Abstract
Activation of Crtc1 (also known as Mect1/Torc1) by a t(11;19) chromosomal rearrangement underlies the etiology of malignant salivary gland tumors. Since LKB1 is a target for mutational inactivation in lung cancer and was recently shown to regulate hepatic Crtc2/CREB transcriptional activity in mice, we now present evidence suggesting disruption of an LKB1/Crtc pathway in cancer. Although Crtc1 is preferentially expressed in adult brain tissues, we observed elevated levels of steady-state Crtc1 in thoracic tumors. In addition, we demonstrate that somatic loss of LKB1 is associated with underphosphorylation of endogenous Crtc1, enhanced Crtc1 nuclear localization, and enhanced expression of the Crtc prototypic target gene, NR4A2/Nurr1. Inhibition of NR4A2 was associated with growth suppression of LKB1-null tumors, but showed little effect on LKB1-wildtype cells. These data strengthen the role of dysregulated Crtc as a bona fide cancer gene, present a new element to the complex LKB1 tumorigenic axis, and suggest that Crtc genes may be aberrantly activated in a wider range of common adult malignancies.
Keywords: Crtc1, LKB1, lung cancer, Mect1/Torc1, NR4A2
Introduction
The Crtc1 (aka Mect1/Torc1) gene was initially isolated as a fusion partner with Maml2 in mucoepidermoid salivary gland and lung tumors that carry a t(11;19) chromosomal rearrangement (Tonon et al., 2003). The Crtc1-Maml2 fusion transcript was subsequently identified in primary thyroid, breast, cervix, lung, and cutaneous sweat gland tumors with clear-cell, mucoepidermoid tumor-like histologic features (Achcar et al., 2009; Behboudi et al., 2005; Camelo-Piragua et al., 2009; Enlund et al., 2004; Kaye, 2009; Kazakov et al., 2007; Lennerz et al., 2008; Tirado et al., 2007) unifying a group of tumors that arise from mucous/serous glands scattered throughout the body. Since Crtc gene members are potent cAMP/CREB co-activators (Conkright et al., 2003; Iourgenko et al., 2003) and the ectopic expression of Crtc1-Maml2 activated a similar group of target genes (Coxon et al., 2005; Wu et al., 2005), a current model proposes that the fusion oncogene transforms cells by aberrantly co-activating specific Crtc1 inducible targets (Kaye, 2006). For example, using a doxycycline inducible Crtc1-Maml2 vector system we identified marked induction of previously known cAMP target genes, such as PEPCK, amphiregulin, NR4A2, and NR4A3 (Coxon et al., 2005), which corresponded to the prototypic gene promoters activated by wildtype Crtc1 (Conkright et al., 2003; Iourgenko et al., 2003). This model is further supported by the observation that Crtc1-Maml2 did not appear to alter expression of Maml2 target genes and that small in-frame deletions within Crtc1 exon 1 abolished both CREB binding and Crtc1-Maml2-mediated tumorigenicity of rat RK3E cells (Conkright et al., 2003; Iourgenko et al., 2003) (Coxon et al., 2005; Wu et al., 2005).
Therefore, while Crtc1-Maml2 is etiologically linked to a diverse group of uncommon human malignancies, we were interested to study if activation of Crtc1 by an alternate mechanism might be an important event in other tumor types. For example, the Crtc2 homolog was shown to regulate gluconeogenesis by integrating calcium and metabolic signaling through calcineurin and AMPK/SIK kinase families (Bittinger et al., 2004; Fu and Screaton, 2008; Screaton et al., 2004; Shaw et al., 2005). In these experiments, Crtc2 transcriptional co-activator function was negatively regulated by phosphorylation at sites including a critical serine171 that mediates cytoplasmic sequestration and/or degradation (Screaton et al., 2004). Conversely, dephosphorylation of Crtc2 at serine171 resulted in nuclear import with activation of several cAMP/CREB target genes including orphan nuclear receptors and anabolic enzymes necessary for glycogen and fatty acid storage (Wang et al., 2008). Previous studies in yeast and mammalian cells had identified LKB1 as an essential activator for the AMPK gene family (Hawley et al., 2003; Hong et al., 2003), and the targeted loss of LKB1 in murine liver was shown to reduce hepatic AMPK activity resulting in enrichment of nuclear, underphosphorylated Crtc2 with induction of gluconeogenesis and elevation of circulating murine blood glucose levels (Shaw et al., 2005). In the case of Crtc1, the homologous serine151 residue (167 in isoform II) was shown to serve as a target for inhibition by AMPK/SIK phosphorylation which might explain the constitutive activity of the Crtc1-Maml2 fusion oncogene which only included the CREB binding Crtc1 aa residues 1–42. Since expression of endogenous Crtc1 proteins have not been extensively tested in human tumor samples and since LKB1 is a frequent target for mutational inactivation in certain tumor subtypes, we examined Crtc1 protein expression, sub-localization, and activation of the prototypic Crtc1 target gene, NR4A2, in a group of tumor cell lines.
Results
Crtc1 protein mobility is associated with LKB1 status in tumor cell lines
Although Crtc1 mRNA was reported to be enriched in brain tissues (Kovacs et al., 2007; Zhou et al., 2006), there had been few studies of protein expression in human tumor samples or cell lines. We, therefore, tested steady-state protein levels on a normal human tissues immunoblot using Crtc1 peptide antisera and observed a strong signal at approximately 85 kDa exclusively in normal brain tissue (data not shown). We validated the antisera by detecting the endogenous 135 kDa Crtc1-Maml2 fusion protein in the lung mucoepidermoid cancer cell line (H3118) as well as recombinant Crtc1-Maml2 fusion product in a lung cancer cell line (H2009) transiently transfected with either the empty parental vector or the fusion oncogene (Figure 1a). In addition to the fusion peptide, however, we detected abundant expression of an endogenous 85 kDa Crtc1 species in both tumor cell lines. To examine the pattern of endogenous Crtc1 expression in cancer, we tested the specificity of the antisera by detecting a signal in an independent source of normal brain and absent expression in skeletal muscle and then tested a series of brain tumor samples (Figure 1b). We detected low levels of steady-state Crtc1 in four high-grade glioma tumors and an increased level of protein expression in three different oligodendroglioma tumor samples. In addition, we observed that the Crtc1 protein species in these tumor samples showed a variable migration pattern with either a broad-based 85kDa signal or slower mobility forms resembling the migration pattern of phosphorylated and underphosphorylated Crtc2 (Screaton et al., 2004). In contrast, when we tested other tumor cell lines that included: three LKB1 wildtype lung cancer cell lines (H522, H2087, and H358), three LKB1-null lung cancer cell lines (H23, H460, H2126), and three other LKB1-null lines (HSY, HSG, ACC3), we noted a correlation between null LKB1 status and the presence of a constitutive discrete, faster mobility endogenous Crtc1 signal (Figure 2a). For example, while we observed that the Crtc1 protein pattern could vary between faster and slower mobility species in LKB wildtype cells harvested at different times, the LKB1 null cells predominantly expressed the faster mobility pattern. This pattern, however, was observed only with the Crtc1 custom peptide antisera (epitope: Crtc1 aa 19–34 with 2 mismatches against Crtc2 and 5 mismatches against Crtc3). In contrast, a pan-Crtc antisera generated in our laboratory against a GST-fusion peptide spanning Crtc1 exon 1 could not discriminate these mobility shifts using the same lung tumor protein lysates (Supplemental Figure 1) suggesting that the aa 19–34 peptide antisera may be selectively detecting a specific Crtc1 isoform. Using the anti-Crtc1 peptide antisera or a specific anti-Crtc2/Torc2 antisera (Cell Signaling Technologies) on duplicate lanes, we observed co-expression of both endogenous Crtc1 and Crtc2 in the same lung tumor cell extract (Supplemental Figure 1), but the faster migration of Crtc2 at 80 kDa demonstrated the specificity for each antisera. The observation that the phosphorylation of a specific isoform of human Crtc1 may depend on LKB1 regulated kinase activity extends recent results from studies with either murine tissues or recombinant protein that demonstrated that the phosphorylation and gel mobility of the Crtc2 homolog was dependent on LKB1 status through the activation of downstream AMPK/SIK-related kinase members (Shaw et al., 2005).
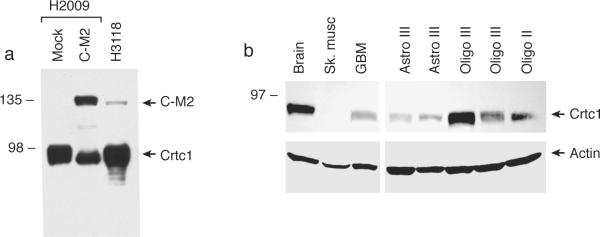
Figure 1. Endogenous Crtc1 in normal and human tumor cells.
a) N-terminal anti-Crtc1 antisera detects the recombinant Crtc1-Maml2 fusion peptide (C-M2) transfected into H2009 lung cancer cells and the endogenous C-M2 fusion peptide (H3118 lung mucoepidermoid cells) at 135 kDa (upper arrow). Steady-state expression of the endogenous 85 kDa Crtc1 is detected in both cell lines (lower arrow). b) Variable expression of endogenous Crtc1 in brain tumor samples. Glioblastoma (GBM), astrocytoma (Astro), oligodendroglioma (Oligo), grade II/III (II/III).
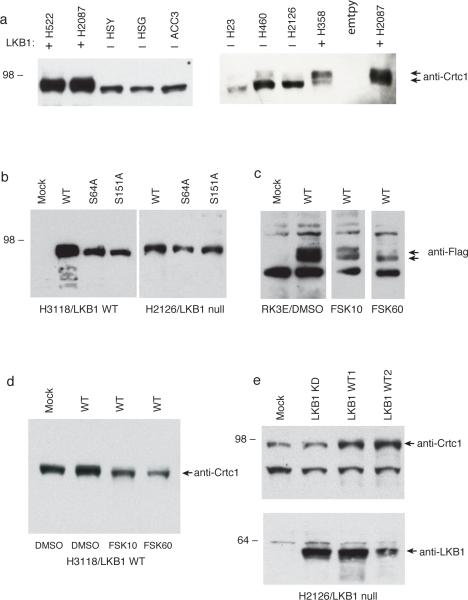
Figure 2. Endogenous and recombinant Crtc1 mobility pattern linked with LKB1 status in human tumor cells.
a) 60 μg protein extracts subjected to imunoblotting using peptide anti-Crtc1 antisera. LKB1 protein status is indicated above each lane and was confirmed by imunoblotting for each sample (data not shown). The samples on the left panel were run on a gradient gel and on the right panel on a 7% acrylamide gel to emphasize the size differences. Arrows depict the faster and slower Crtc1 mobility forms. b) Flag-tagged wildtype (wt) Crtc1 and the indicated point mutant Crtc1 vectors were transfected into either LKB1 wildtype cells (H3118) or LKB1 null cells (H2126) and protein extracts were subjected to immunoblotting with anti-Flag. c) Flag-tagged wt Crtc1 was transfected in LKB1 wildtype cells and subjected to incubation with forskolin 10 microgram/1 hr (FSK10) or 60 microgram/4 hrs (FSK60) treatment as indicated. d) H3118 cells were repeated as in panel c and immunoblotted with anti-Crtc1 antisera. e) LKB1-null H2126 cells were mock-transfected or transfected with either the kinase dead (KD) or wt LKB1 plasmids and protein extracts subjected to imunoblotting with either anti-Crtc1 (top) or anti-LKB1 (bottom).
To test if the Crtc1 immunoblot mobility shift detected by our peptide antisera was related to LKB1 dependent phosphorylation, we performed three separate experiments. First, we generated a wildtype recombinant Flag-tagged Crtc1 (wt), as well as two Crtc1 point mutants corresponding to a S64A mutation at a candidate glycosylation site regulating phosphorylation and at a S151A mutation corresponding to the predicted AMPK/LKB1 phosphorylation site identified by alignment with both human Crtc2/Torc2 and drosophila Crtc/Torc (Wang et al., 2008). We observed a broad-based mobility signal when the wt Flag-Crtc1 plasmid was transfected into LKB1 wt cells (H3118), however, we detected predominantly the faster mobility migration pattern with the S151A Crtc1 mutant in the same LKB1 wt cell line (Figure 2b, left panel). In contrast, when the identical vectors were transfected into the LKB1 null lung cancer cell line (H2126), the wt Crtc1 product co-migrated with the faster mobility S151A mutant (Figure 2b, right panel). These data support the model where LKB1 is a regulator of phosphorylation for both endogenous and recombinant Crtc1. We next transfected FLAG tagged wt Crtc1 into another LKB1wt cell line (RK3E) followed by incubation with forskolin, which has been previously reported to induce Crtc2/Torc2 dephosphorylation (Screaton et al., 2004). Using an anti-FLAG antisera, we observed that the broad-based, wildtype recombinant Crtc1 signal could be resolved into discrete mobility species after 1 hr treatment with 10 μM forskolin which collapsed to predominantly the underphosphorylated signal that co-migrated with the endogenous unphosphorylated signal following 4 hr exposure to 60 μM forskolin (Figure 2c). We confirmed this observation, by transfecting the FLAG-wt Crtc1 vector into H3118 cells followed by treatment with either DMSO alone or forskolin/DMSO at the indicated concentrations. In this experiment, however, we immunoblotted the lysates with the anti-Crtc1 antisera and observed that the mobility of both endogenous and recombinant Crtc1 products were shifted fully to the underphosphorylated form (Figure 2d). Finally, we transfected the H2126 LKB1 null cells with either an empty parental vector, a kinase-dead (KD) LKB1 (K78I) point mutant, or two different subclones of wildtype LKB1 plasmid (Shaw et al., 2004). We confirmed expression of the mutant KD and the two wildtype LKB1 vectors by immunoblotting with anti-LKB1 and subjected the same extracts to analysis with the anti-Crtc1 antisera. Although the efficiency of the transient transfection in H2126 cells was estimated at 30% (data not shown) we could detect a shift in mobility of endogenous Crtc1 in cells transfected with wildtype LKB1 but no effect with the KD control product or vector alone (Figure 2e). In summary, these data support the hypothesis that a subset of lung tumors with somatic mutations for LKB1 show aberrant mobility of an endogenous Crtc1 isoform.
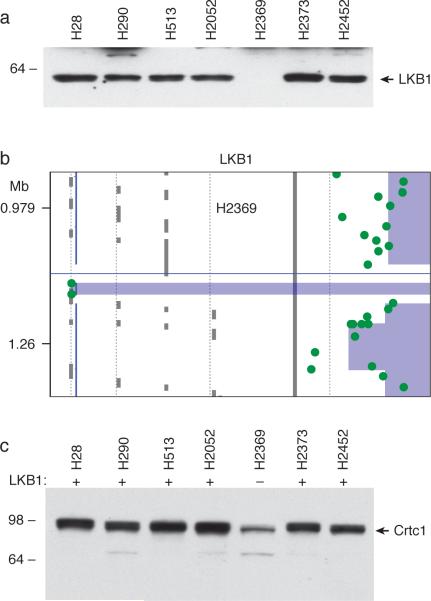
To strengthen the association of Crtc1 activation with loss of LKB1 status, we tested a separate dataset of tumor samples for LKB1 status, rather than screening initially for an abnormal Crtc1 migration pattern. We selected mesothelioma tumor cell lines which had not been previously studied for LKB1 status and found one case (H2369) with null LKB1 protein expression (Figure 3a). In addition, we confirmed the molecular basis for LKB1 loss by detection a discrete internal 40-kilobase homozygous deletion using a high density oligo-based CGH assay (Figure 3b). Subsequent immunoblot analysis of the Crtc1 migration pattern showed a predominantly discrete migrating Crtc1 species in only the tumor cell line that corresponded to LKB1 null status (Figure 3c).
Figure 3. LKB1 null status associated with faster Crtc1 mobility.
a) mesothelioma tumor cells lines were subjected to imunoblotting with anti-LKB1. b) High resolution CGH analysis for LKB1 detects homozygous deletion within LKLB1 locus exclusively for H2369 cells. c) matched tumor cells lines subjected to immunoblotting with anti-Crtc1.
Activation of endogenous Crtc1 in LKB1 null tumor: nuclear localization
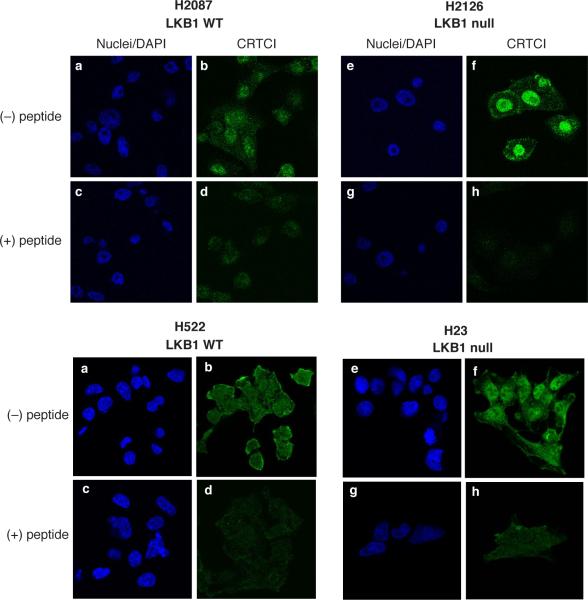
The transcriptional CREB co-activator function of Crtc2/Torc2 has been causally associated with nucleocytoplasmic shuttling in response to changes in AMPK/SIK2 phosphorylation suggesting that loss of LKB1 function in lung cancer samples should also increase the proportion of endogenous Crtc1 within the nuclear compartment of these tumor cells. To test this hypothesis, we subjected LKB wildtype and LKB mutant lung cancer cells to immunofluorescence using anti-Crtc1 in the presence or absence of the competing epitope peptide used in the generation of the rabbit polyclonal antisera. We observed that the LKB1 wildtype lung cancer cell lines H2087 and H522 showed predominantly cytoplasmic staining which was blocked by the addition of the immunogenic Crtc1 peptide used to gnerate the antisera (Figure 4). In contrast, the LKB1 null lung cancer cell lines H2126 and H23, which expressed predominantly the underphosphorylated Crtc1 species, showed increased nuclear staining for endogenous Crtc1 which could be competed by the blocking Crtc1 peptide (Figure 4). These data is also consistent with a recent study of the single drosophila Crtc product which was shown to be regulated by phosphorylation-dependent nuclear export (Wang et al., 2008).
Figure 4. Enhanced nuclear staining of endogenous Crtc1 in LKB1 null cells.
a) H2087 and H522 LKB1 wildtype cells and b) H2126 and H23 LKB1 null cells were stained with DAPI and anti-Crtc1 in the absence or presence of the blocking immunogenic Crtc1 peptide and visualized by immunofluorescence.
Activation of endogenous Crtc1 in LKB1 null tumor: NR4A2/Nurr1 as a downstream target in cancer
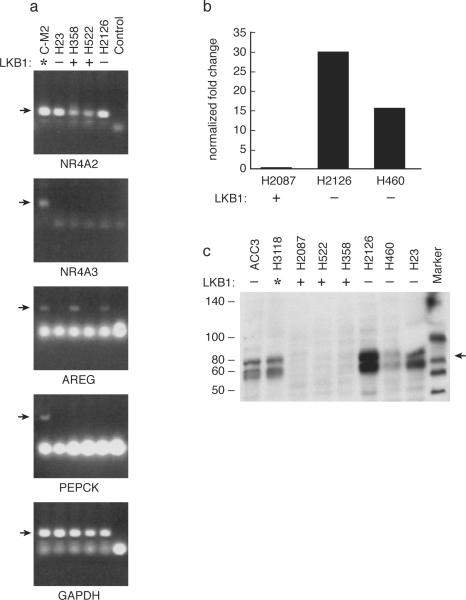
We, and others, had previously shown that the Crtc1-Maml2 fusion oncogene could potently activate a group of CREB inducible genes (Conkright et al., 2003; Coxon et al., 2005; Iourgenko et al., 2003; Wu et al., 2005). These included genes for the gluconeogenic pathway, a series of orphan nuclear receptors that were previously known to be regulated by cAMP/CREB signaling, and other candidate CRE regulated gene products. Since we observed that LKB1 null tumor cells have enhanced Crtc1 nuclear localization, we tested if these tumor cells have also selectively activated a specific set of downstream cAMP/CREB mediators. To test this hypothesis, we harvested RNA from two LKB1 null and two LKB1 wildtype lung tumor cell lines as well as from a Crtc1-Maml2 transfectant clone that we and others had used previously to identify candidate downstream target genes by oligonucleotide-based global microarray profiling (Coxon et al., 2005; Ravnskjaer et al., 2007; Screaton et al., 2004; Wu et al., 2005). We performed RT-PCR using paired primers for four prototypic Crtc1 inducible gene including PEPCK, NR4A2, NR4A3, and amphiregulin. Although we detected expression of each of these genes in the control Crtc1-Maml2 stable transfectant clone (C-M2*), we only detected enhanced expression of NR4A2 in the LKB1 null lung tumor samples as compared with LKB1 wildtype lung cancer samples (Figure 5a). Quantitative RT-PCR with RNA prepared from a different set of LKB1 (+) and (−) tumor cells again showed significant induction of NR4A2 in LBK1 null (H2126 and H460) as compared with LKB1 wildtype (H2087) cell lines. (Figure 5b) In addition, we validated these data by performing immunoblot analysis and observed an elevated level of endogenous steady-state NR4A2 protein in LKB1 null and in the H3118 Crtc1-Maml2 fusion positive tumor lines as compared to LKB1 wildtype cells (Figure 5c).
Figure 5. Activation of the Crtc1/CREB target gene NR4A2/Nurr1 in LKB1 null samples.
a) RT-PCR for the indicated target genes and GAPDH in cells carrying the Crtc1-Maml2 fusion (*C-M2) and LKB1 wildtype (+) and LKB1 null (−) tumor samples. b) Quantitative RT-PCR for NR4A2/Nurr1 in LKB1 wildtype (H2087) and LKB1 null (H2126 and H460) samples. The NR4A2 signal was normalized for GAPDH expression. c) Protein immunoblot analysis for NR4A2 in tumor samples with indicated matched LKB1 status.
Nr4A2 knock-down shows growth suppression in tumors with activated Crtc1
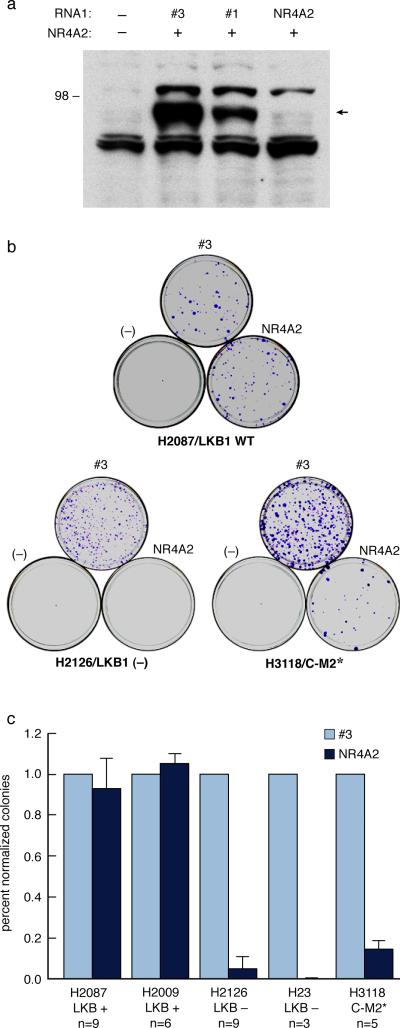
Previous studies had shown that siRNA knock-down of NR4A2 was associated with reduced colony growth or cell death of HeLa cells (Ke et al., 2004) suggesting that Nr4A2 can exhibit cell survival signals. Since HeLa cells are also known to be LKB1 null, we tested if efficient knock-down of NR4A2 might selectively inhibit growth of Crtc1-Maml2 positive or LKB1 null cell lines as compared with LKB1 wt cell lines. Although we attempted to make several different RNAi hairpin plasmids directed against NR4A2, we were successful in only obtaining one sequence that could efficiently (>95%) knock-down recombinant Flag-tagged NR4A2 expression (Figure 6). When we transfected either the NR4A2 RNAi or control RNAi plasmids into the LKB1 wildtype lung cancer cells, H2087 and H2009, we observed no difference in colony growth after 4 weeks in G418 selective media. This data suggests that the suppression of NR4A2 with the pSHAG RNAi vector is not toxic to mammalian cells that do not overexpress the protein. In contrast, transfection of the identical NR4A2 RNAi plasmid into H3118 Crtc1-Maml2 positive cells with deregulated NR4A2 expression showed 85% colony growth suppression. In addition, transfection into the LKB1 null lung tumors, H2126 and H23, showed >90% growth suppression. These data confirm earlier data that NR4A2 suppression can inhibit growth of a subset of cancer cells (Ke et al., 2004; Li et al., 2006) and suggest that NR4A2 may be a useful molecular target for tumor cells with aberrant activation of Crtc target genes.
Figure 6. NR4A2 RNAi colony growth inhibition.
a) NR4A2 immunoblot showing RNAi protein knock-down by either control pSHAG RNAi #3 or pSHAG #1 sequences or by NR4A2 specific pSHAG RNAi vector. b) crystal violet staining of 10 cm dishes with indicated tumor lines grown 4 weeks in G418-containing media following transfection with mock (−), control pSHAG #3 RNAi (#3), or pSHAG NR4A2 RNAi plasmids. c) colony growth suppression data (mean and standard deviation) for pSHAG NR4A2 RNAi as compared with control RNAi transfection on the indicated tumor cell lines.
Discussion
Since the biology of LKB1 is complex and linked with many signaling pathways spanning cell polarity, cellular metabolism, and cell cycle control (Katajisto et al., 2007; Sanchez-Cespedes, 2007; Shah et al., 2008), there remains an imperfect understanding of how somatic or inherited mutations of LKB1 lead to tumorigenesis. We have now identified another element to this pathway by observing that somatic loss of LKB1 in tumor cell lines is associated with enhanced Crtc1 activation resulting in deregulated expression of several cAMP/CREB inducible targets, including NR4A2/Nurr1. We and others had previously shown that Crtc1 is a bona fide cancer gene when activated by a t(11;19) chromosomal translocation resulting in the aberrant transcription of certain Crtc1 inducible genes, including the orphan nuclear receptor NR4A2 (Coxon et al., 2005; Tonon et al., 2003; Wu et al., 2005). Since LKB1 has been identified as a key regulator for Crtc2 transcriptional activity (Katoh et al., 2006; Shaw et al., 2005) and since somatic mutations have been identified in a subset of human lung tumors and cell lines (Ji et al., 2007; Sanchez-Cespedes et al., 2002), our data now suggest that enhanced Crtc gene family activity may participate in the tumorigenic process within LKB1 null tumors that are not otherwise associated with the recurrent t(11;19) rearrangement. Interestingly, overexpression of NR4A2 had been previously reported for HeLa and H460 cell lines, which are both known to be LKB1 null (Katoh et al., 2006; Tiainen et al., 1999). NR4A2 has also been tested as a candidate tumor suppressor gene where siRNA knock-down in HeLa cells was associated with parameters of growth inhibition and apoptosis (Ke et al., 2004). In addition, deregulation of NR4A1-3 family members have been associated with both alterations in glucose metabolism (Pei et al., 2006), as well as cell survival signals (Wallen-Mackenzie et al., 2003) and tumorigenesis (Li et al., 2006). The low or undetectable levels of steady-state NR4A2 expression observed in LKB1 wildtype lung tumor cell lines when grown to confluence is also consistent with prior data from studies on colon cancer cell lines which showed low or undetectable NR4A2 expression under basal growth conditions, but induction of NR4A2 following incubation with the prostaglandin PGE2 (Holla et al., 2006). The authors observed that the PGE2-mediated induction of NR4A2 conveyed pro-survival signals and was dependent on cAMP signaling which is consistent with a potential role for Crtc gene members as intermediaries in a signaling pathway. These data, therefore, suggest that persistent expression of NR4A2 under basal growth conditions may be an important biomarker. In addition, a recent study confirmed that induction of PGE2 by timed incubations with IBOP in serum starved lung cancer cell lines resulted in enhanced NR4A2 expression. Although the authors observed induction in LKB1 wildtype cells, the strongest effect was demonstrated in the LKB1 null cells H23, H460, and A549 and they mapped the NR4A2 promoter activation to a CREB binding site (Li and Tai, 2009), suggesting again that activation of Crtc gene members may participate in an enhanced lability for NR4A2 induction.
Defining the role of a potential LKB1:Crtc pathway, however, will be complicated by potential redundancy and cross-talk in the signaling cascade where LKB1, in concert with upstream nutrient/energy sensing and calcium fluxes, regulate the kinase activity of many different AMPK family members, which in turn phosphorylate and inhibit multiple downstream targets in addition to Crtc gene members (Shaw et al., 2004). While AMPK, Sik2, and Mark2 have been proposed as major proximate regulators for Crtc2 (Dentin et al., 2007; Fu and Screaton, 2008; Hezel and Bardeesy, 2008; Lizcano et al., 2004), Sik1 has also been suggested as an important kinase inhibitor for Crtc1 transcriptional activity (Katoh et al., 2006). Further, SIK1 is a Crtc1/cAMP/CREB target gene whose induction is proposed to phosphorylate and mediate a negative feedback loop on Crtc1 in neuronal cells (Li et al., 2009). Accordingly, we examined Sik1 expression in the lung tumor cell lines and observed a heterozygous genomic Sik1 deletion and absent Sik1 protein expression in the lung tumor cell line that showed the strongest constitutive Crtc1 activity (H2126; Sanger COSMIC database, http://www.sanger.ac.uk/genetics/CGP and data not shown). It is noteworthy that the lung cancer cell line with the strongest Crtc1 nuclear localization pattern (H2126) showed loss of both SIK1 and LKB1 function, however, more work is required to determine if loss of Sik1/2 expression cooperates with LKB1 null status since we did not see a strong correlation of Sik1 protein levels in other tumors samples with defined LKB1 status (data not shown). Sik1, however, was independently identified in a global unbiased RNAi screen for kinases that cooperate with PI3KCA to modulate tumor growth in soft agar, and reduced SIK1 expression was also correlated with a poor prognosis outcome in datasets obtained from patients with breast cancer (Cheng et al., 2009). These observations suggest that SIK1 may participate in mediating LKB1 tumor suppressor activity and members of the Crtc gene family have been proposed as novel candidate downstream targets for SIK kinase signaling (Shaw, 2009).
Finally, while LKB1 can cooperate with mutant K-ras in a mouse lung tumor model, LKB1 mutations are detected in human tumor samples that are either wildtype or mutant for K-ras, p53, and CDKN2a/arf/RB cancer genes suggesting that LKB1 tumorigenesis may not be dependent on these pathways (Ji et al., 2007). These observations suggest a parallel LKB1/AMPK family pathway important for the survival of a substantial subset of human tumors. Targeting Crtc gain-of-function activation, therefore, is a plausible approach for cancer therapeutics since Drosophila mutants carrying homozygous loss of the single Crtc gene were viable and fertile suggesting a reduced potential for normal tissue toxicity (Wang et al., 2008), and there are multiple strategies and commercially available pharmacologic agents that can impact either Crtc/Torc phosphorylation, nuclear/cytoplasmic trafficking, or downstream target signaling (Carling, 2006; Huang et al., 2008; Screaton et al., 2004; Shaw et al., 2005). These data offer another element for the LKB1 axis and suggest that Crtc1 can be activated by chromosomal translocation in uncommon salivary gland tumors or by the loss of LKB1 function in a wider range of common adult malignancies.
Materials and Methods
Cell lines, plasmids, RT-PCR
All tumor cell lines were obtained from the ATCC repository (Manassas, VA) or from the National Naval Medical Center, Bethesda and maintained in RPMI with 10% serum or ACL4 with 10% serum (H2087). The Crtc1 mammalian expression vector was subcloned using a cDNA (GenBank #AY040323) that corresponded to the predominant Crtc1 isoform present in the lung cancer samples and which aligned with the exon splicing pattern reported for the single Drosophila Crtc/Torc cDNA sequence (Wang et al., 2008). Wildtype LKB1 and kinase dead LKB1 point mutant were obtained from Addgene (Cambridge, MA) (Shaw et al., 2004). NR4A2 expression vector was obtained from Genecopoeia (Germantown, MD). Additional single point gene mutation expression vectors were generated using site-directed oligonucleotide mutagenesis as described by the manufacturer (Promega, Madison, WI). RNA was subjected to RTPCR using Superscript Reverse Transcriptase (Invitrogen) and PuRe Taq Ready-to-go beads (GE Healthcare) with indicated paired oligonucleotide primers as previously published (Coxon et al., 2005). GAPDH was amplified as a control for RNA loading. cDNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems).was also subjected to Q-PCR analysis using the NR4A2 Taq Man Gene Expression Assay (Applied Biosystems) for Fast Real-Time PCR (Applied Biosystems 7900HT). NR4A2 expression was normalized to GAPDH expression.
Antibodies
Protein extracts harvested from cell transfection experiments were subjected to imunoblotting using anti-Crtc1 polyclonal rabbit antisera generated against a custom exon 1 N-terminal peptide (aa 19–34, Rockland antibodies, Gilbertsville, PA), anti-LKB1 and anti-Crtc2/Torc2 (Cell Signaling, Danvers, MA), anti-NR4A2 (R&D systems), or anti-Flag (Sigma-Aldrich).
Immunoflourescence
Indicated cell lines were grown to 70% confluency on coverslips in multiwell plates. Cells were washed in PBS, fixed in 3% methanol free formaldehyde (Polysciences, Warrington, PA) for 15 minutes, and washed again in PBS. Cells were then incubated overnight with a 1:1000 dilution of the polyclonal Crtc1 antisera at 4° C. Alternatively, the antisera was pre-incubated with a five-fold excess of the Crtc1 blocking peptide at 4° C for 2 hours prior to overnight incubation. To visualize, cells were washed in PBS and incubated with the Alexafluor 488 fluorochrome-conjugated secondary antibody (Invitrogen) for 2 hours in the dark. Cells were washed in PBS and coverslips were mounted with Prolong Gold antifade reagent with DAPI (Invitrogen) according to the manufacturer's protocol.
RNAi
pSHAG RNAi control and NR4A2 specific vectors were generated and tested as previously published (Komiya et al., 2006; Paddison et al., 2002) using pSHAG design strategies as outlined at the RNAi central website (http://katahdin.cshl.org:9331/homepage/portal/scripts/main2.pl?link=plasmids&content=plasmids.html). For colony growth inhibition assays, plasmids with a neo resistant marker were transfected using FuGENE 6 reagent (Roche Applied Science) into indicated subconfluent tumor cells using 10 cm cell culture plates and incubated with the appropriate G418 concentrations starting 72 hr following transfection. Plates were stained with crystal violet at 4 weeks and scored for colony formation as previously published (Komiya et al., 2006).
Supplementary Material
Supplemental Figure 1 a) Immunoblot of lung cancer samples (60μg whole cell protein lysates) using a rabbit polyclonal pan-Crtc antisera generated against Crtc1 exon 1 aa 2–42. Upper arrowhead depicts 85 kDa species that co-migrates with signal detected with the peptide Crtc1 antisera. Lower arrowhead depicts cross-reactive 65 kDa bands that may represent Crtc splice isoforms or the smaller Crtc3 homolog. b) specificity of the 85 and 65 kDa signals shown by efficient blocking following incubation with the Gst-Crtc1 immunogen. c) Immunoblot analysis showing co-expression of Crtc2 (80 kDa) and Crtc1 (85 kDa) in duplicate lanes of the LKB1 wildtype H2087 lung cancer cell line and showing specificity for each antisera.
Acknowledgements
We would like to thank Jie Li (NINDS) for brain tumor samples. We are grateful to William Meyerson for technical assistance.
References
- Achcar RD, Nikiforova MN, Dacic S, Nicholson AG, Yousem SA. Mammalian mastermind like 2 11q21 gene rearrangement in bronchopulmonary mucoepidermoid carcinoma. Hum Pathol. 2009;40:854–60. doi: 10.1016/j.humpath.2008.11.007. [DOI] [PubMed] [Google Scholar]
- Behboudi A, Winnes M, Gorunova L, van den Oord JJ, Mertens F, Enlund F, et al. Clear cell hidradenoma of the skin-a third tumor type with a t(11;19)--associated TORC1-MAML2 gene fusion. Genes Chromosomes Cancer. 2005;43:202–5. doi: 10.1002/gcc.20168. [DOI] [PubMed] [Google Scholar]
- Bittinger MA, McWhinnie E, Meltzer J, Iourgenko V, Latario B, Liu X, et al. Activation of cAMP response element-mediated gene expression by regulated nuclear transport of TORC proteins. Curr Biol. 2004;14:2156–61. doi: 10.1016/j.cub.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Camelo-Piragua SI, Habib C, Kanumuri P, Lago CE, Mason HS, Otis CN. Mucoepidermoid carcinoma of the breast shares cytogenetic abnormality with mucoepidermoid carcinoma of the salivary gland: a case report with molecular analysis and review of the literature. Hum Pathol. 2009;40:887–92. doi: 10.1016/j.humpath.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Carling D. LKB1: a sweet side to Peutz-Jeghers syndrome? Trends Mol Med. 2006;12:144–7. doi: 10.1016/j.molmed.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Cheng H, Liu P, Wang ZC, Zou L, Santiago S, Garbitt V, et al. SIK1 couples LKB1 to p53-dependent anoikis and suppresses metastasis. Sci Signal. 2009;2:ra35. doi: 10.1126/scisignal.2000369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conkright MD, Canettieri G, Screaton R, Guzman E, Miraglia L, Hogenesch JB, et al. TORCs: transducers of regulated CREB activity. Mol Cell. 2003;12:413–23. doi: 10.1016/j.molcel.2003.08.013. [DOI] [PubMed] [Google Scholar]
- Coxon A, Rozenblum E, Park YS, Joshi N, Tsurutani J, Dennis PA, et al. Mect1-Maml2 fusion oncogene linked to the aberrant activation of cyclic AMP/CREB regulated genes. Cancer Res. 2005;65:7137–44. doi: 10.1158/0008-5472.CAN-05-1125. [DOI] [PubMed] [Google Scholar]
- Dentin R, Liu Y, Koo SH, Hedrick S, Vargas T, Heredia J, et al. Insulin modulates gluconeogenesis by inhibition of the coactivator TORC2. Nature. 2007;449:366–9. doi: 10.1038/nature06128. [DOI] [PubMed] [Google Scholar]
- Enlund F, Behboudi A, Andren Y, Oberg C, Lendahl U, Mark J, et al. Altered Notch signaling resulting from expression of a WAMTP1-MAML2 gene fusion in mucoepidermoid carcinomas and benign Warthin's tumors. Exp Cell Res. 2004;292:21–8. doi: 10.1016/j.yexcr.2003.09.007. [DOI] [PubMed] [Google Scholar]
- Fu A, Screaton RA. Using kinomics to delineate signaling pathways: control of CRTC2/TORC2 by the AMPK family. Cell Cycle. 2008;7:3823–8. doi: 10.4161/cc.7.24.7241. [DOI] [PubMed] [Google Scholar]
- Hawley SA, Boudeau J, Reid JL, Mustard KJ, Udd L, Makela TP, et al. Complexes between the LKB1 tumor suppressor, STRAD alpha/beta and MO25 alpha/beta are upstream kinases in the AMP-activated protein kinase cascade. J Biol. 2003;2:28. doi: 10.1186/1475-4924-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hezel AF, Bardeesy N. LKB1; linking cell structure and tumor suppression. Oncogene. 2008;27:6908–19. doi: 10.1038/onc.2008.342. [DOI] [PubMed] [Google Scholar]
- Holla VR, Mann JR, Shi Q, DuBois RN. Prostaglandin E2 regulates the nuclear receptor NR4A2 in colorectal cancer. J Biol Chem. 2006;281:2676–82. doi: 10.1074/jbc.M507752200. [DOI] [PubMed] [Google Scholar]
- Hong SP, Leiper FC, Woods A, Carling D, Carlson M. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc Natl Acad Sci U S A. 2003;100:8839–43. doi: 10.1073/pnas.1533136100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Wullschleger S, Shpiro N, McGuire VA, Sakamoto K, Woods YL, et al. Important role of the LKB1-AMPK pathway in suppressing tumorigenesis in PTEN-deficient mice. Biochem J. 2008;412:211–21. doi: 10.1042/BJ20080557. [DOI] [PubMed] [Google Scholar]
- Iourgenko V, Zhang W, Mickanin C, Daly I, Jiang C, Hexham JM, et al. Identification of a family of cAMP response element-binding protein coactivators by genome-scale functional analysis in mammalian cells. Proc Natl Acad Sci U S A. 2003;100:12147–52. doi: 10.1073/pnas.1932773100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Ramsey MR, Hayes DN, Fan C, McNamara K, Kozlowski P, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448:807–10. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- Katajisto P, Vallenius T, Vaahtomeri K, Ekman N, Udd L, Tiainen M, et al. The LKB1 tumor suppressor kinase in human disease. Biochim Biophys Acta. 2007;1775:63–75. doi: 10.1016/j.bbcan.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Katoh Y, Takemori H, Lin XZ, Tamura M, Muraoka M, Satoh T, et al. Silencing the constitutive active transcription factor CREB by the LKB1-SIK signaling cascade. FEBS J. 2006;273:2730–48. doi: 10.1111/j.1742-4658.2006.05291.x. [DOI] [PubMed] [Google Scholar]
- Kaye FJ. Emerging biology of malignant salivary gland tumors offers new insights into the classification and treatment of mucoepidermoid cancer. Clin Cancer Res. 2006;12:3878–81. doi: 10.1158/1078-0432.CCR-06-0791. [DOI] [PubMed] [Google Scholar]
- Kaye FJ. Mutation-associated fusion cancer genes in solid tumors. Mol Cancer Ther. 2009;8:1399–408. doi: 10.1158/1535-7163.MCT-09-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazakov DV, Vanecek T, Belousova IE, Mukensnabl P, Kollertova D, Michal M. Skin-type hidradenoma of the breast parenchyma with t(11;19) translocation: hidradenoma of the breast. Am J Dermatopathol. 2007;29:457–61. doi: 10.1097/DAD.0b013e318156d76f. [DOI] [PubMed] [Google Scholar]
- Ke N, Claassen G, Yu DH, Albers A, Fan W, Tan P, et al. Nuclear hormone receptor NR4A2 is involved in cell transformation and apoptosis. Cancer Res. 2004;64:8208–12. doi: 10.1158/0008-5472.CAN-04-2134. [DOI] [PubMed] [Google Scholar]
- Komiya T, Park Y, Modi S, Coxon A, Oh H, Kaye FJ. Sustained expression of Mect1-Maml2 is essential for tumor cell growth in salivary gland cancers carrying the t(11;19) translocation. Oncogene. 2006;25:6128–32. doi: 10.1038/sj.onc.1209627. [DOI] [PubMed] [Google Scholar]
- Kovacs KA, Steullet P, Steinmann M, Do KQ, Magistretti PJ, Halfon O, et al. TORC1 is a calcium- and cAMP-sensitive coincidence detector involved in hippocampal long-term synaptic plasticity. Proc Natl Acad Sci U S A. 2007;104:4700–5. doi: 10.1073/pnas.0607524104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennerz JK, Perry A, Mills JC, Huettner PC, Pfeifer JD. Mucoepidermoid Carcinoma of the Cervix: Another Tumor With the t(11;19)-associated CRTC1-MAML2 Gene Fusion. Am J Surg Pathol. 2008 doi: 10.1097/PAS.0b013e318190cf5b. [DOI] [PubMed] [Google Scholar]
- Li QX, Ke N, Sundaram R, Wong-Staal F. NR4A1, 2, 3--an orphan nuclear hormone receptor family involved in cell apoptosis and carcinogenesis. Histol Histopathol. 2006;21:533–40. doi: 10.14670/HH-21.533. [DOI] [PubMed] [Google Scholar]
- Li S, Zhang C, Takemori H, Zhou Y, Xiong ZQ. TORC1 regulates activity-dependent CREB-target gene transcription and dendritic growth of developing cortical neurons. J Neurosci. 2009;29:2334–43. doi: 10.1523/JNEUROSCI.2296-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Tai HH. Activation of Thromboxane A2 Receptors Induces Orphan Nuclear Receptor Nurr1 Expression and Stimulates Cell Proliferation in Human Lung Cancer Cells. Carcinogenesis. 2009 doi: 10.1093/carcin/bgp161. [DOI] [PubMed] [Google Scholar]
- Lizcano JM, Goransson O, Toth R, Deak M, Morrice NA, Boudeau J, et al. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 2004;23:833–43. doi: 10.1038/sj.emboj.7600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16:948–58. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei L, Waki H, Vaitheesvaran B, Wilpitz DC, Kurland IJ, Tontonoz P. NR4A orphan nuclear receptors are transcriptional regulators of hepatic glucose metabolism. Nat Med. 2006;12:1048–55. doi: 10.1038/nm1471. [DOI] [PubMed] [Google Scholar]
- Ravnskjaer K, Kester H, Liu Y, Zhang X, Lee D, Yates JR, 3rd, et al. Cooperative interactions between CBP and TORC2 confer selectivity to CREB target gene expression. EMBO J. 2007;26:2880–9. doi: 10.1038/sj.emboj.7601715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Cespedes M. A role for LKB1 gene in human cancer beyond the Peutz-Jeghers syndrome. Oncogene. 2007;26:7825–32. doi: 10.1038/sj.onc.1210594. [DOI] [PubMed] [Google Scholar]
- Sanchez-Cespedes M, Parrella P, Esteller M, Nomoto S, Trink B, Engles JM, et al. Inactivation of LKB1/STK11 is a common event in adenocarcinomas of the lung. Cancer Res. 2002;62:3659–62. [PubMed] [Google Scholar]
- Screaton RA, Conkright MD, Katoh Y, Best JL, Canettieri G, Jeffries S, et al. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell. 2004;119:61–74. doi: 10.1016/j.cell.2004.09.015. [DOI] [PubMed] [Google Scholar]
- Shah U, Sharpless NE, Hayes DN. LKB1 and lung cancer: more than the usual suspects. Cancer Res. 2008;68:3562–5. doi: 10.1158/0008-5472.CAN-07-6620. [DOI] [PubMed] [Google Scholar]
- Shaw RJ. Tumor suppression by LKB1: SIK-ness prevents metastasis. Sci Signal. 2009;2:pe55. doi: 10.1126/scisignal.286pe55. [DOI] [PubMed] [Google Scholar]
- Shaw RJ, Bardeesy N, Manning BD, Lopez L, Kosmatka M, DePinho RA, et al. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell. 2004;6:91–9. doi: 10.1016/j.ccr.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Shaw RJ, Lamia KA, Vasquez D, Koo SH, Bardeesy N, Depinho RA, et al. The kinase LKB1 mediates glucose homeostasis in liver and therapeutic effects of metformin. Science. 2005;310:1642–6. doi: 10.1126/science.1120781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiainen M, Ylikorkala A, Makela TP. Growth suppression by Lkb1 is mediated by a G(1) cell cycle arrest. Proc Natl Acad Sci U S A. 1999;96:9248–51. doi: 10.1073/pnas.96.16.9248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirado Y, Williams MD, Hanna EY, Kaye FJ, Batsakis JG, El-Naggar AK. CRTC1/MAML2 fusion transcript in high grade mucoepidermoid carcinomas of salivary and thyroid glands and Warthin's tumors: implications for histogenesis and biologic behavior. Genes Chromosomes Cancer. 2007;46:708–15. doi: 10.1002/gcc.20458. [DOI] [PubMed] [Google Scholar]
- Tonon G, Modi S, Wu L, Kubo A, Coxon AB, Komiya T, et al. t(11;19)(q21;p13) translocation in mucoepidermoid carcinoma creates a novel fusion product that disrupts a Notch signaling pathway. Nat Genet. 2003;33:208–13. doi: 10.1038/ng1083. [DOI] [PubMed] [Google Scholar]
- Wallen-Mackenzie A, Mata de Urquiza A, Petersson S, Rodriguez FJ, Friling S, Wagner J, et al. Nurr1-RXR heterodimers mediate RXR ligand-induced signaling in neuronal cells. Genes Dev. 2003;17:3036–47. doi: 10.1101/gad.276003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Goode J, Best J, Meltzer J, Schilman PE, Chen J, et al. The insulin-regulated CREB coactivator TORC promotes stress resistance in Drosophila. Cell Metab. 2008;7:434–44. doi: 10.1016/j.cmet.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Liu J, Gao P, Nakamura M, Cao Y, Shen H, et al. Transforming activity of MECT1-MAML2 fusion oncoprotein is mediated by constitutive CREB activation. Embo J. 2005;24:2391–402. doi: 10.1038/sj.emboj.7600719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Wu H, Li S, Chen Q, Cheng XW, Zheng J, et al. Requirement of TORC1 for late-phase long-term potentiation in the hippocampus. PLoS ONE. 2006;1:e16. doi: 10.1371/journal.pone.0000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1 a) Immunoblot of lung cancer samples (60μg whole cell protein lysates) using a rabbit polyclonal pan-Crtc antisera generated against Crtc1 exon 1 aa 2–42. Upper arrowhead depicts 85 kDa species that co-migrates with signal detected with the peptide Crtc1 antisera. Lower arrowhead depicts cross-reactive 65 kDa bands that may represent Crtc splice isoforms or the smaller Crtc3 homolog. b) specificity of the 85 and 65 kDa signals shown by efficient blocking following incubation with the Gst-Crtc1 immunogen. c) Immunoblot analysis showing co-expression of Crtc2 (80 kDa) and Crtc1 (85 kDa) in duplicate lanes of the LKB1 wildtype H2087 lung cancer cell line and showing specificity for each antisera.