ABSTRACT
Vaccines play an essential role in controlling the rates of fatality and morbidity. Vaccines not only arrest the beginning of different diseases but also assign a gateway for its elimination and reduce toxicity. This review gives an overview of the possible uses of computational tools for vaccine design. Moreover, we have described the initiatives of utilizing the diverse computational resources by exploring the immunological databases for developing epitope-based vaccines, peptide-based drugs, and other resources of immunotherapeutics. Finally, the applications of multi-graft and multivalent scaffolding, codon optimization and antibodyomics tools in identifying and designing in silico vaccine candidates are described.
KEYWORDS: Vaccine, reverse vaccinology, rational design, antibodyomics tools, epitope prediction, multi-graft scaffolding
1. Introduction
Vaccine design is a complicated process, however, advances in bioinformatics will probably make vaccine design and drug development easy.1 The design of vaccines can be divided into two broad categories: the traditional and the modern approach. The design of traditional vaccines is expensive, time-consuming, and not applicable for antigenically diverse pathogens.2 This is because of the genetic/antigenic diversity of pathogens, insufficient information about the interaction between pathogen and host, absence of a permissive cell line, and lack of successful animal models.3,4 It has been observed that vaccine development for severe diseases, such as smallpox, human immunodeficiency virus causing acquired immunodeficiency syndrome (HIV-AIDS), and tuberculosis (TB) was also affected by these drawbacks.3 On the other hand, vaccines developed by the traditional approach for smallpox, polio, and diphtheria have several drawbacks and faced many problems.5
Due to limitations of the conventional technology, modern technologies have come into existence, including recombinant DNA technology, rational vaccinology, structural biology, conjugate vaccines, next-generation technology and epitope-based vaccine design. With the help of recombinant DNA technology, vaccines developed are regarded as safe, effective and inexpensive as compared to other traditional vaccines and apply for the bulk production of sub-unit vaccines.6 Several in silico tools have been designed for the development of immunotherapy along with peptide-based drugs discovery in the previous two decades. Therefore, it is crucial to develop novel therapeutics with prophylactic vaccines and computational tools against different diseases like malaria, HIV-AIDS, and tuberculosis.7 Practice is required in the field of genomics, structural biology, computational biology, and rational vaccinology to improve the development of vaccines.8 Initiation of sequence analysis and recombinant DNA technology (RDT) opened the way to innovative vaccine design, including the concept of epitope-based vaccine design. The genomic analysis of pathogens also facilitates the classification and recognization of the protective epitope.9
Modern computational design starts as a dynamic force to facilitate structural vaccinology, whereby protein antigens are designed to prepare novel biomolecules with better immunological properties.10 Regular progress in vaccine development and diagnostic fields accelerate the broad application of structural vaccinology (SV), reverse vaccinology (RV) and antigen recognition technology.11 However, systems biology aids in predicting the host-pathogen interactions, and improves adjuvant capability to provide long-lasting immunity.12 In these novel technologies, rational vaccinology is an innovative and functionally applicable approach to design the potent immunogen for the induction of prolonged protective immunity. With the help of this technology, synthetic peptide vaccine was designed for the treatment of asthma.13 The comprehensive vaccines for viral pathogens such as HIV, influenza and hepatitis C virus may be designed through rational vaccinology approach as reported by Burton, 2017.14 Antigen prediction is an important criterion in the process of vaccine development. Vaxi Jen is an online software, based on the alignment-free approach and can directly predict the antigens.15 It is the first online server for alignment-independent prediction of protective antigens. The modern technology of vaccine design also includes reverse vaccinology, which accelerates the process of vaccine development.16 Epitope mapping is also a crucial factor in designing an effective vaccine as it generates vigorous reactions from both B cells and T cells and in silico prediction successfully increases the epitope prediction.17,18 A multi-epitope peptide vaccine was developed to stimulate an effective immune response for the treatment of brucellosis. Ren et al., 201919 prepared a multi-epitope vaccine through bioinformatic tools for evaluating its immune response in mice, and high production of IgG antibodies was observed.
Broadly neutralizing antibodies (bNAbs) is a new term in immuno-informatics and is still in the computational pipeline. It was initially applied to analyze a different class of HIV-1 bNAbs entirely based on 454-sequencing method.20 These antibodies have the feature of targetting only conserved epitopes of the microbes that play a significant role in virulence21 and develops a new area of research to design a vaccine against quickly mutating viruses such as HIV and influenza.22 The proper implementation of computational tools minimizes the various challenges in the field of vaccine development. Computational biology also constitutes side-chain prediction tools to design an antibody and predict its structure.23 Different aspects of multi-graft, multivalent scaffolding, codon optimization, and antibodyomics tools to identify and design potential vaccine candidate are also well described. This review provides relevant information about the latest computational tools that are essential for vaccine design since all of them have a unique feature and applcation according to the need of the situation.
2. Vaccine design: systems biology and structural antigen design
Vaccines not only arrest the beginning of different diseases, but also assign a doorway for its elimination and help in reducing the toxicity.24 Systems biology and structure-based antigen design are novel techniques to develop vaccines. A biological system is thoroughly analyzed via systematically including diverse areas such as genetics, biology, and chemistry. It gives valuable information about the gene, protein, and different metabolic pathway involved in pathogenesis.25 Systems biology collects a massive amount of biological data from the various hierarchical levels. The information about protein expression levels, DNA sequences, RNA, microRNAs, metabolite biology, protein-protein and protein-DNA interactions are obtained from the various biological datasets.26 The data generated will be further integrated and formulated through mathematical models to explain the structure of the system. Additionally, it helps in the analysis of the sequences of genes and proteins involved in the virulence in different microbes. The progress of “omics” technologies such as proteomics, genomics, metabolomics, and transcriptomics offers a comprehensive study of systems biology.27 Proteomics play an essential role in the field of vaccine design including immunogenic techniques along with a genome-based approach and to discover the potent immunogenic protein.28 Proteomic experiments in microorganisms were verified by whole genome sequencing and bioinformatics tools to discover new vaccines.29 Therefore, computational biology is an essential factor to fulfill this approach.
The detection of efficient biomarkers is the primary aim of molecular medicine. Systems biology has significantly identified the biological markers used for the diagnosis of various diseases.30 Groves et al., 201831 reported that systems biology enhances the recognition of a radiation-specific biomarker. Muhammad et al., 201932 showed that a computational approach, including systematic simulation-based meta-analytical framework, successfully predicted biomarkers. Oh et al., 201833 also demonstrated that the systems biology approach could play an essential role in designing potent vaccines against different diseases like Ebola or Zika virus (ZIKV), dengue, avian influenza and thrombocytopenia syndrome (SFTS).
Computational design is emerging as a driving force for structural vaccinology, where protein antigens are designed to produce new biomolecules to enhance immunological properties.34 Structural vaccinology induces a higher protective immune response, thus aiding in discovering novel antigens.35 The production of the vaccine can be enhanced by stabilizing the structure of complex antigens. Kaufmann and Flechtner36 mentioned that Herpes Simplex Virus (HSV) vaccine could be rationally developed as an alternative option for the treatment of the disease. Bajic et al., 201937 reported that VH1-69-encoded antibodies against influenza virus, HIV-1, and HCV. The mentioned antibodies have similar genetic and structural characteristics, and neutralize a wide spectrum of viral strains. Trobaugh et al., 201938 suggested that encephalitis virus vaccine could be designed by the implantation of structural vaccinology approaches, which is also referred to as rational vaccinology.
3. Adjuvants in vaccine design
An adjuvant can be defined as a vaccine component that enhances host immune response and plays an essential role in the development of vaccine.39 Adjuvants made of aluminum were used 90 years ago to enhance the immune response of the vaccines. Different aluminum salt products are used in vaccines that have a special feature of immunopotentiation along with safety records.40 Basically, aluminum hydroxide and aluminum phosphate are the two types of aluminum adjuvants used in specific licensed vaccines.41 They are prepared by vaccine companies and can be easily purchased by manufacturers like Brenntag Chemtrade, Biosector, and SPI Pharma. Generally, they can be simply recognized through their trade names like Alhydrogel, Rehydragel, and Adju-Phos. ImjectTM Alum made of amorphous aluminum hydroxycarbonate and crystalline magnesium hydroxide were used for preclinical and experimental studies.42 Calabro et al., 201343 reported that MF59 is a potent adjuvant that recruits CD11b+ blood mononuclear cells in the mouse muscle. MF59 is used in clinical trials as a component in prophylactic and therapeutic vaccines of infectious disease, cancer, and allergies. It also stimulates different immune cells such as neutrophils, eosinophils, macrophages, and monocytes. Adjuvants can be designed by incorporating various components such as TLR4 agonist, flagellin, and T-helper agonists.44 Kanzler et al, 200745 demonstrated that Toll-like receptors (TLRs) can enhance immune response, and thus can be used as vaccine adjuvant. TLR ligand-based adjuvants produce a robust immune response in the signaling of MyD88 in macrophages.46
Aucouturier et al., 200247 reported that montanide ISA 720 and 51 are used as water-in-oil emulsion adjuvants for human vaccine development. The TLR4 agonist, glucopyranosyl lipid adjuvant (GLA), protected mouse-adapted Ebola virus (ma-EBOV) and was prepared in a stable emulsion (SE) to stimulate immunogen and promote durable protection. Different adjuvants such as virosome, MPL and MF59 are applied in the design of vaccines like Invivac, Fendrix, and Pandemic Influenza vaccines, respectively.48 MPL is the foremost and only TLR ligand in licensed human vaccines, in the form of AS04 used for allergy treatment. This adjuvant is derivative of a liposaccharide that shows a reduced toxicity and maintains major immunostimulatory reaction of lipopolysaccharide.49 No harmful effects of MPL were observed in the rabbits when weekly doses were administered. In addition, it does not show any adverse effect on respiratory function, reproduction or genotoxicity. QS-21 induced antigen-specific antibody responses, including CD8+ T-cell response in mice and maintained a balanced production of IgG1 and IgG2a as compared to aluminum hydroxide that significantly favors IgG1 production.50 It can be used as an efficient adjuvant against feline leukemia virus (FeLV) in the form of a recombinant retroviral sub-unit vaccine. Many databases are available to find the adjuvant, such as Vaxjo (http://www.violinet.org/vaxj).51 This database incorporates approximately 400 vaccines that use an adjuvant and contains more than 100 vaccine adjuvants. Additionally, vaccine adjuvant design includes database development, omics bioinformatics, data analysis, and literature mining.52
4. Rational vaccine design
Rational vaccine design is an innovative approach in the field of vaccinology and is applied to design potent immunogens for the induction of prolonged protective immunity.53 Generally, rational vaccinology is applicable for viral pathogens such as hepatitis C virus, HIV, and influenza.54 The rationally developed vaccine consists of antigens, its delivery systems and an adjuvant to stimulate an immune response against specific epitopes of a particular pathogen. Computational modeling is an efficient tool to design the structure of a protein that can be determined through template-based and free modeling. Template-based modeling is entirely based on the 3D structure of a protein consisting of a selection of templates, sequence alignment, models construction, quality estimation, and structural modification.55 Vaccine design based on a protein structure depends on the conserved sites present on pathogens and the neutralizing antibodies with conserved sites.56 These neutralizing antibodies are adequate to inactivate antibodies and also induces prolonged protective immunity.
Vaxi Jen, the online software for the prediction of the antigens, can perform alignment-independent prediction of protective antigens and allows antigen classification separately based on the physicochemical properties of proteins. The server can be used on its own or in combination with alignment-based.57 TLRs identify the pathogen and are responsible for inducing innate immunity. Peta-flops-scale supercomputers can be used for modulation, screening, and identification of new lead structures for hTLR4. Moreover, they are applied for cancer immunotherapy to design polymeric hybrid micelles.58 With the incorporation of this technology, 12 compounds associated with tryptamine were screened and developed by in silico tools to preserve their molecular geometry while interacting with the hTLR4 binding site.59 RDT is successfully applied in rational vaccinology for the production of chimeric proteins. The classification of these proteins was performed by the help of Expasy ProtParam. A synthetic peptide vaccine was developed to enhance the efficiency of antigen presentation to stimulate the humoral immune response and thus help in the treatment of asthma as reported by Hayman et al., 2018.60
Rational vaccinology has been applied to discover a novel synthetic peptide vaccine for the treatment of asthma. Researchers described that a peptide of interleukin-13 was expressed and upregulated in asthma, and this could serve as a crucial antigen to use for screening of asthma through experimental and computational methods.61
Human astrovirus can cause viral diarrhea, especially in children and immune-compromised patients, and still, there is no vaccine available to prevent this infection.62 The rational vaccine design technology could be carried out for the development of a vaccine against it. Martinez et al., 201763 reported that cancer vaccines derived from MUC1-glycopeptides by implementing rational design technology. Different approaches are mentioned in Figure 1 for the rational vaccine design. The VBRC NERVE is a novel tool used to determine how specific proteins can act as potential vaccine candidates.64 Furthermore, the Promoter Scan can recognize potential epitopes that are suitable for immune response and the expression of the gene.9
Figure 1.
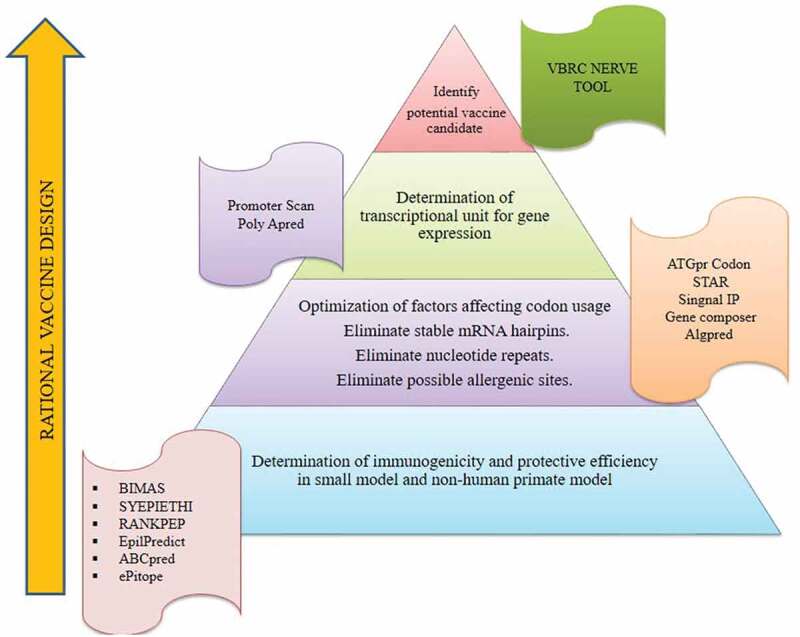
Rational vaccinology and its procedure to design vaccines through the VBRC NERVE tool. Illustration showing the steps involved in the identification of potential vaccine candidates through rational vaccinology. Each step uses different bioinformatics tools which are mentioned.
Bioinformatics approaches, such as structural approaches, MD simulations, and docking are also applied for the development of vaccines. It was reported that Chimeric Simian-Human Immunodeficiency Viruses (SHIVs) act as anti-HIV Env interventions in nonhuman primate (NHP) models and are designed by rational technology.65 Infection by Staphylococcus aureus causes high mortality and morbidity in humans. Additionally, Kailasan et al., 201966 reported that Leukocidin AB could be rationally designed as a toxoid vaccine against this infection. Tai et al., 201967 also reported that Zika virus sub-unit vaccine can be rationally designed with high efficiency in which envelope protein domain III (EDIII) is engineered to be used as a vaccine candidate. Trobaugh et al., 201938 showed that a rational approach is applied for the attenuation of eastern equine encephalitis virus (EEEV), a mosquito-transmitted alpha virus.
5. Computational tools for vaccines development
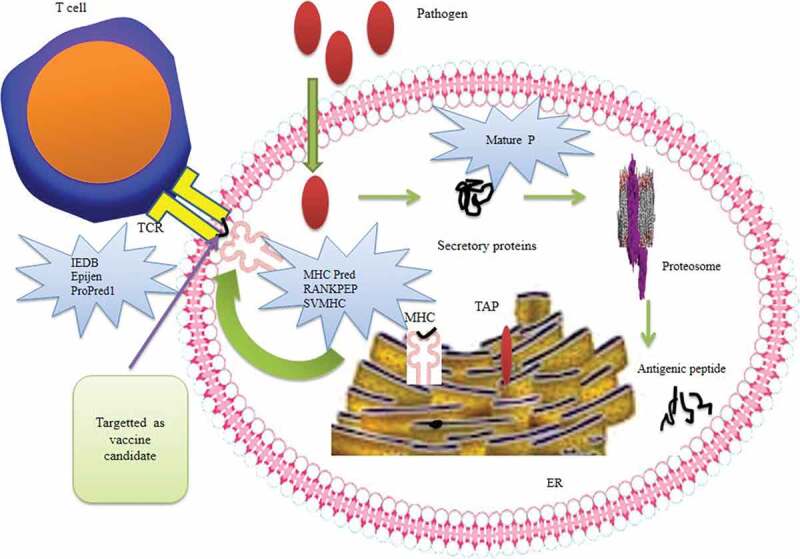
The biological information generated in genetics, biotechnology, and molecular biology is well organized and stored with the help of bioinformatic tools.68 Use of computational tools prior to lab experimentation is more advantageous as they are cost effective and take less time to operate. Immuno-informatics is a novel term applied to the conversion of large-scale immunological data in a compact form through the combination of computational and mathematical approaches.69 These tools are based entirely on statistical and machine learning systems and are well established in analyzing and modeling molecular interactions during antigen presentation and processing. In Figure 2, different computational tools, for the development of vaccines, along with their software, are mentioned.
Figure 2.
Computational strategies for the development of potent vaccine including antigen processing. Illustration showing the processing and screening of the antigen as an immunogen. The pathogen enters the cell and secretes proteins, which can be predicted by the Mature P software. Further processing of the protein is performed by proteosome and the antigenic peptide is released. This peptide, capable of binding MHC, can be predicted by MHCPred, RANKPEP, and SVMHC software. The peptide is then targeted as an epitope by the epitope prediction tools (IEDB, EpiJen, ProPred) and is selected as a potential vaccine candidate.
5.1. Side-chain and backbone modeling tools
Different types of receptors known as side chains are present on the cells and perform their function as gatekeeper of the cells. Each side chain has its own characteristic structure, and only the substance identical to them can enter the cell. The side-chain prediction is an essential constituent of computational biology for designing and predicting antibody structure.70 SCWRL and SCAP are tools which have been applied efficiently for modeling; they are used to determine and analyze mutations in protein side chains in silico. Moreover, these modeling tools can be applied for recognition and optimization of specific antibodies along with its affinity toward a particular target.71 Additionally, a powerful modeling program is available to design glycan epitopes on immunogens.72 Antibody modeling tools can be operated by backbone-dependent rotamer libraries.
Significantly, backbone modeling tools help in the modification of different antibodies. Leem et al., 201873 described position-dependent antibody rotamer swapper (PEARS, http://opig.stats.ox.ac.uk/webapps/pears), a side chain predictor which uses the IMGT position-dependent distribution of rotamers. It performs the side-chain prediction in less than 10 seconds.74 Additionally, DRAGON and GADGET are the antibodies use protein folding programs used to predict the secondary structure and ligand-binding site of the proteins. RAMBLE is an additional bioinformatics analysis tool with different permutations to check the connectivity of disulfide bonds, chain topology, and tryptone side-chain alignment.75 RAPPER is an in silico approach to generate 3D modeling of proteins for comparative analysis with a high degree of accuracy. It can be used to identify target sequences by exploring the conformational structure of a protein.76
5.2. Multi-graft and multivalent scaffolding
Multi-graft and multivalent scaffolding are the prominent approaches for the development of vaccines. A multivalent ligand has multiple copies of ligands which are capable of binding to different sites of the receptor.77 The concept of scaffolding expands the view of vaccine design based on epitope engineering. The implementation of epitopes of interest executed to the scaffolds of heterologous proteins was proven by studying HIV-1, flu, and RSV by Walenskey and Bird, 2010.78 Multivalent scaffolding technology is applied to epitope vaccine design based on the fact that a scaffold is present in the protein of interest. Perhaps it can be considered that multivalent scaffolds presents an antigen in a highly ordered and repetitive manner to induce a strong immune response. Furthermore, Ullah et al., 201979 reported that an inhibitor of the scaffold protein RACK1 (Receptor for Activated C Kinase 1) could inhibit the proliferation of HSV. The virus-like particles (VLPs) are small biological structures consisting of viral proteins similar to virion but not having genetic material and is incapable of stimulating an immune response. Barwal et al., 201680 also defined the virus-like particles as an attractive nano-particulate scaffold to apply in biological science and medicine. Hill et al., 201881 also mentioned that VLPs could be engineered to develop as antigens and thus help in the drug discovery and delivery. Rynda et al., 201482 demonstrated that virus-like particles could be used to make a vaccine candidate by exploring the lung as a site for immuno-stimulation. Prediction of protein structure is performed through composite modeling, which uses multiple templates for the development of multi-graft scaffold immunogen.
Identification of a specific scaffold having multiple epitopes is challenging, antibodies choose these epitopes without any hinderance.83 Küry et al., 201784 demonstrated that a proteosome scaffold subunit functions only during the phase of development of neurogenesis, and also showed that PSMD12 variants might affect neurodevelopment. Hence it can be said that protein engineering helped in the development of novel diagnostics and therapeutic agents. Different protein scaffolds such as DARPins (designed ankyrin repeat proteins) cysteine knots can be used as a scaffold to represent a functional site.85 Multivalent interaction of biological molecules is performed in different biochemical events to enhance the binding affinity, the avidity, and specificity of the ligand to the receptor. Hence multivalent ligands could be an alternate way to treat diseases as reported by Greenspan and Cavacini, 2019.86
A multigraft interface is a novel approach applied to graft epitopes so that antibody binding specificity can be improved and thus potentially affect the nature of antibodies.87 It is also applicable for the engineering of novel epitope scaffolds that exhibit neutralizing antibody 2F5 of HIV-1and also deal with the CDR H3 antibody loop. Gourlay et al., 201788 presented an automated computational tool, SAGE (strategy in alignment and grafting of epitopes), for the insertion of immune-generating epitopes onto a given scaffold. The approach assigns the identification of a graft position on any target antigen with a known three-dimensional structure, which is fast, extensive, and efficient tool. Mishra et al., 201889 reported prime scanning of epitope grafting and studied about computational grafting of malarial epitopes in serum albumin.
5.3. Antibodyomics tools
Antibodyomics is an essential computational tool initially applied for the analysis of the different classes of HIV-1 bNAbs, and based entirely on 454-sequencing.90 Significantly it is an innovative approach for the development of a vaccine against antigenically variable viruses. Antibodyomics tools consist of different phases such as putative germline genes, error correction, and comparison of different bNAbs.91 The variable chains of the antibody have complementary-determining regions (CDR) where these bNAbs specifically bind. Additional information can be obtained to create a standard database through CDR3 analysis. This analysis helps to determine germline precursors and intermediate immunoglobulins from an NGS-derived repertoire. There are two analytical parameters on which it is based; the first is the identification of a sequence to a known bNAb (Y-axis), while the second one includes the analysis of the divergent sequences of putative germline genes (X-axis). The graph plotted between the two axes, and it was observed that closely linked somatic variants form ‘clusters’ which are different from the main sequence population.92 Furthermore, these variants are recognized with the intra-donor phylogenetic analysis and help in searching the sequence that has a similar evolutionary pattern as template bNAbs. It is utilized for the de novo recognition of VRC01-like broad neutralizing antibodies from HIV-1-infected donors.
For the development of epitope vaccines, computational tools like structure-based immunogen design and a wide range analysis of antibody can be used.93 Kwong et al., 201790 reported that neutralizing antibodies can be developed by understanding the information about genetics and immunological processes. Serum neutralization can also be performed to identify and quantify neutralizing antibodies.
5.4. Reverse vaccinology
Reverse vaccinology is a broad term applied to recognize potential vaccine candidates through analyzing the proteome of the pathogen with the help of computational tools. It is an essential technology for the mapping of epitopes and prediction of monovalent peptide vaccines to be used in the therapeutic processes.94 Reverse vaccinology is advantageous as it analyzes the complete genome of the pathogen and specifically chooses proteins that act as a potential antigen. Vaxign is a computational approach used to predict ideal vaccine candidates and develop distinct vaccines against proteins responsible for antibiotic resistance in the pathogens.95 Additionally, this technology is appropriate for the screening of the antigenic peptide in different pathogens like Neisseria meningitides, Group B Streptococcus (GBS), and Porphyromonas gingivalis.96 Functionally, NERVE program is an essential tool used for the development of reverse vaccinology. In this technology, open-reading frames (ORFs) play a significant role and screening of potential ORFs can be performed by the NERVE program.97 The computational analysis helps in the identification and localization of sub-cellular proteins having adhesion-like properties. HensBC can determine and assemble ORFs of different proteins (mitochondrial, cytoplasmic, nuclear, or extracellular) with 80% efficiency to make them a potential vaccine candidate.9
The identification of the pathogenic genome and complete screening of open reading frames (ORFs) is performed to select the specific sequence of the peptides responsible for an immunogenic response. Incorporation of in silico tools such as GLIMMER, ORF-FINDER, and GS-Finder will help scan the whole genome of the pathogenic strain. Following scanning, the identification of therapeutic proteins is carried out by various in silico tools such as ProDom, Pfam, and PROSITE. Initially, reverse vaccinology was applied for the development of Group B meningococcus vaccine by applying different software programs. Yee, 201998 reported that EV-A71 is a next-generation vaccine candidate against Enterovirus A71 which is responsible for causing foot and mouth diseases and severe neurological complications. Implementation of reverse genetics technology can be used to develop an rgEV-A71 strain to act as a potential vaccine candidate. It is difficult to control Ctenocephalides felis and disease risks associated with them, hence Contreras et al., 201899 suggested that the reverse vaccinology approach could be applied for the development of a novel vaccine against it. Moreover, this technique also helps in the identification of MHC Class-II-restricted epitopes from Leishmania donovani against visceral leishmaniasis.100 It can be successfully applied for the designing of novel vaccines against serogroup B Neisseria meningitidis, which is a cause of meningitis.101
Nosocomial infection is mainly caused by Pseudomonas aeruginosa in immuno-compromised patients, and recently three proteins, PSE17-1, PSE41-5, and PSE54, were identified by the application of reverse vaccinology as potential vaccine antigens. These are novel lectins of P. aeruginosa and contribute a significant role in infecting host cells.102 Naz et al., 2019103 made the computational pipeline termed “PanRV” that applied for both pangenome and reverse vaccinology approaches. It includes four functional modules- Pangenome Estimation Module (PGM), Reverse Vaccinology Module (RVM), Functional Annotation Module (FAM), and Antibiotic Resistance Association Module (ARM). Multi-epitope subunit vaccine can be designed by incorporating reverse vaccinology against avian influenza A (H7N9) as reported by Hasan et al., 2019.104 Reverse vaccinology has been successfully applied to categorize novel potential vaccine candidates against Acinetobacter baumannii, which is an evolving pathogen mainly found in intensive care units (ICU). Araujo et al., 201918 also discussed the success of omics and reversed vaccinology for the prediction of novel vaccine target in the whole genome of Corynebacterium pseudotuberculosis, which is an etiological agent of veterinary related diseases.
5.5. Codon optimization and other available software
The expression of proteins can be enhanced with a technology known as codon optimization, an approach in the field of immunoinformatics.105 Different codon optimization algorithms are available for the high production of protein. Codon Optimization OnLine (COOL, http://bioinfo.bti.a-star.edu.sg/COOL/),106 is a new tool that functions to synthesize genes. With the implementation of COOL, different codon optimization parameters including codon pairing, codon adaptation index, and specific codon usage, can be customized. An online application known as OPTIMIZER (http://genomes.urv.es/OPTIMIZER107) is built to optimize codon usage of a gene to increase its expression level.108 However, OPTIMIZER can optimize strongly expressed genes in more than 150 prokaryotic species during the process of translational, thus, it can predict highly expressed genes.
CpG is another technique applied for the optimization of a codon and concerns CG dinucleotides. Studies have shown that the immune response of DNA vaccines could be raised by CpG motif engineering.109 Narum et al., 2001110 reported the protein expression of DNA vaccines could be enhanced by the optimization of gene fragments that code for Plasmodium falciparum merozoite proteins, which ultimately raised immunity in mice. Stachyra et al., 2016111 also showed that codon optimization can be used to make potential DNA vaccines for avian influenza virus H5N1 in chickens and mice. Pattern recognition receptors (PRRs) are the components of the innate immune system used to differentiate DNA of prokaryotes from eukaryotes. They use ‘CpG dinucleotide motifs’ in base specific context and do not disturb codon optimization.
Further modification of the sequence of a protein, DNA and RNA can be performed to predict the codon usage via a combinatory algorithm. Remarkably the optimized codon is used in E. coli for the expression of recombinant TEV-protease.112 Gao et al., 2004113 explained the advanced application of GUI software applied to optimize and break the open reading frame (ORF) of giving DNA into triplets. CodonWizard is an also automatic software program for modification of codon optimization and freely available for scientists.114
5.6. Epitope prediction tools
The epitope is the determinant of the antibody attachment site on the part of antigen and is recognized through the host immune cells. An effective vaccine could be designed by the epitope prediction tools as the epitope stimulates immune reactions from both B cells and T cells.115 Different computational tools such as support vector machines (SVMs), motif-based systems, QSAR (quantitative structure-activity relationship analysis), structure-based, neural networks, and Hidden Markov models (HMMs) approaches are used to analyze the peptide interactions.116 Mapping of B cell and T cell epitopes is known as “epitope fishing”, which can screen the potential epitope in a pathogen. Epitope mapping in the genome of Mycobacterium tuberculosis was reported and analyzed with the help of predictive algorithms.117 Thousands of alleles are present on the A and B loci of HLAs. This group of alleles is termed as super type; they occasionally bind to the same set of peptides and contribute to vaccine design.118 Different potential supertypes could be discovered through the scoring matrix of the position specified by the alignment of MHC-I peptides. Support Vector Machines (SVMs) are applied to discriminate data into two distinct groups, based on the statistical theory: the binders and non-binders.119 However, the Hidden Markov model (HMM) helps find the sequences that have ‘binder-like’ qualities and also identify complicated peptide patterns through the implementation of a Bayesian neural network.
B-cell epitope prediction is performed by different methods such as hydrophilicity profile, flexibility profile, surface probability and HMM.120 Antigen processing and its selection are also important criteria in vaccine design. The development of a potential vaccine candidate with the help of bioinformatics tools are explained in Figure 3. These bioinformatics tools could be successfully implemented for the prediction of protein epitope domains targeted via human CD4+ T-cells.
Figure 3.
A review of applied computational methods for designing vaccines. Illustration showing various computational tools that help in the development of a vaccine. The antibodyomics tools modify the antibody to enhance binding to the antigen. Codon optimization tools such as Codon Optimization Online (COOL), OPTIMIZER, and Codon Wizard optimize the codon for enhanced gene expression. Epimatrix and TEPITOPE SYFPEITHI are scaffolding and epitope prediction tools. Modeling tools such as CHARMm, MacroModel, and MOIL are used for modeling the structure of the protein.
There are different tools available for B-cell epitope prediction.121 ElliPro is used to determine the presence of discontinuous and conformational epitopes. DiscoTope (http://www.cbs.dtu.dk/services/DiscoTope/)122 predicts discontinuous B-cell epitopes by analyzing 3D protein structures and has been used to predict the epitope in Alkhumra hemorrhagic fever virus (AHFV). DiscoTope can also be used for designing drugs and peptide-based vaccine, and in the development of diagnostic kits. The 3D-Epitope-Explorer (3DEX) software allows mapping of conformational epitopes using 3D structures of proteins based on an algorithm. Other prediction tools include CEP (conformational epitope prediction, http://bioinfo.ernet.in/cep.html), Hopp-Woods hydrophilicity, Kyte-Doolittle hydrophilicity, Eisenberg moment, Karplus-Schultz flexibilty, Emini surface probability, and the PROTEAN module of the LASERGENE software (DNASTAR, Inc, USA).121 Poorinmohmmad et al., 2014123 reported that Discotope could be successfully applying for the prediction of the conformational epitope in Alkhumra hemorrhagic fever virus (AHFV). Moreover, it can be utilized for designing of the drug, peptide-based vaccine, and development of the diagnostic kit. The 3D-Epitope-Explorer (3DEX) software allow mapping of conformational epitopes using 3D structures protein based on algorithm.124
The Artificial Neural Network (ANN) and Quantitative Matrices (QM) are the basis of nHLAPred, which is used for the prediction of MHC-I binding peptides. Whether 9-mer peptides would bind an MHC-I molecule or not will be predicted by the Kernel-based Inter-allele peptide binding prediction SyStem (KISS) in SVM. Different databases such as MHCBN, LANL, SYFPEITHI Parker hydrophilicity, BepiPred and Immune Epitope Database (IEDB; www.immuneepitope.org) are additional online tools for the prediction of B-cell epitopes125.
Different types of vaccines, along with their development strategies based on epitope prediction under pipeline, are discussed in Table 1. There has been no successful vaccine against Plasmodium vivax until now. With the help of epitope prediction, the potential epitope on AMA-1 was identified and developed as a highly effective vaccine candidate.148 Ren et al., 201919 developed a multi-epitope vaccine using bioinformatic tools and evaluated its immune response in mice. They observed a high production of IgG antibodies that protect against lethal doses of Acinetobacter baumannii. Trypanosomiasis is a tropical disease that is caused by the genus Trypanosoma and affects domestic animals and humans. Guedes et al., 2019149 used in silico tools to predict and characterize B-cell epitopes for South American and African T. vivax strain to be used in diagnostics.
Table 1.
Different types of vaccines and their developing strategies.
| S.No. | Vaccine type | Pathogen | Disease | Strategy | Reference |
|---|---|---|---|---|---|
| 1. | Multi-epitope based | Kaposi’s sarcoma-associated herpesvirus | Kaposi sarcoma | Immuno-informatics | Chauhan et al., 2019126 |
| 2. | Multi-epitope vaccine | Pseudomonas aeruginosa | Nosocomial infections | Comparative proteomics | Solanki et al., 2019127 |
| 3. | DNA vaccine | Ebola virus | Ebola virus disease | Computer design; gene expression; immunogenicity | Bazhan et al., 2019128 |
| 4. | Multi-epitope Peptide Vaccine | Neisseria gonorrhoeae | Gonorrhea | In-silico hierarchical approach | Jain et al., 2016129 |
| 5. | Subunit vaccine | Marburg virus | Hemorrhagic fever (MHF) | Reverse vaccinology | Hasan et al., 2019130 |
| 6. | Epitope-based | Nairovirus | Crimean-Congo hemorrhagic fever (CCHF) | Molecular docking and dynamics methods | Nosrati et al., 2019131 |
| 7. | Peptide-based vaccine | Providencia stuartii | Purple urine bag syndrome | Reverse Vaccinology (RV) | Asad et al., 2018132 |
| 8. | Multi-epitope vaccine | Human papilloma virus (HPV) | Warts and cancer | Structural vaccinology | Negahdaripour et al., 2018133 |
| 9. | Next generation vaccines | Echinococcus granulosus | Cystic echinococcosis | Systems vaccinology and mathematical/computational modeling | Pourseif et al., 2017134 |
| 10. | Peptide vaccine | Human papillomavirus (HPV) | Cervical cancer | Immunoinformatics and structural vaccinology approaches | Doorbar et al., 2015135 |
| 11. | Multi-epitope peptide vaccine | Brucella spp. | Brucellosis | Immuno-informatics | Saadi et al., 2017136 |
| 12. | Epitope-Based Peptide Vaccine | Mokola Rabies Virus | Meningo-encephalo-myelitis | In silico Approaches | Mohammed et al., 2017137 |
| 13. | T cells with Chimeric Antigen Receptor (CAR) | Acute lymphoblastic leukemia | Acute lymphoblastic leukemia | Genetic engineering | Dokmanović et al., 2017138 |
| 14. | Peptide vaccine | HPV | HPV-associated cancer | Conventional with Immuno-informatics | Atherton et al., 2018139 |
| 15. | Dendritic Cell Vaccines | Cancer antigens | Cancer | Immuno-informatics | Doytchinova et al., 2018140 |
| 16. | DNA Vaccines | Papillomavirus (HPV) | Human Cervical cancer and cervical intraepithelial neoplasia (CIN) | Genetic immuno-therapy, pharmaco-logical tool | Cordeiro et al., 2018141 |
| 17. | Epitopes based | Zika virus | Guillain–Barré syndrome | In silico-predicted immunogenic | Makhluf et al., 2018142 |
| 18. | Epitopes based | Vibrio anguillarum | Vibriosis | Reverse vaccinology | Baliga et al., 2018143 |
| 19. | Multi-epitope subunit vaccine | Chikungunya virus | Chikungunya | Immuno-informatics | Narula, et al., 2018144 |
| 20. | Sub-unit vaccine | Plasmodium falciparum and P. vivax | Malaria | Rational design | Draper et al., 2018145 |
| 21. | Epitope based vaccine | Dengue virus | Dengue | Immuno-informatics and Molecular Docking | Shen et al., 2018146 |
| 22. | Multi-Epitope based vaccine | Staphylococcus aureus | Skin infections and food poisoning | Immuno-informatics and in silico approach | Hajighahramani et al., 2017.147 |
T-cell epitopes are essential for designing the vaccines as they play a vital role in the cellular response. These epitopes can be identified through T cell receptors from various cells including B-cells, CTLs etc.150 Examples of T-cell epitope prediction tools are- BIMAS, IEDB, NetMHC, ProPred, TEPITPE, and CTLpred. Epijen is a freely available online software used for the prediction of T-cell epitopes and predicts epitopes based on quantitative matrices.151 This approach can be applied to develop vaccines against HIV and malaria. T-cell epitopes can be designed by the use of recombinant DNA technology and bioinformatics tools alongside the knowledge of the genetic background of the pathogen and host immune response.152 Recombinant DNA technologies make epitope-based vaccines more efficient, safe, and less expensive. Moutaftsi et al., 2006153 reported that various CD8 + T-cell epitopes can be predicted in a vaccinia virus WR strain. Glanville et al., 2017154 showed that the GLIPH algorithm accelerates the identification of T-cell epitopes by specifying T-cell receptor groups. Gutiérrez et al., 2016155 validated the prediction of T-cell epitopes in the swine influenza model. The function of PigMatrix and its ability to differentiate between immunogenic and non-immunogenic peptides were also validated.
6. Conclusion and future perspective
In summary, vaccine development can be considered as one of the significant factors for global public health. The traditional techniques have several drawbacks for vaccine design, but the implementation of computational tools will overcome these limitations. Immunoinformatics approaches are more beneficial, and thus the demand for modern technologies such as reverse vaccinology, epitope prediction, and structural vaccinology, including rational approaches, are more in demand to develop the potential vaccine candidates. Different tools applied for protein scaffolding, and epitope prediction contribute an essential role in vaccine design. This approach is advantageous as it is accomplished by analyzing the entire genome of the pathogen as well as by recognizing the proteins that act as a potential antigens. This allows flexible analysis that cannot be performed by traditional methods. The use of computational tools is beneficial for vaccine researchers, vaccine recipients as well as for public health policy-makers and epidemiologists. These tools may also be used to design vaccines for new, emerging diseases. The development of vaccines requires sound knowledge of immunology along with integration of the different areas, including cell biology, physical chemistry, and computational science. The combination of these disciplines will enhance the discovery of potential vaccine candidates.
Funding Statement
Authors acknowledge M.D. University, Rohtak, India, for providing infrastructural facilities. SV acknowledges the support as University Research Scholarship by M.D. University, Rohtak, India. The authors wish to acknowledge NIH fellows editorial board for the scientific editing of the manuscript. The authors also acknowledge the infrastructural support from Department of Science and Technology, New Delhi, Govt. of India, through FIST grant (Grant No. 1196 SR/FST/LS-I/ 2017/4).
Disclosure of potential conflicts of interest
The authors declare no known conflict of interest.
References
- 1.Sieber CC, Kiesswetter E, Kwetkat A, Heppner HJ, Schoene D, Freiberger E.. Prevention: public healthcare, nutrition, physical activity, vaccination. In: Roller-Wirnsberger R, Singler K, Polidori MC, editors. Learning geriatric medicine: a study guide for medical students. Cham, Switzerland: Springer International Publishing; 2018. p. 237–62. doi: 10.1007/978-3-319-61997-2_24. [DOI] [Google Scholar]
- 2.Wolf MC, Freiberg AN, Zhang T, Akyol-Ataman Z, Grock A, Hong PW, Li J, Watson NF, Fang AQ, Aguilar HC, et al. A broad-spectrum antiviral targeting entry of enveloped viruses. Proc Natl Acad Sci U S A. 2010;107:3157–62. doi: 10.1073/pnas.0909587107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kharkar PB, Talkar SS, Kadwadkar NA, Patravale VB. Nanosystems for oral delivery of immunomodulators. In: Andronescu E, Grumezescu AM, editors. Nanostructures for Oral Medicine. Elsevier, 2017;Chap 11:295-334. [Google Scholar]
- 4.Arora G, Misra R, Sajid A. Model systems for pulmonary infectious diseases: paradigms of anthrax and tuberculosis. Curr Top Med Chem. 2017;17:2077–99. doi: 10.2174/1568026617666170130111324. [DOI] [PubMed] [Google Scholar]
- 5.Thomas S, Dilbarova R, Rappuoli R. Future challenges for vaccinologists. Methods Mol Biol. 2016;1403:41–55. doi: 10.1007/978-1-4939-3387-7_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sinha S, Kuo CY, Ho JK, White PJ, Jazayeri JA, Pouton CW. A suicidal strain of listeria monocytogenes is effective as a DNA vaccine delivery system for oral administration. Vaccine. 2017;35:5115–22. doi: 10.1016/j.vaccine.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 7.Hotez PJ, Molyneux DH, Fenwick A, Ottesen E, Ehrlich Sachs S, Sachs JD. Incorporating a rapid-impact package for neglected tropical diseases with programs for HIV/AIDS, tuberculosis, and malaria. PLoS Med. 2006;3:e102. doi: 10.1371/journal.pmed.0030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rappuoli R, Aderem AA. vision for vaccines against HIV, tuberculosis and malaria. Nature. 2020;2011:463–69. [DOI] [PubMed] [Google Scholar]
- 9.Davies MN, Flower DR. Harnessing bioinformatics to discover new vaccines. Drug Discov Today. 2007;12:389–95. doi: 10.1016/j.drudis.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 10.Paladino A, Marchetti F, Rinaldi S, Colombo G. Protein design: from computer models to artificial intelligence. Wiley Interdiscip Rev: Comput Mol Sci. 2017;7:e1318. [Google Scholar]
- 11.Soria-Guerra RE, Nieto-Gomez R, Govea-Alonso DO, Rosales-Mendoza S. An overview of bioinformatics tools for epitope prediction: implications on vaccine development. J Biomed Inform. 2015;53:405–14. doi: 10.1016/j.jbi.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 12.Sherman AC, Mehta A, Dickert NW, Anderson EJ, Rouphael N. The future of flu: a review of the human challenge model and systems biology for advancement of influenza vaccinology. Front Cell Infect Microbiol. 2019;9:107. doi: 10.3389/fcimb.2019.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bramwell VW, Perrie Y. The rational design of vaccines. Drug Discov Today. 2005;10:1527–34. doi: 10.1016/S1359-6446(05)03600-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lua LH, Connors NK, Sainsbury F, Chuan YP, Wibowo N, Middelberg AP. Bioengineering virus-like particles as vaccines. Biotechnol Bioeng. 2014;111:425–40. doi: 10.1002/bit.25159. [DOI] [PubMed] [Google Scholar]
- 15.Ali M, Pandey RK, Khatoon N, Narula A, Mishra A, Prajapati VK. Exploring dengue genome to construct a multi-epitope based subunit vaccine by utilizing immunoinformatics approach to battle against dengue infection. Sci Rep. 2017;7:9232. doi: 10.1038/s41598-017-09199-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dalsass M, Brozzi A, Medini D, Rappuoli R. Comparison of open-source reverse vaccinology programs for bacterial vaccine antigen discovery. Front Immunol. 2019;10:113. doi: 10.3389/fimmu.2019.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pyclik M, Gorska S, Brzozowska E, Dobrut A, Ciekot J, Gamian A, Brzychczy-Włoch M.. Epitope mapping of streptococcus agalactiae elongation factor tu protein recognized by human sera. Front Microbiol. 2018;9:125. doi: 10.3389/fmicb.2018.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Araújo CL, Alves J, Nogueira W, Pereira LC, Gomide AC, Ramos R, Azevedo V, Silva A, Folador A. Prediction of new vaccine targets in the core genome of Corynebacterium pseudotuberculosis through omics approaches and reverse vaccinology. Gene. 2019;702:36–45. doi: 10.1016/j.gene.2019.03.049. [DOI] [PubMed] [Google Scholar]
- 19.Ren S, Guan L, Dong Y, Wang C, Feng L, Xie Y. Design and evaluation of a multi-epitope assembly peptide vaccine against acinetobacter baumannii infection in mice. Swiss Med Wkly. 2019;149:w20052. [DOI] [PubMed] [Google Scholar]
- 20.He L, Lin X, de Val N, Saye-Francisco KL, Mann CJ, Augst R, Morris CD, Azadnia P, Zhou B, Sok D, et al. Hidden lineage complexity of glycan-dependent HIV-1 broadly neutralizing antibodies uncovered by digital panning and native-like gp140 trimer. Front Immunol. 2017;8:1025. doi: 10.3389/fimmu.2017.01025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moore PL. The neutralizing antibody response to the HIV-1 env protein. Curr HIV Res. 2018;16:21–28. doi: 10.2174/1570162X15666171124122044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ekiert DC, Wilson IA. Broadly neutralizing antibodies against influenza virus and prospects for universal therapies. Curr Opin Virol. 2012;2:134–41. doi: 10.1016/j.coviro.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skolnick J, Fetrow JS. From genes to protein structure and function: novel applications of computational approaches in the genomic era. Trends Biotechnol. 2000;18:34–39. [DOI] [PubMed] [Google Scholar]
- 24.Shoenfeld Y, Aron-Maor A. Vaccination and autoimmunity-’vaccinosis’: a dangerous liaison? J Autoimmun. 2000;14:1–10. doi: 10.1006/jaut.1999.0346. [DOI] [PubMed] [Google Scholar]
- 25.Zhou J, Miller JH. Microbial genomics–challenges and opportunities: the 9th international conference on microbial genomes. J Bacteriol. 2002;184:4327–33. doi: 10.1128/jb.184.16.4327-4333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gehlenborg N, O’Donoghue SI, Baliga NS, Goesmann A, Hibbs MA, Kitano H, Kohlbacher O, Neuweger H, Schneider R, Tenenbaum D, et al. Visualization of omics data for systems biology. Nat Methods. 2010;7:S56–68. doi: 10.1038/nmeth.1436. [DOI] [PubMed] [Google Scholar]
- 27.Homuth G, Teumer A, Volker U, Nauck M. A description of large-scale metabolomics studies: increasing value by combining metabolomics with genome-wide SNP genotyping and transcriptional profiling. J Endocrinol. 2012;215:17–28. doi: 10.1530/JOE-12-0144. [DOI] [PubMed] [Google Scholar]
- 28.Bambini S, Rappuoli R. The use of genomics in microbial vaccine development. Drug Discov Today. 2009;14:252–60. doi: 10.1016/j.drudis.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Groot AS. Immunomics: discovering new targets for vaccines and therapeutics. Drug Discov Today. 2006;11:203–09. doi: 10.1016/S1359-6446(05)03720-7. [DOI] [PubMed] [Google Scholar]
- 30.Nelma PG, Massimo L. Innovation in metabonomics to improve personalized health care. handbook of biomarkers and precision medicine. CRC Press; 2019. [Google Scholar]
- 31.Groves AM, Williams JP, Hernady E, Reed C, Fenton B, Love T, Finkelstein JN, Johnston CJ. A potential biomarker for predicting the risk of radiation-induced fibrosis in the lung. Radiat Res. 2018;190:513–25. doi: 10.1667/RR15122.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muhammad SA, Fatima N, Paracha RZ, Ali A, Chen JY. A systematic simulation-based meta-analytical framework for prediction of physiological biomarkers in alopecia. J Biol Res (Thessalon). 2019;26:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oh SJ, Choi YK, Shin OS. Systems biology-based platforms to accelerate research of emerging infectious diseases. Yonsei Med J. 2018;59:176–86. doi: 10.3349/ymj.2018.59.2.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Graham BS, Gilman MSA, McLellan JS. Structure-based vaccine antigen design. Annu Rev Med. 2019;70:91–104. doi: 10.1146/annurev-med-121217-094234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lemaire D, Barbosa T, Rihet P. Coping with genetic diversity: the contribution of pathogen and human genomics to modern vaccinology. Braz J Med Biol Res. 2012;45:376–85. doi: 10.1590/s0100-879x2011007500142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaufmann JK, Flechtner JB. Evolution of rational vaccine designs for genital herpes immunotherapy. Curr Opin Virol. 2016;17:80–86. doi: 10.1016/j.coviro.2016.01.021. [DOI] [PubMed] [Google Scholar]
- 37.Bajic G, van der Poel CE, Kuraoka M, Schmidt AG, Carroll MC, Kelsoe G, Harrison SC. Autoreactivity profiles of influenza hemagglutinin broadly neutralizing antibodies. Sci Rep. 2019;9:3492. doi: 10.1038/s41598-019-40175-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trobaugh DW, Sun C, Dunn MD, Reed DS, Klimstra WB, Morrison TE. Rational design of a live-attenuated eastern equine encephalitis virus vaccine through informed mutation of virulence determinants. PLoS Pathog. 2019;15:e1007584. doi: 10.1371/journal.ppat.1007584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tom JK, Albin TJ, Manna S, Moser BA, Steinhardt RC, Esser-Kahn AP. Applications of immunomodulatory immune synergies to adjuvant discovery and vaccine development. Trends Biotechnol. 2019;37:373–88. doi: 10.1016/j.tibtech.2018.10.004. [DOI] [PubMed] [Google Scholar]
- 40.Harandi AM, Medaglini D, Shattock RJ. Working group convened by E. Vaccine Adjuvants: a Priority for Vaccine Research. Vaccine. 2010;28:2363–66. doi: 10.1016/j.vaccine.2009.12.084. [DOI] [PubMed] [Google Scholar]
- 41.HogenEsch H, O’Hagan DT, Fox CB. Optimizing the utilization of aluminum adjuvants in vaccines: you might just get what you want. NPJ Vaccines. 2018;3:51. doi: 10.1038/s41541-018-0089-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hogenesch H. Mechanism of immunopotentiation and safety of aluminum adjuvants. Front Immunol. 2012;3:406. doi: 10.3389/fimmu.2012.00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Calabro S, Tritto E, Pezzotti A, Taccone M, Muzzi A, Bertholet S, De Gregorio E, O’Hagan DT, Baudner B, Seubert A. The adjuvant effect of MF59 is due to the oil-in-water emulsion formulation, none of the individual components induce a comparable adjuvant effect. Vaccine. 2013;31:3363–69. doi: 10.1016/j.vaccine.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 44.Mbow ML, De Gregorio E, Valiante NM, Rappuoli R. New adjuvants for human vaccines. Curr Opin Immunol. 2010;22:411–16. doi: 10.1016/j.coi.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 45.Kanzler H, Barrat FJ, Hessel EM, Coffman RL. Therapeutic targeting of innate immunity with toll-like receptor agonists and antagonists. Nat Med. 2007;13:552–59. doi: 10.1038/nm1589. [DOI] [PubMed] [Google Scholar]
- 46.Saeed AM, Duffort S, Ivanov D, Wang H, Laird JM, Salomon RG, Cruz-Guilloty F, Perez VL, Vavvas D. The oxidative stress product carboxyethylpyrrole potentiates TLR2/TLR1 inflammatory signaling in macrophages. PLoS One. 2014;9:e106421. doi: 10.1371/journal.pone.0106421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aucouturier J, Dupuis L, Deville S, Ascarateil S, Ganne V. Montanide ISA 720 and 51: a new generation of water in oil emulsions as adjuvants for human vaccines. Expert Rev Vaccines. 2002;1:111–18. doi: 10.1586/14760584.1.1.111. [DOI] [PubMed] [Google Scholar]
- 48.Nordly P, Madsen HB, Nielsen HM, Foged C. Status and future prospects of lipid-based particulate delivery systems as vaccine adjuvants and their combination with immunostimulators. Expert Opin Drug Deliv. 2009;6:657–72. doi: 10.1517/17425240903018863. [DOI] [PubMed] [Google Scholar]
- 49.Burt DS, Lowell GH, White GL, Jones D, Rioux C. U.S. Patent No. 7,524,509. Washington, DC: US Patent and Trademark Office; 2009. [Google Scholar]
- 50.Sun H, Zheng Q. Haemolytic activities and adjuvant effect of gynostemma pentaphyllum saponins on the immune responses to ovalbumin in mice. Phytother Res. 2005;19:895–900. doi: 10.1002/ptr.1754. [DOI] [PubMed] [Google Scholar]
- 51.He Y, Xiang Z. Databases and in silico tools for vaccine design. Methods Mol Biol. 2013;993:115–27. doi: 10.1007/978-1-62703-342-8_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He Y. Vaccine adjuvant informatics: from data integration and analysis to rational vaccine adjuvant design. Front Immunol. 2014;5:32. doi: 10.3389/fimmu.2014.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wallis J, Shenton DP, Carlisle RC. Novel approaches for the design, delivery and administration of vaccine technologies. Clin Exp Immunol. 2019;196:189–204. doi: 10.1111/cei.13287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Du L, Tai W, Yang Y, Zhao G, Zhu Q, Sun S, Liu C, Tao X, Tseng C-TK, Perlman S, et al. Introduction of neutralizing immunogenicity index to the rational design of MERS coronavirus subunit vaccines. Nat Commun. 2016;7:13473. doi: 10.1038/ncomms13473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T. Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc. 2009;4:1–13. doi: 10.1038/nprot.2008.197. [DOI] [PubMed] [Google Scholar]
- 56.Scheid J, Nussenzweig M, Bjorkman P, Diskin R. Human immunodeficiency virus neutralizing antibodies and methods of use thereof. Patent number: US20140328862A1. 2014.
- 57.Bazmara H, Rasooli I, Jahangiri A, Sefid F, Astaneh S, Payandeh Z. Antigenic properties of iron regulated proteins in acinetobacter baumannii: an in silico approach. Int J Pept Res Ther. 2019;25:205–13. doi: 10.1007/s10989-017-9665-6. [DOI] [Google Scholar]
- 58.Li H, Li Y, Wang X, Hou Y, Hong X, Gong T, Zhang Z, Sun X. Rational design of polymeric hybrid micelles to overcome lymphatic and intracellular delivery barriers in cancer immunotherapy. Theranostics. 2017;7:4383–98. doi: 10.7150/thno.20745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Honegr J, Dolezal R, Malinak D, Benkova M, Soukup O, Almeida J, Franca T, Kuca K, Prymula R.. Rational design of a new class of toll-like receptor 4 (TLR4) tryptamine related agonists by means of the structure- and ligand-based virtual screening for vaccine adjuvant discovery. Molecules. 2018;23:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hayman CM, Hermans IF, Painter GF. Increased efficacy of NKT cell-adjuvanted peptide vaccines through chemical conjugation. In: Witczak ZJ, Bielski R, editors. Coupling and decoupling of diverse molecular units in glycosciences. Cham, Switzerland: Springer International Publishing; 2018. p. 309–35. doi: 10.1007/978-3-319-65587-1_14. [DOI] [Google Scholar]
- 61.Wills-Karp M. Interleukin-13 in asthma pathogenesis. Immunol Rev. 2004;202:175–90. doi: 10.1111/j.0105-2896.2004.00215.x. [DOI] [PubMed] [Google Scholar]
- 62.Johnson C, Hargest V, Cortez V, Meliopoulos VA, Schultz-Cherry S. Astrovirus pathogenesis. Viruses. 2017:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martinez-Arzate SG, Tenorio-Borroto E, Barbabosa Pliego A, Diaz-Albiter HM, Vazquez-Chagoyan JC, Gonzalez-Diaz H. PTML model for proteome mining of B-cell epitopes and theoretical-experimental study of Bm86 protein sequences from Colima, Mexico. J Proteome Res. 2017;16:4093–103. doi: 10.1021/acs.jproteome.7b00477. [DOI] [PubMed] [Google Scholar]
- 64.Alansari Z, Anuar NB, Kamsin A, Soomro S, Belgaum MR. Computational intelligence tools and databases in bioinformatics. 4th IEEE International Conference on Engineering Technologies and Applied Sciences (ICETAS), AMA International University, Bahrain; 2017. p. 1–6. [Google Scholar]
- 65.O’Brien SP, Swanstrom AE, Pegu A, Ko S-Y, Immonen TT, Del Prete GQ, Fennessey CM, Gorman J, Foulds KE, Schmidt SD, et al. Rational design and in vivo selection of SHIVs encoding transmitted/founder subtype C HIV-1 envelopes. PLoS Pathog. 2019;15:e1007632. doi: 10.1371/journal.ppat.1007632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kailasan S, Kort T, Mukherjee I, Liao GC, Kanipakala T, Williston N, Ganjbaksh N, Venkatasubramaniam A, Holtsberg FW, Karauzum H, Adhikari RP, et al. Rational design of toxoid vaccine candidates for staphylococcus aureus leukocidin AB (LukAB). Toxins (Basel). 2019:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tai W, Chen J, Zhao G, Geng Q, He L, Chen Y, Zhou Y, Li F, Du L, Heise MT. Rational design of zika virus subunit vaccine with enhanced efficacy. J Virol. 2019;93. doi: 10.1128/JVI.02187-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Orozco A, Morera J, Jimenez S, Boza R. A review of bioinformatics training applied to research in molecular medicine, agriculture and biodiversity in Costa Rica and Central America. Brief Bioinform. 2013;14:661–70. doi: 10.1093/bib/bbt033. [DOI] [PubMed] [Google Scholar]
- 69.Dubey KK, Luke GA, Knox C, Kumar P, Pletschke BI, Singh PK, Shukla P. Vaccine and antibody production in plants: developments and computational tools. Brief Funct Genomics. 2018;17:295–307. doi: 10.1093/bfgp/ely020. [DOI] [PubMed] [Google Scholar]
- 70.Goldenzweig A, Fleishman SJ. Principles of protein stability and their application in computational design. Annu Rev Biochem. 2018;87:105–29. doi: 10.1146/annurev-biochem-062917-012102. [DOI] [PubMed] [Google Scholar]
- 71.He L, Zhu J. Computational tools for epitope vaccine design and evaluation. Curr Opin Virol. 2015;11:103–12. doi: 10.1016/j.coviro.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kwong PD, Ofek G, Guenaga J, Wyatt R, Yang Z, Zhou T, Nabel GJ, Tang M, Schief W, Baker D, Epitope-transplant scaffolds and their use. United States Patent Application Publication US 2010/0068217A1. 2010.
- 73.Leem J, Georges G, Shi J, Deane CM. Antibody side chain conformations are position-dependent. Proteins. 2018;86:383–92. doi: 10.1002/prot.25453. [DOI] [PubMed] [Google Scholar]
- 74.Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Taylor WR, Munro REJ, Petersen K, Bywater RP. Ab initio modelling of the N-terminal domain of the secretin receptors. Comput Biol Chem. 2003;27:103–14. [DOI] [PubMed] [Google Scholar]
- 76.Fiser A. Comparative protein structure modelling. In: Daniel J. Rigden, editor, From protein structure to function with bioinformatics. Dordrecht, Netherlands: Springer; 2017. p. 57–90. [Google Scholar]
- 77.Li M-H, Zong H, Leroueil PR, Choi SK, Baker JR Jr.. Ligand characteristics important to avidity interactions of multivalent nanoparticles. Bioconjug Chem. 2017;28:1649–57. doi: 10.1021/acs.bioconjchem.7b00098. [DOI] [PubMed] [Google Scholar]
- 78.Walensky LD, Bird G. Structured viral peptide compositions and methods of use. MA, USA: Dana Farber Cancer Institute, inc. Pub No WO/2010/148335; 2010. [Google Scholar]
- 79.Ullah H, Hou W, Dakshanamurthy S, Tang Q. Host targeted antiviral (HTA): functional inhibitor compounds of scaffold protein RACK1 inhibit herpes simplex virus proliferation. Oncotarget. 2019;10:3209–26. doi: 10.18632/oncotarget.26907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Barwal I, Kumar R, Kateriya S, Dinda AK, Yadav SC. Targeted delivery system for cancer cells consist of multiple ligands conjugated genetically modified CCMV capsid on doxorubicin GNPs complex. Sci Rep. 2016;6:37096. doi: 10.1038/srep37096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hill BD, Zak A, Khera E, Wen F. Engineering virus-like particles for antigen and drug delivery. Curr Protein Pept Sci. 2018;19:112–27. doi: 10.2174/1389203718666161122113041. [DOI] [PubMed] [Google Scholar]
- 82.Rynda-Apple A, Patterson DP, Douglas T. Virus-like particles as antigenic nanomaterials for inducing protective immune responses in the lung. Nanomedicine (Lond). 2014;9:1857–68. doi: 10.2217/nnm.14.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cai C, Lin J, Lu Y, Zhang Q, Wang L. Polypeptide self-assemblies: nanostructures and bioapplications. Chem Soc Rev. 2016;45:5985–6012. doi: 10.1039/c6cs00013d. [DOI] [PubMed] [Google Scholar]
- 84.Küry S, Besnard T, Ebstein F, Khan TN, Gambin T, Douglas J, Bacino CA, Craigen WJ, Sanders SJ, Lehmann A, et al. De novo disruption of the proteasome regulatory subunit PSMD12 causes a syndromic neurodevelopmental disorder. Am J Hum Genet. 2017;100:352–63. doi: 10.1016/j.ajhg.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mouratou B, Behar G, Pecorari F. Artificial affinity proteins as ligands of immunoglobulins. Biomolecules. 2015;5:60–75. doi: 10.3390/biom5010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Greenspan NS, Cavacini LA. 15 - immunoglobulin function. In: Rich RR, Fleisher TA, Shearer WT, Schroeder HW, Frew AJ, Weyand CM, editors. Clinical immunology. 5th ed. London; 2019. p. 223–33.e1. [Google Scholar]
- 87.Azoitei ML, Ban YA, Kalyuzhny O, Guenaga J, Schroeter A, Porter J, Wyatt R, Schief WR. Computational design of protein antigens that interact with the CDR H3 loop of HIV broadly neutralizing antibody 2F5. Proteins. 2014;82:2770–82. doi: 10.1002/prot.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gourlay L, Peri C, Bolognesi M, Colombo G. Structure and computation in immunoreagent design: from diagnostics to vaccines. Trends Biotechnol. 2017;35:1208–20. doi: 10.1016/j.tibtech.2017.06.018. [DOI] [PubMed] [Google Scholar]
- 89.Mishra S, Manish M. Studies on computational grafting of malarial epitopes in serum albumin. Comput Biol Med. 2018;102:126–31. doi: 10.1016/j.compbiomed.2018.09.018. [DOI] [PubMed] [Google Scholar]
- 90.Kwong PD, Chuang G-Y, DeKosky BJ, Gindin T, Georgiev IS, Lemmin T, Schramm CA, Sheng Z, Soto C, Yang A-S, et al. Antibodyomics: bioinformatics technologies for understanding B-cell immunity to HIV-1. Immunol Rev. 2017;275:108–28. doi: 10.1111/imr.12480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Scheepers C. Host factors and broadly neutralizing antibodies in South African women infected with HIV-1 subtype C [Doctoral Thesis]. 2016. [Google Scholar]
- 92.Telenti A, Lippert C, Chang PC, DePristo M. Deep learning of genomic variation and regulatory network data. Hum Mol Genet. 2018;27:R63–R71. doi: 10.1093/hmg/ddy115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Margarit I, Rappuoli R. Microbial genomics: targeted antimicrobial therapy and genome vaccines. Genomic medicine: principles and practice. 2nd ed. Oxford, UK: Oxford University Press; 2015. [Google Scholar]
- 94.Kaushik DK, Sehgal D. Developing antibacterial vaccines in genomics and proteomics era. Scand J Immunol. 2008;67:544–52. doi: 10.1111/j.1365-3083.2008.02107.x. [DOI] [PubMed] [Google Scholar]
- 95.Xiang Z, He Y. Vaxign: a web-based vaccine target design program for reverse vaccinology. Procedia Vaccinol. 2009;1:23–29. doi: 10.1016/j.provac.2009.07.005. [DOI] [Google Scholar]
- 96.He Y, Xiang Z, Mobley HL. Vaxign: the first web-based vaccine design program for reverse vaccinology and applications for vaccine development. J Biomed Biotechnol. 2010;2010:297505. doi: 10.1155/2010/297505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Heinson AI, Woelk CH, Newell ML. The promise of reverse vaccinology. Int Health. 2015;7:85–89. doi: 10.1093/inthealth/ihv002. [DOI] [PubMed] [Google Scholar]
- 98.Yee PTI, Tan SH, Ong KC, Tan KO, Wong KT, Hassan SS, Poh CL. Development of live attenuated enterovirus 71 vaccine strains that confer protection against lethal challenge in mice. Sci Rep. 2019;9:4805. doi: 10.1038/s41598-019-41285-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Contreras M, Villar M, Artigas-Jeronimo S, Kornieieva L, Mуtrofanov S, de la Fuente J. A reverse vaccinology approach to the identification and characterization of ctenocephalides felis candidate protective antigens for the control of cat flea infestations. Parasit Vectors. 2018;11:43. doi: 10.1186/s13071-018-2618-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dikhit MR, Kumar A, Das S, Dehury B, Rout AK, Jamal F, Sahoo GC, Topno RK, Pandey K, Das VNR, et al. Identification of potential MHC class-II-restricted epitopes derived from leishmania donovani antigens by reverse vaccinology and evaluation of their CD4+ T-cell responsiveness against visceral leishmaniasis. Front Immunol. 2017;8:1763. doi: 10.3389/fimmu.2017.01763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Del Tordello E, Rappuoli R, Delany I. Chapter 3 - reverse vaccinology: exploiting genomes for vaccine design. In: Modjarrad K, Koff WC, editors. Human Vaccines. USA: Academic Press; 2017. p. 65–86. [Google Scholar]
- 102.Day CJ, Hartley-Tassell LE, Seib KL, Tiralongo J, Bovin N, Savino S, Masignani V, Jennings MP. Lectin activity of pseudomonas aeruginosa vaccine candidates PSE17-1, PSE41-5 and PSE54. Biochem Biophys Res Commun. 2019;513:287–90. doi: 10.1016/j.bbrc.2019.03.092. [DOI] [PubMed] [Google Scholar]
- 103.Naz K, Naz A, Ashraf ST, Rizwan M, Ahmad J, Baumbach J, Ali A. PanRV: pangenome-reverse vaccinology approach for identifications of potential vaccine candidates in microbial pangenome. BMC Bioinformatics. 2019;20:123. doi: 10.1186/s12859-019-2713-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hasan M, Ghosh PP, Azim KF, Mukta S, Abir RA, Nahar J, Hasan Khan MM. Reverse vaccinology approach to design a novel multi-epitope subunit vaccine against avian influenza A (H7N9) virus. Microb Pathog. 2019;130:19–37. doi: 10.1016/j.micpath.2019.02.023. [DOI] [PubMed] [Google Scholar]
- 105.Chin JX, Chung BK-S, Lee D-Y. Codon Optimization OnLine (COOL): a web-based multi-objective optimization platform for synthetic gene design. Bioinformatics. 2014;30:2210–12. doi: 10.1093/bioinformatics/btu192. [DOI] [PubMed] [Google Scholar]
- 106.Magistrelli G, Poitevin Y, Schlosser F, Pontini G, Malinge P, Josserand S, Corbier M, Fischer N. Optimizing assembly and production of native bispecific antibodies by codon de-optimization. MAbs. 2017;9:231–39. doi: 10.1080/19420862.2016.1267088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Puigbo P, Guzman E, Romeu A, Garcia-Vallve S. OPTIMIZER: a web server for optimizing the codon usage of DNA sequences. Nucleic Acids Res. 2007;35:W126–31. doi: 10.1093/nar/gkm219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Trösemeier J-H, Rudorf S, Loessner H, Hofner B, Reuter A, Schulenborg T, Koch I, Bekeredjian-Ding I, Lipowsky R, Kamp C. Optimizing the dynamics of protein expression. Sci Rep. 2019;9:7511. doi: 10.1038/s41598-019-43857-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Harish N, Gupta R, Agarwal P, Scaria V, Pillai B. DyNAVacS: an integrative tool for optimized DNA vaccine design. Nucleic Acids Res. 2006;34:W264–6. doi: 10.1093/nar/gkl242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Narum DL, Kumar S, Rogers WO, Fuhrmann SR, Liang H, Oakley M, Taye A, Sim BK, Hoffman SL. Codon optimization of gene fragments encoding plasmodium falciparum merzoite proteins enhances DNA vaccine protein expression and immunogenicity in mice. Infect Immun. 2001;69:7250–53. doi: 10.1128/IAI.69.12.7250-7253.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Stachyra A, Redkiewicz P, Kosson P, Protasiuk A, Gora-Sochacka A, Kudla G, Sirko A.. Codon optimization of antigen coding sequences improves the immune potential of DNA vaccines against avian influenza virus H5N1 in mice and chickens. Virol J. 2016;13:143. doi: 10.1186/s12985-016-0512-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Liu B, Kong Q, Zhang D, Yan L. Codon optimization significantly enhanced the expression of human 37-kDa iLRP in Escherichia coli. 3 Biotech. 2018;8:210. doi: 10.1007/s13205-018-1234-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gao W, Rzewski A, Sun H, Robbins PD, Gambotto A. UpGene: application of a web-based DNA codon optimization algorithm. Biotechnol Prog. 2004;20:443–48. doi: 10.1021/bp0300467. [DOI] [PubMed] [Google Scholar]
- 114.Rehbein P, Berz J, Kreisel P, Schwalbe H. “CodonWizard” - an intuitive software tool with graphical user interface for customizable codon optimization in protein expression efforts. Protein Expr Purif. 2019;160:84–93. doi: 10.1016/j.pep.2019.03.018. [DOI] [PubMed] [Google Scholar]
- 115.Kreiter S, Vormehr M, van de Roemer N, Diken M, Löwer M, Diekmann J, Boegel S, Schrörs B, Vascotto F, Castle JC, et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature. 2015;520:692–96. doi: 10.1038/nature14426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kar P, Ruiz-Perez L, Arooj M, Mancera RL. Current methods for the prediction of T-cell epitopes. Pept Sci. 2018;110:e24046. doi: 10.1093/jnci/djx216. [DOI] [Google Scholar]
- 117.Maji A, Misra R, Kumar Mondal A, Kumar D, Bajaj D, Singhal A, Arora G, Bhaduri A, Sajid A, Bhatia S, et al. Expression profiling of lymph nodes in tuberculosis patients reveal inflammatory milieu at site of infection. Sci Rep. 2015;5:15214. doi: 10.1038/srep15214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Purcell AW, McCluskey J, Rossjohn J. More than one reason to rethink the use of peptides in vaccine design. Nat Rev Drug Discov. 2007;6:404–14. doi: 10.1038/nrd2224. [DOI] [PubMed] [Google Scholar]
- 119.Vivona S, Gardy JL, Ramachandran S, Brinkman FSL, Raghava GP, Flower DR, Filippini F. Computer-aided biotechnology: from immuno-informatics to reverse vaccinology. Trends Biotechnol. 2008;26:190–200. doi: 10.1016/j.tibtech.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 120.Zaman R, Chowdhury SY, Rashid MA, Sharma A, Dehzangi A, Shatabda S. HMMBinder: DNA-binding protein prediction using HMM profile based features. Biomed Res Int. 2017;2017:4590609. doi: 10.1155/2017/4590609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ganguly B, Rastogi SK, Prasad S. Computational designing of a poly-epitope fecundity vaccine for multiple species of livestock. Vaccine. 2013;32:11–18. doi: 10.1016/j.vaccine.2013.10.086. [DOI] [PubMed] [Google Scholar]
- 122.Greenbaum JA, Andersen PH, Blythe M, Bui -H-H, Cachau RE, Crowe J, Davies M, Kolaskar AS, Lund O, Morrison S, et al. Towards a consensus on datasets and evaluation metrics for developing B-cell epitope prediction tools. J Mol Recognit. 2007;20:75–82. doi: 10.1002/jmr.815. [DOI] [PubMed] [Google Scholar]
- 123.Poorinmohammad N, Mohabatkar H. Homology modeling and conformational epitope prediction of envelope protein of alkhumra haemorrhagic fever virus. J Arthropod Borne Dis. 2015;9:116–24. [PMC free article] [PubMed] [Google Scholar]
- 124.Sun P, Guo S, Sun J, Tan L, Lu C, Ma Z. Advances in in-silico B-cell epitope prediction. Curr Top Med Chem. 2019;19:105–15. doi: 10.2174/1568026619666181130111827. [DOI] [PubMed] [Google Scholar]
- 125.Rahman MS, Rahman MK, Saha S, Kaykobad M, Rahman MS. Antigenic: an improved prediction model of protective antigens. Artif Intell Med. 2019;94:28–41. doi: 10.1016/j.artmed.2018.12.010. [DOI] [PubMed] [Google Scholar]
- 126.Chauhan V, Rungta T, Goyal K, Singh MP. Designing a multi-epitope based vaccine to combat kaposi sarcoma utilizing immunoinformatics approach. Sci Rep. 2019;9:2517. doi: 10.1038/s41598-019-39299-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Solanki V, Tiwari M, Tiwari V. Prioritization of potential vaccine targets using comparative proteomics and designing of the chimeric multi-epitope vaccine against pseudomonas aeruginosa. Sci Rep. 2019;9:5240. doi: 10.1038/s41598-019-41496-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bazhan SI, Antonets DV, Karpenko LI, Oreshkova SF, Kaplina ON, Starostina EV, Dudko SG, Fedotova SA, Ilyichev AA.. In silico designed ebola virus T-cell multi-epitope DNA vaccine constructions are immunogenic in mice. Vaccines (Basel). 2019:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jain R, Sonkar SC, Chaudhry U, Bala M, Saluja D. In-silico hierarchical approach for the identification of Potential Universal Vaccine Candidates (PUVCs) from neisseria gonorrhoeae. J Theor Biol. 2016;410:36–43. doi: 10.1016/j.jtbi.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 130.Hasan M, Azim KF, Begum A, Khan NA, Shammi TS, Imran AS, Chowdhury IM, Urme SRA. Vaccinomics strategy for developing a unique multi-epitope monovalent vaccine against Marburg marburgvirus. Infect Genet Evol. 2019;70:140–57. doi: 10.1016/j.meegid.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 131.Nosrati M, Behbahani M, Mohabatkar H. Towards the first multi-epitope recombinant vaccine against Crimean-Congo hemorrhagic fever virus: A computer-aided vaccine design approach. J Biomed Inform. 2019;93:103160. doi: 10.1016/j.jbi.2019.103160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Asad Y, Ahmad S, Rungrotmongkol T, Ranaghan KE, Azam SS. Immuno-informatics driven proteome-wide investigation revealed novel peptide-based vaccine targets against emerging multiple drug resistant providencia stuartii. J Mol Graph Model. 2018;80:238–50. doi: 10.1016/j.jmgm.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 133.Negahdaripour M, Nezafat N, Eslami M, Ghoshoon MB, Shoolian E, Najafipour S, Morowvat MH, Dehshahri A, Erfani N, Ghasemi Y. Structural vaccinology considerations for in silico designing of a multi-epitope vaccine. Infect Genet Evol. 2018;58:96–109. doi: 10.1016/j.meegid.2017.12.008. [DOI] [PubMed] [Google Scholar]
- 134.Pourseif MM, Moghaddam G, Saeedi N, Barzegari A, Dehghani J, Omidi Y. Current status and future prospective of vaccine development against echinococcus granulosus. Biologicals. 2018;51:1–11. doi: 10.1016/j.biologicals.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 135.Doorbar J, Egawa N, Griffin H, Kranjec C, Murakami I. Human papillomavirus molecular biology and disease association. Rev Med Virol. 2015;25(Suppl 1):2–23. doi: 10.1002/rmv.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Saadi M, Karkhah A, Nouri HR. Development of a multi-epitope peptide vaccine inducing robust T cell responses against brucellosis using immunoinformatics based approaches. Infect Genet Evol. 2017;51:227–34. doi: 10.1016/j.meegid.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 137.Mohammed AA, Hashim O, Elrahman KAA, Hamdi A, Hassan MA. Epitope-based peptide vaccine design against mokola rabies virus glycoprotein G utilizing in silico approaches. Immunome Res. 2017;13. [Google Scholar]
- 138.Dokmanović L, Milošević G, Perić J, Tošić N, Krstovski N, Janić D, Pavlovic S.. Next generation sequencing as a tool for pharmacogenomic profiling – nine novel potential genetic markers for targeted therapy in childhood acute lymphoblastic leukemia. Srp Arh Celok Lek. 2017;146:407–11. doi: 10.2298/SARH171003194D. [DOI] [Google Scholar]
- 139.Atherton MJ, Stephenson KB, Nikota JK, Hu QN, Nguyen A, Wan Y, Lichty BD. Preclinical development of peptide vaccination combined with oncolytic MG1-E6E7 for HPV-associated cancer. Vaccine. 2018;36:2181–92. doi: 10.1016/j.vaccine.2018.02.070. [DOI] [PubMed] [Google Scholar]
- 140.Doytchinova IA, Flower DR. In silico prediction of cancer immunogens: current state of the art. BMC Immunol. 2018;19:11. doi: 10.1186/s12865-018-0248-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cordeiro MN, De Lima RCP, Paolini F, Melo A, Campos APF, Venuti A, De Freitas AC.. Current research into novel therapeutic vaccines against cervical cancer. Expert Rev Anticancer Ther. 2018;18:365–76. doi: 10.1080/14737140.2018.1445527. [DOI] [PubMed] [Google Scholar]
- 142.Makhluf H, Shresta S. Development of zika virus vaccines. Vaccines (Basel). 2018:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Baliga P, Shekar M, Venugopal MN. Potential outer membrane protein candidates for vaccine development against the pathogen vibrio anguillarum: A reverse vaccinology based identification. Curr Microbiol. 2018;75:368–77. doi: 10.1007/s00284-017-1390-z. [DOI] [PubMed] [Google Scholar]
- 144.Narula A, Pandey RK, Khatoon N, Mishra A, Prajapati VK. Excavating chikungunya genome to design B and T cell multi-epitope subunit vaccine using comprehensive immunoinformatics approach to control chikungunya infection. Infect Genet Evol. 2018;61:4–15. doi: 10.1016/j.meegid.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 145.Draper SJ, Sack BK, King CR, Nielsen CM, Rayner JC, Higgins MK, Long CA, Seder RA.. Malaria vaccines: recent advances and New Horizons. Cell Host Microbe. 2018;24:43–56. doi: 10.1016/j.chom.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Shen WF, Galula JU, Liu JH, Liao MY, Huang CH, Wang YC, Wu HC, Liang JJ, Lin YL, Whitney MT, et al. Epitope resurfacing on dengue virus-like particle vaccine preparation to induce broad neutralizing antibody. eLife 2018;7:e38970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hajighahramani N, Nezafat N, Eslami M, Negahdaripour M, Rahmatabadi SS, Ghasemi Y. Immunoinformatics analysis and in silico designing of a novel multi-epitope peptide vaccine against staphylococcus aureus. Infect Genet Evol. 2017;48:83–94. doi: 10.1016/j.meegid.2016.12.010. [DOI] [PubMed] [Google Scholar]
- 148.Bueno LL, Lobo FP, Morais CG, Mourão LC, de ÁRAM, Soares IS, Fontes CJ, Lacerda MV, Olórtegui CC, Bartholomeu DC, et al. Identification of a highly antigenic linear B cell epitope within plasmodium vivax apical membrane antigen 1 (AMA-1). PLoS One. 2011;6:e21289. doi: 10.1371/journal.pone.0021289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Guedes RLM, Rodrigues CMF, Coatnoan N, Cosson A, Cadioli FA, Garcia HA, Gerber AL, Machado RZ, Minoprio PMC, Teixeira MMG, et al. A comparative in silico linear B-cell epitope prediction and characterization for South American and African Trypanosoma vivax strains. Genomics. 2019;111:407–17. doi: 10.1016/j.ygeno.2018.02.017. [DOI] [PubMed] [Google Scholar]
- 150.Sette A, Fikes J. Epitope-based vaccines: an update on epitope identification, vaccine design and delivery. Curr Opin Immunol. 2003;15:461–70. doi: 10.1016/s0952-7915(03)00083-9. [DOI] [PubMed] [Google Scholar]
- 151.Doytchinova IA, Guan P, Flower DR. EpiJen: a server for multistep T cell epitope prediction. BMC Bioinformatics. 2006;7:131. doi: 10.1186/1471-2105-7-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Dias DS, Ribeiro PAF, Martins VT, Lage DP, Costa LE, Chavez-Fumagalli MA, Ramos FF, Santos TT, Ludolf F, Oliveira JS, et al. Vaccination with a CD4(+) and CD8(+) T-cell epitopes-based recombinant chimeric protein derived from leishmania infantum proteins confers protective immunity against visceral leishmaniasis. Transl Res. 2018;200:18–34. doi: 10.1016/j.trsl.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 153.Moutaftsi M, Peters B, Pasquetto V, Tscharke DC, Sidney J, Bui -H-H, Grey H, Sette A. A consensus epitope prediction approach identifies the breadth of murine T(CD8+)-cell responses to vaccinia virus. Nat Biotechnol. 2006;24:817–19. doi: 10.1038/nbt1215. [DOI] [PubMed] [Google Scholar]
- 154.Glanville J, Huang H, Nau A, Hatton O, Wagar LE, Rubelt F, Ji X, Han A, Krams SM, Pettus C, et al. Identifying specificity groups in the T cell receptor repertoire. Nature. 2017;547:94–98. doi: 10.1038/nature22976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gutiérrez AH, Loving C, Moise L, Terry FE, Brockmeier SL, Hughes HR, Martin WD, De Groot AS, Krammer F. In vivo validation of predicted and conserved T cell epitopes in a swine influenza model. PLoS One. 2016;11:e0159237. doi: 10.1371/journal.pone.0159237. [DOI] [PMC free article] [PubMed] [Google Scholar]


