Abstract
Background
The ongoing COVID-19 pandemic has caused more than 193,825 deaths during the past few months. A quick-to-be-identified cure for the disease will be a therapeutic medicine that has prior use experiences in patients in order to resolve the current pandemic situation before it could become worsening. Artificial intelligence (AI) technology is hereby applied to identify the marketed drugs with potential for treating COVID-19.
Methods
An AI platform was established to identify potential old drugs with anti-coronavirus activities by using two different learning databases; one consisted of the compounds reported or proven active against SARS-CoV, SARS-CoV-2, human immunodeficiency virus, influenza virus, and the other one containing the known 3C-like protease inhibitors. All AI predicted drugs were then tested for activities against a feline coronavirus in in vitro cell-based assay. These assay results were feedbacks to the AI system for relearning and thus to generate a modified AI model to search for old drugs again.
Results
After a few runs of AI learning and prediction processes, the AI system identified 80 marketed drugs with potential. Among them, 8 drugs (bedaquiline, brequinar, celecoxib, clofazimine, conivaptan, gemcitabine, tolcapone, and vismodegib) showed in vitro activities against the proliferation of a feline infectious peritonitis (FIP) virus in Fcwf-4 cells. In addition, 5 other drugs (boceprevir, chloroquine, homoharringtonine, tilorone, and salinomycin) were also found active during the exercises of AI approaches.
Conclusion
Having taken advantages of AI, we identified old drugs with activities against FIP coronavirus. Further studies are underway to demonstrate their activities against SARS-CoV-2 in vitro and in vivo at clinically achievable concentrations and doses. With prior use experiences in patients, these old drugs if proven active against SARS-CoV-2 can readily be applied for fighting COVID-19 pandemic.
Keywords: AI, DNN, COVID-19, SARS-CoV-2, Feline coronavirus, Drug repurposing
At a glance of commentary
Scientific background on the subject
Artificial intelligence (AI) has been applied in drug design through generation of a learning-prediction model and perform quick virtual screening to present accurate biology and chemistry outputs. Together with the strategy of drug repurposing, AI can quickly identify drugs promising to fight against the emergent diseases such as COVID-19 that is in its urgent need of efficacious medicines.
What this study adds to the field
COVID-19 has become pandemic worldwide and lacks of effective therapeutics. The study adopted AI approach to lead quick screening against the marketed drugs to identify drugs potentially with anti-coronavirus activity and possibly for treating COVID-19. Several old drugs and compounds were here identified with in vitro anti-coronavirus activity. The application of AI approach in drug discovery is therefore proven feasible. With prior use experiences in patients, these old drugs, if shown active against SARS-CoV-2, can be readily applied to treat the COVID-19 patients.
The outbreak of a new coronavirus SARS-CoV-2, previously known as the 2019-nCoV, has caused 2.81 million confirmed cases of the infection. There are more than 193,825 patients died of the coronavirus disease 2019 (COVID-19) affecting 210 countries and territories and the death toll is still climbing up sharp. SARS-CoV-2 was first reported found in Wuhan, China and, soon after, a human-to-human transmission is confirmed followed by a global COVID-19 pandemic as WHO declared on the 11th of March. Majority of the patients of COVID-19 present with fever and respiratory symptoms and death in some serious complications [1]. There are a number of clinical studies ongoing for treating COVID-19 with potential old drugs and investigational new drugs. In order to quickly resolve the current pandemic with more medicinal cures, repurpose of old drug is one of the strategies highly appreciated.
We have hereby practiced with an artificial intelligence (AI) system for searching marketed drugs potentially with antiviral activities against coronaviruses. An AI system could be an efficient tool to quickly screen large number of compounds with assigned learning datasets and an intention to find drugs for specific purposes such as for treating COVID-19. The established AI system analyzed the most important descriptors, generated relevant prediction models, performed screening for the marketed drugs to quickly identify those with potential activities inhibiting SARS-CoV-2. An in vitro cell model for feline coronavirus replication was set up to evaluate the AI-identified drugs for antiviral activity verification.
Feline coronavirus is an α-coronavirus and the virus causing enteritis in domestic and wild cats. Approximately 5–15% of infected cats develop feline infectious peritonitis (FIP) which is fatal to cats [2]. The infection by FIP virus in cats presented similar features to the severe acute respiratory syndrome (SARS) infection such as pulmonary lesions in humans [3]. It was documented that both the nucleoside analog GS-441524 and 3C-like protease inhibitor GC376 exhibited antiviral activities against FIP virus in vitro and were effective for treating FIP in cats [4,5]. Remdesivir (GS-5734) is the prodrug of GS-441524 that inhibits viral RNA-dependent RNA polymerase and prevents viral replication in vitro including the middle east respiratory syndrome (MERS) virus, Ebola virus, Lassa fever virus, Junin virus and respiratory syncytial virus [6]. Remdesivir is now an investigational new drug that shows promising outcomes in compassionate uses. The use of FIP virus in vitro replication cell model in the present study is therefore relevant and shall provide a useful screening tool to identify marketed drugs with a broad-spectrum antiviral activity.
Methods
Two different learning datasets were generated in which one is consisted of the compounds reported or proven active against SARS-CoV, SARS-CoV-2, human immunodeficiency virus (HIV), and influenza virus and the other contains the known 3C-like protease inhibitors. An AI-system was established and, based on the learning datasets to predict drugs potentially active against coronavirus out of the marketed drugs. The predicted drugs were then tested for activities against a feline coronavirus in in vitro cell-based assay for a verification. During the AI practices, these assay results were served as feedbacks to the AI system for relearning and thus to generate a modified AI model to search for old drugs.
Generation of datasets and AI models
Three types of molecular descriptors, extended connectivity fingerprint (ECFP), functional-class fingerprints (FCFPs), and octanol–water partition coefficient (ALogP_count), were performed to AI learning. The extended connectivity fingerprints (ECFPs) are generated in a molecule-directed manner by systematically recording the neighborhood of each non-hydrogen atom into multiple circular layers up to a given diameter of that molecule. The functional-class fingerprints (FCFPs) [7] detail circular fingerprints via the pharmacophore identification of atoms, which reports topological pharmacophore fingerprints. The ALogP_count is an array of 120 numbers that correspond to the 120 Ghose and Crippen atom types [8]. The 613 descriptors in total were used for the AI practices and prediction of the promising drugs. All these descriptors were generated by Discovery Studio v18.1/Calculates ligand properties program (BIOVIA Inc., San Diego, CA, USA), including ALogP_count (101 descriptors), ECFP_4 (256 descriptors), and FCFP_4 (256 descriptors) as shown in the supplemental information. The system used a Deep Neural Network (DNN) [9] algorithm to identify the most important descriptors and gave different weightings to generate AI prediction models. We collected information of those compounds and drugs that had been reported with promising activities for treating COVID-19, such as anti-influenza drugs [10], drugs and compounds inhibiting SARS-CoV [11], HIV drugs [12], and some showing activities against SARS-CoV-2 as recently reported [13,14]. The other AI prediction model is generated from a learning dataset of 210 drugs and compounds reported with inhibitory activities on the 3C-like protease of SARS-CoV listed in Table 1 [[15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35]]. The database adopted to search for marketed drugs was the DrugBank at website: https://www.drugbank.ca.
Table 1.
Learning datasets for Al training.
| Dataset for AI Model 1 |
|
|
|
|
| Dataset for AI Model 2 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Dataset for Modified AI Model |
|
|
|
Virus and cells
The serotype II FIP virus of Taiwan isolate NTU156 strain, was a kind gift from National Taiwan University [36]. The virus was propagated and titrated in the feline catus whole fetus-4 (Fcwf-4) cells (ATCC®CRL-2787). Fcwf-4 cells were amplified and maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) with 1% penicillin/streptomycin at 37 °C with 5% CO2.
Nonspecific cytotoxicity assay
Confluent Fcwf-4 cells were seeded in 96-well plates and were treated with various concentrations of testing compounds at up to 50 μM at 37 °C with 5% CO2 for 48 h. Cell cytotoxicity was visualized and measured by crystal violet staining.
Measurement of antiviral effects in FIP virus-infected Fcwf-4 cells
In brief, 3 × 104 Fcwf-4 cells per well were seeded in 96-well plates containing DMEM supplemented with 10% FBS and incubated at 37 °C with 5% CO2. Sixteen hr post inoculation, the cells were infected with NTU156 at 300 TCID50 per well and incubated at 37 °C for 1 h and the supernatant were discarded followed by addition of 0.4, 2, 10, and 50 μM of testing compounds in DMEM medium containing 2% FBS. The culture plates were incubated for additional 48 h at 37 °C with 5% CO2 followed by fixing with 10% formalin and stained with 0.1% crystal violet 20% ethanol (w/v). The cytopathic effect (CPE) caused by FIP virus were measured visually for intensity of crystal violet staining. Back titration of FIP virus were performed parallelly and the TCID50 values were calculated according a Reed and Muench method [37].
Results
Results from AI model 1 approach
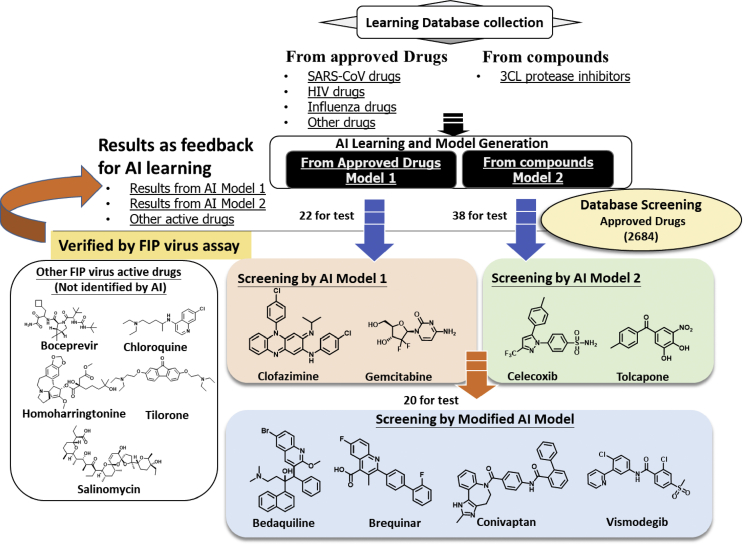
AI prediction model 1 was exercised based on the learning dataset listed in Table 1, containing some of the drugs for influenza, from previous reports showing activities against SARS-CoV, HIV drugs, and drugs and compounds of potential inhibiting SARS-CoV-2 as recently reported as shown in Fig. 1. The AI model identified 22 drugs to be having the potential of antiviral effects. Followed by evaluations on these 22 drugs with the in vitro cell-based FIP virus replication assay, 2 promising drugs clofazimine and gemcitabine were verified with activity against the FIP coronavirus (Table 2).
Fig. 1.
Flow scheme of AI approaches along with antiviral activity verification assays. Two independent datasets were compiled as the learning inputs to generate two AI prediction models of different approaches. Firstly, Model 1 was generated form the known drugs with antiviral activities. Model 2 was generated form the 3C-like protease inhibitors. A database for the market-approved drugs were adopted in which the AI system screens for potential drugs with antiviral activities. The AI-predicted drugs were verified with antiviral activities by a cell-based FIP virus replication assay. Finally, these assay results served as feedbacks for the AI relearning and evolution progress. The Modified AI model was established to screen further and again verified by the FIP virus replication assay. The processing flows are indicated with the arrows.
Table 2.
Assay results of the drugs predicted by AI Model 1.
| AI predicted drugs | Concentration (μM) |
|
|---|---|---|
| Cytotoxicity | Viral Inhibition | |
| 5-Azacytidine | >10 | NA |
| Agomelatine | >50 | >50 |
| Carprofen | >50 | >50 |
| Clofazimine | >50 | >10 |
| Cobicistat | >50 | >50 |
| Cyclofenil | >10 | NA |
| Cytarabine | >2 | NA |
| Diosmetin | >50 | >50 |
| Dolasetron | >50 | >50 |
| Flurbiprofen | >50 | >50 |
| Gemcitabine | >50 | ≧2 |
| Histamine | >50 | >50 |
| Ipriflavone | >50 | >50 |
| Loratadine | >10 | NA |
| Melatonin | >50 | >50 |
| Pexidartinib | >2 | NA |
| Ramosetron | >50 | >50 |
| Ruxolitinib | >10 | NA |
| Sennoside A | >50 | >50 |
| Torsemide | >50 | >50 |
| Triclabendazole | >2 | NA |
| Tropisetron | >50 | >50 |
| GC376 | >50 | 0.4–2 |
Abbreviations: NA: not available to detect due to cytotoxicity;
GC376: a reference compound.
Results from AI model 2 approach
An independent approach from the AI model 1, AI prediction model 2 was practiced based on the learning dataset of 210 inhibitors of the 3C-like protease of SARS-CoV listed in Table 1. The AI model predicted 38 drugs with potential inhibiting the 3C-like protease of SARS-CoV and they were subject to tests in the FIP virus replication assay. Two out of the 38 AI-identified drugs, celecoxib and tolcapone, showed antiviral activities against the FIP coronavirus (Table 3).
Table 3.
Assay results of the drugs predicted by AI Model 2.
| AI predict drugs | Concentration (μM) |
|
|---|---|---|
| Cytotoxicity | Viral Inhibition | |
| ABT-199 | >2 | NA |
| Baricitinib | >50 | >50 |
| Bifonazole | >10 | NA |
| Celecoxib | ≧50 | 50 |
| Chlorothiazide | >50 | >50 |
| Clorsulon | >50 | >50 |
| Dapsone | >50 | >50 |
| Dihydroergotamine | >50 | >50 |
| Dolasetron | >50 | >50 |
| Eletriptan HBr | >50 | >50 |
| Eptifibatide | >50 | >50 |
| Etodolac | >50 | >50 |
| Fenbufen | >50 | >50 |
| Flufenamic acid | >50 | >50 |
| Fomepizole | >50 | >50 |
| Histamine | >50 | >50 |
| Hydrochlorothiazide | >50 | >50 |
| Indapamide | >50 | >50 |
| Naratriptan | >50 | >50 |
| Nimesulide | >50 | >50 |
| Nitazoxanide | >10 | NA |
| Nitisinone | >50 | >50 |
| Quinapril | >50 | >50 |
| Rizatriptan | >50 | >50 |
| Ruxolitinib | >10 | NA |
| Serotonin | >50 | >50 |
| Sorafenib | >50 | >50 |
| Sumatriptan | >50 | >50 |
| Tadalafil | >50 | >50 |
| Thiabendazole | >50 | >50 |
| Tofacitinib | >50 | >50 |
| Tolcapone | >50 | 50 |
| Trichlormethiazide | >50 | >50 |
| Triclabendazole | >2 | NA |
| Triflusal | >50 | >50 |
| Tropisetron | >50 | >50 |
| Vemurafenib | >10 | NA |
| Zinc Pyrithione | >0.4 | NA |
Abbreviation: NA: not available to detect due to cytotoxicity.
Results from the modified AI model
The results from the two AI models 1 and 2 were shown in Table 2, Table 3, respectively, and were served together as a feedback dataset to the AI system for relearning and therefore a modified AI prediction system was established and used for the follow-up prediction exercise. During our integrated efforts for repurposing drugs to tackle COVID-19, there were additional drugs found active in our in vitro assay against the FIP virus (Table 4), such as boceprevir [38], chloroquine [39], homoharringtonine [40], tilorone [41], and salinomycin [42] as they were also reported previously with potential to inhibit SARS-CoV or SARS-CoV-2. These 5 drugs were included as a sub-dataset for the relearning and thus to generate the modified AI prediction system which was used to screen and identify those 20 drugs out of the approved 2684 drugs for tests in the FIP virus replication assay (Table 5). With the AI evolution, the system showed a higher fidelity and 4 drugs (bedaquiline, brequinar, conivaptan, and vismodegib) out of the 20 AI-identified were verified with antiviral activities against the FIP coronavirus (Fig. 2).
Table 4.
Other drugs showing antiviral activities against FIP virus.
| Drugs | Concentration (μM) |
|
|---|---|---|
| Cytotoxicity | Viral Inhibition | |
| Boceprevir | >50 | 50 |
| Chloroquine | >10 | 10 |
| Homoharringtonine | >2 | 2 |
| Salinomycin | >10 | 10 |
| Tilorone | >10 | 10 |
Table 5.
Assay results of drugs predicted by the modified Al model.
| AI predicted drugs | Concentration (μM) |
|
|---|---|---|
| Cytotoxicity | Viral Inhibition | |
| Bedaquiline | >50 | 50 |
| Bifonazole | >10 | NA |
| Brequinar | >50 | 2 |
| Clotrimazole | >2 | NA |
| Conivaptan | >50 | 50 |
| Duvelisib | >50 | >50 |
| Econazole | >10 | NA |
| Fenticonazole | >2 | NA |
| Fumaric acid | >50 | >50 |
| Lapatinib | >10 | NA |
| Miconazole | >2 | NA |
| Miconazole (nitrate) | >10 | NA |
| Pranlukast | >50 | >50 |
| Sertaconazole | >10 | NA |
| Sulconazole | >10 | NA |
| Sulconazole (nitrate) | >10 | NA |
| Tacrolimus | >50 | >50 |
| Telmisartan | >50 | >50 |
| Tipifarnib | >10 | NA |
| Vismodegib | >50 | 50 |
Abbreviation: NA: not available to detect due to cytotoxicity.
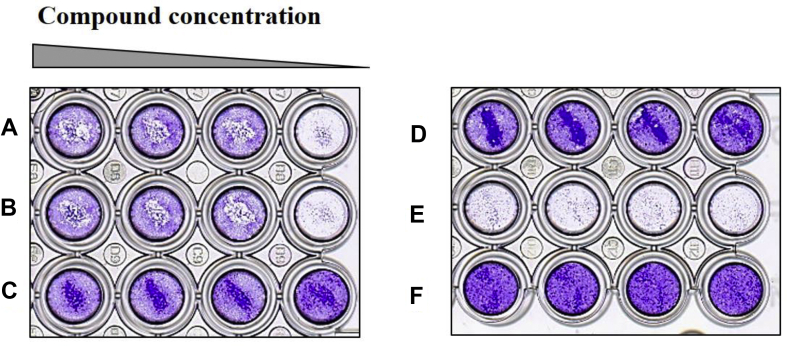
Fig. 2.
Fcwf-4 cells infected with FIP virus showed cytopathic effects, crystal violet staining. FIP virus (NTU156) infected cells treated with brequinar of various concentrations (0.4, 2, 10, 50 μM) in duplicate wells (Row A and B) and cytotoxicity of brequinar at 0.4, 2, 10, 50 μM was also investigated (Row C). FIPV-infected cells treated with a positive control, anti-FIP virus compound GC-376 (Row D) and vehicle only (Row E). Uninfected cells without drugs in parallel treatments were shown (Row F).
Discussion
Predicted by the AI system, 8 drugs were identified with antiviral activities against FIP virus proliferation in the Fcwf-4 cells. Together with the 5 additional drugs identified during the current AI approach and drug repurposing practices, there are 13 drugs with great potentials for further development toward treating COVID-19. Chloroquine showed inhibition activity against FIP virus at 10 μM, which is in agreement with the recent report about its broad-spectrum antiviral effects and it is one of the drugs used to treat COVID-19 patients in China and authorized for emergency use in United States [14]. Remdesivir is a SARS-CoV-2 RNA-dependent RNA polymerase inhibitor and a promising drug candidate for treating COVID-19 [4]; now used in COVID-19 patients in clinical trials in a number of countries including Taiwan. GS-441524 is the active metabolite of remdesivir and showed activity against FIP virus at around 2 μM similar to the previously reported for GS-441524 being developed as a veterinary medicine treating feline coronavirus infections [4]. The results therefore provide solid evidence to support the validity of using cell-based FIP virus infection in vitro model as an antiviral activity assay in the present study. To have the promising drugs further being tested in a SARS-CoV-2 in vitro assay and in vivo animal models is planned to be ongoing.
Among these AI-identified active drugs, four drugs (brequinar, clofazimine, gemcitabine and conivaptan) are of potential in their clinical uses and hereby elaborated further. Brequinar is a synthetic quinolinecarboxylic acid analogue and a dihydroorotate dehydrogenase (DHODH) inhibitor with experiences of patients uses for antitumor activities [43]. It was also used as an immunosuppressant in combination with other drugs such as cyclosporine for synergism [44]. It is of high water solubility and given by IV infusion. The biodistribution studies in rats of brequinar showed that liver and small intestines were the most exposed organs. The elimination rate of brequinar from all tissues were approximately equal to that from the blood circulation indicating no tissue accumulation of the intact drug molecules [45]. The present finding demonstrated that brequinar has an inhibiting activity of 2–10 μM. The systemically exposed plasma concentration range of brequinar in patients are around 73–261 μM for doses of 135–300 m2/kg in 5-day infusion period suggesting that brequinar may be effective against coronaviruses in appropriate dose regimens given alone or in combination with others [44].
Clofazimine is a water-soluble dye developed for the treatment of tuberculosis and used in combination for leprosy. It has a long circulation half-life of 25 days and the serum concentrations remain relatively low with evident tissue accumulation to extremely high levels approximately as 1.5 mg/g in jejunum. After 20 weeks treatment, the peak concentrations in lungs were 106 μg/g and 9020 μg/g in the lungs and spleen, respectively, in mice [46]. Clofazimine exhibited a dose-related relationship of tissue and serum concentrations (1.56–2.23 μM) from an oral dose of 100–300 mg in patients. A long onset of at least two weeks was observed in patients. Taking the advantages of the very slow clearance from body long half-life, and high drug concentration in the lungs [47], clofazimine with minimum adverse effects could be further investigated for treating COVID-19.
Conivaptan is a dual V1A/V2 vasopressin antagonist and given intravenously for the treatment of euvolemic and hypervolemic hyponatremia. The peak concentration C0.5hr of the loading dose at 40 mg/day was about 1.4 μM and the maintenance dose at 40 mg/day for 4-day infusion was around 0.4 μM in patients [48]. Although these concentrations were lower than the inhibitory concentrations at approximately 50 μM, a clinical regimen may be manageable to seek possible efficacy and tolerability in COVID-19 patients with proper study design.
Gemcitabine is an antimetabolite and first-line anticancer drug for pancreatic cancer. It also exhibits activities against breast cancer, ovarian cancer, non-small cell lung cancer, and bladder cancer. Gemcitabine is currently used alone or combined in cancer patients received 600–1200 mg/m2 gemcitabine of 60–120 min i.v. infusion patients in treatment cycles with achievable plasma concentrations of 25–139 μM [49]. Side effects of gemcitabine include pneumonitis, pulmonary fibrosis, pulmonary edema, and adult respiratory distress syndrome. These pulmonary events could lead to fatal respiratory failure in some cases despite discontinuation of gemcitabine therapy 2 weeks ahead. A treatment regimen of low dose and frequency may be explored for single use against COVID-19 in animal models and for combined uses with the other drugs [50]. The present study revealed that the inhibitory activity of gemcitabine on coronavirus FIPV was approximately at 2 μM. A proper combined use with the other drugs in a lower dose of gemcitabine may overcome the pulmonary adverse effects while active inhibiting SARS-CoV-2 for cure of COVID-19 in further preclinical and clinical studies.
Conclusion
AI approach is an efficient way to help quickly identify potential drugs as antiviral therapeutics. Some of the AI-identified drugs show activities against the proliferation of feline coronavirus in vitro. At the time of writing, there are still thousands of people dying of COVID-19 pandemic every day and we hope any of these AI-identified old drugs could soon make it to an effective cure for COVID-19.
Funding
A special government fund to tackle the COVID-19 pandemic from the Ministry of Health and Welfare, Taiwan.
Conflicts of Interest
The authors declare that they have no competing interests.
Acknowledgments
We thank Dr. Kung-Yee Liang for his support and helpful editing on the study and manuscript writing.
Footnotes
Peer review under responsibility of Chang Gung University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bj.2020.05.001.
Contributor Information
Yi-Yu Ke, Email: yiyuke@nhri.edu.tw.
Chiung-Tong Chen, Email: ctchen@nhri.edu.tw.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Hui D.S., E I.A., Madani T.A., Ntoumi F., Kock R., Dar O. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health - the latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pedersen N.C. Serologic studies of naturally occurring feline infectious peritonitis. Am J Vet Res. 1976;37:1449–1453. [PubMed] [Google Scholar]
- 3.Paltrinieri S. Human severe acute respiratory syndrome (SARS) and feline coronaviruses. J Feline Med Surg. 2004;6:131–132. doi: 10.1016/j.jfms.2003.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pedersen N.C., Perron M., Bannasch M., Montgomery E., Murakami E., Liepnieks M. Efficacy and safety of the nucleoside analog GS-441524 for treatment of cats with naturally occurring feline infectious peritonitis. J Feline Med Surg. 2019;21:271–281. doi: 10.1177/1098612X19825701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pedersen N.C., Kim Y., Liu H.W., Kankanamalage A.C.G., Eckstrand C., Groutas W.C. Efficacy of a 3C-like protease inhibitor in treating various forms of acquired feline infectious peritonitis. J Feline Med Surg. 2018;20:378–392. doi: 10.1177/1098612X17729626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sheahan T.P., Sims A.C., Graham R.L., Menachery V.D., Gralinski L.E., Case J.B. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med. 2017;28:eaal3653. doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rogers D., Hahn M. Extended-connectivity fingerprints. J Chem Inf Model. 2010;50:742–754. doi: 10.1021/ci100050t. [DOI] [PubMed] [Google Scholar]
- 8.Ghose A.K., Crippen G.M. Atomic physicochemical parameters for three-dimensional-structure-directed quantitative structure-activity relationships. 2. Modeling dispersive and hydrophobic interactions. J Chem Inf Comput Sci. 1987;27:21–35. doi: 10.1021/ci00053a005. [DOI] [PubMed] [Google Scholar]
- 9.Ma J.S., Sheridan R.P., Liaw A., Dahl G.E., Svetnik V. Deep neural nets as a method for quantitative structure-activity relationships. J Chem Inf Model. 2015;55:263–274. doi: 10.1021/ci500747n. [DOI] [PubMed] [Google Scholar]
- 10.Barik S. New treatments for influenza. BMC Med. 2012;10:104. doi: 10.1186/1741-7015-10-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang Y.W., Yeh T.K., Lin K.T., Chen W.C., Yao H.T., Lan S.J. Pharmacokinetics of anti-SARS-CoV agent niclosamide and its analogs in rats. J Food Drug Anal. 2006;14:329–333. [Google Scholar]
- 12.Chow W.A., Jiang C.L., Guan M. Anti-HIV drugs for cancer therapeutics: back to the future? Lancet Oncol. 2009;10:61–71. doi: 10.1016/S1470-2045(08)70334-6. [DOI] [PubMed] [Google Scholar]
- 13.Ling C.Q. Traditional Chinese medicine is a resource for drug discovery against 2019 novel coronavirus (SARS-CoV-2) J Integr Med. 2020;18:87–88. doi: 10.1016/j.joim.2020.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosa S.G.V., Santos W.C. Clinical trials on drug repositioning for COVID-19 treatment. Rev Panam Salud Publica. 2020;44:e40. doi: 10.26633/RPSP.2020.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blanchard J.E., Elowe N.H., Huitema C., Fortin P.D., Cechetto J.D., Eltis L.D. High-throughput screening identifies inhibitors of the SARS coronavirus main proteinase. Chem Biol. 2004;11:1445–1453. doi: 10.1016/j.chembiol.2004.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen C.N., Lin C.P., Huang K.K., Chen W.C., Hsieh H.P., Liang P.H. Inhibition of SARS-CoV 3C-like protease activity by theaflavin-3,3'-digallate (TF3) Evid Based Complement Alternat Med. 2005;2:209–215. doi: 10.1093/ecam/neh081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen L., Chen S., Gui C., Shen J., Shen X., Jiang H. Discovering severe acute respiratory syndrome coronavirus 3CL protease inhibitors: virtual screening, surface plasmon resonance, and fluorescence resonance energy transfer assays. J Biomol Screen. 2006;11:915–921. doi: 10.1177/1087057106293295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen L.R., Wang Y.C., Lin Y.W., Chou S.Y., Chen S.F., Liu L.T. Synthesis and evaluation of isatin derivatives as effective SARS coronavirus 3CL protease inhibitors. Bioorg Med Chem Lett. 2005;15:3058–3062. doi: 10.1016/j.bmcl.2005.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghosh A.K., Gong G., Grum-Tokars V., Mulhearn D.C., Baker S.C., Coughlin M. Design, synthesis and antiviral efficacy of a series of potent chloropyridyl ester-derived SARS-CoV 3CLpro inhibitors. Bioorg Med Chem Lett. 2008;18:5684–5688. doi: 10.1016/j.bmcl.2008.08.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghosh A.K., Xi K., Grum-Tokars V., Xu X., Ratia K., Fu W. Structure-based design, synthesis, and biological evaluation of peptidomimetic SARS-CoV 3CLpro inhibitors. Bioorg Med Chem Lett. 2007;17:5876–5880. doi: 10.1016/j.bmcl.2007.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacobs J., Grum-Tokars V., Zhou Y., Turlington M., Saldanha S.A., Chase P. Discovery, synthesis, and structure-based optimization of a series of N-(tert-butyl)-2-(N-arylamido)-2-(pyridin-3-yl) acetamides (ML188) as potent noncovalent small molecule inhibitors of the severe acute respiratory syndrome coronavirus (SARS-CoV) 3CL protease. J Med Chem. 2013;56:534–546. doi: 10.1021/jm301580n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jain R.P., Pettersson H.I., Zhang J., Aull K.D., Fortin P.D., Huitema C. Synthesis and evaluation of keto-glutamine analogues as potent inhibitors of severe acute respiratory syndrome 3CLpro. J Med Chem. 2004;47:6113–6116. doi: 10.1021/jm0494873. [DOI] [PubMed] [Google Scholar]
- 23.Kao R.Y., Tsui W.H., Lee T.S., Tanner J.A., Watt R.M., Huang J.D. Identification of novel small-molecule inhibitors of severe acute respiratory syndrome-associated coronavirus by chemical genetics. Chem Biol. 2004;11:1293–1299. doi: 10.1016/j.chembiol.2004.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuo C.J., Liu H.G., Lo Y.K., Seong C.M., Lee K.I., Jung Y.S. Individual and common inhibitors of coronavirus and picornavirus main proteases. FEBS Lett. 2009;583:549–555. doi: 10.1016/j.febslet.2008.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu I.L., Mahindroo N., Liang P.H., Peng Y.H., Kuo C.J., Tsai K.C. Structure-based drug design and structural biology study of novel nonpeptide inhibitors of severe acute respiratory syndrome coronavirus main protease. J Med Chem. 2006;49:5154–5161. doi: 10.1021/jm060207o. [DOI] [PubMed] [Google Scholar]
- 26.Mukherjee P., Desai P., Ross L., White E.L., Avery M.A. Structure-based virtual screening against SARS-3CL(pro) to identify novel non-peptidic hits. Bioorg Med Chem. 2008;16:4138–4149. doi: 10.1016/j.bmc.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thanigaimalai P., Konno S., Yamamoto T., Koiwai Y., Taguchi A., Takayama K. Design, synthesis, and biological evaluation of novel dipeptide-type SARS-CoV 3CL protease inhibitors: structure-activity relationship study. Eur J Med Chem. 2013;65:436–447. doi: 10.1016/j.ejmech.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thanigaimalai P., Konno S., Yamamoto T., Koiwai Y., Taguchi A., Takayama K. Development of potent dipeptide-type SARS-CoV 3CL protease inhibitors with novel P3 scaffolds: design, synthesis, biological evaluation, and docking studies. Eur J Med Chem. 2013;68:372–384. doi: 10.1016/j.ejmech.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsai K.C., Chen S.Y., Liang P.H., Lu I.L., Mahindroo N., Hsieh H.P. Discovery of a novel family of SARS-CoV protease inhibitors by virtual screening and 3D-QSAR studies. J Med Chem. 2006;49:3485–3495. doi: 10.1021/jm050852f. [DOI] [PubMed] [Google Scholar]
- 30.Turlington M., Chun A., Tomar S., Eggler A., Grum-Tokars V., Jacobs J. Discovery of N-(benzo[1,2,3]triazol-1-yl)-N-(benzyl)acetamido)phenyl) carboxamides as severe acute respiratory syndrome coronavirus (SARS-CoV) 3CLpro inhibitors: identification of ML300 and noncovalent nanomolar inhibitors with an induced-fit binding. Bioorg Med Chem Lett. 2013;23:6172–6177. doi: 10.1016/j.bmcl.2013.08.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wen C.C., Kuo Y.H., Jan J.T., Liang P.H., Wang S.Y., Liu H.G. Specific plant terpenoids and lignoids possess potent antiviral activities against severe acute respiratory syndrome coronavirus. J Med Chem. 2007;50:4087–4095. doi: 10.1021/jm070295s. [DOI] [PubMed] [Google Scholar]
- 32.Wu C.Y., Jan J.T., Ma S.H., Kuo C.J., Juan H.F., Cheng Y.S. Small molecules targeting severe acute respiratory syndrome human coronavirus. Proc Natl Acad Sci U S A. 2004;101:10012–10017. doi: 10.1073/pnas.0403596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Q., Chen L., He X., Gao Z., Shen X., Bai D. Design and synthesis of cinanserin analogs as severe acute respiratory syndrome coronavirus 3CL protease inhibitors. Chem Pharm Bull. 2008;56:1400–1405. doi: 10.1248/cpb.56.1400. [DOI] [PubMed] [Google Scholar]
- 34.Zhang J., Huitema C., Niu C., Yin J., James M.N., Eltis L.D. Aryl methylene ketones and fluorinated methylene ketones as reversible inhibitors for severe acute respiratory syndrome (SARS) 3C-like proteinase. Bioorg Chem. 2008;36:229–240. doi: 10.1016/j.bioorg.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J., Pettersson H.I., Huitema C., Niu C., Yin J., James M.N. Design, synthesis, and evaluation of inhibitors for severe acute respiratory syndrome 3C-like protease based on phthalhydrazide ketones or heteroaromatic esters. J Med Chem. 2007;50:1850–1864. doi: 10.1021/jm061425k. [DOI] [PubMed] [Google Scholar]
- 36.Lin C.N., Chang R.Y., Su B.L., Chueh L.L. Full genome analysis of a novel type II feline coronavirus NTU156. Virus Gene. 2013;46:316–322. doi: 10.1007/s11262-012-0864-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reed L.J., Muench H. A simple method of estimating fifty per cent endpoints. Am J Epidemiology. 1938;27:493–497. [Google Scholar]
- 38.Kaeppler U., Stiefl N., Schiller M., Vicik R., Breuning A., Schmitz W. A new lead for nonpeptidic active-site-directed inhibitors of the severe acute respiratory syndrome coronavirus main protease discovered by a combination of screening and docking methods. J Med Chem. 2005;48:6832–6842. doi: 10.1021/jm0501782. [DOI] [PubMed] [Google Scholar]
- 39.Wang M.L., Cao R.Y., Zhang L.K., Yang X.L., Liu J., Xu M.Y. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choy K.T., Yin-Lam Wong A., Kaewpreedee P., Sia S.F., Chen D., Yan Hui K.P. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antivir Res. 2020;178:104786. doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ekins S., Lane T.R., Madrid P.B. Tilorone: a broad-spectrum antiviral invented in the USA and commercialized in Russia and beyond. Pharmaceut Res. 2020;37:71. doi: 10.1007/s11095-020-02799-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang R., Wang L., Zhang N., Deng X., Su M., Su Y. Development of small-molecule MERS-CoV inhibitors. Viruses. 2018;10:721. doi: 10.3390/v10120721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sykes D.B. The emergence of dihydroorotate dehydrogenase (DHODH) as a therapeutic target in acute myeloid leukemia. Expert Opin Ther Targets. 2018;22:893–898. doi: 10.1080/14728222.2018.1536748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arteaga C.L., Brown T.D., Kuhn J.G., Shen H.S.L., Orourke T.J., Beougher K. Phase-I clinical and pharmacokinetic trial of brequinar sodium (Dup 785 Nsc 368390) Canc Res. 1989;49:4648–4653. [PubMed] [Google Scholar]
- 45.Shen H.S.L., Chen S.F., Behrens D.L., Whitney C.C., Dexter D.L., Forbes M. Distribution of the novel anticancer drug candidate brequinar sodium (Dup-785, Nsc-368390) into Normal and tumor-tissues of nude-mice bearing human-colon carcinoma xenografts. Canc Chemother Pharmacol. 1988;22:183–186. doi: 10.1007/BF00273407. [DOI] [PubMed] [Google Scholar]
- 46.Jadhav M.V., Sathe A.G., Deore S.S., Patil P.G., Joshi N.G. Tissue concentration, systemic distribution and toxicity of clofazimine-an autopsy study. Indian J Pathol Microbiol. 2004;47:281–283. [PubMed] [Google Scholar]
- 47.Swanson R.V., Adamson J., Moodley C., Ngcobo B., Ammerman N.C., Dorasamy A. Pharmacokinetics and pharmacodynamics of clofazimine in a mouse model of tuberculosis. Antimicrob Agents Chemother. 2015;59:3042–3051. doi: 10.1128/AAC.00260-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marbury T., Fox J., Kaelin B., Pavliv L. Pharmacokinetics of conivaptan use in patients with severe hepatic impairment. Drug Des Dev Ther. 2017;11:373–382. doi: 10.2147/DDDT.S125459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caffo O., Fallani S., Marangon E., Nobili S., Cassetta M.I., Murgia V. Pharmacokinetic study of gemcitabine, given as prolonged infusion at fixed dose rate, in combination with cisplatin in patients with advanced non-small-cell lung cancer. Canc Chemother Pharmacol. 2010;65:1197–1202. doi: 10.1007/s00280-010-1255-7. [DOI] [PubMed] [Google Scholar]
- 50.Siddall E., Khatri M., Radhakrishnan J. Capillary leak syndrome: etiologies, pathophysiology, and management. Kidney Int. 2017;92:37–46. doi: 10.1016/j.kint.2016.11.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.