Abstract
As in conventional 1H MRI, T1 and T2 relaxation times of hyperpolarized (HP) 13C nuclei can provide important biomedical information. Two new approaches were developed for simultaneous T1 and T2 mapping of HP 13C probes based on balanced steady state free precession (bSSFP) acquisitions: a method based on sequential T1 and T2 mapping modules, and a model-based joint T1/T2 approach analogous to MR fingerprinting. These new methods were tested in simulations, HP 13C phantoms, and in vivo in normal Sprague-Dawley rats. Non-localized T1 values, low flip angle EPI T1 maps, bSSFP T2 maps, and Bloch-Siegert B1 maps were also acquired for comparison. T1 and T2 maps acquired using both approaches were in good agreement with both literature values and data from comparative acquisitions. Multiple HP 13C compounds were successfully mapped, with their relaxation time parameters measured within heart, liver, kidneys, and vasculature in one acquisition for the first time.
Keywords: 13C, hyperpolarized, T1, T2, Mapping, SSFP
Graphical Abstract
1. Introduction
The development of hyperpolarized (HP) 13C imaging with dissolution dynamic nuclear polarization, which provides a >10,000-fold signal enhancement for injected substrates, has enabled monitoring of various physiological processes for a wide range of diseases [1-3]. Probes such as pyruvate and urea inform on metabolism and perfusion [4-6], respectively, with several new probes currently being developed for additional applications [7-9]. Recent phase I and phase II human clinical trials have shown successful translation of [1-13C]pyruvate for monitoring metabolic conversion to [1-13C]lactate, [1-13C]alanine, and [1-13C]bicarbonate in prostate, brain, and liver cancer, and in the heart [10-15].
13C T1’s and T2’s are important parameters for characterizing new probes, pulse sequence optimization, calculations of rate constants such as kPL [16], and could also be used for assessment of healthy versus diseased tissue, as is commonly done in 1H imaging [17-20]. The development of quantitative MRI has indicated the advantage of direct quantitation of these parameters for a wide variety of clinical applications. Estimation of these parameters for 1H has been well studied and can be done either individually [21-26] or simultaneously with MR fingerprinting with the balanced steady-state free precession (bSSFP) sequence [27-29]. High spatial resolution T2 mapping has been recently developed for calculating in vivo 13C T2’s using bSSFP [30-34], while T1 values for 13C compounds, which represent decay of the hyperpolarization towards thermal equilibrium, have been reported on a non-localized basis. Furthermore, mapping the distribution of both parameters in vivo in one acquisition has not been investigated to our knowledge. Simultaneous acquisition of T1 and T2 data is challenging for multiple reasons, such as the limited lifetime of the HP magnetization, conversion of metabolically-active compounds, and flow.
bSSFP has previously been shown to provide high SNR imaging of 13C compounds [30,31,35] in an efficient and rapid fashion and has attractive properties for simultaneous mapping of T1 and T2 by analogy with its use in 1H MR fingerprinting. In this study, we aimed to develop and apply simultaneous in vivo T1 and T2 mapping of multiple hyperpolarized 13C probes using two new approaches based on specialized bSSFP acquisitions: a method based on sequential T1 and T2 mapping modules, and a model-based joint T1/T2 approach analogous to MR fingerprinting. These two different approaches were successfully tested in HP 13C phantoms and normal rats, producing high spatial resolution T1 and T2 maps of heart, liver, kidneys, and vasculature.
2. Methods
2.1. Sample Preparation
[2-13C]pyruvate, [13C]urea, [13C,15N2]urea, and HP001 were prepared as described previously [31,36]. The compounds were individually polarized in a HyperSense system (Oxford Instruments, Abingdon, UK) operating at 1.35 K and 3.35 T to achieve polarizations of ~15-20% for each compound. The compounds were then dissolved in appropriate media: 4.5 mL of 80 mM NaOH/40 mM Tris buffer for [2-13C]pyruvic acid resulting in 80 mM [2-13C]pyruvate (hereafter referred to as C2-pyruvate); 5 mL of 1x phosphate-buffered saline for [13C]urea resulting in 110 mM [13C]urea; 5 mL of 1x phosphate-buffered saline for [13C,15N2]urea resulting in 110 mM [13C,15N2]urea; and 5 mL of 1x phosphate-buffered saline for HP001 resulting in 100 mM HP001.
2.2. Animal Preparation and Hardware
All animal studies were done under protocols approved by the University of California San Francisco Institutional Animal Care and Use Committee (IACUC). Normal female Sprague-Dawley rats (Charles River Laboratories, Wilmington, MA, Age: ~ 1 year old, Average Weight: ~284 g), anesthetized using isoflurane (1.5%, gas flow rate 1 L/min) and inserted with lateral tail vein catheters, were used during the course of these experiments.
All experiments were performed on a GE MR750 3 Tesla clinical MRI scanner (GE Healthcare, Waukesha, WI) using custom dual-tuned 13C/1H quadrature transceiver radiofrequency (RF) coils with an 8 cm diameter. For the duration of the experiments, all animals were placed in a supine position on a heated pad within the coil, centered at the level of the kidneys. A 1 mL enriched [13C]urea vial phantom (6.0 M) was placed adjacent to the abdomen and used for frequency and power calibration.
2.3. MR Experiments
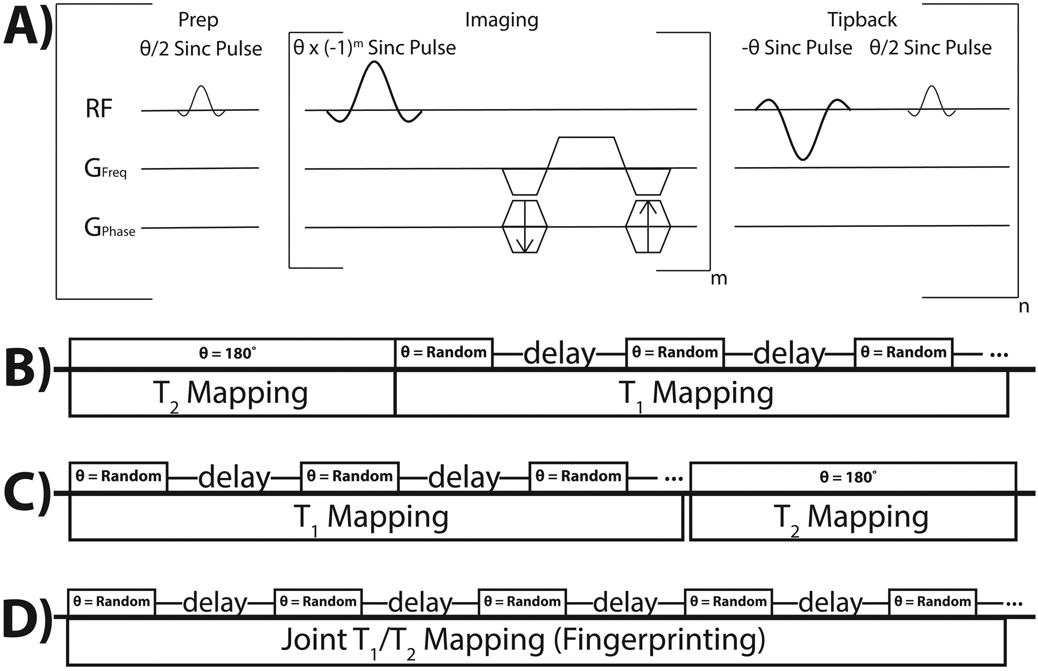
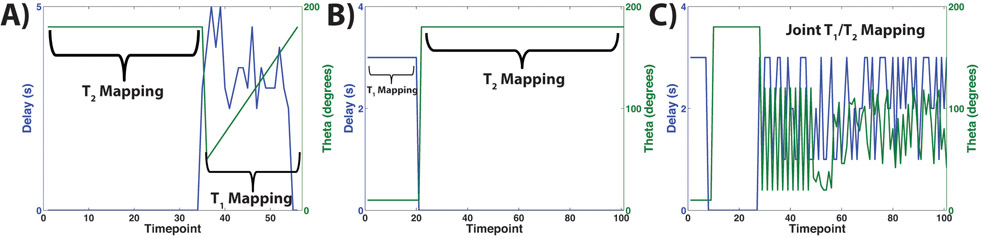
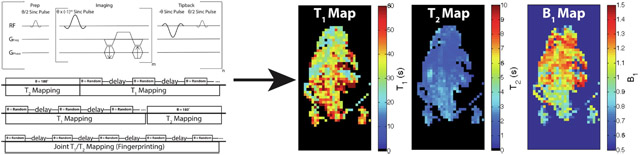
A custom bSSFP sequence was developed for this study, consisting of three portions for dynamic imaging (Figure 1A): preparatory portion consisting of a θ/2-TR/2 pulse for catalyzation, imaging portion consisting of alternating phase sinc pulses and balanced gradients, and a tipback portion consisting of a θ-TR-θ/2-TR/2 sequence for storing the magnetization along the longitudinal axis for imaging multiple time-points. Each acquisition consisted of one of two approaches: either two independent scanning modules (dual module) (Figure 1B and 1C) or modified MR fingerprinting (model-based joint T1T2 approach) (Figure 1D). The dual module approach featured one T2 mapping module and one T1 mapping module (in either order). The T2 mapping module was acquired as described previously [31], The T1 module specifically involved using delays between imaging, whereby the magnetization tipped back onto the longitudinal axis after imaging would decay by T1. Initial studies utilized both constant and variable delays, ranging from 1-10 s, as well as constant and variable flip angles, ranging from 10-180° (θ) during the T1 mapping module, to investigate potential in vivo SNR limitations and consequent signal fitting. Figure 2A shows an example set of delays and flip angles for a T2 followed by T1 mapping acquisition, with variable delays and flip angles for the T1 mapping portion, while Figure 2B shows the delays and flip angles for a T1 followed by T2 mapping acquisition. Initial studies of the modified MR fingerprinting approach involved random delays between imaging, ranging from 1-9 s, and random flip angles, ranging from 10-180° (θ), throughout the entire acquisition, with Figure 2C showing an example set of delays and flip angles for one acquisition. Simulations were performed for the modified MR fingerprinting approach to generate potential combinations of flip angles, delays, and total time-points using the signal equation detailed below. All simulations had added Gaussian noise to mimic in vivo conditions and combinations resulting in calculated T1 T2, and B1 values within 10% of the inputted values were subsequently utilized. The B1 value represents the B1 ratio, where a ratio of 1.0 would represent the exact flip angle intended.
Figure 1:
bSSFP-based methods for simultaneous T1 and T2 mapping of HP 13C probes. (A) Depiction of the bSSFP sequence used in this study, with the preparatory pulse, imaging portion, and tipback pulses for storing the magnetization during the delay portions. (B) Schematic of the dual module approach, with T2 mapping followed by T1 mapping. (C) Schematic of a different iteration of the dual module approach, with T1 mapping followed by T2 mapping. (D) Schematic of the modified MR fingerprinting approach that jointly estimated T1, T2, and B1. The rectangles in B-D represent the image acquisition portions with the depicted θ.
Figure 2:
Depiction of representative sequence delays and flip angles used for each approach. (A) Delay and flip angles for a T2 mapping module followed by a T1 mapping module. The first 16 time-points represent the T2 mapping module and featured no delay and θ = 180°. The rest of the time-points represent the T1 mapping module, which featured random delays between 6 and 10 s, and a ramping variable flip angle scheme. (B) Delay and flip angles for a T1 mapping module followed by a T2 mapping module. The first 20 time-points represent the T1 mapping module and featured a constant 3 s delay and θ = 10°. The rest of the time-points represent the T2 mapping module, which featured no delay and θ = 180°. (C) Delay and flip angles for a modified MR fingerprinting approach. The first 8 time-points featured a constant 3 s delay and θ = 10° and served as a “mini” T1 mapping module. The next 22 time-points featured no delay and θ = 180° and served as a “mini” T2 mapping module. The rest of the time-points featured random delays between 1 and 3 s, and random flip angles between 20° and 120°.
All in vivo acquisitions were acquired as 2D coronal projections (no slice-select gradient) and featured the following parameters: 14 x 7 cm2 FOV, 1.25 x 1.25 - 4 x 4 mm2 in-plane spatial resolution, 1.6 ms sinc pulse with a TBW = 4, 5.6-6.4 ms TR, 40-90 time-points, scans starting at 30 s after start of injection, with ~3 mL injected over 12 s. For comparison, individual T1 maps, T2 maps, and B1 maps were acquired using echo-planar imaging (EPI) [37], bSSFP [31], and Bloch-Siegert B1 mapping [38,39], respectively. Single-shot echo-planar imaging (EPI) T1 maps were also acquired as coronal projections and featured the following parameters: 2.5 x 2.5 mm2 in-plane spatial resolution, 3.2 ms sinc pulse with a TBW = 4, constant 5° flip angle, 1 s TR, 80 time-points, scans starting at 30 s after start of injection. bSSFP T2 maps were acquired with the following parameters: 14 x 7 cm2 FOV, 2.5 x 2.5 mm2 in-plane spatial resolution, 1.6 ms sinc pulse with a TBW = 4, 5.6 ms TR, 100 time-points, scans starting at 30 s after start of injection. Bloch-Siegert B1 maps were acquired in the coronal plane with a single-band spectral-spatial excitation pulse and a single-shot spiral readout and featured the following parameters: 8.5 x 8.5 cm2 FOV, 2.5 x 2.5 mm2 in-plane spatial resolution, 1 cm slice thickness, fermi pulse duration TRF of 12 ms, frequency offset ωRF of ± 4.5 kHz, 10° flip angle, 200 ms TR, scans starting at 15 s after start of injection. Non-localized T1 values were also acquired using a 500 μs hard pulse, 5° flip angle, 3 s TR, and 100 time-points. For anatomic reference, 3D bSSFP proton images (16 x 8 x 4.8 cm, 256 x 128 x 80, 5.1 ms TR, 50° flip angle) were acquired.
2.4. Phantom Experiments
All hyperpolarized phantom ([13C,15N2]urea) acquisitions were also acquired as 2D coronal projections and featured the following parameters: 14 x 7 cm2 FOV, 1 x 1 mm2 in-plane spatial resolution, 1.6 ms sinc pulse with a TBW = 4, 7.7 ms TR, 80-110 time-points, scans starting at 20 s after the syringe was placed into the coil.
2.5. Signal Model and Fitting
All data was reconstructed and analyzed in Matlab (MathWorks, Natick, Massachusetts, USA). The signal train for each approach was modeled using the bSSFP signal equation developed by Scheffler [40], adopted for hyperpolarized 13C imaging (i.e. transient state with negligible T1 recovery):
| [1] |
where θ is the flip angle, E1 = exp(−TR/T1), E2 = exp(−TR/T2), and n is the pulse number (i.e. phase encode). During the delay portion of the T1 mapping module and the modified MR fingerprinting approach, the magnetization was assumed to decay by exp(−delay/T1). Mz,0 corresponded to the initial longitudinal magnetization for each time-point, after accounting for the delay portion.
An initial SNR threshold, which varied from 5-20 depending on the compound, was used to filter out low SNR voxels. Additionally, respiratory motion correction in the superior/inferior direction was performed using rigid translation via mutual information, as described previously [41]. All fitting was done on a voxel-by-voxel basis and only voxels with fits of R2>0.9 were kept. The fitting of the dual module approach acquisitions was done by first fitting the T2 mapping portion to a single exponential fit, and then feeding in the resulting values into the fitting of the T1 mapping portion. The fitting for the T1 mapping portions of the dual module approach, as well as the modified MR fingerprinting, was done using non-linear least squares with the trust-region-reflective algorithm. The initial guesses for the algorithm were based on known non-localized T1 values for each compound, as well as previously acquired mean T2 maps in the case of the modified MR fingerprinting. For comparison for the modified MR fingerprinting, T1 and T2 were fit using dictionary matching via the maximum inner product method [42], where the dictionary was created with a range of T1’s (200 ms increments from 10-100 s), T2’s (100 ms increments from 0.1 to 10 s), and B1’s (2% increments from 50-150%), where B1 is the ratio of the obtained flip angle to the desired flip angle [28].
EPI T1 maps were calculated on a voxel-by-voxel basis by fitting to a mono-exponential curve after respiratory motion and flip angle correction. bSSFP T2 maps were calculated as described previously [31], with the mean T2 maps used for comparison.
3. Results
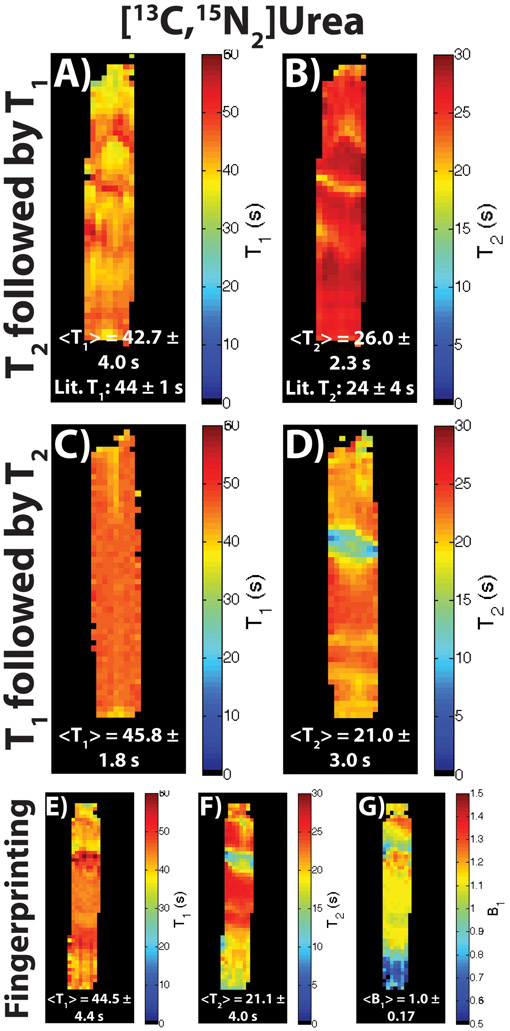
Figure 3 shows the calculated parameter maps for the hyperpolarized [13C,15N2]urea phantom acquisitions of all three approaches. The dual module approach with the T2 module followed by the T1 module is shown in parts A and B, while the dual module approach with the T1 module followed by the T2 module is shown parts C and D. The modified MR fingerprinting approach is shown in parts E-G. The acquisitions were similar to those shown in Figure 2, albeit with more T1 and T2 mapping time-points due to the longer solution state T1 and T2 compared to in vivo for [13C,15N2]urea. The mean and intra-map standard deviation of the T1 and T2 maps from all three acquisitions matched up well among each other and the literature values. The B1 map in part G, with a mean value of 1.0, matched up well with the expected profile of the syringe within the volume coil, with some drop-off near the edge of the coil as seen at the bottom of the syringe. Deviations within the maps can be attributed to some B0 inhomogeneity along the S/I dimension.
Figure 3:
Resulting T1, T2, and B1 maps in a hyperpolarized [13C,15N2]urea phantom for all three types of acquisitions described. (A,B) T1 and T2 maps for a dual module acquisition (T2 mapping module followed by a T1 mapping module). The acquisition was similar to the one depicted in Figure 2A, except with more T2 time-points acquired due to the long solution state T2 of the compound. (C,D) T1 and T2 maps for a different iteration of the dual module acquisition (T1 mapping module followed by a T2 mapping module). The acquisition was similar to the one depicted in Figure 2B, except with more T1 and T2 time-points acquired due to the long solution state T1 and T2 of the compound. (E-G) T1, T2, and B1 maps for a modified MR fingerprinting approach, with the acquisition similar to the one depicted in Figure 2C, except with more T1 and T2 time-points acquired due to the long solution state T1 and T2 of the compound. For all three acquisitions, the resulting mean ± intra-map standard deviation agreed well with the literature value for all the T1 and T2 maps. The B1 also had a mean of 1.0, which is expected with a stationary syringe centered in a volume coil. Deviations in values in the S/I direction for all maps can be attributed to some B0 inhomogeniety, as well as some B1 drop-off at the edge of the coil.
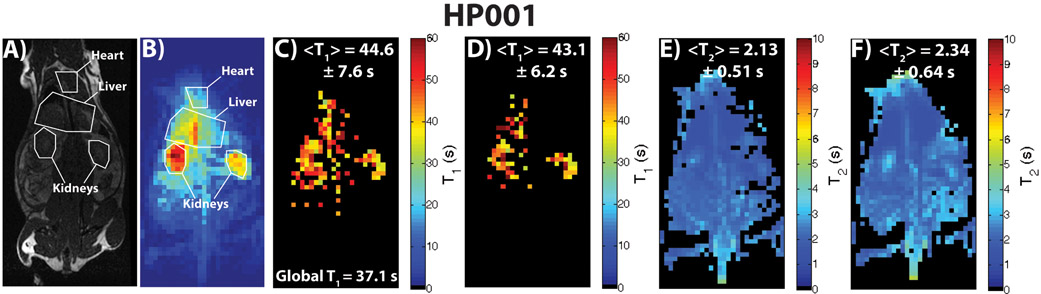
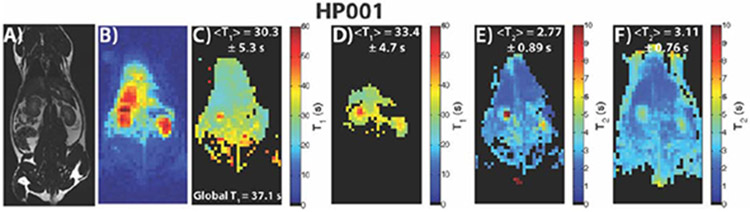
Figure 4 shows results for an HP001 acquisition of the dual module approach (T2 mapping followed by T1 mapping), including a 1H anatomical slice (A), representative time-point (B), T1 map (C), EPI T1 map (D), T2 Map (E), and bSSFP T2 map (F). The T1 mapping module featured variable delays and flip angles, as seen in Figure 2A. The relevant anatomical structures are outlined, with the location being similar in all in vivo acquisitions in the study. The mean and intra-map standard deviation of the T1 map was 44.6 ± 7.6 s, which was close to the non-localized T1 of 37.1 s, with the T1 distribution matching well with the EPI T1 map in the abdomen. The T2 maps also agreed well with each other in terms of distribution and T2’s and mean and intra-map standard deviation. The Bland-Altman plots in Supporting Figure 1A and 1B show good agreement between the dual module and comparative acquisitions on a voxel-by-voxel basis, with most voxels agreeing to within a few seconds (for T1)/less than 0.5 s (for T2) of each other, except for voxels corresponding to long T1’s and T2’s in the kidneys that can be attributed to both renal filtration effects and some low SNR fitting of the dual module approach. Supporting Video 1 shows all the time-points of the acquisition, along with the signal train for a representative voxel within the kidney, where rapid T2 decay occurs first, followed by slower T1 decay.
Figure 4:
Depiction of a 1H anatomical slice (A), representative time-point (B), T1 map (C), EPI T1 Map (D), dual module T2 map (E), and individually acquired T2 map (F) for a dual module (T2 mapping followed by T1 mapping) HP001 acquisition in a normal Sprague-Dawley rat with 2.5 x 2.5 mm2 in-plane spatial resolution (N = 3). The acquisition featured both a variable delay and flip angle during the T1 mapping module, as seen in Figure 2A. The distribution as well as mean and intra-map standard deviation of the dual module T1 map (C) matched up well with the EPI T1 map (D), with the highest T1 values seen within the kidneys. The T2 maps in E and F also matched up well in terms of distribution and mean and intra-map standard deviation.
Figure 5 shows results for an HP001 acquisition of a different iteration of the dual module approach (T1 mapping followed by T2 mapping), including a 1H anatomical slice (A), representative time-point (B), T1 map (C), EPI T1 Map (D), T2 map (E), and bSSFP T2 map (F). The T1 mapping portion was done with a constant delay of 3 s and constant flip angle of 5° as illustrated in Figure 2B. The mean and intra-map standard deviation of the T1 map was 30.3 ± 5.3 s, with the T1 and T2 distribution matching well with the EPI T1 map and bSSFP T2 map, respectively. As with the dual module approach in Figure 4, the Bland-Altman plots in Supporting Figure 1C and 1D show good agreement between this iteration of the dual module approach and the comparative acquisitions on a voxel-by-voxel basis, with most voxels agreeing to within a few seconds (for T1)/less than 0.5 s (for T2) of each other, except for voxels corresponding to long T1’s and T2’s in the kidneys. The T2 maps in particular demonstrate the effects of renal filtration since the dual module T2 map was effectively acquired at 1 minute after the start of the acquisition and compared to the start of the comparative bSSFP T2 map, which gave rise to considerably longer T2 values in the kidneys. Supporting Video 2 shows all the time-points of the acquisition, along with the signal train for a representative voxel within the kidney, where slower T1 decay occurs first, followed by rapid T2 decay.
Figure 5:
Depiction of a 1H anatomical slice (A), representative time-point (B), T1 map (C), EPI T1 Map (D), dual module T2 map (E), and individually acquired T2 map (F) for a dual module (T1 mapping followed by T2 mapping) HP001 acquisition in a normal Sprague-Dawley rat with 2.5 x 2.5 mm2 in-plane spatial resolution (N = 3). The distribution within the T1 maps in parts C and D matched up well with each other within the liver and kidneys. The T2 maps in parts E and F also matched up well, with renal filtration over the course of the acquisition resulting in comparatively longer T2’s in the kidneys of part E.
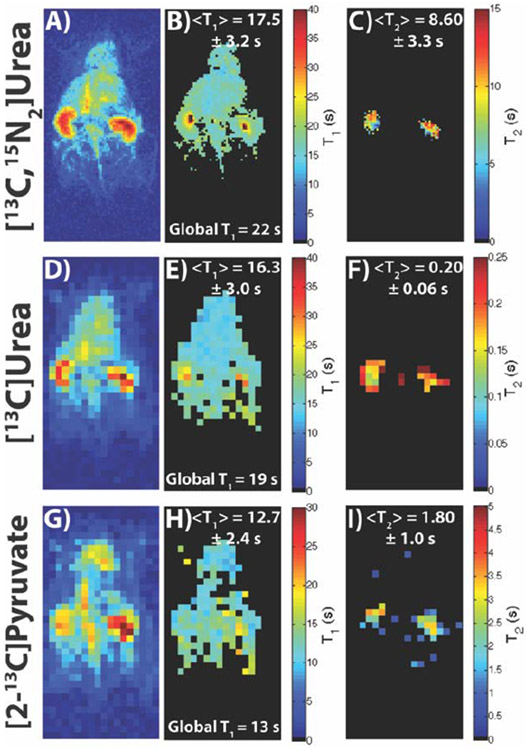
Figure 6 shows the representative time-point, T1 map, and T2 map of [13C,15N2]urea (A-C), [13C]urea (D-F), and [2-13C]pyruvate (G-H) acquired with a dual module acquisition (T1 mapping followed by T2 mapping). The non-localized T1 was close to or within the mean and intra-map standard deviation of the T1 map, and T2 maps matched up with previously acquired maps [30,31,41], although some of the longer T2 values in the kidneys can be attributed to renal filtration over the course of the acquisition. Even with the relatively shorter T1 and T2’s of [2-13C]pyruvate, as well as the short T2’s of [13C]urea, the tailored acquisition resulted in enough high SNR time-points for accurate fitting of both parameters in the kidneys.
Figure 6:
Depiction of a representative time-point, T1 map, and T2 map for [13C,15N2]urea (A-C), [13C]urea (D-F), and [2-13C]pyruvate (G-I) from a dual module acquisition (T1 mapping followed by T2 mapping) (N = 1 for all compounds). The T1 maps matched up well with the global T1 as well as the HP001 T1 maps in Figures 4-5, with the longest T1’s seen in the kidneys. The T2 maps of each compound matched up well with previously acquired maps, with the longest T2 values seen in the kidneys due to renal filtration. Additionally, the [13C,15N2]urea maps showed the capability of 1.25 x 1.25 mm2 spatial resolution for T1/T2 mapping. For 1H anatomical image, see Figure 7A.
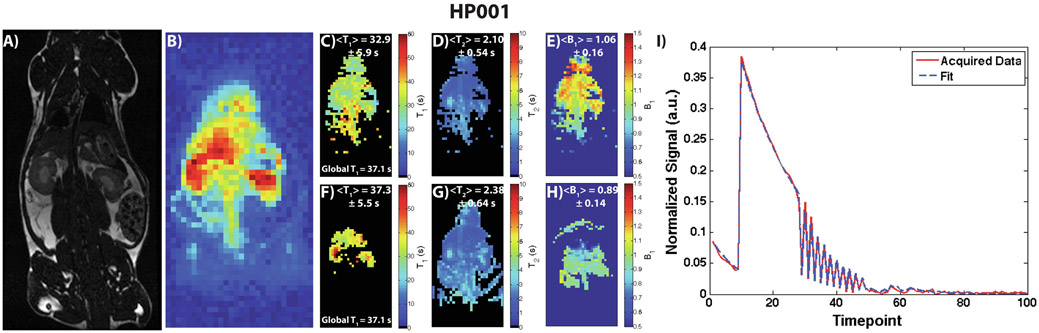
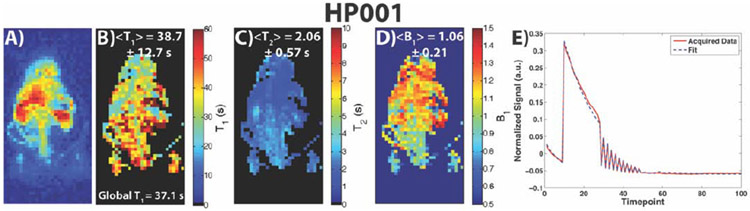
Figure 7 shows results for an HP001 acquisition of the modified MR fingerprinting approach and associated comparison maps, including a 1H anatomical slice (A), representative time-point (B), T1 map (C), T2 map (D), B1 Map (E), EPI T1 map (F), bSSFP T2 map (G), Bloch-Siegert B1 map (H), and an example signal fit from a kidney voxel (I). The mean and intra-map standard deviation of the T1 map was 32.9 ± 5.9 s, which is close to the non-localized T1 of 37.1 s, and matches well with the EPI T1 map. The Bland-Altman plot in Supporting Figure 1E indicates good agreement between the two acquisitions on a voxel-by-voxel basis as well, with most voxels agreeing to within a few seconds of each other, except for voxels corresponding to long T1’s in the kidneys that can be attributed to renal filtration effects. Similarly, the calculated T2 map matched up well with the bSSFP T2 map based on the calculated means and intra-map standard deviations, as well as qualitatively based on the distribution of the T2’s within the anatomy. The Bland-Altman plot in Supporting Figure 1F also indicates good agreement between the two acquisitions on a voxel-by-voxel basis, with most voxels agreeing to less than 0.5 seconds of each other. The additional B1 map calculated with this approach matched up with the expected coil profile for a rat heart/abdomen acquisition and the Bloch-Siegert B1 map within the kidneys, although deviation is visible within the liver due to potential motion effects during the readout of the Bloch-Siegert B1 mapping. Supporting Video 3 shows all the time-points of the acquisition, along with the signal train for a representative voxel within the kidney, with the signal exhibiting initial T1 decay and T2 decay as part of the “mini” modules depicted in Figure 2C, followed by random flip angles and delays for the remaining time-points. Dictionary matching was also able to successfully calculate all three maps (Figure 8), although the spatial smoothness of the T1 map was lower compared to non-linear least squares fit as evidenced by the larger intra-map standard deviation.
Figure 7:
Depiction of a 1H anatomical slice (A), representative time-point (B), T2 map (C), T1 map (D), and B1 Map (E), and for a modified MR fingerprinting HP001 acquisition in a normal Sprague-Dawley rat with 2.5 x 2.5 mm2 in-plane spatial resolution (N = 3). The fitting was done using nonlinear least squares via the trust-region-reflective algorithm. An EPI T1 map (F), bSSFP T2 map (G), and Bloch-Siegert B1 map (H) (resized for display purposes) are shown here for comparison. The distribution in the modified MR fingerprinting T1 and T2 maps match up well with the comparison maps, as well as maps in Figures 4 and 5. The B1 map matches up well with the Bloch-Siegert map and with the expected coil profile of a rat acquisition, although some deviations do exist within the liver and kidneys. An example signal fit from a kidney voxel (I) is also shown here, indicating a good agreement between the acquired signal and least-squares fit.
Figure 8:
Depiction of a representative time-point (A), T2 map (B), T1 map (C), B1 Map (D), and an example signal fit from a kidney voxel (E) for the same acquisition as Figure 7 (HP001 acquisition in a normal Sprague-Dawley rat with 2.5 x 2.5 mm2 in-plane spatial resolution). The fitting was done using dictionary matching via the inner product method. The distribution in the T2 and B1 maps match up well with maps in Figure 7, while the T1 map shows considerable difference from the T1 map in Figure 7, with lower spatial smoothness and overestimation of the T1 values based on the mean and intra-map standard deviation.
Table 1 shows a summary of the T1 and T2 values calculated here for the four compounds studied and the comparative global/gold standard/literature values. We also added values for [1-13C]pyruvate due to its importance in the hyperpolarized 13C community, although we did not use the compound here.
Table 1:
Compilation of T1 and T2 values calculated in this publication (Meas.) and comparative global/literature values (Global/Lit.). These values are for health Sprague-Dawley rats acquired at 3T. We also added literature values of [1-13C]pyruvate due its widespread use in HP 13C research and clinical trials. These values are strongly reflective of kidney parameter values, particularly T2, and some discrepancies may be due to renal filtration (see discussion).
| [1-13C]Pyr | [2-13C]Pyr | HP001 | [13C,14N2]Urea | [13C,15N2]Urea | |
|---|---|---|---|---|---|
| T1 (s) (Meas.) | N/A | 12.7 | 35.9 (mean of Fig. 4C, 5C, and 7C) | 16.3 | 17.5 |
| T1 (s) (Global/Lit.) | ~30 [43] | 13 (Global) | 37.1 (Global) 37.9 (mean of Fig. 4D, 5D, and 7F) 32 [36] | 19 (Global) | 22 (Global) |
| T2 (s) (Meas.) | N/A | 1.8 | 2.3 (mean of Fig. 4E, 5E, and 7D) | 0.20 | 8.6 |
| T2 (s) (Lit.) | ~0.6 [33] | 0.8 [31] | 2.6 (mean of Fig. 4F, 5F, and 7G) | ~0.25 [30] | See [30] for distributions in kidney |
5. Discussion and Conclusion
We have developed two successful approaches for in vivo high spatial resolution simultaneous T1 and T2 mapping using the bSSFP sequence: dual module approach and modified MR fingerprinting. Two iterations of the dual module approach were demonstrated, with individual T1 and T2 mapping portions performed in a sequential manner, and with each portion being acquired in either order. The modified MR fingerprinting successfully extracted T1, T2, and B1 maps by fitting the acquired signal to the analytical bSSFP model after a randomized delay/flip angle acquisition. The distribution of these parameters from several HP 13C compounds, with a wide range of T1’s and T2’s, was shown within the kidneys, liver, heart, and vasculature. The approaches developed here can be used for quantitative 13C MRI by using T1 and T2 relaxometry to monitor various biological processes, such as intracellular versus extracellular uptake [44] and renal urea handling [41]. We were also able to validate the initial implementation of these approaches in vivo using EPI (for T1 values), bSSFP (for T2 values), and Bloch-Siegert (for B1 values).
Each of the described approaches has advantages that can be exploited for mapping specific compounds as well as achieving the highest in vivo distribution of these parameters. The advantage of the dual module approach was the straightforward manner of extracting the T1 and T2 maps since each parameter was sampled individually within different modules of the acquisition. Therefore only one parameter was being fit at one time, making the data analysis much less complicated. Furthermore, the exact timing of each module can be designed prior to the acquisition based on known estimates of the parameters for a given compound, thereby allowing proper sampling of each parameter, i.e. sampling for about 2-3 T1’s and T2’s for accurate fitting. Of the two iterations presented here, the one with the T1 mapping module first via the acquisition detailed in Figure 5 was seen to be the most advantageous with regards to achieving a high and accurate distribution of each parameter at various spatial resolutions for multiple compounds. The magnetization usage of this approach was more optimal compared to acquiring the T2 mapping module first because more magnetization remained for the T2 mapping module as compared to the other iteration where the T2 mapping module would use up considerable magnetization, thereby limiting the SNR of the subsequent T1 mapping module. This is further illustrated in the Bland-Altman plot in Supporting Figure 1A, where some of the T1 map voxels matched up poorly with the comparative EPI T1 map due to some low SNR fits that led to some overfitting. A limitation of the dual module approach was the lack of sensitivity to high spatial resolution B1 mapping since the acquisition did not have the necessary train of flip angles necessary for calculating B1 variation accurately [28]. A variable flip angle T1 mapping module would be a possibility for additionally mapping B1, although we noticed difficulty in fitting both T1 and B1 simultaneously with this approach due to low SNR and limited time-points available for fitting.
The main advantage of the modified MR fingerprinting approach was the ability to map T1, T2, and B1 at a high spatial resolution without any loss in the spatial distribution of any parameters, unlike what was seen in the dual module approach within the module that was acquired second. However, the data fitting was more difficult compared to the dual module approach since three parameters were being fit simultaneously. As with the dual module approach, the choice of temporal resolution between samples is crucial towards being sensitive in detecting the three parameters, with the set of variable flip angles and delays simulated prior to the acquisition. Based on the different schemes tried, the best results were seen with a delay ranging from 1-3 s and flip angles (theta) ranging from 20-120°, which allowed the magnetization to last for ~60-90 time-points or ~120-180 s (Figure 6). This scheme also featured additional “mini” T1 and T2 mapping modules (acquired on the order of one T1 and one T2) (Fig. 2C) that provided better initial estimates, as well as upper and lower bounds, for the non-linear least squares fit. This provided sufficient parameter sensitivity, due to long enough inter-imaging delays relative to the 13C T1, and variation in low and high flip angles that provided additional sensitivity to T1 (low flip angles), as well as T2 (high flip angles) and B1 (variation in the flip angles that induce signal oscillations). This scheme also provided a large amount of high SNR time-points, which was seen to be necessary for accurate fitting of the parameters as too few time-points, which was acquired in a few animals with long (up to 9 s) delay acquisitions or too many low flip angles, resulted in several voxels having visibly inaccurate T1’s and T2’s due to an inability of the algorithm to distinguish an expected fit from an inaccurate one (Supporting Fig. 2). Based on experience and literature, T1 mapping requires ~2-3 T1 of samples spaced ~3 s for accurate fitting. Therefore, design of the acquisition needs to have the required sensitivity to the T1 of the probe via low flip angles and proper sampling timing. However, since SNR can be a limiting factor in all these acquisitions, and T1 takes the longest amount of time to sample (whether in dual module approach or utilizing the “mini” modules), then T1 becomes challenging to accurately measure when also trying to measure T2 (and B1) because the whole acquisition needs to be optimized to have accurate SNR and sampling to measure each parameter. T1 measurement can also be very sensitive to the flip angle chosen as correcting for T1 with an incorrect flip angle (even by a few degrees) can cause a change of ~5-10 s in the T1 calculation. Based on these challenges, further work is needed to balance the length of the “mini” modules with the length of the random train of delays and flip angles to generate enough high SNR time-points for fitting, as well as further optimize the random train to improve B1 sensitivity [45].
Based on the results of all three methods presented here, we believe the most stable approach is using the dual module approach with T1 mapping followed by T2 mapping because of the ease and stability of the subsequent fittings. This approach appeared to be best validated by our gold standards (EPI and bSSFP) compared to the other two. One aspect of future work will focus on improving the model for more accurate fitting of the acquired in vivo data. In general, this could help resolve some of the differences seen between the developed approaches and “gold” standard acquisitions (EPI, bSSFP, Bloch-Siegert) at long T1 and T2 values, as illustrated by via the Bland-Altman plots. The purpose of this study was to demonstrate the feasibility of mapping multiple parameters from one acquisition, and as such we used the simple, but straightforward, bSSFP analytical model for signal fitting and a fully sampled acquisition for easy reconstruction. We did not incorporate B0 into the model because the frequency response of our acquisitions showed minimal variations over ±25 Hz based on the pulse width, time-bandwidth product, TR, and range of flip angles used (Supporting Fig. 3). Future acquisitions could incorporate a linear ramp of ~5 preparation pulses into the “prep” portion of the imaging sequence for additional off-resonance insensitivity [46,47]. B0 can also be added to the model via Bloch simulations as is currently done in 1H MR fingerprinting, at the expense of a more complicated data analysis. Additionally, based on the differences between the modified MR fingerprinting and Bloch-Siegert acquisitions, the B1 term in the model can be considered a “nuisance” parameter that accounts for B1, as well as other effects, such as B0 and motion. The signal fits and parameter maps were better with the B1 term included compared to without, but the calculated values might be slightly off of the true B1 measured by the Bloch-Siegert, such as ~1.1 versus ~1 in a kidney voxel as seen in Figures 7D and 7G. However, incorporation of B0 into the model could lead to an improved B1 estimation by allowing us to tease out the effects of B0 and B1 on the calculation of T1 and T2.
We did not account for metabolism, such as for [2-13C]pyruvate, where conversion to compounds with short T1’s and T2’s, such as [2-13C]lactate, would result in an apparent decrease of the calculated [2-13C]pyruvate T1’s and T2’s, since we utilized an RF pulse with a wide spectral bandwidth. We also did not account for renal filtration, which causes a dynamic elongation of T1 and T2 in the kidneys, particularly in the renal pelvis (Fig. 6), due to a removal of red blood cells and proteins that causes T1 and T2 to lengthen back towards solution state values [30,41]. Metabolism can be accounted for by incorporating a metabolic conversion rate to the T1 portion of the model as is currently done with calculation of kPL [6]. Renal filtration can be accounted for within the kidney voxels by having T1 and T2 not be restricted to one value (representing a constant in vivo value), but to a vector of increasing values resulting in time-varying T1 and T2 maps. In general, the T1’s and T2’s measured here can be considered to be approximate or “apparent” values due to some confounding effects, such as RF transmitter variation and vascular contributions to some voxels [30].
Flow and perfusion are other sources of confounding effects that can cause deviations in relaxation measurements. The bSSFP sequence is first moment flow compensated along the frequency direction, so phase induced by flow along this direction should be fully rephased. However, flow along the phase encode direction will cause some dephasing in the voxel, which would lead to signal loss and a decrease in the apparent relaxation measurement, particularly T2 [30]. Perfusion into tissues over the course of the imaging sequence would cause changes in the local microenvironment for a particular probe. This could have a variety of effects, such as elongation of the T2 for urea due to perfusion within the kidneys, or perfusion into more metabolic regions of the body, which could be accounted for in the model as described above.
The clinical utility in this method can be looked at from two perspectives. One, as mentioned in the introduction, changes in these relaxation parameters can reveal differences in healthy versus diseased tissue, with a high resolution spatial distribution being necessary to identify where any abnormalities area, as demonstrated previously [32,34,41]. Second, we can use the distribution of relaxation parameters to help improve subsequent acquisitions, such as EPI or 3D bSSFP. For example, the spatial parameter information can be used to design an appropriate, on-the-fly, patient-specific, variable flip angle schedule to maximize the spatiotemporal SNR for and/or (if the same flip angle scheme is used for all patients) more accurately calculate other parameters that rely on these relaxation parameters, such as kPL, post-acquisition. However, to obtain this parameter information, an additional injection would be required, which would make widespread use a challenge, although any information from patients would be beneficial for the hyperpolarized 13C community. Additionally, more work would need to be done to adapt our technique to [1-13C]pyruvate and associated metabolites due to these metabolites resonating close to one another at 3T.
Another aspect of future work will focus on adding undersampling to improve the spatial resolution. Undersampling 1H parameter mapping [21,22] and MR fingerprinting acquisitions [27-29] is commonly done to speed up these acquisitions without any loss in pattern recognition and serves as a basis for undersampling HP acquisitions. While HP acquisitions are already fast due to rapid inherent decay of the HP signal, undersampling would reduce the number of phase encodes needed for higher spatial resolution (e.g. 1 x 1 mm2), thereby preserving more magnetization for acquisition of multiple time-points. We have previously showed both 2D and 3D T2 mapping using a locally low rank plus sparse reconstruction that featured matrix sizes of 140x70x20 (2D) and 120x60x18x20 (3D). Several types of reconstructions can be leveraged, with some initial retrospective simulations (Supporting Fig. 4) indicating both a temporal subspace and low rank reconstruction [22] and model-based reconstruction [21] being viable options going forward.
Supplementary Material
Supporting Figure 1: Bland-Altman plots comparing the T1 and T2 maps from the developed approaches and comparison EPI/bSSFP maps. (A,B) Comparison of the dual module approach (T2 mapping followed by T1 mapping) T1 map and EPI T1 map (A), as well as T2 map and bSSFP T2 map (B) is shown here. The maps matched up well with each other in most voxels, although some differences occurred at long T1/T2 values due to renal filtration and some overfitting due to low SNR voxels in the dual module approach. (C,D) Comparison of the dual module approach (T1 mapping followed by T2 mapping) T1 map and EPI T1 map (C), as well as T2 map and bSSFP T2 map (D) is shown here. The maps matched up well with each other in most voxels, although some differences occurred at long T1/T2 values due to renal filtration elongating the T1/T2 over the course of the dual module acquisition. (E,F) Comparison of the modified MR fingerprinting T1 map and EPI T1 map (E), as well as T2 map and bSSFP T2 map (F) is shown here. As with A-D, the maps matched up well with each other in most voxels, although similar differences occurred at long T1/T2 values due to renal filtration and some overfitting due to low SNR voxels in the EPI acquisition for the T1 map. For all plots, the zero line (i.e. no difference between voxels), and the mean ± 1.96*standard deviation (SD) (95% confidence interval) lines are displayed.
Supporting Figure 2: Comparison of fits for a modified MR fingerprinting acquisition of HP001 with long delays (up to 9 s) and only 20 high SNR time-points available for fitting in a kidney voxel, along with resulting parameter maps. Nonlinear least squares was used for fitting. (A) The calculated fit matched up very closely with the acquired signal, although the resulting values for T1 and T2 were lower and higher, respectively, than ideal values, such as those seen in Figures 3 and 4. A plot of the simulated signal trains for combinations of T1 and T2 values show how close all curves match up to one another when only 20 time-points are available for fitting, causing the fitting problem to be poorly conditioned due to lack of signal variation. (B-D) The resulting T2, T1, and B1 maps are shown here, along with the mean and intra-map standard deviation. The mean of the T1 map was considerably lower than the maps found in Figures 3-5, while the T2 map had poor spatial smoothness owing to the high intra-map standard deviation (for comparison, Figure 2E had an intra-map standard deviation of 1.1). The difficulty in fitting illustrated in part A resulted in several voxels in the T2 map with values close 10 s, which was chosen as the upper bound. Consequently, the T1 values were shorter than expected in those voxels, indicating erroneous fits. Acquisition schemes for the modified MR fingerprinting approach with >~60-80 time-points provided fits closer to ideal values and resulted in smoother spatial variation within the parameter maps. Addition of “mini” modules, as depicted in Figure 2C, can help condition the fitting better by providing close initial estimates as well as upper and lower bounds for the fit.
Supporting Figure 3: Depiction of the simulated frequency response for different flip angles using the parameters described in the methods section (1.6 ms TBW = 4 sinc pulse played with a TR of 5.6 ms). For the simulations, θ was set to (A) 10°, (B) 20°, (C) 40°, (D) 60°, (E) 120°, and (F) 180°, which represents the range of the flip angles used in these studies. The variation within ±25 Hz was relatively minimal for both low and high flip angles, allowing us to ignore the contribution of B0 during the signal modeling.
Supporting Figure 4: Comparison of different 50% undersampled reconstructions of a random flip angle and delay acquisition. The first image from the ground truth (A), temporal subspace and low rank recon (B), model-based (via nonlinear conjugate gradient) recon (C), and zero-filled recon (D) can be seen. The dictionary described in the methods section was used for the sparsifying transform. Both recons in B and C had low nRMSE and considerably outperformed the zero-filled recon. Qualitatively, both recons removed the aliasing artifacts present in part D.
Supporting Video 1: An iteration of a T2 mapping followed by T1 mapping acquisition in a normal Sprague-Dawley rat with 2.5 x 2.5 mm2 in-plane spatial resolution is shown here along with the signal evolution for a representative kidney voxel. The delays and flip angles used in this acquisition are shown in Figure 2A and the results are shown in Figure 4. The first 16 time-points represent the T2 mapping module, while the rest of the time-points represent the T1 mapping module.
Supporting Video 2: An iteration of a T1 mapping followed by T2 mapping acquisition in a normal Sprague-Dawley rat with 2.5 x 2.5 mm2 in-plane spatial resolution is shown here along with the signal evolution for a representative kidney voxel. The delays and flip angles used in this acquisition are shown in Figure 2B and the results are shown in Figure 5. The first 20 time-points represent the T1 mapping module, while the rest of the time-points represent the T2 mapping module.
Supporting Video 3: An iteration of a modified MR fingerprinting HP001 acquisition in a normal Sprague-Dawley rat with 2.5 x 2.5 mm2 in-plane spatial resolution is shown here along with the signal evolution for a representative kidney voxel. The delays and flip angles used in this acquisition are shown in Figure 2C and the results are shown in Figures 7 and 8. The randomness of the signal evolution mimics the flip angle train in Figure 2C, with the longest signal lasting in the kidneys due to renal filtration characteristics that elongate the T1 and T2 over time. The first 8 time-points show T1 decay as part of the “mini” T1 mapping module, while the next 19 time-points (3 removed for overranging from original 22) featured show T2 decay as part of the “mini” T2 mapping module.
Highlights.
Two new approaches were developed for simultaneous T1 and T2 mapping of HP 13C probes based on balanced steady state free precession (bSSFP) acquisitions: a method based on sequential T1 and T2 mapping modules, and a model-based joint T1/T2 approach analogous to MR fingerprinting
T1 and T2 maps acquired using both approaches were in good agreement with both literature values and data from comparative acquisitions.
Multiple HP 13C compounds were mapped, with their relaxation time parameters measured within heart, liver, kidneys, and vasculature in healthy rats.
6. Acknowledgements
We gratefully acknowledge Dr. Robert Bok, Romelyn Delos Santos, Lucas Carvajal, and Mark van Criekinge for all their help with the experiments, as well as grant support from NIH P41EB013598, R01EB017449, R01CA183071 and R01EB013427. NIH K01DK099451 supported CVM.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Ardenkjaer-Larsen JH, Fridlund B, Gram A, Hansson G, Hansson L, Lerche MH, Servin R, Thaning M, Golman K, Increase in signal-to-noise ratio of > 10,000 times in liquid-state NMR., Proc. Natl. Acad. Sci. U. S. A. 100 (2003) 10158–10163. doi: 10.1073/pnas.1733835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kurhanewicz J, Vigneron DB, Brindle K, Chekmenev EY, Comment A, Cunningham CH, Deberardinis RJ, Green GG, Leach MO, Rajan SS, Rizi RR, Ross BD, Warren WS, Malloy CR, Analysis of cancer metabolism by imaging hyperpolarized nuclei: prospects for translation to clinical research., Neoplasia. 13 (2011) 81–97. doi: 10.1593/neo.101102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Comment A, Merritt ME, Hyperpolarized Magnetic Resonance as a Sensitive Detector of Metabolic Function, Biochemistry. 53 (2014) 7333–7357. doi: 10.1021/bi501225t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Golman K, Petersson JS, Metabolic Imaging and Other Applications of Hyperpolarized 13C1, Acad. Radiol 13 (2006) 932–942. doi: 10.1016/j.acra.2006.06.001. [DOI] [PubMed] [Google Scholar]
- [5].Von Morze C, Larson PEZ, Hu S, Keshari K, Wilson DM, Ardenkjaer-Larsen JH, Goga A, Bok R, Kurhanewicz J, Vigneron DB, Imaging of blood flow using hyperpolarized [13C]urea in preclinical cancer models, J. Magn. Reson. Imaging 33 (2011) 692–697. doi: 10.1002/jmri.22484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Chen HY, Larson PEZ, Bok RA, Von Morze C, Sriram R, Santos RD, Santos JD, Gordon JW, Bahrami N, Ferrone M, Kurhanewicz J, Vigneron DB, Assessing prostate cancer aggressiveness with hyperpolarized dual-agent 3D dynamic imaging of metabolism and perfusion, Cancer Res. 77 (2017) 3207–3216. doi: 10.1158/0008-5472.CAN-16-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Gallagher FA, Kettunen MI, Day SE, Hu D-E, Ardenkjær-Larsen JH, ‘t Zandt R. in, Jensen PR, Karlsson M, Golman K, Lerche MH, Brindle KM, Magnetic resonance imaging of pH in vivo using hyperpolarized 13C-labelled bicarbonate, Nature. 453 (2008) 940–943. doi: 10.1038/nature07017. [DOI] [PubMed] [Google Scholar]
- [8].Von Morze C, Bok RA, Ohliger MA, Zhu Z, Vigneron DB, Kurhanewicz J, Hyperpolarized [13C]ketobutyrate, a molecular analog of pyruvate with modified specificity for LDH isoforms, Magn. Reson. Med 75 (2016) 1894–1900. doi: 10.1002/mrm.25716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Marco-Rius I, von Morze C, Sriram R, Cao P, Chang G-Y, Milshteyn E, Bok RA, Ohliger MA, Pearce D, Kurhanewicz J, Larson PEZ, Vigneron DB, Merritt M, Monitoring acute metabolic changes in the liver and kidneys induced by fructose and glucose using hyperpolarized [2-13C]dihydroxyacetone, Magn. Reson. Med 77 (2017) 65–73. doi: 10.1002/mrm.26525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Nelson SJ, Kurhanewicz J, Vigneron DB, Larson PEZ, Harzstark AL, Ferrone M, van Criekinge M, Chang JW, Bok R, Park I, Reed G, Carvajal L, Small EJ, Munster P, Weinberg VK, Ardenkjaer-Larsen JH, Chen AP, Hurd RE, Odegardstuen L-I, Robb FJ, Tropp J, Murray JA, Metabolic imaging of patients with prostate cancer using hyperpolarized [1-13C]pyruvate., Sci. Transl. Med 5 (2013) 198ra108. doi: 10.1126/scitranslmed.3006070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chen H-Y, Larson PEZ, Gordon JW, Bok RA, Ferrone M, Van Criekinge M, Carvajal L, Cao P, Pauly JM, Kerr AB, Park I, Slater JB, Nelson SJ, Munster PN, Aggarwal R, Kurhanewicz J, Vigneron DB, Technique development of 3D dynamic CS-EPSI for hyperpolarized 13C pyruvate MR molecular imaging of human prostate cancer, Magn. Reson. Med 80 (2018) 2062–2072. doi: 10.1002/mrm.27179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Gordon JW, Chen H-Y, Autry A, Park I, Van Criekinge M, Mammoli D, Milshteyn E, Bok R, Xu D, Li Y, Aggarwal R, Chang S, Slater JB, Ferrone M, Nelson S, Kurhanewicz J, Larson PEZ, Vigneron DB, Translation of Carbon-13 EPI for hyperpolarized MR molecular imaging of prostate and brain cancer patients, Magn. Reson. Med (2018) 1–8. doi: 10.1002/mrm.27549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Miloushev VZ, Granlund KL, Boltyanskiy R, Lyashchenko SK, Deangelis LM, Mellinghoff IK, Brennan CW, Tabar V, Yang TJ, Holodny AI, Sosa RE, Guo YW, Chen AP, Tropp J, Robb F, Keshari KR, Metabolic Imaging of the Human Brain with Hyperpolarized 13C Pyrucate Demonstrates 13C Lactate Production in Brain Tumor Patients, Cancer Res. 78 (2018) 3755–3761. doi: 10.1158/0008-5472.CAN-18-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Grist JT, Mclean MA, Riemer F, Schulte RF, Deen SS, Zaccagna F, Woitek R, Daniels CJ, Kaggie JD, Matys T, Patterson I, Slough R, Gill AB, Chhabra A, Eichenberger R, Laurent M, Comment A, Gillard JH, Coles AJ, Tyler DJ, Wilkinson I, Basu B, Lomas DJ, Graves MJ, Brindle KM, Gallagher FA, Quantifying normal human brain metabolism using hyperpolarized [1–13C] pyruvate and magnetic resonance imaging, Neuroimage. 189 (2019) 171–179. doi: 10.1016/j.neuroimage.2019.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Cunningham CH, Lau JYC, Chen AP, Geraghty BJ, Perks WJ, Roifman I, Wright GA, Connelly KA, Hyperpolarized 13C Metabolic MRI of the Human Heart: Initial Experience, Circ. Res 119 (2016) 1177–1182. doi: 10.1161/CIRCRESAHA.116.309769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Maidens J, Member S, Gordon JW, Arcak M, Larson PEZ, Optimizing flip angles for metabolic rate estimation in hyperpolarized carbon-13 MRI, IEEE Trans. Med. Imaging 0062 (2016) 1–10. doi: 10.1109/TMI.2016.2574240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bottomley P, Hardy C, Argersinger R, Allen-Moore G, A review of 1H nuclear magnetic resonance relaxation in pathology: Are T1 and T2 diagnostic, Med. Phys 14 (1987) 1–37. doi: 10.1118/1.596111. [DOI] [PubMed] [Google Scholar]
- [18].Damadian R, Tumor Detection by Nuclear Magnetic Resonance, Science (80-.). 171 (1971) 1151–1153. [DOI] [PubMed] [Google Scholar]
- [19].Tofts PS, Quantitative MRI of the brain: measuring change caused by disease, John Wiley and Sons, Chichester, UK, 2003. doi: 10.1002/0470869526. [DOI] [Google Scholar]
- [20].Damadian R, Zaner K, Hor D, DiMaio T, Human Tumors Detected by Nuclear Magnetic Resonance, Proc. Natl. Acad. Sci. U. S. A 71 (1974) 1471–1473. doi: 10.1073/pnas.71.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Doneva M, Börnert P, Eggers H, Stehning C, Sénégas J, Mertins A, Compressed sensing reconstruction for magnetic resonance parameter mapping, Magn. Reson. Med 64 (2010) 1114–1120. doi: 10.1002/mrm.22483. [DOI] [PubMed] [Google Scholar]
- [22].Tamir J.l., Uecker M, Chen W, Lai P, Alley MT, Vasanawala SS, Lustig M, T2 shuffling: Sharp, multicontrast, volumetric fast spin-echo imaging, Magn. Reson. Med 77 (2017) 180–195. doi: 10.1002/mrm.26102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gowland P, Mansfield P, Accurate measurement of T1 in vivo in less than 3 seconds using echo-planar imaging, Magn. Reson. Med 30 (1993) 351–354. doi: 10.1002/mrm.1910300312. [DOI] [PubMed] [Google Scholar]
- [24].Bojorquez JZ, Bricq S, Acquitter C, Brunotte F, Walker PM, Lalande A, What are normal relaxation times of tissues at 3 T?, Magn. Reson. Imaging 35 (2017) 69–80. doi: 10.1016/j.mri.2016.08.021. [DOI] [PubMed] [Google Scholar]
- [25].Taylor AJ, Salerno M, Dharmakumar R, Jerosch-Herold M, T1 Mapping Basic Techniques and Clinical Applications, JACC Cardiovasc. Imaging 9 (2016) 67–81. doi: 10.1016/j.jcmg.2015.11.005. [DOI] [PubMed] [Google Scholar]
- [26].Jezzard P, Duewell S, Balaban RS, MR relaxation times in human brain: measurement at 4 T., Radiology. 199 (1996) 773–779. doi: 10.1148/radiology.199.3.8638004. [DOI] [PubMed] [Google Scholar]
- [27].Ma D, Gulani V, Seiberlich N, Liu K, Sunshine JL, Duerk JL, Griswold M. a., Magnetic resonance fingerprinting, Nature. 495 (2013) 187–192. doi: 10.1038/nature11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Buonincontri G, Sawiak SJ, MR fingerprinting with simultaneous B1 estimation, Magn. Reson. Med 76 (2016) 1127–1135. doi: 10.1002/mrm.26009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hamilton JI, Jiang Y, Chen Y, Ma D, Lo W-C, Griswold M, Seiberlich N, MR fingerprinting for Rapid Quantification of Myocardial T1 , T2 , and Proton Spin Density, Magn. Reson. Med 77 (2017) 1446–1458. doi: 10.1002/mrm.26216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Reed GD, Von Morze C, Bok R, Koelsch BL, Van Criekinge M, Smith KJ, Shang H, Larson PEZ, Kurhanewicz J, Vigneron DB, High Resolution 13C MRI With Hyperpolarized Urea: In Vivo T2 Mapping and 15N Labeling Effects, IEEE Trans. Med. Imaging 33 (2014) 362–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Milshteyn E, Von Morze C, Reed GD, Shang H, Shin PJ, Zhu Z, Chen H, Bok R, Goga A, Kurhanewicz J, Larson PEZ, Vigneron DB, Development of high resolution 3D hyperpolarized carbon-13 MR molecular imaging techniques, Magn. Reson. Imaging 38 (2017) 152–162. doi: 10.1016/j.mri.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Yen YF, Le Roux P, Mayer D, King R, Spielman D, Tropp J, Pauly KB, Pfefferbaum A, Vasanawala S, Hurd R, T2 relaxation times of 13C metabolites in a rat hepatocellular carcinoma model measured in vivo using 13C-MRS of hyperpolarized [1-13C]pyruvate, NMR Biomed. 23 (2010) 414–423. doi: 10.1002/nbm.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Yen Y-F, Le Roux P, Bok R, Tropp J, Chen A, Zhang V, Zierhut M, Albers M, Park I, Nelson S, Vigneron D, Kurhanewicz J, Apparent T2 of 13C-labeled Metabolites In Vivo, in: Proc. 16th Annu. Meet. ISMRM, Toronto, Canada, 2008: p. 1747. [Google Scholar]
- [34].Laustsen C, Nørlinger TS, Hansen DC, Qi H, Nielsen PM, Bertelsen LB, Ardenkjaer-larsen JH, Jørgensen HS, Hyperpolarized 13C Urea Relaxation Mechanism Reveals Renal Changes in Diabetic Nephropathy, Magn. Reson. Med 75 (2016) 515–518. doi: 10.1002/mrm.26036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hansen ESS, Stewart NJ, Wild JM, Stødkilde-jørgensen H, Laustsen C, Hyperpolarized 13C,15N2-Urea MRI for Assessment of the Urea Gradient in the Porcine Kidney, Magn. Reson. Med 76 (2016) 1895–1899. doi: 10.1002/mrm.26483. [DOI] [PubMed] [Google Scholar]
- [36].Von Morze C, Larson PEZ, Hu S, Yoshihara HA, Bok RA, Goga A, Ardenkjaer-Larsen JH, Vigneron DB, Investigating tumor perfusion and metabolism using multiple hyperpolarized 13C compounds: HP001, pyruvate and urea, Magn. Reson. Imaging 30 (2012) 305–311. doi: 10.1016/j.mri.2011.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Gordon JW, Vigneron DB, Larson PEZ, Development of a Symmetric Echo Planar Imaging Framework for Clinical Translation of Rapid Dynamic Hyperpolarized 13C Imaging, Magn. Reson. Med (2016) 1–7. doi: 10.1002/mrm.26123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Sacolick LI, Wiesinger F, Hancu I, Vogel MW, B1 mapping by Bloch-Siegert shift, Magn. Reson. Med 63 (2010) 1315–1322. doi: 10.1002/mrm.22357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lau AZ, Chen AP, Cunningham CH, Integrated Bloch-Siegert B1 mapping and multislice imaging of hyperpolarized 13C pyruvate and bicarbonate in the heart, Magn. Reson. Med 67 (2012) 62–71. doi: 10.1002/mrm.22977. [DOI] [PubMed] [Google Scholar]
- [40].Scheffler K, On the transient phase of balanced SSFP sequences, Magn. Reson. Med 49 (2003) 781–783. doi: 10.1002/mrm.10421. [DOI] [PubMed] [Google Scholar]
- [41].Reed GD, von Morze C, Verkman AS, Koelsch BL, Chaumeil MM, Lustig M, Ronen SM, Bok RA, Sands JM, Larson PEZ, Wang ZJ, Larsen JHA, Kurhanewicz J, Vigneron DB, Imaging Renal Urea Handling in Rats at Millimeter Resolution using Hyperpolarized Magnetic Resonance Relaxometry, Tomography. 2 (2016) 125–137. doi: 10.18383/j.tom2016.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Jiang Y, Ma D, Seiberlich N, Gulani V, Griswold MA, MR Fingerprinting Using Fast Imaging with Steady State Precession ( FISP ) with Spiral Readout, Magn. Reson. Med 74 (2015) 1621–1631. doi: 10.1002/mrm.25559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Bahrami N, Swisher CL, Von Morze C, Vigneron DB, Larson PEZ, Kinetic and perfusion modeling of hyperpolarized 13C pyruvate and urea in cancer with arbitrary RF flip angles., Quant. Imaging Med. Surg 4 (2014) 24–32. doi: 10.3978/j.issn.2223-4292.2014.02.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Karlsson M, Jensen PR, Ardenkjær-Larsen JH, Lerche MH, Difference between Extra- and Intracellular T1 Values of Carboxylic Acids Affects the Quantitative Analysis of Cellular Kinetics by Hyperpolarized NMR, Angew. Chemie - Int. Ed 55 (2016) 13567–13570. doi: 10.1002/anie.201607535. [DOI] [PubMed] [Google Scholar]
- [45].Cohen O, Rosen MS, Algorithm comparison for schedule optimization in MR fingerprinting, Magn. Reson. Imaging 41 (2017) 15–21. doi: 10.1016/j.mri.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Shang H, Sukumar S, Von Morze C, Bok RA, Marco-Rius I, Kerr A, Reed GD, Milshteyn E, Ohliger MA, Kurhanewicz J, Larson PEZ, Pauly JM, Vigneron DB, Spectrally Selective Three-Dimensional Dynamic Balanced Steady-State Free Precession for Hyperpolarized C-13 Metabolic Imaging With Spectrally Selective Radiofrequency Pulses, Magn. Reson. Med (2016). doi: 10.1002/mrm.26480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Le Roux P, Simplified model and stabilization of SSFP sequences, J. Magn. Reson 163 (2003) 23–37. doi: 10.1016/S1090-7807(03)00115-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Figure 1: Bland-Altman plots comparing the T1 and T2 maps from the developed approaches and comparison EPI/bSSFP maps. (A,B) Comparison of the dual module approach (T2 mapping followed by T1 mapping) T1 map and EPI T1 map (A), as well as T2 map and bSSFP T2 map (B) is shown here. The maps matched up well with each other in most voxels, although some differences occurred at long T1/T2 values due to renal filtration and some overfitting due to low SNR voxels in the dual module approach. (C,D) Comparison of the dual module approach (T1 mapping followed by T2 mapping) T1 map and EPI T1 map (C), as well as T2 map and bSSFP T2 map (D) is shown here. The maps matched up well with each other in most voxels, although some differences occurred at long T1/T2 values due to renal filtration elongating the T1/T2 over the course of the dual module acquisition. (E,F) Comparison of the modified MR fingerprinting T1 map and EPI T1 map (E), as well as T2 map and bSSFP T2 map (F) is shown here. As with A-D, the maps matched up well with each other in most voxels, although similar differences occurred at long T1/T2 values due to renal filtration and some overfitting due to low SNR voxels in the EPI acquisition for the T1 map. For all plots, the zero line (i.e. no difference between voxels), and the mean ± 1.96*standard deviation (SD) (95% confidence interval) lines are displayed.
Supporting Figure 2: Comparison of fits for a modified MR fingerprinting acquisition of HP001 with long delays (up to 9 s) and only 20 high SNR time-points available for fitting in a kidney voxel, along with resulting parameter maps. Nonlinear least squares was used for fitting. (A) The calculated fit matched up very closely with the acquired signal, although the resulting values for T1 and T2 were lower and higher, respectively, than ideal values, such as those seen in Figures 3 and 4. A plot of the simulated signal trains for combinations of T1 and T2 values show how close all curves match up to one another when only 20 time-points are available for fitting, causing the fitting problem to be poorly conditioned due to lack of signal variation. (B-D) The resulting T2, T1, and B1 maps are shown here, along with the mean and intra-map standard deviation. The mean of the T1 map was considerably lower than the maps found in Figures 3-5, while the T2 map had poor spatial smoothness owing to the high intra-map standard deviation (for comparison, Figure 2E had an intra-map standard deviation of 1.1). The difficulty in fitting illustrated in part A resulted in several voxels in the T2 map with values close 10 s, which was chosen as the upper bound. Consequently, the T1 values were shorter than expected in those voxels, indicating erroneous fits. Acquisition schemes for the modified MR fingerprinting approach with >~60-80 time-points provided fits closer to ideal values and resulted in smoother spatial variation within the parameter maps. Addition of “mini” modules, as depicted in Figure 2C, can help condition the fitting better by providing close initial estimates as well as upper and lower bounds for the fit.
Supporting Figure 3: Depiction of the simulated frequency response for different flip angles using the parameters described in the methods section (1.6 ms TBW = 4 sinc pulse played with a TR of 5.6 ms). For the simulations, θ was set to (A) 10°, (B) 20°, (C) 40°, (D) 60°, (E) 120°, and (F) 180°, which represents the range of the flip angles used in these studies. The variation within ±25 Hz was relatively minimal for both low and high flip angles, allowing us to ignore the contribution of B0 during the signal modeling.
Supporting Figure 4: Comparison of different 50% undersampled reconstructions of a random flip angle and delay acquisition. The first image from the ground truth (A), temporal subspace and low rank recon (B), model-based (via nonlinear conjugate gradient) recon (C), and zero-filled recon (D) can be seen. The dictionary described in the methods section was used for the sparsifying transform. Both recons in B and C had low nRMSE and considerably outperformed the zero-filled recon. Qualitatively, both recons removed the aliasing artifacts present in part D.
Supporting Video 1: An iteration of a T2 mapping followed by T1 mapping acquisition in a normal Sprague-Dawley rat with 2.5 x 2.5 mm2 in-plane spatial resolution is shown here along with the signal evolution for a representative kidney voxel. The delays and flip angles used in this acquisition are shown in Figure 2A and the results are shown in Figure 4. The first 16 time-points represent the T2 mapping module, while the rest of the time-points represent the T1 mapping module.
Supporting Video 2: An iteration of a T1 mapping followed by T2 mapping acquisition in a normal Sprague-Dawley rat with 2.5 x 2.5 mm2 in-plane spatial resolution is shown here along with the signal evolution for a representative kidney voxel. The delays and flip angles used in this acquisition are shown in Figure 2B and the results are shown in Figure 5. The first 20 time-points represent the T1 mapping module, while the rest of the time-points represent the T2 mapping module.
Supporting Video 3: An iteration of a modified MR fingerprinting HP001 acquisition in a normal Sprague-Dawley rat with 2.5 x 2.5 mm2 in-plane spatial resolution is shown here along with the signal evolution for a representative kidney voxel. The delays and flip angles used in this acquisition are shown in Figure 2C and the results are shown in Figures 7 and 8. The randomness of the signal evolution mimics the flip angle train in Figure 2C, with the longest signal lasting in the kidneys due to renal filtration characteristics that elongate the T1 and T2 over time. The first 8 time-points show T1 decay as part of the “mini” T1 mapping module, while the next 19 time-points (3 removed for overranging from original 22) featured show T2 decay as part of the “mini” T2 mapping module.