Abstract
In order to benefit the public, community workers and scientific community, we hereby present a chronicle of SARS-CoV-2 that leads to the unseen precedent of social distancing and lockdown owing to coronavirus disease (COVID-19). Information on this life-threatening pandemic of COVID-19 is sparse and discrete; and the urgency is such that the dissemination of information is increasing with numerous daily publications on the topic. Therefore, we developed a comprehensive review on various aspects of SARS-CoV-2 and COVID-19. We scientifically compiled published research, news, and reports from various sources to comprehend and summarize the information and findings on Coronaviruses. The review explicitly covers the aspects like genome and pedigree of SARS-CoV-2; epidemiology, prognosis, pathogenesis, symptoms and diagnosis of COVID-19 in order to catalog the right information on transmission route, and influence of environmental factors on virus transmissions, for the robust understanding of right strategical steps for proper COVID-19 management. We have explicitly highlighted several useful information and facts like: i) No established relationship between progression of SARS-CoV-2 with temperature, humidity and/or both, ii) The underlying mechanism of SARS-CoV-2 is not fully understood, iii) Respiratory droplet size determines drop and airborne-based transmission, iv) Prognosis of COVID-19 can be done by its effects on various body organs, v) Infection can be stopped by restricting the binding of S protein and AE2, vi) Hydroxychloroquine is believed to be better than chloroquine for COVID-19, vii) Ivermectin with Vero-hSLAM cells is able to reduce infection by ~5000 time within 2 days, and viii) Nafamostat mesylate can inhibit SARS-CoV-2 S protein-initiated membrane fusion. We have also suggested future research perspectives, challenges and scope.
Keywords: SARS-CoV-2, SARS-CoV, MERS-CoV, COVID-19, Transmission, Pathogenesis
Graphical abstract
Highlights
-
•
Phylogeny of the SARS-CoV-2 is remarkably similar to SARS of pangolin and/or bat origin.
-
•
Transmission depends on respiratory droplet size, environmental condition and immunity.
-
•
COVID-19 progression is highly debated for temperature, humidity, and lifestyle dependence.
-
•
Infection can be restricted through restricting binding of S protein and AE2.
-
•
Increasing neutrophil and lymphocyte ratio (NLR) may give early sign of COVID-19.
1. Introduction
In December 2019, the first outbreak of respiratory contagion owing an uncharted etiology was reported in Wuhan city of China. The World Health Organisation (WHO) used the term ‘novel corona virus 2019’ on 29th December 2019 for the outbreak that affected humans in Wuhan, China, which currently is being referred as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), as proposed by Coronavirus Study Group (CSG) of the International Committee on 11th February 2020 (Guo et al., 2020). Further, the official name of the disease caused by SARS-CoV-2 has been termed as coronavirus disease (COVID-19) by WHO (Wang et al., 2020a; Wu et al., 2020a; Wu et al., 2020b; Zhou et al., 2020b; Zhou et al., 2020a). The genomic characterization revealed close relation of COVID-19 virus with the two bat coronaviruses (SARS-CoV and pangolin coronaviruses) and further the linkage of most of the cases to the Huanan Seafood Wholesale Market, indicating its zoonotic origin (Novel, 2020; Perlman, 2020; ECDC (14/02/2020); WHO (11/03/2020a)). The virus is reported to belong to the group of β – coronavirus (Wang et al., 2020a). The virus is named as coronavirus due to its particular shape resembling a crown with projected spikes made of protein. The coronavirus comprises of a capsid with single-stranded ribonucleic acid (ssRNA), which is allied with a nucleoprotein in matrix protein (Tyrrell and Myint, 1996). Like other SARS virus, the SARS-CoV-2 has three types of hosts (i.e. natural, intermediate, and final), leading to a great amount of difficulty in its prevention and treatment (Wang et al., 2020a). SARS-CoV-2 has caused much greater pandemics than severe acute respiratory syndrome (SARS) in 2002–2004 and Middle East Respiratory Syndrome-related Coronavirus (MERS) in 2012.
As reported by Wang et al. (2020b), the COVID-19 outbreak range is rapid and more extensive than SARS. According to WHO report published on 29th February, as of 12th February 2020, a total number of infected cases were 79,394 cases and 2838 deaths (Wang et al., 2020b) and one month after, on 11th March 2020, COVID-19 was declared as a global pandemic owing to unprecedented global escalation in the affected people, and gravity of likely repercussions. An exponential curve has already been shaped up by mid of the March 2020, for both total infected people and deaths. By the time the revision of the present work is being submitted i.e. till 29th April 2020, WHO reports 3,018,952 worldwide confirmed cases of COVID-19 infection with a death count of 207,973 around the globe, derailing every business sectors, institutions, life, public health, and the world economy (Phelan et al., 2020; Wang et al., 2020a; Wang et al., 2020b; Wang et al., 2020c; Wang et al., 2020d).
The present review work provides a comprehensive platform to understand about COVID-19 from the point view of its: i) virology and genome, ii) Epidemiology, iii) Symptoms and Prognosis, iv) Pathogenesis and Diagnosis, v) Transmission routes, vi) Effect of environmental factors, and vii) existing treatment strategies/management for COVID-19. The present work intend to not only help the scientific community by enlisting probable future research directions pertaining to COVID-19, but will also act as a general awareness guide for the public and policymakers.
2. Virology and genome of SARS-CoV-2
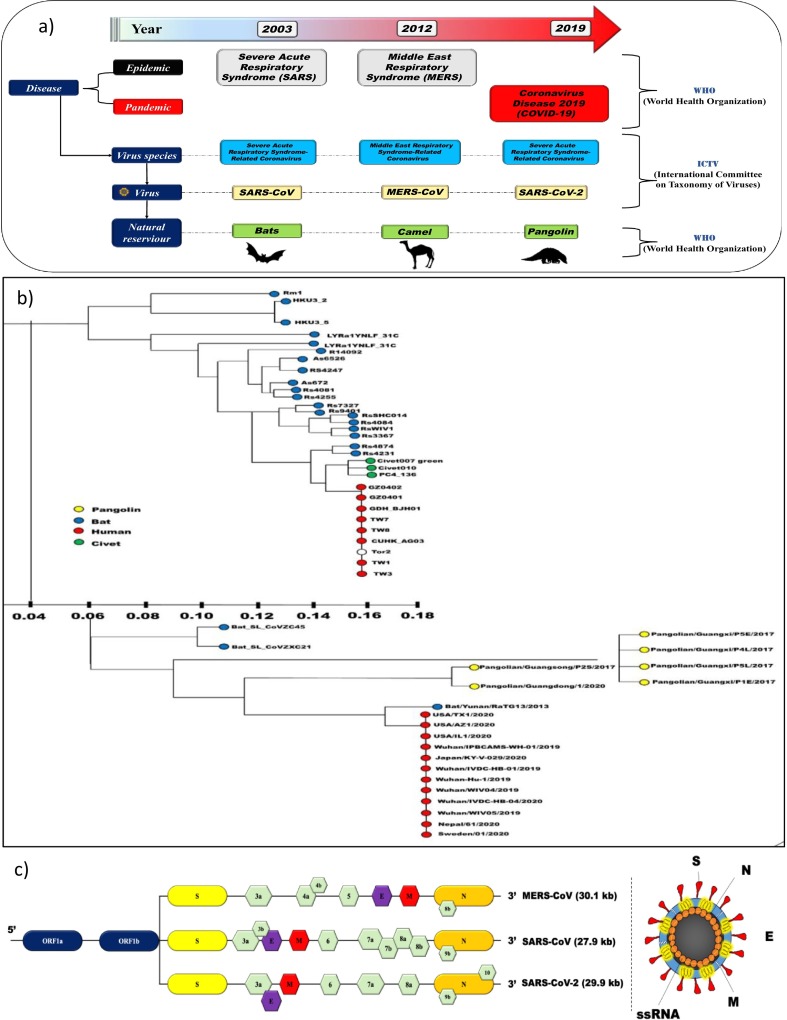
Fig. 1(a) demonstrates the naming of coronavirus with respect to virus taxonomy and diseases caused in a timeline fashioned, and Fig. 1(b) provides a descriptive phylogenetic tree of SARS-like coronaviruses (CoV) i.e. SARS-CoV, MERS-CoV, and SARSCoV-2. According to Li et al., 2020 genome of SARS-CoV-2 has ten open reading frameworks (ORFs) that are similar to other CoV. Fig. 1(c) illustrates that at 1/3rd of the genome, the ORFs encode four main structural proteins: spike (S), envelope (E), nucleocapsid (N), and membrane (M) protein in SARS-CoV-2, SARS-CoV and MERS-CoV. SARS-CoV-2 belongs to a particular class of viruses with enveloped structure, containing single-stranded RNA (Tyrrell and Myint, 1996; Su et al., 2016) and has a base pair (bp) of about 26–32 kilobase (kb) (Su et al., 2016) of diameter 80–120 nm (Wang et al., 2020a). As reported by the studies of Chan et al. (2013), Perlman and Netland (2009) and Su et al. (2016), there are four identified genera of coronavirus i.e. α, β, γ, δ. It was β-CoV that was isolated and identified in the patients of Wuhan city (China) at the onset of outbreak. β-CoV has 88% resemblance with bat-SL-CoVZC45 and bat-SL-CoVZXC21 (Lu et al., 2020), and about 79% of the genomic resemblance of SARS-CoV-2, is reported with SARS-like bat COV-MG772933 (Wu et al., 2020a). The International Committee on Taxonomy of Viruses (ICTV) (C.S.G., 2020) declared isolated β-CoV as “SARS-CoV-2 owing to the genetic similarity of the coronavirus to the virus of SARS outbreak of 2003” (Guo et al., 2020; Su et al., 2016).
Fig. 1.
(a) Naming of coronavirus with respect to taxonomy and disease caused [Modified from Gorbalenya et al., 2020 (Nature Microbiology)]; (b) Phylogenetic tree of SARS-like coronaviruses and (c) Open reading frame (ORF) SARS-like coronaviruses [Figure adopted from Li et al., 2020 (Pharmaceutical Analysis)].
3. COVID-19 outspread: infection and morbidity
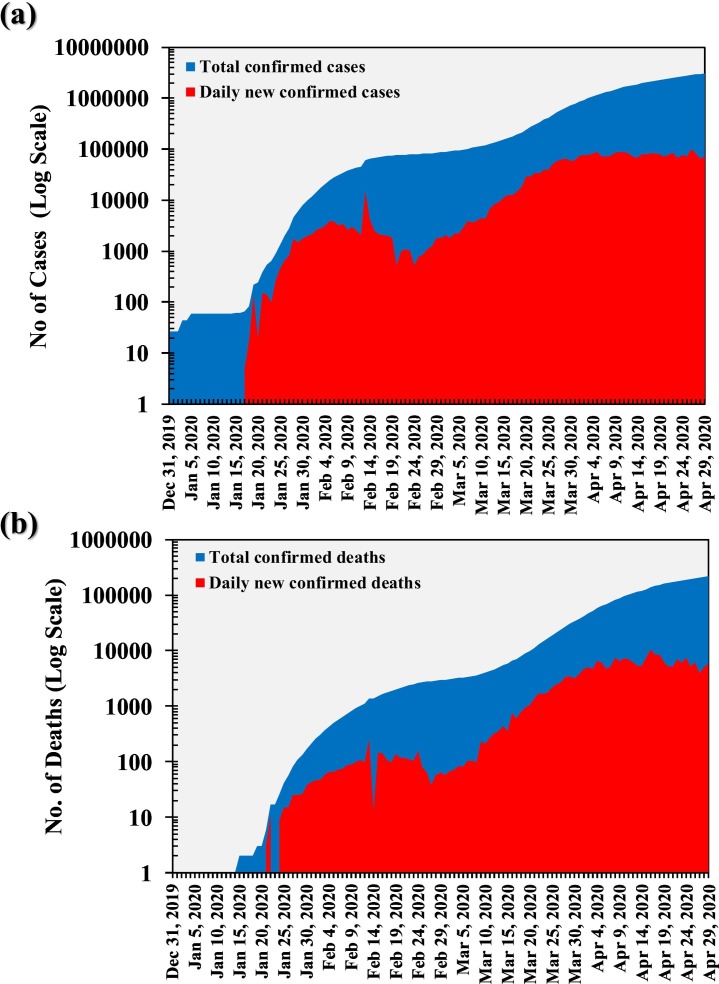
On 9th January 2020, Wuhan witnessed the first confirmed death due to COVID-19 (The New York Times, 23rd January 2020b). The Philippines on 1st February reported first death outside China province (Ramzy and May, 2020), and on 14th February, France reported first death outside Asia (The New York Times, 15th February 2020a). COVID-19 spread has been rapid and random comprising dozens of deaths in Iran, South Korea, and Italy by 28th February (COVID-19 in Korea, 5th April 2020). Around 40 countries over all the continents, except the Antarctica, had reported deaths by 13th March 2020 (Gumbrecht and Howard, 2020). Fig. 2 represents the total and daily new confirmed cases (Fig. 2a) and deaths (Fig. 2b) worldwide till 29th April 2020, by when the daily new cases are ~70,000, and daily reported new deaths are >5000. Since the start of this pandemic till 29th April 2020, WHO reports 3,018,952 confirmed cases worldwide with a death count of 207,973 (WHO Situation Report-100, 2020). However, actual case count is likely to be much higher than the reported count owing to lack of testing, in general; and especially for the asymptomatic patients (Reuters, 2020b). Likewise, the actual death count is much higher because as per the standard official protocols, the death accounted in the report is only of people who were tested positive for COVID-19 (Reuters, 2020a).On the other hand, as of 29th April 2020, total of 991,999 cases have been recovered from COVID-19 across the world as per the data collected by John Hopkin University, USA (COVID-19 Map, 2020).
Fig. 2.
Cumulative variation in total and daily new COVID-19 a) Cases, and b) deaths in the world since December 31st and April, 30th 2020. [Data Source: ourworldindata.org].
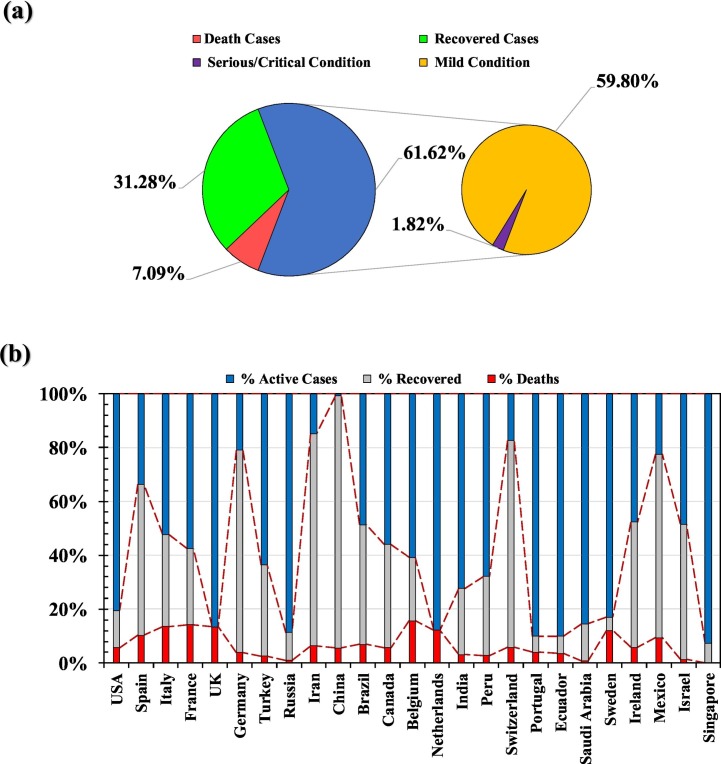
Fig. 3(a) shows the spread of total confirmed cases of COVID-19 in the top 25 countries, in terms of % active cases (current laboratory confirmed cases), % deaths, and % recovered cases, and in comparison to the world cases (Fig. 3(b)). Supplementary Table 1 reports countries and territories with laboratory-confirmed COVID-19 cases and deaths as per the WHO Situation Report-100 (2020). Till 29th April 2020, the USA is the most infected country in terms of confirmed cases (983,457) followed by Spain (210,773), Italy (201,505), United Kingdom (161,149), Germany (157,641), France (125,464), Turkey (114,653), Russia (99,399), Iran (92,584), and China (84,369) (WHO Situation Report-100, 2020). While the order of top ten countries changes for the reported death count where the USA (50,492) remained at the first place but the Italy (27,359) supersedes Spain (23,822), and rest countries follows the order of France (23,627) > United Kingdom (21,678) > Germany (6115) > Iran (5877) > China (4643) > Turkey (2992) > Russia (972) (WHO Situation Report-100, 2020). However the most interesting thing is ratio of reported death versus reported case in these top ten countries that varied between 18.8% to 1.0% with an order of France (18.8%) > Italy (13.6%) > UK (13.5%) > Spain (11.3%) > Iran (6.3%) > China (5.5%) > USA (5.1%) > Germany (3.9%) > Turkey (2.6%) > Russia (1.0%). Such a huge variation in the mortality rate seems to be governed by various factors like population characteristics (sex, age, and overall health), time since the initial outbreak, healthcare system quality, the volume of testing and treatment options (Roser et al., 2020). In general, countries (i.e. China, Iran, Switzerland, Germany, Mexico, Israel, Spain and Ireland) with early outbreak of COVID-19 are showing considerable recovery rate; with china being on the top with ~95% of the recovery from the infection. It looks like that fatality rate is considerably high (>10%) in European countries like Italy, Spain, UK, Netherlands, Belgium, France, Sweden and Mexico. The Supplementary Table 1 reports Countries and territories with laboratory-confirmed COVID-19 cases and deaths (WHO Situation Report-100, 2020). Overall, there seem to exist a globally good correlation between daily reported active cases and total active cases around the world (Fig. 3).
Fig. 3.
Distribution of COVID-19 Cases among active, recovered and deaths in a) the world; and b) top 25 infected countries (By 30th April 2020) [Data Source: worldometers.info].
4. Swinging symptoms of COVID-19
COVID-19 infected patients exhibit mostly symptoms like fever, fatigue, headache, cough, difficulty in breathing, and sore throat (Guan et al., 2020; Huang et al., 2020). Based on 1099 samples of the confirmed cases of COVID-19, till 29th January 2020, the common clinical manifestations were in order of high temperature (89%) > cough (68%) > tiredness (38%) > sputum (33%) > breath shortening (19%) > sore throat (14%) > headache (14%) > vomiting (5%) > diarrhea (4%). These manifestations show consistency with previous analysis of Hubei province, China (Wang et al., 2020a; Wang et al., 2020b; Wang et al., 2020c; Wang et al., 2020d; Huang et al., 2020; Chen et al., 2020). Thus, the dominant symptoms of COVID-19 are fever and cold; and rare symptoms are respiratory and gastrointestinal. Such symptomatic condition implies that SARS-COV-2 exhibits different viral tropism than other influenza causing viruses like MERS-CoV (Assiri et al., 2013), and SARS-CoV (Lee et al., 2003; Wang et al., 2016). The earliest screening may be carried out with the help of neutrophil to lymphocyte ratio (NLR) (Qin et al., 2020), as infection instantly leads to the reduction of lymphocytes which causes an increase in NLR (Guan et al., 2020; Liu et al., 2020b; Liu et al., 2020a).
Overall, WHO defines COVID-19 patient as: i) An individual with acute respiratory illness like fever and/or any sign of respiratory ailments like cough, sore throat, and/or has a travel history to locations reported community COVID-19 spread during the 14 days prior to the onset of symptoms. OR (2) An individual with any acute respiratory illness and/or been in contact with a confirmed or probable COVID-19 individual in the last 14 days. OR (3) An individual with severe acute respiratory illness with unexplained clinical diagnosis of alternate reasons for the problem. The central idea is to consider even a slightest probable case as a suspected patient based on above points with inconclusive or awaiting test results. While, a laboratory confirmed case is a person with a laboratory confirmation i.e. tested positive for COVID-19, irrespective of clinical signs and symptoms (asymptomatic patient). The trickiest feature of COVID-19 has lately been identified as being the asymptomatic (up to 60% in India). The risk is so heavy, spread is so fast and symptoms are of so varying nature that even travelling to a community spread country or being a physical contact of an infected person are being treated as symptoms.
5. Diagnosis of COVID-19 through biological and environmental samples
Diagnostic testing for the SARS-CoV-2 is currently undertaken using two approaches: whole genome sequencing and real-time reverse transcriptase-polymerase chain reaction (rRT-PCR, Nucleic Acid Testing). Table 1 compiles the emerging diagnostics techniques being used/developed for SARS-CoV-2. The sequencing approach was used primarily in the early days of the outbreak, almost all diagnostic testing for SARS-CoV-2 is done using rRT-PCR (Zhou et al., 2020b; Zhou et al., 2020a; Li et al., 2020). Many companies also launched RT-qPCR test kits to assist clinical diagnosis Chu et al. (2020) described two 1-step RTqPCR assays to detect two different regions (ORF1b and N) of the viral genome. Saliva serves as a promising non-invasive specimen for the diagnosis of COVID-19 patients with a high positive rate of 91.7% (11/12) in determining the disease using RT-qPCR (non-probes SYBR based fluorescence signal) (To et al., 2020). Though this technique has a sensitivity of 50%–79% of detection, depending on the protocol used and the sample type; the number of clinical specimens collected (Yam et al., 2003), safety hazards and detection time are some issues. CT scan is suggested as an auxiliary diagnostic method to avoid reporting false results and to increase sensitivity (Li et al., 2020). Early diagnosis and evaluation of disease severity of COVID-19 patients can be performed using high-resolution CT (HRCT) scan of the chest (Pan et al., 2020b; Pan et al., 2020a). However, inability to differentiate from other viral pneumonia makes the interpretation of the hysteresis of abnormal CT imaging tedious and inadequate (Li et al., 2020). Nevertheless, the chest CT had a very low misdiagnosed rate (3.9%, 2/51) in comparison with any other method (Ai et al., 2020).
Table 1.
Emerging diagnostics technological platform with their corresponding sample types and working principles for SARS-CoV-2.
| Platform | Type of technology | Sample type | Working |
|---|---|---|---|
| CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)a | Nucleic acid | Serum | PCR, CRISPR/Ca9-mediated lateral flow nucleic assay. |
| RAT (Rapid Antigen Test)b | Protein | Serum | Gold-coated antibodies produce colorimetric signal on paper in presence of target. |
| RPA (Recombinase Polymerase Amplification)c | Nucleic acid | Fecal and Nasal swabs | Forward and reverse primers blind to DNA and amplify strands at 37 °C. |
| RCA (Rolling Circle Amplification)d | Nucleic acid | Serum | DNA polymerase used to extend a circular primer and repeatedly replicate the sequence. |
| SIMOA (Single-Molecule Enzyme-Linked Immunosorbent Assay)e | Protein | Serum | Digital readout of colored product by enzymatic reaction in presence of target. |
| NASBA (Nucleic Acid Sequence Based Amplification)f | Nucleic acid | Nasal swabs | Transcription based amplification for RNA targets. |
| LAMP (Loop-Mediated Isothermal Amplification)g | Nucleic acid | Throat swabs | Isothermal DNA synthesis using self-recurring strand displacement reactions. |
| RT-LAMP (Reverse Transcription Loop-Mediated Isothermal Amplification)h | Nucleic acid | Nasopharyngeal aspirates | Reverse transcriptase LAMP reaction for RNA targets. |
| ELISA (Enzyme-Linked Immunosorbent Assay)i | Protein | Serum | Enzymatic reaction to produce colored product by enzymatic reaction in presence of target. |
In previous studies on SARS or MERS, lung parenchymal irregularity was reported in the central and bilateral upper lobes of lungs (Das et al., 2016; Paul et al., 2004). However, Li and Xia (2020) showed that the peripheral (sub-pleural), and middle and lower zones of lungs showed an abnormality at the initial stages, followed by affecting the upper lobes, and in severe cases, all five lobes of both lungs were affected. Recently, the Center for disease control and prevention (CDC, 2020) is working on Serologic (Antibody) immunoassays (IAs) testing i.e. Protein testing, for diagnosis of COVID-19 (Vashist, 2020). The automated chemiluminescent IA (CLIA), manual ELISA, and rapid lateral flow IA (LFIA) are most notable IAs, these detects the immunoglobulin M (IgM) and immunoglobulin G (IgG) produced in person in response to SARS-CoV-2 infection. Here the body is looked for antibodies; the positive test shows that the body has antibodies that may have resulted from infection with SARS-CoV-2 or related coronavirus. These serologic diagnostic tests are still in the research stage of development. A report by Biomedomics showed that Abbott ID Now™ COVID-19 test that utilizes isothermal nucleic acid amplification technology to detect viral RNA can detect SARS-CoV-2 in 5 min (Li, 2020). It can take throat, nasal, nasopharyngeal, and oropharyngeal swabs as samples to run a molecular test for the RdRp gene (Vashist, 2020). Further, wastewater based epidemiological (WBE) surveillance for the genetic material of SARS-CoV-19 has drawn loads of attention that can be helpful in early detection as well as resurgence of COVID-19 (Kumar et al., 2019a; Kumar et al., 2019b; Kumar et al., 2020a; Kumar et al., 2020b; Kumar et al., 2020c; Kitajima et al., 2020). WBE surveillance are expected to become handy for the potential eradication of the virus in a community.
6. Entry, replication and pathogenesis of COVID-19
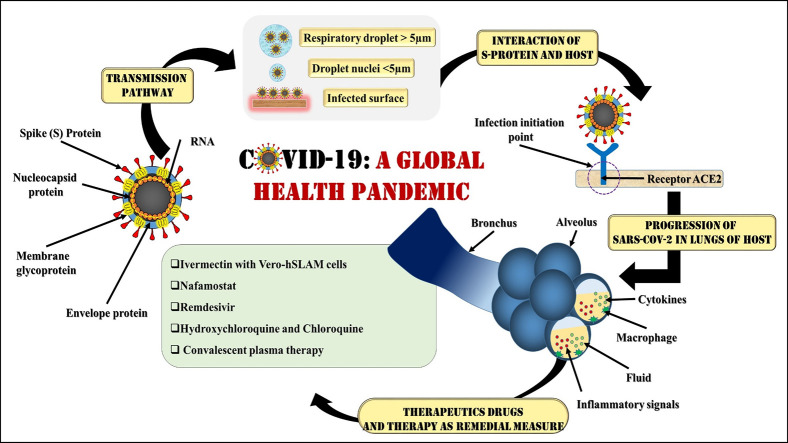
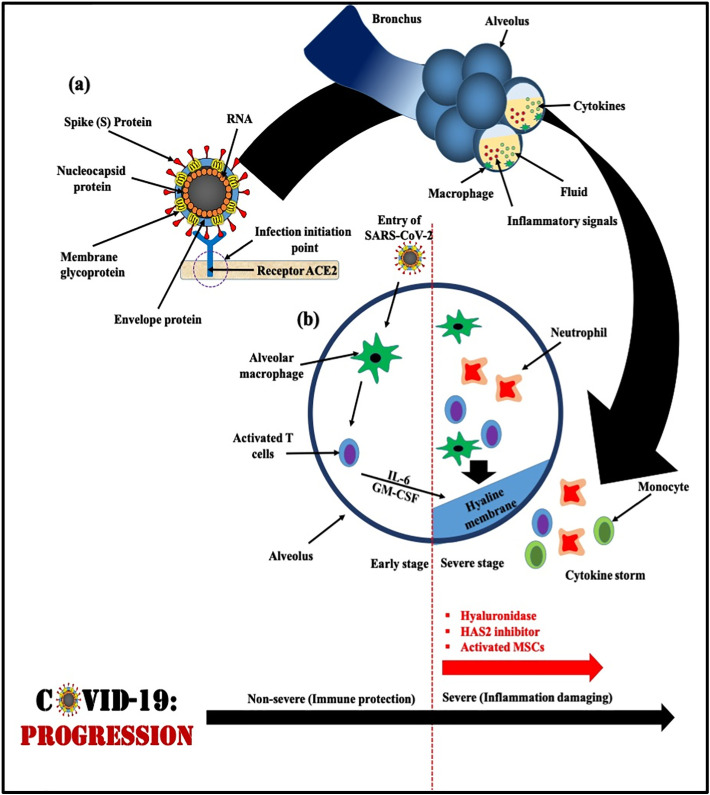
SARS-CoV-2 moves from the back of the throat to the lungs and then ultimately into the blood. All coronaviruses contain specific genes in downstream regions of ORF1 that encode proteins for viral replication, nucleocapsid, and spikes formation (Van Boheemen et al., 2012). SARS-CoV-2 and SARS-CoV binds with a receptor called angiotensin-converting enzyme 2 (ACE-2) found on the respiratory cells with the help of S protein, which helps in entry and replication for further spreading the infection. The receptor-binding domain (RBD) is loosely attached among viruses leading to infection of multiple host (Raj et al., 2013; Perlman and Netland, 2009). Fig. 4(a) demonstrates the interaction of viral S protein with ACE-2 on the host cell surface which is highly significant being an initiation step of infection/pathogenicity. Cryogenic Electronic Microscopic structure analysis revealed that the binding affinity of S protein to ACE-2 is about 10–20 times higher for SARS-CoV-2 than that of SARS-CoV, which is the reason for the higher transmissibility and contagiousness of SARS-CoV-2 as compared to SARS-CoV. After continuous multiplication, when virus reaches the lungs it causes inflammation in the alveoli or lung sacs leading to pneumonia. Lung inflammation makes breathing difficult for an infected person within 8 to 15 days from the onset of the symptoms and the state named acute respiratory distress syndrome (ARDS). While the functional receptor of MERS-CoV is known as Dipeptidyl peptidase 4 (DPP-4) or CD 26 (Li et al., 2020). Further, the S protein invasion process is facilitated by serine protease (TMPRSS211) produced by the host cell. After entering into the host body cell a single-stranded positive RNA is released by the viral genome which translates into viral polyproteins by the translation machinery of host cell protein. These polyproteins are further sliced into effector proteins by viral proteinases 3CLpro and PLpro which results into the immune suppression. Naturally, when the virus enters the cells, it offers antigen to the antigen presentation cells (APC), a central part of the antiviral immunity in the human body (Li et al., 2020), however, there still exists a lack of information about the antigen presentation of SARS-CoV-2. Therefore, till date, no SARS-CoV-2 specific antiviral agents could be discovered for a possible treatment to save lives and to produce an effective vaccine for future prevention.
Fig. 4.
Schematic diagram of COVID-19 progression; (a) Interaction between spike (S) protein and receptor (ACE2), the encircled zone is the initiation point from where infection spreads in host [Modified from_ Liu et al., 2020b; Liu et al., 2020a]; (b) Schematic representation of progression of SARS-CoV-2 inside host body [Modified from Shi et al., 2020 (Cell Death & Differentiation)].
Fig. 4(b) demonstrates the progression of SARS-CoV-2 inside the host body. Inflammatory response of ARDS is mainly caused by the release of large amounts of pro-inflammatory cytokines known as ‘cytokine storm’ (Li et al., 2020). SARS-CoV-2 infection undergoes ‘cytokine storm’ that triggers viral sepsis and inflammatory-induced lung injury leading to complications like pneumonitis, respiratory failure, ARDS, shock, organ failure, and potentially death (Yang et al., 2020; Prompetchara et al., 2020). During the cytokine storm mechanism in the body a wide variety of cytokines (IFN-α, IFN-γ, IL-1β, IL-6, IL-12, IL-18, IL-33, TNF-α, TGFβ, and others) and chemokines (CCL2, CCL3, CCL5, CXCL8, CXCL9, CXCL10, and others) are released (Huang et al., 2020; Cameron et al., 2008). Although, the detailed beneficial role of cytokines is unclear currently due to the recent progression of the disease, there are already clinically tested drugs available which may add value to the current treatment process of cytokine storm. Interleukin-6 inhibitors, or IL-6 inhibitors drugs, which were found to be helpful in patients of France, China, and New York works based on blocking a specific cytokine associated with inflammation. Further, Supplementary Table 2 shows various supporting statements given by doctors and researchers regarding ‘cytokine storm’ mechanisms and response in our body. Overall, understanding of immuno-pathological mechanism of SARS-CoV-2 infection has great potential to develop therapeutic strategies to contain the pandemic.
7. Prognosis: susceptibility and recovery
The prognosis of COVID-19 can be done by its effects on various body organs. The recovery rate from the infection of COVID-19 is varying based on its severity. People with mild condition (with few or no symptoms having a resemblance to some upper respiratory diseases such as common cold) are recovering within two weeks, the recovery duration can go up to six weeks for severe or critical disease cases. Two to eight weeks of duration from the symptoms onset has been observed to death (WHO-China Joint Mission on Coronavirus Disease 2019 (COVID-19) 16–24 February 2020). Various complications and health issues associated with COVID-19 have been reported, like Pneumonia affecting lungs, ARDS which may lead to respiratory and multi-organ failure (Cascella et al., 2020; Heymann and Shindo, 2020), abnormal clotting, sepsis and damage to kidney, liver, and heart (Zhou et al., 2020b; Zhou et al., 2020a; Xu et al., 2020). In order to understand the severity of the virus outbreak, it is crucial to focus on the population groups which are at high risk, which will allow them to direct the resources towards the most vulnerable.
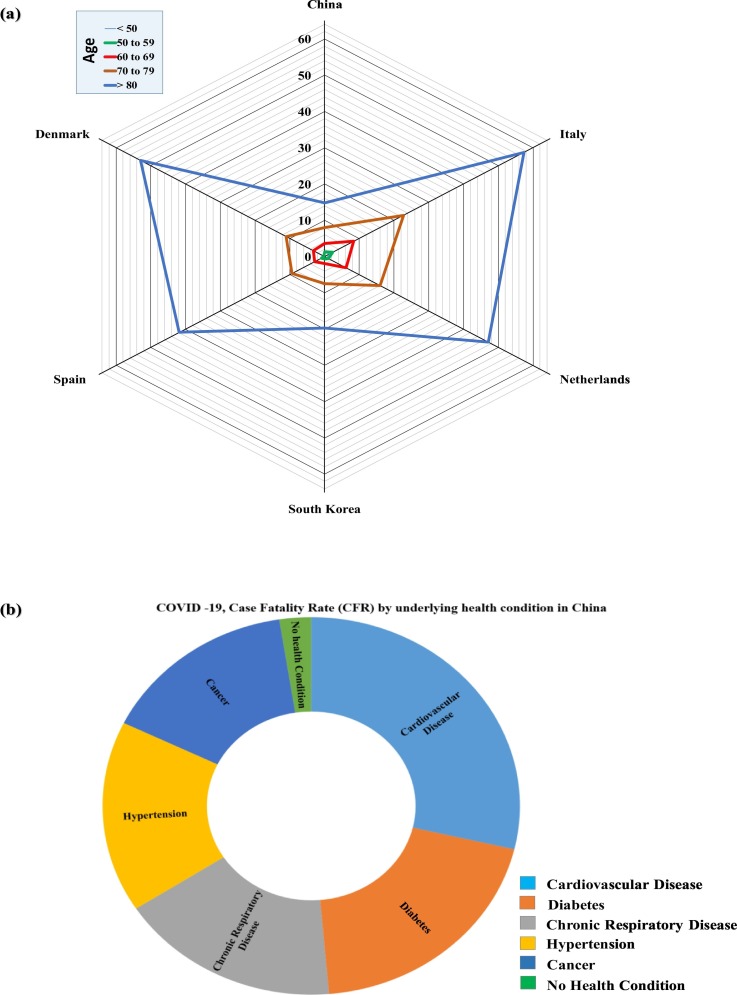
Fig. 5(a) summarises the case fatality rate (CFR) by the age group of various countries (Actualización n° 67, 5th April 2020; COVID-19 (Denmark), 6th April 2020; Epidemia (Rome) COVID-19, 6th April 2020; COVID-19 (Korea), 5th April 2020; Epidemiologische situatie COVID-19, 3rd April 2020; Surveillances, V., 2020). CFR can be defined as the ratio of the number of confirmed deaths and the number of confirmed cases. CFR may not represent the accurate results for the deaths caused by COVID-19, as it depends on several factors such as the outbreak stage of the country, extent of testing and even accuracy of testing. The CFR shows high variance among the age groups. The age group of >80 years is considered a vulnerable age group due to highest death risk associated with it in all the countries. However, the age group below 50 years, and children can have mild symptoms, but the risk of death is low. The CFR caused by COVID-19 is distinctive than the other historical pandemics like malaria and Spanish flu, where the most vulnerable age group was children or young adults. However, in the case of COVID-19, along with the age group the underlying health conditions also have a significant impact on the death risk (Fig. 5(b)). The early-stage CFR data of China shows that people with underlying health conditions such as cardiovascular disease, cancer, hypertension, chronic respiratory disease, and diabetes, are at higher risk (WHO, 23rd January 2020b). The CFR due to COVID-19 was highest for those underlying cardiovascular disease (10.5%), followed by diabetes (7.3%), chronic respiratory disease (6.3%), hypertension (6.0%), and cancer (5.6%). A comparison suggests that people with no previous underlying health condition had ten times lesser CFR (0.9%) and thus the elderly age group with pre-existing health conditions are more. Overall, the susceptibility and recovery of the disease is highly dependent upon the innate immunity of the person.
Fig. 5.
a) Radial diagram illustrating the age correlation with morbidity and b) Pie diagram illustrating Case Fatality Rate (CFR) by age and underling health conditions due to COVID-19. [CFR is based on Actualización n° 67, 5th April 2020; COVID-19 in Denmark, 6th April 2020; Epidemia in Rome COVID-19, 6th April 2020; COVID-19 in Korea, 5th April 2020; Epidemiologische situatie COVID-19, 3rd April 2020; Surveillances, V., 2020].
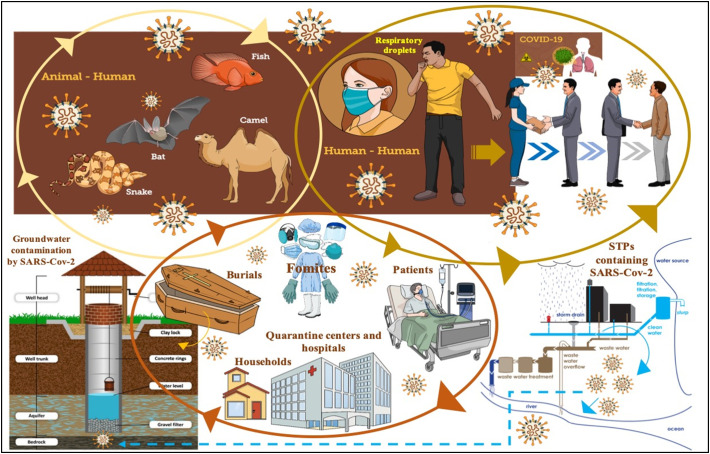
8. Transmission pathway and environmental factors
Fig. 6 presents a transmission pathway of the COVID-19 and their intersections with various environment. The primary mode of transmission of SARS-CoV-2 is between human-to-human via respiratory droplets and contact routes. There are two modes of transmission - (1) Droplet transmission and (2) Airborne transmission. Unlike other respiratory diseases, the mode of transmission is through droplets coming out during coughing and sneezing. As per WHO (2014), the respiratory infection transmission depends on the size of drop particles. If the particle size is >5–10 μm, it is referred to as respiratory droplets, and <5 μm is called droplet nuclei. Also, droplet-based transmission will occur when the uninfected person is within the 1 m radius of an infected person (Galbadage et al., 2020; Tellier et al., 2019) as the droplets can also travel within the range of 6 ft (Cennimo, 2020). While airborne transmission occurs when the microbe is present inside the droplet nuclei (particle size <5 μm) and subjected to the condition that the droplet nuclei can remain in the air for a longer duration and can travel up to a distance of 1 m (WHO, 2014). However, the probability of airborne transmission of COVID-19 is very less and case-specific such as generation of aerosols like endotracheal intubation, bronchoscopy, open suctioning, administration of nebulized treatment, and manual ventilation before intubation, turning the patient to the prone position, disconnecting the patient from the ventilator, non-invasive positive-pressure ventilation, tracheostomy, and cardiopulmonary resuscitation during medical procedures and treatment process.
Fig. 6.
Transmission pathways of the COVID-19 and their intersections with various environment.
Zhang et al. (2020) investigated the presence of SARS-CoV-2 in the feces of an infected person to understand if the SARS-CoV-2 can lead to any kind of gastrointestinal infections and further transmission. However, to date, no such study has been conducted that could establish a relationship between the fecal−oral transmission of the SARS-CoV-2. Zhou et al. (2020a) reported that viral shedding is one of the most significant ways of transmission and observed that the viral shedding was found to last for 37 days in survivors. The amount of viral load contributed to the environment by asymptomatic and symptomatic patients was conducted by Bai et al. (2020), Zou et al. (2020b) and Zou et al. (2020c). The authors reported that transmission was possible even in the case of asymptomatic patients, but in the case of symptomatic patients increase in viral loads was observed.
As of now, the environmental factors that has been studied to understand the COVID-19 transmissions, are the ambient environmental temperature, humidity but still, no authenticity on the relationship is confirmed yet (Wang et al., 2020a; Wang et al., 2020b; Wang et al., 2020c; Wang et al., 2020d; Kumar et al., 2020a; Tobías and Molina, 2020). Apart from climatic conditions (temperature and humidity), the transmission may be further triggered by population and medical facility (Dalziel et al., 2018; Hemmes et al., 1960). Based on the assumption that the SARS-CoV-2 is similar to SARS-CoV, it has been hypothesized that the SARS-CoV-2 may not survive at high temperature and humidity (Chan et al., 2011; Kumar et al., 2020a; Kumar et al., 2020b; Kumar et al., 2020c; Tobías and Molina, 2020) due to breakdown of the lipid membrane at elevated temperature (Schoeman and Fielding, 2019). Bukhari and Jameel (2020) investigated the effect of temperature and humidity on the transmission of COVID-19 disease. For the study, the data until March 22, 2020, were analyzed for sub-tropical countries. It was found that from January 22, 2020, to March 21, 2020, the number of cases increased in the region with a mean temperature range of 4–17 °C and absolute humidity 3-9 g/cm3. However, for temperature and absolute humidity higher than 17 °C and 9 g/cm3 lesser number of cases were reported. For countries with temperatures higher than 18 °C (between March 11 and 19, 2020) like India, Brazil, Indonesia, and Pakistan, the sudden rise in COVID-19 cases may be due to the recent initiation of large-scale COVID-19 testing. Based on the data obtained, it was hypothesized that the lower number of cases in a tropical climate might be due to the warm, and humid climate. The underlying hypothesis of decrease in the spread of viruses during warmer seasons is higher vitamin D levels, resulting in better immune responses (Aranow, 2011; Kumar et al., 2020a; Kumar et al., 2020b; Kumar et al., 2020c). Interestingly, a common man notion is that in cold weather, people tend to stay inside, often in a closed room, at times close to each other, and that enhances the infection probability. A study confirmed a strong relationship does exist between the reported cases, temperature, and absolute humidity, but the basis of this relationship is not lucid. It is difficult to delineate the effect of environmental factors like temperature and humidity, individually and in combination.
van Doremalen et al. (2020) studied the behavior of SARS-CoV-1 and SARS-CoV-2 and observed that SARS-CoV-1 and CoV-2 are very similar in terms of stability in the environment and reported that SARS-CoV-2 remained active on the surface of the plastic and stainless steel - 48–72 h; cardboard - 24 h; copper -4 h; 3 h in aerosols under experimental laboratory conditions but in real situation, this retention time may vary depending on temperature, humidity, ventilation, and the extent of virus deposited on any surface (Kumar et al., 2020a; Tobías and Molina, 2020). Wang et al. (2020a), Wang et al. (2020b), Wang et al. (2020c) and Wang et al. (2020d) estimated an effective reproductive number (R) to understand the severity of infection (Wallinga and Teunis, 2004). The R was developed based on the data obtained from 100 Chinese cities with >40 cases. The study was carried out to understand the effect of climatic factors (temperature and humidity) on R, which will indirectly can tell about the transmission of SARS-CoV-2. The results showed lowering of R value by 0.0225 and 0.0158 with increase in temperature by 1 °C and increase in relative humidity by 1%, respectively. The finding reported by Wang et al. (2020a), Wang et al. (2020b), Wang et al. (2020c) and Wang et al. (2020d) are consistent with the findings for the influenza virus (Park et al., 2020; Steel et al., 2011; Lipsitch and Viboud, 2009; Shaman and Kohn, 2009), where the high temperature and high humidity leads to a significant reduction in transmission of the influenza virus. This might be due to the following reasons: (i) Cold and dry weather might cause weakness in the immunity of the host (Kudo et al., 2019; Eccles, 2002), (ii) unlike influenza virus they might be more stable in lower temperature and within the respiratory droplets (Lowen and Steel, 2014; Tellier, 2009). Also, the observations made for SARS CoV for temperature and humidity (Chan et al., 2011; Yuan et al., 2006) were inconsistent with the findings of Wang et al. (2020a), Wang et al. (2020b), Wang et al. (2020c) and Wang et al. (2020d).
9. Current treatment options for COVID-19
Li et al., 2020 reported at present, there is no proven antiviral agent that can control the COVID-19 outbreak and inactivate SARS-CoV-2. By 29th April 2020, no drug has been approved by United States food and drug administration (2020) that can prevent COVID-19. Currently, management strategies for COVID-19 patients with mild symptoms include infection prevention by using oseltamivir/ intravenous antibiotics, clinical supportive measures for severe patients such as oxygen and mechanical ventilator support (Liu and Li, 2020). Some of the therapeutics drugs and therapy which are under investigation for controlling COVID-19 are: (i) Ivermectin with Vero-hSLAM cells; (ii) Nafamostat; (iii) Remdesivir; (iv) Hydroxychloroquine and Chloroquine; and (v) Convalescent plasma therapy.
FDA-approved Ivermectin as an anti-parasitic and anti-viral in-vitro activity. Ivermectin inhibits IN nuclear import and the human immunodeficiency virus-1 (HIV-1) replication and thus reduces infection by ~5000 times within two days after consumption. It showed a 99.98% reduction in viral RNA (Caly et al., 2020). Another drug, Nafamostat mesylate (Fusan) as demonstrated by Japanese researchers can inhibit the fusion of SARS-CoV-2 (S) protein and initiated membrane at achievable and safe concentrations in the patients (Hannah, 2020). Remdesivir is an antiviral drug that is intravenous and inhibits the synthesis of viral RNA by preventing the replication of RNA by early termination of RNA transcription (Li et al., 2020). Lo et al. (2017) successfully demonstrated that Remdesivir is a potent antiviral towards SARS and MERS-CoV. As per the CDC, “Remdesivir has in-vitro activity against SARS-CoV-2 and in-vitro and in-vivo activity against related beta coronaviruses”.
Hydroxychloroquine and chloroquine are anti-malarial drugs that are taken through oral administration, and both of the drugs belong to the quinolone family. Yazdany and Kim (2020) demonstrated that both the medicines have a potent antiviral property that can control SARS-CoV-2 in-vitro. Roque (2020) reported that the use of hydroxychloroquine is much safer and has more potential of inhibiting SARS-CoV-2. Hydroxychloroquine has been proven more successful than chloroquine (inhibition rate did not exceed 50%) at inhibiting SARS-CoV-2 (Yao et al., 2020). Paton et al. (2011) and Ooi et al. (2006) reported negative results of hydroxychloroquine and chloroquine during random testing for influenza in random patients. Henceforth, there is lack of report, facts and figures to support the use of hydroxychloroquine and chloroquine as an efficient treatment mechanism (Yazdany and Kim, 2020).
Early signs are that convalescent plasma therapy can reduce the mortality rate in SARS-CoV-2 patients (Cheng et al., 2005; Lai, 2005; Soo et al., 2004). Mair-Jenkins et al., 2015 showed recovery from SARS-CoV-2 at early-stage of treatment with convalesced plasma therapy. Unlike SARS-CoV (Mair-Jenkins et al., 2015) and MERS-CoV (Koenig, 2015; Li et al., 2020), many patients are donating plasma with SARS-CoV-2 antibodies to control COVID-19. Duan et al. (2020) demonstrated the potential of convalescent plasma therapy to treat the severe COVID-19 patients. 10 patients were treated with plasma therapy, a 200 ml of convalescent plasma neutralize with antibody titers above 1:640 was given to the patients who showed rapid improvement in symptoms with three days of convalescent plasma transfusion. However, treatment by convalescent plasma therapy is still questionable (Liu and Li, 2020).
10. Conclusions
COVID-19 is a severe global health issue which is caused by SARS-CoV-2. The genomic study revealed that the phylogeny of the SARS-CoV-2 is very similar to SARS-like bat/Pangolin. The disease result in respiratory illness like SARS-CoV and MERS-CoV and may cause death in severe cases. The mortality is significantly higher in the elderly age group, mostly having pre-existing health conditions. At the initial stage the disease may be identified by symptoms such as fever, dry cough, muscle pain and fatigue but challenge of identifying the asymptomatic patient is huge. The transmission is mainly through the respiratory droplets and their diameters, and through direct contact with an infected surface. In the case of airborne transmission, the probability of being infected is very less and case-specific. The higher transmissibility and contagiousness of SARS-CoV-2 may be attributed to the high binding affinity of SARS-CoV-2, S protein to ACE2. The rapid transmission is due to the weak linkage between the receptor-binding domain (RBD) of SARS-CoV2 and the host cell. Some of the profound drugs available to control cytokine storm are Interleukin-6 inhibitors or IL-6 inhibitors, but for older people or people with past medical histories inhibiting the immune system may result in severe health issues. One of the probable ways to curb the infection is to restrict the binding between S protein of SARS-CoV-2 to ACE2. Though the cause and effects are known still the transmission mechanism is not fully understood.
As a control measure, what best can be done and being implemented globally are self-isolation and social distancing. Once there is established virus transmission within the community, proper quarantine of symptomatic may have a meaningful impact to control further spread. Also, more clinical testing apart from laboratory testing should be carried out to validate the proposed drugs and therapy further. Strong ways to hold the COVID-19 outspread are: i) better surveillance plans like wastewater based epidemiological surveillance; ii) local and regional support through government and community collaboration; iii) cooperation to health workers for more clinical testing; iv) awareness through the improvement of health advisories; v) encouragement of clinical testing of the cases with mild symptoms; and vi) better resource allocations to the economic vulnerable groups during the social distancing. Last but not the least, research is the key to bring the breakthrough to contain the COVID-19 spread for which some of the key research aspects may be: i) Development of statistical-based predictive model to understand the future outbreak or re-occurrences of COVID-19 or alike pandemic; ii) Understanding the environmental impacts to curb the COVID-19 through additional ways of adaptation to the local conditions; iii) Development of comprehensive library for SARS-COV-2; iv) Invention of drug and therapy to control the contagion by various scientific communities; v) Pace the global testing of patient to obtain the data to find the troubleshooting of asymptomatic patient identification; vi) Lifestyle enhancement to increase the immunity, and sanitation awareness; vi) Identification of the hotspot areas to target better containment strategies; and vii) Distribution of immunity boosting medication to narrow down risk and vulnerability of the population as a precautionary measure.
CRediT authorship contribution statement
Manish Kumar: Conceptualization, Visualization, Writing - original draft, Supervision, Writing - review & editing. Kaling Taki: Writing - original draft, Visualization, Writing - review & editing. Rohit Gahlot: Formal analysis, Data curation. Ayushi Sharma: Formal analysis, Writing - review & editing. Kiran Dhangar: Formal analysis, Data curation, Writing - review & editing.
Acknowledgments
We acknowledge the moral support of various initiatives started by the administration of IIT Gandhinagar. We also acknowledge the help received from Ms. Payal Mazumder, Dr. Arbind Patel, Mr. Alok K Thakur and anonymous referees.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2020.139278.
Appendix A. Supplementary data
Supplementary tables
References
- Actualización n° 67 . Ministerio de Sanidad, Consumo y Bienestar Social; 2020. Enfermedad por el Coronavirus (COVID-19) (PDF) (Report) (in Spanish) (5th April) [Google Scholar]
- Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020 doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amer H.M., El Wahed A.A., Shalaby M.A., Almajhdi F.N., Hufert F.T., Weidmann M. A new approach for diagnosis of bovine coronavirus using a reverse transcription recombinase polymerase amplification assay. J. Virol. Methods. 2013;193(2):337–340. doi: 10.1016/j.jviromet.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranow C. Vitamin D and the immune system. J. Investig. Med. 2011;59(6):881–886. doi: 10.231/JIM.0b013e31821b8755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assiri A., Al-Tawfiq J.A., Al-Rabeeah A.A., Al-Rabiah F.A., Al-Hajjar S., Al-Barrak A., Flemban H., Al-Nassir W.N., Balkhy H.H., Al-Hakeem R.F., Makhdoom H.Q. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect. Dis. 2013;13(9):752–761. doi: 10.1016/S1473-3099(13)70204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Yao L., Wei T., Tian F., Jin D.Y., Chen L., Wang M. Presumed asymptomatic carrier transmission of COVID-19. Jama. 2020;323(14):1406–1407. doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch I., De Puig H., Hiley M., Carré-Camps M., Perdomo-Celis F., Narváez C.F., Salgado D.M., Senthoor D., O’Grady M., Phillips E., Durbin A. Rapid antigen tests for dengue virus serotypes and Zika virus in patient serum. Sci. Transl. Med. 2017;9(409) doi: 10.1126/scitranslmed.aan1589. (p. eaan1589) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukhari Q., Jameel Y. 2020. Will Coronavirus Pandemic Diminish by Summer? (Available at SSRN 3556998) [Google Scholar]
- Caly L., Druce J.D., Catton M.G., Jans D.A., Wagstaff K.M. The FDA-approved Drug Ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir. Res. 2020:104787. doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron M.J., Bermejo-Martin J.F., Danesh A., Muller M.P., Kelvin D.J. Human immunopathogenesis of severe acute respiratory syndrome (SARS) Virus Res. 2008;133(1):13–19. doi: 10.1016/j.virusres.2007.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascella M., Rajnik M., Cuomo A., Dulebohn S.C., Di Napoli R. Statpearls [Internet] StatPearls Publishing; 2020. Features, evaluation and treatment coronavirus (COVID-19) [PubMed] [Google Scholar]
- Cennimo D.J. 2020. Coronavirus Disease 2019 (COVID-19): Medscape (April 4, 2020) (Accessed 29 April 2020) [Google Scholar]
- Centers for disease control and prevention (CDC) 2020. https://www.cdc.gov/coronavirus/2019-ncov/hcp/therapeutic-options.html (7th April)
- Chan K.H., Peiris J.S., Lam S.Y., Poon L.L.M., Yuen K.Y., Seto W.H. The effects of temperature and relative humidity on the viability of the SARS coronavirus. Advances in virology. 2011;2011 doi: 10.1155/2011/734690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.W., To K.K.W., Tse H., Jin D.Y., Yuen K.Y. Interspecies transmission and emergence of novel viruses: lessons from bats and birds. Trends Microbiol. 2013;21(10):544–555. doi: 10.1016/j.tim.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Yu T. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Wong R., Soo Y.O.Y., Wong W.S., Lee C.K., Ng M.H.L., Chan P., Wong K.C., Leung C.B., Cheng G. Use of convalescent plasma therapy in SARS patients in Hong Kong. Eur. J. Clin. Microbiol. Infect. Dis. 2005;24(1):44–46. doi: 10.1007/s10096-004-1271-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D.K., Pan Y., Cheng S.M., Hui K.P., Krishnan P., Liu Y., Ng D.Y., Wan C.K., Yang P., Wang Q., Peiris M. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin. Chem. 2020;66(4):549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COVID-19 in Danmark . Statens Serum Institut; 2020. Epidemiologisk overvågningsrapport den 6. april 2020 (Report) Danish. (6th April) [Google Scholar]
- CSG of the International The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020:1. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalziel B.D., Kissler S., Gog J.R., Viboud C., Bjørnstad O.N., Metcalf C.J.E., Grenfell B.T. Urbanization and humidity shape the intensity of influenza epidemics in US cities. Science. 2018;362(6410):75–79. doi: 10.1126/science.aat6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das K.M., Lee E.Y., Langer R.D., Larsson S.G. Middle east respiratory syndrome coronavirus: what does a radiologist need to know? Am. J. Roentgenol. 2016;206(6):1193–1201. doi: 10.2214/AJR.15.15363. [DOI] [PubMed] [Google Scholar]
- Duan K., Liu B., Li C., Zhang H., Yu T., Qu J., Zhou M., Chen L., Meng S., Hu Y., Peng C. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc. Natl. Acad. Sci. 2020;117:9490–9496. doi: 10.1073/pnas.2004168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eccles R. An explanation for the seasonality of acute upper respiratory tract viral infections. Acta Otolaryngol. 2002;122(2):183–191. doi: 10.1080/00016480252814207. [DOI] [PubMed] [Google Scholar]
- Epidemia in Rome COVID-19 . Istituto Superiore di Sanità; Rome: 2020. Aggiornamento nazionale 6 aprile 2020 (PDF) (Report) in Italian. (6th April) [Google Scholar]
- Epidemiologische situatie COVID-19 in Nederland . Rijksinstituut voor Volksgezondheid en Milie; Bilthoven: 2020. (PDF) (Report) (in Dutch) (3rd April) [Google Scholar]
- European Centre for Disease Prevention and Control (ECDC) 2020. Outbreak of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): Increased Transmission Beyond China—Fourth Update. (PDF, 14th February) [Google Scholar]
- Galbadage T., Peterson B.M., Gunasekera R.S. Does COVID-19 spread through droplets alone? Faculty Articles & Research. 2020;408 doi: 10.3389/fpubh.2020.00163. https://digitalcommons.biola.edu/faculty-articles/408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaev A.A. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S., Du B. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbrecht J., Howard J. WHO declares novel coronavirus outbreak a pandemic. 2020. https://edition.cnn.com/2020/03/11/health/coronavirus-pandemic-world-health-organization/index.html Available at. (12th March)
- Guo Y.R., Cao Q.D., Hong Z.S., Tan Y.Y., Chen S.D., Jin H.J., Tan K.S., Wang D.Y., Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak–an update on the status. Military Medical Research. 2020;7(1):1–10. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah B. 2020. https://www.drugtargetreview.com/news/58915/nafamostat-inhibits-sars-cov-2-infection-preventing-covid-19-transmission/ (31st March)
- Hemmes J.H., Winkler K., Kool S.M. Virus survival as a seasonal factor in influenza and poliomyelitis. Nature. 1960;188(4748):430–431. doi: 10.1038/188430a0. [DOI] [PubMed] [Google Scholar]
- Heymann D.L., Shindo N. COVID-19: what is next for public health? Lancet. 2020;395(10224):542–545. doi: 10.1016/S0140-6736(20)30374-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M., Ninomiya A., Minekawa H., Notomi T., Ishizaki T., Van Tu P., Tien N.T.K., Tashiro M., Odagiri T. Rapid diagnosis of H5N1 avian influenza virus infection by newly developed influenza H5 hemagglutinin gene-specific loop-mediated isothermal amplification method. J. Virol. Methods. 2007;141(2):173–180. doi: 10.1016/j.jviromet.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Johns Hopkins Coronavirus Resource Center COVID-19 Map. 2020. https://coronavirus.jhu.edu/map.html> [online] Available at.
- Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba P., P C., Hamilton K.A., Haramoto E., Rose J.B. SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig K.L. Identify-isolate-inform: a modified tool for initial detection and management of Middle East Respiratory Syndrome patients in the emergency department. Western Journal of Emergency Medicine. 2015;16(5):619. doi: 10.5811/westjem.2015.7.27915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo E., Song E., Yockey L.J., Rakib T., Wong P.W., Homer R.J., Iwasaki A. Low ambient humidity impairs barrier function and innate resistance against influenza infection. Proc. Natl. Acad. Sci. 2019;116(22):10905–10910. doi: 10.1073/pnas.1902840116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Ram B., Honda R., Poopipattana C., Canh V.D., Chaminda T., Furumai H. Concurrence of antibiotic resistant bacteria (ARB), viruses, pharmaceuticals and personal care products (PPCPs) in ambient waters of Guwahati, India: urban vulnerability and resilience perspective. Sci. Total Environ. 2019;693 doi: 10.1016/j.scitotenv.2019.133640. [DOI] [PubMed] [Google Scholar]
- Kumar M., Chaminda T., Honda R., Furumai H. Vulnerability of urban waters to emerging contaminants in India and Sri Lanka: resilience framework and strategy. APN Science Bulletin. 2019;9:57–66. [Google Scholar]
- Kumar M., Kuroda K., Dhangar K. The most eagerly awaited summer of the Anthropocene: a perspective of SARS-CoV-2 decay and seasonal change. Groundw. Sustain. Dev. 2020;11 doi: 10.1016/j.gsd.2020.100400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Ram B., Sewwandi H., Honda R., Chaminda T. Treatment enhances the prevalence of antibiotic-resistant bacteria and antibiotic resistance genes in the wastewater of Sri Lanka, and India. Environ. Res. 2020;183 doi: 10.1016/j.envres.2020.109179. [DOI] [PubMed] [Google Scholar]
- Kumar M., Chaminda T., Honda R. Seasonality impels the antibiotic resistance in Kelani River of the emerging economy of Sri Lanka. npj Clean Water. 2020;3(12) [Google Scholar]
- Lai S.T. Treatment of severe acute respiratory syndrome. Eur. J. Clin. Microbiol. Infect. Dis. 2005;24(9):583–591. doi: 10.1007/s10096-005-0004-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N., Hui D., Wu A., Chan P., Cameron P., Joynt G.M., Ahuja A., Yung M.Y., Leung C.B., To K.F., Lui S.F. A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 2003;348(20):1986–1994. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- Li James X. Cellex Inc. 2020. https://www.fda.gov/media/136622/ (1st April)
- Li Y., Xia L. Coronavirus disease 2019 (COVID-19): role of chest CT in diagnosis and management. Am. J. Roentgenol. 2020:1–7. doi: 10.2214/AJR.20.22954. [DOI] [PubMed] [Google Scholar]
- Li X., Geng M., Peng Y., Meng L., Lu S. Molecular immune pathogenesis and diagnosis of COVID-19. Journal of Pharmaceutical Analysis. 2020;10:102–108. doi: 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsitch M., Viboud C. Influenza seasonality: lifting the fog. Proc. Natl. Acad. Sci. 2009;106(10):3645–3646. doi: 10.1073/pnas.0900933106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Li S. Convalescent plasma: a valid option in the treatment of COVID-19? Insights in clinical and Cellular Immunology. 2020;4:001–002. doi: 10.29328/journal.icci.1001012. [DOI] [Google Scholar]
- Liu K., Fang Y.Y., Deng Y., Liu W., Wang M.F., Ma J.P., Xiao W., Wang Y.N., Zhong M.H., Li C.H., Li G.C. Clinical characteristics of novel coronavirus cases in tertiary hospitals in Hubei Province. Chin. Med. J. 2020 doi: 10.1097/CM9.0000000000000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Zhou Q., Li Y., Garner L.V., Watkins S.P., Carter L.J., Smoot J., Gregg A.C., Daniels A.D., Jervey S., Albaiu D. 2020. Research and Development on Therapeutic Agents and Vaccines for COVID-19 and Related Human Coronavirus Diseases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo M.K., Jordan R., Arvey A., Sudhamsu J., Shrivastava-Ranjan P., Hotard A.L., Flint M., McMullan L.K., Siegel D., Clarke M.O., Mackman R.L. GS-5734 and its parent nucleoside analog inhibit Filo-, Pneumo-, and Paramyxoviruses. Sci. Rep. 2017;7 doi: 10.1038/srep43395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowen A.C., Steel J. Roles of humidity and temperature in shaping influenza seasonality. J. Virol. 2014;88(14):7692–7695. doi: 10.1128/JVI.03544-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair-Jenkins J., Saavedra-Campos M., Baillie J.K., Cleary P., Khaw F.M., Lim W.S., Makki S., Rooney K.D., Convalescent Plasma Study Group, Nguyen-Van-Tam J.S., Beck C.R. The effectiveness of convalescent plasma and hyperimmune immunoglobulin for the treatment of severe acute respiratory infections of viral etiology: a systematic review and exploratory meta-analysis. J. Infect. Dis. 2015;211(1):80–90. doi: 10.1093/infdis/jiu396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel N., Gomes S.A., Chemin I., Trépo C., Kay A. Improved rolling circle amplification (RCA) of hepatitis B virus (HBV) relaxed-circular serum DNA (RC-DNA) J. Virol. Methods. 2013;193(2):653–659. doi: 10.1016/j.jviromet.2013.07.045. [DOI] [PubMed] [Google Scholar]
- Novel C.P.E.R.E. The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China. Zhonghua liu xing bing xue za zhi= Zhonghua liuxingbingxue zazhi. 2020;41(2):145. doi: 10.3760/cma.j.issn.0254-6450.2020.02.003. [DOI] [PubMed] [Google Scholar]
- Ooi E.E., Chew J.S.W., Loh J.P., Chua R.C. In vitro inhibition of human influenza a virus replication by chloroquine. Virol. J. 2006;3(1):39. doi: 10.1186/1743-422X-3-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Guan H., Zhou S., Wang Y., Li Q., Zhu T., Hu Q., Xia L. Initial CT findings and temporal changes in patients with the novel coronavirus pneumonia (2019-nCoV): a study of 63 patients in Wuhan, China. European radiology, pp. 2020:1–4. doi: 10.1007/s00330-020-06731-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F., Ye T., Sun P., Gui S., Liang B., Li L., Zheng D., Wang J., Hesketh R.L., Yang L., Zheng C. Time course of lung changes on chest CT during recovery from 2019 novel coronavirus (COVID-19) pneumonia. Radiology. 2020 doi: 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.E., Son W.S., Ryu Y., Choi S.B., Kwon O., Ahn I. Effects of temperature, humidity, and diurnal temperature range on influenza incidence in a temperate region. Influenza Other Respir. Viruses. 2020;14(1):11–18. doi: 10.1111/irv.12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paton N.I., Lee L., Xu Y., Ooi E.E., Cheung Y.B., Archuleta S., Wong G., Smith A.W. Chloroquine for influenza prevention: a randomised, double-blind, placebo controlled trial. Lancet Infect. Dis. 2011;11(9):677–683. doi: 10.1016/S1473-3099(11)70065-2. [DOI] [PubMed] [Google Scholar]
- Paul N.S., Roberts H., Butany J., Chung T., Gold W., Mehta S., Konen E., Rao A., Provost Y., Hong H.H., Zelovitsky L. Radiologic pattern of disease in patients with severe acute respiratory syndrome: the Toronto experience. Radiographics. 2004;24(2):553–563. doi: 10.1148/rg.242035193. [DOI] [PubMed] [Google Scholar]
- Perlman S. 2020. Another Decade, another Coronavirus. [Google Scholar]
- Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7(6):439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan A.L., Katz R., Gostin L.O. The novel coronavirus originating in Wuhan, China: challenges for global health governance. Jama. 2020;323(8):709–710. doi: 10.1001/jama.2020.1097. [DOI] [PubMed] [Google Scholar]
- Prompetchara E., Ketloy C., Palaga T. Immune responses in COVID-19 and potential vaccines: lessons learned from SARS and MERS epidemic. Asian Pac. J. Allergy Immunol. 2020;38:1–9. doi: 10.12932/AP-200220-0772. [DOI] [PubMed] [Google Scholar]
- Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., Xie C., Ma K., Shang K., Wang W., Tian D.S. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj V.S., Mou H., Smits S.L., Dekkers D.H., Müller M.A., Dijkman R., Muth D., Demmers J.A., Zaki A., Fouchier R.A., Thiel V. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495(7440):251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramzy A., May T. Philippines reports first coronavirus death outside China. 2020. https://www.nytimes.com/2020/02/02/world/asia/philippines-coronavirus-china.html Available at.
- Report of the WHO-China Joint Mission on Coronavirus Disease 2019 (COVID-19) 2020. https://www.who.int/docs/default-source/coronaviruse/who-china-joint-mission-on-covid-19-final-report.pdf Available at. (16th–24th February)
- Reuters Italy's coronavirus deaths could be underestimated in data: official. 2020. https://www.reuters.com/article/us-health-coronavirus-italy-data/italys-coronavirus-deaths-could-be-underestimated-in-data-official-idUSKBN21I250
- Reuters Special report: Italy and South Korea virus outbreaks reveal disparity in deaths and tactics. 2020. https://www.reuters.com/article/us-health-coronavirus-response-specialre-idUSKBN20Z27P
- Rissin D.M., Kan C.W., Campbell T.G., Howes S.C., Fournier D.R., Song L., Piech T., Patel P.P., Chang L., Rivnak A.J., Ferrell E.P. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat. Biotechnol. 2010;28(6):595. doi: 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roque M.R. Chloroquine and hydroxychloroquine toxicity. Medscape. 20202020 https://emedicine.medscape.com/article/1229016-overview Accessed on 23rd March, 2020. [Google Scholar]
- Roser M., Ritchie H., Ortiz-Ospina E. Our World in Data. 2020. Coronavirus disease (COVID-19)–statistics and research. [Google Scholar]
- Rowe T., Abernathy R.A., Hu-Primmer J., Thompson W.W., Lu X., Lim W., Fukuda K., Cox N.J., Katz J.M. Detection of antibody to avian influenza A (H5N1) virus in human serum by using a combination of serologic assays. J. Clin. Microbiol. 1999;37(4):937–943. doi: 10.1128/jcm.37.4.937-943.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16(1):69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaman J., Kohn M. Absolute humidity modulates influenza survival, transmission, and seasonality. Proc. Natl. Acad. Sci. 2009;106(9):3243–3248. doi: 10.1073/pnas.0806852106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Wang Y., Shao C., Huang J., Gan J., Huang X., Bucci E., Piacentini M., Ippolito G., Melino G. 2020. COVID-19 Infection: The Perspectives on Immune Responses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Nishimura H., Saijo M., Okamoto M., Noda M., Tashiro M., Taguchi F. Diagnosis of human respiratory syncytial virus infection using reverse transcription loop-mediated isothermal amplification. J. Virol. Methods. 2007;139(1):78–84. doi: 10.1016/j.jviromet.2006.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soo Y.O.Y., Cheng Y., Wong R., Hui D.S., Lee C.K., Tsang K.K.S., Ng M.H.L., Chan P., Cheng G., Sung J.J.Y. Retrospective comparison of convalescent plasma with continuing high-dose methylprednisolone treatment in SARS patients. Clin. Microbiol. Infect. 2004;10(7):676–678. doi: 10.1111/j.1469-0691.2004.00956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel J., Palese P., Lowen A.C. Transmission of a 2009 pandemic influenza virus shows a sensitivity to temperature and humidity similar to that of an H3N2 seasonal strain. J. Virol. 2011;85(3):1400–1402. doi: 10.1128/JVI.02186-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Wong G., Shi W., Liu J., Lai A.C., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24(6):490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surveillances, V The epidemiological characteristics of an outbreak of 2019 novel Coronavirus diseases (COVID-19)—China, 2020. China CDC Weekly. 2020;2(8):113–122. [PMC free article] [PubMed] [Google Scholar]
- Tellier R. Aerosol transmission of influenza A virus: a review of new studies. J. R. Soc. Interface. 2009;6(suppl_6):S783–S790. doi: 10.1098/rsif.2009.0302.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellier R., Li Y., Cowling B.J., Tang J.W. Recognition of aerosol transmission of infectious agents: a commentary. BMC Infect. Dis. 2019;19(1):101. doi: 10.1186/s12879-019-3707-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The New York Times Coronavirus Live Updates: First Death Outside Asia Reported in France. 2020. https://www.nytimes.com/2020/02/15/world/asia/coronavirus-china-live-updates.html (15th Febuary)
- The New York Times Coronavirus Death Toll Climbs in China, and a Lockdown Widens. 2020. https://www.nytimes.com/2020/01/23/world/asia/china-coronavirus.html (23rd January)
- The updates on COVID-19 in Korea (Report). Korea Centers for Disease Control and Prevention (KCDC) 2020. https://www.cdc.go.kr/board/board.es?mid=a30402000000&bid=0030&act=view&list_no=366739&tag=&nPage=1 Available at. (5th April)
- To K.K.W., Tsang O.T.Y., Yip C.C.Y., Chan K.H., Wu T.C., Chan J.M.C., Leung W.S., Chik T.S.H., Choi C.Y.C., Kandamby D.H., Lung D.C. Clinical Infectious Diseases: An Official Publication of the Infectious Diseases Society of America. 2020. Consistent detection of 2019 novel coronavirus in saliva. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobías A., Molina T. Is temperature reducing the transmission of COVID-19? Environ. Res. 2020 doi: 10.1016/j.envres.2020.109553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrrell D.A., Myint S.H. 4th edition. University of Texas Medical Branch at Galveston; 1996. Coronaviruses. In Medical Microbiology. [PubMed] [Google Scholar]
- United States food and drug administration. Revised information for investigational COVID-19 convalescent plasma. https://www.fda.gov/vaccines-blood-biologics/investigational-new-drug-ind-or-device-exemption-ide-process-cber/revised-information-investigational-covid-19-convalescent-plasma (3rd April) 2020.
- Van Boheemen S., de Graaf M., Lauber C., Bestebroer T.M., Raj V.S., Zaki A.M., Osterhaus A.D., Haagmans B.L., Gorbalenya A.E., Snijder E.J., Fouchier R.A. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio. 2012;3(6) doi: 10.1128/mBio.00473-12. (pp. e00473–12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., Tamin A., Harcourt J.L., Thornburg N.J., Gerber S.I., Lloyd-Smith J.O. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020;382(16):1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashist S.K. 2020. In Vitro Diagnostic Assays for COVID-19: Recent Advances and Emerging Trends. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallinga J., Teunis P. Different epidemic curves for severe acute respiratory syndrome reveal similar impacts of control measures. Am. J. Epidemiol. 2004;160(6):509–516. doi: 10.1093/aje/kwh255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Xiao X., Lu J., Chen Z., Li K., Liu H., Luo L., Wang M., Yang Z. Factors associated with clinical outcome in 25 patients with avian influenza A (H7N9) infection in Guangzhou, China. BMC Infect. Dis. 2016;16(1):534. doi: 10.1186/s12879-016-1840-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. Jama. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Tang K., Feng K., Lv W. 2020. High Temperature and High Humidity Reduce the Transmission of COVID-19. (Available at SSRN 3551767) [Google Scholar]
- Wang L.S., Wang Y.R., Ye D.W., Liu Q.Q. A review of the 2019 Novel Coronavirus (COVID-19) based on current evidence. Int. J. Antimicrob. Agents. 2020:105948. doi: 10.1016/j.ijantimicag.2020.105948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Xiong E., Tian T., Cheng M., Lin W., Wang H., Zhang G., Sun J., Zhou X. Clustered regularly interspaced short palindromic repeats/Cas9-mediated lateral flow nucleic acid assay. ACS Nano. 2020;14(2):2497–2508. doi: 10.1021/acsnano.0c00022. [DOI] [PubMed] [Google Scholar]
- Wat D., Gelder C., Hibbitts S., Cafferty F., Bowler I., Pierrepoint M., Evans R., Doull I. The role of respiratory viruses in cystic fibrosis. J. Cyst. Fibros. 2008;7(4):320–328. doi: 10.1016/j.jcf.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Situation Report-100 Countries, territories or areas with reported laboratory-confirmed COVID-19 cases and deaths as of 29th April 2020. 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200429-sitrep-100-covid-19.pdf?sfvrsn=bbfbf3d1_2 30th April)
- World Health Organization Geneva: World Health Organization; 2014. Infection prevention and control of epidemic- and pandemic-prone acute respiratory infections in health care. https://apps.who.int/iris/bitstream/handle/10665/112656/9789241507134_eng.pdf?sequence=1 Available from. [PubMed]
- World Health Organization (WHO) 2020. WHO Director-General’s Opening Remarks at the Media Briefing on COVID-19—11 March 2020. (11 March) [Google Scholar]
- World Health Organization (WHO) 2020. WHO Director-General’s Statement on the Advice of the IHR Emergency Committee on Novel Coronavirus. (23rd January) [Google Scholar]
- Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., Sheng J. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Shi L., Wang Y. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. (published online ahead of print February 18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yam W.C., Chan K.H., Poon L.L.M., Guan Y., Yuen K.Y., Seto W.H., Peiris J.S.M. Evaluation of reverse transcription-PCR assays for rapid diagnosis of severe acute respiratory syndrome associated with a novel coronavirus. J. Clin. Microbiol. 2003;41(10):4521–4524. doi: 10.1128/JCM.41.10.4521-4524.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Shen C., Li J., Yuan J., Yang M., Wang F., Li G., Li Y., Xing L., Peng L., Wei J. 2020. Exuberant Elevation of IP-10, MCP-3 and IL-1ra during SARS-CoV-2 Infection Is Associated With Disease Severity and Fatal Outcome. (medRxiv) [Google Scholar]
- Yao X., Ye F., Zhang M., Cui C., Huang B., Niu P., Liu X., Zhao L., Dong E., Song C., Zhan S. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdany J., Kim A.H. Use of hydroxychloroquine and chloroquine during the COVID-19 pandemic: what every clinician should know. Ann. Intern. Med. 2020 doi: 10.7326/M20-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., Yun H., Lan W., Wang W., Sullivan S.G., Jia S., Bittles A.H. A climatologic investigation of the SARS-CoV outbreak in Beijing, China. Am. J. Infect. Control. 2006;34(4):234–236. doi: 10.1016/j.ajic.2005.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chen C., Zhu S., Shu C., Wang D., Song J., Song Y., Zhen W., Zijian F., Wu G., Xu J. Isolation of 2019-nCoV from a stool specimen of a laboratory-confirmed case of the coronavirus disease 2019 (COVID-19) China CDC Weekly. 2020;2(8):123–124. [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D. 2020. Discovery of a Novel Coronavirus Associated With the Recent Pneumonia Outbreak in Humans and Its Potential Bat Origin. (BioRxiv) [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J., Guo Q. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382(12):1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables