Abstract
We survey advances in information extraction from the free-text of electronic medical records as related to the complex domain of oncology. Current models for correlating Electronic Medical Records with –omics data largely ignore the clinical text, which remains an important source of phenotype information for cancer patients. This data convergence has the potential to enable new insights about cancer initiation, progression, metastasis, and response to treatment. Insights from this real-world data will catalyze clinical care, research, and regulatory activities. Natural language processing methods are needed to extract these rich cancer phenotypes from the clinical text. We review the advances of natural language processing and information extraction methods relevant to oncology since the Yim et al, 2016 paper in JAMA Oncology. The current survey is based on publications from PubMed as well as NLP and machine learning conference proceedings. Because of this broad catchment, the survey summarizes the main trends in natural language processing and information extraction for oncology over the last 3 years organized by task and application. Given the interdisciplinary nature of the fields of oncology and information extraction, this survey serves as a critical trail marker on the path to higher fidelity oncology phenotypes from real-world data.
Keywords: Natural Language Processing, Machine Learning, Artificial Intelligence, Oncology, Phenotyping, Electronic Medical Records
Introduction
Data produced during the processes of clinical care and research in oncology are proliferating at an exponential rate. In the past decade, use of electronic medical records (EMRs) has increased significantly in the United States,1 driven at least in part by incentivization from the Health Information Technology for Economic and Clinical Health (HITECH) Act of 2009.2 In parallel, large databases such as the National Cancer Institute’s Surveillance, Epidemiology, and End Results program (SEER),3 the National Cancer Database (NCDB),4 The Cancer Genome Atlas (TCGA)5 and the Human Tumor Atlas Network (HTAN)6 are increasingly important avenues for clinical and translational oncology research. However, significant nuanced phenotype data is locked in clinical free-text, which remains the primary form of documenting and communicating clinical presentations, provider impressions, procedural details, and management decision-making.7 Despite the proliferation of EMR and -omics data, critical and precise phenotype information is often detailed only in these clinical texts. Natural language processing (NLP), broadly defined as the transformation of language structures into computable representations, is key to large-scale extraction of nuanced data within clinical texts. As a subfield of artificial intelligence, clinical NLP (cNLP), which refers to the analysis of clinical or healthcare texts (as opposed to clinical application, per se) has been around for decades. However, only in recent years have compute power and algorithms advanced sufficiently to demonstrate its power towards broadening oncologic investigation.
There are excellent prior review papers of cNLP. Spyns8 covers the period before 1995. Meystre et al9 survey the 1998-2008 developments. Yim et al10 provide an overview with a special emphasis on oncology for the period of 2008-2016. Neveol et al11 offer a first broad overview of cNLP for languages other than English. These surveys capture three distinct methodology phases in NLP – from exclusively rule-based systems through the shift towards probabilistic methods to the dominance of machine learning. Kreimeyer et al 12 review existing cNLP systems. Some popular cNLP systems are MetaMap13,14 (concept mapping), Apache cTAKES15,16 (classic NLP components, concept mapping, entities and attributes, relations, temporality), YTex17 (entity and attributes), OBO annotator18 (concept mapping), TIES19 (linking of pathology reports to tissue bank data), MedLEE20 (entities and attributes, relations), CLAMP21 (entities and attributes), NOBLE (entities and attributes)22.
The mid-2010s mark a transformational milestone for the field where plentiful digitized textual data and hardware advances met powerful mathematical abstractions in a super connected world that led to the explosive interest in general artificial intelligence (e.g. autonomous cars) and NLP in particular (e.g. Google translator™, Apple Inc.’s Siri®, movie recommenders). Herein, we review major recent developments in cNLP methods for cancer since that watershed point. We discuss their applications for translational investigation and future directions. We cover publications since the 2016 review by Yim et al10, which are: (1) focused on cNLP of EMR text related to cancer, (2) peer-reviewed, (3) published in English and use English EMR text, (4) sourced from MEDLINE and major computational linguistics and machine learning venues – the annual conferences of the Association of Computational Linguistics, North American Association of Computational Linguistics, European Association of Computational Linguistics, Empirical Methods for Natural Language Processing, International Conference on Machine Learning, Neural Information Processing Systems Conference, Machine Learning for Healthcare, SemEval, International Conference for High Performance Computing, and IEEE International Conference on Biomedical Health Informatics. Our goal is to highlight recent exceptional papers with implications for the broader cancer research community; thus, this survey is not a systematic meta-review. We acknowledge that much work is taking place outside traditional academic environments (i.e., industry) and we attempt to include it to the extent it meets this survey’s inclusion criteria. For ease of reading, terms and definitions are presented in Table 1.
Table 1:
Terms and definitions
| Term | Definition |
|---|---|
| Accuracy | Where TP is true positive; TN is true negative; FP is false positive; and FN is false negative. |
| Artificial intelligence | A process through which machines mimic “cognitive” functions that humans associate with other human minds, such as language comprehension. |
| Area under the curve (AUC) | A metric of binary classification; range from 0 to 1, 0 being always wrong, 0.5 representing random chance, and 1, the perfect score. |
| Artificial neural network | Computing systems that are inspired by, but not necessarily identical to, the biological neural networks that constitute human brain. |
| Attribute | Facts, details or characteristics of an entity. |
| Autoencoder | A class of artificial neural networks. |
| Concept mapping | A diagram that depicts suggested relationships between concepts. |
| Convolutional neural network | A class of artificial neural networks. |
| Decision tree | A tree-like graph or model of decisions and their possible consequences, including chance event outcomes, resource costs, and utility. |
| Deep learning | A subclass of a broader family of machine learning methods based on artificial neural networks. The designation “deep” signifies multiple layers of the neural network |
| Entities | A person, place, thing or concept about which data can be collected. Examples in the clinical domain include diseases/disorders, signs/symptoms, procedures, medications, anatomical sites |
| F1 score | Values range from 0 to 1 (perfect score) |
| Graphics processing unit | A specialized electronic circuit designed to perform very fast calculations needed for training artificial neural networks. |
| K-nearest neighbors | A non-parametric method used for classification and regression in pattern recognition |
| Latent representation | Word representations that are not directly observed but are rather inferred through a mathematical model |
| Machine learning | The scientific study of algorithms and probabilistic models that computer systems use in order to perform a specific task effectively without using explicit instructions, relying on patterns and inference instead |
| Precision | Where TP is true positive, and FP is false positive. |
| Probabilistic methods | A nonconstructive method, primarily used in combinatorics, for proving the existence of a prescribed kind of mathematical object |
| Recall | Where TP is true positive, and FN is false negative. |
| Recurrent neural network | A class of artificial neural networks |
| Rule-based system | Systems involving human-crafted or curated rule sets. |
| Semantic representation | Ways in which the meaning of a word or sentence is interpreted. |
| Supervised learning | Machine learning method that infers a function from labeled training data consisting of a set of training examples. |
| Support vector machine | Supervised learning models with associated learning algorithms that analyze data used for classification and regression analysis. |
| tensor | A mathematical object analogous to but more general than a vector, represented by an array of components that are functions of the coordinates of a space. |
| Transfer learning | A machine learning technique where a model trained on one task is re-purposed on a second related task. |
| Unsupervised learning | Self-organized Hebbian learning that helps find previously unknown patterns in data set without pre-existing labels. |
| Word embedding | The collective name for a set of language modeling and feature learning techniques in natural language processing (NLP) where words or phrases from the vocabulary are mapped to vectors of real numbers. |
We highlight results measured in either accuracy, harmonic mean of recall/sensitivity and precision/positive predictive value (F1 score), or area under the curve (AUC) (trade-off between true positive and false positive rates). These performance metrics reflect a comparison against human-generated data (referred to as gold-standard annotations), thus they capture agreement between NLP systems and humans. Gold-standard annotations are also used for training algorithms (supervised learning). The inter-annotator agreement (IAA) measures human performance and serves as a system performance target.
Major NLP algorithmic advances
The last three years have shown the development of a variety of methodologies for NLP with a general shift towards a particular machine learning category -- deep learning (DL)23. DL techniques were initially conceived in the 1980’s but not operationalized until the convergence of three critical elements: massive digital text corpora, novel but compute and data intensive algorithms, and powerful, massively parallel computing architectures currently using graphics processing units (GPUs).24 For many tasks, DL is considered state-of-the-art in artificial intelligence.25–27 The key differentiator between DL and feature-rich machine learners is the concept of representation learning.28 Feature-rich algorithms require expert knowledge – linguistic, semantic, biomedical, or world -- to determine the information of interest. Some examples of feature-rich learners are support vector machines (SVM) and random forests (RF).29 In the clinical domain, the engineered features are often guided by biomedical dictionaries, clinical ontologies, or biomedical knowledge from domain experts. Instead, DL models automatically discover mathematically and computationally convenient abstractions from raw data needed for classification without the need for explicitly defined features.23,25 These representations can range from simple word representations and word embeddings30 to complex hierarchies that capture contextual meaning and relationships between words, phrases, and other compositional derivatives. This capability of DL algorithms can potentially unmask unknown relationships buried within large quantities of data, which can be particularly advantageous in cancer research and practice.25 Furthermore, DL algorithms can uniquely take advantage of transfer learning26, the ability to learn from data not in the target domain, and then apply this knowledge to other domains. For example, one DL model may be trained on large, openly available non-medical text data (e.g., Wikipedia), and then this model’s knowledge is applied effectively in cNLP tasks through fine tuning the model’s parameters on smaller but directly relevant clinical text corpora.
Most DL architectures are built on the artificial neural network with interconnected nodes (neurons) arranged in layers.23 The variations in the arrangement and interconnections of these layers result in various elaborate networks, or architectures, suitable for addressing a variety of tasks. The most popular among these include: convolutional neural networks (CNNs), optimal for data where spatial relationships encode critical information; recurrent neural networks (RNNs), advantageous for sequentially ordered data (e.g. time-series data); and autoencoders, suitable for learning problems from noisy data, or data where prior information about data are partially or entirely unknown.23 There is a substantial amount of research in the general (as opposed to clinical) application of DL, demonstrating its potential in NLP31.
Linguistic variability, combined with the abundance of medical terminology, abbreviations, synonyms, jargon, and spelling inconsistencies prevalent in clinical text, make cNLP a particularly challenging problem. DL has shown remarkable results in extracting low- and high-level abstractions from raw text data with semantic and syntactic capabilities. This ability is often accompanied by excellent performance across translational science applications 25,32 and as highlighted below.
Latest cNLP application developments
Task: Extracting temporality and timelines
Longitudinal representations of patients’ cancer journeys are a cornerstone of translational research enabling rich studies across variables (e.g. tumor molecular profile) and outcomes (e.g. treatment efficacy). Extracting timelines from the EMR free-text has become a line of cNLP research on its own. Since 2016, under the auspices of SemEval, Clinical TempEval shared tasks have challenged the NLP research community to establish state-of-the-art methods and results for temporal relation extraction with a focus on oncology. The dataset for these shared tasks consists of 400 cancer patients distributed evenly between colon and brain cancers, each represented by pathology, radiology and clinical notes (the THYME corpus described in 33 and available at 34). The tasks consisted of identifying event expressions, time expressions, and temporal relations (see Fig 1 for an example). The relation between the event and the document creation time is called DocTimeRel with values of BEFORE, OVERLAP, BEFORE-OVERLAP and AFTER which provide a course-level temporal positioning on a timeline.
Figure 1.
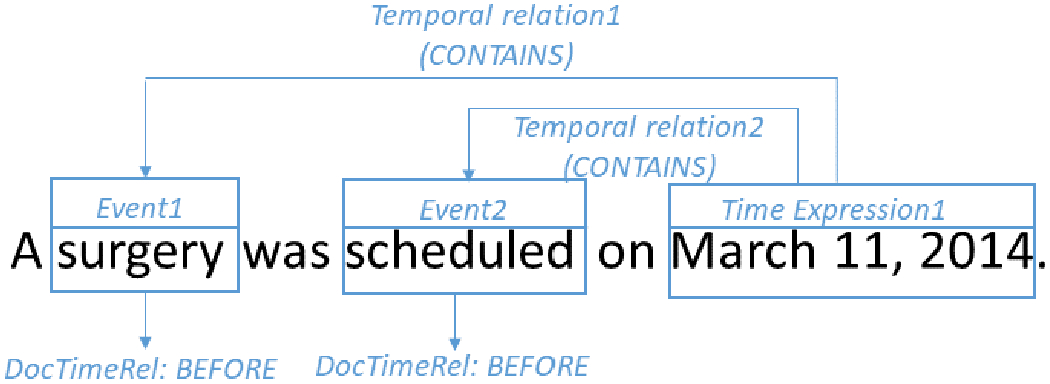
Clinical TempEval example: two events, one time expression, two temporal relations, two relations to the document creation time (DocTimeRel).
Clinical TempEval 201635 focused on developing methods from colon cancer EMR data and testing on colon cancer data (within-domain evaluation). The results suggest that current state-of-the-art systems perform extremely well on most event- and time expression- related tasks -- gap between system performance and IAA (or human performance) < 0.05 F1. However, the temporal relation tasks remained a challenge. Systems that predict DocTimeRel relation lagged about 0.09 F1 behind IAA. For other types of temporal relations, systems lagged about 0.25 F1 behind IAA.
Clinical TempEval 201736 addressed the question of how well systems trained on one cancer medical domain (colon cancer) perform in predicting timelines in another cancer medical domain (brain cancer). The results showed that developing clinical timeline extraction methods that generalize across cancer domains is an open research question. Across the board, there was a 0.20+ F1 drop in performance when systems were trained on colon cancer and tested on brain cancer. Providing a small amount of target domain (brain cancer) training data improved performance.
Methods employed by the Clinical TempEval participants are wide ranging -- from classic methods (logistic regression, conditional random fields, SVMs, pattern matching) to various architectures of latest DL techniques (RNNs, CNNs with inputs of word and character embeddings). Clinical TempEval 2017 showed there was no one specific method that provides the best results, although the combination of various approaches appeared a promising path.
Outside of Clinical TempEval, experimentation with advanced DL architectures and various data streams for timeline extraction of cancer patient EMRs has intensified. Tourille et al. explored neural networks and domain adaptation strategies37. Chen et al.38 and Dligach et al.39 dealt with simplifications of time expression representations in a neural approach. Some latest trends include DL models which combine a small portion of the labeled THYME data with unlabeled publicly available data (Google News30 and social media) to achieve results about 0.02 F1 below IAA40. The current best reported result is 0.684 F141.
Open source systems for timeline extraction include Apache cTAKES temporal module42, Heidel-Time43 (for temporal expressions and their normalization), and rule-based extensions of Stanford CoreNLP44.
The task of extracting temporality from EMR clinical narrative has advanced dramatically since 2016. In the last three years, the performance on the Clinical TempEval test set moved from 0.573 to 0.684 F1 for finer grained temporal relations and reached 0.835 F1 for DocTimeRel. This last result enables exploring select temporally sensitive applications such as outcomes extraction which was pointed out as one of the most challenging yet to be addressed use cases in the 2016 survey paper.
Application: Extracting Tumor and Cancer Characteristics
Information extraction from pathology reports, which have a more consistent structure than other free text EMR documents, presents a tractable challenge to the field of cNLP.45 Since the 2016 survey, the oncology NLP field has moved beyond cancer stage and TNM extraction into the extraction of more comprehensive cancer and tumor attributes. Qiu et al.46 presented a CNN for information abstraction of primary cancer site topography from breast and lung cancer pathology reports from the Louisiana Cancer Registry, reporting 0.72 F1. Using the same corpus, Gao et al.47 boosted performance using a more elaborate DL architecture (hierarchical attention neural network). The authors reported 0.80 F1 for cancer site topography and 0.90 F1 for histological grade. However, the authors noted significant computational demands of their DL solution.
Alawad et al.48 showed that for extraction of cancer primary site, histological grade, and laterality, training CNN to make multiple predictions simultaneously (multi-task learning) outperformed single task models. In a later study, the authors explored the computational demands of CNN cNLP models and the role of high-performance computing for achieving population-level automated coding of pathology documents to achieve near real-time cancer surveillance for cancer registry development.49 Using a corpus of 23,000 pathology reports, they reported 0.84 F1 for primary cancer site extraction across 64 cancer sites using their CNN model, significantly outperforming a random forest classifier with 0.76 F1.
Yala et al.50 used boosting51 to extract tumor information from breast pathology reports and achieved 90% accuracy for extracting carcinoma and atypia categories. Since gold-standard datasets are a necessary but resource-intensive requirement of ML algorithms, this study also investigated the minimum number of annotations needed to maintain at least 0.9 F1 without the system being pretrained. They reported this to be approximately 400. Using similar methods, Acevedo et al.52 found the rate of abnormal findings in asymptomatic patients to be 7%, and to increase with age. These results are higher than previously reported, suggesting the clinical value of these algorithms over current epidemiologic methods to measure cancer incidence and prevalence. In a study of multiple diseases, Gehrmann et al. 25 reported an improvement in F1 score and AUC for advanced cancer using CNNs over rule-based systems.
The open source DeepPhe platform53, 54 is a hybrid system for extracting a number of tumor and cancer attributes. It implements a variety of artificial intelligence approaches – rules, domain knowledge bases, machine learning (feature-rich and DL) – to crawl the entire cancer patient chart (not restricted to pathology notes), extract and summarize the information related to tumors and cancers and their characteristics. The IAA ranged from 0.46 to 1.00 F1, and system agreement with humans ranged from 0.32 to 0.96 F1. System highest result is on primary site extraction (0.96 F1); lowest – PR method extraction (0.32 F1).
Castro et al55 developed an NLP system to annotate and classify all BI-RADS mentions present in a single radiology report which can serve as the foundation for future studies that will leverage automated BI-RADS annotation, providing feedback to radiologists as part of a learning health system loop 56.
Application: Clinical Trials Matching
Clinical trials determine safety and effectiveness of new medical treatments; with the successes of recent years including new classes of therapies (e.g., immunotherapy; CAR-T cells), the clinical trial landscape has exploded. Nevertheless, adult patient participation in clinical trials remains low, especially among underrepresented minorities. This limits trial completion, generalizability, and interpretation of trial findings. Thus, there is a great deal of interest in clinical trial matching. This is not a simple problem, given the need to extract information from trial protocols written in natural language and match the findings with characteristics from individual EMRs.
Since the 2016 survey paper10, researchers have explored DL technology to identify relevant information found in patients’ EMRs to establish eligibility for clinical trials. Bustos et al. developed a CNN, leveraging its representation learning capability, to extract medical knowledge reflecting eligibility criteria from clinical trials57. They reported promising results using CNNs compared to state-of-the-art classification algorithms including FastText58, SVM, and k-Nearest Neighbors (kNN). Shivade et al.59, and Zhang et al.60 developed SVMs to automate the classification of eligibility criteria to facilitate trial matching for specific patient populations.
Yala et al.50 and Osborne et al.61 used Boostexter62 and MetaMap13,14 respectively on rule-based regular expressions to automatically extract relevant patient information from EMRs, predominantly free-text reports, to identify patient cohorts with characteristics of interest for clinical trials or other relevant reporting. There are also a panoply of commercial solutions emerging in this space, but our search did not reveal any publications by these commercial entities.
Application: Pharmacovigilance and Pharmacoepidemiology
Pharmacovigilance, drug-safety surveillance, and factors associated with non-adherence play an important role in improving patient outcomes by personalizing cancer treatments, monitoring and understanding adverse drug events (ADEs) as well as minimizing risks associated with different therapies. The 2016 survey paper identifies outcomes extraction as one of the challenges for cNLP because temporality extraction plays a key role. With the advances in temporality extraction in the last three years (see section Extracting Temporality and Timelines), methods for outcomes extraction have also improved.
A variety of methods have been explored including Logistic Regression, SVM, Random Forest, Decision Tree, and DL to analyze EMR data to predict treatment prescription, quality of care, and health outcomes of cancer patients. Using data from the SEER3 cancer registry as gold-standard for cancer stages, and variables extracted from linked Medicare claims data, Bergquist et al.63 classified lung cancer patients receiving chemotherapy into different stages of severity, with a hybrid method of rules and ensemble ML algorithms. This system achieved 93% accuracy demonstrating its potential applications to study the quality of care for lung cancer patients and health outcomes.
Survival analysis plays an important role for clinical decision support. In oncology care the choice of treatment depends greatly on prognosis, sometimes difficult for physicians to determine. Gensheimer et al.64 proposed a hybrid pipeline that combines semantic data mining with neural embeddings of sequential clinical notes and outputs a probability of >3 months life expectancy.
Yang et al.65 applied a tensorized RNN on sequential clinical records to extract a latent representation from the entire patient history, and used it as the input to an Accelerated Failure Time model to predict the survival time of metastatic breast cancer patients. Yin et al.66 applied word embeddings to discover topics in patient-provider communications associated with an increased likelihood of early treatment discontinuation in the adjuvant breast cancer setting. Overall, treatment toxicity extraction remains an open research area.
Shareable Resources for NLP in Oncology
Recent years have seen cancer cNLP tasks tackled occasionally at mainstream NLP conferences and affiliated workshops (in open-domain NLP, top research is preferentially presented at conferences). While still relatively rare, this has the potential to greatly benefit cancer cNLP research, with a larger community of NLP researchers working directly on these problems in addition to the more specialized cNLP community. The prerequisite for this trend to continue is access to shareable data resources as also pointed out in the 2016 survey paper. The colon and brain cancer THYME corpus was used in several general domain conference and workshop papers,37,38,40,67–69 while a radiology report dataset from a 2007 challenge (available at 70) was used in another,71 and SEER-provided (though unshared thus not available for distribution) corpus was used in yet another.72 Other work using ad hoc resources has been used for methods development but this is a less sustainable model due to the rarity of expertise in both cancer and NLP.73–75 A recently developed resource created gold-standard annotations of the semantics of sentences in notes describing patients with cancer.76 More shared resources, community challenges, and publicity for both, will likely lead to more focused development of new methods for cancer information extraction - a challenge that the community needs to address.
Application at the point of care
The focus of our survey paper is on NLP technologies for cancer translational studies. However, we briefly review the applications of these technologies for direct patient care which has rightfully proceeded with caution given that even small system error rates could lead to harm. Lee et al77 studied concordance of IBM Watson for Oncology®, a commercial NLP-based treatment recommendation system, with the recommendations of local experts and it was 48.9%. Similar results are reported in 78, 79 . Furthermore, such applications are treated as Software as Medical Device (SaMD) by the US Food and Drug Administration which, justifiably, is a high bar to clear. 80,81. Some cautious use cases provide assistance to physicians82, 83 in the form of question-answering and summarization. Voice tools in healthcare, which represent a distinct sub-domain of NLP, are primarily used for (1) documentation, (2) commands, and (3) interactive response and navigation to patients.84
Implications and future directions
As discussed above, NLP technology for cancer has made strides since the 2016 survey paper which states that at that time “oncology-specific NLP is still in its infancy”. Given the breadth and depth of the research we surveyed in the current manuscript, we believe the field has expanded enabled by state-of-the-art methods and abundant digital EMR data. We observe more collaborations between NLPers and oncologists which was one of the take-away lessons from Yim et al.
State-of-the-art machine learning methods require significant amounts of human-labeled data to learn from, which is expensive and time-consuming. This presents a methodological challenge towards learning paradigms from vast unlabeled datasets (lightly supervised or unsupervised methods). Another challenge lies in the portability of the machine learners as they represent the distributions of the data they learned from. If translated to a domain with a different distribution (e.g. colorectal to brain cancer), there is a substantial drop in performance (see section Extracting Temporality and Timelines)). Thus, domain adaptation remains an unsolved and hot scientific problem. Large scale translational science is likely to cross country boundaries and harvest data from EMRs written in a variety of languages. Therefore, the cNLP research community needs to think about multi-lingual machine learning to enable such bold studies. On the hardware side, DL methods require vast computational resources available only to a very few and not necessarily solvable by a cloud computing environment. Last but not least, ethical considerations of the application of these powerful technologies should be discussed, at the bare minimum whether the underlying data on which machine learners are trained represents the whole of human diversity.
In research, real-world big data has great potential to improve cancer care. Gregg et al present a risk stratification research for prostate cancer85. The utilization of real-world big data is a key focus area of the National Cancer Institute.86 SEER and NCDB, the two major cancer registry databases in the United States, have limitations in terms of coverage, accuracy, and granularity that introduce bias. 3,4,87,88,89,90 Currently, database building requires manual annotation of clinical free-text, which is resource intensive and prone to human error. cNLP can support more rapid, large-scale, and standardized database development. Automated, semi-automated and accurate identification of cancer cases will be particularly helpful in studying underrepresented patient populations and rare cancers. Additionally, cNLP can facilitate analysis of unstructured data that are poorly documented in databases but widely accepted to be critical for prognostication and management decision-making, most notably patient-reported outcomes.91 Our hope is that larger, more accurate, and granular clinical databases can be integrated with -omics databases to enable translational research to better understand oncologic phenotype relationships. This data convergence has the potential to enable new insights about cancer initiation, progression, metastasis, and response to treatment.
While NLP has yet to make major inroads in the clinical setting, some of the potential applications are clear. Direct extraction of cancer phenotypes from source data (pathology and radiology reports) could reduce redundancy and prevent ambiguity within a patient’s chart, minimizing confusion and medical errors. Summarization and information retrieval applications can reduce search burden and enable clinicians to spend more time with their patients. Clinical decision support tools could help reduce the increasingly burdensome cognitive load placed on clinicians, although the results reported thus far by efforts such as IBM Watson for Oncology® raise serious concerns about what the bar for accuracy of clinical recommendations should be for routine use. In fact, these results are a cautionary tale of the challenges of domain adaptation – the software was widely reported to have been trained on hypothetical cases at a highly specialized cancer center, leading to incorrect and possibly unsafe recommendations92. At this time, NLP technology is not yet ripe for direct patient care except in carefully observed scenarios.
Conclusion
cNLP has the potential to affect almost all aspects of the cancer care continuum, and multidisciplinary collaboration is necessary to ensure optimal advancement of the field. As there are few individuals with expertise in both oncology and NLP, clinical oncologists, basic and translational scientists, bioinformaticians, and epidemiologists should work with computer scientists to identify and prioritize the most important clinical questions and tasks that can be addressed with this technology. Further, oncology subject matter experts will be needed to create gold datasets. Once an NLP technology is developed, oncologists and cancer researchers should take a primary role in evaluating it to determine its utility for research and their clinical value. While standards for clinical evaluation of software, including artificial intelligence systems, are evolving,93 NLP tools that directly affect management decisions should be considered for evaluation in a trial setting by clinical investigators familiar with the technology and FDA guidelines80. In partnership, computer scientists, oncology researchers, and clinicians can take full advantage of the recent advances in NLP technology to fully leverage the wealth of data stored and rapidly accumulating in our EMRs.
Acknowledgements
The work was supported by funding from 1U24CA184407(NCI), U01CA231840 (NCI), UG3 CA243120 (NCI), R01LM010090 (NLM) and R01GM114355 (NIGMS).
This work has been supported in part by the Joint Design of Advanced Computing Solutions for Cancer (JDACS4C) program established by the U.S. Department of Energy (DOE) and the National Cancer Institute (NCI) of National Institutes of Health. This work was performed under the auspices of the U.S. Department of Energy by Argonne National Laboratory under Contract DE-AC02-06-CH11357, Lawrence Livermore National Laboratory under Contract DE-AC52-07NA27344, Los Alamos National Laboratory under Contract DE-AC5206NA25396, and Oak Ridge National Laboratory under Contract DE-AC05-00OR22725.
This manuscript has been authored by UT-Battelle, LLC under Contract No. DE-AC05-00OR22725 with the U.S. Department of Energy. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a non-exclusive, paid-up, irrevocable, world-wide license to publish or reproduce the published form of the manuscript, or allow others to do so, for United States Government purposes. The Department of Energy will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan (http://energy.gov/downloads/doe-public-access-plan).
Footnotes
Conflict of Interest Statement: none
References
- 1.Cohen MF Impact of the HITECH financial incentives on EHR adoption in small, physician-owned practices. Int. J. Med. Inf 94, 143–154 (2016). [DOI] [PubMed] [Google Scholar]
- 2.H.R. 1 (111th): American Recovery and Reinvestment Act of 2009 -- House Vote #46 -- Jan 28, 2009. GovTrack.us; Available at: https://www.govtrack.us/congress/votes/111-2009/h46. (Accessed: 11th February 2019) [Google Scholar]
- 3.Surveillance, Epidemiology, and End Results Program. SEER; Available at: https://seer.cancer.gov/index.html. (Accessed: 11th February 2019) [Google Scholar]
- 4.National Cancer Database. American College of Surgeons; Available at: https://www.facs.org/quality-programs/cancer/ncdb. (Accessed: 11th February 2019) [Google Scholar]
- 5.The Cancer Genome Atlas Home Page. The Cancer Genome Atlas - National Cancer Institute; (2011). Available at: https://cancergenome.nih.gov/. (Accessed: 11th February 2019) [Google Scholar]
- 6.Human Tumor Atlas Network (HTAN), https://www.cancer.gov/research/key-initiatives/moonshot-cancer-initiative/implementation/human-tumor-atlas.
- 7.Rosenbloom ST et al. Data from clinical notes: a perspective on the tension between structure and flexible documentation. J. Am. Med. Inform. Assoc. JAMIA 18, 181–186 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spyns P Natural language processing in medicine: an overview. Methods Inf. Med 35, 285–301 (1996). [PubMed] [Google Scholar]
- 9.Meystre SM, Savova GK, Kipper-Schuler KC & Hurdle JF Extracting information from textual documents in the electronic health record: a review of recent research. Yearb. Med. Inform 128–144 (2008). [PubMed] [Google Scholar]
- 10.Yim W-W, Yetisgen M, Harris WP & Kwan SW Natural Language Processing in Oncology: A Review. JAMA Oncol. 2, 797–804 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Névéol A, Dalianis H, Velupillai S, Savova G & Zweigenbaum P Clinical Natural Language Processing in languages other than English: opportunities and challenges. J. Biomed. Semant 9, 12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kreimeyer K et al. Natural language processing systems for capturing and standardizing unstructured clinical information: A systematic review. J. Biomed. Inform 73, 14–29 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aronson AR Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc. AMIA Symp. 17–21 (2001). [PMC free article] [PubMed] [Google Scholar]
- 14.Aronson AR & Lang F-M An overview of MetaMap: historical perspective and recent advances. J. Am. Med. Inform. Assoc. JAMIA 17, 229–236 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Savova GK et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J. Am. Med. Inform. Assoc 17, 507–513 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Apache cTAKES; ctakes.apache.org.
- 17.Garla V et al. The Yale cTAKES extensions for document classification: architecture and application. J. Am. Med. Inform. Assoc. JAMIA 18, 614–620 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.OBO Foundry. (2015) http://www.obofoundry.org.
- 19.TIES v5 - Clinical Text Search Engine. http://ties.upmc.com/index.html.
- 20.Friedman C A broad-coverage natural language processing system. Proc. AMIA Symp. 270–274 (2000). [PMC free article] [PubMed] [Google Scholar]
- 21.Soysal E et al. CLAMP - a toolkit for efficiently building customized clinical natural language processing pipelines. J. Am. Med. Inform. Assoc. JAMIA (2017). doi: 10.1093/jamia/ocx132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tseytlin E et al. NOBLE – Flexible concept recognition for large-scale biomedical natural language processing. BMC Bioinformatics 17, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodfellow Ian, Yoshua B, & Courville A Deep Learning. (MIT Press, 2016). [Google Scholar]
- 24.Rumelhart DE, Hinton GE & Williams RJ Learning representations by back-propagating errors. Nature 323, 533 (1986). [Google Scholar]
- 25.Gehrmann S et al. Comparing deep learning and concept extraction based methods for patient phenotyping from clinical narratives. PLOS ONE 13, e0192360 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Young T, Hazarika D, Poria S & Cambria E Recent trends in deep learning based natural language processing. Ieee Comput. Intell. Mag 13, 55–75 (2018). [Google Scholar]
- 27.Goldberg Y A primer on neural network models for natural language processing. J. Artif. Intell. Res 57, 345–420 (2016). [Google Scholar]
- 28.Bengio Y, Courville A & Vincent P Representation Learning: A Review and New Perspectives. ArXiv12065538 Cs (2012). [DOI] [PubMed] [Google Scholar]
- 29.Manning CD, Raghavan P & Schütze H Introduction to Information Retrieval. (Cambridge University Press, 2008). [Google Scholar]
- 30.Mikolov T, Sutskever I, Chen K, Corrado GS & Dean J Distributed Representations of Words and Phrases and their Compositionality in Advances in Neural Information Processing Systems 26 (eds. Burges CJC, Bottou L, Welling M, Ghahramani Z & Weinberger KQ) 3111–3119 (Curran Associates, Inc., 2013). [Google Scholar]
- 31.LeCun Y, Bengio Y & Hinton G Deep learning. Nature 521, 436–444 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Banerjee I et al. Comparative effectiveness of convolutional neural network (CNN) and recurrent neural network (RNN) architectures for radiology text report classification. Artif. Intell. Med (2018). doi: 10.1016/j.artmed.2018.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Styler WF et al. Temporal Annotation in the Clinical Domain. Trans. Assoc. Comput. Linguist 2, 143–154 (2014). [PMC free article] [PubMed] [Google Scholar]
- 34.THYME corpus, available through hNLP Center membership. at https://healthnlp.hms.harvard.edu/center/pages/data-sets.html.
- 35.Bethard S et al. SemEval-2016 Task 12: Clinical TempEval. in Proceedings of the 10th International Workshop on Semantic Evaluation (SemEval-2016) 1052–1062 (Association for Computational Linguistics, 2016). [Google Scholar]
- 36.Bethard S, Savova G, Palmer M & Pustejovsky J SemEval-2017 Task 12: Clinical TempEval. in Proceedings of the 11th International Workshop on Semantic Evaluation (SemEval-2017) 565–572 (Association for Computational Linguistics, 2017). [Google Scholar]
- 37.Tourille J, Ferret O, Neveol A & Tannier X Neural Architecture for Temporal Relation Extraction: A Bi-LSTM Approach for Detecting Narrative Containers. in Proceedings of the 55th Annual Meeting of the Association for Computational Linguistics (Volume 2: Short Papers) 224–230 (Association for Computational Linguistics, 2017). [Google Scholar]
- 38.Lin C, Miller T, Dligach D, Bethard S & Savova G Representations of Time Expressions for Temporal Relation Extraction with Convolutional Neural Networks. in BioNLP 2017 322–327 (Association for Computational Linguistics, 2017). [Google Scholar]
- 39.Dligach D, Miller T, Lin C, Bethard S & Savova G Neural Temporal Relation Extraction. in Proceedings of the 15th Conference of the European Chapter of the Association for Computational Linguistics: Volume 2, Short Papers 746–751 (Association for Computational Linguistics, 2017). [Google Scholar]
- 40.Lin C et al. Self-training improves Recurrent Neural Networks performance for Temporal Relation Extraction. in Proceedings of the Ninth International Workshop on Health Text Mining and Information Analysis 165–176 (Association for Computational Linguistics, 2018). [Google Scholar]
- 41.Lin Chen, Miller Timothy, Dligach Dmitriy, Bethard Steven & Savova Guergana. A BERT-based Universal Model for Both Within- and Cross-sentence Clinical Temporal Relation Extraction. in Clinical NLP Workshop (2019). [Google Scholar]
- 42.Lin C, Dligach D, Miller TA, Bethard S & Savova GK Multilayered temporal modeling for the clinical domain. J. Am. Med. Inform. Assoc. JAMIA 23, 387–395 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strötgen J & Gertz M Multilingual and cross-domain temporal tagging. Lang. Resour. Eval 47, 269–298 (2013). [Google Scholar]
- 44.Manning C et al. The Stanford CoreNLP Natural Language Processing Toolkit. in Proceedings of 52nd Annual Meeting of the Association for Computational Linguistics: System Demonstrations 55–60 (Association for Computational Linguistics, 2014). doi: 10.3115/v1/P14-5010 [DOI] [Google Scholar]
- 45.Liu K, Hogan WR & Crowley RS Natural Language Processing methods and systems for biomedical ontology learning. J. Biomed. Inform 44, 163–179 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qiu JX, Yoon H-J, Fearn PA & Tourassi GD Deep Learning for Automated Extraction of Primary Sites From Cancer Pathology Reports. IEEE J. Biomed. Health Inform 22, 244–251 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Gao S et al. Hierarchical attention networks for information extraction from cancer pathology reports. J. Am. Med. Inform. Assoc. JAMIA (2017). doi: 10.1093/jamia/ocx131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alawad M, Yoon H & Tourassi GD Coarse-to-fine multi-task training of convolutional neural networks for automated information extraction from cancer pathology reports. in 2018 IEEE EMBS International Conference on Biomedical Health Informatics (BHI) 218–221 (2018). doi: 10.1109/BHI.2018.8333408 [DOI] [Google Scholar]
- 49.HPC-Based Hyperparameter Search of MT-CNN for Information Extraction from Cancer Pathology Reports. Available at: https://sc18.supercomputing.org/proceedings/workshops/workshop_pages/ws_cafcw107.html. (Accessed: 12th February 2019)
- 50.Yala A et al. Using machine learning to parse breast pathology reports. Breast Cancer Res. Treat 161, 203–211 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Schapire RE The Boosting Approach to Machine Learning: An Overview. (2002).
- 52.Acevedo F et al. Pathologic findings in reduction mammoplasty specimens: a surrogate for the population prevalence of breast cancer and high-risk lesions. Breast Cancer Res. Treat (2018). doi: 10.1007/s10549-018-4962-0 [DOI] [PubMed] [Google Scholar]
- 53.Savova GK et al. DeepPhe: A Natural Language Processing System for Extracting Cancer Phenotypes from Clinical Records. Cancer Res. 77, e115–e118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Public release of the DeepPhe analytic software. (DeepPhe, 2019). [Google Scholar]
- 55.Castro SM et al. Automated annotation and classification of BI-RADS assessment from radiology reports. J. Biomed. Inform 69, 177–187 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chandran UR et al. TCGA Expedition: A Data Acquisition and Management System for TCGA Data. PLoS ONE 11, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bustos A & Pertusa A Learning Eligibility in Cancer Clinical Trials using Deep Neural Networks. Appl. Sci 8, 1206 (2018). [Google Scholar]
- 58.Joulin A, Grave E, Bojanowski P & Mikolov T Bag of Tricks for Efficient Text Classification. ArXiv160701759 Cs (2016). [Google Scholar]
- 59.Shivade C, Hebert C, Regan K, Fosler-Lussier E & Lai AM Automatic data source identification for clinical trial eligibility criteria resolution. AMIA Annu. Symp. Proc. AMIA Symp. 2016, 1149–1158 (2016). [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang K & Demner-Fushman D Automated classification of eligibility criteria in clinical trials to facilitate patient-trial matching for specific patient populations. J. Am. Med. Inform. Assoc. JAMIA 24, 781–787 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Osborne JD et al. Efficient identification of nationally mandated reportable cancer cases using natural language processing and machine learning. J. Am. Med. Inform. Assoc. JAMIA 23, 1077–1084 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schapire RE & Singer Y BoosTexter: A boosting-based system for text categorization. Mach. Learn 39, 135–168 (2000). [Google Scholar]
- 63.Bergquist SL Classifying Lung Cancer Severity with Ensemble Machine Learning in Health Care Claims Data. 14. [PMC free article] [PubMed]
- 64.Gensheimer MF et al. Automated Survival Prediction in Metastatic Cancer Patients Using High-Dimensional Electronic Medical Record Data. J. Natl. Cancer Inst (2018). doi: 10.1093/jnci/djy178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang Y, Fasching PA & Tresp V Modeling Progression Free Survival in Breast Cancer with Tensorized Recurrent Neural Networks and Accelerated Failure Time Models. 13
- 66.Yin Z et al. The therapy is making me sick: how online portal communications between breast cancer patients and physicians indicate medication discontinuation. J. Am. Med. Inform. Assoc 25, 1444–1451 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lin C, Miller T, Dligach D, Bethard S & Savova G Improving temporal relation extraction with training instance augmentation. in Proceedings of the 15th Workshop on Biomedical Natural Language Processing 108–113 (2016). [Google Scholar]
- 68.Galvan D, Okazaki N, Matsuda K & Inui K Investigating the Challenges of Temporal Relation Extraction from Clinical Text. in Proceedings of the Ninth International Workshop on Health Text Mining and Information Analysis 55–64 (2018). [Google Scholar]
- 69.Leeuwenberg A & Moens M-F Word-Level Loss Extensions for Neural Temporal Relation Classification. in Proceedings of the 27th International Conference on Computational Linguistics 3436–3447 (2018). [Google Scholar]
- 70.ICD-9 radiology corpus, available through hNLP Center membership. at https://healthnlp.hms.harvard.edu/center/pages/data-sets.html.
- 71.Karimi S, Dai X, Hassanzadeh H & Nguyen A Automatic diagnosis coding of radiology reports: a comparison of deep learning and conventional classification methods. BioNLP 2017 328–332 (2017). [Google Scholar]
- 72.Zamaraeva O, Howell K & Rhine A Improving Feature Extraction for Pathology Reports with Precise Negation Scope Detection. in Proceedings of the 27th International Conference on Computational Linguistics 3564–3575 (2018). [Google Scholar]
- 73.Jagannatha A Structured prediction models for RNN based sequence labeling in clinical text. in Proceedings of the 2016 Conference on Empirical Methods in Natural Language Processing 856–865 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jagannatha AN & Yu H Bidirectional RNN for Medical Event Detection in Electronic Health Records. in Proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies 473–482 (Association for Computational Linguistics, 2016). doi: 10.18653/v1/N16-1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shivade C, de Marneffe M-C, Fosler-Lussier E & Lai AM Identification, characterization, and grounding of gradable terms in clinical text. in Proceedings of the 15th Workshop on Biomedical Natural Language Processing 17–26 (2016). [Google Scholar]
- 76.Roberts K, Si Y, Gandhi A & Bernstam E A FrameNet for Cancer Information in Clinical Narratives: Schema and Annotation. in Proceedings of the Eleventh International Conference on Language Resources and Evaluation (LREC-2018) (European Language Resource Association, 2018). [Google Scholar]
- 77.Lee W-S et al. Assessing Concordance With Watson for Oncology, a Cognitive Computing Decision Support System for Colon Cancer Treatment in Korea. JCO Clin. Cancer Inform 2, 1–8 (2018). [DOI] [PubMed] [Google Scholar]
- 78.Kim EJ et al. Early experience with Watson for oncology in Korean patients with colorectal cancer. PloS One 14, e0213640 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Choi YI et al. Concordance Rate between Clinicians and Watson for Oncology among Patients with Advanced Gastric Cancer: Early, Real-World Experience in Korea. Can. J. Gastroenterol. Hepatol 2019, 8072928 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Health C for D. and R. Artificial Intelligence and Machine Learning in Software as a Medical Device. FDA https://www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-software-medical-device, (2019).
- 81.US FDA Proposed Regulation for AI and ML software; https://www.fda.gov/media/122535/download.
- 82.Schuler A, Callahan A, Jung K & Shah NH Performing an Informatics Consult: Methods and Challenges. J. Am. Coll. Radiol. JACR 15, 563–568 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hirsch JS et al. HARVEST, a longitudinal patient record summarizer. J. Am. Med. Inform. Assoc. JAMIA 22, 263–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kumah-Crystal YA et al. Electronic Health Record Interactions through Voice: A Review. Appl. Clin. Inform 9, 541–552 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gregg JR et al. Automating the Determination of Prostate Cancer Risk Strata From Electronic Medical Records. JCO Clin. Cancer Inform. 2017, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.NCI FY 2020 Annual Plan and Budget Proposal Released. National Cancer Institute; (2018). Available at: https://www.cancer.gov/news-events/cancer-currents-blog/2018/sharpless-nci-annual-plan-2020. (Accessed: 11th February 2019) [Google Scholar]
- 87.Giordano SH et al. Limits of observational data in determining outcomes from cancer therapy. Cancer 112, 2456–2466 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Noone A-M et al. Comparison of SEER Treatment Data With Medicare Claims. Med. Care 54, e55–64 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Baldwin L-M et al. Linking physician characteristics and medicare claims data: issues in data availability, quality, and measurement. Med. Care 40, IV-82–95 (2002). [DOI] [PubMed] [Google Scholar]
- 90.Lerro CC, Robbins AS, Phillips JL & Stewart AK Comparison of cases captured in the national cancer data base with those in population-based central cancer registries. Ann. Surg. Oncol 20, 1759–1765 (2013). [DOI] [PubMed] [Google Scholar]
- 91.Hernandez-Boussard T, Tamang S, Blayney D, Brooks J & Shah N New Paradigms for Patient-Centered Outcomes Research in Electronic Medical Records: An Example of Detecting Urinary Incontinence Following Prostatectomy. EGEMS Wash. DC 4, 1231 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.IBM’s Watson recommended ‘unsafe and incorrect’ cancer treatments. STAT; (2018). Available at: https://www.statnews.com/2018/07/25/ibm-watson-recommended-unsafe-incorrect-treatments/. (Accessed: 13th June 2019) [Google Scholar]
- 93.Developing a Software Precertification Program: A Working Model; https://www.fda.gov/downloads/MedicalDevices/DigitalHealth/DigitalHealthPreCertProgram/UCM605685.pdf.


