Antimicrobial-resistant ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) pathogens represent a global threat to human health. The acquisition of antimicrobial resistance genes by ESKAPE pathogens has reduced the treatment options for serious infections, increased the burden of disease, and increased death rates due to treatment failure and requires a coordinated global response for antimicrobial resistance surveillance.
KEYWORDS: Acinetobacter, Enterobacter, Enterobacterales, Enterococcus, Klebsiella, Pseudomonas aeruginosa, Staphylococcus aureus, antibiotic resistance, multidrug resistance
SUMMARY
Antimicrobial-resistant ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) pathogens represent a global threat to human health. The acquisition of antimicrobial resistance genes by ESKAPE pathogens has reduced the treatment options for serious infections, increased the burden of disease, and increased death rates due to treatment failure and requires a coordinated global response for antimicrobial resistance surveillance. This looming health threat has restimulated interest in the development of new antimicrobial therapies, has demanded the need for better patient care, and has facilitated heightened governance over stewardship practices.
INTRODUCTION
The emergence of multidrug-resistant (MDR) bacteria (bacteria resistant to more than three antibiotic classes) (1) has been paralleled by a waning antibiotic development pipeline (2). The U.S. Centers for Disease Control and Prevention (CDC) and the World Health Organization (WHO) categorize antimicrobial-resistant (AMR) pathogens as a looming threat to human health (3, 4). Currently, no systematic international surveillance of AMR exists (3), but available reports estimate that more than 2 million AMR infections with a death toll of 29,000 occur in the United States per annum, at an attributable health care cost of more than $4.7 billion (4). In Europe, over 33,000 deaths and 874,000 disability-adjusted life years are attributed to hospital-acquired (HA) and community-acquired (CA) AMR infections each year, accounting for $1.5 billion in direct and indirect costs (5, 6). In developing nations, where economic loss estimates are not available, communicable diseases remain the leading cause of death, and these are now heightened by emerging and reemerging infectious diseases (7–9). While AMR genes occur naturally in the environment, the use of antibiotics has selected for the presence of AMR genes. The lack of rapid diagnostic methods to identify bacterial pathogens and AMR genes in clinical settings has resulted in the often unnecessary use of broad-spectrum antibiotics (10).
In February 2017, to focus and guide research and development related to new antibiotics, the WHO published its list of pathogens for which new antimicrobial development is urgently needed. Within this broad list, ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) pathogens (11) were designated “priority status” (12).
Through genetic mutation and the acquisition of mobile genetic elements (MGEs) (13), ESKAPE pathogens have developed resistance mechanisms against oxazolidinones, lipopeptides, macrolides, fluoroquinolones, tetracyclines, β-lactams, β-lactam–β-lactamase inhibitor combinations, and antibiotics that are the last line of defense, including carbapenems, glycopeptides, and clinically unfavorable polymyxins (14–19). Comparatively, resistance to lipoglycopeptides is rare and has only recently been documented (20). This may be potentially attributed to the dual action of lipoglycopeptides in inhibiting both peptidoglycan synthesis and destabilizing the bacterial cell membrane. Overall, the constitutive and/or inducible expression of these drug resistance mechanisms has resulted in the increased representation of bacterial species with these mechanisms in hospital-acquired infections (12).
Since the turn of the 1990s, the development and commercialization of novel antibiotics have slowed. Between 2017 and 2019, 11 new antimicrobial therapies were approved by the U.S. Food and Drug Administration (U.S. FDA) (21). Of these 11 antimicrobials, 4 were approved by the European Union European Medicines Agency (E.U. EMA): the meropenem-vaborbactam combination (Vaborem), eravacycline (Xerava), delafloxacin (Baxdela/Quofenix), and the imipenem-cilastatin-relebactam combination (Recarbrio; a positive opinion toward the granting of marketing authorization was recommended in December 2019, and approval was provided in February 2020) (22–25). Apart from these antimicrobials, during this time frame, the E.U. EMA additionally approved ceftobiprole (Zeftera; also approved by the Australian Therapeutic Goods Agency in 2016 and by Health Canada in 2015), whereas the Japanese Pharmaceutical and Medical Devices Agency (PMDA) approved lascufloxacin (Lasvic) (26–29). Global initiatives to deliver new stand-alone antibacterial therapies or complementing alternative therapies are urgently needed. In this review, we assess the current state of AMR in ESKAPE pathogens, with a focus on current and emerging drug development avenues in the response against AMR.
VANCOMYCIN-RESISTANT ENTEROCOCCI
Enterococcus faecium is a prominent cause of health care-associated infections, and hospital-adapted lineages are increasingly resistant to vancomycin (30) (Table 1). The dissemination of Enterococcus in the United States occurred in two separate waves. The first wave began in the 1980s and was associated with the introduction of third-generation cephalosporins driving the emergence of vancomycin- and ampicillin-resistant Enterococcus faecalis (31). The second wave, dominated by vancomycin-resistant E. faecium (VREfm), was hypothesized to have spread from the United States to other parts of the world. Several European countries have now reported increases in VREfm prevalence in hospitalized patients (32, 33). In Australia, 47% of E. faecium blood culture isolates are VREfm, contributing to an incidence rate of vancomycin-resistant enterococci (VRE) which surpasses that of many other high-income nations (34, 35). VREfm multilocus sequence types (ST) pertaining to clonal complex 17 (CC17) are currently responsible for a significant burden of hospital-acquired infection (36). Highly prevalent in the gut microbiome of wild and domesticated animals (37, 38), CC17 strains have been associated with outbreaks in Europe, Asia, South America, and Australia (34, 39–42). Although the zoonotic transfer of CC17 strains from animals to humans is largely attributed to the spread of this complex, fresh food has also been found to be a significant reservoir (36). Despite spread in the community appearing high, community-associated infections caused by CC17 strains are uncommon.
TABLE 1.
Clinical characteristics of ESKAPE pathogensa
| Species | Resistances | Clinical manifestations | Major ST/CC | Mortality rates | Treatments | Key characteristics |
|---|---|---|---|---|---|---|
| Vancomycin-resistant Enterococcus | Vancomycin (184, 471), ampicillin (472), linezolid (35), teicoplanin (473), piperacillin (474), cephalosporins (64), multidrug resistant (184) | Catheter-associated-UTI, vascular catheter-associated bacteremia, intra-abdominal and pelvic infection, endocarditis (51) | E. faecium ST17 (CC17) (35), ST203 (CC17) (475), ST796 (476), ST1421 (35), and CC17 (36); E. faecalis CC2 (477), CC9 (477), ST6 (478), and ST16 (479) | Over 30% for bacteremia (35, 480); 2.5-fold increase in mortality from bacteremia caused by VRE compared to that from bacteremia caused by vancomycin-sensitive bacteria (473) | Nitrofurantoinb (481), fosfomycin (482), linezolid (480), daptomycin (18), chloramphenicol (483), doxycycline (483), high-dose ampicillin and sulbactam (483), omadacycline (396) | 10% of all HA bloodstream infections (484, 485); tolerant to heat, chlorine, and alcohol preparations (486); E. faecium demonstrates significantly higher levels of resistance than E. faecalis (35); commonly encountered as asymptomatic colonization (487) |
| Methicillin-resistant S. aureus | Aminoglycosides (488), β-lactams (489), chloramphenicol (488), trimethoprim (313), macrolides (313), tetracycline (313), fluoroquinolones (64), multidrug resistant (488) | Acute bacterial skin and skin structure infection (490), bacteremia (488), pneumonia (491), osteoarticular infection (492), endocarditis (488) | ST5 (65), ST8 (493), ST22 (35), ST30 (494), ST59 (495), ST72 (CC8) (496), ST80 (70), ST398 (livestock associated) (71, 497) | Greater than 20% for bloodstream infection (76, 77); overall mortality ranges from 15–50% (498) | Vancomycin (488), clindamycin (499), daptomycin (500), linezolid (501), tedizolid (502), dalbavancin (503), tigecycline (504), trimethoprim and sulfamethoxazole (505), pristinamycin (506), omadacycline (396), lefamulin (403) | In Asia, 50% of all S. aureus bloodstream infections are caused by MRSA (507); in USA, HA-MRSA infections have decreased by 54% (508); in Europe, the total proportion of reported MRSA infections among S. aureus infections decreased from 19.6% in 2014 to 16.4% in 2018 (64); 20–40% of the population carries S. aureus as a commensal organism (488) |
| K. pneumoniae | Polymyxins (276), carbapenems (509), fluoroquinolones (64), third-generation cephalosporins (64), aminoglycosides (64), tetracyclines (276), pandrug resistant (276) | Pneumonia (510), pyogenic liver abscesses (511), necrotizing and soft tissue infection (92), bloodstream infection, meningitis (512), endophthalmitis (512), UTI (513) | ST11 (82, 514), ST15 (82, 515), ST17 (516), ST37 (516), ST101 (82, 517), ST147 (518), ST258 (148, 519), ST307 (88, 89), ST405 (520), ST512 (82) | 40% to 70% for CRKP bloodstream infection (509, 521); 40% for CRKP pulmonary infection (521, 522); 25% to 47% for hvKP necrotizing and soft tissue infection (90, 92) | Aminoglycosides (523), polymyxin combination therapy (524), tigecycline (79), meropenem (523), meropenem-vaborbactam (525), ertapenem and meropenem (526), imipenem-cilastatin-relebactam (24), ceftazidime-avibactam (527), plazomicin (393), eravacycline (394) | USA has more than 7,000 HA-CRKP infections per year (80); in Taiwan, 80% of pyogenic liver abscess cases are attributed to hvKP (511) |
| A. baumannii | Carbapenems (103), polymyxins (108), β-lactams (103), tigecycline (103), ceftazidime (103), fourth-generation cephalosporins (103), multidrug resistant (101, 103) | Ventilator-associated pneumonia (528), central line bloodstream infections (528), nosocomial meningitis (529), skin and soft tissue infection (530), catheter-associated UTI (528) | ST195 (CC92) (531), ST457 (CC92) (531), pan-European epidemic clones I, II, and III (532) | 35% for ventilator-associated pneumonia and bloodstream infections (533) | Colistin (534), tigecycline (102), cefiderocol (412), eravacycline (394) | 2% of all HA-infections in USA and Europe (100, 101); high mutation frequency upon desiccation (535); persistence in biofilms during soft tissue infection (536); tolerance to low-ethanol environments and resistance to chlorhexidine-based disinfectants (537, 538) |
| P. aeruginosa | First- and second-generation cephalosporins (110), piperacillin-tazobactam (35, 110), aminoglycosides (110), quinolones (110), carbapenems (35, 110), polymyxins (110), multidrug resistant (539) | UTI (540), bloodstream infection (539), ventilator-associated pneumonia (64), chronic respiratory infection (541), skin and soft tissue infection (542), endocarditis (543) | ST111 (544), ST175 (112, 544), ST233 (544), ST235 (111, 545), ST253 (544), ST292 (114), ST1725 (544) | 67% for MDR bacteremia (539); 33.9% for UTI (540) | Piperacillin-tazobactam (35), ceftolozane-tazobactam (546), ceftazidime (35), meropenem (35), ciprofloxacin (35), ceftazidime-avibactam (527), cefiderocol (412), imipenem-cilastatin-relebactam (24) | High incidence of infection in burn victims (542); 51,000 HA infections in USA per year (547–550) |
| Enterobacter species | Carbapenems (3), fourth-generation cephalosporins (102), fluoroquinolones (102), β-lactams (157), polymyxins (130), multidrug resistant (102), pandrug resistant (130) | UTI (551), bloodstream infection (552), neonatal pneumonia (553), skin and soft tissue infection (554), intra-abdominal infection (555), endocarditis (556), septic arthritis (556) | In K. aerogenes, ST4 (127) and ST93 (127); in E. cloacae, ST66 (557), ST78 (557), ST108 (557), ST144 (557), and ST171 (128) | Exceeds 40% for E. cloacae bloodstream infection (552, 558) | Nitrofurantoinb (35), cefepime (35), ceftriaxone (35), ciprofloxacin (35), gentamicin (35), meropenem (35), piperacillin-tazobactam (35), trimethoprim with or without sulfamethoxazole (35), imipenem-cilastatin-relebactam (24) | E. cloacae is the 3rd most frequent Enterobacterales species causing bloodstream infection (552); infections are prevalent in neonates and elderly individuals (556, 559); clinically relevant E. hormaechei is an important emerging pathogen within the E. cloacae complex (125, 126) |
Abbreviations: ST, sequence type; CC, clonal complex; UTI, urinary tract infection; HA, hospital acquired; CRKP, carbapenem-resistant K. pneumoniae; hvKP, hypervirulent K. pneumoniae.
Nitrofurantoin is prescribed only for uncomplicated urinary tract infections.
Compared to the durations of outbreaks caused by the other ESKAPE pathogens, VREfm outbreaks have a long duration, approximating 11 months, on average (43, 44). The entry of VREfm into the bloodstream of hospitalized patients is typically preceded by antibiotic exposure, enabling VREfm to become the predominant species in the gastrointestinal tract (45, 46). The duration of prior antibiotic exposure is strongly associated with a subsequent risk of VRE infection (47). In a 2016 national survey of 1,058 bloodstream infections caused by Enterococcus in Australia, almost 50% of E. faecium isolates were vancomycin resistant (48). In the United States, the incidence of hospital- and community-acquired VRE infection between 2012 and 2017 significantly decreased (4). The management of patients infected with VRE is complicated by the excess cost and disruption resulting from the need for isolation rooms, contact precautions, and dedicated room cleaning. The treatment of significant infection relies upon second-line antibiotic therapies (e.g., tigecycline and daptomycin), which are often associated with increased cost, diminished efficacy, and a greater risk of toxicity compared with the cost, efficacy, and risk of toxicity of first-line antibiotic therapies (Table 1) (49, 50). Defining the additional risk of a poor outcome attributable to vancomycin resistance in enterococci has been challenging, largely because of the confounding effects of comorbidity (51). Most studies have demonstrated an association of VRE infection with excess mortality, the duration of hospital admission, and treatment costs (52, 53), especially when VRE cause a bloodstream infection (54).
METHICILLIN-RESISTANT STAPHYLOCOCCUS AUREUS
Methicillin resistance was first identified in Staphylococcus aureus in 1961 as a consequence of widespread penicillin usage (55). The introduction of penicillin also heightened the emergence of penicillinase-producing S. aureus. Although methicillin-resistant S. aureus (MRSA) is still a significant burden in U.S. health care settings, the incidence of hospital-acquired MRSA (HA-MRSA) is declining (4, 56) (Table 1). Opposite this finding, the incidence of community-acquired MRSA (CA-MRSA) infections in the same region has significantly increased (56). CA-MRSA infections emerged among the indigenous population of Australia in the 1980s (57) and in otherwise healthy communities of the United States and Canada in the 1990s (58). In North America, where CA-MRSA is prevalent, MRSA epidemics are largely attributed to the emergence of either of two unrelated MRSA clones (59, 60). The MRSA clone USA400, isolated from the pediatric population, initiated the first epidemic wave and remains a common cause of community-onset disease among indigenous populations in Alaska and the Pacific Northwest (61). Since 2001, USA400 has been superseded by an epidemic caused by MRSA USA300 (61, 62) and closely related variants, which are now the most prevalent CA-MRSA isolates in North America and northern parts of South America.
Today, the burden of MRSA across the world varies substantially (4, 63, 64). In China, the prevalence of HA- and CA-MRSA infections wavered remarkably between 2007 and 2018. The prevalence of HA-MRSA clones ST239-t030 and ST239-t037 was significantly reduced (from 20.3% to 1% and 18.4% to 0.5%, respectively), and these have now been replaced by the ST5-t2460 clone (from 0% to 17.3%), which has seen a rapid emergence. Furthermore, the incidence of CA-MRSA clones ST59 and ST398 also increased over the same period (from 1.0% to 5.8% and 1.8% to 10.5%, respectively) (65). In Northern Europe (i.e., the United Kingdom and France), a steady decrease in the prevalence of HA-MRSA was observed between 2015 and 2018 and was largely attributed to improved national infection control programs (64, 66, 67). In comparison, the rates of HA-MRSA in Southern Europe (i.e., Portugal, Spain, Italy, and Greece) remain high (5, 64).
CA-MRSA strains have typically been associated with skin and soft tissue infections, whereas HA-MRSA strains are associated with severe pneumonia and bloodstream infections (68). The division between CA- and HA-MRSA strains is becoming indistinct, with CA-MRSA strains now identified to be a causative agent of bloodstream infections in nosocomial settings. MRSA ST80 is a well-defined agent of CA-MRSA in Europe. Although it is now becoming less prevalent in select European countries (69), CA-MRSA ST80 is now a major contributor of infection in defined health care settings (70). Furthermore, examples of CA-MRSA (e.g., ST398) have been shown to be associated with exposure to livestock (particularly pigs) in Europe (71) (Table 1). Although individuals with direct exposure to livestock are the most at risk from livestock-associated MRSA (LA-MRSA), it has now been reported that LA-MRSA substantially contributes to the burden of nosocomial infection in Europe (72). One of the less-defined and neglected subgroups of S. aureus is borderline oxacillin-resistant S. aureus (BORSA). Found both in community settings and in hospital settings, BORSA is characterized by intermediate resistance to penicillinase-resistant penicillins, with oxacillin MICs being between 1 and 8 μg/ml (73). Lacking the mecA gene, BORSA is not truly either methicillin resistant or methicillin sensitive, and frequent misidentification poses a significant threat to patient treatment and outcome, as severe BORSA infections may be nonresponsive to high doses of oxacillin (74). Overall, MRSA infections carry additional health care burdens in terms of morbidity, length of hospital stay, health care costs, and quality of life (75). The rate of mortality following S. aureus bloodstream infection exceeds 20%, and the presence of methicillin resistance is independently associated with increased mortality (76, 77).
KLEBSIELLA PNEUMONIAE
Cephalosporin- and carbapenem-class antibiotics have been a mainstay of treatment for serious infections caused by Enterobacterales, such as K. pneumoniae, but efficacy has been compromised by the widespread acquisition of genes encoding enzymes, such as extended-spectrum β-lactamases (ESBLs) and carbapenemases, which mediate the respective resistance to these critical drugs (19). High rates of mortality, often exceeding 40%, have been associated with severe infections caused by carbapenem-resistant Enterobacterales (CRE) (78). Effective antimicrobial options are often lacking, and treatment typically requires reliance on drugs with a risk of toxicity (e.g., aminoglycosides, polymyxins) or other safety concerns (e.g., tigecycline) (79) (Table 1). Carbapenem-resistant K. pneumoniae (CRKP) strains are the most clinically prominent CRE (64, 80). In the United States, carbapenemases carried by K. pneumoniae were originally reported in 2001 (81). Since then, the genes encoding these β-lactamases have spread among several Gram-negative bacterial species. Between 2005 and 2010, an increase in CRKP isolates causing invasive infections was reported across Europe (64). The spread of CRKP in Europe has been driven by direct and indirect patient-to-patient transmission in nosocomial settings, largely attributed to ST11, ST15, ST101, and ST258 strains, along with the ST258 derivative ST512 (82) (Table 1). The global burden of CRKP has now been further exacerbated by successive waves of CRKP emerging from several locations across the Indian Ocean rim, the United States, and China (83–87). The global dissemination of CRKP is exemplified by the CRKP clone ST307. The ST307 clone has successfully disseminated across every major continent (88), demonstrating extremely high transmission rates in health care settings (89).
Recent reports suggest that AMR hypervirulent K. pneumoniae (hvKP) strains are also emerging. In Taiwan, hvKP causes as many cases of necrotizing fasciitis as Streptococcus pyogenes and is associated with a higher mortality rate (47% versus 19%) (90). The detection of hvKP is now being reported around the world in both high- and low-income settings (87, 91, 92). An important laboratory feature frequently seen in hvKP strains is the presence of a hypermucoviscous phenotype (in association with the K1 and K2 capsular serotypes) (93).
ACINETOBACTER BAUMANNII
A. baumannii infections typically occur in hospitalized patients or patients with significant contact with the health care system (94). Historically, A. baumannii has been associated with hot and/or humid geographic climates (95, 96). Between 1987 and 1996, the frequency of both community- and hospital-acquired infections across the United States was observed to rise by 50% between the months of July and October (97). Since the 1970s, A. baumannii has become increasingly common in temperate climates, a shift largely attributed to improved environmental persistence mechanisms and MDR development (98). Community-acquired pneumonia due to A. baumannii has been described in tropical regions of Asia and Australia among individuals with a history of alcohol abuse (99). Although A. baumannii infection rates are comparatively low compared to those of other ESKAPE pathogens (100, 101), approximately 45% of all global A. baumannii isolates are considered MDR, with rates exceeding 60% in the United States (4, 101), Latin America, and the Middle East (102). Turkey and Greece have reported MDR rates exceeding 90% (103). These levels of MDR for A. baumannii are over four times higher than those observed in K. pneumoniae and P. aeruginosa (3). A key aspect of A. baumannii physiology is the propensity to develop rapid resistance. From 2011 to 2016, the rate of identification of A. baumannii isolates resistant to carbapenem- and β-lactam-class antibiotics has increased by over 30% globally (103). The spread of MDR and carbapenem-resistant A. baumannii (CRAB) isolates is largely associated with three international clonal lineages: CC1, CC2, and CC3 (104, 105). CC1 is prevalent worldwide, while CC2 and CC3 are highly prevalent in Europe and North America. CC15 and CC79 are also predominant in Central and South America (106, 107). With the emergence of pandrug-resistant isolates, last-resort carbapenem- and polymyxin-class antibiotics are no longer effective (103, 108) (Table 1). Without adequate action via improved epidemiological surveillance and therapeutic development, A. baumannii has the capacity to potentiate a global epidemic.
PSEUDOMONAS AERUGINOSA
Widely present in aquatic environments, P. aeruginosa is a Gram-negative opportunistic human pathogen commonly associated with severe respiratory infections in patients with impaired immunity. While P. aeruginosa is responsible for 10% of all nosocomial infections, there is also increasing acknowledgment of P. aeruginosa as a cause of community-acquired infection.
The plasticity and adaptability of the P. aeruginosa genome, conferred by a repertoire of regulatory genes (>8% of the 6-Mb genome), are key features in the pathogen’s ability to chronically persist in the host and evade antibiotic treatment (109). Intrinsically resistant to a wide array of antimicrobial agents, P. aeruginosa currently displays resistance to multiple classes of antibiotics (6, 110) (Table 1). In the United States, although AMR rates remain high, surveillance suggests a trend toward declining rates of resistance (4). Globally, patterns of P. aeruginosa AMR vary substantially. Today, the highest rates of AMR in P. aeruginosa occur in North, Central, and South America, Western and Central Europe, China, India, and Southeast Asia (7). With an enhanced capacity to acquire and maintain foreign antibiotic resistance elements, P. aeruginosa lineages ST235 and ST175 have emerged as high-risk globally dispersed clones and remain a major contributor of hospital-acquired infection (111, 112). Furthermore, the widespread distribution of P. aeruginosa nosocomial isolates resistant to last-resort polymyxin- and carbapenem-class antibiotics is well documented (7, 113, 114).
Patients with chronic or inherited lung disease, such as bronchiectasis and cystic fibrosis (CF), are highly susceptible to persistent pulmonary infection, with episodic exacerbations requiring hospitalization and intravenous antibiotics, with a subsequent risk of selection for MDR (115). P. aeruginosa has been shown to remain viable in the lungs of patients diagnosed with CF for over a decade (116). P. aeruginosa colonizes moist environments and therefore can be found in many health care settings, especially in the context of chronic wounds, respiratory support, or urinary tract devices, where biofilm formation predisposes for persistence, immune evasion, and antimicrobial resistance (117, 118).
ENTEROBACTER SPECIES
Over the last 35 years, Enterobacter aerogenes (now renamed Klebsiella aerogenes) and Enterobacter cloacae species have presented as significant threats to neonatal wards and patients in intensive care units, particularly those dependent on mechanical ventilation (119). The emergence of these two Enterobacter species as clinically significant MDR pathogens has occurred in concurrent epidemic waves. From the early 1990s to 2003, E. aerogenes was the most clinically prevalent cause of Enterobacter nosocomial infection (119). During this period, the hospital-acquired E. aerogenes infection incidence was high in Western Europe (120, 121), largely attributed to the dispersion of a single epidemic clone (122, 123). In about 2010, E. aerogenes was superseded by E. cloacae as the most common clinically isolated species of the genus (124). It is worth noting that other members of the E. cloacae complex, especially Enterococcus hormaechei, are clinically relevant and are often difficult to discriminate at the species level based on standard phenotypic assays (125, 126).
MDR Enterobacter species are an increasing cause of hospital-acquired infection. In the United States, E. aerogenes ST4 and ST93 currently represent prevalent lineages associated with nosocomial infection (127). For the E. cloacae complex, recent data suggest that carbapenem resistance has directionally spread across the United States due to the dissemination of hospital-associated carbapenem-resistant E. cloacae ST178 and ST78 isolates (128). Prior to 2005, an estimated 99.9% of Enterobacter strains were sensitive to carbapenems (129). Carbapenem resistance is now reported in all WHO health regions (3). Moreover, pandrug-resistant E. aerogenes has also emerged, displaying resistance to the last-resort antibiotic colistin (130) (Table 1). To complicate the treatment of bacterial infections further, E. aerogenes is capable of harboring subpopulations of colistin-resistant bacteria which are undetectable using current diagnostic testing strategies (131).
ESCHERICHIA COLI
Although not formally recognized as part of the ESKAPE group of pathogens, AMR Escherichia coli is identified as a major cause of bloodstream and urinary tract infection (UTI) in both community and health care settings globally (5, 35, 64). Sepsis is one of the most common manifestations of E. coli UTI. In Australian inpatient and emergency department settings, E. coli is the most prevalent Gram-negative bacterial species isolated from both blood and urine cultures (35). Over the past decade, several pandemic clones of MDR uropathogenic E. coli (e.g., ST131 and ST95) have disseminated worldwide (132, 133). Through horizontal gene transfer, E. coli typically acquires resistance genes from other members of the Enterobacterales. High rates of resistance to aminopenicillins, fluoroquinolones, aminoglycosides, and third-generation cephalosporins are noted across Europe (64). Although carbapenem resistance is rare in invasive E. coli strains, the general situation in Europe for CRE, including E. coli, was shown to worsen between 2010 and 2018 (134). Furthermore, in 2016, resistance to the last-resort polymyxin, colistin, was identified in E. coli strains isolated from pig farms in China (135). Although not discussed further in this review, AMR E. coli is currently one of the largest clinical burdens facing both human and animal health. In order not to exacerbate these challenges further, organizations involved in AMR policy, research and development (R&D), and surveillance need to consider this pathogen as a critical public health concern.
ESKAPE PATHOGEN MECHANISMS OF ANTIBIOTIC RESISTANCE
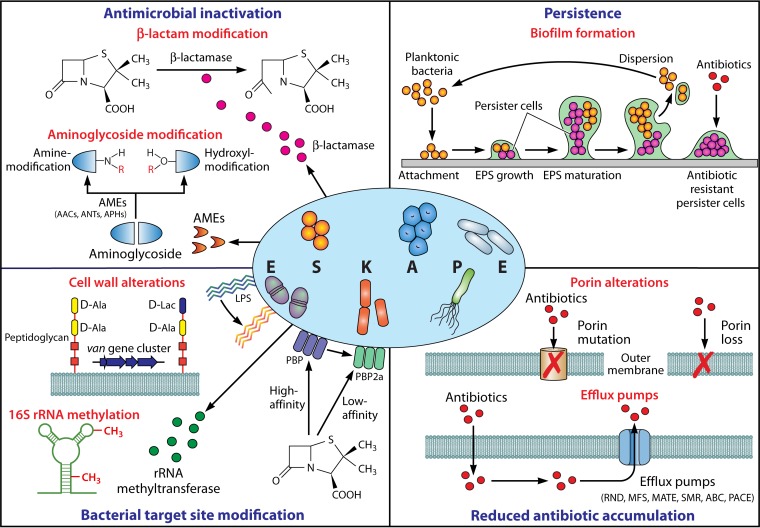
Given the frequency at which ESKAPE organisms are encountered in the clinical setting, it is not surprising that numerous different AMR mechanisms are observed in these pathogens. These can be broadly categorized into four groups, comprising (i) inactivation or alteration of the antimicrobial molecule, (ii) bacterial target site modifications, (iii) reduced antibiotic penetration/accumulation, and (iv) the formation of bacterial biofilms (Fig. 1). Here we explore the most important AMR determinants that have contributed to the success of ESKAPE pathogens in the modern-day clinical setting.
FIG 1.
Mediators of ESKAPE pathogen antimicrobial resistance. Mechanisms facilitating antimicrobial resistance in ESKAPE pathogens can be broadly categorized into four groups: (i) enzyme-mediated antimicrobial inactivation, which either irreversibly destroys the active antibiotic site (e.g., hydrolytic cleavage of the β-lactam ring by β-lactamases) or covalently modifies key structural elements of the drug to hinder the bacterial target site interaction (e.g., aminoglycoside-modifying enzymes that catalyze hydroxyl/amino group modifications); (ii) bacterial target site modification, which prevents the binding or which reduces the affinity of the antibiotic molecule at the cell surface (e.g., LPS modification, PBP2a expression with reduced β-lactam affinity, and van gene cluster-mediated peptidoglycan modification) or intracellularly (e.g., 16S RNA methylation); (iii) reduced antibiotic accumulation through the mutation or loss of outer membrane channels (e.g., OprD in P. aeruginosa, CarO in A. baumannii, and OmpK36 in K. pneumoniae) and expression of efflux systems to actively extrude drugs out of the cell (e.g., RND, MFS, MATE, SMR, ABC, and PACE); and (iv) persistence through biofilm-embedded cells which demonstrate a markedly higher tolerance to antimicrobial agents than planktonic bacteria. AMEs, aminoglycoside-modifying enzymes; AACs, aminoglycoside acetyltransferases; ANTs, aminoglycoside nucleotidyltransferases; APHs, aminoglycoside phosphotransferases; LPS, lipopolysaccharide; PBP, penicillin-binding protein; RND, resistance-nodulation-division; MFS, major facilitator superfamily; MATE, multidrug and toxic compound extrusion; SMR, small multidrug resistance; ABC, ATP-binding cassette; PACE, proteobacterial antimicrobial compound efflux; EPS, extracellular polymeric substance.
Antibiotic Inactivation/Alteration
One of the most common AMR mechanisms employed by ESKAPE pathogens involves the production of enzymes that irreversibly destroy or neutralize antibiotics. Such enzymes are particularly prevalent among the Gram-negative pathogens and comprise those (i) that destroy the active antibiotic site (e.g., hydrolytic cleavage of the β-lactam ring by β-lactamases) or (ii) that covalently modify key structural elements of the drug to hinder bacterial target site interaction (e.g., aminoglycoside-modifying enzymes [AMEs] that catalyze hydroxyl/amino group modifications).
β-Lactamases.
β-Lactamase enzymes were first identified soon after the initial discovery and purification of penicillin (136). Since then, >2,600 unique β-lactamases enabling resistance to one or more β-lactams (i.e., penicillins, cephalosporins, monobactams, and carbapenems) have been described (137). β-Lactamases remain the most important resistance mechanism among Gram-negative ESKAPE pathogens, where they are concentrated within the periplasm, thus hydrolyzing the β-lactam agents prior to reaching the penicillin-binding protein (PBP) target in the cell wall.
β-Lactamase enzymes are typically classified according to their primary molecular structure (i.e., the Ambler scheme [138]) or combined hydrolytic and inhibition functional properties (i.e., the Bush-Jacoby system [139]). Ambler class A enzymes contain serine in their active site and comprise penicillinases, cephalosporinases, narrow- and broad-spectrum β-lactamases, extended-spectrum β-lactamases (ESBLs), and carbapenemases. Overall, they represent the largest cluster of β-lactamase enzymes and collectively are capable of inactivating most β-lactam classes, including the penicillins, early cephalosporins, third-generation oxyimino-cephalosporins, monobactams, cephamycins, and carbapenems. Their susceptibility to inhibition by clavulanic acid and tazobactam is variable, though all are inhibited by novel β-lactamase inhibitor agents, including avibactam, relebactam, and vaborbactam (139, 140).
Ambler class A enzymes comprise various important β-lactamases that are frequently observed in Gram-negative (e.g., TEM, SHV, CTX-M, and KPC) and Gram-positive (e.g., penicillinase) ESKAPE pathogens. Indeed, blaZ-encoded penicillinases that emerged widely and soon after the introduction of penicillin are now detectable in ∼85% of clinically significant S. aureus isolates and some Enterococcus spp. (141–144). Likewise, the narrow-spectrum TEM-type β-lactamases, which readily hydrolyze early cephalosporins and penicillins, are frequently encountered in K. pneumoniae and Enterobacter spp. but have also been reported in A. baumannii and P. aeruginosa. SHV-1, which has a substrate and inhibition profile similar to that of TEM-1, is almost universally found in the progenitor species, K. pneumoniae (144, 145).
Due to a combination of strong selection pressures and the frequency at which AMR determinants are mobilized between organisms, both TEM- and SHV-type enzymes have undergone extensive evolution in recent decades (145). This has resulted in the proliferation and dissemination of numerous plasmid-encoded ESBL variants that can also hydrolyze oxyimino-β-lactams and aztreonam (139, 145). Other class A ESBLs, namely, enzymes of the CTX-M, PER, GES, and VEB families, have also been reported across all Gram-negative ESKAPE pathogens. Characteristically, most class A ESBL enzymes remain susceptible to clavulanic acid, though Bush-Jacoby subgroup 2br and 2ber ESBLs (e.g., TEM-30, SHV-10, and TEM-50) show reduced susceptibility to various β-lactamase inhibitors (146). Concerningly, inhibitor-resistant β-lactamases have also been reported in K. pneumoniae strains harboring KPC serine carbapenemase enzymes (147). Plasmid-encoded KPCs have been associated with major outbreaks worldwide (e.g., the outbreak caused by K. pneumoniae ST258) and hydrolyze virtually all β-lactams, including carbapenems (148). Despite this, there is emerging evidence that infections with KPC-producing organisms can be successfully targeted with various new β-lactamase–β-lactamase inhibitor combinations, including imipenem-cilastatin-relebactam, meropenem-vaborbactam, and ceftazidime-avibactam (149). Unfortunately, the rapid evolution of ceftazidime-avibactam resistance has already been reported in K. pneumoniae ST258 blaKPC-3-harboring isolates and in non-ST258 clonal backgrounds and additional blaKPC variants (17, 150, 151).
Ambler class B metallo-β-lactamases (MBLs) represent another clinically important group of enzymes capable of hydrolyzing most β-lactams, including carbapenems. However, in contrast to other β-lactamases, they require Zn2+ at their active site, display a low affinity for aztreonam, and are inhibited by EDTA (139). The most prominent MBLs encountered in the Gram-negative ESKAPE pathogens (e.g., MBLs of the IMP, VIM, and NDM families) are encoded on conjugative plasmids. IMP- and VIM-type MBLs were first detected in clinical P. aeruginosa isolates (152, 153) but have since been identified in K. pneumoniae, E. cloacae complex isolates, and Acinetobacter spp. (154–157). NDM-type enzymes have also been detected across all Gram-negative ESKAPE bacteria and are of particular concern due to the fact that they are incorporated into transferable genetic elements that also encode determinants for resistance to other antibiotic classes (157, 158).
Group C β-lactamases comprise chromosomally encoded cephalosporinases, such as AmpC, that are found in many Enterobacterales (including Enterobacter spp.), P. aeruginosa, and Acinetobacter spp. (159). They are most active on narrow- to intermediate-spectrum cephalosporins plus aztreonam and are usually resistant to clavulanic acid. The rate of constitutive expression of AmpC is usually low, but clinically relevant resistance is inducible during therapy (139). Plasmid-mediated resistance involving group C enzymes has also been reported widely, including reports of plasmids in organisms, such as K. pneumoniae, that do not normally contain genes encoding these enzymes on their chromosome (159).
β-Lactamases belonging to Ambler class D primarily consist of oxacillin-hydrolyzing enzymes (OXA), which are able to hydrolyze oxacillin and its derivatives, which display ESBL-like substrate properties, and which show variable resistance to β-lactam inhibitors (139). Importantly, some OXA-type β-lactamases, such as OXA-48 and its derivatives, also confer carbapenem resistance. OXA-type enzymes are most frequently found in Acinetobacter spp., where they are often located on the chromosome. However, plasmid-borne OXA-48-like enzymes are now widely distributed in many Enterobacterales species, including K. pneumoniae and Enterobacter spp. (160), many of which express other ESBLs, such as CTX-M-15, and thus provide resistance to most β-lactam agents (161).
Aminoglycoside-modifying enzymes.
The most common aminoglycoside resistance mechanism encountered among ESKAPE pathogens occurs through the production of AMEs. During transportation of the drug across the cytoplasmic membrane, these enzymes covalently catalyze specific hydroxyl or amino group modifications of the aminoglycoside molecule, thus reducing antibacterial activity through diminished bacterial ribosomal subunit binding. Based on their biochemical activity, there are three classes of AMEs (i.e., aminoglycoside acetyltransferases [AACs], aminoglycoside phosphotransferases [APHs], and aminoglycoside nucleotidyltransferases [ANTs]). Enzymes within each class are then further subdivided according to the position of the modification site, resistance profile, and specific protein designation (162). Earlier work has shown that the global distribution of AMEs varies with respect to geography, antibiotic selection pressure, and bacterial species (163, 164). Depending on the specific enzyme and the host organism, genes coding for AMEs are located on plasmids, on transposons, or in the chromosome (162), though the high frequency of these resistance determinants among ESKAPE pathogens is largely attributable to acquisition via horizontal gene transfer (165).
AACs encompass the largest AME class and in an acetyl coenzyme A-dependent manner catalyze the acetylation of specific amino groups present on the antibiotic acceptor molecule. Of the four AAC subclasses, the AAC(1) and AAC(3) enzymes target amino group positions 1 and 3 of the central 2-deoxystreptamine ring, respectively, whereas the AAC(2′) and AAC(6′) subclasses modify the respective 2′ and 6′ amino group positions of the 2,6-dideoxy-2,6-diamino-glucose ring (166). While comprehensive analyses of global AAC epidemiology remain relatively scarce, recent investigations conducted in the United States, Europe, and Asia indicate that Gram-negative ESKAPE pathogens most frequently encode AAC(3) and AAC(6′) enzymes, which collectively confer resistance to gentamicin, tobramycin, and amikacin (165, 167, 168).
APHs comprise the second most abundant class of AMEs, which decrease aminoglycoside binding affinity by catalyzing ATP-dependent phosphorylation of —OH groups on the antibiotic molecule. Of the seven different APH subclasses [i.e., APH(4), APH(6), APH(9), APH(3′), APH(2ʺ), APH(3ʺ), and APH(7ʺ)], APH(3′) is the most widely distributed among clinical isolates, with the aph(3′)-IIIa gene being recognized as a key determinant of plasmid-mediated amikacin resistance in both S. aureus and Enterococcus spp. (165).
The final class of AMEs encompasses the ANTs, which reduce aminoglycoside toxicity via the magnesium-dependent transfer of a nucleoside monophosphate to —OH groups on the antibiotic molecule. Overall, there are five subclasses of ANTs [i.e., ANT(6), ANT(9), ANT(4′), ANT(2ʺ), and ANT(3ʺ)], of which ANT(4′) and ANT(2ʺ) are the most clinically relevant. ANT(4′) enzymes conferring resistance to amikacin and tobramycin have been detected in S. aureus, Enterococcus spp., K. pneumoniae, and P. aeruginosa. ANT(2ʺ), encoded by the ant(2ʺ)-Ia (or aadB) gene, is frequently associated with gentamicin and tobramycin resistance across all the Gram-negative ESKAPE organisms (165).
Most importantly, broad-spectrum aminoglycoside resistance in the ESKAPE pathogens is often conferred through the presence of multiple or bifunctional AMEs. This frequently occurs among Gram-negative organisms, where multiple AMEs result in significantly increased aminoglycoside resistance (169–171). Likewise, expression of the bifunctional AAC(6′)-APH(2″) enzyme, which resides on the common Tn4001 transposon, accounts for high-level gentamicin resistance in both S. aureus and Enterococcus spp. (including MRSA and VRE strains) worldwide (152). More recently, a variant enzyme termed AAC(6′)-Ib-cr, which confers low-level plasmid-mediated aminoglycoside and ciprofloxacin resistance, has been described in K. pneumoniae, Enterobacter spp., A. baumannii, and P. aeruginosa (172–175).
Target Site Modifications
Another common AMR strategy employed by the ESKAPE pathogens is to modify the antibiotic target site, thereby reducing the affinity or preventing the binding of the antibiotic molecule. Specifically, these mechanisms include (i) target enzyme modification, (ii) ribosomal target site alterations, and (iii) cell wall precursor alterations.
Target enzyme modifications.
β-Lactam antibiotics inhibit bacteria by binding to PBP enzymes anchored in the cell wall. In MRSA, resistance to methicillin and other β-lactam antibiotics is mediated through expression of the foreign mecA gene. mecA codes for PBP2a, a modified PBP with a low affinity for β-lactams, which renders most β-lactam agents completely ineffective against MRSA (176). mecA is located within the staphylococcal cassette chromosome mec (SCCmec), which also encodes a two-component regulatory system (TCRS; designated MecI and MecR1), site-specific ccr recombinase genes, as well as three joining (J) regions that can contain additional resistance determinants, mobile genetic elements (MGEs), and regulators (176). Cryptic or low-level mecA-positive MRSA strains displaying oxacillin MICs of ≤2 μg/ml are often misidentified as methicillin-sensitive S. aureus, proving a particular problem in the accurate identification of CA- and LA-MRSA (177, 178).
Thirteen distinct SCCmec types of various sizes and with various genetic contents have been identified thus far in S. aureus (179). Isolates possessing multidrug resistance and larger SCCmec types are typically associated with hospital-acquired MRSA (HA-MRSA) strains (e.g., SCCmec types I to III), whereas community-acquired strains expressing predominantly β-lactam resistance alone are more often associated with smaller SCCmec cassettes (e.g., types IV and V). Interestingly, two other mec gene homologues (designated mecB and mecC) have been recently identified in MRSA (180, 181). Though the frequency of strains expressing mecB is unclear at present, recent studies indicate that mecC-encoding S. aureus strains are predominantly found across the United Kingdom and Europe at a low but variable prevalence across several host species, including livestock and humans (176, 182, 183).
Both E. faecalis and E. faecium also express PBP5, a low-affinity chromosomally encoded ortholog of PBP2a in MRSA, which confers intrinsic low- to moderate-level β-lactam resistance (penicillin MICs are 2 to 8 μg/ml for E. faecalis and 16 to 32 μg/ml for E. faecium). In addition, up to 90% of hospital-associated E. faecium strains show high-level ampicillin resistance (MICs, >128 μg/ml), arising through the overproduction of PBP5 or polymorphisms in PBP5, which further decrease the affinity for β-lactam agents (184, 185). Although uncommonly reported, alterations in A. baumannii PBPs can also contribute to carbapenem resistance (186).
Another important example in which AMR arises in ESKAPE pathogens through modification of enzyme targets is fluoroquinolone resistance. Fluoroquinolones, such as ciprofloxacin and norfloxacin, represent some of the most widely prescribed antimicrobial agents worldwide. These are active against most ESKAPE organisms and target the DNA gyrase and topoisomerase IV enzymes, necessary for bacterial DNA repair and replication. Each of these heterotetrameric topoisomerases consists of two pairs of subunits (A and B) encoded by the gyrA and gyrB genes, respectively (or the parC and parE topoisomerase IV homologues, respectively) (187). Fluoroquinolone resistance most commonly occurs through spontaneous gyrA and parC mutations that give rise to amino acid changes clustered in the 5′ quinolone-binding region of the enzyme (188–190), though there is some evidence to suggest that B-subunit alterations also contribute to reduced susceptibility (191, 192). The level of resistance achieved by single-target mutations is dependent on both the specific agent and the bacterial species (187), while the accumulation of multiple mutations across both target enzymes often leads to the evolution of a high-level fluoroquinolone resistance phenotype (193).
Plasmid-mediated quinolone resistance (PMQR) conferred by Qnr-family proteins represents another fluoroquinolone resistance mechanism in K. pneumoniae and Enterobacter spp. (194, 195). qnr-encoded proteins (e.g., QnrA, QnrB, QnrS) bind directly to the DNA gyrase antibiotic target, thereby providing low-level fluoroquinolone resistance. PMQR is common among ESBL-producing organisms and can augment fluoroquinolone resistance levels arising through other mechanisms (194, 195).
Ribosomal target site alterations.
A major mechanism of resistance to macrolide-lincosamide-streptogramin B (MLSB) antibiotics in S. aureus and Enterococcus spp. is mediated by the erm-encoded rRNA methyltransferases. These enzymes mono- or dimethylate the A2058 residue within the 23S rRNA of the bacterial 50S ribosomal subunit, thus impairing MLSB target binding (196, 197). Expression of erm can be either constitutive or inducible. Constitutively expressing strains display cross-resistance to all MLSB agents. In contrast, inducibly resistant strains show resistance to 14- and 15-member inducer macrolides (e.g., erythromycin, clarithromycin, and azithromycin) but remain susceptible to lincosamides and streptogramin. There are 42 currently described classes of erm genes, many of which are located on mobile genetic elements (MGEs). erm(A) resides on transposon Tn554 as part of the SCCmec II cassette found predominantly in HA-MRSA strains. erm(C) is primarily associated with plasmid-mediated resistance in methicillin-susceptible S. aureus, whereas erm(B) is more commonly found in enterococci (198, 199).
ESKAPE organism resistance to linezolid and aminoglycosides is also mediated at the ribosomal level. Indeed, linezolid resistance in both S. aureus and Enterococcus spp. can arise through mutations in genes encoding 23S rRNA and/or 50S ribosomal subunit proteins or via Cfr-mediated methylation of 23S rRNA at residue A2503 (200). The cfr gene is transferable within MGEs, often in association with other AMR determinants (e.g., erm) (201, 202), and has been detected in staphylococcal strains possessing other linezolid resistance mechanisms (203). The enzymatic methylation of 16S rRNA conferring high-level aminoglycoside resistance (to all aminoglycosides, including plazomicin [described below]) has also recently emerged as an important acquired AMR mechanism in the Gram-negative ESKAPE pathogens (204, 205). To date, 10 different classes of 16S rRNA methyltransferases have been documented worldwide (e.g., ArnA, RmfA to RmfH, and NmpA). Concerningly, these enzymes are often located on plasmids that harbor the genes for other MDR determinants (e.g., blaOXA-23 and blaNDM), thus further reducing the available treatment options (205).
Cell wall precursor alterations.
One of the most important AMR mechanisms that has emerged in Gram-positive ESKAPE organisms in recent decades has been the development of glycopeptide resistance. In susceptible Gram-positive organisms, bacterial cell wall biosynthesis is inhibited by glycopeptides that target outer cell wall d-Ala–d-Ala peptidoglycan precursor residues. Glycopeptide resistance in enterococci involves the acquisition of van gene clusters which coordinate (i) the synthesis of modified peptidoglycan precursors that exhibit subdued glycopeptide binding (i.e., the natural d-Ala–d-Ala termini are replaced with either d-Ala-d-lactate or d-Ala–d-serine) and (ii) the production of d,d-carboxypeptidases that eliminate residual natural d-Ala–d-Ala precursors from the host cell (184, 206). To date, nine distinct van gene clusters have been classified, with the majority of human VRE infections being attributed to E. faecium and E. faecalis isolates carrying vanA and vanB gene clusters. vanA-mediated resistance occurs most frequently and is characterized by high-level resistance to both vancomycin and teicoplanin (206). The vanA gene cluster is typically associated with Tn1546 and related transposons, which can be localized on both plasmids and chromosomal DNA (207). In contrast, the vanB gene cluster confers resistance to only vancomycin and is most often carried by Tn1547/Tn5382 transposons that localize to the chromosome (208, 209).
Since 2002, sporadic cases of vancomycin-resistant S. aureus (termed VRSA) infection have also been reported. This form of vancomycin resistance (MIC, ≥16 μg/ml) is conferred by the vanA gene cluster on Tn1546, which is acquired via conjugative transfer of enterococcal plasmids (210, 211). In such instances, vancomycin resistance is maintained either by retention of the donor enterococcal plasmid within the S. aureus recipient or through transposition of the incoming Tn1546 element onto an endogenous staphylococcal plasmid. Most cases of VRSA infection have been observed among patients with prior/current VRE infections, though the frequency of such detections is low, with less than 20 cases being reported across North America, South America, and Europe to date (212–215). This most likely reflects several factors, including plasmid instability, the relatively low prevalence of donor Enterococcus strains containing compatible plasmids carrying vanA, robust S. aureus restriction modification systems which restrict unmodified DNA from entering the cell, as well as VanA-associated fitness costs (216–219).
A much more commonly encountered issue is the detection of S. aureus isolates that exhibit intermediate resistance to vancomycin (i.e., MICs, 4 to 8 μg/ml; termed vancomycin-intermediate S. aureus [VISA] strains). This form of AMR typically emerges through prolonged exposure to vancomycin, giving rise to an initial heterogeneous VISA (hVISA) phenotype, in which a small subpopulation of cells demonstrates MICs of ≥4 μg/ml (220). The precise mechanisms underlying the hVISA/VISA phenotypes are incompletely understood, though various studies indicate the role of genetic modifications to regulatory genes and global epigenetic changes which lead to cell wall thickening, decreased peptidoglycan cross-linking, and autolytic activity, as well as an excess of d-Ala–d-Ala residues (198, 221–225). As opposed to person-to-person transmission, the vast majority of hVISA and VISA infections arise via in vivo evolution within individual patients and typically involve pandemic HA-MRSA lineages (e.g., ST239 and ST5). However, it should be noted that CA-MRSA clones, including the USA300 clone, can also exhibit this resistance phenotype (146, 226).
Resistance to daptomycin, an agent that has activity against Gram-positive bacteria and that is related to host cationic antimicrobial peptides (AMPs), has also been observed in both S. aureus and enterococci in recent years. The precise mechanisms of resistance are yet to be fully elucidated, but it has been postulated that alterations in cell surface charge, phospholipid composition/metabolism, and membrane stress responses are involved (227). Recent studies also highlight the emergence of acquired polymyxin (another cationic AMP) resistance in K. pneumoniae, A. baumannii, and P. aeruginosa arising from remodeling of outer membrane (OM) lipopolysaccharide (LPS) lipid A structures. These modifications contribute to reduce the net negative charge of the LPS, thus reducing its polymyxin binding efficiency. In K. pneumoniae, loss-of-function mutations of the mgrB gene (a negative feedback regulator of the PhoPQ TCRS), mutations driving the expression of the PhoPQ, PmrAB, and CrrAB TCRS, as well as the acquisition of the plasmid-mediated mcr gene all give rise to resistance-associated lipid A modifications (e.g., addition of 4-amino-4-deoxy-l-arabinose [Ara4N], phosphoethanolamine [PEtN], and 2-hydroxymyristate through increased expression of the pmrHFIJKLM operon, pmrC, and lpxO, respectively) (135, 228–230). Of these, mgrB inactivation has been reported the most frequently and, interestingly, also gives rise to other modifications that collectively promote virulence and that attenuate early host defense responses (228). The primary mechanisms of polymyxin resistance in A. baumannii comprise mutations in the PmrAB TCRS leading to PEtN synthesis and the loss of LPS through inactivation of the lpxA, lpxC, and lpxD lipid A biosynthesis genes (229). Polymyxin resistance in P. aeruginosa is conveyed by five TCRS, including PmrAB, PhoPQ, ParRS, ColRS, and CpsRS, most frequently resulting in the constitutive expression of pmrHFIJKLM and the addition of Ara4N (229).
Reduced Antibiotic Penetration and Accumulation
Porins.
Mutations leading to the downregulation, balance, function, and/or loss of the outer membrane protein channels (porins) also represent important mediators of AMR among Gram-negative ESKAPE pathogens. Hydrophilic agents, such as the β-lactams (including carbapenems) and some fluoroquinolones which rely on porins to penetrate the outer membrane barrier, are particularly affected. Moreover, these mutations can arise during treatment (231, 232) and, importantly, enhance the influence of other resistance mechanisms, such as efflux pumps and degradative enzymes (198). For example, the loss or modification of the P. aeruginosa OprD porin is linked to reduced carbapenem susceptibility (233). Likewise, the loss or inactivation of CarO in A. baumannii is associated with imipenem resistance (234). During antibiotic therapy, it is also recognized that K. pneumoniae and some Enterobacter spp. can sequentially alter the balance of different porins. In some cases, the sorbitol-sensitive Omp35 porin is replaced with Omp36, which has a smaller channel size. These Omp35-deficient, Omp36-producing strains typically exhibit intermediate carbapenem susceptibility profiles, while those lacking both porins show carbapenem resistance (233, 235, 236). Overexpression of the LamB porin in association with strains showing porin deficiency or reduced porin expression can also contribute to reduce β-lactam susceptibility (235, 236). Mutations leading to conformational changes in the eyelet region of the E. aerogenes Omp36 lumen with reduced β-lactam permeability have also been reported (235).
Efflux pumps.
The expression of bacterial efflux pumps, which actively extrude drugs out of the cell, also greatly contributes to AMR. Genes encoding efflux pumps can be located on the chromosome or within MGEs. To date, six major families of efflux pumps have been characterized, comprising the (i) resistance-nodulation-division (RND), major facilitator superfamily (MFS), multidrug and toxic compound extrusion (MATE), small multidrug resistance (SMR), ATP-binding cassette (ABC), and proteobacterial antimicrobial compound efflux (PACE) families (234, 237). All six families are represented within the ESKAPE group, with individual exporters varying in terms of their substrate specificity. Of note, RND-type efflux pump-mediated resistance is of particular concern with respect to AMR among Gram-negative bacteria. For example, the chromosomally encoded MexAB-OprM efflux system in P. aeruginosa exhibits broad substrate specificity and when overexpressed confers fluoroquinolone, aminoglycoside, and β-lactam resistance. Likewise, the overproduction of AcrAB-TolC is characteristic of multidrug-resistant K. pneumoniae and Enterobacter strains. The A. baumannii AdeABC, AdeFGH, and AdeIJK RND-type efflux pumps are also associated with broad-range AMR (234, 238–240). More recently, the chromosomally encoded OqxAB efflux pump, which contributes to reduced quinolone and chloramphenicol susceptibility, has been identified in K. pneumoniae (195, 241). OqxAB homologues have also been observed in some Enterobacter spp., though, aside from tigecycline (242), these elements are not thought to contribute to clinically relevant drug resistance under in vitro conditions (241).
Other Mechanisms and Survival Strategies
Biofilms.
In addition to the aforementioned classical AMR mechanisms, it is now also recognized that growth within biofilms can further impede antimicrobial activity. Biofilms are structured, surface-attached microbial communities encased in an extracellular matrix (ECM) which demonstrate a markedly higher tolerance to antimicrobial agents than nonadherent planktonic cells (118, 243, 244). Most notably, biofilms play a prominent role in chronic infections, such as those involving P. aeruginosa in the airways of patients with cystic fibrosis and indwelling medical device infections caused by S. aureus and A. baumannii (118, 245). The reduced antibiotic susceptibility exhibited by biofilm-embedded cells is thought to be multifactorial and can vary according to the species and genetic makeup of the organism(s), the nature of the antimicrobial agent, the developmental stage of the biofilm, and the environmental conditions (118, 246). More recently, it has been recognized that bacterial aggregation can also give rise to reduced antibiotic susceptibility independent of growth on a surface. Some of the factors attributable to the increased antibiotic recalcitrance of biofilms include (i) restriction of antibiotic penetration by the ECM, (ii) the secretion of antibiotic-modifying enzymes, extracellular DNA, and other macromolecules into the ECM, (iii) the accumulation of filamentous bacteriophages which promote the formation of liquid crystalline structures, (iv) differential metabolic activity, (v) the emergence of persister cells (see below), (vi) biofilm-associated upregulation of bacterial efflux, (vii) enhanced horizontal gene transfer and mutation frequency, and (viii) interactions between different bacterial species within mixed-species biofilms (246–249). A classic example of the last two factors was reported by Weigel and colleagues, who observed that a plasmid carrying a vanA vancomycin resistance gene in a VRSA strain arose from a VRE strain present within the same multispecies biofilm (250).
Antibiotic tolerance and persistence.
Aside from antibiotic resistance, which is characterized by the presence of inheritable resistance-encoding genes or mutations that give rise to an increased MIC, there is increasing evidence that some ESKAPE pathogens are able to overcome treatment through antibiotic tolerance. Antibiotic tolerance enables an entire bacterial population to withstand transient exposures to high doses of bactericidal antibiotics (e.g., β-lactams and quinolones) without a change in the MIC. This occurs in the absence of any genetic resistance factor and is typically associated with an arrested (or dormant) growth state which is reversed upon removal of the antibiotic exposure (251, 252). Antibiotic tolerance can arise from genetic mutations but may also be conferred by stressful external conditions, including nutrient limitation, host factors, temperature, and antibiotic treatment (251). Concerningly, recent studies of MRSA infections in humans also indicate that the evolution of antibiotic tolerance can facilitate the emergence of mutational resistance (253). Quantitative assessment of antibiotic tolerance can be reliably achieved by the minimum duration for killing of 99% of a bacterial population (MDK99), which evaluates the time that it takes to eradicate 99% of a bacterial population at antibiotic concentrations that substantially exceed the MIC (252, 254). In a related phenomenon, antibiotic tolerance can also be observed among subpopulations of bacterial cells termed “persisters.” Antibiotic persistence is frequently associated with biofilm infections and is characterized by a biphasic MDK99.99 killing curve which displays the emergence of a clonal persister subpopulation over time. Persister bacterial cells do not respond to antibiotics, and although they fail to divide in the presence of bactericidal antimicrobials, they are not killed. Upon treatment cessation, these persistent subpopulations are then able to resume growth, thus contributing to relapsing or chronic infection (252, 255, 256).
Intracellular survival.
Another possible factor contributing to AMR among ESKAPE pathogens is the observation that some species can be internalized and then survive for extended periods within host cells. Indeed, recent in vitro studies show that upon engulfment by alveolar macrophages, both K. pneumoniae and E. faecalis are able to survive and persist within unique intracellular vacuolar compartments (257, 258). Likewise, there is accumulating evidence that S. aureus has the capacity to adhere to, enter, and survive within both professional and nonprofessional phagocytes, including macrophages; epithelial, endothelial, and mammary cells; keratinocytes; osteoblasts; and fibroblasts (259, 260). In such instances, it is thus plausible that the microbes are able to not only evade many of the hosts’ immune defenses but also remain insulated from the activity of cell-impermeant antibiotics, thus providing a reservoir for disseminated and/or latent infection. Such a scenario was recently illustrated by Lehar and colleagues, who showed that, compared to extracellular planktonic bacteria, intracellular MRSA isolates exhibit a 100-fold increase in the vancomycin MIC, as well as an enhanced propensity for systemic dissemination in an antibiotic-treated mouse infection model (261).
MOBILE GENETIC ELEMENTS CONFERRING ANTIMICROBIAL RESISTANCE
While bacteria can be intrinsically resistant to certain antibiotics, they may also accumulate AMR genes on MGEs. MGEs are segments of DNA that are capable of capturing genes and mediating their movement within the genome (intracellular mobility) or between different cells (intercellular mobility). In this fashion, MGEs are responsible for much of the observed phenotypic variability in AMR both within and between bacterial species. The association of AMR and MGEs has been extensively reviewed recently (262). Thus, here we summarize those elements most relevant to the ESKAPE pathogens, mainly, plasmids, insertion sequences (IS) and transposons (Tn), integrative and conjugative elements (ICE), and other genomic islands (GI) (Table 2).
TABLE 2.
MGEs associated with carriage of antimicrobial resistance genes in ESKAPE pathogensa
| Species | Resistance(s) | MGE type | MGE | Key characteristics | Reference(s) |
|---|---|---|---|---|---|
| Vancomycin-resistant Enterococcus | Vancomycin | Tn | Tn1546 | A composite transposon (IS16) responsible for dissemination of vancomycin resistance in both enterococci and staphylococci | 560, 561 |
| Vancomycin | ICE | Tn1549-like | vanB is typically associated with Tn1549-like and Tn5382-like transposons, which are closely related; significant role in the global spread of vancomycin resistance | 334, 335 | |
| Chloramphenicol, aminoglycosides, tetracycline, kanamycin, MLS, chloramphenicol, streptomycin, vancomycin | Plasmid | Inc18-type (e.g., pR23 and pEF-01), RepA_N plasmids (e.g., pRUM-like) | Conjugative; broad host range, allowing for transfer into a variety of bacterial species; responsible for introducing vancomycin resistance into MRSA | 262, 562, 563, 564, 565 | |
| Methicillin-resistant S. aureus | β-Lactams | Tn | Tn552-like elements | Tn552 is a complex unit transposon responsible for mobilization and dissemination of β-lactam resistance in staphylococci; associated with a pSK1-like and pSK41-like plasmid | 272 |
| Vancomycin | Tn | Tn1546 | A composite transposon (IS16) responsible for dissemination of vancomycin resistance in both enterococci and staphylococci | ||
| Tetracycline, chloramphenicol, erythromycin | Plasmid | Small (1- to 10-kb) multicopy plasmids | Typically carries a single resistance gene | 314–316 | |
| β-Lactams, aminoglycosides, trimethoprim, antiseptics | Plasmid | pSK1-like plasmids | Associated with carriage of the Tn552-like β-lactam resistance transposons | 317 | |
| Aminoglycosides, β-lactams, vancomycin | Plasmid | pSK41-like plasmids (e.g., pSK41, pGO1, and pLW103) | Conjugative; associated with carriage of the Tn552-like β-lactam resistance transposons; associated with carriage of vanA on the Tn1546 glycopeptide resistance transposon | 318–321 | |
| Methicillin, penicillin, β-lactams | GI | SCCmec element | SCCmec has a limited distribution and is restricted to 11 major clonal lineages of S. aureus from 5 clonal complexes | 336–338 | |
| Methicillin, glycopeptides | IS | IS1182, IS256 | Insertional deactivation, resulting in increased resistance | 566, 567 | |
| Enterobacterales (K. pneumoniae and Enterobacter spp.) | Multidrug: β-lactams, carbapenems, trimethoprim, chloramphenicol, fluoroquinolones | IS | ISEcp1, ISCR | Typically encodes a promiscuous transposase which can mobilize adjacent genes when they fail to identify terminal repeat sequences; ISEcp1 elements are commonly associated with mobilization of β-lactamase genes; ISCR elements are commonly associated with complex class 1 integrons | 273–276, 280, 568, 569 |
| Carbapenems, colistin | IS | Various IS, including ISEcp1 | Insertional deactivation resulting in increased resistance | 276, 281, 282, 570 | |
| Multidrug: β-lactams, aminoglycosides, trimethoprim, antiseptics, carbapenems, colistin, cephalosporins | Plasmid | IncF-type, IncI, IncH (HiI1 and HI2), IncL, IncC, IncN, IncH, IncX3 | Typically, low-copy-number plasmids; broad and narrow host ranges; can act as vehicles for carriage of other mobile resistance elements, e.g., transposons and integrons | 293, 294, 571 | |
| A. baumannii | Multidrug: β-lactams, aminoglycosides chloramphenicol, tetracycline, sulfonamide | Tn | AbaR, AbGR11 | Tn7-like unit transposons | 270, 271 |
| Carbapenems | Tn | Tn2006 | The Tn2006 composite transposon (ISAb1) is responsible for mobilization of the blaOXA-23 carbapenemase | ||
| Carbapenems | IS | ISAb825, ISAb125 | Insertional deactivation of the outer membrane protein carO results in elevated carbapenem MICs | 223 | |
| Carbapenems | IS | ISAba1 | IS insertion upstream of blaOXA-53 drives expression of the gene; IS-mediated constitutive expression of blaOXA-53 confers high-level carbapenem resistance | 283 | |
| Gentamicin, tobramycin | Plasmid | pRAY-like plasmids | Small (6- to 10-kb), widely distributed plasmids | 303, 304 | |
| Multidrug: carbapenems and kanamycin | Plasmid | RepAci6-like plasmids (e.g., pAB-G7-2 and pACICU2), pNDM-BJ01-like plasmids | Kanamycin resistance is associated with carriage of the TnaphA6 transposon; carbapenem resistance is associated with carriage of the Tn2006 transposon; pNDM-BJ01-like plasmids from A. lwoffii carry the blaNDM-1 carbapenemase genes | 302, 305, 306 | |
| P. aeruginosa | Carbapenems, β-lactams | IS | IS21, ISPA26 | Insertional deactivation, resulting in increased resistance | 572, 573 |
| Carbapenems | Plasmid | IncP-2 plasmids | Carbapenemases associated with class 1 integrons carried on IncP-2-type plasmids | 309–311 | |
| Carbapenems, aminoglycosides | ICE | P. aeruginosa pathogenicity island (e.g., PAPI-1, PAGI-2/PAGI-3-like) | Multidrug resistance is associated with carriage of antimicrobial resistance genes on Tn6162 and Tn6163 elements in a genomic island | 333 |
Abbreviations: MGEs, mobile genetic elements; ICE, integrative and conjugative elements; Tn, transposon; IS, insertion sequence; GI, genomic island; MLS, macrolide lincosamide sulfonamides; SCCmec, staphylococcal cassette chromosome mec.
Insertion Sequences and Transposons
IS are small elements (typically, <3 kb) that are capable of self-transposition. The canonical IS unit is composed of one or two genes required for mobility, flanked by terminal inverted repeats (IRs) (263, 264). IS are capable of mobilizing neighboring genes (cargo genes) in structures called composite/compound transposons, where two copies of an identical or related IS mobilize the region between them (265, 266). Classic examples of composite transposons associated with the carriage of AMR genes include Tn9 (IS1; chloramphenicol resistance), Tn10 (IS10; tetracycline resistance), Tn5 (IS50; aminoglycoside and bleomycin resistance) (262, 265), and, more recently, Tn6330 (ISApl1), which is responsible for mobilizing the colistin resistance gene mcr-1 (135, 262, 267). More complex unit transposons can also be found in both Gram-negative and Gram-positive bacteria. Unit transposons are large IS-like elements flanked by terminal IRs with genes (for example, tnpA [transposase] and tnpR [resolvase] in Tn3) that facilitate replicative transposition. In the ESKAPE pathogens, AMR genes are frequently associated with the Tn3 family (Tn1, Tn2, and Tn3) (207, 268, 269), Tn7-like unit transposons (270, 271), and Tn552-like elements (272) (Table 2).
In some cases, single IS elements can also mobilize neighboring genes. ISEcp1 is commonly associated with the β-lactamase genes blaCTX-M (e.g., blaCTX-M-1, blaCTX-M-9, blaCTX-M-15), blaCMY-2, and blaACC and, more recently, with the blaOXA-181 carbapenemase gene (273–276). ISEcp1 elements encode promiscuous transposases that can recognize a variety of different sequences as right IRs (IRr), thereby allowing them to capture adjacent genes. Related IS (IS1247, ISKpn23, and ISEnca1) have also been associated with the mobilization of adjacent resistance genes in a manner similar to that for ISEcp1 (277, 278). Recently, it has been demonstrated that IS26 can produce a circular intermediate consisting of a single copy of IS26 and a DNA segment immediately adjacent to it. This structure, termed a translocatable unit (TU), can then move by a replicative mechanism (279). ISCR elements also form circular intermediates. They move and can capture adjacent genes by rolling-circle replication (280).
IS elements can also impact the evolution of AMR in the host by the transpositional deactivation of genes and by modulating the expression of adjacent genes through the delivery of promoter or terminator sequences (reviewed in reference 264). The insertional deactivation of uptake systems is a common mechanism by which IS elements can affect antibiotic susceptibilities. For example, IS-mediated deactivation of the ompK36 porin in K. pneumoniae results in elevated carbapenem MICs (281). Similarly, insertional inactivation of the mgrB regulatory gene in K. pneumoniae drives the overexpression of the pmrHFIJKLM operon, conferring colistin resistance (276, 282).
Many IS carry strong promoter sequences, and their insertion upstream of chromosomal genes can drive the expression of that gene and influence AMR. This mechanism is clearly evident in A. baumannii, where insertion of an ISAba1 element upstream of the blaOXA-51 gene confers carbapenem resistance (283). Similar mechanisms of IS-mediated constitutive expression of resistance genes have been observed in K. pneumoniae and P. aeruginosa (284, 285). Alternatively, an IS may provide only the −35 region promoter component, which, together with a −10 region donated by an adjacent gene, forms a hybrid promoter to drive the expression of neighboring genes. Hybrid IS promoters have been identified in at least 17 different bacterial species (286), including A. baumannii, K. pneumoniae, and P. aeruginosa (283, 287, 288).
Plasmids
Plasmids are an important vehicle for gene transfer in both Gram-negative and Gram-positive bacteria (289). Typically, plasmids are circular, double-stranded, and self-replicating DNA molecules that are readily vertically inherited in a growing population (290). While it is clear that the MGEs described thus far (IS elements, transposons, etc.) are primarily responsible for mobilizing resistance genes, the dissemination of these genes is mainly attributed to conjugative plasmids. Plasmids are rich in IS and other MGEs carrying AMR genes and facilitate the intra- and interspecies horizontal transfer of these elements (135, 291–293).
MDR Enterobacterales (K. pneumoniae and Enterobacter species) carry plasmids from a wide variety of different incompatibility (Inc) groups (294) (Table 2), but those from Inc group types F (multiple F-type replicons can be found together in multireplicon plasmids), I, H (HI1 and HI2), L, C, and N are most frequently associated with multidrug resistance (293, 294). Of particular concern among the Enterobacterales is the role that these plasmids continue to play in the emergence and dissemination of ESBLs, particularly those of the blaCTX-M type (19, 295, 296); AmpC-type cephalosporinases (blaCMY-2 and blaDHA-1) (297–299); carbapenemase-encoding genes (blaVIM, blaKPC, blaNDM, and blaOXA-48) (161, 300, 301); and plasmid-mediated colistin resistance (mcr) (229). Comparatively little is known about plasmids from A. baumannii compared to what is known about plasmids from the Enterobacterales. However, in 2011, a Europe-wide study of clinical A. baumannii isolates found that resistance plasmids were mainly associated with carriage of the blaOXA (blaOXA-23, blaOXA-58-like, and blaOXA-40) carbapenemase genes (302), kanamycin and amikacin resistance, and gentamicin and tobramycin resistance (303–306). Additionally, plasmids related to pNDM-BJ01 from Acinetobacter lwoffii carry the globally distributed blaNDM-1 carbapenemase gene (307).
In P. aeruginosa, resistance genes are typically found on chromosomal resistance islands rather than plasmids. However, P. aeruginosa may carry large (∼300- to 500-kb), transferable IncP-2 plasmids (308) associated with the carriage of carbapenemase genes, specifically, blaIMP (blaIMP-9 and blaIMP-45) (309, 310) and blaVIM (blaVIM-2) (311) carbapenemases, on class 1 integrons.
AMR plasmids are frequently found in clinical staphylococcal isolates (312, 313). These include small (1- to 10-kb) multicopy plasmids typically encoding a single resistance gene (314–316) and larger (>15-kb) multiresistance plasmids, such as the prototype pSK1 family of multiresistance plasmids (317) (Table 2). A single family of larger (>30-kb) conjugative multiresistance plasmids, including pSK41, pGO1, and pLW1043 (318, 319), can also be found in clinical strains of staphylococci and are credited with the emergence of aminoglycoside, β-lactam, and vancomycin resistance in S. aureus populations (267, 320–326).
In the enterococci, AMR is largely encoded on Inc18 and RepA_N plasmids (262). Both Inc18 and RepA_N plasmids have a broad host range, allowing for their transfer into a variety of bacterial species, and are responsible for introducing vancomycin resistance into MRSA (218).
Genomic Islands and Integrative Conjugative Elements
Genomic islands (GIs) are discrete genomic loci that have been acquired through horizontal gene transfer (HGT) (327). Many different types of GIs exist (328–330), and describing them all is beyond the scope of this review. Here, we only briefly describe the integrative conjugative elements (ICE) and the SCCmec element.
ICE are conjugative elements that integrate into the host chromosome and that are passively replicated along with the host genome during cell division (331, 332). In P. aeruginosa ICE, specifically, the P. aeruginosa pathogenicity/genomic island-type ICE (PAPI-1, PAGI-2/PAGI-3-like), are the most important for acquired AMR genes (333) (Table 2). ICE are also important carriers of AMR genes in enterococci (334). Of particular note is the vanB-associated Tn1549, which has played a significant role in the global spread of vancomycin resistance (335).
In staphylococci, SCCmec is a genomic island carried in the chromosome of MRSA isolates (336). The SCCmec element carries the mecA gene, encoding a low-affinity penicillin-binding protein, PBP2a, that confers resistance to methicillin, penicillin, and other β-lactam antibiotics. Horizontal transfer of the SCCmec element facilitates the movement of the mecA gene and methicillin resistance among the staphylococci. However, despite this capacity for mobility, SCCmec has a limited distribution and appears to be restricted to 11 major clonal lineages of S. aureus from 5 clonal complexes (337, 338).
Contribution of Horizontal Gene Transfer to the Spread of Mobile Genetic Elements
HGT facilitates the movement of MGEs between bacteria and is considered the primary mechanism for the emergence and spread of AMR among pathogenic bacteria (339). Of the three primary mechanisms of HGT, conjugation, transduction, and transformation, the dissemination of MGEs that carry AMR genes is mostly by conjugation. Conjugative plasmids and ICE facilitate the rapid dissemination of AMR genes among bacteria of diverse origins (19, 340–343). Additionally, conjugative plasmids can also mobilize nonconjugative plasmids, including those with broad host ranges (344). Although it has been less well studied, there is currently indirect evidence that transduction can contribute to the spread of AMR genes between members of the same bacterial species. Bacteriophages isolated from hospital-acquired MRSA, Pseudomonas, and Acinetobacter strains have been shown to transduce AMR genes to recipient strains under laboratory conditions (345–348). There is currently no substantial direct evidence to suggest that transformation contributes to the spread of AMR among the ESKAPE pathogens. However, Acinetobacter, Pseudomonas, and Staphylococcus strains are all capable of DNA uptake and natural transformation (349). and the Enterobacterales are predicted to be naturally competent (350, 351). Additionally, natural transformation can facilitate the transfer of transposons and integrons between bacterial species (352). Consequently, the potential role that transformation plays in the spread of AMR genes cannot be overlooked.
Coselection of Antimicrobial Resistance with Detergents and Biocides
Due to the fitness cost imposed by plasmid carriage, plasmids tend to be lost from bacterial populations under conditions in which they provide no selective advantage (353, 354). However, antibiotic stewardship programs and the restricted use of key antibiotics have not been successful in controlling the emergence and spread of AMR. One mechanism which promotes the persistence of AMR genes in bacterial populations is coselection with genes that confer resistance to metals and antimicrobial biocides (355–358). Coselection can result when antibiotic resistance and biocide resistance share a mechanism, for example, modification of the cell wall/membrane preventing entry into the cell or upregulation of efflux pumps to remove unwanted compounds from the cell (359–361). Coselection can also occur when the bacteria carry genes that promote resistance or decreased susceptibility to both types of antimicrobial compounds. The colocation of AMR and biocide resistance genes on plasmids and other MGEs (296–298) is particularly problematic, as exposure to the latter can facilitate the spread of AMR genes by HGT. In 2015, a large-scale study on coselection potential that examined 2,522 bacterial genomes and 4,582 plasmids found that among plasmids carrying genes for AMR and biocide resistance, 57% were conjugative, whereas only 18% of plasmids carrying either AMR or biocide resistance genes were conjugative (357).
Although many different mechanisms of coselection exist, of particular concern in clinical environments is the qac family of biocide resistance genes, specifically, qacEΔ1, a truncated version of the qacE gene associated with low-level resistance to quaternary ammonium compounds (QACs) and other biocides (e.g., biguanides, diamidines, and xanthenes) (362, 363). qacEΔ1 is frequently found as a component of AMR gene cassettes on class 1 integrons, which are more prevalent in bacteria exposed to detergent and other biocides than in bacteria not exposed to these compounds (362, 364, 365). Biocides, such as quaternary ammonium compounds, are heavily used as cleaning agents in hospitals. Consequently, intertwined AMR and QAC resistance mechanisms can represent a long-term selection pressure for the maintenance and spread of AMR in clinical environments (366–369).
Antibiotic stewardship programs are designed to limit the use of key antibiotics to prevent the spread of AMR. However, in order for these programs to be successful, it is also important to consider measures which prevent the accumulation of biocides and other antimicrobial toxicants that can promote coselection.
THERAPEUTIC ADVANCES AGAINST ESKAPE PATHOGENS
Antimicrobial drug discovery is highly challenging, and the current rise in AMR is eroding the efficacy of available antibiotics (370). Since the early 1960s, only 4 new classes of antibiotics have been introduced: quinolones, lincosamides, oxazolidinones, and cyclic lipopeptides. The global financial antibiotic market, estimated at $30 billion, is still dominated by classes of antibiotics discovered over half a century ago (371). Frequently, “novel antibiotics” is a term often used for successive compounds derived from established antibiotic classes (371). The reserved use of such novel drugs as last-resort measures has constrained the profit margins of the pharmaceutical industry, leading organizations to withdraw their research effort from antimicrobial drug discovery (370). Economic barriers surrounding the R&D of antimicrobial drug design are largely instigated by the large operational cost of clinical trials, estimated to be upwards of $130 million, with postapproval follow-on trials amounting to an added $146 million (372). Antimicrobial drug candidates targeting more than one clinical indication require significantly more financial support, further exacerbating the financial hurdle (373). The financial justification for the development and commercialization of new therapies often fails to outweigh the value to public health and foregoing investment (374). Limited by stewardship practices, the net value of an R&D project has been estimated to be €31.5 million, which in turn considerably weakens the economic incentive (375). Today, the majority of pharmaceutical companies which do undertake R&D have few antimicrobial products on the market aimed at the WHO’s priority pathogen list (376). Due to the absence of revenue from sales, these companies are largely dependent on external partnering funding sources, such as the U.S. Biomedical Advanced Research and Development Authority (BARDA), Combating Antibiotic Resistant Bacteria Biopharmaceutical Accelerator (CARB-X), the Wellcome Trust, the National Institute of Allergy and Infectious Diseases (NIAID), and the Innovative Medicines Initiative (IMI; a joint undertaking between the European Union and the European Pharmaceutical Industry), initiatives funded solely and/or in partnership (376–379). Identifying antibiotic-related health priorities, defining appropriate stewardship practices, and developing new sustainable economic models to stimulate antibiotic innovation are key intersecting themes currently requiring a cooperative restructure. Funded by IMI, Driving Reinvestment in Research and Development and Responsible Antibiotic Use (DRIVE-AB) is a pioneering project composed of 15 public and 7 private partners from 12 countries aiming to tackle this issue (380). Through the use of surveillance systems data, antibiotic prescription databases, and published literature, DRIVE-AB aims to develop models which transform the way in which policy makers stimulate and financially incentivize antibiotic innovation (380, 381). Despite collaborative projects like these, the majority of new R&D programs aimed at tackling AMR are currently funded by public and nonprofit partnerships (374). This approach often neglects obstacles, such as licensing, affordability, and stewardship, which affect access to new antimicrobials in low- and middle-income countries. Even though antibiotic drug discovery has reduced and investment from the pharmaceutical industry has receded, promising antimicrobial strategies are still being developed (Table 3) (371, 382–389).
TABLE 3.
Antimicrobial drugs in the clinical pipeline as of February 2020a
| Drug name | Drug class | Drug target | Development stage | Company | Pathogen target | Applicationb | Information of noteb |
|---|---|---|---|---|---|---|---|
| Plazomicin (Zemdri) | Aminoglycoside | 30S subunit of bacterial ribosome | U.S. FDA approved on 26 June 2018 | Achaogen Inc. | E. coli, K. pneumoniae, Proteus mirabilis, E. cloacae, and CRE | cUTI including acute pyelonephritis | Nephrotoxicity has been reported |
| Cefiderocol (Fetroja) | Siderophore–β-lactam (cephalosporin) | PBP | U.S. FDA approved on 14 November 2019 | Shionogi & Co. Ltd. | CRE, CRAB, and CRPA | cUTI | |
| Imipenem + cilastatin + relebactam (Recarbrio) | β-Lactam (carbapenem) + β-lactamase inhibitor (diazabicyclooctane) | PBP, β-lactamase | U.S. FDA approved on 17 July 2019; E.U. EMA approved on 13 February 2020 | Merck & Co., Inc. | CRE and CRPA | cUTI including pyelonephritis, cIAI, and HA and VA bacterial pneumonia | |
| Meropenem- vaborbactam (Vabomere/Vaborem) | β-Lactam (carbapenem) + β-lactamase inhibitor (cyclic boronate) | PBP, β-lactamase | U.S. FDA approved on 30 August 2017; E.U. EMA approved on 20 November 2018 | Rempex Pharmaceuticals Inc. (wholly owned subsidiary of The Medicines Co.) | E. coli, K. pneumoniae, E. cloacae species complex, and CRE | cUTI including pyelonephritis | Anaphylactic and central nervous system reactions have been reported |
| Cefepime + zidebactam (WCK 5222) | β-Lactam (cephalosporin) + β-lactamase inhibitor (diazabicyclooctane) | PBP, β-lactamase | Phase I | Wockhardt Ltd. | ESKAPE pathogens, pneumococci, and streptococci | cUTI and HA and VA bacterial pneumonia | Highly broad-spectrum antimicrobial |
| ETX0282 + cefpodoxime + proxetil (ETX0282CPDP) | β-Lactam (cephalosporin) + β-lactamase inhibitor (diazabicyclooctane) | PBP, β-lactamase | Phase I | Entasis Therapeutics Inc. | MDR A. baumannii and CRAB | UTI | Designated a QIDP by U.S. FDA and awarded fast-track review |
| Meropenem + nacubactam OP0595/RG6080) | β-Lactam (carbapenem) + β-lactamase inhibitor (diazabicyclooctane) | PBP, β-lactamase, PBP2 | Phase I | Meiji Seika Pharma Co. Ltd./Fedora Pharmaceuticals Inc. (Roche licensee) | CRE, CRPA | Bacterial infections | |
| Cefepime + AAI101 | β-Lactam (cephalosporin) + β-lactamase inhibitor (β-lactam) | PBP, β-lactamase | Phase III | Allecra | CRE | cUTI including pyelonephritis, cIAI, and HA and VA bacterial pneumonia | Designated a QIDP by U.S. FDA and awarded fast-track review |
| Cefepime + taniborbactam (VNRX-5133) | β-Lactam (cephalosporin) + β-lactamase inhibitor (cyclic boronate) | PBP, β-lactamase | Phase III | VenatoRx Pharmaceuticals Inc. | MDR enteric organisms and ESBL-, KPC-, OXA-, and NDM-, VIM-producing P. aeruginosa | cIAI and cUTI | Designated a QIDP by U.S. FDA and awarded fast-track review |
| Sulbactam-durlobactam (ETX2514SUL) | β-Lactam (sulbactam) + β-lactamase inhibitor (diazabicyclooctane) | PBP, β-lactamase | Phase III | Entasis Therapeutics Inc. | CRAB | A. baumannii infections | |
| BOS-228 (LYS228) | β-Lactam (monobactam) | PBP | Phase II | Boston Pharmaceuticals (licensed from Novartis AG) | CRE | cUTI and cIAI | May be used in combination with other β-lactam antibiotics |
| Ceftobiprole (Zeftera) | β-Lactam (cephalosporin) | PBP | Phase III; E.U. EMA approved via decentralized procedure; Australian TGA approved for HA and CA pneumonia on 8 February 2016; Health Canada approved for HA and CA pneumonia on 12 October 2015; Health Canada approved for ABSSSI in June 2008 | Basilea Pharmaceutica | S. aureus | ABSSSI and CA and HA bacterial pneumonia | |
| Tebipenem-pivoxil (SPR994/SPR859) | β-Lactam (carbapenem) | PBP | Phase III; Japanese PMDA approved on 22 April 2009 | Spero Therapeutics Inc. | ESBL-producing E. coli and K. pneumoniae | CA bacterial pneumonia and complicated UTI | Oral administration for home-based care |
| Sulopenem/sulopenem-etzadroxil | β-Lactam (carbapenem) | PBP | Phase III | Iterum Therapeutics Ltd. | MDR Gram-negative pathogens | cUTI, uncomplicated UTI, and cIAI | Available as oral and intravenous preparations |
| Cefilavancin (TD-1792) | Glycopeptide–β-lactam (cephalosporin) hybrid | PG chain elongation, PBP | Phase III | R-Pharm/Theravance Biopharma Inc. | ABSSSI | ||
| Delafloxacin (Baxdela/Quofenix) | Fluoroquinolone | Bacterial type II topoisomerase | U.S. FDA approved on 19 June 2017; E.U. EMA approved on 16 December 2019 | Melinta Therapeutics Inc. | ESKAPE pathogens except A. baumannii | ABSSSI | Serious adverse reactions, including tendinitis, tendon rupture, and neuropathy |
| Lascufloxacin (Lasvic) | Fluoroquinolone | Bacterial type II topoisomerase | Japanese PMDA approved on 20 September 2019 | Kyorin Pharmaceutical Co. Ltd. | Klebsiella spp, Enterobacter spp., MRSA, and S. pneumoniae | CA bacterial pneumonia | Activity against MRSA comparable to that of linezolid and vancomycin |
| Alalevonadifloxacin + levonadifloxacin (WCK 771/WCK 2349) | Fluoroquinolone | Bacterial type II topoisomerase | Phase III | Wockhardt Ltd. | MRSA, VRSA, and MDR pneumococci, E. coli, and K. pneumoniae | ABSSSI and HA bacterial pneumonia | Active against MSRA biofilm |
| OPS-2071 | Quinolone | Bacterial type II topoisomerase | Phase II | Otsuka Pharmaceutical Co. Ltd. | C. difficile | C. difficile infection | |
| Nemonoxacin (Tiagexyn) | Quinolone | Bacterial type II topoisomerase | Phase II; approved in Taiwan, Russia, Turkey, China, and Latin America during 2016 | TaiGen Biotechnology Co. Ltd. | S. aureus | CA bacterial pneumonia, diabetic foot infection, and ABSSSI | Designated a QIDP by U.S. FDA and awarded fast-track review |
| Zoliflodacin (ETX0914) | Spiropyrimidinetrione | Bacterial type II topoisomerase | Phase III | Entasis Therapeutics Inc. | N. gonorrhoeae | Uncomplicated N. gonorrhoeae infection | Designated a QIDP by U.S. FDA and awarded fast-track review |
| ACX-362E | DNA polymerase IIIC inhibitor | DNA polymerase IIIC | Phase 1 | Acurx Pharmaceuticals LLC | C. difficile | C. difficile infections | Potential to be applied against all ESKAPE pathogens |
| MGB-BP-3 | Distamycin | DNA minor groove binder | Phase II | MGB Biopharma Ltd. | C. difficile | C. difficile-associated diarrhea | Targets C. difficile in vegetative state before it sporulates |
| Sodium fusidate (ARV-1801) | Fusidane | Elongation factor G | Phase III; approved (for dosing regimens as an alternative to what is currently being tested in clinical trials) for ABSSSI in South Korea, Japan, Canada, the European Union, Australia, New Zealand, Thailand, India, and Taiwan | Arrevus Ltd. | MRSA | ABSSSI and prosthetic joint infections | Granted orphan drug designation by U.S. FDA |
| Omadacycline (Nuzyra) | Tetracycline | 30S subunit of bacterial ribosome | U.S. FDA approved on 2 October 2018; E.U. EMA considered granting marketing authorization | Paratek Pharmaceuticals Inc. | ESKAPE pathogens | CA bacterial pneumonia and ABSSSI | Favorable oral and/or intravenous administration; Paratek Pharmaceuticals withdrew from E.U. EMA marketing authorization approval on 9 October 2019 due to commercial feasibility issues |
| Eravacycline (Xerava) | Tetracycline | 30S subunit of bacterial ribosome | U.S. FDA approved on 27 August 2018; E.U. EMA approved on 9 September 2018 | Tetraphase Pharmaceuticals Inc. | ESKAPE pathogens, including CRE and CRAB | cIAI | Not indicated for treatment of complicated UTI |
| KBP-7072 | Tetracycline | 30S subunit of bacterial ribosome | Phase I | KBP BioSciences Pharmaceutical Technical Co. Ltd. | CA bacterial pneumonia | Designated a QIDP by U.S. FDA and awarded fast-track review | |
| TP-271 | Tetracycline | 30S subunit of bacterial ribosome | Phase I | Tetraphase Pharmaceuticals Inc. | Respiratory pathogens | CA bacterial pneumonia | Designated a QIDP by U.S. FDA and awarded fast-track review |
| TP-6076 | Tetracycline | 30S subunit of bacterial ribosome | Phase I | Tetraphase Pharmaceuticals Inc. | CRE and CRAB | Bacterial infections | CARB-X funded ($4 million) |
| Lefamulin (Xelenta) | Pleuromutilin | 50S subunit of bacterial ribosome | U.S. FDA approved on 19 August 2019; E.U. EMA marketing authorization application submitted in May 2019 (now accepted); Chinese NMPA granted clinical trial application approval on 13 June 2019 | Nabriva Therapeutics AG | MRSA | ABSSSI and CA and HA bacterial pneumonia | Low toxicity profile; available as both intravenous and oral treatments |
| Delpazolid (LCB01-0371) | Oxazolidinone | 50S subunit of bacterial ribosome | Phase II | LegoChem Biosciences Inc. | Bacterial infections and tuberculosis | Granted orphan drug designation, a QIDP, and fast-track review by U.S. FDA | |
| DNV3837 (MCB3837) | Oxazolidinone-quinolone hybrid | 50S subunit of bacterial ribosome, bacterial type II topoisomerase | Phase I | Deinove SA (formerly Morphochem AG) | C. difficile | C. difficile infections | Narrow spectrum for C. difficile, which may reduce gut dysbiosis; designated a QIDP by U.S. FDA and awarded fast-track review |
| Contezolid (MRX-I/MRX-415) | Oxazolidinone | 50S subunit of bacterial ribosome | Phase III | MicuRx Pharmaceuticals Inc. | MRSA and S. pneumoniae | ABSSSI | |
| Nafithromycin (WCK 4873) | Macrolide | 50S subunit of bacterial ribosome | Phase II | Wockhardt Ltd. | MDR pneumococci | CA bacterial pneumonia | High level of lung and alveolar macrophage penetration; low hepatic toxicity |
| SPR206 | Polymyxin | Cell membrane | Phase I | Spero Therapeutics Inc. | CRE, CRPA, CRAB, and XDR strains | cUTI and HA and VA bacterial pneumonia | Reduced nephrotoxicity |
| SPR741 | Polymyxin | Cell membrane | Phase I | Spero Therapeutics Inc. | Gram-negative ESKAPE pathogens | Bacterial infections | Potentiator with reduced nephrotoxicity; antibiotic paring is yet to be announced |
| Afabicin (Debio 1450) | Benzofuran naphthyridine | FabI | Phase II | Debiopharm International SA | Staphylococcus sp. | ABSSI | Low propensity for resistance development |
| CG-549 | Benzyl pyridinone | FabI | Phase II | Crystal Genomics Inc. | MRSA and VRSA | ABSSSI | |
| Iclaprim | 2,4-Diaminopyrimidine | Dihydrofolate reductase | Complete response letter to Investigational New Drug application (U.S. FDA) | Motif Bio PLC | Gram-positive bacteria, including MRSA | ABSSSI and HA and VA bacterial pneumonia | Granted orphan status and designated a QIDP by U.S. FDA; low propensity for resistance development |
| CRS3123 | Diaryldiamine | Methionyl-tRNA synthetase | Phase I | Crestone Inc. | C. difficile | C. difficile infections | Narrow spectrum for C. difficile, which may reduce gut dysbiosis |
| Brilacidin | Defensin mimetic | Cell membrane | Phase II | Innovation Pharmaceuticals Inc. | ABSSSI | New class of antibiotic; designated a QIDP by U.S. FDA and awarded fast-track review | |
| Murepavadin (POL7080) | Antimicrobial peptide mimetic | LptD | Phase III | Polyphor Ltd. | CRPA | HA and VA bacterial pneumonia (caused by P. aeruginosa) | Designated a QIDP by U.S. FDA and awarded fast-track review |
| Ridinilazole (SMT 19969) | Bis-benzimidazole | Inhibition of cell division and reduction of toxin production | Phase III | Summit Therapeutics PLC | C. difficile | C. difficile infection | Narrow spectrum for C. difficile, which may reduce gut dysbiosis; designated a QIDP by U.S. FDA and awarded fast-track review |
The table includes data from reference 390. Abbreviations: CA, community acquired; HA, hospital acquired; VA, ventilator associated; UTI, urinary tract infection; cUTI, complicated UTI; cIAI, complicated intra-abdominal infection; ABSSSI, acute bacterial skin and skin structure infection; QIDP, qualified infectious disease product; U.S. FDA, U.S. Food and Drug Administration; E.U. EMA, European Union European Medicines Agency; Australian TGA, Australian Therapeutic Goods Administration; Chinese NMPA, Chinese National Medical Products Administration; Japanese PMDA, Japanese Pharmaceutical and Medical Devices Agency; BARDA, U.S. Biomedical Advanced Research and Development Authority; PBP, penicillin-binding protein; PG, peptidoglycan; MRSA, methicillin-resistant S. aureus; CRE, carbapenem-resistant Enterobacterales; CRAB, carbapenem-resistant A. baumannii; CRPA, carbapenem-resistant P. aeruginosa; XDR, extremely drug resistant.
As defined by the company website.
Recently Approved Drugs
In 2018, three new antibiotics with the potential to treat serious bacterial infections successfully emerged through clinical trials with either U.S. FDA or E.U. EMA approval (390) (Table 3). All three drug compounds target the 30S subunit of the bacterial ribosome. The aminoglycoside class analog plazomicin (Zemdri; Achaogen Inc.) was approved by the U.S. FDA in June 2018 for the treatment of patients 18 years of age or older for complicated UTI including pyelonephritis. Plazomicin was not approved by the U.S. FDA for the treatment of bloodstream infections due to a lack of effectiveness (390–392). From a randomized trial involving 609 patients, once-daily treatment with plazomicin was shown to be noninferior to treatment with meropenem (treatment every 8 h) for use against complicated UTIs and acute pyelonephritis caused by Enterobacterales, including MDR strains (393). Plazomicin also demonstrated efficacy better than that of meropenem for eradication of aminoglycoside-resistant and ESBL-producing Enterobacterales (393).
Eravacycline (Xerava; Tetraphase Pharmaceuticals Inc.) is a tetracycline analog approved by the U.S. FDA in August 2018 and the E.U. EMA in September 2018 (22, 390, 394) (Table 3). Eravacycline has been developed solely for the treatment of complicated intra-abdominal infections caused by Gram-negative and Gram-positive ESKAPE pathogens, including CRE and CRAB (394). In the IGNITE4 clinical trial, eravacycline was shown to be noninferior to meropenem for the treatment of complicated intra-abdominal infections caused by ESBL-producing Enterobacterales (395). Furthermore, patients treated with eravacycline experienced relatively low rates (3 to 5%) of adverse events, such as nausea, vomiting, and diarrhea (395).
Omadacycline (Nuzyra; Paratek Pharmaceuticals Inc.), a tetracycline-class drug active against Gram-negative and Gram-positive ESKAPE pathogens, was approved by the U.S. FDA in October 2018 for the treatment of CA bacterial pneumonia, acute bacterial skin and skin structure infection (ABSSSI), and complicated and uncomplicated UTI (390, 396) (Table 3). In a recent clinical trial, once-daily administration through interchanging intravenous and oral administration demonstrated that omadacycline was noninferior to the commonly prescribed moxifloxacin for an early clinical response, as defined by survival, against CA bacterial pneumonia (397). Unlike the U.S. FDA, the E.U. EMA recommended granting marketing authorization for omadacycline solely for use in patients with ABSSSI and not in those with CA bacterial pneumonia. As a result, Paratek Pharmaceuticals Inc. withdrew omadacycline from the E.U. EMA marketing authorization application on 9 October 2019, stating that “it would not be commercially feasible to market Nuzyra just for the treatment of skin and skin structure infections” (398). The absence of omadacycline in Europe illustrates the current financial and practical problems facing antimicrobial R&D, i.e., balancing product profitability with appropriate stewardship practices. To combat these obstacles, DRIVE-AB has proposed a delinking initiative which provides a financial reward ($1 billion) per innovative antibiotic, allowing revenues for new antibiotics to be partially or fully delinked from the number of units sold, thus allowing for the revenues to be based upon the value to society (399).
In 2019, five new antimicrobial drug therapies were approved by either the U.S. FDA, the E.U. EMA, or the Japanese PMDA. Of these five therapies, four drugs, imipenem-cilastatin-relebactam (Recarbrio; Merck & Co., Inc.), lefamulin (Xelenta; Nabriva Therapeutics AG), lascufloxacin (Lasvic; Kyorin Pharmaceutical Co. Ltd.), and cefiderocol (Fetroja; Shionogi & Co. Ltd.) demonstrated efficacy against ESKAPE pathogens (note that pretomanid [TB Alliance] was approved for the treatment of MDR tuberculosis) (390) (Table 3). In July 2019, the three-drug combination imipenem, cilastatin, and relebactam was approved by the U.S. FDA for the treatment of complicated UTI and complicated intra-abdominal infection caused by E. coli, E. cloacae, K. pneumoniae, K. aerogenes, and P. aeruginosa) (400). Furthermore, the E.U. EMA adopted a positive opinion for imipenem-cilastatin-relebactam, recommending to grant a marketing authorization for the treatment of infections due to aerobic Gram-negative organisms in adults with limited treatment options (December 2019). Imipenem-cilastatin-relebactam was subsequently approved by the E.U. EMA in February 2020 (24). Imipenem-cilastatin-relebactam, comprised of the existing antibiotics imipenem and cilastatin, contains a novel β-lactamase inhibitor, relebactam, a third new β-lactamase inhibitor after avibactam and vaborbactam (401).
Lefamulin (Xelenta; Nabriva Therapeutics AG), developed for the treatment of CA bacterial pneumonia, was the second therapeutic to be approved by the U.S. FDA in 2019 (August) (402) (Table 3). In the LEAP 2 trial (a phase III, noninferiority trial that compared oral lefamulin to oral moxifloxacin), lefamulin demonstrated noninferiority to moxifloxacin in the management of CA bacterial pneumonia. Lefamulin presents a new oral and intravenous treatment against CA bacterial pneumonia, particularly for pneumonia caused by CA-MRSA (403). In May 2019, Nabriva Therapeutics AG submitted a marketing authorization application (now accepted) to the E.U. EMA for both intravenous and oral formulations of lefamulin for the treatment of CA bacterial pneumonia in adults 18 years of age and older (404, 405).
Lascufloxacin (Lasvic; Kyorin Pharmaceutical Co. Ltd.), a novel fluoroquinolone antimicrobial, was approved by the Japanese PMDA on 20 September 2019 for the treatment of CA bacterial pneumonia caused by a range of bacterial pathogens, including quinolone-resistant Staphylococcus and Klebsiella spp. (406–408). In a phase II clinical trial, oral administration of lascufloxacin (75 mg) demonstrated clinical cure rates of 90%, bacteriological eradication rates of 96%, and an overall incidence of adverse drug reaction of 11.1% (409). Furthermore, during a double-blind phase III clinical trial, lascufloxacin demonstrated noninferiority to orally administered quinolones in the management of CA bacterial pneumonia and sinusitis (410).
Cefiderocol (Fetroja; Shionogi & Co. Ltd.), approved by the U.S. FDA in November 2019, is a novel siderophore-cephalosporin conjugate approved for the treatment of patients 18 years of age or older with complicated UTI including kidney infections caused by susceptible Gram-negative pathogens (i.e., CRE, CRAB, and carbapenem-resistant P. aeruginosa [CRPA]) (21, 411) (Table 3). In a phase II, double-blind, randomized clinical trial, the safety and efficacy of cefiderocol were assessed. The trial found that 72.6% of the patients who were administered cefiderocol but only 54.6% who those who received imipenem-cilastatin had resolution of symptoms and eradication of bacteria at 7 days posttreatment (412).
New Drug Classes in Clinical Trials
The portfolio of compounds currently in clinical trials more often than not consists of derivatives of chemical classes for which there is already an underlying mechanism of resistance. Over the past 24 months, all drugs approved by the U.S. FDA, E.U. EMA, and Japanese PMDA were drug analogs based on established antibiotic classes (390). Murepavadin is one of the few new class of antibiotics which entered into a phase III clinical trial (now temporarily suspended) (Table 3). Murepavadin is selective against the protein transporter LptD, which mediates the transport of LPS to the outer leaflet (413). As the first in class of the outer membrane protein-targeting peptidomimetic antibiotics, murepavadin displays potent activity against carbapenemase-producing and polymyxin-resistant P. aeruginosa strains (413). Murepavadin was under clinical development for the treatment of HA pneumonia and ventilator-associated (VA) pneumonia. Unfortunately, enrollment in phase III studies for the systemic treatment of nosocomial pneumonia has now been temporarily suspended due to higher-than-expected levels of acute kidney injury in participants. Aerosol formulations of murepavadin for the treatment of nosocomial and chronic P. aeruginosa respiratory infections are still being actively pursued (414).
Brilacidin (Innovation Pharmaceuticals Inc.), currently undergoing a phase II clinical trial, is a synthetic new class of defensin mimetic for the treatment of ABSSSI caused by S. aureus (Table 3). As a late-stage antibiotic drug candidate, brilacidin is being advanced to the clinic under the Qualified Infectious Disease Product designation by the U.S. FDA, enabling fast-track review (390).
New Drugs in Clinical Trials—Overcoming Antibiotic Toxicity
The innate toxicity of certain antibiotics, combined with the compromised state of the infected patient, often shapes clinical dosing regimens. The rates of drug-induced nephrotoxicity range from 14% to 26% in adults, and nephrotoxicity is the most common toxicity issue associated with antibiotic prescription (415). The nephrotoxic effects of last-resort polymyxin-class antibiotics and the widespread level of carbapenem resistance in ESKAPE pathogens have severely limited patient treatment options. In a bid to overcome these problems, two new polymyxin analogs, SPR206 and SPR741 (Spero Therapeutics Inc.), have been developed (390, 416) (Table 3). SPR206 is an investigational drug candidate in a phase I clinical trial with broad-spectrum antimicrobial activity against polymyxin-sensitive, Gram-negative pathogens (including CRE, CRPA, and CRAB) associated with complicated UTI, HA pneumonia, and VA pneumonia (416). SPR741, formerly NAB741, is a potentiator molecule that has been successfully evaluated in a phase I clinical trial, demonstrating potent synergy with azithromycin, clarithromycin, erythromycin, fusidic acid, mupirocin, retapamulin, rifampin, and telithromycin (417). SPR741 has the potential to treat a broad range of bacterial disease states, including those caused by CRE, CRPA, and CRAB strains. In contrast to polymyxin B, SPR206 yields an aromatic β-amino acid modification at the fatty acid-AA1 component of polymyxin B. SPR741 lacks two cationic diaminobutyryl residues in the linear portion of the peptide and lacks the 6-methyloctanoyl or 6-methylheptanoyl fatty acid tail found in polymyxin B (418). These respective structural modifications have significantly improved the safety and dose-limiting nephrotoxicity issues associated with polymyxin-class antibiotics.
Alternative Drug Trial Approaches
New drug trial strategies are emerging, focusing on both alleviating the increased demand for last-resort therapies and minimizing further infection complications by reducing patient hospital admission lengths. Such new trial strategies include testing new antibiotic-antibiotic potentiator combinations and reducing hospital-stay periods beyond the critical point of infection to improve treatment outcomes.
Carbapenems are still considered the most effective therapy for select ESKAPE pathogen infections, particularly those caused by ESBL-producing Enterobacterales. Unfortunately, the increased use of carbapenems in clinical settings has created a selection pressure for the emergence of carbapenem resistance. In an effort to define carbapenem-sparing alternatives, a recent drug trial (MERINO trial) examined whether piperacillin-tazobactam combinations could demonstrate efficacy comparable to that of carbapenems (419). Among patients with E. coli and K. pneumoniae bloodstream infections, 30-day survival did not improve upon piperacillin-tazobactam treatment compared to that achieved with meropenem therapy (420). Although results from the MERINO trial did not support the use of piperacillin-tazobactam as a carbapenem-sparing treatment, the trial highlighted the need for carbapenem-sparing strategies and for further study into β-lactam–β-lactamase inhibitor drug combinations.
Reducing the length of hospital stay following the initial stage of infection is associated with a decreased risk of further complications and better outcomes of the patient disease state (421, 422). In 2018, with the objective of reducing patient hospital admission times, the Partial Oral Treatment of Endocarditis (POET) trial evaluated the efficacy of intravenous antibiotics compared with that of oral antibiotics among stable patients with infective endocarditis (423). The premise of this study was to allow patient treatment to take place outside of hospitals, without the requirement for intravenous catheters. The primary outcome from the POET study showed that oral antibiotics were noninferior to intravenous antibiotics at preventing adverse events (i.e., all-cause death, unplanned cardiac surgery, embolic events, or relapse of bacteremia) (423). The impact of this landmark study may significantly minimize the challenges associated with parenteral treatment, including logistics, monitoring, and risks of complications associated with intravenous catheters (i.e., secondary local and systemic infections).
Combinational Drug Therapy
The protracted R&D of effective, novel antimicrobial drug candidates has contributed to the failure to combat the resurgence of AMR bacterial infections. Pairing existing antimicrobials with either other antimicrobials or nonantimicrobial compounds offers a valuable strategy to address the problem of AMR. This strategy is not new (424, 425), and it is well recognized that antibiotic monotherapy is not universally efficacious for all bacterial infections. Adjuvants are often administered in combination with antibiotic therapy, resulting in better patient outcomes. Antibiotic adjuvants can be divided into two classes: class I adjuvants, which act on the pathogen, and class II adjuvants, which act on host properties to potentiate antibiotic action (426).
Blocking resistance mechanisms against existing antibiotics.
(i) Class I adjuvants.
From the available repertoire of class I adjuvants, β-lactamase inhibitors have proven to be the most successful. Clavulanic acid, isolated from Streptomyces clavuligerus (427), inactivates Ser β-lactamases and has been paired with amoxicillin for over 30 years to create the drug Augmentin. Recently, several human clinical trials have examined the efficacy of class A and C β-lactamase inhibitors: avibactam, vaborbactam, and relebactam coformulated with ceftazidime and relebactam paired with imipenem against CRKP (428). In current clinical practice, β-lactamase inhibitors do not inhibit the class B, Zn-dependent MBLs found broadly in Enterobacterales, Pseudomonas, and Acinetobacter species (429). The fungus-derived aspergillomarasmine A has been shown to be efficacious against MBL-producing bacteria in in vivo models of infection (430). Aspergillomarasmine A functions to rescue the antibiotic activity of meropenem expressing NDM-1 and VIM MBLs by sequestering Zn2+ ions essential for catalytic activity. Other antibiotic class I adjuvants which have been investigated include 2-aminoimidazole-based compounds, which disrupt two-component signaling (431); anthracyclines, which potentiate rifampin and linezolid (432); SPR741, a polymyxin-derived molecule designed to minimize nephrotoxicity and potentiate rifampin activity (433); and hydroxyquinoline analogues, among others, which potentiate the activity of multiple classes of antibiotics against a range of Gram-positive AMR pathogens (389, 434).
(ii) Class II adjuvants.
Enhancement of host defense mechanisms offers an alternative set of targets for antibiotic adjuvants. Over the past decade, several innate immune-enhancing peptides developed to treat sepsis (E-5564 [Eisai Pharmaceuticals] and TAK-242 [Takeda Pharmaceuticals]) have reached phase III clinical trials but failed to progress (435). Despite these shortcomings, new immune-stimulating therapies are continuing to be explored, with promising outcomes.
Recent studies have reported on the synthesis of novel immunotherapeutic compounds which successfully mediate the opsonization of E. coli and P. aeruginosa in human serum (436). This technology utilizes polymyxin B-hapten conjugates to facilitate a two-step process whereby the lipid A binding scaffold of polymyxin B decorates the surface of Gram-negative bacteria with antibody-recruiting haptens. Mortality arising from serious infection implies failure of the patient’s innate immune response. To correct and enhance innate immune cell function, pharmacological strategies have been centered around hypoxia-inducible factor 1 (HIF), the central regulator of the cellular response to hypoxic stress (437). HIF has been proposed to be a master regulator of innate immunity (438). HIF functions to both control the recruitment of neutrophils to the site of infection and increase the functional neutrophil life span by inhibiting apoptotic pathways. A series of in vivo infection models has demonstrated that reduced HIF levels diminish neutrophil function and render mice more susceptible to serious group A Streptococcus and P. aeruginosa infections (439, 440). Conversely, elevated levels of HIF have demonstrated enhanced control of MRSA skin infection in mice (441). HIF modulation has the capacity to serve as a complementary therapy to classical anti-infective strategies, making it a significant target for immune-boosting therapy.
Alternative Nondrug Therapies
Preclinical and clinical R&D of new antimicrobials encompasses a wide range of drug discovery pathways: (i) bacteriophage therapy, (ii) drug repurposing, (iii) monoclonal antibody (MAb) therapy, (iv) vaccine development, and (v) fecal microbiota transplantation (FMT) are some of the many novel approaches currently being investigated to control the burden of AMR.
Bacteriophage therapy.
Bacteriophage therapy was first applied in 1917 as an oral preparation to treat dysentery (387). As a result of antibiotic discovery, the use of bacteriophage treatment significantly reduced. Today, bacteriophage therapy again represents a potentially effective strategy for the control of AMR bacteria. Within the past 5 years, a number of phage preparations have undergone clinical trials. Examples include bacteriophage preparations for burn wound infections (ClinicalTrials.gov study identifier NCT02116010) and persistent postoperative respiratory infection (ClinicalTrials.gov study identifier NCT00945087) (387).
Through the utilization of the CRISPR technology, several companies have engineered bacteriophages which demonstrate in vivo efficacy against AMR bacterial infections. These distinct therapies, formulated by Locus and Eligo Bioscience, exploit the Cas3 and Cas9 enzyme systems, respectively, to degrade bacterial DNA, leading to cellular death. Therapies developed by both companies are expected to enter clinical trials within the next 12 months (442).
Although bacteriophages are not currently U.S. FDA approved for human use (due to the uncertainty surrounding the host immune response), the U.S. FDA has committed to facilitating the testing of phage therapy in clinical trials. In 2019, the U.S. FDA accepted an Investigational New Drug application by physician scientists at the University of California, San Diego (443, 444). In collaboration with AmpliPhi Biosciences Corporation, the proposed phase I/II trial will test AB-SA01, an experimental bacteriophage combination for the treatment of ventricular assisted devices infected with MDR S. aureus. The trial will evaluate the safety, tolerability, and efficacy of intravenously administered AB-SA01 bacteriophage therapy in combination with complementing antibiotic therapy (444).
Globally, the lack of an appropriate legal and regulatory framework has been a significant obstacle to the advancement of phage therapy. In 2016, the Belgium government began implementing a magistral phage therapy framework that centers on the magistral preparation (compounded prescription drug product in the United States) of phage medicines tailor-made for the patient, providing for a broader and more structured application of phages in Belgium (445). For the proposed magistral phage strategy process, characterized and quality-tested phages are to be transferred to a hospital pharmacy for possible incorporation in patient-tailored magistral formulas. It is anticipated that other European Union members will adopt this phage therapy framework in the near future, hastening the clinical development of safe-for-human-use phage therapy.
Repurposing existing drugs used for noninfectious disease.
Repurposing existing drugs represents a viable alternative to de novo drug discovery and favorably reduces the time, cost, and risk associated with drug innovation (446). Drug repurposing has already been shown to be efficacious against several Gram-positive and Gram-negative ESKAPE pathogens. Glatiramer acetate (Copaxone), a widely used treatment for multiple sclerosis, displays antibacterial activity against E. coli, A. baumannii, and, notably, P. aeruginosa clinical strains isolated from cystic fibrosis patients (447). Ebselen, a synthetic anti-inflammatory drug, and the oncology drugs adarotene and floxuridine have displayed bactericidal activity against MRSA and VRSA strains in vivo (448, 449). Efforts to repurpose existing drugs as antimicrobial agents or as antibiotic resistance breakers are continually being pursued both in the academic sector and in the industrial sector (371, 389, 450). Due to the marginal incentives of the current pharmaceutical R&D model for antibiotic development and discovery, drug repurposing may present an efficacious solution to the burden of AMR.
Monoclonal antibody therapy.
Largely used as a therapeutic for oncology and rheumatic indications, MAb therapy presents a viable alternative for the treatment of AMR bacterial infections. The bacterial specificity of MAb therapy (i.e., it is not targeted to the host microflora), combined with a low propensity for resistance development, is a key feature which makes MAb therapy well positioned for the treatment of AMR bacterial infections (451). To date, only three MAbs have been approved for clinical use by the U.S. FDA, and none of them has been directed against ESKAPE pathogens. Both raxibacumab (Human Genome Sciences, Inc.; approved in December 2012) (452) and obiltoxaximab (Elusys Therapeutics, Inc.; approved in March 2016) are approved for the treatment of adult and pediatric patients with inhalational anthrax due to Bacillus anthracis (453), while bezlotoxumab (Merck Sharp & Dohme Corp.; approved in October 2016) is approved for the prevention of recurrent Clostridioides difficile infection in high-risk patients (454). Although no existing approved MAb therapy targets ESKAPE pathogens, several MAb therapies have been clinically evaluated for treatment of infections caused by MDR P. aeruginosa. Three MAbs, panobacumab (Adris Pharmaceuticals), KB001 (KaloBios), and MEDI3902 (AstraZeneca Pharmaceutical Company), have been evaluated in an early clinical trial targeting MDR P. aeruginosa. Panobacumab, currently in phase II clinical trials, is an antilipopolysaccharide IgM antibody directed against O-polysaccharide and is under investigation for the treatment of nosocomial pneumonia caused by P. aeruginosa O11 (455, 456). KB001 is a PEGylated MAb fragment directed against PcrV (a protein subunit of the P. aeruginosa type III secretion system) which was evaluated for use in mechanically ventilated CF patients suffering from chronic P. aeruginosa infection (457). KB001 was observed to be well tolerated in patients, reduced the levels of the sputum inflammatory marker interleukin-8, and significantly improved patient lung function (458). Able to facilitate opsonophagocytosis in vitro, MEDI3902 is a bivalent, bispecific MAb targeting PcrV and Psl (an exopolysaccharide involved in colonization and tissue adherence) (459). During a phase I dose-escalation study, subjects administered MEDI3902 demonstrated no serious treatment-related adverse effects, making it appropriate for use in ventilated intensive care unit (ICU) patients (460). In the ongoing EVADE phase II clinical trial, the safety and efficacy of MEDI3902 will be assessed for the prevention of VA pneumonia in adult ICU patients (461).
Vaccine development.
Conceptually, bacterium-targeted vaccine therapies provide a means to decrease the public demand on existing antibiotics, while they also enable protection for those who are both vaccinated and unvaccinated (herd immunity). Unfortunately, vaccines are not currently available for infections caused by ESKAPE pathogens, with many vaccine candidates failing to elicit an immunogenic protective effect in clinical trials (462–464).
Of the limited ESKAPE pathogen-targeted vaccines which have recently undergone clinical trials, the S. aureus 4-antigen vaccine (SA4Ag; Pfizer; which has now ceased enrollment) was one of the few candidates which demonstrated efficacy in the prevention of invasive S. aureus infection in humans (465). SA4Ag consists of capsular polysaccharide serotypes 5 and 8 conjugated to the nontoxic mutant form of diphtheria toxin (CRM197), a recombinant mutant clumping factor A (ClfA), and a recombinant manganese transporter C (MntC) (465). In a phase I/II randomized trial, single-dose vaccination of healthy adults ranging from 65 to 80 years of age with SA4Ag was shown to be well tolerated and induced rapid high levels of bacterium-killing antibodies and sustained immune responses at 12 months postvaccination (465). Unfortunately, in December 2018, the respective clinical trials ceased enrollment due to meeting its prespecified futility criteria at an interim efficacy assessment. Despite this, a 36-month postvaccination serological extension study was still undertaken and recently completed. In this trial of 440 participants, persistent functional immune responses to S. aureus antigens were observed through a 36-month time period (466).
In the face of the limited number of vaccines in the clinical pipeline, encouraging preclinical studies have recently demonstrated the in vivo efficacy of vaccines against CRKP (467), P. aeruginosa (462), and A. baumannii (468). Vaccines have already been shown to be very effective at reducing the incidence of other AMR pathogens, such as Streptococcus pneumoniae and Haemophilus influenzae type b (464). The prospect of new ESKAPE-targeted vaccines which complement existing AMR therapies will significantly aid in mitigating the spread and threat of ESKAPE pathogens.
FMT strategies.
Antibiotic exposure, particularly in long-term acute care hospital patients, can alter the patient microbiota and significantly reduce colonization resistance (469). FMT-based approaches are currently being investigated for protection against AMR bacterial colonization. A recent study demonstrated that blood disorder patients treated with multiple FMT procedures exhibited gastrointestinal decolonization of VRE, ESBL-producing Enterobacterales, CRE, and CRPA in 60% of all cases (470). These data highlight the utility of FMT approaches in reversing the gut dysbiosis that predisposes patients to colonization with AMR ESKAPE pathogens.
OUTLOOK
AMR represents one of the few challenges that unites global interests and concerns for human and animal health and the food and agricultural sectors. Exacerbated by the acquisition of AMR genes, ESKAPE pathogens represent the paradigm for resistance, pathogenesis, and disease transmission in both the community and clinical settings. Although heterogeneous at the genetic level, the general mechanisms surrounding ESKAPE pathogen emergence and persistence are broadly shared. Mediated in part through HGT, resistance strategies encompassing drug inactivation, modification of the antibiotic target site, and a reduction of antibiotic accumulation in the bacterial cell are common strategies shared by all ESKAPE pathogens. Combined with the collective ability to form biofilm on innate and biological surfaces, ESKAPE pathogens remain highly prevalent in clinical settings.
To constrain the spread of ESKAPE pathogens, it is now well recognized that collaborative global and regional efforts are required by policy makers, funders, and those responsible for the treatment and management of ESKAPE pathogens (12, 35, 64). Aside from novel drug development, these collaborative endeavors will require sustainable stewardship practices to reduce the inappropriate use of antibiotics in both the human health and agricultural sectors. Despite efforts to coordinate international and national AMR surveillance, it would be well-advised for AMR policy, drug development, and surveillance efforts to include both ESKAPE pathogens and other serious health threats, such as AMR E. coli, Neisseria gonorrhoeae, and Campylobacter spp. These will require equal attention to avoid selecting for a new group of AMR pathogen threats (12). Improvements in factors encompassing AMR surveillance, diagnostics, patient education, and patient treatment options will together help facilitate the control of AMR.
Biographies

David M. P. De Oliveira is currently a Research Fellow at the Australian Infectious Diseases Research Centre, University of Queensland. He completed his Ph.D. studies in December 2015 at the Illawarra Health and Medical Research Institute at the University of Wollongong. His Ph.D. thesis focused on characterizing novel host-bacterial interactions which contribute to group A streptococcal pathogenesis. His current research themes center around novel infectious disease therapies, with a particular focus on repurposing existing nonantimicrobial therapies into direct-acting or potentiating antimicrobial therapies which target AMR bacterial pathogens.

Brian M. Forde completed a Ph.D. at University College Cork, Ireland, in 2012. He joined The University of Queensland in 2012 as a Postdoctoral Fellow at the School of Chemistry and Molecular Biosciences (SCMB). He is currently a Research Fellow at the SCMB and The University of Queensland Centre for Clinical Research (UQCCR). His research his focused on the use of whole-genome sequencing for the surveillance of multidrug-resistant human pathogens in clinical and public health microbiology, real-time tracking of transmission routes during nosocomial outbreaks, and the implementation of genomic research into clinical practice.

Timothy J. Kidd completed a B.Appl.Sc. in microbiology and biotechnology at the Queensland University of Technology in 1998 and thereafter worked as a senior scientist within various clinical microbiology laboratories located in Queensland, Australia. In 2012, he received his Ph.D. in medical microbiology at The University of Queensland, Australia. Between 2014 and 2019 he secured successive European Respiratory Society RESPIRE2 Marie Sklodowska-Curie Postdoctoral and National Health and Medical Research Council Early Career Fellowships based at Queen’s University of Belfast, United Kingdom, and The University of Queensland. He currently holds a combined research and teaching position within the School of Chemistry and Molecular Biosciences at The University of Queensland. His research interests focus on cystic fibrosis (CF) airway infections and Klebsiella pneumoniae infection biology, in particular, the epidemiology and transmission of CF pathogens, the mechanisms and detection of antimicrobial resistance, as well as the characterization of novel K. pneumoniae virulence determinants.

Patrick N. A. Harris is an Infectious Disease Physician, Medical Microbiologist, and NHMRC Early Career Research Fellow at The University of Queensland Centre for Clinical Research (UQCCR). He completed his Ph.D. in 2018 under the supervision of Prof. David Paterson. He was the lead author on the MERINO trial, reported in JAMA in 2018. His research has a focus on antibiotic-resistant bacteria and the use of randomized clinical trials to define the optimal treatment for these problematic infections as well as the application of bacterial genomics to clinical practice.

Mark A. Schembri is an Australian National Health & Medical Research Council Senior Research Fellow and Deputy Director of the Australian Infectious Diseases Research Centre at The University of Queensland. Dr. Schembri completed his Ph.D. at Monash University and postdoctoral studies at the Technical University of Denmark. In 2001, he was awarded a Fellowship from the Danish Natural Sciences Research Council and appointed to Lecturer. He joined The University of Queensland in 2004 as a Senior Lecturer and was promoted to Reader in 2007 and to Professor in 2010. Dr. Schembri’s research is in the field of molecular microbiology and bacterial pathogenesis. His specialist interest is in the area of uropathogenic Escherichia coli (UPEC), with a focus on the genetics, genomics, and virulence of multidrug-resistant UPEC clones and the role of cell surface factors in UPEC adhesion, aggregation, biofilm formation, and colonization of the urinary tract.

Scott A. Beatson is a Group Leader at The University of Queensland (UQ) and a member of the Australian Infectious Diseases Research Centre. In 2002 Dr. Beatson was awarded a Ph.D. from UQ in the area of bacterial genomics and pathogenesis. He completed Postdoctoral Fellowships in bioinformatics at the University of Oxford and the University of Birmingham before returning to UQ as a National Health and Medical Research Fellow (NHMRC) Howard Florey Research Fellow. In 2008, he established the Microbial Genomics Group at UQ and has since been supported by fellowships from both the Australian Research Council and NHMRC. He uses genomics to investigate transmission, pathogenesis, and antibiotic resistance in bacteria, with a focus on mobile genetic elements. Recent work includes genomic investigations of AMR pathogens causing hospital outbreaks and phylogenomic analyses of the pandemic multidrug-resistant Escherichia coli ST131 clone.

David L. Paterson is a Director at The University of Queensland Centre for Clinical Research. He is also a Consultant Infectious Diseases Physician at the Royal Brisbane and Women's Hospital. He received his medical degree and Ph.D. from The University of Queensland. Dr. Paterson has received research funding from the Centers for Disease Control and Prevention (CDC), Australia's National Health and Medical and Medical Research Council (NHMRC), and the Medical Research Future Fund. A 2008 Frank Fenner Award for Advanced Research in Infectious Diseases by the Australasian Society for Infectious Diseases (ASID) quickly followed, and in 2009 he was awarded a Queensland Health Senior Clinical Research Fellowship and subsequently 2 NHMRC Practitioner Fellowships. He is now an NHMRC investigator grant holder. His research focuses on the molecular and clinical epidemiology of infections with AMR organisms, with the intent of translation of knowledge into optimal prevention and treatment of these infections.

Mark J. Walker is the Director of the Australian Infectious Disease Research Centre, a multidisciplinary network based at The University of Queensland, and the QIMR Berghofer Medical Research Centre. He was awarded an Alexander Von Humboldt Fellowship in 1998 and a Fulbright Senior Scholar Fellowship in 2005. Dr. Walker was elected a Fellow of the American Academy for Microbiology in 2013. He works with a collaborative network of Australian and international researchers to understand the emergence and evolution of AMR in bacterial pathogens, investigate host-pathogen interactions, and translate these research findings into new therapeutics and vaccines.
REFERENCES
- 1.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 2.Sommer MOA, Munck C, Toft-Kehler RV, Andersson DI. 2017. Prediction of antibiotic resistance: time for a new preclinical paradigm? Nat Rev Microbiol 15:689–696. doi: 10.1038/nrmicro.2017.75. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization. 2014. Antimicrobial resistance: global report on surveillance 2014. https://www.who.int/antimicrobial-resistance/publications/surveillancereport/en/. Accessed 26 June 2019.
- 4.Centers for Disease Control and Prevention. 2019. Antibiotic resistant threats in the United States 2019. https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf. Accessed 15 January 2020.
- 5.Cassini A, Högberg LD, Plachouras D, Quattrocchi A, Hoxha A, Simonsen GS, Colomb-Cotinat M, Kretzschmar ME, Devleesschauwer B, Cecchini M, Ouakrim DA, Oliveira TC, Struelens MJ, Suetens C, Monnet DL, Strauss R, Mertens K, Struyf T, Catry B, Latour K, Ivanov IN, Dobreva EG, Tambic Andraševic A, Soprek S, Budimir A, Paphitou N, Žemlicková H, Schytte Olsen S, Wolff Sönksen U, Märtin P, Ivanova M, Lyytikäinen O, Jalava J, Coignard B, Eckmanns T, Abu Sin M, Haller S, Daikos GL, Gikas A, Tsiodras S, Kontopidou F, Tóth Á, Hajdu Á, Guólaugsson Ó, Kristinsson KG, Murchan S, Burns K, Pezzotti P, Gagliotti C, Dumpis U, et al. 2019. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect Dis 19:56–66. doi: 10.1016/S1473-099(18)30605-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weist K, Hogberg LD. 2016. ECDC publishes 2015 surveillance data on antimicrobial resistance and antimicrobial consumption in Europe. Euro Surveill 21(46):pii=30401. doi: 10.2807/1560-7917.ES.2016.21.46.30399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Center for Disease Dynamics Economics & Policy. 2015. The state of the world’s antibiotics, 2015. https://cddep.org/publications/state_worlds_antibiotics_2015/. Accessed 1 November 2019.
- 8.Kayange N, Kamugisha E, Mwizamholya DL, Jeremiah S, Mshana SE. 2010. Predictors of positive blood culture and deaths among neonates with suspected neonatal sepsis in a tertiary hospital, Mwanza-Tanzania. BMC Pediatr 10:39. doi: 10.1186/1471-2431-10-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roca A, Quinto L, Abacassamo F, Morais L, Valles X, Espasa M, Sigauque B, Sacarlal J, Macete E, Nhacolo A, Mandomando I, Levine MM, Alonso PL. 2008. Invasive Haemophilus influenzae disease in children less than 5 years of age in Manhica, a rural area of southern Mozambique. Trop Med Int Health 13:818–826. doi: 10.1111/j.1365-3156.2008.02061.x. [DOI] [PubMed] [Google Scholar]
- 10.Akova M. 2016. Epidemiology of antimicrobial resistance in bloodstream infections. Virulence 7:252–266. doi: 10.1080/21505594.2016.1159366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rice LB. 2008. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J Infect Dis 197:1079–1081. doi: 10.1086/533452. [DOI] [PubMed] [Google Scholar]
- 12.World Health Organization. 2017. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. http://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf?ua=1. Accessed 2 November 2019.
- 13.Beatson SA, Walker MJ. 2014. Microbiology. Tracking antibiotic resistance. Science 345:1454–1455. doi: 10.1126/science.1260471. [DOI] [PubMed] [Google Scholar]
- 14.Bin Zaman S, Hussain MA, Nye R, Mehta V, Mamun KT, Hossain N. 2017. A review on antibiotic resistance: alarm bells are ringing. Cureus 9:e1403. doi: 10.7759/cureus.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naylor NR, Atun R, Zhu N, Kulasabanathan K, Silva S, Chatterjee A, Knight GM, Robotham JV. 2018. Estimating the burden of antimicrobial resistance: a systematic literature review. Antimicrob Resist Infect Control 7:58. doi: 10.1186/s13756-018-0336-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iguchi S, Mizutani T, Hiramatsu K, Kikuchi K. 2016. Rapid acquisition of linezolid resistance in methicillin-resistant Staphylococcus aureus: role of hypermutation and homologous recombination. PLoS One 11:e0155512. doi: 10.1371/journal.pone.0155512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giddins MJ, Macesic N, Annavajhala MK, Stump S, Khan S, McConville TH, Mehta M, Gomez-Simmonds A, Uhlemann AC. 2017. Successive emergence of ceftazidime-avibactam resistance through distinct genomic adaptations in bla(KPC-2)-harboring Klebsiella pneumoniae sequence type 307 isolates. Antimicrob Agents Chemother 62:e02101-17. doi: 10.1128/AAC.02101-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herc ES, Kauffman CA, Marini BL, Perissinotti AJ, Miceli MH. 2017. Daptomycin nonsusceptible vancomycin resistant Enterococcus bloodstream infections in patients with hematological malignancies: risk factors and outcomes. Leuk Lymphoma 58:2852–2858. doi: 10.1080/10428194.2017.1312665. [DOI] [PubMed] [Google Scholar]
- 19.Paterson DL, Bonomo RA. 2005. Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev 18:657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kussmann M, Karer M, Obermueller M, Schmidt K, Barousch W, Moser D, Nehr M, Ramharter M, Poeppl W, Makristathis A, Winkler S, Thalhammer F, Burgmann H, Lagler H. 2018. Emergence of a dalbavancin induced glycopeptide/lipoglycopeptide non-susceptible Staphylococcus aureus during treatment of a cardiac device-related endocarditis. Emerg Microbes Infect 7:202. doi: 10.1038/s41426-018-0205-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.CenterWatch. 2020. FDA approved drugs. https://www.centerwatch.com/directories/1067-fda-approved-drugs/topic/546-bacterial-infections. Accessed 10 January 2020.
- 22.European Medicines Agency. 2020. Xerava. https://www.ema.europa.eu/en/medicines/human/EPAR/xerava. Accessed 13 January 2020.
- 23.European Medicines Agency. 2020. Quofenix. https://www.ema.europa.eu/en/medicines/human/EPAR/quofenix. Accessed 13 January 2020.
- 24.European Medicines Agency. 2020. Recarbrio. https://www.ema.europa.eu/en/medicines/human/EPAR/recarbrio. Accessed 13 January 2020.
- 25.European Medicines Agency. 2020. Vaborem. https://www.ema.europa.eu/en/medicines/human/EPAR/vaborem. Accessed 11 March 2020.
- 26.Giacobbe DR, De Rosa FG, Del Bono V, Grossi PA, Pea F, Petrosillo N, Rossolini GM, Tascini C, Tumbarello M, Viale P, Bassetti M. 2019. Ceftobiprole: drug evaluation and place in therapy. Expert Rev Anti Infect Ther 17:689–698. doi: 10.1080/14787210.2019.1667229. [DOI] [PubMed] [Google Scholar]
- 27.PMDA. 2019. Pharmaceuticals and medical devices agency: list of approved products. https://www.pmda.go.jp/english/review-services/reviews/approved-information/drugs/0002.html. Accessed 10 January 2020.
- 28.Australian Government Therapeutic Goods Administration. 2016. Australian public assessment report for ceftobiprole medocaril sodium. https://www.tga.gov.au/sites/default/files/auspar-ceftobiprole-medocaril-sodium-160208.pdf. Accessed 11 March 2020.
- 29.Government of Canada. 2020. Drug product database. https://www.canada.ca/en/health-canada/services/drugs-health-products/drug-products/drug-product-database.html. Accessed 10 January 2020.
- 30.Lebreton F, van Schaik W, McGuire AM, Godfrey P, Griggs A, Mazumdar V, Corander J, Cheng L, Saif S, Young S, Zeng Q, Wortman J, Birren B, Willems RJ, Earl AM, Gilmore MS. 2013. Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. mBio 4:e00534-13. doi: 10.1128/mBio.00534-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murray BE. 1990. The life and times of the Enterococcus. Clin Microbiol Rev 3:46–65. doi: 10.1128/cmr.3.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pinholt M, Larner-Svensson H, Littauer P, Moser CE, Pedersen M, Lemming LE, Ejlertsen T, Sondergaard TS, Holzknecht BJ, Justesen US, Dzajic E, Olsen SS, Nielsen JB, Worning P, Hammerum AM, Westh H, Jakobsen L. 2015. Multiple hospital outbreaks of vanA Enterococcus faecium in Denmark, 2012–13, investigated by WGS, MLST and PFGE. J Antimicrob Chemother 70:2474–2482. doi: 10.1093/jac/dkv142. [DOI] [PubMed] [Google Scholar]
- 33.Gastmeier P, Schroder C, Behnke M, Meyer E, Geffers C. 2014. Dramatic increase in vancomycin-resistant enterococci in Germany. J Antimicrob Chemother 69:1660–1664. doi: 10.1093/jac/dku035. [DOI] [PubMed] [Google Scholar]
- 34.Coombs GW, Pearson JC, Daley DA, Le T, Robinson OJ, Gottlieb T, Howden BP, Johnson PD, Bennett CM, Stinear TP, Turnidge JD, Australian Group on Antimicrobial Resistance . 2014. Molecular epidemiology of enterococcal bacteremia in Australia. J Clin Microbiol 52:897–905. doi: 10.1128/JCM.03286-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Australian Commission on Safety and Quality in Health Care. 2019. AURA 2019. Third Australian report on antimicrobial use and resistance in human health. https://www.safetyandquality.gov.au/sites/default/files/2019-06/AURA-2019-Report.pdf. Accessed 10 November 2019.
- 36.Lee T, Pang S, Abraham S, Coombs GW. 2019. Antimicrobial-resistant CC17 Enterococcus faecium: the past, the present and the future. J Glob Antimicrob Resist 16:36–47. doi: 10.1016/j.jgar.2018.08.016. [DOI] [PubMed] [Google Scholar]
- 37.Goncalves A, Igrejas G, Radhouani H, Lopez M, Guerra A, Petrucci-Fonseca F, Alcaide E, Zorrilla I, Serra R, Torres C, Poeta P. 2011. Detection of vancomycin-resistant enterococci from faecal samples of Iberian wolf and Iberian lynx, including Enterococcus faecium strains of CC17 and the new singleton ST573. Sci Total Environ 410-411:266–268. doi: 10.1016/j.scitotenv.2011.09.074. [DOI] [PubMed] [Google Scholar]
- 38.Marques C, Belas A, Franco A, Aboim C, Gama LT, Pomba C. 2018. Increase in antimicrobial resistance and emergence of major international high-risk clonal lineages in dogs and cats with urinary tract infection: 16 year retrospective study. J Antimicrob Chemother 73:377–384. doi: 10.1093/jac/dkx401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.da Silva LP, Pitondo-Silva A, Martinez R, da Costa Darini AL. 2012. Genetic features and molecular epidemiology of Enterococcus faecium isolated in two university hospitals in Brazil. Diagn Microbiol Infect Dis 74:267–271. doi: 10.1016/j.diagmicrobio.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 40.Kang M, Xie Y, He C, Chen ZX, Guo L, Yang Q, Liu JY, Du Y, Ou QS, Wang LL. 2014. Molecular characteristics of vancomycin-resistant Enterococcus faecium from a tertiary care hospital in Chengdu, China: molecular characteristics of VRE in China. Eur J Clin Microbiol Infect Dis 33:933–939. doi: 10.1007/s10096-013-2029-z. [DOI] [PubMed] [Google Scholar]
- 41.Strateva T, Sirakov I, Dimov S, Trifonova A, Savov E, Mitov I. 2018. Clonal spread of vanA Enterococcus faecium sequence type 203 in Bulgarian hospitals. Infect Dis (Lond) 50:718–721. doi: 10.1080/23744235.2018.1453946. [DOI] [PubMed] [Google Scholar]
- 42.Gao W, Howden BP, Stinear TP. 2018. Evolution of virulence in Enterococcus faecium, a hospital-adapted opportunistic pathogen. Curr Opin Microbiol 41:76–82. doi: 10.1016/j.mib.2017.11.030. [DOI] [PubMed] [Google Scholar]
- 43.Satilmis L, Vanhems P, Benet T. 2016. Outbreaks of vancomycin-resistant enterococci in hospital settings: a systematic review and calculation of the basic reproductive number. Infect Control Hosp Epidemiol 37:289–294. doi: 10.1017/ice.2015.301. [DOI] [PubMed] [Google Scholar]
- 44.Sood G, Perl TM. 2016. Outbreaks in health care settings. Infect Dis Clin North Am 30:661–687. doi: 10.1016/j.idc.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ubeda C, Taur Y, Jenq RR, Equinda MJ, Son T, Samstein M, Viale A, Socci ND, van den Brink MR, Kamboj M, Pamer EG. 2010. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest 120:4332–4341. doi: 10.1172/JCI43918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buffie CG, Pamer EG. 2013. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol 13:790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gouliouris T, Warne B, Cartwright EJP, Bedford L, Weerasuriya CK, Raven KE, Brown NM, Török ME, Limmathurotsakul D, Peacock SJ. 2018. Duration of exposure to multiple antibiotics is associated with increased risk of VRE bacteraemia: a nested case-control study. J Antimicrob Chemother 73:1692–1699. doi: 10.1093/jac/dky075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coombs GW, Daley DA, Thin Y, Pang S, Collignon P, Bradbury S, Gottlieb T, Robertson G, Branley J, Barbaro D, Huntington P, van Hal S, Beukers A, Iredell J, Ginn A, Givney R, Winney I, Newton P, Hoddle M, Baird R, Hennessy J, McLeod J, Binotto E, Thomsett B, Nimmo G, George N, Maloney S, Curtis C, Horvath R, Martin L, Runnegar N, Douglas J, Robson J, Peachey G, Papanaoum K, Wells N, Warner M, Smith K, Cooley L, Jones D, Kalukottege P, Wilcox K, Spelman D, Bernhard R, Johnson P, Hurren F, Korman T, Kotsanas D, Daley A, Gonis G. 2018. Australian Group on Antimicrobial Resistance (AGAR) Australian Enterococcal Sepsis Outcome Programme (AESOP) annual report 2016. Commun Dis Intell 42:PII=S2209-6051(18)00020-9. [DOI] [PubMed] [Google Scholar]
- 49.Prasad P, Sun JF, Danner RL, Natanson C. 2012. Excess deaths associated with tigecycline after approval based on noninferiority trials. Clin Infect Dis 54:1699–1709. doi: 10.1093/cid/cis270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Silverman JA, Mortin LI, VanPraagh ADG, Li TC, Alder J. 2005. Inhibition of daptomycin by pulmonary surfactant: in vitro modeling and clinical impact. J Infect Dis 191:2149–2152. doi: 10.1086/430352. [DOI] [PubMed] [Google Scholar]
- 51.Cetinkaya Y, Falk P, Mayhall CG. 2000. Vancomycin-resistant enterococci. Clin Microbiol Rev 13:686–707. doi: 10.1128/cmr.13.4.686-707.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prematunge C, MacDougall C, Johnstone J, Adomako K, Lam F, Robertson J, Garber G. 2016. VRE and VSE bacteremia outcomes in the era of effective VRE therapy: a systematic review and meta-analysis. Infect Control Hosp Epidemiol 37:26–35. doi: 10.1017/ice.2015.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chiang HY, Perencevich EN, Nair R, Nelson RE, Samore M, Khader K, Chorazy ML, Herwaldt LA, Blevins A, Ward MA, Schweizer ML. 2017. Incidence and outcomes associated with infections caused by vancomycin-resistant enterococci in the United States: systematic literature review and meta-analysis. Infect Control Hosp Epidemiol 38:203–215. doi: 10.1017/ice.2016.254. [DOI] [PubMed] [Google Scholar]
- 54.DiazGranados CA, Jernigan JA. 2005. Impact of vancomycin resistance on mortality among patients with neutropenia and enterococcal bloodstream infection. J Infect Dis 191:588–595. doi: 10.1086/427512. [DOI] [PubMed] [Google Scholar]
- 55.Eriksen KR. 1961. “Celbenin"-resistant staphylococci. Ugeskr Laeger 123:384–386. (In Danish.) [PubMed] [Google Scholar]
- 56.Kourtis AP, Hatfield K, Baggs J, Mu Y, See I, Epson E, Nadle J, Kainer MA, Dumyati G, Petit S, Ray SM, Emerging Infections Program MRSA Author Group, Ham D, Capers C, Ewing H, Coffin N, McDonald LC, Jernigan J, Cardo D, Program EI. 2019. Vital signs: epidemiology and recent trends in methicillin-resistant and in methicillin-susceptible Staphylococcus aureus bloodstream infections—United States. MMWR Morb Mortal Wkly Rep 68:214–219. doi: 10.15585/mmwr.mm6809e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Udo EE, Pearman JW, Grubb WB. 1993. Genetic analysis of community isolates of methicillin-resistant Staphylococcus aureus in Western Australia. J Hosp Infect 25:97–108. doi: 10.1016/0195-6701(93)90100-e. [DOI] [PubMed] [Google Scholar]
- 58.Diekema DJ, Pfaller MA, Schmitz FJ, Smayevsky J, Bell J, Jones RN, Beach M, SENTRY Participants Group . 2001. Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997–1999. Clin Infect Dis 32:S114–S132. doi: 10.1086/320184. [DOI] [PubMed] [Google Scholar]
- 59.Gilbert M, MacDonald J, Gregson D, Siushansian J, Zhang K, Elsayed S, Laupland K, Louie T, Hope K, Mulvey M, Gillespie J, Nielsen D, Wheeler V, Louie M, Honish A, Keays G, Conly J. 2006. Outbreak in Alberta of community-acquired (USA300) methicillin-resistant Staphylococcus aureus in people with a history of drug use, homelessness or incarceration. CMAJ 175:149–154. doi: 10.1503/cmaj.051565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mulvey MR, MacDougall L, Cholin B, Horsman G, Fidyk M, Woods S, Saskatchewan CA-MRSA Study Group . 2005. Community-associated methicillin-resistant Staphylococcus aureus, Canada. Emerg Infect Dis 11:844–850. doi: 10.3201/eid1106.041146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.David MZ, Rudolph KM, Hennessy TW, Boyle-Vavra S, Daum RS. 2008. Molecular epidemiology of methicillin-resistant Staphylococcus aureus, rural southwestern Alaska. Emerg Infect Dis 14:1693–1699. doi: 10.3201/eid1411.080381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Diep BA, Sensabaugh GF, Somboona NS, Carleton HA, Perdreau-Remington F. 2004. Widespread skin and soft-tissue infections due to two methicillin-resistant Staphylococcus aureus strains harboring the genes for Panton-Valentine leucocidin. J Clin Microbiol 42:2080–2084. doi: 10.1128/JCM.42.5.2080-2084.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lim WW, Wu P, Bond HS, Wong JY, Ni KW, Seto WH, Jit M, Cowling BJ. 2019. Determinants of methicillin-resistant Staphylococcus aureus (MRSA) prevalence in the Asia-Pacific region: a systematic review and meta-analysis. J Glob Antimicrob Resist 16:17–27. doi: 10.1016/j.jgar.2018.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.European Antimicrobial Resistance Surveillance Network. 2019. Surveillance of antimicrobial resistance in Europe. Annual report of the European Antimicrobial Resistance Surveillance Network (EARS-Net) 2018. https://www.ecdc.europa.eu/sites/default/files/documents/surveillance-antimicrobial-resistance-Europe-2018.pdf. Accessed 13 January 2020.
- 65.Dai YX, Liu JL, Guo W, Meng HW, Huang Q, He L, Gao QQ, Lv HY, Liu Y, Wang YN, Wang H, Liu Q, Li M. 2019. Decreasing methicillin-resistant Staphylococcus aureus (MRSA) infections is attributable to the disappearance of predominant MRSA ST239 clones, Shanghai, 2008–2017. Emerg Microbes Infect 8:471–478. doi: 10.1080/22221751.2019.1595161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Duerden B, Fry C, Johnson AP, Wilcox MH. 2015. The control of methicillin-resistant Staphylococcus aureus blood stream infections in England. Open Forum Infect Dis 2:ofv035. doi: 10.1093/ofid/ofv035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Carlet J, Astagneau P, Brun-Buisson C, Coignard B, Salomon V, Tran B, Desenclos JC, Jarlier V, Schlemmer B, Parneix P, Regnier B, Fabry J, Hea F, French National Program for Prevention of Healthcare-Associated Infections and Antimicrobial Resistance . 2009. French National Program for Prevention of Healthcare-Associated Infections and Antimicrobial Resistance, 1992–2008: positive trends, but perseverance needed. Infect Control Hosp Epidemiol 30:737–745. doi: 10.1086/598682. [DOI] [PubMed] [Google Scholar]
- 68.DeLeo FR, Otto M, Kreiswirth BN, Chambers HF. 2010. Community-associated meticillin-resistant Staphylococcus aureus. Lancet 375:1557–1568. doi: 10.1016/S0140-6736(09)61999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dermota U, Jurca T, Harlander T, Kosir M, Zajc U, Golob M, Zdovc I, Kosnik IG. 2016. Infections caused by community-associated methicillin-resistant Staphylococcus aureus European clone (St80) in Slovenia between 2006 and 2013. Zdr Varst 55:121–125. doi: 10.1515/sjph-2016-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Drougka E, Foka A, Liakopoulos A, Doudoulakakis A, Jelastopulu E, Chini V, Spiliopoulou A, Levidiotou S, Panagea T, Vogiatzi A, Lebessi E, Petinaki E, Spiliopoulou I. 2014. A 12-year survey of methicillin-resistant Staphylococcus aureus infections in Greece: ST80-IV epidemic? Clin Microbiol Infect 20:O796–O803. doi: 10.1111/1469-0691.12624. [DOI] [PubMed] [Google Scholar]
- 71.Fluit AC. 2012. Livestock-associated Staphylococcus aureus. Clin Microbiol Infect 18:735–744. doi: 10.1111/j.1469-0691.2012.03846.x. [DOI] [PubMed] [Google Scholar]
- 72.van Alen S, Ballhausen B, Peters G, Friedrich AW, Mellmann A, Kock R, Becker K. 2017. In the centre of an epidemic: fifteen years of LA-MRSA CC398 at the university hospital Munster. Vet Microbiol 200:19–24. doi: 10.1016/j.vetmic.2016.01.021. [DOI] [PubMed] [Google Scholar]
- 73.Hryniewicz MM, Garbacz K. 2017. Borderline oxacillin-resistant Staphylococcus aureus (BORSA)—a more common problem than expected? J Med Microbiol 66:1367–1373. doi: 10.1099/jmm.0.000585. [DOI] [PubMed] [Google Scholar]
- 74.Skinner S, Murray M, Walus T, Karlowsky JA. 2009. Failure of cloxacillin in treatment of a patient with borderline oxacillin-resistant Staphylococcus aureus endocarditis. J Clin Microbiol 47:859–861. doi: 10.1128/JCM.00571-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pada SK, Ding Y, Ling ML, Hsu LY, Earnest A, Lee TE, Yong HC, Jureen R, Fisher D. 2011. Economic and clinical impact of nosocomial meticillin-resistant Staphylococcus aureus infections in Singapore: a matched case-control study. J Hosp Infect 78:36–40. doi: 10.1016/j.jhin.2010.10.016. [DOI] [PubMed] [Google Scholar]
- 76.Cosgrove SE, Sakoulas G, Perencevich EN, Schwaber MJ, Karchmer AW, Carmeli Y. 2003. Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: a meta-analysis. Clin Infect Dis 36:53–59. doi: 10.1086/345476. [DOI] [PubMed] [Google Scholar]
- 77.Kaasch AJ, Barlow G, Edgeworth JD, Fowler VG, Hellmich M, Hopkins S, Kern WV, Llewelyn MJ, Rieg S, Rodriguez-Baño J, Scarborough M, Seifert H, Soriano A, Tilley R, Tőrők ME, Weiß V, Wilson APR, Thwaites GE. 2014. Staphylococcus aureus bloodstream infection: a pooled analysis of five prospective, observational studies. J Infect 68:242–251. doi: 10.1016/j.jinf.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tzouvelekis LS, Markogiannakis A, Psichogiou M, Tassios PT, Daikos GL. 2012. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin Microbiol Rev 25:682–707. doi: 10.1128/CMR.05035-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kaewpoowat Q, Ostrosky-Zeichner L. 2015. Tigecycline: a critical safety review. Expert Opin Drug Saf 14:335–342. doi: 10.1517/14740338.2015.997206. [DOI] [PubMed] [Google Scholar]
- 80.Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats in the United States, 2013. https://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013-508.pdf. Accessed 23 October 2019.
- 81.Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, Alberti S, Bush K, Tenover FC. 2001. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother 45:1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.David S, Reuter S, Harris SR, Glasner C, Feltwell T, Argimon S, Abudahab K, Goater R, Giani T, Errico G, Aspbury M, Sjunnebo S, EuSCAPE Working Group, ESGEM Study Group, Feil EJ, Rossolini GM, Aanensen DM, Grundmann H. 2019. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat Microbiol 4:1919–1929. doi: 10.1038/s41564-019-0492-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hall JM, Corea E, Sanjeewani HD, Inglis TJ. 2014. Molecular mechanisms of beta-lactam resistance in carbapenemase-producing Klebsiella pneumoniae from Sri Lanka. J Med Microbiol 63:1087–1092. doi: 10.1099/jmm.0.076760-0. [DOI] [PubMed] [Google Scholar]
- 84.Hall JM, Ingram PR, O'Reilly LC, Inglis T. 2016. Temporal flux in beta-lactam resistance among Klebsiella pneumoniae in Western Australia. J Med Microbiol 65:429–437. doi: 10.1099/jmm.0.000242. [DOI] [PubMed] [Google Scholar]
- 85.Guh AY, Bulens SN, Mu Y, Jacob JT, Reno J, Scott J, Wilson LE, Vaeth E, Lynfield R, Shaw KM, Vagnone PMS, Bamberg WM, Janelle SJ, Dumyati G, Concannon C, Beldavs Z, Cunningham M, Cassidy PM, Phipps EC, Kenslow N, Travis T, Lonsway D, Rasheed JK, Limbago BM, Kallen AJ. 2015. Epidemiology of carbapenem-resistant Enterobacteriaceae in 7 US communities, 2012–2013. JAMA 314:1479–1487. doi: 10.1001/jama.2015.12480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang R, Chan EWC, Zhou HW, Chen S. 2017. Prevalence and genetic characteristics of carbapenem-resistant Enterobacteriaceae strains in China. Lancet Infect Dis 17:256–257. doi: 10.1016/S1473-3099(17)30072-5. [DOI] [PubMed] [Google Scholar]
- 87.Wei DD, Wan LG, Deng Q, Liu Y. 2016. Emergence of KPC-producing Klebsiella pneumoniae hypervirulent clone of capsular serotype K1 that belongs to sequence type 11 in mainland China. Diagn Microbiol Infect Dis 85:192–194. doi: 10.1016/j.diagmicrobio.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 88.Wyres KL, Hawkey J, Hetland MAK, Fostervold A, Wick RR, Judd LM, Hamidian M, Howden BP, Lohr IH, Holt KE. 2019. Emergence and rapid global dissemination of CTX-M-15-associated Klebsiella pneumoniae strain ST307. J Antimicrob Chemother 74:577–581. doi: 10.1093/jac/dky492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lowe M, Kock MM, Coetzee J, Hoosien E, Peirano G, Strydom KA, Ehlers MM, Mbelle NM, Shashkina E, Haslam DB, Dhawan P, Donnelly RJ, Chen L, Kreiswirth BN, Pitout J. 2019. Klebsiella pneumoniae ST307 with blaOXA-181, South Africa, 2014–2016. Emerg Infect Dis 25:739–747. doi: 10.3201/eid2504.181482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cheng NC, Yu YC, Tai HC, Hsueh PR, Chang SC, Lai SY, Yi WC, Fang CT. 2012. Recent trend of necrotizing fasciitis in Taiwan: focus on monomicrobial Klebsiella pneumoniae necrotizing fasciitis. Clin Infect Dis 55:930–939. doi: 10.1093/cid/cis565. [DOI] [PubMed] [Google Scholar]
- 91.Bernal NP, Latenser BA, Born JM, Liao J. 2012. Trends in 393 necrotizing acute soft tissue infection patients 2000–2008. Burns 38:252–260. doi: 10.1016/j.burns.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 92.Krapp F, Morris AR, Ozer EA, Hauser AR. 2017. Virulence characteristics of carbapenem-resistant Klebsiella pneumoniae strains from patients with necrotizing skin and soft tissue infections. Sci Rep 7:13533. doi: 10.1038/s41598-017-13524-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li W, Sun GZ, Yu YH, Li N, Chen M, Jin RH, Jiao YM, Wu H. 2014. Increasing occurrence of antimicrobial-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in China. Clin Infect Dis 58:225–232. doi: 10.1093/cid/cit675. [DOI] [PubMed] [Google Scholar]
- 94.Munoz-Price LS, Weinstein RA. 2008. Acinetobacter infection. N Engl J Med 358:1271–1281. doi: 10.1056/NEJMra070741. [DOI] [PubMed] [Google Scholar]
- 95.Villegas MV, Hartstein AI. 2003. Acinetobacter outbreaks, 1977–2000. Infect Control Hosp Epidemiol 24:284–295. doi: 10.1086/502205. [DOI] [PubMed] [Google Scholar]
- 96.Ibrahim ME. 2019. Prevalence of Acinetobacter baumannii in Saudi Arabia: risk factors, antimicrobial resistance patterns and mechanisms of carbapenem resistance. Ann Clin Microbiol Antimicrob 18:1. doi: 10.1186/s12941-018-0301-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McDonald LC, Banerjee SN, Jarvis WR, National Nosocomial Infections Surveillance System . 1999. Seasonal variation of Acinetobacter infections: 1987–1996. Clin Infect Dis 29:1133–1137. doi: 10.1086/313441. [DOI] [PubMed] [Google Scholar]
- 98.Harding CM, Hennon SW, Feldman MF. 2018. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat Rev Microbiol 16:91–102. doi: 10.1038/nrmicro.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Davis JS, McMillan M, Swaminathan A, Kelly JA, Piera KE, Baird RW, Currie BJ, Anstey NM. 2014. A 16-year prospective study of community-onset bacteremic Acinetobacter pneumonia: low mortality with appropriate initial empirical antibiotic protocols. Chest 146:1038–1045. doi: 10.1378/chest.13-3065. [DOI] [PubMed] [Google Scholar]
- 100.Lob SH, Hoban DJ, Sahm DF, Badal RE. 2016. Regional differences and trends in antimicrobial susceptibility of Acinetobacter baumannii. Int J Antimicrob Agents 47:317–323. doi: 10.1016/j.ijantimicag.2016.01.015. [DOI] [PubMed] [Google Scholar]
- 101.Magill SS, Edwards JR, Bamberg W, Beldavs ZG, Dumyati G, Kainer MA, Lynfield R, Maloney M, McAllister-Hollod L, Nadle J, Ray SM, Thompson DL, Wilson LE, Fridkin SK, Emerging Infections Program Healthcare-Associated Infections and Antimicrobial Use Prevalence Survey Team . 2014. Multistate point-prevalence survey of health care-associated infections. N Engl J Med 370:1198–1208. doi: 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Giammanco A, Cala C, Fasciana T, Dowzicky MJ. 2017. Global assessment of the activity of tigecycline against multidrug-resistant Gram-negative pathogens between 2004 and 2014 as part of the tigecycline evaluation and surveillance trial. mSphere 2:e00310-16. doi: 10.1128/mSphere.00310-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xie RQ, Zhang XD, Zhao Q, Peng B, Zheng J. 2018. Analysis of global prevalence of antibiotic resistance in Acinetobacter baumannii infections disclosed a faster increase in OECD countries. Emerg Microbes Infect 7:31. doi: 10.1038/s41426-018-0038-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lee HY, Chang RC, Su LH, Liu SY, Wu SR, Chuang CH, Chen CL, Chiu CH. 2012. Wide spread of Tn2006 in an AbaR4-type resistance island among carbapenem-resistant Acinetobacter baumannii clinical isolates in Taiwan. Int J Antimicrob Agents 40:163–167. doi: 10.1016/j.ijantimicag.2012.04.018. [DOI] [PubMed] [Google Scholar]
- 105.Zarrilli R, Pournaras S, Giannouli M, Tsakris A. 2013. Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int J Antimicrob Agents 41:11–19. doi: 10.1016/j.ijantimicag.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 106.Tavares LCB, de Vasconcellos FM, de Sousa WV, Rocchetti TT, Mondelli AL, Ferreira AM, Montelli AC, Sadatsune T, Tiba-Casas MR, Camargo CH. 2019. Emergence and persistence of high-risk clones among MDR and XDR A. baumannii at a Brazilian teaching hospital. Front Microbiol 9:2898. doi: 10.3389/fmicb.2018.02898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Escandón-Vargas K, Reyes S, Gutiérrez S, Villegas MV. 2017. The epidemiology of carbapenemases in Latin America and the Caribbean. Expert Rev Anti Infect Ther 15:277–297. doi: 10.1080/14787210.2017.1268918. [DOI] [PubMed] [Google Scholar]
- 108.Qureshi ZA, Hittle LE, O'Hara JA, Rivera JI, Syed A, Shields RK, Pasculle AW, Ernst RK, Doi Y. 2015. Colistin-resistant Acinetobacter baumannii: beyond carbapenem resistance. Clin Infect Dis 60:1295–1303. doi: 10.1093/cid/civ048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gellatly SL, Hancock R. 2013. Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog Dis 67:159–173. doi: 10.1111/2049-632X.12033. [DOI] [PubMed] [Google Scholar]
- 110.Yayan J, Ghebremedhin B, Rasche K. 2015. Antibiotic resistance of Pseudomonas aeruginosa in pneumonia at a single university hospital center in Germany over a 10-year period. PLoS One 10:e0139836. doi: 10.1371/journal.pone.0139836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Treepong P, Kos VN, Guyeux C, Blanc DS, Bertrand X, Valot B, Hocquet D. 2018. Global emergence of the widespread Pseudomonas aeruginosa ST235 clone. Clin Microbiol Infect 24:258–266. doi: 10.1016/j.cmi.2017.06.018. [DOI] [PubMed] [Google Scholar]
- 112.Cabot G, Lopez-Causape C, Ocampo-Sosa AA, Sommer LM, Dominguez MA, Zamorano L, Juan C, Tubau F, Rodriguez C, Moya B, Pena C, Martinez-Martinez L, Plesiat P, Oliver A, Spanish Network for Research in Infectious Diseases . 2016. Deciphering the resistome of the widespread Pseudomonas aeruginosa sequence type 175 international high-risk clone through whole-genome sequencing. Antimicrob Agents Chemother 60:7415–7423. doi: 10.1128/AAC.01720-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Xipell M, Bodro M, Marco F, Losno RA, Cardozo C, Soriano A. 2017. Clinical experience with ceftazidime/avibactam in patients with severe infections, including meningitis and lung abscesses, caused by extensively drug-resistant Pseudomonas aeruginosa. Int J Antimicrob Agents 49:266–268. doi: 10.1016/j.ijantimicag.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 114.Fan X, Wu Y, Xiao M, Xu ZP, Kudinha T, Bazaj A, Kong F, Xu YC. 2016. Diverse genetic background of multidrug-resistant Pseudomonas aeruginosa from mainland China, and emergence of an extensively drug-resistant ST292 clone in Kunming. Sci Rep 6:26522. doi: 10.1038/srep26522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Langan KM, Kotsimbos T, Peleg AY. 2015. Managing Pseudomonas aeruginosa respiratory infections in cystic fibrosis. Curr Opin Infect Dis 28:547–556. doi: 10.1097/QCO.0000000000000217. [DOI] [PubMed] [Google Scholar]
- 116.LaFayette SL, Houle D, Beaudoin T, Wojewodka G, Radzioch D, Hoffman LR, Burns JL, Dandekar AA, Smalley NE, Chandler JR, Zlosnik JE, Speert DP, Bernier J, Matouk E, Brochiero E, Rousseau S, Nguyen D. 2015. Cystic fibrosis-adapted Pseudomonas aeruginosa quorum sensing lasR mutants cause hyperinflammatory responses. Sci Adv 1:e1500199. doi: 10.1126/sciadv.1500199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Newman JW, Floyd RV, Fothergill JL. 2017. The contribution of Pseudomonas aeruginosa virulence factors and host factors in the establishment of urinary tract infections. FEMS Microbiol Lett 364:fnx124. doi: 10.1093/femsle/fnx124. [DOI] [PubMed] [Google Scholar]
- 118.Hoiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. 2010. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents 35:322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 119.Davin-Regli A, Pagès J-M. 2015. Enterobacter aerogenes and Enterobacter cloacae; versatile bacterial pathogens confronting antibiotic treatment. Front Microbiol 6:392. doi: 10.3389/fmicb.2015.00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Georghiou PR, Hamill RJ, Wright CE, Versalovic J, Koeuth T, Watson DA, Lupski JR. 1995. Molecular epidemiology of infections due to Enterobacter aerogenes—identification of hospital outbreak-associated strains by molecular techniques. Clin Infect Dis 20:84–94. doi: 10.1093/clinids/20.1.84. [DOI] [PubMed] [Google Scholar]
- 121.Grattard F, Pozzetto B, Tabard L, Petit M, Ros A, Gaudin OG. 1995. Characterization of nosocomial strains of Enterobacter aerogenes by arbitrarily primed-PCR analysis and ribotyping. Infect Control Hosp Epidemiol 16:224–230. doi: 10.1086/647094. [DOI] [PubMed] [Google Scholar]
- 122.Bosi C, Davin-Regli A, Bornet C, Mallea M, Pages JM, Bollet C. 1999. Most Enterobacter aerogenes strains in France belong to a prevalent clone. J Clin Microbiol 37:2165–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chevalier J, Mulfinger C, Garnotel E, Nicolas P, Davin-Régli A, Pagès J-M. 2008. Identification and evolution of drug efflux pump in clinical Enterobacter aerogenes strains isolated in 1995 and 2003. PLoS One 3:e3203. doi: 10.1371/journal.pone.0003203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mezzatesta ML, Gona F, Stefani S. 2012. Enterobacter cloacae complex: clinical impact and emerging antibiotic resistance. Future Microbiol 7:887–902. doi: 10.2217/fmb.12.61. [DOI] [PubMed] [Google Scholar]
- 125.Paauw A, Caspers MPM, Schuren FHJ, Leverstein-van Hall MA, Deletoile A, Montijn RC, Verhoef J, Fluit AC. 2008. Genomic diversity within the Enterobacter cloacae complex. PLoS One 3:e3018. doi: 10.1371/journal.pone.0003018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Annavajhala MK, Gomez-Simmonds A, Uhlemann AC. 2019. Multidrug-resistant Enterobacter cloacae complex emerging as a global, diversifying threat. Front Microbiol 10:44. doi: 10.3389/fmicb.2019.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Malek A, McGlynn K, Taffner S, Fine L, Tesini B, Wang J, Mostafa H, Petry S, Perkins A, Graman P, Hardy D, Pecora N. 2019. Next-generation sequencing based hospital outbreak investigation yields insight into Klebsiella aerogenes population structure and determinants of carbapenem resistance and pathogenicity. Antimicrob Agents Chemother 63:e02577-18. doi: 10.1128/AAC.02577-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gomez-Simmonds A, Annavajhala MK, Wang Z, Macesic N, Hu Y, Giddins MJ, O’Malley A, Toussaint NC, Whittier S, Torres VJ, Uhlemann A-C. 2018. Genomic and geographic context for the evolution of high-risk carbapenem-resistant Enterobacter cloacae complex clones ST171 and ST78. mBio 9:e00542-18. doi: 10.1128/mBio.00542-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sader HS, Biedenbach DJ, Jones RN. 2003. Global patterns of susceptibility for 21 commonly utilized antimicrobial agents tested against 48,440 Enterobacteriaceae in the SENTRY Antimicrobial Surveillance Program (1997–2001). Diagn Microbiol Infect Dis 47:361–364. doi: 10.1016/S0732-8893(03)00052-X. [DOI] [PubMed] [Google Scholar]
- 130.Thiolas A, Bollet C, La Scola B, Raoult D, Pagès J-M. 2005. Successive emergence of Enterobacter aerogenes strains resistant to imipenem and colistin in a patient. Antimicrob Agents Chemother 49:1354–1358. doi: 10.1128/AAC.49.4.1354-1358.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Band VI, Crispell EK, Napier BA, Herrera CM, Tharp GK, Vavikolanu K, Pohl J, Read TD, Bosinger SE, Trent MS, Burd EM, Weiss DS. 2016. Antibiotic failure mediated by a resistant subpopulation in Enterobacter cloacae. Nat Microbiol 1:16053. doi: 10.1038/nmicrobiol.2016.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Schembri MA, Ben Zakour NL, Phan MD, Forde BM, Stanton-Cook M, Beatson SA. 2015. Molecular characterization of the multidrug resistant Escherichia coli ST131 clone. Pathogens 4:422–430. doi: 10.3390/pathogens4030422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mathers AJ, Peirano G, Pitout J. 2015. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev 28:565–591. doi: 10.1128/CMR.00116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Brolund A, Lagerqvist N, Byfors S, Struelens MJ, Monnet DL, Albiger B, Kohlenberg A, European Antimicrobial Resistance Genes Surveillance Network (EURGen-Net) Capacity Survey Group . 2019. Worsening epidemiological situation of carbapenemase-producing Enterobacteriaceae in Europe, assessment by national experts from 37 countries, July 2018. Euro Surveill 24(9):pii=1900123. doi: 10.2807/1560-7917.ES.2019.24.9.1900123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 136.Abraham EP, Chain E. 1988. An enzyme from bacteria able to destroy penicillin. 1940. Rev Infect Dis 10:677–678. [PubMed] [Google Scholar]
- 137.Naas T, Oueslati S, Bonnin RA, Dabos ML, Zavala A, Dortet L, Retailleau P, Iorga BI. 2017. Beta-Lactamase Database (BLDB—structure and function. J Enzyme Inhib Med Chem 32:917–919. doi: 10.1080/14756366.2017.1344235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ambler RP. 1980. The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci 289:321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- 139.Bush K, Jacoby GA. 2010. Updated functional classification of beta-lactamases. Antimicrob Agents Chemother 54:969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bush K, Bradford PA. 2019. Interplay between beta-lactamases and new beta-lactamase inhibitors. Nat Rev Microbiol 17:295–306. doi: 10.1038/s41579-019-0159-8. [DOI] [PubMed] [Google Scholar]
- 141.Richter SS, Doern GV, Heilmann KP, Miner S, Tendolkar S, Riahi F, Diekema DJ. 2016. Detection and prevalence of penicillin-susceptible Staphylococcus aureus in the United States in 2013. J Clin Microbiol 54:812–814. doi: 10.1128/JCM.03109-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sarti M, Campanile F, Sabia C, Santagati M, Gargiulo R, Stefani S. 2012. Polyclonal diffusion of beta-lactamase-producing Enterococcus faecium. J Clin Microbiol 50:169–172. doi: 10.1128/JCM.05640-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zscheck KK, Murray BE. 1991. Nucleotide sequence of the beta-lactamase gene from Enterococcus faecalis HH22 and its similarity to staphylococcal beta-lactamase genes. Antimicrob Agents Chemother 35:1736–1740. doi: 10.1128/aac.35.9.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wright G, McArthur AG. 2018. The comprehensive antibiotic resistance database. http://arpcard.mcmaster.ca/. Accessed 1 December 2019.
- 145.Gniadkowski M. 2008. Evolution of extended-spectrum beta-lactamases by mutation. Clin Microbiol Infect 14(Suppl 1):11–32. doi: 10.1111/j.1469-0691.2007.01854.x. [DOI] [PubMed] [Google Scholar]
- 146.Canton R, Morosini MI, Martin O, de la Maza S, de la Pedrosa E. 2008. IRT and CMT beta-lactamases and inhibitor resistance. Clin Microbiol Infect 14:53–62. doi: 10.1111/j.1469-0691.2007.01849.x. [DOI] [PubMed] [Google Scholar]
- 147.Bradford PA, Bratu S, Urban C, Visalli M, Mariano N, Landman D, Rahal JJ, Brooks S, Cebular S, Quale J. 2004. Emergence of carbapenem-resistant Klebsiella species possessing the class A carbapenem-hydrolyzing KPC-2 and inhibitor-resistant TEM-30 beta-lactamases in New York City. Clin Infect Dis 39:55–60. doi: 10.1086/421495. [DOI] [PubMed] [Google Scholar]
- 148.Nordmann P, Cuzon G, Naas T. 2009. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis 9:228–236. doi: 10.1016/S1473-3099(09)70054-4. [DOI] [PubMed] [Google Scholar]
- 149.Vena A, Castaldo N, Bassetti M. 2019. The role of new beta-lactamase inhibitors in Gram-negative infections. Curr Opin Infect Dis 32:638–646. doi: 10.1097/QCO.0000000000000600. [DOI] [PubMed] [Google Scholar]
- 150.Gottig S, Frank D, Mungo E, Nolte A, Hogardt M, Besier S, Wichelhaus TA. 2019. Emergence of ceftazidime/avibactam resistance in KPC-3-producing Klebsiella pneumoniae in vivo. J Antimicrob Chemother 74:3211–3216. doi: 10.1093/jac/dkz330. [DOI] [PubMed] [Google Scholar]
- 151.Zhang P, Shi Q, Hu H, Hong B, Wu X, Du X, Akova M, Yu Y. 2020. Emergence of ceftazidime/avibactam resistance in carbapenem-resistant Klebsiella pneumoniae in China. Clin Microbiol Infect 26:124.e1–124.e4. doi: 10.1016/j.cmi.2019.08.020. [DOI] [PubMed] [Google Scholar]
- 152.Lauretti L, Riccio ML, Mazzariol A, Cornaglia G, Amicosante G, Fontana R, Rossolini GM. 1999. Cloning and characterization of blaVIM, a new integron-borne metallo-beta-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob Agents Chemother 43:1584–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Watanabe M, Iyobe S, Inoue M, Mitsuhashi S. 1991. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 35:147–151. doi: 10.1128/aac.35.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Matsumura Y, Peirano G, Devinney R, Bradford PA, Motyl MR, Adams MD, Chen L, Kreiswirth B, Pitout J. 2017. Genomic epidemiology of global VIM-producing Enterobacteriaceae. J Antimicrob Chemother 72:2249–2258. doi: 10.1093/jac/dkx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Peirano G, Matsumura Y, Adams MD, Bradford P, Motyl M, Chen L, Kreiswirth BN, Pitout J. 2018. Genomic epidemiology of global carbapenemase-producing Enterobacter spp., 2008–2014. Emerg Infect Dis 24:1010–1019. doi: 10.3201/eid2406.171648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lee K, Ha GY, Shin BM, Kim JJ, Kang JO, Jang SJ, Yong D, Chong Y, Korean Nationwide Surveillance of Antimicrobial Resistance (KONSAR) Group . 2004. Metallo-beta-lactamase-producing Gram-negative bacilli in Korean nationwide surveillance of antimicrobial resistance group hospitals in 2003: continued prevalence of VIM-producing Pseudomonas spp. and increase of IMP-producing Acinetobacter spp. Diagn Microbiol Infect Dis 50:51–58. doi: 10.1016/j.diagmicrobio.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 157.Cornaglia G, Giamarellou H, Rossolini GM. 2011. Metallo-beta-lactamases: a last frontier for beta-lactams? Lancet Infect Dis 11:381–393. doi: 10.1016/S1473-3099(11)70056-1. [DOI] [PubMed] [Google Scholar]
- 158.Jovcic B, Lepsanovic Z, Suljagic V, Rackov G, Begovic J, Topisirovic L, Kojic M. 2011. Emergence of NDM-1 metallo-beta-lactamase in Pseudomonas aeruginosa clinical isolates from Serbia. Antimicrob Agents Chemother 55:3929–3931. doi: 10.1128/AAC.00226-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Jacoby GA. 2009. AmpC beta-lactamases. Clin Microbiol Rev 22:161–182. doi: 10.1128/CMR.00036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Pitout JDD, Peirano G, Kock MM, Strydom KA, Matsumura Y. 2020. The global ascendency of OXA-48-type carbapenemases. Clin Microbiol Rev 33:e00102-19. doi: 10.1128/CMR.00102-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Poirel L, Bonnin RA, Nordmann P. 2012. Genetic features of the widespread plasmid coding for the carbapenemase OXA-48. Antimicrob Agents Chemother 56:559–562. doi: 10.1128/AAC.05289-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Shaw KJ, Rather PN, Hare RS, Miller GH. 1993. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol Rev 57:138–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kobayashi N, Alam M, Nishimoto Y, Urasawa S, Uehara N, Watanabe N. 2001. Distribution of aminoglycoside resistance genes in recent clinical isolates of Enterococcus faecalis, Enterococcus faecium and Enterococcus avium. Epidemiol Infect 126:197–204. doi: 10.1017/s0950268801005271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Miller GH, Sabatelli FJ, Hare RS, Glupczynski Y, Mackey P, Shlaes D, Shimizu K, Shaw KJ. 1997. The most frequent aminoglycoside resistance mechanisms—changes with time and geographic area: a reflection of aminoglycoside usage patterns? Aminoglycoside Resistance Study Groups. Clin Infect Dis 24(Suppl 1):S46–S62. doi: 10.1093/clinids/24.supplement_1.s46. [DOI] [PubMed] [Google Scholar]
- 165.Ramirez MS, Tolmasky ME. 2010. Aminoglycoside modifying enzymes. Drug Resist Updat 13:151–171. doi: 10.1016/j.drup.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Krause KM, Serio AW, Kane TR, Connolly LE. 2016. Aminoglycosides: an overview. Cold Spring Harb Perspect Med 6:a027029. doi: 10.1101/cshperspect.a027029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Castanheira M, Davis AP, Serio AW, Krause KM, Mendes RE. 2019. In vitro activity of plazomicin against Enterobacteriaceae isolates carrying genes encoding aminoglycoside-modifying enzymes most common in US Census divisions. Diagn Microbiol Infect Dis 94:73–77. doi: 10.1016/j.diagmicrobio.2018.10.023. [DOI] [PubMed] [Google Scholar]
- 168.Costello SE, Deshpande LM, Davis AP, Mendes RE, Castanheira M. 2019. Aminoglycoside-modifying enzyme and 16S ribosomal RNA methyltransferase genes among a global collection of Gram-negative isolates. J Glob Antimicrob Resist 16:278–285. doi: 10.1016/j.jgar.2018.10.020. [DOI] [PubMed] [Google Scholar]
- 169.Almaghrabi R, Clancy CJ, Doi Y, Hao B, Chen L, Shields RK, Press EG, Iovine NM, Townsend BM, Wagener MM, Kreiswirth B, Nguyen MH. 2014. Carbapenem-resistant Klebsiella pneumoniae strains exhibit diversity in aminoglycoside-modifying enzymes, which exert differing effects on plazomicin and other agents. Antimicrob Agents Chemother 58:4443–4451. doi: 10.1128/AAC.00099-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hasani A, Sheikhalizadeh V, Ahangarzadeh Rezaee M, Rahmati-Yamchi M, Hasani A, Ghotaslou R, Goli HR. 2016. Frequency of aminoglycoside-modifying enzymes and ArmA among different sequence groups of Acinetobacter baumannii in Iran. Microb Drug Resist 22:347–353. doi: 10.1089/mdr.2015.0254. [DOI] [PubMed] [Google Scholar]
- 171.Holbrook SYL, Garneau-Tsodikova S. 2018. Evaluation of aminoglycoside and carbapenem resistance in a collection of drug-resistant Pseudomonas aeruginosa clinical isolates. Microb Drug Resist 24:1020–1030. doi: 10.1089/mdr.2017.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Libisch B, Poirel L, Lepsanovic Z, Mirovic V, Balogh B, Paszti J, Hunyadi Z, Dobak A, Fuzi M, Nordmann P. 2008. Identification of PER-1 extended-spectrum beta-lactamase producing Pseudomonas aeruginosa clinical isolates of the international clonal complex CC11 from Hungary and Serbia. FEMS Immunol Med Microbiol 54:330–338. doi: 10.1111/j.1574-695X.2008.00483.x. [DOI] [PubMed] [Google Scholar]
- 173.Park CH, Robicsek A, Jacoby GA, Sahm D, Hooper DC. 2006. Prevalence in the United States of aac(6')-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob Agents Chemother 50:3953–3955. doi: 10.1128/AAC.00915-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Perilli M, Sabatini A, Pontieri E, Celenza G, Segatore B, Bottoni C, Bellio P, Mancini A, Marcoccia F, Brisdelli F, Amicosante G. 2015. OXA-23 carbapenemase in multidrug-resistant Acinetobacter baumannii ST2 type: first identification in L'Aquila Hospital (Italy). Microb Drug Resist 21:97–101. doi: 10.1089/mdr.2014.0056. [DOI] [PubMed] [Google Scholar]
- 175.Robicsek A, Strahilevitz J, Jacoby GA, Macielag M, Abbanat D, Park CH, Bush K, Hooper DC. 2006. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat Med 12:83–88. doi: 10.1038/nm1347. [DOI] [PubMed] [Google Scholar]
- 176.Lakhundi S, Zhang K. 2018. Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin Microbiol Rev 31:e00020-18. doi: 10.1128/CMR.00020-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Chen FJ, Huang IW, Wang CH, Chen PC, Wang HY, Lai JF, Shiau YR, Lauderdale TLY, TSAR Hospitals . 2012. mecA-positive Staphylococcus aureus with low-level oxacillin MIC in Taiwan. J Clin Microbiol 50:1679–1683. doi: 10.1128/JCM.06711-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Pu WX, Su Y, Li JX, Li CH, Yang ZQ, Deng HP, Ni CX. 2014. High incidence of oxacillin-susceptible mecA-positive Staphylococcus aureus (OS-MRSA) associated with bovine mastitis in China. PLoS One 9:e88134. doi: 10.1371/journal.pone.0088134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Baig S, Johannesen TB, Overballe-Petersen S, Larsen J, Larsen AR, Stegger M. 2018. Novel SCCmec type XIII (9A) identified in an ST152 methicillin-resistant Staphylococcus aureus. Infect Genet Evol 61:74–76. doi: 10.1016/j.meegid.2018.03.013. [DOI] [PubMed] [Google Scholar]
- 180.Becker K, van Alen S, Idelevich EA, Schleimer N, Seggewiß J, Mellmann A, Kaspar U, Peters G. 2018. Plasmid-encoded transferable mecB-mediated methicillin resistance in Staphylococcus aureus. Emerg Infect Dis 24:242–248. doi: 10.3201/eid2402.171074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Garcia-Alvarez L, Holden MT, Lindsay H, Webb CR, Brown DF, Curran MD, Walpole E, Brooks K, Pickard DJ, Teale C, Parkhill J, Bentley SD, Edwards GF, Girvan EK, Kearns AM, Pichon B, Hill RL, Larsen AR, Skov RL, Peacock SJ, Maskell DJ, Holmes MA. 2011. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect Dis 11:595–603. doi: 10.1016/S1473-3099(11)70126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Paterson GK. 13 January 2020. Low prevalence of livestock-associated methicillin-resistant Staphylococcus aureus clonal complex 398 and mecC MRSA among human isolates in North-West England. J Appl Microbiol doi: 10.1111/jam.14578. [DOI] [PubMed] [Google Scholar]
- 183.Paterson GK, Harrison EM, Holmes MA. 2014. The emergence of mecC methicillin-resistant Staphylococcus aureus. Trends Microbiol 22:42–47. doi: 10.1016/j.tim.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Arias CA, Murray BE. 2012. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol 10:266–278. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zapun A, Contreras-Martel C, Vernet T. 2008. Penicillin-binding proteins and beta-lactam resistance. FEMS Microbiol Rev 32:361–385. doi: 10.1111/j.1574-6976.2007.00095.x. [DOI] [PubMed] [Google Scholar]
- 186.Poirel L, Nordmann P. 2006. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin Microbiol Infect 12:826–836. doi: 10.1111/j.1469-0691.2006.01456.x. [DOI] [PubMed] [Google Scholar]
- 187.Hooper DC. 2001. Emerging mechanisms of fluoroquinolone resistance. Emerg Infect Dis 7:337–341. doi: 10.3201/eid0702.010239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Laponogov I, Sohi MK, Veselkov DA, Pan XS, Sawhney R, Thompson AW, McAuley KE, Fisher LM, Sanderson MR. 2009. Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases. Nat Struct Mol Biol 16:667–669. doi: 10.1038/nsmb.1604. [DOI] [PubMed] [Google Scholar]
- 189.Morais Cabral JH, Jackson AP, Smith CV, Shikotra N, Maxwell A, Liddington RC. 1997. Crystal structure of the breakage-reunion domain of DNA gyrase. Nature 388:903–906. doi: 10.1038/42294. [DOI] [PubMed] [Google Scholar]
- 190.Wohlkonig A, Chan PF, Fosberry AP, Homes P, Huang J, Kranz M, Leydon VR, Miles TJ, Pearson ND, Perera RL, Shillings AJ, Gwynn MN, Bax BD. 2010. Structural basis of quinolone inhibition of type IIA topoisomerases and target-mediated resistance. Nat Struct Mol Biol 17:1152–1153. doi: 10.1038/nsmb.1892. [DOI] [PubMed] [Google Scholar]
- 191.Schmitz FJ, Jones ME, Hofmann B, Hansen B, Scheuring S, Luckefahr M, Fluit A, Verhoef J, Hadding U, Heinz HP, Kohrer K. 1998. Characterization of grlA, grlB, gyrA, and gyrB mutations in 116 unrelated isolates of Staphylococcus aureus and effects of mutations on ciprofloxacin MIC. Antimicrob Agents Chemother 42:1249–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Yoshida H, Bogaki M, Nakamura M, Yamanaka LM, Nakamura S. 1991. Quinolone resistance-determining region in the DNA gyrase gyrB gene of Escherichia coli. Antimicrob Agents Chemother 35:1647–1650. doi: 10.1128/aac.35.8.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Komp Lindgren P, Karlsson A, Hughes D. 2003. Mutation rate and evolution of fluoroquinolone resistance in Escherichia coli isolates from patients with urinary tract infections. Antimicrob Agents Chemother 47:3222–3232. doi: 10.1128/aac.47.10.3222-3232.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Robicsek A, Jacoby GA, Hooper DC. 2006. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect Dis 6:629–640. doi: 10.1016/S1473-3099(06)70599-0. [DOI] [PubMed] [Google Scholar]
- 195.Ruiz J. 2019. Transferable mechanisms of quinolone resistance from 1998 onward. Clin Microbiol Rev 32:e00007-19. doi: 10.1128/CMR.00007-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Horinouchi S, Weisblum B. 1980. Posttranscriptional modification of mRNA conformation: mechanism that regulates erythromycin-induced resistance. Proc Natl Acad Sci U S A 77:7079–7083. doi: 10.1073/pnas.77.12.7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Weisblum B. 1995. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother 39:577–585. doi: 10.1128/aac.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Munita JM, Arias CA. 2016. Mechanisms of antibiotic resistance. Microbiol Spectr 4:VMBF-0016-2015. doi: 10.1128/microbiolspec.VMBF-0016-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Murphy E, Huwyler L, de Freire Bastos MDC. 1985. Transposon Tn554: complete nucleotide sequence and isolation of transposition-defective and antibiotic-sensitive mutants. EMBO J 4:3357–3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Shaw KJ, Barbachyn MR. 2011. The oxazolidinones: past, present, and future. Ann N Y Acad Sci 1241:48–70. doi: 10.1111/j.1749-6632.2011.06330.x. [DOI] [PubMed] [Google Scholar]
- 201.Locke JB, Zuill DE, Scharn CR, Deane J, Sahm DF, Denys GA, Goering RV, Shaw KJ. 2014. Linezolid-resistant Staphylococcus aureus strain 1128105, the first known clinical isolate possessing the cfr multidrug resistance gene. Antimicrob Agents Chemother 58:6592–6598. doi: 10.1128/AAC.03493-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Mendes RE, Deshpande LM, Castanheira M, DiPersio J, Saubolle MA, Jones RN. 2008. First report of cfr-mediated resistance to linezolid in human staphylococcal clinical isolates recovered in the United States. Antimicrob Agents Chemother 52:2244–2246. doi: 10.1128/AAC.00231-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Locke JB, Morales G, Hilgers M, Kedar GC, Rahawi S, Jose Picazo J, Shaw KJ, Stein JL. 2010. Elevated linezolid resistance in clinical cfr-positive Staphylococcus aureus isolates is associated with co-occurring mutations in ribosomal protein L3. Antimicrob Agents Chemother 54:5352–5355. doi: 10.1128/AAC.00714-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Cox G, Ejim L, Stogios PJ, Koteva K, Bordeleau E, Evdokimova E, Sieron AO, Savchenko A, Serio AW, Krause KM, Wright GD. 2018. Plazomicin retains antibiotic activity against most aminoglycoside modifying enzymes. ACS Infect Dis 4:980–987. doi: 10.1021/acsinfecdis.8b00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Doi Y, Wachino J-I, Arakawa Y. 2016. Aminoglycoside resistance: the emergence of acquired 16S ribosomal RNA methyltransferases. Infect Dis Clin North Am 30:523–537. doi: 10.1016/j.idc.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Kristich CJ, Rice LB, Arias CA. 2014. Enterococcal infection—treatment and antibiotic resistance In Gilmore MS, Clewell DB, Ike Y, Shankar N (ed), Enterococci: from commensals to leading causes of drug resistant infection. Massachusetts Eye and Ear Infirmary, Boston, MA. [PubMed] [Google Scholar]
- 207.Arthur M, Molinas C, Depardieu F, Courvalin P. 1993. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol 175:117–127. doi: 10.1128/jb.175.1.117-127.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Carias LL, Rudin SD, Donskey CJ, Rice LB. 1998. Genetic linkage and cotransfer of a novel, vanB-containing transposon (Tn5382) and a low-affinity penicillin-binding protein 5 gene in a clinical vancomycin-resistant Enterococcus faecium isolate. J Bacteriol 180:4426–4434. doi: 10.1128/JB.180.17.4426-4434.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Quintiliani R Jr, Courvalin P. 1996. Characterization of Tn1547, a composite transposon flanked by the IS16 and IS256-like elements, that confers vancomycin resistance in Enterococcus faecalis BM4281. Gene 172:1–8. doi: 10.1016/0378-1119(96)00110-2. [DOI] [PubMed] [Google Scholar]
- 210.Perichon B, Courvalin P. 2009. VanA-type vancomycin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 53:4580–4587. doi: 10.1128/AAC.00346-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.McGuinness WA, Malachowa N, DeLeo FR. 2017. Vancomycin resistance in Staphylococcus aureus. Yale J Biol Med 90:269–281. [PMC free article] [PubMed] [Google Scholar]
- 212.Melo-Cristino J, Resina C, Manuel V, Lito L, Ramirez M. 2013. First case of infection with vancomycin-resistant Staphylococcus aureus in Europe. Lancet 382:205. doi: 10.1016/S0140-6736(13)61219-2. [DOI] [PubMed] [Google Scholar]
- 213.Panesso D, Planet PJ, Diaz L, Hugonnet JE, Tran TT, Narechania A, Munita JM, Rincon S, Carvajal LP, Reyes J, Londono A, Smith H, Sebra R, Deikus G, Weinstock GM, Murray BE, Rossi F, Arthur M, Arias CA. 2015. Methicillin-susceptible, vancomycin-resistant Staphylococcus aureus, Brazil. Emerg Infect Dis 21:1844–1848. doi: 10.3201/eid2110.141914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Rossi F, Diaz L, Wollam A, Panesso D, Zhou Y, Rincon S, Narechania A, Xing G, Di Gioia TSR, Doi A, Tran TT, Reyes J, Munita JM, Carvajal LP, Hernandez-Roldan A, Brandão D, van der Heijden IM, Murray BE, Planet PJ, Weinstock GM, Arias CA. 2014. Transferable vancomycin resistance in a community-associated MRSA lineage. N Engl J Med 370:1524–1531. doi: 10.1056/NEJMoa1303359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Division of Healthcare Quality Promotion, Centers for Disease Control and Prevention. 2015. Investigation and control of vancomycin-resistant Staphylococcus aureus (VRSA): 2015 update. https://www.cdc.gov/hai/pdfs/VRSA-Investigation-Guide-05_12_2015.pdf. Accessed 5 February 2020.
- 216.Foucault ML, Courvalin P, Grillot-Courvalin C. 2009. Fitness cost of VanA-type vancomycin resistance in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 53:2354–2359. doi: 10.1128/AAC.01702-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Zhu WM, Clark N, Patel JB. 2013. pSK41-like plasmid is necessary for Inc18-like vanA plasmid transfer from Enterococcus faecalis to Staphylococcus aureus in vitro. Antimicrob Agents Chemother 57:212–219. doi: 10.1128/AAC.01587-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Zhu W, Murray PR, Huskins WC, Jernigan JA, McDonald LC, Clark NC, Anderson KF, McDougal LK, Hageman JC, Olsen-Rasmussen M, Frace M, Alangaden GJ, Chenoweth C, Zervos MJ, Robinson-Dunn B, Schreckenberger PC, Reller LB, Rudrik JT, Patel JB. 2010. Dissemination of an Enterococcus Inc18-like vanA plasmid associated with vancomycin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 54:4314–4320. doi: 10.1128/AAC.00185-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Kos VN, Desjardins CA, Griggs A, Cerqueira G, Van Tonder A, Holden MTG, Godfrey P, Palmer KL, Bodi K, Mongodin EF, Wortman J, Feldgarden M, Lawley T, Gill SR, Haas BJ, Birren B, Gilmore MS. 2012. Comparative genomics of vancomycin-resistant Staphylococcus aureus strains and their positions within the clade most commonly associated with methicillin-resistant S. aureus hospital-acquired infection in the United States. mBio 3:e00112-12. doi: 10.1128/mBio.00112-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Howden BP, Davies JK, Johnson PD, Stinear TP, Grayson ML. 2010. Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications. Clin Microbiol Rev 23:99–139. doi: 10.1128/CMR.00042-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Boyle-Vavra S, Challapalli M, Daum RS. 2003. Resistance to autolysis in vancomycin-selected Staphylococcus aureus isolates precedes vancomycin-intermediate resistance. Antimicrob Agents Chemother 47:2036–2039. doi: 10.1128/aac.47.6.2036-2039.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Cui L, Iwamoto A, Lian JQ, Neoh HM, Maruyama T, Horikawa Y, Hiramatsu K. 2006. Novel mechanism of antibiotic resistance originating in vancomycin-intermediate Staphylococcus aureus. Antimicrob Agents Chemother 50:428–438. doi: 10.1128/AAC.50.2.428-438.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Cui L, Ma X, Sato K, Okuma K, Tenover FC, Mamizuka EM, Gemmell CG, Kim MN, Ploy MC, El-Solh N, Ferraz V, Hiramatsu K. 2003. Cell wall thickening is a common feature of vancomycin resistance in Staphylococcus aureus. J Clin Microbiol 41:5–14. doi: 10.1128/JCM.41.1.5-14.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Renzoni A, Barras C, Francois P, Charbonnier Y, Huggler E, Garzoni C, Kelley WL, Majcherczyk P, Schrenzel J, Lew DP, Vaudaux P. 2006. Transcriptomic and functional analysis of an autolysis-deficient, teicoplanin-resistant derivative of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 50:3048–3061. doi: 10.1128/AAC.00113-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Howden BP, Peleg AY, Stinear TP. 2014. The evolution of vancomycin intermediate Staphylococcus aureus (VISA) and heterogenous-VISA. Infect Genet Evol 21:575–582. doi: 10.1016/j.meegid.2013.03.047. [DOI] [PubMed] [Google Scholar]
- 226.Zhang SS, Sun XX, Chang WJ, Dai YY, Ma XL. 2015. Systematic review and meta-analysis of the epidemiology of vancomycin-intermediate and heterogeneous vancomycin-intermediate Staphylococcus aureus isolates. PLoS One 10:e0136082. doi: 10.1371/journal.pone.0136082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Miller WR, Bayer AS, Arias CA. 2016. Mechanism of action and resistance to daptomycin in Staphylococcus aureus and enterococci. Cold Spring Harb Perspect Med 6:a026997. doi: 10.1101/cshperspect.a026997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Kidd TJ, Mills G, Sa-Pessoa J, Dumigan A, Frank CG, Insua JL, Ingram R, Hobley L, Bengoechea JA. 2017. A Klebsiella pneumoniae antibiotic resistance mechanism that subdues host defences and promotes virulence. EMBO Mol Med 9:430–447. doi: 10.15252/emmm.201607336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Poirel L, Jayol A, Nordmann P. 2017. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev 30:557–596. doi: 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Wright MS, Suzuki Y, Jones MB, Marshall SH, Rudin SD, van Duin D, Kaye K, Jacobs MR, Bonomo RA, Adams MD. 2015. Genomic and transcriptomic analyses of colistin-resistant clinical isolates of Klebsiella pneumoniae reveal multiple pathways of resistance. Antimicrob Agents Chemother 59:536–543. doi: 10.1128/AAC.04037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Quinn JP, Dudek EJ, DiVincenzo CA, Lucks DA, Lerner SA. 1986. Emergence of resistance to imipenem during therapy for Pseudomonas aeruginosa infections. J Infect Dis 154:289–294. doi: 10.1093/infdis/154.2.289. [DOI] [PubMed] [Google Scholar]
- 232.Hasdemir UO, Chevalier J, Nordmann P, Pages JM. 2004. Detection and prevalence of active drug efflux mechanism in various multidrug-resistant Klebsiella pneumoniae strains from Turkey. J Clin Microbiol 42:2701–2706. doi: 10.1128/JCM.42.6.2701-2706.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Pages JM, James CE, Winterhalter M. 2008. The porin and the permeating antibiotic: a selective diffusion barrier in Gram-negative bacteria. Nat Rev Microbiol 6:893–903. doi: 10.1038/nrmicro1994. [DOI] [PubMed] [Google Scholar]
- 234.Li XZ, Plesiat P, Nikaido H. 2015. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin Microbiol Rev 28:337–418. doi: 10.1128/CMR.00117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Davin-Regli A, Lavigne JP, Pages JM. 2019. Enterobacter spp.: update on taxonomy, clinical aspect, and emerging antimicrobial resistance. Clin Microbiol Rev 32:e00002-19. doi: 10.1128/CMR.00002-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Masi M, Winterhalter M, Pages JM. 2019. Outer membrane porins. Subcell Biochem 92:79–123. doi: 10.1007/978-3-030-18768-2_4. [DOI] [PubMed] [Google Scholar]
- 237.Hassan KA, Liu Q, Henderson PJ, Paulsen IT. 2015. Homologs of the Acinetobacter baumannii AceI transporter represent a new family of bacterial multidrug efflux systems. mBio 6:e01982-14. doi: 10.1128/mBio.01982-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Su CC, Morgan CE, Kambakam S, Rajavel M, Scott H, Huang W, Emerson CC, Taylor DJ, Stewart PL, Bonomo RA, Yu EW. 2019. Cryo-electron microscopy structure of an Acinetobacter baumannii multidrug efflux pump. mBio 10:e01295-19. doi: 10.1128/mBio.01295-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Leus IV, Weeks JW, Bonifay V, Smith L, Richardson S, Zgurskaya HI. 2018. Substrate specificities and efflux efficiencies of RND efflux pumps of Acinetobacter baumannii. J Bacteriol 200:e00049-18. doi: 10.1128/JB.00049-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Pagdepanichkit S, Tribuddharat C, Chuanchuen R. 2016. Distribution and expression of the Ade multidrug efflux systems in Acinetobacter baumannii clinical isolates. Can J Microbiol 62:794–801. doi: 10.1139/cjm-2015-0730. [DOI] [PubMed] [Google Scholar]
- 241.Wong MHY, Chan EWC, Chen S. 2015. Evolution and dissemination of OqxAB-like efflux pumps, an emerging quinolone resistance determinant among members of Enterobacteriaceae. Antimicrob Agents Chemother 59:3290–3297. doi: 10.1128/AAC.00310-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Veleba M, Schneiders T. 2012. Tigecycline resistance can occur independently of the ramA gene in Klebsiella pneumoniae. Antimicrob Agents Chemother 56:4466–4467. doi: 10.1128/AAC.06224-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Hill D, Rose B, Pajkos A, Robinson M, Bye P, Bell S, Elkins M, Thompson B, MacLeod C, Aaron SD, Harbour C. 2005. Antibiotic susceptibilities of Pseudomonas aeruginosa isolates derived from patients with cystic fibrosis under aerobic, anaerobic, and biofilm conditions. J Clin Microbiol 43:5085–5090. doi: 10.1128/JCM.43.10.5085-5090.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Nickel JC, Ruseska I, Wright JB, Costerton JW. 1985. Tobramycin resistance of Pseudomonas aeruginosa cells growing as a biofilm on urinary catheter material. Antimicrob Agents Chemother 27:619–624. doi: 10.1128/aac.27.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Percival SL, Suleman L, Vuotto C, Donelli G. 2015. Healthcare-associated infections, medical devices and biofilms: risk, tolerance and control. J Med Microbiol 64:323–334. doi: 10.1099/jmm.0.000032. [DOI] [PubMed] [Google Scholar]
- 246.Hall CW, Mah TF. 2017. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol Rev 41:276–301. doi: 10.1093/femsre/fux010. [DOI] [PubMed] [Google Scholar]
- 247.Schembri MA, Hjerrild L, Gjermansen M, Klemm P. 2003. Differential expression of the Escherichia coli autoaggregation factor antigen 43. J Bacteriol 185:2236–2242. doi: 10.1128/jb.185.7.2236-2242.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Secor PR, Michaels LA, Ratjen A, Jennings LK, Singh PK. 2018. Entropically driven aggregation of bacteria by host polymers promotes antibiotic tolerance in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 115:10780–10785. doi: 10.1073/pnas.1806005115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Secor PR, Sweere JM, Michaels LA, Malkovskiy AV, Lazzareschi D, Katznelson E, Rajadas J, Birnbaum ME, Arrigoni A, Braun KR, Evanko SP, Stevens DA, Kaminsky W, Singh PK, Parks WC, Bollyky PL. 2015. Filamentous bacteriophage promote biofilm assembly and function. Cell Host Microbe 18:549–559. doi: 10.1016/j.chom.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Weigel LM, Donlan RM, Shin DH, Jensen B, Clark NC, McDougal LK, Zhu W, Musser KA, Thompson J, Kohlerschmidt D, Dumas N, Limberger RJ, Patel JB. 2007. High-level vancomycin-resistant Staphylococcus aureus isolates associated with a polymicrobial biofilm. Antimicrob Agents Chemother 51:231–238. doi: 10.1128/AAC.00576-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Brauner A, Fridman O, Gefen O, Balaban NQ. 2016. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat Rev Microbiol 14:320–330. doi: 10.1038/nrmicro.2016.34. [DOI] [PubMed] [Google Scholar]
- 252.Balaban NQ, Helaine S, Lewis K, Ackermann M, Aldridge B, Andersson DI, Brynildsen MP, Bumann D, Camilli A, Collins JJ, Dehio C, Fortune S, Ghigo JM, Hardt WD, Harms A, Heinemann M, Hung DT, Jenal U, Levin BR, Michiels J, Storz G, Tan MW, Tenson T, Van Melderen L, Zinkernagel A. 2019. Definitions and guidelines for research on antibiotic persistence. Nat Rev Microbiol 17:441–448. doi: 10.1038/s41579-019-0196-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Liu JF, Gefen O, Ronin I, Bar-Meir M, Balaban NQ. 2020. Effect of tolerance on the evolution of antibiotic resistance under drug combinations. Science 367:200–204. doi: 10.1126/science.aaj2191. [DOI] [PubMed] [Google Scholar]
- 254.Brauner A, Shoresh N, Fridman O, Balaban NQ. 2017. An experimental framework for quantifying bacterial tolerance. Biophys J 112:2664–2671. doi: 10.1016/j.bpj.2017.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Yan J, Bassler BL. 2019. Surviving as a community: antibiotic tolerance and persistence in bacterial biofilms. Cell Host Microbe 26:15–21. doi: 10.1016/j.chom.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Cohen NR, Lobritz MA, Collins JJ. 2013. Microbial persistence and the road to drug resistance. Cell Host Microbe 13:632–642. doi: 10.1016/j.chom.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Cano V, March C, Insua JL, Aguilo N, Llobet E, Moranta D, Regueiro V, Brennan GP, Millan-Lou MI, Martin C, Garmendia J, Bengoechea JA. 2015. Klebsiella pneumoniae survives within macrophages by avoiding delivery to lysosomes. Cell Microbiol 17:1537–1560. doi: 10.1111/cmi.12466. [DOI] [PubMed] [Google Scholar]
- 258.Zou J, Shankar N. 2016. The opportunistic pathogen Enterococcus faecalis resists phagosome acidification and autophagy to promote intracellular survival in macrophages. Cell Microbiol 18:831–843. doi: 10.1111/cmi.12556. [DOI] [PubMed] [Google Scholar]
- 259.Fraunholz M, Sinha B. 2012. Intracellular Staphylococcus aureus: live-in and let die. Front Cell Infect Microbiol 2:43. doi: 10.3389/fcimb.2012.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Lowy FD. 2000. Is Staphylococcus aureus an intracellular pathogen? Trends Microbiol 8:341–343. doi: 10.1016/s0966-842x(00)01803-5. [DOI] [PubMed] [Google Scholar]
- 261.Lehar SM, Pillow T, Xu M, Staben L, Kajihara KK, Andlen RV, DePalatis L, Raab H, Hazenbos WL, Morisaki JH, Kim J, Park S, Darwish M, Lee BC, Hernandez H, Loyet KM, Lupardus P, Fong RN, Yan DH, Halouni CC, Luis E, Khalfin Y, Plise E, Heong JC, Lyssikatos JP, Strandh M, Koefoed K, Andersen PS, Flygare JA, Tan MW, Brown EJ, Ariathasan SM. 2015. Novel antibody-antibiotic conjugate eliminates intracellular S. aureus. Nature 527:323–328. doi: 10.1038/nature16057. [DOI] [PubMed] [Google Scholar]
- 262.Partridge SR, Kwong SM, Firth N, Jensen SO. 2018. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev 31:e00088-17. doi: 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Siguier P, Gourbeyre E, Varani A, Ton-Hoang B, Chandler M. 2015. Everyman's guide to bacterial insertion sequences. Microbiol Spectr 3:MDNA3-0030-2014. doi: 10.1128/microbiolspec.MDNA3-0030-2014. [DOI] [PubMed] [Google Scholar]
- 264.Vandecraen J, Chandler M, Aertsen A, Van Houdt R. 2017. The impact of insertion sequences on bacterial genome plasticity and adaptability. Crit Rev Microbiol 43:709–730. doi: 10.1080/1040841X.2017.1303661. [DOI] [PubMed] [Google Scholar]
- 265.Haniford DB, Ellis MJ. 2015. Transposons Tn10 and Tn5. Microbiol Spectr 3:MDNA3-0002-2014. doi: 10.1128/microbiolspec.MDNA3-0002-2014. [DOI] [PubMed] [Google Scholar]
- 266.Bennett PM. 2008. Plasmid encoded antibiotic resistance: acquisition and transfer of antibiotic resistance genes in bacteria. Br J Pharmacol 153(Suppl 1):S347–S357. doi: 10.1038/sj.bjp.0707607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Weigel LM, Clewell DB, Gill SR, Clark NC, McDougal LK, Flannagan SE, Kolonay JF, Shetty J, Killgore GE, Tenover FC. 2003. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science 302:1569–1571. doi: 10.1126/science.1090956. [DOI] [PubMed] [Google Scholar]
- 268.Partridge SR. 2015. What's in a name? ISSwi1 corresponds to transposons related to Tn2 and Tn3. mBio 6:e01344-15. doi: 10.1128/mBio.01344-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Nicolas E, Lambin M, Dandoy D, Galloy C, Nguyen N, Oger CA, Hallet B. 2015. The Tn3-family of replicative transposons. Microbiol Spectr 3:MDNA3-0060-2014. doi: 10.1128/microbiolspec.MDNA3-0060-2014. [DOI] [PubMed] [Google Scholar]
- 270.Post V, White PA, Hall RM. 2010. Evolution of AbaR-type genomic resistance islands in multiply antibiotic-resistant Acinetobacter baumannii. J Antimicrob Chemother 65:1162–1170. doi: 10.1093/jac/dkq095. [DOI] [PubMed] [Google Scholar]
- 271.Hamidian M, Hall RM. 2017. Origin of the AbGRI1 antibiotic resistance island found in the comM gene of Acinetobacter baumannii GC2 isolates. J Antimicrob Chemother 72:2944–2947. doi: 10.1093/jac/dkx206. [DOI] [PubMed] [Google Scholar]
- 272.Gregory PD, Lewis RA, Curnock SP, Dyke KG. 1997. Studies of the repressor (BlaI) of beta-lactamase synthesis in Staphylococcus aureus. Mol Microbiol 24:1025–1037. doi: 10.1046/j.1365-2958.1997.4051770.x. [DOI] [PubMed] [Google Scholar]
- 273.Poirel L, Lartigue MF, Decousser JW, Nordmann P. 2005. ISEcp1B-mediated transposition of blaCTX-M in Escherichia coli. Antimicrob Agents Chemother 49:447–450. doi: 10.1128/AAC.49.1.447-450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Karim A, Poirel L, Nagarajan S, Nordmann P. 2001. Plasmid-mediated extended-spectrum beta-lactamase (CTX-M-3 like) from India and gene association with insertion sequence ISEcp1. FEMS Microbiol Lett 201:237–241. doi: 10.1111/j.1574-6968.2001.tb10762.x. [DOI] [PubMed] [Google Scholar]
- 275.Bauernfeind A, Schneider I, Jungwirth R, Sahly H, Ullmann U. 1999. A novel type of AmpC beta-lactamase, ACC-1, produced by a Klebsiella pneumoniae strain causing nosocomial pneumonia. Antimicrob Agents Chemother 43:1924–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Zowawi HM, Forde BM, Alfaresi M, Alzarouni A, Farahat Y, Chong TM, Yin WF, Chan KG, Li J, Schembri MA, Beatson SA, Paterson DL. 2015. Stepwise evolution of pandrug-resistance in Klebsiella pneumoniae. Sci Rep 5:15082. doi: 10.1038/srep15082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Chen YG, Qu TT, Yu YS, Zhou JY, Li LJ. 2006. Insertion sequence ISEcp1-like element connected with a novel aph(2ʺ) allele [aph(2ʺ)-Ie] conferring high-level gentamicin resistance and a novel streptomycin adenylyltransferase gene in Enterococcus. J Med Microbiol 55:1521–1525. doi: 10.1099/jmm.0.46702-0. [DOI] [PubMed] [Google Scholar]
- 278.Partridge SR. 2016. Mobilization of blaBKC-1 by ISKpn23? Antimicrob Agents Chemother 60:5102–5104. doi: 10.1128/AAC.00785-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Harmer CJ, Hall RM. 2016. IS26-mediated formation of transposons carrying antibiotic resistance genes. mSphere 1:e00038-16. doi: 10.1128/mSphere.00038-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Toleman MA, Bennett PM, Walsh TR. 2006. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol Mol Biol Rev 70:296–316. doi: 10.1128/MMBR.00048-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Lee CH, Chu C, Liu JW, Chen YS, Chiu CJ, Su LH. 2007. Collateral damage of flomoxef therapy: in vivo development of porin deficiency and acquisition of blaDHA-1 leading to ertapenem resistance in a clinical isolate of Klebsiella pneumoniae producing CTX-M-3 and SHV-5 beta-lactamases. J Antimicrob Chemother 60:410–413. doi: 10.1093/jac/dkm215. [DOI] [PubMed] [Google Scholar]
- 282.Cannatelli A, Giani T, D'Andrea MM, Di Pilato V, Arena F, Conte V, Tryfinopoulou K, Vatopoulos A, Rossolini GM, COLGRIT Study Group . 2014. MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob Agents Chemother 58:5696–5703. doi: 10.1128/AAC.03110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Chen TL, Wu RC, Shaio MF, Fung CP, Cho WL. 2008. Acquisition of a plasmid-borne blaOXA-58 gene with an upstream IS1008 insertion conferring a high level of carbapenem resistance to Acinetobacter baumannii. Antimicrob Agents Chemother 52:2573–2580. doi: 10.1128/AAC.00393-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Aubert D, Naas T, Heritier C, Poirel L, Nordmann P. 2006. Functional characterization of IS1999, an IS4 family element involved in mobilization and expression of beta-lactam resistance genes. J Bacteriol 188:6506–6514. doi: 10.1128/JB.00375-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Aubert D, Naas T, Nordmann P. 2003. IS1999 increases expression of the extended-spectrum beta-lactamase VEB-1 in Pseudomonas aeruginosa. J Bacteriol 185:5314–5319. doi: 10.1128/jb.185.17.5314-5319.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Depardieu F, Podglajen I, Leclercq R, Collatz E, Courvalin P. 2007. Modes and modulations of antibiotic resistance gene expression. Clin Microbiol Rev 20:79–114. doi: 10.1128/CMR.00015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Naas T, Philippon L, Poirel L, Ronco E, Nordmann P. 1999. An SHV-derived extended-spectrum beta-lactamase in Pseudomonas aeruginosa. Antimicrob Agents Chemother 43:1281–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Lee KY, Hopkins JD, Syvanen M. 1990. Direct involvement of IS26 in an antibiotic resistance operon. J Bacteriol 172:3229–3236. doi: 10.1128/jb.172.6.3229-3236.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Aminov RI. 2011. Horizontal gene exchange in environmental microbiota. Front Microbiol 2:158. doi: 10.3389/fmicb.2011.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Shintani M, Sanchez ZK, Kimbara K. 2015. Genomics of microbial plasmids: classification and identification based on replication and transfer systems and host taxonomy. Front Microbiol 6:242. doi: 10.3389/fmicb.2015.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Guglielmini J, Quintais L, Garcillan-Barcia MP, de la Cruz F, Rocha EP. 2011. The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet 7:e1002222. doi: 10.1371/journal.pgen.1002222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Thomas CM, Nielsen KM. 2005. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol 3:711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 293.Carattoli A. 2009. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother 53:2227–2238. doi: 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Rozwandowicz M, Brouwer MSM, Fischer J, Wagenaar JA, Gonzalez-Zorn B, Guerra B, Mevius DJ, Hordijk J. 2018. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J Antimicrob Chemother 73:1121–1137. doi: 10.1093/jac/dkx488. [DOI] [PubMed] [Google Scholar]
- 295.Pitout J. 2010. Infections with extended-spectrum beta-lactamase-producing Enterobacteriaceae changing epidemiology and drug treatment choices. Drugs 70:313–333. doi: 10.2165/11533040-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 296.Petty NK, Ben Zakour NL, Stanton-Cook M, Skippington E, Totsika M, Forde BM, Phan MD, Moriel DG, Peters KM, Davies M, Rogers BA, Dougan G, Rodriguez-Bano J, Pascual A, Pitout JDD, Upton M, Paterson DL, Walsh TR, Schembri MA, Beatson SA. 2014. Global dissemination of a multidrug resistant Escherichia coli clone. Proc Natl Acad Sci U S A 111:5694–5699. doi: 10.1073/pnas.1322678111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Bauernfeind A, Chong Y, Schweighart S. 1989. Extended broad-spectrum beta-lactamase in Klebsiella pneumoniae including resistance to cephamycins. Infection 17:316–321. doi: 10.1007/bf01650718. [DOI] [PubMed] [Google Scholar]
- 298.Philippon A, Arlet G, Jacoby GA. 2002. Plasmid-determined AmpC-type beta-lactamases. Antimicrob Agents Chemother 46:1–11. doi: 10.1128/aac.46.1.1-11.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Hennequin C, Chlilek A, Beyrouthy R, Bonnet R, Robin F. 2018. Diversity of DHA-1-encoding plasmids in Klebsiella pneumoniae isolates from 16 French hospitals. J Antimicrob Chemother 73:2981–2989. doi: 10.1093/jac/dky285. [DOI] [PubMed] [Google Scholar]
- 300.Deshpande LM, Jones RN, Fritsche TR, Sader HS. 2006. Occurrence and characterization of carbapenemase-producing Enterobacteriaceae: report from the SENTRY antimicrobial surveillance program (2000–2004). Microb Drug Resist 12:223–230. doi: 10.1089/mdr.2006.12.223. [DOI] [PubMed] [Google Scholar]
- 301.Walsh TR, Weeks J, Livermore DM, Toleman MA. 2011. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis 11:355–362. doi: 10.1016/S1473-3099(11)70059-7. [DOI] [PubMed] [Google Scholar]
- 302.Towner KJ, Evans B, Villa L, Levi K, Hamouda A, Amyes SG, Carattoli A. 2011. Distribution of intrinsic plasmid replicase genes and their association with carbapenem-hydrolyzing class D beta-lactamase genes in European clinical isolates of Acinetobacter baumannii. Antimicrob Agents Chemother 55:2154–2159. doi: 10.1128/AAC.01661-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Lean SS, Yeo CC. 2017. Small, enigmatic plasmids of the nosocomial pathogen, Acinetobacter baumannii: good, bad, who knows? Front Microbiol 8:1547. doi: 10.3389/fmicb.2017.01547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Hamidian M, Nigro SJ, Hall RM. 2012. Variants of the gentamicin and tobramycin resistance plasmid pRAY are widely distributed in Acinetobacter. J Antimicrob Chemother 67:2833–2836. doi: 10.1093/jac/dks318. [DOI] [PubMed] [Google Scholar]
- 305.Hamidian M, Holt KE, Pickard D, Dougan G, Hall RM. 2014. A GC1 Acinetobacter baumannii isolate carrying AbaR3 and the aminoglycoside resistance transposon TnaphA6 in a conjugative plasmid. J Antimicrob Chemother 69:955–958. doi: 10.1093/jac/dkt454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Hamidian M, Hall RM. 2014. pACICU2 is a conjugative plasmid of Acinetobacter carrying the aminoglycoside resistance transposon TnaphA6. J Antimicrob Chemother 69:1146–1148. doi: 10.1093/jac/dkt488. [DOI] [PubMed] [Google Scholar]
- 307.Hu H, Hu Y, Pan Y, Liang H, Wang H, Wang X, Hao Q, Yang X, Yang X, Xiao X, Luan C, Yang Y, Cui Y, Yang R, Gao GF, Song Y, Zhu B. 2012. Novel plasmid and its variant harboring both a bla(NDM-1) gene and type IV secretion system in clinical isolates of Acinetobacter lwoffii. Antimicrob Agents Chemother 56:1698–1702. doi: 10.1128/AAC.06199-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Jacoby GA, Sutton L, Knobel L, Mammen P. 1983. Properties of IncP-2 plasmids of Pseudomonas spp. Antimicrob Agents Chemother 24:168–175. doi: 10.1128/aac.24.2.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Liu J, Yang L, Chen D, Peters BM, Li L, Li B, Xu Z, Shirtliff ME. 2018. Complete sequence of pBM413, a novel multidrug resistance megaplasmid carrying qnrVC6 and blaIMP-45 from Pseudomonas aeruginosa. Int J Antimicrob Agents 51:145–150. doi: 10.1016/j.ijantimicag.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 310.Xiong J, Alexander DC, Ma JH, Deraspe M, Low DE, Jamieson FB, Roy PH. 2013. Complete sequence of pOZ176, a 500-kilobase IncP-2 plasmid encoding IMP-9-mediated carbapenem resistance, from outbreak isolate Pseudomonas aeruginosa 96. Antimicrob Agents Chemother 57:3775–3782. doi: 10.1128/AAC.00423-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Botelho J, Grosso F, Quinteira S, Mabrouk A, Peixe L. 2017. The complete nucleotide sequence of an IncP-2 megaplasmid unveils a mosaic architecture comprising a putative novel blaVIM-2-harbouring transposon in Pseudomonas aeruginosa. J Antimicrob Chemother 72:2225–2229. doi: 10.1093/jac/dkx143. [DOI] [PubMed] [Google Scholar]
- 312.Shearer JE, Wireman J, Hostetler J, Forberger H, Borman J, Gill J, Sanchez S, Mankin A, Lamarre J, Lindsay JA, Bayles K, Nicholson A, O'Brien F, Jensen SO, Firth N, Skurray RA, Summers AO. 2011. Major families of multiresistant plasmids from geographically and epidemiologically diverse staphylococci. G3 (Bethesda) 1:581–591. doi: 10.1534/g3.111.000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Malachowa N, DeLeo FR. 2010. Mobile genetic elements of Staphylococcus aureus. Cell Mol Life Sci 67:3057–3071. doi: 10.1007/s00018-010-0389-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Mojumdar M, Khan SA. 1988. Characterization of the tetracycline resistance gene of plasmid pT181 of Staphylococcus aureus. J Bacteriol 170:5522–5528. doi: 10.1128/jb.170.12.5522-5528.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Horinouchi S, Weisblum B. 1982. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol 150:815–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Horinouchi S, Weisblum B. 1982. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibodies. J Bacteriol 150:804–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Jensen SO, Apisiridej S, Kwong SM, Yang YH, Skurray RA, Firth N. 2010. Analysis of the prototypical Staphylococcus aureus multiresistance plasmid pSK1. Plasmid 64:135–142. doi: 10.1016/j.plasmid.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 318.Morton TM, Eaton DM, Johnston JL, Archer GL. 1993. DNA sequence and units of transcription of the conjugative transfer gene complex (trs) of Staphylococcus aureus plasmid pGO1. J Bacteriol 175:4436–4447. doi: 10.1128/jb.175.14.4436-4447.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Liu MA, Kwong SM, Jensen SO, Brzoska AJ, Firth N. 2013. Biology of the staphylococcal conjugative multiresistance plasmid pSK41. Plasmid 70:42–51. doi: 10.1016/j.plasmid.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 320.Archer GL, Johnston JL. 1983. Self-transmissible plasmids in staphylococci that encode resistance to aminoglycosides. Antimicrob Agents Chemother 24:70–77. doi: 10.1128/aac.24.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Goering RV, Ruff EA. 1983. Comparative analysis of conjugative plasmids mediating gentamicin resistance in Staphylococcus aureus. Antimicrob Agents Chemother 24:450–452. doi: 10.1128/aac.24.3.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Littlejohn TG, DiBerardino D, Messerotti LJ, Spiers SJ, Skurray RA. 1991. Structure and evolution of a family of genes encoding antiseptic and disinfectant resistance in Staphylococcus aureus. Gene 101:59–66. doi: 10.1016/0378-1119(91)90224-y. [DOI] [PubMed] [Google Scholar]
- 323.Morton TM, Johnston JL, Patterson J, Archer GL. 1995. Characterization of a conjugative staphylococcal mupirocin resistance plasmid. Antimicrob Agents Chemother 39:1272–1280. doi: 10.1128/aac.39.6.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, Sensabaugh GF, Perdreau-Remington F. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 325.Evans J, Dyke KG. 1988. Characterization of the conjugation system associated with the Staphylococcus aureus plasmid pJE1. J Gen Microbiol 134:1–8. doi: 10.1099/00221287-134-1-1. [DOI] [PubMed] [Google Scholar]
- 326.Bender J, Strommenger B, Steglich M, Zimmermann O, Fenner I, Lensing C, Dagwadordsch U, Kekule AS, Werner G, Layer F. 2015. Linezolid resistance in clinical isolates of Staphylococcus epidermidis from German hospitals and characterization of two cfr-carrying plasmids. J Antimicrob Chemother 70:1630–1638. doi: 10.1093/jac/dkv025. [DOI] [PubMed] [Google Scholar]
- 327.Juhas M, van der Meer JR, Gaillard M, Harding RM, Hood DW, Crook DW. 2009. Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol Rev 33:376–393. doi: 10.1111/j.1574-6976.2008.00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Bellanger X, Payot S, Leblond-Bourget N, Guédon G. 2014. Conjugative and mobilizable genomic islands in bacteria: evolution and diversity. FEMS Microbiol Rev 38:720–760. doi: 10.1111/1574-6976.12058. [DOI] [PubMed] [Google Scholar]
- 329.Mantri Y, Williams KP. 2004. Islander: a database of integrative islands in prokaryotic genomes, the associated integrases and their DNA site specificities. Nucleic Acids Res 32:D55–D58. doi: 10.1093/nar/gkh059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Dobrindt U, Hochhut B, Hentschel U, Hacker J. 2004. Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol 2:414–424. doi: 10.1038/nrmicro884. [DOI] [PubMed] [Google Scholar]
- 331.Burrus V, Pavlovic G, Decaris B, Guedon G. 2002. Conjugative transposons: the tip of the iceberg. Mol Microbiol 46:601–610. doi: 10.1046/j.1365-2958.2002.03191.x. [DOI] [PubMed] [Google Scholar]
- 332.Delavat F, Miyazaki R, Carraro N, Pradervand N, van der Meer JR. 2017. The hidden life of integrative and conjugative elements. FEMS Microbiol Rev 41:512–537. doi: 10.1093/femsre/fux008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Roy Chowdhury P, Scott MJ, Djordjevic SP. 2017. Genomic islands 1 and 2 carry multiple antibiotic resistance genes in Pseudomonas aeruginosa ST235, ST253, ST111 and ST175 and are globally dispersed. J Antimicrob Chemother 72:620–622. doi: 10.1093/jac/dkw471. [DOI] [PubMed] [Google Scholar]
- 334.Cochetti I, Tili E, Mingoia M, Varaldo PE, Montanari MP. 2008. erm(B)-carrying elements in tetracycline-resistant pneumococci and correspondence between Tn1545 and Tn6003. Antimicrob Agents Chemother 52:1285–1290. doi: 10.1128/AAC.01457-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Launay A, Ballard SA, Johnson PD, Grayson ML, Lambert T. 2006. Transfer of vancomycin resistance transposon Tn1549 from Clostridium symbiosum to Enterococcus spp. in the gut of gnotobiotic mice. Antimicrob Agents Chemother 50:1054–1062. doi: 10.1128/AAC.50.3.1054-1062.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Katayama Y, Takeuchi F, Ito T, Ma XX, Ui-Mizutani Y, Kobayashi I, Hiramatsu K. 2003. Identification in methicillin-susceptible Staphylococcus hominis of an active primordial mobile genetic element for the staphylococcal cassette chromosome mec of methicillin-resistant Staphylococcus aureus. J Bacteriol 185:2711–2722. doi: 10.1128/jb.185.9.2711-2722.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Enright MC, Robinson DA, Randle G, Feil EJ, Grundmann H, Spratt BG. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc Natl Acad Sci U S A 99:7687–7692. doi: 10.1073/pnas.122108599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Katayama Y, Zhang HZ, Hong D, Chambers HF. 2003. Jumping the barrier to beta-lactam resistance in Staphylococcus aureus. J Bacteriol 185:5465–5472. doi: 10.1128/jb.185.18.5465-5472.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.von Wintersdorff CJH, Penders J, van Niekerk JM, Mills ND, Majumder S, van Alphen LB, Savelkoul PHM, Wolffs P. 2016. Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front Microbiol 7:173. doi: 10.3389/fmicb.2016.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Conlan S, Thomas PJ, Deming C, Park M, Lau AF, Dekker JP, Snitkin ES, Clark TA, Luong K, Song Y, Tsai YC, Boitano M, Dayal J, Brooks SY, Schmidt B, Young AC, Thomas JW, Bouffard GG, Blakesley RW, NISC Comparative Sequencing Program, Mullikin JC, Korlach J, Henderson DK, Frank KM, Palmore TN, Segre JA. 2014. Single-molecule sequencing to track plasmid diversity of hospital-associated carbapenemase-producing Enterobacteriaceae. Sci Transl Med 6:254ra126. doi: 10.1126/scitranslmed.3009845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Weingarten RA, Johnson RC, Conlan S, Ramsburg AM, Dekker JP, Lau AF, Khil P, Odom RT, Deming C, Park M, Thomas PJ, NISC Comparative Sequencing Program, Henderson DK, Palmore TN, Segre JA, Frank KM. 2018. Genomic analysis of hospital plumbing reveals diverse reservoir of bacterial plasmids conferring carbapenem resistance. mBio 9:e02011-17. doi: 10.1128/mBio.02011-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Grundmann H, Aires-De-Sousa M, Boyce J, Tiemersma E. 2006. Emergence and resurgence of meticillin-resistant Staphylococcus aureus as a public-health threat. Lancet 368:874–885. doi: 10.1016/S0140-6736(06)68853-3. [DOI] [PubMed] [Google Scholar]
- 343.Gardete S, Tomasz A. 2014. Mechanisms of vancomycin resistance in Staphylococcus aureus. J Clin Invest 124:2836–2840. doi: 10.1172/JCI68834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Meyer R. 2009. Replication and conjugative mobilization of broad host-range IncQ plasmids. Plasmid 62:57–70. doi: 10.1016/j.plasmid.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Maslanova I, Stribna S, Doskar J, Pantucek R. 2016. Efficient plasmid transduction to Staphylococcus aureus strains insensitive to the lytic action of transducing phage. FEMS Microbiol Lett 363:fnw211. doi: 10.1093/femsle/fnw211. [DOI] [PubMed] [Google Scholar]
- 346.Chlebowicz MA, Mašlaňová I, Kuntová L, Grundmann H, Pantůček R, Doškař J, van Dijl JM, Buist G. 2014. The staphylococcal cassette chromosome mec type V from Staphylococcus aureus ST398 is packaged into bacteriophage capsids. Int J Med Microbiol 304:764–774. doi: 10.1016/j.ijmm.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 347.Blahova J, Kralikova K, Krcmery V, Jezek P. 2000. Low-frequency transduction of imipenem resistance and high-frequency transduction of ceftazidime and aztreonam resistance by the bacteriophage AP-151 isolated from a Pseudomonas aeruginosa strain. J Chemother 12:482–486. doi: 10.1179/joc.2000.12.6.482. [DOI] [PubMed] [Google Scholar]
- 348.Krahn T, Wibberg D, Maus I, Winkler A, Bontron S, Sczyrba A, Nordmann P, Puhler A, Poirel L, Schluter A. 2016. Intraspecies transfer of the chromosomal Acinetobacter baumannii bla(NDM-1) carbapenemase gene. Antimicrob Agents Chemother 60:3032–3040. doi: 10.1128/AAC.00124-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Johnston C, Martin B, Fichant G, Polard P, Claverys JP. 2014. Bacterial transformation: distribution, shared mechanisms and divergent control. Nat Rev Microbiol 12:181–196. doi: 10.1038/nrmicro3199. [DOI] [PubMed] [Google Scholar]
- 350.Cameron ADS, Redfield RJ. 2006. Non-canonical CRP sites control competence regulons in Escherichia coli and many other gamma-proteobacteria. Nucleic Acids Res 34:6001–6014. doi: 10.1093/nar/gkl734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Palchevskiy V, Finkel SE. 2006. Escherichia coli competence gene homologs are essential for competitive fitness and the use of DNA as a nutrient. J Bacteriol 188:3902–3910. doi: 10.1128/JB.01974-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Domingues S, Nielsen KM, da Silva GJ. 2012. Various pathways leading to the acquisition of antibiotic resistance by natural transformation. Mob Genet Elements 2:257–260. doi: 10.4161/mge.23089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Baltrus DA. 2013. Exploring the costs of horizontal gene transfer. Trends Ecol Evol 28:489–495. doi: 10.1016/j.tree.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 354.Vogwill T, MacLean RC. 2015. The genetic basis of the fitness costs of antimicrobial resistance: a meta-analysis approach. Evol Appl 8:284–295. doi: 10.1111/eva.12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Wiggins AG, LaVoie SP, Wireman J, Summers AO. 2018. Thinking outside the (pill) box: does toxic metal exposure thwart antibiotic stewardship best practices? Plasmid 99:68–71. doi: 10.1016/j.plasmid.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 356.Lin F, Xu Y, Chang Y, Liu C, Jia X, Ling B. 2017. Molecular characterization of reduced susceptibility to biocides in clinical isolates of Acinetobacter baumannii. Front Microbiol 8:1836. doi: 10.3389/fmicb.2017.01836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Pal C, Bengtsson-Palme J, Kristiansson E, Larsson D. 2015. Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genomics 16:964. doi: 10.1186/s12864-015-2153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Canton R, Ruiz-Garbajosa P. 2011. Co-resistance: an opportunity for the bacteria and resistance genes. Curr Opin Pharmacol 11:477–485. doi: 10.1016/j.coph.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 359.McDonnell G, Russell AD. 2001. Antiseptics and disinfectants: activity, action, and resistance. Clin Microbiol Rev 14:227–228. doi: 10.1128/CMR.12.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Schweizer HP. 1998. Intrinsic resistance to inhibitors of fatty acid biosynthesis in Pseudomonas aeruginosa is due to efflux: application of a novel technique for generation of unmarked chromosomal mutations for the study of efflux systems. Antimicrob Agents Chemother 42:394–398. doi: 10.1128/AAC.42.2.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Nikaido H. 1998. Antibiotic resistance caused by Gram-negative multidrug efflux pumps. Clin Infect Dis 27:S32–S41. doi: 10.1086/514920. [DOI] [PubMed] [Google Scholar]
- 362.Paulsen IT, Littlejohn TG, Radstrom P, Sundstrom L, Skold O, Swedberg G, Skurray RA. 1993. The 3' conserved segment of integrons contains a gene associated with multidrug resistance to antiseptics and disinfectants. Antimicrob Agents Chemother 37:761–768. doi: 10.1128/aac.37.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Kazama H, Hamashima H, Sasatsu M, Arai T. 1999. Characterization of the antiseptic-resistance gene qacE delta 1 isolated from clinical and environmental isolates of Vibrio parahaemolyticus and Vibrio cholerae non-O1. FEMS Microbiol Lett 174:379–384. doi: 10.1111/j.1574-6968.1999.tb13593.x. [DOI] [PubMed] [Google Scholar]
- 364.Gillings MR, Duan XJ, Hardwick SA, Holley MP, Stokes HW. 2009. Gene cassettes encoding resistance to quaternary ammonium compounds: a role in the origin of clinical class 1 integrons? ISME J 3:209–215. doi: 10.1038/ismej.2008.98. [DOI] [PubMed] [Google Scholar]
- 365.Gaze WH, Abdouslam N, Hawkey PM, Wellington E. 2005. Incidence of class 1 integrons in a quaternary ammonium compound-polluted environment. Antimicrob Agents Chemother 49:1802–1807. doi: 10.1128/AAC.49.5.1802-1807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Sidhu MS, Heir E, Sorum H, Holck A. 2001. Genetic linkage between resistance to quaternary ammonium compounds and beta-lactam antibiotics in food-related Staphylococcus spp. Microb Drug Resist 7:363–371. doi: 10.1089/10766290152773374. [DOI] [PubMed] [Google Scholar]
- 367.Baker-Austin C, Wright MS, Stepanauskas R, McArthur JV. 2006. Co-selection of antibiotic and metal resistance. Trends Microbiol 14:176–182. doi: 10.1016/j.tim.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 368.Kazama H, Hamashima H, Sasatsu M, Arai T. 1998. Distribution of the antiseptic-resistance gene qacE delta 1 in Gram-positive bacteria. FEMS Microbiol Lett 165:295–299. doi: 10.1111/j.1574-6968.1998.tb13160.x. [DOI] [PubMed] [Google Scholar]
- 369.Liu WJ, Fu L, Huang M, Zhang JP, Wu Y, Zhou YS, Zeng J, Wang GX. 2017. Frequency of antiseptic resistance genes and reduced susceptibility to biocides in carbapenem-resistant Acinetobacter baumannii. J Med Microbiol 66:13–17. doi: 10.1099/jmm.0.000403. [DOI] [PubMed] [Google Scholar]
- 370.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. 2007. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Discov 6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 371.Fischbach MA, Walsh CT. 2009. Antibiotics for emerging pathogens. Science 325:1089–1093. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Renwick M, Mossialos E. 2018. What are the economic barriers of antibiotic R&D and how can we overcome them? Expert Opin Drug Discov 13:889–892. doi: 10.1080/17460441.2018.1515908. [DOI] [PubMed] [Google Scholar]
- 373.Sertkaya A, Eyraud J, Birkenbach A, Franz C, Ackerley N, Overton V, Outterson K. 2014. Analytical framework for examining the value of antibacterial products. https://aspe.hhs.gov/system/files/pdf/76891/rpt_antibacterials.pdf. Accessed 20 January 2020.
- 374.Theuretzbacher U, Outterson K, Engel A, Karlen A. 19 November 2019. The global preclinical antibacterial pipeline. Nat Rev Microbiol doi: 10.1038/s41579-019-0288-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Sharma P, Towse A. 2011. New drugs to tackle antimicrobial resistance: analysis of EU policy options. https://www.ohe.org/publications/new-drugs-tackle-antimicrobial-resistance-analysis-eu-policy-options. Accessed 8 January 2020.
- 376.Access to Medicine Foundation. 2018. Antimicrobial resistance benchmark 2018. https://accesstomedicinefoundation.org/media/uploads/downloads/5bc5edd8367eb_Antimicrobial-Resistance-Benchmark-2018.pdf. Accessed 8 January 2020.
- 377.Innovative Medicines Initiative (IMI). 2017. New drugs for bad bugs. The Innovative Medicines Initiative response to antimicrobial resistance. https://www.imi.europa.eu/sites/default/files/uploads/documents/projects/IMI_AMR_2017_LR.pdf. Accessed 7 January 2020.
- 378.Innovative Medicines Initiative (IMI). 2019. VALUE-Dx. The value of diagnostics to combat antimicrobial resistance by optimising antibiotic use. https://www.imi.europa.eu/projects-results/project-factsheets/value-dx. Accessed 7 January 2020.
- 379.COMBACTE. 2019. COMBACTE-CARE: combatting bacterial resistance in Europe—carbapenem-resistance. https://www.combacte.com/about/about-combacte-care-detail/. Accessed 7 January 2020.
- 380.DRIVE AB. 2014. Driving reinvestment in R&D for antibiotics and their responsible use. http://drive-ab.eu/. Accessed 8 January 2020.
- 381.Ardal C, Baraldi E, Theuretzbacher U, Outterson K, Plahte J, Ciabuschi F, Rottingen JA. 2018. Insights into early stage of antibiotic development in small- and medium-sized enterprises: a survey of targets, costs, and durations. J Pharm Policy Pract 11:8. doi: 10.1186/s40545-018-0135-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Ling LL, Schneider T, Peoples AJ, Spoering AL, Engels I, Conlon BP, Mueller A, Schaberle TF, Hughes DE, Epstein S, Jones M, Lazarides L, Steadman VA, Cohen DR, Felix CR, Fetterman KA, Millett WP, Nitti AG, Zullo AM, Chen C, Lewis K. 2015. A new antibiotic kills pathogens without detectable resistance. Nature 517:455–459. doi: 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Summers WC. 2001. Bacteriophage therapy. Annu Rev Microbiol 55:437–451. doi: 10.1146/annurev.micro.55.1.437. [DOI] [PubMed] [Google Scholar]
- 384.Liu CI, Liu GY, Song Y, Yin F, Hensler ME, Jeng WY, Nizet V, Wang AH, Oldfield E. 2008. A cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulence. Science 319:1391–1394. doi: 10.1126/science.1153018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Vaara M, Vaara T. 1983. Sensitization of Gram-negative bacteria to antibiotics and complement by a nontoxic oligopeptide. Nature 303:526–528. doi: 10.1038/303526a0. [DOI] [PubMed] [Google Scholar]
- 386.Zinkernagel AS, Peyssonnaux C, Johnson RS, Nizet V. 2008. Pharmacologic augmentation of hypoxia-inducible factor-1alpha with mimosine boosts the bactericidal capacity of phagocytes. J Infect Dis 197:214–217. doi: 10.1086/524843. [DOI] [PubMed] [Google Scholar]
- 387.Gill EE, Franco OL, Hancock RE. 2015. Antibiotic adjuvants: diverse strategies for controlling drug-resistant pathogens. Chem Biol Drug Des 85:56–78. doi: 10.1111/cbdd.12478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Theuretzbacher U, Gottwalt S, Beyer P, Butler M, Czaplewski L, Lienhardt C, Moja L, Paul M, Paulin S, Rex JH, Silver LL, Spigelman M, Thwaites GE, Paccaud JP, Harbarth S. 2019. Analysis of the clinical antibacterial and antituberculosis pipeline. Lancet Infect Dis 19:e40–e50. doi: 10.1016/S1473-3099(18)30513-9. [DOI] [PubMed] [Google Scholar]
- 389.Bohlmann L, De Oliveira DMP, El-Deeb IM, Brazel EB, Harbison-Price N, Ong CY, Rivera-Hernandez T, Ferguson SA, Cork AJ, Phan MD, Soderholm AT, Davies MR, Nimmo GR, Dougan G, Schembri MA, Cook GM, McEwan AG, von Itzstein M, McDevitt CA, Walker MJ. 2018. Chemical synergy between ionophore PBT2 and zinc reverses antibiotic resistance. mBio 9:e02391-18. doi: 10.1128/mBio.02391-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.The PEW Charitable Trusts. 2019. Antibiotics currently in global clinical development. https://www.pewtrusts.org/-/media/assets/2019/08/arp_antibiotics_currently_in_global_clinical_development_data_table_v2.pdf. Accessed 18 November 2019.
- 391.Shaeer KM, Zmarlicka MT, Chahine EB, Piccicacco N, Cho JC. 2019. Plazomicin: a next-generation aminoglycoside. Pharmacotherapy 39:77–93. doi: 10.1002/phar.2203. [DOI] [PubMed] [Google Scholar]
- 392.U.S. Food and Drug Administration. 2018. Centre for drug evaluation and research. Application number: 210303Orig1s000. Summary review. https://www.accessdata.fda.gov/drugsatfda_docs/nda/2018/210303Orig1s000SumR.pdf. Accessed 14 January 2020.
- 393.Wagenlehner FME, Cloutier DJ, Komirenko AS, Cebrik DS, Krause KM, Keepers TR, Connolly LE, Miller LG, Friedland I, Dwyer JP, EPIC Study Group . 2019. Once-daily plazomicin for complicated urinary tract infections. N Engl J Med 380:729–740. doi: 10.1056/NEJMoa1801467. [DOI] [PubMed] [Google Scholar]
- 394.Snydman DR, McDermott LA, Jacobus NV, Kerstein K, Grossman TH, Sutcliffe JA. 2018. Evaluation of the in vitro activity of eravacycline against a broad spectrum of recent clinical anaerobic isolates. Antimicrob Agents Chemother 62:e02206-17. doi: 10.1128/AAC.02206-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Solomkin JS, Gardovskis J, Lawrence K, Montravers P, Sway A, Evans D, Tsai L. 2019. IGNITE4: results of a phase 3, randomized, multicenter, prospective trial of eravacycline vs. meropenem in the treatment of complicated intra-abdominal infections. Clin Infect Dis 69:921–929. doi: 10.1093/cid/ciy1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Dougherty JA, Sucher AJ, Chahine EB, Shihadeh KC. 2019. Omadacycline: a new tetracycline antibiotic. Ann Pharmacother 53:486–500. doi: 10.1177/1060028018818094. [DOI] [PubMed] [Google Scholar]
- 397.Stets R, Popescu M, Gonong JR, Mitha I, Nseir W, Madej A, Kirsch C, Das AF, Garrity-Ryan L, Steenbergen JN, Manley A, Eckburg PB, Tzanis E, McGovern PC, Loh E. 2019. Omadacycline for community-acquired bacterial pneumonia. N Engl J Med 380:517–527. doi: 10.1056/NEJMoa1800201. [DOI] [PubMed] [Google Scholar]
- 398.European Medicines Agency. 2019. Withdrawal of application for the marketing authorisation of Nuzyra (omadacycline). https://www.ema.europa.eu/en/documents/medicine-qa/questions-answers-withdrawal-application-marketing-authorisation-nuzyra-omadacycline_en.pdf. Accessed 11 March 2020.
- 399.DRIVE AB. 2018. DRIVE AB report: revitalizing the antibiotic pipeline: stimulatinh innovation while driving sustainable used and global access. http://drive-ab.eu/wp-content/uploads/2018/01/DRIVE-AB-Final-Report-Jan2018.pdf. Accessed 12 March 2020.
- 400.Merck & Co., Inc. 2019. Recarbrio prescribing information. http://bit.ly/2U4x4bP. Accessed 23 November 2019.
- 401.Papp-Wallace KM, Barnes MD, Alsop J, Taracila MA, Bethel CR, Becka SA, van Duin D, Kreiswirth BN, Kaye KS, Bonomo RA. 2018. Relebactam is a potent inhibitor of the KPC-2 beta-lactamase and restores imipenem susceptibility in KPC-producing Enterobacteriaceae. Antimicrob Agents Chemother 62:e00174-18. doi: 10.1128/AAC.00174-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.U.S. Food and Drug Administration. 2019. FDA approves new antibiotic to treat community-acquired bacterial pneumonia. https://www.fda.gov/news-events/press-announcements/fda-approves-new-antibiotic-treat-community-acquired-bacterial-pneumonia. Accessed 3 November 2019.
- 403.Alexander E, Goldberg L, Das AF, Moran GJ, Sandrock C, Gasink LB, Spera P, Sweeney C, Paukner S, Wicha WW, Gelone SP, Schranz J. 2019. Oral lefamulin vs moxifloxacin for early clinical response among adults with community-acquired bacterial pneumonia: the LEAP 2 randomized clinical trial. JAMA 322:1661–1671. doi: 10.1001/jama.2019.15468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Nabriva Therapeutics AG. 2020. Lefamulin. https://www.nabriva.com/pipeline-research. Accessed 11 March 2020.
- 405.European Medicines Agency. 2020. EMEA-002075-PIP01-16-M01. https://www.ema.europa.eu/en/medicines/human/paediatric-investigation-plans/emea-002075-pip01-16-m01. Accessed 11 March 2020.
- 406.Kyorin Pharmaceutical Co. Ltd. 2019. Kyorin pharmaceutical receives marketing approval for oral quinolone antibacterial agent “Lasvic tablets 75mg.” https://www.kyorin-pharm.co.jp/en/news/691640e4c4142c191c8d4061a71c02e299076a4a.pdf. Accessed 11 March 2020.
- 407.Kishii R, Yamaguchi Y, Takei M. 2017. In vitro activities and spectrum of the novel fluoroquinolone lascufloxacin (KRP-AM1977). Antimicrob Agents Chemother 61:e00120-17. doi: 10.1128/AAC.00120-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Furuie H, Tanioka S, Shimizu K, Manita S, Nishimura M, Yoshida H. 2018. Intrapulmonary pharmacokinetics of lascufloxacin in healthy adult volunteers. Antimicrob Agents Chemother 62:e02169-17. doi: 10.1128/AAC.02169-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Kato T, Odajima M, Kohno S. 2017. Phase II clinical study of oral lascufloxacin in community-acquired pneumonia, abstr P-1362. Abstr 27th Congr ECCMID. [Google Scholar]
- 410.Kyorin Pharmaceutical Co. Ltd. 2019. NHI drug price listing and release of oral quinolone antibacterial agent “Lasvic tablets 75mg.” https://www.kyorin-pharm.co.jp/en/news/a329f0ae64024c1173f40660eede0efb37f1cbb0.pdf. Accessed 12 March 2020.
- 411.Bonomo RA. 2019. Cefiderocol: a novel siderophore cephalosporin defeating carbapenem-resistant pathogens. Clin Infect Dis 69:S519–S520. doi: 10.1093/cid/ciz823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Zhanel GG, Golden AR, Zelenitsky S, Wiebe K, Lawrence CK, Adam HJ, Idowu T, Domalaon R, Schweizer F, Zhanel MA, Lagace-Wiens PRS, Walkty AJ, Noreddin A, Lynch JP, Karlowsky JA. 2019. Cefiderocol: a siderophore cephalosporin with activity against carbapenem-resistant and multidrug-resistant Gram-negative bacilli. Drugs 79:271–289. doi: 10.1007/s40265-019-1055-2. [DOI] [PubMed] [Google Scholar]
- 413.Martin-Loeches I, Dale GE, Torres A. 2018. Murepavadin: a new antibiotic class in the pipeline. Expert Rev Anti Infect Ther 16:259–268. doi: 10.1080/14787210.2018.1441024. [DOI] [PubMed] [Google Scholar]
- 414.Bernardini F, Dale GE, Wach A, Obrecht D. 2019. WS01-4 pharmacokinetics and pharmacodynamics of murepavadin (POL7080) in neutropenic lung infection models when evaluated by aerosol administration. J Cyst Fibros 18(Suppl 1):S2. doi: 10.1016/S1569-1993(19)30120-1. [DOI] [Google Scholar]
- 415.Perazella MA. 2018. Pharmacology behind common drug nephrotoxicities. Clin J Am Soc Nephrol 13:1897–1908. doi: 10.2215/CJN.00150118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Spero Therapeutics Inc. 2018. Spero Therapeutics announces positive top-line data for two product candidates from its potentiator platform. https://investors.sperotherapeutics.com/node/7091/pdf. Accessed 21 April 2020.
- 417.Corbett D, Wise A, Langley T, Skinner K, Trimby E, Birchall S, Dorali A, Sandiford S, Williams J, Warn P, Vaara M, Lister T. 2017. Potentiation of antibiotic activity by a novel cationic peptide: potency and spectrum of activity of SPR741. Antimicrob Agents Chemother 61:e00200-17. doi: 10.1128/AAC.00200-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Vaara M, Siikanen O, Apajalahti J, Fox J, Frimodt-Moller N, He H, Poudyal A, Li JA, Nation RL, Vaara T. 2010. A novel polymyxin derivative that lacks the fatty acid tail and carries only three positive charges has strong synergism with agents excluded by the intact outer membrane. Antimicrob Agents Chemother 54:3341–3346. doi: 10.1128/AAC.01439-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Harris PNA, Peleg AY, Iredell J, Ingram PR, Miyakis S, Stewardson AJ, Rogers BA, McBryde ES, Roberts JA, Lipman J, Athan E, Paul SK, Baker P, Harris-Brown T, Paterson DL. 2015. Meropenem versus piperacillin-tazobactam for definitive treatment of bloodstream infections due to ceftriaxone non-susceptible Escherichia coli and Klebsiella spp (the MERINO trial): study protocol for a randomised controlled trial. Trials 16:24. doi: 10.1186/s13063-014-0541-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Harris PNA, Tambyah PA, Lye DC, Mo Y, Lee TH, Yilmaz M, Alenazi TH, Arabi Y, Falcone M, Bassetti M, Righi E, Rogers BA, Kanj S, Bhally H, Iredell J, Mendelson M, Boyles TH, Looke D, Miyakis S, Walls G, Al Khamis M, Zikri A, Crowe A, Ingram P, Daneman N, Griffin P, Athan E, Lorenc P, Baker P, Roberts L, Beatson SA, Peleg AY, Harris-Brown T, Paterson DL, MERINO Trial Investigators and the Australasian Society for Infectious Disease Clinical Research Network (ASID-CRN) . 2018. Effect of piperacillin-tazobactam vs meropenem on 30-day mortality for patients with E. coli or Klebsiella pneumoniae bloodstream infection and ceftriaxone resistance: a randomized clinical trial. JAMA 320:984–994. doi: 10.1001/jama.2018.12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Kehlet H. 2008. Fast-track colorectal surgery. Lancet 371:791–793. doi: 10.1016/S0140-6736(08)60357-8. [DOI] [PubMed] [Google Scholar]
- 422.Khoo CK, Vickery CJ, Forsyth N, Vinall NS, Eyre-Brook IA. 2007. A prospective randomized controlled trial of multimodal perioperative management protocol in patients undergoing elective colorectal resection for cancer. Ann Surg 245:867–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Iversen K, Ihlemann N, Gill SU, Madsen T, Elming H, Jensen KT, Bruun NE, Høfsten DE, Fursted K, Christensen JJ, Schultz M, Klein CF, Fosbøll EL, Rosenvinge F, Schønheyder HC, Køber L, Torp-Pedersen C, Helweg-Larsen J, Tønder N, Moser C, Bundgaard H. 2019. Partial oral versus intravenous antibiotic treatment of endocarditis. N Engl J Med 380:415–424. doi: 10.1056/NEJMoa1808312. [DOI] [PubMed] [Google Scholar]
- 424.Jewetz E, Gunnison JB, Bruff JB, Coleman VR. 1952. Studies on antibiotic synergism and antagonism. Synergism among seven antibiotics against various bacteria in vitro. J Bacteriol 64:29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Bushby SR, Hitchings GH. 1968. Trimethoprim, a sulphonamide potentiator. Br J Pharmacol Chemother 33:72–90. doi: 10.1111/j.1476-5381.1968.tb00475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Tyers M, Wright GD. 2019. Drug combinations: a strategy to extend the life of antibiotics in the 21st century. Nat Rev Microbiol 17:141–155. doi: 10.1038/s41579-018-0141-x. [DOI] [PubMed] [Google Scholar]
- 427.Neu HC, Fu KP. 1978. Clavulanic acid, a novel inhibitor of beta-lactamases. Antimicrob Agents Chemother 14:650–655. doi: 10.1128/aac.14.5.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Wong DR, van Duin D. 2017. Novel beta-lactamase inhibitors: unlocking their potential in therapy. Drugs 77:615–628. doi: 10.1007/s40265-017-0725-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Wright GD. 2016. Antibiotic adjuvants: rescuing antibiotics from resistance. Trends Microbiol 24:862–871. doi: 10.1016/j.tim.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 430.King AM, Reid-Yu SA, Wang WL, King DT, De Pascale G, Strynadka NC, Walsh TR, Coombes BK, Wright GD. 2014. Aspergillomarasmine A overcomes metallo-beta-lactamase antibiotic resistance. Nature 510:503–506. doi: 10.1038/nature13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Thompson RJ, Bobay BG, Stowe SD, Olson AL, Peng LL, Su ZM, Actis LA, Melander C, Cavanagh J. 2012. Identification of BfmR, a response regulator involved in biofilm development, as a target for a 2-aminoimidazole-based antibiofilm agent. Biochemistry 51:9776–9778. doi: 10.1021/bi3015289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Cox G, Koteva K, Wright GD. 2014. An unusual class of anthracyclines potentiate Gram-positive antibiotics in intrinsically resistant Gram-negative bacteria. J Antimicrob Chemother 69:1844–1855. doi: 10.1093/jac/dku057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Zurawski DV, Reinhart AA, Alamneh YA, Pucci MJ, Si YZ, Abu-Taleb R, Shearer JP, Demons ST, Tyner SD, Lister T. 2017. SPR741, an antibiotic adjuvant, potentiates the in vitro and in vivo activity of rifampin against clinically relevant extensively drug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 61:e01239-17. doi: 10.1128/AAC.01239-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Jen FE, Everest-Dass AV, El-Deeb IM, Singh S, Haselhorst T, Walker MJ, von Itzstein M, Jennings MP. 2020. Neisseria gonorrhoeae becomes susceptible to polymyxin B and colistin in the presence of PBT2. ACS Infect Dis 6:50–55. doi: 10.1021/acsinfecdis.9b00307. [DOI] [PubMed] [Google Scholar]
- 435.Hancock REW, Nijnik A, Philpott DJ. 2012. Modulating immunity as a therapy for bacterial infections. Nat Rev Microbiol 10:243–254. doi: 10.1038/nrmicro2745. [DOI] [PubMed] [Google Scholar]
- 436.Feigman MS, Kim S, Pidgeon SE, Yu Y, Ongwae GM, Patel DS, Regen S, Im W, Pires MM. 2018. Synthetic immunotherapeutics against Gram-negative pathogens. Cell Chem Biol 25:1185–1194. doi: 10.1016/j.chembiol.2018.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Bhandari T, Nizet V. 2014. Hypoxia-inducible factor (HIF) as a pharmacological target for prevention and treatment of infectious diseases. Infect Dis Ther 3:159–174. doi: 10.1007/s40121-014-0030-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Zarember KA, Malech HL. 2005. HIF-1 alpha: a master regulator of innate host defenses? J Clin Invest 115:1702–1704. doi: 10.1172/JCI25740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Peyssonnaux C, Datta V, Cramer T, Doedens A, Theodorakis EA, Gallo RL, Hurtado-Ziola N, Nizet V, Johnson RS. 2005. HIF-1 alpha expression regulates the bactericidal capacity of phagocytes. J Clin Invest 115:1806–1815. doi: 10.1172/JCI23865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Berger EA, McClellan SA, Vistisen KS, Hazlett LD. 2013. HIF-1 alpha is essential for effective PMN bacterial killing, antimicrobial peptide production and apoptosis in Pseudomonas aeruginosa keratitis. PLoS Pathog 9:e1003457. doi: 10.1371/journal.ppat.1003457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Okumura CYM, Hollands A, Tran DN, Olson J, Dahesh S, von Kockritz-Blickwede M, Thienphrapa W, Corle C, Jeung SN, Kotsakis A, Shalwitz RA, Johnson RS, Nizet V. 2012. A new pharmacological agent (AKB-4924) stabilizes hypoxia inducible factor-1 (HIF-1) and increases skin innate defenses against bacterial infection. J Mol Med (Berl) 90:1079–1089. doi: 10.1007/s00109-012-0882-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Reardon S. 2017. Modified viruses deliver death to antibiotic-resistant bacteria. Nature 546:587–588. doi: 10.1038/nature.2017.22173. [DOI] [PubMed] [Google Scholar]
- 443.UC San Diego Health. 2019. With OK from FDA, UC San Diego researchers prepare to launch novel phage study. https://health.ucsd.edu/news/releases/Pages/2019-01-08-FDA-okays-uc-san-diego-to-launch-novel-phage-study.aspx. Accessed 13 August 2019.
- 444.Lehman SM, Mearns G, Rankin D, Cole RA, Smrekar F, Branston SD, Morales S. 2019. Design and preclinical development of a phage product for the treatment of antibiotic-resistant Staphylococcus aureus infections. Viruses 11:88. doi: 10.3390/v11010088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Pirnay JP, Verbeken G, Ceyssens PJ, Huys I, De Vos D, Ameloot C, Fauconnier A. 2018. The magistral phage. Viruses 10:64. doi: 10.3390/v10020064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Chong CR, Sullivan DJ. 2007. New uses for old drugs. Nature 448:645–646. doi: 10.1038/448645a. [DOI] [PubMed] [Google Scholar]
- 447.Christiansen SH, Murphy RA, Juul-Madsen K, Fredborg M, Hvam ML, Axelgaard E, Skovdal SM, Meyer RL, Sorensen U, Moller A, Nyengaard JR, Norskov-Lauritsen N, Wang M, Gadjeva M, Howard KA, Davies JC, Petersen E, Vorup-Jensen T. 2017. The immunomodulatory drug glatiramer acetate is also an effective antimicrobial agent that kills Gram-negative bacteria. Sci Rep 7:15653. doi: 10.1038/s41598-017-15969-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Kim W, Zhu WP, Hendricks GL, Van Tyne D, Steele AD, Keohane CE, Fricke N, Conery AL, Shen S, Pan W, Lee K, Rajamuthiah R, Fuchs BB, Vlahovska PM, Wuest WM, Gilmore MS, Gao HJ, Ausubel FM, Mylonakis E. 2018. A new class of synthetic retinoid antibiotics effective against bacterial persisters. Nature 556:103–107. doi: 10.1038/nature26157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Younis W, Thangamani S, Seleem MN. 2015. Repurposing non-antimicrobial drugs and clinical molecules to treat bacterial infections. Curr Pharm Des 21:4106–4111. doi: 10.2174/1381612821666150506154434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Brown D. 2015. Antibiotic resistance breakers: can repurposed drugs fill the antibiotic discovery void? Nat Rev Drug Discov 14:821–832. doi: 10.1038/nrd4675. [DOI] [PubMed] [Google Scholar]
- 451.McConnell MJ. 2019. Where are we with monoclonal antibodies for multidrug-resistant infections? Drug Discov Today 24:1132–1138. doi: 10.1016/j.drudis.2019.03.002. [DOI] [PubMed] [Google Scholar]
- 452.Tsai CW, Morris S. 2015. Approval of raxibacumab for the treatment of inhalation anthrax under the US Food and Drug Administration “animal rule.” Front Microbiol 6:1320. doi: 10.3389/fmicb.2015.01320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Hou AW, Morrill AM. 2017. Obiltoxaximab: adding to the treatment arsenal for Bacillus anthracis infection. Ann Pharmacother 51:908–913. doi: 10.1177/1060028017713029. [DOI] [PubMed] [Google Scholar]
- 454.Kufel WD, Devanathan AS, Marx AH, Weber DJ, Daniels LM. 2017. Bezlotoxumab: a novel agent for the prevention of recurrent Clostridium difficile infection. Pharmacotherapy 37:1298–1308. doi: 10.1002/phar.1990. [DOI] [PubMed] [Google Scholar]
- 455.Adis Insight Drugs. 2019. Panobacumab—Aridis Pharmaceuticals. https://adisinsight.springer.com/drugs/800024401. Accessed 10 January 2020.
- 456.Secher T, Fauconnier L, Szade A, Rutschi O, Fas SC, Ryffel B, Rudolf MP. 2011. Anti-Pseudomonas aeruginosa serotype O11 LPS immunoglobulin M monoclonal antibody panobacumab (KBPA101) confers protection in a murine model of acute lung infection. J Antimicrob Chemother 66:1100–1109. doi: 10.1093/jac/dkr038. [DOI] [PubMed] [Google Scholar]
- 457.Milla CE, Chmiel JF, Accurso FJ, VanDevanter DR, Konstan MW, Yarranton G, Geller DE, for the KB001 Study Group . 2014. Anti-PcrV antibody in cystic fibrosis: a novel approach targeting Pseudomonas aeruginosa airway infection. Pediatr Pulmonol 49:650–658. doi: 10.1002/ppul.22890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Jain R, Beckett VV, Konstan MW, Accurso FJ, Burns JL, Mayer-Hamblett N, Milla C, VanDevanter DR, Chmiel JF, KB001-A Study Group . 2018. KB001-A, a novel anti-inflammatory, found to be safe and well-tolerated in cystic fibrosis patients infected with Pseudomonas aeruginosa. J Cyst Fibros 17:484–491. doi: 10.1016/j.jcf.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 459.DiGiandomenico A, Warrener P, Hamilton M, Guillard S, Ravn P, Minter R, Camara MM, Venkatraman V, MacGill RS, Lin J, Wang Q, Keller AE, Bonnell JC, Tomich M, Jermutus L, McCarthy MP, Melnick DA, Suzich JA, Stover CK. 2012. Identification of broadly protective human antibodies to Pseudomonas aeruginosa exopolysaccharide Psl by phenotypic screening. J Exp Med 209:1273–1287. doi: 10.1084/jem.20120033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Ali SO, Yu XQ, Robbie GJ, Wu Y, Shoemaker K, Yu L, DiGiandomenico A, Keller AE, Anude C, Hernandez-Illas M, Bellamy T, Falloon J, Dubovsky F, Jafri HS. 2019. Phase 1 study of MEDI3902, an investigational anti-Pseudomonas aeruginosa PcrV and Psl bispecific human monoclonal antibody, in healthy adults. Clin Microbiol Infect 25:629.e1–629.e6. doi: 10.1016/j.cmi.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 461.EVADE. 2019. Effort to prevent nosocomial pneumonia caused by Pseudomonas aeruginosa in mechanically ventilated subjects. https://www.combacte.com/trials/evade/. Accessed 10 January 2020.
- 462.Hoggarth A, Weaver A, Pu QQ, Huang T, Schettler J, Chen F, Yuan XF, Wu M. 2019. Mechanistic research holds promise for bacterial vaccines and phage therapies for Pseudomonas aeruginosa. Drug Des Devel Ther 13:909–924. doi: 10.2147/DDDT.S189847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Rello J, Krenn CG, Locker G, Pilger E, Madl C, Balica L, Dugernier T, Laterre PF, Spapen H, Depuydt P, Vincent JL, Bogar L, Szabo Z, Volgyes B, Manez R, Cakar N, Ramazanoglu A, Topeli A, Mastruzzo MA, Jasovich A, Remolif CG, Soria LD, Hernandez MAA, Balart CR, Kremer I, Molnar Z, von Sonnenburg F, Lyons A, Joannidis M, Burgmann H, Welte T, Klingler A, Hochreiter R, Westritschnig K. 2017. A randomized placebo-controlled phase II study of a Pseudomonas vaccine in ventilated ICU patients. Crit Care 21:22. doi: 10.1186/s13054-017-1601-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Jansen KU, Knirsch C, Anderson AS. 2018. The role of vaccines in preventing bacterial antimicrobial resistance. Nat Med 24:10–19. doi: 10.1038/nm.4465. [DOI] [PubMed] [Google Scholar]
- 465.Creech CB, Frenck RW, Sheldon EA, Seiden DJ, Kankam MK, Zito ET, Girgenti D, Severs JM, Immermann FW, McNeil LK, Cooper D, Jansen KU, Gruber W, Eiden J, Anderson AS, Baber J. 2017. Safety, tolerability, and immunogenicity of a single dose 4-antigen or 3-antigen Staphylococcus aureus vaccine in healthy older adults: results of a randomised trial. Vaccine 35:385–394. doi: 10.1016/j.vaccine.2016.11.032. [DOI] [PubMed] [Google Scholar]
- 466.Creech CB, Frenck RW, Fiquet A, Feldman R, Kankam MK, Pathirana S, Baber J, Radley D, Cooper D, Eiden J, Gruber WC, Jansen KU, Anderson AS, Gurtman A. 2020. Persistence of immune responses through 36 months in healthy adults after vaccination with a novel Staphylococcus aureus 4-antigen vaccine (SA4Ag). Open Forum Infect Dis 7:ofz532. doi: 10.1093/ofid/ofz532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Seeberger PH, Pereira CL, Khan N, Xiao GZ, Diago-Navarro E, Reppe K, Opitz B, Fries BC, Witzenrath M. 2017. A semi-synthetic glycoconjugate vaccine candidate for carbapenem-resistant Klebsiella pneumoniae. Angew Chem Int Ed Engl 56:13973–13978. doi: 10.1002/anie.201700964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Ainsworth S, Ketter PM, Yu JJ, Grimm RC, May HC, Cap AP, Chambers JP, Guentzel MN, Arulanandam BP. 2017. Vaccination with a live attenuated Acinetobacter baumannii deficient in thioredoxin provides protection against systemic Acinetobacter infection. Vaccine 35:3387–3394. doi: 10.1016/j.vaccine.2017.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Wang TZ, Kodiyanplakkal RPL, Calfee DP. 2019. Antimicrobial resistance in nephrology. Nat Rev Nephrol 15:463–481. doi: 10.1038/s41581-019-0150-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Bilinski J, Grzesiowski P, Sorensen N, Madry K, Muszynski J, Robak K, Wroblewska M, Dzieciatkowski T, Dulny G, Dwilewicz-Trojaczek J, Wiktor-Jedrzejczak W, Basak GW. 2017. Fecal microbiota transplantation in patients with blood disorders inhibits gut colonization with antibiotic-resistant bacteria: results of a prospective, single-center study. Clin Infect Dis 65:364–370. doi: 10.1093/cid/cix252. [DOI] [PubMed] [Google Scholar]
- 471.Remschmidt C, Schroder C, Behnke M, Gastmeier P, Geffers C, Kramer TS. 2018. Continuous increase of vancomycin resistance in enterococci causing nosocomial infections in Germany—10 years of surveillance. Antimicrob Resist Infect Control 7:54. doi: 10.1186/s13756-018-0353-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Arias CA, Murray BE. 2008. Emergence and management of drug-resistant enterococcal infections. Expert Rev Anti Infect Ther 6:637–655. doi: 10.1586/14787210.6.5.637. [DOI] [PubMed] [Google Scholar]
- 473.O'Driscoll T, Crank CW. 2015. Vancomycin-resistant enterococcal infections: epidemiology, clinical manifestations, and optimal management. Infect Drug Resist 8:217–230. doi: 10.2147/IDR.S54125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Mioljević V, Marković-Denić L, Vidović A, Jovanović M, Tosić T, Tomin D. 2013. Risk factors for vancomycin-resistant Enterococcus colonization in hematologic patients. Vojnosanit Pregl 70:1109–1116. doi: 10.2298/vsp1312109m. [DOI] [PubMed] [Google Scholar]
- 475.Johnson PDR, Ballard SA, Grabsch EA, Stinear TP, Seemann T, Young HL, Grayson ML, Howden BP. 2010. A sustained hospital outbreak of vancomycin-resistant Enterococcus faecium bacteremia due to emergence of vanB E. faecium sequence type 203. J Infect Dis 202:1278–1286. doi: 10.1086/656319. [DOI] [PubMed] [Google Scholar]
- 476.Lister DM, Kotsanas D, Ballard SA, Howden BP, Carse E, Tan K, Scott C, Gillespie EE, Mahony AA, Doherty R, Korman TM, Johnson PDR, Stuart RL. 2015. Outbreak of vanB vancomycin-resistant Enterococcus faecium colonization in a neonatal service. Am J Infect Control 43:1061–1065. doi: 10.1016/j.ajic.2015.05.047. [DOI] [PubMed] [Google Scholar]
- 477.Ruiz-Garbajosa P, Bonten MJM, Robinson DA, Top J, Nallapareddy SR, Torres C, Coque TM, Canton R, Baquero F, Murray BE, del Campo R, Willems R. 2006. Multilocus sequence typing scheme for Enterococcus faecalis reveals hospital-adapted genetic complexes in a background of high rates of recombination. J Clin Microbiol 44:2220–2228. doi: 10.1128/JCM.02596-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.McBride SM, Fischetti VA, LeBlanc DJ, Moellering RC, Gilmore MS. 2007. Genetic diversity among Enterococcus faecalis. PLoS One 2:e582. doi: 10.1371/journal.pone.0000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479.Freitas AR, Novais C, Ruiz-Garbajosa P, Coque TM, Peixe L. 2009. Clonal expansion within clonal complex 2 and spread of vancomycin-resistant plasmids among different genetic lineages of Enterococcus faecalis from Portugal. J Antimicrob Chemother 63:1104–1111. doi: 10.1093/jac/dkp103. [DOI] [PubMed] [Google Scholar]
- 480.Erlandson KM, Sun J, Iwen PC, Rupp ME. 2008. Impact of the more-potent antibiotics quinupristin-dalfopristin and linezolid on outcome measure of patients with vancomycin-resistant Enterococcus bacteremia. Clin Infect Dis 46:30–36. doi: 10.1086/523588. [DOI] [PubMed] [Google Scholar]
- 481.Zhanel GG, Hoban DJ, Karlowsky JA. 2001. Nitrofurantoin is active against vancomycin-resistant enterococci. Antimicrob Agents Chemother 45:324–326. doi: 10.1128/AAC.45.1.324-326.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Falagas ME, Vouloumanou EK, Samonis G, Vardakas KZ. 2016. Fosfomycin. Clin Microbiol Rev 29:321–347. doi: 10.1128/CMR.00068-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Linden PK. 2002. Treatment options for vancomycin-resistant enterococcal infections. Drugs 62:425–441. doi: 10.2165/00003495-200262030-00002. [DOI] [PubMed] [Google Scholar]
- 484.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis 39:309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 485.Pinholt M, Ostergaard C, Arpi M, Bruun NE, Schonheyder HC, Gradel KO, Sogaard M, Knudsen JD, Danish Collaborative Bacteraemia Network (DACOBAN) . 2014. Incidence, clinical characteristics and 30-day mortality of enterococcal bacteraemia in Denmark 2006–2009: a population-based cohort study. Clin Microbiol Infect 20:145–151. doi: 10.1111/1469-0691.12236. [DOI] [PubMed] [Google Scholar]
- 486.Kramer A, Schwebke I, Kampf G. 2006. How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC Infect Dis 6:130. doi: 10.1186/1471-2334-6-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Santajit S, Indrawattana N. 2016. Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed Res Int 2016:2475067. doi: 10.1155/2016/2475067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488.Turner NA, Sharma-Kuinkel BK, Maskarinec SA, Eichenberger EM, Shah PP, Carugati M, Holland TL, Fowler VG. 2019. Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research. Nat Rev Microbiol 17:203–218. doi: 10.1038/s41579-018-0147-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Harkins CP, Pichon B, Doumith M, Parkhill J, Westh H, Tomasz A, De Lencastre H, Bentley SD, Kearns AM, Holden M. 2017. Methicillin-resistant Staphylococcus aureus emerged long before the introduction of methicillin into clinical practice. Genome Biol 18:130. doi: 10.1186/s13059-017-1252-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490.Corey GR, Arhin FF, Wikler MA, Sahm DF, Kreiswirth BN, Mediavilla JR, Good S, Fiset C, Jiang H, Moeck G, Kabler H, Green S, O'Riordan W, SOLO I, SOLO II Investigators . 2016. Pooled analysis of single-dose oritavancin in the treatment of acute bacterial skin and skin-structure infections caused by Gram-positive pathogens, including a large patient subset with methicillin-resistant Staphylococcus aureus. Int J Antimicrob Agents 48:528–534. doi: 10.1016/j.ijantimicag.2016.07.019. [DOI] [PubMed] [Google Scholar]
- 491.Rubinstein E, Kollef MH, Nathwani D. 2008. Pneumonia caused by methicillin-resistant Staphylococcus aureus. Clin Infect Dis 46(Suppl 5):S378–S385. doi: 10.1086/533594. [DOI] [PubMed] [Google Scholar]
- 492.McNeil JC, Vallejo JG, Hulten KG, Kaplan SL. 2018. Osteoarticular infections following open or penetrating trauma in children in the post-community-acquired methicillin-resistant Staphylococcus aureus era: the impact of Enterobacter cloacae. Pediatr Infect Dis J 37:1204–1210. doi: 10.1097/INF.0000000000001991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493.Chuang YY, Huang YC. 2013. Molecular epidemiology of community-associated meticillin-resistant Staphylococcus aureus in Asia. Lancet Infect Dis 13:698–708. doi: 10.1016/S1473-3099(13)70136-1. [DOI] [PubMed] [Google Scholar]
- 494.Liu YD, Wang H, Du N, Shen EH, Chen HB, Niu JQ, Ye HF, Chen MJ. 2009. Molecular evidence for spread of two major methicillin-resistant Staphylococcus aureus clones with a unique geographic distribution in Chinese hospitals. Antimicrob Agents Chemother 53:512–518. doi: 10.1128/AAC.00804-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 495.Zhao CJ, Liu YM, Zhao MZ, Liu YL, Yu Y, Chen HB, Sun QN, Chen HW, Jiang W, Liu YD, Han SM, Xu YC, Chen MJ, Cao B, Wang H. 2012. Characterization of community acquired Staphylococcus aureus associated with skin and soft tissue infection in Beijing: high prevalence of PVL+ ST398. PLoS One 7:e38577. doi: 10.1371/journal.pone.0038577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496.Song JH, Hsueh PR, Chung DR, Ko KS, Kang CI, Peck KR, Yeom JS, Kim SW, Chang HH, Kim YS, Jung SI, Son JS, So TMK, Lalitha MK, Yang YH, Huang SG, Wang H, Lu QA, Carlos CC, Perera JA, Chiu CH, Liu JW, Chongthaleong A, Thamlikitkul V, Van PH, ANSORP Study Group . 2011. Spread of methicillin-resistant Staphylococcus aureus between the community and the hospitals in Asian countries: an ANSORP study. J Antimicrob Chemother 66:1061–1069. doi: 10.1093/jac/dkr024. [DOI] [PubMed] [Google Scholar]
- 497.Sahibzada S, Abraham S, Coombs GW, Pang S, Hernandez-Jover M, Jordan D, Heller J. 2017. Transmission of highly virulent community-associated MRSA ST93 and livestock-associated MRSA ST398 between humans and pigs in Australia. Sci Rep 7:5273. doi: 10.1038/s41598-017-04789-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.van Hal SJ, Jensen SO, Vaska VL, Espedido BA, Paterson DL, Gosbell IB. 2012. Predictors of mortality in Staphylococcus aureus bacteremia. Clin Microbiol Rev 25:362–386. doi: 10.1128/CMR.05022-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 499.Dillon HC, Derrick CW. 1975. Clinical experience with clindamycin hydrochloride: 1. Treatment of streptococcal and mixed streptococcal-staphylococcal skin infections. Pediatrics 55:205–212. [PubMed] [Google Scholar]
- 500.Fowler VG, Boucher HW, Corey GR, Abrutyn E, Karchmer AW, Rupp ME, Levine DP, Chambers HF, Tally FP, Vigliani GA, Cabell CH, Link AS, DeMeyer I, Filler SG, Zervos M, Cook P, Parsonnet J, Bernstein JM, Price CS, Forrest GN, Fätkenheuer G, Gareca M, Rehm SJ, Brodt HR, Tice A, Cosgrove SE, S. aureus Endocarditis and Bacteremia Study Group . 2006. Daptomycin versus standard therapy for bacteremia and endocarditis caused by Staphylococcus aureus. N Engl J Med 355:653–665. doi: 10.1056/NEJMoa053783. [DOI] [PubMed] [Google Scholar]
- 501.Weigelt J, Itani K, Stevens D, Lau W, Dryden M, Knirsch C, Linezolid CSSTI Study Group . 2005. Linezolid versus vancomycin in treatment of complicated skin and soft tissue infections. Antimicrob Agents Chemother 49:2260–2266. doi: 10.1128/AAC.49.6.2260-2266.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Shorr AF, Lodise TP, Corey GR, De Anda C, Fang E, Das AF, Prokocimer P. 2015. Analysis of the phase 3 ESTABLISH trials of tedizolid versus linezolid in acute bacterial skin and skin structure infections. Antimicrob Agents Chemother 59:864–871. doi: 10.1128/AAC.03688-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 503.Boucher HW, Wilcox M, Talbot GH, Puttagunta S, Das AF, Dunne MW. 2014. Once-weekly dalbavancin versus daily conventional therapy for skin infection. N Engl J Med 370:2169–2179. doi: 10.1056/NEJMoa1310480. [DOI] [PubMed] [Google Scholar]
- 504.Tanaseannu C, Bergallo C, Teglia O, Jasovich A, Oliva ME, Dukart G, Dartois N, Cooper CA, Gandjini H, Mallick R, 303 and 318 Study Groups . 2008. Integrated results of 2 phase 3 studies comparing tigecycline and levofloxacin in community-acquired pneumonia. Diagn Microbiol Infect Dis 61:329–338. doi: 10.1016/j.diagmicrobio.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 505.Paul M, Bishara J, Yahav D, Goldberg E, Neuberger A, Ghanem-Zoubi N, Dickstein Y, Nseir W, Dan M, Leibovici L. 2015. Trimethoprim-sulfamethoxazole versus vancomycin for severe infections caused by meticillin resistant Staphylococcus aureus: randomised controlled trial. BMJ 350:h2219. doi: 10.1136/bmj.h2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506.Ng J, Gosbell IB. 2005. Successful oral pristinamycin therapy for osteoarticular infections due to methicillin-resistant Staphylococcus aureus (MRSA) and other Staphylococcus spp. J Antimicrob Chemother 55:1008–1012. doi: 10.1093/jac/dki108. [DOI] [PubMed] [Google Scholar]
- 507.Chen CJ, Huang YC. 2014. New epidemiology of Staphylococcus aureus infection in Asia. Clin Microbiol Infect 20:605–623. doi: 10.1111/1469-0691.12705. [DOI] [PubMed] [Google Scholar]
- 508.Dantes R, Mu Y, Belflower R, Aragon D, Dumyati G, Harrison LH, Lessa FC, Lynfield R, Nadle J, Petit S, Ray SM, Schaffner W, Townes J, Fridkin S, Emerging Infections Program-Active Bacterial Core Surveillance MRSA Surveillance Investigators . 2013. National burden of invasive methicillin-resistant Staphylococcus aureus infections, United States, 2011. JAMA Intern Med 173:1970–1978. doi: 10.1001/jamainternmed.2013.10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509.Tzouvelekis LS, Markogiannakis A, Piperaki E, Souli M, Daikos GL. 2014. Treating infections caused by carbapenemase-producing Enterobacteriaceae. Clin Microbiol Infect 20:862–872. doi: 10.1111/1469-0691.12697. [DOI] [PubMed] [Google Scholar]
- 510.Gu DX, Dong N, Zheng ZW, Lin D, Huang M, Wang LH, Chan EWC, Shu LB, Yu J, Zhang R, Chen S. 2018. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis 18:37–46. doi: 10.1016/S1473-3099(17)30489-9. [DOI] [PubMed] [Google Scholar]
- 511.Hsieh PF, Lin TL, Lee CZ, Tsai SF, Wang JT. 2008. Serum-induced iron-acquisition systems and TonB contribute to virulence in Klebsiella pneumoniae causing primary pyogenic liver abscess. J Infect Dis 197:1717–1727. doi: 10.1086/588383. [DOI] [PubMed] [Google Scholar]
- 512.Siu LK, Yeh KM, Lin JC, Fung CP, Chang FY. 2012. Klebsiella pneumoniae liver abscess: a new invasive syndrome. Lancet Infect Dis 12:881–887. doi: 10.1016/S1473-3099(12)70205-0. [DOI] [PubMed] [Google Scholar]
- 513.Quan JJ, Zhao DD, Liu LL, Chen Y, Zhou JC, Jiang Y, Du XX, Zhou ZH, Akova M, Yu YS. 2017. High prevalence of ESBL-producing Escherichia coli and Klebsiella pneumoniae in community-onset bloodstream infections in China. J Antimicrob Chemother 72:273–280. doi: 10.1093/jac/dkw372. [DOI] [PubMed] [Google Scholar]
- 514.Hu L, Liu Y, Deng L, Zhong Q, Hang Y, Wang Z, Zhan L, Wang L, Yu F. 2016. Outbreak by ventilator-associated ST11 K. pneumoniae with co-production of CTX-M-24 and KPC-2 in a SICU of a tertiary teaching hospital in central China. Front Microbiol 7:1190. doi: 10.3389/fmicb.2016.01190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 515.Ewers C, Stamm I, Pfeifer Y, Wieler LH, Kopp PA, Schønning K, Prenger-Berninghoff E, Scheufen S, Stolle I, Günther S, Bethe A. 2014. Clonal spread of highly successful ST15-CTX-M-15 Klebsiella pneumoniae in companion animals and horses. J Antimicrob Chemother 69:2676–2680. doi: 10.1093/jac/dku217. [DOI] [PubMed] [Google Scholar]
- 516.Navon-Venezia S, Kondratyeva K, Carattoli A. 2017. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev 41:252–275. doi: 10.1093/femsre/fux013. [DOI] [PubMed] [Google Scholar]
- 517.Frasson I, Lavezzo E, Franchin E, Toppo S, Barzon L, Cavallaro A, Richter SN, Palu G. 2012. Antimicrobial treatment and containment measures for an extremely drug-resistant Klebsiella pneumoniae ST101 isolate carrying pKPN101-IT, a novel fully sequenced bla(KPC-2) plasmid. J Clin Microbiol 50:3768–3772. doi: 10.1128/JCM.01892-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 518.D'Andrea MM, Arena F, Pallecchi L, Rossolini GM. 2013. CTX-M-type beta-lactamases: a successful story of antibiotic resistance. Int J Med Microbiol 303:305–317. doi: 10.1016/j.ijmm.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 519.Wyres KL, Holt KE. 2016. Klebsiella pneumoniae population genomics and antimicrobial-resistant clones. Trends Microbiol 24:944–956. doi: 10.1016/j.tim.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 520.López-Camacho E, Paño-Pardo JR, Ruiz-Carrascoso G, Wesselink J-J, Lusa-Bernal S, Ramos-Ruiz R, Ovalle S, Gómez-Gil R, Pérez-Blanco V, Pérez-Vázquez M, Gómez-Puertas P, Mingorance J. 2018. Population structure of OXA-48-producing Klebsiella pneumoniae ST405 isolates during a hospital outbreak characterised by genomic typing. J Glob Antimicrob Resist 15:48–54. doi: 10.1016/j.jgar.2018.06.008. [DOI] [PubMed] [Google Scholar]
- 521.Tumbarello M, Trecarichi EM, De Rosa FG, Giannella M, Giacobbe DR, Bassetti M, Losito AR, Bartoletti M, Del Bono V, Corcione S, Maiuro G, Tedeschi S, Celani L, Cardellino CS, Spanu T, Marchese A, Ambretti S, Cauda R, Viscoli C, Viale P, ISGRI-SITA (Italian Study Group on Resistant Infections of the Società Italiana Terapia Antinfettiva) . 2015. Infections caused by KPC-producing Klebsiella pneumoniae: differences in therapy and mortality in a multicentre study. J Antimicrob Chemother 70:2133–2143. doi: 10.1093/jac/dkv086. [DOI] [PubMed] [Google Scholar]
- 522.Hauck C, Cober E, Richter SS, Perez F, Salata RA, Kalayjian RC, Watkins RR, Scalera NM, Doi Y, Kaye KS, Evans S, Fowler VG, Bonomo RA, van Duin D. 2016. Spectrum of excess mortality due to carbapenem-resistant Klebsiella pneumoniae infections. Clin Microbiol Infect 22:513–519. doi: 10.1016/j.cmi.2016.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 523.Paterson DL, Ko WC, Von Gottberg A, Mohapatra S, Casellas JM, Goossens H, Mulazimoglu L, Trenholme G, Klugman KP, Bonomo RA, Rice LB, Wagener MM, McCormack JG, Yu VL. 2004. Antibiotic therapy for Klebsiella pneumoniae bacteremia: implications of production of extended-spectrum beta-lactamases. Clin Infect Dis 39:31–37. doi: 10.1086/420816. [DOI] [PubMed] [Google Scholar]
- 524.Souli M, Galani I, Antoniadou A, Papadomichelakis E, Poulakou G, Panagea T, Vourli S, Zerva L, Armaganidis A, Kanellakopoulou K, Giamarellou H. 2010. An outbreak of infection due to beta-lactamase Klebsiella pneumoniae carbapenemase 2-producing K. pneumoniae in a Greek university hospital: molecular characterization, epidemiology, and outcomes. Clin Infect Dis 50:364–373. doi: 10.1086/649865. [DOI] [PubMed] [Google Scholar]
- 525.Petty LA, Henig O, Patel TS, Pogue JM, Kaye KS. 2018. Overview of meropenem-vaborbactam and newer antimicrobial agents for the treatment of carbapenem-resistant Enterobacteriaceae. Infect Drug Resist 11:1461–1472. doi: 10.2147/IDR.S150447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 526.El Nekidy WS, Mooty MY, Attallah N, Cardona L, Bonilla MF, Ghazi IM. 2017. Successful treatment of multidrug resistant Klebsiella pneumoniae using dual carbapenem regimen in immunocompromised patient. IDCases 9:53–55. doi: 10.1016/j.idcr.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 527.Shirley M. 2018. Ceftazidime-avibactam: a review in the treatment of serious Gram-negative bacterial infections. Drugs 78:675–692. doi: 10.1007/s40265-018-0902-x. [DOI] [PubMed] [Google Scholar]
- 528.Weiner LM, Webb AK, Limbago B, Dudeck MA, Patel J, Kallen AJ, Edwards JR, Sievert DM. 2016. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011–2014. Infect Control Hosp Epidemiol 37:1288–1301. doi: 10.1017/ice.2016.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 529.Kim B-N, Peleg AY, Lodise TP, Lipman J, Li J, Nation R, Paterson DL. 2009. Management of meningitis due to antibiotic-resistant Acinetobacter species. Lancet Infect Dis 9:245–255. doi: 10.1016/S1473-3099(09)70055-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 530.Sievert DM, Ricks P, Edwards JR, Schneider A, Patel J, Srinivasan A, Kallen A, Limbago B, Fridkin S, National Healthcare Safety Network (NHSN) Team and Participating NHSN Facilities . 2013. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–2010. Infect Control Hosp Epidemiol 34:1–14. doi: 10.1086/668770. [DOI] [PubMed] [Google Scholar]
- 531.Zhou K, Tang X, Wang L, Guo Z, Xiao S, Wang Q, Zhuo C. 2018. An emerging clone (ST457) of Acinetobacter baumannii clonal complex 92 with enhanced virulence and increasing endemicity in south China. Clin Infect Dis 67:S179–S188. doi: 10.1093/cid/ciy691. [DOI] [PubMed] [Google Scholar]
- 532.van Dessel H, Dijkshoorn L, van der Reijden T, Bakker N, Paauw A, van den Broek P, Verhoef J, Brisse S. 2004. Identification of a new geographically widespread multiresistant Acinetobacter baumannii clone from European hospitals. Res Microbiol 155:105–112. doi: 10.1016/j.resmic.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 533.Antunes LCS, Visca P, Towner KJ. 2014. Acinetobacter baumannii: evolution of a global pathogen. Pathog Dis 71:292–301. doi: 10.1111/2049-632X.12125. [DOI] [PubMed] [Google Scholar]
- 534.Lăzureanu V, Poroșnicu M, Gândac C, Moisil T, Bădițoiu L, Laza R, Musta V, Crișan A, Marinescu A-R. 2016. Infection with Acinetobacter baumannii in an intensive care unit in the western part of Romania. BMC Infect Dis 16(Suppl 1):95. doi: 10.1186/s12879-016-1399-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 535.Norton MD, Spilkia AJ, Godoy VG. 2013. Antibiotic resistance acquired through a DNA damage-inducible response in Acinetobacter baumannii. J Bacteriol 195:1335–1345. doi: 10.1128/JB.02176-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 536.Thompson MG, Black CC, Pavlicek RL, Honnold CL, Wise MC, Alamneh YA, Moon JK, Kessler JL, Si YZ, Williams R, Yildirim S, Kirkup BC, Green RK, Hall ER, Palys TJ, Zurawski DV. 2014. Validation of a novel murine wound model of Acinetobacter baumannii infection. Antimicrob Agents Chemother 58:1332–1342. doi: 10.1128/AAC.01944-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 537.Nwugo CC, Arivett BA, Zimbler DL, Gaddy JA, Richards AM, Actis LA. 2012. Effect of ethanol on differential protein production and expression of potential virulence functions in the opportunistic pathogen Acinetobacter baumannii. PLoS One 7:e51936. doi: 10.1371/journal.pone.0051936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 538.Hassan KA, Jackson SM, Penesyan A, Patching SG, Tetu SG, Eijkelkamp BA, Brown MH, Henderson PJF, Paulsen IT. 2013. Transcriptomic and biochemical analyses identify a family of chlorhexidine efflux proteins. Proc Natl Acad Sci U S A 110:20254–20259. doi: 10.1073/pnas.1317052110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 539.Tacconelli E, Tumbarello M, Bertagnolio S, Citton R, Spanu T, Fadda G, Cauda R. 2002. Multidrug-resistant Pseudomonas aeruginosa bloodstream infections: analysis of trends in prevalence and epidemiology. Emerg Infect Dis 8:220–221. doi: 10.3201/eid0802.010121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 540.Lamas Ferreiro JL, Álvarez Otero J, González González L, Novoa Lamazares L, Arca Blanco A, Bermúdez Sanjurjo JR, Rodríguez Conde I, Fernández Soneira M, de la Fuente Aguado J. 2017. Pseudomonas aeruginosa urinary tract infections in hospitalized patients: mortality and prognostic factors. PLoS One 12:e0178178. doi: 10.1371/journal.pone.0178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 541.Marvig RL, Sommer LM, Molin S, Johansen HK. 2015. Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nat Genet 47:57–64. doi: 10.1038/ng.3148. [DOI] [PubMed] [Google Scholar]
- 542.Dou Y, Huan JN, Guo F, Zhou ZD, Shi Y. 2017. Pseudomonas aeruginosa prevalence, antibiotic resistance and antimicrobial use in Chinese burn wards from 2007 to 2014. J Int Med Res 45:1124–1137. doi: 10.1177/0300060517703573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 543.Dapas JI, Rivero C, Burgos P, Vila A. 2016. Pseudomonas aeruginosa infective endocarditis following aortic valve implantation: a note of caution. Open Cardiovasc Med J 10:28–34. doi: 10.2174/1874192401610010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 544.Aguilar-Rodea P, Zúñiga G, Rodríguez-Espino BA, Olivares Cervantes AL, Gamiño Arroyo AE, Moreno-Espinosa S, de la Rosa Zamboni D, López Martínez B, Castellanos-Cruz MDC, Parra-Ortega I, Jiménez Rojas VL, Vigueras Galindo JC, Velázquez-Guadarrama N. 2017. Identification of extensive drug resistant Pseudomonas aeruginosa strains: new clone ST1725 and high-risk clone ST233. PLoS One 12:e0172882. doi: 10.1371/journal.pone.0172882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 545.Maatallah M, Cheriaa J, Backhrouf A, Iversen A, Grundmann H, Do T, Lanotte P, Mastouri M, Elghmati MS, Rojo F, Mejdi S, Giske CG. 2011. Population structure of Pseudomonas aeruginosa from five Mediterranean countries: evidence for frequent recombination and epidemic occurrence of CC235. PLoS One 6:e25617. doi: 10.1371/journal.pone.0025617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 546.Bassetti M, Castaldo N, Cattelan A, Mussini C, Righi E, Tascini C, Menichetti F, Mastroianni CM, Tumbarello M, Grossi P, Artioli S, Carannante N, Cipriani L, Coletto D, Russo A, Digaetano M, Losito AR, Peghin M, Capone A, Nicole S, Vena A, CEFTABUSE Study Group . 2019. Ceftolozane/tazobactam for the treatment of serious Pseudomonas aeruginosa infections: a multicentre nationwide clinical experience. Int J Antimicrob Agents 53:408–415. doi: 10.1016/j.ijantimicag.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 547.Fujii A, Seki M, Higashiguchi M, Tachibana I, Kumanogoh A, Tomono K. 2014. Community-acquired, hospital-acquired, and healthcare-associated pneumonia caused by Pseudomonas aeruginosa. Respir Med Case Rep 12:30–33. doi: 10.1016/j.rmcr.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 548.Sligl WI, Dragan T, Smith SW. 2015. Nosocomial Gram-negative bacteremia in intensive care: epidemiology, antimicrobial susceptibilities, and outcomes. Int J Infect Dis 37:129–134. doi: 10.1016/j.ijid.2015.06.024. [DOI] [PubMed] [Google Scholar]
- 549.Oncul O, Oksuz S, Acar A, Ulkur E, Turhan V, Uygur F, Ulcay A, Erdem H, Ozyurt M, Gorenek L. 2014. Nosocomial infection characteristics in a burn intensive care unit: analysis of an eleven-year active surveillance. Burns 40:835–841. doi: 10.1016/j.burns.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 550.Lund-Palau H, Turnbull AR, Bush A, Bardin E, Cameron L, Soren O, Wierre-Gore N, Alton EWFW, Bundy JG, Connett G, Faust SN, Filloux A, Freemont P, Jones A, Khoo V, Morales S, Murphy R, Pabary R, Simbo A, Schelenz S, Takats Z, Webb J, Williams HD, Davies JC. 2016. Pseudomonas aeruginosa infection in cystic fibrosis: pathophysiological mechanisms and therapeutic approaches. Expert Rev Respir Med 10:685–697. doi: 10.1080/17476348.2016.1177460. [DOI] [PubMed] [Google Scholar]
- 551.Edlin R, Shapiro D, Hersh A, Copp H. 2013. Antibiotic resistance patterns in outpatient pediatric urinary tract infections. J Urol 190:222–227. doi: 10.1016/j.juro.2013.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 552.Chen CH, Huang CC. 2013. Risk factor analysis for extended-spectrum beta-lactamase-producing Enterobacter cloacae bloodstream infections in central Taiwan. BMC Infect Dis 13:417. doi: 10.1186/1471-2334-13-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 553.Wang HJ, Shi H, Zhou W, Hu ZQ, Mu LY, Su M, Jiang YM. 2012. Common pathogens and clinical characteristics of neonatal pneumonia. Zhongguo Dang Dai Er Ke Za Zhi 14:898–902. [PubMed] [Google Scholar]
- 554.Múñez E, Ramos A, Alvarez de Espejo T, Vaqué J, Sánchez-Payá J, Pastor V, Asensio A. 2012. Aetiology of surgical infections in patients undergoing craniotomy. Neurocirugia (Astur) 23:54–59. doi: 10.1016/j.neucir.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 555.Fata F, Chittivelu S, Tessler S, Kupfer Y. 1996. Gas gangrene of the arm due to Enterobacter cloacae in a neutropenic patient. South Med J 89:1095–1096. [DOI] [PubMed] [Google Scholar]
- 556.Sanders WE, Sanders CC. 1997. Enterobacter spp.: pathogens poised to flourish at the turn of the century. Clin Microbiol Rev 10:220–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 557.Izdebski R, Baraniak A, Herda M, Fiett J, Bonten MJ, Carmeli Y, Goossens H, Hryniewicz W, Brun-Buisson C, Gniadkowski M, MOSAR WP2, WP3 and WP5 Study Groups . 2015. MLST reveals potentially high-risk international clones of Enterobacter cloacae. J Antimicrob Chemother 70:48–56. doi: 10.1093/jac/dku359. [DOI] [PubMed] [Google Scholar]
- 558.Lin YC, Chen TL, Ju HL, Chen HS, Wang FD, Yu KW, Liu CY. 2006. Clinical characteristics and risk factors for attributable mortality in Enterobacter cloacae bacteremia. J Microbiol Immunol Infect 39:67–72. [PubMed] [Google Scholar]
- 559.Hunter CJ, Petrosyan M, Ford HR, Prasadarao NV. 2008. Enterobacter sakazakii: an emerging pathogen in infants and neonates. Surg Infect (Larchmt) 9:533–539. doi: 10.1089/sur.2008.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 560.Robredo B, Torres C, Singh KV, Murray BE. 2000. Molecular analysis of Tn1546 in vanA-containing Enterococcus spp. isolated from humans and poultry. Antimicrob Agents Chemother 44:2588–2589. doi: 10.1128/aac.44.9.2588-2589.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 561.Simjee S, White DG, McDermott PF, Wagner DD, Zervos MJ, Donabedian SM, English LL, Hayes JR, Walker RD. 2002. Characterization of Tn1546 in vancomycin-resistant Enterococcus faecium isolated from canine urinary tract infections: evidence of gene exchange between human and animal enterococci. J Clin Microbiol 40:4659–4665. doi: 10.1128/jcm.40.12.4659-4665.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 562.Schwarz FV, Perreten V, Teuber M. 2001. Sequence of the 50-kb conjugative multiresistance plasmid pRE25 from Enterococcus faecalis RE25. Plasmid 46:170–187. doi: 10.1006/plas.2001.1544. [DOI] [PubMed] [Google Scholar]
- 563.Liu Y, Wang Y, Wu C, Shen Z, Schwarz S, Du XD, Dai L, Zhang W, Zhang Q, Shen J. 2012. First report of the multidrug resistance gene cfr in Enterococcus faecalis of animal origin. Antimicrob Agents Chemother 56:1650–1654. doi: 10.1128/AAC.06091-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 564.Lim SK, Tanimoto K, Tomita H, Ike Y. 2006. Pheromone-responsive conjugative vancomycin resistance plasmids in Enterococcus faecalis isolates from humans and chicken feces. Appl Environ Microbiol 72:6544–6553. doi: 10.1128/AEM.00749-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 565.Hegstad K, Mikalsen T, Coque TM, Werner G, Sundsfjord A. 2010. Mobile genetic elements and their contribution to the emergence of antimicrobial resistant Enterococcus faecalis and Enterococcus faecium. Clin Microbiol Infect 16:541–554. doi: 10.1111/j.1469-0691.2010.03226.x. [DOI] [PubMed] [Google Scholar]
- 566.Fujimura T, Murakami K. 2008. Staphylococcus aureus clinical isolate with high-level methicillin resistance with an lytH mutation caused by IS1182 insertion. Antimicrob Agents Chemother 52:643–647. doi: 10.1128/AAC.00395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 567.Maki H, McCallum N, Bischoff M, Wada A, Berger-Bächi B. 2004. tcaA inactivation increases glycopeptide resistance in Staphylococcus aureus. Antimicrob Agents Chemother 48:1953–1959. doi: 10.1128/AAC.48.6.1953-1959.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 568.Partridge SR, Hall RM. 2003. In34, a complex In5 family class 1 integron containing orf513 and dfrA10. Antimicrob Agents Chemother 47:342–349. doi: 10.1128/aac.47.1.342-349.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 569.Toleman MA, Walsh TR. 2008. Evolution of the ISCR3 group of ISCR elements. Antimicrob Agents Chemother 52:3789–3791. doi: 10.1128/AAC.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 570.Mussi MA, Limansky AS, Viale AM. 2005. Acquisition of resistance to carbapenems in multidrug-resistant clinical strains of Acinetobacter baumannii: natural insertional inactivation of a gene encoding a member of a novel family of beta-barrel outer membrane proteins. Antimicrob Agents Chemother 49:1432–1440. doi: 10.1128/AAC.49.4.1432-1440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 571.Wang Y, Tong MK, Chow KH, Cheng VCC, Tse CWS, Wu AKL, Lai RWM, Luk WK, Tsang DNC, Ho PL. 2018. Occurrence of highly conjugative IncX3 epidemic plasmid carrying bla(NDM) in Enterobacteriaceae isolates in geographically widespread areas. Front Microbiol 9:2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 572.Boutoille D, Corvec S, Caroff N, Giraudeau C, Espaze E, Caillon J, Plesiat P, Reynaud A. 2004. Detection of an IS21 insertion sequence in the mexR gene of Pseudomonas aeruginosa increasing beta-lactam resistance. FEMS Microbiol Lett 230:143–146. doi: 10.1016/S0378-1097(03)00882-6. [DOI] [PubMed] [Google Scholar]
- 573.Evans JC, Segal H. 2007. A novel insertion sequence, ISPa26, in oprD of Pseudomonas aeruginosa is associated with carbapenem resistance. Antimicrob Agents Chemother 51:3776–3777. doi: 10.1128/AAC.00837-07. [DOI] [PMC free article] [PubMed] [Google Scholar]