neuron lamination. PlexBEcTM, a truncated PlexB protein lacking its cytoplasmic domain, functions as a dominant negative receptor, efficiently blocking PlexB function in vivo (Wu et al., 2012). We used three different approaches to assess how PlexBEcTM affects targeting of FB neurons, combining PlexBEcTM expression with a reporter gene to visualize P-Fr neuronal-process targeting in the FB (Figures 1A and 2A).
First, we labeled all P-Fr (green) and ExFl2 (red) neurons by using R37G12-GAL4 and R84C10-lexA to drive CD8-GFP and mtdT, respectively (Figure 1B). In control brains, P-Fr and ExFl2 processes are located in adjacent FB layers with minimal overlap (Figures 1Bi and 3Bii). However, when PlexBEcTM is expressed in P-Fr neurons, their processes innervate the dorsal FB layer 6, which is normally occupied by ExFl2 axons (Figures 1Bv and 3Bvi, arrowheads; quantification in Figures 1C and 1D). Some P-Fr processes also innervate the EB when PlexBEcTM is expressed in P-Fr neurons (Figure 1Bviii, arrow). These phenotypes recapitulate what we observed when Sema-2b was knocked down in the mFB (Figure 7D) and thus suggest that PlexB functions as a Sema-2b receptor to control P-Fr process lamination within the mFB.
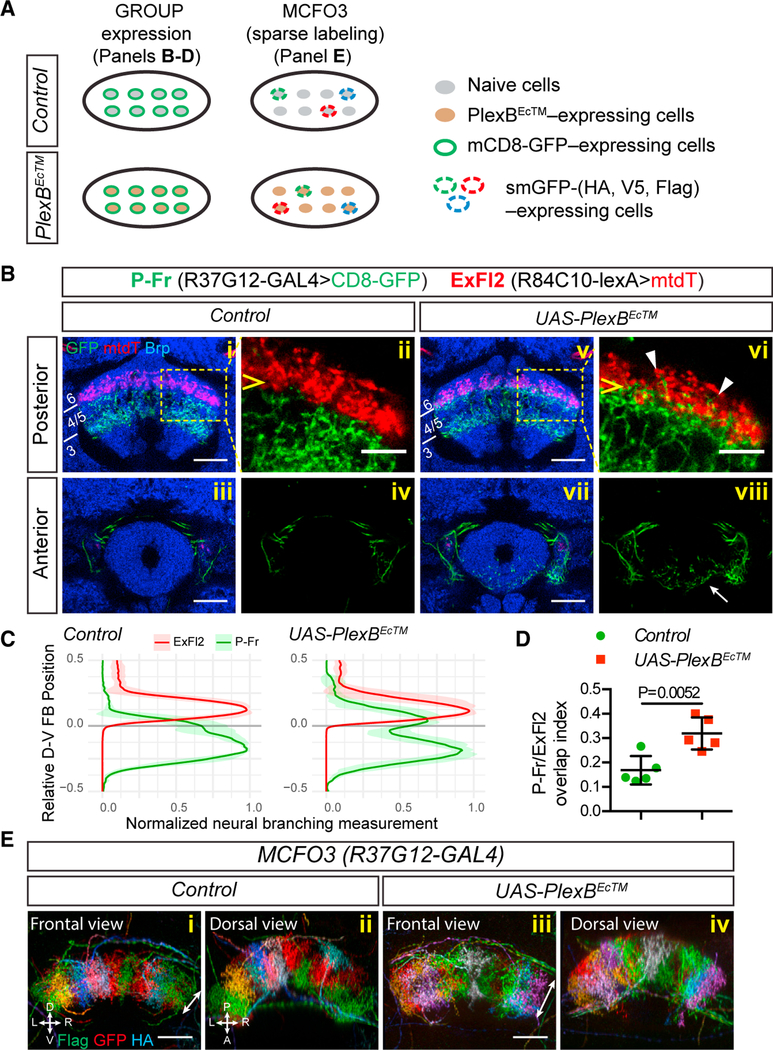
Figure 3. Plexin B Is Required for P-Fr Neuron-Process Lamination.
(A) Schematics showing two different approaches used to express reporter genes in neurons belonging to a single group (“GROUP”) or a subset of neurons within that group (“MCFO”). Lower diagrams show that PlexBEcTM (a dominant-negative truncated PlexB receptor) is mis-expressed in the entire group of P-Fr neurons in both cases. smGFP = spaghetti monster GFP.
(B) P-Fr and ExFl2 neurons were labeled with R37G12-GAL4 driving mCD8-GFP expression and R84C10-lexA driving mtdT expression, respectively. CadN staining (blue) labels adult brain structures. In control animals (i–iv), P-Fr processes (green) and ExFl2 axons (red) are located in different FB layers (P-Fr in layers 3–5 and ExFl2 in layer 6) (i–ii). When PlexBEcTM is expressed in P-Fr neurons (v–viii), their processes extend into FB layer 6 (posterior section, arrowheads in vi) and also mis-project into the EB (anterior section, arrow in viii).
(C) Quantification of P-Fr and ExFl2 process distributions along the dorsal-ventral (D-V) axis in the FB. We measured P-Fr branches at specific D-V positions by counting numbers of voxels that had GFP or mtdT labeling intensities above the chosen thresholds; these values were then normalized to the largest value for each individual brain (X axis). We aligned the lower boundary of ExFl2 axon staining and set it as position 0 and we calculated the relative D-V position by normalizing the absolute distance between each D-V position and position 0 to the total D-V extent of the FB (Y axis). There is expansion of P-Fr processes (green line) into layer 6 (above position 0).
(D) The Overlap index of P-Fr processes with ExFl2 axons is defined by the sum of P-Fr branch measurements above position 0 divided by total measurements across all D-V positions (see Star Methods for additional quantification details; t test was performed for this comparison; n = 5 brains for each genotype).
(E) R37G12-GAL4 and MCFO3 transgenes were combined so that individual P-Fr neurons were labeled in multiple colors. Note that processes from individual P-Fr neurons occupy individual columns orthogonal to the plane of FB lamanae across the total extent of the FB. When PlexBEcTM is expressed in all P-Fr neurons, their processes extend ectopically along the D-V axis (double-headed arrows); this is similar to what we observe with only mCD8-GFP labeling (B). The columnar organization of these processes appears largely intact despite PlexBEcTM expression.
All scale bars represent 20 μm.
Second, we used the multi-colored flip-out (MCFO) technique (Nern et al., 2015) to differentially label individual P-Fr neurons. Relatively dense labeling (through the use of MCFO3) of P-Fr neurons in control brains revealed that P-Fr processes are segregated into multiple columns within the mFB (Figures 1Ei and 3Eii). This columnar organization was largely preserved when P-Fr process lamination was altered after PlexBEcTM expression in all P-Fr neurons (Figures 1Eiii and 3Eiv). We also observed changes in individual P-Fr neuron process lamination when we used sparser MCFO4 single-cell labeling together with PlexBEcTM expression in all P-Fr neurons (Figures 2B and 2C). When we measured the extent of single P-Fr cell processes along the D-V and horizontal axes in Z stack projection images, we found that P-Fr processes expand along the D-V axis (Figure 2F) but not along the horizontal axis (Figure 2G) after PlexBEcTM expression in all P-Fr neurons. The similarity between control and PlexBEcTM groups in P-Fr process extension along the horizontal axis supports the idea that columnar organization of P-Fr processes in the FB is preserved when laminar targeting is disrupted.
Third, we used mosaic analysis with a repressible cell marker (MARCM) to generate single-neuron P-Fr clones and directly assess cell-autonomous requirements for PlexB signaling in P-Fr process development. We labeled individual P-Fr neurons with mCD8-GFP and labeled all P-Fr neurons by using R37G12-lexA to drive mtdT expression (Figure 2D). When PlexBEcTM was expressed in single GFP-labeled P-Fr neurons, their processes extended further dorsally than did mtdT-labeled processes from wild-type P-Fr neurons, which remained confined to layers 3–5 (Figure 2Diii). Individual PlexBEcTM expressing P-Fr neurons extended processes within layer 6; they crossed many single-column boundaries and expanded laterally to occupy a much larger region than in more ventral layers (Figures 2Diii and 4Div; quantification in Figure 2E). When we measured overall process diameters along the D-V and horizontal axes of single-cell P-Fr MARCM clones in Z stack projection images, we found that P-Fr processes expanded further along the D-V axis in the PlexBEcTM group than in the control group; this is similar to what we observed in our MCFO4 experiments (Figure 2F). However, unlike MCFO4-labeled P-Fr cells in the context of PlexBEcTM expression in all P-Fr neurons, MARCM-labeled P-Fr single cells that expressed PlexBEcTM showed much wider horizontal expansion than did the controls (Figures 2D and 2G). This is presumably because single PlexBEcTM-expressing over-extending cells do not confront other normal P-Fr neuronal processes in layer 6, and so they are free to expand horizontally (Figure 2H). These results show that individual P-Fr processes are normally guided by exogenous Sema2b signaling via PlexB to target correct FB laminae. Further, they suggest that neighboring P-Fr processes interact with each other through as-yet-unknown molecules to promote column formation; this indicates that the formation of laminae and columns utilizes independent mechanisms in the FB.
Figure 4. Cell-Autonomous Requirement for PlexB in P-Fr Process Lamination.
(A) Schematics showing two different approachesused here for expressing reporter genes in an individual neuron within a group. Lower diagrams show that PlexBECTM is expressed in the entire group of P-Fr neurons when MCFO4 labeling is used, but it is expressed in a single P-Fr neuron that expresses the reporter gene when MARCM (mosaic analysis with a repressible cell marker) is used.
(B) The MCFO4 fly line, which expresses a less robust flipase (FLP) than does the MCFO3 line, was used for labeling single P-Fr neurons (green). The FB is illuminated with anti-Brp staining (red). When PlexBEcTM is expressed in all P-Fr neurons, GFP-labeled single P-Fr neuron processes exhibit a dorsal expansion but also express preservation of their columnar organization.
(C) Quantification of the normalized distributionof MCFO4-labeled P-Fr neuron processes along the D-V axis in control and PlexBEcTM- expressing conditions. P-Fr processes are measured as in Figure 1C except that only GFP labeling is quantified and the ventral boundary of each P-Fr process is aligned and set to position 0. Note the dorsal shift of P-Fr process extension when PlexBEcTM is overexpressed in all P-Fr neurons. n = 10 brains for each genotype.
(D) MARCM is used for expressing mCD8-GFP and PlexBEcTM in single P-Fr neurons under the control of the R37G12-GAL4 driver; all P-Fr neurons are simultaneously labeled by R37G12-lexA driving mtdT. In control animals, single CD8-GFP-labeled P-Fr neurons elaborate processes within a single column in FB layers 3–5; this is similar to other mtdT-expressing P-Fr neurons ([i] and [ii]). When PlexBEcTM was expressed in single P-Fr neurons, their processes ectopically innervated FB layer 6; they extended dorsally beyond other P-Fr neurons and expanded laterally to cover an area in layer 6 that was larger than their normal lateral columnar extent in more ventral layers ([iii] and [iv]).
(E) Quantification of the normalized distribution of MARCM-labeled P-Fr neuron processes along the D-V axis, as in (C). Note that P-Fr processes have relatively more branches in the dorsal FB than in the ventral FB following PlexBEcTM expression.
(F and G) Comparisons of P-Fr process width (normalized to FB width) along the D-V or lateral axes, as shown in the yellow boxes in B and D, respectively. n = 10 neurons for both MCFO4 control and MCFO4 PlexBEcTM; n = 8 neurons for MARCM control and 11 neurons for MARCM PlexBEcTM. Two-way ANOVA with multiple comparisons was used for (F) and (G).
(H) Schematics summarizing phenotypes exhibited by P-Fr neurons in the experimental conditions presented in this figure and in Figure 1. The cell bodies, and also the axonal and dendritic trajectories, of all P-Fr neurons are not shown in each diagram, with the exception of one P-Fr neuron.
All scale bars represent 20 μm.
P-Fr Neuron Lamination Defects Result in Ectopic Connectivity with ExFl2 Neurons
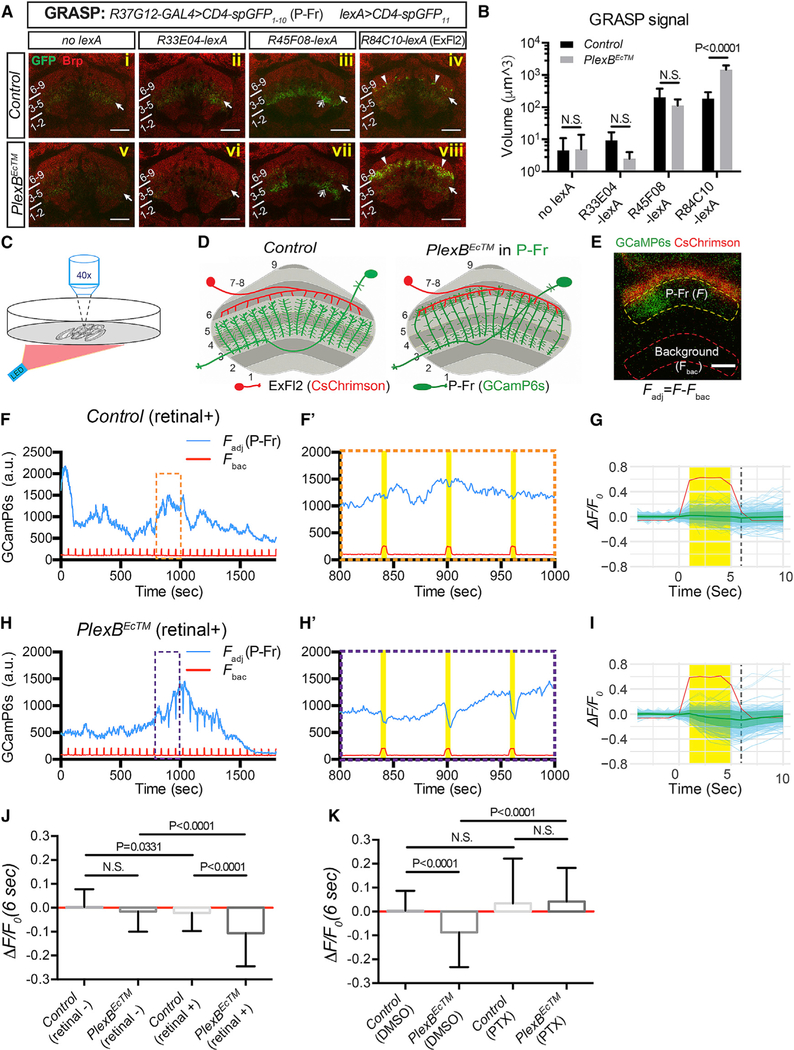
P-Fr neuron processes in the FB harbor both pre- and post-synaptic terminals (Wolff et al., 2015). We found that both types of terminals were still present in P-Fr processes even when P-Fr process lamination was altered by PlexBEcTM expression. This is because we observed equal distributions of GFP-tagged synaptotagmin (Syt-GFP, a synaptic vesicle marker) and DenMark (a dendritic marker) throughout P-Fr processes in both control and PlexBEcTM conditions (Figure S5A). To examine potential P-Fr connections with other FB neurons, and to determine whether these are altered when they express PlexBEcTM, we first employed the GFP reconstitution across synaptic partners (GRASP) approach to assess physical association between P-Fr processes and other FB neurons (Gordon and Scott, 2009; Feinberg et al., 2008). R37G12-GAL4 drives UAS-CD4-spGFP1–10 in P-Fr neurons, and several lexA drivers express lexAop-CD4-spGFP11 in different layer 6 input neurons; for example, such drivers include R84C10-lexA in large-field ExFl2 neurons, and R33E04-lexA and R45F08-lexA in as-yet-uncharacterized small-field neurons (Figure S5B). By using an antibody directed against GFP, we detected weak GFP immunostaining in the mFB in all conditions (Figure 3A, arrows); this result shows that this antibody weakly binds to spGFP1–10. Interestingly, strong GFP staining due to the same antibody, which presumably reveals true GRASP signals, was sparsely detected between FB layer 5 and 6 with only R84C10-lexA driving lexAop-CD4-spGFP11 in ExF12 neurons (Figure 3Aiv, arrowheads). These GRASP signals were significantly stronger within FB layer 6 when PlexBEcTM was expressed in P-Fr neurons (Figure 3Aviii, arrowheads; quantification in Figure 3B). The other two lexA drivers did not generate GRASP signals in the dorsal FB (Figures 3Aii, 5Aiii, 5Avi, and 5Avii). Instead, GRASP signals were detected in FB layers 3 and 4 when R45F08-lexA was used to drive lexAop-CD4-spGFP11 in both control and PlexBEcTM groups, and there was no significant difference between them (Figures 3Aiii and 5Avii, double arrows; quantification in Figure 3B). These GRASP signals are most likely reconstituted GFP between P-Fr processes and FB neurons that innervate FB layers 3 and 4. Together, these results strongly suggest that P-Fr neurons form few contacts with ExFl2 neurons in WT animals, but these contacts are selectively increased when P-Fr processes expand dorsally into layer 6.
Figure 5. P-Fr Process Lamination Defects Lead to Ectopic Inhibitory Synaptic Input from ExFl2 Neuron Axons.
(A) GFP reconstitution across synaptic partners (GRASP) was used to probe potential synaptic connections between P-Fr processes and other neurons that innervate FB layer 6 (R33E04-lexA and R45F08-lexA for small-field neurons and R84C10-lexA for large-field ExFl2 neurons). The CD4-spGFP1–10 was expressed in P-Fr neurons using R37G12-GAL4, and CD4-spGFP11 was expressed in different lexA-expressing neuronal populations. GRASP signals were examined through the use of a rabbit anti-GFP antibody. Weak GFP staining was observed in all conditions with or without lexA drivers (arrows) suggesting that this antibody weakly recognizes spGFP1–10. Stronger GFP immunolabeling was seen in layer 3 when the R45F08-lexA driver was used (double arrows in [iii] and [vii]), showing that this antibody strongly recognizes reconstituted GFP. Strong, punctated, GFP signals were detected near the lower boundary of layer 6 (arrowheads in [iv]) or within layer 6 (arrowheads in [viii]) only when the R84C10-lexA driver was used to express CD4-spGFP11 in ExFl2 neurons. Strong GRASP reconstitution signals in the control conditions are quite sparse (iv) as compared to GRASP signals when PlexBEcTM is expressed in P-Fr neurons (viii).
(B) Quantification of the total volume of all voxels that have GFP immunolabeling above the defined threshold across all brains (see Star Methods) in the FB for GRASP experiments shown in (A) (n = 8 brains for each genotype).
(C) Schematics showing the experimental strategy for ex vivo two-photon Ca2+ imaging (920 nm) paired with optical stimulation (620 nm, 0.338 mW/cm2).
(D) Schematics presenting the experimental design (E–K) for examining synaptic connections between ExFl2 axons (expressing CsChrimson-mtdT) and P-Fr processes (expressing GCaMP6s).
(E) A live imaging snapshot shows CsChrimson-mtdT (red) expression in ExFl2 axons and GCaMP6s (green) in P-Fr processes in the FB. The average GCaMPfluorescence intensity was measured in two identical contours: one inside the FB (within yellow dashed line) for GCaMP6s fluorescence in P-Fr processes (F), and a second outside of the FB (within red dashed line) for background calibration (Fbac). The difference between fluorescence measurements in these two contours gives rise to the adjusted CCaMP6s fluorescence in P-Fr processes (Fadj = F-Fbac). P-Fr processes can be activated in a column-specific fashion, as shown here by the increase of GCaMP fluorescence only on the left side of the FB. All imaging was performed in the presence of TTX.
(F) A representative trace shows dynamic changes of adjusted GCaMP6s fluorescence in P-Fr processes (blue line) and background fluorescence (red line) duringa 30 min imaging period in in a control brain. The pulsed increases in background fluorescence serve as internal indicators of LED stimulation events (on for 5 s per min). Three stimulation events (orange box) are expanded and shown in (F’). No correlation between LED stimulation and GCaMP fluorescence in P-Fr processes was noted in the control (yellow bars).
(G) Individual stimulation events (4 s prior to LED on, 5 s during LED on, and 5 s after LED off) are overlaid and GCaMP fluorescence changes are shown as ΔF/F0 (light blue for individual events and green lines and margins for mean ± SD). Time 0 is the time point just before the LED light is on. F0 is the average of Fadj 4 s (time point−3 to 0 s) before the LED is on. ΔF = Fadj –F0 for each time point is shown. See Star Methods for additional details. The ΔF/F0 at 6 s (dashed black line) is plotted in (J) and (K) for comparisons among different genotypes and conditions. The red line shows the average background fluorescence changes, which include many larger peak amplitudes than P-Fr fluorescence changes (light blue lines), and these do not differ between experimental and control animals. The y axis only represents the scale of P-Fr but not background fluorescence changes. A total of 301 trials from 22 brains is shown here.
(H and I) Examples and cumulative analyses of GCaMP6s fluorescence changes in P-Fr processes in PlexBEc brains (otherwise similar to [F]–[G]). GCaMP6s fluorescence transiently decreases following each LED stimulation. A total of 266 trials from 22 brains is shown in I.
(J) Quantification of ΔF/F0 (6 s) in control and PlexBEcTM groups, without or with all-trans retinal feeding (n = 186 events for control [retinal]; n = 220 for PlexBEcTM [retinal]; n = 301 for control [retinal+]; n = 266 for PlexBEcTM [retinal+]; a total of 7–22 brains was imaged for each condition). LED stimulation results in a negative shift of ΔF/F0 only when PlexBEcTM-expressing flies are fed with retinal. N is the number of LED stimulation events quantified (F0 > 200 a.u.) in(J) and (K).
(K) Quantification of ΔF/F0 (at 6 s post-LED on) in control and PlexBEcTM groups without or with picrotoxin (PTX, 10 μM) in the bath with the imaged brains (n = 214 trials for control [DMSO]; n = 213 for PlexBEcTM [DMSO]; n = 240 for control [PTX]; n = 239 for PlexBEcTM [PTX]; eight brains were imaged for each condition). All flies were fed retinal. PTX treatment abolished the strong negative responses elicited by LED stimulation in the control brains and in the PlexBEcTM group. Two-way ANOVA with multiple comparisons was performed for (B); one-way ANOVA with Tukey’s multiple comparisons was used for (J) and (K).
All scale bars represent 20 μm.
To test for functional synaptic connections between ExFl2 and P-Fr neurons, we employed ex vivo two-photon Ca2+ imaging and optogenetics (Dag et al., 2019). We tested whether activation of ExFl2 neurons can affect P-Fr activity or vice versa (Figure 3C). We first asked whether optical stimulation of ExFl2 neurons activates or inactivates P-Fr neurons, as reported by GCaMP6s fluorescence changes in P-Fr processes (Figure 3D). In control adult brains, LED stimulation of ExFl2 neurons did not change GCaMP6s fluorescence in P-Fr processes (Figures 3F and 3G). However, in flies that expressed PlexBEcTM in P-Fr neurons, optical activation of ExFl2 neurons was associated with transient depression of GCaMP6s fluorescence in P-Fr neurons (Figures 3H and 3I). There was a significant difference between control and PlexBEcTM groups in GCaMP6s fluorescence changes following LED stimulation (ΔF/F0 (6 s)) (Figure 3J). The quantification shown here only includes events with pre-stimulation GCaMP6s fluorescence signals above a predetermined threshold (F0>200 a.u.), since depression of GCaMP6s fluorescence levels can only be detected when Ca2+ activity is above baseline. We calculated baseline GCaMP6s fluorescence metrics (mean and SD of F0, F0 span [max–min] for each brain sample) and found no statistically significant differences between control and experimental groups (data not shown); this suggests that basal Ca2+ levels and dynamics in the PlexBECTM group were similar to those in the corresponding control group.
To further validate the specificity of these inhibitory events, we conducted several additional control experiments. First, these transient depressions of GCaMP6s fluorescence are not seen in the absence of all-trans retinal (a cofactor required for CsChrimson activation) (Figure 3J), and this shows that GCaMP6s depression is specifically due to ExFl2 activation. Second, these events are also eliminated when 10 μM picrotoxin (PTX, a broad-spectrum chloride channel blocker) is present in the bath (Figure 3K). Because a higher concentration of PTX, such as 100 μM PTX, is required for efficiently blocking glutamate-gated chloride channels (Liu and Wilson, 2013), the depression events we observed here are most likely mediated by inhibitory GABAA receptors. Lastly, we did not detect GCaMP fluorescence changes in P-Fr neurons when R33E04 neurons (small-field neurons that target layer 6) were optically stimulated in either control or PlexBEcTM-expression conditions (data not shown). These results are in line with our GRASP data and thus suggest that P-Fr neurons receive specific synaptic inputs from ExFl2 axons in FB layer 6.
Because FB P-Fr processes also contain pre-synaptic terminals (Figure S5A), and because ExFl2 axons form axon-axonal dopaminergic connections (Liu et al., 2012; Ueno et al., 2012), we asked whether we could detect synaptic transmission in the opposite direction, from P-Fr processes to ExFl2 axons (Figures S5C and S5D). We observed that optical activation of P-Fr neurons did not change GCaMP6s fluorescence in ExFl2 axons in flies expressing PlexBEcTM in P-Fr neurons. Quantification of these data reveals no statistical difference between control and experimental groups (Figures S5E–S5I) and thus demonstrates that synaptic transmission from P-Fr processes to ExFl2 axons in the FB is either absent or comparably much weaker than transmission from ExF12 to P-Fr neurons. We could see that this lack of GCaMP6s fluorescence-level depression after P-Fr optogenetic stimulation is not simply due to low baseline Ca2+ levels in ExFl2 because we were able to record robust and dynamic GCaMP6s fluorescence during the duration of imaging epochs (Figures S5E and S5G, and data not shown).
To further demonstrate the significance of these functional imaging experiments and to exclude pseudoreplication effects that can result from acquisition of multiple measurements from the same brains, we performed additional statistical analyses by using a linear mixed effect model (see Star Methods for details) in which different F0 thresholds (0, 50, 500, 1000 a.u.) were investigated (only threshold = 0 a.u. is shown in Figure S6). These analyses show the same effects of LED stimulation in PlexBEcTM (Retinal+) brains independent of chosen thresholds. They are also consistent with data and analyses presented above and lead to the same conclusion: neural activity measured by changes in GCaMP6s fluorescence in P-Fr neurons that express PlexBEcTM are negatively correlated with optical stimulation of ExFl2 neurons.
Taken together, our GRASP and optogenetic/Ca2+ imaging experiments demonstrate that alteration of process lamination within the FB leads to the formation of ectopic connectivity between P-Fr and ExFl2 neurons.
P-Fr Neurons Modulate Arousal Threshold during Nighttime Sleep
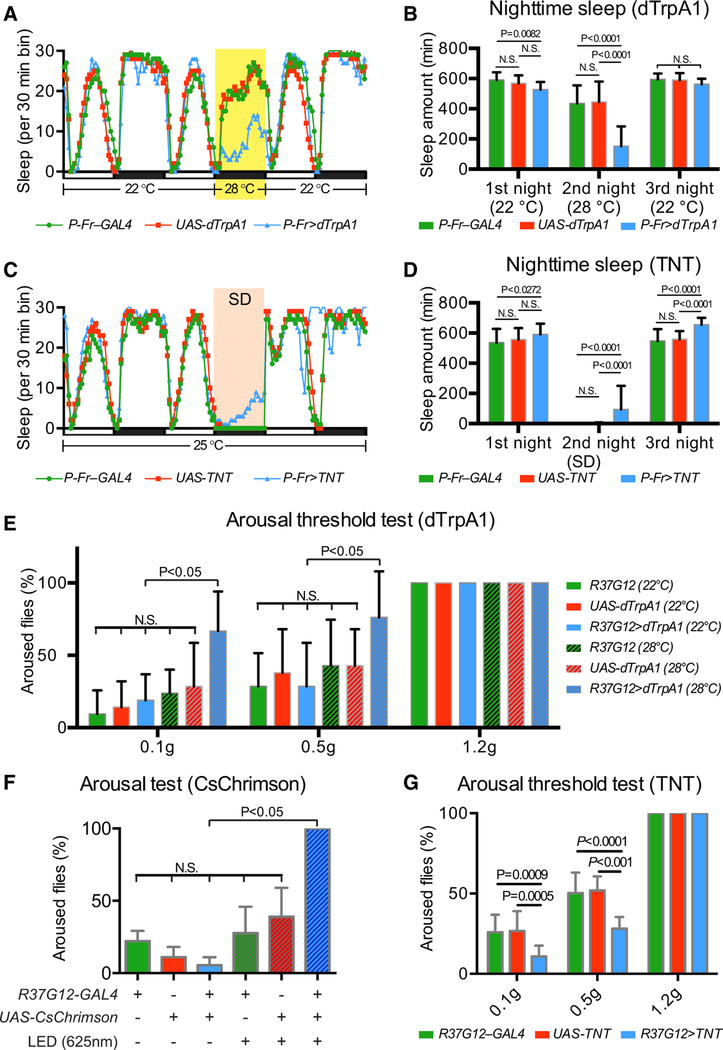
ExFl2 neurons, which function mainly as FB input neurons, have been implicated in regulating adult fly sleep behavior (Liu et al., 2012; Ueno et al., 2012; Qian et al., 2017; Donlea et al., 2011). A pair of dopaminergic neurons promotes fly wakefulness by locally inhibiting ExFl2 neurons that promote sleep. Our results show that P-Fr and ExFl2 neurons normally innervate adjacent layers in the FB. ExFl2 neurons apparently form ectopic inhibitory synapses onto P-Fr neurons after P-Fr lamination defects resulting from perturbation of Sema-2b/PlexB signaling. Therefore, we next wondered whether P-Fr neurons also play a role in regulating sleep behavior and whether alterations in ExFl2/P-Fr connections formed during pupal development affect sleep behavior in the adult. To address this question, we conditionally activated P-Fr neurons expressing the heat-activated cation channel dTrpA1 under the control of P-Fr–GAL4 (P-Fr–specific split-GAL4, see Star Methods for details) and also R37G12-GAL4 (Figures S7A and S7B) by increasing the temperature from 22C to 28 (or 29)°C for 12 h during the night. We measured sleep during this time interval and observed that the temperature shift reduced sleep in control and experimental groups (Figures 4A and S7C). However, select activation of P-Fr neurons led to significantly less total sleep time in these flies than in control flies (P-Fr–GAL4 flies, 434.8 ± 120.7 min of average total second nighttime sleep; UAS-dTrpA1 flies, 446.2 ± 135.0 min; P-Fr > dTrpA1 flies, 152.9 ± 130.7 min) (Figure 4B) (See Figure S7D and Star Methods for R37G12-GAL4 results). These results show that increasing P-Fr neuron activity promotes wakefulness.
To better understand P-Fr neuron functions in sleep, we blocked synaptic transmission in P-Fr neurons by using P-Fr– GAL4 or R37G12-GAL4 to selectively drive tetanus toxin light chain (TNT) expression. TNT-expressing flies showed normal sleep/wake cycles and no change in total duration of nighttime sleep when they were compared to controls. However, when we disrupted fly sleep through the use of a strong mechanical stimulus (Liu et al., 2016), flies expressing TNT (under either GAL4 driver) showed resistance to this sleep deprivation manipulation and slept significantly more than did control flies (Figures 4C, 4D, S7E, and S7F). This sleep loss difference during mechanical deprivation suggests that P-Fr neurons play a role in regulating arousal during sleep.
We next investigated this issue with more precision by conducting sleep arousal threshold analysis, a technique in which mechanical stimuli of differing strengths are used to arouse sleeping flies (Liu et al., 2016). We found that R37G12-GAL4 driving TrpA1 expression or CsChrimson expression in P-Fr neurons, after temperature elevation or light stimulation, respectively, resulted in flies that had a lower arousal threshold (i.e., were more easily aroused by external stimuli) (Figures 4E and 4F); in contrast, TNT-expressing flies had a higher arousal threshold (i.e., were harder to arouse with external stimuli) during nighttime sleep than did controls (Figure 4G). Thus, P-Fr neuron activity is strongly correlated with sleep behavioral changes: higher P-Fr activities decrease arousal threshold, and lower P-Fr activities increase it.
Because we had found that P-Fr neurons most likely receive greater inhibitory input from ExFl2 neurons when they express PlexBEcTM, we next asked whether PlexBEcTM expression in P-Fr neurons produced sleep phenotypes similar to those produced by TNT expression in P-Fr neurons; we tested this by assessing sleep time and arousal thresholds during mechanical sleep deprivation. Indeed, PlexBEcTM-expressing flies also showed resistance to sleep deprivation (Figures 5A and 5B) and had a higher arousal threshold than did controls during nighttime sleep (Figure 5C). To further examine whether mechanosensation or locomotion is affected in these dTrpA1, TNT, or PlexBEcTM expression paradigms, we tested the mechanosensory responses in these flies by using a grooming-based assay (Murphy et al., 2015). Our results show no difference between control and experimental groups (Figure 5D). We also calculated locomotion during the first 12 h day or night periods of our 3 day sleep assays (Figures 4A, 4C, 5A, 5C, S7C, and S7E). The results show that locomotion changes are not correlated with arousal threshold changes observed in these flies (Figure S8).
Together, these observations show that select genetic manipulation of Sema-2b/PlexB signaling in FB neuron subtypes alters connectivity in a sleep circuit and results in significant changes in sleep behavior, underscoring the importance of CX laminar organization.
DISCUSSION
We show here that in the fly brain CX, the secreted semaphorin Sema-2b, signaling via its PlexB receptor, regulates lamination of P-Fr neuron processes in the mFB. Mis-projection of P-Fr neuronsintoanadjacentlaminaresultsintheformationofectopicconnections between P-Fr and dorsal FB-projecting ExFl2 neurons, and this is associated with an increase in arousal threshold during nighttime sleep—these effects resemble those of P-Fr neuron inactivation. These results suggest that laminar organization of CX neuronal processes is critical for constraining synaptic contacts and thereby assuring normal connectivity and behavior.
Our finding that Sema-2b regulates FB lamination is reminiscent of our previous observations on sensory afferent axon targeting in the Drosophila embryonic CNS (Wu et al., 2011), where secreted semaphorins function at short range to locally guide axon targeting and Sema-2b can act as an attractant. Secreted semaphorins are also implicated in gradient-mediated, longer-range guidance events in the Drosophila antennal lobe (Sweeney et al., 2011; Joo et al., 2013). It will be important to determine what controls the local or more diffuse distribution of secreted semaphorins. P-Fr processes are attracted by Sema-2b in the mFB via signaling through the PlexB receptor. Interestingly, overexpression of PlexB in P-Fr neurons results in very similar lamination defects to those we observe following PlexB loss of function (LoF) (data not shown). A similar phenomenon, whereby PlexB GoF and LoF exhibit similar phenotypes, has been observed in fly olfactory circuit formation (Li et al., 2018; Guajardo et al., 2019), and this suggests that correctly tuned PlexB activity is required for normal axonal and dendritic responses to these ligands.
Our experiments show that different cellular and molecular mechanisms govern the formation of laminae in the FB compared to the EB. For example, ring (R) neuron axons in the EB segregate into different layers soon after they reach the midline, but FB neurons exhibit diverse cellular behaviors during FB layer formation. Sema-2b is expressed within the FB, but it is absent in the EB, and it signals through PlexB “forward” signaling to attract neuronal processes. In contrast, in the EB, the transmembrane semaphorin Sema-1a mediates axon lamination of certain R neuron subtypes through “reverse” signaling (Xie et al., 2017).
CX substructures and neural connectivity are compartmentalized along multiple dimensions. Columns are formed perpendicular to laminae in a wide range of neural systems in many organisms, from insects to vertebrates, and the columnar organization of PB and EB circuits in the CX has been implicated in regulating insect navigation behaviors (Heinze and Homberg, 2007; Heinze et al., 2009; Seelig and Jayaraman, 2015; Kim et al., 2017; Green et al., 2017; Turner-Evans et al., 2017). However, the cellular mechanisms and molecules that control column formation, and how column and lamina formation are coordinated during CX development, were not known. Our observations of PlexB LoF lamination phenotypes in FB P-Fr neurons suggest that one mechanism underlying column formation involves homotypic repulsion among small-field neurons that innervate neighboring columns. Homotypic repulsion mediates tiling of axon terminals and dendritic arbors in both invertebrate and vertebrate nervous systems (Lefebvre et al., 2015), and it will be interesting to determine whether the molecular mechanisms that constrain FB column organization are similar to those that regulate neuronal process tiling in sensory systems.
EB and FB neurons are implicated in regulating fly sleep behavior. These include the R5 neurons (formally called R2, which innervate the EB) (Liu et al., 2016; Omoto et al., 2017), the ExFl2 neurons (which innervate the FB) (Donlea et al., 2011; Liu et al., 2012; Ueno et al., 2012), and helicon cells (which connect ExFl2 FB neurons and R5 EB neurons) (Donlea et al., 2018). Our behavioral results show, for the first time, that in addition to these neurons, P-Fr neurons that project to the mFB also modulate sleep behaviors and control arousal in sleep. It is unclear whether P-Fr neurons participate in the ExFl2-helicon-R5 circuit or if they function in parallel to it (Figure 5E). Our functional imaging and behavioral analyses of flies that express PlexBEcTM in P-Fr neurons suggest that FB lamination provides a structural basis for separating distinct streams of information flow along ExFl2 or P-Fr neurons, since lamination defects resulting from disruption of Sema-2b/PlexB signaling disturb this separation and correlate well with changes in sleep behavior. However, we cannot rule out other possible explanations for how loss of PlexB signaling in P-Fr neurons affects their functions and animal behaviors. For example, PlexBEcTM expression in P-Fr neurons might not only result in expansion of their processes into layer 6 but could also cause changes in normal synaptic connections within layers 3–5. Future studies that map P-Fr neuron synaptic partners in the FB will help address this issue.
Understanding the relationship between structural principles underlying circuit development and behavioral output is key for discerning the logic of nervous system organization. The Drosophila CX is a major sensory integration and locomotion control center in the insect brain, and it provides an extremely useful model for studying how neural circuits are formed and how they control animal behavior (Turner-Evans and Jayaraman, 2016). Detailed investigation into CX development, neuronal composition, connectivity, and overall organization continues to provide critical understanding of the circuitry that underlies integration of extrinsic sensory experience and internal brain states and leads to the subsequent generation of appropriate motor output. We can expect that a thorough understanding of CX neural-circuit organization and function will also inform our understanding of analogous, phylogenetically conserved features critical for the execution of complex behaviors.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Alex L. Kolodkin (kolodkin@jhmi.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Fly Strains and Genetics
A detailed list of the genotypes of the flies used in each figure is included in Table S1. All GMR GAL4, lexA and R37G12-p65AD lines were generated at the Janelia Research Campus/HHMI (Pfeiffer et al., 2008; Jenett et al., 2012) and were obtained from the Bloomington Drosophila Stock Center (BDSC) at Indiana University.
The VT036267-GAL4 fly line was obtained from the Vienna Drosophila Resource Center (VDRC). A VT036267-GAL4DBD line was generated from the GAL4 line using the HACK system (Lin and Potter, 2016; Xie et al., 2018).
The following transgenes were used: UAS-CD8::GFP (RRID: BDSC_5130) (Lee and Luo, 1999), 13xlexAop2-IVS-myr-Tomato (RRID: BDSC_52272), UAS-Syt::GFP, UAS-DenMark (RRID: BDSC_33064) (Nicolaï et al., 2010), UAS-CD4::spGFP1–10, lexAopCD4::spGFP11 (RRID: BDSC_57321) (Gordon and Scott, 2009), tub-GAL80ts (RRID: BDSC_7108), UAS-Dicer2 (RRID: BDSC_24644) (Dietzl et al., 2007), UAS-PlexB, UAS-3xMyc-PlexB and UAS-3xMyc-PlexBEcTM (Wu et al., 2011), MCFO3 and MCRO4 (Nern et al., 2015), eyFLP, UASmcd8GFP, FRT19A; sp/CyO; l(3)/TM3,Sb (RRID: BDSC_42728), hsFLP, tubP-GAL80, FRT19A (RRID: BDSC_5132), UAS-dTrpA1 (RRID: BDSC_26263), UAS-TNT (RRID: BDSC_28838), UAS-Stinger-GFP (RRID: BDSC_65402) (Vef et al., 2006). The following lines were kindly shared by Vivek Jayaraman/HHMI: 20xUAS-CsChrimson-mCherry, 13xlexAop-CsChrimson-mtdT, 20xUAS-IVS-GCamP6s and 13xlexAop-IVS-GCamP6s. The following RNAi lines were ordered from the Vienna Drosophila Resource Center (VDRC) (Dietzl et al., 2007): UAS-Sema-2b-RNAi (KK101842), and from BDSC (Perkins et al., 2015): UAS-Sema-2b-RNAi (HM05143, RRID: BDSC_28942).
The following mutant alleles were used for gene expression analysis or loss-of-function experiments: Sema-2bC4 (Wu et al., 2011), PlexBKG00878 (Ayoob et al., 2006), PlexBMI15559 (Venken et al., 2011). PlexBGAL4 was generated from PlexBMI15559 by converting the MiMic cassette to a ‘Trojan’ exon containing T2A-GAL4 components (Diao et al., 2015). Briefly, PlexBMI15559 flies were recombined with yw; sp/CyO; LoxP(Trojan-GAL4.1) (RRID: BDSC_60305) and yw, hs-Cre, vas-dɸC31 (RRID: BDSC_60299) flies through multistep crosses to combine all components required for the MiMic cassette exchange. Candidate progeny flies were crossed to UAS-mCD8::GFP reporter flies for screening.
Genetic Analyses
Flies were reared at 25°C for most experiments. For MARCM analyses, hsFLP was used to generate mosaic clones, as previously described (Lee and Luo, 1999) with minor modifications. To generate small P-Fr neuron clones, middle and late 3rd instar larvae were heat shocked once for 60 min at 37°C, or twice for 60 min at 37°C, on two consecutive days, and then adult female flies were dissected for immunohistochemistry analysis. For RNAi experiments, parental flies were kept at 25°C to lay eggs, and F1 larvae were transferred to and raised at 29°C until F1 pupae were dissected at specific developmental stages.
For adult fly lifespan analysis, 10–5 newly emerging male flies were collected in each vial and were transferred into new vials every 2 days until all flies were dead. The number of live flies in each vial was recorded at the time of transfer. The survival rate was calculated as the number of living flies recorded each day divided by the starting number of flies in each vial.
METHOD DETAILS
Immunohistochemistry
Fly brains were quickly dissected from pupae submerged in cold phosphate buffered saline (PBS, pH 7.4), or if from adult flies, in cold PBS with 0.1% Triton X-100 (0.1% PBT); brains were immediately transferred into fixation buffer (4% paraformaldehyde in 0.1% PBT). Brains were gently rotated in fixation buffer for 20 min at room temperature (RT). After washing with 0.1% PBT, fly brains were incubated with blocking buffer (5% normal goat serum in 0.3% PBT) for 1 h at RT. Then, brains were incubated with primary antibodies for two days at 4°C, or for one day at RT. Brains were washed extensively (20 min, 3 times in 0.3% PBT at RT) after primary antibody incubation. After washing, brains were incubated with secondary antibodies for another two days at 4°C, or in some cases one day at RT. Brains were washed intensively (20 min, 3 times in 0.3% PBT at RT) following secondary antibody incubation. After a final wash, brains were incubated with a drop of Vectashield mounting medium (Vector Laboratories, H-1000) overnight at 4°C. Then, brains were loaded onto glass slides (Superfrost Plus, Fisherbrand) prepared with silicon spacers (120 μm thick) (Grace Biolabs), covered with a glass coverslip (1.5 oz., Fisherbrand), and then used for image analyses.
The following primary antibodies were used: chicken anti-GFP (1:1000 dilution, AVES, RRID: AB_10000240), mouse anti-GFP (1:100, Sigma-Aldrich G6539, RRID: AB_259941), rabbit anti-DsRed (1:1000,Clontech, RRID: AB_10013483), rat anti-HA (1:500, Roche, 3F10, RRID: AB_390915), mouse anti-Flag (1:500, Sigma-Aldrich, RRID: AB_259529), mouse anti-Myc (1:1000, Sigma-Aldrich, RRID: AB_309725), mouse anti-Brp (1:50, Developmental Studies Hybridoma Bank (DSHB), Nc82, RRID: AB_2314868), rat anti-CadN (1:50, DSHB, DN-Ex#8, RRID: AB_2619582), rabbit anti-Sema-2b (RRID: AB_2569774). The secondary antibodies were raised in goat against rabbit, chicken, mouse, and rat antisera (Life Technology) and conjugated to Alexa 488 (1:1000), Alexa 555 (1:1000) or Alexa 647 (1:300). Antibodies were prepared in blocking buffer with 0.02% NaN3; some primary antibodies were reused several times.
A polyclonal rabbit anti-GFP antibody (1:1000, Synaptic Systems, RRID: AB_887725) was used for our GRASP experiments. For two reasons were this rabbit antibody chosen over the monoclonal mouse anti-GFP antibody (Sigma G6539), which was previously used for detecting reconstituted GFP (Gordon and Scott, 2009). First, the rabbit antibody gave stronger signals illuminating reconstituted GFP. Second, the rabbit antibody weakly binds to spGFP1–10, while the mouse antibody does not, so we were able to use these ‘noise’ signals as an internal control for spGFP1–10 expression. The signals for spGFP1–10 and reconstituted GFP are significantly different and were quantitatively separated under different intensity threshold conditions in our analyses.
Image Acquisition
Most of fixed tissue images were taken on a LSM700 confocal microscope (Zeiss) using either a 20X air lens (N.A. 0.8) or a 63X oil immersion lens (N.A. 1.4), except that images in Figure S6, A&B were acquired on a Axio Observer Z1 microscope (Zeiss) using a 20X air lens (N.A. 0.35). Most of the confocal image stacks were taken under 1X zoom, in a 512X512 configuration, and using 1 μm (20X lens) and 0.5 μm (63X lens) Z resolution. Images stacks were processed with Fiji (ImageJ), Adobe Photoshop CS6 and Imaris (Bitplane). Figures were composed using Adobe Illustrator CS6.
Optogenetic Photostimulation and Functional Imaging
All-trans retinal (ATR) (R2500, Sigma) was prepared as a 40 mM stock solution dissolved in ethanol, and this stock was diluted into melted standard fly food to achieve a final concentration of 400 μM. Re-solidified food was used immediately or stored at 4C for no more than 1 week. Flies were raised on ATR food in darkness for 24–48 h before dissection. Fly brains were quickly dissected in Drosophila physiological saline solution (101 mM NaCl, 3 mM KCl, 1 mM CaCl2, 4 mM MgCl2, 1.25mM NaH2PO4, 20.7 mM NaHCO3, and 5 mM glucose; pH 7.2), which was pre-bubbled with 95% O2 and 5% CO2 for at least 10 min. Then, brains were transferred to a 35mm glass bottom Petri dish (FluoroDish, World Precision Instruments, INC.) containing 2 mL of saline solution and 1 mM tetrodotoxin (TTX). Picrotoxin (PTX) stock solution (10 mM) was prepared in DMSO and was diluted at 1:1000 in the saline solution when required. Calcium imaging was conducted using an upright LSM 880 system (Zeiss) with a two-photon laser source (920 nm) and a 40x water-immersion lens (NA 0.8). Time-lapse images were acquired at 1 Hz.
A red LED with peak wavelength at 627 nm (ALK-26M-1UP-KIT with Luexeon Rebel Red LED and wide optic, LED supply) and Buckpuck driver (RapidLED, Randolph, Vermont) was used to stimulate CsChrimson-expressing neurons. The LED was mounted directly underneath the FluoroDish and light was presented at 0.338 mW/mm2 as measured with a light meter (Cal-light 400, the Cooke Corporation). Photostimulation was delivered by 5 s pluses with 55 s intervals and was controlled by a microcontroller board (Aduino Uno).
Beam-break Analyses for Sleep Measurements
Single fly sleep behavior was measured using consolidated locomotor inactivity as previously described (Liu et al., 2016). Flies were reared in the vials containing standard Drosophila medium. 0–2 day old female flies were collected and loaded into glass tubes containing 5% sucrose/2% agarose. One day later, their activities were then monitored using the Drosophila Activity Monitoring System (Trikinetics) in an incubator with a 12 h:12 h light:dark (LD) condition. Activity counts were collected in 1 min bins, and sleep was identified as periods of inactivity lasting at least 5 min. Sleep data were collected for 3 days, after discarding the first day following loading.
Standard Mechanical Sleep Deprivation
For overnight sleep deprivation experiments, flies were mechanically stimulated for 2 s per min for 12 h using a vortexer mounting plate and multi-tube vortexer (Trikinetics) (Liu et al., 2016).
Arousal Threshold Measurement
To quantify arousal threshold, increasing mechanical stimulus intensities at ZT16 (0.1g-force), ZT18 (0.5g-force), and ZT20 (1.2g-force) for 1 s were applied by using a vortexer mounting plate and multi-tube vortexer (Trikinetics) (Liu et al., 2016). Flies inactive for 5 min prior to the stimulus were considered asleep, and only data from flies with a sleep bout >15 min prior to the mechanical stimulus were analyzed. Flies displaying at least 1 count of activity within the 3 min after the stimulus were considered aroused.
Heat Activation
For UAS-dTrpA1 experiments, flies were raised at 22°C and 1 day of baseline sleep was recorded at 22°C. Flies were then shifted to 28C for 12 h during the night (ZT12-ZT24), in order to activate the neurons.
Single Fly Video Analyses
For experiments involving simultaneous measurement of sleep behavior with optogenetic manipulation, individual flies were anesthetized on ice and glued to a 0.025 mm thick stainless steel shim using dental wax. Flies were given ad libitum access to food (rehydrated food flakes, Nutri-Fly Instant, Genesee Scientific), and leg movements were continuously monitored with an IR-sensitive CCD camera (Ailipu Technology) at 2 fps. Inactivity assessed by leg movements was transformed to sleep behavior via a frame-subtraction approach using a noise threshold algorithm, as previously described (Tabuchi et al., 2018). Sleep was identified using a 5 min window for this assay was validated by arousal threshold criteria, as determined by administration of varying strengths of air puffs. 90 min of recording data were used for these analyses, because the first 30 min of the 2 h window were used to allow the fly to acclimate to the preparation.
Single Fly Arousal Measurement
Under the single fly video platform described above, instead of mechanical stimulus, air puffing stimuli with a pressure of 7 psi were used in the presence or absence of optogenetic activation of P-Fr cells. The pressure of air puffing stimulus was controlled and delivered by Picospritzer III (Parker Hannifin).
Optogenetic Stimulation of Living Flies
Flies were fed 1 mM all-trans-retinal (MilliporeSigma), mixed in rehydrated food flakes (Nutri-Fly Instant, Genesee Scientific). A collimated LED light source (625nm, Thorlabs) was used for photostimulation of individual CsChrimson-expressing flies. A fiber optic cannula (Thorlabs) was attached to the LED to converge the light. An Arduino Uno board (Arduino) was connected to a computer running MATLAB and used to control the timing of photostimulation. Photostimulation was delivered by 5ms duration 3 Hz frequency pulses having a poissonally distributed temporal structure, using a MATLAB-connected Arduino interface.
Mechanosensory Response Analyses
To validate the integrity of mechanosensory responses, these were tested as previously described (Murphy et al., 2015). Flies were anesthetized on ice and then were decapitated using microdissection scissors. Headless flies were placed in a closed, moist, environment and incubated overnight for recovery. Then, their mechanosensory responses were tested via eliciting a grooming reflex by deflecting the single scutellar bristle toward the fly body with fine forceps followed by observation of leg movement responses. These responses were quantified by giving a score of one to flies that lift their leg in response to bristle stimulation, and a score of zero to flies that do not move their legs. Five trials spaced 2 min apart were performed for each fly, and the percentage of responsiveness was calculated by obtained score number divided by five.
QUANTIFICATION AND STATISTICAL ANALYSIS
Standard Image Processing
To quantify Sema-2b distribution in the FB (Figure 7A), single optical sections from similar anterior-posterior positions were manually selected, using Sema-2b antibody staining in FB layer 9 as a landmark, from Z stacked images of eight 40 h APF pupal brains. A box of 100X300 pixels (19.85X59.54 mm) was drawn to cover the FB area around the midline (shown in Figure 7A). Grey scale values for Sema-2b immunofluorescence along the dorsal-ventral (D-V) axis (the long-axis of the box) were measured using the ‘Plot Profile’ function in the Fuji (ImageJ) software package. The gray scale value in each D-V position was normalized to the largest value in each brain. The mean of normalized gray scale value in each D-V position from eight brains was plotted against D-V position (‘0’ was set for the dorsal margin of the box and numbers decrease ventrally).
For comparison of Sema-2b immunolabeling in control and RNAi pupal brains, single optical sections from similar anterior-posterior positions were manually selected from Z stacked images of control or RNAi groups (eight 48 h APF pupal brains for each group). The average greyscale value of Sema-2b immunofluorescence was measured within the FB (defined by CadN staining) and was normalized to the average greyscale value for the entire visual field. The ratios were plotted in Figure 7C.
To quantify dorsal FB innervation by P-Fr processes, single optical sections from similar anterior-posterior positions (corresponding to the ‘posterior plane’ in Figure 7D) were manually selected from Z stacked images of adult brains. P-Fr processes were measured by counting pixel areas that had mtdT immunofluorescence greyscale values above a threshold that was automatically determined by the software (Fuji) to exclude background signals. The P-Fr processes located within layer 6 (identified by stronger Brp staining in the dorsal FB) were normalized to total P-Fr processes within the entire FB for each brain, and ratios were plotted in Figure 7E.
Branching Distribution Analysis—MARCM and MCFO4
Because of FB curvature, branch distribution analysis was performed using an FB-centric coordinate system (Figures S4A and S4B). The Dorsal-Ventral axis of the FB (D-VFB) was defined as the axis orthogonal to the FB layers. Note that the orientation of the D-VFB axis was annotated separately for each neural branching distribution in our dataset, since its orientation in the image volume varies along the Left-Right axis of the brain due to the curvature of the FB. The Anterior-Posterior axis of the FB (A-PFB) was defined as the axis orthogonal to the D-VFB axis and the Left-Right brain axis (L-Rbrain). The Left-Right axis of the FB (L-RFB) was defined as the axis tangential to the curvature of the FB; in other words, orthogonal to both the D-VFB and A-PFB axes. The L-RFB axis was manually annotated in the frontal view of the FB, which was obtained from each image volume using ImageJ’s “3D Project” tool. To correct for the rotated orientation of the FB in our imaging volumes, this projection was performed at a 20-degree angle of rotation around the L-Rbrain axis, which was also the x axis of our imaging volumes. Mounting was found to produce a consistent orientation across samples; hence a constant 20-degree rotation was used for all samples. In the MARCM experiments, R37G12-lexA>mtdT staining was used as an anatomical reference for annotating the L-RFB axis. In MCFO4 experiments, staining of each single neuron was used to define the L-RFB axis, assisted by the gross anatomical features of FB background staining. Note that the L-RFB therefore also varies along the Left-Right axis of the brain, remaining tangential to the FB curvature.
Neural branching distributions were computed as a function of position along the D-VFB axis (Figure S4C). These branching measurements were calculated as the total number of fluorescence intensity voxels above a chosen threshold (see below) in each plane that is orthogonal to the D-VFB axis, and hence tangential to the FB layer curvatures. In all these planes corresponding to positions along the D-VFB axis, voxel counts proceeded in two successive steps along the A-PFB and L-RFB axes. First, voxel counts were summed along the A-PFB axis using a modified version of ImageJ’s “3D Project” tool, yielding an intermediate voxel count as a function of D-VFB and L-RFB position. As before, to correct for the rotated orientation of the FB in our imaging volumes, this 3D voxel count projection was performed at a 20-degree angle of rotation around the L-Rbrain axis (Figure S4B). A voxel was counted when its fluorescence level exceeded an arbitrary low threshold (threshold = 7, maximum pixel value = 255). Second, these intermediate voxel counts were summed along the L-RFB axis, yielding a 1-dimensional distribution of voxel counts along the D-VFB axis. For some images, outgoing axons contributed significantly to the fluorescence signal, thus obscuring the start and end of layers 3–5 of the FB. These processes were manually masked where possible.
FB size differed slightly among samples. To correct for this, in each image volume the height (in pixels) along the D-Vbrain axis (y axis in our image volumes) was annotated in the Z slice to show where this height was at its maximum. This pixel height annotation underwent the same 20-degree rotation around the L-Rbrain axis (x axis in our image volumes), thus creating a measure of FB pixel height along the D-VFB axis. All D-VFB axis positions were subsequently normalized to this FB height measurement and computed separately for each sample. The measure of total branching at each D-VFB position was normalized to the maximum branching measured for each sample. In Figures 2C and 2E, thick lines represent mean voxel counts across samples as a function of D-VFB position. The ribbons indicate standard deviations. Other than manual annotation, all data were processed using custom Python (version 3.5) and R (version 3.4) scripts. All data files and custom ImageJ, Python, and R scripts for data processing and plotting, and manual annotations are available in the repository accompanying this paper.
Branching Distribution Analysis—Group Expression Experiment
Neural branching in the group expression experiments (Figure 1C) was analyzed as described above for the MARCM and MCFO4 experiments, with the exception that neurons could no longer be individually labeled. Instead, branching was measured orthogonal to the D-VFB axis, as described above, but for a longer segment of the FB: starting at approximately the leftmost point of the FB and ending closely toward the middle of the FB while maintaining an approximately straight tangent line. All further analyses proceeded as described above for MARCM and MCFO4 experiments. The overlap index of P-Fr and ExFl2 defined the integral of the normalized branch counts within layer 6 compared to total branch counts throughout FB layers. Layer 6 was arbitrarily set as the region beyond (dorsal to) the point at which ExFl2 fluorescence started to exceed 5% of the maximum ExFl2 fluorescence in that sample.
Functional Imaging Analysis
Contours of P-Fr or ExFl2 neuronal processes were manually traced for each sample, guided by GCaMP6s fluorescence. For P-Fr GCamP6s experiments, contours of P-Fr processes excluded layer 6 to reduce bleaching by the strong CsChrimson-mtdT fluorescence from ExFl2 axons. Masks were used to calculate background fluorescence and were computed by positioning manually traced contours of P-Fr or ExFl2 processes in the most ventral position in each image stack, resulting in no overlap between the P-Fr or ExFl2 contours. Fan-shape body and background fluorescence signals were computed using these masks. Fluorescence signals were normalized using the areas of the respective FB and background masks. The difference between raw FB fluorescence (F) and background fluorescence (Fbac) was calculated as the ‘adjusted’ P-Fr or ExFl2 fluorescence (Fadj = F-Fbac).
Because LED stimulation itself substantially increases background fluorescence across the entire imaging field, LED stimulation onset was defined as the derivative of the background fluorescence signal rising above an arbitrarily set threshold (threshold = 50), which was significantly surpassed by most events (mean background fluorescence change of detected events = 131 ± 61) and was far above the baseline fluorescence fluctuations (max fluorescence change during the 10 s before event detection = 25, excluding the frame immediately before event detection), and so reliably detects LED onset. Detected events were confirmed to have the correct period, and only 64 events (or 1.35% of the 4719 total events scored) were found to have remained undetected based on manual inspection of the data. Because LED onset was not synchronized to image frame acquisition, slight fluorescent waveform misalignments resulted from desynchronized acquisition. However, these slight deviations do not affect our results, since fluorescent changes, ΔF(6 s), were measured six seconds after LED onset was detected. Since LED stimulation only lasts five seconds, measuring ΔF(6 s) ensures that the signal is not contaminated by LED excitation. Although all of our figures are based on a six second delay in measuring ΔF, relative to LED onset, the same conclusions hold at delays of five, seven or eight seconds (data not shown). Based on LED stimulations, 31 min time-lapse images were broken down into 30 individual events, each containing: 4 frames proceeding, 5 frames during and 5 frames following the 5 s LED stimulation. Within each event, changes in GCaMP6s fluorescence were computed as ΔF/F0. F0 is, therefore, the average adjusted fluorescence in P-Fr or ExFl2 processes during the four frames preceding LED onset. The fluorescence change, DF, was computed as Fadj-F0 for each frame. We plotted ΔF/F0(6 s) to compare fluorescence changes among different genotypes and treatments. Other than manual annotation of P-Fr and ExFl2 outlines, all data were processed using custom Python (version 3.5) and R (version 3.4) scripts. All data files and custom Python and R scripts for data processing and plotting, and manual annotations are available in the repository accompanying this paper.
Mixed Effect Model Analysis
To model the effect of LED stimulation on each experimental group, we used the following linear mixed effect model to capture all interactions between experimental conditions:
where genotype ˛ε{Control, PlexBEcTM}, solution ˛ {Retinal-, Retinal+, Retinal+/DMSO, Retinal+/PTX}, LED ˛ {ON, OFF} are fixed effects. The variables trial and sample represent the timing of the trial and specific brain sample, respectively, and are treated as random effects. LEDOFF samples were generated by artificially shifting the window over which ΔF/F0(6 s) is computed by 30 s (half the period of trial repetition). LEDON samples correspond to the original (unshifted) ΔF/F0(6 s) analysis window synchronized with LED stimulation. The model was fit using the nlme R package (version 3.1–137) (Pinheiro, Bates, DebRoy, Sarkar, 2013). In the absence of LED stimulation, neither genetic background nor solution contents, nor their interaction, were associated with a depression of activity following LED stimulation, nor was LED stimulation alone (p > 0.5 for all variables). However, LED stimulation in PlexBEcTM (Retinal+) brains (that is, the LED: PlexBEcTM:Retianl+ interaction term in our model) was associated with a marginal 0.1 change in ΔF/F0(6 s) compared to baseline (p = 0.0002, 95% CI = [−0.15, 0.05]). LED stimulation in PlexBEcTM (Retinal+DMSO) brains was associated with a marginal 0.08 change in ΔF/F0(6 s) compared to baseline (p = 0.0088, 95% CI = [−0.14, 0.02]). LED stimulation across Retinal+ brains is associated with a marginal 0.05 change in DF/F0(6 s) (p = 0.01, 95% CI = [0.01, 0.09]). For all other model parameters, p > 0.05.
The ExFl2 experiments were modeled similarly as follows: ΔF/F0(6 s) = 1 + genotype * LED + (1 + trial j sample). No significant effects were observed.
Statistical Analysis
Unless specified, all statistical tests were performed by Prism 6 (GraphPad); methods are listed below. Post hoc power analysis was conducted on existing datasets by G*Power 3.1 (Universität Düsseldorf). All t tests or one-way ANOVA tests that have p < 0.05 also€ have power >0.8. N equals to number of animals in most of cases, except that N represents number of trials in Figures 3J, 3K, S5I, and S6.
Supplementary Material
Figure 6. P-Fr Neurons Regulate Arousal during Sleep.
(A) A split GAL4 fly line (R37G12-p65AD; VT036267GAL4BD, abbreviated as P-Fr–GAL4 here) was generated to specifically target P-Fr neurons (see expression pattern in Figure S6B). Sleep profiles of P-Fr–GAL4-GAL4/+ (green line), UAS-dTrpA1/+ (red line), and P-Fr>dTrpA1 (blue line) flies during a three-day recording period are shown here. Sleep time is plotted in 30 min bins. White and black bars indicate 12 h light and dark periods, respectively. The yellow column indicates 12 h dTrpA1 activation at 28°C.
(B) Quantification of nighttime sleep amount per flyin the first, second, and third 12 h dark periods is shown in (A). The temperature increase (from 22C to 28°C) reduced nighttime sleep overall but P-Fr>TrpA1 flies exhibited significantly more sleep loss than the two control groups. A total of 30–31 flies were tested for each genotype.
(C) Sleep profiles of P-Fr–GAL4/+ (green line), UASTNT/+ (red line), and P-F >TNT (blue line) flies in a three-day recording at 25°C. The yellow column indicates 12 h sleep deprivation with mechanical stimulation (~1.0 g).
(D) Quantification of nighttime sleep amount per fly in the first, second, and third 12 h dark periods shown in (C). P-Fr–GAL4/+ n = 23 flies; UAS-TNT/+ n = 32 flies; P-Fr>TNT n = 32 flies. Mechanical stimulation largely attenuated fly sleep overall, but P-Fr>TNT flies exhibited a significant amount of residual sleep when they were compared to flies in the two control groups.
(E) Sleep arousal threshold tests of flies in which P-Fr neuron activity was tuned through the use of R37G12-GAL4 to express dTrpA1 at two different temperatures (22°C versus 28°C), here showing the percentage of flies aroused by mild (0.1g), moderate (0.5g), or strong (1.2g) mechanical stimuli during nighttime sleep. A total of seven flies were tested for each genotype (g = relative centrifugal force). More R37G12-GAL4>dTrpA1 flies were aroused than in control groups after mild and moderate mechanical stimuli.
(F) Sleep arousal tests of flies in which P-Fr neuron activity was tuned through the use of R37G12GAL4 to express CsChrimson coupled with LED stimulation, here showing the percentage of flies aroused by air-puffing stimuli (7 psi) during sleep. A total of six flies were used for each genotype. Note that all R37G12-GAL4>CsChrimson flies with optical activation were aroused after air puff stimulation, which is significantly different from the results for other control groups.
(G) Sleep-arousal-threshold tests of flies in which P-Fr neuron inactivation was accomplished through the use of TNT expression, here showing the percentage of flies aroused by mild (0.1g), moderate (0.5g), or strong (1.2g) mechanical stimuli during nighttime sleep. A total of 6–10 flies was used for each genotype for tests. In contrast to (E) and (F), fewer R37G12-GAL4>TNT flies were aroused compared to flies in control groups following mild and moderate stimuli. Two-way ANOVA with multiple comparisons was conducted for all panels in this figure except that a one-way ANOVA was used for (F).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Chicken anti-GFP | AVES | RRID: AB_10000240 |
| Mouse anti-GFP | Sigma-Aldrich G6539 | RRID: AB_259941 |
| Rabbit anti-GFP | Systems | RRID: AB_887725 |
| Rabbit anti-DsRed | Clontech | RRID: AB_10013483 |
| Rat anti-HA | Roche | RRID: AB_390915 |
| Mouse anti-Flag | Sigma-Aldrich | RRID: AB_259529 |
| Mouse anti-Myc | Sigma-Aldrich | RRID: AB_309725 |
| Mouse anti-Brp (Nc82) | Developmental Studies Hybridoma Bank | RRID: AB_2314868 |
| Rat anti-CadN (DN-Ex#8) | Developmental Studies Hybridoma Bank | RRID: AB_2619582 |
| Rabbit anti-Sema-2b | Liqun Luo lab, Stanford | RRID: AB_2569774 |
| Goat anti-Chicken IgG (H+L) Alexa Fluor 488 | Thermo Fisher Scientific | Cat#: A32931 |
| Goat anti-Mouse IgG (H+L) Alexa Fluor 488 | Thermo Fisher Scientific | Cat#: A-11029 |
| Goat anti-Rabbit IgG (H+L) Alexa Fluor 488 | Thermo Fisher Scientific | Cat#: A-11034 |
| Goat anti-Mouse IgG (H+L) Alexa Fluor 555 | Thermo Fisher Scientific | Cat#: A-21424 |
| Goat anti-Rabbit IgG (H+L) Alexa Fluor 555 | Thermo Fisher Scientific | Cat#: A-21429 |
| Goat anti-Rat IgG (H+L) Alexa Fluor 555 | Thermo Fisher Scientific | Cat#: A-21434 |
| Goat anti-Mouse IgG (H+L) Alexa Fluor 647 | Thermo Fisher Scientific | Cat#: A-21236 |
| Goat anti-Rat IgG (H+L) Alexa Fluor 647 | Thermo Fisher Scientific | Cat#: A-21427 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Tetrodotoxin citrate | Abcam | Cat#: ab120055 |
| Picrotoxin | Sigma-Aldrich | Cat#: P1675-1G |
| 16% Paraformaldehyde | Electron Microscopy Sciences | Cat#: 15710 |
| All-trans retinal | Millipore Sigma | Cat#: R2500 |
| Nutri-Fly Instant food flakes | Genesee Scientific | Cat#: 66–117 |
| Experimental Models: Organisms/Strains | ||
| Drosophila: R37G12-GAL4 | Bloomington Drosophila Stock Center | RRID: BDSC_49967 |
| Drosophila: R37G12-lexA | Bloomington Drosophila Stock Center | RRID: BDSC_52765 |
| Drosophila: R37G12-p65AD(attP40) | Bloomington Drosophila Stock Center | RRID: BDSC_70661 |
| Drosophila: R84C10-GAL4 | Bloomington Drosophila Stock Center | RRID: BDSC_48378 |
| Drosophila: R84C10-lexA | Bloomington Drosophila Stock Center | RRID: BDSC_54339 |
| Drosophila: R13C03-GAL4 | Bloomington Drosophila Stock Center | RRID: BDSC_48550 |
| Drosophila: R13C03-lexA | Bloomington Drosophila Stock Center | RRID: BDSC_54421 |
| Drosophila: R28C12-lexA | Bloomington Drosophila Stock Center | RRID: BDSC_54622 |
| Drosophila: R45F08-GAL4 | Bloomington Drosophila Stock Center | RRID: BDSC_49565 |
| Drosophila: R45F08-lexA | Bloomington Drosophila Stock Center | RRID: BDSC_61545 |
| Drosophila: R33E04-GAL4 | Bloomington Drosophila Stock Center | RRID: BDSC_49752 |
| Drosophila: R33E04-lexA | Bloomington Drosophila Stock Center | RRID: BDSC_54728 |
| Drosophila: R92D04-GAL4 | Bloomington Drosophila Stock Center | RRID: BDSC_40615 |
| Drosophila: R83H12-GAL4 | Bloomington Drosophila Stock Center | RRID: BDSC_40374 |
| Drosophila: R40E08-lexA | Bloomington Drosophila Stock Center | RRID: BDSC_52779 |
| Drosophila: VT036267-GAL4 | Vienna Drosophila Resource Center | N/A |
| Drosophila: VT036267-GAL4DBD | In this study | N/A |
| Drosophila: UAS-mCD8::GFP | Bloomington Drosophila Stock Center | RRID: BDSC_5130 |
| Drosophila: 13xlexAop2-IVS-myr-Tomato | Bloomington Drosophila Stock Center | RRID: BDSC_52272 |
| Drosophila: UAS-Syt::GFP, UAS-DenMark | Bloomington Drosophila Stock Center | RRID: BDSC_33064 |
| Drosophila: UAS-CD4::spGFP1–10, lexAop-CD4::spGFP11 | Bloomington Drosophila Stock Center | RRID: BDSC_57321 |
| Drosophila: tub-GAL80ts | Bloomington Drosophila Stock Center | RRID: BDSC_7108 |
| Drosophila: UAS-Dicer2 | Bloomington Drosophila Stock Center | RRID: BDSC_24644 |
| Drosophila: UAS-TNT | Bloomington Drosophila Stock Center | RRID: BDSC_28838 |
| Drosophila: 20XUAS-IVS-CsChrimson.mVenus | Bloomington Drosophila Stock Center | RRID: BDSC_55136 |
| Drosophila: UAS-dTrpA1 | Bloomington Drosophila Stock Center | RRID: BDSC_26263 |
| Drosophila: MCFO3 | Rubin lab | N/A |
| Drosophila: MCFO4 | Rubin lab | N/A |
| Drosophila: UAS-PlexB | Kolodkin lab | N/A |
| Drosophila: UAS-Myc-PlexB | Kolodkin lab | RRID: BDSC_66163 |
| Drosophila: UAS-Myc-PlexBEcTM | Kolodkin lab | RRID: BDSC_65744 |
|
Drosophila: eyFLP, UASmcd8GFP, FRT19A; sp/ CyO; l(3)/TM3,Sb |
Bloomington Drosophila Stock Center | RRID: BDSC_42728 |
| Drosophila: hsFLP, tubP-GAL80, FRT19A | Bloomington Drosophila Stock Center | RRID: BDSC_5132 |
| Drosophila: w;; 13xlexAop2-IVS-GCaMP6s, 20xUAS-CsChrimson-mCherry | Jayaraman lab | N/A |
| Drosophila: w;; 20xUAS-GCaMP6s, 13xlexAop2-CsChrimson-mtdT | Jayaraman lab | N/A |
| Drosophila: UAS-Sema-2a | Kolodkin lab | N/A |
| Drosophila: UAS-Sema-2a-TMGFP | Kolodkin lab | RRID: BDSC_65747 |
| Drosophila: UAS-Sema-2b | Kolodkin lab | N/A |
| Drosophila: UAS-Sema-2b-TMGFP | Kolodkin lab | RRID: BDSC_65748 |
| Drosophila: UAS-Sema-2b-RNAi | Vienna Drosophila Resource Center | KK101842 |
| Drosophila: UAS-Sema-2b-RNAi | Bloomington Drosophila Stock Center | HM05143, RRID: BDSC_28942 |
| Drosophila: Sema-2bC4 | Kolodkin lab | RRID: BDSC_66734 |
| Drosophila: PlexBKG00878 | Bloomington Drosophila Stock Center | RRID: BDSC_14579 |
| Drosophila: PlexBMI15559 | Bloomington Drosophila Stock Center | RRID: BDSC_61730 |
| Drosophila: PlexBGAL4 | In this study | N/A |
| Drosophila: yw; sp/CyO; LoxP(Trojan-GAL4.1 | Bloomington Drosophila Stock Center | RRID: BDSC_60305 |
| Drosophila: yw, hs-Cre, vas-dFC31 | Bloomington Drosophila Stock Center | RRID: BDSC_60299 |
| Drosophila: w1118 | Bloomington Drosophila Stock Center | RRID: BDSC_5905 |
| Drosophila: UAS-Stinger-GFP | Bloomington Drosphila Stock Center | RRID: BDSC_65402 |
| Software and Algorithms | ||
| MATLAB R2017b | Mathworks | RRID: SCR_001622 |
| Prism 6 | GraphPad | RRID: SCR_002798 |
| Fiji | https://fiji.sc/ | RRID: SCR_002285 |
| Adobe Photoshop CS6 | Adobe | RRID: SCR_014199 |
| Adobe Illustrator CS6 | Adobe | RRID: SCR_010279 |
| Python 3.6 | Python Software Foundation | RRID: SCR_008394 |
| R 3.5.1 | The R Foundation | RRID: SCR_001905 |
| nlme 3.1 (R package) | CRAN | RRID: SCR_015655 |
| Other | ||
| 8-well silicon spacer | Grace Biolabs | Cat#: 654008 |
| 1-well silicon spacer | Grace Biolabs | Cat#: 654004 |
| 35mm FluoroDish | World Precision Instruments/Fisher Scientific | Cat#: 50-823-005 |
| Dental wax | GC Corporation | 27B2X00008000016 |
| Fiber optic cannula | Thorlabs | CFML21L10 |
| Arduino Uno board | Arduino | A000066 |
| Collimated LED light source (625nm) | Thorlabs | M625F2 |
| LED light source (627nm) | LED supply | ALK-26M-1UP-KIT with Luexeon Rebel Red LED and wide optic |
| Light meter | Cooke Corporation | Cal-light 400 |
| Buckpuck driver | RapidLED | 3023-D-E700 |
| Picospritzer III | Parker Hannifin | 052-0500-900 |
Highlights.
Sema-2b/PlexB-mediated attraction controls fan-shaped body (FB) lamination
Column formation in the FB is independent of lamination
FB lamination selectively regulates inhibitory-synapse formation
FB lamination facilitates development of a sleep circuit
ACKNOWLEDGMENTS
We thank members of the Kolodkin and Wu laboratories for helpful discussions throughout the course of this project. We thank L. Luo for antibodies and fly stocks, V. Jayaraman for fly stocks, and M. Pucak and the NINDS Multi-photon Imaging Core Facility at JHMI (P30 NS50274) for imaging and data analysis. We thank L. Luo for communication of results prior to publication. We also thank the Bloomington Stock Center and the Vienna Drosophila Resource Center for fly stocks. This work was supported by the National Institutes of Health (NIH) (grants 1R01 NS100792 to M.N.W. and K99 NS101065 to M.T.) and the Howard Hughes Medical Institute (HHMI) (A.L.K.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Ayoob JC, Terman JR, and Kolodkin AL (2006). Drosophila Plexin B is a Sema-2a receptor required for axon guidance. Development 133, 2125–2135. [DOI] [PubMed] [Google Scholar]
- Baier H. (2013). Synaptic laminae in the visual system: molecular mechanisms forming layers of perception. Annu. Rev. Cell Dev. Biol 29, 385–416. [DOI] [PubMed] [Google Scholar]
- Dag U, Lei Z, Le JQ, Wong A, Bushey D, and Keleman K. (2019). Neuronal reactivation during post-learning sleep consolidates long-term memory in Drosophila. eLife 8, e42786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao F, Ironfield H, Luan H, Diao F, Shropshire WC, Ewer J, Marr E, Potter CJ, Landgraf M, and White BH (2015). Plug-and-play genetic access to Drosophila cell types using exchangeable exon cassettes. Cell Rep.10, 1410–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. (2007). A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448, 151–156. [DOI] [PubMed] [Google Scholar]
- Donlea JM, Thimgan MS, Suzuki Y, Gottschalk L, and Shaw PJ (2011). Inducing sleep by remote control facilitates memory consolidation in Drosophila. Science 332, 1571–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlea JM, Pimentel D, and Miesenbö ck G. (2014). Neuronal machinery of sleep homeostasis in Drosophila. Neuron 81, 1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlea JM, Pimentel D, Talbot CB, Kempf A, Omoto JJ, Hartenstein V, and Miesenbö ck G. (2018). Recurrent circuitry for balancing sleep need and sleep. Neuron 97, 378–389.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Krishnaswamy A, De la Huerta I, and Sanes JR (2014). Type II cadherins guide assembly of a direction-selective retinal circuit. Cell 158, 793–807. [DOI] [PubMed] [Google Scholar]
- Duan X, Krishnaswamy A, Laboulaye MA, Liu J, Peng YR, Yamagata M, Toma K, and Sanes JR (2018). Cadherin combinations recruit dendrites of distinct retinal neurons to a shared interneuronal scaffold. Neuron 99, 1145–1154.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg EH, Vanhoven MK, Bendesky A, Wang G, Fetter RD, Shen K, and Bargmann CI (2008). GFP reconstitution across synaptic partners (GRASP) defines cell contacts and synapses in living nervous systems.Neuron 57, 353–363. [DOI] [PubMed] [Google Scholar]
- Gordon MD, and Scott K. (2009). Motor control in a Drosophila taste circuit. Neuron 61, 373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J, Adachi A, Shah KK, Hirokawa JD, Magani PS, and Maimon G. (2017). A neural circuit architecture for angular integration in Drosophila.Nature 546, 101–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guajardo R, Luginbuhl DJ, Han S, Luo L, and Li J. (2019). Functional divergence of Plexin B structural motifs in distinct steps of Drosophila olfactory circuit assembly. eLife 8, e48594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J, and Staiger JF (2017). The functioning of a cortex without layers. Front. Neuroanat 11, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanesch UF, Fischbach K-F, and Heisenberg M. (1989). Neuronal architecture of the central complex in Drosophila melanogaster. Cell Tissue Res. 257, 343–366. [Google Scholar]
- Heinze S, and Homberg U. (2007). Maplike representation of celestial E-vector orientations in the brain of an insect. Science 315, 995–997. [DOI] [PubMed] [Google Scholar]
- Heinze S, Gotthardt S, and Homberg U. (2009). Transformation of polarized light information in the central complex of the locust. J. Neurosci 29, 11783–11793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenett A, Rubin GM, Ngo TT, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, et al. (2012). A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2, 991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo WJ, Sweeney LB, Liang L, and Luo L. (2013). Linking cell fate, trajectory choice, and target selection: genetic analysis of Sema-2b in olfactory axon targeting. Neuron 78, 673–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser MS, Yue Z, and Sehgal A. (2014). A critical period of sleep for development of courtship circuitry and behavior in Drosophila. Science 344, 269–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SS, Rouault H, Druckmann S, and Jayaraman V. (2017). Ring attractor dynamics in the Drosophila central brain. Science 356, 849–853. [DOI] [PubMed] [Google Scholar]
- Kolodkin AL, and Hiesinger PR (2017). Wiring visual systems: common and divergent mechanisms and principles. Curr. Opin. Neurobiol 42, 128–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottler B, Bao H, Zalucki O, Imlach W, Troup M, van Alphen B, Paulk A, Zhang B, and van Swinderen B. (2013). A sleep/wake circuit controls isoflurane sensitivity in Drosophila. Curr. Biol 23, 594–598. [DOI] [PubMed] [Google Scholar]
- Lee T, and Luo L. (1999). Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22, 451–461. [DOI] [PubMed] [Google Scholar]
- Lefebvre JL, Sanes JR, and Kay JN (2015). Development of dendritic form and function. Annu. Rev. Cell Dev. Biol 31, 741–777. [DOI] [PubMed] [Google Scholar]
- Li J, Guajardo R, Xu C, Wu B, Li H, Li T, Luginbuhl DJ, Xie X, and Luo L. (2018). Stepwise wiring of the Drosophila olfactory map requires specific Plexin B levels. eLife 7, e39088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CC, and Potter CJ (2016). Editing transgenic DNA components by inducible gene replacement in Drosophila melanogaster. Genetics 203, 1613–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Chuang CC, Hua TE, Chen CC, Dickson BJ, Greenspan RJ, and Chiang AS (2013). A comprehensive wiring diagram of the protocerebral bridge for visual information processing in the Drosophila brain. Cell Rep. 3, 1739–1753. [DOI] [PubMed] [Google Scholar]
- Liu WW, and Wilson RI (2013). Glutamate is an inhibitory neurotransmitter in the Drosophila olfactory system. Proc. Natl. Acad. Sci. USA 110, 10294–10299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Liu S, Kodama L, Driscoll MR, and Wu MN (2012). Two dopaminergic neurons signal to the dorsal fan-shaped body to promote wakefulness in Drosophila. Curr. Biol 22, 2114–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Liu Q, Tabuchi M, and Wu MN (2016). Sleep drive is encoded by neural plastic changes in a dedicated circuit. Cell 165, 1347–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy TP, Luu DD, DeSimone JA, O’Brien TC, Lally CJ, Lindblad JJ, and Webster SM (2015). A behavioral assay for mechanosensation of MARCM-based clones in Drosophila melanogaster. J. Vis. Exp 106, e53537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern A, Pfeiffer BD, and Rubin GM (2015). Optimized tools for multicolor stochastic labeling reveal diverse stereotyped cell arrangements in the fly visual system. Proc. Natl. Acad. Sci. USA 112, E2967–E2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaï LJ, Ramaekers A, Raemaekers T, Drozdzecki A, Mauss AS, Yan J, Landgraf M, Annaert W, and Hassan BA (2010). Genetically encoded dendritic marker sheds light on neuronal connectivity in Drosophila. Proc. Natl. Acad. Sci. USA 107, 20553–20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaou N, and Meyer MP (2015). Lamination speeds the functional development of visual circuits. Neuron 88, 999–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omoto JJ, Keles MF, Nguyen BM, Bolanos C, Lovick JK, Frye MA, and Hartenstein V. (2017). Visual input to the Drosophila central complex by developmentally and functionally distinct neuronal populations. Curr. Biol 27, 1098–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng YR, Tran NM, Krishnaswamy A, Kostadinov D, Martersteck EM, and Sanes JR (2017). Satb1 regulates Contactin 5 to pattern dendrites of a mammalian retinal ganglion cell. Neuron 95, 869–883.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins LA, Holderbaum L, Tao R, Hu Y, Sopko R, McCall K, YangZhou D, Flockhart I, Binari R, Shim HS, et al. (2015). The Transgenic RNAi Project at Harvard Medical School: Resources and validation. Genetics 201, 843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer K, and Homberg U. (2014). Organization and functional roles of the central complex in the insect brain. Annu. Rev. Entomol 59, 165–184. [DOI] [PubMed] [Google Scholar]
- Pfeiffer BD, Jenett A, Hammonds AS, Ngo TT, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, et al. (2008). Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl. Acad. Sci. USA 105, 9715–9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro J, Bates D, DebRoy S, and Sarkar D; R Core Team (2013). nlme: Linear and nonlinear mixed effects models. R package version 3, pp. 1–140. https://CRAN.R-project.org/package=nlme. [Google Scholar]
- Qian Y, Cao Y, Deng B, Yang G, Li J, Xu R, Zhang D, Huang J, and Rao Y. (2017). Sleep homeostasis regulated by 5HT2b receptor in a small subset of neurons in the dorsal fan-shaped body of Drosophila. eLife 6, e23496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes JR, and Yamagata M. (2009). Many paths to synaptic specificity. Annu. Rev. Cell Dev. Biol 25, 161–195. [DOI] [PubMed] [Google Scholar]
- Seelig JD, and Jayaraman V. (2015). Neural dynamics for landmark orientation and angular path integration. Nature 521, 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney LB, Chou YH, Wu Z, Joo W, Komiyama T, Potter CJ, Kolodkin AL, Garcia KC, and Luo L. (2011). Secreted semaphorins from degenerating larval ORN axons direct adult projection neuron dendrite targeting. Neuron 72, 734–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi M, Monaco JD, Duan G, Bell B, Liu S, Liu Q, Zhang K, and Wu MN (2018). Clock-generated temporal codes determine synaptic plasticity to control sleep. Cell 175, 1213–1227.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner-Evans DB, and Jayaraman V. (2016). The insect central complex. Curr. Biol 26, R453–R457. [DOI] [PubMed] [Google Scholar]
- Turner-Evans D, Wegener S, Rouault H, Franconville R, Wolff T, Seelig JD, Druckmann S, and Jayaraman V. (2017). Angular velocity integration in a fly heading circuit. eLife 6, e23496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno T, Tomita J, Tanimoto H, Endo K, Ito K, Kume S, and Kume K. (2012). Identification of a dopamine pathway that regulates sleep and arousal in Drosophila. Nat. Neurosci 15, 1516–1523. [DOI] [PubMed] [Google Scholar]
- Vef O, Cleppien D, Loffler T, Altenhein B, and Technau GM (2006). A new strategy for efficient in vivo sreening of mutagenized Drosophila embryos. Dev. Genes Evol 216, 105–108. [DOI] [PubMed] [Google Scholar]
- Venken KJ, Schulze KL, Haelterman NA, Pan H, He Y, Evans-Holm M, Carlson JW, Levis RW, Spradling AC, Hoskins RA, and Bellen HJ (2011). MiMIC: A highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat. Methods 8, 737–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff T, Iyer NA, and Rubin GM (2015). Neuroarchitecture and neuroanatomy of the Drosophila central complex: A GAL4-based dissection of protocerebral bridge neurons and circuits. J. Comp. Neurol 523, 997–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Sweeney LB, Ayoob JC, Chak K, Andreone BJ, Ohyama T, Kerr R, Luo L, Zlatic M, and Kolodkin AL (2011). A combinatorial semaphorin code instructs the initial steps of sensory circuit assembly in the Drosophila CNS. Neuron 70, 281–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Andreone BJ, and Kolodkin AL (2012). The coordinate regulation of sensory afferent CNS targeting and CNS longitudinal tract organization in Drosophila during neural development. J. Peripher. Nerv. Syst 17 (Suppl 3), 34–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Tabuchi M, Brown MP, Mitchell SP, Wu MN, and Kolodkin AL (2017). The laminar organization of the Drosophila ellipsoid body is semaphorin-dependent and prevents the formation of ectopic synaptic connections. eLife 6, e25328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T, Ho MCW, Liu Q, Horiuchi W, Lin CC, Task D, Luan H, White BH, Potter CJ, and Wu MN (2018). A genetic toolkit for dissecting dopamine circuit function in Drosophila. Cell Rep. 23, 652–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Awasaki T, Yu HH, He Y, Ding P, Kao JC, and Lee T. (2013). Diverse neuronal lineages make stereotyped contributions to the Drosophila locomotor control center, the central complex. J. Comp. Neurol 521, 2645–Spc1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JM, and Armstrong JD (2010). Structure of the adult central complex in Drosophila: organization of distinct neuronal subsets. J. Comp. Neurol 518, 1500–1524. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.